The Arabis alpina APETALA2 ortholog, PERPETUAL FLOWERING2, coordinates the age-dependent response to vernalization and it is required to facilitate the activation of A. alpina FLOWERING LOCUS C after vernalization.
Keywords: APETALA2, AP2, FLOWERING LOCUS C, FLC, juvenility, PEP1, PEP2, perennial, PERPETUAL FLOWERING 1, vernalization
Abstract
The floral repressor APETALA2 (AP2) in Arabidopsis regulates flowering through the age pathway. The AP2 ortholog in the alpine perennial Arabis alpina, PERPETUAL FLOWERING 2 (PEP2), was previously reported to control flowering through the vernalization pathway via enhancing the expression of another floral repressor PERPETUAL FLOWERING 1 (PEP1), the ortholog of Arabidopsis FLOWERING LOCUS C (FLC). However, PEP2 also regulates flowering independently of PEP1. To characterize the function of PEP2, we analyzed the transcriptomes of pep2 and pep1 mutants. The majority of differentially expressed genes were detected between pep2 and the wild type or between pep2 and pep1, highlighting the importance of the PEP2 role that is independent of PEP1. Here, we demonstrate that PEP2 activity prevents the up-regulation of the A. alpina floral meristem identity genes FRUITFUL (AaFUL), LEAFY (AaLFY), and APETALA1 (AaAP1), ensuring floral commitment during vernalization. Young pep2 seedlings respond to vernalization, suggesting that PEP2 regulates the age-dependent response to vernalization independently of PEP1. The major role of PEP2 through the PEP1-dependent pathway takes place after vernalization, when it facilitates PEP1 activation both in the main shoot apex and in axillary branches. These multiple roles of PEP2 in the vernalization response contribute to the A. alpina life cycle.
Introduction
Plant adaptation to the environment requires the modification of developmental traits, among which flowering time is key to ensure successful production of offspring. Alpine habitats in which juvenile survival is very low are mainly dominated by perennial species (Billings and Mooney, 1968). In general, the perennial growth habit relies on the differential behavior of meristems on the same plant so that some will stay vegetative whereas others will initiate flowering (Amasino, 2009; Lazaro et al., 2018). The main environmental cue that promotes flowering in alpine species is the exposure to prolonged cold, a process called vernalization. Alpine environments are characterized by short growing seasons and long periods of snow coverage. Thus, to ensure reproductive success, alpine plants initiate flower buds in response to prolonged cold several months or years before anthesis (Diggle, 1997; Meloche and Diggle, 2001). However, exposure to long periods of cold does not always result in flowering. This is especially true for perennial species, as most of them have a prolonged juvenile phase and are not competent to flower at a young age (Bergonzi and Albani, 2011).
The molecular mechanisms regulating flowering in response to vernalization or to the age of the plant have been mainly studied in the annual model plant Arabidopsis thaliana. The MADS box transcription factor FLOWERING LOCUS C (FLC) is the major regulator of flowering in response to vernalization (Michaels and Amasino, 1999; Sheldon et al., 2000). FLC transcriptionally regulates floral integrator genes, such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and genes involved in the age pathway, suggesting an interplay between these two pathways (Deng et al., 2011; Mateos et al., 2017). Comparative studies between Arabidopsis and the alpine perennial Arabis alpina demonstrated that the FLC ortholog in A. alpina, PERPETUAL FLOWERING1 (PEP1), also controls flowering in response to vernalization. In addition, PEP1 contributes to the perennial growth habit by repressing flowering in a subset of axillary meristems after vernalization (Wang et al., 2009; Lazaro et al., 2018). Flower buds in A. alpina are formed during prolonged exposure to vernalizing conditions. The length of vernalization determines PEP1 reactivation in the inflorescence. After insufficient vernalization, PEP1 mRNA is reactivated and results in the appearance of floral reversion phenotypes such as bracts and vegetative inflorescence branches (Lazaro et al., 2018). In the axillary branches, the duration of vernalization does not influence PEP1 expression, and PEP1 transcript is high irrespective of the duration of vernalization (Lazaro et al., 2018). The fate of these axillary branches is determined by a combined action of the age pathway and PEP1 (Wang et al., 2011; Park et al., 2017).
In Arabidopsis, the age pathway is regulated by two miRNAs and their targets. At a young age high miRNA 156 (miR156) levels prevent flowering. As plants get older miR156 accumulation gradually decreases, whereas miR172 levels follow the opposite pattern and gradually increase during development (Wu et al., 2009). The biological function of miR156 is exerted by its targets that encode members of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKEs (SPLs) transcription factor family (Schwab et al., 2005; Wu and Poethig, 2006; Wu et al., 2009; Xu et al., 2016). From these, SPL9 and SPL15 have been reported to activate the transcription of miRNA172b, which in turn represses the expression of a small subfamily of APETALA2-like transcription factors by a translational mechanism (Aukerman and Sakai, 2003; Chen, 2004; Mathieu et al., 2009; Wu et al., 2009; Hyun et al., 2016). This subfamily includes six members: AP2, TARGET OF EARLY ACTIVATION TAGGED1 to 3 (TOE1–TOE3), SCHLAFMUTZE (SMZ), and SCHNARCHZAPFEN (SNZ) (Aukerman and Sakai, 2003; Schmid et al., 2003; Mathieu et al., 2009; Yant et al., 2010). Arabis alpina has a very distinct juvenile phase and the accession Pajares needs to grow for at least 5 weeks in long days (LDs) before it is able to flower in response to vernalization (Wang et al., 2011; Bergonzi et al., 2013). The role of miR156 is conserved in A. alpina as miR156b-overexpressing lines block flowering in response to vernalization, while mimicry lines (MIM156), used to reduce miRNA activity, flower when vernalized at the age of 3 weeks (Bergonzi et al., 2013). However, the complementary temporal accumulation of miR156 and miR172 during development is uncoupled in A. alpina (Bergonzi et al., 2013). Similar to A. thaliana, the accumulation of miR156 is reduced in the shoot apex of A. alpina plants that get older and acquire competence to flower in long days, but miR172 expression is low (Bergonzi et al., 2013). For flowering to occur and to observe an increase in miR172 levels in the shoot apex, vernalization is required (Bergonzi et al., 2013). However, vernalization is only effective in mature (old) plants but not in juvenile (young) plants that still have high levels of miR156 (Bergonzi et al., 2013). The initiation of flowering during cold in mature plants correlates with the gradual increase in expression of the floral organ identity genes LEAFY (AaLFY), FRUITFUL (AaFUL), and APETALA1 (AaAP1) (Lazaro et al., 2018). In perennials, such as apple and poplar, the homologs of the floral repressor TERMINAL FLOWER1 (TFL1) regulate the juvenile period. Transgenic Malus domestica and Populus trichocarpa lines with decreased expression of TFL1 have a shortened juvenile phase (Kotoda et al., 2006; Mohamed et al., 2010). Similarly, transgenic A. alpina plants in which AaTFL1 expression was reduced can flower even when they are vernalized at a young age (Wang et al., 2011). Interestingly, these lines can flower after exposure to a short (6 weeks instead of 12 weeks) duration of vernalization. These results again suggest an interplay between the age and the vernalization pathways.
In Arabidopsis, AP2 influences a variety of developmental processes, including flowering time through the age pathway and floral development (Yant et al., 2010). Strong recessive ap2 mutant alleles, such as ap2-12, flower early in both LDs and short days (SDs) (Yant et al., 2010). Similarly, lesions in the A. alpina ortholog of AP2, PEP2, has been reported to have a flowering time phenotype (Bergonzi et al., 2013). pep2 mutants flower without vernalization and show compromised perennial traits, similar to pep1-1 mutant plants (Bergonzi et al., 2013). The effect of PEP2 on flowering was first related to the vernalization pathway as it promotes the expression of PEP1 (Bergonzi et al., 2013). In 2-week-old pep2-1 seedlings, PEP1 transcript levels are reduced compared with wild-type plants (Bergonzi et al., 2013). However, PEP2 also has a PEP1-independent role in the regulation of flowering time in A. alpina as flowering is accelerated in the pep1-1 pep2-1 double mutant compared with the single mutants (Bergonzi et al., 2013).
Here, we show that during vernalization PEP2 represses the expression of the floral meristem identity genes AaFUL, AaLFY, and AaAP1. Vernalization accelerates flowering in young pep2-1 plants, indicating that PEP2 regulates the age-dependent response to vernalization. In addition, we report that the PEP1-dependent role of PEP2 takes place after vernalization because PEP2 is required to activate PEP1 after the return to warm temperatures. The involvement of PEP2 in two different aspects of the vernalization response contributes to the perennial life cycle of A. alpina.
Materials and methods
Plant material, growth conditions, and phenotyping
The A. alpina genotypes used in this study were Pajares (wild type), the pep2-1 mutant, and the pep1-1 mutant. The accession Pajares was collected in the Cordillera Cantábrica mountains in Spain at 1400 m altitude (42°59'32''N, 5°45'32''W). Both the pep2-1 and the pep1-1 mutant were isolated from an EMS (ethyl methanesulfonate) mutagenesis in the Pajares background (Wang et al., 2009; Bergonzi et al., 2013; Nordstrom et al., 2013). For the phenotypic analysis, plants were grown in LDs (16 h light and 8 h dark) under temperatures ranging from 20 °C during the day to 18 °C during the night. All vernalization treatments were performed at 4 °C in SD conditions (8 h light and 16 h dark).
Flowering time in the young wild-type, pep2-1, and pep1-1 plants was scored as the number of leaves at flowering and as the number of days to the first open flower after vernalization. Plants were grown for 3 weeks in LD cabinets, vernalized for 12 weeks, and moved back to LDs after cold.
The characterization of flowering time and inflorescence traits with different vernalization durations in the pep2-1 mutant was performed together with the wild type and the pep1-1 mutant in an experiment previously published (fig. 6 in Lazaro et al., 2018). The same data for control wild-type plants were used in Lazaro et al. (2018). Plants were grown for 5 weeks in a LD greenhouse, vernalized for 8, 12, 18, and 21 weeks, and moved back to LD greenhouse conditions on the same day. Flowering time was measured by recording the date on which the first flower opened after vernalization. The number of flowering and vegetative branches and the number of bracts in the inflorescence were measured at the end of flowering, except for plants vernalized for 8 weeks when the measurements took place 14 weeks after vernalization.
The Arabidopsis genotypes used herein were Columbia-0 (Col-0) wild type, ap2-7, and Col FRI San Feliu-2 (Sf-2) (Lee and Amasino, 1995). The ap2-7 mutant was crossed to the Col FRI Sf-2, and the FRI ap2-7 plants were isolated from a selfed F2 progeny that showed ap2 homeotic defects and late flowering.
For the flowering time experiments in Arabidopsis, the total leaf number (rosette and cauline leaves) was scored at the time when the first flower opened.
Construction of plasmids and plant transformation
To obtain the ap2-7 PEP2–VENUS transgenic plant, a 7.4 kb PEP2 genomic region spanning 4 kb upstream of the translational start and 1195 bp downstream of the translational stop was cloned by PCR (NCBI accession number LT669794.1). Subsequently, the VENUS:9Ala coding sequence was inserted either after the ATG or before the STOP codon of PEP2 by employing the polymerase incomplete primer extension (PIPE) method (Klock et al., 2008). Primers used for PIPE cloning are summarized in Supplementary Table S1 at JXB online. The generated recombinant DNA fragments were integrated in the pEarlyGate301 binary vector and transformed into Col through Agrobacterium-mediated floral dip (Clough and Bent, 1998). Selected homozygous lines, Col ProPEP2::VENUS::PEP2 N6-1-3 and Col ProPEP2::PEP2::VENUS C2-1-9, were crossed to ap2-7.
Gene expression analysis
Gene expression analysis was performed on the wild-type, pep1-1, and pep2-1. For pep2-1 samples, homozygous plants were selected after genotyping from a segregating population using a cleaved amplified polymorphic (CAP) marker (Forward primer, CAGCTGCACGGTATGTTTTTC; Reverse primer, GCTTTGTCATAAGCCCTGTG) and NdeI digestion.
For the analysis of the PEP1 expression pattern, the wild type and pep2-1 were grown for 6 weeks in LDs and vernalized for 12 weeks. Main shoot apices were harvested before vernalization, during vernalization, and after vernalization (1, 2, 3, and 4 weeks after the plants returned to warm temperatures). Axillary vegetative apices were harvested from plants growing in LDs 2, 3, and 4 weeks after vernalization. An average of 10 apices were pooled in each sample.
The expression of PEP1, AaSOC1, AaFUL, AaTFL1, AaLFY, and AaAP1 transcripts in the young and adult wild type, pep1-1, and pep2-1 was detected in seedlings grown for 3 weeks (young) or 6 weeks (adult) in LDs and vernalized for 12 weeks. Main shoot apices were harvested before vernalization and during cold, at 4, 8, and 12 weeks in vernalization. For the analysis of AaSPL5, AaSPL9, AaSPL15, and miR156, the main shoot apex was harvested from 3-, 4-, and 6-week old wild-type and pep2-1 plants growing in LDs. An average of 14 apices were pooled in each sample. Expression levels were normalized to both AaPP2A and AaRAN3, except for miR156 which was normalized to SnoR101.
The expression of FLC transcript was analyzed in the shoot apex of FRI and FRI ap2-7 plants grown for 10 d before vernalization, during 40 d of vernalization, and 10 d and 20 d after the return to LD glasshouse conditions. Expression levels were normalized to UBC21. Total plant RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), and a DNase treatment was performed with the Ambion DNA-free kit (Invitrogen) to reduce any DNA contamination. Total RNA (1.5 µg) was used to synthesize cDNA through reverse transcription with SuperScript II Reverse Transcriptase (Invitrogen) and oligo dT(18) as primer. A 2 µl aliquot of a cDNA dilution (1:5) was used as the template for each quantitative PCR (qPCR). For the analysis of miR156 and the SPLs, total RNA was extracted using the miRNeasy® Mini Kit (Qiagen), and a DNase treatment was performed with the Ambion DNA-Free kit (Invitrogen) to reduce DNA contamination. A 200 ng aliquot of RNA was used for reverse transcription of miR156 and SnoR101 using miR156- and SnoR101-specific primers. qPCRs were performed using a CFX96 and CFX384 Real-Time System (Bio-Rad) and the iQ SYBR Green Supermix detection system. Each data point was derived from two or three independent biological replicates and is shown as the mean ±standard deviation.
Primers used for qPCR for PEP1, AaSOC1, AaFUL, AaTFL1, AaLFY, AaAP1, AaSPL5, AaSPL9, AaSPL15, AaPP2A, AaRAN3, miR156, and SnoR101 were described previously (Wang et al., 2009, 2011; Bergonzi et al., 2013; Mateos et al., 2017; Lazaro et al., 2018). Primers used for qPCR for FLC and UBC21 were also described elsewhere (Czechowski et al., 2005; Crevillen et al., 2013).
Statistical analysis
Statistical analyses were performed using the R software. To detect significant differences in gene expression, we controlled for a false discovery rate (FDR) of 0.05 when conducting multiple pairwise comparisons by using Benjamini–Hochberg-corrected P-values. Treatments with significant differences are depicted with letters or asterisks. For the pep2-1 physiological analysis, we conducted multiple pairwise Bonferroni tests (α=0.05) to detect significant differences between the wild type and pep2-1. Here, a non-parametric test could not be conducted due to ties created during rank assignment.
RNAseq analysis
For differential gene expression analysis, we used the RNA sequencing (RNAseq) method on apices from the 3-week-old wild type, and the pep2-1 and pep1-1 mutants. pep2-1 homozygous plants were genotyped from a segregating population using the CAP marker described above. RNA was isolated as described above and total RNA integrity was confirmed on the Agilent BioAnalyzer. The library preparation and sequencing were performed at the Max Planck Genome Center Cologne, Germany (https://mpgc.mpipz.mpg.de/home/). RNAseq was performed with three biological replicates per sample. The libraries were prepared from 1 mg of total RNA using the TruSeq RNA kit (Illumina) and 100 bp single-end reads were sequenced on HiSeq2500 (Illumina). Reads from all samples were mapped on the A. alpina reference genome (Willing et al., 2015) using TopHat (Trapnell et al., 2009) with default parameters. Afterwards, CuffDiff (Trapnell et al., 2010) was used to estimate the mRNA level of each gene by calculating fragments per kilobase of exon model per million reads mapped (FPKM). To calculate the differential gene expression among the samples, FPKM values were used. A log2 fold change (L2FC) ≥1 for up-regulated genes and L2FC ≤ –1 for down-regulated genes, both with a q-value (adjusted P-value) ≤0.05, was used for further analysis.
Gene Ontology (GO) enrichment was performed with the BiNGO plug-in (Maere et al., 2005) implemented in Cytoscape V3.5.1 (Cline et al., 2007). A hypergeometric test was applied to determine the enriched genes, and the Benjamini–Hochberg FDR correction (Benjamini and Hochberg, 1995) was performed in order to limit the number of false positives. The FDR was set up to 0.05.
Sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE117977. Sequences of genes studied can be found in the GenBank/EMBL databases under the following accession numbers: PEP2 (AALP_AA7G245300), cDNA of PEP1 (FJ755930), coding sequence of AaLFY (JF436956), coding sequence of AaSOC1 (JF436957), AaAP1 (KFK41337.1), coding sequence of AaTFL1 (JF436953), and AaFUL (KFK27856.1).
Results
PEP2 influences the expression of genes involved in many plant physiological and developmental responses including flowering
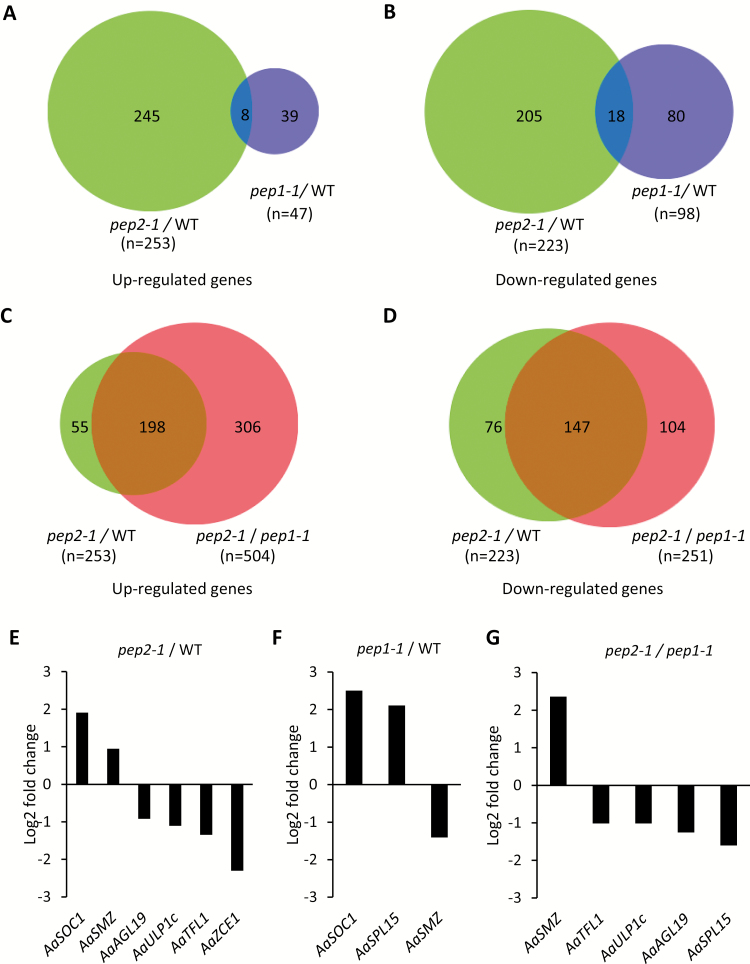
To provide an overview of the role of PEP2 in A. alpina, we performed an RNAseq analysis. We compared the transcriptomes of apices of 3-week-old pep2-1 and pep1-1 mutants with the wild type (Pajares). Three-week-old wild-type and mutant plants are vegetative and have not undergone the transition to flowering (Wang et al., 2011; Bergonzi et al., 2013; Park et al., 2017; Lazaro et al., 2018). Among transcriptomes, the majority of differentially expressed genes were detected in pep2-1. A total of 253 genes were up-regulated and 223 genes were down-regulated in pep2-1 compared with the wild type (Fig. 1A, B; Supplementary Dataset S1). In contrast, only 47 genes were up-regulated and 98 genes were down-regulated in pep1-1 compared with the wild type (Fig. 1A, B; Supplementary Dataset S2). The genes differentially expressed between pep1-1 and the wild type are influenced by PEP1, whereas those differentially expressed between pep2-1 and the wild type are affected by PEP2 through both the PEP1-dependent and PEP1-independent pathway. To identify genes influenced by PEP2 through the PEP1-independent pathway, we compared the transcriptomes of pep2-1 versus pep1-1 (Fig. 1C, D; Supplementary Dataset S3). A total of 504 genes were significantly up-regulated and 251 genes were significantly down-regulated in pep2-1 compared with pep1-1 (Fig. 1C, D). Interestingly, the number of differentially expressed genes detected between pep2-1 and pep1-1 was higher than those detected when single mutants were compared with the wild type. GO analysis demonstrated that the most enriched category for the up-regulated genes in pep2-1 compared with the wild type and in pep2-1 compared with pep1-1 was the biosynthesis of glucosinolates, which are involved in defense against herbivore attack and pathogens (Supplementary Fig. S1) (Keith and Mitchell-Olds, 2017). The overlap in over-represented GO categories in the set of genes up-regulated in pep2-1 in comparison with either the wild type or the pep1-1 mutant was very high, which is to be expected as more genes were up-regulated in pep2-1 compared with the wild type than in pep1-1 compared with the wild type (Fig. 1A; Supplementary Fig. S1A, B). Among the commonly enriched categories for down-regulated genes in pep2-1, we found apoptosis and protein desumoylation (Supplementary Fig. S1C, D).
Fig. 1.
Differentially expressed genes in pep2 and pep1 A. alpina mutants. (A and B) Venn diagram of significantly up-regulated (A) and down-regulated (B) genes in pep2-1 compared with the wild type (WT) and pep1-1 compared with the WT. (C and D) Venn diagram of significantly up-regulated (C) and down-regulated (D) genes in pep2-1 compared with the WT and in pep2-1 compared with pep1-1. (E–G) Flowering time genes differentially expressed in pep2-1 compared with the WT (E), pep1-1 compared with the WT (F), and pep2-1 compared with pep1-1 (G). Expression values are based on RNAseq.
Floral activators and repressors were identified among the differentially expressed genes in pep2-1. For example, the A. alpina ortholog of SOC1 (AaSOC1) was up-regulated in pep2-1 compared with the wild type (Fig. 1E; Supplementary Dataset S1). This effect of PEP2 on AaSOC1 is through PEP1 as AaSOC1 was differentially expressed between pep1-1 and the wild type, but not in pep2-1 versus pep1-1 (Fig. 1F, G; Supplementary Datasets S2, S3; Mateos et al, 2017). The regulation of AaSMZ through PEP2 is different from that through PEP1. AaSMZ was up-regulated in pep2-1 compared with the wild type and down-regulated in pep1-1 compared with the wild type (Fig. 1E, F; Supplementary Datasets S1, S2). In contrast, AaSPL15 was up-regulated in the pep1-1 mutant compared with the wild type and not in pep2-1 compared with the wild type, indicating that PEP2 does not control AaSPL15 expression (Fig. 1E, F; Supplementary Datasets S1, S2). Among the flowering time genes involved in the PEP1-independent role of PEP2 were the floral repressor AaTFL1 and AGAMOUS-LIKE 19 (AaAGL19). AaTFL1 was down-regulated when we compared pep2-1 with both the wild type and pep1-1, suggesting that the effect of PEP2 on AaTFL1 is independent of PEP1 (Fig. 1E–G; Supplementary Datasets S1–S3). Similarly, AGAMOUS-LIKE 19 (AaAGL19) transcripts were down-regulated specifically in the pep2-1 mutant (Fig. 1E–G; Supplementary Datasets S1–S3). We also found the ortholog of AaULP1c (ubiquitin-like protein protease) encoding for a SUMO protease and of CIS-CINNAMIC ACID-ENHANCED 1 (AaZCE1) being differentially expressed specifically in pep2-1 (Fig. 1C–E; Supplementary Datasets S1, S2). Interestingly, both ULP1c and ZCE1 in Arabidopsis control flowering through FLC. Mutations in ULP1c and its homolog, ULP1d, in Arabidopsis cause an early flowering phenotype that can at least partially be due to FLC down-regulation (Conti et al., 2008; Castro et al., 2016). ZCE1 is involved in the regulation of plant growth and development by cis-phenylpropanoids and it has been shown to control bolting time via enhancing FLC expression (Guo et al., 2011).
PEP2 can complement the Arabidopsis ap2 mutant
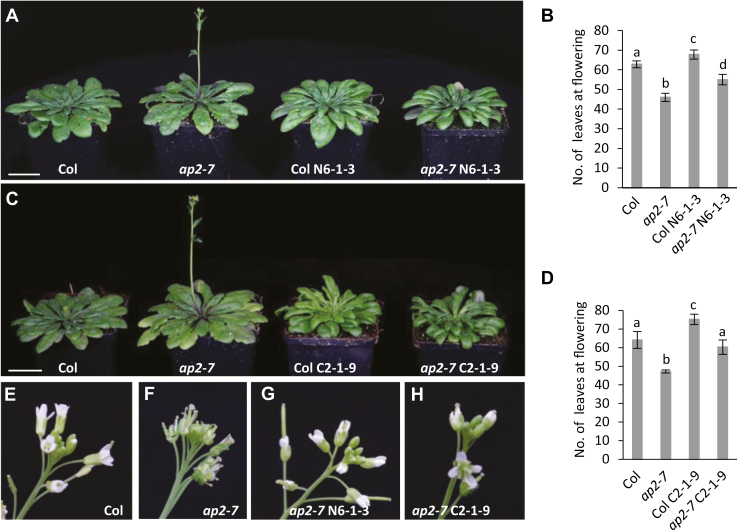
Both the pep2 mutant in A. alpina and the ap2 mutant in Arabidopsis show early flowering and similar floral defects, including the absence of petals and the transformation of sepals to carpels (Bowman et al., 1991; Bergonzi et al., 2013; Nördstrom et al., 2013). To check if both genes have common functions, we expressed PEP2 in the ap2-7 mutant background under the control of its own promoter. We fused a 7.4 kb PEP2 genomic region spanning 4 kb upstream of the translational start and 1.2 kb downstream of the translational stop to the VENUS fluorescent protein, at the N- or C-terminus. Transgenic lines were first obtained in the Col background. Homozygous lines obtained for the N-terminal VENUS (Col ProPEP2::VENUS::PEP2 N6-1-3) and the C-terminal VENUS (Col ProPEP2::PEP2::VENUS C2-1-9) were subsequently crossed to ap2-7. When grown in SDs, the PEP2 constructs complemented the early flowering phenotype of the ap2-7 mutant (Fig. 2A–D). Moreover, the homeotic defects of the ap2 mutant were restored by PEP2, indicating that the A. alpina PEP2 gene controls flowering time and floral organ identity in a similar way to AP2 (Fig. 2E–H).
Fig. 2.
PEP2 can complement the flowering and floral phenotype of the Arabidopsis ap2-7 mutant. (A and B) Phenotypes of Col wild type, the ap2-7 mutant, the Col ProPEP2::VENUS::PEP2 N6-1-3, and the ap2-7 ProPEP2::VENUS::PEP2 N6-1-3 lines grown in SDs (A) and number of leaves at flowering (B). (C and D) Col, the ap2-7 mutant, the Col ProPEP2::PEP2::VENUS C2-1-9, and the ap2-7 ProPEP2::PEP2::VENUS C2-1-9 lines grown in SDs (C) and number of leaves at flowering (D). (A and C) Whole plant pictures were taken 57 days after germination (DAG). Scale bar=3 cm. In (B) and (D), letters stand for significant differences between genotypes determined by multiple pairwise comparisons using Benjamini–Hochberg-corrected P-values (a-value of 0.05). Error bars indicate the standard deviation. (E and F) Inflorescence of Col wild type (E) ap2-7 (F), ap2-7 ProPEP2::VENUS::PEP2 N6-1-3 (G), and ap2-7 ProPEP2::PEP2::VENUS C2-1-9 (H) taken 73 DAG in SDs.
To test whether the effect of PEP2 on PEP1 expression was conserved in Arabidopsis for AP2 and FLC, we combined the ap2-7 mutation with the strong FRI allele from the San Feliu-2 (Sf-2) accession, which enhances Col FLC expression. Although the ap2 mutation accelerated flowering in the FRI Sf-2 background, the expression of FLC was not altered in the apices of these plants at different developmental stages (before, during, or after 40 d of vernalization) (Supplementary Fig. S2). These results indicate that, although the role of AP2 and PEP2 regarding flowering time regulation and floral organ identity is conserved, AP2 does not control FLC expression in a FRI Sf-2 background (Supplementary Fig. S2B).
PEP2 controls the age-dependent response of A. alpina to vernalization
We then investigated whether the PEP1-independent role of PEP2 was similar to that of AP2 in Arabidopsis and, therefore, whether it regulated flowering through the age pathway. We first analyzed the accumulation of miR156 and the transcript level of the A. alpina SPL5, 9, and 15 (AaSPL5, 9, and 15) in the apices of pep2-1 and wild-type seedlings grown for 3, 4, and 6 weeks in LDs (Supplementary Fig. S3). miR156 accumulation in the shoot apex decreased in older seedlings, but a similar pattern was observed in pep2-1 and the wild type (Supplementary Fig. S3A). Transcript levels of AaSPL5, 9, and 15 increased as the plants aged (Supplementary Fig. S3B–D). For AaSPL5 and 15, we observed no significant differences between pep2-1 and the wild type, whereas AaSPL9 mRNA levels differed between the two genotypes only in 6-week-old seedlings (Supplementary Fig. S3B–D). These results are consistent with previous studies in Arabidopsis demonstrating that AP2 controls flowering through the age pathway downstream of miR156 and the SPL genes. As it was previously shown that the age-dependent effect on flowering in A. alpina is only apparent after vernalization (Wang et al., 2011; Bergonzi et al., 2013), we tested whether PEP2 has an age-dependent role in vernalized plants. For this, we vernalized 3-week-old wild-type and pep2-1 seedlings for 12 weeks and measured flowering time after the return to warm temperatures. We also included the pep1-1 mutant in this experiment to rule out a PEP1-dependent effect of PEP2 on flowering time. In accordance with previous studies, under these conditions the wild type did not flower after vernalization and only grew vegetatively (Fig. 3; Wang et al., 2011; Bergonzi et al., 2013). Interestingly, pep2-1 flowered with an average of 18 leaves and 17 d after vernalization, whereas vernalized pep1-1 flowered with 27 leaves similar to non-vernalized pep1-1 plants grown continuously in LDs (Fig. 3; Supplementary Fig. S4; Wang et al., 2009; Bergonzi et al., 2013). These data suggest that vernalization accelerates flowering in young pep2-1 but not in pep1-1 plants. The flowering time phenotype of the mutants is also in contrast to that in LDs when pep1-1 flowers earlier than pep2-1 (Bergonzi et al., 2013). Overall, these results suggest that PEP2 regulates the age-dependent response to vernalization in a PEP1-independent manner.
Fig. 3.
PEP2 regulates the age-dependent response of A. alpina to vernalization. (A) Picture of 3-week-old wild-type (WT), pep1-1, and pep2-1 vernalized for 12 weeks followed by 2 weeks in LDs. Scale bar=5 cm. (B) Flowering time demonstrated as the number of leaves at flowering of 3-week-old WT, pep1-1, and pep2-1 vernalized for 12 weeks. The WT did not flower (NF). The asterisk stands for a significant difference in the total leaf number determined by a Student t-test (P-value <0.01). Error bars indicate the standard deviation.
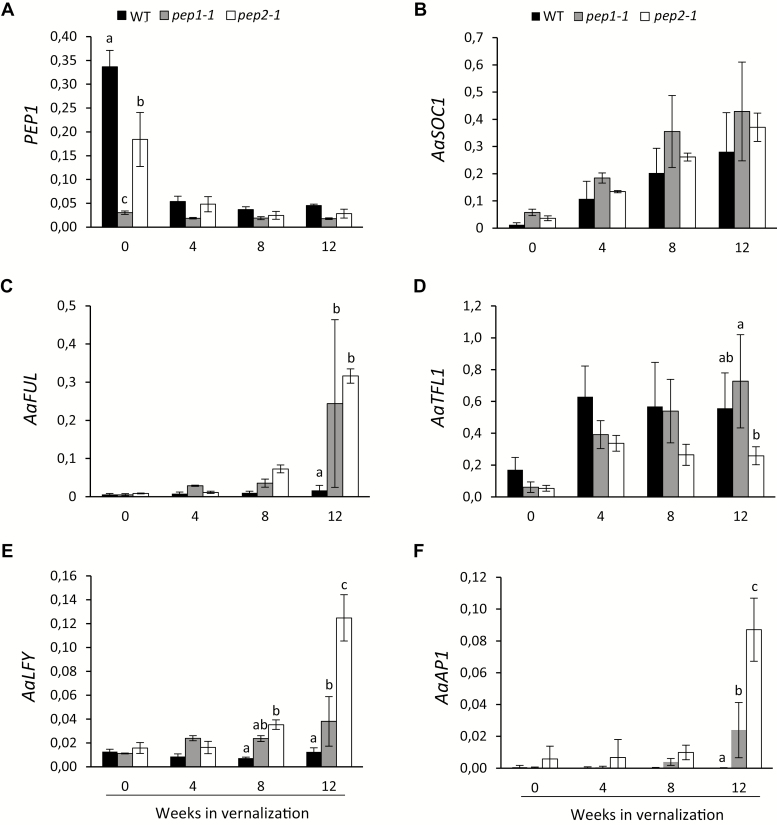
To understand how the young pep2-1 plants accelerate flowering in response to vernalization, we analyzed the expression of PEP1, AaSOC1, AaFUL, AaTFL1, AaLFY, and AaAP1. Three-week-old wild-type, pep2-1, and pep1-1 apices from the main shoot were harvested before and during vernalization at 4, 8, and 12 weeks. In agreement with previous results obtained in seedlings, non-vernalized 3-week-old pep2-1 plants showed lower PEP1 mRNA levels than the wild type (Fig. 4A; Bergonzi et al., 2013). Nevertheless, the PEP1 transcript decreased in a similar way in pep2-1 and wild-type plants, and PEP1 was silenced after 4 weeks in cold (Fig. 4A). These data suggest that, despite the initial difference in PEP1 expression, the lack of PEP2 does not influence PEP1 transcription in young apices during vernalization. The expression of AaSOC1 was gradually up-regulated during vernalization, following the same pattern in the three genotypes (Fig. 4B). In contrast, AaFUL, AaLFY, and AaAP1 showed a differential increase in the wild type and the mutants after 8 and 12 weeks in vernalization (Fig. 4C, E, F). In young wild-type plants AaFUL, AaLFY, and AaAP1 mRNA levels did not rise, indicating that flowering had not been initiated (Fig. 4C, E, F). Moreover, the pep2-1 mutant showed higher levels of AaLFY and AaAP1 than pep1-1 after 12 weeks in vernalization (Figs 3, 4E, F). Interestingly, the pep2-1 mutant also showed reduced expression of AaTFL1 at the end of the cold treatment compared with pep1-1 (Fig. 4D). Taken together, our results indicate that PEP2 activates AaTFL1 and represses AaFUL, AaLFY, and AaAP1 in young apices during vernalization (Fig. 3). This role of PEP2 is independent of PEP1, given that pep1-1 plants vernalized at a young age flowered later than pep2-1 and that PEP1 expression was reduced to the same extent in the wild type and pep2-1 during vernalization (Figs 3, 4A).
Fig. 4.
PEP2 regulates AaFUL, AaTFL1, AaLFY, and AaAP1 expression during vernalization. Relative expression of PEP1 (A), AaSOC1 (B), AaFUL (C), AaTFL1 (D), AaLFY (E), and AaAP1 (F). Three-week-old wild-type (WT), pep1-1, and pep2-1 shoot apices were harvested before and during 12 weeks of vernalization. Letters stand for significant differences between the WT, pep1-1, and pep2-1 at each time point determined by multiple pairwise comparisons using Benjamini–Hochberg-corrected P-values (α-value of 0.05). Graphs with no letters show no significant differences. Error bars indicate the standard deviation.
To investigate whether the regulation of these floral meristem identity genes by PEP2 was also conserved in adult plants, we tested the expression of AaFUL, AaLFY, and AaAP1 during vernalization. Six-week-old wild-type and pep2-1 plants were exposed to 12 weeks of cold and the mRNA levels of AaLFY, AaAP1, and AaFUL were analyzed in the shoot apex before vernalization and 1, 3, 5, 8, and 12 weeks into vernalization. AaFUL mRNA levels were higher in pep2-1 than in the wild type already after 8 weeks of vernalization (Supplementary Fig. S4A). For AaLFY and AaAP1 expression, a significant increase was observed in the pep2-1 mutant only at the end of the 12 weeks of cold (Supplementary Fig. S4B, C). Overall, these results suggest that PEP2 delays flowering by keeping AaFUL, AaLFY, and AaAP1 repressed at the end of the 12 weeks of vernalization, when PEP1 has already been silenced in the apices of both young and adult plants.
PEP2 is required to activate PEP1 expression after vernalization
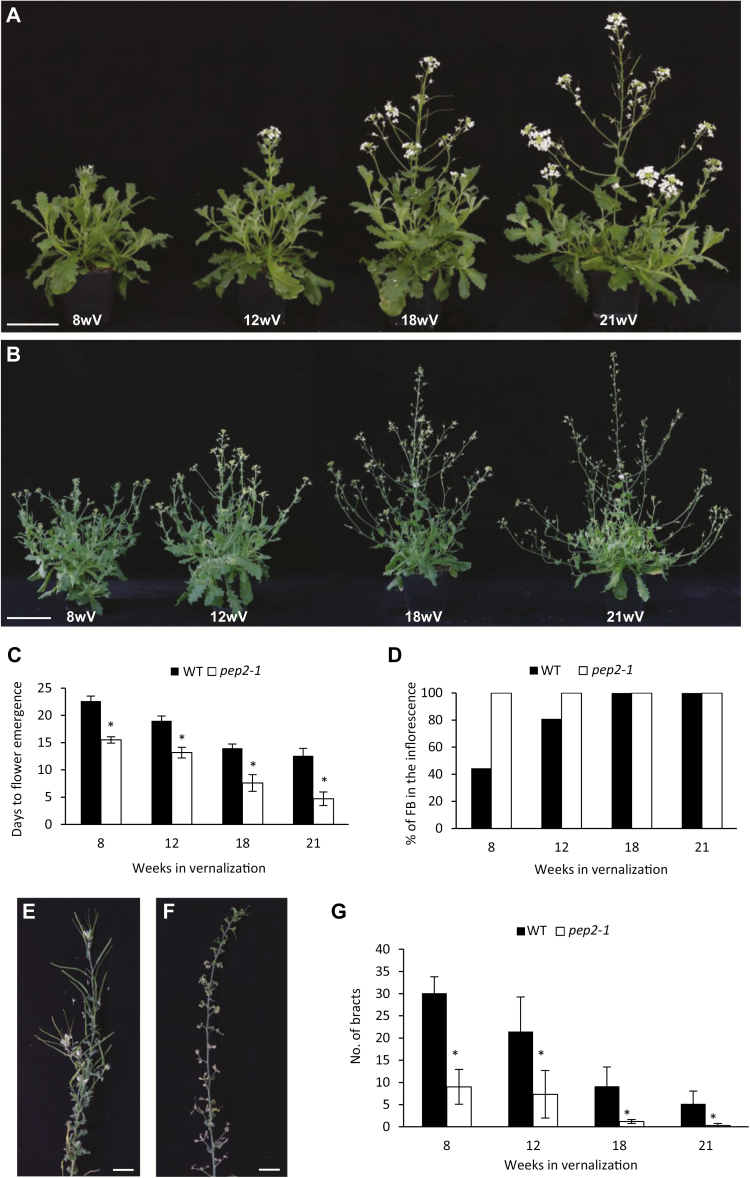
To test the PEP1-dependent role of PEP2, we exposed the pep2-1 mutant and the wild-type plants to different durations of vernalization. Both genotypes were grown for 5 weeks in LDs, vernalized for 8, 12, 18, and 21 weeks, and transferred back to LD greenhouse conditions (Fig. 5A, B). The pep2-1 mutant showed a reduction in the number of days to flower emergence compared with the wild type in all durations of cold (Fig. 5C). In addition, inflorescences in pep2-1 showed reduced floral reversion phenotypes and enhanced commitment of inflorescence branches to flowering (Fig. 5D–G). These results indicate that PEP2 controls flowering time and inflorescence architecture in A. alpina. However, the response of pep2-1 still varied with the duration of vernalization, suggesting that other floral repressors might contribute to flowering in response to vernalization. Also, PEP2 is required to maintain axillary shoots that are located just below the inflorescence in a vegetative state as all axillary branches in the pep2-1 mutant commit to reproductive development (Fig. 5; Bergonzi et al., 2013).
Fig. 5.
The pep2 mutant plants flower earlier than the wild type and show reduced reverted phenotypes. (A) Wild-type (WT) plants exposed to several durations of vernalization (8, 12, 18, and 21 weeks) followed by 3 weeks in LDs. (B) pep2-1 mutant plants exposed to several durations of vernalization (8, 12, 18, and 21 weeks) followed by 3 weeks in LDs. Scale bar=10 cm. (C) Time to flower emergence of WT and pep2-1 plants exposed to different durations of vernalization measured as the number of days to the first open flower. (D) Percentage of flowering inflorescence branches (FB) in the WT and the pep2-1 mutant exposed to 8, 12, 18, and 21 weeks of vernalization at the time the last flower in the inflorescence opened. (E) WT reverted inflorescence in plants vernalized for 8 weeks. (F) pep2-1 mutant inflorescence in plants vernalized for 8 weeks. Scale bar=2 cm. (G) Number of bracts within the inflorescence of the WT and the pep2-1 mutant exposed to 8, 12, 18, and 21 weeks of vernalization at the time the last flower in the inflorescence opened. This experiment was performed together with the pep1-1 mutant in an experiment previously published (Fig. 6 in Lazaro et al., 2018). Data for the WT control is similar between the two papers. Asterisks stand for significant differences between the wild type and the pep2-1 mutant at each time point determined by multiple pairwise Bonferroni tests (α-value of 0.05). Error bars indicate the standard deviation.
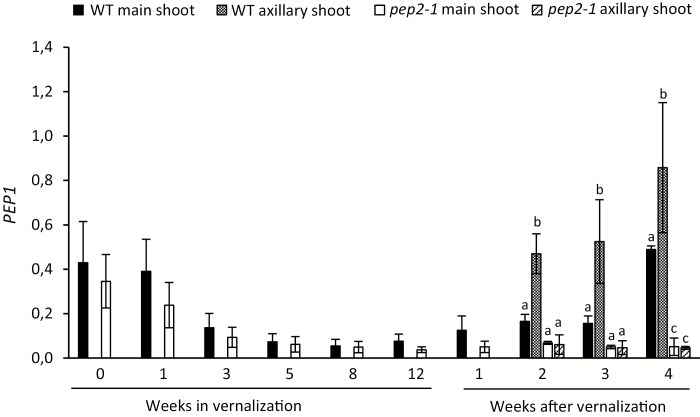
As shown previously in the wild type, PEP1 mRNA is up-regulated in the shoot apical meristem of the main shoot after a non-saturating vernalization (Fig. 6; Wang et al., 2009; Lazaro et al., 2018). This unstable silencing of PEP1 mRNA after cold was abolished in the pep2-1 mutant, suggesting that PEP2 is required to activate PEP1 expression in the shoot apical meristem after insufficient vernalization (Fig. 6). The role of PEP2 in the activation of PEP1 after vernalization is also observed in the axillary branches. All axillary branches in the pep2-1 mutant committed to flowering (Fig. 5B) and showed very low expression of PEP1 when compared with wild-type vegetative branches (Fig. 6). These results suggest that the major contribution of PEP2 is to activate PEP1 transcription after vernalization, both in the shoot apical meristem and in the vegetative axillary branches.
Fig. 6.
PEP2 is required to activate PEP1 expression after vernalization. Relative expression of PEP1 in the shoot apical meristem and in the vegetative axillary meristems of the wild type (WT) and the pep2-1 mutant before, during, and after 12 weeks of vernalization. Letters stand for significant differences between the WT and pep2-1 at each time point determined by multiple pairwise comparisons using Benjamini–Hochberg-corrected P-values (α-value of 0.05). Detailed information on significant differences can be found in Supplementary Table S3. Error bars indicate the standard deviation.
Discussion
Understanding the role of prolonged exposure to low temperatures in flowering is of particular importance in perennial species that will overwinter several times during their life cycle. In temperate perennials, prolonged exposure to cold controls later stages of flowering such as uniform bud break in the spring, whereas in alpine species it ensures floral formation before plants experience favorable environmental conditions for anthesis (Diggle, 1997; Meloche and Diggle, 2001; Lazaro et al., 2018). The maintenance of vegetative development after flowering, which is important for the perennial life strategy, is regulated by the seasonal cycling of floral repressors and the differential response of meristems to flower inductive stimuli due to age-related factors (Wang et al., 2009, 2011; Koskela et al., 2012). Here we characterized the role of the A. alpina floral repressor PEP2, the ortholog of the Arabidopsis AP2. Previous studies had demonstrated that PEP2 controls flowering through a PEP1-dependent and a PEP1-independent pathway (Bergonzi et al., 2013). Our transcriptomic analysis indicated that PEP2 influences the expression of genes involved in several developmental processes. However, many of the identified genes might not be regulated directly by PEP2 but by complex downstream genetic interactions (Fig. 1; Supplementary Fig. S1). We also found both floral promoters and repressors differentially expressed in the pep2 mutant. In Arabidopsis, the AP2 protein was immunoprecipitated from the promoter region of SOC1 (Yant et al., 2010). However, in our study, the effect of PEP2 on AaSOC1 seems to be through PEP1 (Fig. 1). To characterize the PEP1-dependent and the PEP1-independent role of PEP2 in flowering, we also employed physiological analysis and followed the expression of flowering time and meristem identity genes during the A. alpina life cycle. These data indicated that PEP2 controls (i) the age-dependent response to vernalization and (ii) the temporal cycling of the floral repressor PEP1 by ensuring the activation of PEP1 expression after vernalization.
PEP2 controls the age-dependent response to vernalization
PEP2 could rescue the early flowering phenotype of the Arabidopsis ap2-7 mutant, suggesting that its role in flowering time might be conserved (Fig. 2). In Arabidopsis, AP2 is post-transcriptionally regulated by miR172, and miR172b is placed in the age pathway as it is transcriptionally controlled by the miR156 targets SPL9 and SPL15 (Wu et al., 2009; Hyun et al., 2016). AP2 also negatively regulates its own expression by directly binding to its own genomic locus, as well as to the loci of its regulators miR156e, miR172b, and FUL, suggesting that AP2 is transcriptionally regulated by multiple feedback loops (Schwab et al., 2005; Yant et al., 2010; Balanza et al., 2018). The AP2 protein was also immunoprecipitated from the chromatin of floral integrators and genes required for floral meristem development such as SOC1, AGAMOUS (AG), and AP1 (Yant et al., 2010). The transcription of genes such as SOC1 and FUL is also controlled by upstream regulators in the age pathway. SPL9 has been reported to bind to the first intron of SOC1 and SPL15 to FUL and miR172b (Wang et al., 2009; Hyun et al., 2016). Overall, this complex genetic circuit that includes AP2 might contribute to the fast life cycle of Arabidopsis, in which floral transition takes place soon after reproductive competence is acquired. In contrast to Arabidopsis, in A. alpina reproductive competence is uncoupled from flowering initiation. Arabis alpina plants become competent to flower after growing for 5 weeks in LD conditions but only initiate flowering when they are exposed to vernalization (Wang et al., 2009). This suggests that flowering in A. alpina is regulated by a strong interplay between the age and the vernalization pathways. Members of the SPL and AP2 families (e.g. AaSPL15 and AaTOE2) are transcriptionally repressed by PEP1 in addition to the post-transcriptional and post-translational regulation by the miRNAs (Chen, 2004; Bergonzi et al., 2013; Hyun et al., 2016; Xu et al., 2016; Mateos et al., 2017). Although, FLC in Arabidopsis targets a similar set of genes, the strong interplay between the age and the vernalization pathway is most apparent in A. alpina (Deng et al., 2011; Mateos et al., 2017). Vernalization in A. alpina provides the condition where the age effect on flowering is apparent as it silences PEP1. Gradual changes in the accumulation of miR156 and the expression of the SPL genes can be observed in the shoot apex of A. alpina plants that get older in LDs (Bergonzi et al., 2013). However, the accumulation of downstream regulators in the age pathway, such as of miR172, only increases in the shoot apex during vernalization and upon floral transition (Bergonzi et al., 2013). Here we show that the expression of miR156 and of AaSPL5 and 15 is not influenced in pep2 plants grown in LDs (Fig. 1E–G; Supplementary Fig. S3). Given that PEP2 acts partially through PEP1, the lack of an effect in pep2-1 on AaSPL15 can be due either to the residual PEP1 expression in the pep2-1 mutant or to the existence of compensatory genetic mechanisms. Interestingly, AaSPL9 mRNA levels were reduced in 6-week-old pep2-1 seedlings compared with the wild type (Supplementary Fig. S3). This effect of PEP2 on AaSPL9, though, cannot be explained by the feedback loops described in Arabidopsis as AaSPL9 transcript levels would be expected to be higher in pep2-1 than in the wild type (Supplementary Fig. S3; Yant et al., 2010).
The A. alpina ortholog of TFL1 (AaTFL1) has been previously reported to influence the effect of vernalization in an age-dependent manner, although its expression pattern does not differ between juvenile and adult apices before vernalization (Wang et al., 2011). Here we show that vernalization accelerated flowering in young pep2-1 seedlings compared with pep1-1, suggesting that PEP2 also regulates the age-dependent response to vernalization in a PEP1-independent pathway (Fig. 3). Interestingly, in our RNAseq analysis, AaTFL1 transcripts were reduced in the pep2-1 mutant, suggesting that PEP2, together with or through AaTFL1, sets an age threshold for flowering in response to vernalization. One major difference between AaTFL1 and PEP2, though, is that lines with reduced AaTFL1 activity do not flower without vernalization. These results suggest that PEP2 plays additional roles in the regulation of flowering time in A. alpina. Transcriptomic experiments in Arabidopsis also showed that TFL1 mRNA is down-regulated in ap2 inflorescences compared with the wild type (Yant et al., 2010). However, no direct binding of AP2 to the TFL1 locus has been detected by ChIP-Seq and therefore it is unclear whether there is a direct or indirect effect of AP2 on TFL1 transcription (Yant et al., 2010).
PEP2 ensures the activation of PEP1 after vernalization
Previous studies in A. alpina have demonstrated that PEP2 controls flowering in response to vernalization via enhancing the expression of PEP1 (Bergonzi et al., 2013). Here we show that the major role of PEP2 in PEP1 activation takes place after vernalization. PEP1 expression in A. alpina is temporarily silenced during prolonged exposure to cold to define inflorescence fate, while it is highly expressed in axillary branches after vernalization to repress flowering (Wang et al., 2009; Lazaro et al., 2018). We have recently shown that the duration of vernalization influences PEP1 reactivation in the shoot apex after the return to warm temperatures (Lazaro et al., 2018). Phenotypes correlated with high PEP1 mRNA levels after vernalization (e.g. floral reversion and the presence of vegetative axillary branches) were almost absent in the pep2-1 mutant (Fig. 5; Lazaro et al., 2018). Accordingly, PEP1 mRNA levels were reduced in vernalized pep2-1 plants compared with the wild type both in the inflorescence stem and in the axillary branches (Fig. 6; Wang et al., 2009; Lazaro et al., 2018). These results suggest that PEP2 contributes to the perennial life cycle and controls perennial-specific traits via activating PEP1 after vernalization. In Arabidopsis, the introgression of the FRI allele from the Sf-2 accession into Col extends the duration of cold temperatures required to silence FLC (Searle et al., 2006). Northern Arabidopsis accessions such as Lov-1 require several months of vernalization to achieve FLC silencing and, similarly to A. alpina Pajares, a shorter duration of cold temperatures causes FLC reactivation (Shindo et al., 2006). The link between AP2 and FLC is not clear in Arabidopsis. AP2 does not bind to the FLC locus in ChIP-Seq experiments, and in our study FLC expression was not altered in plants where the ap2-7 mutant allele was introgressed into the Col FRI Sf-2 background (Supplementary Fig. S3; Yant et al., 2010). However, as the strongest difference in PEP1 expression in the pep2-1 mutant was observed after vernalization, the effect of AP2 in the Lov-1 accession should be analyzed to rule out a role for AP2 on FLC reactivation after insufficient vernalization.
The unstable silencing of FLC involves changes in the accumulation of the H3 trimethylation at Lys27 (H3K27me3) (Angel et al., 2011; Coustham et al., 2012). The pattern of the H3K27me3 mark at the PEP1 locus also correlates with changes in PEP1 mRNA levels in A. alpina (Wang et al., 2009). PEP1 shows a much higher and broader increase of H3K27me3 during the cold than FLC, and H3K27me3 levels rapidly decrease at PEP1 after short vernalization periods (Wang et al., 2009; Angel et al., 2011; Lazaro et al., 2018). Although the proteins regulating histone modifications at the PEP1 locus are not known, resetting of the epigenetic memory of FLC in Arabidopsis is dependent on the presence of TrxG components and the Jumonji C (JmjC) domain-containing demethylases EARLY FLOWERING 6 (ELF6) and RELATIVE OF EARLY FLOWERING 6 (REF6) (Noh et al., 2004; Yun et al., 2011; Crevillen et al., 2014). It has been shown that AP2 has the ability to interact with a chromatin remodeling factor HISTONE DEACETYLASE 19 (HDA19) to transcriptionally repress one of its targets (Krogan et al., 2012), but AP2 has never been associated with histone demethylases.
We have recently demonstrated that PEP1 is stably silenced in the shoot apical meristem of adult plants that commit to flowering during prolonged exposure to cold (Lazaro et al., 2018). In juvenile plants, a similar duration of vernalization fails to initiate flowering even if PEP1 is silenced during cold (Lazaro et al., 2018). Floral commitment during vernalization is correlated with a higher expression of the floral meristem identity genes, AaFUL, AaLFY, and AaAP1, which are repressed by PEP2 (Lazaro et al., 2018). This is evident by the precocious up-regulation of AaFUL, AaLFY, and AaAP1 mRNA levels in vernalized pep2-1 plants compared with the wild type (Fig. 4; Supplementary Fig. S4). Although, the link between PEP2 and PEP1 resetting is not clear, it seems that the achievement of floral commitment during vernalization is negatively correlated with PEP1 up-regulation after the return to warm temperatures (Lazaro et al., 2018). In Arabidopsis, AP2 is not known to influence FLC transcription. However, AP2 has been reported to be transcriptionally repressed by FUL, and FUL-overexpressing plants show reduced FLC expression (Balanzà et al., 2014, 2018). These results suggest that FUL might regulate FLC transcription independently or through AP2. This might also indicate that in A. alpina the role of PEP2 in PEP1 expression might implicate other flowering time regulators, genes involved in the age pathway and genes ensuring floral commitment during vernalization. However, since PEP1 also transcriptionally regulates genes in these genetic pathways, feedback mechanisms might also occur (Mateos et al., 2017).
Conclusion
Our study demonstrates that PEP2 plays an instrumental role in A. alpina controlling the age-dependent response to vernalization and facilitating the activation of PEP1 after vernalization. As both roles of PEP2 focus on whether floral commitment has been achieved during vernalization, suggests that they might not be completely independent. Upstream regulators of floral meristem identity genes such as PEP2 might control the response to vernalization of individual meristems and contribute to the complex plant architecture of perennials.
Supplementary data
Supplementary data are available at JXB online.
Dataset S1. Transcripts identified as being differentially expressed in pep2-1 compared with the wild type.
Dataset S2. Transcripts identified as being differentially expressed in pep1-1 compared with the wild type.
Dataset S3. Transcripts identified as being differentially expressed in pep2-1 compared with pep1-1.
Fig. S1. GO-enriched categories in the RNAseq experiment.
Fig. S2. AP2 activity does not affect FLC expression in Arabidopsis.
Fig. S3. The expression levels of miR156, AaSPL5, and AaSPL15 do not differ between wild-type and pep2-1 plants growing in long days.
Fig. S4. PEP2 regulates the age-dependent response of A. alpina to vernalization.
Fig. S5. PEP2 controls AaFUL, AaTFL1, AaLFY, and AaAP1 expression during vernalization in adult plants.
Table S1. Primers used for PIPE-cloning of the PEP2 locus.
Table S2. Statistical differences in Supplementary Fig. S2 determined by multiple pairwise comparisons using Benjamini–Hochberg-corrected P-values comparing FLC mRNA levels between FRI and FRI ap2-7 at different developmental stages.
Table S3. Statistical differences in Fig. 6 determined by multiple pairwise comparisons using Benjamini–Hochberg-corrected P-values comparing PEP1 mRNA levels between pep2-1 and the wild type at different developmental stages.
Acknowledgements
We would like to thank SPP1530 and CEPLAS for funding to MCA, SPP1529 for funding to AP and KN, and a Purkyně fellowship from the ASCR to AP. We would also like to thank Margaret Kox for critical reading of the manuscript.
Glossary
Abbreviations
- DAG
days after germination
- LD
long day
- SD
short day.
References
- Amasino R. 2009. Floral induction and monocarpic versus polycarpic life histories. Genome Biology 10, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. 2011. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzà V, Martínez-Fernández I, Ferrándiz C. 2014. Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. Journal of Experimental Botany 65, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzà V, Martínez-Fernández I, Sato S, Yanofsky MF, Kaufmann K, Angenent GC, Bemer M, Ferrándiz C. 2018. Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL–APETALA2 pathway. Nature Communications 9, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B: Methodological 57, 289–300. [Google Scholar]
- Bergonzi S, Albani MC. 2011. Reproductive competence from an annual and a perennial perspective. Journal of Experimental Botany 62, 4415–4422. [DOI] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Ver Loren van Themaat E, Nordström KJ, Wang R, Schneeberger K, Moerland PD, Coupland G. 2013. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340, 1094–1097. [DOI] [PubMed] [Google Scholar]
- Billings WD, Mooney HA. 1968. Ecology of Arctic and Alpine plants. Biological Reviews of the Cambridge Philosophical Society 43, 481–529. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1991. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Castro PH, Couto D, Freitas S, et al. . 2016. SUMO proteases ULP1c and ULP1d are required for development and osmotic stress responses in Arabidopsis thaliana. Plant Molecular Biology 92, 143–159. [DOI] [PubMed] [Google Scholar]
- Chen X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, et al. . 2007. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A. 2008. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. The Plant Cell 20, 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587. [DOI] [PubMed] [Google Scholar]
- Crevillén P, Sonmez C, Wu Z, Dean C. 2013. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO Journal 32, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C. 2014. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WW, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. 2011. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P. 1997. Extreme preformation in alpine Polygonum viviparum: an architectural and developmental analysis. American Journal of Botany 84, 154. [PubMed] [Google Scholar]
- Guo D, Wong WS, Xu WZ, Sun FF, Qing DJ, Li N. 2011. Cis-cinnamic acid-enhanced 1 gene plays a role in regulation of Arabidopsis bolting. Plant Molecular Biology 75, 481–495. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. 2016. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Developmental Cell 37, 254–266. [DOI] [PubMed] [Google Scholar]
- Keith RA, Mitchell-Olds T. 2017. Testing the optimal defense hypothesis in nature: variation for glucosinolate profiles within plants. PLoS One 12, e0180971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock HE, Koesema EJ, Knuth MW, Lesley SA. 2008. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71, 982–994. [DOI] [PubMed] [Google Scholar]
- Koskela EA, Mouhu K, Albani MC, Kurokura T, Rantanen M, Sargent DJ, Battey NH, Coupland G, Elomaa P, Hytönen T. 2012. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca.Plant Physiology 159, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoda N, Iwanami H, Takahashi S, Abe K. 2006. Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. Journal of the American Society for Horticultural Science 131, 74–81. [Google Scholar]
- Krogan NT, Hogan K, Long JA. 2012. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Obeng-Hinneh E, Albani MC. 2018. Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiology 176, 2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM. 1995. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiology 108, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, Krajewski P, Coupland G. 2017. Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proceedings of the National Academy of Sciences, USA 114, E11037–E11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. 2009. Repression of flowering by the miR172 target SMZ. PLoS Biology 7, e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche CG, Diggle PK. 2001. Preformation, architectural complexity, and developmental flexibility in Acomastylis rossii (Rosaceae). American Journal of Botany 88, 980–991. [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed R, Wang CT, Ma C, et al. . 2010. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. The Plant Journal 62, 674–688. [DOI] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS. 2004. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. The Plant Cell 16, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJ, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G, Schneeberger K. 2013. Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nature Biotechnology 31, 325–330. [DOI] [PubMed] [Google Scholar]
- Park JY, Kim H, Lee I. 2017. Comparative analysis of molecular and physiological traits between perennial Arabis alpina Pajares and annual Arabidopsis thaliana Sy-0. Scientific Reports 7, 13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. 2003. Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8, 517–527. [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Development 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. 2000. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proceedings of the National Academy of Sciences, USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes & Development 20, 3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G. 2011. Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. The Plant Cell 23, 1307–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC. 2009. PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427. [DOI] [PubMed] [Google Scholar]
- Willing EM, Rawat V, Maumus F, et al. . 2015. Lack of symmetric CG methylation and long-lasting retrotransposon activity have shaped the genome of Arabis alpina. Nature Plants 1, 14023. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS. 2016. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genetics 12, e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. The Plant Cell 22, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H, Hyun Y, Kang MJ, Noh YS, Noh B, Choi Y. 2011. Identification of regulators required for the reactivation of FLOWERING LOCUS C during Arabidopsis reproduction. Planta 234, 1237–1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.