ABSTRACT
Paneth cells (PCs) are epithelial cells found in the small intestine, next to intestinal stem cells (ISCs) at the base of the crypts. PCs secrete antimicrobial peptides (AMPs) that regulate the commensal gut microbiota. In contrast, little is known regarding how the enteric microbiota reciprocally influences PC function. In this study, we sought to characterize the impact of the enteric microbiota on PC biology in the mouse small intestine. This was done by first enumerating jejunal PCs in germ-free (GF) versus conventionally raised (CR) mice. We next evaluated the possible functional consequences of altered PC biology in these experimental groups by assessing epithelial proliferation, ISC numbers, and the production of AMPs. We found that PC numbers were significantly increased in CR versus GF mice; however, there were no differences in ISC numbers or cycling activity between groups. Of the AMPs assessed, only Reg3γ transcript expression was significantly increased in CR mice. Intriguingly, this increase was abrogated in cultured CR versus GF enteroids, and could not be re-induced with various bacterial ligands. Our findings demonstrate the enteric microbiota regulates PC function by increasing PC numbers and inducing Reg3γ expression, though the latter effect may not involve direct interactions between bacteria and the intestinal epithelium. In contrast, the enteric microbiota does not appear to regulate jejunal ISC census and proliferation. These are critical findings for investigators using GF mice and the enteroid system to study PC and ISC biology.
KEYWORDS: Paneth cell, microbiota, intestinal stem cell, Reg3γ, germ-free, enteroid
Introduction
Originally described in the late 1800’s, Paneth cells (PCs) are highly specialized secretory cells of the small intestinal epithelium.1,2 They are located at the base of the crypts of Lieberkühn, positioned amongst the actively cycling intestinal stem cells (ISCs).3 PCs have a distinct morphology, characterized by large, eosinophilic secretory granules. These cells support an array of homeostatic functions, ranging from innate host defense4 to maintenance of the ISC niche.5 Importantly, PC abnormalities have been implicated in a number of human disease processes, including Crohn’s disease,6 necrotizing enterocolitis,7 and graft-versus-host disease.8 Therefore, there is a strong rationale to understand the factors that regulate the development and function of these cells.
To date, numerous studies have shown that intrinsic host factors are critical for proper PC development and function. Our group has previously shown that background host genetics profoundly impacts PC biology.9 Specifically, we demonstrated that mice on different genetic backgrounds (C57BL/6J and 129/SvEv) show striking differences in PC number and functional antimicrobial peptide (AMP) expression. At a molecular level, several host-signaling pathways have been implicated in PC development. Wnt signaling promotes PC maturation,10 driving the expression of genes required for proper PC development and migration to the base of small intestinal crypts.11 Conversely, Notch signaling blocks secretory cell development (including PCs) by repressing Atoh1, which serves as a master regulator of secretory cell differentiation in the intestine.12,13 Collectively, these studies illustrate the importance of intrinsic host factors that regulate PC development and function.
Despite our increasing understanding of host influences on PC function, the impact of extrinsic luminal factors on PC physiology is less clear. In particular, the intestinal epithelium is intimately situated adjacent to an extensive community of resident microorganisms, collectively termed the gut microbiota. These microbes perform key functions that enhance the health of the host, including modulating caloric utilization,14 regulating mucosal immunity,15 and maintaining epithelial integrity.16 Elegant studies have shown that PCs are able to regulate the ecology of the intestinal microbiota via AMP secretion,17 and that pathogenic bacteria such as Salmonella enterica can influence PC biology.18 However, the impact of commensal gut bacteria on PC development and function is less clear. Indeed, previous work examining the influence of the enteric microbiota on PC biology has yielded conflicting results. Early studies comparing germ-free (GF) and conventionally raised (CR) rodents showed equivalent numbers of PCs in these animals.19,20 However, work by Rodning et al. demonstrated PC hyperplasia in GF laboratory rats,21 while yet other groups have shown reduced PC numbers in GF rodents compared to their CR counterparts.22,23 These discrepant data highlight the gaps in our knowledge of microbial regulation of PC function.
Given the conflicting findings described above, the primary goal of the present study was to thoroughly characterize the impact of the enteric microbiota on PC number and function. To accomplish this, we used flow cytometry and enteroid culture techniques to enumerate jejunal PCs and assess key PC functions, including driving epithelial proliferation, supporting the ISC niche, and producing AMPs. Using these approaches, we describe key differences in PC number and function between CR and GF mice. These findings provide important knowledge regarding the impact of the gut microbiota on PC function, and are critical for investigators using GF mice and enteroid models to study PC and ISC biology.
Results
Germ-free mice have fewer jejunal Paneth cells than conventionally raised mice
PCs were first enumerated using hematoxylin and eosin (H&E)-stained slides from the distal jejunum. Representative histology is shown in Fig. 1A. When quantified in numerous crypts from multiple biological replicates (Fig. 1B), PC numbers were found to be significantly decreased in GF mice relative to CR counterparts (4.1 ± 0.3 versus 6.4 ± 0.3 PCs/crypt respectively). To minimize the potential for sampling error inherent in histologic analyses, we further investigated PC numbers in the entire jejunal epithelium of CR and GF mice via flow cytometry. Using a gating scheme previously described,9 PC numbers were assessed by quantifying the lysozyme-positive (Lyz+) population in jejunal epithelial cell preparations (Fig. 1C). Consistent with our histological data, the percentage of Lyz+ cells in the total jejunum epithelium was significantly lower in GF mice compared to CR animals (0.37 ± 0.11% versus 0.86 ± 0.15% respectively, Fig. 1D). It should be noted that GF mice had longer villi (694 ± 17 versus 490 ± 11 μm) and greater intestinal length (34.2 ± 0.5 versus 30.4 ± 0.5 cm) than their CR counterparts. While this may affect absolute cell numbers, the similar findings in both our histologic and flow cytometry analyses support an overall decrease in jejunal PCs in GF mice.
Figure 1.
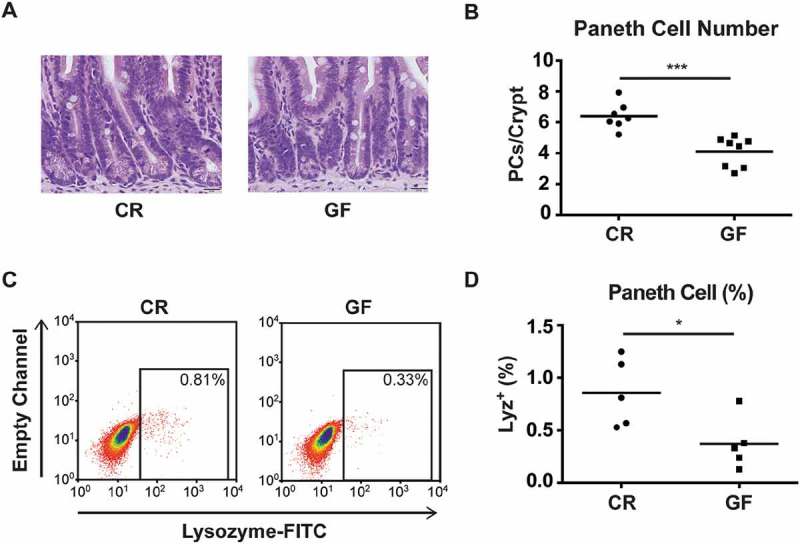
Paneth cell (PC) number is decreased in germ-free (GF) mice. A: Representative H&E images of PCs located at the base of the crypts from the jejunum of conventionally raised (CR) and GF mice (60x magnification, scale bar = 20 μm). At least 2 jejunal sections were prepared per mouse, from 7–8 mice/group. B: Quantitative analysis of PC number per crypt from CR and GF mice. At least 10 crypts/mouse were evaluated (***P < 0.001). C: Representative flow cytometry plots gating Lysozyme (Lyz)+ epithelial cells from CR and GF mice. D: Quantitative analysis of PCs as a percentage of the total epithelium in CR and GF mice (n = 5 pools of mice/group, with 4 mice/pool, *P < 0.05). All quantitative data are represented as scatter plots with a line representing the mean.
Germ-free and conventionally raised mice have similar rates of epithelial proliferation, intestinal stem cell numbers, and cycling status
PCs are known producers of multiple Wnt proteins, which in turn are key drivers of epithelial cell proliferation in the gut.24 Therefore, we hypothesized that the diminished PC numbers observed in GF mice would be associated with decreased proliferation in jejunal crypts. Remarkably, however, 5-ethynyl-2’-deoxyuridine (EdU) immunofluorescence staining showed the proliferative zone of CR and GF crypts to be quite similar (Fig. 2A). This was confirmed quantitatively using flow cytometry (Fig. 2B), which demonstrated a similar percentage of EdU+ cells in the jejunal epithelium of CR and GF mice (3.40 ± 0.63% versus 2.76 ± 0.97% respectively, Fig. 2C).
Figure 2.
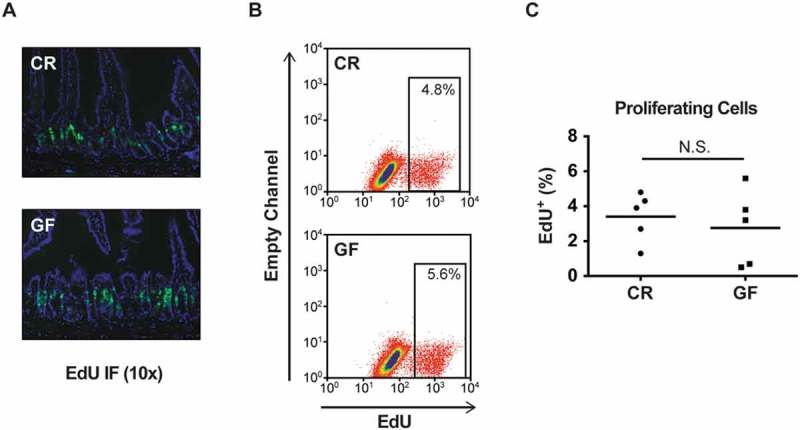
Conventionally raised (CR) and germ-free (GF) mice have similar numbers of total proliferating cells within the intestinal epithelium. A: EdU immunofluorescence (IF) staining from the jejunum of CR and GF mice (10x magnification, single section from 1 mouse/group). B: Representative flow cytometry plots showing total number of proliferating cells (EdU+) in CR and GF mice. C: Quantitative analysis of proliferating cells as a percentage of the total epithelium in CR and GF mice (n = 5 pools of mice/group, with 4 mice/pool, P = 0.5). N.S. = not significant. Quantitative data are represented as scatter plots with a line representing the mean.
Numerous lines of evidence suggest that PCs are an important component of the ISC niche.5,25 Therefore, we next sought to determine if lower PC numbers in GF mice translated to changes in ISC number or activity. This was accomplished by enumerating ISCs using a CD24-staining approach, which offers the advantage of being able to quantitate ISCs in wild-type, non-reporter mice. Previous work has demonstrated that ISCs are enriched in the CD24loLyz– population of intestinal epithelial cells.26 Representative flow cytometry plots gating this CD24loLyz– fraction are shown in Fig. 3A for both CR and GF mice. As demonstrated in Fig. 3B, cumulative analyses from multiple mice revealed that CR and GF animals had similar numbers of ISCs, as measured by the percentage of CD24loLyz– cells within the intestinal epithelium (CR = 1.6 ± 0.4%, GF = 1.3 ± 0.6%). To assess for putative differences in ISC proliferation, EdU staining was used to quantify cycling status. Fig. 3C shows representative flow plots gating on the CD24loLyz–EdU+ cell fraction. When quantified in multiple biological replicates (Fig. 3D), CR and GF mice showed no differences in the percentage of ISCs that were actively cycling (27.4 ± 6.9% versus 24.5 ± 6.8% respectively).
Figure 3.
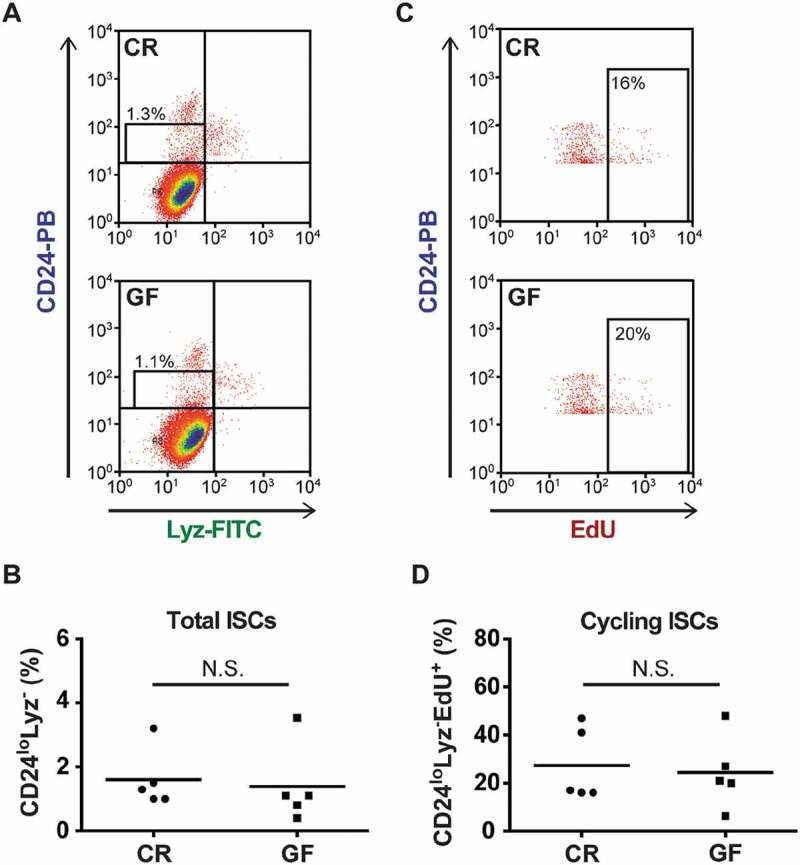
Conventionally raised (CR) and germ-free (GF) mice have similar numbers of intestinal stem cells (ISCs). A: Representative flow cytometry plots showing total ISC numbers (CD24loLyz–) in CR and GF mice. B: Quantitative analysis of ISCs as a percentage of the total epithelium in CR and GF mice (n = 5 pools of mice/group, with 4 mice/pool, P = 0.5). C: Representative flow cytometry plots evaluating the cycling status of ISCs (CD24loLyz–EdU+) in CR and GF mice. Lyz+ cells have been excluded from these gates. D: Quantitative analysis of cycling ISCs as a percentage of total ISCs (n = 5 pools of mice/group, with 4 mice/pool, P = 0.8). N.S. = not significant. Quantitative data are represented as scatter plots with a line representing the mean.
Enteroids from germ-free and conventionally-raised mice demonstrate similar ex vivo crypt survival and proliferation
Because 3-D culture has proven to be a valuable tool to study the behavior of ISCs, we next generated enteroids from GF and CR mice. We then quantified the efficiency of enterosphere and enteroid formation, lateral bud counts, and enteroid area. These parameters have been previously used to assess ISC survival and proliferation.27,28 Representative images of CR and GF enteroids grown in Matrigel for 6 days are shown in Fig. 4A. The efficiency of plated crypts (day 0) in generating enterospheres (day 1) and enteroids (day 6) was quantified in Fig. 4B, and showed no difference between the two experimental groups. This suggests similar ex vivo survival of CR and GF jejunal ISCs. Lateral budding and 2-D area of CR and GF enteroids (day 6) are depicted in Fig. 4C. Again no differences in ex vivo CR and GF ISC proliferation were identified using these parameters.
Figure 4.
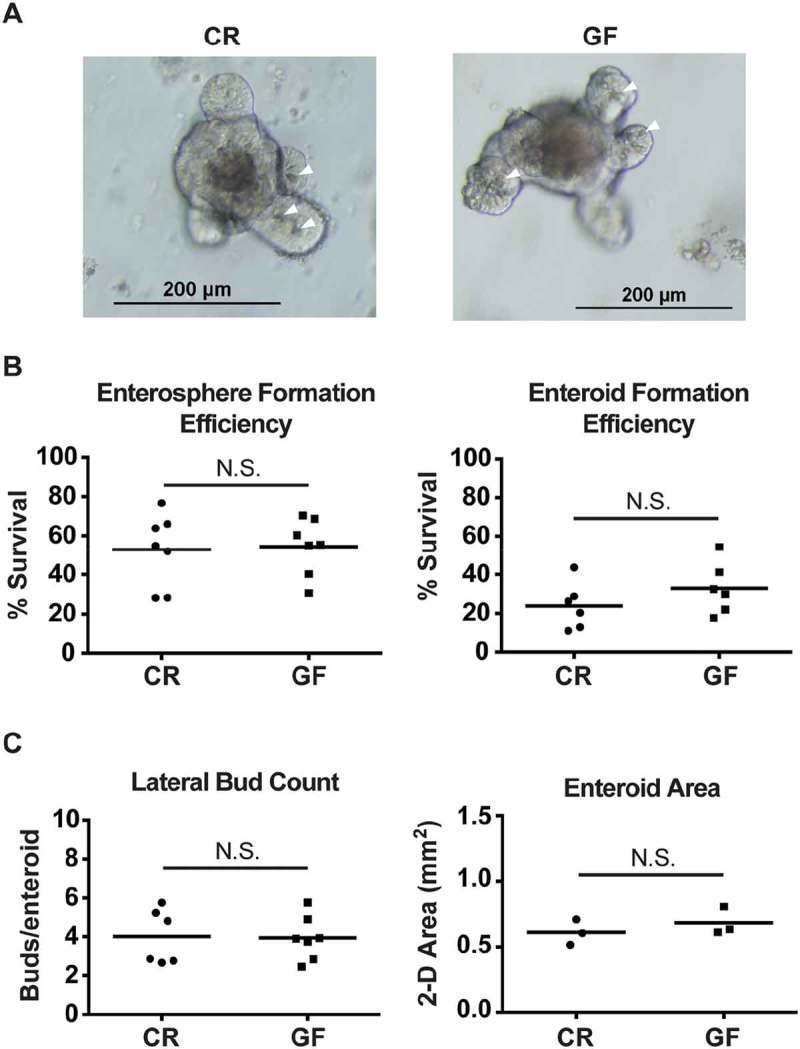
Conventionally raised (CR) and germ-free (GF) jejunal enteroids show no differences in crypt survival or proliferation. A: Representative images of day 6 jejunal enteroids from CR and GF mice. Arrowheads (white) indicate PCs. B: Efficiency of enterosphere (day 1) and enteroid (day 6) formation from cultured CR and GF crypts (n = 6-7 mice/group, P > 0.2 for both measures). Represented as percentage of plated crypts surviving. C: Lateral bud counts (n = 6-7 mice/group) and area (n = 3 mice/group) of CR and GF enteroids on day 6 of culture (P > 0.4 for both measures). For all enteroid studies, 200–400 crypts were plated per mouse. N.S. = not significant. Quantitative data are represented as scatter plots with a line representing the mean.
The enteric microbiota has a limited impact on jejunal Paneth cell antimicrobial peptide transcript expression
In addition to their supporting role within the ISC niche, PCs also contribute to the mucosal defense system of the intestine via the production of AMPs. Although the impact of the microbiota on AMP expression has been examined in ex-GF mice,29-32 to our knowledge this has not been performed in a comprehensive fashion in freshly derived jejunal crypts from mice that have been born and raised in GF versus CR conditions. To accomplish this, we measured GF and CR crypt transcript levels of representative molecules from the primary PC AMP classes expressed in C57BL/6J mice. These include lysozyme (Lyz), C-type lectins (Reg3γ), angiogenin 4 (Ang4), α-defensins (PanCrp), and cryptdin-related sequence peptides (Crs1c). These results are shown in Fig. 5. As indicated, Reg3γ was the only AMP that was significantly regulated by the microbiota in this system. This reduction of Reg3γ expression in the GF state is consistent with previous studies.29,30
Figure 5.
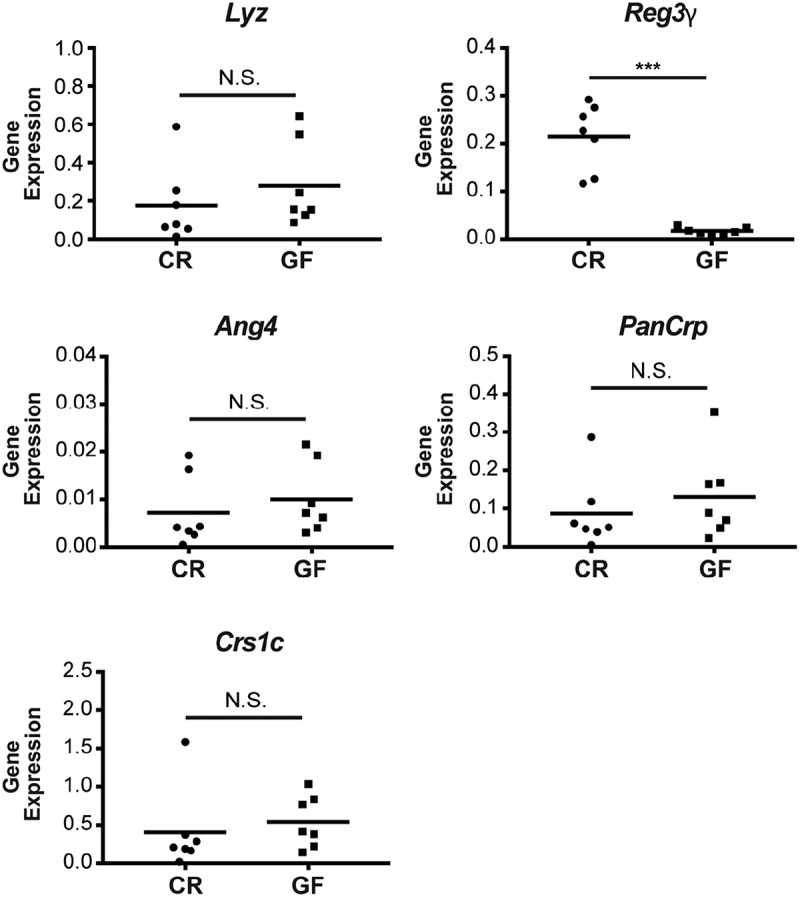
The enteric microbiota selectively regulates jejunal Paneth cell (PC) Reg3γ expression. Transcript levels of the primary antimicrobial peptide classes in C57BL/6J mice were measured in conventionally raised (CR) and germ-free (GF) jejunal crypts, including: lysozyme (Lyz), regenerating islet-derived protein 3 gamma (Reg3γ), angiogenin 4 (Ang4), global α-defensins (PanCrp), and cryptdin-related sequence 1c (Crs1c). Data are shown as target gene expression normalized to β-actin (scatter plot with mean, ***P < 0.001). N.S. = not significant.
Enteroid culture fails to recapitulate in vivo differences of Paneth cell function
While numerous studies have attested that Matrigel-based 3D enteroid cultures are an acceptable in vitro model to study the behavior of ISCs, investigators have only just begun using this system to analyze PC function.33 Given the robust differences in vivo of Reg3γ expression in CR versus GF intestinal crypts shown in Fig. 5, we sought to use enteroid cultures to explore the hypothesis that these differences reflect crypt autonomous signaling from luminal microbiota. Interestingly, the baseline endogenous Reg3γ expression of CR crypts was rapidly lost in culture. The data in Fig. 6 show that by day 6, levels of Reg3γ mRNA in enteroids from CR and GF mice were equivalent to those in the original crypts from GF mice. The obvious explanation for this observation would be that the lack of bacteria and/or bacterial products in the sterile culture system results in loss of the signaling responsible for elevated Reg3γ expression in CR crypts. To assess this, we attempted to re-induce Reg3γ mRNA expression in CR enteroids by stimulating them with a panel of microbial products, including heat-killed bacteria, lipopolysaccharide (LPS), flagellin, and muramyl dipeptide (MDP). No induction of Reg3γ expression was detected upon exposure of CR enteroids to these ligands (data not shown). Similar findings were observed when enteroids were disrupted prior to ligand exposure, to allow for interaction with the apical surface of the epithelial cells.
Figure 6.
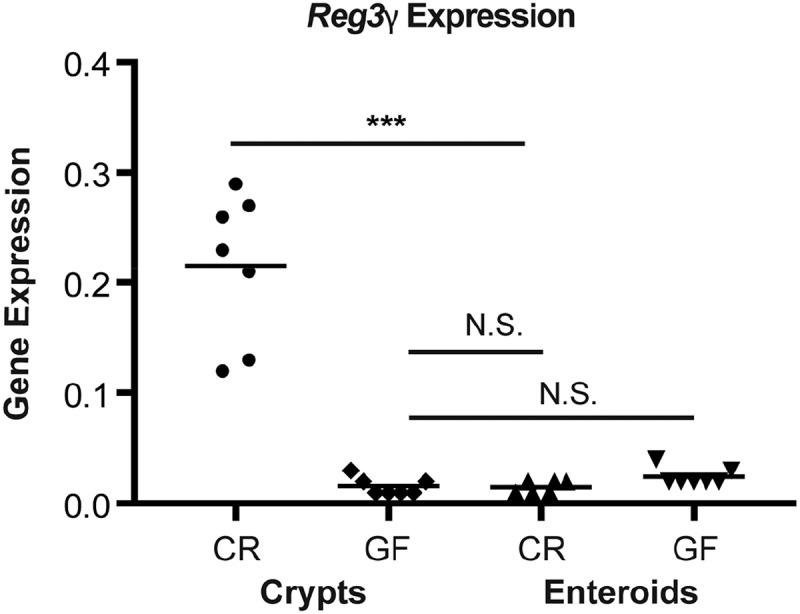
Elevated Reg3γ expression of conventionally raised (CR) crypts is rapidly lost in culture. Scatter plot (with mean) shows Reg3γ transcript expression of freshly isolated CR and germ-free (GF) crypts, and corresponding enteroids harvested after 6 days of culture. Data are depicted as target gene expression normalized to β-actin. Two-way ANOVA showed highly significant main effects of microbial status (P < 0.0001) and culture (P < 0.0001) as well as the interaction of the two variables (P < 0.0001). Pairwise Mann-Whitney comparisons showed the Reg3γ mRNA in CR enteroids to be significantly less than those in CR crypts (***P < 0.002) and to be not significantly (N.S.) different from those in GF crypts (P > 0.94).
Discussion
In this study, we have sought to define the impact of the enteric microbiota on PC biology in the murine small intestine. To this end, we have completed a comprehensive, three-dimensional analysis of PC census within the jejunum of mice lacking an endogenous microbiota, as compared to mice raised in conventional housing conditions. We have also characterized the putative downstream consequences of altered PC function in these experimental groups. In doing so, we have established that the commensal microbiota does indeed regulate PC biology in this region of the intestinal tract. This is evidenced by increased PC numbers and transcript expression of the AMP Reg3γ in the jejuna of CR mice, as compared to GF animals.
Despite the increased PC census in CR mice relative to their GF counterparts, we found no differences in the overall proliferative status of the jejunal epithelium in these experimental groups. These observations extended to the ISC compartment, in which ISC numbers and cycling status were similar between CR and GF mice. This suggests that decreased PC numbers do not directly associate with diminished epithelial proliferation or maintenance of the ISC niche in the jejunum of GF animals. Although PCs are able to support the ISC compartment,5,25 our findings are consistent with work demonstrating that PCs are not essential for the survival and proliferation of active ISCs.34 Specifically, the intestinal stroma is able to serve as a niche for ISCs, even in the absence of epithelial-derived Wnt ligands.35 Such compensatory signaling may allow GF ISCs to develop appropriately, despite a reduction in PC numbers.
The similar rates of epithelial proliferation, ISC numbers, and cycling status between CR and GF mice also indicate that the enteric microbiota does not influence these parameters through PC-independent mechanisms. This is consistent with early studies that showed similar crypt depth and mitotic indices in the jejuna of CR and GF Wistar rats.36 More recently, work by Peck et al. revealed comparable numbers of jejunal Sox9-EGFPLow cells in CR and GF mice.37 Sox9-EGFPLow cells are enriched for active ISCs,38 thereby supporting our results. In contrast, several studies have demonstrated CR mice have increased cell division (versus GF mice) in other segments of the small intestine, including the duodenum39 and ileum.36, 40 This raises the possibility that different regions of the small intestine may display distinct responses to the enteric microbiota. Ultimately, this would require testing in a single study examining ISCs in different small intestinal regions from the same mice. Adding to this complexity, individual bacteria also possess differential abilities to stimulate epithelial proliferation. For example, the bacterial strain Lactobacillus plantarum is able to induce intestinal epithelial proliferation by stimulating host reactive oxygen species (ROS) production.41 In contrast, this effect is not observed with Erwinia carotovora, which does not promote ROS generation. These findings highlight the importance of considering both host and microbial factors when evaluating the impact of the microbiota on epithelial homeostasis in the intestinal tract.
In addition to the complexity of the host and microbial factors described, ISCs themselves comprise a heterogeneous group of cells. Specifically, active ISCs (identified by markers such as Lgr5, Ascl2, and Olfm4) are believed to support baseline homeostasis of the intestinal epithelium; conversely, a highly diverse group of reserve ISCs (marked by Bmi1, mTert, Dclk1, and Lrig1) can be recruited to respond to epithelial injury.42 Importantly, the CD24loLyz– ISC population is enriched for active ISCs, as roughly 85% of Lgr5+ cells fall within this fraction.26 Therefore, it is possible that differences in reserve ISC communities between CR and GF mice could be undetected using a CD24-based analysis. Intriguingly, the study by Peck et al. described above demonstrated increased numbers of Sox9-EGFPHigh cells in GF mice compared to CR counterparts.37 Although the Sox9-EGFPHigh population represents a heterogeneous group of cells, high levels of Sox9 have been linked to the reserve ISC state.43 Hence it is possible that the microbiota has a suppressive effect on reserve ISCs. Future studies using ISC injury models in CR and GF mice could provide valuable insight into the functional relevance of these findings.
While the enteric microbiota does not appear to impact PC influences on the ISC compartment, we did identify alterations in the PC antimicrobial program of GF mice. Specifically, transcript levels of Reg3γ were significantly reduced in GF jejunal crypts, relative to those derived from CR mice. Although Reg3γ can be expressed broadly in intestinal epithelial cells, it is highly abundant in PCs, as evidenced by gene expression profiling of laser-captured PCs, as well as immunogold electron microscopy.29 This previous study also demonstrated an induction of Reg3γ after colonizing GF mice with a complex microbiota, and Reg3γ is known to be regulated by MyD88 signalling,44,45 supporting the concept that bacterial ligands can stimultate its expression. Interestingly, the other major classes of mouse AMPs (Lyz, Ang4, Crs1c, and the α-defensins) were not influenced by the microbiota in the mouse jejunum. Again, it is possible that small intestinal location may impact these findings. Although additional studies are needed, this is supported by evidence that mouse PC AMPs display substantial regional variation along the length of the intestinal tract,30 and work from our group demonstrating that the microbiota does induce mRNA expression of Lyz, Ang4, Reg3γ and the α-defensins in the mouse ileum.9
To mechanistically study how the microbiota regulates jejunal Reg3γ expression, we turned to the enteroid system, which provides a more reductionist approach to study this question. Enteroids derived from CR and GF crypts both contained all 4 differentiated lineages of the small intestine, including PCs. Moreover, we found no differences in the efficiency of enteroid formation, lateral bud counts, or area between enteroids generated from CR and GF mice. Strikingly, when Reg3γ transcripts were measured in these enteroids, the marked in vivo differences in expression between CR and GF mice were abrogated. Specifically, by 6 days after plating, both CR and GF enteroids expressed low levels of Reg3γ that were similar to GF mice. These findings demonstrate that PCs in the enteroid culture system display different biological characteristics to those in vivo, and serve as a caution to investigators using classic enteroid culture to study PC function.
A key difference between jejunal enteroids and in vivo crypts is the lack of a commensal microbiota in the culture system. To determine if bacterial engagement with the intestinal epithelium is required to induce Reg3γ expression, we stimulated CR enteroids with a variety of bacterial ligands at both their basolateral and apical surfaces. None of the ligands tested, including heat-killed bacteria, were able to induce Reg3γ expression in the enteroid system. Similarly, previous work by Davies et al. has shown that the bacterial ligands LPS and Pam3CSK4 do not impact the morphology or survival of jejunal enteroids.28 However, this study did demonstrate that the percentage of Lyz+ PCs was reduced in enteroids treated with the viral mimetic Poly I:C. Emerging data suggest that non-bacterial components of the microbiota, such as viruses, fungi, and archea, play key roles in modulating host physiology.46 Although the impact of these non-bacterial organisms on PC function is beyond the scope of our current study, it will be interesting to examine such influences in future investigations. It should also be noted that ex vivo enteroids lack enteroids also lack influences from the underlying stroma. Therefore, it is possible that microbial regulation of Reg3γ expression may occur indirectly through underlying mesenchymal cells. In future studies, co-culturing enteroids with specific stromal components will allow us to determine if non-epithelial cell types are required to transmit signals from the enteric microbiota to the PC compartment.
Ultimately, the findings presented in this study have important clinical implications, as PC dysfunction is believed to contribute to a wide variety of gastrointestinal disorders. For example, PC-specific disruption of the Crohn’s disease (CD) risk allele Xbp1 leads to spontaneous enteritis, suggesting these cells may be important regulators of intestinal inflammation in specific patients.47 Indeed, several CD risk alleles have been associated with abnormal PC morphology and activity,48 and PC defects in CD patients appear to promote intestinal dysbiosis.49 In addition to the chronic inflammatory processes observed in CD, PCs are also involved in the pathogenesis of more acute intestinal injury. Specifically, PCs appear to be a key target in intestinal graft-versus-host disease.50 This leads to significant disruption of intestinal homeostasis and the enteric microbiota, and is associated with increased patient mortality.51 Finally, PC disruption has also been implicated in the pathogenesis of necrotizing enterocolitis, demonstrating an important role for these cells in the developing intestine.52, 53 The findings presented in the current study represent an important step towards understanding how the microbiota regulates PC function. This knowledge will be necessary to ultimately develop microbial modulation strategies (i.e. probiotics, antibiotics, fecal microbiota transplantation) as novel therapies for these diseases.
In conclusion, our findings demonstrate that the enteric microbiota regulates jejunal PC function by increasing PC numbers and inducing Reg3γ expression. In contrast, the enteric microbiota does not appear to regulate jejunal ISC census and proliferation. We believe these findings are important considerations for investigators using GF mice and the enteroid culture system to study PC and ISC biology. Moreover, they set the stage for future work focused on developing specific microbial-based strategies to enhance PC function that may be of critical importance for a spectrum of gastrointestinal disorders.
Materials and Methods
Mice
All animal experiments were approved by the University of North Carolina Institutional Animal Care and Use Committee, and performed per guidelines dictated by the American Association for Laboratory Care and Research. Adult (8-16 week old), female, wild-type, C57BL/6J mice were housed in either CR or GF conditions under a 12:12-h light-dark cycle. CR mice were born and raised for a minimum of three generations in specific pathogen-free conditions. GF mice were derived and maintained in sterile conditions at the National Gnotobiotic Rodent Resource Center at UNC Chapel Hill.
Histologic analyses
Paneth cell quantification: The small intestine, excluding the duodenum, was procured by isolating intestinal tissue from the ligament of Treitz to the ileo-cecal junction. The jejunum was defined as the proximal one-half of this resected specimen. The distal 1.5 cm of this jejunal segment was removed for histology and fixed in 10% phosphate buffered formalin for 48 hours. The tissue was then embedded longitudinally in paraffin and cut in 5 μm sections for H&E staining. PC number was assessed by counting the granulated eosinophilic cells at the base of crypts. Only crypts that possessed an intact lumen from the base of the crypt to the lumen of the intestine were considered for analysis. A minimum of 10 crypts was analyzed per animal, allowing for the calculation of an average number of PCs/crypt from each mouse. All histological measurements were performed by a blinded scorer.
Proliferation: Mice were euthanized following a 1 h pulse of EdU (100 µg) delivered intraperitoneally. The intestine was removed, flushed, and placed in 4% paraformaldehyde overnight at 4°C, and then transferred to a 30% sucrose solution overnight at 4°C. The tissue was then embedded in optimal cutting temperature (Tissue-Tek catalog no. 4583). 8 µm sections were cut, and EdU was detected using the Alexa 488 azide included in the Imaging Kit (Thermo Fisher catalog no. C10337).
Flow cytometry analyses
Flow cytometry was used to enumerate the following cell populations: 1) total PCs (Lyz+ cells as a percentage of total epithelium); 2) total cycling cells (EdU+ cells as a percentage of total epithelium); 3) total ISCs (CD24loLyz– cells as a percentage of total epithelium); and 4) cycling ISCs (CD24loLyz–EdU+ cells as a percentage of total ISCs). Specific cell staining techniques and gating strategies for each of these populations were performed per published protocols.9,26 For all flow cytometry studies, whole jejuna from 4 mice were pooled, and 5 biological replicates were run for each group (20 mice/group). Jejunal epithelial cells were isolated from CR and GF mice using an established ethylenediaminetetraacetic acid/dispase dissociation method,54 followed by staining and analysis of 1 × 106 cells. The following antibodies were used for cell staining: FITC-conjugated anti-lysozyme (Dako catalog no. F0372), Pacific Blue anti-CD24 (BioLegend catalog no. 101819), EdU-647 Flow Cytometry Kit (Thermo Fisher catalog no. C10424). In all flow cytometry studies, CD45+ hematopoietic cells and debris were excluded based on their established location on bivariate forward scatter versus side scatter plots, and doublets were excluded using pulse width versus forward scatter analysis.26 All gates were established using fluorescence minus one controls to identify labeled cells.55 Further details of this approach are described in Suppl. Fig. 1.
Crypt culture experiments
Crypt isolation and culture: Crypts were isolated from 10 cm jejunal tissue as previously described.56 Specific reagents and methods for crypt culture are described in detail by Sato et al. and are based largely on this protocol.57 Briefly, crypts were plated at a density of 60–100 crypts/well (3-4 wells/mouse) in hESC-qualified Matrigel (BD catalog no. 354277). After solidification of the Matrigel, 200 μl of culture media was added/well. This media was comprised of Advanced DMEM/F12 (Gibco catalog no. 12634-028) containing: L-GlutaMAX (1:100, Gibco catalog no. 35050–061), gentamicin/kanamycin (1:100), HEPES (10 mM, Gibco catalog no. 15630-080), N2 (1:100, Gibco catalog no. 17502-048), B27 (1:50, Gibco catalog no. 17504-044), Y27632 (10 μΜ, Sigma-Aldrich catalog no. Y0503), epidermal growth factor (50 ng/mL, R&D Systems catalog no. 2028-EG-200), noggin (100 ng/mL, R&D Systems catalog no. 1967-NG-025/CF), and R-spondin-1 (500 ng/mL, R&D Systems catalog no. 4645-RS). Media was changed every 6 days.
Enteroid analyses: Analyses were performed using bright field microscopy. Efficiency measurements were calculated by quantifying total number of enterospheres (day 1) or enteroids (day 6) divided by the total number of plated crypts (day 0). These structures were defined as recommended by the Intestinal Stem Cell Consortium.58 Enteroid budding was enumerated by counting total number of lateral buds per enteroid, while area was assessed using ImageJ (https://imagej.nih.gov/ij/). All measurements evaluated total structures from a minimum of four wells per mouse. Each biological replicate is represented as the average of these technical replicates.
Enteroid stimulation studies: Enteroids were grown in culture for 6 days as described above. At this time, they were stimulated with bacterial ligands that had been added to fresh media. Specific ligands included: heat-killed bacteria (Listeria monocytogenes and Salmonella enterica serovar Typhimurium), LPS (10 or 100 μg/mL), flagellin (10 ng/mL), or MDP (10 μg/mL). Multiple stimulation times were assessed, including 1, 6, 24, and 48 hours. To enhance apical exposure of epithelial cells to these ligands, the described conditions were also applied to enteroids that had been physically disrupted by repeat pipetting through a 21 gauge needle and syringe.
Antimicrobial peptide transcript expression analyses
RNA extraction: For each biological replicate, RNA was extracted from ~2000 jejunal crypts or ~150-200 6-day-old enteroids. For crypts, tissues were deposited in 600 μL Buffer RLT (Qiagen catalog no. 79216) supplemented with 10 μL/mL of β-ME, vortexed, and frozen at -80 °C. For enteroids, media from each well was first replaced with 200 μL PBS. The Matrigel was then scraped from the well and deposited with the PBS into a 1.5 mL microfuge tube. This was centrifuged, and the PBS was replaced with 600 μl RLT + β-ME. Samples were then vortexed and frozen as described above. For RNA extraction, all samples were thawed and immediately homogenized by centrifuging through QIAshredder columns (Qiagen catalog no. 79654). Total RNA was then isolated using an RNeasy Mini Kit (Qiagen catalog no. 74104), per manufacturer instructions. Complementary DNA was generated using SuperScript II reverse transcriptase (Thermo Fisher catalog no. 18064014).
Gene expression analyses: quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) was performed using TaqMan Gene Expression Master Mix (Thermo Fisher catalog no. 4369016) or SYBR Green PCR Master Mix (Thermo Fisher catalog no. 4309155), per manufacturer instructions. Specific primer/probe sets were obtained from Applied Biosystems as follows: Actb (Mm02619580_g1), Lyz (Mm00727183_s1), Ang4 (Mm03647554_g1), Reg3γ (Mm00441127_m1). Forward and reverse SYBR Green primers for Crs1c and PanCrp were generated through the Nucleic Acids Core Facility (University of North Carolina, Chapel Hill) based on previously published sequences.59 Target gene expression relative to the housekeeping gene Actb was determined for each biological replicate using the ∆CT method.60 The following equation was utilized for all gene expression calculations: 2 –(ΔCT) = 2 –(CTAMP – CTActb).
Statistical Analyses
GraphPad Prism software version 6 was used to analyze experimental groups. Results are expressed as scatter plots with the mean indicated as a line. Differences between groups were compared using an unpaired Mann-Whitney test. P < 0.05 was considered significant. To determine effects of microbial status and culture conditions on Reg3γ expression, two-by-two tables were generated using qRT-PCR data for Reg3γ and subjected to two-way ANOVA, with the α set at P < 0.05.
Supplementary Material
Funding Statement
This work was supported by the National Institutes of Health under grants K08 DK095917 and R03 DK104005 to ASG, U01 DK085547 to SJH, P40 OD010995 to R. Balfour Sartor, and P30 DK034987 to Robert Sandler.
Acknowledgements
The authors wish to thank Dr. Sayanty Roy for her assistance preparing the figures for this study, and Dr. Allison Rogala for her expert advice and critical review of the manuscript. We also wish to acknowledge assistance from the gnotobiotic (Maureen Bower) and histology cores (Carolyn Suitt) at the UNC Center for Gastrointestinal Biology and Disease.
Disclosure of Interest
The authors report no conflict of interest.
References
- 1.Paneth J. Ueber die secernirenden Zellen des Dünndarm-Epithels. Arch Mikrosk Anat. 1888;31:113–91. doi: 10.1007/BF02955706. [DOI] [Google Scholar]
- 2.Schwalbe G. Beiträge zur Kentniss der Drüsen in den Darmwandungen, insbesondere der Brunnerschen Drüsen. Arch Mikrosk Anat. 1872;8:92–140. doi: 10.1007/BF02955835. [DOI] [Google Scholar]
- 3.Clevers HC, Bevins CL.. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744 PMID:23398152. [DOI] [PubMed] [Google Scholar]
- 4.Ouellette AJ. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010;26:547–53. doi: 10.1097/MOG.0b013e32833dccde PMID:20693892. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H.. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–8. doi: 10.1038/nature09637 PMID:21113151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stappenbeck TS, McGovern DP.. Paneth cell alterations in the development and phenotype of crohn’s disease. Gastroenterology. 2017;152:322–6. doi: 10.1053/j.gastro.2016.10.003 PMID:27729212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McElroy SJ, Underwood MA, Sherman MP.. Paneth cells and necrotizing enterocolitis: a novel hypothesis for disease pathogenesis. Neonatology. 2013;103:10–20. doi: 10.1159/000342340 PMID:23006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriguchi Y, Takashima S, Oka H, Shimoji S, Nakamura K, Uryu H, Shimoda S, Iwasaki H, Shimono N, Ayabe T, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood. 2012;120:223–31. doi: 10.1182/blood-2011-12-401166 PMID:22535662. [DOI] [PubMed] [Google Scholar]
- 9.Gulati AS, Shanahan MT, Arthur JC, Grossniklaus E, von Furstenberg RJ, Kreuk L, Henning SJ, Jobin C, Sartor RB.. Mouse background strain profoundly influences Paneth cell function and intestinal microbial composition. PLoS One. 2012;7:e32403. doi: 10.1371/journal.pone.0032403 PMID:22384242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Es JH, Jay P, Gregorieff A, ME van Gijn, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–6. doi: 10.1038/ncb1240 PMID:15778706. [DOI] [PubMed] [Google Scholar]
- 11.Batlle E, Henderson JT, Beghtel H, MMW van den Born, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–63. doi: 10.1016/S0092-8674(02)01015-2 PMID:12408869. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY.. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–8. doi: 10.1126/science.1065718 PMID:11739954. [DOI] [PubMed] [Google Scholar]
- 13.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY.. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–88. doi: 10.1053/j.gastro.2007.03.047 PMID:17570220. [DOI] [PubMed] [Google Scholar]
- 14.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI.. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–84. doi: 10.1073/pnas.0605374104 PMID:17210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda K, Littman DR.. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848 PMID:27383982. [DOI] [PubMed] [Google Scholar]
- 16.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R.. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002 PMID:15260992. [DOI] [PubMed] [Google Scholar]
- 17.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825 PMID:19855381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez Rodriguez NR, Eloi MD, Huynh A, Dominguez T, Lam AH, Carcamo-Molina D, Naser Z, Desharnais R, Salzman NH, Porter E.. Expansion of Paneth cell population in response to enteric Salmonella enterica serovar Typhimurium infection. Infect Immun. 2012;80:266–75. doi: 10.1128/IAI.05638-11 PMID:22006567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klockars M. Concentration and immunohistochemical localization of lysozyme in germ-free and conventionally reared rats. Acta Pathol Microbiol Scand A. 1974;82:675–82.PMID:4447004. [DOI] [PubMed] [Google Scholar]
- 20.Bry L, Falk P, Huttner K, Ouellette A, Midtvedt T, Gordon JI.. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci U S A. 1994;91:10335–9. doi: 10.1073/pnas.91.22.10335 PMID:7937951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodning CB, Erlandsen SL, Wilson ID, Carpenter AM.. Light microscopic morphometric analysis of rat ileal mucosa: II. Component quantitation of Paneth cells. Anat Rec. 1982;204:33–8. doi: 10.1002/ar.1092040105 PMID:7149281. [DOI] [PubMed] [Google Scholar]
- 22.Satoh Y. Ultrastructure of Paneth cells in germ-free rats, with special reference to the secretory granules and lysosomes. Arch Histol Jpn. 1984;47:293–301. doi: 10.1679/aohc.47.293 PMID:6497592. [DOI] [PubMed] [Google Scholar]
- 23.Satoh Y, Vollrath L.. Quantitative electron microscopic observations on Paneth cells of germfree and ex-germfree Wistar rats. Anat Embryol (Berl). 1986;173:317–22. doi: 10.1007/BF00318915 PMID:3963410. [DOI] [PubMed] [Google Scholar]
- 24.Gregorieff A, Clevers H.. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–90. doi: 10.1101/gad.1295405 PMID:15833914. [DOI] [PubMed] [Google Scholar]
- 25.Gracz AD, Williamson IA, Roche KC, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza RJ, et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17:340–9. doi: 10.1038/ncb3104 PMID:25664616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Furstenberg RJ, Gulati AS, Baxi A, Doherty JM, Stappenbeck TS, Gracz AD, Magness ST, Henning SJ.. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G409–17. doi: 10.1152/ajpgi.00453.2010 PMID:21183658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller MK, Faulk DM, Sundaram N, Shroyer NF, Henning SJ, Helmrath MA.. Intestinal crypts reproducibly expand in culture. J Surg Res. 2012;178:48–54. doi: 10.1016/j.jss.2012.03.037 PMID:22564827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies JM, Santaolalla R, von Furstenberg RJ, Henning SJ, Abreu MT.. The viral mimetic polyinosinic: polycytidylicacid alters the growth characteristics of small intestinal and colonic crypt cultures. PLoS One. 2015;10:e0138531. doi: 10.1371/journal.pone.0138531 PMID:26414184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cash HL, Whitham CV, Behrendt CL, LV Hooper. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119 PMID:16931762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlsson J, Putsep K, Chu H, Kays RJ, Bevins CL, Andersson M.. Regional variations in paneth cell antimicrobial peptide expression along the mouse intestinal tract. BMC Immunol. 2008;9:37. doi: 10.1186/1471-2172-9-37 PMID:18637162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue R, Tsuruta T, Nojima I, Nakayama K, Tsukahara T, Yajima T.. Postnatal changes in the expression of genes for cryptdins 1-6 and the role of luminal bacteria in cryptdin gene expression in mouse small intestine. FEMS Immunol Med Microbiol. 2008;52:407–16. doi: 10.1111/j.1574-695X.2008.00390.x PMID:18328077. [DOI] [PubMed] [Google Scholar]
- 32.Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, Andersson M.. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. 2000;275:40478–82. doi: 10.1074/jbc.M007816200 PMID:11010975. [DOI] [PubMed] [Google Scholar]
- 33.Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG.. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015;8:352–61. doi: 10.1038/mi.2014.72 PMID:25118165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TH, Escudero S, Shivdasani RA.. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc Natl Acad Sci U S A. 2012;109:3932–7. doi: 10.1073/pnas.1113890109 PMID:22355124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, Edison Aliyev J, Wu Y, Bunte R, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–15. doi: 10.1242/dev.104976 PMID:24821987. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa K, Satoh Y, Tanaka H, Ono K.. Influence of conventionalization on small-intestinal mucosa of germ-free Wistar rats: quantitative light microscopic observations. Acta Anat (Basel). 1986;127:296–302. doi: 10.1159/000146301 PMID:3811822. [DOI] [PubMed] [Google Scholar]
- 37.Peck BC, Mah AT, Pitman WA, Ding S, Lund PK, Sethupathy P.. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem. 2017;292:2586–600. doi: 10.1074/jbc.M116.770099 PMID:28053090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST.. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108–18. doi: 10.1152/ajpgi.00004.2009 PMID:19228882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesher S, Walburg HE Jr, Sacher GA Jr.. Generation cycle in the duodenal crypt cells of germ-free and conventional mice. Nature. 1964;202:884–6. doi: 10.1038/202884a0 PMID:14190081. [DOI] [PubMed] [Google Scholar]
- 40.Khoury KA, Floch MH, Hersh T.. Small intestinal mucosal cell proliferation and bacterial flora in the conventionalization of the germfree mouse. Exp Med J. 1969;130:659–70. doi: 10.1084/jem.130.3.659 PMID:4896909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–28. doi: 10.1038/emboj.2013.224 PMID:24141879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henning SJ, von Furstenberg RJ.. GI stem cells - new insights into roles in physiology and pathophysiology. J Physiol. 2016;594:4769–79. doi: 10.1113/JP271663 PMID:27107928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H, Magness ST.. SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology. 2015;149:1553–63e10. doi: 10.1053/j.gastro.2015.07.004 PMID:26170137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG.. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–900. doi: 10.1084/jem.20070563 PMID:17635956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV.. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–63. doi: 10.1073/pnas.0808723105 PMID:19075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sartor RB, Wu GD.. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–39. doi: 10.1053/j.gastro.2016.10.012 PMID:27769810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–6. doi: 10.1038/nature12599 PMID:24089213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanahan MT, Carroll IM, Gulati AS.. Critical design aspects involved in the study of Paneth cells and the intestinal microbiota. Microbes Gut. 2014;5:208–14. doi: 10.4161/gmic.27466 PMID:24637592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu TC, Gurram B, Baldridge MT, Head R, Lam V, Luo C, Cao Y, Simpson P, Hayward M, Holtz ML, et al. Paneth cell defects in crohn’s disease patients promote dysbiosis. JCI Insight. 2016;1:e86907. doi: 10.1172/jci.insight.86907 PMID:27699268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teshima T, Reddy P, Zeiser R.. Acute graft-versus-host disease: novel biological insights. Biol Blood Marrow Transplant. 2016;22:11–6. doi: 10.1016/j.bbmt.2015.10.001 PMID:26453971. [DOI] [PubMed] [Google Scholar]
- 51.Levine JE, Huber E, Hammer ST, Harris AC, Greenson JK, Braun TM, Ferrara JL, Holler E.. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood. 2013;122:1505–9. doi: 10.1182/blood-2013-02-485813 PMID:23760615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White JR, Gong H, Pope B, Schlievert P, McElroy SJ.. Paneth cell disruption-induced necrotizing enterocolitis requires live bacteria and occurs independent of TLR4 signaling. Dis Model Mech. 2017;10:727–36. doi: 10.1242/dmm.028589 PMID:28450472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang C, Sherman MP, Prince LS, Bader D, Weitkamp JH, Slaughter JC, McElroy SJ.. Paneth cell ablation in the presence of Klebsiella pneumoniae induces necrotizing enterocolitis (NEC)-like injury in the small intestine of immature mice. Dis Model Mech. 2012;5:522–32. doi: 10.1242/dmm.009001 PMID:22328592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magness ST, Puthoff BJ, Crissey MA, Dunn J, Henning SJ, Houchen C, Kaddis JS, Kuo CJ, Li L, Lynch J, et al. A multicenter study to standardize reporting and analyses of fluorescence-activated cell-sorted murine intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2013;305:G542–51. doi: 10.1152/ajpgi.00481.2012 PMID:23928185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3%3c194::AID-CYTO1163%3e3.0.CO;2-C PMID:11746088. [DOI] [PubMed] [Google Scholar]
- 56.Seiler KM, Schenhals EL, von Furstenberg RJ, Allena BK, Smith BJ, Scaria D, Bresler MN, Dekaney CM, Henning SJ.. Tissue underlying the intestinal epithelium elicits proliferation of intestinal stem cells following cytotoxic damage. Cell Tissue Res. 2015;361:427–38. doi: 10.1007/s00441-015-2111-1 PMID:25693894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato T, Clevers H.. Primary mouse small intestinal epithelial cell cultures. Methods Mol Biol. 2013;945:319–28. doi: 10.1007/978-1-62703-125-7_19 PMID:23097115. [DOI] [PubMed] [Google Scholar]
- 58.Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, Lynch J, Li L, Magness ST, Martin MG, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–63. doi: 10.1152/ajpgi.00493.2011 PMID:22461030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shanahan MT, Carroll IM, Grossniklaus E, White A, von Furstenberg RJ, Barner R, Fodor AA, Henning SJ, Sartor RB, Gulati AS.. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut. 2013;63:903–10. doi: 10.1136/gutjnl-2012-304190 PMID:23512834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmittgen TD, Livak KJ.. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73 PMID:18546601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


