CRISPR/Cas9-mediated mutagenesis is used for the first time in an octoploid species, the cultivated strawberry, to functionally characterize the role of the B-class MADS box gene FaTM6 in flower development
Keywords: AP3, CRISPR/Cas9, flower development, Fragaria, ananassa, Fragaria vesca, genome editing, MADS-box transcription factor, octoploid, strawberry, TM6
Abstract
The B-class of MADS-box transcription factors has been studied in many plant species, but remains functionally uncharacterized in Rosaceae. APETALA3 (AP3), a member of this class, controls petal and stamen identities in Arabidopsis. In this study, we identified two members of the AP3 lineage in cultivated strawberry, Fragaria × ananassa, namely FaAP3 and FaTM6. FaTM6, and not FaAP3, showed an expression pattern equivalent to that of AP3 in Arabidopsis. We used the CRISPR/Cas9 genome editing system for the first time in an octoploid species to characterize the function of TM6 in strawberry flower development. An analysis by high-throughput sequencing of the FaTM6 locus spanning the target sites showed highly efficient genome editing already present in the T0 generation. Phenotypic characterization of the mutant lines indicated that FaTM6 plays a key role in anther development in strawberry. Our results validate the use of the CRISPR/Cas9 system for gene functional analysis in F. × ananassa as an octoploid species, and offer new opportunities for engineering strawberry to improve traits of interest in breeding programs.
Introduction
In the years that have passed since the formulation of the classic ABC model for floral organ identity (Coen and Meyerowitz, 1991), our understanding of the molecular mechanisms controlling floral organ development has progressed significantly. The activities of the B-class proteins, APETALA3 (AP3) and PISTILLATA (PI), specify petal and stamen identities when combined with the activities of A- and C-class proteins, respectively (Krizek and Meyerowitz, 1996). AP3 and PI arose from an ancestral duplication event, which it is suggested occurred before the diversification of the angiosperms (Kramer et al., 1998). Later, AP3 experienced a second duplication before the diversification of the higher eudicots in the AP3 lineage, resulting in two paralogous lineages, euAP3 and Tomato MADS box gene6 (TM6), which differ in their C-terminal sequence motifs (Pnueli et al., 1991; Kramer et al., 1998). Although there are species that have lost one of the lineages, such as Arabidopsis and Antirrhinum, which lack TM6, and papaya, where euAP3 is absent (Causier et al., 2010), most species posses both euAP3 and TM6 genes, which have functionally diversified. euAP3 genes, such as the Arabidopsis AP3, are mainly involved in both petal and stamen development (Jack et al., 1992). By contrast, TM6-like genes have a predominant role in stamens (de Martino et al., 2006; Rijpkema et al., 2006; Roque et al., 2013).
In strawberry (Fragaria), flowers differ from those of Arabidopsis in several aspects. The most striking difference is the presence of hundreds of independent carpels located on an enlarged stem tip, the receptacle, which expands upon carpel fertilization to generate the fleshy part of the berry, surrounded by the true fruits, the achenes (Nitsch, 1950; Hollender et al., 2012). A role in the ABC model of flower development has not yet been functionally determined for any of the homeotic genes in strawberry, and it is not yet known whether or how these genes contribute to the development of this particular type of flower.
Strawberry possesses a wide range of ploidy levels, varying from diploid, such as the ancestral species F. vesca (woodland strawberry, 2n=2x=14 chromosomes), to decaploid, such as F. iturupensis (2n=10x=70). The cultivated strawberry, F. × ananassa, is an octoploid species (2n=8x=56). Recent studies have proposed that the complex origin of the F. × ananassa genome was the result of hybridization of three or four different species with different levels of ploidy (Tennessen et al., 2014; Sargent et al., 2016). Recently, a virtual reference genome of F. × ananassa has been established after sequencing some wild relatives (Hirakawa et al., 2014); however, a whole-genome sequence for this species has not yet been published. As an alternative, the genome of the diploid F. vesca is commonly used as the reference (Shulaev et al., 2011; Tennessen et al., 2013, 2014; Li et al., 2017; Edger et al., 2018).
Reverse-genetics strategies employed to characterize gene function in strawberry are based on gene down-regulation via post-transcriptional gene-silencing by RNA interference (RNAi) (Guidarelli and Baraldi, 2015). Although it is an essential tool for gene functional studies, the RNAi approach has two main drawbacks: (1) it requires active repression, which may result in incomplete and/or temporary knockdown effects and therefore to an undesirable background expression of the gene (Barrangou et al., 2015); and (2) it triggers gene-silencing by small interfering RNAs (siRNAs), which can lead to non-specific or off-target effects that are widespread and unpredictable (Birmingham et al., 2014). Recent progress in genome editing methods has opened up new possibilities for reverse-genetics studies. In particular, the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-associated 9 endonuclease (Cas9) technology (hereafter CRISPR/Cas9) has become established as a very powerful tool for the introduction of desired mutations due to its simplicity, efficiency, and stability. CRISPR/Cas9-mediated mutagenesis has been widely applied to plant research in the last few years, not only in Arabidopsis, (Jiang et al., 2013, 2014; Li et al., 2013; Feng et al., 2014; Gao and Zhao, 2014) but also in Rosaceae species, such as apple (Malnoy et al., 2016; Nishitani et al., 2016) and, recently, the diploid wild strawberry F. vesca (Zhou et al., 2018). CRISPR/Cas9 has also been used in crops with high ploidy levels such as citrus (triploid), potato, oilseed rape, cotton (tetraploids), and bread wheat (hexaploid) (Weeks, 2017). However, the functionality of this genome editing system has yet to be tested in an octoploid species such as F. × ananassa.
In this study, we used CRISPR/Cas9 to functionally characterize the role of a homeotic gene, FaTM6, in F. × ananassa. Mutations of this gene affect the development of petals, anthers, and pollen grains, and subsequently of berries. This work demonstrates that FaTM6 plays a role equivalent to AP3 in Arabidopsis, and that the CRISPR/Cas9 system can be a suitable tool for functional analyses and molecular breeding in the cultivated strawberry species, which may have important economic implications.
Material and methods
Alignment and phylogenetic tree of AP3- and TM6-like proteins
The Arabidopsis thaliana AP3 protein sequence was used as a query in a BLAST search against translated protein sequences of the strawberry genome (v. 4.0.a1) (Edger et al., 2018) in the Genome Database for Rosaceae (https://www.rosaceae.org/) to obtain Fragaria vesca TM6 (FveTM6) and AP3 (FveAP3) protein sequences. Multiple sequence alignments of AP3- and TM6-like proteins were performed using MUSCLE with the SeaView version 4 program (Gouy et al., 2010). The phylogenetic tree was inferred by the neighbor-joining method. A total of 1000 bootstrap pseudo-replicates were used to estimate the reliability of internal nodes. Evolutionary distances were computed using the Poisson correction method. The tree was rooted using four PI-like sequences, namely At-PI (A. thaliana), TPI (Solanum lycopersicum), FvePI-1, and FvePI-2 (F. vesca). Tree inference was performed using MEGA version 7 (Kumar et al., 2016). The dataset comprised 68 previously reported AP3- and TM6-like genes from gymnosperms, monocots, basal angiosperms, basal eudicots, and core eudicots obtained from GenBank.
Plant material, transient and stable transformation
Fragaria vesca (cv. Reine des Vallées) and F. × ananassa Duch. (cv. Camarosa) plants were grown and maintained under greenhouse conditions (IHSM, Málaga, Spain). Adult tm6 mutant plants were grown under the same conditions but in an independent and isolated greenhouse module to avoid cross-contamination with wild-type pollen. Transient expression of the sgRNA1-2/Cas9 binary vector was performed by infiltration of a suspension of Agrobacterium tumefaciens (strain AGL-0) into fruits at the green stage of development of F. vesca as previously described (Hoffmann et al., 2006). Fruits were collected 10 d after the infiltration when they reached the red stage. For stable transformation of F. × ananassa cv. Camarosa, plants were micropropagated in N30K medium supplemented with 2.20 μM kinetin. Transformation was performed according to the protocol described by Barceló et al. (1998). Leaf discs were transformed with A. tumefaciens (strain LBA4404) carrying a pCAMBIA2300 plasmid that contained the kanamycin resistance gene nptII and the sgRNA1-2/Cas9 cassette. Regenerated shoots were selected in the same medium supplemented with 50 mg l−1 kanamycin and 500 mg l−1 carbenicillin. Resistant plants were transferred to the greenhouse at 20–30 weeks post-transformation.
Design of sgRNAs and vector constructs
The genomic sequence of FveTM6 (FvH4_1g12260), previously annotated as gene14896-v1.0-hybrid, was obtained from the reference genome of Shulaev et al. (2011). Single-guide (sg)RNAs were designed using the ATUM CRISPR/gRNA tool (https://www.atum.bio/eCommerce/cas9/input) with FveTM6 CDS as the input sequence. We performed a BLAST search of FveTM6 CDS against the F. vesca reference genome to select those candidate sgRNAs that were specific to the target gene. Two sgRNAs located in exon 1 and exon 2, and separated by 198 bp (from PAM to PAM) were selected. The CRISPOR web-tool (http://crispor.org) (Haeussler et al., 2016) was used to validate the quality of the selected sgRNAs and to identify putative off-targets.
Two different vectors were used to generate the final sgRNA1-2/Cas9 construct, namely pAtU6:sgRNA and 35S:hSpCas9 (Mao et al., 2013). We cloned both sgRNA1 and sgRNA2 into pAtU6:sgRNA vectors using BbsI. pAtU6:sgRNA1 was cloned into the 35S:hSpCas9 vector using Acc65I and SalI. pAtU6:sgRNA2 was amplified to include the Cfr9 and XbaI restriction sites and cloned into the pAtU6:sgRNA1/35S:hSpCas9 vector, generating a construct with Cas9 and the two sgRNAs cassettes. The final binary vector was obtained by cloning the sgRNA1-2/Cas9 cassette into pCAMBIA2300 using KpnI and XbaI sites. The binary vector was introduced into A. tumefaciens strain AGL-0 for transient expression and into LBA4404 for stable transformation.
Mutation identification
Genomic DNA was isolated using the CTAB method from fruits of the transiently transformed plants, and from leaves of the stably transformed plants. The presence of the transformation cassette was tested by PCR using primers P248 and P249 to amplify Cas9 (Supplementary Table S2 at JXB online). CRISPR/Cas9-mediated indels were detected by agarose gel electrophoresis after PCR using primers flanking both sgRNAs (P180/P181 for transient assay, and P445/P446 for stable lines; Supplementary Table S2). For transient expression experiments, the PCR amplicons were cloned into the pGEMT-Easy vector system (Promega, Madison, USA) and transformed into E. coli DH5α. Single colonies were picked to identify mutations by Sanger sequencing.
Amplicon sequencing and sequence analysis
cDNA from petals and stamens was amplified using P445 and P475, and genomic DNA from leaves was amplified using P445 and P446 (Supplementary Table S2) for high-throughput amplicon sequencing. Resulting amplicons were reamplified for indexing. Libraries were purified with Agencourt® AMPure® XP beads (Beckman Coulter) using the manufacturer’s recommendations, and their quality was verified using a TapeStation 4200 HS DNA (Agilent). Libraries were quantified using real-time PCR, and they were pooled in equimolar ratios before paired-end sequencing (2 × 250 cycles) using a MiSeq system (Illumina).
For the sequence analysis, the paired-end reads were collapsed using the FLASH algorithm developed by Magoč and Salzberg (2011), using a Phred quality score of 33 as a filter. The resulting clusters were then transformed to the FASTA format using a custom Python script. In order to reduce noise, 35 nucleotides in the 5′ and 3′ positions were trimmed using Trimmomatic (Bolger et al., 2014). Custom Python scripts were designed for sequence identification and quantification. For the quantification process, no similarity threshold for sequence clusters was applied. Sequences with at least 1% prevalence were selected as possible allelic variants. However, PCR bias resulted in some missing alleles. In these rare cases, a fingerprint strategy based on SNPs exclusive for the missing allele, allowing variations among the target sites, was designed using custom Python scripts.
The sequences of two putative off-targets located within the coding sequences (off-targets #1 and #3 of the sgRNA1; Supplementary Table S1) were analysed by amplifying the spanning region of both loci with the oligonucleotides P546/P547 and P544/P545, respectively (Supplementary Table S2). The resulting PCR products were cloned into pGEM-T Easy vectors (Promega). For each line, 40 and 45 clones were selected from the constructs for off-targets #1 and #3, respectively, and these were analysed by Sanger sequencing.
Phenotypic analyses
For control plants and tm6 mutant lines, flowers at the pre-anthesis stage were marked so that all phenotypic analyses could be performed at the same developmental stage. SEM visualization of stamens was performed using flowers at 2 d post-anthesis, and that of carpels at the pre-anthesis stage. Stamens and carpels were visualized without processing using a JEOL JSM-6490LV electron microscope under low-vacuum conditions (30 MPa). To examine the pollen morphology, anthers at the dehiscence stage were incubated in absolute ethanol for 3 h, air-dried, and coated with gold in a sputtering Quorum Q150R ES. Gold-coated pollen grains were examined using a MEB JEOL 840 microscope.
Quantification and germination of pollen grains
To quantify the pollen content and analyse their germination in control and tm6 mutant lines, three flowers per genotype were collected at 2 d post-anthesis. For the quantification assay, anthers were removed and incubated with 10% sucrose and 1% acetocarmine, which stains nuclei and is therefore used as a marker for pollen viability (Chang and Neuffer, 1989). Pollen grains were quantified using a Neubauer chamber under a stereomicroscope Multizoom AZ-100 (Nikon). The pollen germination assay was performed as previously described by Ariza et al. (2006). The samples were visualized under Eclipse E800 microscope (Nikon).
Emasculation and cross-pollination
Emasculation was performed by removing all of the stamens in control flowers at the pre-anthesis stage. In order to avoid cross-pollination, the emasculated flowers were covered with cotton. To analyse the functionality of the carpels in tm6 lines, tm6-7 flowers were pollinated using wild-type pollen. Briefly, the anthers of the mutant flowers were removed manually at pre-anthesis to avoid any possible self-fertilization. A small paintbrush was then used to pollinate the tm6 stigmas with wild-type pollen. In order to avoid cross-contamination, cross-pollinated flowers were covered with cotton.
Accession numbers
Most of the protein sequences were obtained from GenBank: MASAKO B3 (Rosa rugosa; AB055966), PaTM6 (Prunus avium; AB763909), MdMADS13 (Malus × domestica; AJ251116), MdTM6 (M. × domestica; AB081093), HmTM6 (Hydrangea macrophylla; AF230703), GDEF1 (Gerbera hybrida; AJ009724), VvTM6 (Vitis vinifera; DQ979341), BalTM6 (Balanophora fungosa; JQ613232), PhTM6 (Petunia × hybrida; DQ539417), LeTM6 (Solanum lycopersicum; X60759), NbTM6 (Nicotiana benthamiana; AY577817), PtAP3-2 (Pachysandra terminalis; AF052871), PtAP3-1 (P. terminalis; AF052870), Gu.ti. AP3-5 (Gunnera tinctoria; AY337757), Gu.ti. AP3-4 (G. tinctoria; AY337756), FavAP31.1 (Fragaria × ananassa; AY429427), MASAKO euB3 (R. rugosa; AB099875), GDEF2 (G. hybrida; AJ009725), HpDEF2 (Hieracium piloselloides; AF180365), HpDEF1 (H. piloselloides; AF180364), AtAP3 (Arabidopsis thaliana;AF115814), CMB2 (Dianthus caryophyllus; L40405), SLM3 (Silene latifolia; X80490), RAD2 (Rumex acetosa; X89108), RAD1 (R. acetosa; X89113), JrAP3 (Juglans regia; AJ313089), RfAP3-2 (Ranunculus ficaria; AF130870), RfAP3-1 (R. ficaria; AF052854), HmAP3 (H. macrophylla; AF230702), NMH7 (Medicago sativa; L41727), VvAP3 (V. vinifera; EF418603), CitMADS8 (Citrus unshui; AB218614), DEF (Antirrhinum majus; X52023), NtDEF (Nicotiana tabacum; X96428), PhDEF (P. × hybrida; DQ539416), StDEF (Solanum tuberosum; X67511), TAP3 (S. lycopersicum; DQ674532), LeAP3 (S. lycopersicum; AF052868), RfAP3-1 (R. ficaria; AF052854), RbAP3-1 (Ranunculus bulbosus; AF052876), RfAP3-2 (R. ficaria; AF130870), RbAP3-2 (R. bulbosus; AF130869), PnAP3-1 (Papaver nudiculae; AF052873), PapsAP3-1 (Papaver somniferum; EF071993), PcAP3 (Papaver californicum; AF052872), LtAP3 (Liriodendron tulipifera; AF052878), MpMADS7 (Magnolia praecocissima; AB050649), Pe.am.AP3 (Persea americana; AY337748), CfAP3-1 (Calycanthus floridus; AF230699), CfAP3-2 (C. floridus; AF230700), PeMADS2 (Phalaenopsis equestris; AY378149), PeMADS5 (P. equestris; AY378148), OsMADS16 (Oryza sativa; AF077760), SILKY1 (Zea mays; AF181479), PeMADS4 (P. equestris; AY378147), LMADS1 (Lilium longiflorum; AF503913), LRDEF (Lilium regale; AB071378), CryMADS1 (Cryptomeria japonica; AF097746), CryMADS2 (C. japonica; AF097747), GnegGGM2 (Gnetum gnemon; AJ132208), GnegGGM13 (G. gnemon; AJ132219), DAL13-1 (Picea abies; AF158543), PrDGL (Pinus radiata; AF120097), GnegGGM15 (G. gnemon; AJ251555), TPI (S. lycopersicum; DQ674531)
FveTM6 (Fragaria vesca; FvH4_1g12260), FveAP3 (F. vesca; FvH4_2g38970). AtPI (A. thaliana; At5g20240), FvePI-1 (F. vesca; FvH4_2g27860.1), FvePI-2 (F. vesca; FvH4_2g278270.1).
Results
Identification and phylogenetic analysis of AP3 lineage genes in F. vesca
To identify genes belonging to the AP3 lineage in strawberry, a BLAST search was performed using the AP3 protein sequence from Arabidopsis (AtAP3) against the reference genome of the diploid F. vesca (cv. Hawaii 4). Two genes with high homology were obtained, FvH4_2g38970 and FvH4_1g12260, sharing 55.45% and 50.43% of amino acid identity with AtAP3, respectively. To place these two genes in a phylogenetic context, we performed a phylogenetic analysis using neighbor-joining of AP3- and TM6-like proteins from gymnosperms to core eudicots (Supplementary Fig. S1). The phylogenetic analysis showed that FvH4_2g38970 (hereafter named FveAP3) and FvH4_1g12260 (hereafter named FveTM6) belong to the euAP3 and TM6 lineages, respectively, indicating that F. vesca, unlike Arabidopsis, contains both AP3 lineages.
Expression analysis of AP3 lineage genes in F. × ananassa
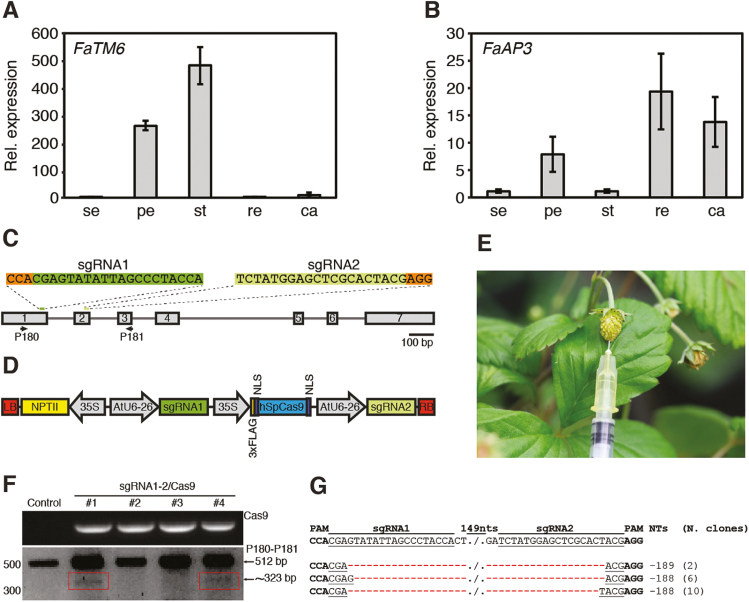
To further investigate the role of the AP3 lineage genes in cultivated strawberry, we first analysed their expression using quantitative real-time PCR (qRT-PCR) in sepals, petals, stamens, receptacles, and carpels of F. × ananassa flowers at stage 12 (Hollender et al., 2012). As shown in Fig. 1A, FaTM6 was expressed in both petals and stamens, with the latter having the higher expression level. In contrast, FaAP3 was expressed mainly in receptacles, followed by carpels and petals, with very little expression in stamens and sepals (Fig. 1B). This suggested that FaTM6, and not FaAP3, is the gene with homologous function to AP3 in Arabidopsis. Hence, we selected FaTM6 to study its role in flower development by CRISPR/Cas9-mediated mutagenesis.
Fig. 1.
Expression of AP3 and TM6 genes in the cultivated strawberry (Fragaria × ananassa), construct design, and evaluation of CRISPR-Cas9-based editing of the wild strawberry, Fragaria vesca. Relative expression of the F. × ananassa TM6 gene (FaTM6) (A) and AP3 gene (FaAP3) (B) in sepals (se), petals (pe), stamens (st), receptacles (re), and carpels (ca) of F. × ananassa flowers at stage 12 as determined by qRT-PCR. Data are means (±SD) of three biological replicates with three technical replicates each. (C) The F. vesca TM6 (FveTM6) locus, including exons (boxes) and introns (lines). sgRNAs 1 and 2 are represented in green and PAM sequences in orange. The primers used for characterization of CRISPR/Cas9 editing are indicated. (D) Schematic representation of the two sgRNAs and Cas9 expression cassettes in a single binary vector pCMABIA2300 (sgRNA1-2/Cas9 vector). (E) Green fruit agro-infiltrated with the sgRNA1-2/Cas9 vector. (F) PCR to detect indels in FveTM6. Top panel: PCR of Cas9 in four fruits that transiently expressed the vector. Bottom panel: PCR with P180 and P181 (Supplementary Table S2) showing a ~323-bp band (red boxes) in fruits #1 and #4 in addition to the wild-type band (512 bp). (G) Alignment of sequences obtained after the purification, cloning, and Sanger sequencing of the ~323 bp band from fruits #1 and #4. Fragments with 189- and 188-bp deletions resulting from simultaneous DSBs in both target sites were detected in two and 16 clones, respectively.
Identification of FaTM6 targets, characterization of FaTM6 alleles, and construct design
We first searched for candidate sgRNAs to edit FaTM6 using the available sequence of FveTM6 from the F. vesca reference genome (cv. Hawaii 4). Two target sites for FveTM6 were selected in order to generate a dual sgRNA construct, allowing us to create a large deletion and/or increase the efficiency of the mutagenesis (Belhaj et al., 2013). The sgRNAs, located within exon 1 (sgRNA1) and exon 2 (sgRNA2), were selected based on their high specificity score and minimum potential off-target activities (Fig. 1C, Supplementary Table S1). sgRNA1 spanned the carboxyl-end of the M-domain and the amino-terminal of the ‘intervening’ (I) region of the FveTM6 protein, while sgRNA2 was located within the I region (Supplementary Fig. S2).
Next, we evaluated the suitability of the two guides designed using the reference genome for editing the two genotypes used in this study [F. vesca cv. Reine des Vallées (RV) and F. × ananassa cv. Camarosa]. In order to detect any polymorphisms in the reference genome that might affect the CRISPR/Cas9-mediated editing, we amplified and cloned the genomic regions of TM6 spanning the two target sites (Fig. 1C, Supplementary Table S2) and performed Sanger sequencing. While F. vesca cv. RV did not show any variation in the TM6 sequence compared to the reference genome, five different alleles were identified in F. × ananassa cv. Camarosa (Supplementary Fig. S3). These alleles contained indels in the first and second introns, and four synonymous and eight non-synonymous SNPs within the coding region. None of these alleles had polymorphisms in the region targeted by sgRNA2, but allele #5 contained a G168T substitution within the PAM-proximal region of sgRNA1 (Supplementary Fig. S3), which might decrease the cleavage efficiency (Xu et al., 2017). To determine which alleles were expressed in petals and stamens, we generated cDNA from these tissues and performed high-throughput amplicon sequencing for the region spanning sgRNA1 and sgRNA2. Our data indicated that at least four of the five FaTM6 alleles identified were expressed in both petals and stamens (Supplementary Table S3). Specifically, we detected alleles #3, #4, #5, and a sequence that might correspond to either allele #1 or #2, which are indistinguishable within the sequenced CDS region. Given this information and with our dual sgRNA strategy, sgRNA2 would probably ensure editing events due to the lack of polymorphisms, while sgRNA1 would allow us to assess the effect of the mismatch in allele #5 on editing efficiency. This design would also result in large deletions if both sites were simultaneously cleaved by Cas9, probably producing non-functional alleles. We designed a single binary vector harboring the two sgRNAs under AtU6-26 promoters, and the Cas9 nuclease under the 35SCaMV promoter, sgRNA1-2/Cas9 (Fig. 1D).
Functionality test of the sgRNA1-2/Cas9 vector by transient transformation of F. vesca fruits
Since the transformation and establishment of stable transgenic plants requires 6–9 months, we first tested the functionality of our dual sgRNA/Cas9 editing construct by transient transformation of diploid F. vesca cv. RV fruits. The sgRNA1-2/Cas9 vector was agro-infiltrated in the receptacle of fruits at the green stage (Fig. 1E), and genomic DNA was extracted at 10 d post-infiltration. PCR amplification was performed with primers spanning the target sites (Supplementary Table S2) in order to detect CRISPR/Cas9-mediated editing events as evidenced by amplicon sizes that were different from the wild-type allele (Fig. 1C). The PCR results confirmed that two of the four fruits infiltrated with the sgRNA1-2/Cas9 vector showed a smaller amplicon in addition to the wild-type (Fig. 1F). Cloning and Sanger sequencing of the smaller amplicon from these two plants confirmed the presence of a deletion of ~190 bp between the two target sites in 18 clones, validating the functionality of the sgRNA1-2/Cas9 vector in the diploid strawberry species (Fig. 1G).
Targeted mutagenesis of FaTM6 in stable transgenic F. × ananassa plants
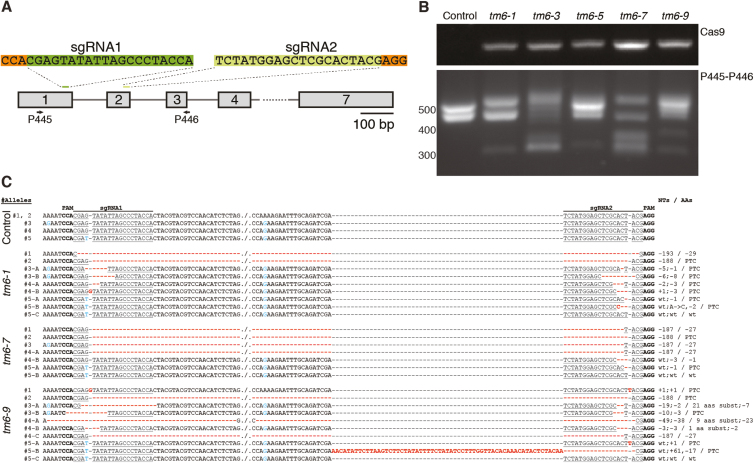
We generated stable transgenic plants of F. × ananassa cv. Camarosa with the same sgRNA1-2/Cas9 vector. We obtained and micropropagated five independent lines, (tm6 lines), and used PCR to examine them for editing events, as described for the transient assays (Fig. 2A; Supplementary Table S2). Different amplicon patterns were obtained between the tm6 lines and the untransformed wild-type plant, indicating the generation of various indels by the CRISPR/Cas9 complex (Fig. 2B). These results confirmed the CRISPR/Cas9-mediated mutagenesis of FaTM6 in F. × ananassa.
Fig. 2.
Identification of CRISPR/Cas9-induced mutations in the Fragaria × ananassa TM6 allele (FaTM6). (A) Schematic representation of the positions of the sgRNAs in the F. vesca TM6 (FveTM6) locus. The primers used for the analysis in agarose gels and for deep sequencing are represented (P445 and P446; Supplementary Table S2). (B) Top panel: identification of the Cas9 gene in transgenic tm6 lines. Bottom panel: detection of mutations at the FaTM6 locus using the P445 and P446 primers. (C) Sequence alignment obtained by high-throughput amplicon sequencing in control and tm6 mutant lines. PAM sequences are marked in bold; sgRNAs are underlined; blue indicates distinctive SNPs among FaTM6 alleles; bold red indicates mutations induced by CRISPR/Cas9-mediated editing. PTC, premature termination codon.
In order to analyse the CRISPR/Cas9-induced mutations at the FaTM6 locus at the molecular level, we selected three transgenic lines, tm6-1, tm6-7, and tm6-9, based on their amplicon patterns. Using high-throughput amplicon sequencing and de novo assembly, nine, seven, and 10 different alignment groups were obtained for tm6-1, tm6-7, and tm6-9, respectively (Fig. 2C; Supplementary Table S3). Allele #1 showed a single editing event in all three lines, while allele #2 had the same deletion in all of them. However, different editing events were obtained for alleles #3, #4, and #5 within each transgenic line, except for tm6-7, which had only one modification in allele #3 (Fig. 2C). As expected, most of the CRISPR/Cas9-induced mutations occurred downstream of the PAM sequences, although a deletion including the whole PAM-sgRNA1 region was also observed in tm6-9.
All sequences obtained for alleles #1 through to #4 showed mutations within the region targeted by sgRNA1 (Fig. 2C). However, no editing was observed in this target site in allele #5, most likely due to the mismatch present in the sgRNA1 seed sequence. For the region targeted by sgRNA2, the sequencing analysis showed that all five alleles were edited, although wild-type sequences were also detected for this target, but only in allele #5.
As expected from the amplicon pattern obtained for these three lines, all of them contained large deletions generated by the simultaneous double-strand breaks (DSBs) in both target sites. These deletions of 187 and 193 nts resulted in a deletion of 27 or 29 amino acids, respectively, or in the generation of a premature stop codon (–188 nts) (Fig. 2C, Supplementary Fig. S4). Most of the editing events generated frameshift mutations, especially in the tm6-1 line, which resulted in the production of seven truncated proteins (#2, #3A, #3B, #4A, #4B, #5A, and #5B) out of the nine allelic variants detected (Supplementary Fig. S4). In addition to the generation of these truncated proteins, shorter amino acid deletions and substitutions were also obtained in tm6-7 and tm6-9.
Off-target analysis
Seven putative off-targets were predicted for sgRNA1 and sgRNA2 (Supplementary Table 1) using the CRISPOR web tool (http://crispor.org) (Haeussler et al., 2016). Out of these putative off-targets, only #1 and #3 were located at CDSs, containing four and five mismatches within the sgRNA1-PAM sequence, respectively. On the other hand, these two genes were very weakly expressed in petals and anthers of F. vesca (Supplementary Fig. S5; Hawkins et al., 2017). Thus, we did not expect any homeotic effect due to possible off-target activities affecting these two genes. Nevertheless, to confirm this hypothesis, we amplified and sequenced the spanning region of both off-targets in the wild-type and tm6-9 mutant lines. As expected, we did not find any sequence variations in the off-target sequences of tm6-9 compared to that of the control (Supplementary Fig. S6).
FaTM6 plays a key role in anther development
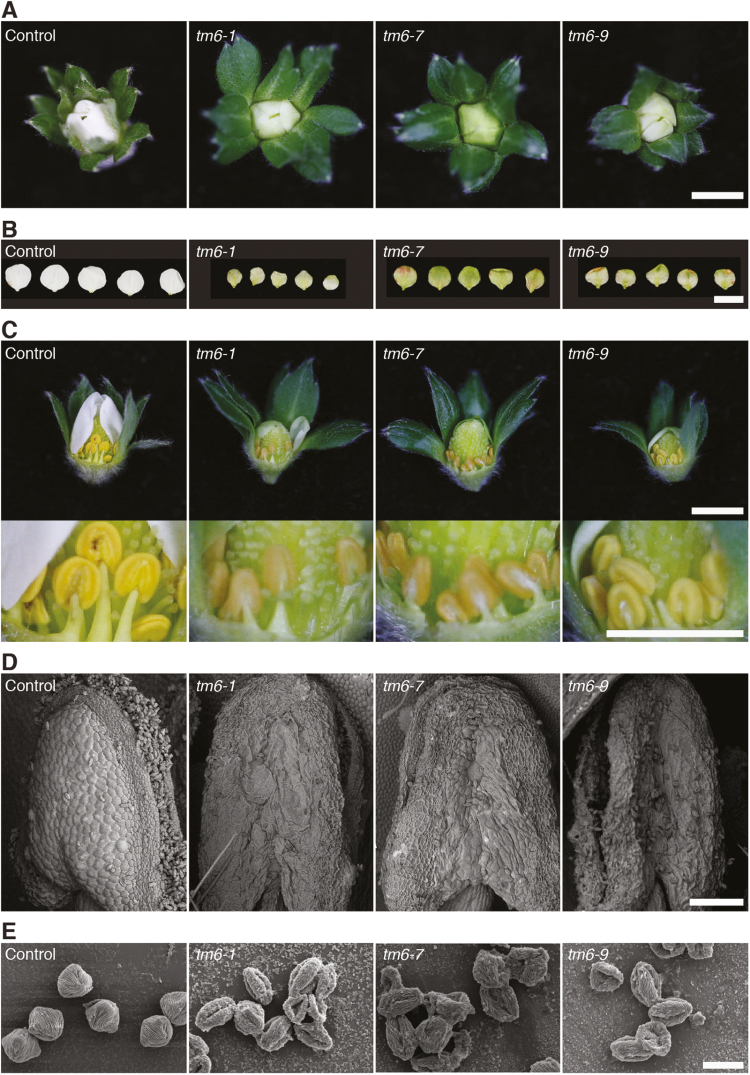
To determine the function of FaTM6, we analysed the flower phenotype of the wild-type and the tm6-1, tm6-7, and tm6-9 mutant lines. At the pre-anthesis stage, petals in the mutant lines were shorter and slightly greener compared to those of the control flowers (Fig. 3A, B). More severe defects were observed in the anthers, which were smaller and darker than that of the wild-type (Fig. 3C). A more detailed anatomical analysis of the anthers at the dehiscence stage and the pollen grains was performed using SEM. Wild-type anthers displayed the typical four-lobed structure, with a very well-defined epidermal layer, and with pollen grains visible at the stomium rupture site (Fig. 3D). However, the anthers of the tm6 mutant lines displayed morphological differences, showing clear defects in the epidermal cell layer and a reduced number of pollen grains at the stomium. This apparent difference in pollen content was quantified as a 10-fold reduction in tm6-1 and tm6-7, and a 50-fold reduction in tm6-9 compared to the control (Supplementary Fig. S7A, B). Moreover, most of the pollen grains from the mutant lines showed aberrant and collapsed structures (Fig. 3E). To test pollen grain viability, acetocarmine staining and pollen germination assays were performed. While acetocarmine successfully stained nuclei in the mutant pollen grains (Supplementary Fig. S7A), we failed to detect any germination in the tm6 mutant lines, in contrast to the wild-type (Supplementary Fig. S7C), indicating that the viability had been strongly impaired in the mutant lines.
Fig. 3.
Phenotypic effects of mutations in Fragaria × ananassa TM6 (FaTM6) in flowers. (A) Flowers of control and three independent tm6 lines at the pre-anthesis stage. (B) Petals of tm6 lines appear smaller and greener. (C) Top panel: flowers at pre-anthesis with some petals removed. Bottom panel: higher magnification to show details of the morphology of the stamens. (D, E), SEM of the structures of the anthers at the dehiscence stage (D) and of the pollen grains (E). Scale bars: (A–C) 1 cm; (D) 200 µm; (E) 20 µm.
Since ovule fertilization and proper development of embryos and achenes are necessary for normal receptacle development (Nitsch, 1950), we assessed fruit formation in the tm6 lines compared with emasculated controls. The emasculation of wild-type flowers caused a complete abortion in receptacle development (Fig. 4A). Consistent with the impaired pollen grain formation, the tm6 mutant lines also showed arrested development of the receptacles (Fig. 4A, B). Nevertheless, a few fruits (5.7 and 2.2% in tm6-7 and tm6-9, respectively) showed a local enlargement of the receptacle around some achenes, indicating that some pollen grains must have retained their viability, allowing a residual pollination (Supplementary Fig. S8).
Fig. 4.
Phenotypic effects of mutations in Fragaria × ananassa TM6 (FaTM6) in fruits and in a complementation experiment. (A) Wild-type flowers emasculated at the pre-anthesis stage phenocopy aborted flowers in tm6 mutant lines. Two representative flowers per genotype are shown. (B) Top panels: adult plants of control and tm6 mutant lines. Bottom panels: control plants develop wild-type berries, but tm6 flowers abort. (C) SEM of the structures of carpels at the pre-anthesis stage. (D) A fruit developed from a tm6-7 flower emasculated and pollinated with wild-type pollen. Scale bars: (A) 1 cm; (C) 200 µm.
Consistent with the lack of FaTM6 expression in carpels, these organs showed normal development (Fig. 4C). In order to confirm the carpel viability in the mutant lines, we pollinated carpels of the tm6-7 line using wild-type pollen. As shown in Fig. 4D a wild-type receptacle was fully developed, indicating that lack of pollination was responsible for the aborted fruit phenotype in the tm6 lines. These results taken together indicate an essential role of FaTM6 in anther and pollen formation during flower development of F. × ananassa.
Discussion
FaTM6 is involved in petal and stamen development
Here, we report that strawberry, unlike Arabidopsis, has maintained both an euAP3 and a TM6 gene. While FveAP3 contains the euAP3 motif in the C-terminal domain of the protein, FveTM6 possesses a motif more similar to those of the paleoAP3 genes (Supplementary Fig. S2) (Kramer et al., 1998). A transcriptome study during floral development of F. vesca reported that FveTM6 (gene14896-v1.0-hybrid) is strongly expressed in anthers (Hollender et al., 2014), consistent with our results in the octoploid strawberry. However, in that study, FveTM6 was mis-annotated as FveAP3, since our phylogenetic analysis showed that this gene belongs instead to the TM6 lineage (Supplementary Fig. S1). Moreover, expression analysis of FaAP3 and FaTM6 in flowers of F. × ananassa showed that FaTM6 and not FaAP3 was the gene with the typical expression pattern of the B-class type (Fig. 1A, B), which is consistent with previous studies in Rosaceae (Hibino et al., 2006).
Sequencing analyses of the region spanning the CRISPR target sites for TM6 indicated that the diploid F. vesca cultivar used in this study was homozygous at this locus (Supplementary Fig. S3). However, the octoploid strawberry F. × ananassa cv. Camarosa showed high heterozygosity at the FaTM6 locus, consistent with the genetically complex genome of this species. At least five alleles of FaTM6 among the four pairs of homoeologous chromosomes were detected. However, we cannot discount an even more complex scenario involving more FaTM6 alleles due to the possible presence of additional polymorphisms outside the region covered in this study. Furthermore, deep sequencing of cDNA from petals and sepals showed that at least four of the five FaTM6 were expressed in both organs, with expression of alleles #3, #4, and #5 confirmed, and one of either allele #1 or allele #2, since these are indistinguishable within the sequenced CDS region (Supplementary Table S3).
The role of euAP3 genes in petal and stamen specification has been well established (Jack et al., 1992; Schwarz-Sommer et al., 1992; de Martino et al., 2006; Roque et al., 2013). However, there have been fewer reports on the role of TM6-like transcription factors, which play a more important role in stamen identity (Rijpkema et al., 2006; Roque et al., 2013). Our results are also consistent with a predominant role for FaTM6 in stamen development since the anthers in the tm6 mutant lines were severely affected, and they showed a drastic reduction in pollen content and viability (Fig. 3C–E, Supplementary Fig. S7), while the petals of the tm6 mutants showed more modest defects in overall size and color (Fig. 3B). This is consistent with the phenotype reported in TM6i (RNAi) lines of tomato, which develop smaller petals that are attributed to a decrease in cell proliferation (de Martino et al., 2006). Similarly, PTD, the Populus trichocarpa TM6 ortholog, has been postulated to play a role in regulating cell proliferation (Sheppard et al., 2000)
Auxin transport from fertilized carpels to the floral receptacle is essential for the latter to grow into a fleshy and edible fruit (Nitsch, 1950; Kang et al., 2013). The high percentage of fruit abortions observed in the tm6 mutant lines, which phenocopy emasculated wild-type flowers, supports the tight coupling of flower and fruit development (Supplementary Fig. S8, Fig. 4A, B). The presence of a small number of aberrant fruits with enlarged portions of receptacle around developed achenes (Supplementary Fig. S8) indicated that some residual viable pollen was formed despite our failure to detect pollen germination (Supplementary Fig. S7C). tm6 mutants developed anatomically normal pistils (Fig. 4C), consistent with the lack of FaTM6 expression in this organ (Fig. 1A). In fact, carpels in the tm6 lines were functional since fruit development was restored using wild-type pollen. All of these findings indicate that the defect in pollen formation rather than gynoecium development caused the fruit abortions in the tm6 lines.
CRISPR/Cas9 is an efficient tool for gene functional analysis in octoploid strawberry
In this study, we designed a dual sgRNA system that efficiently edited the FaTM6 gene in octoploid strawberry. Moreover, we developed a quick and easy validation in vivo of the sgRNA efficiency by performing a transient assay in fruits of the diploid F. vesca.
A deep sequence analysis in three independent transgenic CRISPR lines in F. × ananassa showed a high efficiency of sgRNA1, which drove the editing of four of the five FaTM6 alleles in all lines examined (Fig. 2C, Supplementary Table S3). Only allele #5 remained totally unedited at this target site, probably because of the mismatch in the seed sequence, which has been reported to decrease the cleavage efficiency (Xu et al., 2017). Although Cas9 can still tolerate mismatches within the target site (Hsu et al., 2013; Braatz et al., 2017), our results indicated that the mismatch in the seed sequence of allele #5 totally prevented the Cas9 activity (Fig. 2C, Supplementary Table S3). None of the alleles had mismatches with sgRNA2, and this led to the editing of all the five FaTM6 alleles. Interestingly, an unedited variant for allele #5 was also detected with a low prevalence in each line (#5-C, #5-B, and #5-C in tm6-1, tm6-7, and tm6-9 respectively; Supplementary Table S3). It is possible that the SNP present at the sgRNA1 target site for allele #5 also affected editing at the sgRNA2 site. All of these results show that a preliminary sequence analysis is essential to optimize the efficiency of the CRISPR/Cas9 system in a polyploid and highly heterozygous species such as F. × ananassa, especially when a reference genome sequence is not available. Moreover, when designing CRISPR/Cas9 experiments to edit a polymorphic locus of F. × ananassa, we recommend designing constructs containing multiple sgRNAs against the different allelic variants.
Importantly, and as expected by their number of mismatches to the TM6 locus, no editing was found in the two putative off-target sites located within CDSs (Supplementary Fig. S6). This result provides strong support for the tm6 mutations being responsible for the aberrant development of anthers and petals in these mutant lines.
Despite the octoploid nature of strawberry, our analysis showed that the tm6-1 and tm6-9 lines contained more than eight allelic variants, indicating that these two lines are chimeras. These genetic mosaics are common in CRISPR/Cas9 T0 generations of plants obtained from somatic tissue due to the activity of Cas9 during later stages of shoot development (Liu et al., 2017). Due to the high heterozygosity of cultivated strawberry, breeding lines can only be maintained and propagated clonally by runners. Therefore, even though a full knock-out was not achieved in the T0 generation, continuous clonal propagation of the transgenic lines containing the CRISPR/Cas9 may enable the eventual mutation of all eight homeologs.
Conclusions
In summary, we have characterized the role of a homeotic gene in strawberry, FaTM6, for the first time. We have shown that it is primarily involved in anther development, but also has a role in petal formation. In obtaining these results, we have demonstrated that CRISPR/Cas9 is a powerful tool for gene functional studies in commercial strawberry that can overcome the drawbacks of RNAi-based approaches, such as incomplete and/or unstable knockdown effects and unpredictable off-targets. Moreover, by confirming the feasibility of genome-editing in F. × ananassa, we have shown the potential of this approach for future generation of lines with traits of agronomic interest, despite the high ploidy of this species.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Phylogenetic analysis of TM6 and euAP3 lineage proteins.
Fig. S2. Alignment of AP3- and TM6-like proteins.
Fig. S3. Alignment of TM6 sequences from F. vesca and F. × ananassa.
Fig. S4. Alignment of TM6 predicted amino acid sequences in the tm6 mutant lines.
Fig. S5. Expression analysis of two putative off-targets for sgRNA1.
Fig. S6. Sequence analyses of two putative off-targets for sgRNA1.
Fig. S7. Pollen yield quantification in the tm6 mutant lines.
Fig. S8. Quantification of the tm6 fruit phenotype.
Table S1. Off-target analysis for sgRNA1 and sgRNA2.
Table S2. List of oligonucleotides used in this study.
Table S3. Analysis of high-throughput sequencing of amplicons of TM6 cDNA and genomic DNA.
Acknowledgments
This work and CM-P and DP were supported by the European Research Council (ERC-2014-StG 638134, C.M.-P.) and the Ramón y Cajal program RYC 2013-1269 (MINECO-Universidad de Málaga, Spain, DP). We thank Beth Rowan, Catharina Merchante, and Michael Baldwin for their suggestions to improve the manuscript, and especially Miguel Ángel Botella and Victoriano Valpuesta for helpful discussions. We thank José Sánchez-Sevilla, Alicia Esteban, and José Duarte for technical assistance, and Jian-Kang Zhu for providing the CRISPR/Cas9 vectors.
References
- Ariza MT, López Aranda JM, Soria C. 2006. Optimization of a liquid medium for germination of strawberry pollen. Acta Horticulturae 708, 531–534. [Google Scholar]
- Barceló M, Mansouri El I, Mercado J-Á, Quesada MA, Pliego-Alfaro F. 1998. Regeneration and transformation via Agrobacterium tumefaciens of the strawberry cultivar Chandler. Plant Cell, Tissue and Organ Culture 54, 29–36. [Google Scholar]
- Barrangou R, Birmingham A, Wiemann S, Beijersbergen RL, Hornung V, Smith Av. 2015. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Research 43, 3407–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. 2013. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Kaufmann A, Kozak K. 2014. RNAi and off-target effects. In: Ralph Tripp A, Karpilow JM eds. Frontiers in RNAi, vol. 1. United Arab Emirates: Bentham Science Publishers, 3–20. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braatz J, Harloff H-J, Mascher M, Stein N, Himmelbach A, Jung C. 2017. CRISPR-Cas9 induced mutations in polyploid oilseed rape. Plant Physiology 174, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Castillo R, Xue Y, Schwarz-Sommer Z, Davies B. 2010. Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Molecular Biology and Evolution 27, 2651–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Neuffer MG. 1989. A simple method for staining nuclei of mature and germinated maize pollen. Stain Technology 64, 181–184. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. 2006. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. The Plant Cell 18, 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, VanBuren R, Colle M, et al. 2018. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 7, gix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, et al. 2014. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. 2014. Specific and heritable gene editing in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 4357–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27, 221–224. [DOI] [PubMed] [Google Scholar]
- Guidarelli M, Baraldi E. 2015. Transient transformation meets gene function discovery: the strawberry fruit case. Frontiers in Plant Science 6, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, et al. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Caruana J, Li J, Zawora C, Darwish O, Wu J, Alkharouf N, Liu Z. 2017. An eFP browser for visualizing strawberry fruit and flower transcriptomes. Horticulture Research 4, 17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino Y, Kitahara K, Hirai S, Matsumoto S. 2006. Structural and functional analysis of rose class B MADS-box genes ‘MASAKO BP, euB3, and B3’: Paleo-type AP3 homologue ‘MASAKO B3’ association with petal development. Plant Science 170, 778–785. [Google Scholar]
- Hirakawa H, Shirasawa K, Kosugi S, et al. 2014. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Research 21, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W. 2006. RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. The Plant Journal 48, 818–826. [DOI] [PubMed] [Google Scholar]
- Hollender CA, Geretz AC, Slovin JP, Liu Z. 2012. Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 235, 1123–1139. [DOI] [PubMed] [Google Scholar]
- Hollender CA, Kang C, Darwish O, Geretz A, Matthews BF, Slovin J, Alkharouf N, Liu Z. 2014. Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiology 165, 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jiang W, Yang B, Weeks DP. 2014. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PloS ONE 9, e99225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. 2013. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Research 41, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z. 2013. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. The Plant Cell 25, 1960–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. 1998. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM. 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. 2013. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nature Biotechnology 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dai C, Hu C, Liu Z, Kang C. 2017. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. The Plant Journal 90, 164–176. [DOI] [PubMed] [Google Scholar]
- Liu X, Xie C, Si H, Yang J. 2017. CRISPR/Cas9-mediated genome editing in plants. Methods 121-122, 94–102. [DOI] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Nagamangala Kanchiswamy C. 2016. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Frontiers in Plant Science 7, 1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK. 2013. Application of the CRISPR-Cas system for efficient genome engineering in plants. Molecular Plant 6, 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani C, Hirai N, Komori S, Wada M, Okada K, Osakabe K, Yamamoto T, Osakabe Y. 2016. Efficient genome editing in apple using a CRISPR/Cas9 system. Scientific Reports 6, 31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP. 1950. Growth and morphogenesis of the strawberry as related to auxin. American Journal of Botany 37, 211–215. [Google Scholar]
- Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E. 1991. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. The Plant Journal 1, 255–266. [PubMed] [Google Scholar]
- Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. 2006. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. The Plant Cell 18, 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque E, Serwatowska J, Cruz Rochina M, Wen J, Mysore KS, Yenush L, Beltrán JP, Cañas LA. 2013. Functional specialization of duplicated AP3-like genes in Medicago truncatula. The Plant Journal 73, 663–675. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Yang Y, Šurbanovski N, Bianco L, Buti M, Velasco R, Giongo L, Davis TM. 2016. HaploSNP affinities and linkage map positions illuminate subgenome composition in the octoploid, cultivated strawberry (Fragaria × ananassa). Plant Science 242, 140–150. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. 1992. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. The EMBO Journal 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard LA, Brunner AM, Krutovskii KV, Rottmann WH, Skinner JS, Vollmer SS, Strauss SH. 2000. A DEFICIENS homolog from the dioecious tree black cottonwood is expressed in female and male floral meristems of the two-whorled, unisexual flowers. Plant Physiology 124, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, et al. 2011. The genome of woodland strawberry (Fragaria vesca). Nature Genetics 43, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Govindarajulu R, Ashman TL, Liston A. 2014. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biology and Evolution 6, 3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Govindarajulu R, Liston A, Ashman TL. 2013. Targeted sequence capture provides insight into genome structure and genetics of male sterility in a gynodioecious diploid strawberry, Fragaria vesca ssp. bracteata (Rosaceae). G3: Genes, Genomes, Genetics 3, 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DP. 2017. Gene editing in polyploid crops: wheat, camelina, canola, potato, cotton, peanut, sugar cane, and citrus. Progress in Molecular Biology and Translational Science 149, 65–80. [DOI] [PubMed] [Google Scholar]
- Xu X, Duan D, Chen SJ. 2017. CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: from physical mechanism to off-target assessment. Scientific Reports 7, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang G, Liu Z. 2018. Efficient genome-editing of wild strawberry genes, vector development, and validation. Plant Biotechnology Journal 16, 1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.