Abstract
Tumor HPV status is a prognostic factor for oropharyngeal cancer, but classification methods are not standardized. Here we validate HPV classification methods used in United States cooperative group trials. Tumor DNA and RNA purified from 240 paraffin-embedded oropharyngeal cancers diagnosed from 2000–2009 were scored as evaluable if positive for DNA and mRNA controls by quantitative-PCR. Eighteen high-risk (HR)-HPV types were detected in tumors by consensus PCR followed by HR-HPV E6/7 oncogene expression analysis by quantitative reverse-transcriptase PCR. Sensitivity (S), specificity (SP), positive (PPV) and negative predictive value (NPV) of p16 expression detected by immunohistochemistry (IHC) and HPV16 detected by in situ hybridization (ISH) were evaluated in comparison to HR-HPV E6/7 oncogene expression. Inter-rater agreement among three pathologists was evaluated by kappa statistic. Of 235 evaluable tumors, 158 (67%, 95%CI 61.2–73.3) were positive for HR-HPV E6/7 oncogene expression [HPV type 16 (92%), 18 (3%), 33 (3%), 35 (1%) or 58 (1%)]. p16 IHC had high sensitivity [S 96.8%, SP 83.8%, PPV 92.7%, NPV 92.5%] whereas HPV16 ISH had high specificity [S 88.0%, SP 94.7%, PPV 97.2%, NPV 78.9%] for HR-HPV oncogene expression. Inter-rater agreement was excellent for p16 (kappa=0.95–0.98) and HPV16 ISH (kappa 0.83–0.91). Receiver-operating-curve analysis determined the cross-product of p16 intensity score and percent tumor staining to optimally discriminate HR-HPV E6/7-positive and negative tumors. p16 IHC and HPV16 ISH assays have excellent performance, with high sensitivity and specificity, respectively. Appropriate assay choice depends upon clinical implications of a false-positive or negative test.
Keywords: HPV, oropharyngeal carcinoma, cancer, p16
Introduction
Oropharyngeal squamous cell carcinomas (OSCC) are etiologically heterogeneous, with one subset attributable to sexually acquired human papillomavirus (HPV) infection and another to chronic tobacco and alcohol use.8 In addition to distinct risk-factor profiles, differences extend to prognosis.2,7,21,13,20 Tumor HPV status is now considered the single greatest predictor of survival for patients with local-regionally advanced OSCC. Relative to HPV-negative patients, HPV-positive patients have an approximate 60% reduction in risk of death, corresponding to an absolute survival difference of approximately 30% at five years. 2,7,21,13,20
Despite these clinical advances, there are no commercially available, validated and universally accepted tests for the determination of tumor HPV status. Methods commonly utilized include HPV detection by PCR, in situ hybridization (ISH), or use of a surrogate for the function of the high-risk (HR) HPV E7 protein, the detection of p16 by immunohistochemistry (IHC).5 Available algorithms in the literature for the research laboratory combine all three of these assays28,29, but PCR detection of HPV in DNA purified from tumors is not likely to be feasible in most clinical laboratory settings.
A centralized laboratory evaluated tumor HPV status by p16 IHC and HPV16 ISH for the Eastern Cooperative Oncology Group (ECOG)7 and Radiation Therapy Oncology Group (RTOG) trials2 that helped to establish HPV as an important prognostic factor. The same laboratory is determining eligibility for ongoing trials in both cooperative groups. The laboratory methods used for p16 IHC and HPV16 ISH have shown strong agreement3, demonstrated similar prognostic value2, utilize formalin-fixed, paraffin-embedded (FFPE) material, and are feasible in a clinical pathology laboratory.27 Here, we evaluate assay performance in comparison to HR-HPV E6/7 oncogene expression in tumors, the gold standard for categorizing a tumor as caused by HPV.
Materials and methods
Case selection and pathology review
A total of 240 FFPE biopsies from consecutive patients diagnosed with OSCC and available paraffin-embedded tumor in the hospital pathology archives were obtained from the Ohio State University, University of Chicago, University of California San Francisco and Princess Margaret Hospitals (60 cases per site). Four micron sections were cut onto adherent slides that also contained tissue microarrays created from human tumor xenografts of cell lines with low (SiHa, Cat. No. HTB-35, ATCC, Manassas, VA) and high (CaSki, Cat. No. CRL-1550, ATCC, Manassas, VA) copy number HPV16 and an HPV-negative cell line (C33A, Courtesy of Ventana Inc, Tucson, AZ).
DNA and RNA Isolation
A study-specific standard operating procedure was used by all sites for serial sectioning of paraffin embedded tumor blocks. New blades were used for each tumor sample. Sectioning included: H&E verification of tumor in the specimen; four micron sections X 10 mounted on adherent slides and; 10 micron section “paraffin curls” X two placed in eppendorf tubes for DNA and RNA isolation. DNA was isolated from each specimen using proteinase K digestion, phenol–chloroform extraction and ethanol precipitation as previously described.8 Total RNA was extracted using High Pure RNA Paraffin Kits (Roche, Mannheim, Germany) following the manufacturer’s instructions. DNA and RNA quantity and purity were evaluated with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Inc, Wilmington, DE). After DNase treatment, 0.2 to 0.3 μg of total RNA was reverse transcribed to cDNA by using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Carlsbad, California) in 20 μl of reaction volume. Controls with no reverse transcriptase were performed in parallel for each sample.
HPV DNA detection and genotyping
HPV DNA detection and typing was performed by use of the Inno-LiPA HPV Genotyping kits (Innogenetics, Gent, Belgium) following the manufacturer’s instructions. The test is base on PCR amplification of a 65-bp fragment within the L1 open reading frame of HPV genome using the broad spectrum SFP10 biotinylated primers. Biotinylated amplicon are denatured and hybridized with HPV type-specific oligonucleotide probes that are immobilized as parallel lines on membrane strips. The strip contains 28 HPV types (high-risk 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 82; low-risk types 6, 11, 40, 43, 44, 54, 69, 70, 71, 72). After hybridization and stringent washing, streptavidin-conjugated phosphatase is added and bound to any biotinylated hybrids previously formed. Incubation with BCIP/NBT chromogen yields a purple precipitation and the results can be visually interpreted. Inno-LiPA HPV Genotyping hybridization was performed by AutoBlot 3000H machine (MedTec Inc., Chapel Hill, NC).
HPV type-specific quantitative real-time PCR and reverse transcriptase PCR
HPV viral load in tumor tissue for type 16 was measured in all DNA samples (because of its frequent association with OSCC). HPV viral load and E6/7 RNA transcripts reverse transcribed to cDNA for type 18, 26, 31, 33, 35, 39, 51, 52 and 58 were measured in tumors positive by Inno-LiPA HPV genotyping for corresponding HPV type(s) and for any samples positive for p16 but negative by Inno-LiPa, to account for the possibility of deletion of the L1 region in tumors. HPV type-specific PCR for types 31, 33 and 35 were performed using TaqMan quantitative real-time PCR (qPCR) in ABI’s 7300 real-time PCR systems (Applied Biosystems, Foster City, CA) as previously described.8 Additionally, primers and probes for amplification for HPV types 16, 18, 26, 39, 51, 52 and 58 were designed to target on the E6 and/or E7 region. The primers and probes were synthesized by Integrated DNA Technologies (Coralville, IA). Probes were labeled with 6-carboxy-fluorescein (FAM) at the 5′ end and with Black Hole Quencher-1 (BHQ1) at the 3′ end. The sequences for primers (forward and reverse) and probe were as follows: HPV-16 E6 (bp 94–174, GenBank accession no. K02718): 5’-GAGAACTGCAATGTTTCAGGACC-3’, 5’-TGTATAGTTGTTTGCAGCTCTGTGC-3’, and 5’−56-FAM/CAGGAGCGACCCAGAAAGTTACCACAGTT-3BHQ1–3’; HPV-18 E7 (bp 686–775, Genbank accession no. GQ180792): 5’-GTGTGAAGCCAGAATTGAGC-3’, 5’-ACAAAGGACAGGGTGTTCAG-3’, and 5’−56-FAM/ACGACCTTCGAGCATTCCAGCA-3BHQ1–3’; HPV-26 E7 (bp 559–628, Genbank accession no. NC_001583): 5’-TTTGACAGCTCAGATGAGGA-3’, 5’-CTTCTTGTCCAGCTTGTCT-3’, and 5’−56-FAM/ATAATATGCGTGACCAGCAGGC-3BHQ1–3’; HPV-39 E7 (bp 608–676, GenBank accession no. M62849): 5’-ACCCGACCATGCAGTTAATC-3’, 5’-ATTGTGTGACGCTGTGGTTC-3’, and 5’−56-FAM/CCAACATCAACTACTAGCCAGACGGGA-3BHQ1–3’; HPV-51 E6 (bp 378–462, Genbank accession no. M62877): 5’-TGAAATAGCGGGACGTTG-3’, 5’-GCTTTACACTTGGGTTTCG-3’, and 5’−56-FAM/TGC TGG CAA CGT ACA CGA CAA CG-3BHQ1–3’; HPV-52 E7 (bp 641–703, GenBank accession no. GQ472848): 5’-ACAGCTCAGATGAGGAGGA-3’, 5’-TGGCTTGTTTCTGCTTGTCC-3’, and 5’−56-FAM/ACAGATGGTGTGGACCGGCCA-3BHQ1–3’; HPV-58 E6 (bp 489–548, GenBank accession no. GQ472850): 5’-ATATTTCGGGTCGTTGGA-3’, 5’-TTTGTCTAGGTCGGGG-3’, and 5’−56-FAM/CGCTGTGCAGTGTGTTG-3BHQ1–3’. Standard curves for amplification reactions were generated in duplicate by using a fivefold dilution series (from 250,000 to 3.2 copies) of pUC57 vector (GenScript, Piscataway, NJ) containing the complete type-specific E6 and E7 region in a background of human placental DNA (5 ng/μL). Each PCR reaction contained 1X TaqMan universal PCR master mixes (Applied Biosystems, Branchbury, NJ), 0.1 μmol of probe, 0.2 μmol of each primer, and 2 μL of purified tumor DNA or tumor RNA reverse transcribed cDNA. Amplification conditions included one cycle of 2-minute incubation at 50°C (degradation of the uracil containing DNA) and 10-minute incubation at 95°C, followed by 50 cycles of 15 seconds at 95°C and 60 seconds at 60°C. For all TaqMan real-time PCR assays, the cycle threshold (CT) of unknown samples was determined from an equation derived from a linear regression through the log CT of the standard curve according to the manufacturer’s recommendation. Samples above the lower limit of reproducibility of the assays (for all, ≥ three copies) were considered positive.
An estimate of diploid genome equivalents (e.g. cell number) in each sample was determined by TaqMan quantitative real-time PCR targeting on a single-copy human gene on chromosome 7, human endogenous retrovirus 3 (ERV3)1, The 58-bp ERV3 fragment was amplified for 240 samples and the reaction condition were as previously published.8 Briefly, 2 μL of purified tumor tissue DNA was analyzed. A standard curve was generated in duplicate from a fivefold dilution series (from 150,000 to 1.92 cells) of a diploid human cell line, CCD-18LU (ATCC, Manassas, VA). Results were reported as the number of human diploid genome equivalents of purified genomic DNA from tumor samples that were evaluated for HPV type(s) DNA by Q-PCR.
For RNA normalization, human ribosomal protein large, P0 (RPLP0) was chosen as a reference gene for quantitative reverse transcriptase mediated-PCR (qRT-PCR). Pilot studies determine RPLPO expression to be most stable (out of 40 potential control genes) during histopathological progression of cervical cancer (data not shown). The 73-bp RPLP0 fragment was amplified for 240 samples. The primers and probe sequence were as follows: 5’-ACGGGTACAAACGAGTCCTG-3’, 5’-GCCTTGACCTTTTCAGCAAG-3’, 5’−56-FAM/CCTTGTCTGTGGAGACGGAT-3BHQ1–3’. Standard curves for amplification reactions were generated in duplicate from a fivefold dilution series (from 100,000 to 1.28 copies) of pOTB7 vector (Open Biosystems Products, Huntsville, AL) containing 1170 base pairs of RPLP0 mRNA fragment (Gene Bank BC019014) in a background of Salmon Sperm DNA (6 ng/μL, Invitrogen, CARLSBAD, CA ). Each PCR reaction contained 1 × TaqMan universal PCR master mixes, 0.4 μmol of probe, 0.5 μmol of each primer, and 2 μL of cDNA. Amplification conditions included one cycle of 2-minute incubation at 50°C and 10-minute at 95°C, followed by 50 cycles of 15 seconds at 95°C and 60 seconds at 62°C. Results were reported as HPV type 16, 18, 26, 31 33, 35, 39, 52, 58 mRNA expression level normalized to RPLP0 mRNA expression level as evaluated by real-time PCR.
p16 mRNA expression analysis by quantitative reverse transcriptase PCR
Samples were evaluated for mRNA expression of p16 by qRT-PCR with primers designed to amplify transcript variant 1 of the p16 gene (Genbank accession no. NM_000077.4): 5’-TGCCTTTTCACTGTGTTGGA-3’ and 5’-AAATGCCCACATGAATGTGC-3’. The p16 probe sequence was 5’-(FAM)-AGGGCGTGAGTGCTCACTCCA–(BHQ1)-3’. Each PCR reaction contained 1 × TaqMan universal PCR master mixes, 0.1 μmol of probe, 0.2 μmol of each primer, and 2 μL of cDNA with the following reaction conditions: 50°C for 2 minutes to active AmpErase UNG enzyme, 95°C for 12 minutes to active AmpliTaq Gold enzyme, then 50 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Standard curves for amplification reactions were generated in duplicate by using a fivefold dilution series (from 150,000 to 1.92 cells) of CCD-18Lu human lung cell line (ATCC). Results were reported as p16 mRNA expression level normalized to RPLP0 mRNA expression level as evaluated by qRT-PCR.
p16 immunohistochemistry
Automated p16 immunohistochemistry (IHC) stain using the monoclonal anti-p16INK4a (MTM Laboratories, Heidelberg, Germany) was carried out in the BenchMark XT (Ventana, Tucson, AZ) according to the manufacturer’s IHC staining protocol. Details of p16 staining are provided in the supplemental materials. Slides were counterstained using hematoxylin and bluing reagent (Ventana, Tucson, AZ), and mounted under coverslips.
HPV16 in situ hybridization
HPV16 DNA was detected in paraffin-embedded tumor samples by use of the GenPoint catalyzed in situ hybridization signal amplification system for biotinylated probes (Dako, Carpinteria, CA). This signal amplification system can detect a single integrated copy of HPV16 DNA.10 Briefly, tissue sections mounted onto slides were subjected to deparaffinization, heat-induced target retrieval with the use of a steamer, and digestion with proteinase K (20 ug/mL; Roche Diagnostics, Indianapolis, IN) at room temperature. The slide was hybridized at 37ºC overnight with a biotinylated DNA probe that was specific for HPV16 (code Y1407, Dako) and then subjected to low- and high-stringency washes followed by signal amplification with the use of a Tyramide Signal Amplification System kit (code K0620; Dako). The signal was developed by adding diaminobenzidine to the slide for 3–5 minutes and monitoring color change by light microscopy. The sections were counterstained with hematoxylin and mounted under coverslips.
Interpretation of histopathology
H&E stained slides were used to confirm presence of tumor in the sample and to aid in assay interpretation. Interpretation of slides was independently performed by three pathologists (RJ, ML, BPO) in batches containing 40 cases with H&E and corresponding p16 IHC or H&E with corresponding HPV16 ISH. Interpretation was recorded using a web-based reporting system. p16 IHC was scored as evaluable if strong and diffuse positivity was observed in the tissue microarray positive control mounted on each slide. The highest intensity of p16 staining present in the tumor was scored by each pathologist on an ordinal score of 0–3, relative to the intensity of the positive (score 3) and negative (score 0) tissue microarray controls mounted on each slide. The percent of tumor staining at the highest intensity was also estimated within 5% increments. Pathologists also scored tumor as staining positive or negative for p16 based upon the current standard of strong and diffuse nuclear and cytoplasmic staining in ≥70% of the tumor. The H score was derived from the cross product of the intensity score (0 to 3) and the percent of tumor staining at the highest intensity (0–100%).
HPV16 ISH stained slides were scored as evaluable if punctuate nuclear (SiHa) and diffuse nuclear (CaSki) staining was present in the positive control and absent in the negative control tissue microarrays mounted on the slide. Tumors were scored on the following scale: 3+ (multiple confluent nuclear dots); 2+ (multiple punctate dots in tumor nuclei); 1+ (single punctate dots in tumor cells); 0 (no staining). Cases were considered HPV16 positive for cases scoring 1+ or greater.7
After independent interpretation, slides with discrepant categorical interpretation as p16 or HPV16 positive or negative were resolved by joint review for comparison to HPV E6/7 expression analysis.
Statistical analysis
The study was powered to evaluate the inter-reader agreement among three pathologists for a dichotomous categorization of an oropharyngeal tumor as positive or negative for p16 IHC or HPV16 ISH. A sample size of 206 cases provided 80% power to detect Cohen’s kappa of 0.80 or higher, assuming a 0.50 probability of a positive test and a type I error rate of .0167 (Bonferroni adjustment to account for the three pairs of raters). The sample size was adjusted by 10% to 226 to account for the possibility of non-evaluable samples, and therefore each participating institution provided 60 cases (N=240).
Differences between type-specific gold standard HPV-positive and -negative samples in demographic and clinical characteristics were analyzed using contingency table chi-square tests. Nonparametric Mann-Whitney tests were used to determine equality of medians in laboratory testing values. Cohen’s kappa statistic was used to analyze inter-rater agreement for p16 IHC and HPV16 ISH testing among the three pathologists for all pair-wise combinations and to analyze inter-assay agreement. Contingency tables were used to determine sensitivity, specificity, and positive and negative predictive values (and 95% confidence intervals [CI]) for final p16 and HPV16 classification compared to gold standard testing. All reported p values were two-sided.
A receiver operating curve analysis was conducted to examine and compare the classification accuracy on tumor HPV status among three measures, including P16 IHC maximum tumor intensity, percent staining, and their cross product (H-score). For each of the three classification measures, the area under the curve (AUC) was calculated for each individual rater, and the optimal cut-point was determined by the average sensitivities and specificities among the three raters. Stata 10.1 software (StataCorp, College Station, TX) was used for all analyses.
Results
Study population
A total of 240 cases of incident OSCC diagnosed from 2000–2009 were obtained for analysis by request to the collaborating pathologists and shipped by FedEx to the testing center. The demographic and clinical characteristics of the 235 cases that were both eligible and evaluable for HR-HPV E6/7 expression are shown in Table 1. Three cases were found to be ineligible because of a diagnosis of recurrent (rather than incident) cancer (n=1) or a primary tumor located in the oral cavity (n=2). Two cases were not evaluable for viral expression analysis (see below).
Table 1.
Demographic and clinical characteristics of the cases
| HR Type-specific Gold Standard HPV | X2 P-value | |||
|---|---|---|---|---|
| N=235 | n=77 | n=158 | ||
| Factor | N (%) | Neg. (%) | Pos. (%) | |
| Institution* | ||||
| OSU | 58 (24.7) | 20 (26.0) | 38 (24.1) | |
| UCSF | 60 (25.5) | 15 (19.5) | 45 (28.5) | |
| UC | 57 (24.3) | 20 (26.0) | 37 (23.4) | |
| PMH | 60 (25.5) | 22 (28.6) | 38 (24.1) | 0.518 |
| Gender | ||||
| Female | 58 (24.7) | 26 (33.8) | 32 (20.3) | |
| Male | 177 (75.3) | 51 (66.2) | 126 (79.7) | 0.024 |
| Anatomic subsite | ||||
| Base of tongue | 86 (36.6) | 34 (44.2) | 52 (32.9) | |
| Pharyngeal wallsa | 7 (3.0) | 6 (7.8) | 1 (0.6) | |
| Soft palate | 4 (1.7) | 4 (5.2) | 0 (0) | |
| Tonsil/tonsillar pillar | 138 (58.7) | 33 (42.9) | 105 (66.5) | <0.001 |
| Staging type | ||||
| Clinical | 163 (69.4) | 55 (71.4) | 108 (68.4) | |
| Pathological | 71 (30.2) | 22 (28.6) | 49 (31.0) | |
| Missing | 1 (0.4) | 0 (0) | 1 (0.6) | 0.840 |
| AJCC tumor stage | ||||
| T1 | 75 (31.9) | 16 (20.8) | 59 (37.3) | |
| T2 | 87 (37.0) | 27 (35.1) | 60 (38.0) | |
| T3 | 32 (13.6) | 14 (18.2) | 18 (11.4) | |
| T4 | 36 (15.3) | 19 (24.7) | 17 (10.8) | |
| Missing | 5 (2.1) | 1 (1.3) | 4 (2.5) | 0.009 |
| AJCC nodal stage | ||||
| N0 | 55 (23.4) | 31 (40.3) | 24 (15.2) | |
| N1 | 38 (16.2) | 7 (9.1) | 31 (19.6) | |
| N2 | 109 (46.4) | 31 (40.3) | 78 (49.4) | |
| N3 | 8 (3.4) | 1 (1.3) | 7 (4.4) | |
| Nx | 22 (9.4) | 5 (6.5) | 17 (10.8) | |
| Missing | 3 (1.3) | 2 (2.6) | 1 (0.6) | <0.001 |
| AJCC metastasis stage | ||||
| M0 | 115 (48.9) | 37 (48.1) | 78 (49.4) | |
| M1 | 9 (3.8) | 6 (7.8) | 3 (1.9) | |
| Mx | 86 (36.6) | 24 (31.2) | 62 (39.2) | |
| Missing | 25 (10.6) | 10 (13.0) | 15 (9.5) | 0.110 |
Calendar period of case diagnosis: OSU, 2009; UCSF, 2000–2009; UC, 2005–2009; PMH, 2004–2009
Lateral and posterior
HPV DNA and RNA expression analysis
An analysis of the DNA quantity and quality purified from tumor specimens determined 233 tumors to be evaluable by Q-PCR for the ERV3 gene. By ERV3, a median of 34,206 cells (interquartile range [IQR]: 17.179–62,556) per sample were evaluated for HPV DNA. Median DNA yield was 5.74 ug (IQR 1.92–13.78) per 10 micrometer paraffin section with a median 260/280nm ratio of 1.62 (IQR 1.59–1.66).
When evaluated for presence of HPV DNA by consensus primer PCR targeted to the viral capsid gene L1, 184 of 233 (78.3%, 95% CI 73.0–83.6) cases were positive. HPV16 was detected in the majority of positive samples (n=170, 90.8%). Additional HR-HPV types detected included: 18 (n=6), 33 (n=4), 35 (n=2), 51 (n=1), and 58 (n=2). Two tumors were positive for more than one HR-HPV type and two were positive for low-risk HPV type 6 or 11 and were not further evaluated for HPV E6/7 expression.
Expression of HR-HPV oncogenes E6/7 remains the gold standard for categorizing a tumor as caused by HPV. When samples were analyzed for expression of the endogenous control gene RPLPO by qRT-PCR, 235 of 237 (99.2%) were positive and therefore evaluable. The quantity and quality of total RNA extracted from tumor samples was good, with a median RNA yield of 3.39 ug (IQR:1.33–9.38) per 10 micrometer paraffin section and a median 260/280nm ratio of 2.04 (IQR 1.96–2.08) by spectrophotometry.
When evaluable samples were analyzed for expression of HPV16 E6/7 by qRT-PCR, 146 of 235 (62.1%, 95%CI: 55.9–68.4) were positive, including two samples negative for HPV16 by consensus PCR (consistent with deletion of the L1 region in tumors). Additionally, 12 of 15 cases positive for HR-HPV DNA other than type 16 were positive for E6/7 oncogene expression, yielding a final total of 67.2% (95% CI 61.2–73.3) positive tumors by the gold standard test. HPV type distribution for E6/7 expression-positive samples was: 16, 93.4%; 18, 2.5%; 33, 2.5%; 35, 1.3%; 58, 1.3%. Median E6/7 expression levels were lower for type 16 positive tumors than for other HR-HPV types (205.7 vs. 746.7 copies per 1000 RPLPO), thus confirming true positives for non-16 types. Overall, only 85.9% (158 of 184) of HPV DNA-positive tumors were confirmed as positive for E6/7 oncogene expression (Supplementary Table 2). Viral copy number per cell was significantly higher among cases positive versus negative for viral oncogene expression (median 15.7 vs. 0 copies per cell, p<0.001).
When compared to cases negative for HR-HPV E6/7 expression, positive cases were significantly more common among men and were more likely to be of tonsillar origin, early tumor stage (T1–2), and advanced nodal stage (N2–3) (Table 1).
p16 IHC and HPV16 ISH interpretation
Following review 231 of 235 cases assessed by p16 IHC had tumor present, had evaluable controls and were considered evaluable (Figure 1). As shown in Table 2, inter-rater agreement on interpretation of p16 expression as positive or negative in tumors was very high in all pair-wise comparisons for the three raters (for all, kappa>0.90). After resolution by joint review of tumors with discrepant interpretation, 70.2% (95%CI 64.3–76.1) were p16-positive.
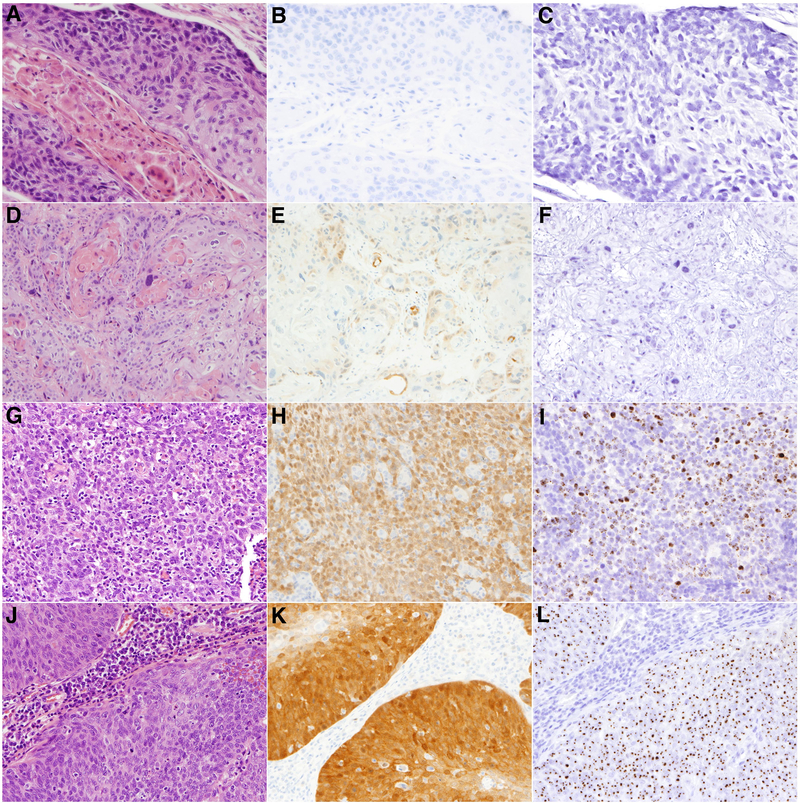
Figure 1. Representative cases of p16 immunohistochemistry (IHC) and HPV16 in situ hybridization (ISH).
Shown are cases of oropharyngeal squamous cell carcinoma evaluated by hematoxylin and eosin staining (panels A, D, G, J), p16 expression by IHC (panels B, E, H, K) and for HPV16 presence by ISH (panels C, F, I, L). Row 1: An example of a p16 negative (B) and HPV16 ISH negative case (C). Row 2: An example of a case with an H-score equal to 15 [p16 IHC intensity score 1 × 15% positive (E)] and negative HPV16 ISH (F). Row 3: An example of a case with an H-score of 160 [p16 intensity score 2 × 80% positive] (H) and positive HPV16 ISH score 3 (I) with multiple confluent brown signal reactions in tumor nuclei. Row 4: An example of a case with an H-score of 285 [p16 intensity score 3 × 95% positive] (K) and positive HPV16 ISH score 2 (L) with single and multiple brown signal reactions in tumor nuclei.
Table 2.
Inter-rater agreement, p16 IHC and HPV16 ISH results
| Rater 2 | Kappa stat | Rater 3 | Kappa stat | |||
|---|---|---|---|---|---|---|
| Site IHC | Neg. | Pos. | (95% CI) | Neg. | Pos. | (95% CI) |
| p16 IHCa | ||||||
| Rater 1 | ||||||
| Negative | 67 | 2 | 0.979 | 63 | 7 | 0.926 |
| Positive | 0 | 162 | (0.950, 1.0) | 0 | 162 | (0.873, 1.0) |
| Rater 2 | ||||||
| Negative | 63 | 4 | 0.957 | |||
| Positive | 0 | 164 | (0.916, 0.999) | |||
| HPV16 ISHb | ||||||
| Rater 1 | ||||||
| Negative | 90 | 1 | 0.964 | 87 | 3 | 0.973 |
| Positive | 3 | 139 | (0.929, 0.999) | 0 | 142 | (0.942, 1.0) |
| Rater 2 | ||||||
| Negative | 86 | 6 | 0.936 | |||
| Positive | 1 | 139 | (0.890, 0.983) | |||
Due to rater evaluation of tumor missing on IHC slide: Rater 1/2, n=231; Rater 1/3, n=232; Rater 2/3, n=231
Due to rater evaluation of tumor missing on ISH slide: Rater 1/2, n=233; Rater 1/3, n=232; Rater 2/3, n=232
All tumors were evaluated for p16 mRNA expression. Median values for p16 transcript expression were significantly higher for tumors positive vs. negative for p16 IHC after consensus review (median 92.0 vs. 4.1 copies per 1000 RPLPO) and for HR-HPV E6/7 expression (median 93.5 vs. 5.8 copies per 1000 RPLPO).
Upon review, 232 of 235 cases evaluated by HPV16 ISH had tumor present on the slide, had evaluable positive and negative controls on the slide, and were therefore considered evaluable for this analysis (Figure 1). As shown in Table 2, inter-rater agreement on interpretation of HPV16 ISH was extraordinarily high in all pair-wise comparisons for the three raters (for all, kappa>0.90). After resolution by joint review of tumors with discrepant interpretation, 60.9% (95%CI 54.6–67.1) were HPV16 ISH positive. Agreement between the p16 IHC and HPV16 ISH assays was very good (kappa=0.70, Supplementary Table).
Comparison to the gold standard of high-risk HPV oncogene expression
We evaluated the performance of p16 IHC and HPV16 ISH in comparison to the gold standard test of HPV oncogene expression for type 16 alone and for all high-risk types (Table 3). The p16 IHC assay had very high sensitivity for HPV16 E6/7 expression. Specificity was 72.1% for HPV16 E6/7 expression, but increased to 83.8% when compared to HR-HPV E6/7 expression. The HPV16 ISH assay had very high sensitivity, specificity, positive and negative predictive value in comparison to HPV16 E6/7 expression (Table 3). Assay sensitivity declined from 96.6% for HPV16 E6/7 expression to 88.0% in comparison to HR-HPV E6/7 expression.
Table 3.
p16 IHC/HPV16 ISH sensitivity analyses
| Gold Standard HPV Test Resultsc | Sensitivity | Specificity | PPV | NPV | Area under curve | ||
|---|---|---|---|---|---|---|---|
| Test | Neg. | Pos. | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) |
| HPV16-type specific | |||||||
| p16 IHC (n=232) | |||||||
| Negative | 62 | 5 | 96.6% | 72.1% | 85.5% | 92.5% | 0.84 |
| Positive | 24 | 141 | (92.2, 98.9) | (61.4, 81.2) | (79.1, 90.5) | (83.4, 97.5) | (0.79, 0.89) |
| HPV16 ISH (n=233) | |||||||
| Negative | 80 | 10 | 93.2% | 92.0% | 95.1% | 88.9% | 0.93 |
| Positive | 7 | 136 | (87.8, 96.7) | (84.1, 96.7) | (90.2, 98.0) | (90.5, 94.5) | (0.89, 0.96) |
| IHC/ISH Combined (n=232) | |||||||
| Both Positivea | |||||||
| Negative | 81 | 13 | 91.1% | 94.2% | 96.4% | 86.2% | 0.93 |
| Positive | 5 | 133 | (85.3, 95.2) | (87.0, 98.1) | (91.7, 98.8) | (77.5, 92.4) | (0.89, 0.96) |
| Either Positiveb | |||||||
| Negative | 60 | 2 | 98.6% | 69.8% | 84.7% | 96.8% | 0.84 |
| Positive | 26 | 144 | (95.1, 99.8) | (58.9, 79.2) | (78.4, 89.8) | (88.8, 99.6) | (0.79, 0.89) |
| HR-HPV Type specific | |||||||
| p16 IHC (n=232) | |||||||
| Negative | 62 | 5 | 96.8% | 83.8% | 92.7% | 92.5% | 0.90 |
| Positive | 12 | 153 | (92.8, 99.0) | (73.4, 91.3) | (87.6, 96.2) | (83.4, 97.5) | (0.86, 0.95) |
| HPV16 ISH (n=233) | |||||||
| Negative | 71 | 19 | 88.0% | 94.7% | 97.2% | 78.9% | 0.91 |
| Positive | 4 | 139 | (81.9, 92.6) | (86.9, 98.5) | (93.0, 99.2) | (69.0, 86.8) | (0.88, 0.95) |
| IHC/ISH Combined (n=232) | |||||||
| Both Positivea | |||||||
| Negative | 72 | 22 | 86.1% | 97.3% | 98.6% | 76.6% | 0.92 |
| Positive | 2 | 136 | (79.7, 91.1) | (90.6, 99.7) | (94.9, 99.8) | (66.7, 84.7) | (0.88, 0.95) |
| Either Positiveb | |||||||
| Negative | 60 | 2 | 98.7% | 81.1% | 91.8% | 96.8% | 0.90 |
| Positive | 14 | 156 | (95.5, 99.8) | (70.3, 89.3) | (86.6, 95.4) | (88.8, 99.6) | (0.85, 0.94) |
Sensitivity analyses for combined ICH/ISH test results set both positive as a positive test and all others (either one or both test negative) as a negative test
Sensitivity analyses for combined ICH/ISH test results set either positive as a positive test and both negative as a negative test
The gold standard test for HPV status is defined as either positive for HPV16 E6/7 expression (top) or for high-risk HPVE6/7 expression (bottom, including types 16, 18, 31, 33, 35, 52).
When p16 IHC and HPV16 ISH testing were evaluated in combination (Table 3), HPV E6/7 expression was present in 135 (98.5%) of 137 cases positive for both, 18 (60%) of 30 p16-positive/HPV16 ISH-negative, 3 (50%) of 6 p16-negative/HPV16-positive and 2 (3.3%) of 61 negative-negative samples. Therefore, a combination of both HPV16 ISH positive and p16 positive had highest specificity in comparison to the gold standard test, with a false positive rate of ~3%. By contrast, use of a combination of either p16 IHC or HPV16 ISH positive will result in the highest sensitivity, but will result in a false-positive rate of ~19%.
Of the 27 (11.4%) cases with discrepant p16 IHC and HPV16 ISH analysis, 12 were found to be HR-HPV E6/7 positive. Analysis of the remaining 15 p16-positive tumors for HPV DNA or RNA revealed all were negative for HPV 16, 18, 31, 33, 35, 45, 52, and 58.
ROC analysis was performed to determine optimal cut-points for p16 IHC interpretation in comparison to gold standard HR-HPV oncogene expression (Table 4). For all three raters, the AUC for percent staining was higher than those for intensity score, indicating that percent staining as a single classification measure was better at discriminating the tumor HPV status. Significant differences in AUC were observed for both intensity score and percent staining among pathologists (for both, p-value equality of areas < 0.02). After averaging among the three raters, a p16 intensity score cut-point of 2 on a scale of 0–3 was most sensitive and percent staining cut-point of 35% on a scale of 0–100% was most specific for HR-HPV E6/7 expression (Table 4). Use of the cross-product of these measures (H-score) resulted in a better clarification of tumor HPV status. An optimal H-score cut-point of 60 on a scale of 0–300 yielded an average sensitivity of 91.6% and specificity of 90.4% for HR-HPV oncogene expression.
Table 4.
Receiver operator characteristic curve analysis for p16 intensity and staining in comparison to HR-HPV E6/7 analysis
| Classification Variable | Area Under Curve (95% CI) | Optimal Cutoff | ||||
|---|---|---|---|---|---|---|
| R1 | R2 | R3 | Cutoff Point | Average Sensitivity (95% CI) | Average Specificity (95% CI) | |
| P16 IHC Maximum Tumor Intensity | 0.9243 (0.8847, 0.9639) | 0.9423 (0.9077, 0.9769) | 0.9193 (0.8777, 0.9609) | 2 | 94.94% (89.95%, 97.60%) | 84.93% (74.24%, 91.85%) |
| P16 IHC Percent at Maximum Tumor Intensity | 0.9412 (0.9084, 0.9741) | 0.9590 (0.9301, 0.9879) | 0.9295 (0.8881, 0.9710) | 35% | 87.98% (81.98%, 92.03%) | 90.09% (80.49%, 95.34%) |
| H-Score | 0.9491 (0.9183, 0.9799) | 0.9570 (0.9288, 0.9852) | 0.9318 (0.8919, 0.9716) | 60 | 91.56% (85.93%, 95.07%) | 90.41% (80.70%, 95.69%) |
Discussion
The p16 IHC and HPV16 ISH assays used in past and ongoing cooperative cancer group trials in the United States for classification of tumor HPV status have excellent assay performance, with high sensitivity and specificity, respectively, for expression of HR-HPV E6/7 oncogenes and excellent inter-rater agreement on interpretation. The sensitivity of HPV16 ISH is limited by the presence of a small proportion of HR-HPV types other than HPV16 in tumors and the specificity of p16 is limited by the presence of p16-positive tumors that are without evidence of HPV DNA or E6/7 expression.
Determination of HPV status for OSCC is rapidly becoming the standard of care, with the majority of pathologists reporting current1 or future intent26 to evaluate all tumors. Tumor HPV status is now accepted as a strong, independent prognostic factor for oropharynx cancer2,9 is predictive of response to treatment with cisplatin induction chemotherapy7,35 and radiotherapy5, and can aid in the differential diagnosis of cystic neck lesions6 and localization of an unknown primary.3,16 The importance of HPV testing is underscored by the increasing number of organizations recommending its use, including the National Comprehensive Cancer Network (NCCN)19, The College of American Pathologists, and The Collaborative Stage Data Collection System, utilized by associations such as American Joint Committee on Cancer and Surveillance Epidemiology and End Results program. Unfortunately, there is no current standard for testing or interpretation of HPV detection assays, and each assay has technical limitations. However, p16 IHC and HPV16 ISH are currently preferred for prognostication, because these methods have been used in the clinical trials that established HPV as an important prognostic factor for these cancers.3,7 Additionally, these assays provide comparable prognostic value to detection of HPV16 E6/7 expression.25
Previous analyses have reported that as many as 50% of HPV-DNA positive tumors are negative for E6/7 mRNA expression.34 In our analysis, 14% of HPV DNA-positive tumors were negative for HR-HPV E6/7 expression and had very low HPV DNA viral load, arguing against use of PCR alone for classification of HPV status. The HPV DNA PCR-positive/p16 IHC-negative tumor once considered a biologically unique class of tumor is likely explained by these false-positive tests.34
HPV ISH using either type-specific or probe “cocktails” can be performed on clinical FFPE and localizes virus topographically to tumor. Although our HPV16 ISH assay has single-copy sensitivity and excellent assay performance in comparison to HPV16 E6/7 expression, it is type-specific, non-automated, technically difficult to perform and not commercially available. Our findings are difficult to compare to other recent analyses of HPV ISH assay performance due to differences in laboratory methodology and use by other investigators of a gold standard of HPV DNA detection by PCR alone.8,29,32 However, as recently reported by Schlecht and colleagues,23 commercially available assays by Ventana (INFORM HPV-III Fam16B) and Dako (HPV16/18) appeared to have less impressive assay performance when compared to HPV16 E6/7 expression (AUC 0.48–0.69).
An increasingly common alternative to HPV ISH is determination of p16 protein expression by IHC. In HPV-associated cancers, p16 is frequently over-expressed due to inactivation of pRB by the HR-HPV E7 oncoprotein and consequent release of pRb-mediated negative regulation of p16.15 In the histopathological progression of cervical cancer, p16 expression increases with severity of dysplasia and cancer and is a possible adjunct to HPV testing for triage of mild vs. moderate or greater dysplasia.11,24 However, as is the case for oropharynx cancers, lack of standardization of testing and interpretation of p16 IHC has in part delayed introduction into the clinic.31
Comparable assay performance for p16 to that observed here has been previously reported23,29, even when a different monoclonal antibody to p16 was utilized. Similarly high agreement on inter-rater interpretation has also been reported23,29, indicating the familiarity of pathologists with interpretation of IHC assays. The currently recommended cut-point for defining a positive p16 IHC assay - strong and diffuse nuclear and cytoplasmic staining of 70% or greater of tumor -was largely experientially determined.3 In this report, the optimal cut-point was evaluated based upon the area under the receiver operating characteristics curves in comparison to HR-HPV E6/7 expression. Our cut-point for staining intensity agreed with the empirically determined cut-point of 2 or greater. However, the optimal cut-point for percent staining was lower at 35% or greater. The H-score cut-point of 60 indicates that a tumor with diffuse low-intensity nuclear and cytoplasmic p16 staining in the majority of the tumor is a true positive, likely because of the effect of highly variable tumor fixation on the intensity of staining. The effect of this alternate cut-point for interpretation of p16 IHC on survival analyses remains to be evaluated.
Test results for p16 IHC and HPV16 ISH had very good agreement. As previously reported2,4,27 approximately 15% of cases have discordant test results and are most frequently p16-positive and HPV ISH-negative. Our data indicate that half of these are attributable to HPV types other than type 16. The resulting misclassification caused by the type-specificity of our ISH assay explains the relative increase in absolute survival difference for p16-positive vs. negative as compared to HPV16-positive vs. negative patients reported in the analysis of RTOG 0129 by Ang et al. 2 As noted in the discussion of the paper by Ang et al., the wide-spectrum HPV ISH assay used in that analysis has unknown sensitivity for non-16 HPV types. These data have, unfortunately, been misinterpreted as evidence that p16 expression has prognostic significance independent of tumor HPV status for OSCC. Indeed, a recent report that p16 had independent prognostic import was complicated by the fact that subsequent testing revealed the majority of p16-positive and HPV-negative tumors to have HPV E6/7 expression.14,18,32 P16 expression has also been recommended as a means by which to discriminate tumors in which HPV does and does not play a biologically meaningful role.34 Although only comprising ~3% of OCSS overall, here we demonstrate that HPV ISH-positive/p16-negative tumors have HR-HPV E6/7 expression and are therefore etiologically associated with HPV. The specific molecular alterations that underlie p16 overexpression in the absence of demonstrable HPV expression as well as lack of p16 expression in the presence of HR-HPV-E6/7 expression are not yet defined, and the clinical outcome of patients with true discordant results remains unknown18 due to small subsets in correlative studies within clinical trials to date. Of note, p16 expression is being evaluated as a possible prognostic factor in cancers at numerous anatomic sites, including lung20, oral cavity cancers17 and prostate cancers12, among others.22 Our data should not be used to guide interpretation of p16 IHC testing at non-oropharyngeal sites.
The appropriate assay to use either singly or in combination will depend upon the clinical implications of a false-positive or false-negative test. Our analyses indicate that p16 IHC or HPV16 ISH alone may result in misclassification of approximately 17–19% of tumors, with the majority for each test being false-positive and false-negative tests, respectively. If used for clinical trial eligibility, p16 IHC testing alone will facilitate enrollment of patients without a true diagnosis of HPV-associated cancer, and the effect of the inclusion of these patients on clinical trial design and outcomes is unpredictable. For clinical trials evaluating “de-intensification” strategies for patients with HPV-associated OSCC, combined p16 IHC and HPV16 ISH is necessary to provide the high specificity required to avoid possible under treatment of patients without a diagnosis of HPV-associated OSCC. Combined testing also aids in prognostication, given the outcome of patients with concordant tests is clear, whereas either test alone may result in erroneous communication regarding the patient’s true prognosis. By contrast, either assay can be used for stratification of patients within clinical trials to assure arms are balanced by p16 or HPV status.
In conclusion, the laboratory methods currently used to determine eligibility for RTOG and ECOG trials have high sensitivity and specificity for OSCC caused by HPV. A validated, commercial assay is clearly desperately needed, as these assays are increasingly being used for decision making for the individual patient. The optimal assay would combine the sensitivity of p16 IHC with the specificity of HPV ISH, with an expanded HPV probe cocktail to account for HR-HPV types other than HPV16.
Supplementary Material
Acknowledgements
Funding provided by The Ohio State University Comprehensive Cancer Center and the National Institute of Dental and Craniofacial Research (DE016631). Ventana Inc., Tucson, AZ, kindly provided some research reagents. The industry sponsor had no role in study design, execution, analysis or writing of the manuscript. The authors thank Andrea Inman for her assistance in study management.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
(Disclosures: Maura Gillison was the recipient of loaned equipment and reagents from Ventana Corp. that assisted with the study. The other authors have no conflicts of interest or funding to disclose)
References
- 1.Ahmed A, Cascarini L, Sandison A, Clarke P. Survey of the use of tests for human papilloma virus and epidermal growth factor receptor for squamous cell carcinoma of the head and neck in UK head and neck multidisciplinary teams. Br J Oral Maxillofac Surg 2011; 3:1–5 [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. New Eng J Med 2010;363:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clin Cancer Res 2003;9:6469–6475. [PubMed] [Google Scholar]
- 4.Begum S, Gillison ML, Nicol TL, Westra WH. Detection of human papillomavirus-16 in fine-needle aspirates to determine tumor origin in patients with metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res 2007;13:1186–1191. [DOI] [PubMed] [Google Scholar]
- 5.Cantley RL, Gabrielli E, Montebelli F, Cimbaluk D, Gattuso P, Petruzzelli G. Ancillary studies in determining human papillomavirus status of squamous cell carcinoma of the oropharynx: a review. Patholog Res Int 2011;2011:138469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao D, Begum S, Ali SZ, Westra WH. Expression of p16 in benign and malignant cystic squamous lesions of the neck. Hum Pathol 2010;41:535–539. [DOI] [PubMed] [Google Scholar]
- 7.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008;100:261–269 [DOI] [PubMed] [Google Scholar]
- 8.Gillison ML, D’Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst 2008;100:407–420. [DOI] [PubMed] [Google Scholar]
- 9.Gillison M HPV and its effect on head and neck cancer prognosis. Clin Adv Hematol Oncol 2010;8:680–682. [PubMed] [Google Scholar]
- 10.Huang CC, Qiu JT, Kashima ML, Kurman RJ, Wu TC. Generation of type-specific probes for the detection of single-copy human papillomavirus by a novel in situ hybridization method. Mod Pathol 1998;11:971–977. [PubMed] [Google Scholar]
- 11.Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 2001;92:276–284 [DOI] [PubMed] [Google Scholar]
- 12.Kudahetti SC, Fisher G, Ambroisine L, et al. Immunohistochemistry for p16, but not Rb or p21, is an independent predictor of prognosis in conservatively treated, clinically localised prostate cancer. Pathology 2010;42:519–523. [DOI] [PubMed] [Google Scholar]
- 13.Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol 2009;27:1992–1998. [DOI] [PubMed] [Google Scholar]
- 14.Lewis JS Jr., Thorstad WL, Chernock RD, et al. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol 2010;34:1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Nakano J, Ueno M, et al. A useful protocol for analyses of mutations of the epidermal growth factor receptor gene. Oncol Rep 2006;15:1503–1505. [PubMed] [Google Scholar]
- 16.Park JM, Jung CK, Choi YJ, et al. The use of an immunohistochemical diagnostic panel to determine the primary site of cervical lymph node metastases of occult squamous cell carcinoma. Hum Pathol 2010;41:431–437. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Sayans M, Suarez-Penaranda JM, Gayoso-Diz P, Barros-Angueira F, Gandara-Rey JM, Garcia-Garcia A. p16(INK4a)/CDKN2 expression and its relationship with oral squamous cell carcinoma is our current knowledge enough? Cancer Lett 2011;306:134–141. [DOI] [PubMed] [Google Scholar]
- 18.Perrone F, Gloghini A, Cortelazzi B, Bossi P, Licitra L, Pilotti S. Isolating p16-positive/HPV-negative oropharyngeal cancer: an effort worth making. Am J Surg Pathol 2011;35:774–777. [DOI] [PubMed] [Google Scholar]
- 19.Pfister DG, Ang KK, Brizel DM, et al. Head and neck cancers. J Natl Compr Canc Netw 2011;9:596–650. [DOI] [PubMed] [Google Scholar]
- 20.Posner MR, Lorch JH, Goloubeva O, et al. Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann Oncol 2011;22:1071–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rischin D, Young RJ, Fisher R, et al. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol 2010;28:4142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romagosa C, Simonetti S, Lopez-Vicente L, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 2011;30:2087–2097. [DOI] [PubMed] [Google Scholar]
- 23.Schlecht NF, Brandwein-Gensler M, Nuovo GJ, et al. A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer. Modern Pathol 2011;24:1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt D, Bergeron C, Denton KJ, Ridder R. p16/ki-67 dual-Stain cytology in the triage of ASCUS and LSIL papanicolaou cytology: Results from the european equivocal or mildly abnormal papanicolaou cytology study. Cancer Cytopathol 2011;119:158–166. [DOI] [PubMed] [Google Scholar]
- 25.Shi W, Kato H, Perez-Ordonez B, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol 2009;27:6213–6221. [DOI] [PubMed] [Google Scholar]
- 26.Shoushtari AN, Rahimi NP, Schlesinger DJ, Read PW. Survey on human papillomavirus/p16 screening use in oropharyngeal carcinoma patients in the United States. Cancer 2010;116:514–519. [DOI] [PubMed] [Google Scholar]
- 27.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer 2010;116:2166–2173. [DOI] [PubMed] [Google Scholar]
- 28.Smeets SJ, Hesselink AT, Speel EJ, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer 2007;121:2465–2472. [DOI] [PubMed] [Google Scholar]
- 29.Thavaraj S, Stokes A, Guerra E, et al. Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol 2011;64:308–312. [DOI] [PubMed] [Google Scholar]
- 30.Tong J, Sun X, Cheng H, et al. Expression of p16 in non-small cell lung cancer and its prognostic significance: A meta-analysis of published literatures. Lung Cancer 2011; 74(2):155–163. [DOI] [PubMed] [Google Scholar]
- 31.Tsoumpou I, Arbyn M, Kyrgiou M, et al. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev 2009;35:210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS Jr. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2011;35:1343–1350. [DOI] [PubMed] [Google Scholar]
- 33.van Houten VM, Snijders PJ, van den Brekel MW, et al. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer 2001;93:232–235. [DOI] [PubMed] [Google Scholar]
- 34.Weinberger PM, Yu Z, Haffty BG, et al. Molecular classification identifies a subset of human papillomavirus--associated oropharyngeal cancers with favorable prognosis. J Clin Oncol 2006;24:736–747. [DOI] [PubMed] [Google Scholar]
- 35.Worden FP, Kumar B, Lee JS, et al. Chemoselection as a strategy for organ preservation in advanced oropharynx cancer: response and survival positively associated with HPV16 copy number. J Clin Oncol 2008;26:3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.