Voltage-gated sodium channels have a conserved multiphase slow-inactivation process. Gamal El-Din et al. show that the early phase involves conformational changes at a critical threonine in the S6 segment, which are followed by a late phase mediated by the intracellular C-terminal domain.
Abstract
Homotetrameric bacterial voltage-gated sodium channels share major biophysical features with their more complex eukaryotic counterparts, including a slow-inactivation mechanism that reduces ion-conductance activity during prolonged depolarization through conformational changes in the pore. The bacterial sodium channel NaVAb activates at very negative membrane potentials and inactivates through a multiphase slow-inactivation mechanism. Early voltage-dependent inactivation during one depolarization is followed by late use-dependent inactivation during repetitive depolarization. Mutations that change the molecular volume of Thr206 in the pore-lining S6 segment can enhance or strongly block early voltage-dependent inactivation, suggesting that this residue serves as a molecular hub controlling the coupling of activation to inactivation. In contrast, truncation of the C-terminal tail enhances the early phase of inactivation yet completely blocks late use-dependent inactivation. Determination of the structure of a C-terminal tail truncation mutant and molecular modeling of conformational changes at Thr206 and the S6 activation gate led to a two-step model of these gating processes. First, bending of the S6 segment, local protein interactions dependent on the size of Thr206, and exchange of hydrogen-bonding partners at the level of Thr206 trigger pore opening followed by the early phase of voltage-dependent inactivation. Thereafter, conformational changes in the C-terminal tail lead to late use-dependent inactivation. These results have important implications for the sequence of conformational changes that lead to multiphase inactivation of NaVAb and other sodium channels.
Introduction
Voltage-gated sodium (NaV) channels generate and propagate action potentials, which can trigger neurotransmission, endocrine secretion, and contraction of cardiac and skeletal muscle (Hille, 2001). They respond to membrane depolarization by rapidly opening to initiate the regenerative depolarizing phase of the action potential, and then they undergo fast inactivation within a few milliseconds, thereby beginning repolarization and contributing to termination of the action potential. Prolonged depolarizing pulses or trains of repetitive depolarizations induce a separate, multiphase, slow-inactivation process, which shapes the length and frequency of trains of action potentials, protects against excitotoxicity, and plays an important role in encoding and transmitting neural information (Cantrell and Catterall, 2001; Vilin and Ruben, 2001; Carr et al., 2003; Chen et al., 2006). Elucidation of the molecular mechanisms underlying the process of multiphasic slow inactivation would be an important advance in understanding NaV channel function and its role in encoding information in the nervous system.
Eukaryotic NaV channels have single pore-forming subunits composed of four homologous domains, whereas the ancestral bacterial NaV channels are composed of four identical polypeptides homologous to one domain in eukaryotic NaV channels (Catterall, 2000; Ren et al., 2001; Catterall and Zheng, 2015). Each subunit of a bacterial NaV channel has a voltage-sensing module and a pore-forming module (Ren et al., 2001; Payandeh et al., 2011), and they share the major functional and biophysical features of their eukaryotic counterparts. The ability to crystallize bacterial NaV channels like NaVAb, NaVRh, NaVMs, NaVAe, and NaVSp1 (Payandeh et al., 2011, 2012; Zhang et al., 2012; Bagnéris et al., 2013; Arrigoni et al., 2016; Sula et al., 2017) and analyze their structure at high resolution makes them invaluable for studying the structural basis of NaV channel functions that are conserved from prokaryotes to mammals.
Slow inactivation is functionally conserved from bacterial to mammalian NaV channels (Vilin and Ruben, 2001; Pavlov et al., 2005; Gamal El-Din et al., 2018), and it has been extensively studied by mutagenesis of mammalian NaV channels and by x-ray crystallography of NaVAb. Bacterial NaV channels show three phases of slow inactivation. Early decay of sodium current in 10 msec to 100 msec during single depolarizing pulses and intermediate, reversible inactivation during long pulses are observed in many bacterial NaV channels (Ren et al., 2001; Pavlov et al., 2005; Payandeh et al., 2012; Bagnéris et al., 2013; Gamal El-Din et al., 2013, 2014; Arrigoni et al., 2016). In contrast, late, very slowly reversible, use-dependent inactivation is prominent in NaVAb but not other bacterial NaV channels (Gamal El-Din et al., 2013). The first two phases of bacterial NaV channel slow inactivation may be homologous to eukaryotic NaV slow inactivation, whereas the third phase of late use-dependent inactivation may be unique to the NaVAb channel (Scheuer, 2014). NaVAb activates at very negative membrane potentials, and its multiphase inactivation processes are engaged at negative membrane potentials relative to other NaV channels, perhaps providing a rationale for its unique third phase of inactivation. We use the terms “early” and “late” to distinguish the first two phases of inactivation during single pulses in NaVAb from the third phase, because all three phases have time courses characteristic of eukaryotic NaV channel slow inactivation, making use of the terms “fast” and “slow” potentially confusing. Mutation of Asn49 to Lys in the S2 segment in the extracellular negative cluster of the voltage sensor shifts the voltage dependence of activation and prevents late use-dependent inactivation (Gamal El-Din et al., 2013), but the structural basis for multiphase inactivation of the pore remains unknown. Here, we have probed the molecular and structural basis for multiphase inactivation of NaVAb using a combination of mutagenesis, electrophysiological recording, and structure determination. Our results reveal that conformational changes involving Thr206 in the S6 segment are required for early slow inactivation, but the long C-terminal tail mediates late use-dependent inactivation. The requirement for the long C-terminal tail of NaVAb in late use-dependent inactivation suggests a potential functional role for this large intracellular domain.
Materials and methods
Molecular biology, protein expression, and protein purification
NaVAb mutations and truncations for both structural biology and electrophysiology were constructed by the introduction of mutations at the desired positions (QuikChange, Agilent) in the genetic background of NaVAb WT and confirmed by sequencing. Recombinant baculovirus was generated using the Bac-to-Bac system (Invitrogen), and Trichoplusia ni insect cells (High Five) were infected for protein production and electrophysiology. Protein was produced and purified as previously described (Payandeh et al., 2012).
Electrophysiology
T. ni cells were grown on 35-mm Petri dishes in Grace’s insect medium (Gibco) supplemented with FBS (10%) and antibiotics (100 µg/ml streptomycin and 100 U/ml penicillin). Cells were infected by replacing the incubation medium with a medium containing the virus encoding NaVAb constructs (10 µg/ml). After 1 h, 2 ml incubation medium was added to the virus-containing medium. Cells were maintained at 25–27°C for at least 24 h before study (Gamal El-Din et al., 2013).
All constructs showed high-level expression that enabled us to measure ionic current 24–48 h after infection. Whole-cell sodium currents were recorded using an amplifier (Axopatch 200; Molecular Devices) with glass micropipettes (2–4 MΩ). Capacitance was subtracted and series resistance was compensated using internal amplifier circuitry; 85–90% of series resistance was compensated. For ionic current measurements, the intracellular pipette solution contained (in mM) 35 NaCl, 105 CsF, 10 EGTA, and 10 HEPES, pH 7.4 (adjusted with CsOH). The extracellular solution contained (in mM) 140 NaCl, 2 CaCl2, 2 MgCl2, and 10 HEPES, pH 7.4 (adjusted with NaOH). The standard voltage-clamp protocol for measuring ionic currents consisted of steps from a holding potential of −180 mV to voltages ranging from −180 mV to +50 mV in 10-mV steps. Use-dependent inactivation was induced by depolarizing from −180 mV to 0 mV or to the indicated potentials at 0.2 Hz or 1 Hz. Voltage-clamp pulses were generated and currents were recorded using Pulse software controlling an Instrutech ITC18 interface (HEKA). Data were analyzed using Igor Pro 6.2 software (WaveMetrics).
NaVAb crystallization and data collection
NaVAb/WT and mutants were reconstituted into DMPC/CHAPSO bicelles (Anatrace) as previously described, mixed in a 1:2 ratio, and set up in a hanging-drop format over well solutions containing 1.8 M ammonium sulfate and 100 mM sodium acetate, pH 4.8. Crystals appeared in 3–5 d and grew to full size within 1 wk (∼50 µm by 100 µm). They were cryoprotected by slow addition of well solution supplemented with increasing concentrations of glucose up to 30%. Crystals were harvested in nylon loops and plunged into liquid nitrogen for data collection. Data were collected at Advanced Light Source (Beamlines BL821 and BL822). As crystals were highly radiation sensitive, exposure times were minimized to limit decay during data collection. Data reported herein were collected from single crystals.
Structure determination and refinement
X-ray diffraction data were integrated and scaled with DENZO/SCALEPACK (Otwinowski and Minor, 1997) and processed with PHENIX (Adams et al., 2010). We investigated multiple space groups and determined the reported space group (I422) by comparison of merging statistics, as well as fully refined structures determined in C2, I222, I4, and I422 with and without inclusion of weakly recorded reflections. The NaVAbΔ28 structure was solved with molecular replacement using the NaVAb/I217C structure (PDB accession no. 3RVY), modified by removal of the S4–S5 linker and base of S5 helix (amino acids 115–137) plus the S6 helix as the search model. Following initial phases, models of both proteins were manually rebuilt based on resulting electron density maps. Electron density was easily interpretable throughout the S6, S4–S5 linker, and C-terminal domain NaVAbΔ28. Simulated annealing omit maps were used to confirm the placement of the S6 at the activation gate, as well as the position of the S4–S5 linker. Lipids and waters were added to both models near the end of refinement. Geometry, B-factors, and electron density fits were assessed with POLYGON (Urzhumtseva et al., 2009).
Online supplemental material
Fig. S1 presents a 3-D view of the amino acid residues involved in the early phase of voltage-dependent inactivation of NaVAb and in slow inactivation of NaV1.4, as listed in Table S1. Fig. S2 presents results on the steady-state inactivation of mutants at the position of Thr206. Fig. S3 presents high-resolution views of the fits of our molecular models for the region around Thr206 and for the C-terminal tail coiled-coil domain to the underlying electron density maps. Table S1 presents a comparison of amino acid residues that are involved in the early phase of voltage-dependent inactivation in the bacterial sodium channel NaVAb and in slow inactivation of the mammalian sodium channel NaV1.4. Table S2 presents the mean data and statistical analysis for the new structural models derived from our x-ray crystallography studies.
Results
Block of early voltage-dependent inactivation by a mutation in S6
The rate of inactivation of NaVAb and other bacterial NaV channels during a single depolarization is ∼10- to 50-fold slower than fast inactivation of mammalian NaV channels (Ren et al., 2001; Ito et al., 2004; Pavlov et al., 2005; Gamal El-Din et al., 2013), and movements of the selectivity filter and S6 segment are implicated in slow inactivation in mammalian sodium channels (Vilin et al., 2001) and NaChBac (Zhao et al., 2004a,b; Pavlov et al., 2005). Amino acid residues that are implicated in slow inactivation both by mutagenesis studies of mammalian NaV channels and by mutagenesis and structural studies of bacterial NaV channels are listed in Table S1 and illustrated in structural format in Fig. S1. The extensive overlap in the amino acid residues implicated in slow inactivation of mammalian and bacterial NaV channels suggest a similar underlying mechanism for slow inactivation of mammalian NaV channels and the early voltage-dependent phase of inactivation of NaVAb.
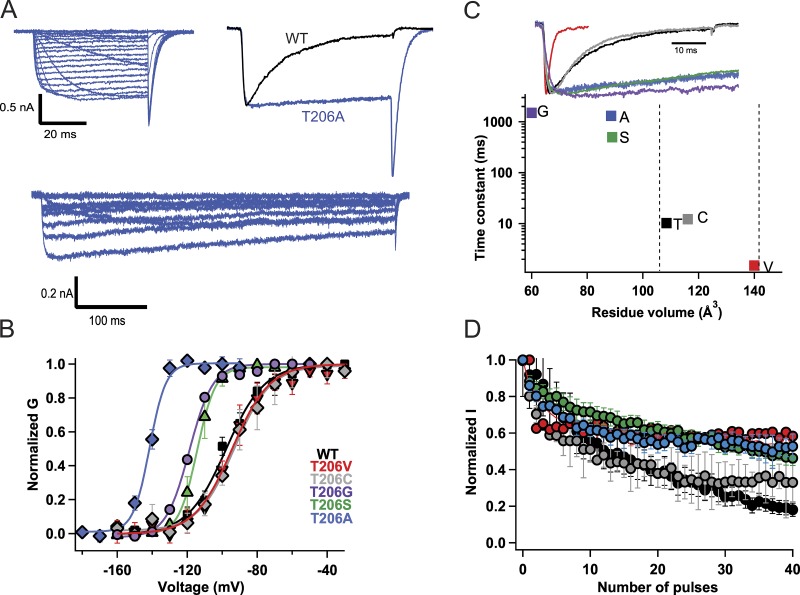
We hypothesized that the gating movements of the S6 segment leading to opening of the pore (Zhao et al., 2004a,b; Pavlov et al., 2005) may also begin the multiphase process of inactivation of NaVAb. Structural and functional experiments have suggested that Thr206 in the S6 segment is involved in both activation and inactivation gating in NaVAb. This residue is in the position homologous to the Gly hinges identified in prokaryotic potassium channels and NaChBac (Zhao et al., 2004a,b; Cuello et al., 2010; Lee et al., 2012). We mutated Thr206 to Ala (Fig. 1) in order to determine whether it would disrupt the early phase of inactivation during the test pulse. Indeed, this mutation abolished inactivation during the pulse (Fig. 1 A, upper traces). Even during long 500-ms depolarizing pulses, little decay of the sodium current was observed (Fig. 1 A, lower traces). The T206A mutation also caused partial loss of steady-state inactivation (Fig. S2 A), and it shifted the activation curve of NaVAb by ∼25 mV in the negative direction (Fig. 1 B). These results indicate that this mutation stabilizes the open state of the channel and allows more complete activation at each test potential (Fig. 1 B) yet uncouples this enhanced voltage-dependent activation from the early phase of slow inactivation.
Figure 1.
Size-dependent effects of mutations of Thr206 on the early phase of inactivation. (A) Top left: Representative current traces of NaVAb/T206A. Cells were held at −180 mV, and 50-ms depolarizing pulses were applied in 10-mV steps from −180 mV to +50 mV. Top right: Comparison of NavAb/T206A inactivation during the pulse (blue) to NaVAb/WT (black). The 100-ms pulse was applied from a holding potential of −180 mV to −70 mV. Bottom: Representative current traces of NaVAb/T206A recorded during 500-ms depolarizing pulses from −180 mV to +50 mV. (B) Voltage dependence of NaVAb/WT and mutants: NaVAb/WT, V1/2 = −97 ± 1.4 mV (black circles; n = 7); NaVAb/T206A, V1/2 = −121 ± 2 mV (blue; n = 5); NaVAb/T206S, V1/2 = −115 ± 1 mV, (green); NaVAb/T206C, V1/2 = −95 ± 4 mV (gray; n = 7); NaVAb/T206V, V1/2 = −95 ± 0.9 mV (red; n = 8). The activation curves were constructed from I–V curves in which 500-ms depolarizing pulses were applied. (C) Top: Representative current traces showing inactivation kinetics of NaVAb Thr206 mutants compared with NaVAb WT. Bottom: Time constants for early voltage-dependent inactivation plotted versus the volume of the amino acid residue at position 206. (D) Peak inward currents measured during each pulse of 1-Hz trains of depolarizations and normalized to the current produced by the first depolarization of the train for NaVAb/WT (black), T206C (gray), T206S (green), T206V (red), T206G (purple), and T206A (blue). Error bars represent SEM.
Mutation of Thr to Ala both prevents hydrogen bond formation and reduces side-chain size (Thr: V = 118.3 Å3; Ala: V = 87.8 Å3; Perkins, 1986). To separate these two effects, we mutated Thr to different residues that would either remove the hydroxyl group of Thr and thereby prevent hydrogen bonding or change the side-chain volume. We found that the T206S mutation abolishes the early phase of inactivation similar to T206A. This suggests that hydrogen bond formation is not sufficient to retain early phase inactivation, because Ser has a hydroxyl group like Thr but removes inactivation during the pulse (Fig. 1 C, green; Tang et al., 2016). However, Ala and Ser have similar molecular volumes (87.8 vs. 91.7 Å3, respectively). Therefore, to test whether side-chain volume is the main factor in tuning early phase inactivation kinetics, we mutated Thr206 to a bulky residue Val (V = 138.8 Å3). To our surprise, the NaVAb/T206V mutation greatly increased the rate of early voltage-dependent inactivation (Fig. 1 C, red). To further test the role of side-chain volume as a determinant of the kinetics of early phase inactivation, we mutated Thr206 to Cys (V = 105.5 Å3) and Gly (V = 59.9 Å3). Indeed, T206C exhibited intermediate inactivation kinetics, whereas T206G inactivated even more slowly than T206A and T206S (Fig. 1 C). Overall, these results demonstrate a striking correlation of inactivation rate with molecular size of the residue at position 206 over a 1,000-fold range of inactivation rates (Fig. 1 C).
All of the Thr206 mutations that reduce side-chain size caused negative shifts in the voltage dependence of activation (Fig. 1 B), whereas they had comparatively small effects on the use-dependent inactivation profile (Fig. 1 D). These results reveal a significant correlation of negative shifts in the voltage dependence of activation with reduction in the molecular volume of the side chain of Gly, Cys, Ser, and Val substituted for Thr206 (Fig. S2 B; r = 0.86), and substitution of Ala has even greater effects (Fig. S2 B). Thus, reducing molecular volume correlates with both negative shifts of activation and dramatic slowing of the rate of inactivation (Fig. 1 C). We hypothesize that Thr206 serves as a molecular hub coupling conformational changes important for activation and the early phase of inactivation; therefore, mutations that reduce side-chain size uncouple activation from inactivation.
Role of the C-terminal tail in early voltage-dependent inactivation
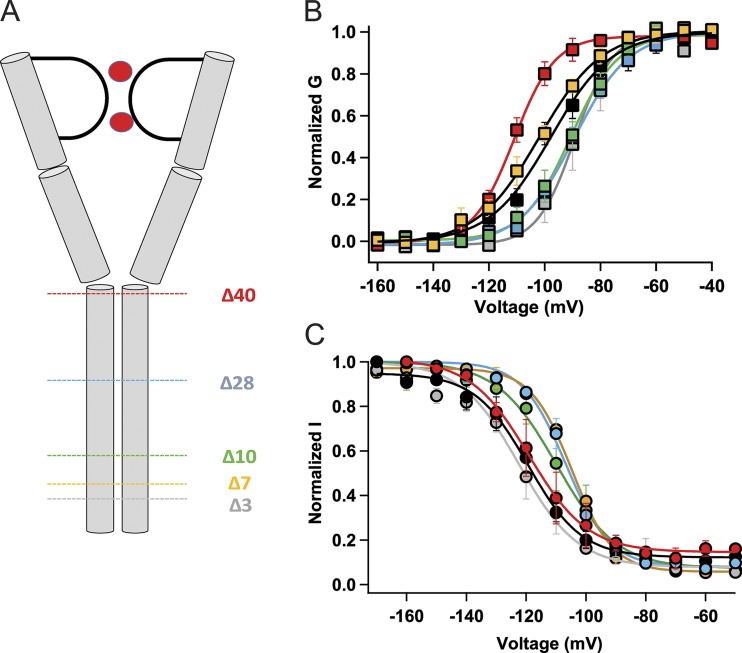
The S6 transmembrane helix continues uninterrupted as an intracellular helix in the C-terminal tail in some bacterial NaV channels (e.g., McCusker et al., 2012; Bagnéris et al., 2013), and we wondered whether this extension of the S6 helix could modulate inactivation kinetics. To address this question, we constructed a series of truncation mutants of the C-terminal tail (NaVAbΔ40, Δ28, Δ10, Δ7, and Δ3; Fig. 2 A). The construct named NaVAbΔ40 encodes the same protein form as NaVAb/1–226, whose structure we determined in an open state previously (Lenaeus et al., 2017). All of these constructs conducted robust voltage-dependent sodium currents (Fig. 2 B). Complete truncation of the C-terminal tail in NaVAbΔ40 facilitates activation, as revealed by an approximately −14-mV shift of the G–V curve compared with NaVAb/WT (V1/2 = −110.7 ± 2.1 for NaVAbΔ40, red; V1/2 = −97 ± 1.4 mV for NaVAb/WT, black; P < 0.001; Fig. 2 B; Lenaeus et al., 2017). On the other hand, this truncation did not affect the steady-state inactivation profile (Fig. 2 C; NaVAbΔ40, V1/2 = −116.1 ± 4.4, red; NaVAb/WT, V1/2 = −119 ± 1.1, black, NS). NaVAbΔ28, Δ10, and Δ3 showed positive shifts in their G–V curves compared with NaVAb/WT (Fig. 2 B; NaVAbΔ28, V1/2 = −90.1 ± 1.5, P < 0.001; blue; NaVAb/Δ10, V1/2 = −90.7 ± 0.8, P < 0.05, green; NaVAbΔ3, V1/2 = −89.1 ± 0.8, gray, P < 0.001). In contrast, NaVAbΔ7 did not affect voltage-dependent activation significantly (Fig. 2 B; NaVAbΔ7; V1/2 = −104.1 ± 2, yellow; NaVAb/WT, V1/2 = −97 ± 1.4 mV, black). Steady-state inactivation measurements showed a positive shift in V1/2 for all constructs except NaVAbΔ40 and NaVAbΔ3 (Fig. 2 C; NaVAbΔ28, V1/2 = −106.7 ± 0.6, P < 0.001, blue; NaVAbΔ10, V1/2 = −109.7 ± 0.6 mV, green, P < 0.001; NaVAbΔ7, V1/2 = −104.7 ± 0.4, yellow, P < 0.001; NaVAbΔ3, V1/2 = −121.7 ± 0.4, gray, NS). The differential effects of these truncations on the activation and inactivation profiles suggest distinct functions of the proximal versus distal C-terminal tail.
Figure 2.
Effects of C-terminal truncation on the voltage dependence of activation and inactivation. (A) A cartoon of NaVAb showing the different truncations of the C-terminal tail domain. (B) G–V curves of NaVAb/WT and C-terminal truncated constructs: NaVAb/WT, V1/2 = −97 ± 1.4 mV (black, n = 7); NaVAbΔ40, V1/2 = −110.7 ± 2.1 mV (red, n = 5); NaVAbΔ28, V1/2 = −90.1 ± 1.5 mV (blue, n = 8); NaVAbΔ10, V1/2 = −90.7 ± 0.8 mV (green, n = 4); NaVAb/Δ7, V1/2 = -104 ± 2 mV (gold, n = 5); and NaVAbΔ3, V1/2 = −89.1 ± 0.8 mv (gray, n = 4) (C) Steady-state inactivation curves of the constructs. NaVAb/WT, V1/2 = −119 ± 1.1 mV (black, n = 5); NaVAbΔ40, V1/2 = −116.1 ± 4.4 mV (red, n = 5); NaVAbΔ28, V1/2 = −106.7 ± 0.6 (blue, n = 3); NaVAbΔ10, V1/2 = −109.7 ± 0.6 mV (green, n = 5); NaVAbΔ7, V1/2 = −104.7 ± 0.4 mV (gold, n = 3); and NaVAbΔ3, V1/2= −121.7 ± 0.4 (gray, n = 5). Error bars represent SEM.
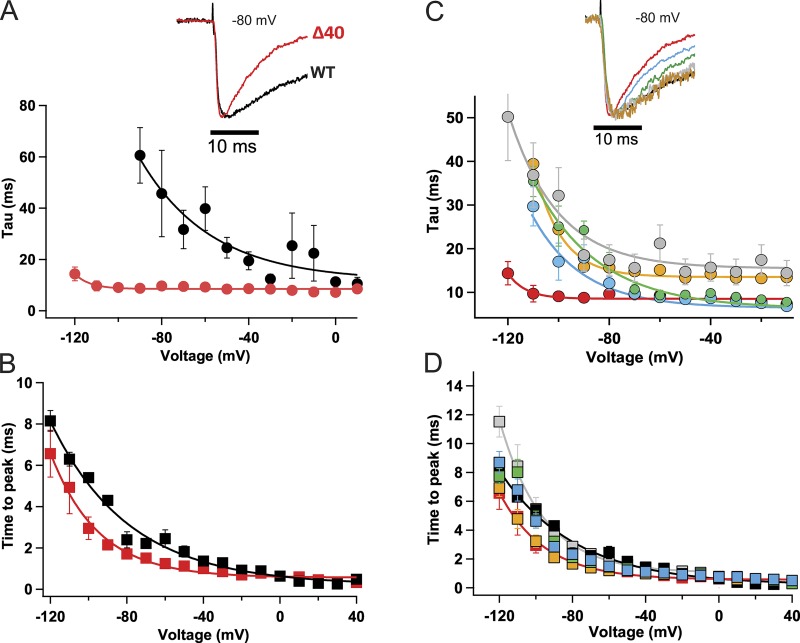
Deletion of the entire C-terminal tail in NaVAbΔ40 accelerated the early phase of inactivation compared with NaVAb/WT (Fig. 3 A; Gamal El-Din et al., 2013). Remarkably, this truncation removed the voltage dependence of current decay and rendered the rate of inactivation during the pulse voltage insensitive (Fig. 3 A, red). However, the kinetics of activation, measured as time to peak current, did not change greatly compared with NaVAb/WT (Fig. 3 B). We analyzed the kinetics of inactivation during the pulse for NaVAbΔ28, Δ10, Δ7, and Δ3. These progressively truncated constructs showed progressively faster kinetics of decay of the sodium current during single depolarizations (Fig. 3 C); however, there were only minor effects on activation kinetics (Fig. 3 D). Therefore, these results demonstrate a specific effect of the C-terminal tail of NaVAb on the rate and voltage dependence of the early phase of voltage-dependent slow inactivation.
Figure 3.
Truncation of the C-terminal tail accelerates the early phase of inactivation. (A) Cells were held at −180 mV, and 50-ms depolarizing pulses were applied in 10-mV steps from −180 mV to +50 mV. Top: Comparison of NaVAbΔ40 inactivation during the pulse (red) to NaVAb/WT (black) during a depolarizing pulse from a holding potential of −180 mV to −80 mV. Bottom: Time constant of the decay of current during depolarizations to the indicated potentials for NaVAbΔ40 (red, n = 4) and NaVAb/WT (black, n = 7). The NaVAb/WT inactivation time constants were adapted from previous work (Gamal El-Din et al., 2013) for ease of comparison. (B) Time to peak current for NaVAbΔ40 (red) and NaVAb/WT (black; n = 5–7). (C) Top: Representative normalized current traces during a depolarizing pulse from a holding potential of −180 mV to −80 mV. Time constant of inactivation for NaVAbΔ40 (red), NaVAbΔ28 (blue), NaVAbΔ10 (green), NaVAbΔ7 (gold), and NaVAbΔ3 (gray); n = 4–11. The inactivation time constants were estimated using the equation yo + A(exp(−t/τ). (D) Time to peak current of the C-terminal truncated constructs, color coded as in C (n = 6–7). Error bars represent SEM.
Control of late use-dependent inactivation of NaVAb by its C-terminal tail
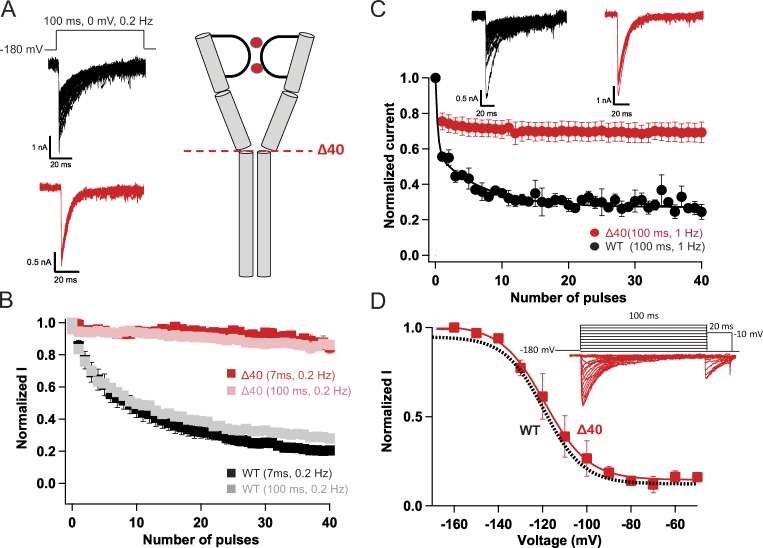
NaVAb/WT activates at very negative potentials and shows a dramatic, very slowly reversible form of use-dependent inactivation (Payandeh et al., 2012; Gamal El-Din et al., 2013). Repetitive depolarizations of NaVAb/WT to 0 mV for either 7 ms or 100 ms at 0.2 Hz resulted in reduction of current amplitude to ∼20% of its initial value after 40 pulses (Fig. 4, A and B, black and gray). In sharp contrast, NaVAbΔ40 shows no use-dependent inactivation in either pulse protocol (Fig. 4, A and B, red). Applying higher frequency repetitive pulses resulted in ∼20% decline during the first few pulses followed by stable current compared with continuous decline for NaVAb/WT (Fig. 4 C, red and black, respectively). By the end of 40 pulses, NaVAbΔ40 lost only 25% of its initial current amplitude compared with a 75% loss in case of NaVAb/WT. In contrast to its loss of late use-dependent inactivation, NaVAbΔ40 showed no effect on early steady-state inactivation during single depolarizations compared with NaVAb/WT (Fig. 4 D). These results indicate that the molecular mechanism of early steady-state inactivation is different from that of late use-dependent inactivation.
Figure 4.
Truncation of the C-terminal tail abolishes late use-dependent inactivation. (A) Representative current traces showing late use-dependent inactivation of NaVAb WT (black) and loss of late use-dependent inactivation in NaVAbΔ40 (red). (B) Peak inward currents recorded during each pulse in trains of depolarizations at 0.2 Hz and normalized to the current produced by the first depolarization of the train for NaVAbΔ40 (7 ms, red, n = 7; 100 ms, pink, n = 5) and NaVAb/WT (7 ms, black, n = 7; 100 ms, gray n = 11). (C) Top: Representative currents recorded during each pulse in trains of depolarizations at 1 Hz. Bottom: Normalized currents during each depolarizing pulse for NaVAbΔ40 (red, n = 3) and NaVAb/WT (black, n = 5). (D) Top: Representative currents for NaVAb40 (red) during 100-ms conditioning pulses followed by 20-ms test pulses. Bottom: Voltage dependence of inactivation for NaVAb40 (red) compared with NaVAb/WT (black), as reported previously (Lenaeus et al., 2017). Error bars represent SEM.
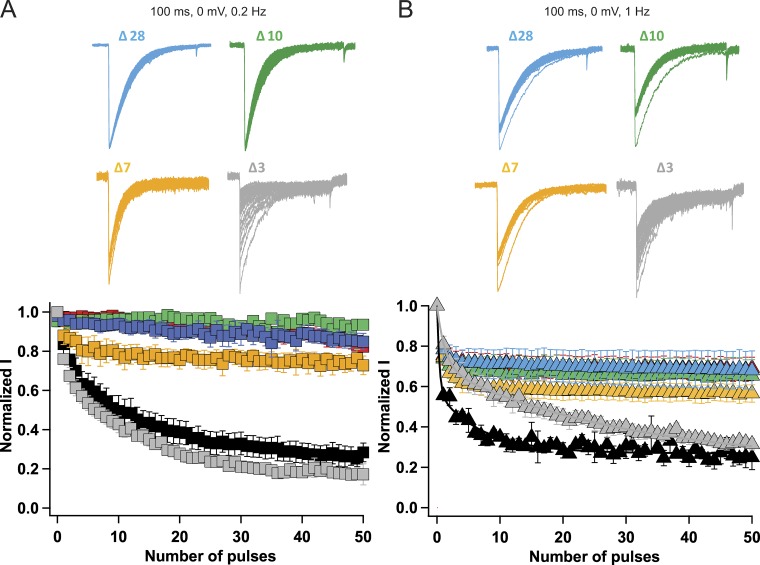
Graded effects of progressive truncation of the C-terminal tail
How many residues must be cut from the C-terminal tail to block late use-dependent inactivation of NaVAb? We found that progressive deletions of the NaVAb C-terminal tail at Δ28, Δ10, Δ7, and Δ3 caused graded effects on use-dependent inactivation (Fig. 5). However, deletion of only 10 residues was sufficient to abolish nearly all of the late use-dependent inactivation of NaVAb (Fig. 5, A and B). At 0.2 Hz, NaVAbΔ28 and NaVAbΔ10 showed no use-dependent inactivation. NaVAbΔ7 showed ∼20–25% current decay during the first few pulses, after which the current stayed relatively constant (Fig. 5 A). On the other hand, NaVAbΔ3 had use-dependent inactivation similar to NaVAb WT (Fig. 5 A). Application of 100-ms depolarizing pulses to 0 mV at 1 Hz resulted in ∼30% current decay for NaVAbΔ28 and NaVAbΔ10, 40% current decay for NaVAbΔ7, and ∼70% current decay for NaVAbΔ3, but with slow kinetics compared with NaVAb/WT. Overall, these results lead to the surprising conclusion that the most distal segment of the C-terminal tail is required for late use-dependent inactivation of NaVAb.
Figure 5.
Graded effects of C-terminal tail truncations on late use-dependent inactivation. (A) Top: Current traces elicited by applying 100-ms depolarizing pulses from a holding potential of −180 mV to 0 mV at 0.2 Hz for different truncated constructs. Bottom: Peak inward currents measured during each train of pulses and normalized to the current produced by the first depolarization of the train for NaVAb/WT (black, n = 7), NaVAb 40 (red, n = 7), NaVAb 28 (blue, n = 12), NaVAb 10 (green, n = 5), NaVAb 7 (yellow, n = 9), and NaVAb 3 (gray, n = 5). (B) Top: Current traces elicited by 100-ms depolarizations from a holding potential of −180 mV at 1 Hz with different constructs. Bottom: Use-dependent inactivation profile of the different constructs during application of 100-ms repetitive pulses applied at 1 Hz. NaVAb WT (black, n = 7), NaVAbΔ40 (red, n = 11), NaVAbΔ28 (blue, n = 6), NaVAbΔ10 (green, n = 5), NaVAbΔ7 (yellow, n = 6), and NaVAbΔ3 (gray, n = 5). Error bars represent SEM.
Structural basis for multiphase inactivation of NaVAb
As a first step toward understanding the structural basis for the striking effects of mutations of Thr206, the S6 segment, and the C-terminal tail in multiphase inactivation of NaVAb, we set out to analyze the structures of the NaVAb mutants with progressively truncated C-terminal tail domains using x-ray crystallography. We were not able to solve the structures of crystals formed by NaVAbΔ10, NaVAbΔ7, and NaVAbΔ3. However, the high-resolution structure of NaVAbΔ28 (Fig. 6 A and Table S2) has given key insights into the conformations of the S6 inactivation gate, the C-terminal tail, and the region of Thr206 and nearby residues.
Figure 6.
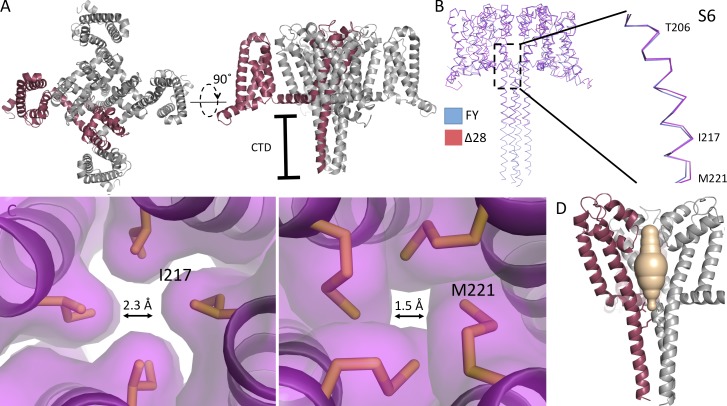
The structure of NaVAbΔ28. (A) The overall fold of NaVAbΔ28 is shown as cartoon helices, with one of the four monomers highlighted in red. Left: Top view. Right: Side view. (B) Ribbon overlay of NaVAbΔ28 (dark red) and NaVAb/T206F/V213Y (PDB accession no. 5VB8, blue). The inset shows a close-up view of the overlay in the region of the S6 helix and activation gate. (C) Side view of the pore domain of NaVAbΔ28, with one subunit highlighted as in A and one subunit removed for clarity. The solvent-accessible volume derived from the program MOLE is shown in wheat color to illustrate the closed activated gate at the level of Ile217 (left) and Met221 (right), which are shown in stick format. Arrows are shown to demonstrate the diameter of the activation gate at the level of the sidechain of I217 or M221, respectively. (D) The four S6 helices are shown at the level of the activation gate residues: Ile217 (left) and Met221 (right). The contours illustrate these amino acid residues in space-filling format. From the contours, we estimate that the orifice of the activation gate has a diameter of 2.3 Å at Ile217 and 1.5 Å at Met221. CTD, C-terminal domain; FY, NaVAb/FY; Δ28, NaVAb/Δ28.
Structure of NaVAbΔ28 with a closed pore
The structure of the S6 segments of NaVAbΔ28 precisely overlays the closed-state structure of NaVAb/FY through the level of the truncation at the intracellular surface of the membrane (Fig. 6 B), leading us to assign it as a putative closed state as demonstrated directly for NaVAb/FY (Lenaeus et al., 2017). The activation curve of NaVAbΔ28 (Fig. 2 B) is shifted ~21 mV compared with NaVAbΔ40, which was crystallized in a putative open state (Lenaeus et al., 2017), indicating that the closed state is preferred in NaVAbΔ28. Analysis with MOLE also indicates that the pore is closed at the activation gate (Fig. 6 C, tan) at the positions of Ile271 and Met221 (Fig. 6 C, sticks). The closed conformation of the activation gate of NaVAbΔ28 is further illustrated by the surface representation of the structure at the levels of Ile217 and Met221 at the intracellular ends of the four S6 segments, which interact to seal the activation gate nearly completely (Fig. 6 D). The diameters of the pore estimated from the contours of the amino acid residues shown in Fig. 6 D are 2.1 Å at I217 and 0.8 Å at M221, both far smaller than hydrated Na+. Because NaVAbΔ28 is fully functional, unlike NaVAb/FY (Lenaeus et al., 2017), it provides a valuable additional structural model of the closed state of the pore.
C-terminal tail
Previously published crystal structures of NaVAb have omitted the C-terminal domain because of poorly resolved electron density in this region (Payandeh et al., 2011, 2012), except for a study of a constitutively closed NaVAb mutant, NaVAb/FY, which resolved clear electron density for the C-terminal domain and allowed determination of its structure at high resolution (Lenaeus et al., 2017). In that structure, the C-terminal tail of NaVAb formed a long four-helix bundle, as has been observed in other bacterial NaV channels (McCusker et al., 2012; Bagnéris et al., 2013; Shaya et al., 2014). Similarly, we found that the C-terminal tail of NaVAbΔ28 also forms a four-helix bundle (Fig. 6, A and B). Its structure precisely overlaps the C-terminal of NaVAb/FY, even though it is missing the last 28 amino acid residues. The intersubunit hydrogen-bonding pattern that stabilizes the four-helix bundle is completely conserved, as illustrated for Glu228, Glu229, and His231 in Fig. S3 (B and C). The striking structural similarity between NaVAbΔ28 and NaVAb/FY, despite truncation of 28 residues in the C terminus, suggests a limited range of conformations for this domain. Moreover, the structure of NaVAbΔ28 shows that even large truncations of the C-terminal tail do not disrupt its four-helix bundle structure and therefore support the conclusion that more subtle molecular changes must be responsible for the functional effects of NaVAbΔ28, NaVAbΔ10, and NaVAbΔ7 in the early and late phases of voltage-dependent inactivation.
Thr206
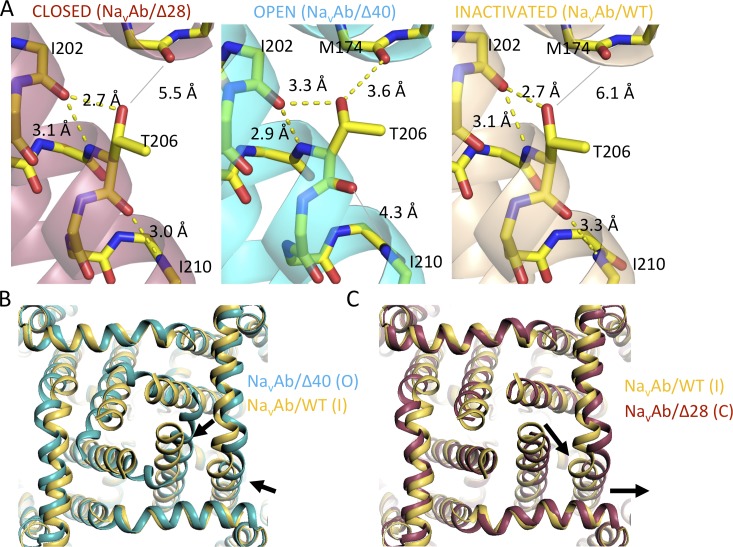
To search for structural changes that accompany the striking functional effects of mutations of Thr206, we compared the relevant regions of the structures of NaVAb/WT, NaVAb/T206A, and NaVAb/T206V determined in NaVAb constructs with different functional states of the pore: NaVAbΔ40 (open, equivalent to NaVAb/1–226; Lenaeus et al., 2017), NaVAb/WT (inactivated), and NaVAbΔ28 (closed). The structures surrounding Thr206 in these three states were quite similar (Fig. 7 A; RMSD 1.05 Å between NaVAbΔ40 and NaVAb/WT; RMSD 1.20 Å between NaVAb/WT and NaVAbΔ28). Although the local conformations surrounding Thr206 in these three states are quite similar, significant changes in the distance of the hydroxyl group of Thr206 relative to Met174 at the inner end of the ion selectivity filter are observed: 5.5 Å in the putative closed state structure, 3.6 Å in the putative open state structure, and 6.1 Å in the putative inactivated state structure (Fig. 7 A). In addition, a hydrogen bond is observed between Thr206 and Ile210 in the closed state, it is lost in the open state, and it is reformed but at greater distance and more oblique angle in the inactivated state (Fig. 7 A). The movement of Thr206 within 3.6 Å of Met174 in the open state may allow formation of a weak hydrogen bond, which is lost upon inactivation (Fig. 7 A). In contrast to the open state, in which the S6 helix is kinked and the side chain of Thr206 has moved closer to underside of the pore helix (Lenaeus et al., 2017; Fig. 7 A), the S6 helix is linear in the closed state, allowing the hydrogen bond to Ile210 to form again and the side chain of Thr206 to move significantly farther away from the pore helix (Figs. 7 A and S3 A). When examined at high resolution, the close fit of the electron density to the model in a simulated annealing omit map confirms the important conformational differences at this key site of bending in the S6 helix as the channel moves to the inactivated state (Fig. S3 A).
Figure 7.
Conformational changes accompanying gating of NaVAb. (A) Close-up views of the S6 helix near Thr206 during the closed, open, and inactivated states of NaVAbΔ28, NaVAb/1–226 (cyan, open; equivalent to NaVAbΔ40) and NaVAb/WT (yellow, inactivated). The main chain of each model is shown in stick format, as is the side chain of Thr206. All other side chains have been removed for clarity. Possible hydrogen bonds are shown as black dashes, and a broken hydrogen bond between the carbonyl of Thr206 and amide of Ile210 is shown in gray in the middle panel. (B) An overlay of NaVAb/1–226 (cyan, open; equivalent to NaVAb 40) and NaVAb/WT (yellow, inactivated) with the view as if one is standing below the membrane and looking upward along the permeation pathway. Voltage sensors have been removed for clarity, and arrows show the conformational changes associated with the transition from the open to the inactivated state. (C) An overlay (as in B) showing a comparison between NaVAb/WT (yellow, inactivated) and NaVAbΔ28 (magenta, closed). As in B, arrows highlight the conformational changes associated with the transition between states.
This series of transitions among functional states also induces clear structural changes in the intracellular activation gate. The distal portions of the S4–S5 linker helix and S6 helix move toward the pore axis during the transition between the putative open and inactivated states (Fig. 7 B, arrows). This movement creates the partial asymmetric collapse of the S6 segments in the inactivated state, with two S6 segments moving toward the axis of the pore and two moving away (Payandeh et al., 2012; Zhang et al., 2012). Similar movements of these segments away from the axis of the pore are observed in the transition from the inactivated state to the closed state (Fig. 7 C, arrows). A crucial result of this movement is to return the activation gate to a fourfold symmetric square conformation in readiness for eventual reopening in a concerted transition to the conducting state. Collectively, these data support earlier conclusions regarding asymmetric pore collapse in the transition from the open state to the inactivated state (Payandeh et al., 2012; Zhang et al., 2012) and further show that the pore conformation returns to fourfold symmetric configuration in the transition from the inactivated state to the closed state in preparation for the next pore opening. The fourfold symmetric conformation of the closed activation gate in our original structure of NaVAb/I217C in the preopen state (Payandeh et al., 2011) fits closely with this proposed series of conformational transitions that control movement of Na+ through the activation gate.
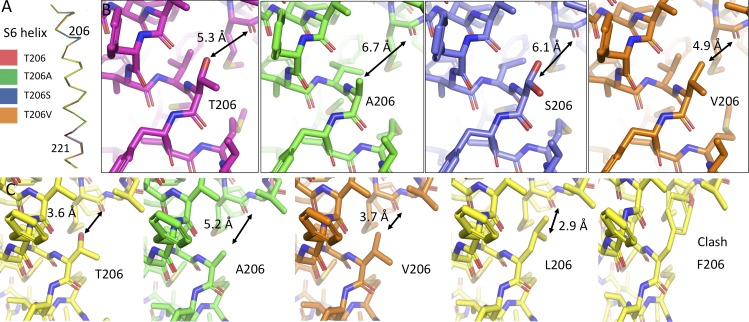
The structures of the S6 helices of NaVAb/T206A, NaVAb/T206S, and NaVAb/T206V were similar to WT when studied in the closed state in NaVAbΔ28 (Fig. 8, A and B). Close inspection of the structures at position 206 helps explain the relationship between side-chain volume and channel gating (Fig. 8 B). The relatively large side chains of Thr206 and Val206 leave a distance of ∼5 Å between the nearest S6 residue and the underside of the pore helix, when measured from the center of the closest nonhydrogen atom at position 206 and the center of the main chain carbonyl oxygen of Met174. The smaller side chains of Ala206 and Ser206 leave a larger distance between the same points (in the range of 6.1–6.7 Å), perhaps explaining the effect of side-chain volume on activation and inactivation gating. To further study this point, we made models of the mutants at position 206 in the open-state structure of NaVAb (Fig. 8 C). Each model was created by mutating the side chain of T206 into the most frequent rotamer of the substituted amino acid. Native Thr206 (Fig. 8 C, T206) shows a distance of 3.6 Å between the hydroxyl oxygen of Thr206 and the main-chain carbonyl of Met174, while mutations to Ala (A206) or Val (V206) have distances of 5.2 Å and 3.7 Å, respectively. In contrast, introducing Leu (L206) resulted in a distance of 2.9 Å between the side chain and the carbonyl of Met174 and introducing Phe (F206) gave a distance of 1.4 Å. These distances are both less than the sum of the van der Waals radii of the nearest atoms (3.2 Å for the terminal carbon of Leu and the carbonyl of Met174, and 3.4 Å for the carbon at the edge of the phenyl ring of Phe and the carbonyl of Met174). Thus, these larger side chains create a steric clash between the S6 helix in the open conformation and the underside of pore helix. These apparent structural clashes are consistent with our finding that these mutants can be expressed as protein but do not activate (Lenaeus et al., 2017; unpublished data for T206L). We speculate that these larger side chains exceed the available space and thereby prevent activation. It would be interesting to have structures of NaVAb constructs with these substitutions in the open state. Unfortunately, we could not solve the structures of any T206 mutations in the NaVAb/Δ40 construct that allows determination of the open-state structure.
Figure 8.
The structures of Thr206 mutants. (A) Side view of the isolated S6 helices of NaVAbΔ28 (dark red), NaVAbΔ28/T206A (green), NaVAbΔ28/T206S (blue), and NaVAbΔ28/T206V (orange). (B) Stick models are shown for each of the mutations in A, with focus on the 206 position of the S6 helix and the nearby portion of the pore helix. Coloring is as in A. Distances between the centers of the atoms of the side chain at position 206 and the main-chain carbonyl of Met174 are displayed in black in order to illustrate the differences among these mutations. Two rotamers are shown for T206S (blue), as determined in the crystal structure. (C) Hypothetical positions of Thr206 mutations in the open state (PDB accession no. 5VB2), with (from left to right) Thr206 (WT), T206A, T206V, T206L, and T206F. In each case, the model was created by substituting the Thr206 position with the amino acid shown, without energy minimization or any other rotamer selection. Rotamers selected are the most commonly encountered in the PDB and are meant to be illustrative of space constraints.
Discussion
A hinge region in the S6 segment initiates activation and early voltage-dependent inactivation
It has been suggested that slow inactivation of bacterial NaV channels occurs through a C-type inactivation mechanism involving conformational changes in the pore (Pavlov et al., 2005). Previous crystallographic studies revealed asymmetric pore collapse as the likely structural mechanism for inactivation (Payandeh et al., 2012; Zhang et al., 2012). However, it is unknown how voltage-dependent activation of the channel is linked to voltage-dependent inactivation and asymmetric collapse of the pore. Structure–function studies suggested that both activation and inactivation of bacterial NaV channels occur via twisting and bending movements of the S6 pore-lining helix at a Gly hinge, such as Gly219 in NaChBac (Zhao et al., 2004b). The same mechanism has been proposed for the bacterial K+ channels KcsA and MthK (Cuello et al., 1998; Perozo et al., 1999; Jiang et al., 2002a,b). In NaChBac, mutation of the neighboring Thr residue (Thr220) also greatly slows the early phase of slow inactivation (Zhao et al., 2004a,b; Lee et al., 2012). Here, we have found through mutagenesis and structural studies that Thr206 is part of a voltage-dependent coupling hub in NaVAb, as indicated by the strong reciprocal shifts in the kinetics and voltage dependence of activation versus inactivation in the mutants T206A/S/G compared with T206V. These results suggest that this segment of the S6 segment undergoes two conformational changes, one that initiates opening of the activation gate and a second that couples activation to the multiphase process of inactivation. Our structural studies provide a potential rationale for this mechanism. As illustrated in Fig. 7 (A and B), we find that the S6 helix kinks at the level of Thr206 during activation and then unkinks during inactivation. The kinking and unkinking of S6 is accompanied by a change in the position and potential binding partners of the side chain of Thr206, with the activated, kinked conformation having its Thr206 side chain closer to the underside of the pore helix and the unkinked conformation of S6 having its Thr206 side chain within hydrogen-bonding distance of the main chain of the S6 segment. Side-chain volume is the critical factor in determining how the hinge at Thr206 position interacts with the bottom of the selectivity filter. The early voltage-dependent phase of inactivation may be initiated by repulsion between Thr206 in the S6 helix and Met174 on the underside of the pore helix, which would provide a potential mechanism for larger side chains in this position to accelerate inactivation and smaller side chains to delay inactivation.
The C-terminal tail opposes inactivation during single depolarizations
Asymmetric collapse during inactivation alters the conformation along the full length of the pore from the extracellular vestibule and selectivity filter to the intracellular activation gate (Payandeh et al., 2012; Zhang et al., 2012). Therefore, conformational changes in the C-terminal tail extending beyond the activation gate might also affect the early phase of inactivation. We found that C-terminal truncations modulate early voltage-dependent inactivation during single depolarizations. Progressive deletions from the C terminus cause graded increases in the rate of the early voltage-dependent inactivation process, without major effects on the rate of activation. These increases in the rate of the early phase of inactivation of NaVAb with progressive truncation of the C-terminal tail reflect loss of voltage dependence, as if truncation of the C-terminal tail removes a voltage-dependent brake on the early phase of the inactivation process. A similar effect has been shown previously in the KcsA channel, where truncation of the C-terminal tail enhanced inactivation kinetics (Cuello et al., 2010). In contrast, previous studies found that deleting the C-terminal tail of NaVSulP and NaVMs slowed the early phase of inactivation during single depolarizations, and point mutations that disrupt the four-helix bundle also caused that effect (Irie et al., 2012; Bagnéris et al., 2013). Similarly, deleting the C-terminal domain or mutating specific residues within it is a prerequisite for function of the bacterial sodium channel NaVAe (Arrigoni et al., 2016). These results suggest that the C-terminal domain can modulate the voltage dependence and kinetics of the early phase of voltage-dependent inactivation of different NaV channels over a wide range, from locking the channel in an inactivated state to more subtle modulation of the rate of inactivation in different directions, depending on the molecular context.
The C-terminal tail is required for late use-dependent inactivation
Truncation of the C-terminal tail of NaVAb/WT removes the late use-dependent phase of inactivation, which seems to be unique to NaVAb among bacterial NaV channels studied to date. Cutting 10 residues from the distal end of the tail was sufficient to abolish use-dependent inactivation at a low frequency (0.2 Hz); however, at a higher frequency (1 Hz), ∼20–30% use-dependent inactivation occurs during the first few pulses, and then the peak current stays constant for up to 50 pulses. The distal 20 residues of the tail, comprising a coiled-coil region, are hydrophobic (9 hydrophobic residues out of 20) compared with proximal part that is mostly hydrophilic (4 hydrophobic residues out of 20). These hydrophobic interactions in the coiled coil likely help to stabilize the distal part of the tail in the four-helix bundle structure. However, our crystal structure of NaVAbΔ28 indicates that the distal C-terminal tail is not required to maintain the four-helix bundle. Evidently, the strong effects of the Δ7, Δ10, and Δ28 mutations on late use-dependent inactivation do not require unwinding of the four-helix bundle.
Arrigoni et al. (2016) showed that the proximal part of the C-terminal domain (the “neck” domain) is subject to temperature-dependent unfolding transitions during gating, while the coiled-coil part stays intact. They found that only the neck has a major effect on activation, with just a minor role for the coiled coil. Their experiments did not reveal a major effect of the C-terminal domain on the rate of inactivation, again illustrating that control of late use-dependent inactivation by the C-terminal domain depends on its molecular context and may serve different functions in different bacterial NaV channels.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health research grants R01 NS15751 (W.A. Catterall) and R01 HL122808 (W.A. Catterall and N. Zheng) and the Howard Hughes Medical Institute (N. Zheng).
The authors declare no competing financial interests.
Author contributions: All authors designed experiments, T.M. Gamal El-Din, M.J. Lenaeus, and K. Ramanadane carried out experiments and analyzed data. All authors contributed to writing the paper.
Kenton J. Swartz served as editor.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni C., Rohaim A., Shaya D., Findeisen F., Stein R.A., Nurva S.R., Mishra S., Mchaourab H.S., and Minor D.L. Jr. 2016. Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell. 164:922–936. 10.1016/j.cell.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnéris C., Decaen P.G., Hall B.A., Naylor C.E., Clapham D.E., Kay C.W., and Wallace B.A.. 2013. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat. Commun. 4:2465 10.1038/ncomms3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell A.R., and Catterall W.A.. 2001. Neuromodulation of Na+ channels: an unexpected form of cellular plasticity. Nat. Rev. Neurosci. 2:397–407. 10.1038/35077553 [DOI] [PubMed] [Google Scholar]
- Carr D.B., Day M., Cantrell A.R., Held J., Scheuer T., Catterall W.A., and Surmeier D.J.. 2003. Transmitter modulation of slow, activity-dependent alterations in sodium channel availability endows neurons with a novel form of cellular plasticity. Neuron. 39:793–806. 10.1016/S0896-6273(03)00531-2 [DOI] [PubMed] [Google Scholar]
- Catterall W.A. 2000. Structure and regulation of voltage-gated Ca2+ channels. Annu. Rev. Cell Dev. Biol. 16:521–555. 10.1146/annurev.cellbio.16.1.521 [DOI] [PubMed] [Google Scholar]
- Catterall W.A., and Zheng N.. 2015. Deciphering voltage-gated Na+ and Ca2+ channels by studying prokaryotic ancestors. Trends Biochem. Sci. 40:526–534. 10.1016/j.tibs.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yu F.H., Surmeier D.J., Scheuer T., and Catterall W.A.. 2006. Neuromodulation of Na+ channel slow inactivation via cAMP-dependent protein kinase and protein kinase C. Neuron. 49:409–420. 10.1016/j.neuron.2006.01.009 [DOI] [PubMed] [Google Scholar]
- Cuello L.G., Romero J.G., Cortes D.M., and Perozo E.. 1998. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 37:3229–3236. 10.1021/bi972997x [DOI] [PubMed] [Google Scholar]
- Cuello L.G., Jogini V., Cortes D.M., Pan A.C., Gagnon D.G., Dalmas O., Cordero-Morales J.F., Chakrapani S., Roux B., and Perozo E.. 2010. Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature. 466:272–275. 10.1038/nature09136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamal El-Din T.M., Martinez G.Q., Payandeh J., Scheuer T., and Catterall W.A.. 2013. A gating charge interaction required for late slow inactivation of the bacterial sodium channel NavAb. J. Gen. Physiol. 142:181–190. 10.1085/jgp.201311012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamal El-Din T.M., Scheuer T., and Catterall W.A.. 2014. Tracking S4 movement by gating pore currents in the bacterial sodium channel NaChBac. J. Gen. Physiol. 144:147–157. 10.1085/jgp.201411210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamal El-Din T.M., Lenaeus M.J., and Catterall W.A.. 2018. Structural and functional analysis of sodium channels viewed from an evolutionary perspective. Handb. Exp. Pharmacol. 246:53–72. 10.1007/164_2017_61 [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ionic Channels of Excitable Membranes. Third edition Sinauer Associates, Sunderland, MA. [Google Scholar]
- Irie K., Shimomura T., and Fujiyoshi Y.. 2012. The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate. Nat. Commun. 3:793 10.1038/ncomms1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Xu H., Guffanti A.A., Wei Y., Zvi L., Clapham D.E., and Krulwich T.A.. 2004. The voltage-gated Na+ channel NaVBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc. Natl. Acad. Sci. USA. 101:10566–10571. 10.1073/pnas.0402692101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., and MacKinnon R.. 2002a Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. 10.1038/417515a [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., and MacKinnon R.. 2002b The open pore conformation of potassium channels. Nature. 417:523–526. 10.1038/417523a [DOI] [PubMed] [Google Scholar]
- Lee S., Goodchild S.J., and Ahern C.A.. 2012. Local anesthetic inhibition of a bacterial sodium channel. J. Gen. Physiol. 139:507–516. 10.1085/jgp.201210779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaeus M.J., Gamal El-Din T.M., Ing C., Ramanadane K., Pomès R., Zheng N., and Catterall W.A.. 2017. Structures of closed and open states of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 114:E3051–E3060. 10.1073/pnas.1700761114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker E.C., Bagnéris C., Naylor C.E., Cole A.R., D’Avanzo N., Nichols C.G., and Wallace B.A.. 2012. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 3:1102 10.1038/ncomms2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., and Minor W.. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Pavlov E., Bladen C., Winkfein R., Diao C., Dhaliwal P., and French R.J.. 2005. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys. J. 89:232–242. 10.1529/biophysj.104.056994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Gamal El-Din T.M., Scheuer T., Zheng N., and Catterall W.A.. 2012. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 486:135–139. 10.1038/nature11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S.J. 1986. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur. J. Biochem. 157:169–180. 10.1111/j.1432-1033.1986.tb09653.x [DOI] [PubMed] [Google Scholar]
- Perozo E., Cortes D.M., and Cuello L.G.. 1999. Structural rearrangements underlying K+-channel activation gating. Science. 285:73–78. 10.1126/science.285.5424.73 [DOI] [PubMed] [Google Scholar]
- Ren D., Navarro B., Xu H., Yue L., Shi Q., and Clapham D.E.. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. 10.1126/science.1065635 [DOI] [PubMed] [Google Scholar]
- Scheuer T. 2014. Bacterial sodium channels: models for eukaryotic sodium and calcium channels. Handb. Exp. Pharmacol. 221:269–291. 10.1007/978-3-642-41588-3_13 [DOI] [PubMed] [Google Scholar]
- Shaya D., Findeisen F., Abderemane-Ali F., Arrigoni C., Wong S., Nurva S.R., Loussouarn G., and Minor D.L. Jr. 2014. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J. Mol. Biol. 426:467–483. 10.1016/j.jmb.2013.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sula A., Booker J., Ng L.C., Naylor C.E., DeCaen P.G., and Wallace B.A.. 2017. The complete structure of an activated open sodium channel. Nat. Commun. 8:14205 10.1038/ncomms14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Gamal El-Din T.M., Swanson T.M., Pryde D.C., Scheuer T., Zheng N., and Catterall W.A.. 2016. Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature. 537:117–121. 10.1038/nature19102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzhumtseva L., Afonine P.V., Adams P.D., and Urzhumtsev A.. 2009. Crystallographic model quality at a glance. Acta Crystallogr. D Biol. Crystallogr. 65:297–300. 10.1107/S0907444908044296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilin Y.Y., and Ruben P.C.. 2001. Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem. Biophys. 35:171–190. 10.1385/CBB:35:2:171 [DOI] [PubMed] [Google Scholar]
- Vilin Y.Y., Fujimoto E., and Ruben P.C.. 2001. A single residue differentiates between human cardiac and skeletal muscle Na+ channel slow inactivation. Biophys. J. 80:2221–2230. 10.1016/S0006-3495(01)76195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., et al. 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134. 10.1038/nature11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Scheuer T., and Catterall W.A.. 2004a Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc. Natl. Acad. Sci. USA. 101:17873–17878. 10.1073/pnas.0408270101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yarov-Yarovoy V., Scheuer T., and Catterall W.A.. 2004b A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron. 41:859–865. 10.1016/S0896-6273(04)00116-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.