Abstract
BioGears is an open‐source, lumped parameter, full‐body human physiology engine. Its purpose is to provide realistic and comprehensive simulations for medical training, research, and education. BioGears incorporates a physiologically based pharmacokinetic/pharmacodynamic (PK/PD) model that is designed to be applicable to a diversity of drug classes and patients and is extensible to future drugs. In addition, BioGears also supports drug interactions with various patient insults and interventions allowing for a realistic research framework and accurate dose‐patient responses. This tutorial will demonstrate how the generic BioGears PK/PD model can be extended to a new substance for prediction of drug administration outcomes.
The development of increasingly accurate physiological modeling and simulation tools represents a critical opportunity for advancement in the medical research and training fields. Training and educational applications require physiology models capable of characterizing, among other outcomes, cardiopulmonary interactions, pharmacokinetic/pharmacodynamic (PK/PD) behavior, and homeostatic feedback responses. Many of these models have been developed in some detail but have not been integrated within a supported framework. BioGears rectifies this shortcoming by introducing a whole‐body physiology model with a lumped parameter backbone that uses conservation principles to drive systemic flow and manages validated feedback responses. This simulation environment, developed in C++, is offered as open‐source software to encourage cooperative development in the medical research community and to lower the barrier to creating medical training content. The PK/PD model defined in BioGears operates within this whole‐body framework. As a compartmental, physiology‐based model, BioGears leverages physiochemical data and data from in vivo studies to determine the time evolution of drug distribution and clearance on an organ‐specific level. Such detailed distribution tracking is crucial for developing patient‐specific dosing regimens,1, 2 determining optimal sampling times,3 designing drug–drug interaction studies,4 and establishing PD end points.5
Other excellent open‐source tools exist for the pharmacological community to perform in silico modeling following or coupled to in vitro analysis. PK‐Sim provides an excellent compartmental framework for analyzing protein‐drug binding and drug metabolism and degradation in the body.6 Pharmacological applications of PK‐Sim include studies of hormone regulation,7 gastrointestinal absorption,8 computational oncology,9 and many others.10, 11 The BioGears compartmental PK model builds on this work by considering whole‐body distribution of molecules and their transport into the tissues.12, 13 BioGears computes whole‐body drug effects using standard maximum effect (Emax) models that other studies have implemented using the MoBi framework. Although robust, MoBi is a separate framework and puts much of the burden of data management and model construction on the user. In BioGears, these processes are integrated into the overall design. The time‐course of a drug in the body can be directly influenced by conditions, such as respiratory impairment and dynamic insults, such as asthma attack and hemorrhage. BioGears achieves this level of interaction by embedding the traditional compartmental PK/PD structure within a physics‐based lumped‐parameter circuit representation of the cardiopulmonary system that considers fluid dynamics and physiological feedback. This tight integration gives BioGears its uniqueness and promotes simulation of complex medical scenarios requiring dynamic drug response.
We, therefore, wish to demonstrate how a new substance can be modeled in BioGears for the purpose of predicting drug outcomes in silico. The tutorial begins with an overview of the BioGears engine, focusing primarily on substance definition and the underlying PK/PD model. After introducing the BioGears Toolkit, we show how to create a new substance file and design scenarios to validate the PK/PD behavior predicted by BioGears. Finally, we highlight how the interaction of the PK/PD model with the entirety of the BioGears engine supports simulation of complex physiological scenarios that arise in medical research.
Overview of biogears
Model and data structure
BioGears uses electrical circuit analogs augmented by physics‐based models and feedback mechanisms to simulate physiologic behavior. Circuit abstractions of the cardiovascular system, known as zero‐D lumped‐parameter models, were first postulated by Otto Frank.14 His model, which included a capacitor in parallel with a resistance element, effectively related blood pressure to vessel elasticity.15 Improvements upon this initial model introduced time‐varying compliance elements, which captured more complex cardiac mechanics—such as pulsatile flow behavior—and accurately tracked pressure‐volume relationships over a cardiac cycle.16, 17, 18, 19, 20 Lumped‐parameter models of the systemic vasculature have since been integrated with full cardiopulmonary models,21, 22 which BioGears has expanded upon by introducing circuit elements representing all major organs.
Although not capable of reproducing the spatial accuracy achieved by finite element methods,23 the BioGears full‐body, zero‐D lumped parameter circuit provides an excellent modeling scaffold at a fraction of the computational cost. BioGears models linked to the circuit are organized into systems with data stored hierarchically. For instance, the Cardiovascular System assigns relevant data to compartments that represent major organs, which are further partitioned into vascular (blood), tissue, and extravascular spaces (Figure 1). A Common Data Model (CDM) facilitates data transfer between systems and provides generic algorithms for unit conversion and data tracking. CDM algorithms also calculate that state of the BioGears circuit at each simulation time step and update substance transport between compartments and systems. With this approach, BioGears can conduct accurate simulations at approximately six times real time with accurate accounting of mass and momentum conservation.
Figure 1.
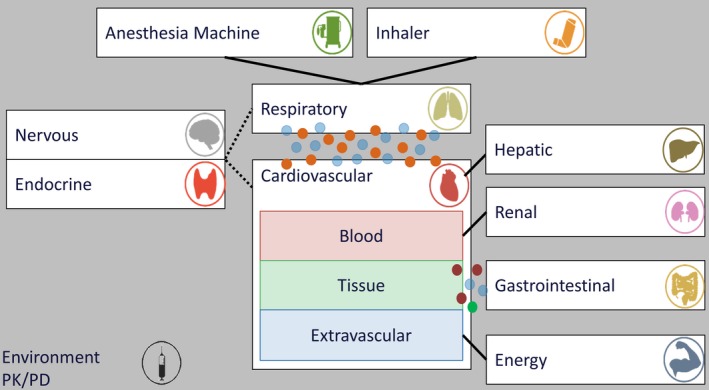
BioGears consists of several unique systems built upon a cardiopulmonary lumped parameter model. Solid lines indicate direct data transfer between systems; dotted lines indicate feedback communication; circles indicate substance diffusion between systems. Substances transfer also occurs among the three subcompartments of the cardiovascular system (blood, tissue, and extravascular). Equipment may be optionally connected to the system (anesthesia machine and inhaler). The environment and pharmacokinetic/pharmacodynamic (PK/PD) are shown set apart because they can affect all systems.
BioGears may be used as a standalone application or integrated with simulators, sensor interfaces, and other existing models. Each instance of the BioGears engine models the physiology of a single patient. Patients are defined by basic parameters, such as gender, height, and weight. The relative health of a patient can be adjusted prior to beginning a simulation by applying a condition, such as heart failure or diabetes. Once a simulation begins, users may subject the patient to a variety of actions that can be broadly categorized as acute insults, interventions, and environmental changes. BioGears also supports interactions between the patient and supported medical equipment, including mechanical ventilators, electrocardiography machines, and inhalers.
Substance definitions
As of the latest BioGears release, the substance library contains over 40 entries spanning such biochemical families as hormones, electrolytes, metabolites, and drugs (Table S1 ). These substances, like all data structures in BioGears, are defined using the extensible markup language (XML. Properties in XML schema can be simple types that accept a single argument or complex types that encapsulate multiple simple subelements. Schema elements defined in BioGears obey the following conventions. A string value or enumeration is declared as:
<Property Name>Value</Property Name>
Note that if the property is a complex type, the enumeration value will be another schema element. Scalar and double elements are declared using the structure.
<Property Name value=“value” unit=“unit”/>
Declaring the unit allows the CDM to store the value such that it can be easily converted to other units throughout the course of a simulation. If a value is unitless, the unit tag is omitted.
With the exception of its name and physical state, the properties required to completely define a BioGears Substance depend on its function. A drug will not be properly processed by the PK/PD system unless its schema definition also includes clearance, PKs, and PDs data. Each of these additional data elements are complex types with numerous subelements, as elucidated by Figure 2.
Figure 2.
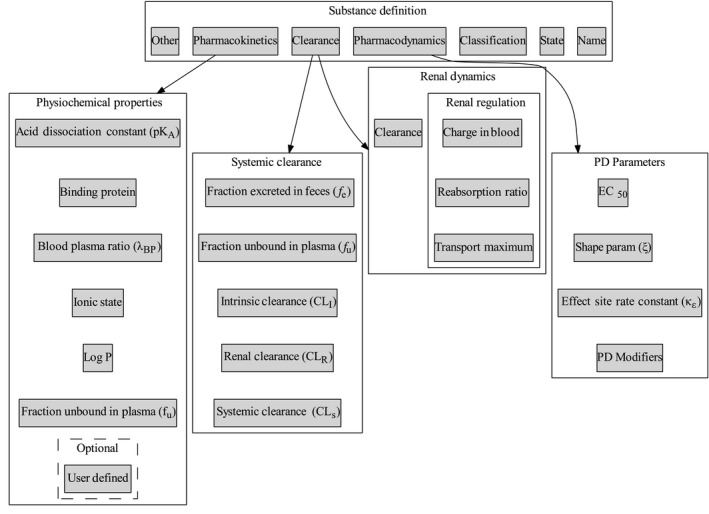
Organization of drug‐specific parameters in BioGears substance schema. Dashed lines are optional data entries for a given substance.xml definition. The Toolkit folder (introduced later) has numerous examples of completed substance definitions. EC50, half‐maximum concentration; PD, pharmacodynamic.
Although not required, it is recommended to provide a drug classification, if relevant. Doing so allows BioGears to organize drugs with similar mechanisms of action. For example, in the current library, morphine and fentanyl belong to the opioid class because they predominantly target similar receptors in the central nervous system. Existing drug classifications include anesthetic, opioid, reversal agent (currently naloxone reversal of opioid overdose), and sedative. If a drug does not fall into one of these categories, then the tag may be omitted.
Clearance data are partitioned between systemic and renal dynamics because a substance may enter one of two renal clearance models. The first is a detailed mechanistic reabsorption model that requires additional physiochemical data (see Renal Regulation in Figure 2), whereas the second is a generic model that uses a lumped clearance term (clearance). Currently, the mechanistic model mostly governs ions and electrolytes (e.g., sodium, potassium, and lactate) and has not been extended to drugs. Therefore, a drug substance schema should only specify clearance under the renal dynamics data type. This value must be duplicated exactly to the renal clearance data field nested under the systemic type. Doing so notifies the renal system that the generic clearance model should be preferred. The systemic type contains another duplicated parameter in the form of fraction unbound in plasma (fU), although the rationale for the duplication is much simpler. BioGears enforces system‐level data encapsulation and the drug and renal models, which both require access to fU, and must, therefore, each store a separate copy.
The PK schema field is usually completed by physiochemical data from which BioGears estimates tissue:plasma partition coefficients according to the methods outlined in the Distribution section. However, in cases when the resulting model does not meet the validated literature, the generated coefficients may be overwritten. Ketamine and morphine currently use the latter definition, if an example is sought. For the sake of brevity, a complete list of PD modifiers is deferred to Table S2 . Likewise, Table S3 elaborates upon additional optional parameters that, while not strictly required by the PK/PD model, may be of interest.
PK model
Absorption/administration
The BioGears PK model supports bolus and continuous infusions of liquid substances, as well as inhalation of gases and aerosols. Boluses may be given intravenously, intra‐arterially, or intramuscularly. The model parses these options by adding the bolus dose to the appropriate BioGears compartment (vena cava, aorta, or muscle tissue, respectively). In the event of intramuscular dosing, the muscle tissue:plasma partition coefficient determines the rate at which the drug diffuses into the bloodstream. Continuous infusions enter the cardiovascular model exclusively via the vena cava. Inhaled substances are introduced via the interaction of the BioGears respiratory system with the inhaler model (in the case of albuterol) or the environmental conditions model (in the case of carbon monoxide). More information regarding inhaled substance modeling—which is outside the scope of this tutorial—is available in the BioGears online documentation (https://www.biogearsengine.com/systems).
BioGears does not currently support oral administration. Until recently, the absence of a detailed gastrointestinal model had been the primary roadblock in this respect. Now that the BioGears gastrointestinal system has been validated, we expect to begin work on oral drug PKs in the near future.
Distribution
The BioGears fluid circuit methodology dictates drug transport throughout the cardiovascular system. Within each vascular compartment, mass exchange with the tissue operates according to the principles of perfusion‐limited diffusion, that is,
| (1) |
In Eq. (1), m represents the mass of the drug, Q B the rate of blood flow into the vascular compartment, C B and C T the respective drug concentrations in the vascular and tissue compartments, and K TB the tissue:blood partition coefficient.24 BioGears generates estimates of K TB for each compartment using a slight modification of the methods reported by Rodgers et al. and Rodgers and Rowland.12, 13 For moderate to strong bases, K TB is given by
| (2) |
Multiplication by the ratio of f u to λBP converts Eq. (2) from a tissue:plasma basis (as derived by Rogers and Rowland) to the desired tissue:blood basis. Likewise, for acids, neutral compounds, and weak bases
| (3) |
The parameters X and Y depend on the state of the drug according to
| (4) |
| (5) |
The PKs data type associated with a drug (Figure 2) contains the bulk of the information required to evaluate these expressions. The parameters f u, λBP, P, and pKa are self‐evident. Determining which of Eqs. (2) and (3) to use—as well as how to evaluate Eqs. (4) and (5), if needed—requires the Ionic State specified by the schema. Furthermore, the identity of the binding protein plays a role in establishing K Bind.
Aside from the plasma pH (pHp) and the intracellular pH (pHIW), which are calculated at each time step of a simulation, the remaining parameters are tissue‐dependent values stored within BioGears. These values include extracellular and intracellular water fractions (f EW and f IW), neutral lipid and phospholipid fractions (f NL and f NP), and binding protein partition coefficients (K Bind). Currently supported binding proteins include albumin, lipoprotein, and alpha‐1‐acid glycoprotein. Table S4 contains a complete list of these parameters organized by tissue compartment.
Clearance
Clearance occurs predominantly by hepatic metabolism and renal elimination. The model calculates hepatic clearance using the relationship
| (6) |
Q H represents the rate of blood supplied to the liver by the portal vein (which receives blood from the small intestine, large intestine, and splanchnic compartments). Both the intrinsic clearance (ClI)—which is normalized by body weight—and f u are specified in the substance schema (Figure 2). The mass of drug metabolized and removed from the liver compartment at a given time t is, therefore:
| (7) |
C L(t) is the concentration of drug in the liver vasculature at time t, whereas Δt is the time step size. Multiplication by body weight (BW) renders the total hepatic clearance (ClH) on a volume‐per‐time basis. The metabolized mass (m H) is assumed to be completely inactive from a PD standpoint, as BioGears does not currently consider the activity of active metabolites.
As stated previously, a dynamic renal filtration model regulates the elimination of many BioGears substances. The model reacts to fluctuations in pressure and osmolality to estimate the glomerular filtration rate and determine the ratio of substance filtration to reabsorption. This model, however, has not yet been extended to drug substances. Instead, drugs enter a generic model analogous to Eq. (7) but parameterized by the drug concentration in the kidneys and a renal clearance normalized by BW (ClR in Figure 2). Half of the calculated mass eliminated is taken from each kidney compartment.
BioGears supports additional elimination mechanisms. If a significant proportion of a substance is excreted in feces, that fraction may be indicated in the substance schema. Mass excreted in the feces is taken from the liver compartment. Likewise, if clearance occurs by an alternate pathway (i.e., not hepatic or renal), the total systemic clearance (ClS in Figure 2) can be adjusted. The systemic clearance collectively represents all possible routes of clearance; thus, the rate at which mass is cleared by “other” processes is the difference between ClS and the sum of ClH and ClR. Any alternate clearance process is assumed to take place in the bloodstream; therefore, any mass removed in this manner is taken from the vena cava compartment.
PDs
A major challenge associated with computational pharmacology is accounting for drug onset latency and duration of action. BioGears accounts for the potential for counter‐clockwise hysteresis by introducing an effect compartment that qualitatively captures the delay incurred as a drug is transported from the plasma to its site of action.25 The time‐evolution of this effect‐site concentration is governed by a first‐order rate constant, as indicated in Eq. (8).26
| (8) |
PD response is assumed to be governed by a typical Hill‐type relationship dependent upon the effect‐site concentration, as stated in Eq. (9). The half‐maximum concentration and shape parameter (EC50 and γ, respectively) are unique to each drug. Drugs are also assigned a series of fractional‐effect modifiers (E m), each one corresponding to a specific physiologic response (e.g., heart rate, respiration rate, etc.). Changes in states (ΔE) are accomplished by multiplying the relevant modifier (E m) and baseline value (E bl) by the dimensionless Hill function. Currently supported PD responses (and their range of accepted inputs) are covered fully in Table S2 . This subset of patient responses will be expanded in the coming years.
| (9) |
Running biogears
Development packages
BioGears is being developed continuously with a wide range of user groups in mind. As such, three deployment packages are available (all available for Windows, Mac, and Linux operating systems at https://www.biogearsengine.com/download). The first package is a Toolkit intended for researchers, educators, and students who wish to execute existing BioGears actions while monitoring the physiological state of the virtual patient. A graphical user interface (GUI) accompanies the Toolkit, allowing scenarios to be executed without writing code. The GUI also plots requested data in real time and stores the plots automatically when the scenario has finished execution.
The BioGears Software Development Kit offers an intermediate option. The Software Development Kit users enjoy more control over scenario execution and data extraction than the Toolkit allows. At the same time, inclusion of prebuilt libraries and headers, as well as numerous “How‐To” sample files, reduces the burden of development incurred by working directly with the source code. This balance makes the Software Development Kit the ideal platform through which to integrate the BioGears engine with existing software applications.
Finally, advanced users have the option of downloading the BioGears source code (which can also be pulled from the BioGears Github page, https://github.com/BioGearsEngine). Developers can view implementation, contribute improvements, and integrate their own models with existing BioGears systems. Working with the source code requires functional experience with C++ and may also necessitate extension of the CDM. Developers working with the source code who require technical assistance can reach out to the BioGears team via the Github page or our community forum (https://www.biogearsengine.com/forums/). This tutorial, however, will focus entirely on using the Toolkit.
Toolkit overview
To download the Toolkit, navigate to https://www.biogearsengine.com/download and select the appropriate platform from the available selections (located in the green banner heading the page). The three download options will become visible, as will links to extensive documentation and validation. Choosing the Toolkit option will download a zipped file titled Biogears‐7.0.0_[OS]‐Toolkit, where OS is the selected platform. After extracting the folder, the top level directory will be biogears‐[OS]. Selecting the BioGearsGUI batch (or shell) file will launch the GUI, producing the view show in Figure 3.
Figure 3.
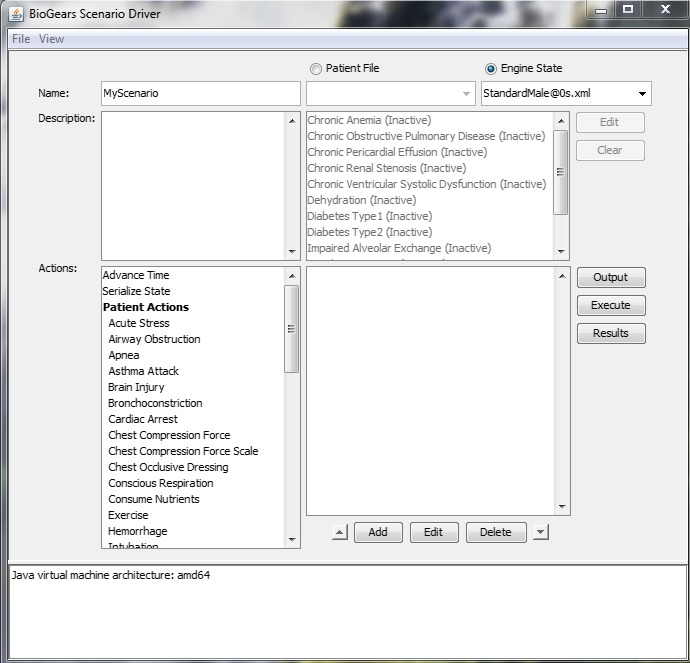
The main window of the BioGears graphical user interface.
Name and Description are fields that users may optionally update to better organize their scenarios. The results file and plot folder will assume the input to Name; thus, by default, the GUI writes output to MyScenario.txt and plots data in a folder called MyScenarioGraphs.
Virtual patient physiology is established by selecting either a Patient File or an Engine State. The distinction between these two options lies in the fact that BioGears usually must undergo a stabilization period before executing a scenario. During stabilization, the engine runs until it reaches a steady state within an acceptable tolerance of the patient parameters defined in a Patient File. It is possible, though, to serialize the state of the engine after it has stabilized. This saved Engine State can be loaded directly to a scenario at a later time, bypassing the need for stabilization. Available Patient Files and their dual Engine States can be viewed in the Patients and States folders, respectively. In most cases, loading an Engine State is the most expedient option. The exception to this rule occurs when modeling a patient with a condition (i.e., diabetes type 1). Because conditions constitute deviation from normal patient parameters, the engine must be put through stabilization. Conditions, therefore, can only be run using a Patient File (which is why the list of conditions is shown as unavailable in Figure 3).
Scenarios can be initiated in one of two ways. Users can explore scenarios designed by the BioGears team for validation purposes. These scenarios can be accessed via Files‐>Open‐>Scenarios. The Scenarios folder is further organized into categories such as Drug, Energy Environment, and Patient. Opening any one of the scenario.xml files will populate the scenario field of the GUI main window (the blank field below the list of Conditions in Figure 3) with a list of actions to be executed. Although not visible from the main window, data requests will be populated at the same time.
Alternatively, novel scenarios can be constructed using the Actions menu. Chosen actions will appear in the scenario field, and there is no limit to the number of actions that may be applied. Note that the passage of time must be explicitly set using the Advance Time action. After constructing a scenario in this way, at least one data request must be defined. Selecting the Output option will generate a new menu where the user can specify data returns with the appropriate units. At that point, the scenario is ready and may be initialized using the Run button. A new window will track the progress of the simulation. When it has exited, the Results button may be selected to view plots of the data that were requested via the Output function.
PK/PD Toolkit scenarios
We will now demonstrate the process for using the BioGears Toolkit to create a new substance definition, capture its PK/PD behavior, and model its action in a complex virtual patient setting. We elect to create arginine vasopressin (AVP), also known as vasopressin and antidiuretic hormone. AVP affects multiple systems (cardiovascular and renal) and is currently of interest in shock therapy research. Thus, our example will prove both relevant and capable of demonstrating the spectrum physiologic feedback systems modeled in BioGears. Although a naturally occurring endogenous compound, AVP does not present itself in significant levels in well‐hydrated individuals (1–2 pg/mL plasma concentration).27 We, therefore, do not expect the absence of baseline AVP levels to confound the results of our scenarios.
Substance parameters
Within the top‐level Toolkit folder resides the substances directory, which contains all of the currently supported substance schema files. These files are excellent sources to consult when constructing a new substance. We create a new xml in this location entitled “Vasopressin.xml” and note that the file name must match the name in the substance schema exactly. Using available literature and the schema structure detailed in Figure 2, we define the general, clearance, and PK properties of vasopressin (see Model‐Code S1 ). PD modifiers are left at 0 for the PK validation scenario. The lone exception is the parameter EC50, which must be assigned a value to prevent the engine from generating an error.
PK validation
The scenario used to assess the PK model performance derives from a study conducted by Glänzer.28 Healthy male volunteers were administered AVP intravenously for 60 minutes at a rate of 160 pmol/minute (0.174 μg/minute). Clearance was monitored for an hour postinfusion. To model this scenario, we navigate to the Actions menu in the GUI main window and select Substance Infusion. This action requires a concentration, a rate of infusion, and a substance. We set a concentration of 0.174 μg/mL and an infusion rate of 1.0 mL/minute to achieve the desired 0.174 μg/minute dose regimen. We then select Vasopressin from the Substance dropdown menu (Figure 4 , top left). (Note: If the substance does not appear in the menu, first ensure that the name of the substance xml file is identical to the Name element given in the schema and then confirm that the PK and PDs elements are defined correctly, as the drug will not be processed without them.)
Figure 4.
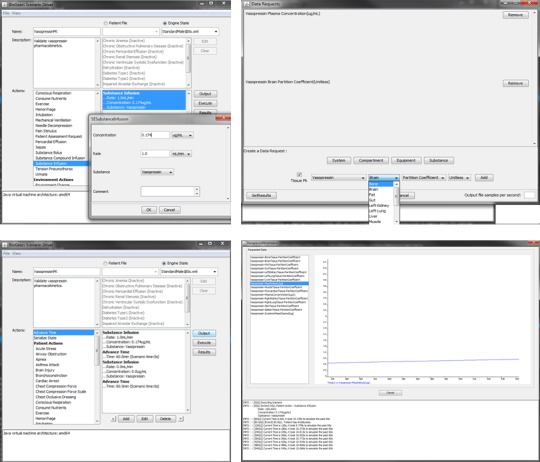
GUI implementation. Vasopressin pharmacokinetic scenario (top left). Creating the substance infusion action (bottom left). The complete scenario with all actions defined (top right). Creating data requests using the “Output” button (bottom right). Scenario view of real time results after selecting “Execute.”
Time advancement must be explicitly stated as an action. We create an Advance Time action and assign it a value of 60 minutes. We then create a new Substance Infusion action—again with respect to vasopressin—and set both the concentration and rate to 0. Doing so signals the model to discontinue the initial vasopressin infusion. After creating another Advance Time action (60 minutes), our scenario description is complete (Figure 4 , bottom left).
Scenarios cannot be run without creating at least one data request. We select Output from the main window, which generates a new window labeled Data Requests (Figure 4 , top right). Requests are organized by System (e.g., cardiovascular, respiratory), Compartment (e.g., brain, heart), Equipment, and Substance. In this scenario, all of the required outputs can be found beneath Substance. A new list of options will appear with fields for substance, property, and unit. We set these fields to Vasopressin, Plasma Concentration, and μg/mL, respectively, and then click Add. The request will appear in the area above the menu (Figure 4 , top right). Next, we select the Tissue PK box to the left of the substance name, which brings up an alternate menu with a list of tissues. We query the partition coefficient from each tissue and then return to the main menu.
At this point, we may begin our scenario by clicking the Execute button in the main window. Doing so will bring up a new window that tracks the progress of the simulation (Figure 4 , bottom right). The scenario data requests will appear in the left‐most area. Navigating between the requests will bring up a corresponding plot, which is updated continuously until the end of the scenario. The field at the bottom of the window will track the progress of the simulation, logging actions (like the Substance Infusion) and patient events that the model detects. When the scenario has finished, the log will indicate that data plots are being generated, as well as the location in which these plots are stored. Alternatively, plots can be viewed by selecting the Results button from the main menu.
After completing our scenario, we validate the results against those reported by Glänzer. Because we cannot project the literature results onto the plot generated by the GUI (Figure 5 a), we use a Python script to plot the results written to VasopressinPKResults.txt against the literature (Figure 5 b). We see that the time evolution of the Vasopressin plasma concentration predicted by BioGears compares very favorably with the results reported by Glänzer. Table S5 gives the tissue partition coefficients associated with this fit.
Figure 5.
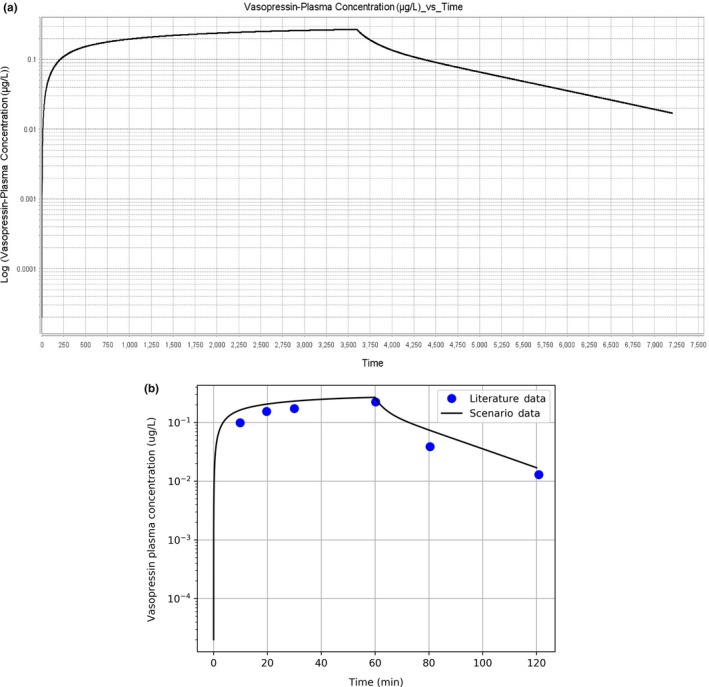
Vasopressin pharmacokinetic curve. (a) Vasopressin plasma concentration plot output by BioGears graphical user interface. (b) Vasopressin plasma concentration predicted by BioGears (black) compared to literature data (blue circles).
PD validation
Validation scenarios
Vasopressin exhibits vasoconstrictive and antidiuretic activity; therefore, we require values for the diastolic pressure, systolic pressure, and (renal) tubular permeability modifiers (Table S2 ). Because parameter estimation cannot currently be performed in BioGears, these parameters must be qualitatively estimated from available studies. As such, we will create cardiovascular and renal validation scenarios. The cardiovascular scenario is adopted from the work of Ebert et al.,29 who infused healthy male subjects with vasopressin at variable rates over 55‐minute intervals. Similarly, the renal scenario recreates an experiment that monitored urine production rate and urine osmolality over a range of hour‐long vasopressin infusions.30 Figures S1 and S2 demonstrate how these two scenarios may be defined using the BioGears GUI.
Before executing these scenarios, we must also define parameters that complete the Emax model described in Eqs. (8) and (9). Vanner et al.31 estimated the EC50 associated with vasopressin‐induced constriction of submucosal arterioles to be 1.2 nM (1.3 × 10−3 μg/mL). Another study performed with respect to human cerebral arteries obtained a similar result of 0.72 nM (7.8 × 10−4 μg/mL).32 Although a useful starting point, we must consider that these results were obtained in isolated tissue and may not be reflective of in vivo conditions. We must also take into account the renal activity of vasopressin, which is known to be significant at plasma concentrations that elicit minimal cardiovascular response.33 In fact, symptoms mimicking dehydration (i.e., low urine flow and high urine osmolality) can be induced at vasopressin plasma levels of 20 pM (2.1 × 10−5 μg/mL).30 To consolidate these observations, we test median EC50 values with respect to the values and adjust the magnitude of the effect modifiers accordingly.
None of the aforementioned studies reported a value for the shape parameter, γ. However, when Möhring et al.34 plotted the mean arterial blood pressure (MAP) increase for three subjects as a function of vasopressin plasma concentration, the shape of the curve was consistent with γ = 1.0. Finally, no sources reporting hysteresis could be found; therefore, the effect site rate constant is set to 0 (this flags the model that plasma and effect site concentration are equal). Model‐Code S1 shows the finalized Vasopressin substance file after tuning.
Cardiovascular validation
BioGears predicts a cardiovascular response consistent with the data published by Ebert et al.29 The model performs particularly well at low infusion rates. Estimated AVP plasma concentration (Figure 6 a) and percent deviation in mean arterial pressure and heart rate (Figure 6 b) all fall within the margin of error of the corresponding literature values. However, the quality of the fit declines at higher doses. Even though BioGears somewhat overestimates the AVP concentration at the second infusion level, the MAP does not respond accordingly (Figure 6 c).
Figure 6.
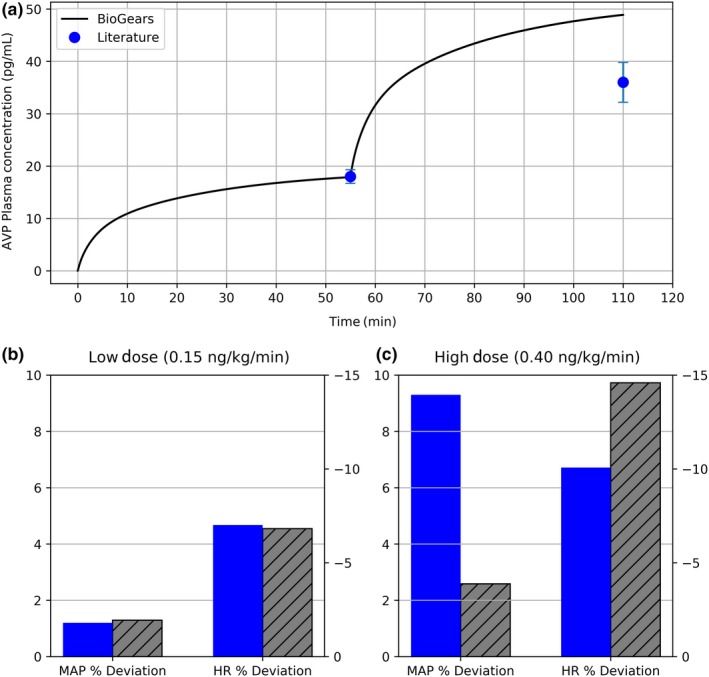
Validation of vasopressin cardiovascular pharmacokinetic/pharmacodynamic. (a) The plasma concentration predicted over the two infusion levels (0.15 and 0.40 ng/kg/minute) compares favorably with that reported by Ebert et al.29 (b) At low infusion doses, the percent increase in mean arterial pressure (MAP) and percent decrease in heart rate (HR) predicted by BioGears (gray) are in line with experimental values of Ebert (blue). (c) As infusion dose is increased, the BioGears baroreflex over‐aggressively opposes MAP elevation, causing an exaggerated bradycardia. AVP, arginine vasopressin.
This muted response can be attributed to the BioGears baroreceptor model. Physiologically speaking, baroreceptors initiate a negative feedback cascade that regulates blood pressure. BioGears models this system according to the work of Ottesen et al.35 The model counters deviations in MAP from its baseline by adjusting the heart elastance, systemic resistance, systemic compliance, and heart rate. This mechanism ex2plains why vasopressin induces—albeit indirectly—a heart rate response even though no heart rate modifier was defined. In any case, the validation literature would suggest that the BioGears baroreflex becomes overly aggressive at AVP plasma concentrations above ~20 pg/mL.
Renal validation results
Validation of renal metrics proved to be more qualitative due to the drastic influence patient hydration has on experimental results. In the study consulted for validation data,30 patients were water‐loaded prior to vasopressin infusion. Baseline urinary flow rates were greater and urine osmolality was lower in these studies compared to BioGears initial values. The output from BioGears mimics what should be expected of an antidiuretic. The urine production rate predicted by BioGears reaches a minimum on par with reported values29 (Figure 7 a). Predicted urine osmolality, while trending in the right direction (Figure 7 b), does not reach the expected range of 875‐1,000 mOsm/kg.29
Figure 7.
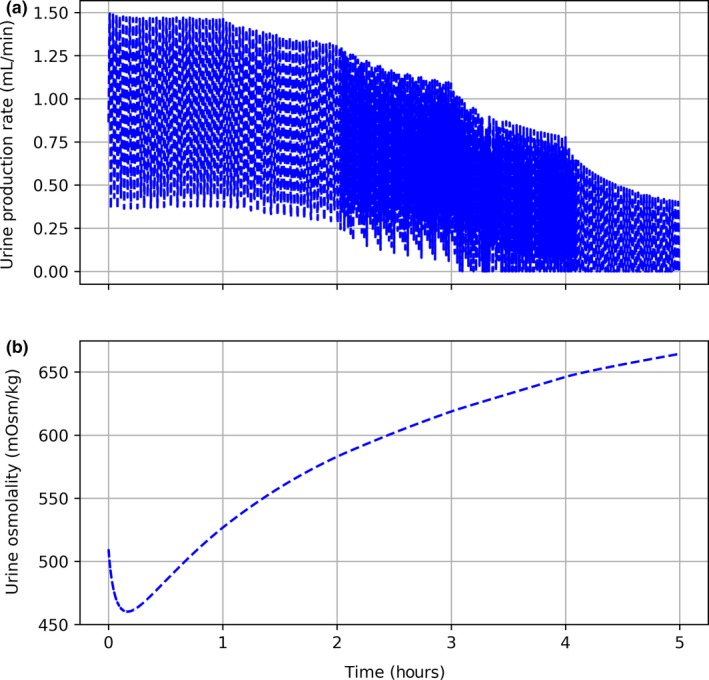
Vasopressin renal validation. Each hour, the infusion rate of vasopressin is increased. (a) Urine production rate progressively declines. (b) Decrease in urine production is accompanied by an increase in urine osmolality.
Applications to current research efforts
Vasopressin in shock therapy
Although useful in isolation, the BioGears PK/PD model is meant to contribute toward accurate simulation of more complex scenarios arising in medical research. Vasopressin presents an interesting case because of its potential role in promoting recovery from multiple forms of shock, including distributive shock (such as septic shock and neurogenic shock) and hypovolemic shock (such as hemorrhagic shock).36 With particular attention to hemorrhagic shock, it has been observed that the early stages of bleeding are characterized by massive increases in plasma vasopressin levels.37 The AVP concentration subsequently diminishes quickly, implicating vasopressin as a key component in the rapid‐onset, early physiological response to hemorrhage. The primary goal in this initial response phase is to maintain adequate perfusion pressure. Thus, it is postulated that blood pressure in the latter stages of hemorrhagic shock can be sustained by administering exogenous vasopressin. Furthermore, animal studies of uncontrolled intra‐abdominal hemorrhage have indicated that this blood pressure rescue does not exacerbate blood loss, as AVP's actions include redistribution of blood flow away from the gut, skin, and muscle toward the heart and brain.38, 39, 40 Thus, vasopressin might simultaneously salvage blood pressure and decrease the amount of fluids required for resuscitation.
Although some human case studies have evaluated the efficacy of vasopressin therapy in hemorrhagic shock,41 few rigorous clinical trials exist. One large effort, the Vasopressin in Traumatic Shock (VITRIS) study, did not garner enough participants to be more than exploratory. Another comprehensive study that showed great promise was initiated at the University Hospital in San Antonio.42 Trauma patients meeting the study criteria received one of two fluid resuscitation treatments: saline only or vasopressin in saline. The primary outcomes monitored were total fluids administered over 5 hours and patient mortality over 24 hours and 30 days. Only 78 patients were enrolled, though, which reduced the statistical power of the study. Recruitment issues notwithstanding, studies involving traumatic injuries are subject to multiple uncontrolled variables. Injury severity, injury location, transport time, and treatment received before admittance to the hospital can all confound results.
In such cases in which human studies are either difficult to perform or to replicate, in silico experiments can be quite informative. BioGears supports hemorrhagic insults, thus, we can customize scenarios in which we compare resuscitation therapies both with and without vasopressin. The outcome of this simulation will not, of course, generate actionable conclusions. Our hope, though, is to demonstrate that BioGears can be used as an exploratory tool to drive more advanced model development and research in this particular field.
BioGears hemorrhage scenario
A hemorrhage action in BioGears is defined by a wound location and initial bleeding rate. The action activates a path on the cardiovascular circuit connecting the wound site to ground (i.e., outside the body). The specified bleeding rate and the pressure in the compartment at the time of hemorrhage initiation determine the flow resistance assigned to this path. By using a resistance element instead of a flat, continuous bleeding rate, the model captures pulsatile flow behavior and allows the bleeding rate to fluctuate as a function of blood pressure.
We create a hemorrhage object in the vena cava compartment with an initial bleeding rate of 350 mL/minute. After 15 minutes, at which point nearly 50% of blood volume has been lost, we stop the bleeding (performed by setting the bleed rate to 0.0). We then administer a rescue bolus of saline (1.0 L) and begin either the control (saline only) or experimental (0.2 unit/minute of vasopressin in saline, assuming 2 U = 5 μg43) treatment. Although the vasopressin in saline treatment could be defined by instantiating two separate infusions, we instead create a new BioGears Substance Compound called VasopressinInSaline (Model‐Code S2 ) and save it in the same substances folder in which we defined the Vasopressin Substance. The concentration of vasopressin specified in this compound and the experimental infusion rate derive from guidelines published with respect to management of gastrointestinal and variceal bleeding.44 A second rescue bolus of saline is administered 15 minutes after treatment initiation. At 30 minutes and 40 minutes posthemorrhage termination, we administer one unit of packed red blood cells. Figure S3 compares the two treatment definitions as defined in the BioGears GUI.
Discussion
BioGears closely replicates the naturally occurring cardiovascular response to massive hemorrhage. The first line of defense is to increase the heart rate to help preserve blood pressure, cardiac output, and tissue perfusion. Given the magnitude of blood loss, however, the compensatory tachycardia incompletely counteracts the decrease in blood pressure, which diminishes further. The renal system responds to this decline in blood pressure by decreasing the glomerular filtration rate to retain fluid.
After initiation of the experimental protocol, plasma levels of AVP rapidly increase after simulated exogenous administration (Figure 8 a). A substantial increase in MAP occurs almost immediately, well beyond that demonstrated with saline infusion alone (Figure 8 b). Recovery stagnates shortly thereafter, likely due to fluid leaking from the vasculature to the extravascular space. Indeed, blood pressure recovery resumes following the second bolus of saline (t = 30). The second rescue bolus also improves the condition of the control patient, although not to the same degree. This observation supports the hypothesis that vasopressin—while not sufficient in isolation to promote full recovery—augments resuscitation by reducing the amount of fluid loading required to stabilize a patient. Secondary effects of blood pressure stabilization include tachycardia reversal (Figure 8 c) and heightened glomerular filtration rate (Figure 8 d), the latter of which indicates improved end‐organ perfusion.
Figure 8.
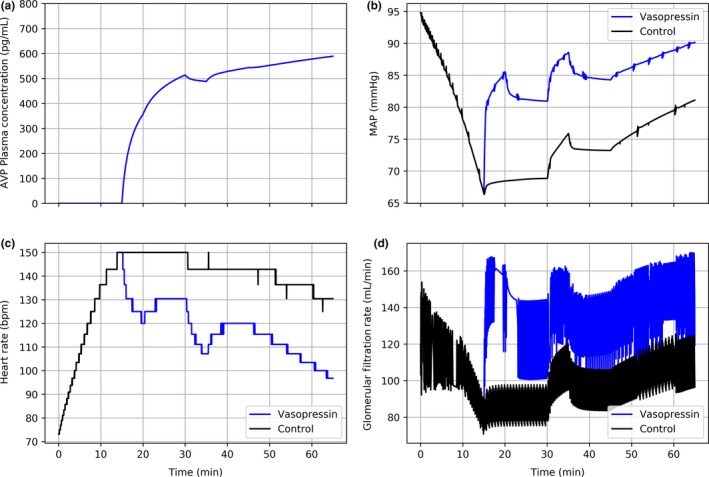
Comparison of control (black) and experimental vasopressin (blue) therapies in virtual patient with severe hemorrhage. Hemorrhage is initiated at t = 0 and terminated at t = 15. Rescue bolus of 1.0 L of normal saline is administered at t = 15. Control and experimental treatments are begun at t = 15 and continued through the end of the simulation. A second 1.0 L bolus of normal saline is given at t = 30. One unit of packed red blood cells are infused at t = 45 and t = 55: (a) arginine vasopressin plasma concentration, (b) mean arterial pressure, (c) heart rate, (d) glomerular filtration rate. AVP, arginine vasopressin; MAP, mean arterial pressure.
Although certainly pronounced, the rise in blood pressure seen in this simulation underestimates that observed in comparable human case studies. This discrepancy probably stems from the fact that the BioGears baroreceptor model is constantly active throughout the hemorrhage. In reality, there is evidence to support baroreceptor suppression after ~1 L of blood loss.46 Increased perfusion of the capillary beds is also not simulated in BioGears during hemorrhage. Thus, during in vivo administration, AVP would be expected to act for the most part unopposed. Only minor model adjustments—such as introducing blood volume dependence to the sympathetic and parasympathetic responses—would be required to address this issue.
The generic BioGears Emax PD model further limits the physiologic accuracy of blood pressure response. In the current implementation, drug‐specific blood pressure modifiers induce an identical resistance change across every pathway on the cardiovascular circuit (except the brain). That is, the model assumes that vasoconstriction and vasodilation are systemic. Vasopressin, however, promotes localized vasoconstriction. The cardioregulatory receptors that AVP targets (V1 receptors) reside primarily in the liver, smooth muscle, and peripheral tissue.33 As a result, the skin, skeletal muscle, and mesenteric vessels experience the greatest degree of vasoconstriction at elevated plasma AVP concentrations. Thus, blood flow is preferentially shunted toward other organs, most notably the brain. Given the importance of maintaining cerebral perfusion under shock conditions, this feature would greatly enhance the fidelity of the hemorrhage scenario. Extending the BioGears PK/PD model to include site‐of‐action effects will, therefore, be a priority going forward.
Conclusion
BioGears is a robust open‐source human physiology modeling platform with myriad potential applications, including enhancement of healthcare simulation, research, and medical training. Comprehensive pharmacologic properties are included in the physiology engine, grounded in existing literature values, allowing for realistic modeling of patient‐drug relationships. Implications include exploratory modeling of complex pharmacologic scenarios that are prohibitively difficult or impossible to evaluate with human studies. In addition, simulated medical training scenarios can be rapidly developed and modified to fit trainer and learner objectives.
Funding
This research and development project was conducted by Applied Research Associates and is made possible by a cooperative agreement that was awarded and administered by the US Army Medical Research & Materiel Command (USAMRMC) and the Telemedicine and Advanced Technology Research Center (TATRC), at Fort Detrick, MD, under Contract Number W81XWH‐13‐2‐0068.
Conflict of Interest
The authors declared no competing interests for this work.
Supporting information
Table S1. BioGears substance library.Table S2. Pharmacodynamic effect modifiers currently supported by BioGears.Table S3. Additional optional substance parameters. Parameter names are written exactly as they appear in the substance schema.Table S4. Water and lipid volume fractions and tissue to plasma protein ratios for BioGears compartments.Table S5. Tissue:plasma partition coefficients for Vasopressin predicted by BioGears PK/PD model.Model‐Code S1. Vasopressin extensible markup language (XML) schema definition.Model‐Code S2. VasopressinInSaline Substance Compound XML definition.Figure S1. BioGears GUI scenario window defining the cardiovascular pharmacodynamic scenario.Figure S2. BioGears GUI scenario window defining the renal pharmacodynamic scenario.Figure S3. BioGears GUI scenario windows comparing two treatment regimens for resuscitation from massive hemorrhage: saline only (left) and saline with vasopressin (right).
References
- 1. Di, L. et al A perspective on the prediction of drug pharmacokinetics and disposition in drug research and development. Drug Metab. Dispos. 41, 1975–1993 (2013a). [DOI] [PubMed] [Google Scholar]
- 2. Sager, J.E. , Yu, J. , Raguenau‐Majlessi, I. & Isoherranen, N. Physiologically based pharmacokinetic (PBPK) modeling and simulation approaches: a systematic review of published models, applications and model verification. Drug Metab. Dispos. 43, 1823–1837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rowland, M. , Peck, C. & Tucker, G. Physiologically‐based pharmacokinetics in drug development and regulatory science. Annu. Rev. Pharmacol. Toxicol. 51, 45–73 (2011). [DOI] [PubMed] [Google Scholar]
- 4. Shardlow, C. , Generaux, G. , Patel, A. , Tai, G. , Tran, T. & Bloomer, J. Impact of physiologically‐based pharmacokinetic modelling and simulation in drug development. Drug Metab. Dispos. 41, 1994–2003 (2013). [DOI] [PubMed] [Google Scholar]
- 5. Lötsch, J. Pharmacokinetic–pharmacodynamic modeling of opioids. J. Pain Symptom Manage. 29, 90–103 (2005). [DOI] [PubMed] [Google Scholar]
- 6. Niederalt, C. et al A generic whole body physiologically based pharmacokinetic model for therapeutic proteins in PK‐Sim. J. Pharmacokinet. Pharmacodyn. 45, 235–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schaller, S. et al A generic integrated physiologically based whole‐body model of the glucose‐insulin‐glucagon regulatory system. CPT Pharmacometrics Syst. Pharmacol. 2, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thelen, K. et al Evolution of a detailed physiological model to simulate the gastrointestinal transit and absorption process in humans, part 1: oral solutions. J. Pharm. Sci. 100, 5324–5345 (2011). [DOI] [PubMed] [Google Scholar]
- 9. Block, M. et al Physiologically‐based PK/PD modeling for oncology: applications for antibody drug conjugates. J. Pharmacokinet. Pharmacodyn. 41, S58–S59 (2014). [Google Scholar]
- 10. Eissing, T. , Lippert, J. & Willmann, S. Pharmacogenomics of codeine, morphine, and morphine‐6‐glucuronide. Mol. Diagn. Ther. 16, 43–53 (2012). [DOI] [PubMed] [Google Scholar]
- 11. Eissing, T. et al A computational systems biology software platform for multiscale modeling and simulation: integrating whole‐body physiology, disease biology, and molecular reaction networks. Front. Physiol. 2, 4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodgers, T. , Leahy, D. & Rowland, M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate‐to‐strong bases. J. Pharm. Sci. 94, 1259–1276 (2005). [DOI] [PubMed] [Google Scholar]
- 13. Rodgers, T. & Rowland, M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J. Pharm. Sci. 95, 1238–1257 (2006). [DOI] [PubMed] [Google Scholar]
- 14. Frank, O. Die grundform des arteriellen pulses. Erste Abhandlung Mathematische Analyse Z. Biol. 37, 483–526 (1899). [Google Scholar]
- 15. Burton, A.C. On the physical equilibrium of small blood vessels. Am. J. Physiol. Legacy Content 164, 319–329 (1951). [DOI] [PubMed] [Google Scholar]
- 16. Masuzawa, T. , Fukui, Y. & Smith, N.T. Cardiovascular simulation using a multiple modeling method on a digital computer—simulation of interaction between the cardiovascular system and angiotensin II. J. Clin. Monit. 8, 50–58 (1992). [DOI] [PubMed] [Google Scholar]
- 17. Sunagawa, K. Models of ventricular contraction based on time‐varying elastance. Crit. Rev. Biomed. Eng. 7, 193–228 (1982). [PubMed] [Google Scholar]
- 18. Ursino, M. , Fiorenzi, A. & Belardinelli, E. The role of pressure pulsatility in the carotid baroreflex control: a computer simulation study. Comput. Biol. Med. 26, 297–314 (1996). [DOI] [PubMed] [Google Scholar]
- 19. Grodins, F.S. Integrative cardiovascular physiology: a mathematical synthesis of cardiac and blood vessel hemodynamics. Q. Rev. Biol. 34, 93–116 (1959). [DOI] [PubMed] [Google Scholar]
- 20. Stergiopulos, N.I.K.O.S. , Meister, J.J. & Westerhof, N.I.C.O. Determinants of stroke volume and systolic and diastolic aortic pressure. Am. J. Physiol. Heart Circ. Physiol. 270, H2050–H2059 (1996). [DOI] [PubMed] [Google Scholar]
- 21. Lu, K. , Clark, J.W. Jr , Ghorbel, F.H. , Ware, D.L. & Bidani, A. A human cardiopulmonary system model applied to the analysis of the Valsalva maneuver. Am. J. Physiol. Heart Circ. Physiol. 281, H2661–H2679 (2001). [DOI] [PubMed] [Google Scholar]
- 22. Lu, K. et al Cerebral autoregulation and gas exchange studied using a human cardiopulmonary model. Am. J. Physiol. Heart Circ. Physiol. 286, H584–H601 (2004). [DOI] [PubMed] [Google Scholar]
- 23. Kerckhoffs, R.C. , Neal, M.L. , Gu, Q. , Bassingthwaighte, J.B. , Omens, J.H. & McCulloch, A.D. Coupling of a 3D finite element model of cardiac ventricular mechanics to lumped systems models of the systemic and pulmonic circulation. Ann. Biomed. Eng. 35, 1–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khalil, F. & Laer, S. Physiologically based pharmacokinetic modeling: methodology, applications, and limitations with a focus on its role in pediatric drug development. J. Biomed. Biotechnol. 2011, 907461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Louizos, C. , Yáñez, J.A. , Forrest, L. & Davies, N.M. Understanding the hysteresis loop conundrum in pharmacokinetic/pharmacodynamic relationships. J. Pharm. Pharm. Sci. 17, 34 (2014). [PMC free article] [PubMed] [Google Scholar]
- 26. Upton, R.N. & Mould, D.R. Basic concepts in population modeling, simulation, and model‐based drug development: part 3—Introduction to pharmacodynamic modeling methods. CPT Pharmacometrics Syst. Pharmacol. 3, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilman, A.G. Goodman and Gilman's The Pharmacological Basis of Therapeutics 11th edn (McGraw‐Hill, New York, 2006). [Google Scholar]
- 28. Glänzer, K. , Prüßing, B. , Düsing, R. & Kramer, H.J. Hemodynamic and hormonal responses to 8‐arginine‐vasopressin in healthy man: effects of indomethacin. Klin. Wochenschr. 60, 1234–1239 (1982). [DOI] [PubMed] [Google Scholar]
- 29. Ebert, T.J. , Cowley, A.W. & Skelton, M. Vasopressin reduces cardiac function and augments cardiopulmonary baroreflex resistance increases in man. J. Clin. Investig. 77, 1136–1142 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morton, J.J. , Padfield, P.L. & Forsling, M.L. A radioimmunoassay for plasma arginine‐vasopressin in man and dog: application to physiological and pathological states. J. Endocrinol. 65, 411–424 (1975a). [DOI] [PubMed] [Google Scholar]
- 31. Vanner, S. , Jiang, M.M. , Brooks, V.L. & Surprenant, A. Characterization of vasopressin actions in isolated submucosal arterioles of the intestinal microcirculation. Circ. Res. 67, 1017–1026 (1990). [DOI] [PubMed] [Google Scholar]
- 32. Martinez, M.C. , Aldasoro, M. , Vila, J.M. , Medina, P. & Lluch, S. Responses to vasopressin and desmopressin of human cerebral arteries. J. Pharmacol. Exp. Ther. 270, 622–627 (1994). [PubMed] [Google Scholar]
- 33. Treschan, T.A. & Peters, J. The vasopressin system: physiology and clinical strategies. Anesthesiology 105, 599–612 (2006). [DOI] [PubMed] [Google Scholar]
- 34. Möhring, J. et al Greatly enhanced pressor response to antidiuretic hormone in patients with impaired cardiovascular reflexes due to idiopathic orthostatic hypotension. J. Cardiovasc. Pharmacol. 2, 367–376 (1980). [DOI] [PubMed] [Google Scholar]
- 35. Ottesen, J.T. , Olufsen, M.S. & Larsen, J.K. Applied Mathematical Models in Human Physiology (Society for Industrial and Applied Mathematics, Philadelphia, PA, 2004). [Google Scholar]
- 36. Cohn, S.M. , Blackbourne, L.H. , Landry, D.W. , Proctor, K.G. , Walley, K.R. & Wenzel, V. San Antonio vasopressin in shock symposium report. Resuscitation 81, 1473–1475 (2010). [DOI] [PubMed] [Google Scholar]
- 37. Tsuneyoshi, I. , Onomoto, M. , Yonetani, A. & Kanmura, Y. Low‐dose vasopressin infusion in patients with severe vasodilatory hypotension after prolonged hemorrhage during general anesthesia. J. Anesth. 19, 170–173 (2005). [DOI] [PubMed] [Google Scholar]
- 38. Voelckel, W.G. et al Arginine vasopressin, but not epinephrine, improves survival in uncontrolled hemorrhagic shock after liver trauma in pigs. Crit. Care Med. 31, 1160–1165 (2003). [DOI] [PubMed] [Google Scholar]
- 39. Feinstein, A.J. , Cohn, S.M. , King, D.R. , Sanui, M. & Proctor, K.G. Early vasopressin improves short‐term survival after pulmonary contusion. J. Trauma Acute Care Surg. 59, 876–883 (2005). [DOI] [PubMed] [Google Scholar]
- 40. Morales, D. et al Reversal by vasopressin of intractable hypotension in the late phase of hemorrhagic shock. Circulation 100, 226–229 (1999). [DOI] [PubMed] [Google Scholar]
- 41. Krismer, A.C. et al Employing vasopressin as an adjunct vasopressor in uncontrolled traumatic hemorrhagic shock. Der Anaesthesist 54, 220–224 (2005). [DOI] [PubMed] [Google Scholar]
- 42. Cohn, S.M. , McCarthy, J. , Stewart, R.M. , Jonas, R.B. , Dent, D.L. & Michalek, J.E. Impact of low‐dose vasopressin on trauma outcome: prospective randomized study. World J. Surg. 35, 430–439 (2011). [DOI] [PubMed] [Google Scholar]
- 43. Leng, G. & Sabatier, N. Measuring oxytocin and vasopressin: bioassays, immunoassays and random numbers. J. Neuroendocrinol. 28 (2016). doi: 10.1111/jne.12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ready, J.B. , Robertson, A.D. & Rector, W.G. Effects of vasopressin on portal pressure during hemorrhage from esophageal varices. Gastroenterology 100, 1411–1416 (1991). [PubMed] [Google Scholar]
- 45. Gleason, E. et al Standard infusion concentrations: critical care medications. Hartford Hospital <portal.hartfordanesthesia.net/Portals/25/Content/Standard%20Infusions.pdf> (2013). Accessed February 7, 2018.
- 46. Foex, B.A. Systemic responses to trauma. Br. Med. Bull. 55, 726–743 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. BioGears substance library.Table S2. Pharmacodynamic effect modifiers currently supported by BioGears.Table S3. Additional optional substance parameters. Parameter names are written exactly as they appear in the substance schema.Table S4. Water and lipid volume fractions and tissue to plasma protein ratios for BioGears compartments.Table S5. Tissue:plasma partition coefficients for Vasopressin predicted by BioGears PK/PD model.Model‐Code S1. Vasopressin extensible markup language (XML) schema definition.Model‐Code S2. VasopressinInSaline Substance Compound XML definition.Figure S1. BioGears GUI scenario window defining the cardiovascular pharmacodynamic scenario.Figure S2. BioGears GUI scenario window defining the renal pharmacodynamic scenario.Figure S3. BioGears GUI scenario windows comparing two treatment regimens for resuscitation from massive hemorrhage: saline only (left) and saline with vasopressin (right).


