Abstract
Background
The prevalence of Helicobacter pylori resistance to metronidazole and clarithromycin is high in Indonesia. Moreover, the increasing levofloxacin resistance rates in the absence of bismuth treatment in Indonesia has led to the use of other antibiotics as alternative regimens.
Methods
We determined the minimum inhibitory concentrations (MICs) of five alternative antibiotics for H. pylori (rifaximin, rifabutin, furazolidone, garenoxacin, and sitafloxacin) using the agar dilution method and assessed mutations associated with antibiotic resistance using next-generation sequencing.
Result
Analysis of 106 strains isolated from 1039 adult dyspeptic patients revealed that none of the strains were furazolidone-resistant. All strains were also sensitive to rifabutin and sitafloxacin. In contrast, the rates of resistance to rifaximin and garenoxacin were high (38.9% and 6.7%, respectively). The strains isolated from patients on Java Island had the highest resistance rates to garenoxacin and rifaximin. In addition, the resistance was distributed evenly among the ethnic groups, ranging between 25.0% and 69.2%. Except for rifaximin, for which the resistance rate was 38.9%, the other four antibiotics could be successfully employed to eradicate levofloxacin- and metronidazole-resistant H. pylori infections in vitro. Interestingly, garenoxacin-sensitive strains were found in regions with high clarithromycin resistance rates such as Bali and Papua Islands. In contrast, rifaximin might not be considered as an alternative antibiotic in regions with high clarithromycin resistance. There was an inconsistent association between gyrA and gyrB mutations and garenoxacin resistance. We confirmed that the I837V (replacement of isoleucine at position 837 with valine), A2414T/V, Q2079K and K2068R were the predominant rpoB point mutations. There was an association between vacA genotypes of H. pylori and rifaximin resistance (P = 0.048).
Conclusion
furazolidone-, rifabutin-, and sitafloxacin-based therapies might be considered as alternative regimens to eradicate H. pylori in Indonesia, including regions with high metronidazole and clarithromycin resistance rates. Moreover, sitafloxacin but not garenoxacin should be considered for eradication of levofloxacin-resistant strains.
Keywords: Indonesia, drug resistance, Helicobacter pylori, antibiotics
Introduction
Helicobacter pylori eradication has led to a significant decrease in the incidence of gastric cancer and can prevent its progression.1,2 The H. pylori eradication regimens established in the Asia-Pacific region and three countries in East Asia (Japan, South Korea, and China) have been summarized in the recent guidelines.3–6 Nevertheless, resistance to clarithromycin, which is included in the first-line therapy for H. pylori, has recently emerged in several regions across the globe.7–10 In addition, resistance to alternative regimens including metronidazole was significantly associated with their frequent use.9,11 Moreover, high levofloxacin resistance was reported in several countries in Asia, and this even reached a rate of up to 67%.9,11–15 Based on the Maastricht Consensus V, a suitable first-line regimen is considered to be effective against H. pylori if the cure rate is >90%,16 and thus, it can prevent secondary antibiotic resistance. However, further investigation is warranted to assess the antibiotic sensitivity of H. pylori to overcome the multiple treatment failures, with H. pylori eradication failure in >20% of cases, in specific countries to determine the best rescue treatment regimens.17
Indonesia, located in Southeast Asia, is the fourth most populous country in the world, with a total population of ~260 million in 2017, which is composed of various ethnic groups. Java, Sumatra, Papua, Kalimantan, and Sulawesi Island are the five main islands, with half of the total population living on Java Island. Similar to other regions in Indonesia, we previously reported high resistance to clarithromycin (21.4%) on Java Island, the rate of which is more than the limit of 15% recommended by the Maastricht consensus.18 In addition, the resistance rates to metronidazole and levofloxacin in Indonesian H. pylori strains are high (46.8% and 31.2%, respectively). Importantly, the prevalence of H. pylori infection in Indonesians, particularly among the major ethnic group of Javanese, is low (2.4%),19 highlighting the difficulties in isolating strains and conducting clinical trials on H. pylori eradication in Indonesia. In addition, although dyspepsia is the fifth most common symptom in an inpatient setting in Indonesia, the availability of gastrointestinal endoscopy is limited, and it is predominantly utilized on Java Island.20
Among the several antibiotics proposed as alternative regimens for H. pylori is furazolidone, a synthetic nitrofuran with broad-spectrum antimicrobial activity that blocks bacterial metabolism by interfering with bacterial oxidoreductase activity.21–25 Furthermore, in a study, the sensitivity of H. pylori to rifabutin and the utility of rifabutin as a rescue regimen following treatment failure with other antibiotics were reported in >50% of the subjects.26 Rifabutin is an antituberculosis agent which acts on DNA-directed RNA polymerase and inhibits transcription in H. pylori.27–29 Rifaximin is a semisynthetic derivate of rifamycin with antimicrobial activities against a broad spectrum of organisms, including H. pylori, is not absorbed in the gastrointestinal tract, and associated with mutations in rpoB.30 Conversely, garenoxacin and sitafloxacin, two novel quinolones, were proposed to treat H. pylori-resistant strains harboring gyrA mutation.31 In this study, we examined the resistance profile of H. pylori to several antibiotics used as alternative regimens in a geographical area with a high prevalence of clarithromycin-and metronidazole-resistant H. pylori strains. Our findings suggest several potential regimens that might overcome the hurdle of clarithromycin and metronidazole resistance, and the results might be of value not only for Indonesia but also for countries worldwide. Furthermore, we identified several point mutations in H. pylori that might confer rifaximin resistance.
Materials and methods
Patients and H. pylori
This nationwide study included 1,039 adult dyspeptic patients who underwent endoscopic biopsy between August 2012 and February 2016 in 18 cities on eight Indonesian islands. Among these 1,039 patients, 752 were reported in a previous study.18 Gastric biopsy specimens of the remaining 287 patients used in the current study (Figure 1) were obtained from the following regions: Cimacan (n=22) and Surabaya (n=22) on Java Island; Padang (n=33), Palembang (n=38), and Dolok Sanggul (n=47) on Sumatera Island; Gunungsitoli (n=32) on Nias Island; Kolaka (n=50) on Sulawesi Island; and Merauke (n=43) on Papua Island. There were 599 males (age range, 17–88 years; mean, 46.14±13.63 years) and 439 females (age range, 14–80 years; mean, 47.79±14.4 years). Patients with bleeding due to esophageal varices, those with a history of partial gastric resection, and those with a history of successfully eradicated H. pylori infection were excluded. All procedures applied in this work complied with the ethical standards of the relevant national and institutional committees on human experimentation and with the Declaration of Helsinki of 1975, as revised in 2008 and 2013. Peptic ulcer disease was diagnosed by endoscopic examination, whereas the diagnosis of gastritis was based on histologic examination. The review board or the ethics committee of the following institutions reviewed and approved the study protocol: Dr. Cipto Mangunkusumo Teaching Hospital (Jakarta, Indonesia), Dr. Soetomo Teaching Hospital (Surabaya, Indonesia), Dr. Wahidin Sudirohusodo Teaching Hospital (Makassar, Indonesia), and Oita University Faculty of Medicine (Yufu, Japan). All study participants agreed to follow the study protocol and provided written informed consent. For the participants who were <18 years old, the parents or legal guardian provided written informed consent.
Figure 1.
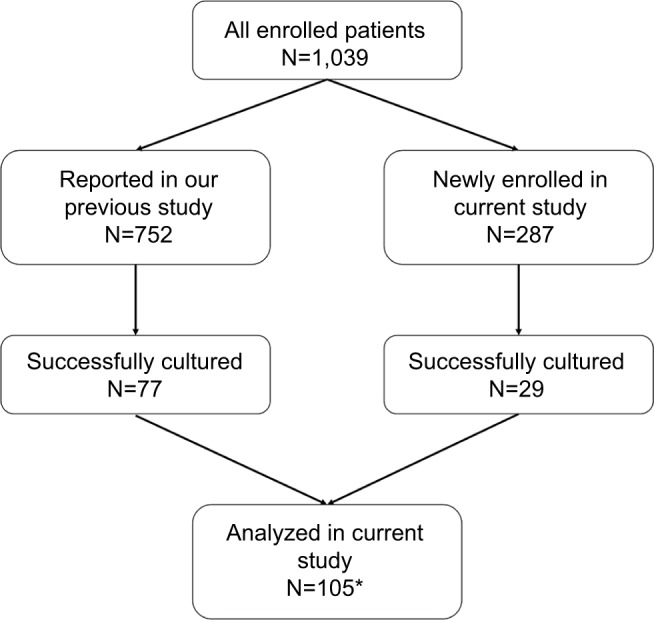
The chart showing the enrollment of patients in the current study.
Note: *One strain (Malang1) could not grow well, and hence, we excluded it from the study.
H. pylori was isolated from homogenized antral biopsy specimens by inoculating onto selective agar plates and incubating the plates for up to 10 days in microaerophilic environment (10% O2, 5% CO2, and 85% N2) at 37°C. The colonies that grew were subcultured onto antibiotic-free Mueller–Hinton II agar (Beckton Dickinson, Franklin Lakes, NJ, USA) supplemented with 10% horse blood under the same microaerophilic conditions. H. pylori isolates were confirmed based on colony morphology and Gram staining as well as oxidase, catalase, and urease test results. The isolates were stored in Brucella broth (Difco, Franklin Lakes, NJ, USA) supplemented with 10% dimethyl sulfoxide and 10% horse serum at −80°C.
Antibiotic susceptibility testing
The twofold agar dilution method was used to determine minimum inhibitory concentrations (MICs) of furazolidone (Tokyo Chemical Company, Tokyo, Japan), rifaximin (Tokyo Chemical Company), rifabutin (Sigma-Aldrich Co., St. Louis, MO, USA), garenoxacin (Sigma-Aldrich Co.), and sitafloxacin (Haoyuan Chemexpress, Shanghai, China) according to M07-A9 version of methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically (approved standard, Clinical and Laboratory Standard Institute). Briefly, the isolates were subcultured on Mueller–Hinton II agar supplemented with 10% horse blood. The bacteria were diluted in Brucella broth and adjusted to be equivalent to a McFarland opacity standard of 0.5. The prepared bacterial suspension (1 µL) was then inoculated using 2 mm-pin inoculator (Tokken Inc. Chiba, Japan) on Mueller–Hinton II agar supplemented with 5% horse blood. The MICs were determined after a 72-hour incubation. An H. pylori strain from American Type Culture Collection (catalog # 43504) was used as the quality control. Resistance breakpoints were determined based on an MIC of >4 mg/L for furazolidone and rifaximin and >1 mg/L for rifabutin, garenoxacin, and sitafloxacin, as described previously.32–35 The final concentrations of furazolidone and rifaximin ranged from 0.25 to 32 µg/mL, while those of rifabutin, garenoxacin, and sitafloxacin ranged from 0.064 to 8 µg/mL.
Detection of virulence factors and resistant strains
H. pylori DNA was extracted using the commercially available DNeasy® kit (Qiagen, Hilden, Germany) and stored at −20°C until further analysis. Data on the gyrA and gyrB mutations in H. pylori were available for the 752 patients who were reported in our previous publication.18 In addition, mutation analyses were performed for gyrA and gyrB mutation status in the remaining 287 specimens. Furthermore, next-generation sequencing (MiSeq next-generation sequencer; Illumina, San Diego, CA, USA) was used to analyze all specimens for full-length rpoB, oipA status (“on” or “off ”), and the presence of vacA (s1 or s2; m1 or m2; and i1, i2, or i3), iceA (iceA1 or iceA2), jhp0562, and b-(1,3)galT genotypes of the Indonesian strains. The BLAST algorithm implemented in the CLC Genomics Workbench software (ver. 11; Qiagen NV, Venlo, Netherlands) was used for the analysis. The sequences of hp0701, hp0501, hp1198, and hp0638 of the strain 26695 (GenBank accession number AE000511.1) were used as queries to obtain the gyrA, gyrB, rpoB, and oipA sequences, respectively, from the Indonesian next-generation sequencing data. The variants related to antibiotic resistance were predicted by comparing all the rpoB sequences of resistant strains and five random sensitive strains with the rpoB sequence of the strain 26695 for rifaximin resistance and gyrA and gyrB for garenoxacin and sitafloxacin resistance. Briefly, after obtaining the rpoB, gyrA, and gyrB sequences and confirming the absence of insertions or deletions leading to frameshift mutations, the sequences were aligned at the codon level using the MAFFT software (http://mafft.cbrc.jp/alignment/server/). Subsequently, each codon of the resistant and sensitive strains was compared to the reference sequence using our original PERL script and confirmed by visual inspection. Variants found in both the resistant and the sensitive strains were considered as normal variants and were excluded from further analysis. Variants found in the resistant strains but not in the sensitive ones were considered as variants related to antibiotic resistance.
Statistical analyses
Discrete variables were analyzed by the chi-squared test, whereas interval/ratio variables were analyzed using Student’s t-test or the Mann–Whitney U test. P values of <0.05 were considered statistically significant. All statistical analyses were performed using the SPSS statistical software package version 18.0 (SPSS Inc., Chicago, IL, USA).
Results
Resistance of H. pylori to alternative antibiotics
Twenty-nine H. pylori strains were isolated from 287 patients including 1, 1, 10, 1, 7, and 9 strains from Surabaya, Palembang, Dolok Sanggul, Gunungsitoli, Kolaka, and Merauke, respectively. No H. pylori strains could be isolated from the specimens obtained from the patients in Cimacan and Padang. We also reanalyzed the 77 strains that were assessed for their sensitivity to clarithromycin, amoxicillin, metronidazole, tetracycline, and levofloxacin in our previous study.18 However, one of these strains (Malang1) did not grow properly. Therefore, a total of 105 strains were analyzed in the current study.
Overall, more than half of the strains (61/105, 58.1%) were sensitive to all five antibiotics examined in this study. Forty strains were resistant to rifaximin (38.9%; Table 1). In addition, the rate of garenoxacin resistance was 6.7% (7/105). In contrast, none of the examined strains exhibited resistance to furazolidone, rifabutin, or sitafloxacin. Four strains were resistant to two antibiotics. The rates of resistance to rifaximin and garenoxacin were higher in males than in females (28/67 [42.4%] vs 12/38 [31.5%] and 5/67 [7.6%] vs 2/38 [5.2%], respectively), although these differences were not statistically significant (P=0.28 and P=0.65, respectively). The antibiotic-resistant strains were more frequent among those older than 30 years of age, albeit in the absence of a significant association. Overall, 95, 1, and 9 antibiotic-resistant strains were isolated from patients with chronic gastritis, gastric cancer, and peptic ulcer, respectively. The rate of garenoxacin resistance was higher in the patients with chronic gastritis than in those with peptic ulcer (6/95 [6.3%] vs 0/9 [0.0%]; P=0.001)
Table 1.
Rates of resistance to alternative antibiotics in Helicobacter pylori strains isolated in Indonesia
| Characteristics | N | Resistance (%)
|
||||
|---|---|---|---|---|---|---|
| Furazolidone | Sitafloxacin | Garenoxacin | Rifaximin | Rifabutin | ||
|
| ||||||
| Total | 105 | 0 (0.0) | 0 (0.0) | 7 (6.7) | 40 (38.9) | 0 (0.0) |
| Sex | ||||||
| Male | 67 | 0 (0.0) | 0 (0.0) | 5 (7.6) | 28 (42.4) | 0 (0.0) |
| Female | 38 | 0 (0.0) | 0 (0.0) | 2 (5.2) | 12 (31.5) | 0 (0.0) |
| Age (years) | ||||||
| 17–30 | 12 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (25.0) | 0 (0.0) |
| 31–40 | 13 | 0 (0.0) | 0 (0.0) | 1 (7.6) | 5 (38.4) | 0 (0.0) |
| 41–50 | 28 | 0 (0.0) | 0 (0.0) | 2 (7.1) | 12 (42.8) | 0 (0.0) |
| 51–60 | 34 | 0 (0.0) | 0 (0.0) | 3 (8.8) | 13 (38.2) | 0 (0.0) |
| >60 | 18 | 0 (0.0) | 0 (0.0) | 1 (5.5) | 6 (33.3) | 0 (0.0) |
| Clinical outcome | ||||||
| Gastritis | 95 | 0 (0.0) | 0 (0.0) | 6 (6.3) | 34 (35.7) | 0 (0.0) |
| PUD | 9 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (33.3) | 0 (0.0) |
| Cancer | 1 | 0 (0.0) | 0 (0.0) | 1 (100) | 1 (100) | 0 (0.0) |
Abbreviation: PUD, peptic ulcer disease.
Rates of antibiotic resistance according to location and ethnicity
The rate of garenoxacin resistance was highest among the H. pylori strains obtained from Java Island compared to those from the other regions (15.4% vs 10.0%, 6.2%, and 4.7% from the Sumatera, Papua, and Sulawesi Island, respectively; Table 2). The garenoxacin resistance was not detected in any of the strains from Kalimantan, Timor, and Bali. In contrast, more than half of the strains isolated from the specimens of patients from Kalimantan, Sulawesi, and Bali Islands had rifaximin resistance (60.0%, 52.4%, and 50.0%, respectively). Finally, the rate of rifaximin resistance was at least 20% in all the study locations.
Table 2.
Rates of resistance to alternative antibiotics in Helicobacter pylori strains isolated in specific regions of Indonesia
| Region | N | Resistance (%)
|
||||
|---|---|---|---|---|---|---|
| Furazolidone | Sitafloxacin | Garenoxacin | Rifaximin | Rifabutin | ||
|
| ||||||
| Bali | 6 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (50.0) | 0 (0.0) |
| Java | 13 | 0 (0.0) | 0 (0.0) | 2 (15.4) | 4 (30.7) | 0 (0.0) |
| Kalimantan | 5 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (60.0) | 0 (0.0) |
| Papua | 16 | 0 (0.0) | 0 (0.0) | 1 (6.2) | 6 (37.5) | 0 (0.0) |
| Sulawesi | 21 | 0 (0.0) | 0 (0.0) | 1 (4.7) | 11 (52.4) | 0 (0.0) |
| Sumateraa | 30 | 0 (0.0) | 0 (0.0) | 3 (10.0) | 7 (23.3) | 0 (0.0) |
| Timor | 14 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (42.8) | 0 (0.0) |
Note:
Strains obtained from patients from Nias Island were combined with those of Sumatera Island due to the low sample number.
The analysis of the rates of antibiotic resistance according to ethnicity revealed that the garenoxacin resistance rate of 20% was higher in the strains isolated from the Chinese Indonesian patients than in those isolated from the Bataknese, Buginese, and Papuan patients (9.6%, 7.7%, and 6.2%, respectively; Table 3); however, this difference was not statistically significant (P=0.44, P=0.33, and P=0.31, respectively). None of the strains isolated from the Ambonese, Balinese, Dayak, Javanese, Minahasanese, and Timor patients exhibited garenoxacin resistance, indicating that the bacterial strains in the patients belonging to these ethnic groups were sensitive to furazolidone, rifabutin, garenoxacin, and sitafloxacin but not rifaximin. Only one strain from a Javanese patient (only one strain was isolated) was sensitive to all five antibiotics. Rifaximin resistance was distributed evenly among ethnic groups (Table 3), with the rates ranging between 25.0% and 69.2% (P=0.81). Within the ethnic groups with the highest prevalence of H. pylori in Indonesia,19 all the strains were resistant to garenoxacin and rifaximin, with the higher resistance rate to rifaximin observed in the Buginese patients compared with that in the Papuan and Bataknese patients (9/13 [69.2%] vs 6/16 [37.5%], P=0.01 and 8/31 [25.8%], P=0.001, respectively).
Table 3.
Prevalence of antibiotic resistance in Helicobacter pylori isolates based on ethnicity
| Ethnicity | Island | N | Resistance (%)
|
||||
|---|---|---|---|---|---|---|---|
| FUR | SIT | GAR | RFX | RIF | |||
|
| |||||||
| Ambonese | Java | 4 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (50.0) | 0 (0.0) |
| Bataknese | Sumatera and Java | 31 | 0 (0.0) | 0 (0.0) | 3 (9.6) | 8 (25.8) | 0 (0.0) |
| Balinese | Bali | 6 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (50.0) | 0 (0.0) |
| Buginese | Sulawesi | 13 | 0 (0.0) | 0 (0.0) | 1 (7.7) | 9 (69.2) | 0 (0.0) |
| Chinese | Java and Kalimantan | 10 | 0 (0.0) | 0 (0.0) | 2 (20.0) | 3 (30.0) | 0 (0.0) |
| Dayak | Kalimantan | 2 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) |
| Javanese | Java | 1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Minahasanese | Sulawesi | 8 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (25.0) | 0 (0.0) |
| Papuan | Papua | 16 | 0 (0.0) | 0 (0.0) | 1 (6.2) | 6 (37.5) | 0 (0.0) |
| Timor | Timor | 14 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (42.8) | 0 (0.0) |
Abbreviations: FUR, furazolidone; GAR, garenoxacin; RFX, rifaximin; RIF, rifabutin; SIT, sitafloxacin.
Comparison of alternative and standard antibiotic regimens
We determined the rates of resistance of the 76 H. pylori strains reported in our previous study14 to the five antibiotics and compared them with the rates of resistance to the standard antibiotics used for H. pylori infection (Table 4). Our findings above indicated that, except for rifaximin with a resistance rate of 35.5% (27/76), there was a possibility that the remaining four antibiotics might overcome the high rate of resistance to levofloxacin and metronidazole. Interestingly, H. pylori in the regions with high rates of clarithromycin resistance, such as Bali and Papua Islands (1/6, 16.7% and 1/7, 14.3%, respectively), was still sensitive to garenoxacin, although this finding could be due to the low number of strains with clarithromycin resistance. In contrast, the isolate from Java Island with the highest rate of clarithromycin resistance also exhibited a high rate of rifaximin resistance. Thus, we suggest that rifaximin should not be considered as an alternative in areas with high clarithromycin resistance. Garenoxacin may combat H. pylori in regions with high amoxicillin resistance such as Papua Island (0.0% vs 14.3% of resistance rate for garenoxacin and amoxicillin, respectively) but not in regions with high tetracycline resistance such as Java Island (both 15.4% of resistance rate) (Table 4).
Table 4.
Comparison of the five alternative antibiotics with the standard regimens as reported by Miftahussurur et al18
| Island | n | Resistant regimens (%)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAM | AMX | MNZ | TCN | LVX | FUR | SIT | GAR | RFX | RIF | ||
|
| |||||||||||
| Total | 76 | 7 (9.1) | 4 (5.2) | 36 (46.7) | 2 (2.6) | 24 (31.2) | 0 (0.0) | 0 (0.0) | 5 (6.5) | 27 (35.5) | 0 (0.0) |
| Bali | 6 | 1 (16.7) | 0 (0.0) | 2 (33.3) | 0 (0.0) | 1 (16.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (50.0) | 0 (0.0) |
| Java | 13 | 3 (23.0) | 0 (0.0) | 7 (46.1) | 2 (15.4) | 7 (53.8) | 0 (0.0) | 0 (0.0) | 2 (15.4) | 4 (30.7) | 0 (0.0) |
| Kalimantan | 5 | 0 (0.0) | 0 (0.0) | 1 (20.0) | 0 (0.0) | 1 (20.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (60.0) | 0 (0.0) |
| Papua | 7 | 1 (14.3) | 1 (14.3) | 3 (42.9) | 0 (0.0) | 2 (28.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (28.5) | 0 (0.0) |
| Sulawesi | 13 | 1 (7.7) | 1 (7.7) | 4 (30.8) | 0 (0.0) | 2 (15.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (46.1) | 0 (0.0) |
| Sumatera | 18 | 1 (5.6) | 1 (5.6) | 16 (88.9) | 0 (0.0) | 8 (44.4) | 0 (0.0) | 0 (0.0) | 3 (16.7) | 3 (16.7) | 0 (0.0) |
| Timor | 14 | 0 (0.0) | 1 (7.1) | 3 (21.4) | 0 (0.0) | 3 (21.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (42.8) | 0 (0.0) |
Abbreviations: AMX, amoxicillin; CAM, clarithromycin; FUR, furazolidone; GAR, garenoxacin; LVX, levofloxacin; MNZ, metronidazole; RFX, rifaximin; RIF, rifabutin; SIT, sitafloxacin; TCN, tetracycline.
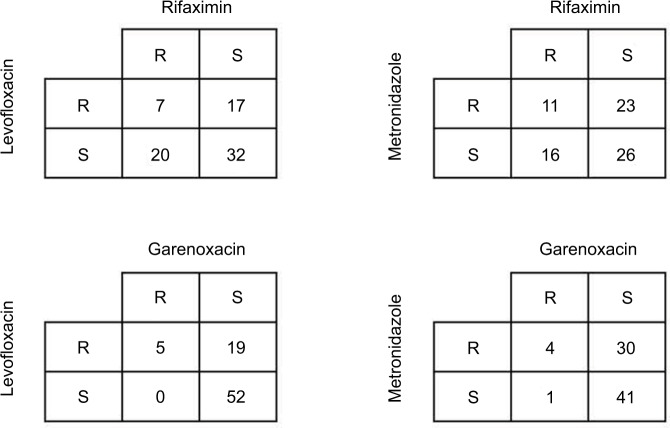
To further analyze the associations among resistance rates of metronidazole, levofloxacin, garenoxacin, and rifaximin, we created a two-by-two table (Figure 2). Only seven strains (9.2%) exhibited resistance to both rifaximin and levofloxacin. Furthermore, the percentage of the levofloxacin-resistant/ rifaximin-sensitive strains was lower than that of the levofloxacin-sensitive/rifaximin-resistant strains (17/76 [22.4%] vs 20/76 [26.3%]). In contrast, the percentage of the metronidazole-resistant/rifaximin-sensitive strains was higher than that of the metronidazole-sensitive/rifaximin-resistant strains (23/76 [30.3%] vs 16/76 [21.1%]). Conversely, the percentages of the metronidazole-resistant/garenoxacin-sensitive and the levofloxacin-resistant/ garenoxacin-sensitive strains were higher than those of the metronidazole-sensitive/garenoxacin-resistant and levofloxacin-sensitive/garenoxacin-resistant strains (19/76 [25.0%] and 30/76 [39.5%] vs 0/76 [0.0%] and 1/76 [1.3%], respectively).
Figure 2.
The associations among rates of resistance to metronidazole, levofloxacin, garenoxacin, and rifaximin.
Mutations associated with garenoxacin resistance
We analyzed the gyrA and gyrB mutations from the two strains with high MICs for garenoxacin identified in the current study (Merauke20 and Kolaka72; Table 5) together with those identified in our previous study.18 Four strains with the highest MICs for garenoxacin (2 mg/L) were associated with a high MIC for levofloxacin (>32 mg/L). In addition, three of these strains had an amino acid substitution at Asp91 or Asn87 in the GyrA, which were predominantly associated with the highest MICs for levofloxacin.18 However, the 13 garenoxacin-sensitive strains with low MIC values (<0.063–0.5 mg/L) were also associated with those mutations, suggesting an inconsistent effect of these mutations. Moreover, none of the garenoxacin-resistant strains harbored a substitution at Arg484 or Ser479 of the gyrB. Finally, none of the strains harbored parC or parE, the two important genes associated with quinolone resistance in other bacteria.
Table 5.
Mutations associated with quinolones
| No. | Strains | gyrA mutation | gyrB mutation | MIC of LVX (mg/L) | MIC of GAR (mg/L) | MIC of SIT (mg/L) |
|---|---|---|---|---|---|---|
| 1 | Jayapura1 | N87K | None | >32 | 0.25 | 0.063 |
| 2 | Jayapura21 | N87K | None | >32 | 0.125 | 0.063 |
| 3 | Kupang2 | D91N, A129T | S479G | 4 | 0.125 | <0.063 |
| 4 | Kupang11 | D91Y | None | >32 | 0.25 | <0.063 |
| 5 | Kupang23 | A129T | S479G | >32 | 0.125 | <0.063 |
| 6 | Kupang41 | D91N | R484K | 8 | 0.5 | <0.063 |
| 7 | Malang1a | D91N | None | 16 | n.a. | n.a. |
| 8 | Manado18 | None | None | 8 | 0.5 | <0.063 |
| 9 | Manado20 | D91Y | None | 8 | 0.25 | <0.063 |
| 10 | Medan3 | N87I | None | >32 | 2 | 0.25 |
| 11 | Medan10 | None | None | 25 | <0.063 | <0.063 |
| 12 | Medan15 | R140K, D192N | None | >32 | 2 | 0.5 |
| 13 | Medan17 | D34N | None | 16 | 0.5 | <0.063 |
| 14 | Medan18 | D91G, D161N | None | 4 | <0.063 | 0.063 |
| 15 | Medan22 | D91N | None | >32 | 0.5 | 0.125 |
| 16 | Medan23 | D34Y, R140K | None | 4 | 0.063 | <0.063 |
| 17 | Medan30 | D91N | None | >32 | 1 | 0.125 |
| 18 | Pontianak50 | D91G | None | >32 | 0.063 | <0.063 |
| 19 | Surabaya71 | D91N | None | >32 | 2 | 0.25 |
| 20 | Surabaya79 | N87Y | R484K | >32 | 0.5 | <0.063 |
| 21 | Surabaya137 | N87K | None | >32 | 0.5 | <0.063 |
| 22 | Surabaya151 | N87K | None | >32 | 0.5 | 0.125 |
| 23 | Surabaya283 | D91Y | None | >32 | 2 | <0.063 |
| 24 | Surabaya304 | D91G | None | >32 | 0.5 | 0.063 |
| 25 | Merauke20b | E103G | None | na | 1 | <0.063 |
| 26 | Kolaka72b | D91G | None | na | 1 | <0.063 |
Notes: An MIC >1 mg/L was used as a resistance breakpoint for levofloxacin, garenoxacin, and sitafloxacin.
One of the previously isolated strains could not sustain growth.
The new strains with high MIC of garenoxacin that were not reported in our previous study.
Abbreviation: GAR, garenoxacin; LVX, levofloxacin; MIC, minimum inhibitory concentration; na, not available; SIT, sitafloxacin.
Mutations associated with rifaximin resistance
We analyzed full-length rpoB from 40 rifaximin-resistant strains based on the next-generation sequencing data, with an average sequencing coverage ranging from 82.43x to 560.85x and a Q30 score percentage ranging from 80.59% to 96.31% (Table S1). Five random rifaximin-sensitive strains were used for comparison. Pairwise alignment identified that the garenoxacin-sensitive strains shared 95.7%–97.8% identity with the reference strain 26695. Using the strain 26695 and the garenoxacin-sensitive control strains, DNA sequence analysis of rpoB from all rifaximin-sensitive strains revealed intact reading frames that lacked nonsense mutations. Among all 2,890 codons of rpoB, 1,010 had non-synonymous substitutions, indicating a change of nucleotide without a change in the amino acid (silent mutations). In contrast, majority of the rifaximin-resistant strains (39/40 [97.5%]) contained missense mutations (Table 6). We confirmed that the predominant point mutations of rpoB were the replacement of isoleucine at position 837 with valine amino acid (8/40 [20%]), alanine at position 2,414 with valine or threonine (8/40 [20%]), glutamine at position 2,079 with lysine (7/40 [17.5%]), and lysine at position 2,068 with arginine (7/40 [17.5%]).
Table 6.
Mutations associated with rifaximin resistance
| No. | Strain name | MIC (mg/L) | rpoB mutation |
|---|---|---|---|
| 1 | Surabaya47 | 4 | S355Y, I741V, T2002M, Q2079K |
| 2 | Jayapura21 | 4 | V1125I, A2454V |
| 3 | Jayapura06 | 8 | L547F, K786R, I837V, A964T, V1275I, A1533S, P1623S, D1697N, G1908E, A2099T, S2640Y |
| 4 | Jakarta9 | 4 | I64V, L295I, S355Y, V657I, T1023I, S1197A, Q2042R, K2068R |
| 5 | Kupang10 | 4 | G523C, I832V, E877K, K1006E, E1528D, Q1666H, N1944S, A2255V, G2512S, S2619I, V2774M |
| 6 | Kupang23 | 4 | L169S, S355H, A693T, I837V, K854R, L977I, I1351T, N1999H, K2068R, Q2079K, S2415N, I2481V, V2528L, P2679S, M2696T |
| 7 | Kupang26 | 4 | L169A, S355H, I837V, L977I, N1999H, Q2079K, S2415N, A2472V, I2481V, V2528L, P2679S, M2696T |
| 8 | Kupang29 | 4 | K42R, I748V, T773I, A958T, E969D, S986G, A1025V, V1052I, V1122I, A2414V, S2619I |
| 9 | Kupang30 | 4 | S355H, S627N, I837V, A1025V, D1162N, K1165R, L1401I, R1711H, Q2079K, A2234T, D2380E |
| 10 | Kupang34 | 4 | S355H, A732V, I837V, V955I, L977I, V1028A, N1999H, K2068R, Q2079K, S2415N, I2481V, V2528L, P2679S, M2696T |
| 11 | Bangli42 | 4 | I141V, E163D, N642D, R954W, E996G, M1264I, E1407K, Y2275C, K2462E, V2469I, G2480R, T2539A |
| 12 | Bangli47 | 4 | L314F, E1151K, I1190T, R1711H, K2068R, Y2326C, K2418Q, V2528L, T2536A, F2537L, K2538S, K2557R, V2561M, A2570T, S2734G |
| 13 | Bangli64 | 4 | R63H, E162K, L295I, I336T, M667I, A735T, P816S, I837V, V867A, R973H, T975I, Q1010R, E1572Q, A1691V, A2346T, S2390G, G2491D, A2541V |
| 14 | Manado29 | 4 | D255N, P259S, A1168V, A1181V, A1533T, L1765F, V1939I, A1950T, L2328I, R2694H, I2824V |
| 15 | Manado31 | 8 | K307E, R984H, V1028A, A1533T, A2255V, Y2326C, A2494T |
| 16 | Merauke20 | 4 | K786R, A964T, R2313H, G2512S |
| 17 | Merauke21 | 4 | A1181V, G2180S, A2472T, P2545S, V2664M |
| 18 | Merauke27 | 4 | I140V, A1533T, S1701N, A2505T, V2664M |
| 19 | Merauke37 | 4 | S78P, P95H, M313L, K786R, A964T, L977F, E1014K, M1242I, D1379N, A1533S, G2180S, A2414T, R2477H, A2494T, I2730V |
| 20 | Kolaka72 | 4 | R274I, S355Y, V538I, T635A, R1248H, G2403S, A2541, N2602D, R2641K, D2788G |
| 21 | Kolaka79 | 4 | – |
| 22 | Kolaka96 | 4 | R708K, L982S, E1161K, N1709D, E2382K, V2469I, R2477Y, V2528L, T2536A, F2537L, K2538S, K2557R, V2561M, R2882K |
| 23 | Kolaka98 | 4 | P259S, A735T, S743A, I837V, T975S, A1025S, K1165R, D1379Y, K2068R, Q2079K, K2421R |
| 24 | Kolaka99 | 4 | L295I, S355Y, A473V, M1175I, R1711H, Q2079K, A2454V, P2612S, S2675G |
| 25 | Makasar31 | 8 | L295I, I336T, G615D, A735V, I837V, Q1010R, D1379Y, V1491I, H1985Y, V2237M, A2317V, D2380E, R2506C |
| 26 | Makasar45 | 4 | E1161K, A2414V |
| 27 | Makasar52 | 4 | E1161K, A2414V |
| 28 | Makasar55 | 4 | P931S, A1643M, A1950T, K2068R, D2380E |
| 29 | Medan56 | 4 | V303I, K1540N, A2414V, A2454V |
| 30 | Medan67 | 4 | A497T, A958T, N1598H, T2002M, R2313C, A2414V |
| 31 | Medan75 | 4 | I512V, M667I, A964V, T1402M, A2414V, E2599A, V2638I, S2791G |
| 32 | Padang42 | 4 | H153Y, P931S, H1985Y, E2183K, A2414V, E2604G |
| 33 | Pontianak44 | 4 | A487T, E969D, V1291I, A2255V, V2447I |
| 34 | Pontianak50 | 4 | S355Y, S627N, E969K, R1563K, M1627I, A1676V, S1794N, N1999H, V2037I, D2449N, A2459T, T2533M, M2696T, E2859G |
| 35 | Pontianak5 | 4 | V2802L |
| 36 | Surabaya283 | 4 | P259S, T440A, A497T, E1232D, N1944S, V2037I, I2428V, I2564V, Y2740H, K2889R |
| 37 | Surabaya304 | 8 | S78A, S355Y, K398R, V657I, E1486D, D2226S, L2328I, A2357V |
| 38 | Medan15 | 4 | A473V, Q991R, E1059G, S1197A, K2068R |
| 39 | Medan22 | 32 | I66V, L295I, S355Y, I586L, E655K, V657I, G1620S, A2541T, L2881I |
| 40 | Medan25 | 8 | V52I, I66V, E106G, V657I, A756V, D2380E, K2482R |
Note: S355Y means tyrosine replaced serine amino acid in the position 355.
Abbreviation: MIC, minimal inhibitory concentration.
Virulence factors and antibiotic resistance types
In addition to the data on virulence factors that we reported previously,18 we analyzed virulence factors in the 29 newly identified H. pylori strains including cagA, vacA, iceA, jhp0562/b-(1,3)galT, and oipA. There was an association between the vacA genotype of H. pylori with rifaximin resistance (P=0.048). The genotypes s2m1 and s1m1 of vacA tended to be more frequent in the garenoxacin-resistant strains compared with the vacA s1m2 and s2m2 genotypes (2/2 [100.0%], 32/74 [43.2%], 6/26 [23.1%], and 0/2 [0.0%], respectively; P=0.051). There were no significant associations between other virulence factors and antibiotic resistance.
Nucleotide sequencing
The nucleotide sequences were deposited in the DDBJ under accession numbers LC420353–LC420380 (vacA), LC420381–LC420408 (oipA), LC420409–LC420436 (jhp0562 and jhp0563), LC420437–LC420462 (iceA), LC420463–LC420466 (gyrA and gyrB), and LC420467– LC420511 (rpoB).
Discussion
The current study revealed that none of the H. pylori strains isolated from Indonesian patients were resistant to furazolidone, suggesting that furazolidone might be considered as an alternative H. pylori treatment regimen in Indonesia, especially in regions with high prevalence of strains exhibiting dual resistance to clarithromycin and metronidazole.36 Our results are in agreement with those reported by a study from a neighboring country, Malaysia, which also found that all the isolated strains were sensitive to furazolidone.37 Furazolidone use has been proposed in recent guidelines for H. pylori management in developing countries, due to its efficacy, low rate of primary bacterial resistance, and lack of alternative and low-cost therapies.38,39 To improve the H. pylori cure rates, bismuth should be added to therapy. For example, the addition of bismuth to quadruple therapy including furazolidone has been successful in China, with cure rates reaching 92.26% with minimal side effects.40 However, bismuth is unavailable in certain regions41 due to potential bismuth-associated carcinogenic effects, including mutagenicity and genotoxicity in in vitro and animal models,42,43 and it was classified as a type III carcinogen for humans in 1997 by the International Agency on Research on Cancer. Furthermore, there are currently no standardized rescue therapies available for patients who fail the initial furazolidone-based treatment.
Our finding of all isolated strains exhibiting rifabutin sensitivity provides support for rifabutin as a potential alternative antibiotic against H. pylori. The concentrations of rifabutin in the gastric juice were reported to be 10–17 times higher than in peripheral blood.44 The antibacterial activity of rifabutin, which is not affected by the low pH environment in the stomach, is higher than that of rifampicin.27 Importantly, its target is different from that of clarithromycin. Therefore, its efficacy in strains with primary clarithromycin resistance, even in those who are also resistant to metronidazole, is high,45,46 although the majority of the clinical trials that define these differences were conducted in Western countries. Nonetheless, the adverse effects of rifabutin such as myelotoxicity should be considered. Furthermore, the increased use of rifabutin in Indonesia, a country with high tuberculosis prevalence, might lead to rifabutin resistance of Mycobacterium tuberculosis. Several studies reported substantial in vitro cross-resistance to rifampicin, a main component of the tuberculosis therapy regimens, although rifabutin resistance in H. pylori in in vitro was rarely reported.28 A history of rifampicin treatment should be taken into consideration before prescribing rifabutin for H. pylori eradication to reduce the possibility of failure of tuberculosis treatment and H. pylori eradication. Importantly, rifabutin use in combination with clarithromycin should be avoided, based on evidence showing the inhibition of rifabutin metabolism by clarithromycin in liver microsomes,47 which suggests that potential toxicity might arise with combination use.
Compared with the other fluoroquinolones, sitafloxacin is a more potent inhibitor of DNA gyrase and topoisomerase IV, which play important roles in bacterial DNA repair, transcription, replication, and recombination.48 Sitafloxacin improves the efficacy of quinolone-based rescue therapy by virtue of its ability to eradicate H. pylori strains with gyrA mutations.49 However, limited access and availability are the main concerns regarding sitafloxacin. Currently, Japan and Thailand are the only countries that provide sitafloxacin in their health care system, and clinical trials for sitafloxacin are underway in Western countries.50 In contrast, although garenoxacin was also reported to eradicate H. pylori strains with gyrA mutations,51 there were several strains that were resistant to this antibiotic in the current study. Interestingly, the MIC value of levofloxacin was not associated with MIC value of garenoxacin. For example, although all seven garenoxacin-resistant strains exhibited the highest MICs for levofloxacin (>32 mg/L), all ten strains with the highest levofloxacin MIC were sensitive to garenoxacin. The lower antibacterial activity of garenoxacin against H. pylori compared with that of sitafloxacin might be associated with the high affinity of sitafloxacin to DNA gyrase.52 Due to high levofloxacin resistance rates in Indonesia, our finding should be instrumental in formulating second-line regimen guidelines to eradicate H. pylori.
One study found that double mutations in gyrA were associated with a sevenfold increase in sitafloxacin MIC compared with the pretreatment MICs and that double mutations in gyrA, including the mutations at Asp91 and Asn87, were associated with eradication failure.53 We found that double mutations were not associated with an increase in the MIC of sitafloxacin, although none of the strains harbored both Asp91 and Asn87 mutations. Similar to sitafloxacin, none of the single or double mutations in GyrA or GyrB were associated with garenoxacin resistance. Although parC and parE are important genes associated with quinolone resistance, none of the isolated H. pylori strains exhibited the presence of expressed these genes, as previously described.54 Our results suggest that genes other than gyrA or gyrB were associated with resistance to sitafloxacin and garenoxacin, which should be investigated in future studies.
Among the several alternative drugs tested in the current study, the rate of resistance was highest to rifaximin; this finding is in agreement with a previous study showing that rifaximin-based triple therapy did not achieve acceptable H. pylori cure rates.55,56 However, rifaximin is a promising H. pylori drug due to poor absorbance in the blood, which can minimize adverse effects, and its higher bioavailability in the gastrointestinal tract than that of other antibiotics.57 The poor eradication rates might be due to a failure in achieving sufficient therapeutic concentrations under and within the gastric mucosal layer, which is a frequent site of H. pylori colonization.58 Therefore, well-designed clinical trials are necessary to evaluate rifaximin efficacy against H. pylori, including high-dose regimens of longer duration, additional bioadhesive formulations, and combinations with mucolytic agents for persistent coverage of the gastric mucosa.58
Although rpoB mutations were reported to play a role in rifaximin resistance in other bacteria such as Escherichia coli,59 Clostridium difficile,60,61 Staphylococcus,62 and M. tuberculosis,63 only one study found an association between codons 524–545 and 585 of rpoB with rifabutin resistance in H. pylori.30 In the current study, we found numerous missense mutations in rpoB of rifaximin-resistant strains, including novel and predominant mutations: 837I, 2414A, 2079K, and 2068K. Although the mechanism remains unclear, the risk for horizontal transmission of the rpoB mutations is lower than that of a resistance gene located on a plasmid or transposon; however, certain yet-to-be determined conditions and improper prescription or usage of antibiotic could still facilitate the rapid transmission of such mutations.62 Strict control should be practiced to prevent rifaximin failure in H. pylori eradication.
The major limitation of this study was the relatively small number of samples, and when we divided the samples based on regions, it yielded very low sample number in each region. Therefore, it may be difficult to represent H. pylori strains in whole Indonesia. Further study with a bigger sample size is necessary. However, these samples, obtained from 1,039 endoscopic patients, comprised the biggest cohort of H. pylori strains isolated in Indonesia thus far. Indonesia is a wide country and consists of many ethnic groups. Among those, some ethnic groups had a much higher prevalence of H. pylori infection than the others; however, the overall gastric cancer risk in Indonesia is low, suggesting that Indonesia may become the best example for Asian enigma similar to South Asia. In addition, only a fraction of the genomic changes that were related to drug resistance, among a total of 1,600 genes of H. pylori, were examined in the current study. Although sitafloxacin is a potent drug for H. pylori, it has not been approved by the Indonesian National Agency of Drug and Food Control. Thus, sitafloxacin-based regimens cannot be currently prescribed in Indonesia.
Conclusion
Furazolidone-, rifabutin-, and sitafloxacin-based therapies should be considered as alternative regimens to eradicate H. pylori in Indonesia, including regions with high rates of metronidazole and clarithromycin resistance. Moreover, sitafloxacin but not garenoxacin could inhibit the levofloxacin-resistant H. pylori strains.
Supplementary material
Table S1.
Mutations associated with rifaximin resistance
| No. | Strain ID | Average coverage | Q30 percentage | Rifaximin |
|---|---|---|---|---|
| 1 | Manado1 | 326.27 | 91.21 | S |
| 2 | Surabaya71 | 346.57 | 90.99 | S |
| 3 | Surabaya47 | 264.51 | 88.23 | R |
| 4 | Surabaya68 | 408.21 | 91.31 | S |
| 5 | Surabaya69 | 504.26 | 88.53 | S |
| 6 | Surabaya71 | 560.21 | 88.06 | S |
| 7 | Surabaya79 | 554.35 | 94.39 | S |
| 8 | Jayapura1 | 570.45 | 95.95 | S |
| 9 | Jayapura3 | 560.85 | 96.31 | S |
| 10 | Jayapura6 | 589.56 | 95.97 | R |
| 11 | Jayapura8 | 231.21 | 84.54 | S |
| 12 | Jayapura15 | 489.67 | 96.36 | S |
| 13 | Jayapura16 | 509.21 | 95.14 | S |
| 14 | Jayapura21 | 502.21 | 96.49 | R |
| 15 | Jakarta9 | 115.74 | 91.23 | R |
| 16 | Medan17 | 142.34 | 80.59 | S |
| 17 | Medan23 | 166.24 | 90.67 | S |
| 18 | Medan27 | 91.58 | 80.76 | S |
| 19 | Medan3 | 93.57 | 80.88 | S |
| 20 | Medan10 | 85.96 | 80.44 | S |
| 21 | Medan11 | 278.2 | 87.68 | S |
| 22 | Medan15 | 260.45 | 80.59 | R |
| 23 | Medan19 | 176.28 | 83.32 | S |
| 24 | Medan20 | 164.28 | 82.27 | S |
| 25 | Medan22 | 115.51 | 82.74 | R |
| 26 | Medan23 | 264.07 | 81.06 | S |
| 27 | Medan25 | 219.52 | 80.97 | R |
| 28 | Medan28 | 169.54 | 80.8 | S |
| 29 | Medan30 | 237.04 | 80.42 | S |
| 30 | Makasar31 | 87.65 | 81.19 | R |
| 31 | Makasar45 | 156.35 | 89.51 | R |
| 32 | Makasar47 | 85.35 | 80.68 | S |
| 33 | Makasar52 | 87.07 | 83.13 | R |
| 34 | Makasar55 | 89.29 | 83.49 | R |
| 35 | Makasar56 | 82.43 | 82.46 | S |
| 36 | Pontianak63 | 82.77 | 88.97 | S |
| 37 | Pontianak75 | 85.13 | 86.89 | R |
| 38 | Pontianak20 | 191.58 | 89.57 | S |
| 39 | Pontianak44 | 128.24 | 89.45 | R |
| 40 | Pontianak50 | 99.51 | 91.01 | R |
| 41 | Manado5 | 171.08 | 89.76 | S |
| 42 | Manado18 | 136.04 | 90.18 | S |
| 43 | Manado20 | 110.47 | 87.37 | S |
| 44 | Manado26 | 151.85 | 86.67 | S |
| 45 | Manado28 | 175.98 | 93.44 | S |
| 46 | Manado29 | 90.53 | 93.63 | R |
| 47 | Manado31 | 102.87 | 93.18 | R |
| 48 | Kupang2 | 156.23 | 82.79 | S |
| 49 | Kupang5 | 132.2 | 85.25 | S |
| 50 | Kupang6 | 120.2 | 83.27 | S |
| 51 | Kupang10 | 105.54 | 82.16 | R |
| 52 | Kupang11 | 196.38 | 80.2 | S |
| 53 | Kupang15 | 192.96 | 82.36 | S |
| 54 | Kupang23 | 125.05 | 84.94 | R |
| 55 | Kupang26 | 98.87 | 89.29 | R |
| 56 | Kupang28 | 146.88 | 86.58 | S |
| 57 | Kupang29 | 144.7 | 81.55 | R |
| 58 | Kupang30 | 85.85 | 85.47 | R |
| 59 | Kupang33 | 158.02 | 83.55 | S |
| 60 | Kupang34 | 238.16 | 87.82 | R |
| 61 | Kupang35 | 240.73 | 85.3 | S |
| 62 | Kupang41 | 112.69 | 85.57 | S |
| 63 | Kupang42 | 86.98 | 81.43 | R |
| 64 | Kupang47 | 142.12 | 85.5 | R |
| 65 | Kupang64 | 171.52 | 82.59 | R |
| 66 | Kupang73 | 134.33 | 88.07 | S |
| 67 | Kupang83 | 152.03 | 85.08 | S |
| 68 | Medan18 | 201.24 | 89.67 | S |
| 69 | Medan31 | 162.6 | 90.39 | S |
| 70 | Medan32 | 264.33 | 88.11 | S |
| 71 | Medan33 | 368.99 | 86.63 | S |
| 72 | Nias9 | 123.05 | 84.92 | S |
| 73 | Medan36 | 97.32 | 84.74 | S |
| 74 | Medan37 | 95.96 | 85.43 | S |
| 75 | Medan40 | 125.98 | 85.11 | S |
| 76 | Medan49 | 163.37 | 83.54 | S |
| 77 | Medan50 | 146.38 | 83.51 | S |
| 78 | Medan56 | 192.02 | 83.51 | R |
| 79 | Medan67 | 223.14 | 85.69 | R |
| 80 | Medan68 | 137.93 | 84.11 | S |
| 81 | Medan73 | 224.19 | 83.87 | S |
| 82 | Medan75 | 194.47 | 83.86 | R |
| 83 | Padang42 | 157.75 | 88.29 | R |
| 84 | Surabaya106 | 84.06 | 80.06 | S |
| 85 | Surabaya137 | 104.09 | 86.49 | S |
| 86 | Surabaya151 | 96.79 | 82.68 | S |
| 87 | Surabaya192 | 106.55 | 85.02 | S |
| 88 | Surabaya283 | 107.28 | 87.04 | R |
| 89 | Surabaya304 | 112.26 | 86.21 | R |
| 90 | Merauke3 | 243.44 | 85.05 | S |
| 91 | Merauke5 | 219.32 | 84.41 | S |
| 92 | Merauke7 | 225.09 | 83.16 | S |
| 93 | Merauke8 | 269.32 | 86.05 | S |
| 94 | Merauke12 | 191.03 | 85.7 | S |
| 95 | Merauke20 | 145.23 | 88.24 | R |
| 96 | Merauke21 | 338.84 | 87.53 | R |
| 97 | Merauke27 | 185.99 | 80.84 | R |
| 98 | Merauke37 | 304.88 | 86.56 | R |
| 99 | Kolaka56 | 219.48 | 85.48 | S |
| 100 | Kolaka72 | 196.53 | 86.86 | R |
| 101 | Kolaka79 | 343.49 | 86.15 | R |
| 102 | Kolaka94 | 256.4 | 83.82 | S |
| 103 | Kolaka96 | 190.85 | 82.58 | R |
| 104 | Kolaka98 | 206.04 | 83.99 | R |
| 105 | Kolaka99 | 279.25 | 81.86 | R |
Abbreviations: R, resistant; S, sensitive.
Acknowledgments
The authors would like to thank Dr OK Yulizal, Dr Abdul Rahman, Dr Kanserina Esthera Dachi, Dr Fardah Akil, Dr Willi Brodus Uswan, Dr David Simanjuntak, Dr Jimmy Bradley Waleleng, Dr Alexander Michael Joseph Saudale, Dr Fauzi Yusuf, Dr Syifa Mustika, Dr Pangestu Adi, Dr Hasan Maulahela, and Prof. Maria Inge Lusida for their kind help in the sample acquisition process. They would also like to thank Dr Dalla Doohan, Dr Kartika Afrida Fauzia, Dr Phawinee Subsomwong, and Dr Junko Akada for their excellent technical assistance. This study was funded by grants from the National Institutes of Health (DK62813) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (221S0002, 16H06279, 15H02657, and 16H05191) (YY). The study was also supported by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Core-to-Core Program: B. Africa-Asia Science Platform (YY). LAW is a doctoral student who was supported by the MEXT Scholarship Program for 2015. In addition, the Ministries of Research, Technology and Higher Education in the World Class Professor Program (123.4/D2.3/KP/2018) also supported this research (MM).
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yoon SB, Park JM, Lim CH, Cho YK, Choi MG. Effect of Helicobacter pylori eradication on metachronous gastric cancer after endoscopic resection of gastric tumors: a meta-analysis. Helicobacter. 2014;19(4):243–248. doi: 10.1111/hel.12146. [DOI] [PubMed] [Google Scholar]
- 2.Ford AC, Forman D, Hunt RH, Yuan Y, Moayyedi P. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. BMJ. 2014;348:g3174. doi: 10.1136/bmj.g3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fock KM, Katelaris P, Sugano K, et al. Second asia-pacific consensus guidelines for Helicobacter pylori infection. J Gastroenterol Hepatol. 2009;24(10):1587–1600. doi: 10.1111/j.1440-1746.2009.05982.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim SG, Jung HK, Lee HL, et al. Guidelines for the diagnosis and treatment of Helicobacter pylori infection in Korea, 2013 revised edition. Korean J Gastroenterol. 2013;62(1):3–26. doi: 10.4166/kjg.2013.62.1.3. [DOI] [PubMed] [Google Scholar]
- 5.Asaka M. A new approach for elimination of gastric cancer deaths in Japan. Int J Cancer. 2013;132(6):1272–1276. doi: 10.1002/ijc.27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinese Society of Gastroenterology, Chinese Study Group on Helicobacter pylori. Liu WZ, Xie Y, et al. Fourth Chinese national consensus report on the management of Helicobacter pylori infection. J Dig Dis. 2013;14(5):211–221. doi: 10.1111/1751-2980.12034. [DOI] [PubMed] [Google Scholar]
- 7.Ghotaslou R, Leylabadlo HE, Asl YM. Prevalence of antibiotic resistance in Helicobacter pylori: a recent literature review. World J Methodol. 2015;5(3):164–174. doi: 10.5662/wjm.v5.i3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro-Jarabo JM, Fernández-Sánchez F, Fernández-Moreno N, et al. Prevalence of primary resistance of Helicobacter pylori to clarithromycin and levofloxacin in Southern Spain. Digestion. 2015;92(2):78–82. doi: 10.1159/000435949. [DOI] [PubMed] [Google Scholar]
- 9.Kuo YT, Liou JM, El-Omar EM, et al. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2017;2(10):707–715. doi: 10.1016/S2468-1253(17)30219-4. [DOI] [PubMed] [Google Scholar]
- 10.Binh TT, Shiota S, Nguyen LT, et al. The incidence of primary antibiotic resistance of Helicobacter pylori in Vietnam. J Clin Gastroenterol. 2013;47(3):233–238. doi: 10.1097/MCG.0b013e3182676e2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aftab H, Miftahussurur M, Subsomwong P, Ahmed F, Khan AK, Yamaoka Y. Helicobacter pylori antibiotic susceptibility patterns in Bangladesh: emerging levofloxacin resistance. J Infect Dev Ctries. 2016;10(3):245–253. doi: 10.3855/jidc.7713. [DOI] [PubMed] [Google Scholar]
- 12.Miftahussurur M, Shrestha PK, Subsomwong P, Sharma RP, Yamaoka Y. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol. 2016;16(1):256. doi: 10.1186/s12866-016-0873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thung I, Aramin H, Vavinskaya V, et al. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43(4):514–533. doi: 10.1111/apt.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peretz A, Paritsky M, Dinisman-Zavulunov E, Pastukh N, Glyatman T, On A. Susceptibility of Helicobacter pylori to levofloxacin and rifampicin in Israel. Microb Drug Resist. 2015;21(4):448–451. doi: 10.1089/mdr.2014.0250. [DOI] [PubMed] [Google Scholar]
- 15.Liang CM, Cheng JW, Kuo CM, et al. Levofloxacin-containing second-line anti-Helicobacter pylori eradication in Taiwanese real-world practice. Biomed J. 2014;37(5):326–330. doi: 10.4103/2319-4170.125650. [DOI] [PubMed] [Google Scholar]
- 16.Megraud F, Coenen S, Versporten A, et al. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62(1):34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 17.Gisbert JP. “Rescue” regimens after Helicobacter pylori treatment failure. World J Gastroenterol. 2008;14(35):5385–5402. doi: 10.3748/wjg.14.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miftahussurur M, Syam AF, Nusi IA, et al. Surveillance of Helicobacter pylori antibiotic susceptibility in indonesia: different resistance types among regions and with novel genetic mutations. Plos One. 2016;11(12):e0166199. doi: 10.1371/journal.pone.0166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Syam AF, Miftahussurur M, Makmun D, et al. Risk factors and prevalence of Helicobacter pylori in five largest islands of indonesia: a preliminary study. PLoS One. 2015;10(11):e0140186. doi: 10.1371/journal.pone.0140186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdullah AA, Abdullah M, Fauzi A, Syam AF, Simadibrata M, Makmun D. The effectiveness of endoscopic retrograde cholangiopancreatography in the management of patients with jaundice at Cipto Mangunkusumo Hospital, Jakarta. Acta Med Indones. 2012;44(4):298–303. [PubMed] [Google Scholar]
- 21.Xiao SD, Liu WZ, Xia DH, et al. The efficacy of furazolidone and metronidazole in the treatment of chronic gastritis associated with Helicobacter (Campylobacter) pylori - a randomized double-blind placebo-controlled clinical trial. Hepatogastroenterology. 1990;37(5):503–506. [PubMed] [Google Scholar]
- 22.Buzás GM, Józan J. Nitrofuran-based regimens for the eradication of Helicobacter pylori infection. J Gastroenterol Hepatol. 2007;22(10):1571–1581. doi: 10.1111/j.1440-1746.2007.05082.x. [DOI] [PubMed] [Google Scholar]
- 23.Ali BH. Pharmacology and toxicity of furazolidone in man and animals: some recent research. Gen Pharmacol. 1989;20(5):557–563. doi: 10.1016/0306-3623(89)90085-2. [DOI] [PubMed] [Google Scholar]
- 24.Kwon DH, Lee M, Kim JJ, et al. Furazolidone- and nitrofurantoin-resistant Helicobacter pylori: prevalence and role of genes involved in metronidazole resistance. Antimicrob Agents Chemother. 2001;45(1):306–308. doi: 10.1128/AAC.45.1.306-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuge L, Wang Y, Wu S, Zhao RL, Li Z, Xie Y. Furazolidone treatment for Helicobacter pylori infection: a systematic review and meta-analysis. Helicobacter. 2018;23(2):e12468. doi: 10.1111/hel.12468. [DOI] [PubMed] [Google Scholar]
- 26.Gisbert JP, Castro-Fernandez M, Perez-Aisa A, et al. Fourth-line rescue therapy with rifabutin in patients with three Helicobacter pylori eradication failures. Aliment Pharmacol Ther. 2012;35(8):941–947. doi: 10.1111/j.1365-2036.2012.05053.x. [DOI] [PubMed] [Google Scholar]
- 27.Kunin CM. Antimicrobial activity of rifabutin. Clin Infect Dis. 1996;22(Suppl_1):S3–S14. doi: 10.1093/clinids/22.supplement_1.s3. [DOI] [PubMed] [Google Scholar]
- 28.Brogden RN, Fitton A, Rifabutin FA. Rifabutin. A review of its antimicrobial activity, pharmacokinetic properties and therapeutic efficacy. Drugs. 1994;47(6):983–1009. doi: 10.2165/00003495-199447060-00008. [DOI] [PubMed] [Google Scholar]
- 29.Akada JK, Shirai M, Fujii K, Okita K, Nakazawa T. In vitro anti-Helicobacter pylori activities of new rifamycin derivatives, KRM-1648 and KRM-1657. Antimicrob Agents Chemother. 1999;43(5):1072–1076. doi: 10.1128/aac.43.5.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heep M, Beck D, Bayerdörffer E, Lehn N. Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob Agents Chemother. 1999;43(6):1497–1499. doi: 10.1128/aac.43.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miftahussurur M, Yamaoka Y. Appropriate first-line regimens to combat Helicobacter pylori antibiotic resistance: an Asian perspective. Molecules. 2015;20(4):6068–6092. doi: 10.3390/molecules20046068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogata SK, Gales AC, Kawakami E. Antimicrobial susceptibility testing for Helicobacter pylori isolates from Brazilian children and adolescents: comparing agar dilution, E-test, and disk diffusion. Braz J Microbiol. 2014;45(4):1439–1448. doi: 10.1590/s1517-83822014000400039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adachi JA, Dupont HL. Rifaximin: a novel nonabsorbed rifamycin for gastrointestinal disorders. Clin Infect Dis. 2006;42(4):541–547. doi: 10.1086/499950. [DOI] [PubMed] [Google Scholar]
- 34.Nishizawa T, Suzuki H, Matsuzaki J, et al. Helicobacter pylori resistance to rifabutin in the last 7 years. Antimicrob Agents Chemother. 2011;55(11):5374–5375. doi: 10.1128/AAC.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami K, Okimoto T, Kodama M, et al. Sitafloxacin activity against Helicobacter pylori isolates, including those with gyrA mutations. Antimicrob Agents Chemother. 2009;53(7):3097–3099. doi: 10.1128/AAC.01552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence consensus report. Gut. 2017;66(1):6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 37.Goh KL, Navaratnam P. High Helicobacter pylori resistance to metronidazole but zero or low resistance to clarithromycin, levofloxacin, and other antibiotics in Malaysia. Helicobacter. 2011;16(3):241–245. doi: 10.1111/j.1523-5378.2011.00841.x. [DOI] [PubMed] [Google Scholar]
- 38.Hunt RH, Xiao SD, Megraud F, et al. Helicobacter pylori in developing countries. world gastroenterology organisation global guideline. J Gastrointestin Liver Dis. 2011;20(3):299–304. [PubMed] [Google Scholar]
- 39.Coelho LG, León-Barúa R, Quigley EM. Latin-American consensus conference on Helicobacter pylori infection. Latin-American national gastroenterological societies affiliated with the Inter-American Association of Gastroenterology (AIGE) Am J Gastroenterol. 2000;95(10):2688–2691. doi: 10.1111/j.1572-0241.2000.03174.x. [DOI] [PubMed] [Google Scholar]
- 40.Xie Y, Zhu Y, Zhou H, et al. Furazolidone-based triple and quadruple eradication therapy for Helicobacter pylori infection. World J Gastroenterol. 2014;20(32):11415–11421. doi: 10.3748/wjg.v20.i32.11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Francesco V, Ierardi E, Hassan C, Zullo A. Is furazolidone therapy for Helicobacter pylori effective and safe? Dig Dis Sci. 2009;54(10):2298–2299. doi: 10.1007/s10620-009-0748-x. [DOI] [PubMed] [Google Scholar]
- 42.Jin X, Tang S, Chen Q, et al. Furazolidone induced oxidative DNA damage via up-regulating ROS that caused cell cycle arrest in human hepatoma G2 cells. Toxicol Lett. 2011;201(3):205–212. doi: 10.1016/j.toxlet.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 43.Ali BH. Pharmacological, therapeutic and toxicological properties of furazolidone: some recent research. Vet Res Commun. 1999;23(6):343–360. doi: 10.1023/a:1006333608012. [DOI] [PubMed] [Google Scholar]
- 44.Koudriakova T, Iatsimirskaia E, Tulebaev S, et al. In vivo disposition and metabolism by liver and enterocyte microsomes of the antitubercular drug rifabutin in rats. J Pharmacol Exp Ther. 1996;279(3):1300–1309. [PubMed] [Google Scholar]
- 45.Pilotto A, Franceschi M, Rassu M, Furlan F, Scagnelli M. In vitro activity of rifabutin against strains of Helicobacter pylori resistant to metronidazole and clarithromycin. Am J Gastroenterol. 2000;95(3):833–834. doi: 10.1111/j.1572-0241.2000.01900.x. [DOI] [PubMed] [Google Scholar]
- 46.Toracchio S, Capodicasa S, Soraja DB, Cellini L, Marzio L. Rifabutin based triple therapy for eradication of H. pylori primary and secondary resistant to tinidazole and clarithromycin. Dig Liver Dis. 2005;37(1):33–38. doi: 10.1016/j.dld.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Gisbert JP, Calvet X. Review article: rifabutin in the treatment of refractory Helicobacter pylori infection. Aliment Pharmacol Ther. 2012;35(2):209–221. doi: 10.1111/j.1365-2036.2011.04937.x. [DOI] [PubMed] [Google Scholar]
- 48.Okumura R, Hirata T, Onodera Y, Hoshino K, Otani T, Yamamoto T. Dual-targeting properties of the 3-aminopyrrolidyl quinolones, DC-159a and sitafloxacin, against DNA gyrase and topoisomerase IV: contribution to reducing in vitro emergence of quinolone-resistant Streptococcus pneumoniae. J Antimicrob Chemother. 2008;62(1):98–104. doi: 10.1093/jac/dkn136. [DOI] [PubMed] [Google Scholar]
- 49.Matsuzaki J, Suzuki H, Nishizawa T, et al. Efficacy of sitafloxacin-based rescue therapy for Helicobacter pylori after failures of first- and second-line therapies. Antimicrob Agents Chemother. 2012;56(3):1643–1645. doi: 10.1128/AAC.05941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujita K, Fujita M, Ito Y, et al. Preliminary evaluation of a sitafloxacin-containing regimen for relapsed or refractory pulmonary Mycobacterium avium complex disease. Open Forum Infect Dis. 2016;3(3):ofw147. doi: 10.1093/ofid/ofw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki H, Nishizawa T, Muraoka H, Hibi T. Sitafloxacin and garenoxacin may overcome the antibiotic resistance of Helicobacter pylori with gyrA mutation. Antimicrob Agents Chemother. 2009;53(4):1720–1721. doi: 10.1128/AAC.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akasaka T, Kurosaka S, Uchida Y, Tanaka M, Sato K, Hayakawa I. Antibacterial activities and inhibitory effects of sitafloxacin (DU-6859a) and its optical isomers against type II topoisomerases. Antimicrob Agents Chemother. 1998;42(5):1284–1287. doi: 10.1128/aac.42.5.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori H, Suzuki H, Matsuzaki J, Masaoka T, Kanai T. Acquisition of double mutation in gyrA caused high resistance to sitafloxacin in Helicobacter pylori after unsuccessful eradication with sitafloxacin-containing regimens. United European Gastroenterol J. 2018;6(3):391–397. doi: 10.1177/2050640617737215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambur OH, Davidsen T, Frye SA, et al. Genome dynamics in major bacterial pathogens. FEMS Microbiol Rev. 2009;33(3):453–470. doi: 10.1111/j.1574-6976.2009.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramas M, Donday MG, McNicholl AG, Gisbert JP. Efficacy and safety of rifaximin associated with standard triple therapy (omeprazole, clarithromycin and amoxicillin) for H. pylori eradication: a phase IV pilot clinical trial. Gastroenterol Hepatol. 2017;40(10):658–662. doi: 10.1016/j.gastrohep.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 56.Gasbarrini A, Gasbarrini G, Pelosini I, Scarpignato C. Eradication of Helicobacter pylori: are rifaximin-based regimens effective? Digestion. 2006;73(Suppl 1):129–135. doi: 10.1159/000089788. [DOI] [PubMed] [Google Scholar]
- 57.Scarpignato C, Pelosini I. Rifaximin, a poorly absorbed antibiotic: pharmacology and clinical potential. Chemotherapy. 2005;51(Suppl 1):36–66. doi: 10.1159/000081990. [DOI] [PubMed] [Google Scholar]
- 58.Song M, Ang TL. Second and third line treatment options for Helicobacter pylori eradication. World J Gastroenterol. 2014;20(6):1517–1528. doi: 10.3748/wjg.v20.i6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kothary V, Scherl EJ, Bosworth B, et al. Rifaximin resistance in Escherichia coli associated with inflammatory bowel disease correlates with prior rifaximin use, mutations in rpoB, and activity of Phe-Arg-β-naphthylamide-inhibitable efflux pumps. Antimicrob Agents Chemother. 2013;57(2):811–817. doi: 10.1128/AAC.02163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Connor JR, Galang MA, Sambol SP, et al. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother. 2008;52(8):2813–2817. doi: 10.1128/AAC.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huhulescu S, Sagel U, Fiedler A, et al. Rifaximin disc diffusion test for in vitro susceptibility testing of Clostridium difficile. J Med Microbiol. 2011;60(Pt 8):1206–1212. doi: 10.1099/jmm.0.028571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang JY, Kim SE, Kim TH, et al. Emergence of rifampin-resistant staphylococci after rifaximin administration in cirrhotic patients. PLoS One. 2017;12(10):e0186120. doi: 10.1371/journal.pone.0186120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams DL, Spring L, Collins L, et al. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42(7):1853–1857. doi: 10.1128/aac.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Mutations associated with rifaximin resistance
| No. | Strain ID | Average coverage | Q30 percentage | Rifaximin |
|---|---|---|---|---|
| 1 | Manado1 | 326.27 | 91.21 | S |
| 2 | Surabaya71 | 346.57 | 90.99 | S |
| 3 | Surabaya47 | 264.51 | 88.23 | R |
| 4 | Surabaya68 | 408.21 | 91.31 | S |
| 5 | Surabaya69 | 504.26 | 88.53 | S |
| 6 | Surabaya71 | 560.21 | 88.06 | S |
| 7 | Surabaya79 | 554.35 | 94.39 | S |
| 8 | Jayapura1 | 570.45 | 95.95 | S |
| 9 | Jayapura3 | 560.85 | 96.31 | S |
| 10 | Jayapura6 | 589.56 | 95.97 | R |
| 11 | Jayapura8 | 231.21 | 84.54 | S |
| 12 | Jayapura15 | 489.67 | 96.36 | S |
| 13 | Jayapura16 | 509.21 | 95.14 | S |
| 14 | Jayapura21 | 502.21 | 96.49 | R |
| 15 | Jakarta9 | 115.74 | 91.23 | R |
| 16 | Medan17 | 142.34 | 80.59 | S |
| 17 | Medan23 | 166.24 | 90.67 | S |
| 18 | Medan27 | 91.58 | 80.76 | S |
| 19 | Medan3 | 93.57 | 80.88 | S |
| 20 | Medan10 | 85.96 | 80.44 | S |
| 21 | Medan11 | 278.2 | 87.68 | S |
| 22 | Medan15 | 260.45 | 80.59 | R |
| 23 | Medan19 | 176.28 | 83.32 | S |
| 24 | Medan20 | 164.28 | 82.27 | S |
| 25 | Medan22 | 115.51 | 82.74 | R |
| 26 | Medan23 | 264.07 | 81.06 | S |
| 27 | Medan25 | 219.52 | 80.97 | R |
| 28 | Medan28 | 169.54 | 80.8 | S |
| 29 | Medan30 | 237.04 | 80.42 | S |
| 30 | Makasar31 | 87.65 | 81.19 | R |
| 31 | Makasar45 | 156.35 | 89.51 | R |
| 32 | Makasar47 | 85.35 | 80.68 | S |
| 33 | Makasar52 | 87.07 | 83.13 | R |
| 34 | Makasar55 | 89.29 | 83.49 | R |
| 35 | Makasar56 | 82.43 | 82.46 | S |
| 36 | Pontianak63 | 82.77 | 88.97 | S |
| 37 | Pontianak75 | 85.13 | 86.89 | R |
| 38 | Pontianak20 | 191.58 | 89.57 | S |
| 39 | Pontianak44 | 128.24 | 89.45 | R |
| 40 | Pontianak50 | 99.51 | 91.01 | R |
| 41 | Manado5 | 171.08 | 89.76 | S |
| 42 | Manado18 | 136.04 | 90.18 | S |
| 43 | Manado20 | 110.47 | 87.37 | S |
| 44 | Manado26 | 151.85 | 86.67 | S |
| 45 | Manado28 | 175.98 | 93.44 | S |
| 46 | Manado29 | 90.53 | 93.63 | R |
| 47 | Manado31 | 102.87 | 93.18 | R |
| 48 | Kupang2 | 156.23 | 82.79 | S |
| 49 | Kupang5 | 132.2 | 85.25 | S |
| 50 | Kupang6 | 120.2 | 83.27 | S |
| 51 | Kupang10 | 105.54 | 82.16 | R |
| 52 | Kupang11 | 196.38 | 80.2 | S |
| 53 | Kupang15 | 192.96 | 82.36 | S |
| 54 | Kupang23 | 125.05 | 84.94 | R |
| 55 | Kupang26 | 98.87 | 89.29 | R |
| 56 | Kupang28 | 146.88 | 86.58 | S |
| 57 | Kupang29 | 144.7 | 81.55 | R |
| 58 | Kupang30 | 85.85 | 85.47 | R |
| 59 | Kupang33 | 158.02 | 83.55 | S |
| 60 | Kupang34 | 238.16 | 87.82 | R |
| 61 | Kupang35 | 240.73 | 85.3 | S |
| 62 | Kupang41 | 112.69 | 85.57 | S |
| 63 | Kupang42 | 86.98 | 81.43 | R |
| 64 | Kupang47 | 142.12 | 85.5 | R |
| 65 | Kupang64 | 171.52 | 82.59 | R |
| 66 | Kupang73 | 134.33 | 88.07 | S |
| 67 | Kupang83 | 152.03 | 85.08 | S |
| 68 | Medan18 | 201.24 | 89.67 | S |
| 69 | Medan31 | 162.6 | 90.39 | S |
| 70 | Medan32 | 264.33 | 88.11 | S |
| 71 | Medan33 | 368.99 | 86.63 | S |
| 72 | Nias9 | 123.05 | 84.92 | S |
| 73 | Medan36 | 97.32 | 84.74 | S |
| 74 | Medan37 | 95.96 | 85.43 | S |
| 75 | Medan40 | 125.98 | 85.11 | S |
| 76 | Medan49 | 163.37 | 83.54 | S |
| 77 | Medan50 | 146.38 | 83.51 | S |
| 78 | Medan56 | 192.02 | 83.51 | R |
| 79 | Medan67 | 223.14 | 85.69 | R |
| 80 | Medan68 | 137.93 | 84.11 | S |
| 81 | Medan73 | 224.19 | 83.87 | S |
| 82 | Medan75 | 194.47 | 83.86 | R |
| 83 | Padang42 | 157.75 | 88.29 | R |
| 84 | Surabaya106 | 84.06 | 80.06 | S |
| 85 | Surabaya137 | 104.09 | 86.49 | S |
| 86 | Surabaya151 | 96.79 | 82.68 | S |
| 87 | Surabaya192 | 106.55 | 85.02 | S |
| 88 | Surabaya283 | 107.28 | 87.04 | R |
| 89 | Surabaya304 | 112.26 | 86.21 | R |
| 90 | Merauke3 | 243.44 | 85.05 | S |
| 91 | Merauke5 | 219.32 | 84.41 | S |
| 92 | Merauke7 | 225.09 | 83.16 | S |
| 93 | Merauke8 | 269.32 | 86.05 | S |
| 94 | Merauke12 | 191.03 | 85.7 | S |
| 95 | Merauke20 | 145.23 | 88.24 | R |
| 96 | Merauke21 | 338.84 | 87.53 | R |
| 97 | Merauke27 | 185.99 | 80.84 | R |
| 98 | Merauke37 | 304.88 | 86.56 | R |
| 99 | Kolaka56 | 219.48 | 85.48 | S |
| 100 | Kolaka72 | 196.53 | 86.86 | R |
| 101 | Kolaka79 | 343.49 | 86.15 | R |
| 102 | Kolaka94 | 256.4 | 83.82 | S |
| 103 | Kolaka96 | 190.85 | 82.58 | R |
| 104 | Kolaka98 | 206.04 | 83.99 | R |
| 105 | Kolaka99 | 279.25 | 81.86 | R |
Abbreviations: R, resistant; S, sensitive.