Abstract
Achieving adequate margins during tumor margin resection is critical to minimize the recurrence rate and maximize positive patient outcomes during skin cancer surgery. Although Mohs micrographic surgery is by far the most effective method to treat nonmelanoma skin cancer, it can be limited by its inherent required infrastructure, including time-consuming and expensive on-site histopathology. Previous studies have demonstrated that Raman spectroscopy can accurately detect basal cell carcinoma (BCC) from surrounding normal tissue; however, the biophysical basis of the detection remained unclear. Therefore, we aim to explore the relevant Raman biomarkers to guide BCC margin resection. Raman imaging was performed on skin tissue samples from 30 patients undergoing Mohs surgery. High correlations were found between the histopathology and Raman images for BCC and primary normal structures (including epidermis, dermis, inflamed dermis, hair follicle, hair shaft, sebaceous gland and fat). A previously developed model was used to extract the biochemical changes associated with malignancy. Our results showed that BCC had a significantly different concentration of nucleus, keratin, collagen, triolein and ceramide compared to normal structures. The nucleus accounted for most of the discriminant power (90% sensitivity, 92% specificity – balanced approach). Our findings suggest that Raman spectroscopy is a promising surgical guidance tool for identifying tumors in the resection margins.
1. Introduction
Nonmelanoma skin cancer is by far the most common malignancy worldwide. Among more than 5 million new cases diagnosed annually in the US, approximately 80% are basal cell carcinoma (BCC) [1]. Currently, Mohs micrographic surgery (Mohs) is the most effective method for the treatment of BCC. The 5-yr recurrence rate of Mohs (1 – 3% for primary BCC and 5 – 7% for recurrent BCC) is much lower than standard surgical excision (3 – 10% in primary BCC and >17% in recurrent BCC) [2]. Mohs involves iterative excision of surgical margins of each stage, followed by frozen section histopathology. If the histopathological diagnosis indicates tumor still exists, further tissue layers will be removed until the margins are clear.
Although effective, Mohs has several limitations, including time, expense, training requirements and lab infrastructure. Most tumors require 1 to 3 stages (sometimes as many as 5 – 6) for complete removal, with patients waiting under local anesthesia between each stage [3]. The total time for Mohs surgery can be anywhere from one to five hours. Infrastructure requirements may also be limiting, including the building and maintenance of histology lab, staff training and physician training. As these requirements pose significant barriers to its use, Mohs is used in less than half (~40%) of cases, and less effective treatments such as standard surgical excision are more frequently employed [4]. Moreover, disparities in access to care, such as in among different geographic regions and racial and ethnic groups, leads to substantial underutilization of Mohs in underserved populations [4,5]. Therefore, a critical unmet need exists for low-infrastructure technologies that would enable general dermatologists to perform resections with high accuracy.
Raman spectroscopy is a nondestructive and label-free optical technique, and has demonstrated great clinical merits for tumor margin assessment in numerous types of cancer, including skin [6], brain [7,8], oral cavity [9,10], breast [11,12], and stomach [13]. Those studies either use fiber-optical Raman probe for single point sampling or Raman microscopy (also called Raman microspectroscopy) for optical imaging. One major advantage of Raman microscopy is that it has high resolution and sectioning comparable with that of conventional histology. Moreover, it provides an objective diagnosis with minimal tissue processing. Previous studies have demonstrated Raman microscopy can discriminate BCC from normal skin tissues with sensitivity of 90 – 100% and specificity of 85 – 93%. Nijssen et al. was among the first to discriminate BCC from surrounding normal tissue using Raman spectroscopy and reached 100%/93% sensitivity/specificity (15 patients, 59 spectra) [14]. Lieber et al. developed an in vivo Raman microscopy and achieved 100%/91% sensitivity/specificity (19 patients, 42 spectra) in classifying BCC and squamous cell carcinoma from paired normal skin tissues [15]. Larraona-Puy et al. demonstrated the ability of Raman microscopy in discriminating BCC from surrounding normal tissue with 90%/85% sensitivity/specificity (20 patients, 329 spectra) [16]. Kong et al. combined Raman microscopy with autofluorescence imaging to increase acquisition speed and achieved 100% sensitivity and 92.9% specificity for discriminating BCC [6].
Prior studies mostly utilized statistical algorithms to extract the spectral differences between BCC and normal tissue, such as principal component analysis [6,14], linear discriminant analysis [17] and maximum representation and discrimination feature [15]. Although such methods provided high diagnostic accuracy, they did not elucidate the nature and biochemical processes responsible for the spectral differences. Understanding the biophysical basis of the discriminatory power of Raman spectroscopy would increase fundamental knowledge of cancer processes as well as lay the groundwork for improving the diagnostic performance of the technology [18]. Therefore, our aim in this study is to obtain biophysically relevant markers from Raman spectra of BCC and surrounding normal tissue, and then build diagnostic model to guide BCC tumor margin delineation.
Our group recently proposed a biophysical human skin cancer model, an inverse model that infers the skin’s biochemical makeup from its Raman spectrum [19]. Different from previous studies that selected a number of Raman bands as “fingerprints” to discriminate between healthy skin and tumor regions [16,20], our method is based on the fitting of pure spectral components. We validated the model using previous in vivo human skin cancer screening data [21], and demonstrated the feasibility of Raman spectroscopy to capture relevant biophysical changes accounting for the in vivo diagnosis [22]. Later, we presented a preliminary study of BCC tumor margin detection using the biophysical model based on a small data set from 14 patients [23]. This study demonstrated the feasibility of detecting biophysical changes between BCC and five primary normal structures (epidermis, dermis, hair follicle, sebaceous gland and fat), but has several limitations: firstly, the number of patients is small; secondly, inflamed dermis was not included in the study, which may be confused with BCC in histopathological diagnosis [6,16]; finally, a more comprehensive analysis is needed to link our biophysical approach with molecular vibrational approaches [24,25].
Here, we demonstrate that Raman spectroscopy is highly sensitive in capturing the biochemical differences between BCC and surrounding normal skin structures. Based on these biochemical differences, we can develop diagnostic algorithms to accurately discriminate BCC in Mohs excisions, which supports the future development of intraoperative assessment of tumor margins.
2. Materials and methods
2.1 Patients and sample preparation
This study was approved by the Institutional Review Board (IRB) at The University of Texas at Austin and the Seton Healthcare Family. A total of 30 frozen tissue blocks were collected from 30 patients who had undergone Mohs at Austin Dermatologic Surgery Center. 18 samples were found to have both BCC tumors and surrounding normal tissue, and 12 samples contained only normal tissue. Before the Raman experiment, 20μm tissue sections were sliced from frozen tissue blocks at −22C and transferred onto low Raman background glass slides (MgF2 substrates). Serial sections were transferred onto microscope slides for hematoxylin and eosin (H&E) staining. The H&E technique stains cell nuclei in purple (hematoxylin), and intracellular or extracellular protein in red (eosin). Tissue structure identification and histopathological diagnosis were provided by a board-certified dermatologist.
2.2 Raman imaging experiment and data preprocessing
Raman imaging was performed using a custom-built confocal Raman microscope. We used a longer wavelength laser (830nm) to minimize tissue autofluorescence. Reflectance confocal and bright-field images are also collected simultaneously. A detailed system description can be found elsewhere [19]. The lateral, axial and spectral resolution of the system was approximately 1 μm, 8 μm and 8 cm−1, respectively. The power delivered to the sample was approximately 45mW. Raman images were collected from “tissue-level” regions varying from 60 × 60 μm2 to 100 × 100 μm2 (2μm steps, 2s per step). The “tissue-level” regions consist of ~10 – 100 cells, approximating the resolution of a dermatologist reading an H&E slide.
Raw Raman spectra underwent wavenumber calibration, dark noise removal, cosmic ray removal, smoothing, and a fifth-order polynomial fitting [26] to remove tissue fluorescence background. Spectral response calibration was conducted using a tungsten halogen lamp (LS-1-CAL, Ocean Optics, FL, USA). The effective spectral range was 800 to 1790 cm−1. Data were normalized to the area under curve.
2.3 Clustering analysis
Raman pseudo-color images were generated by k-means clustering. K-means is an unsupervised algorithm for cluster analysis and can easily handle large amounts of Raman spectroscopy data for cell [27] and tissue [14] imaging. The first 100 principal components accounting for 95% - 99% of the variation in the data set served as input for K-means. The number of clusters was determined by visual comparison of the pseudo-color image and histopathology. Each cluster was represented by a centroid Raman spectrum and assigned a different color. To eliminate spectral outliers, any spectrum that belonged to a cluster that was more than three times the standard deviation from the mean of that cluster was omitted [14]. We then annotated the centroid Raman spectrum of each cluster as either BCC or normal skin structures: inflamed dermis (Inf), epidermis (Epi), dermis (Der), hair follicle (HF), hair shaft (HS), sebaceous gland (SG), and fat. HS is a long filament in the center of the follicle extended above the surface of epidermis (also called hair). HF is the sheath of cells and connective tissue that surrounds the root of a hair. HF and HS were separated because they had heterogenous biochemical composition [17]. While muscle tissue can be present in Mohs sections and has been studied in other Raman studies [6,20], it generally appears in a small minority of cases; thus, we have excluded it from this study.
2.4 Raman biophysical model
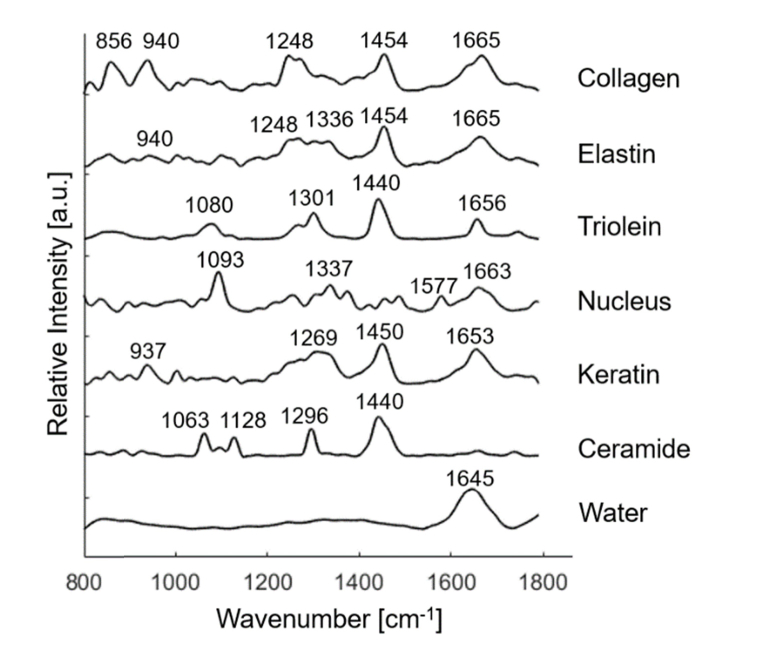
A previously developed biophysical model [19] was used to extract the biochemical composition from the centroid Raman spectra of BCC and normal tissue structures. The model consists of seven Raman active components: collagen, elastin, triolein, keratin, nucleus, ceramide, and water, as shown in Fig. 1. Melanin was not included because melanin played a minor role in fitting the spectra of normal tissue and non-pigmented BCC. All the model components were extracted from human skin sections in situ and contained rich biochemical and structural information. For instance, collagen and elastin were the major constituents in dermal extracellular matrix, while keratin was rich in epidermal extracellular matrix. Nucleus represented the nuclear material in the cell. Ceramide was important constituent in epidermal lipid, while triolein existed in small amount in sebaceous lipid and large amount in subcutaneous fat. Peak positions of the main Raman bands are displayed in Table 1 [19,28].
Fig. 1.
Raman active components in the biophysical model of skin, including collagen, elastin, triolein, nucleus, keratin, ceramide, and water. Peak positions of the main Raman bands are labeled.
Table 1. Peak positions of the main Raman bands in the Raman active components.
| Raman peaks [cm−1] | Band assignments | Components |
|---|---|---|
| 835 | DNA backbone: O-P-O/tyrosine | nucleus |
| 855 | CCH bending (aromatic) of protein | elastin, keratin |
| 856 | C-C vibration of the collagen backbone | collagen |
| 937 | C-C stretching of proline and valine and protein backbone | keratin |
| 940 | C-C stretching of protein backbone | collagen, elastin |
| 1003 | C-C vibration of phenyl ring | collagen, elastin, keratin |
| 1063 | C-C asymmetric skeletal stretching of lipids (trans-conformation) | ceramide |
| 1080 | C-C skeletal stretching in lipids | triolein |
| 1093 | O-P-O symmetric stretching vibration of the DNA backbone | nucleus |
| 1128 | C-C symmetric skeletal stretching | ceramide |
| 1248 | Amide III (β-sheet and random coil conformations) | collagen, elastin |
| 1254 | β sheet/ thymine/ cytosine (DNA base/ DNA & RNA base) | nucleus |
| 1269 | Amide III (α-helix conformation), C-N stretching, N-H in-plane bending | collagen, elastin, keratin |
| 1301 | C-H modes (CH2 twisting and wagging) of lipids; CH2/CH3 bands | triolein |
| 1336 | Amide III, C-N stretching, N-H in-plane bending | elastin |
| 1337 | adenine, guanine (DNA & RNA base) | nucleus |
| 1440 | CH2/CH3 bands | triolein, ceramide |
| 1450 | C-H bending of proteins | keratin |
| 1454 | C-H stretching, C-H asymmetric deformation | collagen, elastin |
| 1577 | Adenine/ guanine (DNA base) | nucleus |
| 1645 | O-H bending mode of liquid water | water |
| 1653 | C-O stretching model of amide I | keratin |
| 1656 | C-C lipids | triolein |
| 1663 | Thymine/ guanine (DNA base) | nucleus |
| 1665 | C-O amide I vibration | collagen, elastin |
2.5 Model fitting and statistical analysis
Each centroid Raman spectrum was described as a linear combination of the model components according to a non-negative linear least-squares fitting criteria. The fit coefficients were then visualized using scatter plots. The variation of the fit coefficient is represented by the error bar generated by the 95% confidence interval. The fit coefficients of BCC and individual normal tissue structures were statistically compared. To account for dependencies in the data inherent from measuring multiple skin structures per patient, linear fixed-effects models were employed with the skin structures (epidermis, dermis, etc.) treated as a fixed effect and the patient treated as a random effect. The models were fitted using restricted maximum likelihood and p-values were derived from t-tests using Satterthwaite approximations [29,30].
2.6 Diagnostic algorithm
A logistic regression classifier was built to discriminate BCC from normal tissue structures based on their fit coefficients. Leave-one-patient-out receiver operator characteristic (ROC) analysis was used to determine the optimum number of input model components., i.e., the models were trained using a subset of 29 patients and tested on the remaining one patient. To avoid overfitting, the number of input model components was in all cases no more than 4. Leave-one-spectrum-out ROC analysis was also performed for inspection of the misclassified spectra. The area under the ROC curve (AUC) was calculated to measure the discriminatory power of the classification model. The combination of sensitivity and specificity with of greatest clinical value was obtained from the ROC curve. Sensitivity determines the ability of the model to correctly identify the positive group, whereas specificity is the ability of the model to correctly identify the negative group. Positive predictive value (PPV) and negative predictive value (NPV) were derived from Eq. (1) and (2), respectively. PPV is the probability that the positive group (BCC) identified by the model is truly positive. NPV is the probability that the negative group (normal tissue) identified by the model is truly negative.
| (1) |
| (2) |
3. Results
3.1 Annotated tissue spectra database
In total, we obtained 223 centroid Raman spectra from 30 patients, including 50 spectra from BCC, and 173 spectra from normal structures (including inflamed dermis (N = 19), epidermis (N = 26), dermis (N = 47), hair follicle (N = 31), hair shaft (N = 18), sebaceous gland (N = 22), and fat (N = 10)).
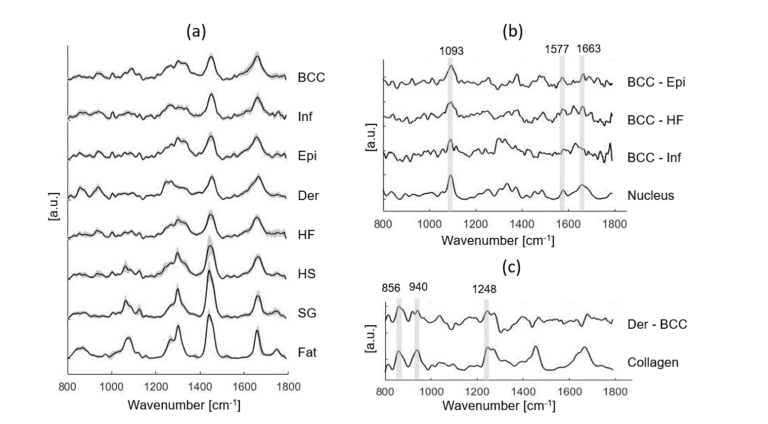
Figure 2 shows a typical example of Raman experiment. Good visual correspondence was observed between Raman pseudo-color images and H&E images of the serial section. Figure 3 shows the mean Raman spectra of BCC and normal structures. The main differences between BCC and epidermis/hair follicle/inflamed dermis can be found at 1093, 1577, 1663 cm−1 (assigned to nucleus), while the main differences between dermis and BCC can be found at 856, 940, 1248 cm−1 (assigned to collagen).
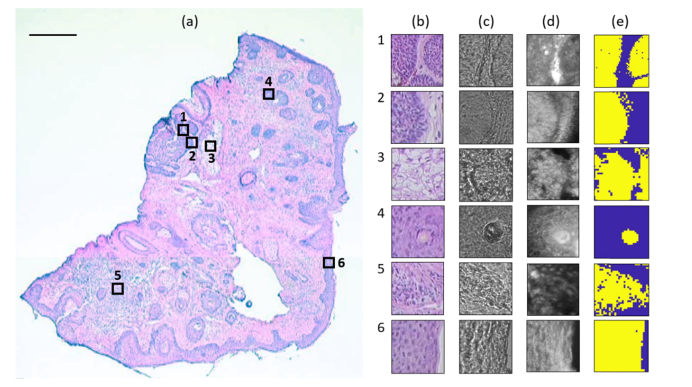
Fig. 2.
Raman experiment on a typical skin tissue section. (a) H&E image shows six measured regions of 100 × 100 μm2, being represented as empty squares. Scale bar: 500 μm. (b) H&E image of the serial section. (c) Bright-field image. (d) Reflectance confocal images. (e) Raman pseudo-color image generated by k-means. Region 1 and 2 contains BCC (yellow) and dermis (blue), region 3 contains sebaceous gland (yellow) and MgF2 substrate (blue), region 4 contains hair shaft (yellow) and hair follicle (blue), region 5 contains inflamed dermis (yellow) and dermis (blue), and region 6 contains epidermis (yellow) and MgF2 substrate (blue).
Fig. 3.
(a) Mean Raman spectra ± SD of all individual tissue structures, including BCC, Inf (inflamed dermis), Epi (epidermis), Der (dermis), HF (hair follicle), HS (hair shaft), SG (sebaceous gland) and fat. (b) Spectral differences of mean spectra of BCC minus Epi, BCC minus HF, and BCC minus Inf are compared with the basis spectrum of nucleus. (c) Spectral difference of mean spectra of dermis minus BCC is compared with the basis spectrum of collagen. Peak positions of the main Raman bands are labeled.
3.2 Biophysical model fitting results
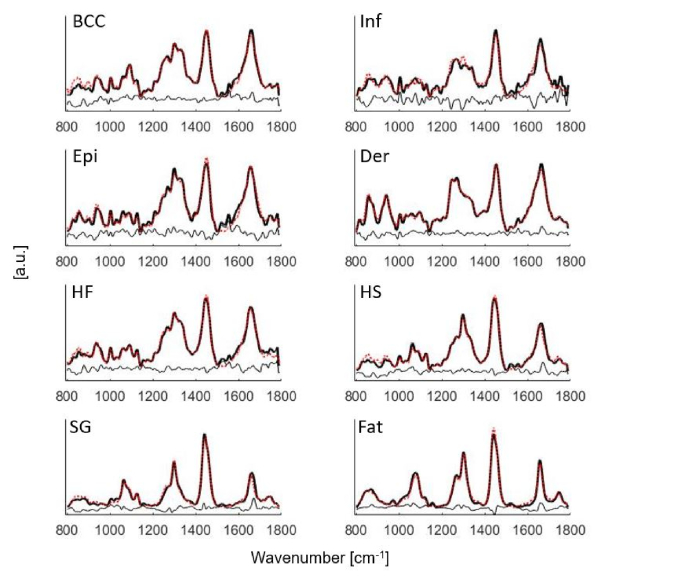
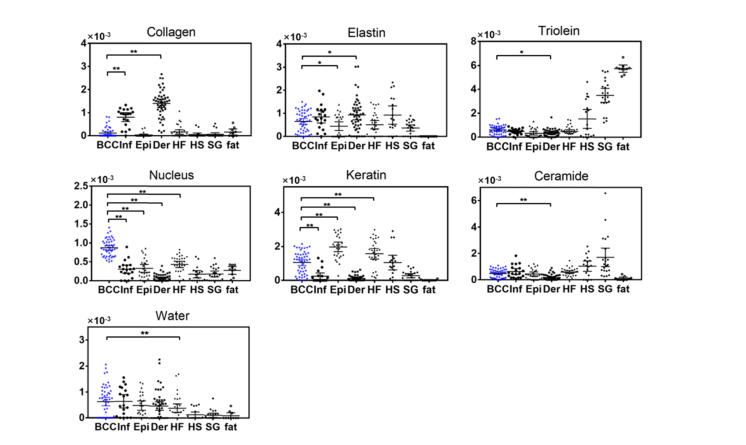
Figure 4 shows the mean Raman spectra fit to the model components in Fig. 1. The fit coefficients of the model components computed from the biophysical model were visualized in Fig. 5. Statistical significance for BCC versus inflamed dermis, BCC versus epidermis, BCC versus dermis, and BCC versus hair follicle was demonstrated in Fig. 5. A complete list of statistical comparison between BCC and individual normal structures was displayed in Table 2.
Fig. 4.
Mean Raman spectra of BCC, Inf (inflamed dermis), Epi (epidermis), Der (dermis), HF (hair follicle), HS (hair shaft), SG (sebaceous gland) and fat fit to the model components in Fig. 1. Black solid lines: mean tissue spectra. Red dotted lines: model fits. Residuals are also plotted on the bottom.
Fig. 5.
Fit coefficients of the biophysical markers for BCC (N = 50), Inf (inflamed dermis, N = 19), Epi (epidermis, N = 26), Der (dermis, N = 47), HF (hair follicle, N = 31), HS (hair shaft, N = 18), SG (sebaceous gland, N = 22) and fat (N = 10). Each point represents a spectrum data. Statistical significance of BCC versus Inf, BCC versus Epi, BCC versus Der, and BCC versus HF are labeled. *p≤0.05, **p≤0.01.
Table 2. Statistical comparison between BCC and individual normal structures (*p≤0.05, **p≤0.01).
| Collagen | Elastin | Triolein | Nucleus | Keratin | Ceramide | Water | ||
|---|---|---|---|---|---|---|---|---|
| BCC vs. | Inflammation | ** | ** | ** | ||||
| Epidermis | * | ** | ** | |||||
| Dermis | ** | * | * | ** | ** | ** | ||
| Hair follicle | ** | ** | ** | |||||
| Hair shaft | ** | ** | ** | |||||
| Sebaceous gland | ** | ** | ** | ** | ** | ** | ||
| Fat | ** | ** | ** | ** | * |
3.3 Discrimination between BCC and normal tissue structures
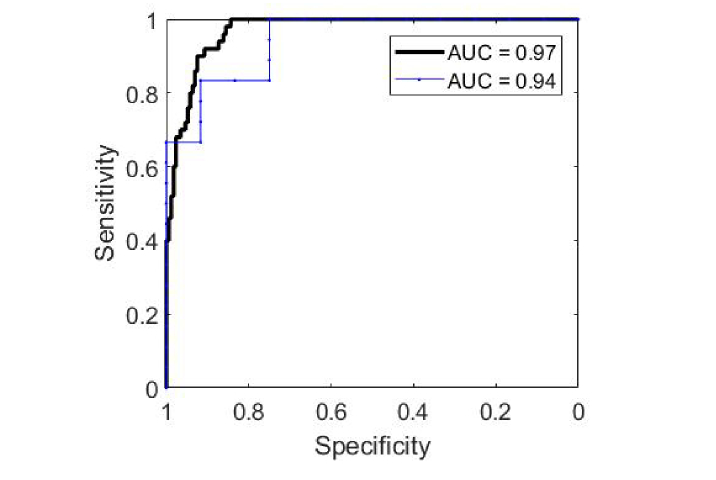
The optimum results to classify BCC from all normal structures were reached by using the fit coefficient of nucleus alone, leading to an AUC of 0.94 for leave-one-patient-out ROC analysis, and 0.97 for leave-one-spectrum-out ROC analysis (Fig. 6). Additional inclusion of keratin, triolein, and collagen in the classification model did not further improve the diagnostic performance (Table 3). A summary of misclassifications is displayed in Table 4. One may see that hair follicle and epidermis are more commonly misclassified as BCC compared to other normal structures. The discrimination threshold is chosen to prioritize either high sensitivity or high specificity, or a balanced combination of sensitivity and specificity (both ≥90%). By prioritizing high sensitivity, classification result achieved 100% sensitivity, 84% specificity, 65% PPV, and 100% NPV; while by prioritizing high specificity, classification result reached 52% sensitivity, 99% specificity, 93% PPV, and 88% NPV.
Fig. 6.
ROC analysis for classifying BCC from all normal structures. Black thick line: leave-one-spectrum-out ROC curve. Blue thin line: leave-one-patient-out ROC curve.
Table 3. Discriminating between BCC and all normal structures using optimum combination of components.
| # components | Optimum combinations | Leave-one-patient-out ROC AUC |
Leave-one-spectrum-out ROC AUC |
|---|---|---|---|
| 1 | [nucleus] | 0.94 | 0.97 |
| 2 | [nucleus, keratin] | ||
| [nucleus, triolein] | |||
| [nucleus, collagen] | 0.94 | 0.97 | |
| 3 | [nucleus, triolein, keratin] | ||
| [nucleus, triolein, collagen] | 0.94 | 0.97 | |
| 4 | [nucleus, keratin, triolein, collagen] | 0.93 | 0.96 |
Table 4. Summary of misclassifications by prioritizing high sensitivity or specificity.
| Sensitivity/ Specificity (%) |
PPV/ NPV (%) |
BCC | Normal tissue structures |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Inf | Epi | Der | HF | HS | SG | Fat | ||||
| Prioritizing high sensitivity | 100/84 | 65/100 | 0/50 | 3/19 | 6/26 | 0/47 | 15/31 | 2/18 | 1/22 | 0/10 |
| Prioritizing high specificity | 52/99 | 93/88 | 24/50 | 1/19 | 1/26 | 0/47 | 0/31 | 0/18 | 0/22 | 0/10 |
| Balanced sensitivity and specificity | 90/92 | 76/97 | 5/50 | 2/19 | 5/26 | 0/47 | 6/31 | 1/18 | 0/22 | 0/10 |
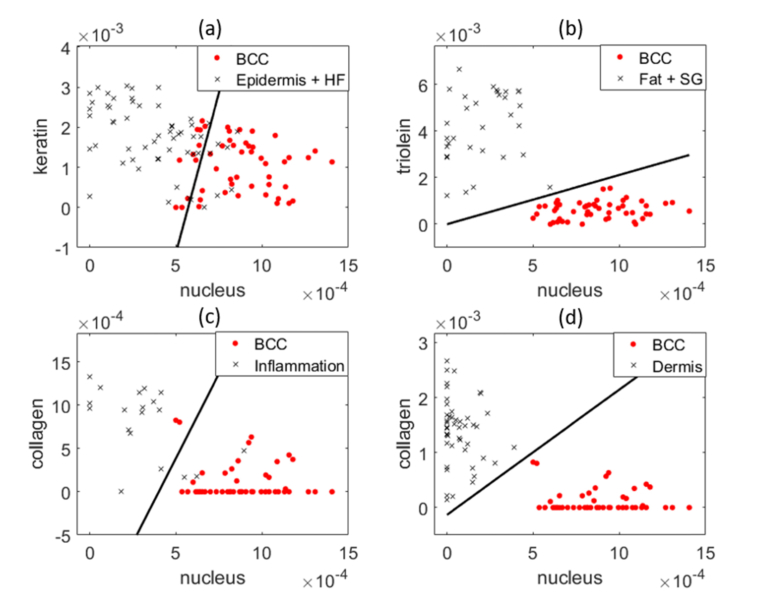
Figure 7 represents separating BCC from different categories of normal structures using the fit coefficients of two primary model components. Figure 7(a) shows that epidermis and hair follicle have the largest overlap with BCC compared to other normal structures. The overlap occurs when epidermis and hair follicle have comparable level of keratin and nucleus content as BCC. Figure 7(b) shows that fat and sebaceous gland can be easily separated from BCC because they have distinct nucleus and lipid content. Figure 7(c) (d) demonstrate inflamed dermis/dermis can also be separated from BCC using their distinct differences in nucleus and collagen.
Fig. 7.
Scatter plots demonstrates the performance of two primary model components in discriminating BCC from normal structures. (a) Nucleus and keratin content of BCC, epidermis and HF. (b) Nucleus and triolein content of BCC, fat and SG. (c) Nucleus and ceramide content of BCC and inflamed dermis. (d) Nucleus and collagen content of BCC and dermis. Red dots: BCC. Black crosses: normal tissue structures. Each point represents a spectrum data. The black line is the decision line drawn by logistic regression.
4. Discussion
In this study, we evaluated the accuracy of Raman to discriminate between BCC and normal skin structures in excised Mohs skin sections. In contrast to previous studies, our discriminatory model was built upon the biochemical differences of Raman active components extracted from a previously developed model [19].
Our results show markedly different biochemical and structural compositions between BCC and normal tissue (Fig. 5 and Table 2). One important finding is that the spectra of BCC has statistically significantly larger contribution of nucleus material compared to normal tissue structures, which is consistent with previous Raman studies [14,16,31]. As seen in Fig. 3(b), mean BCC spectra have stronger contribution from 1093, 1577 and 1663 cm−1 assigned to nucleus compared to epidermis, hair follicle and inflamed dermis. This is also consistent with the H&E image in Fig. 2(b), where there are smaller amounts of cytoplasm and higher density of cells present in the BCC tumor. Other studies have pointed out similar or increased nucleic contributions in inflamed regions according to the presence of specific Raman bands [6,20].
Another important finding is that BCC has significantly larger contribution from keratin compared to inflamed/normal dermis, and lower contribution from keratin than epidermis and HF. Keratin is a fibrous structural protein produced by keratinocytes and is abundant in normal epidermis. Keratinocytes are the predominant cells originating in the basal layer between epidermis and dermis, which then move towards the skin surface in a process of maturation and differentiation. The outermost skin layer, stratum corneum, consists about 80% of keratin in dry weight [25]. HF is also rich in keratin. The epithelium of HF forms a cylinder with different concentric layers, including the inner and outer root sheath, with each one expressing a distinct pattern of keratin [32]. Because keratin expression is closely related to differentiation of tumors, it plays a significant role in identifying the origin of BCC. Several studies have discovered that BCC may arise from germinative cells within the basal layer of epidermis or follicular structures [33,34].
Furthermore, BCC has significantly smaller amounts of collagen and elastin compared to dermis. The difference spectra in Fig. 3(c) shows that BCC spectra have weaker contribution from 856, 940, 1248 cm−1 assigned to the vibrational modes of collagen type I and elastin. This is likely the result of the epidermal origin of the BCC, high in cellular content and low in connective tissue. We also found collagen in inflamed dermis is higher than BCC, but lower than normal dermis. This confirms the observation of an early study that a dense inflammatory infiltrate appears to have less collagen than normal dermis [14].
Finally, BCC has significantly larger amount of triolein and ceramide compared to dermis, and larger amount of triolein compared to epidermis. The difference spectrum of BCC and dermis also shows higher contribution at 1080, 1128, 1440 cm−1 associated with lipids. A previous biophysical model built upon nonmelanoma skin cancer found an increased contribution of triolein to BCC spectra [35]. Another biophysical model also found an increase in triolein in urological carcinoma lesions [36]. Those studies suggested that maintaining sufficient lipid levels may be necessary to sustain fast tumor growth.
Some normal structures have high biochemical similarity. For instance, the biochemical compositions of epidermis and HF are highly similar, which agrees with the fact that HF is an invagination of normal dermis [17]. As a result, a previous study grouped epidermis and HF together for clustering analysis [6]. The biochemical composition of HS may resemble either HF or SG. This is because HS consists of terminally differentiated keratinocytes that are produced by HF [32], but it is sometimes coated by the sebum secreted by SG [37]. This also explains why the fit coefficients of HS have larger variation compared to the other components. Fat and SG both have low nucleus and high triolein content, so they can be easily discriminated from BCC (Fig. 7(b)).
We evaluated the diagnostic performance by prioritizing either high sensitivity or high specificity (Table 4). Achieving high specificity is clinically significant in tissue-conserving surgeries such as Mohs, when preserving normal tissue is a critical concern. Appropriate use criteria of Mohs include tumor location (such as “mask areas” of face), size, and patient type [38]. On the other hand, achieving high sensitivity is more clinically significant when the primary goal is to capture the entire margin based on aggressive growth histology rather than preserving normal tissue.
By prioritizing high sensitivity, we achieved 100% sensitivity and 84% specificity in discriminating BCC from all normal tissue structures. Our results show that that nucleus accounts for most of the discriminant ability. By using the fit coefficient of nucleus alone, 100% of the spectra annotated as dermis (40/40) and fat (10/10) were correctly classified. Most of the spectra annotated as SG (95%, 21/22) and HS (89%, 16/18) were also correctly classified. The misclassification in these latter two categories may be due to unknown tissue structures grouped as the same cluster as SG or HS, leading to high fitting error. On the contrary, HF, epidermis and inflamed dermis were the normal structures that were more easily misclassified as BCC. 16 out of 31 spectra annotated as HF were correctly classified (52%), 20 out of 26 spectra annotated as epidermis were correctly classified (77%), and 16 out of 19 spectra annotated as inflamed dermis were correctly classified (84%).
HF and epidermis were most easily misclassified as BCC. Figure 7(a) demonstrates that those misclassifications occur due to high nucleus content in some of HF and epidermis. The main reason is that HF may have abundant basal cells in inner and outer root sheath layer, whereas epidermis is rich in basal keratinocyte stem cells in stratum basale layer and polyhedral keratinocytes in stratum spinosum layer. About 16% of inflamed dermis was also misclassified as BCC. Corresponding with the H&E images indicated that inflamed dermis regions have higher number of nucleus than normal dermis, so their spectra were more similar to BCC than normal dermis.
By prioritizing high specificity, we achieved 99% specificity, 52% sensitivity and 93% PPV. High specificity indicates a region has high risk, so a dermatologist could remove more tissue from the corresponding region with high confidence of it being tumor [39]. In a Mohs guidance setting, this approach could reduce the number of skin samples processed for histopathology. The dermatologist would still make a histopathological diagnosis on the final stage to ensure all the tumors are entirely removed; thus, one may be able to tolerate lower specificity. Ultimately, this system would need to be tested in an intraoperative setting to determine its impact on reducing the number of tissue samples needing histology processing. Our results showed 24 out of 50 spectra annotated as BCC were misclassified as normal structures. All the spectra annotated as dermis, HF, HS, SG and fat were classified correctly. Most of the spectra annotated as inflamed dermis (95%, 18/19) and epidermis (96%, 25/26) were correctly classified.
Although we have demonstrated Raman microscopy is highly accurate in evaluating skin tumor surgical margin, one major limitation is lengthy acquisition time. To raster scan a tissue 1mm2 in size, it would take around 10 – 20 hours, making it unpractical for intraoperative use. To overcome this limitation, several wide-field imaging techniques could be employed. Kong et al. integrated Raman microscopy with tissue autofluorescence imaging and achieved one or two orders of magnitude faster speed [6]. Karen et al. developed fluorescence confocal mosaicking microscopy and proved its potential for rapid assessment of BCC margins during Mohs [40]. Flores et al used fluorescence confocal mosaicking microscopy to enable rapid detection of residual tumor directly in the surgical wounds on patients [41]. Further directions for this work include speeding up Raman acquisition without losing molecular specificity, and combining Raman spectroscopy with wide-field imaging technique for fast intraoperative surgical guidance.
Acknowledgments
We would like to thank the Seton Healthcare family and the Austin Dermatological Surgical Center for collaboration. We appreciate the time and help of all the staff, physician assistants, and doctors. We would like to thank Sandra Esparza and Leandra Turner for preparing samples, Professor Aaron Baker for providing the histology facility, Greg Lyness and Esther Maier for H&E staining, and UT Statistics & Data Science Department for providing statistical consulting.
Funding
Cancer Prevention & Research Institute of Texas (RP130702).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Society A. C., Cancer facts & figures 2018 (American Cancer Society, Altanta, 2018). [Google Scholar]
- 2.Mosterd K., Krekels G. A., Nieman F. H., Ostertag J. U., Essers B. A., Dirksen C. D., Steijlen P. M., Vermeulen A., Neumann H., Kelleners-Smeets N. W., “Surgical excision versus Mohs’ micrographic surgery for primary and recurrent basal-cell carcinoma of the face: a prospective randomised controlled trial with 5-years’ follow-up,” Lancet Oncol. 9(12), 1149–1156 (2008). 10.1016/S1470-2045(08)70260-2 [DOI] [PubMed] [Google Scholar]
- 3.Smeets N. W., Kuijpers D. I., Nelemans P., Ostertag J. U., Verhaegh M. E., Krekels G. A., Neumann H. A., “Mohs’ micrographic surgery for treatment of basal cell carcinoma of the face--results of a retrospective study and review of the literature,” Br. J. Dermatol. 151(1), 141–147 (2004). 10.1111/j.1365-2133.2004.06047.x [DOI] [PubMed] [Google Scholar]
- 4.Viola K. V., Jhaveri M. B., Soulos P. R., Turner R. B., Tolpinrud W. L., Doshi D., Gross C. P., “Mohs micrographic surgery and surgical excision for nonmelanoma skin cancer treatment in the Medicare population,” Arch. Dermatol. 148(4), 473–477 (2012). 10.1001/archdermatol.2011.2456 [DOI] [PubMed] [Google Scholar]
- 5.Shareef M. S., Hussain W., “The Mohs histotechnician: a review of training and practice within 29 centres in the UK,” Clin. Exp. Dermatol. 38(6), 589–593 (2013). 10.1111/ced.12021 [DOI] [PubMed] [Google Scholar]
- 6.Kong K., Rowlands C. J., Varma S., Perkins W., Leach I. H., Koloydenko A. A., Williams H. C., Notingher I., “Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy,” Proc. Natl. Acad. Sci. U.S.A. 110(38), 15189–15194 (2013). 10.1073/pnas.1311289110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jermyn M., Mok K., Mercier J., Desroches J., Pichette J., Saint-Arnaud K., Bernstein L., Guiot M. C., Petrecca K., Leblond F., “Intraoperative brain cancer detection with Raman spectroscopy in humans,” Sci. Transl. Med. 7(274), 274ra19 (2015). 10.1126/scitranslmed.aaa2384 [DOI] [PubMed] [Google Scholar]
- 8.Kirsch M., Schackert G., Salzer R., Krafft C., “Raman spectroscopic imaging for in vivo detection of cerebral brain metastases,” Anal. Bioanal. Chem. 398(4), 1707–1713 (2010). 10.1007/s00216-010-4116-7 [DOI] [PubMed] [Google Scholar]
- 9.Behl I., Kukreja L., Deshmukh A., Singh S. P., Mamgain H., Hole A. R., Krishna C. M., “Raman mapping of oral buccal mucosa: a spectral histopathology approach,” J. Biomed. Opt. 19(12), 126005 (2014). 10.1117/1.JBO.19.12.126005 [DOI] [PubMed] [Google Scholar]
- 10.Cals F. L. J., Bakker Schut T. C., Caspers P. J., Baatenburg de Jong R. J., Koljenović S., Puppels G. J., “Raman spectroscopic analysis of the molecular composition of oral cavity squamous cell carcinoma and healthy tongue tissue,” Analyst (Lond.) 143(17), 4090–4102 (2018). 10.1039/C7AN02106B [DOI] [PubMed] [Google Scholar]
- 11.Haka A. S., Volynskaya Z., Gardecki J. A., Nazemi J., Lyons J., Hicks D., Fitzmaurice M., Dasari R. R., Crowe J. P., Feld M. S., “In vivo margin assessment during partial mastectomy breast surgery using raman spectroscopy,” Cancer Res. 66(6), 3317–3322 (2006). 10.1158/0008-5472.CAN-05-2815 [DOI] [PubMed] [Google Scholar]
- 12.Keller M. D., Vargis E., de Matos Granja N., Wilson R. H., Mycek M. A., Kelley M. C., Mahadevan-Jansen A., “Development of a spatially offset Raman spectroscopy probe for breast tumor surgical margin evaluation,” J. Biomed. Opt. 16(7), 077006 (2011). 10.1117/1.3600708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teh S. K., Zheng W., Ho K. Y., Teh M., Yeoh K. G., Huang Z., “Diagnostic potential of near-infrared Raman spectroscopy in the stomach: differentiating dysplasia from normal tissue,” Br. J. Cancer 98(2), 457–465 (2008). 10.1038/sj.bjc.6604176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nijssen A., Bakker Schut T. C., Heule F., Caspers P. J., Hayes D. P., Neumann M. H. A., Puppels G. J., “Discriminating basal cell carcinoma from its surrounding tissue by Raman spectroscopy,” J. Invest. Dermatol. 119(1), 64–69 (2002). 10.1046/j.1523-1747.2002.01807.x [DOI] [PubMed] [Google Scholar]
- 15.Lieber C. A., Majumder S. K., Ellis D. L., Billheimer D. D., Mahadevan-Jansen A., “In vivo nonmelanoma skin cancer diagnosis using Raman microspectroscopy,” Lasers Surg. Med. 40(7), 461–467 (2008). 10.1002/lsm.20653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Larraona-Puy, A. Ghita, A. Zoladek, W. Perkins, S. Varma, I. H. Leach, A. A. Koloydenko, H. Williams, and I. Notingher, “Development of Raman microspectroscopy for automated detection and imaging of basal cell carcinoma,” J Biomed Opt 14, 054031–054010 (2009). 10.1117/1.3251053 [DOI] [PubMed]
- 17.Larraona-Puy M., Ghita A., Zoladek A., Perkins W., Varma S., Leach I., Koloydenko A., Williams H., Notingher I., “Discrimination between basal cell carcinoma and hair follicles in skin tissue sections by Raman micro-spectroscopy,” J. Mol. Struct. 993(1-3), 57–61 (2011). 10.1016/j.molstruc.2010.10.046 [DOI] [Google Scholar]
- 18.Pence I., Mahadevan-Jansen A., “Clinical instrumentation and applications of Raman spectroscopy,” Chem. Soc. Rev. 45(7), 1958–1979 (2016). 10.1039/C5CS00581G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng X., Moy A. J., Nguyen H. T. M., Zhang J., Fox M. C., Sebastian K. R., Reichenberg J. S., Markey M. K., Tunnell J. W., “Raman active components of skin cancer,” Biomed. Opt. Express 8(6), 2835–2850 (2017). 10.1364/BOE.8.002835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boitor R., Kong K., Shipp D., Varma S., Koloydenko A., Kulkarni K., Elsheikh S., Schut T. B., Caspers P., Puppels G., van der Wolf M., Sokolova E., Nijsten T. E. C., Salence B., Williams H., Notingher I., “Automated multimodal spectral histopathology for quantitative diagnosis of residual tumour during basal cell carcinoma surgery,” Biomed. Opt. Express 8(12), 5749–5766 (2017). 10.1364/BOE.8.005749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim L., Nichols B., Migden M. R., Rajaram N., Reichenberg J. S., Markey M. K., Ross M. I., Tunnell J. W., “Clinical study of noninvasive in vivo melanoma and nonmelanoma skin cancers using multimodal spectral diagnosis,” J. Biomed. Opt. 19(11), 117003 (2014). 10.1117/1.JBO.19.11.117003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng X., Moy A. J., Nguyen H. T. M., Zhang Y., Zhang J., Fox M. C., Sebastian K. R., Reichenberg J. S., Markey M. K., Tunnell J. W., “Raman biophysical markers in skin cancer diagnosis,” J. Biomed. Opt. 23(5), 1–10 (2018). 10.1117/1.JBO.23.5.057002 [DOI] [PubMed] [Google Scholar]
- 23.Feng X., Moy A. J., Nguyen H. T., Zhang Y., Fox M. C., Sebastian K. R., Reichenberg J. S., Markey M. K., Tunnell J. W., “Raman spectroscopy reveals biophysical markers in skin cancer surgical margins,” in Biomedical Vibrational Spectroscopy 2018: Advances in Research and Industry, (International Society for Optics and Photonics, 2018), 104900B. [Google Scholar]
- 24.Barry B., Edwards H., Williams A., “Fourier transform Raman and infrared vibrational study of human skin: assignment of spectral bands,” J. Raman Spectrosc. 23(11), 641–645 (1992). 10.1002/jrs.1250231113 [DOI] [Google Scholar]
- 25.Caspers P. J., Lucassen G. W., Wolthuis R., Bruining H. A., Puppels G. J., “In vitro and in vivo Raman spectroscopy of human skin,” Biospectroscopy 4(5), S31–S40 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Lieber C. A., Mahadevan-Jansen A., “Automated method for subtraction of fluorescence from biological Raman spectra,” Appl. Spectrosc. 57(11), 1363–1367 (2003). 10.1366/000370203322554518 [DOI] [PubMed] [Google Scholar]
- 27.Miljković M., Chernenko T., Romeo M. J., Bird B., Matthäus C., Diem M., “Label-free imaging of human cells: algorithms for image reconstruction of Raman hyperspectral datasets,” Analyst (Lond.) 135(8), 2002–2013 (2010). 10.1039/c0an00042f [DOI] [PubMed] [Google Scholar]
- 28.Prescott B., Steinmetz W., Thomas G. J., Jr., “Characterization of DNA structures by laser Raman spectroscopy,” Biopolymers 23(2), 235–256 (1984). 10.1002/bip.360230206 [DOI] [PubMed] [Google Scholar]
- 29.D. Bates, M. Mächler, B. Bolker, and S. Walker, “Fitting linear mixed-effects models using lme4,” arXiv preprint arXiv:1406.5823 (2014).
- 30.Luke S. G., “Evaluating significance in linear mixed-effects models in R,” Behav. Res. Methods 49(4), 1494–1502 (2017). 10.3758/s13428-016-0809-y [DOI] [PubMed] [Google Scholar]
- 31.M. A. Short, H. Lui, D. McLean, H. Zeng, A. Alajlan, and X. K. Chen, “Changes in nuclei and peritumoral collagen within nodular basal cell carcinomas via confocal micro-Raman spectroscopy,” J. Biomed Opt 11, 034004–034009 (2006). 10.1117/1.2209549 [DOI] [PubMed]
- 32.Schneider M. R., Schmidt-Ullrich R., Paus R., “The hair follicle as a dynamic miniorgan,” Curr. Biol. 19(3), R132–R142 (2009). 10.1016/j.cub.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 33.Miller S. J., “Biology of basal cell carcinoma (Part I),” J. Am. Acad. Dermatol. 24(1), 1–13 (1991). 10.1016/0190-9622(91)70001-I [DOI] [PubMed] [Google Scholar]
- 34.Yoshikawa K., Katagata Y., Kondo S., “Biochemical and immunohistochemical analyses of keratin expression in basal cell carcinoma,” J. Dermatol. Sci. 17(1), 15–23 (1998). 10.1016/S0923-1811(97)00065-0 [DOI] [PubMed] [Google Scholar]
- 35.Silveira L., Jr., Silveira F. L., Bodanese B., Zângaro R. A., Pacheco M. T. T., “Discriminating model for diagnosis of basal cell carcinoma and melanoma in vitro based on the Raman spectra of selected biochemicals,” J. Biomed. Opt. 17(7), 077003 (2012). 10.1117/1.JBO.17.7.077003 [DOI] [PubMed] [Google Scholar]
- 36.Stone N., Hart Prieto M. C., Crow P., Uff J., Ritchie A. W., “The use of Raman spectroscopy to provide an estimation of the gross biochemistry associated with urological pathologies,” Anal. Bioanal. Chem. 387(5), 1657–1668 (2007). 10.1007/s00216-006-0937-9 [DOI] [PubMed] [Google Scholar]
- 37.Lee W.-S., “Integral hair lipid in human hair follicle,” J. Dermatol. Sci. 64(3), 153–158 (2011). 10.1016/j.jdermsci.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 38.Connolly S. M., Baker D. R., Coldiron B. M., Fazio M. J., Storrs P. A., Vidimos A. T., Zalla M. J., Brewer J. D., Begolka W. S., Berger T. G., Bigby M., Bolognia J. L., Brodland D. G., Collins S., Cronin T. A., Jr., Dahl M. V., Grant-Kels J. M., Hanke C. W., Hruza G. J., James W. D., Lober C. W., McBurney E. I., Norton S. A., Roenigk R. K., Wheeland R. G., Wisco O. J., American Academy of Dermatology. American College of Mohs Surgery. American Society for Dermatologic Surgery Association. American Society for Mohs Surgery. Ad Hoc Task Force , “AAD/ACMS/ASDSA/ASMS 2012 appropriate use criteria for Mohs micrographic surgery: a report of the American Academy of Dermatology, American College of Mohs Surgery, American Society for Dermatologic Surgery Association, and the American Society for Mohs Surgery,” Dermatol. Surg. 38(10), 1582–1603 (2012). 10.1111/j.1524-4725.2012.02574.x [DOI] [PubMed] [Google Scholar]
- 39.Shipp D. W., Rakha E. A., Koloydenko A. A., Macmillan R. D., Ellis I. O., Notingher I., “Intra-operative spectroscopic assessment of surgical margins during breast conserving surgery,” Breast Cancer Res. 20(1), 69 (2018). 10.1186/s13058-018-1002-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karen J. K., Gareau D. S., Dusza S. W., Tudisco M., Rajadhyaksha M., Nehal K. S., “Detection of basal cell carcinomas in Mohs excisions with fluorescence confocal mosaicing microscopy,” Br. J. Dermatol. 160(6), 1242–1250 (2009). 10.1111/j.1365-2133.2009.09141.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flores E. S., Cordova M., Kose K., Phillips W., Rossi A., Nehal K., Rajadhyaksha M., “Intraoperative imaging during Mohs surgery with reflectance confocal microscopy: initial clinical experience,” J. Biomed. Opt. 20(6), 61103 (2015). 10.1117/1.JBO.20.6.061103 [DOI] [PMC free article] [PubMed] [Google Scholar]