ABSTRACT
The micro-environment of cancer cells in the body is mechanically stiffer than that of normal cells. We cultured three breast cell lines of MCF10A-normal, MCF7-noninvasive, and MDA-MB-231-invasive on PDMS substrates with different elastic moduli and different cellular features were examined.Effects of substrate stiffness on cell behavior were evident among all cell lines. Cancerous cells were more sensitive to substrate stiffness for cell behaviors related to cell motility and migration which are necessary for invasion. The invasive cancerous cells were the most motile on substrates with moderate stiffness followed by non-invasive cancerous cells. Gene markers alterations were generally according to the analyzed cell movement parameters. Results suggest that alterations in matrix stiffness may be related to cancer disease and progression.
KEYWORDS: Substrate Stiffness, mammary cancer cell lines, different invasiveness, motility, morphology, proliferation, gene expression
Introduction
Breast cancer is the most prevalent cancer worldwide which is usually diagnosed by the physical palpation of a rigid mass situated on the soft surrounding [1]. The mechanisms by which tumours are formed and progress and later metastasize to other tissues have been vastly studied, however the role of mechanical cues in modulating the growth and progression of the breast cancer cells remain poorly defined. It was demonstrated that an estimate of breast tissue stiffness was associated with breast cancer risks [2]. Furthermore, the atomic force microscopy (AFM) measurements of breast tissues extracted from patients showed that they had a broad range of elastic modulus and usually were stiffer than normal tissues in extracellular matrix regions while cancer cells were softer than normal cells [3], indicating that breast cancer cells develop in a niche stiffer than a normal breast tissue.
In addition to genetic parameters associated with progression of cancer cells, external structural factors have been suggested to be effective [4], such as stiffening of the extracelluar matrix (ECM) that is shown to be associated with tumour development. An in vitro study showed that matrix stiffness altered expression of some genes in hepatocellular carcinoma especially markers that were related to the cell-cell or cell-substrate adhesions. Moreover, matrix stiffening decreased effects of medication to the cancerous cells and increased growth rate [5]. Epithelial cells connect to the ECM via integrin adhesions and it was suggested that ECM stiffening altered cytoskeletal tension through this connection promoting malignant transformation [6]. Furthermore, it was shown that glioblastoma migration and proliferation [7], and the ability of smooth muscle cells [8] and fibrosarcoma cells [9] to migrate highly depends on the substrate stiffness. Interestingly, cell proliferation was also regulated by ECM rigidity since cells were dividing more rapidly on rigid ECMs compared to compliant ones [7,10]. The mechanism of stiffness sensing is guided by intracellular tension through mechanotransductive cascades. Whenever substrate stiffens cell contractile forces rise in order to establish an equilibrium [11]. In this context, the behaviours of cancer cells such as proliferation, invasion, metastasis and their responses to the chemo treatments are highly modified by their surrounding environments including mechanical properties.
Effects of substrate stiffness on cell behaviour are well dependent on cell phenotype. Results of effects of substrate stiffness on three cell types showed different trends. Fibroblasts and endothelial cells were influenced by their substrate stiffness, since their spread area and integrin α5 gene expression and stress fibres were enhanced on the stiff substrates. On the other hand, the behaviour of neutrophils was independent from the substrate stiffness [12]. While the cell area, proliferation, motility and forming stress fibres of macrophage cells were enhanced due to the stiffening of the substrate [13], hepatocyte cells were more functional and well-differentiated on soft substrates. Additionally, they were able to maintain their phenotype better on the soft substrates [14]. Furthermore, the differentiation of mesenchymal stem cells was related to their substrate stiffness. Cells cultured on soft substrates differentiated to the chondrocytes, whereas those cultured on the stiff substrates expressed smooth muscle cells markers [15].
Analysis of effects of substrate stiffness on the behaviour of cancer cells can be assistive in study of cancer invasion and new prognostic tools. Effects of substrate stiffness on cancerous cells have been addressed, although such effects depended on the cell type. It was shown that the variation of stiffness affected proliferation index and spread area of two hepatocellular carcinoma cell lines (Huh7 and HepG2), however such effects were more profound among HepG2 cells [5]. In another study cancer cell lines were categorised as rigidity-dependent and rigidity-independent types whether their proliferation after 5 days culture depended on the substrate stiffness or not. The rate of this effect was different even among rigidity dependent cancer cell lines [16].
Effects of substrate stiffness on a wide range of cell behaviours of cancer cells with different invasive potentials have not been fully investigated. Cancer cells, including breast cancer cells with differing invasiveness have different physical and structural properties as determinants of their behaviour in different stages of cancer [17]. It is of great interest to investigate if such properties are altered by substrate stiffness which might further address novel methods in cancer suppression. In this contribution, we examined effects of substrate stiffness on the major behaviours of breast cancer cell lines with different degrees of invasiveness. MCF10A, MCF7 and MDA-MB-231 cell lines were studied as the normal, non-invasive and invasive breast cancer cell lines respectively. Samples of poly dimethylsiloxane (PDMS) with three elastic moduli were synthesised by changing the ratio of monomer to cross-linker as the substrates of three breast cancer cell lines. We analysed alterations in cell morphology, cytoskeletal architecture, cell proliferation, cell motility, and expression of a wide range of specific genes of cells seeded on the substrates to quantify effects of substrate elastic properties on breast cancer cell behaviours.
Results
Mechanical properties of substrates
Three breast cancer cell lines were cultured on the prepared collagen coated PDMS substrates with varying stiffness for 48 h. Different monomer to cross linker ratios (10:1, 50:1 and 75:1) led to different elastic moduli. Utilizing tensile tests, the elastic moduli of PDMS substrates were obtained 640kpa, 40kpa and 10kpa for 10:1, 50:1 and 75:1 ratios respectively.
Substrate rigidity alters morphology and actin structure of breast cell lines
Analysis of cell area revealed that this parameter increased 59 percent and 173 percent by culturing MCF10A breast cell lines on soft substrate to substrate with moderate stiffness and from substrate with moderate stiffness to stiff substrate respectively. Furthermore, these enhancements were 43 percent and 77 percent for MCF7 and 50 percent and 92 percent for MDA-MB-231 respectively (Figure 1(a)). One-way ANOVA revealed that substrate stiffness significantly altered cell morphological spreading among all tested cell lines (P < 0.0001). Furthermore, Bonferroni’s multiple comparison was performed to compare paired between groups. Results were indicative of significant differences as described in Figures. Both SEM and confocal images demonstrated that the average cell area decreased dramatically by decreasing substrate stiffness among all three cell lines (Figures 1(d) and 2). Cells were well-spread and showed evident and elongated lammelopodia on the stiffest substrates. By substrate softening, cells became semi rounded and their spreading area decreased considerably. Moreover, the number and the length of the lammelopodia decreased intensely by decreasing substrate stiffness. On softer substrates, the long lammelopodia were replaced by numerous shorter filopodia that enhanced cell motility.
Figure 1.
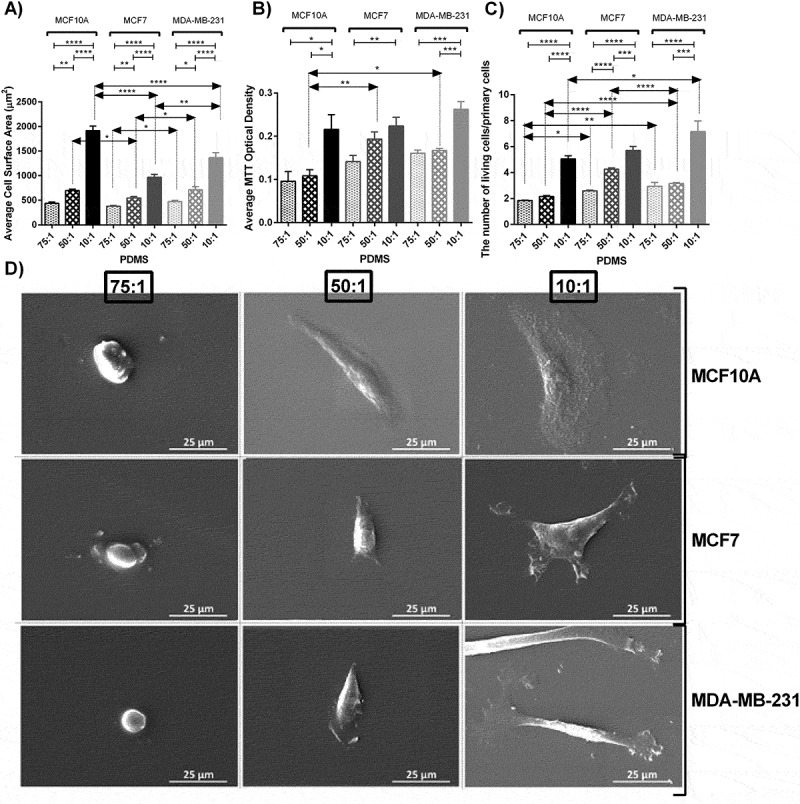
Morphological analysis, proliferation assay and SEM images of MC10A, MCF7 and MDA-MB-231 cell lines cultured on substrates with different elastic moduli. A) Average cellular surface area of different breast cell lines cultured on substrates with different rigidities using Image J software. B) Average cellular proliferation index of different breast cell lines cultured on substrates with different rigidities using MTT assay. C) Direct living cell counting using trypan blue. The number of cells after 48 h culture was divided by the number of primary cells (50,000 cells in each well of 6 well plates in the first day of culture) . D) Scanning electron microscopic (SEM) images of different breast cell lines cultured on substrate with different rigidities. In each case, values represent the mean (± SEM) of measurements from three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
Figure 2.
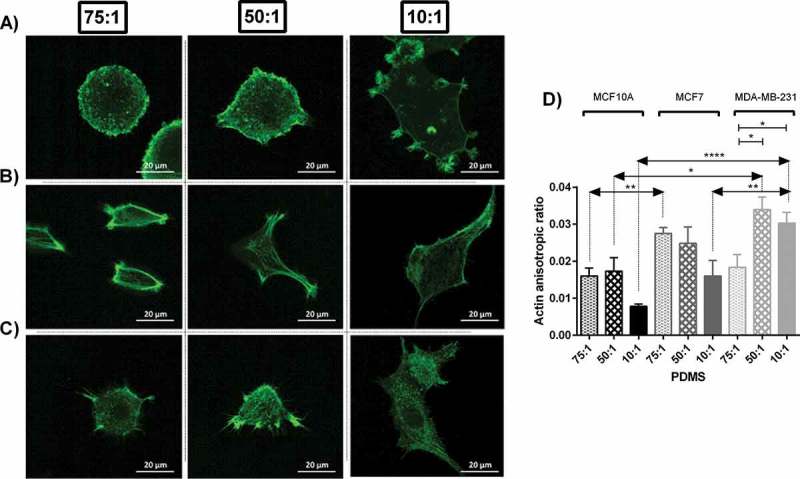
Actin cytoskeletal staining of MC10A, MCF7 and MDA-MB-231 cell lines cultured on substrates with different stiffness values. (A, B, C) Represents confocal images of cells on each substrate stained by phalloidin-FITC. (A) MCF10A. (B) MCF7. (C) MDA-MB-231. (D) Actin anisotropic ratio of breast cell lines cultured on substrates with different stiffness values.
Different morphological features were observed for cells seeded on each substrate among three cell lines (Figure 1(a,d)). The differences among cell area between all cell lines were more prominent on stiff substrates (P < 0.0001) compared to the moderate and soft stiffness (P < 0.05) Although there was a significant difference between morphology of MCF10A and MCF7 cell lines cultured on the moderate stiffness, the difference was not evident when they were cultured on the soft substrate, indicating that by substrate softening, the difference in morphological features such as the surface area became less detectable. In general, among all substrates, the normal breast cell line (MCF10A) had higher surface areas compared to other cell lines. On stiff substrates, the average surface area of MCF10A cells was 1.4 and 2 folds higher than MDA-MB-231 and MCF7 cells respectively. Furthermore, MDA-MB-231 cells were more elongated and had more noticeable lammelopodia than MCF7 cells which is an indication of higher invasion ability of this cell line especially on the stiff substrates (Figure 1(c)).
Among cells which were quantified, normal breast cell lines (MCF10A) and invasive one (MDA-MB-231) demonstrated similar trend in variation of actin structure and isotropic levels. Richer actin structure was observed among cells cultured on substrates with moderate stiffness. For these cells results of actin anisotropic ratio showed least polarisation on stiff substrates and highest on substrates with moderate stiffness (Figure 2(d)). Noninvasive cell lines (MCF7) shows similar trend when was cultured on the soft substrate. Moreover, the average values of actin polarisation were significantly different for cell lines cultured in different stiffness (P < 0.05 for MCF10A and P < 0.01 for MDA-MB-231).
In each substrate, there was a statistical difference between actin polarisation of three cell lines (P < 0.01, P < 0.05 and P < 0.0001 for soft and moderate and stiff substrates respectively). The differences among actin polarisation values are indicators of different actin structure among cell lines. On average, the degree of actin polarisation was higher for cancerous cells compared to normal cells (Figure 2(d)) with the highest values for invasive cells (MDA-MB-231) which means the arrangement of actin fibres in cancer cells was less isotropic (Figure 2), most likely due to rich parallel fibrils within the cortex of cells. Moreover, such index was altered by change in substrate stiffness. In general, substrate with moderate stiffness followed by soft substrates enhanced actin rich cell cortex which is related to stress fibers necessary for cell motility. Normal and highly invasive breast cell lines, MCF10A and MDA-MB-231 respectively, formed richer stress fibres on substrates with moderate stiffness, whereas noninvasive breast cell line, MCF7, showed more noticeable stress fibres when they were cultured on soft substrates (Figure. 2).
Substrate stiffening increases proliferation of breast cell lines
Data demonstrated that substrate stiffening led to higher proliferation measured by MTT assay and direct cell counting among all cell lines (Figure 1(b,c)). One-way ANOVA revealed evident statistical differences for all cell lines cultured on substrates with different rigidities for both MTT assay (MCF10A P < 0.01, MCF7 P < 0.05 and MDA-MB-231 P < 0.0001) and direct cell counting method (P < 0.0001 for all cell lines). The differences among cell proliferation between all cell lines seeded on substrates with different stiffness values were strongly significant for all cell lines for both MTT assay (soft P < 0.05, moderate P < 0.01 and MDA-MB-231 P < 0.05) and direct cell counting (soft P < 0.01, moderate P < 0.0001 and MDA-MB-231 P < 0.01). MTT assay revealed that the average cell proliferation rose 13 percent and 100 percent by culturing MCF10A breast cells on soft substrate to moderate substrate and from moderate substrate to stiff substrate respectively. Furthermore, such increases were 37 percent and 16 percent for MCF7 and 4 percent and 57 percent for MDA-MB-231 accordingly. Such trend was confirmed by direct cell counting.
Substrate rigidity alters cell movement parameters
Cell motility is among major features determining cellular invasion in cancer. To measure effects of substrate rigidity on cell movement, we monitored selected cells cultured on each prepared substrate for 10 hours using time lapse microscopy and calculated three parameters as the determinants of cell motility including cell track length, cell velocity, and cell effective distance as the length of the straight line between the initial and last positions of cells. Generally, effects of stiffness variation on the cell movement parameters were more noticeable among cancerous cells compared to normal cells. Results indicated marked alterations in cell track length in response to change in stiffness of substrates specifically among invasive cancerous cells (Figure 3(a)). MDA-MB-231 cells were highly motile in the substrate with moderate stiffness compared to other substrates. Statistical analyzes revealed significant differences among both motility and velocity caused by substrate stiffness among cancerous cell lines (P < 0.0001). Further post-hoc analysis for comparison of each pair of test groups was performed and results are presented in Figures. Among three breast lines, all cells moved less on the stiffest substrates. For normal and invasive lines, cells were more motile on substrates with moderate stiffness, while for non-invasive cancer cells the longest track length was achieved on soft substrates. The average track length of normal and invasive cells on substrates with moderate stiffness were almost 1.3 and 3 folds of those cultured on the stiff substrate respectively, whereas the average track length of non-invasive cancer cells cultured on the soft substrates was 1.8 folds of those cultured on the stiff substrate.
Figure 3.
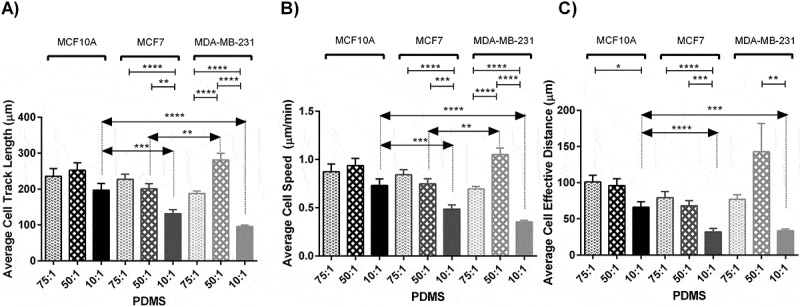
Analyzing cell movement parameters of MC10A, MCF7 and MDA-MB-231 cell lines cultured on substrates with different stiffness values. (A) Represents average cell track length of different breast cancer cell lines cultured on substrates with different rigidities. (B) Represents average cell speed of different breast cancer cell lines cultured on substrates with different rigidities. (C) Represents average cell effective distance of different breast cancer cell lines cultured on substrates with different rigidities. In each case, values represent the mean (± SEM) of measurements from three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
The trend for the velocity of track length was the same as the track length, since cell velocity is obtained through dividing the track length by time. Again, the variation was not significant for normal cells (P > 0.05). The speeds of migration of MCF10A and MDA-MB-231 cells were higher on substrates with moderate stiffness, whereas MCF7 cells obtained highest speed on the soft substrate as it was gradually decreased as the substrate was stiffened (Figure 3(b)). Detailed movies generated during 10 hours monitoring revealed that cells on stiff substrates migrated with developed and broad lamellipodia, whereas the majority of the cells on the soft substrates showed round morphology and thinner and shorter lamellipodia (See Supplementary movie file 2–10). On the other hand, the number of lamellopodia developed on stiff substrates was lower than those on other substrates (Figures1(d) and 2). This might be the reason for higher motility on less stiff substrates. The average cell speed decreased by 22 percent for MCF10A cells on substrates with moderate to high stiffness although not significant (P > 0.05). For MCF7 cells the decreases in cell speed by gradual substrate stiffening were 11 percent and 35 percent. The speed of MDA-MB-231 cells was enhanced significantly on the substrate with moderate stiffness, indicating 50 percent and 192 percent higher values than those cultured on the soft and stiff substrates respectively (P < 0.0001). In order to better understand the role of substrate stiffness on cell motility, the snap shots of the surveyed cells cultured on each substrate captured during 120 min with 15 min intervals are presented in Supplementary Information Figure S3 to S11.
To further study cell motility, cell effective distance was measured over 10 hours of culture on different substrates. For all cell lines, the differences between effective distance of cells cultured on the soft and moderate stiffness were not significant, whereas significant difference was obtained for cells seeded on stiff substrates. One-way ANOVA revealed significant differences among them (MCF10A P < 0.05, MCF7 < 0.01 and MDA-MB-231 < 0.01) (Figure 3(c)).
Compared to cancerous cells, cell motility parameters of normal cells were less under influence of substrate stiffness. On moderate substrates, the motility parameters of invasive breast cell lines were significantly more than other cell lines (p < 0.05).
Substrate rigidity alters gene expression
Effects of substrate stiffness on gene expression after 48 hours culture on substrates with different stiffness values were quantified. The expression levels of Integrin β1 marker increased by substrate stiffening for all cell lines (MCF10A P < 0.0001, MCF7 P < 0.0001 and MDA-MB-231 P < 0.01) (Figure 4(a)). The expression of Integrin β3 decreased for MCF7 cells by stiffening of the substrate (P < 0.0001), although did not change significantly for MCF10A cells (P > 0.05) and increased gradually for MDA-MB-231 cells by substrate stiffening (P < 0.01) (Figure 4(b)). The expression of E-cadherin marker increased gradually for all cell lines by substrate stiffening (P < 0.0001) (Figure 4(c)). The expression of N-cadherin marker decreased gradually by substrate stiffening for MCF10A and MCF7 (P < 0.05 and P < 0.01 respectively) but for MDA-MB-231 cell line expressed more noticeably on the substrates with moderate stiffness (P < 0.0001) (Figure 4(d)).
Figure 4.
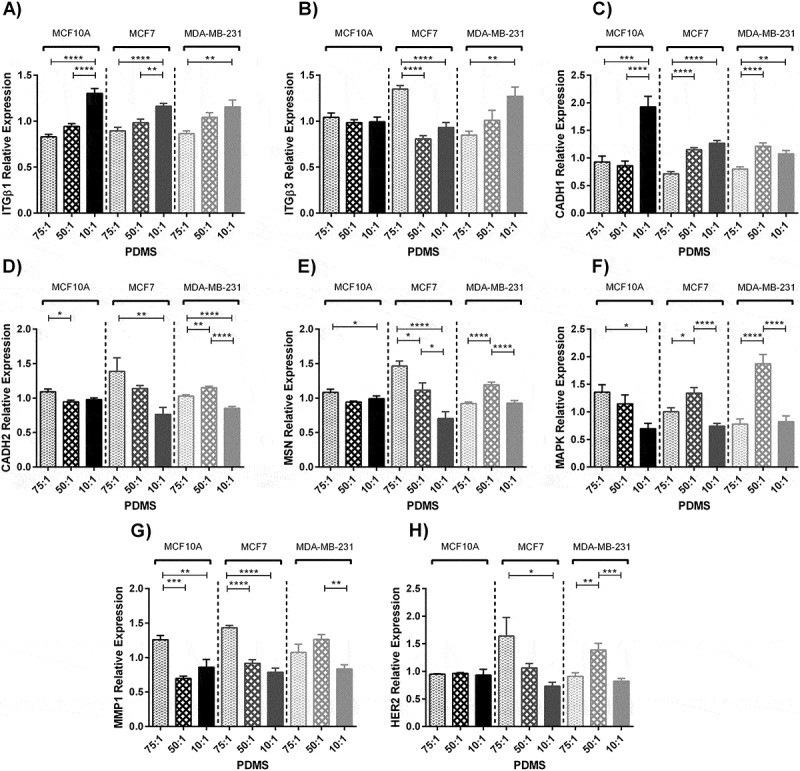
Quantification of gene expression of three breast cell lines cultured on prepared substrates with different rigidities for 2 days using real time PCR. (A, B, C, D, E, F, G, H) Represents relative gene expression of different breast cancer cell lines cultured on substrates with different elastic moduli. (A) Integrin β1. (B) Integrin β3. (C) E-cadherin. (D) N-cadherin. (E) Moesin. (F) Mitogen-activated protein kinase 1. (G) Matrix metalloproteinase1. (H) Human epidermal growth factor receptor 2. In each case, values represent the mean (± SEM) of quantification in three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
Moesin, localising in membranous protrusions, was reduced by substrate stiffening for MCF10A and MCF7 cells (P < 0.05 and P < 0.0001 respectively), although this expression was higher for MDA-MB-231 cells when cultured on the substrates with moderate stiffness compared to other stiffness values (P < 0.0001) (Figure 4(e)). Mitogen-activated protein kinase 1 (MAPK1) expression decreased gradually for MCF10A cells by substrate stiffening (P < 0.05). On the other hand, it was expressed markedly in the cells seeded on substrates with moderate stiffness for MCF7 and MDA-MB-231 cell lines (P < 0.0001) (Figure 4(f)). Although, the matrix metalloproteinase gene expression (MMP1) decreased for MCF10A and MCF7 cells on the substrates with moderate and stiff substrates compared to the soft one (P < 0.001 and P < 0.0001 respect/ively), For MDA-MB-231 cells this expression was boosted when cultured on the substrates with moderate stiffness compared to other stiffness values (P < 0.01) (Figure 4(g)). Human epidermal growth factor receptor 2 (HER2), one of the well-known genes which is up regulated due to the breast cancer diseases, did not change for MCF10A after change in substrate stiffness (P > 0.05) but was over expressed for MCF7 and MDA-MB-231 cells cultured on the soft and moderate substrates respectively (P < 0.05 and P < 0.001 respectively) (Figure 4(h)).
Discussion
Biological cells interact with a highly dynamic environment by which their homeostatic state is defined. Cells receive chemical and physical stimuli in their micro-environment and interact with them through signalling cascades. In vitro studies use custom made substrates to investigate how different physical, chemical and biological characteristics of the substrate influence functionality of cells seeded on them. Among different micro-environmental stimuli, elastic properties of substrates provide a mechanical cue by which cell behaviours are engineered. Cells apply traction forces and create local deformation on the substrates and the neighbouring cells receive these mechanical signals and respond accordingly. Hence a strain contour is generated which depends on the mechanical properties of substrate and location and number of cells [18]. It is of great importance to notice that when effects of mechanical properties of the substrates on cultured cells are surveyed, the cell-substrate interaction depends on other parameters. As an example, when cells are confluent, cell-cell contact is the dominant factor and cells receive lesser signals from neighbouring cells through the substrate; hence the cell-substrate is a secondary issue. In current study, such effect was observed when the density of cells was high enough, i.e. there was lesser impact of substrate stiffness on cell behaviour (data not shown). On the other hand, if cell density is low, cells are isolated and the long distances between cells cause cells to receive weak signals from other cells. This is especially important when substrate is highly stiff and substrate is less deformed under conventional traction forces. In such case, neighbouring cells virtually play no role and cells remodel their structure solely by interaction to the substrate.
There has been a wide range of substrate elastic modulus investigated by researchers from kPa to MPa. In addition to the diversity among elastic modulus within the biological tissues, such variation might be due to the fact that the micro-mechanical environment of cells of the target tissues can be considered both in micro- and macro-scales. Cells synthesise the ECM and the synthesised fibres adjacent to cells appear softer compared to the organised tissue level structure. Hence, regardless of the cell phenotype and the tissue that cells reside in, different in vitro studies have applied different ranges of elastic moduli for the cell substrates. Furthermore, there is a broad range of elastic moduli assigned to the biological tissues, from kPa for soft tissues to GPa for hard tissues depending on the testing methods [19]. Breast tissue is considered as a soft tissue in normal conditions. However, with tumour development, the micro-environmental physical forces are altered [20], and the breast tissue markedly stiffens [21]. In general, most of studies that evaluated effects of substrate stiffness on the behaviour of different cell lines have used similar monomer to cross liker ratios for PDMS substrates [22,23].
Substrate induced cell remodelling is through application of traction forces on the substrate through focal adhesion sites and the interactive force caused by substrate elastic response that is received by adhesion molecules and transfers to the cell body through mechanotransductive pathways which in turn causes cytoskeletal remodelling [24]. In current study, we believe that the Young’s modulus of stiff substrates was high enough that cells could not cause noticeable deformation and the behaviour of cells was less influenced by neighbouring cells. However, for substrates with moderate and low stiffness values, cells received signals from neighbouring cells and respond accordingly. In this case cells feel stiffer medium on substrates with moderate stiffness and softer medium on substrates with low stiffness, hence responded differently. The mentioned phenomenon might be the reason that some parameters were more influenced by stiff substrates while other parameters were more affected by substrates with moderate stiffness. For example, for stiff substrates parameters which are directly influenced by cell-substrate such as morphology and proliferation were significantly affected. On the other hand, cell motility which is guided by neighbouring cells through the substrate deformation was highly manifested on substrates with moderate stiffness and soft substrates. For motility of invasive MDA-MB-231cells, there were clear differences (p < 0.0001) among three groups with the highest motility parameters on substrates with moderate stiffness. Same trend was observed for MCF10A cells although the differences were not significant. An exception was observed for the effective distance parameter. It should be noted that a lower value of effective distance does not mean less motility. It might be due to more fluctuations in the trajectory while the track length is still higher. For MCF-7cells, there were no statistical differences among motility related parameters between two softer substrates. Hence, we concluded that in general, motility parameters are lower in stiff substrates compared to other two substrates, while for other two groups there are no statistical differences between moderate and soft substrates with favour for moderate substrates (Figure 3).
Although numerous studies have described biochemical signals which trigger invasion among cancer cells, fewer studies have investigated biomechanical cues that regulate cancer cell behaviour, among which mechanical microenvironments of cells are of special interest [25–27]. Breast tumours provide rigid environments that are noticeably stiffer than normal breast tissue. Breast cancer cells influence their surroundings through a combination of proteolysis degradation of existing matrix components, secretion of matrix components, induction of strain-stiffening and contractility-dependent bundling, and alignment of ECM fibrils that lead to stiffer ECM [28,29]. To thoroughly study the role of stiffness of surrounding matrix on cancer cell behaviours in vitro, here we examined how substrate elastic modulus, as an important mechanical stimulus, modulates a wide range of behaviours of two breast cell lines with different stages of invasion as well as normal cells. Alterations in morphology, proliferation and motility which are central metastatic features were investigated and gene expression was further measured to identify molecular mechanisms which are responsible for alternation of these behaviours. Our novel results demonstrated that vital behaviours of breast cancer cell lines depended upon the substrate stiffness.
Effects of substrate stiffness on different cells have been investigated. It is identified that fibroblast cells have a tendency to migrate from soft to stiff substrates, a phenomenon known as durotaxis [30]. Furthermore it has been shown that cell migration speed is related to the substrate stiffness, although such effect further depends strongly on the cell phenotype [8,31]. The expression and activity of some proteins highly depend on substrate stiffness especially those that are responsible for regulation of cell contractility such as Rho and ROCK [6]. The functions of these proteins are prominent in many solid tumours [32]. ROCK inhibition promotes responses of cancer cells to treatments [33,34]. Interestingly, it was demonstrated that stiff substrates reduce responses to the treatments similar to the condition of the cancer cells in vivo [5]. Hence molecular mechanism that is responsible for resistance of cancer cells to the treatment may be related to the Rho and ROCK up regulation due to enhancement in the elasticity of tumour microenvironment. Furthermore it was indicated that inhibition of NMMII which is responsible for cytoskeleton assembly and contractility eliminated sensitivity of gioblastoma cells to the stiffness [7]. Alternations in different cellular features as observed in this study may partly be related to the cell contractility which is according to the model of cellular mechanical receptors. In this model cells sense their substrate stiffness mainly by integrins which are transmembrane receptors [35]. Integrins are connected to the cell cytoskeleton through a network of intermediate proteins. Cells contract in order to spread, move or even proliferate through applying traction forces by the cytoskeleton on the substrates through integrins. Such contractility is sensed by the substrate through substrate deformation based on its mechanical properties and in return affects cell function [36]. The cell-substrate interaction leads to cell remodelling by which cell cytoskeleton and subsequently cell behaviours are defined. Cells cannot pull and deform stiff substrates efficiently, hence they generate more contraction forces [12,24,30], and highly spread on the substrate and adapt more surface areas (Figure 1(a)). In order to clarify the effect of substrate stiffness on cellular morphology, another cell line was also studied. MDA-MB-468 is an invasive breast cancer cell line with specific round morphology when adhere to its substrate [37]. Surprisingly, these cells adapted triangular shape and spread visibly on the stiff substrate. The surface area of these cells was also enhanced noticeably by substrate stiffening (Supplementary Figure S2). This confirms a more comprehensive conclusion to show that cancer invasion is sensitive to substrate stiffness. Moreover, an interesting feature of this cell line is its round morphology which is kept in different conditions. It was of interest to observe that even for such cell line the effects of substrate stiffness on cell morphology (and proliferation) still persists with the same order of magnitude.
Cell cytoskeleton structure was analysed by phalloidin staining among cells cultured on each substrate. Significant differences were observed for polarisation ratio of actin structure among cell lines and also between cells seeded on different substrates. Previous researches have mainly focused on changes in actin content of cells when subjected to environmental cues. Here we believe that actin arrangement and structure play a key role in cell behaviour in addition to actin content. It is likely that in specific conditions physical content of actin decreases but the arrangement of actin fibres changes in a way that cells produce higher traction forces due to rearrangement of actin fibres such as generation of stress fibres. As an example, it is well known that cancerous cells are softer, but there are supporting evidences that suggest cancer cells have higher traction forces compared to the normal cells [3,38–40]. The cell softening might be related to lower actin content, while higher traction forces might be due to enhanced actin structure and generation of stress fibees in cortex region of cells which is the major site for cell motility. Hence the higher polarisation is not necessarily equivalent to more actin fibres, whereas altered structure of actin might contribute as well. Here we found that the arrangement of actin structure as it is measured by anisotropic ratio was more anisotropic for cancerous cells with high invasiveness, i.e. more parallel fibres (leading to more anisotropic fibrous structure). Cells seeded on soft and moderate substrates which enhance cell motility formed stress fibres in cell cortex close to the membrane and focal adhesion sites (Figure 2). While an isotropic actin structure within the cell body with generation of stress fibres enhances cell traction force, a cell body with improved actin structure near the membrane and in adherent points indicates enhanced cell motility (Figure 3).
Effects of substrate stiffening on cell behaviour depend on the type of cells, since different cell phenotypes produce different traction forces depending on their cytoskeleton structure. Previous data have shown that substrate stiffening reduced cell motility of normal fiboblast cells [41], and modulated the motility of smooth muscle cells and MDA-MB-231 cells in the biphasic trend [8,42]. According to the recent model of cell migration, it has been shown that intermediate matrix stiffness is optimal for cell invasion [43], similar to our results as we obtained higher motility for MDA-MB-231 cells when they were cultured on substrates with moderate stiffness (Figure 3). It has been indicated that cells on stiff substrates spread more and hence form stable focal adhesions leading to reduction in the cell velocity [42].
There is a model of the cell translocation based on five steps among which cells move from one location to another [44]. These five steps are cell polarisation and cytoplasm membrane protrusion at the front, formation of strong adhesions with the substrate, cell body translocation and detachment of the rear part of the cell [41]. On the soft substrate, these phenomena are weakened resulting in reduced cell motility (Figure 3), hence in general, substrates with moderate stiffness provide higher motility. On the soft substrate, there is a tendency of the cells to move towards each other as we observed in the live cells imaging [30]. Detailed cell movements in each substrate are presented in supplementary information and movies.
Cell proliferation was another parameter depending on the stiffness as it increased evidently on the stiffer substrates (Figure 1(b,c)). Stiff substrates with almost no deformability under traction forces lead to a more proliferative phenotype, however substrates with moderate stiffness and soft substrates result in higher motility with lower proliferation [45,46]. Cells do not tend to keep track on the stiff substrate according to the previous data, hence this leads to shorter effective distance on the stiff substrate [41] (Figure 3(c)). Higher proliferation on stiffer substrates has also been observed in other cell types [16,23]. It seems that substrate stiffening enhances cell cycle progression during mitosis [47]. Cell cycle progression in mammalian cells is strictly regulated by integrin-mediated adhesion to the extracellular matrix [48], as it was confirmed by gene expression analysis (Figure 4), and cell spreading through enhanced focal adhesion sites (Figures 1(d) and 2). The increased Induction of cell proliferation mainly occurs by activating extracellular signal-regulated kinase (ERK), RhoGTPase, and NMMII-based contractility by increasing substrate stiffness [7]. These mitogen pathways are partly inhibited on soft substrates.
Effects of substrate stiffness on cell behaviour were evident among all examined cell lines. Cancerous cells were more sensitive to substrate stiffness for cell behaviours which were related to cell motility and migration which are necessary for invasion. The invasive cancerous cells have the highest sensitivity to all three motility parameters followed by non-invasive cancerous cells. This was further confirmed by actin structure in which more distinct structures were observed for cancerous cells seeded on different substrates. Each substrate with a defined stiffness results in a specific actin polarisation [49] as was confirmed by our results among all three cell lines using Fibril tool. Rich actin content in the cell cortex and focal adhesion sites, together with more organised fibres in the cell body necessary for cell movement [50] and invasion were observed on substrates with optimised stiffness for cancerous cells, specifically for invasive cells. The highly invasive MDA-MB-231 cells developed broader lamellipodia than non-invasive MCF7 cells (Figures 1(a,d) and 2). The marked variation of motility of MDA-MB-231 cells on the substrates with different stiffness values indicates that this cell line is highly responsive to the substrate stiffness. It has been suggested that the strong signalling between neighbouring cells through the substrate caused actin organisation and orientation [12]. The normal cells were least sensitive to substrate stiffness for motility related parameters, however they were more sensitive to cell spreading as an indication of firm adhesion of MCF10A normal breast cell lines compared to cancerous cells [51].
The expression of adhesion markers, ITGB1, CADH1, which regulate adhesion of cells to the substrate and neighbouring cells was gradually enhanced by substrate stiffening (Figure 4). This is probably related to higher spreading which elevates the number of cell-substrate adhesion bonds, and increased cell traction forces that together enhance cell adhesion ability to the substrate [52,53]. Additionally cell-cell tension forces is proportional to cell-ECM traction forces [54]. Increase in expression of adhesion genes leads to higher proliferation because of marked effects on cell cycles (Figure 1(b,c)) [48,55].
Cadherin family provides adhesion between neighbouring epithelial cells. E-cadherin provides more cell-cell adhesion forces than N-cadherin [17,56]. In cancer development, E-cadherins are usually suppressed and N-cadherins are expressed more [17]. The reduced expression of E-cadherin and increased expression of N-cadherin in the substrate with different stiffness values (Figure 4(c,d)) accelerates cell disconnection and invasion of malignant cells, which is required for the invasive cells during metastatic phase [56,57].
Moesin gene produce a protein which connects the plasma membrane and actin structure. Meosin localising in filopodia is required for cell–cell connection and migration [58,59]. As it was demonstrated in SEM and confocal images, cells on soft substrates and substrates with moderate stiffness developed shorter, yet numerous filopodia which are necessary for cell motility. Hence it is expected that MSN and moesin relative expression are enhanced on the soft substrate (Figure 4(e)). Moesin is also part of a gene expression signature associated with cell motility and invasion [60], here our results confirm lower motility of cells on stiff substrates while express less moesin (Figure 3). It has also been demonstrated that decreased level of MSN gene increases patient survival in the cancer disease and has an established role in cancer cell chemosensitivity, metastasis and in epithelial-mesenchymal transition of breast cancer cells [61].
The mechanotransductive cascades responsible for stiffness sensing have been suggested to be through a FAK–Rho–ERK signalling network. Extracellular signal-regulated kinase (ERK) also known as MAP kinases regulate cellular responses to mechanical stimuli [62]; and influence cell migration [63]. They are expressed at higher levels in various tumours of epithelial origin compared to corresponding normal tissues [64]. The MAPK1 gene expression was gradually reduced by substrate stiffening for normal breast cell line (MCF10A), although for cancerous cells the highest expressions occurred on substrates with moderate stiffness (Figure 4(f)). Matrix metalloproteinases (MMPs) have a direct implication in carcinogenesis. Moreover, they have a pivotal role in cancer development and metastasis and are targets for the future therapy [65]. MMP1 especially direct breast cancer cell line metastasis to the lung and bone [66,67] and its inhibition suppresses the migration capacity of the cells [68,69]. This gene expression varied in breast cell lines by stiffening of the substrate (Figure 4(g)). This might be another mechanism for higher motility and speed of surveyed cells on soft substrates compared to the stiff one (Figure 3). HER2 (human epidermal growth factor receptor 2) plays a role in the development of breast cancer and is amplified in 25 to 30 percent of breast cancers and in these cases the encoded protein exists in abnormally high levels in the malignant cells. Laboratory studies specify that HER2 overexpression has a direct role in the pathogenesis of breast cancer [70], and patients that overexpress HER2 have an invasive type of the disease with meaningfully shortened life survival [71]. The fluctuation of the expression of this marker (Figure 4(h)) was according to the motility responses of MCF7 and MDA-MB-231 cell lines to the substrate stiffness variation (Figure 3). Interestingly for studied genes, MCF10A as the normal breast cell lines was less sensitive to the substrate stiffness variation rather than cancerous cell types.
Finally comparing cell lines, it was shown that substrate stiffness influences cell behaviour among healthy and cancerous breast cells. Although normal cells were treated with a more nutrient media, cancerous cells were more prone to the influence of substrate stiffness. When a cell line is cultured in a nutrient media, it become fresher and stay healthier, this lead to be more responsive to its microenvironment. Considering that the major aim of this study was to compare cancer cell lines with different invasive potentials, even assuming a more nutrient media for MCF10A cells, their response to substrate stiffness was virtually weaker than cancerous cells.
Conclusions
In conclusion, here we compared a wide range of cell behaviours between normal and cancerous breast cells on substrates with different elastic moduli. In all measured parameters, we observed that the effects of substrate stiffness on invasion related cell behaviours such as cell motility and migration were more dominant for cancerous breast cell lines compared to normal cell lines. Aside from different culture media, this difference may be related to the fact that substrate sensing approach for cancer cells are different from normal ones [72].
Cancer cells function in tissues with different micromechanical environment to form tumorus and they are less prone to structural remodelling in this phase until they detach from the tumour and journey in metastatic phase. To invade to other tissues, they undergo marked structural changes to achieve cell deformability and high motility necessary for extravasation and intravasation. Such alterations are through a highly distributed actin structure within cell body with lesser stress fibres and diverse focal adhesions (Figure 2). Cancer cells, especially invasive cells form lamellipodia which consist of focal adhesions as the main sites of mechanotransduction (Figure 1(c)). In normal cells podosomes are main structure for adhesion, as they consist of focal adhesions and proteins [73]. Although podosomes and lamellipodia contain lots of common molecular components, podosomes consist of more adhesive molecules than lamellipodia [51]. These differences between normal and cancerous breast cell lines may be contributed to the fact that they were less sensitive to their microenvironment stiffness.
Normal epithelial cells were less sensitive to substrate stiffness for cell motility behaviours even when they were provided a more nutrient medium. Hence it can be inferred that that cancerous cells were more sensitive to substrate stiffness in that manner. Although normal cells are less sensitive to substrate stiffening, our results indicated that alterations in mechanical properties of substrates still cause statistically significant changes in some cell behaviours. Results can assist in combination with traditional cancer treatment strategies to enhance efficacy of treatment methods.
Methods/materials
To examine effects of substrate stiffness on the behaviour of adhesive cells, it is important to use a substrate with tunable stiffness without changing other chemical and physical properties. By controlling the ratio of polymer to crosslinking agent in poly dimethylsiloxane substrates (PDMS), a wide range of elastic modulus can be achieved with negligible changes in other properties [41]. Here we fabricated PDMS substrates with three elastic moduli to study the behaviour of normal and two cancer cell lines with different invasive potentials. Substrates were synthesised and were coated by a thin layer of collagen type I to provide appropriate cell adherence. To make sure that cells sense the stiffness of substrates, the coated layer was manipulated to be thin enough. After 48 hours of culture on PDMS substrates a wide range of cell behaviors including morphology and cytoskeleton structure, proliferation, and gene expression were examined for all three cell lines. Furthermore, cell motility, a behaviour highly influential in cancer invasion was thoroughly investigated.
Synthesis and characterisation of PDMS substrates
Previously established method was followed for fabricating PDMS substrates with different elastic moduli [74]. Substrates were prepared using sylgard 184 (Dow Corning, USA) with different mixing base:curing agent ratios of 10:1, 50:1, 75:1. The mixtures were degassed in 10 mbar for 10 minutes, followed by baking for 48h at 90°C. The cured mixtures were exposed to UV for 30 minutes. The prepared substrates were functionalized with collagen type 1 (Sigmaaldrich, USA) with density of 0.5 mg/mL to achieve a coated surface with a thin layer of collagen which increases cellular adhesive ability to the PDMS surface while does not affect its surface mechanical characteristics [75]. The collagen coating was performed according to the published research [75]. Briefly, on each substrate, collagen type one diluted with PBS with density of 0.5 mg/mL was deposited and incubated at room temperature for 24h. After incubation, the excessive collagen solution was removed and washed three times with PBS abundantly. This led to surface modification of PDMS substrate which enhanced cell viability [75,76], while the thin coating layer did not affect the stiffness of the substrate.
Characterisation of PDMS substrates
Stiffer substrates are more resistive to deformation when exposed to external loads. Young’s elastic modulus (E) of materials, as a determinant of such resistance, is measured by applying a uniaxial tensile force to a standard sample with specific cross-sectional area and measuring the relative change in the sample length (strain). If the relation of tensile stress (force per area) and tensile strain is relatively linear, E is calculated by the slope of the fitted line relating stress to strain. To evaluate elastic modulus of PDMS substrate, a universal testing machine was used for obtaining elastic modulus of each mixing ratio (10:1, 50:1, 75:1) according to ASTM D3185-06 [77,78]. The different ratios of monomer to cross linker, 10:1 and 50:1 and 75:1, lead to the different elasticity of 640 kPa, 40 kPa and 10 kPa respectively.
Cell culture
Human mammary cell lines MCF10A, MCF7, and MDA-MB-231 were provided from National Cell Bank of Iran (Pasteur Institute of Iran, Tehran, Iran) and used for culturing on substrates with different elastic moduli. MCF10A cells were cultured in DMEM/F12 medium (Invitrogen, USA). Horse serum (Invitrogen, USA), epidermal growth factor (EGF) (Sigma, USA), hydrocortisone (Sigma, USA), cholera toxin (Sigma, USA), insulin (Sigma, USA) and pen/strep (Invitrogen, USA) were added to the medium according to the available protocol [79].
MCF7 and MDA-MB-231 cells were cultured in RPMI medium (Gibco, USA) supplemented by 10% foetal bovine serum (FBS) (Invitrogen, USA), penicillin/streptomycin (Invitrogen, USA) and L-glutamine (Sigma, USA).
The selection of different culture media is according to the fact that the behaviour of cells differs when their culturing media change. This effect is prominent when cells are cultured in the different media of other cell lines which yields to influencing enzyme activity and metabolic rates [80]. Here we kept the basal culture medium constant (RPMI-1640, FBS10%), however according to prescribed protocols [7, 8], for ordinary function of MCF10A cells, extra ingredients were provided. This is due to the fact that if MCF10A cell line is cultured on RPMI media, it becomes old and its viability decreases. This results in becoming less responsive to the microenvironment.
MCF10A cells are normal breast cell lines, and MCF7 and MDA-MB-231 cells are categorised as non-invasive and invasive cancerous cells respectively [81]. After adequate cell growing in T25 flasks (about a confluence of 80%), cells were removed by trypsinization under 3 minutes (Sigma, USA) and were transferred to the prepared substrate coated with collagen type 1. The behaviours of cell lines were investigated after 48h culturing on each PDMS substrate.
To better investigate morphological alternation due to the substrate stiffening, another invasive breast cell line, MDA-MB-468, was also studied [37]. These cells were also cultured in RPMI medium (Gibco, USA) supplemented by 10% foetal bovine serum (FBS) (Invitrogen, USA), penicillin/streptomycin (Invitrogen, USA) and L-glutamine (Sigma, USA).
Optical microscopy and morphometric analysis
Morphology assessment was carried out after 48 h of cell seeding with non-confluent density on collagen coated PDMS substrates with different elastic moduli. An inverted Nikon TE2000-E2 microscope equipped with a digital microscope camera was used to capture at least 100 images of each type of cells on each substrate. Using Image J software (http://rsb.info.nih.gov/ij/, NIH), the area of at least ten cells in each image was quantified. A uniform background subtraction to the entire image and subsequent adjustments to the brightness and contrast were applied as necessary.
Scanning electron microscopy (SEM)
To analyse alternations in the morphology of cells due to substrate elastic properties more accurately, cells were cultured on synthesised substrates at non-confluent density for 48 h and then were fixed by 4% glutaraldehyde (Sigma, USA) diluted with phosphate buffer saline (PBS) (Gibco, USA) for approximately 30 min at the room temperature. The images were captured by scanning electron microscope (Seron Technologies, AIS2300C, South Korea). Samples were coated by gold and the voltage was set at 20 kV.
Actin cytoskeletal staining
To examine cytoskeleton structure of cells, actin staining was performed on samples cultured on substrates with different elastic moduli. Cells were cultured on prepared substrates for 48h and then they were fixed by 4% paraformaldehyde for approximately 20 min at the room temperature. Then Cells were washed three times with PBS and placed in 0.1% Triton-X100 (Merck, Germany) in PBS for 20 minutes to create permeability in cells. Cells were then washed three times with PBS and immersed in 1% BSA in PBS for 30 minutes. Phalloidin-FITC (Invitrogen, USA) staining was placed on each sample for 1h. After actin staining, cells were further washed three times with PBS. Finally, cell images were recorded by confocal microscope equipped with TCS SPS microscope (Leica Microsystem GmbH, Germany).
To further quantify actin structure of cells, actin anisotropy was quantified using Fibril tool as a NIH ImageJ plugin. The periphery of at least 15 cells were outlined as the region of interest on each substrate and actin polarisation was surveyed according to the published protocol [82]. The actin polarisation ratio was calculated as a number between 0 and 1, in which 1 describes highly polarised anisotropic structure, i.e. more parallel actin fibres (leading to more anisotropic fibrous structure) compared to random distributed and mesh like actin fibres indicating isotropic network.
Cell proliferation analysis
To examine proliferation of cancer cell lines on different substrates, a calorimetric method of MTT was used [83]. Cells were cultured at a non-confluent density of 5000 for 48 h in each prepared substrate. Then, the culture medium was removed and substrates were rinsed in PBS followed by addition of 1 mL MTT reagent on each substrate, enough to cover the surface of culturing bed. Cells were incubated for 4h until purple precipitate formed, representing living cells. To solve purple precipitate, the remaining colour was extracted and isopropanol alcohol was added on the surface of each substrate for 20 minutes at room temperature. Finally, absorbance length was recorded at 570 nm.
Since substrate stiffness may have direct effects on metabolic activity as MTT assay detects cell metabolic activity, we further performed direct cell counting with the use of trypan blue (Gibco, USA) on each substrate after 48h of cell culture. The proliferation index was obtained by dividing number of cells after culture period to the number of primary cells at the time zero. Cells were cultured with the number of 50,000 cells in each well of 6 well plates in the first day of culture.
Analysis and quantification of cell motility
To analyse cell motility new parameters were introduced to show how far and with what pattern cells migrated on different substrates. Since this requires live cell imaging, a mini-fluorescent microscope capable of operating inside a CO2 incubator equipped with a 4x objective lens (Ziess, Germany) and a digital microscope camera was designed and manufactured. Cells were cultured on synthesised substrates for 24h before imaging. Phase contrast time-lapse images were acquired on optional time intervals over 5h. To monitor cell movement, for each field of view at least 40 cells were specified and the periphery of each cell was used as an object. The coordinates of the mass centre of each cell body was located using Image J software. The coordinates of cell mass centres throughout the time sequence were calculated and were used for obtaining cell movement parameters. Three different parameters were introduced and calculated from the resultant images. To quantify cellular track length, the trajectory of each cell was monitored and the displacement of each moving cell was collected every 15 minutes over 5 hours. To obtain speed of motility, cell track length within 5 hours was divided by the time as the average velocity of cell motility. The ‘effective distance’ was introduced as the direct distance between the initial and last location of cells, indicating how far cells scattered from the initial location. Hence the initial location at time zero and the end location after 5 hours were specified as the primary and secondary locations respectively.
cDNA synthesis and real-time PCR
To examine alterations in gene expression among breast cancer cell lines as a result of the change in substrate stiffness, cells were cultured on each prepared substrate for 48h after the primary adhesion. Then, the total RNA was extracted with the RNeasy plus mini kit (Qiagen, USA). After determining optical density of extracted RNA, they were reversely transcribed to cDNA (Qiagen, USA). A total of nine genes of interest and one housekeeping gene (GAPDH) were analysed.
Integrins are cell surface adhesion receptors which connect the cellular cytoskeleton and the connecting signalling pathways to molecules of the extra-cellular matrix (ECM). The expressions of ITGB1 (integrin β1) and ITGB3 (integrin β3), as the major markers that demonstrate cellular adhesion capability to the substrate [84], were analysed. Cadherins are a family of markers that represent cell-cell adhesion among which the expression levels of E-cadherin (CADH1) and N-cadherin (CADH2) as major markers [85], were quantified. Other quantified genes were further related to the cancer invasiveness and metastasis. Moesin is localised in filopodia and other membranous protrusions that are important for cell-cell recognition and signalling, and cell movement [86]. Mitogen-activated protein kinase 1 (MAPK1) is a gene that encodes a protein that belongs to the MAP kinase family. MMP1 encodes a member of the peptidase M10 family of matrix metalloproteinase (MMPs). These proteins have the specific role to decompose the extracellular matrix in normal biological flow and disease development [65]. Further markers with functions in several cell signalling processes such as proliferation, differentiation, transcriptional regulation, and cellular development were examined [87], among which HER2 (human epidermal growth factor receptor 2) plays an essential role in the development of breast cancer. Normally, HER2 receptors are involved in control of healthy breast cell growth, division, and repair. However, among some breast cancers, malfunction of HER2 marker is observed, a condition known as HER2 gene amplification in which the extra HER2 agents are responsible for extra HER2 receptors and subsequently HER2 overexpression. This causes uncontrolled growth and division of breast cells [88].
The SYBR green primers sequence were either designed using the primer express software v3.0.1 (Applied Biosystems, USA) or obtained from available data and were checked by the Primerblast software (http://ncbi.nlm.nih.gov/tools/primer-blast, NCBI). The forward and reverse sequences with their references are presented in details in Table 1. The comparative cycle threshold method was applied to quantify expression levels. For each cell line cultured on each substrate at least three experiments were performed in duplicates.
Table 1.
Forward and reverse primer sequences.
| Genes | Forward primers | Reverse primers | Reference |
|---|---|---|---|
| GAPDH | GGTGTGAACCATGAGAAGTATGA | GAGTCCTTCCACGATACCAAAG | Design |
| ITGβ1 | CAGTTCAGTTTGCTGTGTGTTTGCTC | CCCACAATTTGGCCCTGCTTGTAT | Design |
| ITGβ3 | CATTACTCTGCCTCCACTACCA | AACGGATTTTCCCATAAGCA | [89] |
| CADH1 | TACACTGCCCAGGAGCCAGA | TGGCACCAGTGTCCGGATTA | [90] |
| CADH2 | TGAAGGTTTGCCAGTGTGACT | CAGCACAAGGATAAGCAGGATGAT | [91] |
| MSN | GAAATTTGTCATCAAGCCCATTG | CCATGCACAAGGCCAAGAT | [92] |
| MAPK | TCTGGCAGGCAGGCAGGCAAT | TGACCGGGAGGAGGAAGGAAGA | Design |
| MMP1 | CTGGCCACAACTGCCAAATG | CTGTCCCTGAACAGCCCAGTACTTA | [93] |
| HER-2 | AGACGAAGCATACGTGA | GTACGAGCCGCACATC | [94] |
Statistical analysis
Data were presented as mean ± standard error. Each parameter was measured in three independent experiments. For analysing cell surface area, actin staining and cell movement parameters at least fifteen cells were examined in each independent experiment related to each substrate stiffness. To examine effects of substrate stiffness among groups with three different substrates and different cell lines one-way ANOVA was performed for verification of statistical significance. Bonferroni’s multiple comparison test in addition to multi variable ANOVA was performed in order to compare variables for each cell line on different substrates and for each substrate between three cell lines. An exception was for real time gene expression which is described as relative for each cell line. The significant alteration in parameters was reported when the calculated p value was less than 0.05.
Ethics approval and consent to participate
Not applicable.
Acknowledgments
We really appreciate Shahram Azari (National Cell Bank of Iran (NCBI), Pasteur Institute of Iran), Saeed Samani (Department of Tissue Engineering & Applied Cell Sciences, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences), Elahe Kashi (School of Allied Medical Sciences, Islamic Azad University Tehran Medical Branch), and Adam J. Engler (University of California, San Diego) for their valuable advice and excellent technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data can be accessed here.
References
- [1].Senkus E.et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26,v8–v30. [DOI] [PubMed] [Google Scholar]
- [2].Boyd NF, Li Q, Melnichouk O, et al. Evidence that breast tissue stiffness is associated with risk of breast cancer. PloS One. 2014;9:e100937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ansardamavandi A, Tafazzoli-Shadpour M, Omidvar R, et al. Quantification of effects of cancer on elastic properties of breast tissue by atomic force microscopy. J Mech Behav Biomed Mater. 2016;60:234–242. [DOI] [PubMed] [Google Scholar]
- [4].Ingber DE. Can cancer be reversed by engineering the tumor microenvironment? Seminars in cancer biology: Elsevier, 2008356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schrader J, Gordon‐Walker TT, Aucott RL, et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53:1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Paszek MJ, Zahir N, Johnson KR, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. [DOI] [PubMed] [Google Scholar]
- [7].Ulrich TA, de Juan Pardo EM, Kumar S.. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. [DOI] [PubMed] [Google Scholar]
- [9].Hansen TD, Koepsel JT, Le NN, et al. Biomaterial arrays with defined adhesion ligand densities and matrix stiffness identify distinct phenotypes for tumorigenic and non-tumorigenic human mesenchymal cell types. Biomater Sci. 2014;2:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hopp I, Michelmore A, Smith LE, et al. The influence of substrate stiffness gradients on primary human dermal fibroblasts. Biomaterials. 2013;34:5070–5077. [DOI] [PubMed] [Google Scholar]
- [11].Kourouklis AP, Kaylan KB, Underhill GH. Substrate stiffness and matrix composition coordinately control the differentiation of liver progenitor cells. Biomaterials. 2016;99:82–94. [DOI] [PubMed] [Google Scholar]
- [12].Yeung T, Georges PC, Flanagan LA, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. [DOI] [PubMed] [Google Scholar]
- [13].Hayenga HN, Adlerz K, Aranda-Espinoza H. Substrate stiffness regulates the behavior of human monocyte-derived macrophages. Biophys J. 2015;108:306a–7a. [DOI] [PubMed] [Google Scholar]
- [14].Natarajan V, Berglund EJ, Chen DX, et al. Substrate stiffness regulates primary hepatocyte functions. RSC Adv. 2015;5:80956–80966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Park JS, Chu JS, Tsou AD, et al. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials. 2011;32:3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tilghman RW, Cowan CR, Mih JD, et al. Matrix rigidity regulates cancer cell growth and cellular phenotype. PloS One. 2010;5:e12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Omidvar R, Tafazzoli-Shadpour M, Shokrgozar MA, et al. Atomic force microscope-based single cell force spectroscopy of breast cancer cell lines: an approach for evaluating cellular invasion. J Biomech. 2014;47:3373–3379. [DOI] [PubMed] [Google Scholar]
- [18].Reinhart-King CA, Dembo M, Hammer DA. Cell-cell mechanical communication through compliant substrates. Biophys J. 2008;95:6044–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Levental I, Georges PC, Janmey PA. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. [DOI] [PubMed] [Google Scholar]
- [20].Yu H, Mouw JK, Weaver VM. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol. 2011;21:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Samani A, Bishop J, Luginbuhl C, et al. Measuring the elastic modulus of ex vivo small tissue samples. Phys Med Biol. 2003;48:2183. [DOI] [PubMed] [Google Scholar]
- [22].Shukla V, Higuita‐Castro N, Nana‐Sinkam P, et al. Substrate stiffness modulates lung cancer cell migration but not epithelial to mesenchymal transition. J Biomed Mater Res. 2016;104:1182–1193. [DOI] [PubMed] [Google Scholar]
- [23].Prauzner-Bechcicki S, Raczkowska J, Madej E, et al. PDMS substrate stiffness affects the morphology and growth profiles of cancerous prostate and melanoma cells. J Mech Behav Biomed Mater. 2015;41:13–22. [DOI] [PubMed] [Google Scholar]
- [24].Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. [DOI] [PubMed] [Google Scholar]
- [25].Yoshii T, Geng Y, Peyton S, et al. Biochemical and biomechanical drivers of cancer cell metastasis, drug response and nanomedicine. Drug Discov Today. 2016;21:1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Insua-Rodríguez J, Oskarsson T. The extracellular matrix in breast cancer. Adv Drug Deliv Rev. 2016;97:41–55. [DOI] [PubMed] [Google Scholar]
- [27].Kai F, Laklai H, Weaver VM. Force matters: biomechanical regulation of cell invasion and migration in disease. Trends Cell Biol. 2016;26:486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wolf K, Wu YI, Liu Y, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. [DOI] [PubMed] [Google Scholar]
- [29].Provenzano PP, Inman DR, Eliceiri KW, et al. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95:5374–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lo C-M, Wang H-B, Dembo M, et al. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lin P-Y, Chu Y-S, Lai Y-T, et al. Interplay between substrate stiffness and TGF-b1 on cellular traction force and migration of MDA-MB-231 cells. FASEB J. 2016;30:1300–1305.26631482 [Google Scholar]
- [32].Itoh K, Yoshioka K, Akedo H, et al. An essential part for Rho–associated kinase in the transcellular invasion of tumor cells. Nat Med. 1999;5:221–225. [DOI] [PubMed] [Google Scholar]
- [33].Ader I, Delmas C, Bonnet J, et al. Inhibition of Rho pathways induces radiosensitization and oxygenation in human glioblastoma xenografts. Oncogene. 2003;22:8861–8869. [DOI] [PubMed] [Google Scholar]
- [34].Rattan R, Giri S, Singh A, et al. Rho/ROCK pathway as a target of tumor therapy. J Neurosci Res. 2006;83:243–255. [DOI] [PubMed] [Google Scholar]
- [35].Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Bio. 2014;15:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mohammadi H, Sahai E. Mechanisms and impact of altered tumour mechanics. Nature cell biology. 2018;20:766–774. [DOI] [PubMed] [Google Scholar]
- [37].Rizwan A, Cheng M, Bhujwalla ZM, et al. Breast cancer cell adhesome and degradome interact to drive metastasis. NPJ Breast Cancer. 2015;1:15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lekka M, Pogoda K, Gostek J, et al. Cancer cell recognition–mechanical phenotype. Micron. 2012;43:1259–1266. [DOI] [PubMed] [Google Scholar]
- [39].Li Z, Persson H, Adolfsson K, et al. Cellular traction forces: a useful parameter in cancer research. Nanoscale. 2017;9:19039–19044. [DOI] [PubMed] [Google Scholar]
- [40].Kraning-Rush CM, Califano JP, Reinhart-King CA. Cellular traction stresses increase with increasing metastatic potential. PloS One. 2012;7:e32572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tzvetkova-Chevolleau T, Stéphanou A, Fuard D, et al. The motility of normal and cancer cells in response to the combined influence of the substrate rigidity and anisotropic microstructure. Biomaterials. 2008;29:1541–1551. [DOI] [PubMed] [Google Scholar]
- [42].Lang NR, Skodzek K, Hurst S, et al. Biphasic response of cell invasion to matrix stiffness in three-dimensional biopolymer networks. Acta Biomater. 2015;13:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ahmadzadeh H, Webster MR, Behera R, et al. Modeling the two-way feedback between contractility and matrix realignment reveals a nonlinear mode of cancer cell invasion. Proceedings of the National Academy of Sciences 2017; 114:E1617–E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 1998;8:51–54. [DOI] [PubMed] [Google Scholar]
- [45].Pelham RJ, Wang Y-L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings of the National Academy of Sciences 1997; 94:13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Giese A, Loo MA, Tran N, et al. Dichotomy of astrocytoma migration and proliferation. Intl J of Cancer. 1996;67:275–282. [DOI] [PubMed] [Google Scholar]
- [47].Wells RG. The role of matrix stiffness in regulating cell behavior. Hepatology. 2008;47:1394–1400. [DOI] [PubMed] [Google Scholar]
- [48].Schwartz MA, Assoian RK. Integrins and cell proliferation. J Cell Sci. 2001;114:2553–2560. [DOI] [PubMed] [Google Scholar]
- [49].Zemel A, Rehfeldt F, Brown A, et al. Optimal matrix rigidity for stress-fibre polarization in stem cells. Nat Phys. 2010;6:468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. [DOI] [PubMed] [Google Scholar]
- [52].Schwarz US, Soiné JR. Traction force microscopy on soft elastic substrates: a guide to recent computational advances. Biochim Biophys Acta (BBA)-Mol Cell Res. 2015;1853:3095–3104. [DOI] [PubMed] [Google Scholar]
- [53].Müller C, Pompe T. Distinct impacts of substrate elasticity and ligand affinity on traction force evolution. Soft Matter. 2016;12:272–280. [DOI] [PubMed] [Google Scholar]
- [54].Maruthamuthu V, Sabass B, Schwarz US, et al. Cell-ECM traction force modulates endogenous tension at cell–cell contacts. Proceedings of the National Academy of Sciences 2011; 108:4708–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Seguin L, Desgrosellier JS, Weis SM, et al. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Panorchan P, Thompson MS, Davis KJ, et al. Single-molecule analysis of cadherin-mediated cell-cell adhesion. J Cell Sci. 2006;119:66–74. [DOI] [PubMed] [Google Scholar]
- [57].Okegawa T, Pong R-C, Li Y, et al. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim Pol-English Ed-. 2004;51:445–458. [PubMed] [Google Scholar]
- [58].Wang CC, Liau JY, Lu YS, et al. Differential expression of moesin in breast cancers and its implication in epithelial–mesenchymal transition. Histopathology. 2012;61:78–87. [DOI] [PubMed] [Google Scholar]
- [59].Wu M, Liu D-Y, Yuan X-R, et al. The expression of moesin in astrocytoma: correlation with pathologic grade and poor clinical outcome. Med Oncol. 2013;30:1–6. [DOI] [PubMed] [Google Scholar]
- [60].Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. [DOI] [PubMed] [Google Scholar]
- [61].Masoumi S, Harisankar A, Gracias A, et al. Understanding cytoskeleton regulators in glioblastoma multiforme for therapy design. Drug Des Devel Ther. 2016;10:2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Provenzano PP, Inman DR, Eliceiri KW, et al. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK–ERK linkage. Oncogene. 2009;28:4326–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhan Y, Kim S, Izumi Y, et al. Role of JNK, p38, and ERK in platelet-derived growth factor–induced vascular proliferation, migration, and gene expression. Arterioscler Thromb Vasc Biol. 2003;23:795–801. [DOI] [PubMed] [Google Scholar]
- [64].Kiyokawa E, Takai S, Tanaka M, et al. Overexpression of ERK, an EPH family receptor protein tyrosine kinase, in various human tumors. Cancer Res. 1994;54:3645–3650. [PubMed] [Google Scholar]
- [65].Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. [DOI] [PubMed] [Google Scholar]
- [66].Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. [DOI] [PubMed] [Google Scholar]
- [67].Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nishimura T, Nakamura K, Kishioka Y, et al. Inhibition of matrix metalloproteinases suppresses the migration of skeletal muscle cells. J Muscle Res Cell Motil. 2008;29:37–44. [DOI] [PubMed] [Google Scholar]
- [69].Zaman MH, Trapani LM, Sieminski AL, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proceedings of the National Academy of Sciences 2006; 103:10889–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Pietras RJ, Arboleda J, Reese DM, et al. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10:2435–2446. [PubMed] [Google Scholar]
- [71].Browne B, O’Brien N, Duffy M, et al. HER-2 signaling and inhibition in breast cancer. Curr Cancer Drug Targets. 2009;9:419–438. [DOI] [PubMed] [Google Scholar]
- [72].Alexander NR, Branch KM, Parekh A, et al. Extracellular matrix rigidity promotes invadopodia activity. Curr Biol. 2008;18:1295–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gimona M, Buccione R, Courtneidge SA, et al. Assembly and biological role of podosomes and invadopodia. Curr Opin Cell Biol. 2008;20:235–241. [DOI] [PubMed] [Google Scholar]
- [74].Fuard D, Tzvetkova-Chevolleau T, Decossas S, et al. Optimization of poly-di-methyl-siloxane (PDMS) substrates for studying cellular adhesion and motility. Microelectron Eng. 2008;85:1289–1293. [Google Scholar]
- [75].Wang L, Sun B, Ziemer KS, et al. Chemical and physical modifications to poly (dimethylsiloxane) surfaces affect adhesion of Caco‐2 cells. J Biomed Mater Res Part. 2010;93:1260–1271. [DOI] [PubMed] [Google Scholar]
- [76].Lee JN, Jiang X, Ryan D, et al. Compatibility of mammalian cells on surfaces of poly (dimethylsiloxane). Langmuir. 2004;20:11684–11691. [DOI] [PubMed] [Google Scholar]
- [77].Park JY, Yoo SJ, Lee E-J, et al. Increased poly (dimethylsiloxane) stiffness improves viability and morphology of mouse fibroblast cells. BioChip J. 2010;4:230–236. [Google Scholar]
- [78].Shin BK, Lee HS, Jeong YK. Mechanical properties and molecular structure of rubber materials with different hardnesses for vibration isolation of elevator cabins. 대한금속재료학회지. 2013;51:713–718. [Google Scholar]
- [79].Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. [DOI] [PubMed] [Google Scholar]
- [80].Gstraunthaler G, Seppi T, Pfaller W. Impact of culture conditions, culture media volumes, and glucose content on metabolic properties of renal epithelial cell cultures. Cell Physiol and Biochem. 1999;9:150–172. [DOI] [PubMed] [Google Scholar]
- [81].Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Boudaoud A, Burian A, Borowska-Wykręt D, et al. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc. 2014;9:457–463. [DOI] [PubMed] [Google Scholar]
- [83].Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. [DOI] [PubMed] [Google Scholar]
- [84].Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adh Migr. 2011;5:421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nollet F, Berx G, Van Roy F. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol Biol Res Commun. 1999;2:77–85. [DOI] [PubMed] [Google Scholar]
- [86].Zhu X, Morales F C, Agarwal NK, et al. Moesin is a glioma progression marker that induces proliferation and Wnt/β-catenin pathway activation via interaction with CD44. Cancer Res. 2013;73:1142–1155. [DOI] [PubMed] [Google Scholar]
- [87].Boutros T, Chevet E, Metrakos P. Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase regulation: roles in cell growth, death, and cancer. Pharmacol Rev. 2008;60:261–310. [DOI] [PubMed] [Google Scholar]
- [88].Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Partheen K, Levan K, Österberg L, et al Four potential biomarkers as prognostic factors in stage III serous ovarian adenocarcinomas. Int J Cancer. 2008;123:2130–2137. [DOI] [PubMed] [Google Scholar]
- [90].Yan L, Zhou J, Gao Y, et al Regulation of tumor cell migration and invasion by the h19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076–3084. [DOI] [PubMed] [Google Scholar]
- [91].Mink SR, Vashistha S, Zhang W, et al Cancer-associated fibroblasts derived from EGFR-TKI–resistant tumors reverse RGFR pathway inhibition by EGFR-TKIs. Mol Cancer Res. 2010;8:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kopp S, Slumstrup L, Corydon TJ, et al Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the random positioning machine. Sci Rep. 2016;6:26887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Naz S, Bashir M, Ranganathan P, et al Protumorigenic actions of S100A2 involve regulation of PI3/Akt signaling and functional interaction with Smad3. Carcinogenesis. 2014;35:14–23. [DOI] [PubMed] [Google Scholar]
- [94].Benoit V, Relic B, de Leval X, et al Regulation of HER-2 oncogene expression by cyclooxygenase-2 and prostaglandin E2. Oncogene. 2004;23:1631–1635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


