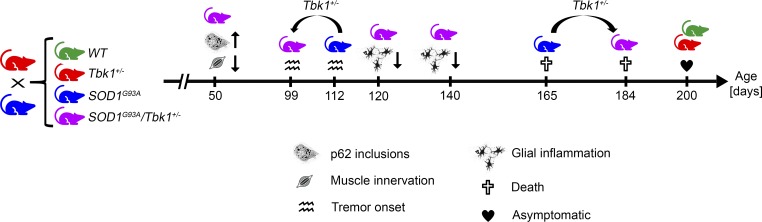
This study shows that mice with an ALS-linked heterozygous Tbk1 deletion are normal until at least 200 d of age. However, proteostatic and neuroinflammatory challenge by additional expression of mutant SOD1 unmasks a two-edged role of TBK1 in motoneuron disease.
Abstract
Heterozygous loss-of-function mutations of TANK-binding kinase 1 (TBK1) cause familial ALS, yet downstream mechanisms of TBK1 mutations remained elusive. TBK1 is a pleiotropic kinase involved in the regulation of selective autophagy and inflammation. We show that heterozygous Tbk1 deletion alone does not lead to signs of motoneuron degeneration or disturbed autophagy in mice during a 200-d observation period. Surprisingly, however, hemizygous deletion of Tbk1 inversely modulates early and late disease phases in mice additionally overexpressing ALS-linked SOD1G93A, which represents a “second hit” that induces both neuroinflammation and proteostatic dysregulation. At the early stage, heterozygous Tbk1 deletion impairs autophagy in motoneurons and prepones both the clinical onset and muscular denervation in SOD1G93A/Tbk1+/− mice. At the late disease stage, however, it significantly alleviates microglial neuroinflammation, decelerates disease progression, and extends survival. Our results indicate a profound effect of TBK1 on brain inflammatory cells under pro-inflammatory conditions and point to a complex, two-edged role of TBK1 in SOD1-linked ALS.
Graphical Abstract
Introduction
Some 5–10% of patients with the fatal neurodegenerative motoneuron (MN) disease amyotrophic lateral sclerosis (ALS) self-report a positive family history (familial ALS [fALS]). Using whole exome sequencing of fALS patients, we and others recently found an exome-wide, highly significant enrichment of mono-allelic TANK-binding kinase 1 (TBK1) loss-of-function mutations in fALS and frontotemporal dementia (FTD) patients (Cirulli et al., 2015; Freischmidt et al., 2015).
TBK1 is a pleiotropic kinase consisting of a catalytic kinase domain and three accessory regulatory domains. Its best characterized biological functions are the regulation of selective autophagy (Weidberg and Elazar, 2011; Wild et al., 2011; Pilli et al., 2012) and modulation of IFN signaling and inflammatory responses (Hemmi et al., 2004; Jin et al., 2012; Hasan et al., 2015, 2017; Yu et al., 2015). In addition, TBK1 has been implicated in energy metabolism (Reilly et al., 2013, 2015; Everts et al., 2014; Zhao et al., 2018), microtubule dynamics (Pillai et al., 2015), and tumorigenesis (Ou et al., 2011).
Pointing to impaired autophagy and consequently reduced clearance of pathological protein oligomers/aggregates, brains of ALS/FTD patients with pathogenic TBK1 mutations exhibit cytoplasmatic (p)TDP-43– and p62-positive inclusions (Freischmidt et al., 2015; Pottier et al., 2015). Mouse studies with deletion of different autophagy-linked Atg genes indicate an ambiguous role of autophagy in MNs (Hara et al., 2006; Nassif et al., 2014; Tokuda et al., 2016; Rudnick et al., 2017).
Neuroinflammation, including activation of microglia and astrocytes, substantially contributes to the exacerbation and progression of the disease in mutant human SOD1 transgenic mouse models of ALS (Beers et al., 2006; Boillée et al., 2006; Yamanaka et al., 2008) and most likely in patients. Further, heterozygous deletion of the α-IFN receptor Ifnar1 significantly prolongs the life span of SOD1G93A mice (Wang et al., 2011). Intriguingly, TBK1 is a well-known inducer of the IFN type I response (Trinchieri, 2010; Ahmad et al., 2016). By contrast, global heterozygous deletion of Tbk1 in combination with selective heterozygous deficiency of Tak1 in the myeloid lineage was recently shown to cause cortical neurodegeneration, microgliosis, and TDP-43 inclusions in 6-mo-old mice (Xu et al., 2018).
Taken together, TBK1 is a central regulator of both selective autophagy and inflammatory responses via IFN type I signaling. Both pathways are suggested to influence the disease course of human ALS and have been shown to modulate disease in transgenic ALS mouse models. Consequently, our study sought to answer which pathways downstream of Tbk1 haploinsufficiency are the most ALS relevant.
Results and discussion
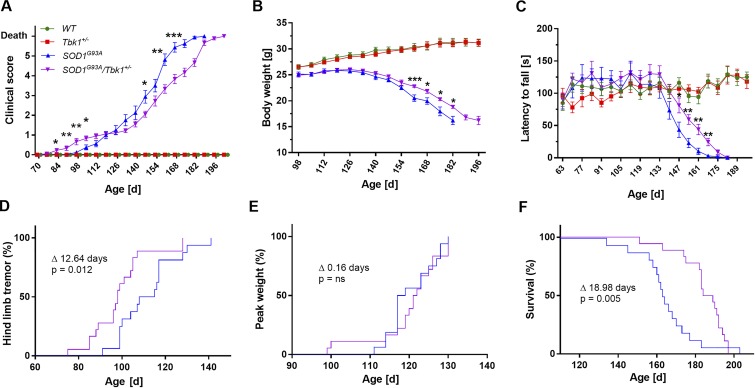
We aimed to determine a possible neurological phenotype of heterozygous Tbk1 knockout mice (Tbk1+/− mice). While homozygous loss of Tbk1 is embryonically lethal in mice (Bonnard et al., 2000), loss of one Tbk1 allele mirrors the genetic defect causing ALS/FTD in humans. In addition, we asked if and how a ubiquitous heterozygous Tbk1 knockout alters the phenotype of SOD1G93A transgenic mice. In this ALS model, the overexpression of (human) SOD1G93A leads to MN degeneration and neuroinflammation and represents a challenge of the proteostatic system (Philips and Rothstein, 2015; Picher-Martel et al., 2016). Thus, we crossed Tbk1+/− mice with SOD1G93A transgenic mice. All four resulting genotypes (WT, Tbk1+/−, SOD1G93A, and SOD1G93A/Tbk1+/− siblings) were subjected to weekly rotarod testing, as well as assessment of the global phenotype progression, weight, and survival. Tbk1+/− and SOD1G93A/Tbk1+/− mice were phenotypically indistinguishable from WT or SOD1G93A siblings at birth. Tbk1+/− mice did not develop motor symptoms or experience weight loss or premature death during the study period of 200 d (Fig. 1, A–C). As expected, SOD1G93A mice developed hind limb tremor (clinical score of 1, see methods section), which became apparent to a blinded investigator at a mean age of 111.8 ± 3.3 d. Remarkably, heterozygous knockout of Tbk1 in addition to SOD1G93A overexpression (SOD1G93A/Tbk1+/− mice) preponed the onset of hind limb tremor to 99.1 ± 3.1 d (Δ12.6 d; P = 0.012; Fig. 1, A and D). However, the age of onset of manifest gait disturbance (score of 2), peak weight, and peak rotarod performance did not significantly differ between SOD1G93A/Tbk1+/− and SOD1G93A siblings (Fig. 1, A-C and E; and Fig. S1 B), suggesting an attenuated progression of symptoms in SOD1G93A/Tbk1+/− mice. During the later disease course, SOD1G93A/Tbk1+/− mice indeed showed a further slowed decline in clinical score, weight, and rotarod performance when compared with SOD1G93A siblings (Fig. 1, A–C; and Fig. S1, A and C; SOD1G93A vs. SOD1G93A/Tbk1+/−: 10% weight loss: 148.5 ± 2.3 vs. 154.9 ± 1.5 d; Δ6.4 d; P = 0.044; half-maximal latency to fall in the rotarod test: 138.9 ± 2.6 vs. 142.3 ± 5.4 d; Δ3.4 d; P = 0.018). Finally, this also led to a significantly extended survival and overall disease duration of SOD1G93A mice with heterozygous Tbk1 knockout compared with single transgenic SOD1G93A siblings (despite the earlier appearance of first symptoms; Fig. 1 F and Fig. S1 D; SOD1G93A vs. SOD1G93A/Tbk1+/−: survival, 165.2 ± 3.7 vs. 184.2 ± 2.8 d; Δ19.0 d; P = 0.005; duration, 44.8 ± 3.0 vs. 63.9 ± 3.4 d; Δ19.1 d; P < 0.001).
Figure 1.
Heterozygous Tbk1 deletion prepones early motor symptoms but slows disease progression and prolongs survival in the SOD1G93A ALS mouse model. (A) Progression of the clinical score at group level. SOD1G93A/Tbk1+/− mice show a bi-phasic, first accelerated and then slowed, disease progression compared with SOD1G93A siblings. (B) Weight curve at group level. SOD1G93A/Tbk1+/− mice show a slowed progression of weight loss compared with SOD1G93A siblings. (C) Performance in the rotarod test at group level over time. SOD1G93A/Tbk1+/− mice show a slowed progression of motor decline compared with SOD1G93A siblings. (D) Kaplan-Meier plot of the fraction of mice with hind limb tremor (score of 1). SOD1G93A/Tbk1+/− mice present with a significantly earlier onset of hind limb tremor than SOD1G93A siblings. (E) Kaplan-Meier plots of the fraction of mice having reached their weight peak. SOD1G93A/Tbk1+/− and SOD1G93A siblings exhibit a similar onset of weight loss. (F) As demonstrated by Kaplan-Meier survival curves, heterozygous deletion of Tbk1 significantly prolongs survival of SOD1G93A mice. n = 16–18 male mice per group in all graphs. Data in A–C are presented as means ± SEM and were analyzed by one-way ANOVA followed by Tukey's multiple comparisons post hoc test. Kaplan-Meier plots were analyzed using the log-rank (Mantel-Cox) test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Considering the unexpected, inverse effect of heterozygous Tbk1 knockout on clinical onset and disease course of SOD1G93A transgenic mice, we next sought to validate this observation at histological and biochemical levels and determine its molecular and cell biological correlates.
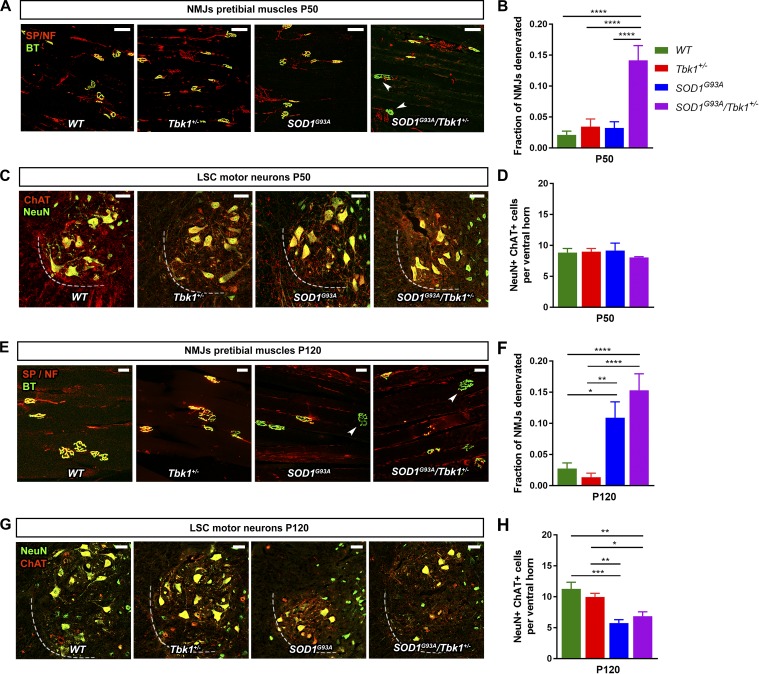
First, we analyzed the innervation of neuromuscular junctions (NMJs) of the pretibial muscles at the age of postnatal day (P) 50 (presymptomatic) and P120 (early symptomatic) in all four genotypes. Consistent with the lack of clinical symptoms during the study period, Tbk1+/− mice did not show NMJ denervation or MN loss at either time point compared with WT siblings (Fig. 2, A–H). By contrast, SOD1G93A/Tbk1+/− mice exhibited a significantly higher denervation of NMJs at P50 than the other groups including SOD1G93A siblings (Fig. 2, A and B), matching the significantly preponed detection of first clinical symptoms, whereas the number of lumbar spinal cord (LSC) MNs was still equal in all genotypes at P50 (Fig. 2, C and D). NMJ denervation and MN loss did not differ between SOD1G93A/Tbk1+/− mice and SOD1G93A siblings at P120 (Fig. 2, E–H). Conclusively, the kinetics of NMJ denervation and MN degeneration of SOD1G93A/Tbk1+/− mice are in agreement with a preponed symptom onset but subsequently decelerated disease progression (Fig. 1).
Figure 2.
Tbk1 haploinsufficiency enhances NMJ denervation in the early phase of the SOD1G93A ALS mouse model. (A) Representative images of NMJs of pretibial muscles at P50 stained with α-bungarotoxin (BT; presynaptic, green), anti-Synaptophysin (SP), and anti–pan-Neurofilament (NF; postsynaptic, red). Completely denervated NMJs are indicated by arrowheads. Scale bar, 100 µm. (B) SOD1G93A/Tbk1+/− mice show significant NMJ denervation as compared with the other genotypes at P50. (C) Representative images of MNs in the anterior horn of the LSC at P50. Scale bar, 50 µm. (D) MN counts in the LSC at P50 do not significantly differ among the four genotypes. (E) Representative images of NMJs of pretibial muscles at P120. Completely denervated NMJs are indicated by arrowheads. Scale bar, 50 µm. (F) No significant difference in the level of NMJ denervation was observed between SOD1G93A/Tbk1+/− and SOD1G93A mice at P120. (G) Representative images of MNs in the anterior horn of the LSC at P120. Scale bar, 50 µm. (H) MN counts in the LSC at P120 do not significantly differ among Tbk1+/− and WT mice or between SOD1G93A/Tbk1+/− and SOD1G93A siblings. n = 5–7 female mice per group in all graphs. Data are presented as means ± SEM. Data presented in B and F were analyzed by Kruskal-Wallis test followed by Dunn's multiple comparisons post hoc test. Data presented in D and H was analyzed by one-way ANOVA followed by Tukey's multiple comparisons post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
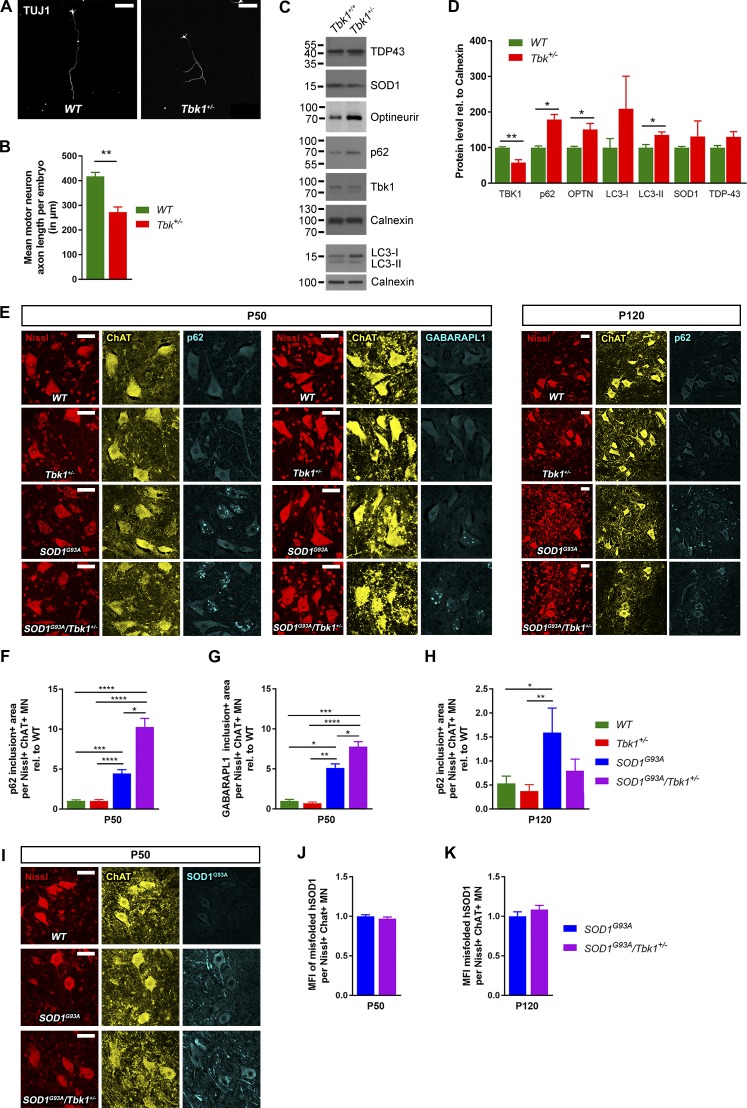
Regulation of autophagy is one of the best established biological functions of TBK1. Similar to the results of our study based on the heterozygous knockout of Tbk1, MN-selective homozygous knockout of the autophagy protein Atg7 preponed the onset of NMJ denervation and hind limb tremor in SOD1G93A mice (Rudnick et al., 2017). This indicates a possible neuroprotective effect of autophagy in the early disease of SOD1G93A transgenic mice. Hence, we sought to answer whether the heterozygous loss of Tbk1 impairs autophagy in LSC MNs in vitro and in vivo. We isolated primary MNs from the spinal cord of embryonic day (E) 12.5 WT and Tbk1+/− embryos using immunopanning (Wiese et al., 2010). Although we did not detect a central nervous system (CNS) phenotype in Tbk1+/− mice during the observation period of 200 d, embryonic Tbk1+/− MNs show impaired axonal elongation after 7 d in culture (Fig. 3, A and B), which points to a possible role of TBK1 in axon growth during development. Alternatively, the observation reflects an MN deficit demasked by cell culture stress as a “second hit,” analogous to the (accentuated) Tbk1-associated axon terminal/NMJ phenotype we observed only in SOD1G93A/Tbk1+/− double-mutant mice compared with SOD1G93A mice (Fig. 2, A and B).
Figure 3.
Tbk1 haploinsufficiency induces markers of impaired autophagy in cultured MNs and in SOD1G93A mice. (A) Representative images of primary MNs from E12.5 mouse embryos 1 wk in culture stained against TUJ1. Scale bar, 100 µm. (B) Heterozygous Tbk1 deletion significantly impairs axonal outgrowth of primary MNs. (C) Representative Western blots of lysate from primary MNs stained against TBK1, p62, OPTN, LC3, SOD1, and TDP-43. (D) Quantification of Western blots from C. TBK1 was reduced by 50% in Tbk1+/− MNs. The autophagy proteins p62, OPTN, and LC3-II significantly accumulated in primary MNs upon heterozygous Tbk1 deletion. n = 3; each n is a pool of three embryos. (E) Representative images of LSC MNs of P50 and P120 mice stained against p62 or GABARAPL1. While p62 aggregates are located within large MNs at P50, the gross of aggregated p62 is located outside of ChAT+ MN somata at P120. Scale bar, 50 µm. (F and G) At P50, heterozygous Tbk1 deletion increases the area of p62+ (F) and GABARAPL1+ (G) round-body inclusions in the LSC of SOD1G93A mice. (H) At P120, the area of intracellular p62+ inclusions per MN does not differ between SOD1G93A/Tbk1+/− and SOD1G93A siblings. (I) Representative images of mouse LSC MNs at P50, stained against human misfolded SOD1G93A (with antibody B8H10). Scale bar, 50 µm. (J and K) Heterozygous Tbk1 deletion does not alter the mean fluorescence intensity of misfolded human SOD1G93A in LSC MNs of P50 and P120 mice. n = 5–7 female mice per group in all in vivo graphs. Data are presented as means ± SEM. Data presented in B, D, J, and K were analyzed by Student’s t test. Data presented in F–H were analyzed by Kruskal-Wallis test followed by Dunn's multiple comparisons post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Furthermore, we observed significantly increased levels of p62, the TBK1-binding autophagy adaptor protein optineurin (OPTN) and LC3-II in Tbk1+/− primary MNs after 7 d in culture, suggesting an impairment of autophagy (Fig. 3, C and D). SOD1 and TDP-43 levels were not affected by Tbk1 haploinsufficiency (Fig. 3, C and D).
Consequently, we extended our analysis of autophagy markers to the in vivo situation. LSC lysates and slices from P50 and P120 mice of all four genotypes were analyzed. The autophagy markers LC3–I, LC3–II, and p62 were unaltered at the age of P50 and P120 in all four genotypes, according to Western blotting of spinal cord protein lysates (Fig. S2, A–D). However, single cell–based analysis using immunohistochemical staining revealed an increased accumulation of aggregated p62 forming round bodies in the cytoplasm of LSC MNs of SOD1G93A/Tbk1+/− mice at P50 compared with SOD1G93A siblings, while Tbk1+/− and WT mice showed essentially no p62+ inclusions (Fig. 3, E and F). We could reproduce this finding using an alternative autophagy marker, GABARAPL1, an Atg8 homologue highly expressed in MNs (Le Grand et al., 2013; Fig. 3, E and G).
As shown in Fig. S2, G and H, the mostly round p62+ inclusions in both SOD1G93A transgenic groups largely colocalized with GABARAPL1 as well as polyubiquitin at P50. We hardly detected round p62 inclusions within ChAT+ neurons at P120, most probably because the majority of large MNs containing these round p62 inclusions at P50 belonged to the more susceptible MN subpopulation and had degenerated between P50 and P120 (as also described previously; e.g., Rudnick et al., 2017). Instead of round p62 inclusions the remaining smaller neurons partially exhibited “skein-like” p62 inclusions at P120 (Fig. S2, K and arrowhead in M), which is consistent with previous findings (Rudnick et al., 2017). The majority of p62 aggregates, however, were not located inside but were outside of the MN somata at P120 and overlapped with GABARAPL1 and polyubiquitin (Fig. S2, I and J). Co-staining of Nissl, Neurofilament, GFAP and Iba1 with p62 suggested that the p62 aggregates outside MN somata colocalized with axons and astrocytes, but hardly with microglia (Fig. S2, K–M). Quantitative analyses revealed that the levels of aggregated p62 inside the remaining ChAT+ MNs at P120 differed significantly between non-SOD1G93A-transgenic and SOD1G93A mice, but not between SOD1G93A mice and SOD1G93A/Tbk1+/− mice, despite a nonsignificant trend toward a reduction in SOD1G93A/Tbk1+/− mice (P = 0.19; Fig. 3 H).
Taken together, the above evidence suggests that at P120 the decelerated disease progression starts to outmatch the disadvantage of Tbk1 haploinsufficiency observed at P50.
TBK1 and phospho-S177–OPTN colocalize with SOD1G93A aggregates in MNs of SOD1G93A mice, suggesting that SOD1G93A aggregates undergo TBK1-directed selective autophagy (Korac et al., 2013). Pointing in the same direction, heterozygous deletion of the autophagy protein Becn1 (Atg6) enhances aggregation of mutant SOD1G93A in SOD1G93A mice (Tokuda et al., 2016). However, we did not find differences in the quantity of either endogenous mouse (WT) SOD1 or human misfolded SOD1G93A in LSC by Western blotting or immunohistochemistry between mutant SOD1G93A mice with or without heterozygous Tbk1 knockout (Fig. 3, I–K; and Fig. S2, E and F). This indicates that the preponed onset of motor symptoms and NMJ denervation in SOD1G93A/Tbk1+/− mice is unlikely to be a direct consequence of increased SOD1G93A levels and suggests other proteins or organelles to be the relevant substrates of disturbed autophagy that are responsible for the effect of heterozygous Tbk1 knockout in the SOD1G93A model.
Beyond its role as an autophagy regulator, abundant evidence connects TBK1 to inflammation. TBK1 is a well-known inducer of the IFN type I response (Trinchieri, 2010). Notably, heterozygous deletion of Ifnar1 significantly prolongs the life span of SOD1G93A mice (Wang et al., 2011) to an extent similar to that observed in the SOD1G93A/Tbk1+/− mice of this study. Moreover, reactive astrocytes and microglia have been repeatedly demonstrated to accelerate disease progression in mutant human SOD1 transgenic mice (Beers et al., 2006; Boillée et al., 2006; Yamanaka et al., 2008).
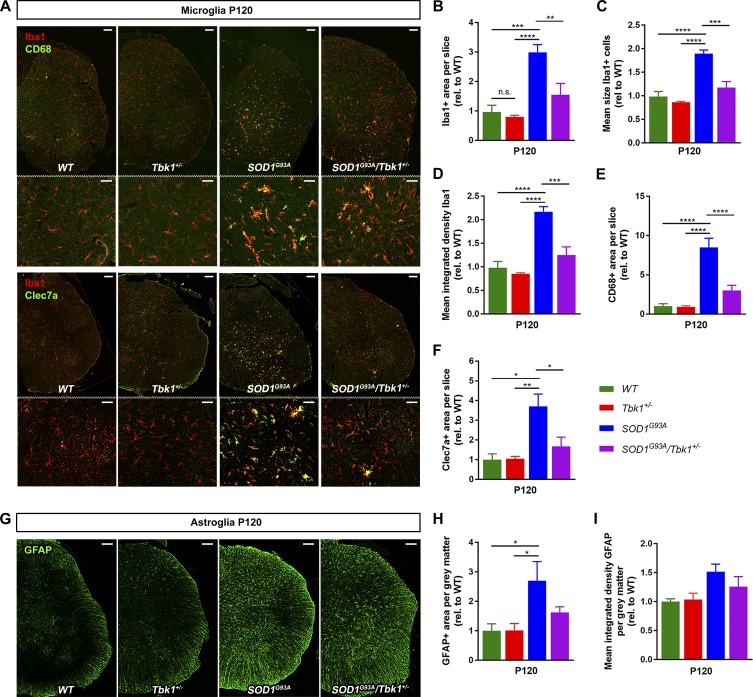
Hence, we hypothesized that Tbk1 haploinsufficiency, beyond autophagy regulation and its role in peripheral leukocytes, may also alleviate CNS inflammation and thereby extend the life span in the mutant SOD1 model. Consequently, we quantified the microglial and astrocytic response at the age of P120, when robust neuroinflammation is found in the SOD1G93A model. Of note, we did not detect differences in the abundances of microglia and astrocytes in the LSC between Tbk1+/− mice and WT mice. By contrast, we found a significantly alleviated microglial activation in SOD1G93A/Tbk1+/− mice compared with SOD1G93A siblings. Immunohistochemical analysis of the LSC of SOD1G93A/Tbk1+/− mice showed a smaller area and reduced integrated density of the Iba1 immunoreactivity, as well as a reduced mean Iba1+ cell size, compared with SOD1G93A siblings (Fig. 4, A–D). In addition, we observed a reduced immunopositive area of the microglial activation markers CD68 and Clec7a (Holtman et al., 2015; Krasemann et al., 2017) in SOD1G93A/Tbk1+/− mice compared with SOD1G93A siblings (Fig. 4, A, E, and F). Clec7a has been described as a marker for the neurodegeneration-associated microglia profile in various models, including SOD1G93A mice (Holtman et al., 2015; Krasemann et al., 2017).
Figure 4.
Tbk1 haploinsufficiency mitigates microglial neuroinflammation in SOD1G93A mice. (A) Representative photomicrographs of LSC hemispheres stained for the microglia markers Iba1, CD68, and Clec7a. Scale bars, 100 µm; 50 µm in magnified images. (B–F) SOD1G93A/Tbk1+/− mice showed a reduced Iba+, CD68+, and Clec7a+ area, reduced integrated density, and reduced mean size of Iba+ microglia, compared with SOD1G93A siblings. (G) Representative photomicrographs of astrogliosis in the gray matter of the LSC at the age of P120. Scale bar, 100 µm. (H and I) A nonsignificant trend toward reduced GFAP+ area (I) and mean integrated density of GFAP (H) in SOD1G93A/Tbk1+/− mice compared with SOD1G93A siblings was observed. n = 5–7 female mice per group. Data are presented as means ± SEM. Data were analyzed by one-way ANOVA followed by Tukey's multiple comparisons post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We furthermore observed a trend toward reduced astrogliosis in the LSC of SOD1G93A/Tbk1+/− mice compared with SOD1G93A siblings at P120, based on GFAP area or integrated GFAP immunoreactivity signal (P = 0.17 and P = 0.52, respectively; Fig. 4, G–I). There was no microgliosis or astrogliosis detectable in LSCs in any of the four genotypes at P50 (Fig. S2, N–P).
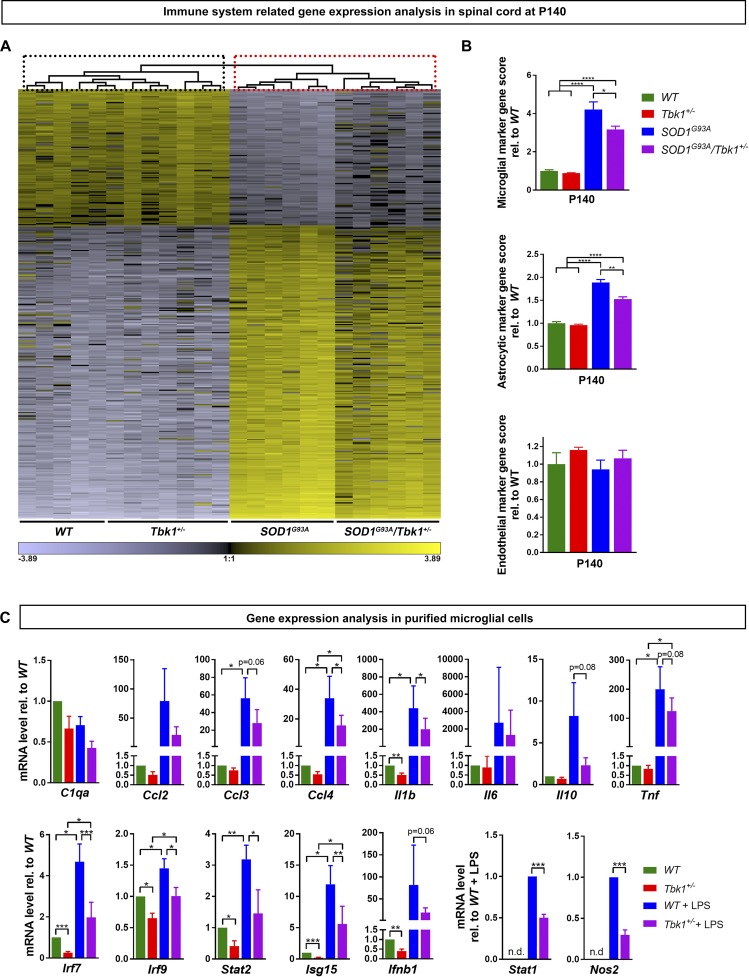
To corroborate the results from these immunohistochemical studies, we next aimed to obtain a more comprehensive picture of neuroinflammation in vivo. To that end, we performed a quantitative NanoString nCounter mRNA expression profiling of a large inflammation-related gene panel (800 genes) of the spinal cord of symptomatic mice and control siblings at P140. In line with the behavioral and immunohistochemical experiments, only the expression of 1 of 800 genes (Ceruloplasmin; [Cp]), beyond Tbk1 itself (which was reduced due to loss of one gene copy), was significantly altered in Tbk1+/− animals compared with WT mice after correction for multiple testing. Consistent with the known strong inflammatory response in ALS mice, we measured 542 inflammation related genes to be significantly altered in SOD1G93A mice compared with WT siblings (mostly induced or increased expression). Upon heterozygous Tbk1 deletion, 96 inflammation-related genes showed significantly less strong induction (among them, the microglial neurodegenerative phenotype markers Axl, Lilrb4, Clec7a, Csf1, and Trem2, previously described in Krasemann et al., 2017), indicating an attenuated inflammatory response in SOD1G93A/Tbk1+/− mice (supplemental dataset). Further supporting that Tbk1 haploinsufficiency attenuates the pro-inflammatory effect of the mutant SOD1 transgene, an unbiased hierarchical cluster analysis did not separate Tbk1+/− from WT animals, but completely separated SOD1G93A/Tbk1+/− mice from their SOD1G93A siblings (Fig. 5 A).
Figure 5.
Tbk1 haploinsufficiency reduces glial activation gene profiles in SOD1G93A mice and in purified microglia. (A) Heat map diagram of unbiased average linkage hierarchical cluster analysis based on 366 significantly altered transcripts identified by one-way ANOVA across all four genotypes (P value threshold: 6.25 × 10−5; corresponding to a P value of 0.05, Bonferroni corrected for multiple testing). The cluster analysis does not separate Tbk1+/− from WT animals (highlighted by dotted black box), but completely separates SOD1G93A/Tbk1+/− mice from their SOD1G93A siblings (highlighted by dotted red box). n = 5–7 mice per group. (B) Scores of cell-specific marker gene expression. Expression of microglial and astroglial marker genes was significantly reduced in SOD1G93A/Tbk1+/− mice compared with SOD1G93A siblings. n = 5–7 mice per group. (C) RT-qPCR of lysates from purified primary microglia untreated or stimulated with LPS (50 ng/ml for 6 h). Ccl4, Il1b, and Nos2 and the IFN-I signaling cascade components Irf7, Irf9, Stat1, Stat2, Isg15, and Ifnb were significantly down-regulated in Tbk1+/− microglia already under basal conditions and/or after activation with LPS. n = 6; each n is a pool of two to three pups. n.d., not detectable. Data are presented as means ± SEM. Data in B were analyzed by one-way ANOVA followed by Tukey's multiple comparisons post hoc test. Data in C were analyzed by one-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Furthermore, the comprehensive NanoString nCounter mRNA expression dataset allowed us to estimate the cell type–specific marker gene signatures described by Danaher et al. (2017) in the analyzed spinal cord tissue. This analysis revealed a significantly attenuated expression of inflammatory genes normally associated with microglial and/or astrocytic responses in SOD1G93A/Tbk1+/− compared with SOD1G93A siblings (Fig. 5 B). Endothelial cell type marker analysis served as a negative control and did not differ among the four genotypes (Fig. 5 B).
Overall, our results show that a global heterozygous deletion of Tbk1 reduces the neuroinflammatory response and attenuates the neurodegeneration-associated differentiation profile of microglia in SOD1G93A mice. To address the question of whether Tbk1 haploinsufficiency has a direct impact on microglial activation, we analyzed the mRNA levels of a subset of genes representing microglial activation markers and/or Tbk1 downstream targets in purified primary microglia cultures. Consistent with our in vivo data, several classically rather pro-inflammatory genes (e.g., Ccl4, Il1b, Nos2) were less induced in LPS-treated Tbk1+/− compared with WT microglial cells, pointing to an attenuated inflammatory response in vitro (Fig. 5 C). Under the cell culture condition without LPS stimulation, which nevertheless represents a less physiological situation than in vivo, there were also some changes in neuroinflammatory gene expression due to heterozygous Tbk1 deletion (Fig. 5 C). Our in vivo results support the hypothesis that microglia are among the primary cell types mediating the modulation of inflammation by Tbk1 haploinsufficiency in the spinal cord of ALS mice.
Taken together, we provide evidence that TBK1 has opposing effects in the early and late phases of disease in the mutant SOD1G93A mouse model of ALS. At the early stage, heterozygous Tbk1 deletion impairs autophagy in MNs and prepones hind limb tremor and NMJ denervation. At the late disease stage, heterozygous Tbk1 deletion substantially alleviates glial activation. Considering our in vitro and in vivo data, the attenuation of neuroinflammation represents the most likely mechanism responsible for the decreased disease progression rate and extended survival of SOD1G93A transgenic mice. In this context, the slowed disease progression observed in SOD1G93A/Tbk1+/− mice would thus represent a non–cell-autonomous effect, in full agreement with abundant previous evidence connecting neuroinflammation and disease progression in the mutant SOD1 ALS mouse model (Beers et al., 2006; Boillée et al., 2006; Yamanaka et al., 2008). However, given the pleiotropic nature of TBK1, multiple alternative cellular pathways may contribute to the alleviated progression and prolonged life span of the SOD1G93A transgenic mice. Cell-type selective deletion of Tbk1 in nonneuronal cells will be necessary for the final confirmation of this claim. In contrast, considering both our in vivo results and the effects of heterozygous Tbk1 deletion on purified cultured MNs, accelerated onset of symptoms and increased NMJ denervation in mutant SOD1G93A mice with heterozygous Tbk1 deletion are more likely MN-autonomous effects. Nevertheless, despite the abundant cell biological data linking TBK1 and autophagy, a caveat must be expressed because the loss of a Tbk1 allele could also indirectly lead to enhanced autophagy impairment in MNs, triggered, for example, by a primary damage to Schwann cells, muscle cells, or macrophages with subsequent damage to motor axons.
Although the heterozygous deletion of Tbk1 in mice corresponds exactly to the (haploinsufficient) Tbk1 loss-of-function mutations that are associated with ALS in humans, we could not detect a motoneuronal phenotype at the behavioral or histological level in Tbk1+/− mice compared with WT mice until the end of the study at an age of 200 d. Of note, in humans Tbk1 loss-of-function mutations show a reduced penetrance and somewhat later age at onset compared with most other fALS mutations (Freischmidt et al., 2015). Thus, we speculate that ALS-linked Tbk1 loss-of-function mutations may require one or more specific genetic or other cofactors representing “second hits” that are present in only a fraction of the normal population and not in mice. This hypothesis is in agreement with our observation that only a second genetic stressor, i.e., mutant SOD1 overexpression, demasks the effects of heterozygous Tbk1 deletion in the CNS of mice.
In summary, our study demonstrates that a heterozygous deletion of Tbk1 is sufficient to affect the function of multiple CNS cell types—at least neurons, astrocytes, and microglia. Our study is thus an important basis for further research on the complex and, depending on the disease stage, antipodal roles of TBK1 in ALS and other CNS disorders.
Material and methods
Animals
B6.Cg-Tg(SOD1*G93A)1Gur/J mice (stock no. 004435; high transgene copy number; https://www.jax.org/strain/004435; hereafter referred to as SOD1G93A mice) were purchased from Jackson Laboratories. Heterozygous B6.129P2-Tbk1tm1Aki mice (hereafter referred to as Tbk1+/− mice; http://www.informatics.jax.org/allele/key/29752; Hemmi et al., 2004) were kindly provided by Shizuo Akira. SOD1G93A mice were bred with Tbk1+/− mice to obtain SOD1G93A/Tbk1+/− mice and respective control siblings. Both strains were bred on the same C57Bl/6 genetic background, and only F1 offspring were analyzed. Tbk1+/− mice had been backcrossed with C57BL/6J mice for >10 generations. Mice were maintained at 22°C with a 14/10-h light/dark cycle and had food and water ad libitum. All animal experiments were performed in accordance with institutional guidelines of the University of Ulm and were approved by the local authority (Regierungspräsidium Tübingen, Germany; animal permission no. 1253).
Genotyping
Genotyping of the SOD1G93A transgene was performed according to the Jackson Laboratory standard PCR protocol (https://www.jax.org/strain/004435). Genotyping of Tbk1-KO was performed as previously described (Möser et al., 2015).
Scoring and motor testing
Twice a week, male mice were subjected to weighing and disease scoring. The mice were evaluated for signs of motor deficit using the following five-stage point scoring system adapted from Weydt et al. (2003): stage 1, manifest hind limb tremor when suspended by the tail; stage 2, manifest gait abnormalities; stage 3, manifest paralysis of one hind limb; stage 4, manifest paralysis of both hind limbs; and stage 5, end stage. The onset (stage 1) was defined retrospectively as the earliest time when the mice showed symptoms for at least two consecutive weeks. The end-stage was determined as the inability to rise within 30 s after being placed on its side. The latency to fall off the rotarod was tested weekly from P50 until end stage by a blinded experimenter using a Rotarod for five mice with tactile user interface (#LE8205; BIOSEB). Rotarod testing consisted of consecutive three rounds per mouse; only the maximum value per day was considered. The start speed of the rod was adjusted to 4 rpm and was accelerated to the maximum speed of 40 rpm during 300 s.
Tissue preparation
At the described time points, mice were deeply anesthetized by i.p. injection of a ketamine/Rompun mixture and were transcardially perfused with 20 ml PBS and 20 ml of 4% paraformaldehyde for fixation. Spinal cords and muscles were fixed overnight with 4% paraformaldehyde, then dehydrated in 30% sucrose (Sigma-Aldrich) in PBS for 48 h at 4°C, embedded in Tissue-Tek O.C.T. Compound (Sakura), and stored at −80°C until use. Embedded spinal cords were sectioned into 12-µm coronal slices using a cryotome (Leica). Serial sections covering the whole LSC were obtained from each animal. Every 10th section was chosen for quantification of anterior horn MNs (total eight sections). Every 20th section was used for quantification of microglia and astrocytes (total four sections each). Pretibial muscles were sectioned into 25-µm longitudinal slices. At least 300 NMJs were recorded per genotype. Mice whose tissue was used for protein analysis were killed by decapitation. The extracted tissue was immediately transferred to liquid nitrogen and stored at −80°C until use.
Immunohistochemistry
Transverse sections of the spinal cord (12 µm thick) were cut using a cryotome. Sections were blocked for 1 h using a permeabilization/blocking solution containing Tris-buffered saline (TBS) with 5% FCS, and 0.25% Triton X-100 (Sigma-Aldrich). After washing once with TBS, sections were stained with combinations of mouse anti-NeuN (1:500; Millipore), goat anti-ChAT (1:100; Millipore), NeuroTrace 640/660 Deep-Red Fluorescent Nissl Stain (1:100; Invitrogen), mouse anti-p62 (1:500; Abcam), rabbit anti-p62 (1:2,000; MBL), rabbit anti-GABARAPL1 (1:1,000; Proteintech), mouse anti-polyubiquitin (1:500; Enzo), rat anti-Clec7a (1:30; InvivoGen), mouse anti-misfolded human SOD1 clone B8H10 (1:250; Medimabs), rabbit anti-Iba1 (1:500; Wako), goat anti-Iba1 (1:1,000; Abcam), rat anti-CD68 (1:100; Bio-Rad), rabbit anti-GFAP (1:750; Abcam), chicken anti-GFAP (1:1,000; Abcam), α-bungarotoxin 488 (Invitrogen; 1:1,000), mouse anti-Neurofilament marker (SMI-32; 1:1,000; BioLegend), and anti-Synaptophysin (1:1,000; Abcam). Antibodies were diluted in TBS containing 0.25% Triton X-100 and 5% horse serum. Sections were incubated with the primary antibody for 12–72 h at 4°C, washed three times with TBS, and incubated with the secondary antibodies in TBS containing 0.25% Triton X-100 and 5% horse serum for 1 h at room temperature while protected from light. Secondary antibodies used for immunofluorescence were donkey anti-rat/rabbit/mouse Alexa Fluor 488/546/647 (1:750; Invitrogen). Sections were then washed three times with TBS and coverslipped in Fluoromount G (Southern Biotech).
Image and data analysis
Sections stained against MNs were recorded with an Axio Observer.A1 microscope (Zeiss). Immunofluorescent muscle and spinal cord sections were recorded with a TCS SP8 confocal laser scanning microscope (Leica), using the same acquisition settings for every section. For stereological analysis, investigators were blind to the genetic background of the animals. Nissl+ ChAT+ cells on sections were counted manually with the ImageJ Cell Counter Plugin (National Institutes of Health). For analysis of Iba1, CD68, Clec7a, and GFAP staining, the Iba1/CD68/Clec7a/GFAP-positive area within the LSC was measured with the ImageJ Threshold Color Plugin and “Analyze Particles” function. For analysis of p62/GABARAPL1+ aggregates, the p62/GABARAPL1+ area per ChAT+ MN was measured using the ImageJ “ROI Manager” function and the ImageJ Threshold Color Plugin. For analysis of misfolded human SOD1G93A, the mean fluorescence intensity of ChAT+ MN was measured using the ImageJ ROI Manager and Analyze Particles functions. For analysis of NMJs, 25 µm z-stacks (3-µm step size) of the pretibial muscles were recorded. The innervation of NMJs was assessed manually by a blinded investigator by comparing the overlap of the presynapse (neurofilaments, synaptophysin) and the postsynapse (α-bungarotoxin; fully innervated, partially innervated, fully denervated) using the ImageJ “Maximum projection” function. NMJs were assessed as “denervated” when presynaptic synaptophysin/neurofilament staining was completely absent.
Primary MN culture
Murine embryonic spinal MNs were isolated and cultured as previously described (Wiese et al., 2010). Briefly, after dissection of the ventrolateral part of E12.5 embryos, spinal cord tissues were incubated for 15 min in 0.05% trypsin in HBSS. Cells were triturated and incubated in Neurobasal medium (Invitrogen), supplemented with 1× Glutamax (Invitrogen) on Nunclon plates (Nunc) precoated with antibodies against the p75 NGF receptor (MLR2; kind gift of Robert Rush, Flinders University, Adelaide, Australia) for 45 min. Plates were washed with Neurobasal medium, and the remaining MNs were recovered from the plate with depolarization solution (0.8% NaCl, 35 mM KCl and 2 mM CaCl2) and collected in full medium (2% horse serum and 1× B27 in Neurobasal medium with 1× Glutamax). After counting, the cell number was adjusted to 1,000 in 100 µl, and 1,000 cells were plated per well on four-well dishes (Greiner; Cellstar) precoated with poly-ornithine/laminin (Invitrogen). Cells were cultured in the presence of brain-derived neurotrophic factor. Axon length was quantified after 7 d in vitro.
Primary astrocyte and microglia culture
Primary cells were prepared from P1-4 Tbk1+/− pups and WT siblings. Microglia were prepared as previously described (Wiesner et al., 2013). Substances and solutions were from Gibco or Sigma-Aldrich. In brief, for microglia, forebrains were digested and dissociated. All cells were seeded in T75 cell culture flasks in supplemented DMEM (Gibco). After 7–10 d in culture, microglia were manually shaken off the astrocyte layer and seeded on 6-well plates with 6 × 105 cells. Microglia cells were harvested up to three times from the astrocyte co-culture. Subsequently, astrocytes were washed from remaining microglia cells and seeded on 6-well plates with 105 cells. Microglia were stimulated with LPS 50 ng/ml for 6 h.
Western blotting
Urea lysis buffer (8 M urea, 10 mM Tris, and 50 mM NaH2PO4, pH 8.0) and TissueLyser II (Qiagen) were used to extract protein from spinal cord tissue. Immunoblotting was performed according to standard procedures, using a total protein amount of 20–30 µg per sample and the XCell II Blot Module system (Thermo Fisher Scientific). The following antibodies were used: rabbit anti-GAPDH (1:1,000; Proteintech), rabbit anti-β-actin (1:1,000; Cell Signaling), rabbit anti-LC3B (1:1,000; Cell Signaling), rabbit anti-LC3B (1:1,000; MBL), mouse anti-SQSTM1/p62 (1:1,000; Abcam), rabbit anti-TBK1 (1:1,000; Thermo Fisher Scientific), rabbit anti-SOD1 (1:1,000, Enzo Life Science), goat anti-mouse-HRP (1:1,000, Life Technologies), and goat anti-rabbit-HRP (1:1,000, Life Technologies). Membranes were developed using the Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and the FUSION SOLO S (Peqlab) system.
Real-time PCR
Total RNA was isolated from primary glial cultures using the RNeasy Plus Micro kit (#73404; Qiagen). Reverse transcription reactions were performed with the QuantiTect Reverse Transcription Kit (#205311; Qiagen), according to the manufacturer`s instructions. Subsequent PCR reactions were run in duplicate on a CFX96 real-time system (Bio-Rad), using the QuantiTect SYBR Green PCR Kit (#204143; Qiagen) and QuantiTect Primer Assays (Qiagen). The following QuantiTect primer assays were used: Ywhaz (#QT00105350), Tbp (#QT00198443), C1qa (#QT01660778), Ccl2 (#QT00167832), Ccl3 (#QT02589426), Ccl4 (#QT00154616), Ilb1 (#01048355), Il6 (#QT00098875), Il10 (#QT00106169), Tnf (#QT00104006), Irf7 (#QT00245266), Irf9 (QT01048698, Stat2 (#QT00160216), Isg15 (#QT01772876), Ifnb1 (#QT00249662), Stat1 (#QT00162183), and Nos2 (#QT00100275). The resulting Ct values were normalized to two housekeeping genes (Ywhaz and Tbp) using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Quantitative NanoString nCounter mRNA expression analysis
Total RNA was isolated from spinal cords of P140 mice using the RNeasy Lipid Tissue Mini Kit (Qiagen), according to the manufacturer’s instructions. Multiplexed mRNA analysis of 800 genes (nCounter Mouse Neuroinflammation Panel plus 30 additional genes [mainly known ALS genes]) was performed using the nCounter platform. The Cell Type Profiling Module of the nCounter Advanced Analysis 2.0 plugin for the nSolver software (NanoString) was used to measure the abundance of various cell populations according to Danaher et al. (2017). The method quantifies cell populations using marker genes that are expressed stably and specifically in given cell types (nCounter Advanced Analysis 2.0 User Manual MAN-10030-03).
Statistics
For comparison of multiple groups, the statistical significance of endpoints was evaluated by one-way ANOVA followed by Tukey’s multiple comparisons post hoc test when data were normally distributed. When data were not normally distributed, statistical significance of endpoints was evaluated by Kruskal-Wallis test followed by Dunn’s multiple comparisons post hoc test. For comparison of two groups and normal distribution of data, the unpaired two-tailed Student’s t test was used. Kaplan-Meier plots were analyzed using the log-rank (Mantel-Cox) test. See the analysis scheme in the supplemental dataset for statistical methods used for analysis of the NanoString nCounter mRNA data. Data are presented as means ± SEM in bar graphs. Statistical significance was reported by the P value of the statistical test procedures and was assessed as significant (*, P < 0.05), strongly significant (**, P < 0.01), or highly significant (***, P < 0.001; ****, P < 0.0001). All statistical analyses were performed with Prism software (version 7.02; GraphPad Software).
Online supplemental material
Fig. S1 shows Kaplan-Meier plots for weight loss and maximum as well as half-maximum latency to fall in the rotarod test and a bar graph displaying disease duration of SOD1G93A/Tbk1+/− compared with SOD1G93A mice. Fig. S2 shows analysis of protein levels of SOD1 and autophagy markers in spinal cord lysates and shows representative pictures of intra- and extramotorneural protein inclusions at P50 and P120. Furthermore, it shows quantification of astrocytes and microglia at P50.
Acknowledgments
We thank Christine V. Möser and Ellen Niederberger (Pharmazentrum Frankfurt/Zentrum für Arzneimittelforschung, Entwicklung und–Sicherheit, Institut für Klinische Pharmakologie, Klinikum der Goethe-Universität, Frankfurt, Germany) for sharing of mice and protocols.
This work was funded by the Baustein-Programm of the Medical Faculty of the University of Ulm (LSBR.0030 to D. Brenner); by The Bruno and Ilse Frick Foundation for ALS Research (award 2015 to J.H. Weishaupt); and by the Association pour la Recherche sur la Sclérose latérale amyotrophique et autres maladies du motoneurone (ARSla, France) and l’Aide à la Recherche des Maladies du Cerveau (ARMC, France) (to C. Lobsiger and S. Boillée).
The authors declare no competing financial interests.
Author contributions: D. Brenner designed and performed experiments and wrote the manuscript. P. Lüningschrör, E. Buck, and A. Freischmidt designed and performed experiments and revised the manuscript. K. Sieverding, C. Bruno, S. Mungwa, L. Fischer, C. Bliederhäuser, J. Ulmer, S. Brockmann, and C. Philibert performed experiments. B. Mayer performed the statistical analyses. T. Satoh and S. Akira created and provided the B6.129P2-Tbk1tm1Aki mice. K. Danzer, S. Boillée, M. Sendtner, and A. Ludolph designed the experiments and revised the manuscript. J. Weishaupt and C. Lobsiger designed the experiments and wrote the manuscript.
References
- Ahmad L., Zhang S.-Y., Casanova J.-L., and Sancho-Shimizu V.. 2016. Human TBK1: A Gatekeeper of Neuroinflammation. Trends Mol. Med. 22:511–527. 10.1016/j.molmed.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers D.R., Henkel J.S., Xiao Q., Zhao W., Wang J., Yen A.A., Siklos L., McKercher S.R., and Appel S.H.. 2006. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 103:16021–16026. 10.1073/pnas.0607423103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillée S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., and Cleveland D.W.. 2006. Onset and Progression in Inherited ALS Determined by MNs and Microglia. Science. 312:1389–1392. [DOI] [PubMed] [Google Scholar]
- Bonnard M., Mirtsos C., Suzuki S., Graham K., Huang J., Ng M., Itié A., Wakeham A., Shahinian A., Henzel W.J., et al. 2000. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19:4976–4985. 10.1093/emboj/19.18.4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E.T., Lasseigne B.N., Petrovski S., Sapp P.C., Dion P.A., Leblond C.S., Couthouis J., Lu Y.-F., Wang Q., Krueger B.J., et al. 2015. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 347:1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaher P., Warren S., Dennis L., D’Amico L., White A., Disis M.L., Geller M.A., Odunsi K., Beechem J., and Fling S.P.. 2017. Gene expression markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer. 5:18 10.1186/s40425-017-0215-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B., Amiel E., Huang S.C.-C., Smith A.M., Chang C.-H., Lam W.Y., Redmann V., Freitas T.C., Blagih J., van der Windt G.J.W., et al. 2014. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15:323–332. 10.1038/ni.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A., Müller K., Zondler L., Weydt P., Mayer B., von Arnim C.A.F., Hübers A., Dorst J., Otto M., Holzmann K., et al. 2015. Serum microRNAs in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 36:2660.e15–2660.e20. 10.1016/j.neurobiolaging.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hasan M., Dobbs N., Khan S., White M.A., Wakeland E.K., Li Q.Z., and Yan N.. 2015. Cutting Edge: Inhibiting TBK1 by Compound II Ameliorates Autoimmune Disease in Mice. J. Immunol. 195:4573–4577. 10.4049/jimmunol.1500162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M., Gonugunta V.K., Dobbs N., Ali A., Palchik G., Calvaruso M.A., DeBerardinis R.J., and Yan N.. 2017. Chronic innate immune activation of TBK1 suppresses mTORC1 activity and dysregulates cellular metabolism. Proc. Natl. Acad. Sci. USA. 114:746–751. 10.1073/pnas.1611113114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H., Takeuchi O., Sato S., Yamamoto M., Kaisho T., Sanjo H., Kawai T., Hoshino K., Takeda K., and Akira S.. 2004. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650. 10.1084/jem.20040520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtman I.R., Raj D.D., Miller J.A., Schaafsma W., Yin Z., Brouwer N., Wes P.D., Möller T., Orre M., Kamphuis W., et al. 2015. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol. Commun. 3:31 10.1186/s40478-015-0203-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Xiao Y., Chang J.-H., Yu J., Hu H., Starr R., Brittain G.C., Chang M., Cheng X., and Sun S.-C.. 2012. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-κB signaling. Nat. Immunol. 13:1101–1109. 10.1038/ni.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korac J., Schaeffer V., Kovacevic I., Clement A.M., Jungblut B., Behl C., Terzic J., and Dikic I.. 2013. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 126:580–592. 10.1242/jcs.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand J.N., Bon K., Fraichard A., Zhang J., Jouvenot M., Risold P.-Y., Boyer-Guittaut M., and Delage-Mourroux R.. 2013. Specific distribution of the autophagic protein GABARAPL1/GEC1 in the developing and adult mouse brain and identification of neuronal populations expressing GABARAPL1/GEC1. PLoS One. 8:e63133 10.1371/journal.pone.0063133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. [DOI] [PubMed] [Google Scholar]
- Möser C.V., Stephan H., Altenrath K., Kynast K.L., Russe O.Q., Olbrich K., Geisslinger G., and Niederberger E.. 2015. TANK-binding kinase 1 (TBK1) modulates inflammatory hyperalgesia by regulating MAP kinases and NF-κB dependent genes. J. Neuroinflammation. 12:100 10.1186/s12974-015-0319-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif M., Valenzuela V., Rojas-Rivera D., Vidal R., Matus S., Castillo K., Fuentealba Y., Kroemer G., Levine B., and Hetz C.. 2014. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 10:1256–1271. 10.4161/auto.28784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y.-H., Torres M., Ram R., Formstecher E., Roland C., Cheng T., Brekken R., Wurz R., Tasker A., Polverino T., et al. 2011. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell. 41:458–470. 10.1016/j.molcel.2011.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T., and Rothstein J.D.. 2015. Rodent models of amyotrophic lateral sclerosis. Curr. Protoc. Pharmacol. 69:5.67.1–5.67.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picher-Martel V., Valdmanis P.N., Gould P.V., Julien J.-P., and Dupré N.. 2016. From animal models to human disease: a genetic approach for personalized medicine in ALS. Acta Neuropathol. Commun. 4:70 10.1186/s40478-016-0340-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S., Nguyen J., Johnson J., Haura E., Coppola D., and Chellappan S.. 2015. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat. Commun. 6:10072 10.1038/ncomms10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M.A., Dupont N., Ornatowski W., Jiang S., Bradfute S.B., et al. 2012. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 37:223–234. 10.1016/j.immuni.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier C., Bieniek K.F., Finch N., van de Vorst M., Baker M., Perkersen R., Brown P., Ravenscroft T., van Blitterswijk M., Nicholson A.M., et al. 2015. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 130:77–92. 10.1007/s00401-015-1436-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly S.M., Chiang S.-H., Decker S.J., Chang L., Uhm M., Larsen M.J., Rubin J.R., Mowers J., White N.M., Hochberg I., et al. 2013. An inhibitor of the protein kinases TBK1 and IKK-ɛ improves obesity-related metabolic dysfunctions in mice. Nat. Med. 19:313–321. 10.1038/nm.3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly S.M., Ahmadian M., Zamarron B.F., Chang L., Uhm M., Poirier B., Peng X., Krause D.M., Korytnaya E., Neidert A., et al. 2015. A subcutaneous adipose tissue-liver signalling axis controls hepatic gluconeogenesis. Nat. Commun. 6:6047 10.1038/ncomms7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick N.D., Griffey C.J., Guarnieri P., Gerbino V., Wang X., Piersaint J.A., Tapia J.C., Rich M.M., and Maniatis T.. 2017. Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc. Natl. Acad. Sci. USA. 114:E8294–E8303. 10.1073/pnas.1704294114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda E., Brännström T., Andersen P.M., and Marklund S.L.. 2016. Low autophagy capacity implicated in motor system vulnerability to mutant superoxide dismutase. Acta Neuropathol. Commun. 4:6 10.1186/s40478-016-0274-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207:2053–2063. 10.1084/jem.20101664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Yang B., and Zhang D.. 2011. Activation of interferon signaling pathways in spinal cord astrocytes from an ALS mouse model. Glia. 59:946–958. 10.1002/glia.21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., and Elazar Z.. 2011. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci. Signal. 4:pe39 10.1126/scisignal.2002355 [DOI] [PubMed] [Google Scholar]
- Weydt P., Hong S.Y., Kliot M., and Möller T.. 2003. Assessing disease onset and progression in the SOD1 mouse model of ALS. Neuroreport. 14:1051–1054. 10.1097/01.wnr.0000073685.00308.89 [DOI] [PubMed] [Google Scholar]
- Wiese S., Herrmann T., Drepper C., Jablonka S., Funk N., Klausmeyer A., Rogers M.-L., Rush R., and Sendtner M.. 2010. Isolation and enrichment of embryonic mouse motoneurons from the lumbar spinal cord of individual mouse embryos. Nat. Protoc. 5:31–38. 10.1038/nprot.2009.193 [DOI] [PubMed] [Google Scholar]
- Wiesner D., Merdian I., Lewerenz J., Ludolph A.C., Dupuis L., and Witting A.. 2013. Fumaric acid esters stimulate astrocytic VEGF expression through HIF-1α and Nrf2. PLoS One. 8:e76670 10.1371/journal.pone.0076670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., Richter B., Korac J., Waidmann O., Choudhary C., et al. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 333:228–233. 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Jin T., Zhu H., Chen H., Ofengeim D., Zou C., Mifflin L., Pan L., Amin P., Li W., et al. 2018. TBK1 Suppresses RIPK1-Driven Apoptosis and Inflammation during Development and in Aging. Cell. 174:1477–1491.e19. 10.1016/j.cell.2018.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., and Cleveland D.W.. 2008. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 11:251–253. 10.1038/nn2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhou X., Chang M., Nakaya M., Chang J.-H., Xiao Y., Lindsey J.W., Dorta-Estremera S., Cao W., Zal A., et al. 2015. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 6:6074 10.1038/ncomms7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Wong K.I., Sun X., Reilly S.M., Uhm M., Liao Z., Skorobogatko Y., and Saltiel A.R.. 2018. TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell. 172:731–743.e12. 10.1016/j.cell.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]