Summary
Global investigation of histone marks in acute myeloid leukemia (AML) remains limited. Analyses of 38 AML samples through integrated transcriptional and chromatin mark analysis exposes 2 major subtypes. One subtype is dominated by patients with NPM1 mutations or MLL-fusion genes, shows activation of the regulatory pathways involving HOX-family genes as targets, and displays high self-renewal capacity and stemness. The second subtype is enriched for RUNX1 or spliceosome mutations, suggesting potential interplay between the 2 aberrations, and mainly depends on IRF family regulators. Cellular consequences in prognosis predict a relatively worse outcome for the first subtype. Our integrated profiling establishes a rich resource to probe AML subtypes on the basis of expression and chromatin data.
Introduction
As a typical hematopoietic neoplasm, acute myeloid leukemia (AML) is frequently a fatal disease (Döhner et al., 2015). It is genetically and clinically heterogeneous (Grimwade et al., 2016), mainly due to the combinations of distinct driver mutations. Epigenetic modifiers are frequently mutated in AML (Wouters and Delwel, 2016) and affect gene transcription by the addition or removal of histone modification, chromatin accessibility, and DNA methylation. Due to highly flexible adaptation to environmental exposures, these epigenetic changes have the potential to improve the prediction of drug responses and targeted treatment using specific inhibitors (Jones et al., 2016). Numerous studies have focused on mapping epigenetic perturbations in AML, mainly DNA methylation, and found some pivotal regulators shaping the AML epigenome and leukemia development (Ley et al., 2013; Cauchy et al., 2015; Figueroa et al., 2010; Li et al., 2016a; McKeown et al., 2017). These studies largely focused on single epigenomic features, which could not reveal systematic chromatin modifications and crosstalk among different epigenetic marks in AMLs. Further characterization by integrating multi-layer datasets, especially histone chromatin immunoprecipitation sequencing (ChIP-seq), would shed more light on epigenetic dynamics in response to AML progression.
Results
AML Classification and Subtype-Specific Features
To comprehensively interrogate the epigenetic signatures and cellular consequences driving the classification of AML subtypes, we combined high-quality ChIP-seq, RNA-seq, DNaseI-seq, and whole-genome bisulfite sequencing (WGBS) profiling on a selection of 38 AMLs representing the abundant genetic heterogeneity (Figures S1A–S1H; Tables S1, S2, S3, and S4). Based on the combination of 6 histone marks, 12 chromatin states (Figure 1A), including 7 active states (states 1–7) and 5 repressed states (states 8–12), were defined. Genomic distribution, DNA accessibility, and methylation levels for each chromatin state in our study is similar to previous findings in normal cell types (Figures S2A and S2B) (Kasowski et al., 2013; Kundaje et al., 2015). In line with a previous study (Glass et al., 2017), AML-associated enhancer regions (EnhS and EnhW) displayed greater differential methylation levels (Figure S2C), while methylation profiles were more similar at promoters (TssF and TssA).
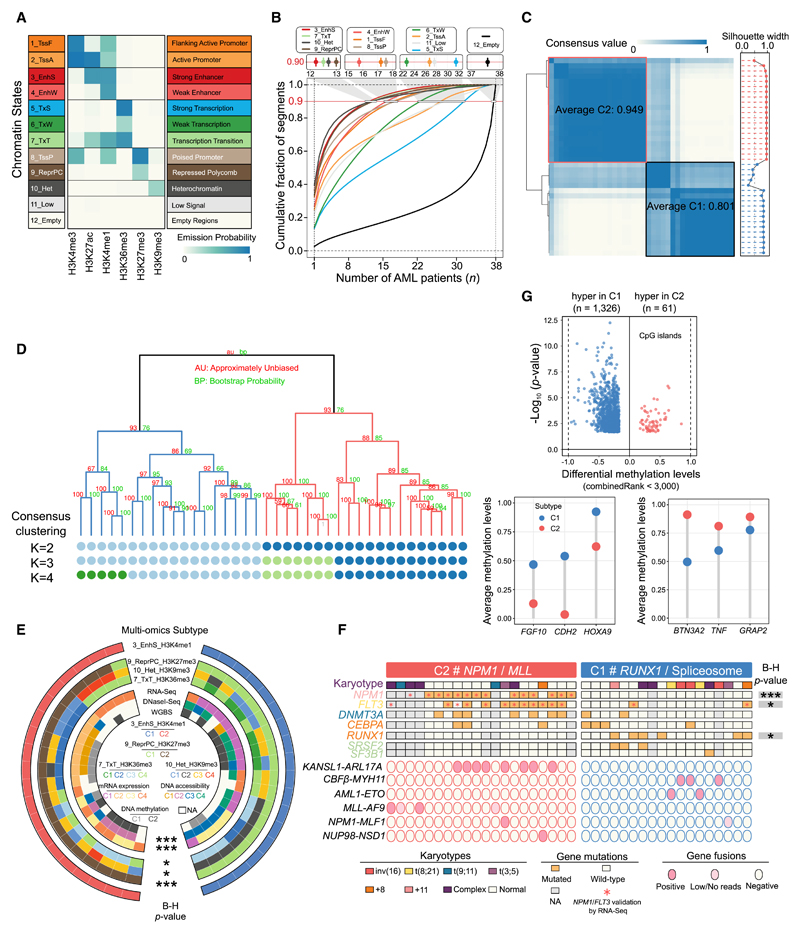
Figure 1. Chromatin State Definition and Subtype Assignment across AMLs.
(A) Combinatorial patterns of 6 histone marks in a 12-state model. The emission probability was learned from ChromHMM based on spatial patterns of histone modifications in chromatin and used to define the chromatin state. A darker shade of blue indicates greater enrichment of the profiled histone marks in a particular state.
(B) Dynamic patterns of chromatin states. The lines indicate genome coverage fraction of each state consistently labeled with that state in at most n (n = 1–38) samples. Approximately 90% of genomic bins with the EnhS state could be found in a small subset of at most 12 samples, and only 1% were commonly shared among 24 or even more samples. In contrast, the quiescent regions labeled as Empty state were the most constitutive, with 65% consistently marked among ≥30 samples, and the fraction of this state uniquely detected in 1 sample was <3%.
(C) Consensus matrices (left) and silhouette scores (right) of 38 AML samples using H3K4me1 signals in strong enhancer state. Consensus values range between 0 (highly dissimilar profiles) and 1 (highly similar profiles), colored white to dark blue. The average consensus score for each subtype is shown in the box.
(D) Hierarchical epigenome clustering of the same data using the pvclust package. Values on the branch indicate bootstrap support scores >1,000 samplings using 2 different approaches. The dots below the dendrogram reveal consensus clusters for k = 2–4. The 3-cluster classification did not split the intermediate box (in Figure 1C) but another subgroup from C2, which could be matched with cluster results from the pvclust method.
(E) Comparison of AML classification on the basis of different datasets. Statistical significance of overlap among multiple clusters is assessed by Fisher’s exact test, followed by the Benjamini-Hochberg (B-H) correction. ***p < 0.001 and *p < 0.05.
(F) Mutation profiles in the 2 AML subtypes. All of the patients with NPM1 insertion and FLT3-ITD were validated by RNA-seq data, as well as all of the translocation fusion products. The strength of association between each mutation and subtypes is assessed by Fisher’s exact test followed by the B-H correction. *p < 0.05, **p < 0.01, and ***p < 0.001.
(G) Differential methylation profiles at global CpG islands between 2 subtypes. A CpG site with combined rank score <3,000 was considered significant.
See also Figures S1, S2, and S3 and Tables S1, S2, S3, S4, and S5.
Similarly, the strong enhancer state (EnhS) dominated by H3K4me1 and H3K27ac exhibited the most sample-specific pattern based on cumulative fraction curves (Figure 1B). This triggered the exploration of AML classification based on the H3K4me1 signal in EnhS regions. Consensus clustering revealed a separation in 2 major molecular clusters (C1 and C2) displaying high consensus values (0.801 and 0.949) and silhouette width profiles (0.724 and 0.932) (Figure 1C), while the separation in 3, 4, or more clusters did not provide significantly better classification. We further estimated approximately unbiased p values for all clusters and found high values (>0.90) for both subtypes (Figure 1D). The comparison between consensus matrices (k = 2–4) and the pvclust dendrogram indicated that the 2 results were identical, supporting the robustness of the partitioning into 2 groups. Finally, clustering of H3K27ac, which is also enriched in the EnhS state, revealed that the samples assigned to the same subtypes by H3K4me1 were always in close proximity (Figure S2D), again validating the H3K4me1-based clustering results.
Principal-component analysis with EnhS H3K4me1 density showed clear separation into 2 groups lacking subtype-specific distribution in the patients’ age, gender, and disease status (Figure S2E). Using H3K27ac for differential analyses at the defined strong enhancer state, a total of 3,629 and 4,400 regions were identified as C1- or C2-specific active enhancers, respectively. Examining the local epigenetic landscape at these enhancers confirmed increased H3K4me1 and H3K27ac and revealed reduced repressive marks, as well as a positive correlation with gene expression (Figures S2F–S2H). Moreover, C2-specific signature genes were upregulated in NPM1 mutated and mixed lineage leukemia (MLL) fusion AMLs, while C1-specific signature genes overlapped with those expressed in t(8;21) AMLs (Figure S2H).
We performed the same clustering analyses using other epigenetic data and evaluated their consistency between subtype identifications. Two major groups with high silhouette values were detected by the H3K27me3-established ReprPC state (Figure S2I), representing almost the same cluster as the H3K4me1-derived state (adjusted p < 0.001; Figures 1E and S2J). Hierarchical clustering based on gene expression and DNA accessibility characterized 4 clusters and showed significant similarity with the 2 EnhS-based subtypes. Our results reveal the identification of 2 clear epigenetic subgroups of AML, despite the genetic heterogeneity of the AML samples.
We determined which mutated genes are subtype specific via Fisher’s exact test. Within our AML cohort, NPM1 mutations were found only in subtype C2 (adjusted p < 0.001) (Figures 1F and S3A; Table S5), while 2 other commonly mutated genes, FLT3-ITD and DNMT3A, also revealed C2-specific patterns. Given this enrichment of the DNA methyltransferase DNMT3A in the C2 subtype, we explored differential methylation levels between the 2 subtypes. This revealed more hypomethylated CpG islands in the C2 group (Figure 1G) and exposed several differentially methylated genes such as HOXA9. The lower methylation levels in the C2 subtype may be related to deficient DNMT3A function or suggest a more unrestricted chromatin structure conferring stronger stemness property for the C2 group. In contrast to C2, patients with mutated RUNX1 or 2 alternative splicing genes (SRSF2 and SF3B1) were specifically allocated in the C1 subtype, while no significant differences between the 2 subtypes in mutational occurrences of the myeloid differentiation factor CEBPA were found. This suggests that epigenetic patterns in AML blasts with CEBPA mutations are dominated by other co-occurring leading aberrations such as NPM1, FLT3, and structural variants, rather than by CEBPA mutation.
We also found that some types of cytogenetic abnormalities seemed subtype specific. For instance, patients with 2 chromosomal variations functionally involving the RUNX1 gene, t(8;21) and inv(16), clustered together in the C1 subtype, while the t(9;11)-associated MLL-AF9 fusion event was found in the C2 subtype (Figures 1F, S3B, and S3C). Given these findings, we named C1 the RUNX1/spliceosome group and C2 the NPM1/MLL group.
Comparing our data with those from The Cancer Genome Atlas (TCGA) (Ley et al., 2013) revealed that our chromatin-based clustering is highly reminiscent of the CpG sparse based subtypes defined by TCGA (Figure S3D). We also examined the mutational spectrum in the AML subtypes inferred from the clustering of other marks (Figures S3E–S3I, top). This revealed that while our enhancer-based clustering congregated samples with the same mutations (Figure 1F), this was not shown when clustering was based on H3K36me3 or H3K9me3. These results revealed that our epigenetic signature partly recapitulates the intrinsic subtypes from other large-scale population studies, but it also exposes a previously unidentified view on cellular conditions and phenotypic plasticity that defines 2 epigenetic subgroups of AML.
The Super-Enhancer Landscape of AML
To explore super-enhancer domains across AML patients, we used our H3K27ac data and assigned each putative superenhancer (SE) to its nearest gene up to 1 Mb away. The SEs exhibited larger size, higher H3K27ac signal, and stronger upregulation in transcriptional levels than defined EnhS and EnhW (Figures 2A, 2B, and S4A) (Cauchy et al., 2015; Li et al., 2016b). Of 4,100 defined SEs, 186 have significantly different H3K27ac enrichment between the 2 AML subtypes identified above and showed a strong positive correlation with the expression levels of the nearest deregulated genes (r = 0.777; Figure 2C), like HOXA/HOXB gene clusters and their cofactors MEIS1 and PBX3. This correlation was not dependent on subtype, as clustering based on other marks revealed a consistent correlation between the presence of an SE and gene activity (Figures S3E–S3I, bottom).
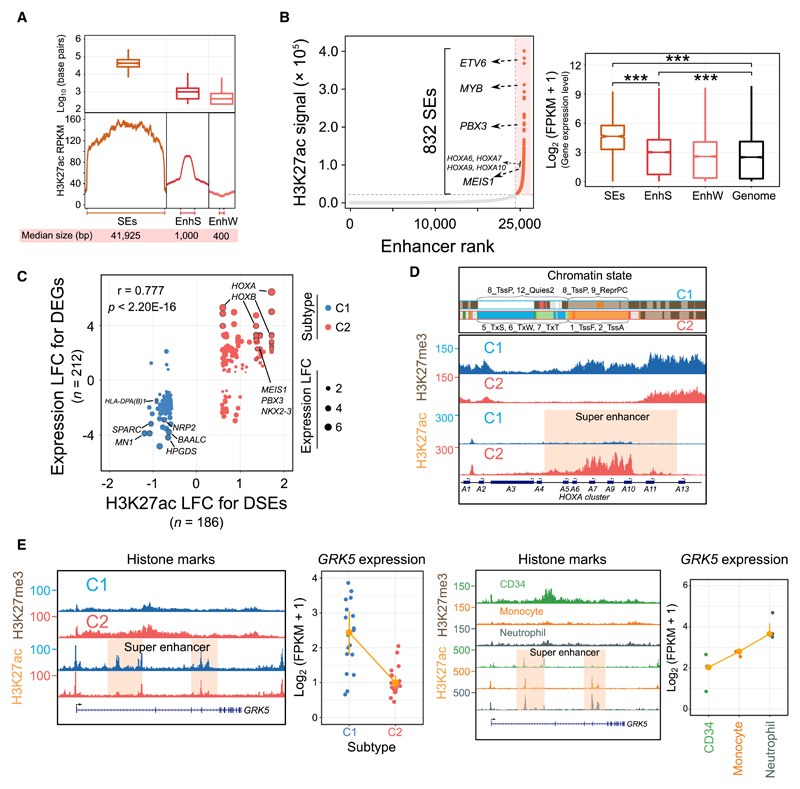
Figure 2. Genome-Wide Landscape and Subtype-Specific Features of Super-Enhancers.
(A) The genomic length and H3K27ac intensity of SEs in AML. EnhS, strong enhancer state; EnhW, weak enhancer state; SE, super-enhancer.
(B) Ranked enhancer plots and their regulatory effects in a representative sample. SEs are orange, and several known AML-associated genes proximal to SEs are highlighted. The figure at right shows the expression differences of genes associated with different types of enhancers. ***p < 0.001.
(C) Correlation between differentially expressed genes (DEGs) and differentially regulatory SEs (DSEs). A total of 212 deregulated genes were assigned to 186 DSEs within a 1-Mb distance. LFC, log2 fold change.
(D) Chromatin states H3K27ac and H3K27me3 signal at the HOXA cluster loci in 2 AML subtypes. For each 200-bp bin from ChromHMM segmentation analysis, we selected the most frequent chromatin state to show in each subtype. All y axis scales are reads per kilobase million (RPKM)-normalized units.
(E) Dynamic H3K27ac and H3K27me3 intensity and expression levels of the GRK5 gene in AML patients, normal CD34+ progenitors, monocytes, and neutrophils.
See also Figure S4.
The HOXA gene cluster was covered by SEs specifically in the NPM1/MLL (C2) group, displaying significantly higher H3K27ac occupancy and lower H3K27me3 signal, as compared to the C1 subtype (Figure 2D). We next examined the expression patterns of HOXA and HOXB genes and found that almost all HOX genes were more abundantly expressed in subtype C2 (Figure S4B). Allocating 179 AML patients from TCGA (Ley et al., 2013) into 2 groups based on the subtype-specific mutations landscape identified in the present study, we also found highly similar expression patterns for HOX genes (Figure S4B). Finally, to compare the epigenetic and transcriptomic features at HOXA regions in AML to normal cell types, we included CD34+ progenitor cell, monocyte, and neutrophil data. We found that normal CD34+ cells were enriched for C2-specific SEs and higher HOXA expression than monocyte and neutrophil cells (Figure S4C). Similarly, the GRK5 gene in the C1 subtype was occupied by high H3K27ac and low H3K27me3 signals, similar to the 2 differentiated cells (Figure 2E). These results suggest that AMLs in the RUNX1/spliceosome cluster (C1) represent more differentiated cells, while those in the NPM1/MLL group (C2) may display a more progenitor-like cellular phenotype.
Transcriptomic Changes in the 2 Epigenetic Subtypes
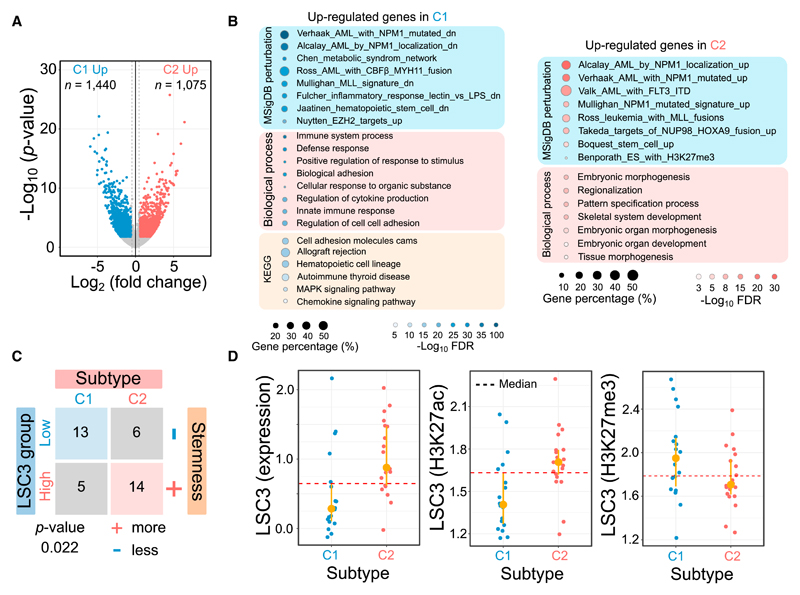
We identified a total of 2,515 significant differentially expressed genes (DEGs) between the RUNX1/spliceosome (C1) and the NPM1/MLL (C2) subtypes (Figure 3A). Examining the expression patterns of these genes revealed intra-group homogeneity with average Spearman correlations of 0.912 in C1 and 0.909 in C2 (Figure S4D). The C2 upregulated gene set represented enriched expression signatures of genetic perturbations induced by NPM1, MLL, and NUP98 defects and contained many genes that are essential for the proliferative properties of stem cells and development (Figure 3B). For C1 upregulated genes, we found enrichment for perturbation pathways related to CBF and MYH11 fusion events and genes increased in the inflammatory, immune, and differentiation properties of AML cells (Figure 3B), again suggesting that C1 represents more differentiated AMLs, while C2 characterizes an earlier stage. Our results also showed strong positive correlation of global gene expression patterns with the TCGA dataset at a Pearson coefficient of 0.716 (Figure S4E). In addition, many epigenetic factors such as homeobox gene families and cofactors, transcription factors, and epigenetic complex showed differential expression between the 2 subtypes (Figure S4F; Table S6). As a key factor in epigenetic programming, CEBPA showed increased expression levels in AMLs and monocytic cells, but no significant difference between C1 and C2 (Figure S4G), suggesting that CEBPA is not the main driving factor in establishing the epigenomic subtypes, which is in line with mutations in CEBPA being present in both.
Figure 3. Subtype-Specific Transcriptomic Signatures.
(A) Volcano plot of gene expression changes between 2 subtypes.
(B) Functional enrichment analysis for differentially expressed genes. The dot size represents the percentage of identified genes against background.
(C) Contingency table comparing putative AML subtypes with different cellular consequences in prognosis predicted from a 3-gene signature model. All AML samples were partitioned into 2 groups based on the LSC3 median cutoff. Samples with higher LSC3 score have greater stemness properties.
(D) Leukemic stem cell score derived from 3 signature genes using gene expression, H3K27ac, and H3K27me3 for the 2 subtypes. The H3K27me3-dependent LSC3 score shows opposite trend due to negative regulation with gene expression.
To assess the cellular consequence for each subtype, we calculated leukemia stem cell (LSC) scores by using a 3-gene signature model (LSC3) (Ng et al., 2016). Based on a median cutoff in predicted LSC3 scores, all of the AML samples could be discretized into high and low groups. We found that the predicted prognosis status of patients showed a significant association with AML subtype C1 or C2 (p = 0.022, Fisher’s exact test) (Figure 3C). The patients with adverse outcomes were more frequently located in the C2 subtype (73.7%), and 68.4% of cases in the favorable group belonged to the C1 subtype, which was validated by several independent predictors such as MSI2 and PBX3 (Figures 3C and S4H) (Byers et al., 2011; Li et al., 2013, 2016b). To explore epigenetic biomarkers for prognosis prediction, we also calculated the LSC3 values using H3K27ac and H3K27me3 signals in promoters and H3K36me3 signals in the gene body for the 3 gene signatures. We found that the C2 subtype possessed a significantly higher percentage of samples belonging to the high-LSC3 group than C1 from both transcriptomic and epigenetic data, in which the lower LSC3 score inferred from H3K27me3 indicated higher stemness due to its negative correlation with gene expression (Figures 3D and S4I).
Also, when only focusing on the 19 AML samples with normal karyotypes, we found that almost all of the samples in C2 have larger LSC3 values inferred from gene expression, while epigenetic marks indicated clear differences (Figure S4J). Our results reveal that patients belonging to group C1 harbor more differentiated AMLs and have relatively favorable prognoses compared to patients in group C2, and suggest that, despite the small sample size used in this study, epigenetic patterns in the 3-gene model may have predictive value.
Relation between Mutations in RUNX1 and Splicing Factors
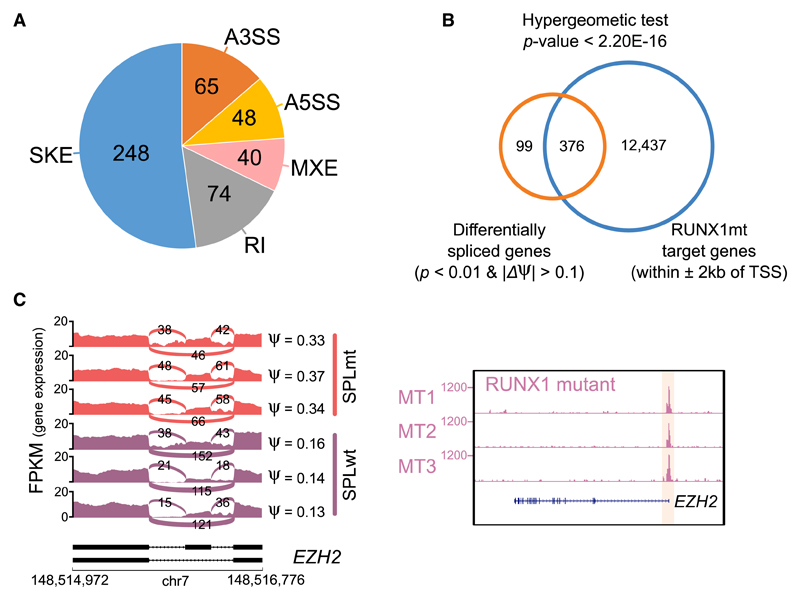
Given that AMLs carrying mutated RUNX1 or splicing factors are specifically in the C1 subtype, we speculated that mutated RUNX1 protein could deregulate the same genes targeted by mutated spliceosome factors (Dvinge et al., 2016). We performed RUNX1 ChIP-seq in the RUNX1 mutant (RUNX1mt) expressing AMLs and found that RUNX1 peaks showed significant enrichment in promoter regions; they also colocalized with active epigenetic marks, especially DNaseI hypersensitive sites (Figures S5A–S5C). Subsequently, we identified 475 genes linked to differential splicing events (Figure 4A) by comparing patients carrying mutations in spliceosome factors with other patients in the C1 subtype. These differentially spliced hits were then compared with the gene list, the promoters of which were occupied by RUNX1 in RUNX1mt AMLs. We found significant overlap between the 2 datasets by hypergeometric testing using all RefSeq genes as background (p < 2.20 × 10−16; Figure 4B). Among the overlapping genes, EZH2, an important component of the PRC2 complex, was found to have higher exon usage in patients with splicing factor mutations (Figure 4C), which is in line with a recent study (Kim et al., 2015). Usage of this exon could lead to a truncated protein product due to a premature stop codon by open reading frame prediction. We also found that its promoter showed the presence of high-affinity binding of RUNX1 in samples with the aberrant RUNX1 gene (Figure 4C, right). Our results suggest that the effects of mutating RUNX1 or splicing factors may converge on the epigenome.
Figure 4. Potential Interplay between Aberrant RUNX1 Protein and Spliceosome Complex.
(A) Differentially spliced events between splicing factor mutant and other AMLs in C1. A3SS/A5SS, alternative 3′/5′ splice sites; MXE, mutually exclusive exons; RI, retained introns; SKE, skipped exons.
(B) Overlap between differentially spliced hits and target genes bound by mutated RUNX1.
(C) Skipped exons and RUNX1 binding patterns at EZH2 loci. Different exon usage between mutated splicing factors and 3 selected other samples (left). Occupancy of RUNX1 at the EZH2 promoter (right).
See also Figure S5.
Mixture Deconvolution and Gene Regulatory Network in 2 Subtypes
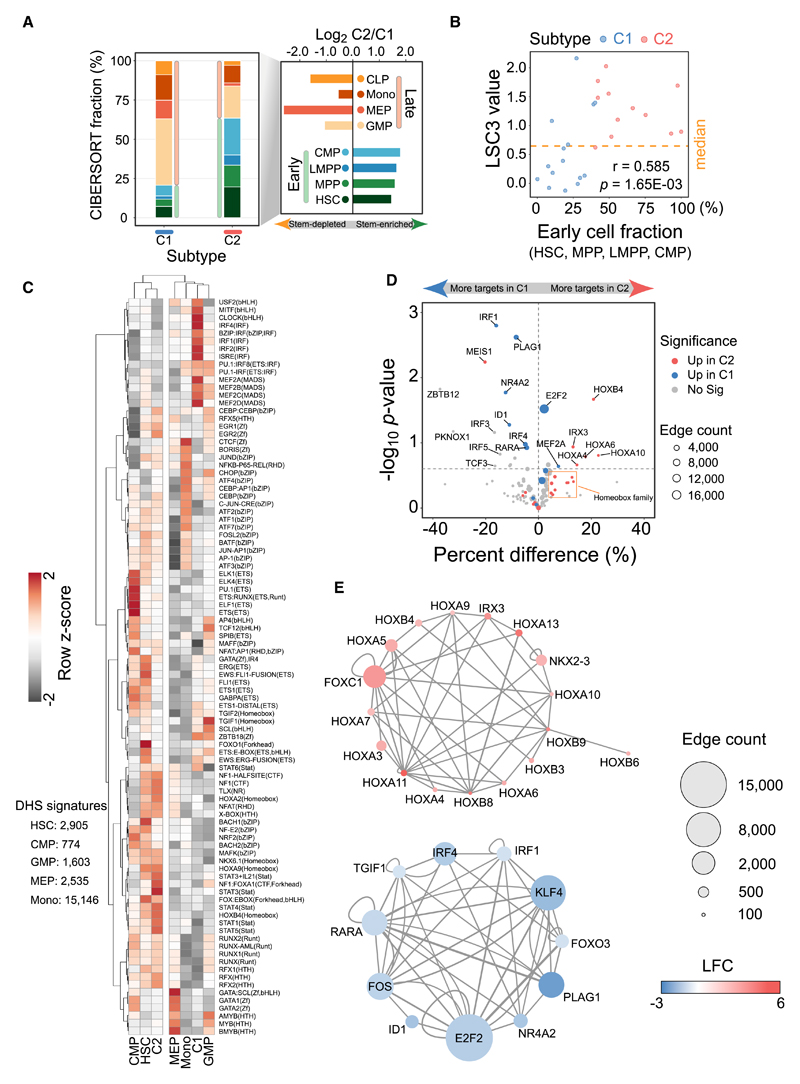
We estimated the attributable fraction of each cell type to quantify their contributions in our AML samples using assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq) and DNaseI-seq data. First, we compared DNaseI-seq data with ATAC-seq data from monocyte cells and showed high concordance between the profiles (average Spearman correlation r = 0.818; Figure S6A), suggesting that these datasets can be directly compared. We found a total of 783 C1-open and 3,676 C2-open DNaseI hypersensitive sites (DHSs) (Figure S6B). Overlap analysis in conjunction with DNA accessibility signatures from 5 different cell types indicates that the RUNX1/spliceosome (C1) subtype is more similar to late-stage cell types, while the NPM1/MLL (C2) subtype maintains signatures from early precursor cells. Second, we performed a deconvolution analysis based on DHSs marked as strong enhancers in AML to define cell subpopulations by integrating ATAC-seq data from 8 other normal cell types (Corces et al., 2016). A cell-mixture decomposition approach predicted that the C2 subtype compromised an average of 63.46% early cells (mainly hematopoietic stem cells [HSCs], multipotent progenitors [MPPs], and common myeloid progenitors [CMPs]), suggesting a more stem-enriched property. In contrast, most cell types (79.27%) in the C1 subtype were from late-stage, differentiated cells (mainly granulocyte-monocyte progenitor [GMP] and monocytes [Mono]) (Figure 5A). A moderate positive correlation (r = 0.585) between the estimated LSC3 score and fractions of early cells showed that patients with the C2 subtype generally have higher early cell percentages and more stem cell properties (Figure 5B).
Figure 5. Distinct Cell Populations, Motif Enrichment, and Regulatory Network between 2 AML Subtypes.
(A) Predicted average fractions of 8 different lineages for 2 AML subtypes. Early cell types include common myeloid progenitor (CMP), hematopoietic stem cell (HSC), lymphoid-primed multipotent progenitor cell (LMPP), and multipotent progenitor cell (MPP). Late cell types include common lymphoid progenitor (CLP), granulocyte-macrophage progenitor cell (GMP), megakaryocyte-erythroid progenitor cell (MEP), and monocyte (Mono).
(B) Consistency of stemness property between mixture deconvolution and 3-gene signature model. The higher fraction of early cell populations shows larger leukemic stem cell values.
(C) Motif enrichment based on unique open chromatin sites in 2 AML subtypes and 5 cell types. Row Z score for each motif is calculated to assess relative enrichment in each cell type.
(D) Differentially connected transcription factors between the 2 subtypes. Upregulated genes in a subtype are prone to modulate more targets in that subtype.
(E) Key deregulated hubs in the subtype-specific regulatory network. Node size denotes the total number of targets regulated by each node.
See also Figure S6.
To explore the key transcription factors (TFs) driving the 2 different AML subtypes, the subtype-specific DHSs and cell type-unique open regions (HSC, CMP, GMP, megakaryocyte-erythroid progenitor [MEP], and Mono) were used for motif discovery (Figure 5C). In addition, for the enriched motifs, we examined the expression levels of the corresponding transcription factors (Figure S6C). Hierarchical cluster results by enrichment degree confirmed the earlier cell stage and C2 correlation (Figure 5C). Specifically, we found that the MEF2 and interferon regulatory factor (IRF) motif families, as well as motifs with a basic helix-loop-helix (bHLH) binding domain, were more enriched in the C1 subtype. In contrast, in the C2 subtype sequence motifs for key hematopoietic regulators such as RUNX1 and homeobox genes were overrepresented.
Moreover, we found many high-quality footprints that could be inspected by the average DNaseI activity profiles, such as the well-known CCCTC-binding factor (CTCF) footprint with strong protection from DNaseI cleavage (Nakahashi et al., 2013) (Figure S6D). We linked these putative footprints to their potential target genes based on the footprint purity score and distance and then inferred subtype-specific gene regulatory networks. Subsequently, the connection number of TFs was compared between the 2 subtypes to identify differentially connected regulators. We found that most upregulated TFs in one subtype also tended to have more target genes and high motif enrichment in the same group, like homeobox genes and IRF families in C2 and C1, respectively (Figure 5D).
We next scrutinized subtype-specific networks from deregulated TFs to explore the interactions between a core set of key regulators in each subtype. For each network, these highly connected TF hubs also presented tight interactions between them (Figure 5E). In the C2-specific network diagram, we found that homeobox genes were major components and have an average of 310 targets, although the hub with the most connections is FOXC1 (1,950 connections). In contrast, the pivotal TFs in the C1 network could regulate relatively more genes, mainly dominated by E2F2, KLF4, and IRF families (Figure 5E). Another 2 TFs, RARA and PLAG1, functionally involved in cell development and differentiation, also showed important regulatory roles in the C1 subtype (Grimwade et al., 2016; Singh et al., 2017). These results revealed the core transcriptional network that drives epigenetic regulation in the 2 subtypes of AML.
Discussion
A comprehensive knowledge of epigenetic signatures is of importance, as in general, these better reveal cellular conditions and phenotypic plasticity than transcriptomic (or genomic) markers alone. RNA-seq data generally suffer from differences in RNA stability, high variation in gene expression levels, and substantial contributions to the overall transcriptome by minor (polluting) cell populations. In contrast, the epigenetic status of, especially, enhancers can better demarcate differentiation trajectories and the clonal composition of the cell population (Corces et al., 2016), which is generally heterogeneous in AML samples. Hence, subtype classification based on the epigenome has the potential to converge patients with similar response to external exposure, such as drugs, into the same group, allowing the identification of clinical indicators for early diagnosis and prognosis.
Here, clustering analysis of H3K4me1 or H3K27me3 uncovers almost the same AML classification patterns to reveal 2 major epigenomic subtypes, C1 and C2. As an enhancer mark, H3K4me1 seems cell type and disease specific and captures cell identity as well as cluster purity, as suggested previously (Kasowski et al., 2013; Kundaje et al., 2015). Similarly, H3K27me3 has also been suggested to contribute to maintaining cell identity, at least in part by regulating lineage-specific TF expression (Conway et al., 2015). The substantial overlap among clustering results from different datasets (ChIP-seq, RNA-seq, DNaseI-seq) points to the robustness of our clustering, and it demonstrated that most epigenetic signatures are strongly interrelated and share a cooperative effect on AML pathogenesis. In contrast, some other marks such as H3K9me3 seem less informative and are more granular. For these marks, increasing their sample size seems warranted.
Our data suggest that the NPM1/MLL (C2) subtype has greater stemness phenotypes owing to the higher enrichment of LSCs, implying that the C2 subtype is likely to be more aggressive and resistant to therapy than the C1 subtype. This finding is supported by the mutational status, as almost all of the samples in the C2 subtype carry FLT3-internal tandem duplication (ITD), IDH1, or t(9;11) aberrations, and abnormally high expression levels of HOXA9, both of which are generally associated with poor prognosis (Bond et al., 2016; Collins and Hess, 2016; Golub et al., 1999; Jung et al., 2015; Li et al., 2012). The 3-gene LSC signature (Ng et al., 2016) using gene expression, H3K27ac, and H3K27me3 suggest inferior cellular consequences in clinical outcomes for the C2 subtype. In addition, the C2 subtype shows epigenomic signatures observed in normal early progenitor cells, suggesting that this subtype largely maintains the epigenetic status of the progenitor lineage. In contrast, the epigenetic signature of the RUNX1/spliceosome (C1) group is characterized by increased repressive marks and a closed chromatin state, likely representing late-stage cells.
In summary, using epigenomic signatures, 2 major AML subtypes are proposed that exhibit distinct mutational characteristics and regulatory mechanisms and that confer different stemness properties. Our findings facilitate a better molecular understanding of the ontogeny of AML, and may ultimately help to improve therapy decision making by designing certain specific epidrugs to reprogram local epigenetic patterns of target genes.
Star★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| H3K4me1 | Diagenode | C15410194; RRID:AB_2637078 |
| H3K4me3 | Diagenode | C15410003-50; RRID:AB_2616052 |
| H3K9me3 | Diagenode | C15410193; RRID:AB_2616044 |
| H3K27ac | Diagenode | C15410196; RRID:AB_2637079 |
| H3K27me3 | Diagenode | C15410195; RRID:AB_2753161 |
| H3K36me3 | Diagenode | C15410192; RRID:AB_2744515 |
| RUNX1 | Abcam | ab23980; RRID:AB_2184205 |
| Critical Commercial Assays | ||
| KAPA library preparation kit | Kapa Biosystems | KK8400 |
| riboZero gold rRNA removal kit | Illumina | MRZG12324 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1031 |
| TruSeq SBS KIT v3 - HS (50 cycles) | Illumina | FC-401-3002 |
| NextSeq 500/550 High Output v2 kit (75 cycles) | Illumina | FC-404-2005 |
| NEBNext High-Fidelity 2 × PCR Master Mix | New England Biolabs | M0541 |
| Second Strand Buffer | Life Technologies | 10812-014 |
| Superscript III Reverse Transcriptase | Life Technologies | 18080-044 |
| DNase I | QIAGEN | 79254 |
| Qubit RNA HS assay kit | Life Technologies | Q32852 |
| Ribozero Gold Kit | Illumina | MRZG12324 |
| Rneasy Mini Kit | QIAGEN | 74106 |
| Deposited Data | ||
| Raw data files for histone ChIP sequencing | This paper | EGAD00001002340, EGAD00001002418, EGAD00001002935 |
| Raw data files for RUNX1 ChIP sequencing | This paper; Mendeley Data | GSE111821; 10.17632/99vfrzcbhm.1 |
| Raw data files for RNA sequencing | This paper | EGAD00001002443, EGAD00001002465, EGAD00001002962, EGAD00001002968 |
| Raw data files for DNaseI sequencing | This paper | EGAD00001002355 |
| Raw data files for WGBS sequencing | This paper | EGAD00001002333, EGAD00001002419 |
| Raw data files for ATAC sequencing | Corces et al., 2016 | GSE74912 |
| Software and Algorithms | ||
| ConsensusClusterPlus | Wilkerson and Hayes, 2010 | http://bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html |
| pvclust | Suzuki and Shimodaira, 2006 | http://stat.sys.i.kyoto-u.ac.jp/prog/pvclust/ |
| BWA | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| Picard | N/A | http://broadinstitute.github.io/picard/ |
| PhantomPeakQualTools | N/A | https://code.google.com/archive/p/phantompeakqualtools/ |
| MACS2 | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| deepTools | Ramírez et al., 2016 | https://github.com/deeptools/deepTools |
| ChromHMM | Ernst and Kellis, 2012 | http://compbio.mit.edu/ChromHMM/ |
| ChromDiff | N/A | http://compbio.mit.edu/ChromDiff/ |
| ROSE | Whyte et al., 2013 | http://younglab.wi.mit.edu/super_enhancer_code.html |
| BEDTools | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| STAR-Fusion | Haas et al., 2017 | https://github.com/STAR-Fusion |
| DESeq2 | Love et al., 2014 | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Cufflinks | Trapnell et al., 2010 | http://cole-trapnell-lab.github.io/cufflinks/ |
| MISO | Katz et al., 2010 | https://miso.readthedocs.io/en/fastmiso/ |
| F-Seq | Boyle et al., 2008 | https://github.com/aboyle/F-seq |
| GEM | Marco-Sola et al., 2012 | http://dat.cnag.cat/wiki/The_GEM_library |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/motif/ |
| PIQ | Sherwood et al., 2014 | https://bitbucket.org/thashim/piq-single |
| Cytoscape | Shannon et al., 2003 | https://cytoscape.org/ |
| bwtool | Pohl and Beato, 2014 | https://github.com/CRG-Barcelona/bwtool |
| CIBERSORT | Newman et al., 2015 | https://cibersort.stanford.edu/ |
| IGV | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
| Other | ||
| EpiFactors database | Medvedeva et al., 2015 | http://epifactors.autosome.ru/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joost H.A. Martens (j.martens@ncmls.ru.nl).
Experimental Model and Subject Details
Patients Acquisition
A total of 38 samples with AML and 2 APLs (acute promyelocytic leukemia) were selected, given that the sample composition in this study should well represent the complex mutational landscape of AML, and all samples should contain enough materials of high quality for multi-omics profiling. Each sample was subjected to ChIP-Seq (six histone marks: H3K4me1, H3K4me3, H3K9me3, H3K27ac, H3K27me3 and H3K36me3) and strand-specific total RNA-Seq, while for the majority of samples DNaseI-Seq (n = 29), targeted mutational analysis (n = 29) and WGBS (n = 21) were also conducted (Table S1). The clinical and biological characteristics of the samples are detailed in Table S1. The study and sample usage were approved by the ethics committees of the contributing institutions. Leukemic samples were either obtained from bone marrow or peripheral blood for subsequent processing. To obtain relative pure cell populations and the largest fraction of leukemic cells (~10 million cells are needed to perform all the experiments) we used fluorescence-activated cell sorting (FACS) based on expression of cell surface markers CD33 or CD34 (Figure S1A; Table S1). For the majority of samples, CD33 enrichment was used and the detailed purification method is listed in Table S1. The cytogenetic information of all subjects was determined at the time of disease diagnosis. Most of samples have undergone mutational analyses by a custom 21-gene sequencing-based assay to assess for frequently mutated genes in AMLs like NPM1, FLT3 and DNMT3A. As APL patients are a separate entity and are treated differently, these patients (n = 2) were excluded from the analysis and processed separately (de Thé, 2015; Petraglia et al., 2018; Singh et al., 2018).
Method Details
Mutation Spectrum Analyses
Genomic DNA was extracted, amplified and subjected to a custom 21-gene sequencing-based assay as previously described (Berger et al., 2017). The 21 target genes (Table S2) are common driver genes with a mutation frequency > 5% in AML, and those well-known variants for each gene were probed in our study. The identified mutation results are shown in Table S3. Considering that the 4-bp insertion in NPM1 and the internal tandem duplication (ITD) in FLT3 are relatively easy to detect, we also confirmed these variants through visual inspection of the RNA-Seq tracks by Integrative Genomics Viewer (IGV) tool (Robinson et al., 2011), and also predicted their status in samples without genomic information. For four genes with high frequency in our study, we used Fisher’s exact test to identify gene pairs with significant exclusivity and co-occurrence.
ChIP-Sequencing
A total of six histone marks were selected for ChIP-Seq, including H3K4me1, H3K4me3, H3K9me3, H3K27ac, H3K27me3 and H3K36me3 (Diagenode C15410194, C15410003-50, C15410193, C15410196, C15410195 and C15410192). Chromatin harvest and sequencing experiments were carried out based on the standard Blueprint protocol (http://www.blueprint-epigenome.eu). For ChIP of each histone mark, around 1 million cells were collected. Purified cells were first cross-linked using 1% formaldehyde (Sigma), and then sonicated to obtain DNA fragments of about 200-300 bp by a Diagenode Bioruptor. Sheared chromatin was incubated with specific antibodies against the six histone markers. After immunoprecipitation, the protein-DNA cross-links were reversed, and the isolated DNA was used for quantitative PCR and sequencing analysis. Meanwhile, a portion of chromatin was processed under the same conditions but without immunoprecipitation step, as a control dataset (input DNA). For each sample, an Illumina library was prepared with the Kapa Hyper Prep Kit, and then subjected to 42 bp single-end sequencing on the Illumina HiSeq 2000 machine.
The RUNX1 ChIP-Seq was performed as described (Mandoli et al., 2016) using RUNX1 antibody (Abcam ab23980) which recognizes both wild-type and mutated RUNX1 protein. After the regular ChIP procedure, four AML samples carrying RUNX1 mutation were sequenced on the HiSeq 2000 platform with 42 bp paired-end reads.
DNaseI-Sequencing
DNaseI-Seq data was generated using the standard protocol of the Blueprint Consortium. Leukemic cells per donor were collected, and nuclei were isolated using Buffer A [15mM NaCl, 60mM KCl, 1mM EDTA (pH 8.0), 0.5mM EGTA (pH 8.0), 15mM Tris-HCl (pH 8.0) and 0.5mM Spermidine] supplemented with 0.015% IGEPAL CA-630 detergent. Nuclei were incubated for 3 minutes at 37°C during DNaseI treatment. The reaction was terminated with stop buffer [50mM Tris-HCl (pH 8.0), 100mM NaCl, 0.10% SDS, 100mM EDTA (pH 8.0), 1mM Spermidine and 0.3mM Spermine]. The sample was subsequently fractionated via 9% Sucrose gradient for 24 hours at 25,000 rpm at 16°C. Fractions containing fragments smaller than 1 kb were purified and further processed according to the Illumina library preparation protocol. After quality assessment, the eligible library was sequenced by Illumina HiSeq 2000 machine and generated 42 bp single-end reads.
Whole Genome Bisulfite Sequencing
Detailed WGBS protocols were conducted as previously described (Kulis et al., 2015). Genomic DNA was sonicated to 50-500 bp using a Covaris E220 and fragments of size 150-300 bp were selected using AMPure XP beads (Agencourt Bioscience). We constructed DNA libraries using the Illumina TruSeq Sample Preparation kit (Illumina Inc., San Diego, CA, USA) based on the Illumina standard protocol. And the DNA underwent two rounds of bisulfite conversion using the EpiTexy Bisulfite kit (QIAGEN). The treated DNA fragments were enriched through seven cycles of PCR using the PfuTurboCx Hotstart DNA polymerase (Stratagene). The quality of library was assessed using the Agilent 2100 BioAnalyzer (Agilent), and the concentration of viable sequencing fragments (molecules carrying adaptors at both extremities) was determined using quantitative PCR with the Library Quantification kit from KAPA Biosystem. Then paired-end DNA sequencing (two reads of 100 bp each) was carried out using the Illumina HiSeq 2000 instrument.
Strand-specific RNA Sequencing
Total RNA was isolated from leukemic cells using the RNeasy RNA extraction kit (QIAGEN, Netherlands) with on-column DNaseI treatment. Ribosomal RNA was removed using the Ribo-Zero rRNA Removal kit (Illumina) following the manufacturer’s recommendations. The RNA concentration was monitored with a Qubit Flurometer (Invitrogen), and the RNA quality was evaluated by the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) prior to library preparation. First strand cDNA synthesis was performed using SuperScript III (Life Technologies), followed by synthesis of the second cDNA strand. Then, strand-specific cDNA library with around 200 bp insert size was constructed using the TruSeq Stranded RNA Sample Preparation kit (Illumina) based on the manufacturer’s instructions. For each library, paired-end sequencing (76 nucleotides each end) was then performed on an Illumina HiSeq 2000 machine.
AML Subtype Classification
Subtype discovery was conducted by ConsensusClusterPlus package (Wilkerson and Hayes, 2010) with top 1% variable peaks or genes (use top 1,000 when under 1,000) which were chosen according to interquartile range (IQR) of normalized peak density or gene expression. ConsensusClusterPlus was run with 1,000 iterations, 80% sample resampling from 2 to 12 clusters (k = 2 to 12) using hierarchical clustering based on Euclidean distance metric and Ward.D2 linkage method. The consensus clustering is a resampling-based method for evaluating stability of the clustering, and the consensus value is the proportion of times (n = 0 ~ 1 inclusive) that the pair’s items (samples) are clustered together across the resampling iterations (i = 1,000 in the present study). We also computed silhouette score to assess the coherence of clusters by evaluating the similarity of patients within or between subtypes. In parallel to this approach, we also used another R package pvclust (Suzuki and Shimodaira, 2006) with the 1,000 iterations of boot-strapping to check the significance and robustness of the clustering based on the same datasets and methods. The optimal number of AML subtypes was mainly determined by consensus cluster and silhouette value changes. The same clustering analyses were done based on histone marks, gene expression, DNA accessibility and methylation level, to compare the consistency of different datasets.
ChIP-Seq Data Analysis
Sequenced reads were aligned against the UCSC human reference genome (GRCh37/hg19) with Burrows-Wheeler Aligner (BWA) program (Li and Durbin, 2009) with default parameters. Each sample with higher read coverage than matched input data was randomly subsampled using Picard DownsampleSam command (http://broadinstitute.github.io/picard/), to increase peaks detection specificity (Chen et al., 2012). The resultant BAM files were subjected to removal of potential PCR and optical duplicates using Picard MarkDuplicates option. Fragment length and quality measurement for each dataset were determined using PhantomPeak-QualTools based on strand cross-correlation approach (https://code.google.com/archive/p/phantompeakqualtools/). Two metrics named normalized strand cross-correlation coefficient (NSC) and relative strand cross-correlation coefficient (RSC) were used for data quality assessment. Peak calling was performed using MACS2 (Zhang et al., 2008) with the estimated fragment size. All peaks were called with input data as the background control. H3K4me1, H3K9me3, H3K27me3 and H3K36me3 peaks were detected using the broad setting (–broad) with a q-value of 0.05, while H3K4me3 and H3K27ac were called using the narrow setting (default) with a q-value of 0.01. For RUNX1 transcription factor, the binding sites were detected using default parameters except for a p-value cutoff of 1 × 10−6. Peaks overlapping with the consensus excludable ENCODE blacklist and on sex chromosomes were discarded to avoid confounding by repetitive regions and gender-specific bias. All alignment files were extended to the estimated fragment length and scaled to RPKM-normalized read coverage files using deepTools (Ramírez et al., 2016) for visualization.
To characterize chromatin states for each individual epigenome, the six histone marks were integrated by applying ChromHMM hidden Markov model (HMM) algorithm (Ernst and Kellis, 2012). ChromHMM was run with default parameters and using input control as background. We trained 13 models ranging from 8 to 20 states, and decided a 12-state model since it could capture the major biologically meaningful combinations. The 12 chromatin states were subsequently defined based on the co-occurrence frequency of individual features. To explore the overall variability of the 12 states across AML patients, we first calculated the number of each 200-bp bin labeled with that state in at least one patient for each state, and the corresponding cumulative fraction in at most n patients (n = 1–38) was computed (Kundaje et al., 2015). The chromatin state with faster cumulative frequency changes means this state is more variable than others.
Super enhancers (SEs) in each sample were predicted by the ROSE algorithm (Whyte et al., 2013) using H3K27ac as the surrogate mark. Briefly, all H3K27ac peaks within ± 2.0 kb around transcription start sites (TSSs) were first excluded. The remaining peaks closer than default distance of 12.5 kb were stitched together, and subsequently ranked by normalized H3K27ac level corrected by input background. Finally, SEs were separated from typical enhancers based on the inflection point of H3K27ac signal curve. Differential SEs between AML subtypes were identified using DESeq2 (Love et al., 2014) with an adjusted p-value less than 0.1 and absolute fold change greater than 1.5. Super enhancer assignment to the nearest genes was determined by BEDTools (Quinlan and Hall, 2010).
RNA-Seq Data Analysis
For expression analyses, the hg19 reference genome index was first generated using STAR aligner (Dobin et al., 2013) with UCSC gene annotation. Paired-end reads were mapped to the indexed genome in two-pass mode with default parameters, to increase alignment accuracy and sensitivity. Stranded gene-level read counts were enumerated at the same time, and used as input for DESeq2 package (Love et al., 2014) to distinguish differential expressed genes among different AML subtypes. Only autosomal genes were analyzed and these greater than 1.5 fold changed at adjusted p-value < 0.1 were considered significantly deregulated. Expression quantification for each RefSeq gene was performed by Cuffnorm function in Cufflinks (Trapnell et al., 2010), to estimate Fragments Per Kilobase per Million aligned reads value (FPKM).
Besides normal mapping, we also turned on detection of chimeric alignments with –chimSegmentMin 20 option, in order to identify genome-wide fusion genes. We used the STAR-Fusion pipeline (https://github.com/STAR-Fusion/STAR-Fusion) to predict recurrent fusion genes based on junction files from the STAR aligner. Only those fusion genes with sum of junction reads and spanning fragments greater than nine were retained, to ensure true positives of predictions.
In addition, we used MISO suites (Katz et al., 2010) with default options to detect alternative splicing events in our study. The MISO annotations contained five types of events: skipped exons (SKE), alternative 3′/5′ splice sites (A3SS, A5SS), mutually exclusive exons (MXE) and retained introns (RI). We first computed percentage splicing index (PSI) value of each event and inferred differentially spliced genes by pairwise comparisons between groups using t test. Significant differences were only considered for p-value < 0.01 and absolute difference in PSI mean of the groups ≥ 0.1.
In order to evaluate clinical outcomes for each AML patient, we used a linear combination of expression value of validated gene signatures and computed a leukemia stem cell (LSC) score (Ng et al., 2016). A reweighted 3-gene signature model was used because this model is more optimal to capture survival differences within small populations. The signature scores (LSC3) were calculated using log2-transformed FPKM after incrementing by 1, and a high LSC3 value suggests a greater fraction of leukemia blasts that conferred resistance to standard AML therapy. As suggested, a median threshold in our data was used to classify scores into high and low groups, in which above- and below-median scores were linked to adverse and favorable outcomes, respectively.
DNaseI-Seq Data Analysis
All DNaseI-Seq reads were mapped to the hg19 reference genome using BWA (Li and Durbin, 2009) with default settings. Non-uniquely mapped reads and PCR duplicates were removed. From these filtered mapped reads, we used F-Seq tool (Boyle et al., 2008) to identify candidate DNaseI hypersensitive sites (DHSs) using default parameters except for a 300 bp feature length and the threshold parameter of 6. To alleviate artificial differences due to genome mappability, a custom 42 bp background track was constructed using GEM tools (Derrien et al., 2012; Marco-Sola et al., 2012) and bffBuilder program (provided by F-Seq) as control. After peak calling, we fitted the DNaseI signal data to a gamma distribution to calculate p-value for each peak, and significant DHSs were determined at a loose p-value of 0.05 cutoff. All DHSs on sex chromosomes were removed from the analysis to allow for comparison across both male and female patients.
Differentially accessible regions were detected using DESeq2 package (Love et al., 2014) with the same cutoffs as in previous analyses, after removing peaks with the total read counts less than five. Motif discovery in the differential DHSs was employed by the findMotifsGenome function in HOMER tool (Heinz et al., 2010) with random background and other default parameters. To compare motif signatures between different subtypes and cell types, we calculated fold change defined as the percentage of target sequence with this motif divided by percentage of background sequences with the same motif.
Transcription factor (TF) footprints were detected using PIQ package (Sherwood et al., 2014) based on the input motifs set from JASPAR database and other collections (Mathelier et al., 2016; Matys et al., 2006). We concatenated all DNaseI alignment files from the same subtypes to provide sufficient sequencing depth for footprinting analyses. Purity scores for the genomic occupation of each TF were predicted to evaluate TF binding affinity. Only those transcription factors with at least 500 high-purity (>0.7) binding sites within a DHS were kept for the following analyses. To infer transcriptional regulatory networks, we conducted similar analyses based on these putative footprint sites as previous studies (Qu et al., 2015; Rendeiro et al., 2016). We computed an interaction score between each transcription factor and each gene, based on its footprint purity score and the distance from a nearby gene. Only those interactions greater than 1.0 were kept and used as edge weight to construct network. The gene regulatory network for each AML subtype was visualized by Cytoscape software (Shannon et al., 2003). To examine differences between AML subtype networks, we focused on these source-target interactions comprising at least one differentially expressed gene (TF or target gene). Subsequently, we divided the degree of each node by the total number of edges in each network, and calculated the percentage difference between two subtypes for the same TF.
WGBS Data Analysis
The common set of called CpG sites including methylation signal and coverage were provided by Centro Nacional de Análisis Genómico (CRG-CNAG). The detailed analyses protocol was performed as described previously (Kulis et al., 2015). WGBS read alignments were generated using GEM software (Marco-Sola et al., 2012) with respect to a converted hg38 reference genome. Only read pairs mapped to the same chromosome with the consistent orientation and reasonable edit distance from the reference were selected for the following analyses. Estimation of genotype and cytosine methylation levels were carried out using software developed at the CNAG, taking into account the observed bases, base quality scores and the strand origin of each read pair. For each genomic position, we generated estimates of the most likely genotype and the methylation proportion (for genotypes containing a C on either strand). A Phred-scaled likelihood ratio for the confidence in the genotype call was estimated for the called genotype at each position. For each sample, CpG sites were selected where both bases were called as homozygous CC followed by GG with a Phred score of at least 20, corresponding to an estimated genotype error level of ≤1%. Sites with coverage greater than 500× were filtered to avoid repetitive regions (centromere/telomere). After quality control procedure, a common set of called CpG sites for all analyzed samples was generated, and used for all downstream analyses.
To investigate the differential DNA methylation levels between AML subtypes, the obtained CpG sites were formatted as inputs for RnBeads package (Assenov et al., 2014). The significance of differential methylation in each site or region was determined by a combined rank score with a threshold of 3,000. All preliminary analyses were done based on hg38 reference genome, so we converted all methylation coordinate results to hg19 assembly using bwtool software (Pohl and Beato, 2014) for data integration and visualization.
AML Deconvolution
To systematically map the compositional difference of blood cells in bulk AML samples, we applied the CIBERSORT (Cell type Identification By Estimating Relative Subsets Of known RNA Transcripts) deconvolution method (Newman et al., 2015) to DNA accessibility data resource. This approach can quantify the relative fractions of cell-type-specific signatures in bulk tumors based on machine learning approach. We downloaded ATAC-seq data for eight primary human blood cells from GSE74912 accession (Corces et al., 2016). The dataset comprised four early cell types, hematopoietic stem cell (HSC), multipotent progenitor cell (MPP), lymphoid-primed multipotent progenitor cell (LMPP), common myeloid progenitor (CMP), and four late cell types, granulocyte-macrophage progenitor cell (GMP), megakaryocyte-erythroid progenitor cell (MEP), monocyte cell (Mono) and common lymphoid progenitor (CLP). We first evaluated the correlation between ATAC-Seq and our DNaseI data. We only focused on DHSs overlapped with strong enhancer (EnhS) from ChromHMM because of high individual variability and cell type specificity of these distal regulatory elements. The RPKM value for each predicted DHSs was calculated to be the input for AMLs deconvolution analyses using 1,000 permutation tests. Only samples with an empirical p-value less than 0.1 were included in the following analyses.
Gene Ontology and Pathway Analysis
To assess the regulatory functions for those subtype-specific elements, we downloaded C2 and C5 collections from Molecular Signatures Database (MSigDB) and performed gene set enrichment analysis (Subramanian et al., 2005). Functional annotation was determined using the hypergeometric test in R by investigating the overlap of genes in identified gene list with genes in archived gene sets. Multiple testing correction was conducted using the Benjamini-Hochberg method, and only those terms with corrected p-values less than 0.01 were called significantly over-represented.
Quantification and Statistical Analysis
The statistical significance of the overlap among any peak or gene sets was determined by the hypergeometric test. Comparison between different groups was tested using Fisher’s exact test for dichotomous variables and Mann-Whitney test for continuous variables.
Supplementary Material
Supplemental Information includes 6 figures and 6 tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.098.
Highlights.
Systematic exploration of the chromatin landscape and regulatory basis of AMLs
Identification of 2 main AML subtypes based on chromatin states
Distinct genetic mutations converge at the chromatin level
Chromatin signatures reveal diverse stemness phenotypes and cellular consequences
Acknowledgments
We thank all of the patients for their sample donations used in this study, and we acknowledge Dr. Eva van den Berg for assistance with the experiments. This work was supported by the BLUEPRINT project (European Union’s Seventh Framework Programme grant agreement no. 282510), the Dutch Children Cancer-Free Foundation (KIKA, project 311), the Netherlands, and the Italian Association Against Cancer ([AIRC] grant no. 17217).
Footnotes
Data and Software Availability
The custom codes used in this study are available at https://github.com/eleven919/JMartensLab.
Author Contributions
J.H.A.M., H.G.S., E.V., and L.A. conceived the study and designed the project; G.Y., A.T.J.W., F.P., and P.N. performed the bioinformatics analyses and data interpretation; E.M.J.-M., A. Mandoli, A. Merkel, K.B., B.K., F.M., A.A.S., E.H., K.H.M.P., A.B. Mulder, J.H.J., L.C., S.H., B.A.v.d.R., P.F., M.-L.Y., I.G., and C.B. contributed to the data collection, reagents usage, and setup of the experiments; G.Y., C.B., J.J.S., L.A., E.V., H.G.S., and J.H.A.M. wrote the manuscript. All of the authors read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
References
- Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, van den Berg E, Sikkema-Raddatz B, Abbott KM, Sinke RJ, Bungener LB, Mulder AB, Vellenga E. Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia. 2017;31:520–522. doi: 10.1038/leu.2016.310. [DOI] [PubMed] [Google Scholar]
- Bond J, Marchand T, Touzart A, Cieslak A, Trinquand A, Sutton L, Radford-Weiss I, Lhermitte L, Spicuglia S, Dombret H, et al. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. Haematologica. 2016;101:732–740. doi: 10.3324/haematol.2015.141218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers RJ, Currie T, Tholouli E, Rodig SJ, Kutok JL. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118:2857–2867. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- Cauchy P, James SR, Zacarias-Cabeza J, Ptasinska A, Imperato MR, Assi SA, Piper J, Canestraro M, Hoogenkamp M, Raghavan M, et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015;12:821–836. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim TK, He HH, Zieba J, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods. 2012;9:609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CT, Hess JL. Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene. 2016;35:1090–1098. doi: 10.1038/onc.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42–48. doi: 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Thé H. Lessons taught by acute promyelocytic leukemia cure. Lancet. 2015;386:247–248. doi: 10.1016/S0140-6736(15)61278-8. [DOI] [PubMed] [Google Scholar]
- Derrien T, Estellé J, Marco Sola S, Knowles DG, Raineri E, Guigó R, Ribeca P. Fast computation and applications of genome mappability. PLoS One. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JL, Hassane D, Wouters BJ, Kunimoto H, Avellino R, Garrett-Bakelman FE, Guryanova OA, Bowman R, Redlich S, Intlekofer AM, et al. Epigenetic Identity in AML Depends on Disruption of Nonpromoter Regulatory Elements and Is Affected by Antagonistic Effects of Mutations in Epigenetic Modifiers. Cancer Discov. 2017;7:868–883. doi: 10.1158/2159-8290.CD-16-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B, Dobin A, Stransky N, Li B, Yang X, Tickle T, Bankapur A, Ganote C, Doak T, Pochet N, et al. STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq. bioRxiv. 2017 doi: 10.1101/120295. [DOI] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- Jung N, Dai B, Gentles AJ, Majeti R, Feinberg AP. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat Commun. 2015;6 doi: 10.1038/ncomms9489. 8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, Boyle AP, Zhang QC, Zakharia F, Spacek DV, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, Li Y, Chung YR, Micol JB, Murphy ME, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Merkel A, Heath S, Queirós AC, Schuyler RP, Castellano G, Beekman R, Raineri E, Esteve A, Clot G, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, Baty JD, et al. The Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V, Mansmann U, Radmacher MD, Maharry KS, Sun M, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, Patel J, Dillon R, Vijay P, Brown AL, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016a;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H, et al. PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute Myeloid Leukemias Characterized by a Core Transcriptome of the MLL-Rearranged Disease. Cancer Res. 2016b;76:619–629. doi: 10.1158/0008-5472.CAN-15-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandoli A, Singh AA, Prange KHM, Tijchon E, Oerlemans M, Dirks R, Ter Huurne M, Wierenga ATJ, Janssen-Megens EM, Berentsen K, et al. The Hematopoietic Transcription Factors RUNX1 and ERG Prevent AML1-ETO Oncogene Overexpression and Onset of the Apoptosis Program in t(8;21) AMLs. Cell Rep. 2016;17:2087–2100. doi: 10.1016/j.celrep.2016.08.082. [DOI] [PubMed] [Google Scholar]
- Marco-Sola S, Sammeth M, Guigó R, Ribeca P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9:1185–1188. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MR, Corces MR, Eaton ML, Fiore C, Lee E, Lopez JT, Chen MW, Smith D, Chan SM, Koenig JL, et al. Superenhancer Analysis Defines Novel Epigenomic Subtypes of Non-APL AML, Including an RARα Dependency Targetable by SY-1425, a Potent and Selective RARα Agonist. Cancer Discov. 2017;7:1136–1153. doi: 10.1158/2159-8290.CD-17-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva YA, Lennartsson A, Ehsani R, Kulakovskiy IV, Vorontsov IE, Panahandeh P, Khimulya G, Kasukawa T, Drablos F. Epi-Factors: a comprehensive database of human epigenetic factors and complexes. Database (Oxford) 2015;2015 doi: 10.1093/database/bav067. bav067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahashi H, Kieffer Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- Petraglia F, Singh AA, Carafa V, Nebbioso A, Conte M, Scisciola L, Valente S, Baldi A, Mandoli A, Petrizzi VB, et al. Combined HAT/EZH2 modulation leads to cancer-selective cell death. Oncotarget. 2018;9:25630–25646. doi: 10.18632/oncotarget.25428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A, Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;30:1618–1619. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K, Zaba LC, Giresi PG, Li R, Longmire M, Kim YH, Greenleaf WJ, Chang HY. Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 2015;1:51–61. doi: 10.1016/j.cels.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendeiro AF, Schmidl C, Strefford JC, Walewska R, Davis Z, Farlik M, Oscier D, Bock C. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nat Commun. 2016;7 doi: 10.1038/ncomms11938. 11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AA, Mandoli A, Prange KH, Laakso M, Martens JH. AML associated oncofusion proteins PML-RARA, AML1-ETO and CBFB-MYH11 target RUNX/ETS-factor binding sites to modulate H3ac levels and drive leukemogenesis. Oncotarget. 2017;8:12855–12865. doi: 10.18632/oncotarget.14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AA, Petraglia F, Nebbioso A, Yi G, Conte M, Valente S, Mandoli A, Scisciola L, Lindeboom R, Kerstens H, et al. Multi-omics profiling reveals a distinctive epigenome signature for high-risk acute promyelocytic leukemia. Oncotarget. 2018;9:25647–25660. doi: 10.18632/oncotarget.25429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127:42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.