ABSTRACT
Bacteria, Archaea, Eukarya and viruses coexist in the human gut, and this coexistence is functionally balanced by symbiotic or antagonistic relationships. Antagonism is often characterized by the production of antimicrobials against other organisms occupying the same environmental niche. Indeed, close co-evolution in the gut has led to the development of specialized antimicrobials, which is attracting increased attention as these may serve as novel alternatives to antibiotics and thereby help to address the global problem of antimicrobial resistance. The gastrointestinal (GI) tract is especially suitable for finding novel antimicrobials due to the vast array of microbes that inhabit it, and a considerable number of antimicrobial producers of both wide and narrow spectrum have been described. In this review, we summarize some of the antimicrobial compounds that are produced by bacteria isolated from the gut environment, with a special focus on bacteriocins. We also evaluate the potential therapeutic application of these compounds to maintain homeostasis in the gut and the biocontrol of pathogenic bacteria.
KEYWORDS: antimicrobial, bacteriocin, gastrointestinal tract, microbiome, biocontrol, antibiotic resistance, probiotic, homeostasis
1. Introduction
Antimicrobials are compounds that kill or inhibit the growth of microorganisms. The availability of antimicrobials, and antibiotics in particular, has improved the quality of life and increased life expectancy. However, antibiotic resistance has become a major threat. Indeed, since the golden era of antibiotic discovery (1940-1960), when many compounds were discovered, especially from soil-derived actinomycetes, few new classes of antibiotics have been discovered, and the majority of new compounds have resulted from chemical modifications of existing ones. In an effort to address this challenge, different approaches have been proposed, like the use of discovery platforms,1 but there has been insufficient investment globally in developing novel procedures/techniques to identify new antimicrobials, and the characterization of these compounds thereafter.
Traditionally, the production of antimicrobials has been considered as an expression of competition and antagonism in bacteria, both inter- and intra-species, whose niches overlap.2 However, it should also be noted that some antimicrobials, and antimicrobial peptides in particular, might have a social purpose, serving to facilitate signaling between individual strains, although not necessarily an indication of cooperation.3 Antimicrobial activity has been proposed to promote biodiversity, as reflected by the differences in metabolic input of antimicrobial producers, sensitive and resistant species and in the ecological fitness that these bacteria show in a shared environment.4,5 More specifically, production of antimicrobials might confer an advantage to the producer against sensitive bacteria, but this production is the result of a metabolic effort, as is the maintenance of immunity among resistant bacteria, which requires less metabolic input than antimicrobial production, but more than that of sensitive bacteria. This has been referred to as the rock-scissors-paper (RSP) dynamic game that ultimately leads to a balanced co-existence in microbial communities.4,5 Different studies, both theoretical and experimental, have been carried out to understand the dynamics of the microbial communities and the role that antibiotics play in this context. These considerations further support the premise that natural sources containing a wide diversity of microorganisms continue to be an underexploited source of new antimicrobials that can be identified and developed at an enhanced rate with the aid of new ‘omics’ technologies, computational advances and development of new analytical tools.6
The human microbiome represents an excellent example of such an environmental niche and contains trillions of microorganisms that have co-evolved with the human host, leading to the view that humans can be considered to be super-organisms, constituted by a vast number of symbiotic relationships. Arguably, the most important microbial communities in humans can be found in the GI tract (the colon is estimated to contain over 70% of all the microbes in the human body7), in the mouth, skin, and urogenitalia. Microbial communities can be also found in, for example, lungs, breast milk and eyes.8 These communities are dynamic and change over time, due to characteristic processes of ecological succession, where microbial communities are substituted by new ones and can be affected by changes of environmental conditions. In the GI tract, the first microbial species that are established in infants are succeeded by new species to form a more mature and complex community and the balance can be altered by the use of antibiotics, dietary changes or introduction of exogenous pathogens.9,10
As the human microbiome is such a dense and diverse compilation of different microbial ecosystems that has not yet been fully explored, it is not difficult to regard it as a promising source of novel metabolites. In 2015, Donia and Fischbach reviewed the chemical spectrum of metabolites produced by the human microbiota and concluded that its diversity was as broad as any other microbial ecosystem, but that these metabolites were highly specific for interacting within the human host.11 More recently, Mousa et al12 (2017) extensively reviewed the same topic, highlighting the variety of specialized metabolic compounds produced by this human microbiota and the effects within the human host. These metabolites include lipids and glycolipids, oligosaccharides, terpenoids, polyketides, amino acids, non-ribosomal peptides and ribosomally synthesized post-translationally modified peptides (RiPPs). This chemical variety also reflects a variety of functions: immunomodulatory, cytotoxic, antioxidant and, of course, antimicrobial, although the function of many of these compounds is not yet identified. New compounds and functions continue to be discovered and characterized. Among the vast number of relevant papers are two recent examples that have shown that a human nasal commensal produces a novel peptide antibiotic that is able to limit Staphylococcus aureus colonization and maintain homeostasis13 and that an antimicrobial produced by skin commensal bacteria has potential for the treatment of atopic dermatitis.14 In the latter case, the observations point to dysbiosis of the skin microbiome as the origin of the problem and a rebalancing of the situation as a potential solution.
Given the aforementioned observations, the variety of species that are coexisting in the human microbiome, most densely in the GI tract, and the number of antimicrobials with a human microbiome origin that have already been described, we can assume that the human gut can continue to be a promising source of new antimicrobials that can help to address specific aetiologies and complex systemic conditions such as metabolic syndrome, inflammatory bowel disease, colorectal cancer and diabetes, that have been started to be associated with an imbalance in gut microbiota communities.15
Here we review the variety of antimicrobials produced by bacteria present in the human GI tract and, in the process, highlight the importance of a targeted approach to discover new effective antimicrobials that can help to maintain good health in humans and to address the problem of antimicrobial resistance.
2. Gut microbiota composition and distribution
Gut microbiota composition is heterogeneous across the GI tract as a result of variations in a broad variety of parameters including fluctuations in temperature, pH, water activity and gas composition as well as differences in physiology throughout the gut. Nutrients are a very important factor because they are the main source of energy for gut bacteria. Their chemical and physical nature determines flow rates in the gut and, therefore, the process of adhesion and washing out of microorganisms. Surface tension can also be affected, having an effect on the physiology of bacteria. Further information on gut conditions and how they influence composition has been extensively reviewed previously.10,16
It is important to consider that the gut microbiota composition is also not static. As noted earlier, alteration of the microbiome structure starts from the moments after birth and varies significantly initially.17,18 However, it stabilizes after the first7 or third10 year of life, maintaining a relatively consistent composition that tends to recover even after acute changes, such as a single exposure to antibiotics.16,17 More sustained changes, like dietary alterations over an extended period can, however, induce longer lasting structural changes.7
Observations of variation among individuals, throughout their intestinal tracts and across their life span, has made it clear that it is challenging to describe what constitutes a healthy gut microbiota.8 However, it has been argued that a functional approach to categorize the gut microbiota may more accurately reflect its ‘health’. This is based on the fact that the microbiota is capable of carrying out a variety of metabolic and molecular functions. This functionality is somewhat redundant and thus remains relatively even in situations where changes in taxonomy are evident.10
There have been considerable advances in approaches to determine microbial composition and the identification of their functional attributes. High throughput new generation DNA sequencing platforms in particular have helped to identify, and partially characterize microbes, most of which cannot be cultured. Care needs to be taken however as the vast majority of investigations relating to the human gut microbiota have been performed using stool samples and, as pointed out by Bajaj et al19 and others, fecal communities do not reflect the gut mucosal microbiota or microbial communities at locations in the upper sections of the GI tract.16
In general, the dominant bacterial phyla in the gut are Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Verrucomicrobia.16 Differences in physiological conditions result in a heterogeneous distribution of bacteria across the GI tract. The presence of bile acids, lower pH, higher oxygen levels and antimicrobial compounds (of both host and bacterial origin) favors colonization of the small intestine by groups of facultative anaerobes, such as Firmicutes (Lactobacillales) and Proteobacteria (Enterobacteriales). In the proximal part of the small intestine, the extreme environmental conditions result in it having a lower microbial density. In the distal part of the small intestine, near the ileum, these conditions become less limiting. Microbial density is highest in the colon where the transit time is slower, and the absence of easily digestible nutrients facilitates the proliferation of fermentative organisms that are able to degrade more complex compounds, including dietary fiber. These organisms include, for example, representatives of the Bacteroidetes (Bacteroidaceae, Prevotellaceae and Rikenellaceae) in the lumen and Firmicutes (Lachnospiraceae and Ruminococcaceae) in the inter-fold regions.16
Despite the fact that bacteria have been most extensively studied, there are other groups of microbes in the human GI tract that have not been as well researched but have an important role in the ecological dynamics, like archaea, viruses and eukaryotes.10,20 Archaea is represented by Euryarcheota and Crenarchaeota. Methanobrevibacter smithii has been identified as the dominant human methanogen where it is estimated to be present in 50% of the adult population21 and the concept of archaebiotics has been proposed, as probiotics of archaeal origin.22 Eukarya presence includes amoebae and fungi. The concept ‘mycobiome’ has been proposed to cover the diversity and dynamics of fungi in the human body.23 It has been suggested that mycobiome and microbiome are intimately regulated, influencing each other in the colonization of the gut niche and having a crucial effect on the overall health status of the host. Recent reviews describe the human mycobiome and its implications in health and disease.24,25 “Virome” is another recent concept to define the community of viruses that can be found within the human host.26 There are also eukaryotic viruses, plant derived viruses that have a dietary origin, giant viruses and bacteriophages.26 Antimicrobials produced by representatives of these groups in the gut have not been identified and characterized yet, to the best of our knowledge. However, Archaea, Eukarya and viruses are already known for their antimicrobial capabilities. Fungi are an especially important source of antibiotics27 and Archaea have also been reported to show antimicrobial activity.28,29 Some bacteriophages produce lytic enzymes that can kill bacteria,30 and the use of the phages derived from the human gut has been proposed as a potential therapeutic to modulate gut composition (phage therapy).26,31 Further examination of these groups might reveal novel antimicrobials and modulation strategies.
3. Non-peptide antimicrobial activity in the gut
Bacteria develop different antagonism strategies to gain ecological advantages over other bacteria in the gut. Some of them are direct strategies, like competitive removal of essential substrates, accumulation of D-amino acids, lowering the oxidation-reduction potential and co-aggregation.32 Another strategy involves the production of substances of metabolic origin that have the ability to limit the growth of surrounding bacteria. Such activity can be specific or non-specific, where the regulatory agent does not address a specific target, hydrogen peroxide (H2O2) being a good example of a chemical with such activity. H2O2 is well known for its antimicrobial properties that are mediated through oxidizing effects on bacterial cells and their molecular structures.33 It has been suggested that H2O2 contributes to maintain a healthy microbiota.34 H2O2 can function synergistically with other compounds produced by bacteria, such as lactic acid,35 but its presence has also been linked to chronic inflammatory lesions in colon.36 Lactic acid and other organic acids are believed to function by virtue of their undissociated form crossing the lipid membrane and its subsequent dissociation in the neutral pH once inside, producing ions that cause stress in the cell. Lactic acid is produced by the carbohydrate metabolism of the so called lactic acid bacteria (LAB), while the important group of short chain fatty acids (SCFA), such as formate, acetate, propionate and butyrate, are produced by bacteria fermenting starches, dietary fiber, sugars, proteins and amino acids37 and their antimicrobial activity is due to acidification of the environment.38 There are other compounds of bacterial origin, like diacetyl (2,3-butadione),39,40 ethanol, CO2, ammonia or phenolic compounds, which have also been associated with antimicrobial activity,11 but, as with the previous examples, their non-specific activity and other side effects,41 make them unsuitable for clinical applications and they will not be discussed further in this review.
4. Peptidic antimicrobial activity in the gut
Bacteria are also capable of producing antimicrobial peptide compounds that are considered to be target-specific. Peptides with antimicrobial properties are classified based on their biosynthesis, which can be ribosomal or non-ribosomal. In general, antimicrobial peptides consist of 10–50 amino acids and their ability to kill bacteria depends on their interaction with bacterial membranes and cell walls. Selectivity depends on membrane or cell wall composition.42 The levels of production and accumulation of these compounds in the gut depend on the producer strains, their chemical structure, bioavailability and physical conditions within the surrounding environment, making the identification and direct isolation of those compounds difficult. Recently, a bioinformatics tool, ClusterFinder, has been used to identify thousands of biosynthetic gene clusters from human metagenomic samples, encoding among other compounds, antimicrobial peptides, highlighting the potentially underexploited therapeutics derived from the human microbiome.43,44
4.1. Non-ribosomal peptides (NRPs)
NRPs are secondary metabolite peptides synthesized by multienzyme complexes of multifunctional peptide synthetases.45 NRPs constitute a large class of bacterial products in several environments and their study has provided many of the antibacterial compounds currently in use, such as penicillins, vancomycin or polymyxin that are considered as peptide derived.45 However, there are very few NRPs characterized from the human microbiota.11 Although analysis of the Antimicrobial Peptide Database (APD3) did not show any NRPs of human gut origin,29 Donia and Fischbach listed some NRPs that had been characterized from bacterial producers isolated from the human microbiome.11 The peptides cereulide, zwittermicin and tilivalline were isolated from pathogens from the human gut although they are not normally present in human gut microbiome.11 Their activity is described as cytotoxic, alongside with colibactin activity, produced by Escherichia coli.46 Zwittermicin activity has been reported as antimicrobial47 but resistance genes for it have also been identified.48 Therefore, the use of such NRPs as therapeutics would be limited.
4.2. Ribosomally synthesized peptides (Bacteriocins)
Bacteriocins are ribosomally synthesized peptides that exhibit antimicrobial activity. The first bacteriocin was discovered by Gratia in 192549 and named “colicin”. Since then, many more bacteriocins have been isolated and characterized, mostly based on their bioactivity, in a wide variety of environments and from many different bacterial species. Bacteriocins have been proposed as mediators in population dynamics and their ecological role in this regard has been studied in recent decades.5, 50, 51 A considerable number of these peptides have originated from bacterial isolates from foods, though many important bacteriocins are produced by bacteria that inhabit the GI tract of humans and animals. Indeed, given the density and diversity of microbial populations present in the gut, it may represent a source of bacteriocin producers that is, as yet, relatively underexplored. Bacteriocins address some of the problems described for traditional antibiotics and that is why their range of applications is expanding towards human and veterinary therapeutics.52,53 They have low toxicity, making them suitable for use as food and beverage preservatives, since traditional antibiotics are prohibited for use in foods. Low toxic effects have been proven for many of the bacteriocins in different studies,54,55 although a small percentage have shown some cytotoxic activity.56 Bioinformatics is also helping to identify putative anti-tumoral activity of bacteriocins, as has been described recently.57,58 Antiviral and antifungal properties of bacteriocins represent other potential application of these peptides.59,60
Some bacteriocins can exhibit broad-spectrum while others exhibit narrow-spectrum antimicrobial activity. The former have the possibility of being used as a traditional therapeutical antibiotic, while the narrow spectrum antimicrobials are more suitable for targeting specific harmful microorganisms without altering the natural populations,61 contributing to maintaining a balance in microbial populations. As a general rule, bacteriocins produced by Gram-positive bacteria show better activity against Gram-positive pathogens and Gram-negative bacteriocins work better on Gram-negative pathogens, but with purified bacteriocins and bioengineered derivatives it has been possible to cross this barrier.62 Furthermore, their activity can be enhanced by the presence of other substances, such as lactic acid, that allows the permeabilization of the bacterial membrane. One of their most attractive features is that resistance is not easily developed against them. In order to do that, target cells would have to alter membrane composition63 or the receptors.64,65 Transporters have also been pointed as a mechanism to develop resistance,66 and although they are not very common, expression of resistance proteins have also been reported.67,68 Bacteriocin combination or using strains that produce more than one, in order to maximize efficacy is another possibility,69 and they can also be combined with other compounds to act synergistically.70,71 In addition, bioactivity is in a nano- to micromolar range, while an absence of taste, color or odor, makes them suitable for industrial and clinical purposes.63
Another asset of such peptides is that they offer the possibility of being bioengineered, either by gene manipulation or partial or complete chemical synthesis, to improve properties such as specific activity, stability and host range. Rational design might be the key to obtain more potent bacteriocins, based on the structure-function relationship;72 and bacteriocins could be modified to have broader activity against new pathogen targets,62 or narrowed activity to target specific pathogens, to treat biofilms and resistant bacteria,73,74 or increase protease resistance against chymotrypsin or trypsin, which can aid their survival in the gut. Key residues have been identified, along with the mechanisms that control activity and regulation that have been studied using natural variants and the generation of peptide derivatives.75 Delivery matrices have been examined to maintain bacteriocin stability and in this regard, nanotechnology has been used as one of the most important approaches, with the use of nanoliposomes or nanoparticles of different composition.76 Improvement of delivery strategies might offer new therapeutic possibilities for bacteriocins, like new targets or controlled delivery.77,78
4.2.1. Identification of bacteriocins
Identification of new bacteriocins originally relied on methods to identify antimicrobial or antagonistic activity of a bacterial strain on a target bacterium. Bioassays for detecting inhibitory activity can present variations for adapting to specific needs.79 One technique is to inoculate media with the specific indicator strain and add a spot of the bacteriocin producing strain or its cell free supernatant (spot test). A common variation is the growth on agar of the producer strain and subsequent overlay with indicator-inoculated soft agar (overlay assay). In both cases the activity is estimated by the inhibition of growth of the indicator strain (Fig. 1). Well diffusion assays and disc assays both involve placement of the cell free supernatant of the producer strain in a well or on a paper disc in indicator inoculated agar followed by incubation, with the bacteriocin producing a zone of inhibition which can be used to measure specific activity (Fig. 1).79 The indicator strain can also be co-cultured with the producer strain or its supernatant and growth assessed by measurement of optical density.80 Despite the extended use of these techniques and their variations, this approach has a limitation, as just a small percentage of bacteria are able to grow in culture and in vitro growth may not provide the appropriate conditions to induce the biosynthesis of bacteriocins.81 Some of the bacteriocin-like compounds identified required the presence of a trigger. For example, the presence of trypsin has proven to be effective as an antimicrobial inducer in human gut isolates,82 and both cell density and carbon source were necessary to induce bacteriocin production in Streptococcus pneumoniae,83 reinforcing the idea that the environment plays an important role in the production of these compounds. Understanding all the factors and interactions that take place in the environment of the potential producing bacteria and the development of strategies to mimic these interactions can be a gargantuan task. A recently developed peptidogenomics strategy attempts to establish a mass spectral molecular network of microbes grown in a plate and integrate this information with genomic and phenotypic information.84 This could be a good first step towards a holistic approach to microbial interactions that could lead to the identification of novel compounds produced from these simultaneous and multilateral interactions in a more realistic and complex environment such as the GI tract.
Figure 1.
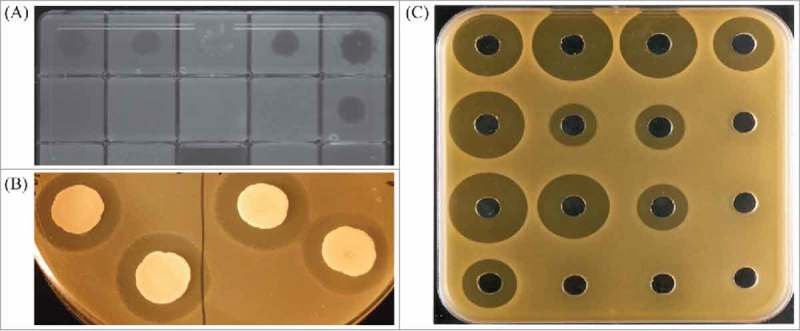
Examples of assays performed to identify antimicrobial activity. (a) spot test; (b) overlay; (c) well diffusion.
The development of molecular tools removes the requirement for functionality influenced by culture conditions and induction. This has allowed the identification of putative bacteriocin clusters in genomes, both in culturable and unculturable species. There are public software and databases to identify putative bacteriocin clusters in sequenced genomes and also offer access to bacteriocin peptide sequences. BAGEL with 482 sequences85 and BACTIBASE with 345 sequences86 are the most commonly used. Broader databases are available to screen for potential new bacteriocin clusters in the gut, like the Human Microbiome Project's reference genome database.87,88 An analysis of this database by Walsh et al identified 74 putative bacteriocin-encoding gene clusters in different phyla.87 A more accurate analysis of the distribution and frequency of specific bacteriocin types can be addressed by the use of Hidden Markov Model, as performed for LanB homologues, allowing the identification of seven new lantibiotic producers.88 Drissi et al developed an extensive database for bacteriocins, BUR (“Bacteriocins of the URMITE database”), where they combined BAGEL, BACTIBASE and NCBI databases for bacteriocin published sequences and also retrieved 641 available genomes from the GI tract that they analyzed in order to incorporate as much bacteriocin data as possible.89 The study of this database provided some interesting observations. First, it showed differences between bacteriocins produced by gut bacteria and bacteria from other environments. These differences were found in the amino acid composition and peptide length, with those for gut bacteria generally being shorter and having less positively charged and less hydrophobic residues, lower percentages of aspartic acid, leucine, arginine and glutamic acid, and higher percentages of lysine and methionine.89 The most predominant class was class I, with 44% of bacteriocins produced in the gut, while class III was less represented (17.3%). This is also a significant difference with bacteriocins from other environments, where class III represents 59.2% and class I only 13%. Different bacterial groups generate different classes of bacteriocins: Actinobacteria (58%), Proteobacteria (94%) and Bacteroidetes (exclusively) were identified mainly as producers of class III bacteriocins, while Firmicutes showed production of bacteriocins of all classes more evenly (20% class I, 45% class II and 35% class III).89
In Tables 1, 2 and 3 we have compiled the known bacteriocins isolated from the human gut based on the analysis of BAGEL3, BACTIBASE and APD3 databases.
Table 1.
Class I bacteriocins produced by bacteria originally isolated from the human gut.
| Class I (modified) | |||||
|---|---|---|---|---|---|
| Group | Distinctive feature | Bacteriocin from human gut isolate | Gut Producer first isolated from human | Activity against | Reference |
| Microcins | Microcin L | E. coli LR05 |
Shigella sonnei E. coli Pseudomonas aeruginosa |
150 | |
| Microcin M | E. coli Nissle 1917 | E. coli, Salmonella | 151 | ||
| Microcin V (ColicinV) | E. coli | E. coli | 49 | ||
| Microcin H47 (MccH47) | E. coli H47 | E. coli, Salmonella, Enterobacter, Shigella, Klebsiella, Proteus spp. | 152 | ||
| Microcin E492 | Klebsiella pneumoniae RYC492 | Escherichia, Klebsiella, Salmonella, Citrobacter, Enterobacter | 153 | ||
| Lasso peptide | Lasso knot structure | Microcin J25 | E. coli AY25 | E. coli, Salmonella sp, Shigella flexneri | 96 |
| Sactibiotics | Sulphur-α-carbon linkages | Thuricin CD | Bacillus thuringiensis DPC 6431 | Clostridium difficile, Bacillus cereus, Bifidobacterium firmus, Listeria monocytogenes | 154 |
| Lantibiotics | Lanthionine bridges | Ruminococcin A | Ruminococcus gnavus | Bacteroides sp, Clostridium sp, Bifidobacterium sp, B. cereus | 116 |
| Nisin O | Blautia obeum A2-162 | Clostridium perfringens, C. difficile, Lactococcus lactis | 118 | ||
| Cytolysin ClyLl and ClyLs | Enterococcus faecalis | Gram-positive bacteria | 114 | ||
| BLD 1648 | Bifidobacterium longum (strain DJO10A) | 117 | |||
| Linear azole-or azoline-containing peptides | Heterocycles but no other modifications | Microcin B17 | E. coli Nissle 1917 | E. coli, Salmonella, Shigella | 151 |
Examples of Class I (Linardins, Proteusins, Patellamide-like cyanobactins, Anacyclamide-like cyanobactins, Thiopeptides, Bottromycins and Glycocins) have not yet been described from human gut isolates. Classification of bacteriocin is based on Cotter et al, 2013.
Table 2.
Class II bacteriocins produced by bacteria originally isolated from the human gut.
| Class II (unmodified or cyclic) | |||||
|---|---|---|---|---|---|
| Group | Distinctive feature | Bacteriocin from human gut isolate | Gut Producer first isolated from human | Activity against | Reference |
| IIa peptides (pediocin PA-1-like bacteriocins) | Conserved YGNGV motif (N represents any amino acid) | Bac43 | Enterococcus faecium VRE82 | E. faecalis, E. faecium, Enterococcus hirae, Enterococcus durans, and L. monocytogenes | 122 |
| Bacteriocin RC714 | E. faecium strain RC714 | E. faecalis, E. faecium, L. monocytogenes, Listeria innocua, Listeria murrayi, Listeria grayi, Lactobacillus paracasei, Lactobacillus plantarum, Leuconostoc sp., and Pediococcus pentosaceus | 124 | ||
| Bacteriocin 31 | E. faecalis YI717 | E. hirae 9790, E. faecium, and L. monocytogenes | 155 | ||
| IIb peptides | Two unmodified peptides are required for activity | ABP-118 | Lactobacillus salivarius UCC-118 | L. monocytogenes | 127 |
| Acidocin J1132 | Lactobacillus acidophilus JCM1132 | Lactobacillus sp | 126 | ||
| Gassericin T | Lactobacillus gasseri SBT 2055 | Lactobacillus delbrueckii subsp. bulgaricus | 130 | ||
| Lactacin F | Lactobacillus johnsonii | Lactobacillus sp, E. faecalis | 156 | ||
| IIc peptides | Cyclic peptides | Gassericin A | L. gasseri LA39 | Bacillus, Clostridium, Lactobacillus spp, Lactococcus lactis, Leuconostoc mesenteroides, Listeria spp, Pediococcus cerevisiae, S. aureus, Streptococcus agalactiae | 157 |
| Reutericin 6 | Lactobacillus reuteri LA 6 | L. acidophilus, L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis | 158 | ||
| AS-48 | E. faecalis AS-48 | Gram-positive and Gram-negative | 159 | ||
| IId peptides | Unmodified, linear, non-pediocin-like, single-peptide bacteriocins | Microcin S | E. coli G3/10 | E. coli G3/10 | 136 |
| Rhamnosin A | Lactobacillus rhamnosus strain 68 | Micrococcus lysodeikticus ATCC 4698 | 138 | ||
| Bac32 | Vancomycin-resistant E. faecium (VRE) 200 | E. faecium, E. hirae, and E. durans | 139 | ||
Examples of Class II (IIe) have not yet been described from human gut isolates. Classification of bacteriocins is based on Cotter et al, 2013.
Table 3.
Class III bacteriocins produced by bacteria originally isolated from the human gut.
| Class III | |||||
|---|---|---|---|---|---|
| Group | Distinctive feature | Bacteriocin from human gut isolate | Gut Producer first isolated from human | Activity against | Reference |
| Bacteriolysins | Colicins | E. coli | Enterobacteria | 49 | |
| Bacteriocin 28b | Serratia marcescens | E. coli | 141 | ||
| Non-lytic bacteriocins | Bacteriocin helveticin J | L. acidophilus NCFM | 142 | ||
4.2.2. Classification of bacteriocins
Due to the heterogeneous nature of the bacteriocins, there have been different criteria to classify them (e.g. by primary structure, molecular weight, mode of action, heat stability, genetic properties). Currently the most accepted is that based on their structure. This review will follow the classification scheme proposed by Cotter et al,56 to highlight the diversity of bacteriocins produced by members of the gut microbiota. Examples of bacteriocin structures and gene operon organization are shown in Figs 2 and 3.
Figure 2.
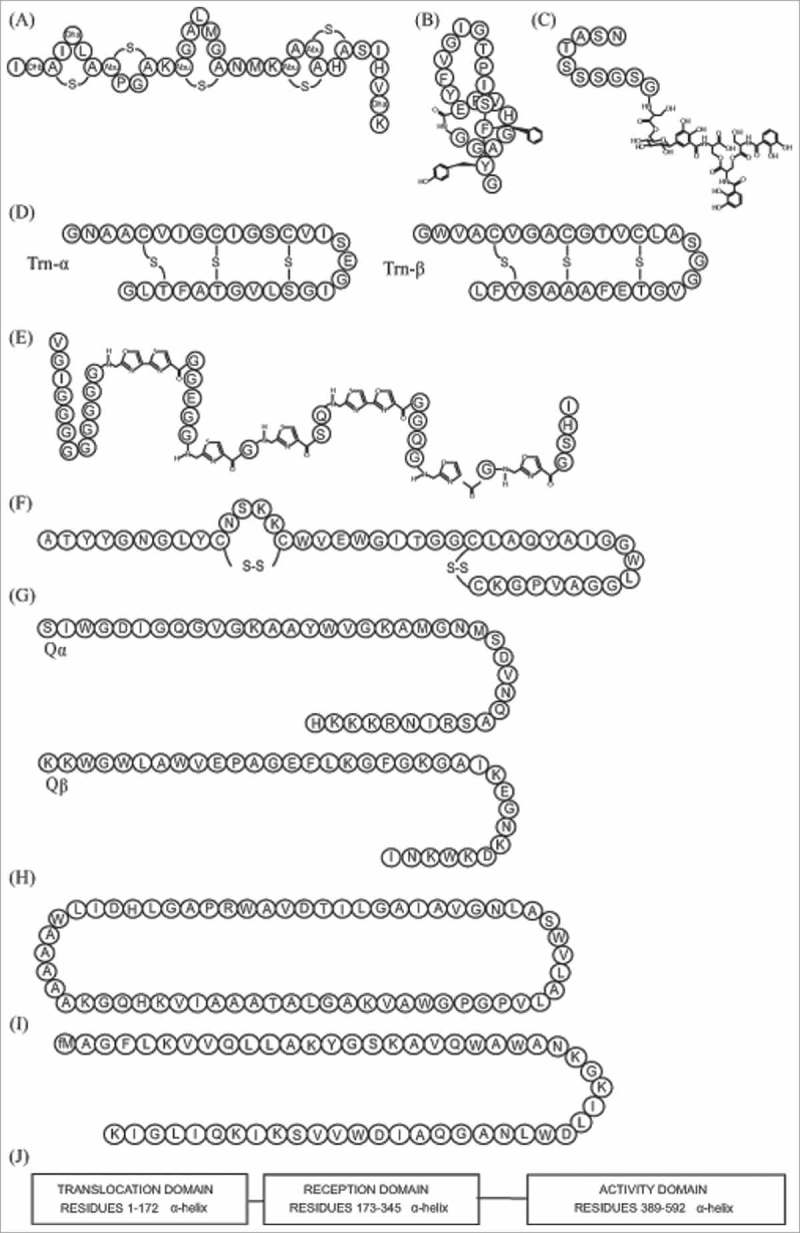
Examples of bacteriocin structures. (a) nisin A1, lantibiotic160; (b) microcin J25, lasso peptide161; (c) microcin E-492, microcin162; (d) thuricin CD subunits, sactipeptide154; (e) microcin B17, linear azole or azoline containing peptides163; (f) enterocin NKR-5-3C1, class IIa164; (g) lactococcin Q1, class IIb164; (h) lactocyclicin Q1, class IIc164; (i) lacticin Q1, class IId164; (j) colicin A1, class III.49 1Note that these bacteriocins have not been described as of human gut origin.
Figure 3.
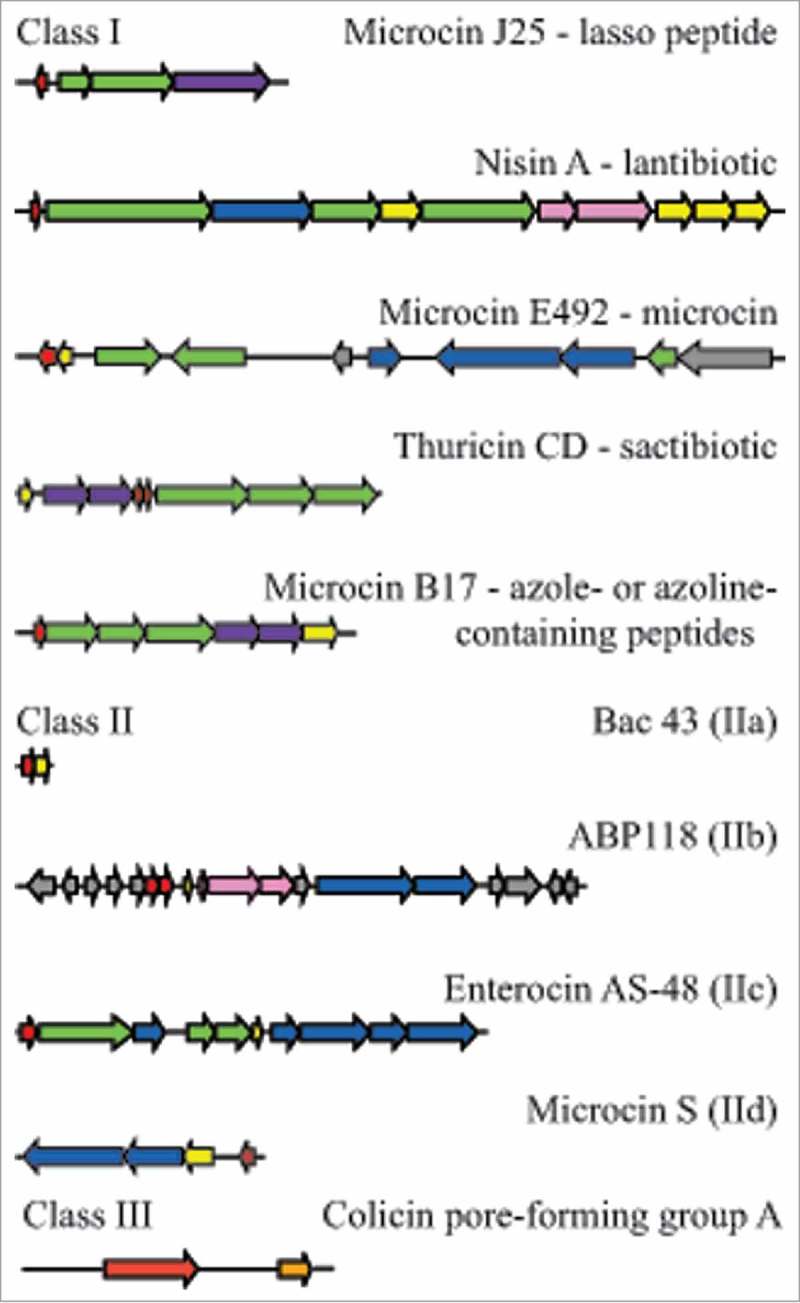
Examples of bacteriocin cluster organization among the different bacteriocin classes. Genes are colored according to the function of their products: red, precursor peptides; green, post-translational modifications; blue, export; yellow, immunity; pink, regulation; purple, export and immunity; orange, lysis; grey, unknown function. Class I: microcin J2591; nisin A109; microcin E-49291; thuricin CD100; microcin B1791; class IIa bacteriocin 43122; class IIb ABP-118127; class IIc AS-48165; class IId microcin S137; class III colicin.49
4.2.2.1. Class I bacteriocins
This class contains different subgroups, which are all small heat-stable peptides of less than 10 kDa and share the process of enzymatic modification during their biosynthesis. The use of characteristic amino acids and structures is distinctive and will have an influence on their future function. The most studied group is lantibiotics, but the development and implementation of more accurate and advanced techniques is broadening our understanding of other modifications discovered more recently. Here we discuss the subgroups of this peptide class whose producers were isolated initially from the human gut microbiota.
4.2.2.1.1. Microcins
Microcins are peptides produced by Enterobacteriaceae90 (Fig. 2). They are differentiated from colicins, also produced by Enterobacteriaceae, by their smaller molecular mass (microcins are smaller than 10 kDa, while colicins are larger than 20 kDa). Microcin operons are constituted by genes encoding a precursor, secretion proteins and elements for self-immunity (Fig. 3). Microcins are subdivided in two groups, based on molecular masses and presence of disulphide bonds and post-translational modifications.90 Thus, class I microcins are encoded on plasmids, smaller than 5 kDa and have extensive post-translational modifications. Class II microcins (between 5–10kDa) can be further classified into class IIa, encoded by plasmids, with one, two or no disulphide bonds and no post-translational modifications, and class IIb, encoded on chromosomes and a non-mandatory post-translational modification.90 They are considered to be hydrophobic and have heat, pH and protease stability.91 Microcins are known for having a precursor peptide that needs to be cleaved during their transport from the producer cells.92 Currently there are at least 16 microcins identified, but only eight of them have been characterized structurally.92 The mode of action of microcins is described as a “Trojan horse”,90 mimicking essential compounds for nutrition, for example an iron-siderophore complex, that the target cell will detect using outer-membrane receptors and will incorporate. They can also be secreted as a non-active compound and be processed to the bioactive form by the target cell. Once inside the cell, microcins bind enzymes or interact with the inner membrane.90,93
4.2.2.1.2. Lasso peptides
These were first described in 1991 by the discovery of anantin.94 They are RiPPs of bacterial origin characterized by their distinctive structure as a knot, including a macrolactam ring crossed through by the C-terminal tail. This structure is sustained by steric interactions and disulphide bridges and it is very stable. There are three classes of lasso peptides described so far, and the criterion followed is the number of disulphide bridges: class I has two disulphide bonds, class II no bonds and class III just one.94 They have been reported to have inhibitory activity against enzymes, receptor antagonistic behavior and a few of them display antimicrobial activity.95 Although during the first years their discovery was activity-based, increased application of genome mining has resulted in a rapid increase of discovery of lasso peptide biosynthetic gene clusters. In a recent review by Heggeman et al95 a total of 38 known lasso peptides were identified. Currently almost all lasso peptides have been isolated from soil, sludge and water bacteria,95 with the exception of zucinodin, isolated from an erythroleukemia cell line, and microcin J25, isolated from baby feces.96 They have also attracted attention because they have been proposed to be used as new chemical scaffolds, due to their stability.
Microcin J25 is the paradigm of lasso peptides and has been studied as a typical example of its class. It has 21 amino acids and is produced by E. coli AY25 that was isolated from newborn feces. Typically, it forms a ring by a lactam linkage between the first glycine and the glutamate at position 8 (Fig. 2B). The remaining thirteen residues form the tail, with the phenylalanine in position 19 and tyrosine at position 20 acting as plugs.97 The biosynthetic cluster has four genes: a precursor (mcjA), cleavager of leader peptide (mcjB), a macrolactam ring formation gene (mcjC) and a gene that encodes the ATP-binding cassette transporter (mcjD) (Fig. 3). Despite their genomic identification, biosynthesis of lasso peptides has not been fully elucidated.98 MccJ25 antimicrobial activity is based on inhibiting the RNA polymerase of Gram-negative bacteria but another activity has been described, depolarizing cell membranes in Salmonella and E. coli species that overexpress fhuA.95
4.2.2.1.3. Sactibiotics
Sactipeptides are a general group of peptides characterized by having an intramolecular bridge between a cysteine sulphur and an α-carbon and do not always show inhibitory activity; the term sactibiotic is used to describe sactipeptides with antimicrobial activity. Many of them have been identified by genome mining99 and currently there are four sactibiotics described: subtilosin A, propionicin F, thuricin H and thuricin CD.100 Among them, only thuricin CD has been isolated from a bacterium from the human gut. Produced by Bacillus thuringiensis DPC6431, it is particularly important because it has been shown to have very specific activity against Clostridium difficile, Bacillus cereus, Bifidobacterium firmus and Listeria monocytogenes.101 It is composed of two subunits, Trnα and Trnβ (Fig. 2D). Its gene cluster contains structural genes, transport, immunity and post translational modification genes (Fig. 3). The presence of genes encoding members of the radical S-adenosulmethionine superfamily of proteins is also a specific characteristic. Thuricin CD's mode of action has just started to be elucidated. Mathur et al showed that both subunits act on the cell membrane, irreversibly collapsing the membrane potential and suggested that the peptides are inserted in the membrane of the cell, forming a pore that leaks ions and ultimately leading to cell lysis.102 The effect of thuricin CD was similar to vancomycin and metronidazole, both routinely used to treat C. difficile infections.
4.2.2.1.4. Linear azole-or azoline-containing peptides
These peptides, also called LAPs, are characterized by presenting different combinations of heterocyclic rings of thi-azole and (methyl)oxazole. These are produced by a cyclodehydration of cysteine and serine or threonine, and a flavin mononucleotide dependent dehydrogenation reaction.103 Their mode of action is not yet fully understood. Microcin B17 is a LAP produced by E. coli isolated from baby feces104 (Fig. 2E). Its cellular target is the DNA gyrase and it has been extensively studied to discover the link between its structure and function, in order to obtain information to design new antimicrobials.105 There is an increasing interest in studying the inhibition of topoisomerases and gyrases as targets for designing antimicrobials, and the study of microcin B17 can offer further insights into the structure and functional analysis of this antimicrobial target.106 In vivo studies have already demonstrated their ability to control infections in infants107 and cattle.108
4.2.2.1.5. Lantibiotics
The term “lantibiotic” comes from “lan-thionine containing antibiotic”. Similar to sactibiotics, lantibiotics are embedded in a bigger group, lantipeptides, not all of which show antimicrobial activity. This subgroup is constituted by small peptides (<5 kDa) with presence of modified amino acids such as dehydroalanine (Dha) and dehydrobutyrine (Dhb), among others.109 Dha and Dhb are derived from the dehydration of Ser and Thr residues respectively, the process followed in order to obtain 2,3-didehydroalanine and (Z)-2,3-didehydrobutyrine; an additional modification is the stereospecific intramolecular addition of a cysteine residue to form lanthionine (Lan) or methyllanthionine (MeLan) bridges. In total up to 17 uncommon amino acids have been described.110 The function of these modified amino acids is not known, but it has been suggested that they interact with free sulfhydryl groups on the cell envelopes of target organisms.109
Genes encoding lantibiotics are organized in clusters which are often highly conserved and have similar functions. A basic lantibiotic cluster contains genes for a precursor peptide, known as LanA, modification enzymes (LanB and LanC or LanM), usually a protease to remove the leader peptide (LanP); an ABC transporter (LanT) that translocates the peptide and sometimes also encodes the protease activity; a two-component regulatory system (LanR and LanK), and proteins responsible for self-immunity (LanI and LanFEG) (Fig. 3). Defined steps in the production and maturation of lantibiotics involve synthesis of the prelantibiotic, dehydration and linkage reactions, removal of the leader sequence, and secretion. Depending on the enzymes present during the maturation process four groups have been differentiated: class I, LanBC-modified (involves LanB, with dehydratase function and LanC, a cyclase); class II, LanM-modified (LanM encodes both dehydratase and cyclase functions); class III uses LanKC and class IV uses LanL, but only class I and II show antimicrobial activity.111
The most common mechanism of action of lantibiotics is through lipid II binding. Lipid II plays an important role in the formation of cell wall and interfering with its function is fatal for the cell. The two ways of disrupting the cell wall are by inhibiting its synthesis or by pore formation.111 Nisin, the most studied lantibiotic (Fig. 2A), has both functions, but not all lantibiotics have the same mode of action. This activity on the cell wall is the reason that lantibiotics are not very effective on Gram negative bacteria, due to their outer membranes that present a barrier for entry to the bacterial cell wall. It has been a common belief that lantibiotics are only produced by Gram-positive bacteria, but the recent identification of pinensins, lantibiotic type bacteriocins produced by the Gram-negative bacteria Chitinophaga pinensis, that also show antifungal activity,112 is an example of how broad metabolic diversity is and that there are functions still to be discovered. At the moment, the front line in lantibiotic research is focused on the transference of the in vitro efficacy to in vivo tests, and the use of bioengineering to improve mode of action and efficiency of delivery systems.113 There are many lantibiotics that are being tested in vivo for a variety of health applications, especially targeting bacteria of clinical relevance, and some are now in pre-clinical development.113
Some of the lantibiotics that can be found in the human gut are synthesized by gut pathogens, like the two-component cytolysin, produced by Enterococcus faecalis.114 This cytolysin is one factor contributing to its virulence and facilitates antibiotic resistant infections,115 and it is highly efficient against other Gram-positive bacteria. Cytolysin is composed of two subunits, large (L) and small (S); the gene cluster also encodes proteins for post translational modification, export, and a protease for activation and immunity. Its activity is regulated by a two-component system that functions via quorum-sensing autoinduction. The products of two genes, cylR1 and cylR2, repress transcription of the bacteriocin components so that production is low. When cell density increases, the small subunit of cytolysin reaches levels high enough to overcome the repression and induce transcription of the bacteriocin components.114 Other lantibiotics are produced by commensal gut microbes. Ruminococcin A, produced by Ruminococcus gnavus, was the first characterized bacteriocin from a strict anaerobe isolated from a human fecal sample and requires trypsin for antimicrobial activity,116 while genomic analysis of B. longum DJO10A identified the presence of a novel lantibiotic gene cluster with activity against a related strain.117 Recently, a new variant of nisin (nisin O), has been identified from the human gut bacterium B. obeum A2-162.118 This showed activity against important gut pathogens C. difficile and C. perfringens.118
4.2.2.2. Class II bacteriocins
Class II constitutes the most extensive group of bacteriocins. They are characterized by being small heat stable peptides (<10 kDa) that do not undergo post translational modifications. According to their specific relationships between structure and function, they can be classified into four subgroups. There have been different criteria to the establishment of these subgroups. Here we will use the one proposed by Cotter et al.56
4.2.2.2.1. Class IIa
Also known as pediocin-like bacteriocins, these contain an N-terminal consensus sequence YGNGVXaaCXaaK/NXaaXaaC which is highly conserved (Fig. 2F). This includes the “pediocin box” (YGNGV). They are characterized also by having two cysteines in the conserved region which form a disulfide bond required for antimicrobial activity.119 The C-terminus is responsible for specificity and their mode of action is via permeabilization of the bacterial cell wall. The sugar transporter mannose phosphotransferase system (Man-PTS) is the target receptor, present in Gram-positive and Gram-negative bacteria,120 and they have particularly strong activity against Listeria spp.119 Biosynthesis of the peptide commonly requires at least four genes: a structural gene encoding a precursor, and the genes encoding an immunity protein, an ATP-binding cassette transporter and a protein for extracellular translocation. They are regulated by quorum sensing, and the regulatory system is composed of three genes, encoding an inducer peptide, a membrane associated histidine protein kinase, and a cytoplasmic response regulator.121 Bacteriocin 43 was isolated and characterized from a vancomycin-resistant clinical isolate Enterococcus faecium (VRE). Its sequence showed high homology with bacteriocin 31 from E. faecalis YI717 and bacteriocin RC714 from E. faecium strain RC714,122 both clinical isolates too.123,124 It is unusual in requiring only 2 genes, one structural and one for immunity122 (Fig. 3).
4.2.2.2.2. Class IIb
This class is represented by bacteriocins that need two different peptides to act synergistically in order to exert their antimicrobial activity125 (Fig. 2G). Normally their genes are located on the same operon and they are expressed simultaneously (Fig. 3). The killing mechanism involves membrane permeabilization. The peptides that constitute the bacteriocin have similar characteristics to one peptide bacteriocins, being usually cationic and hydrophobic or amphiphilic, with a length of 30 to 50 amino acids. Their synthesis includes a double-glycine leader type sequence in the N terminus that it is cleaved off during secretion of the peptide to the outside of the cell.
A range of bacteriocins of this class originating from bacteria of human gut origin are identified in BAGEL3, BACTIBASE and APD3 databases. Acidocin J1132 was produced by from L. acidophilus JMC 1132, isolated from human feces;126 it was found that its production relied on the pH of the medium not being above 7, with maximum production at pH 5. The mode of action against other taxonomically close strains showed a bactericidal activity.126 ABP-118 was the first bacteriocin to be isolated and characterized from a bacterium that was already considered a probiotic.127 L. salivarius UCC118, isolated from the ileal-caecal region of the human GI tract was proven to produce ABP-118 in vivo and it has been tested in order to assess its potential activity, resulting in the control of L. monocytogenes infection in mice.128 Its activity has also been tested as a gut microbiota regulator, leading to population shifts by increasing Bacteroidetes and Proteobacteria and decreasing Actinobacteria.129
Gassericin T is another representative of this class. Produced by L. gasseri SBT 2055 isolated from human faeces, it is part of the lactacin F family, sharing a 60% similarity with lactacin F.130 Lactacin F, another class IIb bacteriocin produced by a gut bacterium, requires two hydrophobic peptides LafA and LafX for its antimicrobial activity and it has been shown to be effective in combination with nisin.131
4.2.2.2.3. Class IIc
These are cyclic peptides that contain a covalent bond between the C and N termini (Fig. 2H). They are generally cationic and have some degree of hydrophobicity. Initially their mode of action was identified as disruption of the cell membrane allowing permeabilization of the bacterial cell membrane to small molecules, leading to cell death. However, inhibition of nucleic acid, protein, cell wall synthesis and enzyme activity have also been reported as additional mechanisms of inhibition.132 Their structure is believed to confer a 3-dimensional stability and makes them more resistant to proteolysis.
Some bacteriocins of human gut origin could be found in the databases. Among them, the paradigm of a circular bacteriocin is considered to be AS-48, an α-helical and cationic peptide first isolated from E. faecalis. It is extensively distributed among different Enterococcus species from clinical to food isolates, and a number of different derivatives, both engineered and natural are available (e.g., AS-48J, isolated from goat cheese133). It maintains stability and solubility through a broad range of pH and temperature, and the most attractive feature is its broad spectrum of activity, affecting both Gram-positive and Gram-negative bacteria. Its applications as a food preservative is well documented, but new functions arise from the implementation of new strategies, like liposome-encapsulation.134 The successful treatment against S. aureus from bovine mastitis opens the possibility of veterinary applications. Other applications being investigated include its use as a leishmanicidal agent135 or against bacterial biofilms.134
Gassericin A is produced by an isolate from the human gut L. gasseri LA39. It has the same primary amino acid sequence as reutericin 6 produced by L. reuteri LA6, both isolated from feces of the same baby,132 but differs in one D-alanine residue, which confers on gassericin A a broader spectrum of activity. In fact, gassericin A can inhibit the growth of L. reuteri LA6, but not the other way around. The activity of reutericin 6 is also influenced by the presence of substances on the surface of the target cells and low pH values, highlighting once more the importance of the secondary and tertiary structures of bacteriocins and the close relationship between this and the mode of action and efficacy.
4.2.2.2.4. Class IId
These are unmodified, linear, non-pediocin like, single peptides (Fig. 2I). This group was proposed as a miscellaneous collection to group different bacteriocins of class II that did not fit elsewhere. There were a few representatives of this class identified from human gut in databases. Microcin S is a bacteriocin produced by E. coli G3/10 that is part of the probiotic marketed as Symbioflor, constituted by six different E. coli genotypes, and used for gastrointestinal disorder treatments, especially intestinal bowel syndrome (IBS).136 Microcin S inhibits the adhesion of the enteropathogenic E. coli E238/6 to intestinal epithelial cells.137 Rhamnosin A138 and Bac 32 were also assigned to this subclass of bacteriocins.139 Bac 32 is encoded by a plasmid and has been identified as present in a wide number of clinical isolates of E. faecium, having a narrow sprectrum of action that only includes other Enterococci.139
4.2.2.3. Bacteriocins class III
These are large (> 30 kDa), heat-labile proteins, usually with different domains (Fig. 2J). Bacteriocins of this group are not very well characterized. They are classified into two groups depending on their mode of action: class IIIa or bacteriolysins, which lyse the cells, and class IIIb or non-lytic bacteriocins, which disrupt the membrane potential. Colicins are the most important type of bacteriocin representing class IIIa.49 The first colicin (colicin V) was originally isolated from E. coli, and is now classified as a microcin, but many colicins have been isolated from other enteric bacteria. They have a narrow spectrum of action and they act by binding to specific receptors on the surface of the target cells. They have different modes of action: inhibiting macromolecular synthesis, causing DNA breakdown and stopping protein synthesis. Colicins can be further divided into two subgroups based on their encoding plasmids: group A are on small plasmids and excreted and group B are encoded by large plasmids and are not excreted, although some of them have mixed characteristics. Colicin operons are formed, as a general trend, by a structural gene, an immunity gene and a lysis protein gene. Bacteriocin 28b, isolated from many biotypes of Serratia marcescens also belongs to class III. Its structural analysis showed similarities to colicins and it is active against E. coli.140 The different biotypes of S. marcescens that produce bacteriocin 28b were clinical isolates, driven by a study of nosocomial infections caused by this bacterium.141 A helveticin J bacteriocin has also been identified from the genome of L. acidophilus NCFM, a probiotic strain that is widely commercialized.142
5. Applications of gut produced antimicrobials
The use of broad spectrum antibiotics has associated problems of increasing antibiotic resistance and concomitant disturbance of the beneficial commensal microbiota. Natural antimicrobials produced by bacteria from the human GI tract are attracting interest in their potential to offer specific solutions to health problems. Bacteriocins can be considered not only as antimicrobials but also, when produced in a mixed microbial community, as regulatory elements to balance the ratio between desirable and undesirable bacteria, offering competitive advantage to beneficial bacteria against bacteria that cause dysbiosis. In an effort to establish the microbiota alterations associated with certain pathologies, correlations between proportions and presence or absence of certain bacterial groups and functionality have been studied. This has revealed dysbiotic relationships between gut microbiota and host pathologies. The disruption of the balance of the microbiota in the gut has been associated with metabolic disease, type 2 diabetes, colorectal cancer or IBD.50 Rational application of bacteriocins, either directly or produced by bacteria in situ as probiotics, is being studied as a method to modulate the structure and function of the gut microbiota and to control these acute and chronic pathologies. Guinane and Cotter (2013) summarized the microbial associations with chronic intestinal diseases.15 Others focus on the probiotic use of bacteriocins and their use to modulate the gut microbiota.143 A strategy of production of many bacteriocins with low activity and a few specific highly active antimicrobials points towards a functionality focused on maintaining homeostasis and providing defensive strategies against elements that could cause dysbiosis. There is an increasing amount of research in this area, but the complexity of GI tract microbial ecology requires further understanding. For example, recent evidence in a tilapia model suggests that abrupt interruption of a probiotic administration can cause gut dysbiosis by increasing host susceptibility to pathogens and other alterations in the intestinal epithelium and metabolites.144 It needs to be taken into consideration whether these results can be extended to humans when designing probiotic treatments.
Bacteriocins can control and protect from specific infections. Microcins have recently proved their ability to act as narrow-spectrum antimicrobials to limit the proliferation of Enterobacteriaceae during intestinal inflammation by giving E. coli Nissle 1917, a probiotic originally isolated from the human gut, a competitive advantage.145 This is the first evidence of the ecological role that microcins might play in the gut and opens a door to further therapeutical applications.
An example of the regulatory potential of bacteriocins in gut microbial populations is bactofencin A. It is produced from a L. salivarius isolated from porcine intestine and, when tested in a distal colon model, although it did not have a direct inhibitory activity against Clostridium, Bacteroides, Bifidobacterium and Fusobacterium, their populations were altered in a positive way.146 More recently, in an experiment where water administered to mice was supplemented with different bacteriocin producer strains, initial 16S rDNA analysis at phylum level suggested that gut microbiota populations were not affected. But a deeper analysis at lower taxonomic levels showed that potential problematic bacteria, like Clostridium spp. or Staphylococcus spp., were inhibited.147 The consensus is that we do not see large changes in in vivo experiments at the level of higher taxonomy level, but there are certainly beneficial and subtle changes that are favorable for the host without major disturbances in the gut, which is a desirable feature. Also, these studies teach us the importance of the scale of analysis to understand effects on the gut microbiome.
One of the main challenges for bacteriocin production in situ is that the producer, in order to be effective, needs to survive and colonize the human gut and the antimicrobial to be active in the gut environment. Thuricin CD illustrates this assertion. Despite its efficacy against C. difficile, when delivering B. thuringiensis DPC6431 directly through the GI tract of mice, results showed that this approach was not a good delivery option.148 However, alternative routes were found to be effective, like rectal administration.148 Also, other possibilities can be explored, like synbiotics and delivery systems to help protect the producer strain or the compound from harsh acidic and enzymatic environments of the GI tract. Tablets of nisin with a pectin / HPMC polymer mixture are an example of this delivery strategy.149
6. Concluding remarks
The search for new antimicrobials has become one of the most urgent tasks to address the challenges of antibiotic resistance. Understanding the environment where problematic bacteria grow and searching for the strategies used by their natural enemies to control them has proven successful. Translating this idea into practical solutions against acute infections and systemic syndromes will be supported by the identification of different antimicrobials of human microbiota origin. Bacteriocins are highly promising, due to the variety of advantages that they present. Current limitations in the identification and isolation of these compounds can be addressed by incorporating new techniques like genomic and bioinformatics tools, genome mining and metagenomics, but there is the need to translate the genomic information into the laboratory, and ultimately to achieve efficacy in vivo. In addition, further understanding of the microbial ecology of the system under study may lead to the development of more effective treatments, both for targeted intervention and to maintain gut homeostasis.
Funding Statement
This work was supported by the Teagasc, 2015066, Science Foundation Ireland (SFI), SFI/12/RC/2273, Science Foundation Ireland (SFI), SFI/11/PI/1137, Biotechnology and Biological Sciences Research Council (BBSRC), BB/J004529/1
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for funding from Walsh Fellowship Project 2015066 (EG-G) and BBSRC Institute Strategic Programme Grant BB/J004529/1 (Quadram Institute Biosciences, MJM and AN). Research in the Cotter lab is supported by SFI; PI award “Obesibiotics” (SFI/11/PI/1137) and centre grant “APC Microbiome Institute” (SFI/12/RC/2273).
Authors contributions
EG-G, MM, PC, and AN designed the manuscript; EG-G wrote the manuscript; MJM, PC and AN critically revised the manuscript and approved the final version.
References
- 1.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–87. doi: 10.1038/nrd3975. PMID:23629505. [DOI] [PubMed] [Google Scholar]
- 2.Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–73. doi: 10.1146/annurev-genet-111212-133307. PMID:24016192. [DOI] [PubMed] [Google Scholar]
- 3.Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, van Wezel GP, Rozen DE. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci U S A. 2015;112:11054–9. doi: 10.1073/pnas.1504076112. PMID:26216986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelsic ED, Zhao J, Vetsigian K, Kishony R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–9. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czárán TL, Hoekstra RF, Pagie L. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci U S A. 2002; 99:786–90. doi: 10.1073/pnas.012399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedorenko V, Genilloud O, Horbal L, Marcone GL, Marinelli F, Paitan Y, Ron EZ. Antibacterial discovery and development: from gene to product and back. Biomed Res Int. 2015;2015:591349. doi: 10.1155/2015/591349. PMID:26339625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut Microbiota in Health and Disease. Physiological Reviews. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 8.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Medicine. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045–00015. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mousa WK, Athar B, Merwin NJ, Magarvey NA. Antibiotics and specialized metabolites from the human microbiota. Nat Prod Rep. 2017;34:1302–31. doi: 10.1039/c7np00021a. [DOI] [PubMed] [Google Scholar]
- 13.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, et al.. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–6. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 14.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, et al.. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378):eaah4680. doi: 10.1126/scitranslmed.aah4680. PMID:28228596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therapeutic Advances in Gastroenterology. 2013;6:295–308. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, et al.. The composition of the gut microbiota throughout life, with an emphasis on early life. Microbial Ecology in Health and Disease. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, et al.. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. American Journal of Physiology – Gastrointestinal and Liver Physiology. 2012;303:G675–85. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère J-F. Archaea and the human gut: new beginning of an old story. World Journal of Gastroenterology. 2014;20:16062–78. doi: 10.3748/wjg.v20.i43.16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brugère J-F, Borrel G, Gaci N, Tottey W, O'Toole PW, Malpuech-Brugère C. Archaebiotics: Proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes. 2014;5:5–10. doi: 10.4161/gmic.26749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huseyin CE, Rubio RC, O'Sullivan O, Cotter PD, Scanlan PD. The fungal frontier: a comparative analysis of methods used in the study of the human gut mycobiome. Front Microbiol. 2017;8:1432. doi: 10.3389/fmicb.2017.01432. PMID:28824566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sam HQ, Chang WM, Chai YL. The fungal mycobiome and its interaction with gut bacteria in the host. Int J Mol Sci. 2017;18:330. doi: 10.3390/ijms18020330. PMID:28165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huseyin CE, Cotter PD, Scanlan PD. Forgotten fungi—the gut mycobiome in human health and disease. FEMS Microbiol Rev. 2017;41:479–511. doi: 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- 26.Scarpellini E, Ianiro G, Attili F, Bassanelli C, De Santis A Gasbarrini A. The human gut microbiota and virome: Potential therapeutic implications. Digestive and Liver Disease. 2015;47:1007–12. doi: 10.1016/j.dld.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Current biology. 2009;19:R437–41. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atanasova NS, Pietilä MK, Oksanen HM. Diverse antimicrobial interactions of halophilic archaea and bacteria extend over geographical distances and cross the domain barrier. MicrobiologyOpen. 2013;2:811–25. doi: 10.1002/mbo3.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–93. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaca AO, Gilmore MS. A lysin to kill. eLife. 2016;5:e16111. doi: 10.7554/eLife.16111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next generation sequencing applied to human gut phage populations. Nat Rev Microbiol. 2012;10:607–17. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno I, Lerayer ALS, de Freitas Leitão MF. Detection and characterization of bacteriocin-producing Lactococcus lactis strains. Revista de Microbiologia. 1999;30:130–6. doi: 10.1590/S0001-37141999000200008. [DOI] [Google Scholar]
- 33.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. In: Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertzberger R, Arents J, Dekker HL, Pridmore RD, Gysler C, Kleerebezem M, de Mattos MJT. H2O2 production in species of the Lactobacillus acidophilus group: a central role for a novel NADH-dependent flavin reductase. Appl Environ Microbiol. 2014;80:2229–39. doi: 10.1128/AEM.04272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atassi F, Servin AL. Individual and co-operative roles of lactic acid and hydrogen peroxide in the killing activity of enteric strain Lactobacillus johnsonii NCC933 and vaginal strain Lactobacillus gasseri KS120.1 against enteric, uropathogenic and vaginosis-associated pathogens. FEMS Microbiol Lett. 2010;304:29–38. doi: 10.1111/j.1574-6968.2009.01887.x. PMID:20082639. [DOI] [PubMed] [Google Scholar]
- 36.Strus M, Gosiewski T, Fyderek K, Wedrychowicz A, Kowalska-Duplaga K, Kochan P, Adamski P, Heczko PB. A role of hydrogen peroxide producing commensal bacteria present in colon of adolescents with inflammatory bowel disease in perpetuation of the inflammatory process. J Physiol Pharmacol. 2009;60:49–54. [PubMed] [Google Scholar]
- 37.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jay JM. Antimicrobial properties of diacetyl. Appl Environ Microbiol. 1982;44:525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langa S, Martín-Cabrejas I, Montiel R, Landete JM, Medina M, Arqués JL. ombined antimicrobial activity of reuterin and diacetyl against foodborne pathogens. Journal of Dairy Science. 2014;97:6116–21. doi: 10.3168/jds.2014-8306. [DOI] [PubMed] [Google Scholar]
- 41.Perry RJ, Peng L, Barry NA, Cline GW, Zhang DY, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–7. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L-j Zhang, RL Gallo. Antimicrobial peptides. Current Biology. 2016;26:R14–9. doi.org/ 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–14. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, et al.. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–21. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hancock REW, Chapple DS. Peptide Antibiotics. Antimicrob Agents Chemother. 1999;43:1317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balskus EP. Colibactin: understanding an elusive gut bacterial genotoxin. Nat Prod Rep. 2015;32:1534–40. doi: 10.1039/c5np00091b. [DOI] [PubMed] [Google Scholar]
- 47.Kevany BM, Rasko DA, Thomas MG. Characterization of the complete Zwittermicin A biosynthesis gene cluster from Bacillus cereus . Appl Environ Microbiol. 2009;75:1144–55. doi: 10.1128/AEM.02518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milner JL, Stohl EA, Handelsman J. Zwittermicin A resistance gene from Bacillus cereus . J Bacteriol. 1996;178:4266–72. doi: 10.1128/jb.178.14.4266-4272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. Colicin Biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gillor O, Etzion A, Riley MA. The dual role of bacteriocins as anti- and probiotics. Appl Microbiol Biotechnol. 2008;81:591–606. doi: 10.1007/s00253-008-1726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riley MA, Gordon DM. The ecological role of bacteriocins in bacterial competition. Trends in Microbiology. 1999;7:129–33. doi: 10.1016/S0966-842X(99)01459-6. [DOI] [PubMed] [Google Scholar]
- 52.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LMT. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018;49:23–28. doi: 10.1016/j.copbio.2017.07.011. PMID:28787641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobson A, Cotter PD, Ross RP, Hill C. Bacteriocin production: a probiotic trait? Appl Environ Microbiol. 2012;78:1–6. doi: 10.1128/AEM.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castiglione F, Cavaletti L, Losi D, Lazzarini A, Carrano L, Feroggio M, Ciciliato I, Corti E, Candiani G, Marinelli F, et al.. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp . Biochemistry. 2007;46:5884–95. doi: 10.1021/bi700131x. [DOI] [PubMed] [Google Scholar]
- 55.Maher S, McClean S. Investigation of the cytotoxicity of eukaryotic and prokaryotic antimicrobial peptides in intestinal epithelial cells in vitro . Biochemical Pharmacology. 2006;71:1289–98. doi: 10.1016/j.bcp.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 56.Cotter PD, Ross RP, Hill C. Bacteriocins – a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen C, Nguyen VD. Discovery of Azurin-Like anticancer bacteriocins from human gut microbiome through homology modeling and molecular docking against the tumor suppressor p53. Biomed Res Int. 2015;8490482, 12 pages. doi: 10.1155/2016/8490482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaur S, Kaur S. Bacteriocins as Potential Anticancer Agents. Frontiers in Pharmacology. 2015;6:272. doi: 10.3389/fphar.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al Kassaa I, Hober D, Hamze M, Chihib NE, Drider D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob Proteins. :177–85. doi: 10.1007/s12602-014-9162-6. PMID:24880436. [DOI] [PubMed] [Google Scholar]
- 60.Drider D, Bendali F, Naghmouchi K, Chikindas ML. Bacteriocins: not only antibacterial agents. Probiotics Antimicrob Proteins. 2016;2014;8:177–82. doi: 10.1007/s12602-016-9223-0. [DOI] [PubMed] [Google Scholar]
- 61.Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Medicine. 2016;8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuwano K, Tanaka N, Shimizu T, Nagatoshi K, Nou S, Sonomoto K. Dual antibacterial mechanisms of nisin Z against Gram-positive and Gram-negative bacteria. Int J Antimicrob Agents. 2005;26:396–402. doi: 10.1016/j.ijantimicag.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Perez RH, Perez MTM, Elegado FB. Bacteriocins from lactic acid bacteria: a review of biosynthesis, mode of action, fermentative production, uses, and prospects. International Journal of Philippine Science and Technology. 2015;8:61–67. [Google Scholar]
- 64.Martinez RCR, De Martinis ECP. Antilisterial activity of a crude preparation of Lactobacillus sakei 1 bacteriocin and its lack of influence on Listeria monocytogenes haemolytic activity. Food Control 2. 005a;16:429–33. doi: 10.1016/j.foodcont.2004.05.002. [DOI] [Google Scholar]
- 65.Martinez RCR, De Martinis ECP. Evaluation of bacteriocin-producing Lactobacillus sakei 1 against Listeria monocytogenes 1/2a growth and haemolytic activity. Brazilian Journal of Microbiology 2. 005b;36:83–87. doi: 10.1590/S1517-83822005000100016. [DOI] [Google Scholar]
- 66.Collins B, Curtis N, Cotter PD, Hill C, Ross RP. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics. Antimicrob Agents Chemother. 2010;54:4416–23. doi: 10.1128/AAC.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khosa S, Frieg B, Mulnaes D, Kleinschrodt D, Hoeppner A, Gohlke H, Smits SHJ. Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae . Sci Rep 2. 016a;6:18679. doi: 10.1038/srep18679. PMID:26727488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khosa S, Lagedroste M, Smits SHJ. Protein defense systems against the lantibiotic nisin: function of the immunity protein NisI and the resistance protein NisR. Front Microbiol 2. 016b;7:504. doi: 10.3389/fmicb.2016.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parada JL, Caron CR, Medeiros ABP, Soccol CR. Bacteriocins from lactic acid bacteria: purification, properties and use as biopreservatives. Brazilian Archives of Biology and Technology. 2007;50:512–42. doi: 10.1590/S1516-89132007000300018. [DOI] [Google Scholar]
- 70.Field D, Daly K, O'Connor PM, Cotter PD, Hill C, Ross RP. Efficacies of nisin A and nisin V semipurified preparations alone and in combination with plant essential oils for controlling Listeria monocytogenes . Appl Environ Microbiol 2. 015b;81:2762–9. doi: 10.1128/AEM.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathur H, Field D, Rea MC, Cotter PD, Hill C, Ross RP. 2017a. Bacteriocin-antimicrobial synergy: a medical and food perspective. Front Microbiol. 8:1205. doi: 10.3389/fmicb.2017.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nes IF, Johnsborg O. Exploration of antimicrobial potential in LAB by genomics. Curr Opin Biotechnol. 2004;15:100–4. doi.org/ 10.1016/j.copbio.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 73.Field D, O'Connor R, Cotter PD, Ross RP, Hill C. In vitro activities of nisin and nisin derivatives alone and in combination with antibiotics against Staphylococcus biofilms. Front Microbiol. 2016;7:508. doi: 10.3389/fmicb.2016.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Field D, Gaudin N, Lyons F, O'Connor PM, Cotter PD, Hill C, Ross RP. A bioengineered nisin derivative to control biofilms of Staphylococcus pseudintermedius . PloS ONE. 2015;19:10. doi: 10.1371/journal.pone.0119684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Field D, Begley M, O'Connor PM, Daly KM, Hugenholtz F, Cotter PD, Hill C, Ross RP. Bioengineered nisin A derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PloS ONE. 2012;7(10):e46884. doi.org/ 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fahim HA, Khairalla AS, El-Gendy AO. Nanotechnology: a valuable strategy to improve bacteriocin formulations. Front Microbiol. 2016;7:1385. doi: 10.3389/fmicb.2016.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heunis T, Bshena O, Klumperman B, Dicks L. Release of bacteriocins from nanofibers prepared with combinations of poly(d,l-lactide) (PDLLA) and poly(ethylene oxide) (PEO). Int J Mol Sci. 2011;12:2158–73. doi: 10.3390/ijms12042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Torres NI, Noll KS, Xu S, Li J, Huang Q, Sinko PJ, Wachsman MB, Chikindas ML. Safety, formulation, and in vitro antiviral activity of the antimicrobial peptide subtilosin against herpes simplex virus type 1. Probiotics Antimicrob Proteins. 2013;5:26–35. doi: 10.1007/s12602-012-9123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoover DG, Harlander SK. 1993. Chapter 2 – Screening Methods for Detecting Bacteriocin Activity. In: Bacteriocins of Lactic Acid Bacteria. San Diego, California: Academic Press; 23–39. doi: 10.1016/B978-0-12-355510-6.50010-5. [DOI] [Google Scholar]
- 80.Fijan S. Antimicrobial effect of probiotics against common pathogens, probiotics and prebiotics in human nutrition and health. InTech. Ch. 2016;10. doi: 10.5772/63141. [DOI] [Google Scholar]
- 81.Chen H, Hoover DG. Bacteriocins and their Food Applications. Comprehensive Reviews in Food Science and Food Safety. 2003;2:82–100. doi: 10.1111/j.1541-4337.2003.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 82.Hatziioanou D, Mayer MJ, Duncan SH, Flint HJ, Narbad A. A representative of the dominant human colonic Firmicutes, Roseburia faecis M72/1, forms a novel bacteriocin-like substance. Anaerobe. 2013;23:5–8. doi: 10.1016/j.anaerobe.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 83.Hoover SE, Perez AJ, Tsui H-CT, Sinha D, Smiley DL, DiMarchi RD, Winkler ME, Lazazzera BA. A new quorum sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Molecular Microbiology. 2015;97:229–43. doi: 10.1111/mmi.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, et al.. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A. 2012;109:E1743–52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41:W448–53. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hammami R, Zouhir A, Le Lay C Ben Hamida J, Fliss I. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiology. 2010;10:22–22. doi: 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walsh CJ, Guinane CM, Hill C, Ross RP, O'Toole PW, Cotter PD. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project's reference genome database. BMC Microbiol. 2015;15:183. doi: 10.1186/s12866-015-0515-4. PMID:26377179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walsh CJ, O'Toole PW and Cotter PD. A Profile Hidden Markov Model to investigate the distribution and frequency of LanB-encoding lantibiotic modification genes in the human oral and gut microbiome. PeerJ. 2017;5:e3254 doi: 10.7717/peerj.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drissi F, Buffet S, Raoult D, Merhej V. Common occurrence of antibacterial agents in human intestinal microbiota. Front Microbiol. 2015;6:441. doi: 10.3389/fmicb.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rebuffat S. Microcins in action: amazing defence strategies of Enterobacteria. Biochemical Society Transactions. 2012;40:1456–62. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 91.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–34. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 92.Zhao Z, Orfe LH, Liu J, Lu S-Y, Besser TE, Call DR. Microcin PDI regulation and proteolytic cleavage are unique among known microcins. Sci Rep. 2017;7:42529. doi: 10.1038/srep42529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ran R, Zeng H, Zhao D, Liu R, Xu X. The novel property of heptapeptide of microcin C7 in affecting the cell growth of Escherichia coli . Molecules. 2017;22:432. doi: 10.3390/molecules22030432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maksimov MO, Pan SJ, James Link A. Lasso peptides: structure, function, biosynthesis, and engineering. Nat Prod Rep. 2012;29:996–1006. doi: 10.1039/c2np20070h. [DOI] [PubMed] [Google Scholar]
- 95.Hegemann JD, Zimmermann M, Xie X, Marahiel MA. Lasso peptides: an intriguing class of bacterial natural products. Acc Chem Res. 2015;48:1909–19. doi: 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- 96.Salomón RA, Farías RN. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli . J Bacteriol. 1992;174:7428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson K-A, Kalkum M, Ottesen J, Yuzenkova J, Chait BT, Landick R, Muir T, Severinov K, Darst SA. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J Am Chem Soc. 2003;125:12475–83. doi: 10.1021/ja036756q. [DOI] [PubMed] [Google Scholar]
- 98.Zhu S, Fage CD, Hegemann JD, Mielcarek A, Yan D, Linne U, Marahiel MA. The B1 protein guides the biosynthesis of a lasso peptide. Sci Rep. 2016;6:35604. doi: 10.1038/srep35604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murphy K, O'Sullivan O, Rea MC, Cotter PD, Ross RP, Hill C. Genome mining for radical SAM protein determinants reveals multiple Sactibiotic-like gene clusters. PloS ONE. 2011;6:e20852. doi: 10.1371/journal.pone.0020852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mathur H, Rea MC, Cotter PD, Hill C, Ross RP. The Sactibiotic subclass of bacteriocins: an update. Current Protein & Peptide Science. 2015;16:549–58. doi: 10.2174/1389203716666150515124831. [DOI] [PubMed] [Google Scholar]
- 101.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile . Proc Natl Acad Sci U S A. 2010; 107:9352–7. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mathur H, Fallico V, O'Connor PM, Rea MC, Cotter PD, Hill C, Ross RP. Insights into the mode of action of the sactibiotic thuricin CD. Front Microbiol. 2017b;8:696. doi: 10.3389/fmicb.2017.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al.. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–60. doi: 10.1039/c2np20085f. PMID:23165928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Asensio C, Pérez-Díaz JC, Martínez MC, Baquero F. A new family of low molecular weight antibiotics from enterobacteria. Biochem Biophys Res Commun. 1976;69:7–14. doi: 10.1016/S0006-291X(76)80264-1. [DOI] [PubMed] [Google Scholar]
- 105.Collin F, Thompson RE, Jolliffe KA, Payne RJ, Maxwell A. Fragments of the bacterial toxin microcin B17 as gyrase poisons. PloS ONE. 2013;8:e61459. doi: 10.1371/journal.pone.0061459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl Microbiol Biotechnol. 2011;92:479–97. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Henker J, Laass M, Blokhin BM, Bolbot YK, Maydannik VG, Elze M, Wolff C, Schulze J. The probiotic Escherichia coli strain Nissle 1917 (EcN) stops acute diarrhoea in infants and toddlers. Eur J Pediatr. 2007;166:311–8. doi: 10.1007/s00431-007-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.von Buenau R Jaekel L, Schubotz E, Schwarz S, Stroff T, Krueger M. Escherichia coli strain Nissle 1917: significant reduction of neonatal calf diarrhea. J. Dairy Sci. 2005;88:317–23. doi: 10.3168/jds.S0022-0302(05)72690-4. [DOI] [PubMed] [Google Scholar]
- 109.McAuliffe O, Ross RP, Hill C. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. 25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 110.Bierbaum G, Sahl HG. Lantibiotics: mode of action, biosynthesis and bioengineering. Current Pharmaceutical Biotechnology. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 111.Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 2016;100:2939–51. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mohr KI, Volz C, Jansen R, Wray V, Hoffmann J, Bernecker S, Wink J, Gerth K, Stadler M, Müller R. Pinensins: the first antifungal lantibiotics. Angew Chem Int Ed. 2015;54:11254–8. doi: 10.1002/anie.201500927. [DOI] [PubMed] [Google Scholar]
- 113.Field D, Cotter PD, Hill C, Ross RP. Bioengineering lantibiotics for therapeutic success. Front Microbiol. 2015;6:1363. doi: 10.3389/fmicb.2015.01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002;415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 115.Queck SY, Khan BA, Wang R, Bach T-HL, Kretschmer D, Chen L, Kreiswirth BN, Peschel A, DeLeo FR, Otto M. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathogens. 2009;5:e1000533. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, Nardi M, Ladiré M, Girardin H, Marcille F, Gomez A, et al.. Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol. 2001;67:4111–8. doi: 10.1128/AEM.67.9.4111-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee J-H, Karamychev V, Kozyavkin S, Mills D, Pavlov A, Pavlova N, Polouchine N, Richardson P, Shakhova V, Slesarev A, et al.. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hatziioanou D, Gherghisan-Filip C, Saalbach G, Horn N, Wegmann U, Duncan SH, Flint HJ, Mayer MJ, Narbad A. Discovery of a novel lantibiotic nisin O from Blautia obeum A2-162, isolated from the human gastrointestinal tract. Microbiology. 2017;163: 1292–305. doi: 10.1099/mic.0.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cui Y, Zhang C, Wang Y, Shi J, Zhang L, Ding Z, Qu X, Cui H. Class IIa bacteriocins: diversity and new developments. Int J Mol Sci. 2012;13:16668–703. doi: 10.3390/ijms131216668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kjos M, Borrero J, Opsata M, Birri DJ, Holo H, Cintas LM, Snipen L, Hernandez PE, Nes IF, Diep DB. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology. 2011;157:3256–67. doi: 10.1099/mic.0.052571-0. [DOI] [PubMed] [Google Scholar]
- 121.Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. The continuing story of Class IIa bacteriocins. Microbiol Mol Biol Rev. 2006;70:564–82. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Todokoro D, Tomita H, Inoue T, Ike Y. Genetic analysis of bacteriocin 43 of vancomycin-resistant Enterococcus faecium . Appl Environ Microbiol. 2006;72:6955–64. doi: 10.1128/AEM.00934-06. PMID:17088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol. 1996;178:3585–93. PMID:8655558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.del Campo R, Tenorio C, Jiménez-Díaz R, Rubio C, Gómez-Lus R, Baquero F, Torres C. Bacteriocin production in vancomycin-resistant and vancomycin-susceptible Enterococcus isolates of different origins. Antimicrob Agents Chemother. 2001;45:905–12. doi: 10.1128/AAC.45.3.905-912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nissen-Meyer J, Oppegård C, Rogne P, Haugen HS, Kristiansen PE. Structure and mode-of-action of the two-peptide (Class-IIb) bacteriocins. Probiotics Antimicrob Proteins. 2010;2:52–60. doi: 10.1007/s12602-009-9021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tahara T, Oshimura M, Umezawa C, Kanatani K. Isolation, partial characterization, and mode of action of acidocin J1132, a two-component bacteriocin produced by Lactobacillus acidophilus JCM 1132. Appl Environ Microbiol. 1996;62:892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Flynn S, van Sinderen D, Thornton GM, Holo H, Nes IF, Collins JK. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology. 2002;148:973–84. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- 128.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CGM. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007; 104:7617–21. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Walsh MC, Gardiner GE, Hart OM, Lawlor PG, Daly M, Lynch B, Richert BT, Radcliffe S, Giblin L, Hill C, et al.. Predominance of a bacteriocin-producing Lactobacillus salivarius component of a five-strain probiotic in the porcine ileum and effects on host immune phenotype. FEMS Microbiology Ecology. 2008;64:317–27. doi: 10.1111/j.1574-6941.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- 130.Kawai Y, Saitoh B, Takahashi O, Kitazawa H, Saito T, Nakajima H, Itoh T. Primary amino acid and DNA sequences of gassericin T, a lactacin F-family bacteriocin produced by Lactobacillus gasseri SBT2055. Bioscience, Biotechnology, and Biochemistry. 2000a;64:2201–8. doi: 10.1271/bbb.64.2201. [DOI] [PubMed] [Google Scholar]
- 131.Dalmau M, Maier E, Mulet N, Viñas M, Benz R. Bacterial membrane injuries induced by lactacin F and nisin. Int Microbiol. 2002;5:73–80. doi: 10.1007/s10123-002-0063-2. PMID:12180783. [DOI] [PubMed] [Google Scholar]
- 132.Maqueda M, Sánchez-Hidalgo M, Fernández M, Montalbán-López M, Valdivia E, Martínez-Bueno M. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:2–22. doi: 10.1111/j.1574-6976.2007.00087.x. [DOI] [PubMed] [Google Scholar]
- 133.Abriouel H, Lucas R, Ben Omar N, Valdivia E, Maqueda M, Martínez-Cañamero M, Gálvez A. Enterocin AS-48RJ: a variant of enterocin AS-48 chromosomally encoded by Enterococcus faecium RJ16 isolated from food. Syst Appl Microbiol. 2005;28:383–97. doi: 10.1016/j.syapm.2005.01.007. PMID:16094865. [DOI] [PubMed] [Google Scholar]
- 134.Burgos MJG, Pulido RP, Aguayo MDL, Galvez A, Lucas R. The cyclic antibacterial peptide enterocin AS-48: isolation, mode of action, and possible food applications. Int J Mol Sci. 2014;15:22706–27. doi: 10.3390/ijms151222706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.MÁ Abengózar, R Cebrián, Saugar JM, Gárate T, Valdivia E, Martínez-Bueno M, Maqueda M, Rivas L. The enterocin AS-48 as an evidence for the use of bacteriocins as new leishmanicidal agents. Antimicrob Agents Chemother. 2017;61(4):e02288-16. doi: 10.1128/AAC.02288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Beimfohr C. A review of research conducted with probiotic E. coli marketed as Symbioflor. Int J Bacteriol. 2016;2016:3535621. doi: 10.1155/2016/3535621. PMID:27995179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zschüttig A, Zimmermann K, Blom J, Goesmann A, Pöhlmann C, Gunzer F. Identification and characterization of microcin S, a new antibacterial peptide produced by probiotic Escherichia coli G3/10. PLoS ONE. 2012;7:e33351. doi: 10.1371/journal.pone.0033351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dimitrijević R, Stojanović M, Živković I, Petersen A, Jankov RM, Dimitrijević L, Gavrović-Jankulović M. The identification of a low molecular mass bacteriocin, rhamnosin A, produced by Lactobacillus rhamnosus strain 68. J Appl Microbiol. 2009;107:2108–15. doi: 10.1111/j.1365-2672.2009.04539.x. PMID:19796123. [DOI] [PubMed] [Google Scholar]
- 139.Inoue T, Tomita H, Ike Y. Bac 32, a novel bacteriocin widely disseminated among clinical isolates of Enterococcus faecium . Antimicrob Agents Chemother. 2006;50:1202–12. doi: 10.1128/AAC.50.4.1202-1212.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guasch JF, Enfedaque J, Ferrer S, Gargallo D, Regué M. Bacteriocin 28b, a chromosomally encoded bacteriocin produced by most Serratia marcescens biotypes. Res Microbiol. 1995;146:477–83. doi: 10.1016/0923-2508(96)80293-2. [DOI] [PubMed] [Google Scholar]
- 141.Grimont PA, Grimont F. Biotyping of Serratia marcescens and its use in epidemiological studies. J Clin Microbiol. 1978;8:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bull M, Plummer S, Marchesi J, Mahenthiralingam E. The life history of Lactobacillus acidophilus as a probiotic: a tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol Lett. 2013;349:77–87. doi: 10.1111/1574-6968.12293. [DOI] [PubMed] [Google Scholar]
- 143.Walsh CJ, Guinane CM, O'Toole PW, Cotter PD. Beneficial modulation of the gut microbiota. FEBS Lett. 2014;588:4120–30. doi: 10.1016/j.febslet.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 144.Liu Z, Liu W, Ran C, Hu J, Zhou Z. Abrupt suspension of probiotics administration may increase host pathogen susceptibility by inducing gut dysbiosis. Sci Rep. 2016;6:23214. doi: 10.1038/srep23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sassone-Corsi M, Nuccio S-P, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–3. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guinane CM, Lawton EM, O'Connor PM, Ó O'Sullivan, Hill C, Ross RP, Cotter PD. The bacteriocin bactofencin A subtly modulates gut microbial populations. Anaerobe. 2016;40:41–49. doi: 10.1016/j.anaerobe.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 147.ÖCO Umu, C Bäuerl, Oostindjer M, Pope PB, Hernández PE, Pérez-Martínez G, Diep DB. The potential of class II bacteriocins to modify gut microbiota to improve host health. PloS ONE. 2016;11:e0164036. doi: 10.1371/journal.pone.0164036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rea MC, Alemayehu D, Casey PG, O'Connor PM, Lawlor PG, Walsh M, Shanahan F, Kiely B, Ross RP, Hill C. Bioavailability of the anti-clostridial bacteriocin thuricin CD in gastrointestinal tract. Microbiology. 2014;160:439–45. doi: 10.1099/mic.0.068767-0. [DOI] [PubMed] [Google Scholar]
- 149.Ugurlu T, Turkoglu M, Gurer US, Akarsu BG. Colonic delivery of compression coated nisin tablets using pectin/HPMC polymer mixture. Eur J Pharm Biopharm. 2007;67:202–10. doi: 10.1016/j.ejpb.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 150.Gaillard-Gendron S, Vignon D, Cottenceau G, Graber M, Zorn N, van Dorsselaer A, Pons A-M. Isolation, purification and partial amino acid sequence of a highly hydrophobic new microcin named microcin L produced by Escherichia coli . FEMS Microbiol Lett. 2000;193:95–98. doi: 10.1111/j.1574-6968.2000.tb09408.x. [DOI] [PubMed] [Google Scholar]
- 151.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology. 2003;149:2557–70. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 152.Laviña M, Gaggero C, Moreno F. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli . J Bacteriol. 1990;172:6585–8. doi: 10.1128/jb.172.11.6585-6588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.de Lorenzo V. Isolation and characterization of microcin E 492 from Klebsiella pneumoniae . Archives of Microbiology. 1984;139:72–75. doi: 10.1007/BF00692715. [DOI] [PubMed] [Google Scholar]
- 154.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile . Proc Natl Acad Sci U S A. 2010;107:9352–7. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol. 1996;178:3585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abee T, Klaenhammer TR, Letellier L. Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration complexes in the cytoplasmic membrane. Appl Environ Microbiol. 1994;60:1006–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kawai Y, Saito T, Kitazawa H, Itoh T. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N and C terminal ends. Bioscience, Biotechnology, and Biochemistry. 1998;62:2438–40. doi: 10.1271/bbb.62.2438. [DOI] [PubMed] [Google Scholar]
- 158.Toba T, Samant SK, Yoshioka E, Itoh T. Reutericin 6, a new bacteriocin produced by Lactobacillus reuteri LA 6. Letters in Applied Microbiology. 1991;13:281–6. doi: 10.1111/j.1472-765X.1991.tb00629.x. [DOI] [Google Scholar]
- 159.Gálvez A, Maqueda M, Valdivia E, Quesada A, Montoya E. Characterization and partial purification of a broad spectrum antibiotic AS-48 produced by Streptococcus faecalis . Canadian Journal of Microbiology. 1986;32:765–71. doi: 10.1139/m86-141. [DOI] [PubMed] [Google Scholar]
- 160.Kaletta C, Entian KD. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. Journal of Bacteriology. 1989;171:1597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Clarke DJ, Campopiano DJ. Maturation of McjA precursor peptide into active microcin MccJ25. Organic & Biomolecular Chemistry. 2007;5:2564–6. doi: 10.1039/B708478A. [DOI] [PubMed] [Google Scholar]
- 162.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–93. doi: 10.1039/B801747F. PMID:18663394. [DOI] [PubMed] [Google Scholar]
- 163.Desriac F, Defer D, Bourgougnon N, Brillet B, Le Chevalier P Fleury Y. Bacteriocin as weapons in the marine animal-associated bacteria warfare: inventory and potential applications as an aquaculture probiotic. Marine Drugs. 2010;8:1153–77. doi: 10.3390/md8041153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Perez RH, Zendo T, Sonomoto K. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Fact. 2014;13 (Suppl 1):S3. doi: 10.1186/1475-2859-13-S1-S3. PMID:25186038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Belkum MJ, Martin-Visscher LA, Vederas JC. Structure and genetics of circular bacteriocins. Trends Microbiol. 2011;19:411–8. doi: 10.1016/j.tim.2011.04.004. PMID:21664137. [DOI] [PubMed] [Google Scholar]


