Head and neck cancer patients taking NSAIDs with PIK3CA tumor alterations demonstrate improved survival. Studies in relevant preclinical models implicate signaling via COX2-mediated production of PGE2 as an underlying mechanism for this survival benefit.
Abstract
PIK3CA is the most commonly altered oncogene in head and neck squamous cell carcinoma (HNSCC). We evaluated the impact of nonsteroidal anti-inflammatory drugs (NSAIDs) on survival in a PIK3CA-characterized cohort of 266 HNSCC patients and explored the mechanism in relevant preclinical models including patient-derived xenografts. Among subjects with PIK3CA mutations or amplification, regular NSAID use (≥6 mo) conferred markedly prolonged disease-specific survival (DSS; hazard ratio 0.23, P = 0.0032, 95% CI 0.09–0.62) and overall survival (OS; hazard ratio 0.31, P = 0.0043, 95% CI 0.14–0.69) compared with nonregular NSAID users. For PIK3CA-altered HNSCC, predicted 5-yr DSS was 72% for NSAID users and 25% for nonusers; predicted 5-yr OS was 78% for regular NSAID users and 45% for nonregular users. PIK3CA mutation predicted sensitivity to NSAIDs in preclinical models in association with increased systemic PGE2 production. These findings uncover a biologically plausible rationale to implement NSAID therapy in PIK3CA-altered HNSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a common epithelial malignancy worldwide with high morbidity and mortality. Primary risk factors for HNSCC include tobacco, alcohol, and human papillomavirus (HPV) infection. Regular use of aspirin was found to be protective against HNSCC development in a large cohort study of patients participating in the National Cancer Institute’s Prostate, Lung, Colorectal, and Ovarian (PLCO) randomized screening trial, as well as in other studies (Gohagan et al., 2000; Jayaprakash et al., 2006; Ahmadi et al., 2010; Wilson et al., 2013). Additional investigations, however, have reported inconsistent findings regarding the potential survival benefit of nonsteroidal anti-inflammatory drugs (NSAIDs) in HNSCC (Rafei et al., 2017; Kim et al., 2018). Moreover, while the PLCO study provided important epidemiological data for a role for NSAIDs in HNSCC consistent with findings from colorectal cancer (CRC) and breast cancer (Baron et al., 2003; Holmes et al., 2010), candidate biomarkers of response and molecular mechanisms were not identified.
Activating mutations in PIK3CA, the gene encoding p110α, the catalytic subunit of phosphoinositide-(3)-kinase α (PI3Kα), are common in HNSCC, with 174 of 504 (35%) tumors harboring PIK3CA mutation or amplification (Cancer Genome Atlas Network, 2015). Signaling through the PI3K pathway induces phosphorylation/activation of AKT (protein kinase B) and production of prostaglandin E2 (PGE2) via cyclooxygenase-2 (COX2) enzyme, a target of NSAIDs.
A correlation between regular NSAID use and improved survival in patients harboring canonical PIK3CA mutations was first described in CRC, an adenocarcinoma, although the mechanism responsible for the therapeutic benefit of NSAIDs was not determined (Liao et al., 2012; Domingo et al., 2013; Elwood et al., 2016). Commonly observed alterations in the PIK3CA gene result from either PIK3CA mutation or gene amplification, which occur at nearly equal frequency in HNSCC. Herein, we tested the hypothesis that regular NSAID use is associated with improved disease-specific survival (DSS) and overall survival (OS) in patients with PIK3CA-mutated or -amplified HNSCC. We also investigated PGE2 secretion as a potential mediator of response to NSAID therapy in PIK3CA-altered preclinical models, hypothesizing a unifying mechanism across cancers.
Results
Clinical and pathological information for our 266-patient cohort was obtained under the auspices of a prospective HNSCC biobanking protocol approved by the Institutional Review Board at the University of Pittsburgh. A detailed description of the construction of our cohort is provided in the Materials and methods section. To investigate the interaction between NSAID use, alteration of PIK3CA, and HNSCC survival, we determined the mutational and amplification status of PIK3CA in these 266 HNSCC tumors and extracted NSAID use from the electronic medical record (EMR). Patients were classified as regular NSAID users if the EMR supported ≥6 mo of “regular use,” defined as ≥2 d/wk (Chan et al., 2005); never users if the EMR documented no prescription or self-reported use; and intermittent users if neither definition was met. Table S1 details the baseline clinical and pathological characteristics of this cohort according to PIK3CA alteration status and NSAID use, and Table S2 details the adjuvant chemoradiation therapies delivered. The majority (67%) of our cohort received adjuvant chemotherapy, radiotherapy, or both (CRT). CRT was equally distributed among patients with PIK3CA mutation/amplification (66% received CRT) and among chronic NSAID users (69% received CRT). DSS and OS did not differ by the administration of CRT (log rank P = 0.3 and 0.5, respectively) suggesting that the administration of adjuvant chemoradiotherapy neither affected outcome directly nor confounded the conclusion regarding the impact of PIK3CA mutation/amplification and chronic NSAID use. 84% of the patient cohort reported a positive smoking history. Smoking history was tested and found to have no effect on DSS (P = 0.301) or OS (P = 0.289). 75 tumors (28%) harbored mutation and/or amplification of PIK3CA. 37 of 266 (14%) contained only mutation, 29 of 266 (11%) had only amplification, and 9 of 266 (3%) harbored both mutation and amplification. In tumors that harbored a PIK3CA mutation, 33 of 46 (72%) had single canonical mutations (at residues E542, E545, and H1047), 12 of 46 (26%) had single noncanonical mutations, and 1 of 46 (2%) harbored both a canonical and a noncanonical mutation (H1047R and D743N). Mutations at E545 in the helical domain were the most common (14 E545K mutations and 1 E545G mutation), followed by mutations at H1047 in the kinase domain (10 H1047R mutations and 3 H1047L mutations) and E542 in the helical domain (six E542K mutations). Other mutations included two R115L mutations and one each of R38C, N345K, G363A, E476Q, D743N, Y890C, C971R, R975S, D1017H, T1025N, and H1048R. Regular NSAID use was identified in 33% (25 of 75) of patients with PIK3CA-altered tumors and 39% (74 of 191) of those with WT, unamplified PIK3CA, and was independent of PIK3CA status (chi-square, P = 0.48). We observed no association between either regular NSAID use or PIK3CA status and any clinical or pathological variable except age (regular NSAID users were on average 4 yr older) and disease status (regular NSAID users with mutant (MT) or amplified PIK3CA were more likely to present with recurrent disease; P = 0.27). The median DSS was not reached but was at least 6 yr. Median OS was 66 mo, with a median follow-up of 40 mo (range 3–106 mo) among survivors. Aspirin was a component of the NSAID regimen in 93% of regular users, and 73% took aspirin exclusively; 75% of aspirin-exclusive users took daily, low-dose (81 mg) drug (details regarding specific NSAID use in the cohort are provided in Table S3). Most regular users (86%) initiated NSAID therapy after receiving their HNSCC diagnosis. There was no indication that the diagnosis of HNSCC informed the decision to take NSAIDs.
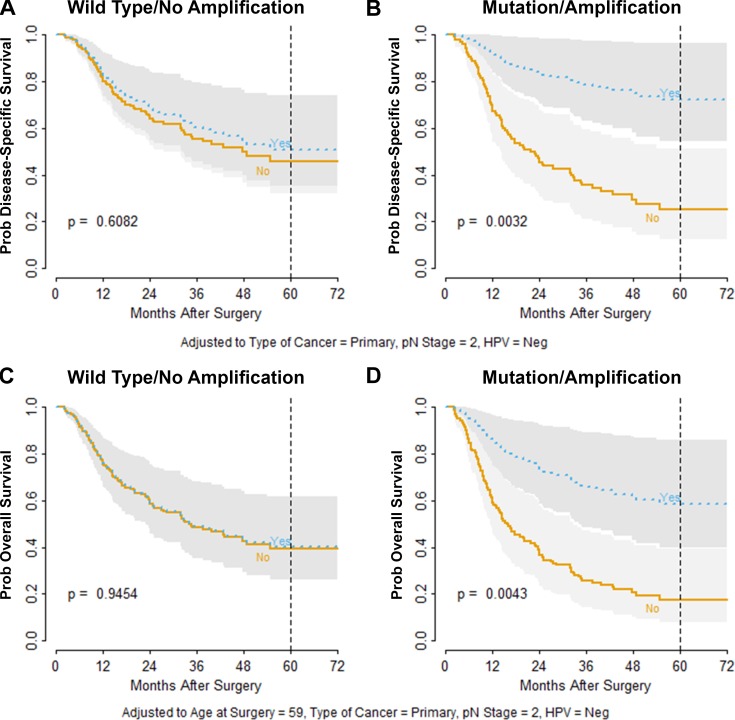
The final multivariate proportional hazard model for DSS was adjusted for cancer type (primary versus recurrence), pathological N (nodal) stage, and HPV status. In addition, we evaluated the interaction between PIK3CA status and regular NSAID use. DSS among patients with WT, unamplified PIK3CA was unaffected by regular NSAID use (Fig. 1 A; P = 0.61). By contrast, regular NSAID use was strongly associated with improved DSS for patients with MT and/or amplified PIK3CA (Fig. 1 B; P = 0.0032). Among patients with altered PIK3CA, regular NSAID users had an absolute 5-yr DSS advantage of 47% (72% in regular users versus 25% for nonregular users; Fig. 1 B; hazard ratio [HR] 0.24; 95% confidence interval [CI] 0.09–0.62; P = 0.0032). However, in the context of unaltered PIK3CA, the disease-specific HR for regular versus nonregular NSAID use was not significant (0.86; 95% CI 0.48–1.54; Table S4). It should also be noted that PIK3CA alteration in nonregular NSAID users was associated with worse DSS (P = 0.047), consistent with the known oncogenic properties of PIK3CA.
Figure 1.
Treatment benefit of regular NSAID exposure for DSS and OS. (A) Model-predicted DSS probability indicates no survival difference between regular users (Yes, blue) versus never or occasional users (No, orange) for HNSCC patients with unaltered PIK3CA. (B) Patients harboring mutated or amplified PIK3CA exhibited a survival advantage with regular NSAID use (Yes, blue). (C) Model-predicted OS probability indicates no survival difference between regular users (Yes, blue) versus never or occasional users (No, orange) for HNSCC patients with unaltered PIK3CA. (D) Patients harboring mutated or amplified PIK3CA exhibited a survival advantage with regular NSAID use (Yes, blue). Dotted vertical reference lines indicate 5 yr of follow-up. P values are based on Z statistics for testing contrasts derived from the design matrix of the selected regression model and are adjusted for HPV status, type of cancer (primary versus recurrence), pathological N stage, and age at surgery (for OS only). Gray bands indicate 95% confidence intervals.
For OS, the final multivariate proportional hazard model was adjusted for PIK3CA status, primary versus recurrence, pathological N stage, HPV status, and age at surgery. In addition, interaction between regular NSAID use and PIK3CA mutation/amplification status was evaluated. As with DSS, regular NSAID users with unaltered PIK3CA had equivalent OS to that of nonregular users (Fig. 1 C; P = 0.95). However, among patients with altered PIK3CA, regular NSAID users had an absolute 5-yr OS advantage of 33% (78% in regular users versus 45% for nonregular users; Fig. 1 D; P = 0.0043). The overall HR for regular NSAID users with PIK3CA mutation and/or amplification was 0.31 (95% CI 0.14–0.69), compared with 0.98 (95% CI 0.60–1.62) for regular users with unaltered PIK3CA (Table S5). Interaction between mutation/amplification status and regular NSAID use was considered significant for the models (Fig. 1) for both DSS (P = 0.025) and OS (P = 0.017). PIK3CA alteration was also associated with worse OS among subjects who were nonregular NSAID users (P = 0.014). The present study is the first to demonstrate a significant advantage in both DSS and OS among HNSCC patients taking regular NSAIDs that is dependent on the presence of a somatic PIK3CA alteration.
HPV status is recognized as a significant prognostic, but not predictive, biomarker in HNSCC, as patients with HPV-positive disease have improved outcomes with any standard-of-care therapy (Buckley et al., 2016). When the model for DSS (Fig. 1) was segregated by HPV status, PIK3CA status, and NSAID use, we observed that for nonregular NSAID users, PIK3CA alteration was associated with worse survival probability irrespective of HPV status (Fig. S1). The diminished DSS probability predicted by PIK3CA alteration was qualitatively reversed for patients taking regular NSAIDs, where PIK3CA alteration predicted significantly improved survival probability in both HPV-positive and HPV-negative disease (Fig. S1). Among HPV-positive patients taking regular NSAIDs (n = 23), no disease-specific deaths were observed with median follow-up of 36 mo (range, 5–97). Kaplan–Meier plots of unadjusted survival estimates also demonstrated the essential interaction between PIK3CA status and NSAID use (data not shown). These findings identify PIK3CA alteration as a molecular predictive and prognostic biomarker relevant to both HPV-positive and HPV-negative HNSCC.
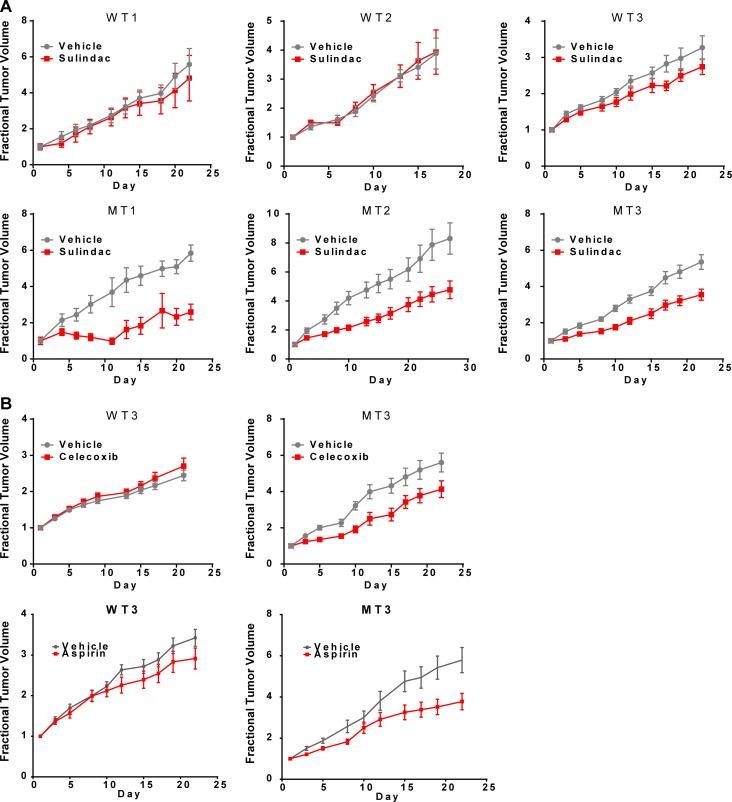
To test our findings using in vivo HNSCC models, mice harboring six distinct patient-derived xenografts (PDXs) endogenously expressing either MT or WT PIK3CA were treated with saline or the NSAIDs sulindac, celecoxib, or aspirin followed by assessment of tumor growth (see Table S6 for PIK3CA mutation details). A mixed linear model was fit to the log-transformed tumor volume data. Sulindac, celecoxib, or aspirin treatment reduced the growth rates of MT PIK3CA tumors compared with vehicle-treated tumors (P < 0.0001; Fig. 2, A and B). In contrast, the growth rates of PIK3CA WT tumors treated with NSAIDs did not differ compared with vehicle-treated controls (Fig. 2, A and B). These data support the potential benefit of regular NSAID use in HNSCC patients with PIK3CA MT tumors.
Figure 2.
HNSCC PDXs harboring PIK3CA mutation are sensitive to NSAID treatment. PDXs were implanted into the flanks of NSG mice and allowed to grow until palpable (n = 4–10 tumors per group). (A) Mice were then treated with vehicle (saline) or sulindac (30 mg/kg by i.p. injection every other day). Three distinct PIK3CA-MT PDX models exhibit significant growth inhibition upon sulindac treatment relative to vehicle control. By contrast, three PIK3CA-WT PDX models do not exhibit sulindac sensitivity. In an analysis combining all PDXs, the interaction of day and treatment group was significant (linear P = 0.016; quadratic P < 0.0001), consistent with differential growth rates. (B) One PIK3CA-MT PDX and one PIK3CA-WT PDX (MT3 and WT3, respectively) were inoculated into mice (n = 10 tumors per group) as above and treated five times per week by oral gavage with vehicle (0.5% methylcellulose, 0.025% Tween 20), celecoxib (40 mg/kg), or aspirin (100 mg/kg). Only the PIK3CA-MT PDX exhibits growth inhibition upon celecoxib or aspirin treatment relative to vehicle (interaction test: linear P < 0.0001; quadratic P < 0.0001). Data are presented as mean ± SEM.
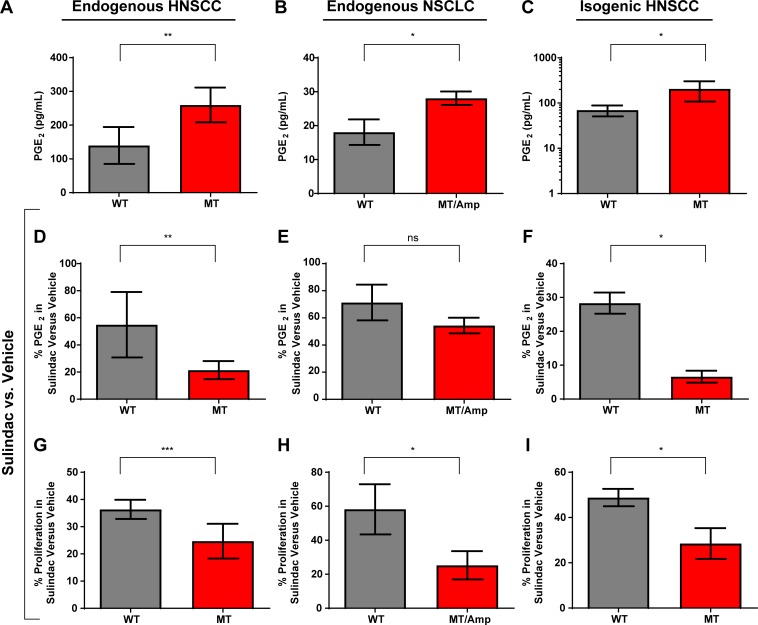
Because nearly all patients in our analysis initiated NSAID treatment following curative therapy (85 of 99), and the specimens interrogated for this study consisted solely of baseline tumor-derived DNA from formalin-fixed, paraffin-embedded material, it is not possible to assess the proposed mechanism in human samples from this cohort, or any human cohort that we know to exist. Furthermore, we are unable to assess circulating levels of PGE2 in these patients, since baseline and/or post-NSAID samples are unavailable. Therefore, we sought to identify a plausible biological mechanism underlying our finding of improved survival in regular NSAID users with PIK3CA-altered tumors by investigating HNSCC preclinical models (Scheper et al., 2007; Katoumas et al., 2015). We first identified a panel of HNSCC cell lines that endogenously express either WT or MT PIK3CA. PIK3CA MT HNSCC cell lines secreted significantly higher basal levels of PGE2 than did WT cells, indicating activation of COX2 signaling (Fig. 3 A). Non–small cell lung cancer (NSCLC) cells with MT/Amp PIK3CA also secreted more PGE2 than did WT NSCLC cells, indicating that PIK3CA may activate COX signaling across cancer types and histologies (Fig. 3 B). We next engineered a PIK3CA WT HNSCC cell line (PCI-52) to stably overexpress WT PIK3CA or a representative PIK3CA mutant (H1047R). Overexpression of MT PIK3CA resulted in phosphorylation of AKT and the downstream target PRAS40 (Fig. S2). Similar findings were observed in a validation cohort of 165 HNSCC tumors, in which mutation and/or amplification of PIK3CA was significantly associated with elevated phosphorylation of AKT at T308 (P = 0.036) and S473 (P = 0.01) relative to WT/unamplified tumors (Fig. S3). Notably, cells with endogenous PIK3CA mutations and cells engineered to overexpress MT PIK3CA expressed markedly elevated levels of COX2 enzyme (Fig. S2) and secreted significantly higher levels of PGE2 (Fig. 3, A–C). Induction of COX2 by MT PIK3CA was due, at least in part, to transcriptional activation (data not shown). The enhanced PGE2 secretion observed in HNSCC cell lines with endogenous PIK3CA mutations was markedly attenuated by treatment with the NSAID sulindac (Fig. 3 D; 79% reduction in MT versus 45% in WT cells, on average; P = 0.0046), as was also the case in NSCLC lines with endogenous PIK3CA mutation or amplification (Fig. 3 E; 46% reduction in MT/Amp versus 29% in WT, on average). These results were confirmed in our engineered, isogenic HNSCC model, where sulindac treatment led to significantly greater reduction in PGE2 secretion in cells overexpressing MT versus WT PIK3CA (Fig. 3 F; average 93% reduction in PCI-52 exogenously expressing MT PIK3CA versus 72% in vector-transfected PCI-52 cells; P = 0.041). Similar findings were observed following treatment of cells with indomethacin (Fig. S4). Absolute PGE2 levels are shown in Fig. S5.
Figure 3.
Cancer cell lines with PIK3CA alteration are more sensitive to NSAID treatment. (A–C) Basal PGE2 secretion (measured via ELISA) is higher in (A) HNSCC (P = 0.0052) cells harboring endogenous PIK3CA mutations (Cal33, Detroit562, and HSC-2) or (B) NSCLC (P = 0.017) cells harboring endogenous PIK3CA mutation or amplification (H460, PIK3CA MT; H3255, PIK3CA amplification) compared with cell lines containing WT, unamplified PIK3CA (Cal27, FaDu, and PE/CA-PJ34.12 for HNSCC; H358 and H1437 for NSCLC), and in (C) isogenic HNSCC cells (PCI-52) engineered to overexpress MT PIK3CA (P = 0.0374). For HNSCC (A, D, and G), three distinct cell lines were used for each genotype, MT or WT; for NSCLC (B, E, and H), two MT/Amp cell lines and two WT cell lines were used. (D–F) Sulindac treatment (48 h) leads to greater reduction of PGE2 secretion in (D) HNSCC (P = 0.0046) but not (E) NSCLC cells harboring endogenous PIK3CA mutations/amplification (P = 0.24), and in (F) isogenic HNSCC cells overexpressing MT PIK3CA (P = 0.041). (G–I) Sulindac treatment (48 h) more potently inhibits cell proliferation in (G) HNSCC (P = 0.0009) or (H) NSCLC (P = 0.036) cells harboring endogenous PIK3CA mutations/amplification, and in (I) isogenic HNSCC cells overexpressing MT PIK3CA (P = 0.041). All panels are depicted as mean + SD (error bars). All experiments were performed in triplicate and repeated once. Data were analyzed by two-way analysis of variance with interaction in which cell type (mutated versus WT) was crossed with treatment (vehicle, sulindac). Specific hypotheses were evaluated by t test with pooled estimates of SE. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.001. ns, not significant.
We next tested cell proliferation as a phenotypic indicator of NSAID efficacy in our panels of HNSCC (Fig. 3 G) and NSCLC (Fig. 3 H) cells with endogenous WT or MT PIK3CA, and in our isogenic HNSCC cells (Fig. 3 I). Sulindac treatment led to significantly reduced cell proliferation in PIK3CA MT cells relative to WT cells in each model tested. We again detected significant interactions between sulindac treatment and PIK3CA status (P = 0.0009 for endogenous HNSCC models, P = 0.036 for endogenous NSCLC models, and P = 0.041 for isogenic HNSCC models), indicating differential sensitivity to sulindac in PIK3CA WT versus MT models with respect to inhibition of cell proliferation. Importantly, exogenous expression of MT PIK3CA in a second isogenic HNSCC cell line model (Cal27) confirmed significantly enhanced growth inhibition following sulindac treatment in cells expressing MT versus WT PIK3CA (P = 0.0029; data not shown). Similar levels of growth inhibition and reduction of PGE2 were seen in HNSCC cells with MT PIK3CA following treatment with either sulindac or celecoxib (not shown). Similarly, sulindac treatment enhanced induction of apoptosis in cells expressing MT versus WT PIK3CA (data not shown). Sulindac also significantly inhibited colony formation in soft agar in PIK3CA MT HNSCC cells compared with WT HNSCC models (data not shown), providing a plausible mechanism for the improved DSS in the human HNSCC cohort.
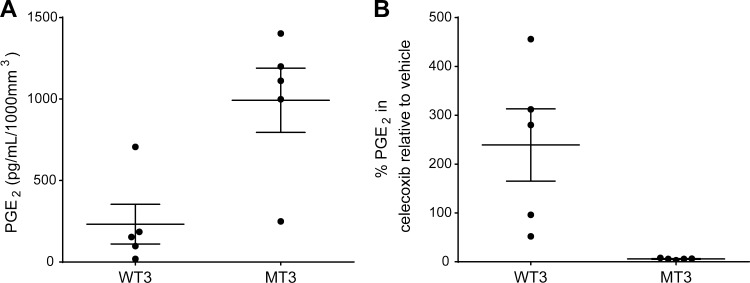
We further observed that mice harboring PDX tumors with MT PIK3CA exhibited elevated levels of circulating PGE2 compared with mice with WT PDX tumors (Fig. 4 A). Moreover, treatment of the PDX-bearing mice with celecoxib, a COX2-selective NSAID, resulted in significantly lower circulating PGE2 in mice with MT PIK3CA tumors compared with those with WT tumors (Figs. 4 B). Similar results were observed with sulindac or aspirin treatment (Fig. S5). Taken together, our findings indicate that PIK3CA mutation or amplification activates PI3K pathway signaling in HNSCC tumors and preclinical models, likely resulting in elevated PGE2 secretion via activation of COX2 enzyme, leading to enhanced NSAID sensitivity.
Figure 4.
PGE2 levels are elevated in PIK3CA MT HNSCC PDX models and are significantly reduced by celecoxib treatment. Mouse serum was extracted immediately after sacrifice following treatment with vehicle or celecoxib as described in Fig. 2. Serum PGE2 levels were measured in triplicate for each mouse using a PGE2 ELISA kit per the manufacturer’s instructions and normalized to the tumor volume per mouse. (A) PGE2 levels in vehicle control treated mice. (B) Percent PGE2 levels in celecoxib-treated mice relative to vehicle control. Significantly lower serum PGE2 levels were observed in mice harboring PIK3CA-MT PDXs treated with celecoxib relative to PGE2 levels in vehicle-treated control mice harboring PIK3CA-WT PDXs. Each data point represents the mean of triplicate measurements for each mouse, while whiskers represent the overall mean ± SEM.
Discussion
Epidemiological studies to date in CRC have highlighted an interaction between canonical PIK3CA mutation, aspirin use, and patient survival, where the underlying biological mechanism remains incompletely understood. Our experimental evidence is in line with the hypothesis that genomic alteration of PIK3CA up-regulates PGE2 secretion, which in turn stimulates cellular proliferation. This autocrine regulatory pathway results in differential susceptibility to COX targeting by NSAIDs. While our results do not rule out molecular mechanisms in addition to PGE2 secretion, they are consistent with an interaction between the PI3K and COX pathways in HNSCC, which may contribute to an oncogene addiction phenotype. In addition, the COX–PGE2 pathway has been implicated as a major immune-inhibitory pathway in HNSCC (Mandapathil et al., 2010). The precise role of COX2 in mediating the impact of NSAIDs on PIK3CA MT cancer across different tumor lineages remains incompletely understood. While an enhanced benefit of adjuvant aspirin was reported among patients with COX2-overexpressing CRC (Chan et al., 2009), a recent study found that the impact of aspirin in PIK3CA MT breast cancer preclinical models was COX2 independent (Henry et al., 2017). Unfortunately, current methodologies are unable to assess PGE2 expression or secretion in formalin-fixed, paraffin-embedded specimens, limiting the analysis of our human specimens. Furthermore, NSAID use in this cohort generally followed curative intent treatment and therefore we are not able to determine if NSAID use is associated with lower COX2 expression in the tumor.
It is noteworthy that the prior report in CRC only analyzed the interaction of the three canonical PIK3CA mutations (exons 9 and 20) with NSAID use and survival. In HNSCC, 64% of PIK3CA mutations are canonical, while 36% are noncanonical (Domingo et al., 2013). Our study sequenced all PIK3CA exons and assessed the impact of both canonical and noncanonical PIK3CA mutations in addition to determining the contribution of gene amplification, thus implicating activation of PI3K signaling as the primary mechanism for NSAID benefit. In addition, CRC is an adenocarcinoma that is distinct from HNSCC with respect to risk factors, prevalence, and known carcinogenic mechanisms. Therefore, our finding that NSAID use in the context of PIK3CA mutation or amplification enhances DSS in both HPV-positive and HPV-negative HNSCC (which are also clearly distinct with respect to risk factors), as well as our results in PIK3CA MT NSCLC cells, suggests that adjuvant NSAID administration may significantly improve outcome in many PIK3CA-altered cancers. The implication of our findings may provide a dramatic impact on human health.
The present study was conducted using a cohort of convenience and, as such, has some inherent limitations. Regular use of NSAIDs was defined through interrogation of the EMR by a combination of prescription refill records and patient self-reporting, which by themselves do not necessarily ensure an accurate estimation of actual use. Regular NSAID users by definition had to survive at least 6 mo, which introduces a potential, although limited, bias. Limitations in cohort size, particularly among subgroups, coupled with inconsistencies in the type, timing, and dosages of NSAID therapy taken by patients in this study limits our ability to make definitive statements regarding specific therapeutic recommendations. To minimize the possibility of index event bias, we carefully adjusted for confounding variables such as disease stage and HPV status (Tables S3 and S4).
We conclude that regular NSAID use likely confers a statistically and clinically significant advantage in DSS as well as OS in patients with PIK3CA-altered HNSCC, irrespective of HPV status, through direct interaction between the PI3K and COX pathways. The magnitude of this apparent advantage, especially given the marked morbidity and mortality of this disease, warrants further study in a prospective randomized clinical trial.
Materials and methods
Cohort construction
Clinical and pathological information was obtained under the auspices of a prospective HNSCC biobanking protocol approved by the Institutional Review Board of the University of Pittsburgh and curated by the HNSCC Specialized Program of Research Excellence (P50CA097190). Of 1,725 patients with surgically resected HNSCC at the University of Pittsburgh Medical Center between May 2008 and March 2013, 807 provided written informed consent for inclusion of their tumors on prospective tissue microarrays. At the time of this study, 216 of these cases were available for targeted PIK3CA sequencing. An additional 141 cases previously characterized by whole-exome sequencing were identified (Stransky et al., 2011; Lui et al., 2013; Cancer Genome Atlas Network, 2015). These 357 cases were filtered according to the following inclusion criteria: determinate PIK3CA mutation and amplification testing; >2 mo of follow-up; surgery within 6 mo of diagnosis; and known cause of death. 91 cases were excluded for nonsquamous histology (10), indeterminate PIK3CA status (51), <2 mo of follow-up (4), delay between diagnosis and surgery (23), or unknown cause of death (3). In total, 266 cases were included for analysis. All patients were treated with curative-intent surgery and standard adjuvant therapy, including radiation and chemotherapy as indicated by clinicopathologic features.
Statistics and model generation
DSS and OS were defined as elapsed time from the date of resection until death due to cancer or death from any cause, respectively. Patients alive at last date of follow-up were censored. Multivariate, proportional hazards regression models for DSS and OS were constructed. Covariates considered for association within each model included gender, age at surgery, year of surgery, tumor site, tumor stage, nodal stage, HPV status, cancer type (primary or recurrent), PIK3CA status (dichotomized as altered by mutation/amplification versus WT), and NSAID use (dichotomized as regular versus nonregular use). Two-way interactions among these variables were also evaluated. DSS and OS models were evaluated with the Akaike and Bayesian information criteria to balance fit and complexity. Final models for DSS and OS were found to meet proportional hazards assumptions using Schoenfeld residuals (Grambsch and Therneau, 1994). Survivor functions were estimated from final proportional hazards models by the discrete hazard method of Kalbfleisch and Prentice (2011). Tests of differences between groups were based on forming contrasts derived from the design matrix of the selected regression model. Computations were made using the rms package (Harrell, 2015) for R (R Team, 2014) statistical software. In vitro data were analyzed by two-way analysis of variance with interaction in which cell type (mutated versus WT) was crossed with treatment (vehicle, sulindac). Expression of PGE2 was natural log–transformed before analysis; survival percentage in proliferation experiments was untransformed. Specific hypotheses were evaluated by a t test with pooled estimates of standard error. Tumor volumes in PDX experiments were analyzed by longitudinal mixed effects linear models applied to log-transformed volume measurements.
Animals
Animal procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco. 5–6-wk old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from the University of California, San Francisco Breeding Core Facility. PDXs were implanted into the flanks of NSG mice under anesthesia and allowed to grow until palpable (∼100 mm3). Mice were treated with vehicle control (saline) or sulindac sulfide i.p. (30 mg/kg; Sigma-Aldrich) every other day. For experiments involving treatment with celecoxib, mice were treated with vehicle control (0.5% methylcellulose + 0.025% Tween 20) or celecoxib by oral gavage (40 mg/kg; Sigma-Aldrich) five times per week. For experiments involving treatment with aspirin, mice were treated with vehicle control (0.5% methylcellulose + 0.025% Tween 20) or aspirin by oral gavage (100 mg/kg; Sigma-Aldrich) five times per week (O’Connor et al., 2004; Scheper et al., 2007; Yip-Schneider et al., 2007; Tury et al., 2016). Tumors were measured every other day by Vernier caliper in two dimensions, and volume was calculated as (larger measurement) × (smaller measurement)2/2. Mixed linear models were applied to log-transformed tumor volumes. Specifically, we fit random coefficient models to verify the interaction between the four groups (formed by crossing mutation status and NSAID exposure) and day of tumor measurement.
Clinicopathologic data and biospecimen analysis
Clinicopathologic variables including age, sex, stage, histopathologic features, and treatment were entered into the clinically annotated database of the University of Pittsburgh Head and Neck Cancer Specialized Program of Research Excellence (SPORE) at the time of informed consent to the biobanking protocol. DSS and OS outcomes are updated monthly and cross-referenced to the Social Security Death Index and the Cancer Registry of the State of Pennsylvania at least annually. The database is linked to the University of Pittsburgh Medical Center EMR where prescription medications and self-reported over-the-counter medications are recorded by date. Use of aspirin and other NSAIDs was extracted from the EMR for each case, including dates of exposure and dose when available. Patients were classified as regular NSAID users if the EMR supported ≥6 mo of regular use, where “regular use” was defined as ≥2 d/wk (Chan et al., 2005); never users if the EMR documented no prescription or self-reported use; and intermittent users if neither definition was met. PIK3CA mutation and amplification status for the whole-exome sequencing cases were generated as previously described (Stransky et al., 2011; Lui et al., 2013; Cancer Genome Atlas Network, 2015). For new cases, formalin-fixed, paraffin-embedded tumor cores, microdissected under guidance from hematoxylin/eosin-stained sections, were obtained for genomic DNA extraction, and next generation sequencing was performed for all PIK3CA exons. DNA extraction, sequencing, coverage analysis, and variant detection were performed by Genewiz. Using a custom primer set, the Ion AmpliSeq Library Preparation Kit 2.0, Ion 316 chips, the Ion Personal Genome Machine, and the Torrent Suite program 4.0, VCF (variant call format) files for each sample were generated and aligned to the reference sequence hg19. Variants were considered valid and reported if they achieved a Phred-based quality score >100 (base call accuracy >99.99%). Fluorescence in situ hybridization was performed on tissue microarrays to assess PIK3CA copy number, where amplification was defined as PIK3CA:CEP3 > 2. HPV status was assessed by p16 immunohistochemistry and HPV in situ hybridization (ISH) in 240 cases, using standard techniques. p16 positivity was defined as strong, diffuse cytoplasmic, and nuclear p16 expression in >70% of tumor cells as previously described (Chiosea et al., 2013). Oropharynx cases positive for p16 and HPV ISH, or p16 only, were classified as HPV positive. Non-oropharynx cases positive for both p16 and HPV ISH were also classified as HPV positive. The clinicopathologic database was constructed independently from the database of PIK3CA alterations.
Cell lines and PDXs
A panel of HNSCC cells with or without endogenous PIK3CA mutation was identified using the Cancer Cell Line Encyclopedia. All of the HNSCC cell lines that were used which harbor endogenous PIK3CA mutations harbor the canonical H1047R mutation. Cal27 (RRID: CVCL_1107), FaDu (RRID: CVCL_1218), and Detroit 562 (RRID: CVCL_1171) cells were obtained from the American Type Culture Collection. PE/CA-PJ34 (clone C12; RRID: CVCL_2679) cells were obtained from Sigma-Aldrich. Cal33 (RRID: CVCL_1108) were obtained from Gerard Milano at the Centre Antoine Lacassagne (Nice, France). HSC-2 (RRID: CVCL_1287) cells were obtained from the Health Science Research Resources Bank. The PCI-52 (RRID: CVCL_RJ99) cell line was obtained from Dr. Theresa Whiteside at the University of Pittsburgh (Pittsburgh, PA). Cal27, FaDu, Detroit 562, Cal33, HSC-2, and PCI-52 cells were maintained in DMEM (Corning) containing 10% FBS (Gemini Bio-Products) and penicillin/streptomycin (Life Technologies). PE/CA-PJ34 cells were maintained in Iscove’s DMEM (Corning) containing 10% FBS, 2 mM l-glutamine (Life Technologies), and penicillin/streptomycin. Lung cancer cell lines were obtained from American Type Culture Collection and maintained in RPMI-1640 (Life Technologies) containing 10% FBS and penicillin/streptomycin. Cell line identities were verified by short tandem repeat profiling performed by Genetica DNA Laboratories or the DNA Sequencing Facility at the University of California, Berkeley, most recently on February 9, 2018. All cells were maintained in a humidified incubator at 37°C and 5% CO2 for ≤6 mo before retesting or starting new cultures from previously tested stocks stored in liquid nitrogen. Mycoplasma status was not routinely assessed. PDXs were developed from human HNSCC tumors as described previously.
Plasmids, lentiviral production, and stable cell line selection
pHAGE-PIK3CA WT and pHAGE-PIK3CA H1047R plasmids were a gift from Dr. Gordon Mills (MD Anderson Cancer Center, Houston, TX) and were cloned into the doxycycline-inducible pLVX-Puro vector (Clontech) using the gateway cloning system (Life Technologies). All vectors were sequence validated. Lentivirus was produced according to the Lenti-X Tet-One Inducible Expression Systems Manual (Clontech) and subsequently used to establish PCI-52 stable cell lines after selection with puromycin (0.5 µg/ml) for several weeks. Validation of each stable clone was performed 48 h after incubation with doxycycline (1 µg/ml) and subsequently examined by immunoblot for Flag tag expression (Sigma-Aldrich).
PGE2 ELISA
Secreted PGE2 was measured using the human PGE2 detection kit from Cayman Chemical Company according to the manufacturer’s instructions. For baseline measurements, cells were plated in triplicate at 10,000 cells per well (100,000 for engineered cells) in 24-well dishes. After 24 h, media were replaced with serum-free media followed by an additional 48-h incubation and subsequent PGE2 measurement. For treatment studies, cells were treated with 200 µM sulindac, 200 µM indomethacin, or 100 µM celecoxib in serum-free media for 48 h followed by PGE2 measurement. For inducible, engineered cell lines, 1 µM doxycycline was added to the medium for 48 h before the experiment.
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay
Cells were plated at 10,000 cells per well (100,000 for engineered cells) in 24-well plates 24 h before treatment with 200 µM sulindac or indomethacin, or 100 µM celecoxib, for 48 h. MTT assays were performed by replacing media with 5 mg/ml MTT (Sigma-Aldrich) in PBS followed by permeabilization and dissolution of formazan product in dimethyl sulfoxide. Absorbance at 570 nm was detected with an Epoch plate reader (BioTek) and considered a surrogate for cell viability/survival.
Immunoblotting
Whole-cell lysates were harvested using radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, and 0.5% NP-40) supplemented with protease and phosphatase inhibitor cocktails. Samples were sonicated for 10 s and then centrifuged at 15,000 g for 10 min at 4°C. Bradford assay was used to determine protein concentrations (Bio-Rad Laboratories). Equal amounts of protein were fractionated by SDS-PAGE, transferred to a polyvinylidene fluoride membrane (Bio-Rad Laboratories), and analyzed by incubation with the corresponding primary antibodies from Cell Signaling Technology: p-AKT-S473 (4060), AKT (4685), p-PRAS40-T246 (13175), PRAS40 (2691), COX2 (12282), and GAPDH (5174). Anti-Flag M2 (Sigma) was used to detect PI3K overexpression. β-Tubulin (Abcam) was used as an alternative loading control. Proteins were detected via incubation with HRP-conjugated secondary antibodies and enhanced chemiluminescence Western Blotting Substrate (Santa Cruz Biotechnology).
Online supplemental material
Fig. S1 illustrates the impact of NSAID use on DSS according to HPV status. Fig. S2 shows activation of PI3K signaling and COX2 expression in HNSCC cells expressing MT PIK3CA. Fig. S3 demonstrates correlation of PIK3CA mutation or amplification with increased PI3K signaling in an independent HNSCC patient cohort. Fig. S4 shows that cancer cells with altered PIK3CA are more sensitive to indomethacin treatment. Fig. S5 shows that cancer cells or PDXs with altered PIK3CA demonstrate a greater reduction of PGE2 secretion upon NSAID treatment. Table S1 lists the baseline characteristics of this cohort. Table S2 lists adjuvant therapies used in this cohort. Table S3 lists specific NSAIDs used in this cohort among 99 regular users. Table S4 lists HRs for DSS by PIK3CA status and regular NSAID exposure. Table S5 lists HRs for OS by PIK3CA status and regular NSAID exposure. Table S6 lists PIK3CA mutations in HNSCC PDX models.
Acknowledgments
We thank the following colleagues who made valuable contributions to this work: Gerard Milano, Theresa Whiteside, and Neil Jeremiah Kelly.
Research reported in this publication was supported by the National Institutes of Health (F30CA180235) and a pilot grant from the American Head and Neck Society to M.L. Hedberg; V Foundation for Cancer Research to J.E. Bauman; the National Institutes of Health (R01DE024728) to D.E. Johnson; the National Institutes of Health (R01CA098372, R01DE023685, and P50CA097190) and the American Cancer Society to J.R. Grandis; and the Department of Veterans’ Affairs Career Development Award Biomedical Laboratory Research & Development to U. Duvvuri. This project used the University of Pittsburgh Cancer Institute Biostatistics Facility that is supported in part by the National Institutes of Health award P30CA047904.
The authors declare no competing financial interests.
Author contributions: M.L. Hedberg, W.E. Gooding, and N.D. Peyser designed and performed experiments and discussed and analyzed data; H. Li, N.E. Bhola, T.R. Zhu, Y. Zeng, T.M. Brand, and V. Olivas helped with performing experiments and data analysis; R.C.K. Jordan, S. VandenBerg, T.G. Bivona, S.I. Chiosea, L. Wang, G.B. Mills, J.T. Johnson, U. Duvvuri, R.L. Ferris, and P. Ha helped supervise research and discussed data; W.E. Gooding, J.E. Bauman, and M.-O. Kim discussed and analyzed data; and M.L. Hedberg, N.D. Peyser, J.R. Grandis, and D.E. Johnson wrote the manuscript.
References
- Ahmadi N., Goldman R., Seillier-Moiseiwitsch F., Noone A.M., Kosti O., and Davidson B.J.. 2010. Decreased risk of squamous cell carcinoma of the head and neck in users of nonsteroidal anti-inflammatory drugs. Int. J. Otolaryngol. 2010:424161 10.1155/2010/424161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron J.A., Cole B.F., Sandler R.S., Haile R.W., Ahnen D., Bresalier R., McKeown-Eyssen G., Summers R.W., Rothstein R., Burke C.A., et al. . 2003. A randomized trial of aspirin to prevent colorectal adenomas. N. Engl. J. Med. 348:891–899. 10.1056/NEJMoa021735 [DOI] [PubMed] [Google Scholar]
- Buckley L., Gupta R., Ashford B., Jabbour J., and Clark J.R.. 2016. Oropharyngeal cancer and human papilloma virus: evolving diagnostic and management paradigms. ANZ J. Surg. 86:442–447. 10.1111/ans.13417 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2015. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 517:576–582. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.T., Giovannucci E.L., Meyerhardt J.A., Schernhammer E.S., Curhan G.C., and Fuchs C.S.. 2005. Long-term use of aspirin and nonsteroidal anti-inflammatory drugs and risk of colorectal cancer. JAMA. 294:914–923. 10.1001/jama.294.8.914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.T., Ogino S., and Fuchs C.S.. 2009. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 302:649–658. 10.1001/jama.2009.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosea S.I., Grandis J.R., Lui V.W., Diergaarde B., Maxwell J.H., Ferris R.L., Kim S.W., Luvison A., Miller M., and Nikiforova M.N.. 2013. PIK3CA, HRAS and PTEN in human papillomavirus positive oropharyngeal squamous cell carcinoma. BMC Cancer. 13:602 10.1186/1471-2407-13-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Church D.N., Sieber O., Ramamoorthy R., Yanagisawa Y., Johnstone E., Davidson B., Kerr D.J., Tomlinson I.P.M., and Midgley R.. 2013. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. J. Clin. Oncol. 31:4297–4305. 10.1200/JCO.2013.50.0322 [DOI] [PubMed] [Google Scholar]
- Elwood P.C., Morgan G., Pickering J.E., Galante J., Weightman A.L., Morris D., Kelson M., and Dolwani S.. 2016. Aspirin in the Treatment of Cancer: Reductions in Metastatic Spread and in Mortality: A Systematic Review and Meta-Analyses of Published Studies. PLoS One. 11:e0152402 10.1371/journal.pone.0152402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohagan J.K., Prorok P.C., Hayes R.B., and Kramer B.S.. Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial Project Team . 2000. The Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial of the National Cancer Institute: history, organization, and status. Control. Clin. Trials. 21(Suppl 6):251S–272S. 10.1016/S0197-2456(00)00097-0 [DOI] [PubMed] [Google Scholar]
- Grambsch P.M., and Therneau T.M.. 1994. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika. 81:515–526. 10.1093/biomet/81.3.515 [DOI] [Google Scholar]
- Harrell F.2015. rms: Regression modeling strategies. R package 4.5-0. http://CRAN.R-project.org/package=rms.
- Henry W.S., Laszewski T., Tsang T., Beca F., Beck A.H., McAllister S.S., and Toker A.. 2017. Aspirin Suppresses Growth in PI3K-Mutant Breast Cancer by Activating AMPK and Inhibiting mTORC1 Signaling. Cancer Res. 77:790–801. 10.1158/0008-5472.CAN-16-2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes M.D., Chen W.Y., Li L., Hertzmark E., Spiegelman D., and Hankinson S.E.. 2010. Aspirin intake and survival after breast cancer. J. Clin. Oncol. 28:1467–1472. 10.1200/JCO.2009.22.7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash V., Rigual N.R., Moysich K.B., Loree T.R., Nasca M.A., Menezes R.J., and Reid M.E.. 2006. Chemoprevention of head and neck cancer with aspirin: a case-control study. Arch. Otolaryngol. Head Neck Surg. 132:1231–1236. 10.1001/archotol.132.11.1231 [DOI] [PubMed] [Google Scholar]
- Kalbfleisch J.D., and Prentice R.L.. 2011. The statistical analysis of failure time data. John Wiley & Sons, Marblehead, MA. [Google Scholar]
- Katoumas K., Nikitakis N., Perrea D., Dontas I., and Sklavounou A.. 2015. In Vivo Antineoplastic Effects of the NSAID Sulindac in an Oral Carcinogenesis Model. Cancer Prev. Res. (Phila.). 8:642–649. 10.1158/1940-6207.CAPR-14-0447 [DOI] [PubMed] [Google Scholar]
- Kim S.A., Roh J.L., Kim S.B., Choi S.H., Nam S.Y., and Kim S.Y.. 2018. Aspirin use and head and neck cancer survival: an observational study of 11,623 person-years follow-up. Int. J. Clin. Oncol. 23:52–58. 10.1007/s10147-017-1165-3 [DOI] [PubMed] [Google Scholar]
- Liao X., Lochhead P., Nishihara R., Morikawa T., Kuchiba A., Yamauchi M., Imamura Y., Qian Z.R., Baba Y., Shima K., et al. . 2012. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N. Engl. J. Med. 367:1596–1606. 10.1056/NEJMoa1207756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui V.W., Hedberg M.L., Li H., Vangara B.S., Pendleton K., Zeng Y., Lu Y., Zhang Q., Du Y., Gilbert B.R., et al. . 2013. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 3:761–769. 10.1158/2159-8290.CD-13-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandapathil M., Szczepanski M.J., Szajnik M., Ren J., Jackson E.K., Johnson J.T., Gorelik E., Lang S., and Whiteside T.L.. 2010. Adenosine and prostaglandin E2 cooperate in the suppression of immune responses mediated by adaptive regulatory T cells. J. Biol. Chem. 285:27571–27580. 10.1074/jbc.M110.127100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor R., Heenan M., Connolly L., Larkin A., and Clynes M.. 2004. Increased anti-tumour efficacy of doxorubicin when combined with sulindac in a xenograft model of an MRP-1-positive human lung cancer. Anticancer Res. 24(2A):457–464. [PubMed] [Google Scholar]
- Rafei H., Lumley C., Han J., Kaffenberger T.M., and Maxwell J.H.. 2017. Aspirin use and survival in head and neck squamous cell cancer (HNC) patients. J. Clin. Oncol. 35:6042 10.1200/JCO.2017.35.15_suppl.6042 [DOI] [Google Scholar]
- R Team 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. http://www.R-project.org.
- Scheper M.A., Nikitakis N.G., Chaisuparat R., Montaner S., and Sauk J.J.. 2007. Sulindac induces apoptosis and inhibits tumor growth in vivo in head and neck squamous cell carcinoma. Neoplasia. 9:192–199. 10.1593/neo.06781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N., Egloff A.M., Tward A.D., Kostic A.D., Cibulskis K., Sivachenko A., Kryukov G.V., Lawrence M.S., Sougnez C., McKenna A., et al. . 2011. The mutational landscape of head and neck squamous cell carcinoma. Science. 333:1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tury S., Becette V., Assayag F., Vacher S., Benoist C., Kamal M., Marangoni E., Bièche I., Lerebours F., and Callens C.. 2016. Combination of COX-2 expression and PIK3CA mutation as prognostic and predictive markers for celecoxib treatment in breast cancer. Oncotarget. 7:85124–85141. 10.18632/oncotarget.13200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.C., Murray L.J., Hughes C.M., Black A., and Anderson L.A.. 2013. Non-steroidal anti-inflammatory drug and aspirin use and the risk of head and neck cancer. Br. J. Cancer. 108:1178–1181. 10.1038/bjc.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip-Schneider M.T., Wu H., Ralstin M., Yiannoutsos C., Crooks P.A., Neelakantan S., Noble S., Nakshatri H., Sweeney C.J., and Schmidt C.M.. 2007. Suppression of pancreatic tumor growth by combination chemotherapy with sulindac and LC-1 is associated with cyclin D1 inhibition in vivo. Mol. Cancer Ther. 6:1736–1744. 10.1158/1535-7163.MCT-06-0794 [DOI] [PubMed] [Google Scholar]