Abstract
Chronic lymphocytic leukemia (CLL) is a frequent hematological neoplasm in which underlying epigenetic alterations are only partially understood. Here we analyze the reference epigenome of seven primary CLLs and the regulatory chromatin landscape of 107 primary cases in the context of normal B-cell differentiation. We identify that the CLL chromatin landscape is largely influenced by distinct dynamics during normal B-cell maturation. Beyond this, we define extensive catalogues of regulatory elements de novo reprogrammed in CLL as a whole and in its major clinico-biological subtypes classified by IGHV somatic hypermutation levels. We uncover that IGHV-unmutated CLLs harbor more active and open chromatin than IGHV-mutated cases. Furthermore, we show that de novo active regions in CLL are enriched for NFAT, FOX and TCF/LEF transcription factor family binding sites. Although most genetic alterations are not associated with consistent epigenetic profiles, CLLs with MYD88 mutations and trisomy 12 show distinct chromatin configurations. Furthermore, we observe that non-coding mutations in IGHV-mutated CLLs are enriched in H3K27ac-associated regulatory elements outside accessible chromatin. Overall, this study provides an integrative portrait of the CLL epigenome, identifies extensive networks of altered regulatory elements and sheds light on the relationship between the genetic and epigenetic architecture of the disease.
Introduction
Over the last three decades, alterations in the epigenomic landscape have gradually emerged as an essential molecular feature of cancer cells, with implications in the pathogenesis, evolution, clinical behavior and therapy of virtually every tumor type1. Out of the broad variety of marks that make up the epigenetic portfolio2, DNA methylation has been the most widely studied in cancer1. In addition, few recent studies have started to analyze genome-wide maps of other marks such as histone modifications and chromatin accessibility3–9. However, the reference epigenome, as defined by the standards of the International Human Epigenome Consortium (IHEC, http://ihec-epigenomes.org/research/reference-epigenome-standards), of purified tumor cells from cancer patients has not been reported yet. Furthermore, given the essential link between the genome and epigenome in cancer development10,11, a comprehensive analysis of (non-)coding somatic mutations and the reference epigenome within the same cancer samples is needed to decipher their mutual relationships. Here, we present an integrative analysis of whole-genome maps of the DNA methylome, six histone modifications with non-overlapping functions (i.e. H3K4me3, H3K4me1, H3K27ac, H3K36me3, H3K9me3 and H3K27me3), chromatin accessibility, three-dimensional chromatin architecture, transcriptome and genome of chronic lymphocytic leukemia (CLL).
CLL is the most frequent leukemia in Western countries and is characterized by heterogeneous molecular features and clinical behaviour12,13. Overall, two major molecular subtypes can be distinguished based on the mutational status of the immunoglobulin variable region loci (IGHV), with those CLL patients having low mutation levels or unmutated IGHV (U-CLL) showing a more aggressive behavior than those with mutated IGHV (M-CLL)14,15. Similar to other neoplasms, the molecular portrait of CLL has mostly been characterized as individual layers of information, such as the genome, transcriptome, DNA methylome and chromatin accessibility8,16–22. Here, we have thoroughly analyzed the epigenome of CLL by sequencing the full reference epigenome of seven CLLs and the chromatin regulatory landscape of 100 additional cases, which were previously characterized by whole-genome and/or whole-exome sequencing (WGS/WES), RNA-seq and DNA methylation microarrays in the context of the International Cancer Genome Consortium (ICGC)20,23. This comprehensive dataset has allowed us to reveal novel insights into the biology and clinical behavior of CLL, and provides a rich resource for researchers studying gene regulation, cell differentiation, and cancer (epi)genomics.
Results
Reference epigenomes of CLL and normal B cells
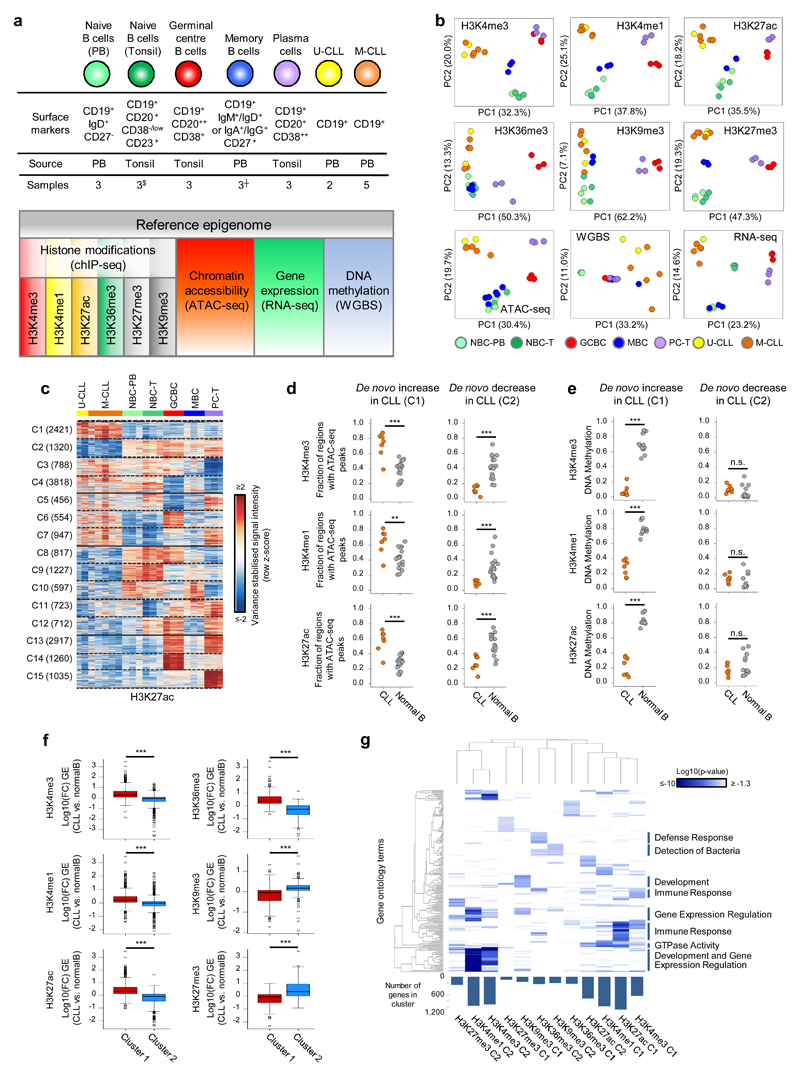
We have generated reference epigenomes, consisting of genome-wide maps of six histone marks, DNA accessibility, DNA methylation and gene expression, of seven representative CLLs, two U-CLL and five M-CLL cases, and five normal mature B-cell subpopulations covering different stages of the differentiation program (Fig. 1a). We confirmed sample identity by comparing the genetic fingerprint of each patient obtained by SNP arrays with genotypes extracted from ChIP-seq, ATAC-seq, WGBS and RNA-seq data. Patient characteristics can be found in Supplementary Table 1. Unsupervised analyses of each layer of the reference epigenome revealed differences both between neoplastic CLLs and B cells, and within normal B cell subpopulations, which showed maturation stage-specific epigenomic profiles (Fig. 1b). We further characterized the dynamics of the six histone modifications and DNA accessibility in CLL and the five normal B-cell subpopulations by K-means clustering. Overall, we identified a mean of 2,729 regions (ranging from 533 to 8,444 depending on the mark, representing from 4.8 to 19.3% of all regions) whose levels were stable in normal B cells and either increased (cluster 1, C1) or decreased (cluster 2, C2) specifically in CLL as a whole (Fig. 1c, Supplementary Fig. 1-2 and Supplementary Table 2). This finding indicates that CLL cells show a global de novo reconfiguration of their chromatin, affecting histone marks with non-overlapping functions as well as chromatin accessibility. In addition, as previously reported16,19, we observed that de novo DNA hypomethylation is more frequent than DNA hypermethylation in the studied CLLs (Supplementary Fig. 3 and Supplementary Table 3). Beyond these findings, we also observed that the CLL chromatin landscape can be linked to different modulation patterns occurring during the normal B-cell differentiation process (Fig. 1c, Supplementary Fig. 2 and Supplementary Table 2). These included regions with similarities to naive (NBCs) and memory B cells (MBCs), which have been proposed as potential cells of origin of CLL12, and regions showing unexpected associations with germinal centre B cells (GCBCs) and plasma cells (PCs), which have not been described to share molecular features with CLL (e.g. C6 and C7 in Fig. 1c). As expected based on the epigenetic patterns shown above, we also observed de novo increase and decrease of gene expression in CLL as well as different modulation patterns of gene expression levels in relation to normal B cells (Supplementary Fig. 4 and Supplementary Table 2). To provide insights into the interplay between histone marks and other layers of the reference epigenome, next we analyzed chromatin accessibility, DNA methylation and gene expression levels of protein coding genes in regions undergoing de novo changes of each histone mark in CLL (Fig. 1d-f, Supplementary Fig. 5, Supplementary Table 4). Regions with de novo increase (C1) of histone marks related to promoters and enhancers (H3K4me3, H3K4me1 and H3K27ac) showed a corresponding increase of chromatin accessibility, decreased DNA methylation and increased expression of the associated genes in CLLs (Fig. 1d-f and Supplementary Fig. 1a). Regions with de novo decrease of these marks (C2) showed an expected decrease in accessibility and gene expression in CLL (Fig. 1d, f), whereas DNA methylation levels were consistently low in all normal and leukemic samples in these regions (Fig. 1e and Supplementary Fig. 1b). Thus, those regulatory regions becoming inactive in CLL do not gain DNA methylation but maintain an imprint of their past activity, supporting the concept that DNA methylation is mostly an accumulative trait24, holding cellular memory of past activity. In contrast, the chromatin configuration of regulatory elements is more dynamic and closely related to transcriptional changes.
Fig. 1. CLL reference epigenomes.
(a) Overview of analyzed CLL and normal B-cell samples (upper panel) for the nine layers of the reference epigenome (lower panel). $no whole-genome bisulfite sequencing data available; ┼six instead of three biologically independent samples analyzed for chromatin accessibility. (b) Unsupervised principal component analysis for the nine layers of the reference epigenome. Number of datapoints analyzed to generate the PCAs: H3K4me3 (n=38,499 independent genomic regions), H3K4me1 (n=37,871 independent genomic regions), H3K27ac (n=47,191 independent genomic regions), H3K36me3 (n=15,561 independent genomic regions), H3K9me3 (n=27,371 independent genomic regions), H3K27me3 (n=12,878 independent genomic regions), ATAC-seq (n=91,671 independent genomic regions), WGBS (n=15,825,190 independent CpGs), RNA-seq (n=36,190 independent genes). Sample sizes were for U-CLL: n=2 biologically independent samples (all nine layers), for M-CLL: n=5 biologically independent samples (all nine layers), for NBC-PB, GCBC and PC-T: n=3 biologically independent samples (all nine layers), for NBC-T: n=3 biologically independent samples (all layers except WGBS that does not include NBC-T), for MBC: n=3 biologically independent samples (all layers except ATAC-seq for which 6 biologically independent samples were used). (c) K-means clustering of independent genomic regions showing differences in the dynamics of H3K27ac levels in CLL and normal B cells. For each cluster (C1-C15) the number of independent genomic regions is indicated in brackets. C1 and C2 respectively represent regions with de novo increase and de novo decrease in CLL. (d) Fraction of regions in CLL (n=7 biologically independent samples) and normal B cells (n=15 biologically independent samples) harboring ATAC-seq peaks in regions with de novo increase (C1) or de novo decrease (C2) in CLL of H3K4me3 (respective P-values 5.5 x 10-4 and 4.2 x 10-6), H3K4me1 (respective P-values 6.1 x 10-3 and 2.9 x 10-5) and H3K27ac (respective P-values 5.5 x 10-4 and 1.9 x 10-4). P-values were calculated using a Wilcoxon rank sum test (two-sided). (e) Median DNA methylation levels in CLL (n=7 biologically independent samples) and normal B cells (n=15 biologically independent samples) of regions with de novo increase (C1) or de novo decrease (C2) in CLL of H3K4me3 (respective P-values 4.5 x 10-4 and 1.6 x 10-1), H3K4me1 (respective P-values 4.5 x 10-4 and 1.6 x 10-1) and H3K27ac (respective P-values 4.5 x 10-4 and 4.2 x 10-1). P-values were calculated using a Wilcoxon rank sum test (two-sided). (f) Boxplots of log10 transformed fold changes (FC) in gene expression (GE) levels in CLL versus normal B cells of all genes located within regions with de novo increase (cluster 1, C1) or de novo decrease (cluster 2, C2) in CLL. For each gene the mean log10 transformed GE levels of CLL (n=7 biologically independent samples) and normal B cells (n=15 biologically independent samples) were calculated and subtracted to obtain the log10 transformed FC between CLL and normal B cells. H3K4me3 (P-value 8.2 x 10-77, mean, minimum, 25th, 50th and 75th percentile and maximum log10(FC) and number of datapoints (= independent genes) C1: 0.43, -1.85, 0.09, 0.29, 0.65, 3.47, 624 and C2: -0.15, -3.62, -0.33, -0.04, 0.10, 1.41, 911), H3K4me1 (P-value 3.9 x 10-50, mean, minimum, 25th, 50th and 75th percentile and maximum log10(FC) and number of datapoints (= independent genes) C1: 0.29, -1.42, 0.05, 0.21, 0.49, 3.47, 971 and C2: -0.05, -2.09, -0.23, -0.02, 0.10, 2.27, 952), H3K27ac (P-value 5.3 x 10-137, mean, minimum, 25th, 50th and 75th percentile and maximum log10(FC) and number of datapoints (= independent genes) C1: 0.44, -1.05, 0.12, 0.32, 0.64, 3.47, 1,081 and C2: -0.25, -2.42, -0.46, -0.09, 0.09, 1.63, 713), H3K36me3 (P-value 1.1 x 10-52, mean, minimum, 25th, 50th and 75th percentile and maximum log10(FC) and number of datapoints (= independent genes) C1: 0.52, -0.65, 0.19, 0.34, 0.72, 3.47, 233 and C2: -0.37, -2.32, -0.68, -0.26, 0.01, 1.13, 235), H3K9me3 (P-value 3.3 x 10-10, mean, minimum, 25th, 50th and 75th percentile and maximum log10(FC) and number of datapoints (= independent genes) C1: -0.16, -1.73, -0.44, -0.04, 0.07, 1.32, 160 and C2: 0.16, -1.91, 0.06, 0.17, 0.30, 1.74, 206) and H3K27me3 (P-value 3.0 x 10-17, mean, minimum, 25th, 50th and 75th percentile and maximum log10(FC) and number of datapoints (= independent genes) C1: -0.22, -2.32, -0.51, -0.06, 0.12, 0.98, 92 and C2: 0.52, -0.93, 0.00, 0.35, 0.93, 3.47, 262). P-values were calculated using a Student's t-test (two-sided). (g) Heatmap of p-values of gene ontology (GO) terms (rows, n= 190 independent GO terms, only the top 20 terms per cluster were included) that were significantly enriched (p-value < 0.05) among the genes overlapping with regions with de novo increase (C1) or de novo decrease (C2) of the six histone marks in CLL. The GO term enrichment and significance were calculated per cluster separately. The number of independent genes per cluster used in this calculation is indicated below the heatmap, their exact numbers were: H3K4me3 (C1: 624, C2: 911), H3K4me1 (C1: 971, C2: 952), H3K27ac (C1: 1,081, C2: 713), H3K36me3 (C1: 233, C2: 235), H3K9me3 (C1: 160, C2: 206) and H3K27me3 (C1: 92, C2: 262). U-CLL, CLL with unmutated IGHV; M-CLL, CLL with mutated IGHV; NBC-PB, naive B cell from peripheral blood; NBC-T, naive B cell from tonsil; GCBC, germinal centre B cell; MBC, memory B cell; PC-T, plasma cell from tonsil; GE, gene expression.
In terms of functional categories, the genes showing de novo increase or decrease of specific histone marks in CLL were involved in different functions, i.e. genes with increased levels of H3K27ac, H3K4me3 and H3K4me1 were related to immune response mechanisms and GTPase activity, while those with decreased levels of H3K4me3 and H3K4me1, tended to be involved in organism development and gene expression regulation (Fig. 1g and Supplementary Table 5).
Chromatin state transitions from normal B cells to CLL
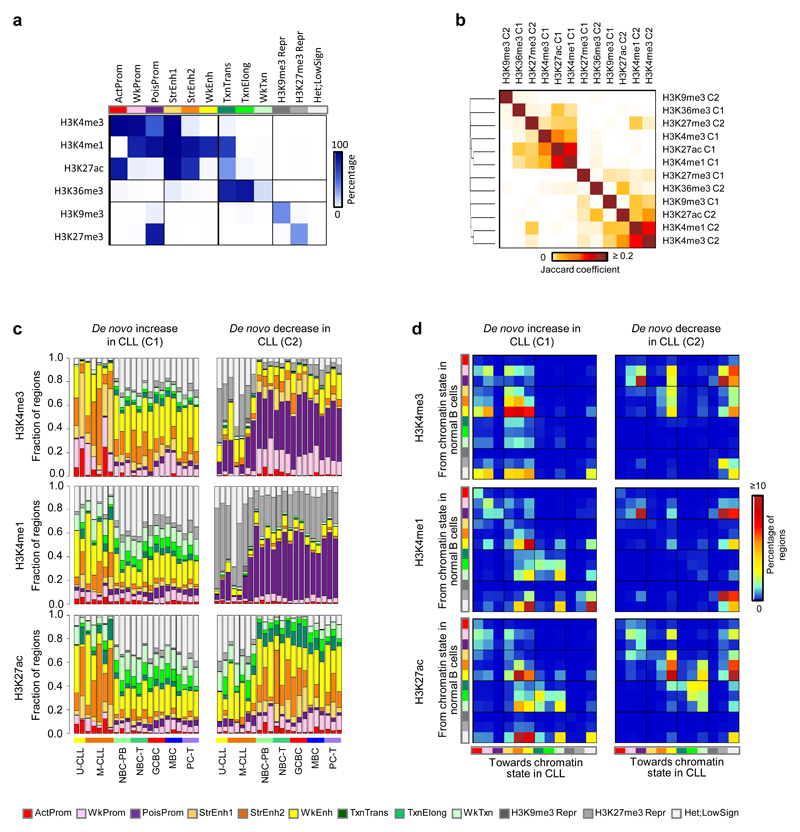
The previous results revealed an extensive modification of the CLL chromatin landscape as compared to normal B cells. To capture overlapping and mutually-exclusive patterns of the different histone modifications25, we generated a chromatin state model specific for B cells using chromHMM26 (Fig. 2a). First of all, with this model we studied the overall relationship between CLL and normal B cells based on the integrative chromatin landscape using the percentage of overlap among chromatin states. As observed previously for the separate histone mark layers of the reference epigenome (Fig. 1b), CLL overall shows the highest resemblance to normal naive and memory B cells (Supplementary Fig. 6). Next, we analyzed the regions with CLL-specific increased or decreased histone mark levels in an integrative manner. We observed that regions with gains of H3K4me3, H3K4me1 and H3K27ac in CLL tended to coincide with each other and to a lesser extent with regions with increased H3K36me3 and decreased H3K27me3 levels. Furthermore, decrease of H3K4me3 and H3K4me1 co-occurred and partially coincided with the loss of H3K27ac and the gain of H3K9me3 (Fig. 2b). Next, we used the chromatin state model to analyze the impact of CLL-specific histone mark, DNA accessibility and DNA methylation alterations on chromatin states (Fig. 2c, Supplementary Fig. 7 and Supplementary Table 6). Globally, we observed that increase or decrease of H3K27ac in CLL was associated with a corresponding increase or decrease of active enhancers and promoters (Fig. 2c). Similarly, increased H3K4me1 and H3K4me3 levels were related to an increase of enhancers and promoters in CLL, respectively (Fig. 2c). Mapping specific chromatin state transitions from normal B cells to CLL (Fig. 2d and Supplementary Fig. 8), we observed that the gain of active enhancers in CLL, upon the increase of H3K4me3, H3K4me1 and H3K27ac, mainly originated from regions classified as weak enhancers or heterochromatin-low signal in normal B cells (Fig. 2d). These data suggest that some fully activated enhancers in CLL are primed in normal B cells, while others become enhancers de novo upon malignant transformation.
Fig. 2. Chromatin states and its transitions in CLL.
(a) Emissions of the generated chromatin state model. Represented are the percentages of regions assigned to a specific chromatin state (columns) that contain a specific histone mark (rows). (b) Jaccard coefficients of genomic regions that show de novo increase (C1) or de novo decrease (C2) of the six different histone marks in CLL. Number of regions analyzed: H3K4me3 C1 (n=1,170 independent regions), H3K4me3 C2 (n=1,423 independent regions), H3K4me1 C1 (n=1,418 independent regions), H3K4me1 C2 (n=1,198 independent regions), H3K27ac C1 (n=2,421 independent regions), H3K27ac C2 (n=1,320 independent regions), H3K36me3 C1 (n=285 independent regions), H3K36me3 C2 (n=251 independent regions), H3K9me3 C1 (n=344 independent regions), H3K9me3 C2 (n=293 independent regions), H3K27me3 C1 (n=208 independent regions), H3K27me3 C2 (n=325 independent regions). (c) Distribution of the different chromatin states in all analyzed samples separately (seven CLLs and 15 normal B cells) at regions with de novo increase (C1) or de novo decrease (C2) of H3K4me3, H3K4me1 and H3K27ac in CLL. (d) Chromatin state transitions from B cells to CLL. Percentages of regions with de novo increase (C1) or de novo decrease (C2) of H3K4me3, H3K4me1 and H3K27ac in CLL that harbor a specific chromatin state in normal B cells (rows, n=15 biologically independent samples) and the same (diagonal, no change of chromatin state) or another state (chromatin state switch) in CLL (columns, n=7 biologically independent samples). The total matrix represents 100 percent of the regions. U-CLL, CLL with unmutated IGHV; M-CLL, CLL with mutated IGHV; NBC-PB, naive B cell from peripheral blood; NBC-T, naive B cell from tonsil; GCBC, germinal centre B cell; MBC, memory B cell; PC-T, plasma cell from tonsil. ActProm, Active Promoter; WkProm, Weak Promoter; PoisProm, poised Promoter; StrEnh1, Strong Enhancer 1; StrEnh2, Strong Enhancer 2; WkEnh, Weak Enhancer; Txn_Trans, Transcription Transition; Txn_Elong, Transcription Elongation; Wk_Txn, Weak Transcription; H3K9me3_Repr, H3K9me3 Repressed; H3K27me3_Repr, H3K27me3 Repressed; Het;LowSign, Heterochromatin;Low Signal.
We also observed that a decrease in H3K4me3 and H3K4me1 in CLL did not alter active regulatory elements, but rather led to a major decrease of poised promoters (Fig. 2c), which mostly became H3K27me3-repressed chromatin in CLL (Fig. 2d). In addition, CLL-specific decrease of H3K27me3 also lead to loss of poised state in a low percentage of the regions, either becoming active (i.e. changing towards weak or active promoters) or inactive (i.e. changing towards heterochromatin-low signal) in CLL (Supplementary Fig. 8). Loss of the poised promoter state seems a general phenomenon in CLL, as a significantly lower percentage of the genome was covered by this chromatin state in CLL as compared to normal B cells (0.008-0.399% vs. 0.232-0.610%, P < 1 x 10-3, two-sided Wilcoxon rank sum test). The transition from poised promoters in normal B cells into stably repressed chromatin in CLL may represent a loss of epigenetic plasticity in CLL without an apparent impact on gene activity. This was for example reflected by the fact that a significantly larger number of the genes decreasing H3K4me3 (n=406 out of 911, 44.6%, P < 1 x 10-3, Fisher’s exact test) or H3K4me1 levels (n=509 out of 952, 53.5%, P < 1 x 10-3, Fisher’s exact test) were neither expressed in CLL nor in normal B cells, as compared to the total number of protein coding genes showing this gene expression pattern (6,186 out of 21,257 genes, 29.1%) (Supplementary Fig. 9). An additional observation supporting the loss of plasticity at these regions in CLL was the fact that they were associated with genes enriched for various gene ontology terms related to organism development (Fig. 1g), which are inactive but remain poised in mature B cells.
Identification of altered regulatory regions involved in CLL pathogenesis
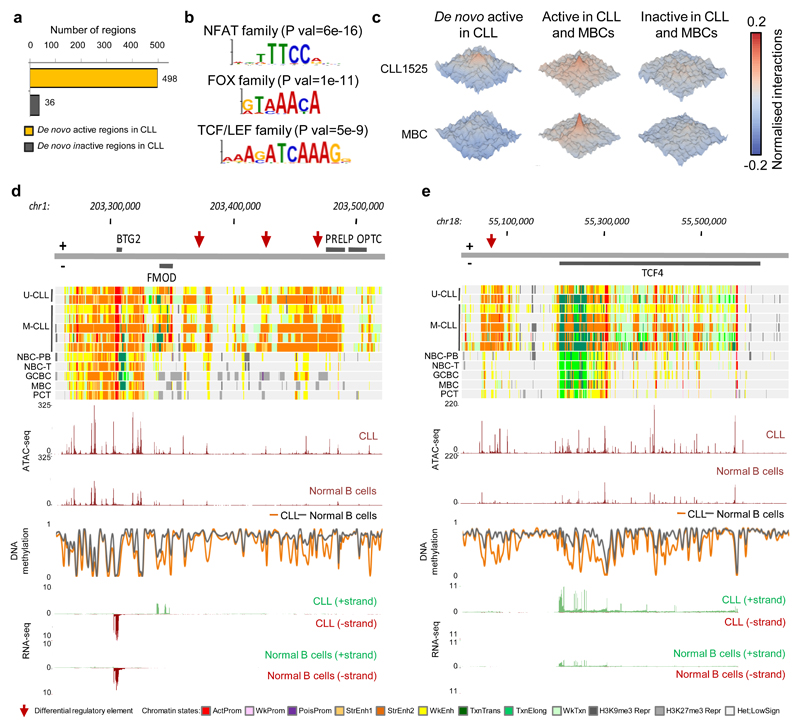
We next designed a stringent approach (Supplementary Fig. 10) to distil, from the previous global analyses, a set of altered regulatory regions that may play an important role in CLL pathogenesis. Using this approach, we detected 534 genomic regions that consistently gained or lost regulatory activity in all seven CLLs as compared to all normal B cells. The majority of these regions (n=498, 93.3%) showed a de novo activation of regulatory elements (Fig. 3a and Supplementary Table 7), which were significantly enriched in super-enhancers27 (n=51 super-enhancers out of 498 regions (10.2%) as compared to the background of n=350 super-enhancers out of 7,121 regions (0.5%), P < 1 x 10-3, Fisher’s exact test). In contrast, we only identified one super-enhancer showing loss of activity in CLL, located within the CLL-silenced gene EBF128. To explore whether de novo changes in chromatin are mediated by specific transcription factors (TF), we mined the regions of interest for TF binding sites. Remarkably, we observed that de novo active chromatin regions were highly enriched for binding motifs of NFAT, FOX and TCF/LEF TF families (Fig. 3b and Supplementary Table 8). These data indicate that chromatin activation, in particular affecting super-enhancers, is an epigenetic feature of CLL, and seems to be mediated by specific TF families. Furthermore, as regions with higher chromatin activity tend to have a higher number of local three-dimensional (3D) chromatin interactions29, we generated in situ HiC-seq30 data in one out of the seven CLLs and MBCs to study this phenomenon. De novo active regions in CLL showed higher levels of local 3D interactions in CLL as compared to MBCs, indicating that chromatin activation in CLL also involves a reconfiguration of the local 3D architecture (Fig. 3c).
Fig. 3. CLL specific regulatory landscape.
(a) Number of independent genomic regions with de novo gain or loss of regulatory elements in CLL. (b) Binding motifs of NFAT, FOX and TCF/LEF transcription family members, which are highly enriched in the accessible loci of the de novo active regions (n=934 independent genomic loci) versus the background (n=1,868 independent genomic loci). Statistical significance was determined using the one-tailed Wilcoxon rank-sum test and the p-values were adjusted using the Bonferroni correction. Out of the list of all enriched TF motifs (Supplementary Table 8), we considered only those expressed in the seven CLLs with reference epigenomes. (c) Normalized interaction frequencies of 3D chromatin interactions within a 100kb window in CLL1525 (upper row) and memory B cells (MBCs, lower row) in regions that are de novo active in CLL (left panels), active in CLL and MBCs (middle panels) and inactive in both (right panels). (d and e) Examples of identified de novo active regions in CLL (red arrows), targeting FMOD (d) and TCF4 (e). Indicated are in the upper panels the chromatin states in all seven biologically independent CLLs and representative samples of each of the normal B-cell subpopulations and below this the median ATAC-seq, DNA methylation and RNA-seq levels of the seven biologically independent CLLs and 15 biologically independent normal B cells. U-CLL, CLL with unmutated IGHV; M-CLL, CLL with mutated IGHV; NBC-PB, naive B cell from peripheral blood; NBC-T, naive B cell from tonsil; GCBC, germinal centre B cell; MBC, memory B cell; PC-T, plasma cell from tonsil. ActProm, Active Promoter; WkProm, Weak Promoter; PoisProm, poised Promoter; StrEnh1, Strong Enhancer 1; StrEnh2, Strong Enhancer 2; WkEnh, Weak Enhancer; Txn_Trans, Transcription Transition; Txn_Elong, Transcription Elongation; Wk_Txn, Weak Transcription; H3K9me3_Repr, H3K9me3 Repressed; H3K27me3_Repr, H3K27me3 Repressed; Het;LowSign, Heterochromatin;Low Signal.
Next, we linked the detected regions to their target genes by a multi-step approach using both linear and 3D proximity, measured by promoter capture Hi-C of one of the seven CLL cases (generated within this study) and normal B cells (previously published)31 and consequent correlation with gene expression (Supplementary Figure 11a). A total of 275 target genes were assigned to the 534 detected regions (Supplementary Figure 11b and Supplementary Table 7). Globally, those genes related to de novo active regions are involved in surface receptor signalling, response to bacteria/lippopolysaccharide, lymphoid organ development as well as cell adhesion and activation (Supplementary Figure 11c and Supplementary Table 5). More specifically, the list of 275 target genes included 11 out of 14 genes (e.g. EBF1, FMOD and LEF1) whose differential expression has been shown to be specific for CLL as compared to other B cell neoplasms32,33. Therefore, we have identified the genome-wide regulatory regions that control the specific transcriptional program of CLL, and distinguish the disease from normal B-cell differentiation. This information represents a solid background to investigate the onco-epigenetic mechanisms underlying leukemic transformation.
The potential role of the 534 identified regions in distant gene regulation, which is a distinctive feature of enhancers, became apparent from the fact that 41.8% (n=223 out of 534) were assigned to one or more distant target genes. For two of these distant target genes, FMOD, a bona fide gene whose expression has diagnostic power in CLL32,33 and TCF4, which encodes a transcription factor involved in the WNT signalling pathway reported to be over-expressed in CLL34, we exemplary show the identified regulatory elements (Fig. 3d, e and Supplementary Fig. 12). Both for the target gene locus and for the regulatory elements, higher chromatin accessibility (ATAC-seq) and lower DNA methylation levels were observed in CLL as compared to normal B cells. Furthermore, by 4C-seq in two CLL cases, we observed that these distant super-enhancers showed 3D interactions with the FMOD and TCF4 promoter (Supplementary Fig. 12), further confirming that these are their target genes in CLL. Interestingly, an upstream TCF4 super-enhancer has been identified in plasmacytoid dendritic cell neoplasms35, while the CLL-associated super-enhancer is located downstream of the gene. These findings suggest the existence of disease-specific enhancer deregulation leading to similar downstream transcriptional effects (e.g. TCF4) or disease-specific transcriptional deregulation (e.g. FMOD).
The regulatory chromatin landscape of clinico-biological CLL subgroups
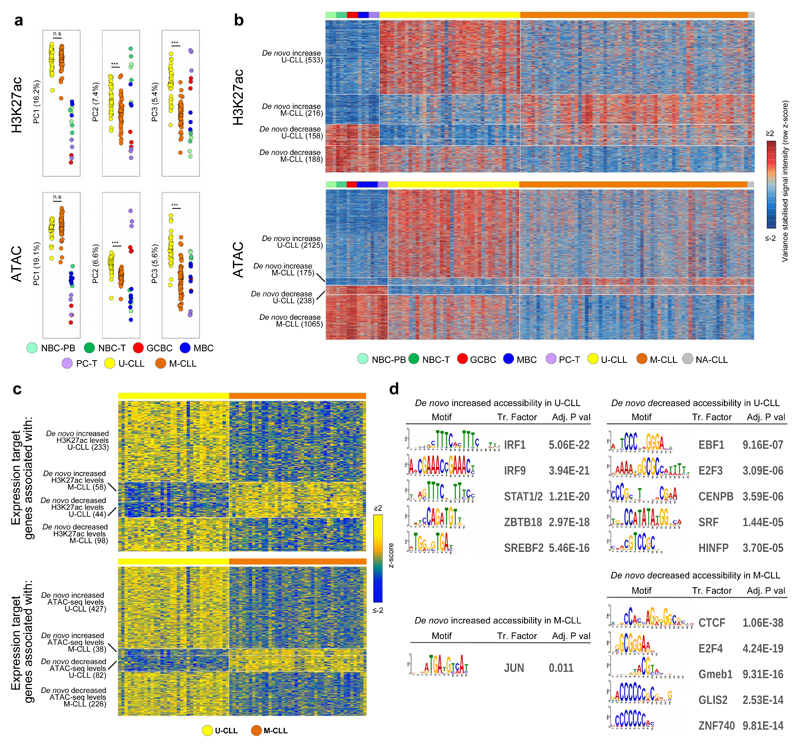
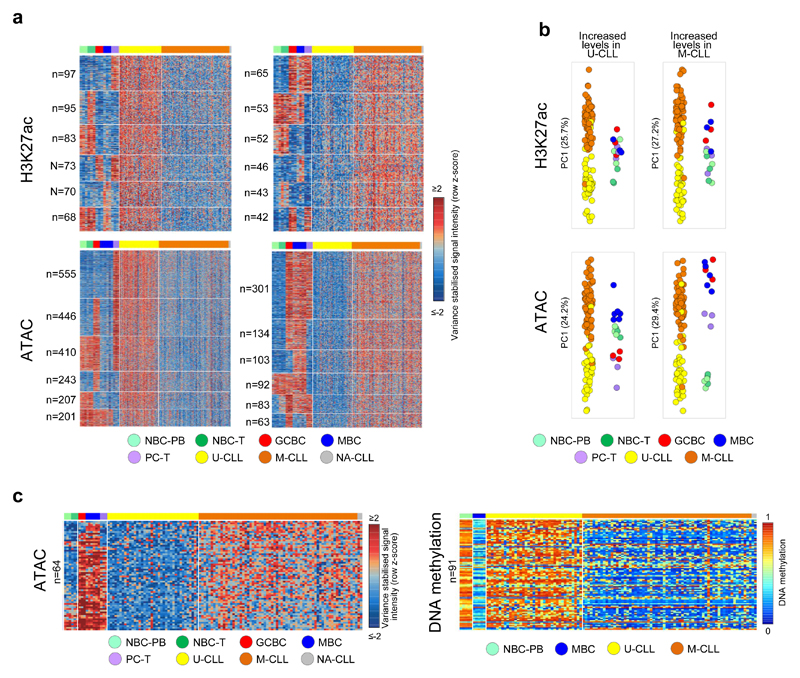
The previous analyses did not have sufficient power to distinguish specific epigenetic modifications that may drive the clinico-biological heterogeneity of CLL, specifically of the two molecular subtypes U-CLL and M-CLL14,15. Therefore, we performed ChIP-seq for H3K27ac and ATAC-seq in 100 additional CLL cases (37 U-CLLs, 61 M-CLLs and two CLLs with unknown IGHV mutation status), bringing the total sample size for these marks to 107 cases. In line with the validation analysis performed in the seven CLLs with reference epigenomes, we also confirmed sample identity of the 100 additional cases. Patient characteristics can be found in Supplementary Table 1. This CLL cohort was extensively characterized previously in the context of the ICGC using RNA-seq (n=78), DNA methylation arrays (n=105), copy number arrays (n=105) and WES and/or WGS (n=105)20. Unsupervised principal component analysis of H3K27ac and ATAC-seq data confirmed that the main source of variability was the difference between CLL as a whole and normal B cells (Fig. 4a). In contrast, the second and third component showed significant differences between U-CLL and M-CLL (Fig. 4a), indicating that a major fraction of chromatin variability is associated with the clinical heterogeneity in CLL patients. Next, we compared U-CLL and M-CLL, and identified 2,818 and 8,803 significant differential regions for H3K27ac and ATAC-seq, respectively (Supplementary Table 9). Overall, the majority of these regions showed higher levels of these marks in U-CLLs, suggesting that clinical aggressiveness in CLL is associated with a more accessible and active chromatin. In addition to the immunogenetic classification of CLL, we also compared the chromatin profiles of a DNA methylation-based CLL classification comprising three clinico-biological entities named NBC-like, MBC-like and intermediate CLLs16,19,36. We observed that the chromatin landscapes of MBC-like and intermediate CLLs (both M-CLL) were distinct from NBC-like CLLs (i.e. U-CLL) but similar to each other (Supplementary Fig. 13), reflecting that the IGHV mutation status is a strong determinant of the regulatory chromatin landscape of CLL.
Fig. 4. De novo chromatin activity and accessibility changes in an extended CLL cohort.
(a) Unsupervised principal component analysis (first three components) of the extended CLL cohort. Number of datapoints analyzed to generate the PCAs: H3K27ac (n=58,790 independent genomic regions) and ATAC-seq (n=115,352 independent genomic regions). Respective P-values for H3K27ac between U-CLL (n=39 biologically independent samples) and M-CLL (n=63 biologically independent samples) of PC1, PC2 and PC3 were 8.4 x 10-1, 6.5 x 10-6 and 4.3 x 10-16) and for ATAC-seq between U-CLL (n=38 biologically independent samples) and M-CLL (n=66 biologically independent samples) of PC1, PC2 and PC3 were 1.5 x 10-1, 9.5 x 10-10 and 5.2 x 10-16). P-values were calculated using a Student's t-test (two-sided). (b) Heatmap of signal intensities of H3K27ac and ATAC-seq in regions that show a de novo change in levels of these marks in U-CLL and M-CLL. Signal intensities are indicated as row z-scores. On the left the number of independent regions per cluster is indicated. (c) Heatmap of gene expression levels of target genes associated with regions that show de novo change in H3K27ac (activity) or ATAC-seq (accessibility) levels in U-CLL and M-CLL. Gene expression levels are indicated as row z-scores. On the left the number of independent target genes is indicated. (d) Top five enriched transcription factor binding sites in regions that show a de novo change in ATAC-seq levels in U-CLL and M-CLL. Out of the list of all enriched TF motifs (Supplementary Table 8), we considered only those expressed in the CLL subgroup with higher accessibility levels. Number of regions analyzed vs. background were: de novo increased accessibility in U-CLL (n= 2,125 vs. 4,250 independent genomic regions) or M-CLL (n=175 vs. 350 independent genomic regions) and de novo decreased accessibility in U-CLL (n=238 vs. 476 independent genomic regions) or M-CLL (n=1,065 vs. 2,130 independent genomic regions). Statistical significance was determined using the one-tailed Wilcoxon rank-sum test and the p-values were adjusted using the Bonferroni correction. U-CLL, CLL with unmutated IGHV; M-CLL= CLL with mutated IGHV; NBC-PB, naive B cell from peripheral blood; NBC-T, naive B cell from tonsil; GCBC, germinal centre B cell; MBC, memory B cell; PC-T, plasma cell from tonsil.
To properly interpret the pathogenic relevance of the differences between U-CLL and M-CLL, we analyzed them in the context of the normal B-cell differentiation. We observed that 38.9% of the differences in H3K27ac (n=1,095 out of 2,818) and 40.9% of the differentially accessible regions (n=3,603 out of 8,803) were stable during B-cell differentiation (Fig. 4b and Supplementary Table 9). Hence, these regions represented subtype-specific epigenetic alterations with de novo increase or decrease of regulatory activity in U-CLL or M-CLL. Using the previously explained strategy (Supplementary Fig. 11a), we identified the target genes of the de novo changes of activity/accessibility in U-CLL and M-CLL (Fig. 4c), which were enriched for distinct biological functions (Supplementary Table 5). Notably, de novo altered chromatin accessibility in U-CLL and M-CLL was associated with markedly different TF motifs (Fig. 4d and Supplementary Table 8). Regions gaining accessibility in U-CLL were enriched in binding sites of multiple TFs including the IRF TF family, whereas regions losing accessibility in M-CLL were highly enriched for CTCF binding sites, suggesting that U-CLL and M-CLL may show differential 3D architectures.
In addition to the regions de novo changing in U-CLL or M-CLL, we identified that the activity/accessibility of 60% of all differential regions was extensively modulated during normal B-cell differentiation. From the DNA methylation perspective, differences in U-CLL and M-CLL have previously been assigned to an epigenetic imprint of their cell of origin, i.e. GC-inexperienced and GC-experienced cells, respectively16. From the chromatin perspective, however, we observed a more complex scenario and we categorized the regions with differential chromatin into 30 patterns based on the similarities of U-CLL or M-CLL to different dynamics during normal B-cell differentiation (Fig. 5a with results of six main patterns and Supplementary Table 9). B-cell dynamic regions with differential H3K27ac showed various patterns without a clear bias of CLLs towards particular normal subpopulations (Fig. 5b). In contrast, the first principal component of B-cell dynamic regions with higher accessibility in M-CLL showed expected cell of origin-based similarities, i.e. U-CLLs derive from cells that have matured outside the germinal center and still maintain a naive-like chromatin accessibility whereas M-CLL stem from GC-experienced cells and thus show similarities to GCBC, MBCs and PCs (Fig. 5b). These differentially accessible regions partially overlapped with the previously identified CLL cell of origin DNA methylation signature16, and showed concordant higher levels of ATAC-seq and lower levels of DNA methylation in M-CLL in comparison with U-CLL (Fig. 5c and Supplementary Table 10). These results imply that both CLL subtypes retain a DNA methylation and chromatin accessibility imprint of their differential cellular origins.
Fig. 5. B cell related chromatin activity and accessibility signatures in the extended CLL cohort.
(a) Heatmap of the signal intensities of H3K27ac and ATAC-seq at differential regions between U-CLL and M-CLL that show dynamic modulation of these marks in normal B cells. Signal intensities are indicated as row z-scores. For each change (up in U-CLL (left panels) or down in U-CLL (right panels)) and each mark the six main (out of the 30 possible) dynamic patterns are shown. On the left the number of independent regions per cluster is indicated. (b) Principal component analysis of all regions that show differential changes in U-CLL versus M-CLL and dynamic modulation in normal B cells. In this case, all regions of all 30 dynamic patterns were included in the analysis, number of datapoints analyzed to generate the PCAs: H3K27ac (n=1,723 independent genomic regions) and ATAC-seq (n=5,200 independent genomic regions). Sample sizes: U-CLL (n=39 biologically independent samples for H3K27ac and 38 for ATAC-seq), M-CLL (n=63 biologically independent samples for H3K27ac and 66 for ATAC-seq), NBC-PB, NBC-T, GCBC and PC-T (n=3 biologically independent samples for H3K27ac and ATAC-seq), MBC (n=3 biologically independent samples for H3K27ac and 6 for ATAC-seq). (c) (left panel) Heatmap of signal intensities of ATAC-seq in the 64 independent genomic regions that show differential higher levels in M-CLL compared to U-CLL that overlap with the previously defined 1,649 CpG signature. Signal intensities are indicated as row z-scores. (right panel) Heatmap of DNA methylation estimates of the 91 independent CpGs that overlap with the ATAC-seq regions represented in the left panel. U-CLL, CLL with unmutated IGHV; M-CLL= CLL with mutated IGHV; NBC-PB, naive B cell from peripheral blood; NBC-T, naive B cell from tonsil; GCBC, germinal centre B cell; MBC, memory B cell; PC-T, plasma cell from tonsil.
Interestingly, the analysis of the B-cell dynamic regions with higher ATAC-seq levels in U-CLL uncovered a relationship between U-CLL and PCs and GCBCs, while M-CLL resembled more NBCs and MBCs (Fig. 5b), which also became apparent in our initial unsupervised analysis (Fig. 4a). The gene ontology analysis of the target genes of active and accessible regions shared by U-CLL, PCs and GCBCs, suggests that the similarities between these cells may be related, among others, to cell cycle regulation and (wnt) signalling (Supplementary Table 5). One example of a gene showing this activation pattern is GFI1 (Supplementary Fig. 14), which encodes a protein involved in cell cycle regulation and becomes up regulated in mature B cells upon antigen stimulation and shows oncogenic activity in aggressive T cell leukemias37,38. These findings suggest that beyond linking chromatin patterns of U-CLL and M-CLL to their cellular origins, chromatin variability in CLL subtypes and normal B cells also seem to reflect different biological behaviors. For instance, U-CLL activates genes operative in proliferative B cell subpopulations such as GCBCs and tonsillar PCs. This phenomenon suggests that U-CLLs may exploit molecular mechanisms present in specific normal B cell subpopulations to achieve higher proliferation39.
Linking somatic genetic changes and the chromatin landscape in CLL
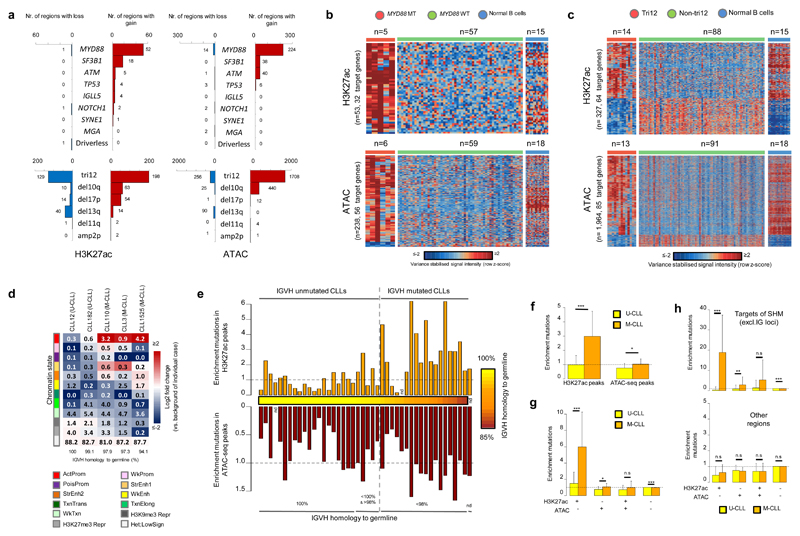
Our thoroughly characterized CLL samples20 provide an opportunity to shed light onto the relationship between chromatin activity/accessibility and somatic genetic changes in CLL. First of all, we investigated whether alterations in 14 common driver genes and copy number variants in CLL (selected based on the presence in at least five cases in our series) were related to specific chromatin signatures by comparing affected vs. non-affected cases (Fig. 6a and Supplementary Table 11). MYD88 mutations showed a consistent pattern of de novo chromatin activation or accessibility associated with over expression of a total of 67 unique target genes, including genes encoding proteins previously linked to NF-kappaB signalling, such as CBLB, PIM1, TNFRSF13B and TNFRSF2140–43 (Fig. 6b and Supplementary Table 12). Similarly, cases with trisomy 12 showed extensive changes in chromatin patterns as compared to unaffected cases, but were intriguingly similar to normal B cells (Fig. 6c and Supplementary Table 12). The broad spectrum of genetic features in CLL also includes driver-less cases, which are CLLs lacking recognized genetic drivers (mainly M-CLLs)18,20. In our series, driver-less (n=15) cases did not show any specific chromatin pattern as compared to other M-CLLs, but rather displayed a pattern consistent with their mutated IGHV status (Fig. 6a and Supplementary Fig. 15). Collectively, these findings suggest that, although few genetic alterations in CLL are associated with particular chromatin profiles, the overall CLL-specific regulatory chromatin landscape does not seem to be established by genetic alterations. Instead, it may be mostly influenced by other factors such as antigen stimulation, B-cell receptor conformation and the microenvironment13,44.
Fig. 6. Somatic genetic alterations in relation to chromatin activity and accessibility.
(a) Number of regions with significant gain or loss of H3K27ac or ATAC-seq levels in CLLs with somatic genetic alterations in the indicated genes/regions as compared to CLL cases without these alterations or in driver-less CLLs as compared to CLLs with mutations in driver genes. Regions with gain/loss within the investigated structural variant were excluded. Statistical significance was determined using the two-sided nbinomWaldTest in the DEseq2 package, corrected for multiple testing (Benjamini-Hochberg). Sample sizes: MYD88-MT vs. MYD88-WT (H3K27ac: n=5 vs. 57, ATAC-seq: n=6 vs. 59 biologically independent samples), SF3B1-MT vs. SF3B1-WT (H3K27ac: n=7 vs. 95, ATAC-seq: n=7 vs. 97 biologically independent samples), ATM-MT vs. ATM-WT (H3K27ac: n=10 vs. 28, ATAC-seq: n=10 vs. 27 biologically independent samples), TP53-MT vs. TP53-WT (H3K27ac: n=5 vs. 97, ATAC-seq: n=5 vs. 99 biologically independent samples), IGLL5-MT vs. IGLL5-WT (H3K27ac: n=6 vs. 56, ATAC-seq: n=7 vs. 58 biologically independent samples), NOTCH1-MT vs. NOTCH1-WT (H3K27ac: n=9 vs. 29, ATAC-seq: n=9 vs. 28 biologically independent samples), SYNE1-MT vs. SYNE1-WT (H3K27ac: n=6 vs. 96, ATAC-seq: n=6 vs. 98 biologically independent samples), MGA-MT vs. MGA-WT (H3K27ac: n=5 vs. 33, ATAC-seq: n=5 vs. 32 biologically independent samples), driverless vs. with mutations in driver genes (H3K27ac: n=15 vs. 47, ATAC-seq: n=15 vs. 50 biologically independent samples), tri12 vs. non-tri12 (H3K27ac: n=14 vs. 88, ATAC-seq: n=13 vs. 91 biologically independent samples), del10q vs. non-del10q (H3K27ac: n=5 vs. 97, ATAC-seq: n=5 vs. 99 biologically independent samples), del17p vs. non-del17p (H3K27ac: n=6 vs. 96, ATAC-seq: n=6 vs. 98 biologically independent samples), del13q vs. non-del13q (H3K27ac: n=45 vs. 57, ATAC-seq: n=46 vs. 58 biologically independent samples), del11q vs. non-del11q (H3K27ac: n=8 vs. 30, ATAC-seq: n=8 vs. 29 biologically independent samples), amp2p vs. non-amp2p (H3K27ac: n=5 vs. 33, ATAC-seq: n=5 vs. 32 biologically independent samples). (b) Heatmap of signal intensities of regions up and down regulated for H3K27ac and ATAC-seq levels in MYD88 mutated CLLs. Signal intensities are indicated as row z-scores. (c) Heatmap of signal intensities of regions up and down regulated for H3K27ac and ATAC-seq levels in CLLs with trisomy 12. Regions with gain of H3K27ac or ATAC-seq levels in chromosome 12 in the trisomy12 cases were excluded. Signal intensities are indicated as row z-scores. (d) Percentage of mutations in specific CLL cases falling into regions with the different chromatin states in the exact same cases. (e) Enrichment of somatic mutations in regions with ATAC-seq and/or H3K27ac in the exact same case (indicated are the ratios of observed versus expected number of mutations in these regions). (f) Mean enrichment in U-CLL (H3K27ac: n=25, ATAC-seq: n=24 biologically independent samples) and M-CLL (H3K27ac: n=17, ATAC-seq: n=18 biologically independent samples) of somatic mutations in regions with H3K27ac (mean U-CLL: 0.99, mean M-CLL: 2.98, P-value 2.7 x 10-5) or ATAC-seq (mean U-CLL: 0.76, mean M-CLL: 1.04, P-value 2.3 x 10-2) in the exact same case (indicated are ratios of observed versus expected number of mutations in these regions). Error bars indicate standard deviations. P-values were calculated using a Wilcoxon rank sum test (two-sided). (g) Mean enrichment in U-CLL (n=24 biologically independent samples) and M-CLL (n=17 biologically independent samples) of somatic mutations in regions with ATAC-seq and/or H3K27ac in the exact same case (indicated are the ratios of observed versus expected number of mutations in these regions). Respective means U-CLL: 1.47, 0.77, 0.74, 1.00, respective means M-CLL: 5.97, 1.08, 0.99, 0.99, and respective P-values: 8.5 x 10-5, 1.7 x 10-2, 3.5 x 10-1 and 1.0 x 10-4. Error bars indicate standard deviations. P-values were calculated using a Wilcoxon rank sum test (two-sided). (h) Mean enrichment in U-CLL (n=24 biologically independent samples) and M-CLL (n=17 biologically independent samples) of somatic mutations in regions with ATAC-seq and/or H3K27ac in the exact same case (indicated are the ratios of observed versus expected number of mutations in these regions) in loci that are known targets of the SHM machinery (upper panel, excluding IG loci, respective means U-CLL: 0.39, 0.80, 1.39, 1.00, respective means M-CLL: 18.87, 2.91, 5.25, 0.92, and respective P-values: 5.3 x 10-6, 8.2 x 10-3, 1.0 x 10-1 and 8.5 x 10-6) and other regions (lower panel, respective means U-CLL: 0.44, 0.75, 0.69, 1.00, respective means M-CLL: 0.62, 0.71, 0.69, 1.00, and respective P-values: 1.6 x 10-1, 8.0 x 10-1, 9.3 x 10-1 and 8.8 x 10-2). Error bars indicate standard deviations. P-values were calculated using a Wilcoxon rank sum test (two-sided). MT, mutated; WT, wild type; tri12, trisomy 12; del, deletion; amp, amplification; U-CLL, CLLs with unmutated IGHV; M-CLL, CLLs with mutated IGHV; SHM, somatic hypermutation; NBC-PB, naive B cell from peripheral blood; NBC-T, naive B cell from tonsil; GCBC, germinal centre B cell; MBC, memory B cell; PC-T, plasma cell from tonsil. ActProm, Active Promoter; WkProm, Weak Promoter; StrEnh1, Strong Enhancer 1; StrEnh2, Strong Enhancer 2; WkEnh, Weak Enhancer; Wk_Txn, Weak Transcription; Het;LowSign, Heterochromatin;Low Signal.
Secondly, we investigated the relationship between all somatic mutations (mostly non-coding) detected by WGS and the chromatin landscape in each of the five cases with reference epigenomes available. Although, as earlier reported in cancer45, most mutations were located in heterochromatin, we also identified a bias of the mutations towards regulatory elements such as promoters and enhancers in M-CLLs (Fig. 6d). A more exhaustive analysis matching somatic mutations detected by WGS to H3K27ac or ATAC-seq peaks in the exact same cases (n=44 CLLs), revealed that the percentage of mutations in active or accessible chromatin per case was respectively ranging from 0.05% to 2.85% and from 0.15% to 1.40%. Nevertheless, we detected a three-fold enrichment of somatic mutations in H3K27ac-associated regions in M-CLLs (Fig. 6e, f). Notably, these mutations mostly occurred in H3K27ac-associated regions lacking ATAC-seq peaks (six-fold enrichment, Fig. 6g). The exclusive presence of this enrichment in M-CLL cases suggested that it was mediated by the somatic hypermutation (SHM) machinery. Indeed, separating SHM targets as previously defined20 (Supplementary Table 13) from non-targets, we observed a 19-fold enrichment of somatic mutations in M-CLLs in H3K27ac-positive/ATAC-seq-negative regions in the former and a depletion (fold enrichment of 0.4) in the latter regions (Fig. 6h), suggesting that accessible regions are protected from the SHM machinery.
Thirdly, we investigated whether particular somatic mutations, mostly in the non-coding fraction, were associated with a local change in chromatin activity and accessibility, representing thus potential non-coding drivers in CLL. To address this issue, we combined the somatic mutations of the 44 CLL cases with their H3K27ac and ATAC-seq signals (Supplementary Figure 16). Out of 106,137 somatic mutations detected in these 44 CLLs, only 114 (0.11%) were associated with a local change in H3K27ac or ATAC-seq signal in the affected CLL case (excluding the immunoglobulin loci), a number consistent with the expected number by chance after performing a random permutation test (a mean of 106.3 random mutations were found that were associated with a local change in H3K27ac or ATAC-seq signal with a standard deviation of 10.9). Hence, with the number of cases available in this study, we did not observe a significant association between somatic mutations and local quantitative changes in genomic activity/accessibility in CLL. We cannot exclude, however, that they may exist if larger series of patients were investigated.
Discussion
In this study we provide an extensive epigenomic characterization of CLL samples and normal B-cell subpopulations, which extends previous studies of the reference epigenome of cancer cell lines46 with detailed information on primary tumors. The identity of all CLL samples studied was validated by genetic fingerprinting. This frequently underestimated quality control step is emerging as an important issue in large-scale sequencing studies47. The strategy of analyzing the CLL epigenome in the context of the entire mature B-cell differentiation program has led to new insights into CLL pathogenesis and clinical behavior. We observe that the epigenomic configuration of CLL as a whole and of its clinico-biological subtypes can be divided into three different types of patterns. First, U-CLL and M-CLL cases show imprints of their cellular origin, i.e. GC-inexperienced and experienced B cells, respectively. Intriguingly, this pattern is only evident for DNA methylation, as previously shown16, and for chromatin accessibility, but not for active regulatory regions marked with H3K27ac. This suggests that not all epigenetic marks seem to hold epigenetic memory, and that the different cellular origins of M-CLL and U-CLL cannot directly be translated into differential chromatin activation. Based on previous findings this may be expected as cell of origin-related differential DNA methylation in M-CLL and U-CLL is not related to differential expression of the target genes36. Second, the CLL chromatin landscape can also be linked to other, more complex, dynamics during the normal B-cell differentiation process, including sets of regions that relate CLL as a whole, M-CLLs or U-CLLs to a variety of combinatorial patterns in NBCs, GCBCs, MBCs and PCs. Although these patterns and their implications in CLL biology deserve further investigation, they already reveal interesting insights. For instance, U-CLLs, although derived from germinal center-inexperienced B cells, acquire chromatin features of proliferative GCBCs, a fact that may partially be associated with the higher proliferation of U-CLLs as compared to M-CLLs39. Third, CLLs also reconfigure their chromatin landscape independently of B-cell differentiation. We provide detailed maps of de novo reprogrammed regulatory elements shared in all CLL samples or present specifically in its clinico-biological subtypes (U-CLL and M-CLL). The former may represent onco-epigenetic events essential for the neoplastic transformation whereas the latter may determine the specific biological features and clinical behavior of CLL subtypes. Interestingly, it seems that extensive chromatin activation may be a feature of worse clinical behavior in CLL, as U-CLLs show more de novo accessible regions and active regulatory elements than M-CLL. De novo chromatin alterations in CLL as a whole, U-CLL and M-CLL seem to be mostly mediated by specific TF families. In particular, NFAT, FOX and TCF/LEF TF families are associated with the de novo active regions in CLL as a whole. Thus, their inhibition may revert chromatin activation and represent rational therapeutic options for CLL. In fact, in the case of NFAT and TCF/LEF, previous studies have highlighted their functional and therapeutic potential in CLL19,34,48,49. Furthermore, in light of the emerging importance of pharmacological agents inhibiting specific epigenetic marks50, the observed alterations in the chromatin landscape of CLL may also represent potential therapeutic targets. In this context, de novo chromatin reprogramming of CLL is marked by the transition from inactive regions in normal B cells to super-enhancers, which have been already shown to be targets for selective pharmacological inhibition in cancer51.
The large number of de novo chromatin changes homogeneously present in CLL or CLL subtypes contrasts with the vast genetic heterogeneity of the disease and the paucity of driver genes mutated in more than 5% of the cases18,20. In terms of the link between genetic and epigenetic changes in CLL, our dataset with both extensive genetic and chromatin characterization of CLL samples allowed us to identify that cases with MYD88 mutations or trisomy 12 represent distinct molecular subgroups from the chromatin perspective, highlighting the specific clinico-biological features of these CLL subtypes52,53. In the case of MYD88, chromatin activation seems to be a direct effect, as the associated genes are downstream effectors of the toll-like receptor pathway. The specific chromatin signature of trisomy 12 CLLs, however, is intriguing. This signature, which is similar between trisomy 12 cases and normal B cells, is derived from the acquisition of chromatin changes in the heterogeneous group of CLLs lacking trisomy 12 rather than from a direct chromatin reprogramming mediated by trisomy 12. More globally, we observe that the mutational landscape of M-CLLs is enriched in regulatory elements, which may constitute potential non-coding drivers20,54. Intriguingly, these mutations in M-CLL are highly enriched in regions associated with H3K27ac-containing nucleosomes outside ATAC-seq peaks, as initially observed for a mutated PAX5 enhancer in M-CLL20. This finding suggests that, although the SHM machinery overall targets active regulatory regions55, it seems that transcription factor binding sites in accessible regions are protected, possibly by blocking access to the SHM machinery or by a higher DNA repair rate. Lastly, we observe that within our CLL series non-coding mutations do not change the activity or accessibility of genomic regions in a quantitative way. Instead, potential non-coding driver mutations may modulate the regulatory potential of already existing promoter and enhancer elements by other means.
In conclusion, this study presents a comprehensive description of the epigenome of CLL samples with complete genetic characterization, and samples spanning the normal B-cell maturation process. The findings derived from the primary analysis of the dataset improve our understanding of the biological basis and clinical behavior of CLL. We identify de novo reprogrammed regulatory regions specifically associated with the development of CLL and its major clinical subtypes, which harbor diagnostic, prognostic and potential therapeutic value. This so far unique dataset also represents a valuable resource for researchers both working in CLL and broader fields such as gene regulation, cell differentiation and neoplastic transformation, and to study the link between genetic variants (somatic and germline) and the epigenome in the context of disease development.
Online methods
Please be referred to the Life Sciences Reporting summary for further details that complement the sections below.
Patients
The clinical and biological characteristics of the 107 patients are shown in Supplementary Table 1. Cases were defined as IGHV-MUT when the identity of immunoglobulin genes was less than 98%. The tumor samples were obtained before administration of any treatment. All patients gave informed consent for their participation in the study following the International Cancer Genome Consortium (ICGC) guidelines and the ICGC Ethics and Policy committee23, and this study was approved by the clinical research ethics committee of the Hospital Clinic of Barcelona.
Collection and preparation of patient and normal samples
Tumor samples were obtained from fresh or cryopreserved mononuclear cells. The CLL fraction was only purified when the tumor content was <85% as assessed by immunostaining of CD19, CD20, CD5 and CD45 followed by flow cytometry. If the tumor content was <85%, CLL cells were purified by selecting CD19 positive cells using AutoMACS (Miltenyi Biotec), until a tumor content of >85% was reached (which was usually obtained after one round of AutoMACS purification). Normal B cell fractions were collected and isolated as previously described, using the indicated surface markers (Fig. 1a)24.
ChIP-seq, ATAC-seq, RNA-seq, WGBS, in situ Hi-C, promoter capture Hi-C, 4C-seq and WGS data generation
ChIP-seq of the six different histone marks and ATAC-seq data were generated as described (http://www.blueprint-epigenome.eu/index.cfm?p=7BF8A4B6-F4FE-861A-2AD57A08D63D0B58). Catalog numbers of antibodies (Diagenode) used are H3K27ac: C15410196/pAb-196-050 (LOT: A1723-0041D), H3K4me1: C15410194/pAb-194-050 (LOT: A1863-001P), H3K4me3: C15410003-50/pAb-003-050 (LOT: A5051-001P), H3K36me3: C15410192/(pAb-192-050 (LOT: A1847-001P), H3K9me3: C15410193/pAb-193-050 (LOT: A1671-001P), H3K27me3: C15410195/pAb-195-050 (LOT: A1811-001P).
Single stranded RNA-seq data of the reference epigenomes was generated as previously described56. Briefly, RNA was extracted using TRIZOL (Life Technologies) and libraries were prepared using a TruSeq Stranded Total RNA Kit with Ribo-Zero Gold (Illumina). Adapter-ligated libraries were amplified and sequenced using 100-bp single-end reads. Fastq files of (non-stranded) RNA-seq data of 78 CLL cases were mined22.
WGBS of the reference epigenomes was generated as previously described24. Briefly, 1–2 μg of DNA was sheared and fragments of 150–300 bp were selected using AMPure XP beads (Agencourt Bioscience). After adaptor ligation (Illumina TruSeq Sample Preparation kit), DNA was treated with sodium bisulfite using the EpiTexy Bisulfite kit (Qiagen). Two rounds of bisulfite conversion were performed to ensure a conversion rate of over 99%. Enrichment for adaptor-ligated DNA was carried out through seven PCR cycles and paired-end DNA sequencing (2 × 100 bp) was then performed using the Illumina HiSeq 2000 platform. Methylation estimates of 105 CLL cases, analyzed by the 450k Human Methylation Array (Illumina), were mined20.
Promoter capture Hi-C interactions of normal B cells31 as well as in situ Hi-C data of GM1287830 were mined. In situ Hi-C of one CLL case and MBCs and promoter capture Hi-C of one CLL case were performed as previously described30,31. 4C templates were prepared for two CLL patients and the JVM-2 cell line as previously described57,58 using 107 cells per 4C library. First and second restriction enzymes per region were for the FMOD enhancer: NlaIII, BfaI; the TCF4 enhancer: DpnII, Csp6I; the FMOD promoter; NlaIII, Csp6I and the TCF4 promoter: DpnII, Csp6I. RE1 and RE2 primers per region were for the FMOD enhancer: 5'- AGGGAAGGCAGGGAAACATG-3', 5'-TACACGCTCATTAACACTGC-3'; the TCF4 enhancer: 5'-TAACTAGAAATGGGGTGATC-3', 5'- AAAAGTGTCAACCTGGAGAA-3'; the FMOD promoter: 5’-GCTGTCCCTTGTCATTCATG-3’, 5’-CTGTGTCCTACCCATTTCAC-3’; and the TCF4 promoter: 5’- TCGGAAAAGTTGAATCGATC-3’, 5’-TTTGATTAAAAAGCGAGTGG-3’.
For 42 CLL patients, WGS data were mined20. WGS data of two CLL patients was generated as previously described20.
Read mapping and data processing
Fastq files of ChIP-seq data were aligned to genome build GRCh38 (using bwa 0.7.7, picard and samtools) and wiggle plots were generated (using PhantomPeakQualTools) as described (http://dcc.blueprint-epigenome.eu/#/md/methods).
Peaks of the histone mark data were called as described (http://dcc.blueprint-epigenome.eu/#/md/methods) using MACS2 (version 2.0.10.20131216). As for many CLL samples (87 out of 107) no input data was available, for all samples H3K27ac peaks were also called without input control. ATAC-seq fastqs were aligned to genome build GRCh38 using bwa 0.7.759 (parameters: -q 5, -P, -a 480) and SAMTOOLS v1.3.160 (default settings). BAM files were sorted and duplicates were marked using PICARD tools v2.8.1 (http://broadinstitute.github.io/picard, default settings). Finally, low quality and duplicates reads were removed using SAMTOOLS v1.3.160 (parameters: -b, -F 4, -q 5, -b, -F 1024). ATAC-seq peaks were determined using MACS2 (v2.1.1.20160309, parameters: -g hs -q 0.05 --keep-dup all -f BAM – nomodel –shift -96 –extsize 200) without input control. For downstream analysis peaks with p-values <1e-5 (H3K36me3, H3K9me3 and H3K27me3) or <1e-9 (H3K4me3, H3K4me1, H3K27ac, ATAC-seq) were included. For each mark a set of consensus peaks, only including regions on chromosome 1-22, present in the normal B cells (n=15 biologically independent samples for histone marks and n=18 biologically independent samples for ATAC-seq) and in the CLL samples (n=7 biologically independent samples for the reference epigenomes, n=104 biologically independent samples for the extended H3K27ac series and n=106 biologically independent samples for the extended ATAC-seq series) was generated by merging the locations of the separate peaks per individual sample. To generate the consensus peak file for the reference epigenomes, only peaks with input were used except for ATAC-seq for which peaks without input were used; for the extended H3K27ac series peaks with (20 CLLs and 15 normal B cells) and without input (104 CLLs and 15 normal B cells) were used; and for the extended ATAC-seq series only peaks without input (106 CLLs and 18 normal B cells) were used. For the histone marks the number of reads per sample per consensus peak were calculated using the genomecov function of bedtools. For the ATAC-seq the number of insertions of the Tn5 transposase per sample per consensus peak were calculated by first determining the estimated insertion sites (shifting the start of the first mate 4bp downstream), followed by the genomecov function of bedtools. Using DEseq261, variance stabilized transformed (vst) values were calculated for all consensus peaks (H3K27ac and ATAC-seq data of extended CLL series) or for the peaks that were present in >1 sample (reference epigenome data). The number of consensus peaks for the reference epigenome analyzes for which vst values were calculated were: 38,499 (H3K4me3); 37,871 (H3K4me1); 47,191 (H3K27ac); 15,561 (H3K36me3); 27,371 (H3K9me3); 12,878 (H3K27me3); and 91,671 (ATAC-seq), and for the extended CLL series: 100,640 (H3K27ac); and 143,668 (ATAC-seq). For the extended CLL series, we corrected the vst values for the consensus SPOT score, i.e., the percentage of total number of reads that fall within the consensus peaks, using the ComBat function from the sva R package62. To that purpose, the cell condition (CLL and the different normal B-cell subtypes) was assigned to each sample and samples were clustered in 20 bins of 5% according to their consensus SPOT score. The bins on the extremes which contained less than five samples were joined with neighboring bins, to ensure that each bin contained at least five samples. PCAs were generated with the prcomp function in R using the (corrected) vst values of all peaks that were present in >1 sample.
RNA-seq data of the reference epigenomes and the fastq files of the 78 samples mined from a previous study22 were aligned to genome build GRCh38, signal files were produced and gene quantifications (gencode 22, 60,483 genes) were calculated as described (http://dcc.blueprint-epigenome.eu/#/md/methods) using the GRAPE2 pipeline with the STAR-RSEM profile (adapted from the ENCODE Long RNA-Seq pipeline). The expected counts and FPKM estimates were used for downstream analysis. The PCA of the RNA-seq data was generated with the prcomp function in R using log10 transformed FPKM (+0.01 pseudocount) data of 36,190 genes with an FPKM standard deviation of >0 in the 22 analyzed samples.
Mapping and determination of methylation estimates were performed as described (http://dcc.blueprint-epigenome.eu/#/md/methods) using GEM3.0. Per sample, only methylation estimates of CpGs with 10 or more reads were used for downstream analysis. The PCA of the DNA methylation data was generated with the prcomp function in R using methylation estimates of 15,825,190 CpGs (chr1-22) with available methylation estimates in all 19 analyzed samples.
Processing of the promoter capture Hi-C data was performed as previously described31. The CHIGAGO software63 was used to determine interacting fragments (CHICAGO score > 5). Hi-C data was processed using TADbit64 for read quality control, read mapping, interaction detection, interaction filtering, and matrix normalization. First, the quality of the experiments was assessed using a Hi-C specific FastQC protocol (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) implemented in TADbit64. Next, a fragment-based strategy in TADbit was used for mapping the paired-end reads to the reference genome (GRCh38) (similar protocol as described65). Mapping resulted in around 65% of reads mapped uniquely to the genome. Next, non-informative contacts between two reads were filtered out, including self-circles, dangling-ends, mapping-errors, random breaks and duplicates as previously described65,66. The final interaction matrices resulted in 83 and 119 million of valid interactions for the CLL and MBC sample, respectively. Assignment of topologically associated domains (TADs) in GM12878 (hg19) was performed using TADbit64 on the GSE63525_GM12878_combined_intrachromosomal_contact_matrices.tar.gz dataset30, followed by liftOver to GRCh38. 4C-seq analysis was performed using the pipeline 4cseqpipe (http://compgenomics.weizmann.ac.il/tanay/?page_id=367) and r3Cseq67. For the 4cseqpipe, default settings were used. For r3Cseq, default settings were used, mapping read counts and interactions using 5,000 base pair windows. Both for 4cseqpipe and r3Cseq, reads corresponding to self-ligated or non-digested fragments were removed.
Somatic mutations present in the two newly sequenced CLL patients were defined as previously described20. Of the 106,197 somatic mutations (chr1-22, genome build hg19) in the 44 CLL patients, 106,137 were successfully lifted over to genome build GRCh38 and were used for the downstream analysis.
Data quality and donor, normal B cell and histone mark identity
The data quality measures of all epigenetic data generated within this study (ChIP-seq and input of all histone marks, ATAC-seq, WGBS and RNA-seq (reference epigenomes only) can be found in Supplementary Table 14. To confirm that all data generated within this study correspond to the correct patient sample, genotypes extracted from each mark of the reference epigenome were matched with the genotype fingerprints of the patients detected by copy number arrays20. For H3K27ac, Input DNA and ATAC-seq genotypes were called using BaseRecalibrator, PrintReads and HaplotypeCaller68 and only positions with Phred score >= 20 were used for the analysis. In the case of WGBS, SNP genotypes with Phred score >= 20 were extracted from the VCF files generated by bs_call in the standard methylation calling pipeline69. For RNA-seq, SNPs were called on the RNA-Seq data using FreeBayes70. Only positions that passed the FreeBayes default filters were used for the analysis. Sample genotype calls were compared with respect to the genotype from the SNP array using an IBS (Identity by State) based statistic. For two sets of genotype calls, SNP positions genotyped in both sets were scored as 0, 1 or 2 according to whether they shared 0, 1 to 2 alleles IBS. This score was then averaged across all such loci to give an average sharing statistic for a pair. Genotype call sets from the same individual would be expected to have an IBS sharing statistic close to 2, while non-matching sets should be in the range 1.2 – 1.6. For the normal B cell subpopulations, snapshots of chromatin states of subpopulation-specific genes and gene expression levels were investigated to confirm that the data correspond to the correct B cell differentiation stages (Supplementary Fig. 17). Read profiles of the different histone marks and ATAC-seq data around transcription start sites (TSS) and gene bodies (GB) were generated to verify the nature of these different layers (Supplementary Fig. 18 and 19). To that end, for the TSS profiles bins of 100 bp around the TSS of all protein coding genes on chromosome 1-22 (50 bins in total, spanning -2500bp to +2500bp) were assigned, while for the GB profiles of the same genes 80 bins were assigned: 15 bins of 100bp (-1500 until TSS), 50 bins each corresponding to 2% of the gene body and 15 bins of 100 bp each (transcriptional termination site until +1500bp). The mean number of reads per bin per sample per mark for the 22 reference epigenome samples (25 in case of ATAC-seq) was calculated using the genomcov function of bedtools and corrected for the total number of mapped reads.
K-means clustering, jaccard coefficients and detection of differentially methylated CpGs and regions
For individual histone marks and ATAC-seq data, only consensus regions present in at least three and in a maximum of 19 out of the 22 samples (22 out of 25 for the ATAC-seq data) were used, i.e., excluding individual specific and constitutive regions. For the RNA-seq dataset, only genes that were expressed (FPKM values equal or greater than 0.1) in at least three out of the 22 samples were included to exclude individual specific genes. Of the included consensus peaks/genes, those differential among the six different subgroups (CLL and five normal B-cell subpopulations) were defined using the likelihood ratio test (FDR <0.01) of the DEseq2 package61. When performing K-means clustering the absolute vst levels (which are dependent on the size of the regions/genes) affect the clustering, while we were only interested in relative differences. Therefore, z-scores are necessary to correct for this phenomenon. Hence, K-means clustering was performed using the z-scores of the vst values of the differential regions/genes. For each, 20 clusters were assigned, which were merged based on pattern similarity.
Pairwise jaccard coefficients of the regions with de novo increase or decrease of the different histone marks in CLL were assigned by calculating the number of base pairs that overlap among the regions divided by the total number of base pairs covered by these regions. The dissimilarity matrix (1-jaccard coefficient) was used for clustering. Differentially methylated CpGs (DMCs) and regions (DMRs) were calculated using methilene71 version 0.2-7. Firstly, from the 15,825,190 CpGs (chr1-22) with available methylation estimates in all 19 analyzed samples, only the ones that were not modulated during normal B cell differentiation (maximum pairwise difference in methylation among normal B cells was 0.25) were selected. Next from this subset of CpGs, DMCs and DMRs were assigned that showed an absolute difference in methylation of at least 0.25 comparing CLL versus normal B cells using default settings in the metilene pipeline. Furthermore, for the detection of DMRs, a minimum number of 3 CpGs and a maximum distance between 2 CpGs of 100 basepairs were used.
Linking histone mark clusters with chromatin accessibility, DNA methylation and gene expression
Per histone mark region the overlapping consensus ATAC-seq peaks of the reference epigenome data were selected. Next, per region per sample was determined whether an ATAC-seq peak was present (1) or absent (0). If no overlapping peaks were found, chromatin accessibility was considered absent (0) in all samples. If more than one consensus peak was found in the histone mark region the mean of present (1) and absent (0) peaks was calculated per sample. Next, for all regions in one cluster a mean of present and absent peaks was calculated per sample.
Median methylation levels of all CpGs within the histone mark regions per cluster were calculated per sample.
Per host gene mean log10(FPKM + 0.01 pseudocount) RNA-seq levels were calculated for CLL, the five different normal B cells separate and normal B cells all together (seven values in total). Boxplots of log10(fold changes) of all genes located in the analyzed regions were generated subtracting the mean log10(FPKM + 0.01 pseudocount) expression levels of normal B cells from the mean expression of CLLs per gene. Finally, if the log10(FPKM + 0.01 pseudocount) expression of a gene was lower than -1 in the CLLs and the five different B-cell subpopulations subgroups it was considered neither expressed in B cells nor in CLL.
Chromatin states and chromatin state transitions
A B-cell specific chromatin state model with 12 emission states was generated using the chromHMM software26 using the six histone marks in the 15 normal B cells, corrected for their corresponding input. Next, this model was used to assign chromatin states in the seven CLL cases. Chromatin states were assigned per 200bp window.
To calculate the overall similarity between CLL and normal B cells based on chromatin states, all regions with differential histone marks among the normal B cells samples (i.e. all the regions of all the 6 histone mark k-means clusters from cluster 3 onwards) were included. From all the included regions (2,167,103 windows of 200 base pairs), the chromatin states were taken and the pair wise fraction of overlap between samples were calculated. The dissimilarity matrix (1-fraction of overlap) was used to cluster the samples.
For all individual regions with de novo increase or decrease of the individual histone marks in CLL, the percentage of each of the 12 chromatin states was counted per sample. Per sample, all percentages were added up to calculate the overall distribution of chromatin states in these regions. In this way, each region, independent of the size, equally contributed to the final distribution.
To calculate chromatin state transitions, each region was divided into 200bp windows. Per 200bp window, the percentages of the 12 chromatin states in 15 normal B cells were calculated as well as the percentages in the seven CLL samples. These vectors were multiplied, generating a 12x12 matrix (rows = normal B cells, columns = CLLs). All matrices of all 200bp windows per region were summed up and corrected for the total number of 200bp windows within the region. In this way, the corrected matrix for each region, independent of the size, had a total value of one. Corrected matrices of all regions per cluster were added up and divided by the total number of regions to calculate the final transition matrix.
Defining de novo (in)active regulatory elements in CLL and their local chromatin interactions
A graphical representation of the strategy is shown in Supplementary Fig. 10. All 8,950 peaks with de novo increase or decrease of H3K27ac, H3K4me3 and H3K4me1 were merged into 7,121 peaks. For each peak the percentage of base pairs covered by active regulatory elements (active promoter + strong enhancer 1 + strong enhancer 2) and inactive chromatin (poised promoter + H3K7me3/H3K9me3 repressed + heterochromatin;low signal) were calculated in normal B cells (n=15 biologically independent samples) and CLLs (n=7 biologically independent samples). Regions were assigned as de novo active regions in CLL if: (i) no significant difference in the percentage of active regulatory elements was observed in normal B cells (Kruskal-Wallis test, p-value <0.1 and in at least one pairwise comparison a difference of >10%), (ii) the percentage of active regulatory elements in CLL was significantly higher than in normal B cells (Wilcoxon rank sum test (two-sided), FDR-value <0.01 and minimal difference of 25%) and (iii) the percentage of inactive chromatin in CLL was significantly lower than in normal B cells (Wilcoxon rank sum test (two-sided), FDR-value <0.01 and minimal difference of 25%). Regions were assigned as de novo inactive regions in CLL if: (i) no significant difference in the percentage of active regulatory elements was observed in normal B cells (Kruskal-Wallis test, p-value <0.1 and in at least one pairwise comparison a difference of >10%), (ii) the percentage of active regulatory elements in CLL was significantly lower than in normal B cells (Wilcoxon rank sum test (two-sided), FDR-value <0.01 and minimal difference of 25%) and (iii) the percentage of inactive chromatin in CLL was significantly higher than in normal B cells (Wilcoxon rank sum test (two-sided), FDR-value <0.01 and minimal difference of 25%). De novo (in)active regulatory elements with a size of more than 10,000 base pairs were considered super-enhancers.
Local chromatin interactions of the de novo active regions in CLL were calculated by using the valid interactions (normalized by one round of ICE66 and by genomic decay) to generate genome-wide interaction maps to perform a meta-analysis of selected regions by merging individual local sub-matrices at 10 kb resolution in a similar fashion as previously published72.
Assignment of target genes and GO analysis
A graphical representation of the assignment of target genes strategy is shown in Supplementary Fig. 11a. Potential protein coding target genes of regulatory regions (de novo active and inactive regions in CLL) and active and accessible chromatic regions (extended CLL series) were assigned by taking the union of (i) the host gene, (ii) the most proximal up- and downstream gene on the positive and negative strand and (iii) genes interacting in 3D space as defined by promoter capture Hi-C. To avoid false positives, per regulatory element, only genes located within the topologically associated domain (TADs) of GM12878 were considered. A potential target gene was assigned to the final list of target genes when a significant difference in expression was observed between the compared groups (DEseq2 package, nbinomWaldTest, FDR < 0.05 (CLL vs. normal B cells or patients with vs. without mutations/copy number variants) or FDR < 0.01 (U-CLL vs. M-CLL)), and only when (i) the gene was expressed in at least one of the compared subgroups (mean(log10(fpkm +0.01)) > -1.0) and (ii) the group with the presence of the regulatory element or the highest H3K27ac or ATAC-seq levels showed higher expression levels.
GO enrichment was performed using the GOstats R package73. As the universe, all GENCODE22 annotated protein coded genes were used. The statistical analysis was conditioned based on the GO structure.
Transcription factor analysis
For the analysis in the 534 de novo regions, reference GRCh38 sequences were extracted from the overlapping consensus ATAC-seq peaks enriched in at least two CLL samples (for the 498 de novo active regions) or in at least two B cell samples (for the 36 de novo inactive regions). In the case of the comparison of U-CLL versus M-CLL, reference GRCh38 sequences were extracted from the differentially enriched peaks in the de novo clusters. The AME tool from MEME suite74 was used for the enrichment analysis of known motifs from the non-redundant vertebrate 2016 Jaspar database75 using a one-tailed Wilcoxon rank-sum test with the maximum score of the sequence, a 0.05 FDR cutoff and a background formed by reference GRCh38 sequences extracted from the consensus ATAC-seq peaks enriched in at least two samples.
Defining differential chromatin activity and accessibility in U-CLL vs. M-CLL and their dynamics in normal B cells
Differential enrichment of H3K27ac/ATAC-seq levels of the consensus regions (extended CLL series) in U-CLL and M-CLL was calculated using DESeq261. The proper condition (U-CLL, M-CLL or normal B cell) per sample and the consensus SPOT (see read mapping and data processing) were introduced into the model. We performed the analysis by contrasting U-CLL and M-CLL samples using the nbinomWaldTest in DEseq2. Next, peaks that were constitutively present in all CLLs or peaks that were not present in at least 10% of any of the two compared subgroups were removed after which the FDR was calculated. Regions with an FDR < 0.001 were considered significantly enriched.
By calculating, for each differential region, whether the mean z-score of the vst value of each normal B-cell subpopulation (five in total) was closer to the mean z-score of U-CLL or M-CLL, 32 patterns (25) of dynamics in normal B cells could be assigned. Two of these patterns, i.e., when all normal B cells are closer to U-CLL or all normal B cells are closer to M-CLL represented de novo changes in respectively M-CLL and U-CLL, while all other patterns represented modulation of H3K27ac or ATAC-seq levels in these regions in normal B cells.
Defining differential chromatin activity and accessibility in patients with mutations in driver genes or copy number variants
Patients compared for these analyses are indicated in Supplementary Table 11. Differential enrichment of H3K27ac/ATAC-seq levels of the consensus regions (extended CLL series) in samples with and without mutations/copy number variants (CNAs) was performed using DESeq261. The proper condition (mutated (MT), wild type (WT), loss, gain or normal B cell) per sample and the consensus SPOT (see read mapping and data processing) were introduced into the model. We performed the analysis by contrasting mutated vs. WT (mutations) or loss/gain vs. WT (CNAs) using the nbinomWaldTest in DEseq2. Next, peaks that were constitutively present in all CLLs or peaks that were not present in at least 10% of any of the two compared subgroups (with a minimum of two samples) were removed after which the FDR was calculated. Regions with an FDR < 0.001 were considered significantly enriched. To exclude any bias due to differences in number of reads, regions covering the copy number alterations were filtered out in case a positive correlation between the copy number change (gain/loss) and the H3K27ac/ATAC-seq signal was found. For example, regions on chromosome 12 were filtered out in the comparison of tri12-positive vs tri12-negative CLLs if they had a higher H3K27ac/ATAC-seq signal intensity in tri12-positive cases.
Enrichment of mutations in H3K27ac, ATAC-seq peaks and chromatin states
Per case, the percentage of mutations within H3K27ac peaks (n=43 cases), ATAC-seq peaks (n=43 cases) and/or the 12 chromatin states (n=5 cases) in the exact same case were calculated. For the H3K27ac and ATAC-seq data, only peaks called without correction for input were used, to avoid a potential bias between samples for which the corresponding input was present and those for which this was absent. To calculate the enrichment of these mutations within these regions, the calculated percentages were divided by the total percentage of the genome that was covered by H3K27ac or ATAC-seq peaks or the specific chromatin states in the exact same case.
Association of somatic mutations with local chromatin changes
A schematic representation of the approach is shown in Supplementary Fig. 16. Consensus H3K27ac and ATAC-seq peaks of the extended CLL series that harbored a somatic mutation in at least one of the 44 CLL cases were included for this analysis (the immunoglobulin loci were excluded). Regions for which somatic mutations were considered to be associated with a local increase in H3K27ac/ATAC-seq levels were assigned if: (i) one or more of the patients with somatic mutations had an H3K27ac/ATAC-seq peak in this region and (ii) one or more of the same patient(s) had a z-score of H3K27ac/ATAC-seq levels of >2, using the mean and standard deviation of CLLs without the somatic mutation and normal B cells. Regions for which somatic mutations were considered to be associated with a local decrease in H3K27ac/ATAC-seq levels were assigned if: (i) at least 10% of the patients without somatic mutations in this region had an H3K27ac/ATAC-seq peak and (ii) one or more of the patients with somatic mutations had a z-score of H3K27ac/ATAC-seq levels of < -2, using the mean and standard deviation of CLLs without the somatic mutation and normal B cells. Next, the mutations per case were permutated (i.e. each patient got assigned the somatic mutations of another case) to calculate how many associating mutations were found by chance.
Supplementary Material
Acknowledgements
This work was funded by the European Union’s Seventh Framework Programme through the Blueprint Consortium (grant agreement 282510), the International Cancer Genome Consortium (Chronic Lymphocytic Leukemia Genome consortium to E.C. and C.L-O), the European Hematology Association (Non-Clinical Advanced Research Fellowships to J.I.M.-S.), the World Wide Cancer Research Foundation Grant No. 16-1285 (to J.I.M-S.), Spanish Ministerio de Economía y Competitividad (MINECO), Grant No. SAF2015-64885-R (to E.C.) and Grant No. PMP15/00007, part of Plan Nacional de I+D+I and co-financed by the ISCIII-Sub-Directorate General for Evaluation and the European Regional Development Fund (FEDER-"Una manera de Hacer Europa") (to E.C.), the Generalitat de Catalunya Suport Grups de Recerca AGAUR 2014-SGR-795 (to E.C.), the CERCA Programme/Generalitat de Catalunya and CIBERONC. R.B was supported by fellowships from the EU (Marie Skłodowska-Curie Inter European Fellowship) and the Lady TATA Memorial Trust (International Award), N.R by the Acció instrumental d’incorporació de científics i tecnòlegs PERIS 2016 from the Generalitat de Catalunya, and M.K. by an AOI grant of the Spanish Association Against Cancer. E.C. is an Academia Researcher of the "Institució Catalana de Recerca i Estudis Avançats" (ICREA) of the Generalitat de Catalunya. This work was partially developed at the Centro Esther Koplowitz (CEK, Barcelona, Spain). We are indebted to the HCB-IDIBAPS Biobank-Tumor Bank and Hematopathology Collection for sample procurement.
Footnotes
Data availability and Accession Code Availability Statements
All the raw data included in this study has been deposited and released, as part of the BLUEPRINT epigenome project, at the European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted at the European Bioinformatics Institute (EBI). They can be found under the unifying EGA accession number EGAD00001004046. Furthermore, we have created a website (http://resources.idibaps.org/paper/the-reference-epigenome-and-regulatory-chromatin-landscape-of-chronic-lymphocytic-leukemia) that includes the large processed data matrices and a link to a genome browser session displaying the generated data.
Author contributions
The Chronic Lymphocytic Leukemia Genome consortium and the BLUEPRINT consortium contributed to this study respectively as part of the International Cancer Genome Consortium and the International Human Epigenome Consortium. Investigator contributions were as follows: T.B., J.D., A.L-G., D.M-G, S.B., M.P., M.A., M.Ku., N.V-D., X.A. and F.P. contributed to sample collection (CLL and normal B cells) as well as to their biological and clinical annotation; M.P., N.V-D., M.G. and I.G. contributed to WGS data generation; N.R., N.V-D., J.H.A.M., H.G.S., J.I.M-S., M.G., I.G., and M-L.Y. contributed to histone mark, ATAC-seq, methylome and transcriptome data generation; R.V-B., J.B., M.G., I.G., J.I.M-S., B.M.J., P.Fr., N.V-D., A.E., A.C.Q and R.B. contributed to In Situ HiC, Promoter Capture HiC and 4C-seq data generation; X.S.P. and C.L-O. contributed to WGS data analysis; R.B., V.C., J.H.A.M., M.D-F., M.Ku., G.Cl., G.Ca., A.M., S.H., A.V., S.U., E.P., R.G., R.R., M.P., D.T., A.D., E.L., M.Ko., M.R., L.C., P.Fl and J.I.M-S contributed to histone mark, ATAC-seq, methylome and transcriptome data analysis; R.V-B., F.S., M.A.M-R., S.W.W., B.M.J., P.Fr., and R.B. contributed to In Situ HiC, Promoter Capture HiC and 4C-seq data analysis; C.L-O, E.C., H.G.S. and R.S. participated in the study design and data interpretation together with R.B. and J.I.M-S.; R.B. and J.I.M-S. directed the research and wrote the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhtar-Zaidi B, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiziev P, et al. Systematic Epigenomic Analysis Reveals Chromatin States Associated with Melanoma Progression. Cell Rep. 2017;19:875–889. doi: 10.1016/j.celrep.2017.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin CY, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muratani M, et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat Commun. 2014;5 doi: 10.1038/ncomms5361. 4361. [DOI] [PubMed] [Google Scholar]
- 7.Queiros AC, et al. Decoding the DNA Methylome of Mantle Cell Lymphoma in the Light of the Entire B Cell Lineage. Cancer Cell. 2016;30:806–821. doi: 10.1016/j.ccell.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rendeiro AF, et al. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nat Commun. 2016;7 doi: 10.1038/ncomms11938. 11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun HJ, et al. Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer Cell. 2016;29:394–406. doi: 10.1016/j.ccell.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khurana E, et al. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 11.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16:145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- 13.Kipps TJ, et al. Chronic lymphocytic leukaemia. Nat Rev Dis Primers. 2017;3 doi: 10.1038/nrdp.2016.96. 16096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damle RN, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 15.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 16.Kulis M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 17.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landau DA, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oakes CC, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48:253–264. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 21.Cahill N, et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia. 2013;27:150–158. doi: 10.1038/leu.2012.245. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira PG, et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014;24:212–226. doi: 10.1101/gr.152132.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Cancer Genome C et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulis M, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seifert M, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012;209:2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javierre BM, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384. e1319. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy BA, et al. A seven-gene expression panel distinguishing clonal expansions of pre-leukemic and chronic lymphocytic leukemia B cells from normal B lymphocytes. Immunol Res. 2015;63:90–100. doi: 10.1007/s12026-015-8688-3. [DOI] [PubMed] [Google Scholar]
- 33.Navarro A, et al. Improved classification of leukemic B-cell lymphoproliferative disorders using a transcriptional and genetic classifier. Haematologica. 2017 doi: 10.3324/haematol.2016.160374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutierrez A, Jr, et al. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood. 2010;116:2975–2983. doi: 10.1182/blood-2010-02-269878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceribelli M, et al. A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancer Cell. 2016;30:764–778. doi: 10.1016/j.ccell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Queiros AC, et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia. 2015;29:598–605. doi: 10.1038/leu.2014.252. [DOI] [PubMed] [Google Scholar]
- 37.Khandanpour C, et al. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell. 2013;23:200–214. doi: 10.1016/j.ccr.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moroy T, Khandanpour C. Growth factor independence 1 (Gfi1) as a regulator of lymphocyte development and activation. Semin Immunol. 2011;23:368–378. doi: 10.1016/j.smim.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Murphy EJ, et al. Leukemia-cell proliferation and disease progression in patients with early stage chronic lymphocytic leukemia. Leukemia. 2017;31:1348–1354. doi: 10.1038/leu.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmaier K, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 41.Nihira K, et al. Pim-1 controls NF-kappaB signalling by stabilizing RelA/p65. Cell death and differentiation. 2010;17:689–698. doi: 10.1038/cdd.2009.174. [DOI] [PubMed] [Google Scholar]
- 42.Kasof GM, et al. Tumor necrosis factor-alpha induces the expression of DR6, a member of the TNF receptor family, through activation of NF-kappaB. Oncogene. 2001;20:7965–7975. doi: 10.1038/sj.onc.1204985. [DOI] [PubMed] [Google Scholar]
- 43.Xia XZ, et al. TACI is a TRAF-interacting receptor for TALL-1, a tumor necrosis factor family member involved in B cell regulation. J Exp Med. 2000;192:137–143. doi: 10.1084/jem.192.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minici C, et al. Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia. Nat Commun. 2017;8 doi: 10.1038/ncomms15746. 15746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 46.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen BS, Quinlan AR. Who's Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am J Hum Genet. 2017;100:406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf C, et al. NFATC1 activation by DNA hypomethylation in chronic lymphocytic leukemia correlates with clinical staging and can be inhibited by ibrutinib. Int J Cancer. 2018;142:322–333. doi: 10.1002/ijc.31057. [DOI] [PubMed] [Google Scholar]
- 49.Wu W, et al. High LEF1 expression predicts adverse prognosis in chronic lymphocytic leukemia and may be targeted by ethacrynic acid. Oncotarget. 2016;7:21631–21643. doi: 10.18632/oncotarget.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 51.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riches JC, et al. Trisomy 12 chronic lymphocytic leukemia cells exhibit upregulation of integrin signaling that is modulated by NOTCH1 mutations. Blood. 2014;123:4101–4110. doi: 10.1182/blood-2014-01-552307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Trillos A, et al. Clinical impact of MYD88 mutations in chronic lymphocytic leukemia. Blood. 2016;127:1611–1613. doi: 10.1182/blood-2015-10-678490. [DOI] [PubMed] [Google Scholar]
- 54.Burns A, et al. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia. 2017 doi: 10.1038/leu.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ecker S, et al. Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types. Genome biology. 2017;18:18. doi: 10.1186/s13059-017-1156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Werken HJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nature methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 58.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nature methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic acids research. 2014;42 doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome biology. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serra F, Baù D, Filion G, Marti-Renom MA. Structural features of the fly chromatin colors revealed by automatic three-dimensional modeling. bioRxiv. 2016:1–29. [Google Scholar]
- 65.Ay F, et al. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC genomics. 2015;16:121. doi: 10.1186/s12864-015-1236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thongjuea S, Stadhouders R, Grosveld FG, Soler E, Lenhard B. r3Cseq: an R/Bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic acids research. 2013;41:e132. doi: 10.1093/nar/gkt373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merkel A, et al. GEMBS — high through-put processing for DNA methylation data from Whole Genome Bisulfite Sequencing (WGBS) bioRxiv. 2017 [Google Scholar]
- 70.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. ArXiv e-prints. 2012;1207 [Google Scholar]
- 71.Juhling F, et al. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26:256–262. doi: 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Wit E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 73.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 74.McLeay RC, Bailey TL. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathelier A, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic acids research. 2016;44:D110–115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.