Abstract
Introduction
Detection of new Actinobacteria is significant to discover new antibiotics because development of new antibiotics is connected to the characterization of novel bacterial taxa. This study has focused on the identification and isolation of antibiotic-producing Actinobacteria from the sediment and the water of Ma’in thermal springs (48-59°C) situated in the center area of Jordan.
Methods
Samples of sediment and water were transferred to glucose yeast malt agar medium and Actinobacteria were cultivated, isolated and identified according to scanning electron microscopy and 16S rRNA gene analysis. Antibacterial activities of the isolates were then tested against different test bacteria by agar well diffusion method.
Results
Three different species of Actinobacteria were isolated (M1-1, M2-2, M3-2) from sediment samples. Based on 16S rRNA gene analysis, isolate M1-1 was found to have only 90% identity percentage with Nocardiopsis sp., however, isolates M2-2 and M3-2 were found to be closely related Streptomyces sp. (97%) and Nocardioides luteus (99%), respectively. The antibacterial activity showed that strain M1-1 is active against P. aeruginosa ATCC 2785 (inhibition zone, 9 mm). Strain M2-2 was found to be active against S. aureus ATCC 29213 (12 mm), B. cereus ATCC 11778 (11 mm), and E. coli ATCC 25922 (9 mm). In respect to strain M3-2, it was found to be active against S. aureus ATCC 29213 (14 mm) and B. cereus ATCC 11778 (9 mm). There were no actinobacterial isolates obtained from water samples despite their significant diversity revealed by our previous metagenomic analysis, which showed the presence of 13 different species dominated by Arthrobacter (an Actinobacterium belonging to family Actinomycetales).
Conclusion
There were 17 different Actinobacteria that could be detected in Ma’in thermal springs (13 unculturable species and 3 culturable species). The culturable Actinobacteria were found to have some antimicrobial activity. Further chemical analysis of the bioactive compounds is recommended.
Keywords: Actinobacteria, antibiotics, Ma’in thermal springs, Jordan
Introduction
The phylum Actinobacteria represents one of the major taxonomic groups in the domain Bacteria with five subclasses, six orders, and fourteen suborders.1 Most species of this phylum are Gram-positive with few exceptions.2,3 Most species in this phylum are aerobic and chemoheterotrophic. In respect to morphology, various morphologies of Actinobacteria are present. However, the growth of some Actinobacteria is associated with the formation of hyphae of bacterial dimensions, less than half the size of fungal hyphae.4,5
Actinobacteria are a well recognized type of bacteria due to their ability to produce antibiotics.4 Examples of antibiotics from Actinobacteria include streptomycin, streptothricin, and actinomycin.4,6 In addition to their antimicrobial activity, previous research indicated that Actinobacteria are a good source of promising compounds with herbicidal, antitumor, antifungal and anthelminthic activities.1
Species of Actinobacteria are common inhabitants of soil.4 However, it is now clear that they are more broadly dispersed in nature and they are not restricted to specific habitats. For instance, they can be found as common inhabitants of plant material,4 aquatic ecosystems including freshwater and marine ecosystems,1,7 and they are also found as harmless commensals in the human body, for example the oral cavity, gastrointestinal tract, and the genitourinary tract.8 In some cases, Actinobacteria can be pathogenic to humans and the example here is Mycobacterium tuberculosis.1
Moreover, Actinobacteria are also known to exist under extreme environments such as those with high temperatures.2 Actinobacteria surviving and growing at high temperature are referred to as thermophilic Actinobacteria. Thermophilic Actinobacteria are known to live at high temperatures between 40 and 80°C.2 Several studies have focused on the microbial ecology of Actinobacteria in common environments; conversely, data about the microbial ecology of thermophilic Actinobacteria in thermal environments, especially those that are endemic to thermal springs, are still limited.7
Ma’in thermal springs (48-59°C),9 located in central Jordan, represent an interesting habitat for different lineages of thermophilic organisms as revealed by earlier studies.10-12 Recently, we have analyzed the microbial diversity in Ma’in thermal springs using culture-independent methods.9 Our findings indicated that the microbial communities in Ma’in thermal springs are very diverse but dominated by Bacteria while Archaea represent just a minor fraction.
In this current study, we studied Actinobacteria from the sediments and the water of Ma’in thermal springs and examined their ability to produce antibacterial agents. Additionally, unpublished data from our previous metagenomic analysis that reflect the diversity of Actinobacteria in the studied habitat were presented.9 The results of this study are expected to broaden our knowledge about thermophilic Actinobacteria present in the thermal springs in Jordan and to establish an initial record of the potential of this environment to yield novel antimicrobials.
Studying the diversity of Actinobacteria inhabiting thermal environments like thermal springs is important to detect new species that could be a promising source of new antibiotics. Taxonomy-based studies help us to reveal novel bacterial taxa and their capabilities of novel antibiotic production.13 Analysis of the published literature shows very scarce information available about the antibiotic-producing Actinobacteria inhabiting Jordanian thermal springs and this study is expected to be a groundwork study on culturable and unculturable Actinobacteria inhabiting the thermal springs in Jordan.
Methods
Collection of sediment and water samples
This study was done on sediment and water obtained from Ma’in thermal springs (Jordan). Three samples of sediments were collected from three different sites at Ma’in Thermal Springs (M1, M2 and M3) that are close to each other. Sediment samples were collected in clean plastic bags. Alternatively, water samples were obtained from three different Ma’in thermal springs (HS1, HS2, HS3) in clean sterile glass bottles as described before.9
Chemical testing of sediment samples
Several chemical properties of the sediments used for the isolation of Actinobacteria were examined. The chemical characteristics of sediment samples (pH, total dissolved solids (TDS), electrical conductivity (EC), salinity, concentration of NO3-, concentration of Cl-, and concentration of F-) were carried out as described previously.9,12 The physical and chemical properties of water samples from HS1, HS2, and HS3 were tested in a previous study by our group.9
Isolation of Actinobacteria
Sediment samples were dried before enrichment by spreading the sediment in a sterile Petri dish and incubating at 50°C for three days. Water samples were not pre-treated. After that, serial dilutions were carried out using normal saline to dilute the samples to 10-1, 10-2, and 10-3. A volume of 200 µL of both sample types was transferred to glucose yeast malt agar medium,14 and incubation was done at 30°C for several days. The resulting bacterial colonies were then compared morphologically based on colonial properties such as shape, color, texture, height, and margins and transferred several times to fresh plates to obtain pure cultures.15 Cultures were stored as glycerol stocks at -20°C.
Scanning electron microscopy
To examine the micro-morphology of the newly isolated Actinobacteria, the Actinobacteria were prepared as mentioned previously,16 and viewed under scanning electron microscope (FEI quanta 200, Thermo Fisher Scientific, Oregon, USA).
DNA extraction
DNA extraction was done using fresh and axenic actinobacterial cultures for the purpose of identification using 16S rRNA gene analysis. Extraction was conducted as described previously.12
Molecular identification and phylogenetic tree construction
The extracted genomic DNA was subjected to 16S rRNA gene amplification and sequencing as described previously.12 The generated sequences were compared to available sequences by BLAST (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi). Phylogenetic analysis was done using maximum likelihood method based on the Tamura-Nei model.17 The phylogenetic trees were built by using MEGA7 Software.18
Extraction of antibacterial agents and evaluating their activity
Extraction of antibacterial agents was done as described previously.14 Briefly, isolates were cultured in medium 5294.14 After 7 days of incubation at 30°C with shaking (160 rpm), twenty milliliters of cultures were mixed with equal volume of ethylacetate for 10 minutes and shaken by a rotary shaker (20 rpm). Then, samples were centrifuged and the upper phase was transferred to new flasks. Ethyl acetate was evaporated and the remaining was solved in ethylacetate:acetone:methanol (1:1:1, v:v:v:). Screening of the antimicrobial activity was carried out as mentioned previously.12 Antibacterial activity of the extract was evaluated against four test bacteria: Pseudomonas aeruginosa ATCC 2785 and Escherichia coli ATCC 25922 (as representatives of Gram-negative bacteria) as well as Staphylococcus aureus ATCC 29213 and Bacillus cereus ATCC 11778 (as representatives of Gram-positive bacteria). The antibacterial activity tests were done by the agar well diffusion method.12
Detection of actinobacterial diversity in water samples
The data about actinobacterial diversity presented in this study are derived from our previous metagenomic analyses carried out by our group on Ma’in thermal springs (HS1, HS2, HS3).9 However, the metagenomic data presented in the results of this study were not published before and they are presented and discussed for the first time.
Results
Chemical properties of sediments
The examined chemical properties of sediments samples include pH, TDS, EC, salinity, concentration of nitrate, concentration of chloride ions, and concentration of fluoride ions. The sediment samples were nearly neutral as indicated by their pH values. Other chemical characteristics of the sediment samples are shown in Table 1. In respect to water samples (HS1, HS2, and HS3), their measured temperature ranged from 48 to 59°C and the pH ranged from 7.44 to 7.76 as revealed previously.9
Table 1. The chemical properties of sediment samples from three Ma’in hot springs sites (M1, M2, and M3) used for the isolation of Actinobacteria .
| M1 | M2 | M3 | |
|---|---|---|---|
| pH | 7.6 | 6.96 | 6.84 |
| Total dissolved solids (mg Kg-1) | 2970 | 7005 | 6005 |
| Electrical conductivity (mS cm-1) | 0.66 | 16.15 | 13.05 |
| Salinity (ppm) | 3 | 7.5 | 6 |
| Nitrate (mg Kg-1) | 1.59 | 4.43 | 3.02 |
| Chloride (mg Kg-1) | 833.35 | 2488 | 2224.75 |
| Fluoride (mg Kg-1) | 3.45 | 2.30 | 2.86 |
Isolation and identification of Actinobacteria from sediments
Five different bacterial isolates were obtained from the sediment samples in this study. However, further analysis by scanning electron microscopy and 16S rRNA gene indicated that only 3 isolates belonged to the phylum Actinobacteria and the other two isolates belonged to the phylum Firmicutes (mainly to the genus Bacillus, data are not shown). The three actinobacterial isolates were assigned as M1-1, M2-2, and M3-2. On the basis of 16S rRNA gene, strain M1-1 was found to have only 90% identity percentage with Nocardiopsis synnemataformans (sequence length is 1072). Identity percentage may suggest that this strain represents a new species in the genus Nocardiopsis based on the similarity that is lower than 97%. Figure 1 shows the morphology of isolate M1-1 and its relationship to the closest relatives.
Figure 1. Scanning electron microscopic image of strain M1-1 and its phylogenetic relationship to the related relatives.
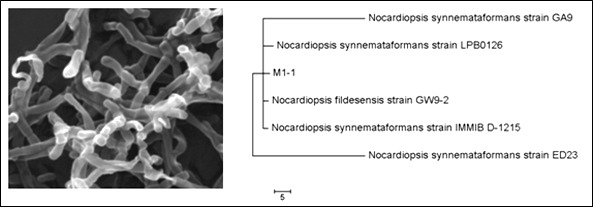
Analysis was carried out by Maximum Likelihood method and the tree was constructed using MEGA7.
In respect to strain M2-2, it was found to be closely related to Streptomyces sp. (identity percentage = 97%; sequence length is 1136 nucleotides). Figure 2 shows the cellular morphology of strain M2-2 under scanning electron microscope and its phylogenetic relationship to the closest relatives.
Figure 2. Scanning electron microscopic image of strain M2-2 and its phylogenetic relationship to the mostly related relatives.
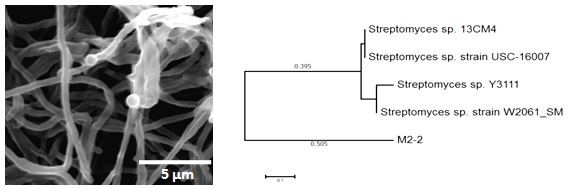
Analysis was carried out by Maximum Likelihood method and the tree was constructed using MEGA7.
Moreover, strain M3-2 was found to be closely related to Nocardioides luteus (identity percentage = 99%; sequence length is 1105 nucleotides). Figure 3 shows cellular morphology of strain M3-2 under scanning electron microscope and its phylogenetic relationship to the closest relatives.
Figure 3. Scanning electron microscopic image of strain M3-2 and its phylogenetic relationship to the mostly related relatives.
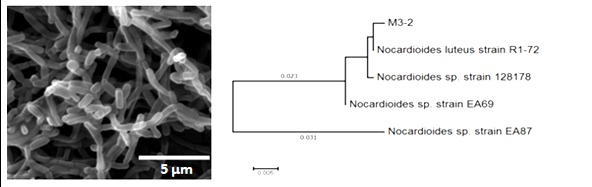
Analysis was carried out by Maximum Likelihood method and the tree was constructed using MEGA7.
Antimicrobial activity of the new isolates
The antibacterial activity of the newly isolated Actinobacteria was examined against P. aeruginosa ATCC 2785, E. coli ATCC 25922, S. aureus ATCC 29213 and B. cereus ATCC 11778. Strain M1-1 was found to be active against P. aeruginosa ATCC 2785 (inhibition zone, 9 mm). Strain M2-2 was found to be active against S. aureus ATCC 29213 (12 mm), B. cereus ATCC 11778 (11 mm), and E. coli ATCC 25922 (9 mm). In respect to strain M3-2, it was found to be active against S. aureus ATCC 29213 (14 mm) and B. cereus ATCC 11778 (9 mm) – Table 2.
Table 2. Diameter of inhibition zones (mm) ± SD formed around the crude extract obtained from antibacterial cultures.
| M1-1 | M2-2 | M3-2 | |
|---|---|---|---|
| P. aeruginosa ATCC 2785 | 9 ± 0.58 | NA | NA |
| E. coli ATCC 25922 | NA | 9 ± 2.3 | NA |
| S. aureus ATCC 29213 | NA | 12 ± 2 | 14 ± 2 |
| B. cereus ATCC 11778 | NA | 11 ± 1.5 | 9 ± 1 |
Actinobacterial diversity in water samples
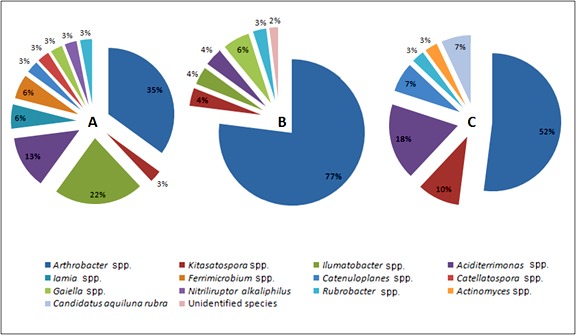
Actinobacteria from water samples could not be isolated by applying the same methods used for sediment samples. However, our unpublished data from a previous metagenomic analysis of Ma’in thermal springs water have shown that the water of Ma’in thermal springs includes a variety of actinobacterial species (13 different species) that are dominated by the genus Arthrobacter. The diversity of Actinobacteria in Ma’in thermal springs is shown in Figure 4.
Figure 4. The major actinobacterial species detected in the three studied Ma’in thermal springs: HS1 (A), HS2 (B) and HS3 (C).
Discussion
Exploring the diversity of Actinobacteria inhabiting thermal environments like thermal springs is important to detect new isolates that could be a promising supply of novel antibiotics.13 The need for new antibiotics is important due to the emergence of pandrug-resistant bacteria and new pathogens.19 Reports indicate that microbial diseases remain the second main cause of mortality around the world despite the vast number of utilized antibiotics. In numbers, microbial infections cause about 17 million deaths per annum.20
Actinobacteria represent a significant source of antibiotics and continue to be a promising target for innovation and discovery. Today, most antibiotics are derived from Actinobacteria and most importantly the Actinomycetes.20 Moreover, extremophilic Actinobacteria represent an important antibiotic source due to several reasons related to their high growth rate and the tendency of their mycelium for rapid breakage and lysis.21
This study revealed the diversity of culturable and unculturable Actinobacteria in the sediments and the water of Ma’in thermal springs (located in Jordan) and reported isolates that are able to produce antimicrobial compounds. Previous studies have documented the isolation of a range of antibiotic-producing species of Actinobacteria originating from thermal springs that are different from our studied springs either in geographical location or the physical and chemical aspects such as temperature and pH. Local and regional studies have frequently documented the isolation of species from thermal habitats belonging to Actinobacteria, like the genus Streptomyces. For instance, Abussaud et al.22 have isolated 30 actinobacterial strains from water and soil of four Jordanian thermal springs (Alshouneh, Waggas, Al-Mansheyah, and Deirallah thermal springs). All isolated strains were identified as Streptomyces species. In the study, the capability of novel isolates to make antimicrobial compounds against E. coli and S. aureus was noticed among 20% and 26% of isolates, respectively.22 At the regional level, Al-Dhabi et al. have recently isolated a new strain of Streptomyces (Streptomyces sp. Al-Dhabi-1) originating from Tharban thermal springs soil located in the southwestern part of Saudi Arabia.23 The strain has also shown a good antimicrobial activity against Streptococcus agalactiae and Klebsiella pneumoniae in addition to some fungal species.23
Thermophilic actinobacterial species with autolytic characteristics were also isolated from the thermal springs in Yunnan, China.21 More diverse actinobacterial populations were also described somewhere else. For instance, Chaudhary and Prabhu (2016) have described the presence of five different genera of Actinobacteria in sediment samples obtained from water springs in Mumbai, India.24 These isolates belong to genera Streptomyces, Micromonospora, Actinomadura, Saccharomonospora, and Thermoactinomyces.24 The isolated strains have a prominent enzymatic activity.24
Based on these studies, it can be concluded that the number and the type of Actinobacteria differ according to the sampling site, physical and chemical properties of the sampling sites, and the isolation strategy. In our study, we were able to detect the frequently isolated genus Streptomyces with a clear antibacterial activity. This agrees with the findings of previous Jordanian and some regional studies that showed Streptomyces as an important genus in such habitats.22-24 Nevertheless, we report in this study the isolation of two additional actinobacterial species: Nocardiopsis sp. and Nocardioides luteus. Both species possess an apparent antibacterial activity.
In respect to water samples, Actinobacteria from water samples could not be isolated even though our metagenomic analysis provided us with the evidence that there is a great diversity of Actinobacteria in Ma’in thermal springs. This can be explained by the isolation method utilized (the medium and the set of incubation conditions), which was not suitable for isolating the detected species. Therefore, we need to apply new isolation strategies to obtain the native Actinobacteria from water samples because this and earlier studies have already indicated that the isolated bacteria obtained in the laboratory represent a minor fraction of what is existing in nature.25 The limitations in this study are variable. As in any isolation study, the success in isolation is dependent on providing the suitable nutritional requirements and how to replicate the in situ physicochemical conditions in the lab and how to suppress the competing groups during isolation.
Conclusion
The following antibiotic-producing Actinobacteria were isolated from the sediment of Ma’in thermal springs (Jordan): Nocardiopsis sp., Streptomyces sp. and Nocardioides luteus. The isolates showed a clear inhibitory activity against E. coli ATCC 25922, P. aeruginosa ATCC 2785, B. cereus ATCC 11778, and S. aureus ATCC 29213. Nevertheless, no actinobacterial isolates could be obtained from water samples despite their significant diversity as revealed by our previous metagenomic analysis, which also showed the dominance of Arthrobacter. New isolation strategies and further chemical analysis of the bioactive compounds is recommended.
Footnotes
Authors’ contributions statement: All authors contributed equally to this study.
Conflicts of interest: All authors – none to disclose.
Funding: None to declare.
References
- 1.Barka EA, Vatsa P, Sanchez L, et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shivalata L, Satyanarayana T. Thermophilic and alkaliphilic Actinobacteria: biology and potential applications. Front Microbiol. 2015;6:1014. doi: 10.3389/fmicb.2015.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventura M, Canchaya C, Tauch A, et al. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madigan M, Martinko J, Stahl D, Clark D. Brock Biology of Microorganisms. 12th Ed. Pearson/Benjamin Cummings; 2009 [Google Scholar]
- 5.Maier RM, Pepper IL, Gerba CP. Environmental Microbiology. 2nd Ed. Academic Press; 2009 [Google Scholar]
- 6.Bérdy J. Microorganisms Producing Antibiotics. In: Sánchez S, Demain Al, editors. Antibiotics. Caister Academic Press; 2015. [Google Scholar]
- 7.Valverde A, Tuffin M, Cowan DA. Biogeography of bacterial communities in hot springs: a focus on the actinobacteria. Extremophiles. 201216;669-79:[Crossref]. doi: 10.1007/s00792-012-0465-9. [DOI] [PubMed] [Google Scholar]
- 8.Engelkirk P, Duben-Engelkirk J. Burton’s Microbiology for the Health Sciences. 9th Ed. Lippincott Williams and Wilkins; 2011 [Google Scholar]
- 9.Hussein EI, Jacob JH, Shakhatreh MAK, Abd Al-Razaq MA, Juhmani AF, Cornelison CT. Exploring the microbial diversity in Jordanian hot springs by comparative metagenomic analysis. MicrobiologyOpen. 2017;6:e521. doi: 10.1002/mbo3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil A, Salim M, Sallal A. Enumeration of thermotolerant bacteria from recreational thermal ponds in Jordan. Cytobios. 1998;96:57–63. [Google Scholar]
- 11.Al-Batayneh K, Jacob J, Hussein E. Isolation and molecular identification of new thermophilic bacterial strains of Geobacillus pallidus and Anoxybacillus flavithermus. Int J Integ Biol. 2011;11:39–43. [Google Scholar]
- 12.Shakhatreh MAK, Jacob JH, Hussein EI, et al. Microbiological analysis, antimicrobial activity, and heavy-metals content of Jordanian Ma’in hot-springs water. J Infect Public Health. 2017;10:789–93. doi: 10.1016/j.jiph.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Müller R, Wink J. Future potential for anti-infectives from bacteria – how to exploit biodiversity and genomic potential. Int J Med Microbiol. 2013;304:3–13. doi: 10.1016/j.ijmm.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Wink J. Methods for the taxonomic description of the Actinobacteria. Compendium of Actinobacteria. [Accessed on: 01 June 2018]. Available at: http://www.dsmz.de/fileadmin/Bereiche/Microbiology/Dateien/Bacterial_Nomenclature_uptodate/Actinomethods.pdf.
- 15.Jacob J, Wink J, Wink M. Antimicrobial activity of actinobacteria from a hypersaline area of the Dead Sea. Proceedings of the Fourth Kuwait Conference of Chemistry Kuwait-Chemistry and Life Sciences; 2016:March 20‐22, Kuwait city, Kuwait. [Google Scholar]
- 16.Muhaidat R, Al-Qudah MA, Samir O, et al. Phytochemical investigation and in vitro antibacterial activity of essential oils from Cleome droserifolia (Forssk.) Delile and C. trinervia Fresen. (Cleomaceae). S Afr J Bot. 2015;99:21–8. [Google Scholar]
- 17.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis. Ver. 1. 0, The Pennsylvania State University, University Park, PA; 1993 [Google Scholar]
- 18.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolain JM, Abat C, Jimeno MT, Fournier PE, Raoult D. Do we need new antibiotics? Clin Microbiol Infect. 2016;22:408–15. doi: 10.1016/j.cmi.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Procópio R, Silva IR, Martins MK, Azevedo JL, Araújo JM. Antibiotics produced by Streptomyces. Braz J Infect Dis. 2012;16:466–71. doi: 10.1016/j.bjid.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Xu LH, Tiang YQ, Zhang YF, Zhao LX, Jiang CL. Streptomyces thermogriseus, a new species of the genus Streptomyces from soil, lake and hot-spring. Int J Syst Bacteriol. 1998;48 Pt 4:1089–93. doi: 10.1099/00207713-48-4-1089. [DOI] [PubMed] [Google Scholar]
- 22.Abussaud MJ, Alanagreh L, Abu-Elteen K. Isolation, characterization and antimicrobial activity of Streptomyces strains from hot spring areas in the northern part of Jordan. Afr J Biotechnol. 2013;12:7124–32. [Google Scholar]
- 23.Al-Dhabi NA, Esmail GA, Duraipandiyan V, Valan Arasu M, Salem-Bekhit MM. Isolation, identification and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles. 2016;20:79–90. doi: 10.1007/s00792-015-0799-1. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhary N, Prabhu S. Thermophilic actinomycetes from hot water spring capable of producing enzymes of industrial importance. Int J Res Stud Biosci. 2016;4:29–35. [Google Scholar]
- 25.Stewart E. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–60. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


