3K3A-APC exerts beneficial therapeutic effects in models of neurological disorders and is currently in development for human stroke. The authors show that 3K3A-APC inhibits BACE1 amyloidogenic pathway in mice, suggesting 3K3A-APC is an effective antiamyloid prevention therapy for Alzheimer’s disease.
Abstract
3K3A-activated protein C (APC), a cell-signaling analogue of endogenous blood serine protease APC, exerts vasculoprotective, neuroprotective, and anti-inflammatory activities in rodent models of stroke, brain injury, and neurodegenerative disorders. 3K3A-APC is currently in development as a neuroprotectant in patients with ischemic stroke. Here, we report that 3K3A-APC inhibits BACE1 amyloidogenic pathway in a mouse model of Alzheimer’s disease (AD). We show that a 4-mo daily treatment of 3-mo-old 5XFAD mice with murine recombinant 3K3A-APC (100 µg/kg/d i.p.) prevents development of parenchymal and cerebrovascular amyloid-β (Aβ) deposits by 40–50%, which is mediated through NFκB–dependent transcriptional inhibition of BACE1, resulting in blockade of Aβ generation in neurons overexpressing human Aβ-precursor protein. Consistent with reduced Aβ deposition, 3K3A-APC normalized hippocampus-dependent behavioral deficits and cerebral blood flow responses, improved cerebrovascular integrity, and diminished neuroinflammatory responses. Our data suggest that 3K3A-APC holds potential as an effective anti-Aβ prevention therapy for early-stage AD.
Introduction
Activated protein C (APC) is an endogenous blood serine protease with anticoagulant and cytoprotective activities mediated by the activation of protease-activated receptor 1 (PAR1) via noncanonical cleavage (Griffin et al., 2018). APC and its cytoprotective analogues exert beneficial effects in preclinical rodent models of stroke (Cheng et al., 2003, 2006; Liu et al., 2004; Zlokovic et al., 2005; Thiyagarajan et al., 2008; Guo et al., 2009a,b; Wang et al., 2009, 2012, 2013, 2016; Sinha et al., 2018), brain trauma (Petraglia et al., 2010; Walker et al., 2010), multiple sclerosis (MS; Han et al., 2008), amyotrophic lateral sclerosis (ALS; Zhong et al., 2009), and systemic models of sepsis; ischemia–reperfusion injury of the heart, kidney, and liver; and diabetes, organ transplants, wound healing, and total body radiation (Griffin et al., 2015, 2018).
3K3A-APC (Lys191–193Ala), a recombinant variant of APC in which three Lys residues (KKK191–193) were replaced with alanine, was engineered to lower APC-associated bleeding risk by reducing APC’s anticoagulant activity by >90% (Mosnier et al., 2004) while retaining normal cytoprotective and cell-signaling activities (Mosnier et al., 2007; Guo et al., 2013; Wang et al., 2016). 3K3A-APC has beneficial therapeutic effects in models of stroke (Guo et al., 2009a,b; Wang et al., 2012, 2013; Sinha et al., 2018), brain trauma (Walker et al., 2010), ALS (Zhong et al., 2009), and MS (Han et al., 2008). It directly protects neurons from divergent inducers of apoptosis via PAR1 and PAR3 (Guo et al., 2009a) similar to wild-type APC (Guo et al., 2004). Cell-signaling APC analogues, including 3K3A-APC, also protect brain endothelium and blood–brain barrier (BBB) integrity from different types of injury by inhibiting apoptosis of endothelial cells and promoting Rac1-dependent stabilization of the endothelial cytoskeleton, which requires PAR1 and endothelial protein C receptor (Guo et al., 2009a,b; Zlokovic and Griffin, 2011; Amar et al., 2018; Griffin et al., 2018). Additionally, APC and 3K3A-APC have potent anti-inflammatory activity (Griffin et al., 2015, 2018). In the nervous system, APC’s cytoprotective analogues suppress microglia activation via PAR1 and inhibit expression of proinflammatory cytokines (Zhong et al., 2009; Zlokovic and Griffin, 2011; Griffin et al., 2018).
3K3A-APC successfully meets the Stroke Therapy Academic Industry Roundtable criteria for preclinical drug assessment for all studied parameters (Zlokovic and Griffin, 2011) and has an established safety and pharmacokinetic profile in human volunteers (Williams et al., 2012; Lyden et al., 2013). Moreover, a recent phase 2a RHAPSODY trial in ischemic stroke patients found that 3K3A-APC is safe, well tolerated, and tends to reduce intracerebral bleeding (Lyden et al., 2016, 2018), consistent with its vasculoprotective effects in animal models (Griffin et al., 2015, 2016; Amar et al., 2018).
Because of its neuroprotective, vasculoprotective, and anti-inflammatory activities in multiple models of neurological disorders, we investigated whether 3K3A-APC can also protect the brain from toxic effects of Alzheimer’s amyloid-β (Aβ) toxin in a mouse model of Alzheimer’s disease (AD). Thus, we administered 3K3A-APC daily (100 µg/kg/d i.p.) for 4 mo in 3-mo-old 5XFAD mice, which overexpress five autosomal dominant AD mutations in neurons, including Swedish, London, and Florida Aβ-precursor protein (APP) mutations and presenilin 1 (PSEN1) M146L and L286V mutations (Oakley et al., 2006). 5XFAD mice begin depositing Alzheimer’s Aβ very early, at 1–2 mo of age, which quickly leads to cognitive dysfunction (Oakley et al., 2006), BBB breakdown (Kook et al., 2012; Giannoni et al., 2016; Park et al., 2017), and neuroinflammation (Oakley et al., 2006; Giannoni et al., 2016) and later in life to loss of neurons and neurodegenerative changes (Oakley et al., 2006). Surprisingly, we found that 3K3A-APC prevents development of amyloid deposits by inhibiting β-secretase 1 (BACE1), the rate-limiting enzyme in the amyloidogenic pathway (Vassar, 2014), in neurons, which slowed down generation of Aβ. Consistent with diminished Aβ pathology, we also found that 3K3A-APC normalized hippocampus-dependent behavioral deficits and cerebral blood flow (CBF) responses, improved cerebrovascular integrity, and diminished neuroinflammatory responses.
Results and discussion
3K3A-APC prevents development of Aβ pathology in 5XFAD mice
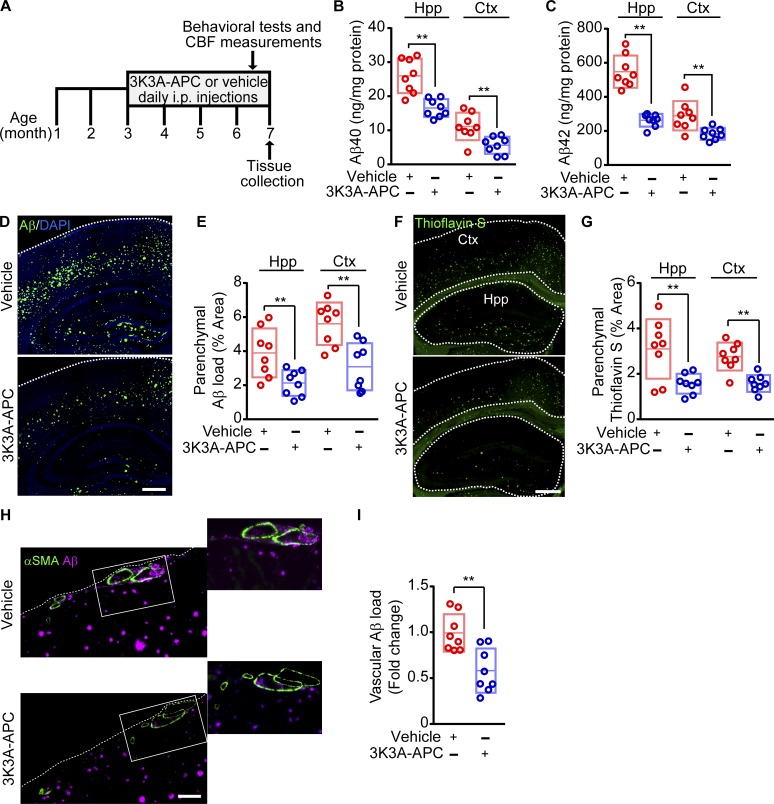
We treated 3-mo-old 5XFAD mice with murine recombinant 3K3A-APC daily (100 µg/kg/d i.p.) or vehicle (control) for 4 mo (Fig. 1 A). The 3K3A-APC dosing regimen was within a range of previously reported treatment with APC’s cell-signaling analogues in models of other neurodegenerative disorders such as ALS (Zhong et al., 2009; Winkler et al., 2014) and MS (Han et al., 2008). First, we examined whether 3K3A-APC treatment influences the development of Aβ pathology. We found that 3K3A-APC compared with vehicle effectively inhibited parenchymal accumulation of Aβ40 (36% and 51% reductions) and Aβ42 (52% and 36% reductions) in the hippocampus and cortex, respectively (Fig. 1, B and C) and reduced by 40–50% total Aβ load (Fig. 1, D and E) and thioflavin S (ThS)–positive amyloid load (Fig. 1, F and G) in the hippocampus and cortex. Compared with vehicle, 3K3A-APC also reduced Aβ load in the vessel wall of the cortical pial arteries and penetrating arterioles by >40%, suggesting delayed development of cerebrovascular Aβ pathology and cerebral amyloid angiopathy (CAA; Fig. 1, H and I).
Figure 1.
3K3A-APC prevents development of Aβ pathology in 5XFAD mice. (A) Dosing paradigm. 3K3A-APC (100 µg/kg/d i.p.) or vehicle (saline) was administered to 5XFAD mice for 4 mo beginning at 3 mo of age. Behavioral tests, CBF responses to whisker stimulation, and tissue collection were performed as indicated. (B and C) Aβ40 (B) and Aβ42 (C) levels in the hippocampus (Hpp) and cortex (Ctx) in vehicle-treated and 3K3A-APC–treated 5XFAD mice. (D and E) Representative images of Aβ immunostaining (coronal sections, D), and quantification of Aβ load in the hippocampus and cortex (E) in 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 500 µm (D). (F and G) Representative images of ThS-positive amyloid deposits (coronal sections; F) and quantification of ThS amyloid load in the hippocampus and cortex (G) in 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 500 µm (F). (H and I) Representative images of Aβ vascular load in small pial arteries and penetrating cortical arterioles (≥20 µm) expressed as Aβ-positive area of αSMA-positive area of the vessel wall (H), and quantification of vascular Aβ load (I) in 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 100 µm (H). In B, C, E, G, and I, data are shown as single points per mouse with boxplots representing means ± SD; n = 8 mice per group. **, P < 0.01 (statistical significance by two-tailed Student’s t test).
Previous work in 5XFAD mice showed lower Aβ load in the hippocampus compared with cortex (Jawhar et al., 2012) despite higher Aβ levels (Kim et al., 2018), consistent with the present findings. It is not entirely clear why Aβ load is lower in the hippocampus when total Aβ levels after guanidine extraction are higher than in cortex. It is possible, however, that the type of Aβ deposits in these two regions can explain this discrepancy; namely, 5XFAD mice have been shown to develop an even number of diffuse and dense Aβ plaques in the hippocampus, whereas in the cortex, the number of diffuse Aβ deposits with lower Aβ content compared with the dense Aβ plaques is twofold higher (Crouzin et al., 2013). After guanidine extraction, a greater ratio of dense compared with diffuse Aβ deposits could yield higher Aβ peptide levels in the hippocampus than in the cortex, consistent with our ELISA measurements (Fig. 1, B and C).
3K3A-APC inhibits amyloidogenic BACE1 pathway in 5XFAD neurons
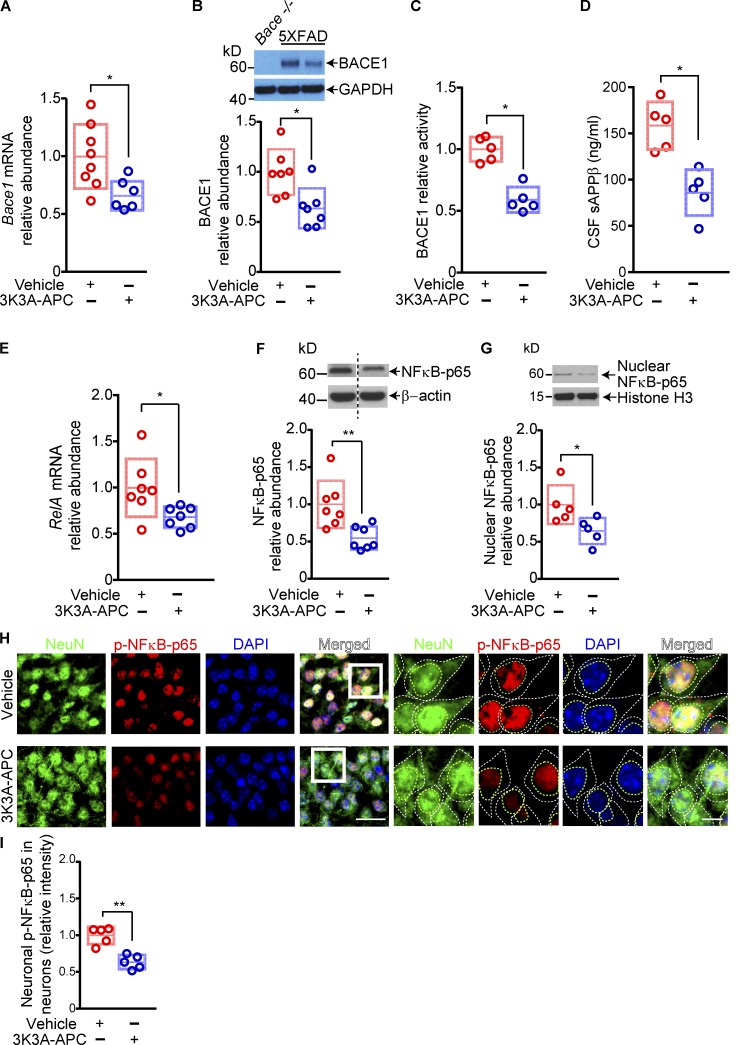
Next, we asked how 3K3A-APC influences development of Aβ pathology. To address this question, we studied whether 3K3A-APC affects APP processing, Aβ clearance, or both. First, we showed that 3K3A-APC treatment slightly, but not significantly (P > 0.05), increased APP levels but did not alter γ-secretase activity, mRNA and protein levels of a disintegrin and metalloproteinase domain–containing protein 10 (ADAM10), an α-secretase responsible for the ectodomain shedding of APP in the mouse brain (Jorissen et al., 2010; Kuhn et al., 2010; Suh et al., 2013), or the Aβ-degrading enzymes neprilysin and insulin-degrading enzyme (IDE), and it only slightly, but not significantly (P > 0.05), increased the levels of Aβ major clearance receptor at the BBB, the low-density lipoprotein receptor–related protein 1 (LRP1; Shibata et al., 2000; Deane et al., 2004; Fig. S1, A–H). Thus, none of the studied possible pathways above gave us a lead to explain the robust antiamyloidogenic effect of 3K3A-APC that we observed. Unexpectedly, we next found that 3K3A-APC substantially reduced brain cortical mRNA and protein levels of BACE1 (Fig. 2, A and B), which was accompanied by an ∼50% decrease in BACE1 cortical activity and a decrease in sAPPβ (the N-terminal BACE1-cleaved soluble fragment of APP; Vassar, 2014) levels in the cerebrospinal fluid (CSF; Fig. 2, C and D). Altogether, these data suggest that 3K3A-APC transcriptionally inhibits BACE1.
Figure 2.
3K3A-APC inhibits BACE1 and NFκB in 5XFAD mice. (A–C) Relative abundance of Bace1 mRNA (A), BACE1 protein levels (B), and BACE1 activity (C) in the cortex of 5XFAD mice treated with vehicle or 3K3A-APC. The relative abundance of BACE1 mRNA (A) and protein (B) was normalized by housekeeping gene Gapdh mRNA levels or GAPDH protein. The specificity of BACE1 antibody was tested on brain homogenates from Bace−/− (null) mice (B). (D) CSF levels of sAPPβ in 5XFAD mice treated with vehicle or 3K3A-APC. (E–G) Relative abundance of RelA (NFκB-p65 subunit) mRNA (E), total NFκB-p65 (F), and nuclear NFκB-p65 (G) protein levels in the cortex of 5XFAD mice treated with vehicle or 3K3A-APC. In E, RelA (NFκB-p65 subunit) mRNA levels were normalized by Gapdh mRNA. In F and G, β-actin and histone 3 were used as loading controls, respectively. (H and I) Representative images of NeuN-positive nuclei, p-NFκB-p65, and DAPI nuclear staining (H) and quantification of p-NFκB-p65 nuclear immunostaining (I) in hippocampal neurons (CA1 region) of 5XFAD mice treated with vehicle or 3K3A-APC. Bar in H, 25 µm (left) and 5 µm (right). In all panels, data are shown as single points per mouse with boxplots representing means ± SD. In A and E, n = 6–8 mice per group; in B and F, n = 7 mice per group; and in C, D, G, and I, n = 5 mice per group. *, P < 0.05; **, P < 0.01 (statistical significance by two-tailed Student’s t test). See Fig. S3 for full gels for Western blot images used for quantification in B, F, and G.
To understand the molecular mechanism underlying the observed BACE1 transcriptional inhibition by 3K3A-APC, based on literature search and previous findings, we investigated whether 3K3A-APC could influence the relationship between the transcription factor NFκB and BACE1. Namely, some earlier reports found that APC transcriptionally inhibits NFκB (Riewald and Ruf, 2005; Cheng et al., 2006) and also blocks NFκB nuclear translocation (White et al., 2000; Joyce et al., 2001; Yuksel et al., 2002; Riewald and Ruf, 2005; Cheng et al., 2006), and other studies have shown that NFκB transcriptionally regulates BACE1 expression (Buggia-Prevot et al., 2008; Chami and Checler, 2012; Chen et al., 2012). Consistent with these reports, we found that 3K3A-APC treatment of 5XFAD mice greatly reduced cortical levels of RelA mRNA, which encodes the NFκB-p65 subunit (Zhang et al., 2017; Fig. 2 E), as well as total and nuclear NFκB-p65 protein levels by 46% and 35%, respectively (Fig. 2, F and G). Immunohistochemical analysis for phosphorylated NFκB-p65 (p-NFκB-p65), the predominant form of NFκB in the nuclei (Zhang et al., 2017), neuronal marker NeuN (neuronal nuclear antigen), and nuclear staining, indicated a substantial decrease in p-NFκB-p65 nuclear levels in neurons of 3K3A-APC–treated mice compared with vehicle-treated 5XFAD mice, as illustrated in the hippocampus (Fig. 2, H and I). Collectively, these data suggest that 3K3A-APC likely inhibited the NFκB–BACE1 pathway regulating Aβ production in neurons.
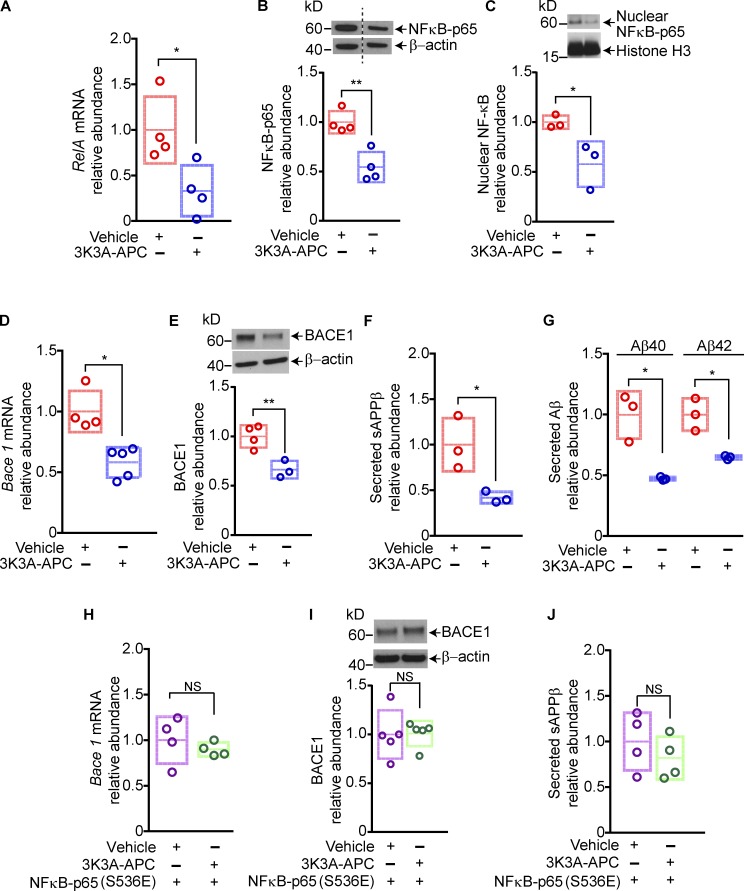
To further validate these findings, we studied the effects of 3K3A-APC on the NFκB–BACE1 pathway in primary cortical neurons isolated from 5XFAD embryos. In these experiments, 3K3A-APC was studied at 5 nM, a concentration moderately higher than the EC50 concentration of 3K3A-APC (3 nM) reported to directly protect cultured mouse cortical neurons from excitotoxic injury (Guo et al., 2009a,b). In agreement with in vivo data, 3K3A-APC suppressed NFκB-p65 expression in 5XFAD primary neurons both at the transcriptional and protein level by 60% and ∼50%, respectively (Fig. 3, A and B), and inhibited nuclear translocation of NFκB-p65 by 45% (Fig. 3 C). Furthermore, 3K3A-APC diminished BACE1 expression at the mRNA and protein level by 35–40% (Fig. 3, D and E), suppressed sAPPβ generation by ∼50%, and substantially reduced neuronal production of Aβ40 and Aβ42 by 53% and 35%, respectively, as shown by reduced levels of secreted Aβ isoforms in the culture medium (Fig. 3, F and G). Finally, we showed that overexpression of the dominant–active NFκB-p65 (S536E) mutant, a phosphomimetic form of NFκB-p65 (Hu et al., 2004), abolished 3K3A-APC–mediated suppression of BACE1 at the mRNA and protein level, as well as its effect on reducing sAPPβ levels (Fig. 3, H–J). Collectively, these in vitro data confirmed that 3K3A-APC inhibited the amyloidogenic Aβ pathway via NFκB-dependent transcriptional inhibition of BACE1.
Figure 3.
3K3A-APC inhibits the amyloidogenic pathway in cortical neurons from 5XFAD mice via NFκB–dependent transcriptional inhibition of BACE1. (A–C) Relative abundance of RelA mRNA (A), total NFκB-p65 (B), and nuclear NFκB-p65 (C) protein levels in primary cortical neurons from 5XFAD mice treated with vehicle or 3K3A-APC (5 nM) for 24 h. In A, RelA (NFκB-p65 subunit) mRNA levels were normalized by Gapdh mRNA; in B and C, β-actin and histone 3 were used as loading controls, respectively. (D–G) Relative abundance of Bace1 mRNA (D), BACE1 protein levels (E), secreted sAPPβ levels (F), and Aβ40 and Aβ42 levels (G) in the culture medium of primary cortical neurons from 5XFAD mice treated with vehicle or 3K3A-APC (5 nM) for 24 h. (H–J) Relative abundance of Bace1 mRNA (H), BACE1 protein (I), and secreted sAPPβ levels in the culture medium (J) in primary cortical neurons from 5XFAD mice transfected with dominant active phosphomimetic mutant form of NFκB-p65 (S536E) and treated with vehicle or 3K3A-APC (5 nM) for 24 h. Cortical neurons were maintained for 14 d in culture before all treatments. In all panels, data are shown as single points per individual culture, with boxplots representing means ± SD; n = 3–5 independent cultures. *, P < 0.05; **, P < 0.01 (statistical significance by two-tailed Student’s t test). See Fig. S3 for full gels for Western blot images used for quantification in B, C, E, and I.
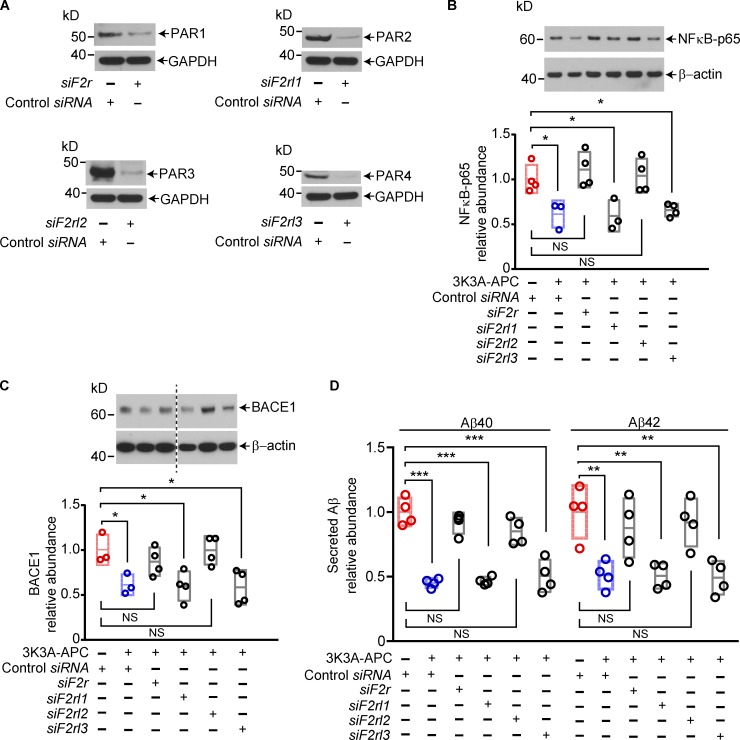
It is well established that PAR1 is critical for APC and 3K3A-APC’s protective effects on cells, including in vivo and in vitro neuroprotective effects (Guo et al., 2004, 2009a, 2013; Wang et al., 2013; Nazir et al., 2017; Griffin et al., 2018; Sinha et al., 2018). Recent studies have shown that APC’s biased signaling following R46, but not R41, cleavage of PAR1 is central to its beneficial protective effects on cells (Mosnier et al., 2014; Nazir et al., 2017), including neuroprotection (Sinha et al., 2018). After silencing PAR1, PAR2, PAR3, and PAR4 in 5XFAD cortical neurons, as previously reported (Guo et al., 2013), we confirmed that PAR1 and PAR3 are required for 3K3A-APC’s inhibition of the NFκB–BACE1 pathway and blockade of Aβ40 and Aβ42 production (Fig. 4), consistent with previous work showing that PAR1 and PAR3 mediate APC and 3K3A-APC’s protective effects on neurons (Guo et al., 2004, 2009a, 2013).
Figure 4.
PAR1 and PAR3 are required for 3K3A-APC's inhibition of the amyloidogenic pathway in cortical neurons from 5XFAD mice. (A) Western blots for PAR1, PAR2, PAR3, and PAR4 in primary cortical neurons from 5XFAD mice 72 h after transfection with siRNA oligonucleotides targeting PAR1 (siF2r), PAR2 (siF2rl1), PAR3 (siF2rl2), and PAR4 (siF2rl3). GAPDH was used as a loading control. (B and C) Relative abundance of total NFκB-p65 (B) and BACE1 (C) protein levels in primary cortical neurons from 5XFAD mice, determined after treatment with vehicle or 3K3A-APC at 5 nM for 24 h after PAR1, PAR2, PAR3, or PAR4 silencing. (D) Secreted Aβ40 and Aβ42 levels in the culture medium from primary cortical neurons from 5XFAD mice after treatment with vehicle or 3K3A-APC at 5 nM for 24 h after PAR1, PAR2, PAR3, or PAR4 silencing. Cortical neurons were maintained for 14 d in culture before silencing PARs. In B and C, β-actin was used as a loading control. In all panels, data are shown as single points per individual culture with boxplots representing means ± SD, n = 3–4 independent cultures. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 (statistical significance by ANOVA). P values were determined by Dunnett’s multiple-comparisons post hoc test (B-D). See Fig. S3 for full gels for Western blot images used for quantification in B and C.
3K3A-APC exerts beneficial functional effects in 5XFAD mice
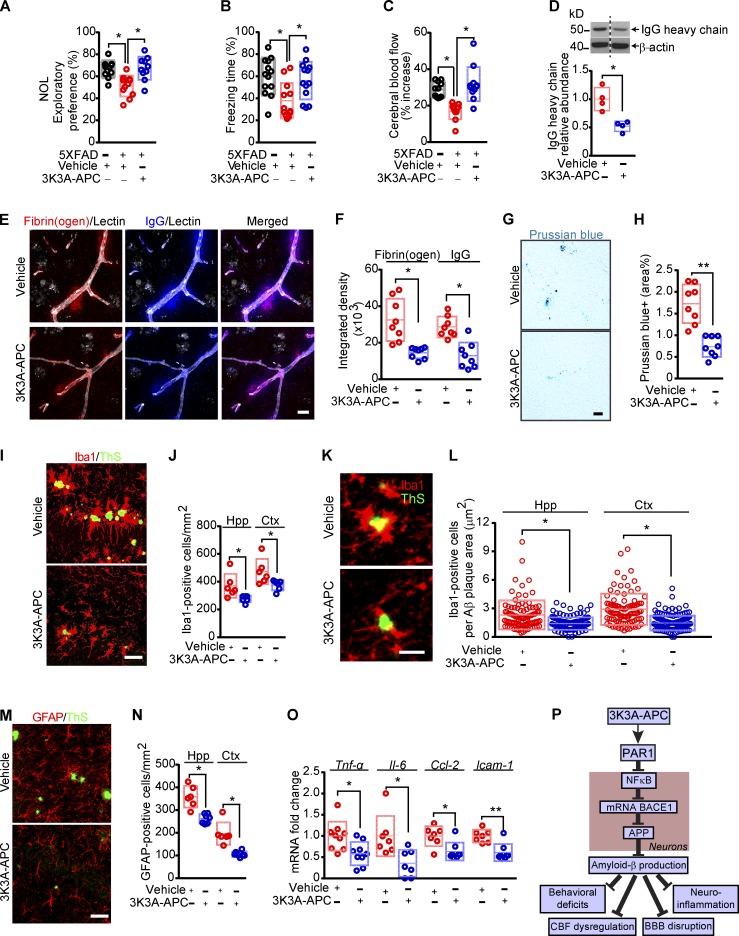
Next, we analyzed the effects of 3K3A-APC on different functional parameters, including hippocampal-dependent behavior, cerebrovascular function, and neuroinflammatory responses. We found that 3K3A-APC completely normalized the performance of 5XFAD mice on novel object location and fear conditioning tests back to control values, as seen in littermate controls (Fig. 5, A and B). Additionally, 3K3A-APC treatment substantially improved CBF responses to whisker stimulation back to control values as in nontransgenic littermates (Fig. 5 C) and improved BBB and cerebrovascular integrity, as shown by 50–60% lower perivascular accumulation of blood-derived immunoglobulin G, fibrinogen, and hemosiderin deposits (Fig. 5, D–H). Consistent with reduced Aβ pathology, 3K3A-APC treatment compared with vehicle suppressed the neuroinflammatory response, as indicated by significantly (P < 0.05) lower numbers of ionized calcium-binding adapter molecule 1 (Iba1)–positive microglia in the hippocampus and cortex tissue (Fig. 5, I and J). Consistent with previously shown APC’s anti-inflammatory activity (Griffin et al., 2018) and reduction in microglia in an ALS model (Zhong et al., 2009), we also found that 3K3A-APC compared with vehicle lowered the microglia count normalized to Aβ plaque load by 39% and 51% in the hippocampus and cortex, respectively (Fig. 5, K and L), suggesting that 3K3A-APC reduces inflammation beyond the reduction in plaque load. 3K3A-APC also reduced the number of glial fibrillary acidic protein (GFAP)–positive astrocytes (Fig. 5, M and N) in the hippocampus and cortex and mRNA levels of proinflammatory cytokines, tumor necrosis factor α (Tnf-α), interleukin 6 (Il-6), chemokine (C-C motif) ligand 2 (Ccl-2), and intercellular adhesion molecule 1 (Icam-1), by 46%, 60%, 36%, and 44%, respectively (Fig. 5 O).
Figure 5.
3K3A-APC beneficial functional effects in 5XFAD mice. (A–C) Novel object location (NOL; A), fear conditioning (B), and CBF responses to whisker stimulation (C) in 5XFAD mice treated with vehicle or 3K3A-APC, compared with nontransgenic littermate controls (black circles). (D) Western blot analyses of IgG heavy chain in cortical homogenates of 5XFAD mice treated with vehicle or 3K3A-APC. (E and F) Confocal representative images (E) and quantification of fibrinogen and IgG extravascular leakage (F) in the somatosensory cortex (layers 2 and 3) of 5XFAD mice treated with 3K3A-APC or vehicle. White, staining for endothelial lectin. Bar, 20 µm (E). (G and H) Representative images of Prussian blue staining (G) and quantification of extravascular Prussian blue–positive hemosiderin deposits (H) in the somatosensory cortex (layers 2 and 3) of 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 20 µm (G). (I and J) Representative images of Iba1-positive microglia (red) and ThS amyloid plaques (green) in the hippocampus CA1 region (I) and quantification of microglia numbers per mm2 in the hippocampus (Hpp) and cortex (Ctx; J) of 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 50 µm (I). (K and L) Representative images of Iba1-positive microglia (red) and ThS-positive amyloid plaques (green) in the hippocampus CA1 region (K) and quantification of microglia numbers expressed per Aβ plaque area in the hippocampus and cortex (L) of 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 25 µm (K). (M and N) Representative images of GFAP-positive astrocytes (red) and ThS-positive amyloid plaques (green) in the hippocampus CA1 region (M) and quantification of astrocyte numbers per mm2 in the hippocampus and cortex (N) of 5XFAD mice treated with vehicle or 3K3A-APC. Bar, 50 µm (M). (O) Relative abundance of Tnf-α, Il-6, Ccl-2, and Icam-1 mRNA levels normalized to the housekeeping Gapdh gene in the cortex of mice treated with vehicle or 3K3A-APC. (P) Diagram illustrating 3K3A-APC inhibition of the amyloidogenic Aβ pathway via NFκB–dependent transcriptional inhibition of BACE1. In A–D, F, H, J, N, and O, data are shown as single points per mouse with boxplots representing means ± SD. In L, individual points are Iba1-positive microglia per μm2 of Aβ plaque area determined from 20 regions of interest per animal from six animals per group, as explained in Materials and methods. In A–C, n = 10–13 mice per group; in D, n = 4 mice per group; in F and H, n = 8 mice per group; in J, L, and N, n = 5–7 mice per group; and in O, n = 6–9 mice per group. *, P < 0.05; **, P < 0.01 (statistical significance by ANOVA or two-tailed Student’s t test). P values were determined by Tukey’s multiple-comparisons post hoc test (A) or Dunn’s multiple-comparisons post hoc test (B and C). See Fig. S3 for full gel for Western blot image used for quantification in D.
Mice treated with 3K3A-APC did not show changes in body weight, physiological parameters (e.g., heart rate, arterial blood pressure, pO2, pCO2, respiration rate, and blood pH), and/or systemic biochemical parameters such as plasma glucose, cholesterol and albumin levels, and liver and kidney analyses (e.g., levels of total protein, alanine aminotransferase, and blood urea nitrogen; Fig. S2).
Discussion of key findings
Our data show that 3K3A-APC blocks neuronal production of Aβ42 and Aβ40 in 5XFAD mice by inhibiting BACE1 amyloidogenic pathway, which prevented development of Aβ parenchymal and cerebrovascular pathology by 40–50% resulting in normalization of hippocampus-dependent memory tasks and CBF responses to neuronal stimulation, improved BBB and cerebrovascular integrity, and suppressed neuroinflammatory responses (Fig. 5 P). This novel mechanism of 3K3A-APC anti-amyloidogenic action adds to the spectrum of its multiple beneficial neuroprotective, vasculoprotective, and anti-inflammatory effects in the nervous system previously shown in models of acute brain injury, such as stroke (Guo et al., 2009a,b; Wang et al., 2012, 2013; Sinha et al., 2018) and brain trauma (Walker et al., 2010), and chronic neurodegenerative disorders such as ALS (Zhong et al., 2009) and MS (Han et al., 2008). Since 3K3A-APC is safe and well tolerated in humans (Williams et al., 2012; Lyden et al., 2013) including stroke patients (Lyden et al., 2018), the present data support that 3K3A-APC holds potential as an effective anti-Aβ therapy for early-stage AD in humans.
The present study did not find that 3K3A-APC treatment affected any other pathway implicated in APP processing and/or Aβ clearance besides BACE1 inhibition. An earlier study reported that wild-type human APC at higher concentrations can increase ADAM10 levels in human neuroblastoma cell line (Li et al., 2014), which may inhibit Aβ production through α-secretase activity (Jorissen et al., 2010; Kuhn et al., 2010; Suh et al., 2013). This finding, however, has not been replicated in the present study. The reason for the difference between previous and the present study is not clear, but might reflect differential species-dependent activities between wild-type anticoagulant human plasma-derived APC used in a previous study, and a recombinant cell-signaling specific murine 3K3A-APC used in the present study.
BACE1 is the rate-limiting enzyme in the amyloidogenic pathway generating synaptotoxic Aβ species and therefore one of the prime drug targets for reducing the levels of Aβ in the AD brain (Vassar, 2014). Genetic studies have shown that complete BACE1 knockout in 5XFAD mice protects from Aβ42 accumulation, prevents neuron loss, and rescues memory deficits in 18-mo-old mice (Ohno et al., 2007). Even deleting a single copy of the BACE1 gene in 5XFAD mice reduced accumulation of Aβ42 and Aβ40 by 50–60% and completely normalized hippocampus-dependent memory tasks in 6-mo-old mice (Kimura et al., 2010). Similarly, sequential and increased deletion of BACE1 in 5XFAD adult mice was capable of reversing amyloid deposition and improving cognitive functions (Hu et al., 2018). A 2-mo treatment of 4-mo-old 5XFAD mice with BACE1 inhibitor led to ∼40% reduction in Aβ42 levels and normalized behavior, but the same treatment of 10-mo-old 5XFAD mice had only modest effects on Aβ pathology and no effect on cognitive function (Devi et al., 2015). A short 2-wk treatment of 1.5-mo-old APPSw/0/PSEN1 mice with BBB-permeable BACE1 inhibitor led to a 75–80% decrease in soluble Aβ levels, whereas 3-mo treatment of 4-mo-old APPSw/0/PSEN1 mice slowed down the formation of new plaques but did not have any effect on existing plaques (Peters et al., 2018). Collectively, these studies suggest that the optimal timing for treatment of Aβ pathology with BACE1 inhibitors is early in the disease course, before widespread Aβ plaque formation occurs.
The transcription factor NFκB is widely expressed in the central nervous system (Zhang et al., 2017), and its increased levels have been shown in AD brains in the vicinity of Aβ plaques associated with increased BACE1 levels (Chen et al., 2012). Studies in APP-expressing cell lines have shown that some NFκB inhibitors can decrease Aβ40 and Aβ42 production, sAPPβ and APP-CTFβ (BACE1 cleaved APP C-terminal fragment) levels, and BACE1 levels, suggesting that they inhibit BACE1 cleavage of APP (Paris et al., 2007, 2010), consistent with reports showing that NFκB transcriptionally regulates BACE1 expression (Buggia-Prevot et al., 2008; Chami and Checler, 2012; Chen et al., 2012). In agreement with findings that APC transcriptionally inhibits NFκB expression (Riewald and Ruf, 2005; Cheng et al., 2006) and blocks NFκB nuclear translocation (White et al., 2000; Joyce et al., 2001; Yuksel et al., 2002; Riewald and Ruf, 2005; Cheng et al., 2006), we showed that 3K3A-APC inhibited expression and nuclear translocation of NFκB in neurons of 5XFAD mice, as demonstrated in both in vivo and cortical neuron cultures from 5XFAD mice. We also showed that 3K3A-APC transcriptionally blocks BACE1 expression in neurons and Aβ production, which is NFκB dependent, as indicated by the lack of 3K3A-APC transcriptional inhibition of BACE1 in neurons overexpressing a dominant-active phosphorylation mimic form of NFκB-p65 (S536E; Hu et al., 2004).
Consistent with findings showing that BACE1 inhibition causing an ∼50% reduction in Aβ pathology completely normalized hippocampus-dependent behavior in early-disease-stage 5XFAD mice (Kimura et al., 2010; Devi et al., 2015), we also found that a similar 40–50% reduction of Aβ deposits by 3K3A-APC fully normalized hippocampus-dependent behavior in 7-mo-old 5XFAD mice back to values seen in nontransgenic controls. As lowering Aβ levels in blood vessels improves CBF responses in APP models (Niwa et al., 2000; Sagare et al., 2007), it is not surprising that reduced Aβ cerebrovascular pathology by 3K3A-APC normalized CBF response to neuronal stimulation in 5XFAD mice. Although, direct neuroprotective effects of 3K3A-APC (Guo et al., 2009a,b; Wang et al., 2009) against Aβ toxicity cannot be ruled out as contributing to normalization of cognitive and CBF responses, 5XFAD mice at this stage still do not develop an overt neuron loss (Oakley et al., 2006). Moreover, previous work in early-disease-stage 5XFAD and APPSw/0/PSEN1 mice (Kimura et al., 2010; Devi et al., 2015; Peters et al., 2018) suggests that BACE1 inhibition in neurons at the level achieved by the current 3K3A-APC treatment can reduce Aβ pathology to a degree sufficient on its own to normalize behavior. Since Aβ vasculotoxic effects in APP models depend on Aβ load (Montagne et al., 2017) and Aβ and amyloid load highly correlate with glial responses in 5XFAD mice (Hu et al., 2018), it is not surprising that 3K3A-APC improved cerebrovascular integrity and suppressed neuroinflammatory responses. As with cognition, the present data, however, cannot rule out possible direct contributions of 3K3A-APC on stabilizing BBB integrity and/or controlling neuroinflammatory response, as reviewed elsewhere (Zlokovic and Griffin, 2011; Griffin et al., 2018).
At present, pharmacological BACE1 inhibition failed to rescue the cognitive decline in mild-to-moderate AD patients (Egan et al., 2018), which indicates that treatment at the symptomatic stage might be too late (Vassar, 2014). Based on animal model findings, ongoing trials with BACE1 inhibitors in individuals with mild cognitive impairment and early-stage AD and prevention trials before the onset of clinical symptoms (https://clinicaltrials.gov/ct2/show/NCT03131453) will likely have a better outcome compared with trials in mild-to-moderate–stage AD (Vassar, 2014). However, the outcome of these trials remains to be seen. In this regard, the present results indicate that 3K3A-APC is likely to be an effective anti-Aβ treatment for early-stage AD and mild cognitive impairment. Whether 3K3A-APC’s direct neuroprotective, vasculoprotective, and anti-inflammatory effects independent of its antiamyloidogenic activity can improve cognitive outcome in late-stage APP models in which BACE1 inhibitors were ineffective (Devi et al., 2015; Peters et al., 2018), and by extension in AD patients at the symptomatic stage, is an intriguing question that remains to be addressed by future studies.
Materials and methods
Animals
Transgenic mice with five familial AD mutations (5XFAD) used in this study were purchased from The Jackson Laboratory (34840-JAX) and were on B6SJL genetic background. 5XFAD mice carry K670N/M67L (Swedish), I716V (Florida), and V717I (London) mutations in human APP (695) and M146L and L286V mutations in the human PSEN1 gene in the brain (Oakley et al., 2006). Both transgenes are regulated by neuronal mouse Thy1 promoter and express transgenes exclusively in neurons (Oakley et al., 2006). Our preliminary data indicated that sex did not influence the development of Aβ pathology in 5XFAD mice at a disease stage that we studied, consistent with several previous reports in 5XFAD mice (Oakley et al., 2006; Devi et al., 2015; Devi and Ohno, 2015; Mariani et al., 2017). Therefore, both female and male mice were used in the study. Treatment groups were composed of gender-balanced littermates randomly assigned to treatment using random number generator. The animals were housed in plastic cages on a 12-h light cycle with ad libitum access to water and a standard laboratory diet. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Southern California following National Institutes of Health guidelines.
Treatment with 3K3A-APC
Murine recombinant 3K3A-APC (KKK192-194AAA) was prepared as described previously (Mosnier et al., 2004). 3K3A-APC diluted in saline (100 µg/kg/d) or saline (vehicle) was administered daily by intraperitoneal injections in 3-mo-old 5XFAD mice for 4 mo.
Behavior
Novel object location
The novel object location test was performed as previously reported, with modifications (Bell et al., 2010; Sagare et al., 2013). Animals were habituated in a 30-cm3 box for 10 min on 3 consecutive d. On the third day, after 10 min of habituation, animals were placed in the box with two identical ∼5 × 5–cm objects placed in the top left and right corners of the testing area. Animals were allowed to explore the objects for 5 min and then returned to their home cages. After 1.5 h, one of the objects was relocated diagonally (novel), and the animals were reintroduced to the testing area and allowed to explore the area for 3 min. After each trial, the testing area and the objects were thoroughly cleaned with 70% ethanol solution. All the trials, including habituation, were recorded with a high-resolution camera, and the amount of time each animal spent exploring the objects was analyzed and presented as the percentage of time spent with novel objects over summarized time spent with novel and old objects. Any animal that showed a preference for either of the two identical objects before replacement with the novel location or stayed in the corner during the habituation was eliminated from the analysis.
Contextual fear conditioning
Contextual fear conditioning test was adapted from previously described protocols (Kimura et al., 2010; Montagne et al., 2018). The experiments were performed using a standard conditioning chamber equipped with a stainless-steel grid floor attached to a shock source that was controlled by FreezeFrame software (Coulbourn Instruments), and a digital camera was mounted on the ceiling to monitor behavior. During training, mice were placed in the conditioning chamber for 5 min and received four footshocks (0.4 mA, duration 1 s) 1 min apart starting after 2 min. Contextual memory was tested in the same chamber the next morning without footshock. The automated FreezeFrame system was used to score the percentage of total freezing time with a threshold set at 10% and minimal bout duration of 0.25 s.
CBF studies
CBF responses to vibrissal stimulation in anesthetized mice (∼0.4% isoflurane) were determined using laser Doppler flowmetry, as described previously (Deane et al., 2012). Briefly, the skin was cut to reveal the skull, and the tip of the laser Doppler probe (Transonic Systems Inc.) was stereotaxically placed 0.5 mm over the whisker barrel cortex area. The contralateral vibrissae were cut to 5 mm and stimulated by gentle stroking at 3–4 Hz for 60 s with a cotton-tip applicator. Rectal temperature was maintained at 37°C using a heated blanket (Homeothermic Blanket; Harvard Apparatus). CBF was recorded during stimulation, and the percentage CBF increase was obtained by subtracting the baseline from the maximum CBF value reached during stimulus. A total of three trials were averaged for each mouse with 10-min recovery periods between trials.
Immunohistochemistry (IHC)
Mice were anesthetized i.p. with 100 mg/kg ketamine and 10 mg/kg xylazine and transcardially perfused with ice-cold 0.01 M PBS, pH 7.4, containing 5 mM EDTA. Brains were rapidly removed, and one hemisphere was embedded in optimal cutting temperature compound (Tissue-Tek); the other hemisphere was saved for protein and RNA analyses. Frozen brains were serially sectioned in a sagittal plane on a cryostat (20 µm) and postfixed with 4% paraformaldehyde for 10 min. After washing with PBS, the sections were blocked in 5% normal donkey serum (Vector Laboratories)/0.3% Triton-X/0.01 M PBS for 1 h and incubated with primary antibodies diluted in blocking solution overnight at 4°C. We used the following antibodies: rabbit anti-human Aβ (to detect Aβ deposits); mouse FITC-conjugated α-smooth muscle actin (αSMA; to visualize vascular smooth muscle cells); rabbit polyclonal anti-human NFκB-p65 (phospho S536), which cross-reacts with mouse NFκB-p65 (phospho-S536); mouse anti-mouse NeuN (to visualize the phosphorylated form of NFκB-p65 staining in neurons); rabbit polyclonal anti-human fibrinogen, which cross-reacts with mouse fibrinogen (Montagne et al., 2018) and recognizes both the monomeric form of fibrinogen and fibrinogen-derived fibrin polymers (to detect extravascular fibrinogen deposits); 647-conjugated donkey anti-mouse IgG (to detect extravascular IgG deposits); rabbit polyclonal anti-mouse Iba1 (to visualize microglia); and rabbit polyclonal anti-bovine GFAP, which cross-reacts with mouse GFAP (to visualize astrocytes; Montagne et al., 2018). To visualize brain vessels, sections were incubated with Dylight 488–conjugated Lycopersicon esculentum lectin together with primary antibodies. After incubation in primary antibodies, sections were incubated with fluorophore-conjugated secondary antibodies (see Table S1 for detailed information on antibodies used). For detection of Aβ plaques, after paraformaldehyde fixation, the sections were incubated in 1% aqueous ThS (T1892; Sigma) for 5 min and rinsed in 80% ethanol, 95% ethanol, and distilled water. To visualize hemosiderin deposits, an Iron stain kit (HT20-1KT; Sigma) was used following the manufacturer’s instructions. All the slides were mounted with DAPI Fluoromount (0100–20; Southern Biotech) or ImmunoHistoMount (sc-45086; Santa Cruz).
Aβ and ThS deposits
Aβ-positive and ThS-positive areas were determined using ImageJ software (National Institutes of Health). Briefly, the images were taken on a BZ9000 fluorescent microscope in single plain at 20× (640 × 480 µm image size) and subjected to threshold processing (Otsu) using ImageJ, and the percent area occupied by the signal in the image area was measured as described previously (Sagare et al., 2013). In each animal, five randomly selected fields from the cortex and hippocampus were imaged and analyzed in four nonadjacent sections (∼100 µm apart). Eight animals in each group were analyzed.
CAA
To visualize CAA, brain sections were double stained with αSMA and Aβ antibody and imaged on a BZ9000 fluorescence microscope in single plain at 10× magnification. In each animal, three sections and five images per section containing pial arteries and penetrating arterioles (≥20 µm) were analyzed as described previously (Cortes-Canteli et al., 2010; Lin et al., 2016), with modifications. Briefly, on merged images containing both (αSMA and Aβ) channels, the outline around the vessels that fits predefined criteria was made using freehand selection tool and the signal was cleared outside using ImageJ software. After thresholding both channels using Otsu, the percent area was calculated for both colors within the outline made previously. Percent area occupied with Aβ was divided by percent area occupied with αSMA to obtain percent Aβ vascular load reflecting development of CAA. A total of 10–15 vessels per animal and 5–7 animals per group were analyzed.
p-NFκB-p65
To visualize the phosphorylated form of NFκB-p65 in neurons, brain sections were stained with NFκB-p65 (phospho S536) and NeuN antibodies. Quantification was performed from images taken on a Nikon confocal microscope in single plain with standardized gain, digital offset, and laser intensity, and these conditions were kept identical for both groups. Briefly, the p-NFκB-p65 relative signal intensity in the NeuN- and DAPI-positive nuclei was measured using ImageJ after Otsu threshold processing, as described previously (Cheng et al., 2006). In each animal, three randomly selected fields from the hippocampus CA1 region were analyzed in three nonadjacent sections (∼100 µm apart). Five animals per group were analyzed.
Extravascular fibrin(ogen) and IgG deposits
Quantification was performed from maximum projections of 10-µm-thick Z-stack images taken on a BZ9000 fluorescent microscope subjected to threshold processing (Otsu) using ImageJ, as described previously (Montagne et al., 2018). Briefly, the amount of perivascular deposits of fibrin(ogen) and IgG was determined as integrated density of the deposits on the abluminal side of the lectin-positive vessels. Representative images were taken on a Nikon confocal microscope with standardized gain, digital offset, and laser intensity, and these conditions were kept identical for both groups. In each animal, five randomly selected fields from the cortex and hippocampus were analyzed in four nonadjacent sections (∼100 µm apart). Seven animals per group were analyzed.
Hemosiderin deposits
Hemosiderin deposits in brain sections were detected by Prussian blue staining, as previously described (Zhong et al., 2008). Percent area occupied by Prussian blue–positive deposits was quantified using ImageJ. In each animal, five randomly selected fields from the cortex and hippocampus were analyzed in four nonadjacent sections (∼100 µm apart), as we reported.
Quantification of Iba1-positive microglia and GFAP-positive astrocytes
Quantification was performed from maximum projections of 10-µm-thick Z-stack images taken on a BZ9000 fluorescence microscope at 20× magnification subjected to threshold processing (Otsu) using ImageJ, as described previously (Montagne et al., 2018). Briefly, the number of Iba1-positive microglia and GFAP-positive astrocytes that also demonstrated DAPI-positive nuclear staining was quantified using the ImageJ Cell Counter analysis tool. In each animal, the numbers of microglia and astrocytes in the cortex and hippocampus were determined in five randomly selected fields from four nonadjacent sections (∼100 µm apart). Six to seven animals per group were analyzed. Representative images were taken on a Nikon confocal microscope with standardized gain, digital offset, and laser intensity, and these conditions were kept identical for both groups.
In a separate experiments, we performed additional analysis of microglia counts per Aβ plaque area using a similar approach that we recently reported for microglia counts normalized to fibrin deposits (Montagne et al., 2018). Briefly, we selected 100 × 100–µm regions of interest (ROIs) with ThS and Iba1 double-stained sections from 3K3A-APC–treated and vehicle-treated 5XFAD mice and assessed the size of each plaque in the ROI and the number of the Iba1-positive cells around each plaque. We then expressed the number of Iba1-positive cells per plaque area defined as number of Iba1-positive cells/µm2 of Aβ plaque area. Twenty randomly selected ROIs per mouse from four sections 100 µm apart were used in these calculations.
Primary neuronal cell culture
Primary cortical neurons were isolated from brains of 5XFAD mouse embryos (embryonic day 15.5) as previously described (Guo et al., 2009a). Isolated neuronal cells from each embryo were separately plated on poly-D-lysine–coated plates at a density of 106 cells per well of 6-well plates. Neuronal cultures were grown for 2 wk in a humidified 5% CO2 incubator at 37°C. A small piece of tissue from each embryo was used for 5XFAD genotyping using primers (5′-AGGACTGACCACTCGACCAG-3′ and 5′-CGGGGGTCTAGTTCTGCAT-3′). Cortical neurons that were positive for 5XFAD genotype and their culture medium was used in further experiments, as described below. All experiments were run in duplicates of at least three independent cultures. Each 5XFAD embryo represented one culture.
Transfection of cortical neurons
2-wk-old differentiated cortical neurons were transfected with T7-RelA (S536E) plasmid (plasmid 24156; Addgene) using Lipofectamine 3000 and following the manufacturer’s instructions (L3000008; Thermo Fisher). Cells were treated with vehicle or 3K3A-APC (5 nM) for 24 h. Neuronal cells were collected for analysis of NFκB-p65 and BACE1 with immunoblotting, and relative abundance of RelA and Bace1 mRNA by real-time quantitative PCR (RT-qPCR). Neuronal culture medium was collected for analysis of human Aβ40, Aβ42, and Swedish sAPPβ levels with ELISA, as described below.
Silencing of PARs
2-wk-old differentiated cortical neurons from 5XFAD mice were transfected with the scrambled or one of the following siRNAs from Dharmacon: F2r (E-054176-00-0010), F2rl1 (E-061445-00-0010), F2rl2 (E-055946-00-0010), and F2rl3 (E-062092-00-0010), in order to silence expression of PAR1, PAR2, PAR3 and PAR4, respectively, following the manufacturer’s instructions and as previously described (Guo et al., 2013). After 72 h, cells were treated with vehicle or 3K3A-APC (5 nM) for 24 h. Neuronal cells were collected for analysis of NFκB-p65 and BACE1 by immunoblotting, and neuronal culture medium was collected for analysis of secreted Aβ40 and Aβ42 levels with ELISA, as described below.
Protein and mRNA quantification
Cortex and hippocampus were isolated and snap frozen from one brain hemisphere from transcardially perfused mice (see the Immunohistochemistry (IHC) section for details) and were used for protein and RNA analyses.
Human Aβ40- and Aβ42-specific ELISA
Hippocampi and cortices were homogenized in ice-cold guanidine buffer (5 M guanidine hydrochloride/50 mM Tris HCl, pH 8), as described previously (Sagare et al., 2007). Hippocampi and cortices, as well as culture medium collected from 5XFAD primary neuronal cultures, were used for measurement of human Aβ40 and Aβ42 levels by using a Meso Scale Discovery assay (K15200E-1).
sAPPβ assay
CSF collected from cisterna magna of anesthetized 5XFAD animals and culture medium collected from 5XFAD primary neuronal cultures (concentrated by 10-fold) were used to determine the levels of sAPPβ released from the Swedish variant of APP by Meso Scale Discovery assay (K151BUE-1).
Immunoblotting
Cortical brain samples, cortical microvessels, or primary cortical neurons were lysed in RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, and Roche protease inhibitor cocktail). After sonication, the samples were centrifuged at 20,000 g for 20 min, and supernatants were used for protein quantification (23228; Thermo Fisher). Nuclear protein extraction of brain samples or cortical neurons was prepared according to the manufacturer’s instructions (78833; Thermo Fisher). Samples were prepared with lithium dodecyl sulfate sample buffer (Invitrogen), and proteins (5–15 µg) were separated by electrophoresis on NuPAGE Novex Bis-Tris precast 4–12% gradient gels (Thermo Fisher). After electrophoretic transfer, nitrocellulose membranes (Bio-Rad) were blocked with blocking buffer (37536; Thermo Fisher) and incubated overnight at 4°C with primary antibodies diluted in blocking solution. After washing with Tris-buffered saline containing 0.1% Tween 20, membranes were incubated with HRP-conjugated donkey anti-rabbit secondary antibody for 1 h at room temperature (see Table S2 for details on primary and secondary antibodies), washed again in Tris-buffered saline containing 0.1% Tween 20, and treated for 5 min with Super Signal West Pico chemiluminescent substrate (Thermo Fisher). Membranes were exposed to CL-XPosure film (Thermo Fisher) and developed in X-OMAT 3000 RA film processer (Kodak). The intensity of blots was determined using ImageJ. The intensity of protein bands was normalized with respective loading control bands.
RT-qPCR
Relative mRNA abundance of human APP (hAPP), mouse β-secretase (Bace1), mouse NFκB-p65 subunit (RelA) in brain homogenates and primary neurons, and of the neuroinflammatory murine cytokines and chemokines, mouse Il6, mouse Icam-1, mouse Tnfα, and mouse Ccl2 in brain homogenates was determined by RT-qPCR. Total RNA from the cortical brain samples or primary cortical neurons was prepared using a RNeasy kit (74104; QIAGEN), and RT-qPCR amplification was performed using a one-step SYBR green qPCR kit (95087; QuantaBio). Relative abundance was calculated using the ΔΔCt method normalized to the housekeeping mouse gene Gapdh, as described previously (Montagne et al., 2018). The primers used are listed in Table 1.
Table 1. List of primers used to determine gene expression levels.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Bace1 | 5′-GATGGTGGACAACCTGAG-3′ | 5′-CTGGTAGTAGCGATGCAG-3′ |
| RelA | 5′-CTTCCTCAGCCATGGTACCTCT-3′ | 5′-CAAGTCTTCATCAGCATCAAACTG-3′ |
| hAPP | 5′-CCAACCAGTGACCATCCAGAACTG-3′ | 5′-GCACTTGTCAGGAACGAGAAGGG-3′ |
| Tnf-α | 5′-CTTCTGTCTACTGAACTTCGGG-3′ | 5′-TGATCTGAGTGTGAGGGTCTG-3′ |
| Il-6 | 5′-CAAAGCCAGAGTCCTTCAGAG-3′ | 5′-GTCCTTAGCCACTCCTTCTG-3′ |
| Ccl2 | 5′-CATCCACGTGTTGGCTCA-3′ | 5′-GATCATCTTGCTGGTGAATCAGT-3′ |
| Icam-1 | 5′-AAGGAGATCACATTCACGGTG-3′ | 5′-TTTGGGATGGTAGCTGGAAG-3′ |
| Adam10 | 5′-GGGAAGAAATGCAAGCTGAA-3′ | 5′-CTGTACAGCAGGGTCCTTGAC-3′ |
| Gapdh | 5′-ACCACAGTCCATGCCATCAC-3′ | 5′-TCCACCACCCTGTTGCTGTA-3′ |
β-Secretase activity assay
β-Secretase 1 (BACE1) activity was measured in brain homogenates using a fluorogenic β-secretase activity assay kit (565785; Calbiochem), according to the manufacturer’s instructions. The enzymatic product was measured on a fluorescence plate reader (Wallac Victor V2; PerkinElmer).
Analysis of physiological and biochemical parameters
Heart rate and systolic and diastolic arterial blood pressure were monitored using a CODA Monitor (Kent Scientific). For respiration rate, the number of breaths was averaged from three individual 60-s trials. Blood gases and pH were determined from a small sample (∼90 µl) of arterial blood, collected from the cannulated right femoral artery, using the i-STAT CG8+ moderately complex panel (03P88-25; Abbott). For analysis of liver and kidney function, ∼200 µl EDTA-plasma was collected and sent to IDEXX BioResearch for screening (test codes 60405 and 60406, respectively).
Statistical analysis
Sample sizes were calculated using nQUERY, assuming a two-sided α level of 0.05, 80% power, and homogenous variances for the two samples to be compared, with the means and common SD for different parameters predicted from published work and our previous studies. For comparison between two groups, an F test was conducted to determine similarity in the variances between the groups that are statistically compared, and statistical significance was analyzed by Student’s t test or Mann-Whitney U test. For multiple comparisons, one-way ANOVA followed by Tukey’s post hoc or Kruskal-Wallis test were used. For all analyses, Shapiro-Wilk normality test was used to test normality of the data and appropriate test to determine statistical significance was applied using GraphPad Prism 7.0 software. P < 0.05 was considered to be significant. All data are shown as scatterplots, with single points per mouse and/or culture and boxplots representing means ± SD, as indicated in the figure legends.
Online supplemental material
Fig. S1 shows no changes in APP mRNA and APP, Notch 1 intracellular domain, ADAM10, IDE, neprilysin, and LRP1 protein levels after 3K3A-APC treatment. Fig. S2 shows that 3K3A-APC treatment does not affect systemic physiological and biochemical parameters. Fig. S3 shows full blots of Western blot data. Table S1 shows primary and secondary antibodies used for IHC. Table S2 shows primary and secondary antibodies used for immunoblotting.
Acknowledgments
We thank J.A. Fernandez, X. Xu, E. Zuniga, E.J. Lawson, K. Kisler, and Y. Wang for technical assistance with some of the experiments; M.D. Sweeney, K. Kisler, Z. Zhao, and A.R. Nelson for helpful discussions; and M.D. Sweeney for critical reading of the manuscript.
This research was supported in part by National Institutes of Health grants NS090904 (B.V. Zlokovic) and HL052246 (J.H. Griffin) for development of APC variants and PAR1 mimetic peptides for stroke and by University of Southern California start-up funds to B.V. Zlokovic.
B.V. Zlokovic is a scientific founder and chairs the scientific advisory board of ZZ Biotech LLC, a biotechnology company with a mission to develop APC and its functional mutants for the treatment of stroke and other neurological disorders. J.H. Griffin is a consultant for ZZ Biotech LLC and inventor of some uses of 3K3A-APC. The remaining authors declare no competing financial interests.
Author contributions: D. Lazic and A.P. Sagare designed and performed experiments and analyzed data. A.M. Nikolakopoulou performed glial data analyses. J.H. Griffin provided murine 3K3A-APC and commented on the text. R. Vassar provided Bace1-knockout brain homogenates and commented on the text. B.V. Zlokovic designed all experiments, analyzed data, and wrote the manuscript.
References
- Amar A.P., Sagare A.P., Zhao Z., Wang Y., Nelson A.R., Griffin J.H., and Zlokovic B.V.. 2018. Can adjunctive therapies augment the efficacy of endovascular thrombolysis? A potential role for activated protein C. Neuropharmacology. 134(Pt B):293–301. 10.1016/j.neuropharm.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R.D., Winkler E.A., Sagare A.P., Singh I., LaRue B., Deane R., and Zlokovic B.V.. 2010. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 68:409–427. 10.1016/j.neuron.2010.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggia-Prevot V., Sevalle J., Rossner S., and Checler F.. 2008. NFkappaB-dependent control of BACE1 promoter transactivation by Abeta42. J. Biol. Chem. 283:10037–10047. 10.1074/jbc.M706579200 [DOI] [PubMed] [Google Scholar]
- Chami L., and Checler F.. 2012. BACE1 is at the crossroad of a toxic vicious cycle involving cellular stress and β-amyloid production in Alzheimer’s disease. Mol. Neurodegener. 7:52 10.1186/1750-1326-7-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Zhou W., Liu S., Deng Y., Cai F., Tone M., Tone Y., Tong Y., and Song W.. 2012. Increased NF-κB signalling up-regulates BACE1 expression and its therapeutic potential in Alzheimer’s disease. Int. J. Neuropsychopharmacol. 15:77–90. 10.1017/S1461145711000149 [DOI] [PubMed] [Google Scholar]
- Cheng T., Liu D., Griffin J.H., Fernández J.A., Castellino F., Rosen E.D., Fukudome K., and Zlokovic B.V.. 2003. Activated protein C blocks p53-mediated apoptosis in ischemic human brain endothelium and is neuroprotective. Nat. Med. 9:338–342. 10.1038/nm826 [DOI] [PubMed] [Google Scholar]
- Cheng T., Petraglia A.L., Li Z., Thiyagarajan M., Zhong Z., Wu Z., Liu D., Maggirwar S.B., Deane R., Fernández J.A., et al. 2006. Activated protein C inhibits tissue plasminogen activator-induced brain hemorrhage. Nat. Med. 12:1278–1285. 10.1038/nm1498 [DOI] [PubMed] [Google Scholar]
- Cortes-Canteli M., Paul J., Norris E.H., Bronstein R., Ahn H.J., Zamolodchikov D., Bhuvanendran S., Fenz K.M., and Strickland S.. 2010. Fibrinogen and beta-amyloid association alters thrombosis and fibrinolysis: a possible contributing factor to Alzheimer’s disease. Neuron. 66:695–709. 10.1016/j.neuron.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzin N., Baranger K., Cavalier M., Marchalant Y., Cohen-Solal C., Roman F.S., Khrestchatisky M., Rivera S., Féron F., and Vignes M.. 2013. Area-specific alterations of synaptic plasticity in the 5XFAD mouse model of Alzheimer’s disease: dissociation between somatosensory cortex and hippocampus. PLoS One. 8:e74667 10.1371/journal.pone.0074667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R., Wu Z., Sagare A., Davis J., Du Yan S., Hamm K., Xu F., Parisi M., LaRue B., Hu H.W., et al. 2004. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 43:333–344. 10.1016/j.neuron.2004.07.017 [DOI] [PubMed] [Google Scholar]
- Deane R., Singh I., Sagare A.P., Bell R.D., Ross N.T., LaRue B., Love R., Perry S., Paquette N., Deane R.J., et al. 2012. A multimodal RAGE-specific inhibitor reduces amyloid β-mediated brain disorder in a mouse model of Alzheimer disease. J. Clin. Invest. 122:1377–1392. 10.1172/JCI58642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L., and Ohno M.. 2015. Effects of BACE1 haploinsufficiency on APP processing and Aβ concentrations in male and female 5XFAD Alzheimer mice at different disease stages. Neuroscience. 307:128–137. 10.1016/j.neuroscience.2015.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L., Tang J., and Ohno M.. 2015. Beneficial effects of the β-secretase inhibitor GRL-8234 in 5XFAD Alzheimer’s transgenic mice lessen during disease progression. Curr. Alzheimer Res. 12:13–21. 10.2174/1567205012666141218125042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.F., Kost J., Tariot P.N., Aisen P.S., Cummings J.L., Vellas B., Sur C., Mukai Y., Voss T., Furtek C., et al. 2018. Randomized Trial of Verubecestat for Mild-to-Moderate Alzheimer’s Disease. N. Engl. J. Med. 378:1691–1703. 10.1056/NEJMoa1706441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoni P., Arango-Lievano M., Neves I.D., Rousset M.-C., Baranger K., Rivera S., Jeanneteau F., Claeysen S., and Marchi N.. 2016. Cerebrovascular pathology during the progression of experimental Alzheimer’s disease. Neurobiol. Dis. 88:107–117. 10.1016/j.nbd.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Griffin J.H., Zlokovic B.V., and Mosnier L.O.. 2015. Activated protein C: biased for translation. Blood. 125:2898–2907. 10.1182/blood-2015-02-355974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J.H., Mosnier L.O., Fernández J.A., and Zlokovic B.V.. 2016. 2016 Scientific Sessions Sol Sherry Distinguished Lecturer in Thrombosis: Thrombotic Stroke: Neuroprotective Therapy by Recombinant-Activated Protein C. Arterioscler. Thromb. Vasc. Biol. 36:2143–2151. 10.1161/ATVBAHA.116.308038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J.H., Zlokovic B.V., and Mosnier L.O.. 2018. Activated protein C, protease activated receptor 1, and neuroprotection. Blood. 132:159–169. 10.1182/blood-2018-02-769026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Liu D., Gelbard H., Cheng T., Insalaco R., Fernández J.A., Griffin J.H., and Zlokovic B.V.. 2004. Activated protein C prevents neuronal apoptosis via protease activated receptors 1 and 3. Neuron. 41:563–572. 10.1016/S0896-6273(04)00019-4 [DOI] [PubMed] [Google Scholar]
- Guo H., Singh I., Wang Y., Deane R., Barrett T., Fernández J.A., Chow N., Griffin J.H., and Zlokovic B.V.. 2009a Neuroprotective activities of activated protein C mutant with reduced anticoagulant activity. Eur. J. Neurosci. 29:1119–1130. 10.1111/j.1460-9568.2009.06664.x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guo H., Wang Y., Singh I., Liu D., Fernández J.A., Griffin J.H., Chow N., and Zlokovic B.V.. 2009b Species-dependent neuroprotection by activated protein C mutants with reduced anticoagulant activity. J. Neurochem. 109:116–124. 10.1111/j.1471-4159.2009.05921.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhao Z., Yang Q., Wang M., Bell R.D., Wang S., Chow N., Davis T.P., Griffin J.H., Goldman S.A., and Zlokovic B.V.. 2013. An activated protein C analog stimulates neuronal production by human neural progenitor cells via a PAR1-PAR3-S1PR1-Akt pathway. J. Neurosci. 33:6181–6190. 10.1523/JNEUROSCI.4491-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Hwang S.-I., Roy D.B., Lundgren D.H., Price J.V., Ousman S.S., Fernald G.H., Gerlitz B., Robinson W.H., Baranzini S.E., et al. 2008. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 451:1076–1081. 10.1038/nature06559 [DOI] [PubMed] [Google Scholar]
- Hu J., Nakano H., Sakurai H., and Colburn N.H.. 2004. Insufficient p65 phosphorylation at S536 specifically contributes to the lack of NF-kappaB activation and transformation in resistant JB6 cells. Carcinogenesis. 25:1991–2003. 10.1093/carcin/bgh198 [DOI] [PubMed] [Google Scholar]
- Hu X., Das B., Hou H., He W., and Yan R.. 2018. BACE1 deletion in the adult mouse reverses preformed amyloid deposition and improves cognitive functions. J. Exp. Med. 215:927–940. 10.1084/jem.20171831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhar S., Trawicka A., Jenneckens C., Bayer T.A., and Wirths O.. 2012. Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Aβ aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging. 33:196.e29–196.e40. 10.1016/j.neurobiolaging.2010.05.027 [DOI] [PubMed] [Google Scholar]
- Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L., Snellinx A., Craessaerts K., Thathiah A., Tesseur I., et al. 2010. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 30:4833–4844. 10.1523/JNEUROSCI.5221-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce D.E., Gelbert L., Ciaccia A., DeHoff B., and Grinnell B.W.. 2001. Gene expression profile of antithrombotic protein c defines new mechanisms modulating inflammation and apoptosis. J. Biol. Chem. 276:11199–11203. 10.1074/jbc.C100017200 [DOI] [PubMed] [Google Scholar]
- Kim W., Ma L., Lomoio S., Willen R., Lombardo S., Dong J., Haydon P.G., and Tesco G.. 2018. BACE1 elevation engendered by GGA3 deletion increases β-amyloid pathology in association with APP elevation and decreased CHL1 processing in 5XFAD mice. Mol. Neurodegener. 13:6 10.1186/s13024-018-0239-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R., Devi L., and Ohno M.. 2010. Partial reduction of BACE1 improves synaptic plasticity, recent and remote memories in Alzheimer’s disease transgenic mice. J. Neurochem. 113:248–261. 10.1111/j.1471-4159.2010.06608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook S.-Y., Hong H.S., Moon M., Ha C.M., Chang S., and Mook-Jung I.. 2012. Aβ1−42-RAGE interaction disrupts tight junctions of the blood-brain barrier via Ca2+-calcineurin signaling. J. Neurosci. 32:8845–8854. 10.1523/JNEUROSCI.6102-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P.-H., Wang H., Dislich B., Colombo A., Zeitschel U., Ellwart J.W., Kremmer E., Rossner S., and Lichtenthaler S.F.. 2010. ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. EMBO J. 29:3020–3032. 10.1038/emboj.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Yu D., and Xu Z.. 2014. Activated protein C inhibits amyloid β production via promoting expression of ADAM-10. Brain Res. 1545:35–44. 10.1016/j.brainres.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Lin B., Hasegawa Y., Takane K., Koibuchi N., Cao C., and Kim-Mitsuyama S.. 2016. High-Fat-Diet Intake Enhances Cerebral Amyloid Angiopathy and Cognitive Impairment in a Mouse Model of Alzheimer’s Disease, Independently of Metabolic Disorders. J. Am. Heart Assoc. 5:e003154 10.1161/JAHA.115.003154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Cheng T., Guo H., Fernández J.A., Griffin J.H., Song X., and Zlokovic B.V.. 2004. Tissue plasminogen activator neurovascular toxicity is controlled by activated protein C. Nat. Med. 10:1379–1383. 10.1038/nm1122 [DOI] [PubMed] [Google Scholar]
- Lyden P., Levy H., Weymer S., Pryor K., Kramer W., Griffin J.H., Davis T.P., and Zlokovic B.. 2013. Phase 1 safety, tolerability and pharmacokinetics of 3K3A-APC in healthy adult volunteers. Curr. Pharm. Des. 19:7479–7485. 10.2174/1381612819666131230131454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden P., Weymer S., Coffey C., Cudkowicz M., Berg S., O’Brien S., Fisher M., Haley E.C., Khatri P., Saver J., et al. 2016. Selecting Patients for Intra-Arterial Therapy in the Context of a Clinical Trial for Neuroprotection. Stroke. 47:2979–2985. 10.1161/STROKEAHA.116.013881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden P., Pryor K.E., Coffey C.S., Cudkowicz M., Conwit R., Jadhav A., Sawyer R.N. Jr., Claassen J., Adeoye O., Song S., et al. NeuroNEXT Clinical Trials Network NN104 Investigators . 2018. Final Results of the RHAPSODY trial: A multi-center, Phase 2 trial using a continual reassessment method to determine the safety and tolerability of 3K3A-APC, a Recombinant Variant of Human Activated Protein C, in combination with tissue plasminogen activator, mechanical thrombectomy or both in moderate to severe acute ischemic stroke. Ann. Neurol. 10.1002/ana.25383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani M.M., Malm T., Lamb R., Jay T.R., Neilson L., Casali B., Medarametla L., and Landreth G.E.. 2017. Neuronally-directed effects of RXR activation in a mouse model of Alzheimer’s disease. Sci. Rep. 7:42270 10.1038/srep42270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne A., Zhao Z., and Zlokovic B.V.. 2017. Alzheimer’s disease: A matter of blood-brain barrier dysfunction? J. Exp. Med. 214:3151–3169. 10.1084/jem.20171406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne A., Nikolakopoulou A.M., Zhao Z., Sagare A.P., Si G., Lazic D., Barnes S.R., Daianu M., Ramanathan A., Go A., et al. 2018. Pericyte degeneration causes white matter dysfunction in the mouse central nervous system. Nat. Med. 24:326–337. 10.1038/nm.4482 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mosnier L.O., Gale A.J., Yegneswaran S., and Griffin J.H.. 2004. Activated protein C variants with normal cytoprotective but reduced anticoagulant activity. Blood. 104:1740–1744. 10.1182/blood-2004-01-0110 [DOI] [PubMed] [Google Scholar]
- Mosnier L.O., Yang X.V., and Griffin J.H.. 2007. Activated protein C mutant with minimal anticoagulant activity, normal cytoprotective activity, and preservation of thrombin activable fibrinolysis inhibitor-dependent cytoprotective functions. J. Biol. Chem. 282:33022–33033. 10.1074/jbc.M705824200 [DOI] [PubMed] [Google Scholar]
- Mosnier L.O., Zlokovic B.V., and Griffin J.H.. 2014. Cytoprotective-selective activated protein C therapy for ischaemic stroke. Thromb. Haemost. 112:883–892. 10.1160/th14-05-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir S., Gadi I., Al-Dabet M.M., Elwakiel A., Kohli S., Ghosh S., Manoharan J., Ranjan S., Bock F., Braun-Dullaeus R.C., et al. 2017. Cytoprotective activated protein C averts Nlrp3 inflammasome-induced ischemia-reperfusion injury via mTORC1 inhibition. Blood. 130:2664–2677. 10.1182/blood-2017-05-782102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa K., Younkin L., Ebeling C., Turner S.K., Westaway D., Younkin S., Ashe K.H., Carlson G.A., and Iadecola C.. 2000. Abeta 1-40-related reduction in functional hyperemia in mouse neocortex during somatosensory activation. Proc. Natl. Acad. Sci. USA. 97:9735–9740. 10.1073/pnas.97.17.9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Cole S.L., Yasvoina M., Zhao J., Citron M., Berry R., Disterhoft J.F., and Vassar R.. 2007. BACE1 gene deletion prevents neuron loss and memory deficits in 5XFAD APP/PS1 transgenic mice. Neurobiol. Dis. 26:134–145. 10.1016/j.nbd.2006.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris D., Patel N., Quadros A., Linan M., Bakshi P., Ait-Ghezala G., and Mullan M.. 2007. Inhibition of Abeta production by NF-kappaB inhibitors. Neurosci. Lett. 415:11–16. 10.1016/j.neulet.2006.12.029 [DOI] [PubMed] [Google Scholar]
- Paris D., Ganey N.J., Laporte V., Patel N.S., Beaulieu-Abdelahad D., Bachmeier C., March A., Ait-Ghezala G., and Mullan M.J.. 2010. Reduction of beta-amyloid pathology by celastrol in a transgenic mouse model of Alzheimer’s disease. J. Neuroinflammation. 7:17 10.1186/1742-2094-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-C., Baik S.H., Han S.-H., Cho H.J., Choi H., Kim H.J., Choi H., Lee W., Kim D.K., and Mook-Jung I.. 2017. Annexin A1 restores Aβ1-42 -induced blood-brain barrier disruption through the inhibition of RhoA-ROCK signaling pathway. Aging Cell. 16:149–161. 10.1111/acel.12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters F., Salihoglu H., Rodrigues E., Herzog E., Blume T., Filser S., Dorostkar M., Shimshek D.R., Brose N., Neumann U., and Herms J.. 2018. BACE1 inhibition more effectively suppresses initiation than progression of β-amyloid pathology. Acta Neuropathol. 135:695–710. 10.1007/s00401-017-1804-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraglia A.L., Marky A.H., Walker C., Thiyagarajan M., and Zlokovic B.V.. 2010. Activated protein C is neuroprotective and mediates new blood vessel formation and neurogenesis after controlled cortical impact. Neurosurgery. 66:165–171, discussion :171–172. 10.1227/01.NEU.0000363148.49779.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewald M., and Ruf W.. 2005. Protease-activated receptor-1 signaling by activated protein C in cytokine-perturbed endothelial cells is distinct from thrombin signaling. J. Biol. Chem. 280:19808–19814. 10.1074/jbc.M500747200 [DOI] [PubMed] [Google Scholar]
- Sagare A., Deane R., Bell R.D., Johnson B., Hamm K., Pendu R., Marky A., Lenting P.J., Wu Z., Zarcone T., et al. 2007. Clearance of amyloid-beta by circulating lipoprotein receptors. Nat. Med. 13:1029–1031. 10.1038/nm1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagare A.P., Bell R.D., Srivastava A., Sengillo J.D., Singh I., Nishida Y., Chow N., and Zlokovic B.V.. 2013. A lipoprotein receptor cluster IV mutant preferentially binds amyloid-β and regulates its clearance from the mouse brain. J. Biol. Chem. 288:15154–15166. 10.1074/jbc.M112.439570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Yamada S., Kumar S.R., Calero M., Bading J., Frangione B., Holtzman D.M., Miller C.A., Strickland D.K., Ghiso J., and Zlokovic B.V.. 2000. Clearance of Alzheimer’s amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J. Clin. Invest. 106:1489–1499. 10.1172/JCI10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R.K., Wang Y., Zhao Z., Xu X., Burnier L., Gupta N., Fernández J.A., Martin G., Kupriyanov S., Mosnier L.O., et al. 2018. PAR1 biased signaling is required for activated protein C in vivo benefits in sepsis and stroke. Blood. 131:1163–1171. 10.1182/blood-2017-10-810895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J., Choi S.H., Romano D.M., Gannon M.A., Lesinski A.N., Kim D.Y., and Tanzi R.E.. 2013. ADAM10 missense mutations potentiate β-amyloid accumulation by impairing prodomain chaperone function. Neuron. 80:385–401. 10.1016/j.neuron.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiyagarajan M., Fernández J.A., Lane S.M., Griffin J.H., and Zlokovic B.V.. 2008. Activated protein C promotes neovascularization and neurogenesis in postischemic brain via protease-activated receptor 1. J. Neurosci. 28:12788–12797. 10.1523/JNEUROSCI.3485-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R. 2014. BACE1 inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimers Res. Ther. 6:89 10.1186/s13195-014-0089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C.T., Marky A.H., Petraglia A.L., Ali T., Chow N., and Zlokovic B.V.. 2010. Activated protein C analog with reduced anticoagulant activity improves functional recovery and reduces bleeding risk following controlled cortical impact. Brain Res. 1347:125–131. 10.1016/j.brainres.2010.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Thiyagarajan M., Chow N., Singh I., Guo H., Davis T.P., and Zlokovic B.V.. 2009. Differential neuroprotection and risk for bleeding from activated protein C with varying degrees of anticoagulant activity. Stroke. 40:1864–1869. 10.1161/STROKEAHA.108.536680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang Z., Chow N., Davis T.P., Griffin J.H., Chopp M., and Zlokovic B.V.. 2012. An activated protein C analog with reduced anticoagulant activity extends the therapeutic window of tissue plasminogen activator for ischemic stroke in rodents. Stroke. 43:2444–2449. 10.1161/STROKEAHA.112.658997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao Z., Chow N., Rajput P.S., Griffin J.H., Lyden P.D., and Zlokovic B.V.. 2013. Activated protein C analog protects from ischemic stroke and extends the therapeutic window of tissue-type plasminogen activator in aged female mice and hypertensive rats. Stroke. 44:3529–3536. 10.1161/STROKEAHA.113.003350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao Z., Rege S.V., Wang M., Si G., Zhou Y., Wang S., Griffin J.H., Goldman S.A., and Zlokovic B.V.. 2016. 3K3A-activated protein C stimulates postischemic neuronal repair by human neural stem cells in mice. Nat. Med. 22:1050–1055. 10.1038/nm.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B., Schmidt M., Murphy C., Livingstone W., O’Toole D., Lawler M., O’Neill L., Kelleher D., Schwarz H.P., and Smith O.P.. 2000. Activated protein C inhibits lipopolysaccharide-induced nuclear translocation of nuclear factor kappaB (NF-kappaB) and tumour necrosis factor alpha (TNF-alpha) production in the THP-1 monocytic cell line. Br. J. Haematol. 110:130–134. 10.1046/j.1365-2141.2000.02128.x [DOI] [PubMed] [Google Scholar]
- Williams P.D., Zlokovic B.V., Griffin J.H., Pryor K.E., and Davis T.P.. 2012. Preclinical safety and pharmacokinetic profile of 3K3A-APC, a novel, modified activated protein C for ischemic stroke. Curr. Pharm. Des. 18:4215–4222. 10.2174/138161212802430413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.A., Sengillo J.D., Sagare A.P., Zhao Z., Ma Q., Zuniga E., Wang Y., Zhong Z., Sullivan J.S., Griffin J.H., et al. 2014. Blood-spinal cord barrier disruption contributes to early motor-neuron degeneration in ALS-model mice. Proc. Natl. Acad. Sci. USA. 111:E1035–E1042. 10.1073/pnas.1401595111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuksel M., Okajima K., Uchiba M., Horiuchi S., and Okabe H.. 2002. Activated protein C inhibits lipopolysaccharide-induced tumor necrosis factor-alpha production by inhibiting activation of both nuclear factor-kappa B and activator protein-1 in human monocytes. Thromb. Haemost. 88:267–273. 10.1055/s-0037-1613197 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Lenardo M.J., and Baltimore D.. 2017. 30 Years of NF-κB: A Blossoming of Relevance to Human Pathobiology. Cell. 168:37–57. 10.1016/j.cell.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Deane R., Ali Z., Parisi M., Shapovalov Y., O’Banion M.K., Stojanovic K., Sagare A., Boillee S., Cleveland D.W., and Zlokovic B.V.. 2008. ALS-causing SOD1 mutants generate vascular changes prior to motor neuron degeneration. Nat. Neurosci. 11:420–422. 10.1038/nn2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Ilieva H., Hallagan L., Bell R., Singh I., Paquette N., Thiyagarajan M., Deane R., Fernandez J.A., Lane S., et al. 2009. Activated protein C therapy slows ALS-like disease in mice by transcriptionally inhibiting SOD1 in motor neurons and microglia cells. J. Clin. Invest. 119:3437–3449. 10.1172/JCI38476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic B.V., and Griffin J.H.. 2011. Cytoprotective protein C pathways and implications for stroke and neurological disorders. Trends Neurosci. 34:198–209. 10.1016/j.tins.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic B.V., Zhang C., Liu D., Fernandez J., Griffin J.H., and Chopp M.. 2005. Functional recovery after embolic stroke in rodents by activated protein C. Ann. Neurol. 58:474–477. 10.1002/ana.20602 [DOI] [PubMed] [Google Scholar]