Abstract
Mesenchymal stem cells’ differentiation into several lineages is coordinated by a complex of transcription factors and co-regulators which bind to specific gene promoters. The Chromatin-Related Mesenchymal Modulator, CHD9 demonstrated in vitro its ability for remodeling activity to reposition nucleosomes in an ATP-dependent manner. Epigenetically, CHD9 binds with modified H3-(K9me2/3 and K27me3). Previously, we presented a role for CHD9 with RNA Polymerase II (Pol II)-dependent transcription of tissue specific genes. Far less is known about CHD9 function in RNA Polymerase I (Pol I) related transcription of the ribosomal locus that also drives specific cell fate. We here describe a new form, the nucleolar CHD9 (n-CHD9) that is dynamically associated with Pol I, fibrillarin, and upstream binding factor (UBF) in the nucleoli, as shown by imaging and molecular approaches. Inhibitors of transcription disorganized the nucleolar compartment of transcription sites where rDNA is actively transcribed. Collectively, these findings link n-CHD9 with RNA pol I transcription in fibrillar centers. Using chromatin immunoprecipitation (ChIP) and tilling arrays (ChIP– chip), we find an association of n-CHD9 with Pol I related to rRNA biogenesis. Our new findings support the role for CHD9 in chromatin regulation and association with rDNA genes, in addition to its already known function in transcription control of tissue specific genes.
Summary
We report several novel and important findings: (i) CHD9 recombinant protein repositions nucleosomes in an ATP-dependent manner and enables nucleosome sliding. (ii) CHD9 bind to histone H3 methylated at lysine 9 and 27 (K9me2/3 and K27me3) in vitro. (iii) The new discovered nucleolar CHD9 (n-CHD9) form shows a dynamic association within the nucleolar organization and during the cell cycle, is localized at sites of ribosomal RNA biogenesis and undergoes a major reorganization within nucleolar transcriptional centers when Pol I transcription is inhibited. The new findings presented here strongly implicate n-CHD9 localized to the nucleoli as a regulator of ribosomal gene transcription which plays also a role in stromal cells fate and differentiation.
The cell fate and tissue specific differentiation need well-coordinated machinery that acts to regulate gene transcription. The dynamic alterations in chromatin structure allow the interaction of transcription factors and RNA polymerases to bind regulatory elements that determine cell fate. Nucleosomes structural modifications rely on the concerted action of ATP-dependent chromatin–remodeling complexes and histone-modifying enzymes which generate chromatin states that are either permissive or incompatible with transcription (Percipalle and Farrants, 2006; Murawska and Brehm, 2011).
Chromatin remodeling proteins of the SWI2/SNF2 protein super family have been classified into four subfamilies based on homologies within the ATPase subunit: SWI2/SNF2, ISWI, INO80, and CHDs (Narlikar et al.,2002; Lusserand Kadonaga, 2003). The CHD family is named for the three domains found in all of its nine members Chromo, Helicase, and DNA-binding domains. All CHD (1–9) proteins contain two tandem chromo domains which are localized at the N-terminus and serve as methylated lysine recognition modules (Yap and Zhou, 2011). The CHD family is heterogeneous based on additional domains and features and thus divided into three sub-families: CHD1–2, CHD3–5, and CHD 6–9 (Benayahu et al., 2007).
The role of chromatin remodelers in transcription regulation is getting new insights that bring some understanding for their tissue specific activities. CHD 6–8 that belong to the third subfamily were shown to have a role in various disease states. CHD6 has been shown to be involved in hematopoietic stem cell disorders and is linked to motor coordination problems in mice (Lathrop et al., 2010). CHD7 has been identified in patients with CHARGE syndrome that is associated with congenital abnormalities in the central nervous system, retina, heart, inner ear, and nasal regions (Aramaki et al., 2006; Ellison, 2008; Zentner et al., 2010). CHD8 plays a role in neurological disorders (Aramaki et al., 2006; Ellison, 2008; Zentner et al., 2010; Ronan et al., 2013) and autism during early development (Bernier et al., 2014).
Our study is focused on CHD9 (CReMM, Chromatin Related Mesenchymal Modulator) that has been shown to be expressed during the differentiation of osteoprogenitors in vivo (Shur and Benayahu, 2005; Shur et al., 2006a) and in vitro (Marom et al., 2006; Shur et al., 2006b). In previous studies, we have demonstrated that CHD9 interacts with A/T-rich sites within promoters of genes that play a role in osteoblast maturation, such as CBFA1/RUNX2, biglycan, and osteocalcin, and this binding was also modulated by hormones (Shur et al., 2006a). Thereby, relating a role for CHD9 in cellular differentiation and bone growth via RNA Pol II transcription. CHD9 sequence contains LXXLL signature motifs that mediate interaction with nuclear receptors: glucocorticoid receptor (Marom etal., 2006) ERalpha and RXRand transcription factors PPARs (Surapureddi et al., 2006).
In this study, we explore the CHD9 protein function in repositioning nucleosome in vitro and its interaction with methylated histones. We also identify the “ two faces” of CHD9 protein that differ in their phosphorylation profile. An un-phosphorylated form is localized to the nucleoli protein and is associated with rDNA genes. This brings a novel function for CHD9 that is linked to Pol I transcription and ribosomal biogenesis in addition to its previously described role in facilitating transcription of Pol II regulated genes (Shur et al., 2006a,b). Pol I transcription takes place within nucleoli at tandem repeats of rDNA genes. Using imaging and molecular approaches, we had demonstrated the co-localization of CHD9 with Pol I, fibrillarin, and upstream binding factor (UBF) at sites of active rDNA transcription in the nucleolus. We examined the expression of CHD9 during the cell cycle and following the use of Pol I specific transcriptional inhibitors demonstrated the interruption of dynamic relationship of CHD9 with the nucleolar compartment. The unique findings we present reveal that CHD9 can serve as a multi-functional chromatin remodeling participating in regulatory processes and transcription systems. This suggests CHD9 to be a useful tool for following the molecular mechanism of differentiation which determines the fate of mesenchymal cells.
Materials and Methods
Recombinant protein production
CHD9 recombinant protein was cloned into a pFastbac-1 vector containing N-terminal sequences coding for the Flag epitope. The FB-CHD9 (C5–2397AA) clone was transformed into competent DH10Bac Escherichia coli cells selected on LB agar plates with kanamycin, gentamicin, and tetracycline. Isolated recombinant bacmid DNA was transfected into insect cells and viral supernatant was collected and high-titer stock generated using the Bac-to-Bac Baculovirus Expression System (Invitrogen, Carlsbad, CA) and used to infect SF9 insect cells.
ATPase activity
This was determined in an assay containing 33 ng recombinant protein, 20 μM ATP, 0.25 mCi/μl of (γ−32P)ATP (3,000Ci/mmol), 10 mM MgCl2, and 0.75 μl HEMGN buffer (25 mM HEPES pH 7.6, 1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.05% NP-40, and 0.3 M KCl) in a 5 μl reaction. Where indicated, 15 ng of purified oligo-nucleosomes were added (gift Akhilesh Nagaich, NIH). Reaction mixtures were incubated for 30min at 26°C and products were spotted onto polyethyleneimine-cellulose thin-layer chromatography plates and resolved with 0.5 M LiCl in 1 M formic acid. Dried plates were imaged and quantitated on a phosphoimager (Tsukiyama et al., 1999; Aalfs et al., 2001).
Chromatin remodeling assays
To generate chromatin for the nucleosome mobilization assay, mononucleosomes were reconstituted on a 216-bp DNA fragment amplified from the pGEM3z-601 plasmid by using a 32P-end-labeled primer. Labeled DNA was gel-purified, mixed with histone octamers in 2 M salt and subjected to serial dilution for assembly. Reconstituted nucleosomes were further purified on a glycerol gradient. The nucleosome sliding assay was carried out at 30°C in “sliding” buffer (20mM HEPES pH 7.9, 50mM KCl, 0.5mM PMSF, 2mM DTT, 0.05% Nonidet P-40, 10% glycerol, 100μg/ml BSA, 10mM MgCl2,4mM ATP or ATP-γ-S) with 10 fmol of labeled mononucleosomes, 300 f mol of cold nucleosomes and ~100 fmol of recombinant protein. The reaction was stopped by the addition of a competitor mix (750 ng of calf thymus DNA and 500 ng of oligonucleosomes). The total reaction was directly loaded onto a 5% native polyacrylamide gel (37.5:1) and run for 4.5 h at 4°C or was further digested by DNaseI, processed, precipitated, and run on denaturing 8% polyacrylamide-urea gels (37.5:1, Aalfs et al., 2001).
Peptide binding assays
Recombinant CHD9 protein (500 ng) was incubated with 2 mg peptides 21-amino acid-biotinylated (trimethylated or dimethylated Lys 4, Lys 9, Lys 27 or unmodified histone H3 N-terminal peptides (Upstate Biotechnology, Lake Placid, NY) and 5 μl streptavidin magnetic beads (Dynal) in binding buffer (150mM NaCl, 50mM HEPES, 0.1% Tween, 10%glycerol, 2 mg/ml pepstatin, 2mg/ml leupeptin, 1mM phenylmethyl sulphonyl fluoride (PMSF), and 0.5 mM dithiothreitol [DTT]) for 2 h at 4°C with rotation. The beads were collected by separation on a magnet, washed three times with binding buffer. Reaction mixtures were separated on 3–8% Tris-glycine gels and immunoblotted with an anti-Flag antibody to detect rCHD9 (Pray-Grant et al., 2005).
Tissue culture
MBA-15 mouse stromal cells (Benayahu et al., 1989) and COS-7 cells were cultured in Dulbecco’s modified essential medium (DMEM) growth media with the addition of 10% heat-inactivated fetal bovine serum (FBS), 1% glutamine, 1% penicillin–streptomycin and maintained in 5% CO2 at 37°C.
Inhibitors of RNA polymerase transcription
MBA-15 cells were treated with RNA Polymerase inhibitors. Actinomycin D (0.04μg/ml) or DRB (5,6-dichloro-1-D-ribofuranosylbenzimidazole, 100μg/ml) was added into the growth media for 3 or 6 h, respectively, and analyzed by immunofluorescence or ChIP assays.
Heterokaryon fusion assay
Fusion assays were performed in MBA-15 cells seeded at 2 × 105/ml on glass coverslips. The following day COS-7 cells (CHD9-null) were overlaid at 2 × 105/ml and allowed to adhere for 4 h. Coverslips were then inverted and incubated for 2 min in polyethyleneglycol 6000 (PEG6000, Sigma–Aldrich, St. Louis, MO) dissolved 1:1 in PBS (w/o Ca and Mg). Coverslips were re-inverted, washed in PBS w/o Ca and Mg and incubated overnight in DMEM containing 10% FBS and 10 mM cytosinarabinoside (Sigma), to prevent overgrowth of non-fused cells. Cells were fixed in 4% paraformaldehyde (PFA, Sigma) in PBS for 20 min, and taken for immunofluorescence staining. Counter-staining with Dapi dye allowed identification of nuclei of different origins (Borer et al., 1989).
Antibodies
Three CHD9 antibodies were utilized: OSB antibody was generated using a non-phosphorylated synthetic peptide of the protein (residues 2,585–2,595); DB-16 antibody was made to an expressed polypeptide (residues 2,332–2,481) (Shur and Benayahu, 2005), 11,424 antibody was generated to a synthetic peptide that encompassed the OSB peptide; anti-UBF (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, cat. sc-13125); anti-fibrillarin (72B9, kindly provided by J. Gall); anti-Polymerase I (Santa Cruz cat. sc-190); anti-Polymerase II (gift from Kevin Gardner, NIH), B23/nucleophosmin a nucleolar marker (FC-61991, Invitrogen), and IgG (Millipore) were used for various applications (IF, Western blots and ChIP assays).
Immunofluorescence (IF) staining
Cultured cells were seeded at 1 × 105/ml on coverslips and incubated overnight. Cells were fixed in 4% paraformaldehyde (PFA, Sigma) in PBS for 20 min and blocked in 0.1% Triton X100, 10% FBS in PBS for 30 min at room temperature. OSB antisera, mouse anti-fibrillarin, and mouse anti-UBF were used as primary antibodies. Secondary antibodies used were anti-rabbit Texas Red-FITC and anti-mouse FITC-Cy5. Antibodies were diluted in blocking media and applied on coverslips for 1 h each at room temperature. Coverslips were mounted onto slides by ProLong anti-fade mounting media (Molecular Probes, Eugene, OR). In competition experiments, OSB or H3K4 peptides were preincubated with OSB antisera overnight and then applied to coverslips.
SDS-PAGE gels and Western blot analysis
Cells were scraped in ice cold PBS and the pellet was suspended in M-PER protein extraction reagent (Pierce, Rockford, IL) containing 150mM NaCl and protease inhibitor cocktail (Roche, Indianapolis, IN). Proteins were separated on 6.5% SDS–PAGE gels or 3–8% radiant NuPage gels for 2 h, then transferred to nitrocellulose and blocked in 5% low-fat milk for 1 hr at room temperature. Blots were probed with anti-OSB or anti-DB-16 diluted in blocking media, overnight at 4°C, followed by anti-rabbit HRP-conjugated antibody (Jackson Labs, West Grove, PA) diluted in blocking media for 2 h at room temperature. Westerns were developed using Super Signal Western blot reagent (Pierce).
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed using OSB, UBF, Pol I, and Pol II antibodies. Immunoprecipitated DNA was analyzed by real-time quantitative PCR analysis using primer pairs from the promoter region of the rDNA transcription unit: (sense) 5′: GAGACAGAATGAGTGAGTGA and (anti-sense) 5′: GTACGACCTCCTTGTTAGAG. qPCR was analyzed by SyBr green Supermix (Bio-Rad, Hercules, CA) on a MyiQ single-color qPCR platform (Bio-Rad). All PCRs were performed in technical duplicates from three biological replicates.
Tilling array ChIP–chip
The protocol used is as described by NimbleGen. Briefly, for each ChIP-chip experiment, 1 to 2 × 108 cells were cross-linked with 1% formaldehyde for 10 min at 37°C, harvested and rinsed with 1 × PBS. Cell nuclei were isolated, pelleted, and sonicated. DNA fragments were enriched by immunoprecipitation with OSB antibody. After heat reversal of the cross-linking, the enriched DNA was amplified by ligation-mediated PCR (LM-PCR) and then fluorescently labeled by using Klenow polymerase and Cy5-labeled dUTP (Amersham Biosciences, Piscataway, NJ). A sample of DNA that was not enriched by immunoprecipitation was subjected to LM-PCR and labeled with Cy3-dUTP. ChIP-enriched and unenriched (input) labeled samples were co-hybridized to microarrays. Microarrays were hybridized 18–20 h at 45°C, washed all according to the protocol (http://www.nimblegen.com/products/chip/index.html.) and scanned using an Agilent Technologies (Santa Clara, CA) microarray scanner (Scacheri et al., 2006).
Results
CHD9 is a nucleosome stimulated, ATP-dependent chromatin remodeling protein
CHD9 contains multiple domains; chromo, helicase, and DNA binding domains (A/T hook) and additionally the SANT domain, found in chromatin remodeling and modifying proteins. CDH9 harbors multiple sites for phosphorylation with an unusually long stretch of 60-serine residues near the C-terminus of the protein (Supplementary Fig. S1A; Shur and Benayahu, 2005). A recombinant form of CHD9 (r-CHD9) was prepared and used for in vitro assay of chromatin remodeling activity using a thin layer chromatography based ATPase assay that measures γ 32P[ATP] hydrolysis. We demonstrated that rCHD9 hydrolyze ATP in the absence of any additional substrates, and this activity was significantly enhanced in the presence of purified oligonucleosomes (Fig. 1A, lane 3 vs. 5). A measure of the activity of a chromatin remodeling protein can be gauged by its ability to slide or mobilize nucleosomes on a fragment of DNA containing a well-positioned nucleosome. To address this, high affinity, strong nucleosome positioning sequences (601) were reconstituted with histone octamers (Lowary and Widom, 1998). Reconstituted nucleosomes adopted a unique position at the 3′ end of the fragment. ATP-dependent repositioning of the nucleosome to a central position on the fragment was detected by a change in electrophoretic mobility of the mononucleosome on a native poly-acrylamide gel (Fig. 1B, lane 2 vs. 1 and lane 4 vs. 3). Remodeler-dependent changes in histone-DNA interactions can also be monitored by altered sensitivities to the minor-groove nuclease, DNaseI (Stockdale et al., 2006). We find that rCHD9 both suppresses and enhances the DNaseI digestion pattern of a positioned nucleosome (Fig. 1C, red arrows). This ability of CHD9 to redistribute nucleosomes is consistent with observations made with other chromatin remodeling proteins (Tong et al., 1998; Stockdale et al., 2006; Thompson et al., 2008).
Fig. 1.
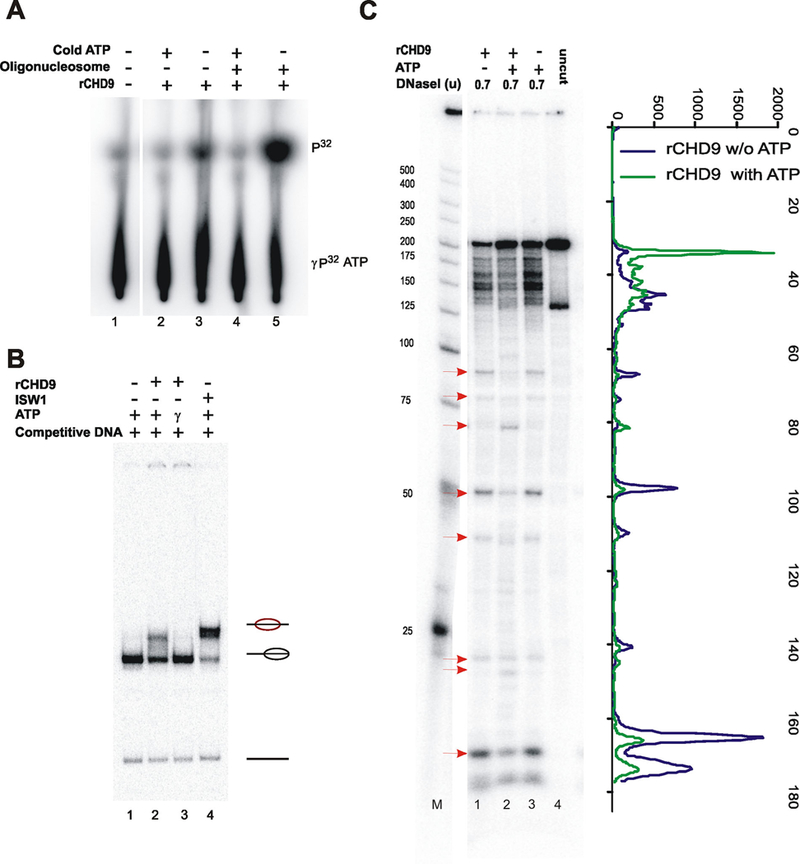
rCHD9 is a nucleosome stimulated, ATP-dependent chromatin remodeling protein. A: rCHD9 in the presence of (γ−32P)ATP and in the presence or absence of oligonucleosomes. Reaction products were separated on PEI-cellulose. B: Mononucleosomes (radiolabeled 601 fragment) were incubated with rCHD9 with ATP or non-hydrolyzable ATP-γ-S and nucleosome mobilization was monitored. Reactions were separated on a native 5% polyacrylamide gel, dried and exposed on a phosphorimager. The positions of nucleosomes (and their relative position on the fragment) and the free DNA are shown to the side. ISWI is used as a positive control in the sliding assay. C: Reconstituted nucleosomes were treated (as in B) followed by DNaseI digestion, with the indicated amounts, for 5 min. Reaction products were separated on 6% polyacrylamide-urea gels. Active chromatin remodeling proteins alter the pattern of DNaseI cutting on positioned nucleosomes. Red arrows reflect sites of increased or decreased cutting as a result of ATP-dependent chromatin remodeling.
CHD9 contains two highly conserved neighboring chromodomains (Benayahu et al., 2007) that have the potential to interact with methylated histone H3 tails (Fig. 2A). We, therefore, examined the in vitro interactions between rCHD9 and biotinylated-H3 peptides that were either methylated or un-methylated at various residues; first by immunoprecipitating with streptavidin-beads followed by Western blotting with Flag antibodies to detect rCHD9. The pull-down assay revealed higher interaction between rCHD9 and K27me3, K9me2/3 and K4me2 peptides that was significantly higher when compared to interactions with unmodified H3 peptide or methylated K4me3 and K27me2 peptides (Fig. 2B). The specificity and stringency of this binding was tested in a competition assay using the rCHD9 protein and target peptides in the presence of an excess of unmodified H3. rCHD9 binding of K27me3 or K9me2 peptides were largely unaffected in the presence of an excess of unmodified H3 peptides as seen on the Western blot (Fig. 2C). The ability of rCHD9 to mobilize nucleosomes in an ATP-dependent manner (Fig. 1) in conjunction with its specificity for methylated H3 histone tails (Fig. 2) provide an evidence for CHD9 being a bona fidé member of the CHD family of chromatin remodeling proteins.
Fig. 2.
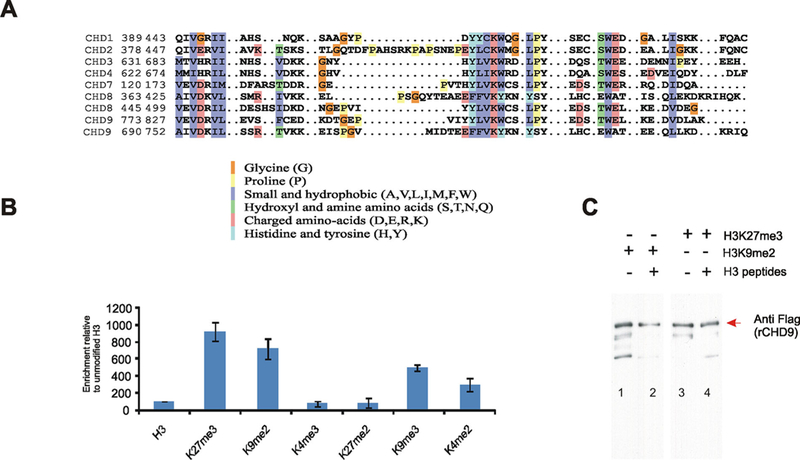
CHD9 binds to methylated H3 peptides in vitro. A: Multiple alighnment of high degree of conservation of chromodomains of CHD proteins. Color scheme refers to the nature of conserved amino acids (e.g., charged and hydrophobic). B: Binding assay analyzed the interaction of rCHD9 with biotinylated H3 methylated peptides was analyzed in vitro by immunoprecipitation with streptavidin beads followed by western blotting with Flag antibodies. C: Competition assay was analyzed on Western blot between K27me3 and K9me2 peptides displaying a preference for binding rCHD9; even in presence of excess of unmodified H3, the competition did not alter the intensity of binding to rCHD9.
n-CHD9 is enriched in the nucleolus
We have previously demonstrated that CHD9 is expressed in marrow stromal osteoprogenitors in vivo and in vitro (Marom et al., 2006; Shur et al., 2006a,b). Currently, we have demonstrated the sub-cellular localization of CHD9 using three CHD9-specific antisera (OSB, DB-16, and 11424; Supplementary Figs. S1 and S2). Western blots recognized the expressed recombinant protein rCHD9 (Supplementary Fig. S1B, lanes 2 and 3) and whole cell lysates from marrow stromal (MBA-15; Supplementary Fig. S1C) probed with OSB or DB-16 antibodies resulted in the identification of the endogenous CHD9. CHD9 protein contains multiple serine phosphorylation sites, and the phosphorylated form was detected by DB-16 antibody (Supplementary Fig. S1B, lanes 3 and 4). The CHD9 form that is recognized by DB-16 shows a reduced reactivity to the antibody when cell lysates were treated with CIP phosphatase (Supplementary Fig. S1C, lane 3 vs. 4). The sub-cellular distribution of endogenous CHD9 was analyzed by immunofluorescence (IF) in MBA-15 cells; IF with OSB antibody showed a significant localization of the CHD9 protein in the nucleolus (Figs. 3–6, Supplementary Fig. S1D left part). This form of the protein is designated as n-CHD9 (nucleolar-CHD9). IF analysis with DB-16 antibody detected the protein throughout the nucleus and cytoplasm (Supplementary Fig. S1D, right part; Marom et al., 2006). The variable sub-cellular distribution may be ascribed to the phosphorylation status of CHD9 protein as was similarly reported for Nopp 140, Nopp 130, and La proteins (Miau et al., 1997; Thiry et al., 2009). For example, the non-phosphorylated La protein is localized in the nucleolus while the phosphorylated form resides in the nucleoplasm (Intine et al., 2004).
Fig. 3.
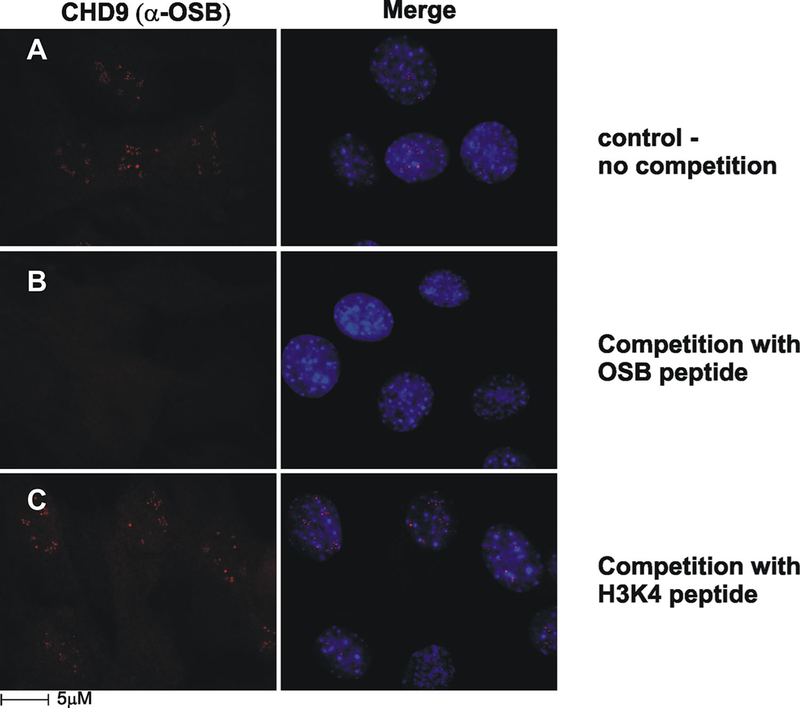
n-CHD9 is highly enriched in the nucleolus observed with α-OSB antisera. A: Immunofluorescence of MBA-15 cells with α-OSB marked the nucleoli. B: α-OSB was incubated with the immunizing OSB peptide resulted in depletion of nucleoli staining. C: α-OSB was incubated with H3K4 peptide. The nucleoli staining pattern was unaffected.
Fig. 6.
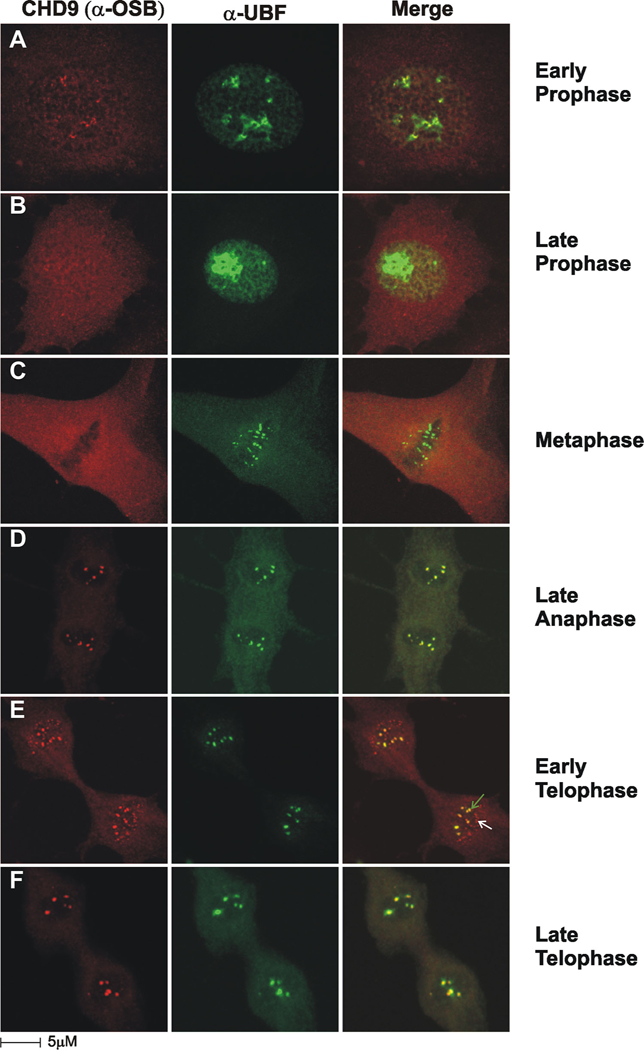
Dynamic association of n-CHD9 with the nucleolus during the cell cycle. A–C: During mitosis pre-rRNA processing factors are separated from the RNA Pol I transcription apparatus, which remains attached to nucleolar organizing regions (NORs) on the metaphase plate. D: During late anaphase both n-CHD9 and upstream binding factor (UBF) are present in nucleolar structures. E,F: At early telophase n-CHD9 and UBF are present in mature nucleolar structures (green arrow), but n-CHD9 is also uniquely present in pre-nucleolar bodies (white arrow). During late telophase both n-CHD9 and UBF are assembled into mature nucleoli.
The nucleoli enrichment of n-CHD9 was validated with an additional antibody (α−11424) that was developed to identify a distinct epitope within CHD9 (Supplementary Fig. S2A and B). The co-localization of n-CHD9 (based on α−11424) with nucleolar markers; B-23/nucleophosmin, UBF, and pol I are shown on (Supplementary Fig. S2A). Immunofluorescence-competition assays in presence of rCHD9, we shown a complete abolish of the 11424 antisera that confirmed the specificity in recognition of the n-CHD9 (Supplementary Fig. 2SB).
The specificity of the nucleoli signal obtained with either OSB or 11,424 antibodies was confirmed in competition experiments using an excess of either the immunizing peptide (Fig. 3B vs. C) or the recombinant protein (Supplementary Fig. S2B). Collectively, the profiles obtained with multiple antibodies revealed that CHD9 cellular distribution varies with post-translation modification of the protein.
Furthermore, a heterokaryon experiment was performed to demonstrate the nucleolar targeting of CHD9 protein. We induced cell fusion between MBA-15 cells (CHD9 expressing) and COS-7 cells (CHD9 null) and followed IF staining with OSB antisera. The CHD9 protein in the heterokaryon was identified by pericentromeric heterochromatic regions characteristic of DAPI staining of MBA-15 cells (Fig. 4A and C). In MBA-15 cells, CHD9 is located in nucleoli and is absent from COS-7 cells (null; Fig. 4B), post fusion CHD9 is also detected in COS-7 nuclei (dotted outline in Fig. 4D) specifically to the nucleoli within these cells. Thus, OSB antisera identified the n-CHD9 form that retains distinctive nucleoli localization when artificially introduced into the heterologous cells. The new evidence here that n-CHD9 localized to the nucleoli, is indicative of a contributory role in RNA pol I transcription
Fig. 4.
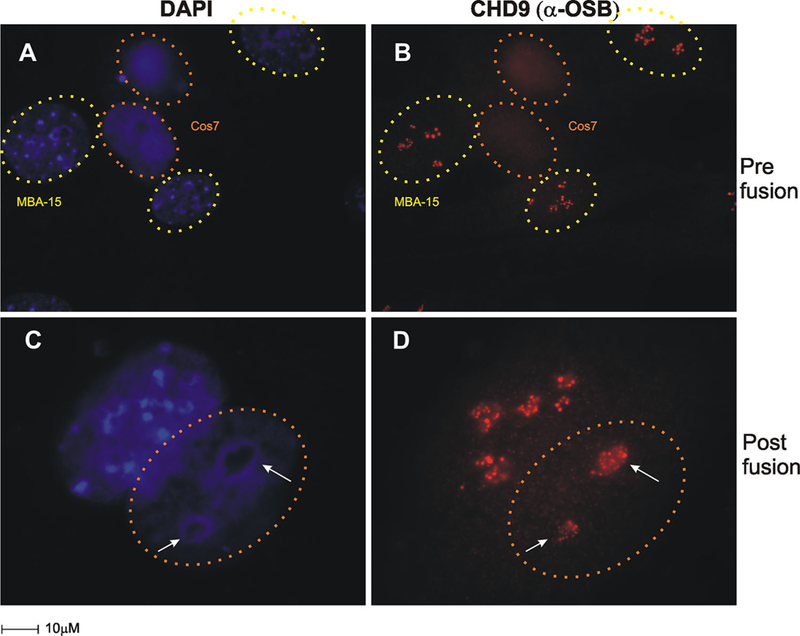
IF of n-CHD9 targeted to nucleoli in CHD9 null cells. Heterokaryon fusion assays were performed between COS-7 (CHD9-null) and MBA-15 (CHD9 expressing). A,B: Cells Pre-fusion; (A) COS-7 cells and MBA-15 cells have distinct heterochromatin staining patterns (MBA-15 cells, identified by pericentric heterochromatin DAPI staining pattern, yellow dotted circles; COS-7 cells, identified by orange dotted circles) (B) IF for n-CHD9 staining patterns (red) were identified in MBA-15 cells prior to cell fusion. C,D: Cells post-fusion; co-culturing for 24 h under cell fusion conditions, resulted in re-localization of n-CHD9 in nucleolar structures in MBA-15 /COS-7 fused cells. Dotted orange circles mark COS-7 cells after fusion expressing n-CHD9.
n-CHD9 localization correlates with centers of ribosomal transcription
The localization of n-CHD9 at the nucleoli was examined along with proteins known to be associated with ribosomal transcriptional centers by imaging (Figs. 5 and 6) and molecular approaches (Fig. 7).
Fig. 5.
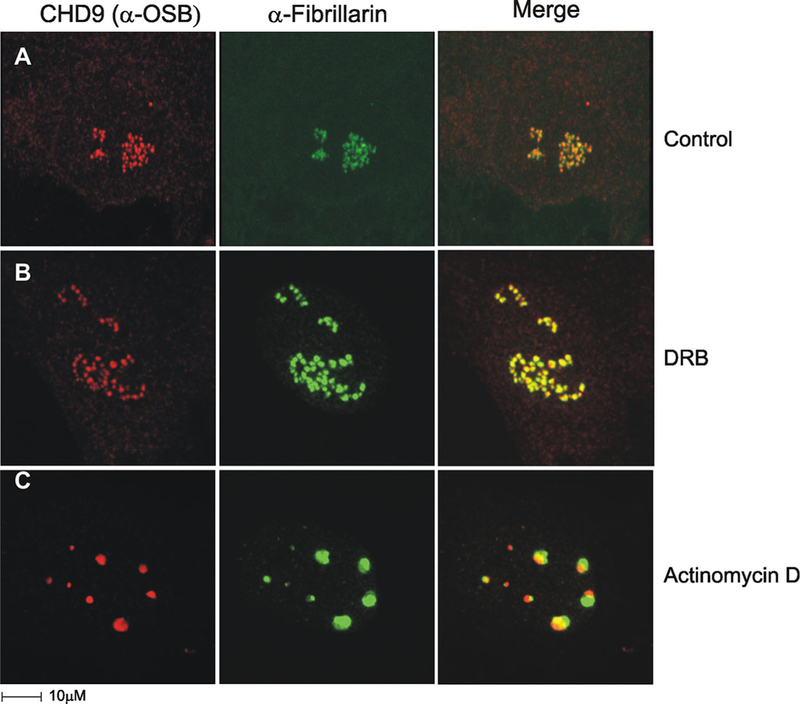
n-CHD9 co-localizes with centers of ribosomal transcription. MBA-15 cells were stained with α-OSB (red) and α-fibrillarin (green), a methylase and processing factor in rRNA maturation. A: Control cell shows an overlay of n-CHD9 and fibrillarin signals indicates partial co-localization (yellow) in the inner portions of fibrillarin positive structures, the dense fibrillar centers (DFC), marking that n-CHD9 localization is limited to fibrillar centers (FCs). B: Transcriptional inhibitor DRB induces redistribution of rDNA genes into “nucleolar necklaces.” The “beads” are known to comprise FCs connected by “strings” of DFCs. C: Actinomycin D inhibits RNA Poly I transcription and induces segregation of the nucleolar components.
Fig. 7.
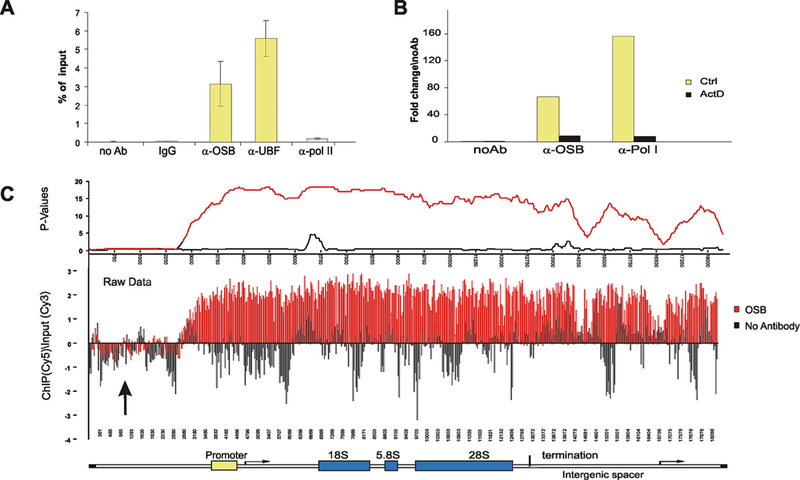
n-CHD9 is binding and broadly distributed across the ribosomal locus. A: ChIP assays were performed with α-OSB, α-UBF, α-Pol I, α-Pol II, IgG, or no antibody. n-CHD9 interacts with rDNA promoter along with the expected interactions of UBF and Pol I. B: As actinomycin D induces dispersal and segregation of FCs, the association of both CHD9 and Pol I are reduced at rDNA. C: ChIP–chip experiments using either α-OSB or no antibody show an enrichment of CHD9 (red) at both promoter and transcribed regions of rDNA in contrast to the no antibody negative control (black). Both raw and calculated P-value data are shown.
Imaging and co-localization with additional proteins allow dissecting the anatomical compartments where transcription of ribosomal genes is performed. In the nucleolus there are three compartments: the fibrillar centers (FC), the site where ribosomal genes are localized and transcribed at its periphery; the dense fibrillar component (DFC), which contains newly synthesized pre-ribosomal particles and the granular component (GC), the site of assembly of pre-ribosomal particles (Carmo-Fonseca et al., 2000; Dundr et al., 2000).
As shown in Figure 5A, IF staining and overlay of n-CHD9 and fibrillarin (a marker of DFC), reveals partial co-localization with the inner portions of fibrillarin positively labeled DFC. This suggests that n-CHD9 localization is restricted to fibrillar centers and implies its potential involvement in rDNA transcription rather than pre-rRNA processing. Adding transcription inhibitors affects the organization of proteins within the DFC and confirms the protein localization at these sites and consequently, their function at the nucleoli. One inhibitor used is the 5,6-dichloro-1b-D-ribofuranosylbenzimidazole (DRB) that inhibits transcription and induces redistribution of rDNA genes into “nucleolar necklaces.” These loose structures are thought to be comprised FCs connected by strings of DFCs (Panse et al., 1999). Upon DRB treatment of MBA-15 cell, we observed redistribution of fibrillarin and similar dispersed appearance for n-CHD9 into necklace staining pattern (Fig. 5B). Actinomycin D (ActD) is a second inhibitor that affects RNA Pol I, and induces segregation of nucleolar components into distinct entities. Following addition of ActD to MBA-15 cells, we have noted by IF (Fig. 5C) that both fibrillarin and n-CHD9 distributions are severely disorganized with clearly separated localizations. n-CHD9 is localized in the nuclear cap which is reminiscent of fibrillar centers, where transcription of rDNA occurs, while fibrillarin is present in segregated dense fibrillar centers, where pre-rRNA processing takes place (Carmo-Fonseca et al., 2000; Dundr et al., 2000). Collectively, these findings strongly link n-CHD9 with active RNA pol I transcription in fibrillar centers.
Dynamic expression and localization of n-CHD9 during cell division
We examined the interaction of n-CHD9 with nucleolar structures for the localization and expression of n-CHD9 during mitosis in conjunction with UBF, a sequence-specific DNA binding protein that has been shown to interact with the core and upstream control elements of the rDNA promoter. In early prophase, nucleoli are disrupted and both n-CHD9 and UBF are associated with nucleoli remnants (Fig. 6A). As overall transcription is shutdown in late prophase (Fig. 6B), UBF remains associated with residual nucleoli structures, however, n-CHD9 is absent from these elements. At metaphase, UBF is associated with nucleolar organizing regions (NORs) on the metaphase plate (Fig. 6C) and as chromosomes decondense. During late anaphase, NORs become transcriptionally active and n-CHD9 and UBF both become present in the newly formed nucleoli (Fig. 6D). During early and late telophase, n-CHD9 and UBF are both present in forming nucleoli and show a high degree of co-localization (green arrow, Fig. 6E and F). In addition n-CHD9 is also present in pre-nucleolar bodies which fuse with transcriptionally active NORs to form nucleoli (white arrow, Fig. 6E). Thus, n-CHD9 displays a unique and dynamic pattern of organization into nucleolar structures that is highly suggestive of a role in rDNA transcription.
n-CHD9 occupancy at the ribosomal locus
We shows on IF the co-localization of n-CHD9 with fibrillarin (Fig. 5), UBF (Fig. 6 or Supplementary Fig. S2) and Pol I (Supplementary Fig. S2) in the nucleoli, all indicates a role in ribosomal gene transcription. Complementary to the IF experiment, interactions between n-CHD9 and the ribosomal gene locus were directly demonstrated in ChIP assay with several antibodies: anti-n-CHD9 (anti-OSB), anti-UBF and antibodies to RNA pol I and pol II. ChIPed DNA was amplified with primers specific to the rDNA promoter showing n-CHD9 was enriched at rDNA promoters (Fig. 7A and B) that is also parallel as expected for UBF (Fig. 7A) and RNA pol I (Fig. 7B) [while RNA pol II was comparable to non-specific IgG or no antibody control (Fig. 7A)]. Treatment of cells with Act D resulted in the disruption of fibrillar centers (Fig. 5C), and consistent with this re-organization of FCs, ChIP experiments showed an abolished binding of RNA Pol I and n-CHD9 at the rDNA promoter (Fig. 7B). rDNA promoter ChIP–chip experiment was extended on a tiling array that spanned the rDNA locus (non-transcribed upstream, promoter, 18S, 5.8S, 28S, termination and intergenic spacer regions; one 50-mer oligo per 100 bp). The ChIP-chip assay revealed an enrichment of n-CHD9 throughout the ribosomal locus (Fig. 7C, compare anti-OSB, red, vs. no antibody, black). The broad distribution of n-CHD9 across the rDNA locus suggests its role rRNA transcription. We have demonstrated a dynamic association of n-CHD9 with ribosomal genes that was confirmed by IFs (Figs. 5 and 6 and Supplementary Fig. S2) and molecular assays (Fig. 7) are indicating a new function for n-CHD9 and a chromatin remodeling role in active rRNA biogenesis.
Discussion
The mesenchymal stem cells’ fate and differentiation is determined by the coordinated transcription regulation of the compacted DNA. The cell differentiation involves the interaction of transcription factors with chromatin remodelers that dictate their binding to promoters region. The chromatin varies in structural hierarchies from arrays of nucleosomes to the highly compacted state at metaphase chromosome during mitosis (Redner et al., 1999), thus there is a necessity for an activity of cell specific chromatin remodeling proteins that govern and facilitate the transcription factor access to packaged DNA (Clapier and Cairns, 2009).
We study the CHD9 remodeling protein, a member of a CHDs remodelers that are divided into three sub-families based on domain features. CHD9 belongs to the third subfamily containing SANT and/or BRK domains. The BRK motif is found in CHD6–9 and also in BRG1 and BRM (SWI/SNF ATPases, Benayahu et al., 2007). The SANT domain has been found in the ISWI family of remodeling proteins and is thought to be required for their interaction with chromatin (Pinskaya et al., 2009). CHD proteins have been shown to play a role during differentiation and development: CHD1 affects gene expression during embryonic development (Gaspar-Maia etal., 2009), CHD7 and CHD8 have been implicated in multi-potent neural crest formation (Benayahu et al., 2007; Bajpai et al., 2010; Jones et al., 2015; Marcos et al., 2014; McCarthy et al., 2014).
We focus on CHD9 and previous in vivo studies revealed its expression during development where it is primarily expressed in osteoprogenitor cells and is absent in mature osteoblasts (Shur and Benayahu, 2005). Consistent with this, CHD9 has been shown to interact with regulatory regions of promoters involved in osteoblastogenesis in vivo and in vitro (Shur et al., 2006a). Ex vivo studies in mesenchymal stem cells show a strong correlation of CHD9 expression with cellular differentiation (Shur and Benayahu, 2005; Shur et al., 2006a). Taken together, this suggests a role for CHD9 as a chromatin remodeler associated with the transcriptional regulation of Pol II controlled genes. It was also shown to interact with nuclear receptors and some transcription factors (Shur and Benayahu, 2005; Marom et al., 2006; Shur et al., 2006a).
In the current study, we extended the knowledge regarding CHD9 structure and function. The chromatin remodeling activity of the protein was demonstrated by assays on r-CHD9 in vitro, which demonstrated the hydrolysis of ATP that was significantly enhanced in the presence of purified oligonucleosomes (Fig. 1). This is consistent with the previous observation of ATPase activity of endogenouse immunipercipitated protein from cell lysates (Shur and Benayahu, 2005). We have demonstrated that rCHD9 was able to perform the remodeling activity in sliding and nucleosome positioning sequences (601) assays with reconstituted histone octamers as in SWI/SNF, ISWI, and CHD’s (Lowary and Widom, 1998). ATP-dependent repositioning of the nucleosome to a central position on the fragment was detected by a change in electrophoretic mobility of the mononucleosome on a native poly-acrylamide gel (Fig. 1). Remodeler-dependent changes in histone-DNA interactions can also be monitored by altered sensitivities to the minor-groove nuclease, DNaseI as shown by others for the ATP dependent family members remodelers such as CHD1 (Stockdale et al., 2006). We found that rCHD9 both suppresses and enhances the DNaseI digestion pattern of a positioned nucleosome. Thus, the ability of CHD9 to redistribute nucleosomes is consistent with observations made with other chromatin remodeling proteins such as CHD1, CHD8 (Tong et al., 1998; Stockdale et al., 2006; Thompson et al., 2008).
CHD9 contains two highly conserved neighboring chromodomains (Shur and Benayahu, 2005; Benayahu et al., 2007) that have the potential to interact with methylated histone H3 tails. The interactions between rCHD9 and H3 peptides that were either methylated or un-methylated at various residues were analyzed by pull-down assay, which revealed an interaction between r-CHD9 and K27me3, K9me2/3 and K4me2 peptides, as compared to no interaction with unmodified H3 peptides or un-methylated. The highest affinity was measured for K9me2/3 and K27me3 peptides (Fig. 2). The ability of r-CHD9 to mobilize nucleosomes in an ATP-dependent fashion (Fig. 1) in conjunction with its’ specificity for methylated H3 histone tails (Fig. 2) provide evidence for CHD9 being a bona fidé member of the CHD family of chromatin remodeling proteins.
Following the biochemical activity analysis on r-CHD9, we revealed that the endogenous protein in MBA-15 cells exists as two alternative forms based on post-traslational modification. The new form described here is n-CHD9 with IF showing unambiguously that CHD9 also localized to the nucleoli and is intimately associated with sites of active rDNA transcription (Figs. 3–6, Supplementary Fig. S1 and 2). The co-localization of n-CHD9 with fibrillarin or UBF is consistent with a connection between n-CHD9 and active ribosomal gene transcription (Fig. 7). Disruption of ribosomal transcription by specific transcriptional inhibitors (DRB and ActD) altered the organization of n-CHD9 nucleolar localization to foci enriched with pol I transcription components (Fig. 5). The allocated sites of fibrillar centers (where transcription of rDNA occurs) as compared with fibrillarin that is present in segregated dense fibrillar centers, where pre-rRNA processing takes place (Carmo-Fonseca et al., 2000; Dundr et al., 2000). Collectively, these findings strongly link n-CHD9 with active RNA pol I transcription in fibrillar centers. Other reports have linked chromatin remodelers also to the transcription regulation of ribosomal genes. In yeast, the ribosomal locus is shown to be associated with both histones and chromatin remodeling proteins Chd1p, Isw1p, and Isw2p (Jones et al., 2007). In mammalian cells, chromatin remodeling enzymes have been implicated in both silencing (NoRC) and activation (B-WICH) of ribosomal genes (Santoro et al., 2002; Zhou et al., 2002; Percipalle and Farrants, 2006). CHD7 which is related to CHARGE syndrome was shown to be a positive regulator of ribosomal biogenesis (Zentner et al., 2010; Kita et al., 2012). We assayed the direct binding of n-CHD9 to rDNA genes by ChiP and ChIP–chip assays that proven the protein to be physically associated and broadly distributed across the ribosomal locus (Fig. 7). We have shown that the extensive binding pattern of n-CHD9 across this locus suggests CHD9 in rDNA transcription of rRNA. The co-localization of n-CHD9 with UBF on IF and the association of n-CHD9 across the gene locus (by ChIP–chip) similar to UBF (Prieto and McStay, 2005; McStay and Grummt, 2008) support for the role n-CHD9 in rRNA elongation. UBF was reported to be an “architectural” factor required in early elongation structures that was enriched at the promoter and across the transcribed portion of the 45S rRNA gene. UBF binding to rRNA repeats was found to be necessary for maintenance of the open chromatin structure found in active NORs (McStay and Grummt, 2008; Sanij and Hannan, 2009).
We have presented that CHD9 has “two faces,” it was shown previously for its’ ability to bind to promotors’ of tissue specific genes (Shur et al., 2006a; Benayahu et al., 2007) and the second form is a protein appears at the nucleoli, n-CHD9 presented in the current study. The n-CHD9 form appears in the nucleolar fibrillar centers, is coincident with the assembly of Nucleolar Organizer Regions during and after cell division (Figs. 5 and 6).
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, and Center for Cancer Research. We would like to acknowledge the assistance of Tatiana Karpova, manager of the LRBGE Optical Imaging Facility.
Contract grant sponsor: NIH.
Contract grant sponsor: National Cancer Institute.
Contract grant sponsor: Center for Cancer Research.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
Conflict of interest: The authors indicate no potential conflicts of interest.
Literature Cited
- Aalfs JD, Narlikar GJ, Kingston RE. 2001. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H.J Biol Chem 276:34270–34278. [DOI] [PubMed] [Google Scholar]
- Aramaki M, Udaka T, Kosaki R, Makita Y, Okamoto N, Yoshihashi H, Oki H, Nanao K, Moriyama N, Oku S, Hasegawa T, Takahashi T, Fukushima Y, Kawame H, Kosaki K. 2006. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J Pediatr 148:410–414. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. 2010. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463:958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayahu D, Kletter Y, Zipori D, Wientroub S. 1989. A bone marrow derived stromal cell line expressing osteoblastic phenotype in vitro and osteogenic capacity in vivo. J Cell Physiol 140:1–7. [DOI] [PubMed] [Google Scholar]
- Benayahu D, Shacham N, Shur I. 2007. Insights on the functional role of chromatin remodelers in osteogenic cells. Crit Rev Eukaryot Gene Expr 17:103–113. [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers LE, Francescatto L, Mefford HC, Rosenfeld JA, Bakken T, O’Roak BJ, Pawlus M, Moon R, Shendure J, Amaral DG, Lein E, Rankin J, Romano C, de Vries BB, Katsanis N, Eichler EE. 2014. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158:263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer RA, Lehner CF, Eppenberger HM, Nigg EA. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379–390. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Mendes-Soares L, Campos I. 2000. To be or not to be in the nucleolus. Nat Cell Biol 2:E107–E112. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO. 2000. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150:433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison J 2008. Gene symbol: CHD7. Disease: CHARGE syndrome. Hum Genet 124:323. [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E. Ramalho-Santos M (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460:863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intine RV, Dundr M, Vassilev A, Schwartz E, Zhao Y, Zhao Y, Depamphilis ML, Maraia RJ. 2004. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol 24:10894–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ. 2007. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol 14:123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Saric N, Russell JP, Andoniadou CL, Scambler PJ, Basson MA. 2015. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita Y, Nishiyama M, Nakayama KI. 2012. Identification of CHD7S as a novel splicing variant of CHD7 with functions similar and antagonisticto those of the full-length CHD7L. Genes Cells 17:536–547. [DOI] [PubMed] [Google Scholar]
- Lathrop MJ, Chakrabarti L, Eng J, Rhodes CH, Lutz T, Nieto A, Liggitt HD, Warner S, Fields J, Stoger R, Fiering S. 2010. Deletion of the Chd6 exon 12 affects motor coordination. Mamm Genome 21:130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276:19–42. [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25:1192–1200. [DOI] [PubMed] [Google Scholar]
- Marcos S, Sarfati J, Leroy C, Fouveaut C, Parent P, Metz C, Wolczynski S, Gerard M, Bieth E, Kurtz F, Verier-Mine O, Perrin L, Archambeaud F, Cabrol S, Rodien P, Hove H, Prescott T, Lacombe D, Christin-Maitre S, Touraine P, Hieronimus S, Dewailly D, Young J, Pugeat M, Hardelin JP, Dode C. 2014. The prevalence of CHD7 missense versus truncating mutations is higher in patients with Kallmann syndromethan in typical CHARGE patients. J Clin Endocrinol Metab 99:E2138–E2143. [DOI] [PubMed] [Google Scholar]
- Marom R, Shur I, Hager GL, Benayahu D. 2006. Expression and regulation of CReMM, a chromodomain helicase-DNA-binding (CHD), in marrow stroma derived osteoprogenitors. J Cell Physiol 207:628–635. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, Kelleher E, O’Brien C, Donohoe G, Gill M, Morris DW, McCombie WR, Corvin A. 2014. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry 19:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I.2008The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24:131–157. [DOI] [PubMed] [Google Scholar]
- Miau LH, Chang CJ,Tsai WH, Lee SC. 1997. Identification and characterization of a nucleolar phosphoprotein, Nopp140, as a transcription factor. Mol Cell Biol 17:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M, Brehm A. 2011. CHD chromatin remodelers and the transcription cycle. Transcription 2:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487. [DOI] [PubMed] [Google Scholar]
- Panse SL, Masson C, Heliot L, Chassery JM, Junera HR. Hernandez-Verdun D.1999. 3-D organization of ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J Cell Sci 112:2145–2154. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Farrants AK. 2006. Chromatin remodelling and transcription: Be-WICHed by nuclear myosin 1. Curr Opin Cell Biol 18:267–274. [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Nair A, Clynes D, Morillon A, Mellor J. 2009. Nucleosome remodeling and transcriptional repression are distinct functions of Isw1 in Saccharomyces cerevisiae. Mol Cell Biol 29:2419–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR. Grant PA (2005). Chdi chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434–438. [DOI] [PubMed] [Google Scholar]
- Prieto JL, McStay B. 2005. Nucleolar biogenesis: The first small steps. Biochem Soc Trans 33:1441–1443. [DOI] [PubMed] [Google Scholar]
- Redner RL, Wang J, Liu JM. 1999. Chromatin remodeling and leukemia: New therapeutic paradigms. Blood 94:417–428. [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. 2013. From neural development to cognition: Unexpected roles for chromatin. Nat Rev Genet 14:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Hannan RD. 2009. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 4:374–382. [DOI] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32:393–396. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS. 2006. Genome-wide analysis of menin binding provides insights into MENI tumorigenesis. PLoS Genet 2:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shur I, Benayahu D. 2005. Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. J Mol Biol 352:646–655. [DOI] [PubMed] [Google Scholar]
- Shur I, Socher R, Benayahu D. 2006. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J Cell Physiol 207:374–378. [DOI] [PubMed] [Google Scholar]
- Shur I, Solomon R, Benayahu D. 2006. Dynamic interactions of chromatin-related mesenchymal modulator, a chromodomain helicase-DNA-binding protein, with promoters in osteoprogenitors. Stem Cells 24:1288–1293. [DOI] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. 2006. Analysis of nucleosome repositioning by yeast ISWI and Chdi chromatin remodeling complexes. J Biol Chem 281:16279–16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. 2006. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochem Biophys Res Commun 343:535–543. [DOI] [PubMed] [Google Scholar]
- Thiry M, Cheutin T, Lamaye F, Thelen N, Meier UT, O’Donohue MF, Ploton D. 2009. Localization of Nopp140 within mammalian cells during interphase and mitosis. Histochem Cell Biol 132:129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Tremblay V, Lin G, Bochar DA. 2008. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol 28:3894–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. I998.Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917–921. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. I999.Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev 13:686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Zhou MM. 2011. Structure and mechanisms of lysine methylation recognition bythe chromodomain in gene transcription. Biochemistry 50:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, Scacheri PC. 2010. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet 19:3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I. 2002. The chromatin remodeling complex NoRC targets HDACI to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21:4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


