In this review, Zheng and Cantley provide a historical perspective of the folate metabolism field, delve into folate chemistry that is often overlooked, and point out various missing links and underdeveloped areas in folate metabolism for future exploration.
Abstract
Folate metabolism is crucial for many biochemical processes, including purine and thymidine monophosphate (dTMP) biosynthesis, mitochondrial protein translation, and methionine regeneration. These biochemical processes in turn support critical cellular functions such as cell proliferation, mitochondrial respiration, and epigenetic regulation. Not surprisingly, abnormal folate metabolism has been causally linked with a myriad of diseases. In this review, we provide a historical perspective, delve into folate chemistry that is often overlooked, and point out various missing links and underdeveloped areas in folate metabolism for future exploration.
Introduction
In 1931, Lucy Wills reported that an unusual form of anemia, macrocytic anemia, could be cured by yeast extracts, suggesting that the disease is a nutritional deficiency (Carpenter, 2003a,b). Soon thereafter, many groups were racing to identify the active nutrient, later named folic acid or folate. This was at a time when modern chromatography techniques were yet to be developed, yet folic acid was successfully isolated by two independent groups using ingenious methods, including using activated charcoal to adsorb and concentrate folic acid and using a microbiological assay to rapidly detect its activity (Hutchings et al., 1941; Mitchell et al., 1941). These intense efforts culminated in 1948, when E.L. Robert Stokstad and colleagues at Lederle Laboratories elucidated the chemical structure of folic acid by chemical degradation and total synthesis (Stokstad and Jukes, 1987).
Having learned of the discovery of folic acid, Sidney Farber recruited a cohort of leukemic children to test whether folic acid might be able to restore normalcy to the blood cells in leukemia, just as it does in macrocytic anemia (Farber et al., 1947). Although this trial was quickly stopped, as folic acid was found to actually exacerbate the disease, it led to a revised idea that folic acid antagonists might stop leukemia. This revised idea was borne out in Farber’s next trial. As described in his landmark paper in 1948 (Farber and Diamond, 1948), one of the tested folic acid antagonists, aminopterin, produced temporary remission in children with acute lymphoblastic leukemia. Thus, the era of cancer chemotherapy had begun.
What also ensued was a “golden era” for enzymology in general (1950–1960; Huennekens, 1996), during which many enzymes involved in folate metabolism were purified from crude cell or tissue extracts and characterized, and their reactions merged into coherent pathways. Among these enzymes, dihydrofolate reductase (DHFR) was identified as the major target of aminopterin and its more widely used close analogue, methotrexate (Werkheiser, 1961; Huennekens, 1994). In the following decades, rapid development of molecular biology techniques allowed cloning of the genes encoding the enzymes, and Robert Schimke discovered that amplification of the DHFR gene confers tumor resistance to methotrexate. Remarkably, this was the first known case of gene amplification in mammalian somatic cells (Schimke, 1989).
Many excellent reviews on folate metabolism have appeared over the past decade, some of which cover metabolic compartmentation (Tibbetts and Appling, 2010; Scotti et al., 2013), folate transport (Zhao et al., 2011), and the roles of folates in redox homeostasis (Ducker and Rabinowitz, 2017), embryonic development (Momb and Appling, 2014), cancers (Locasale, 2013; Yang and Vousden, 2016), and plants (Hanson and Gregory, 2011).
It is sometimes claimed in the current literature that all the components in the metabolic pathways have been identified, and the challenge now is to understand how the metabolic fluxes are regulated. While we wholeheartedly agree with the latter point, we share with others the opinion that many components in mammalian folate metabolism remain to be identified, and their functions remain to be understood. Recently introduced disciplines, such as comparative genomics (Gabaldón and Koonin, 2013), functional genomics (Wang et al., 2014; Shalem et al., 2015; Horlbeck et al., 2016), and organelle proteomics (Chapel et al., 2013; Pagliarini and Rutter, 2013; Calvo et al., 2016), have opened up new opportunities for solving these remaining puzzles. Thus, in this review, we emphasize missing links and underdeveloped areas and discuss potential approaches to illuminate them. We begin with a survey of folate chemistry, an important aspect of folate metabolism that is sometimes overlooked.
Structure and chemistry of folates
Nomenclature
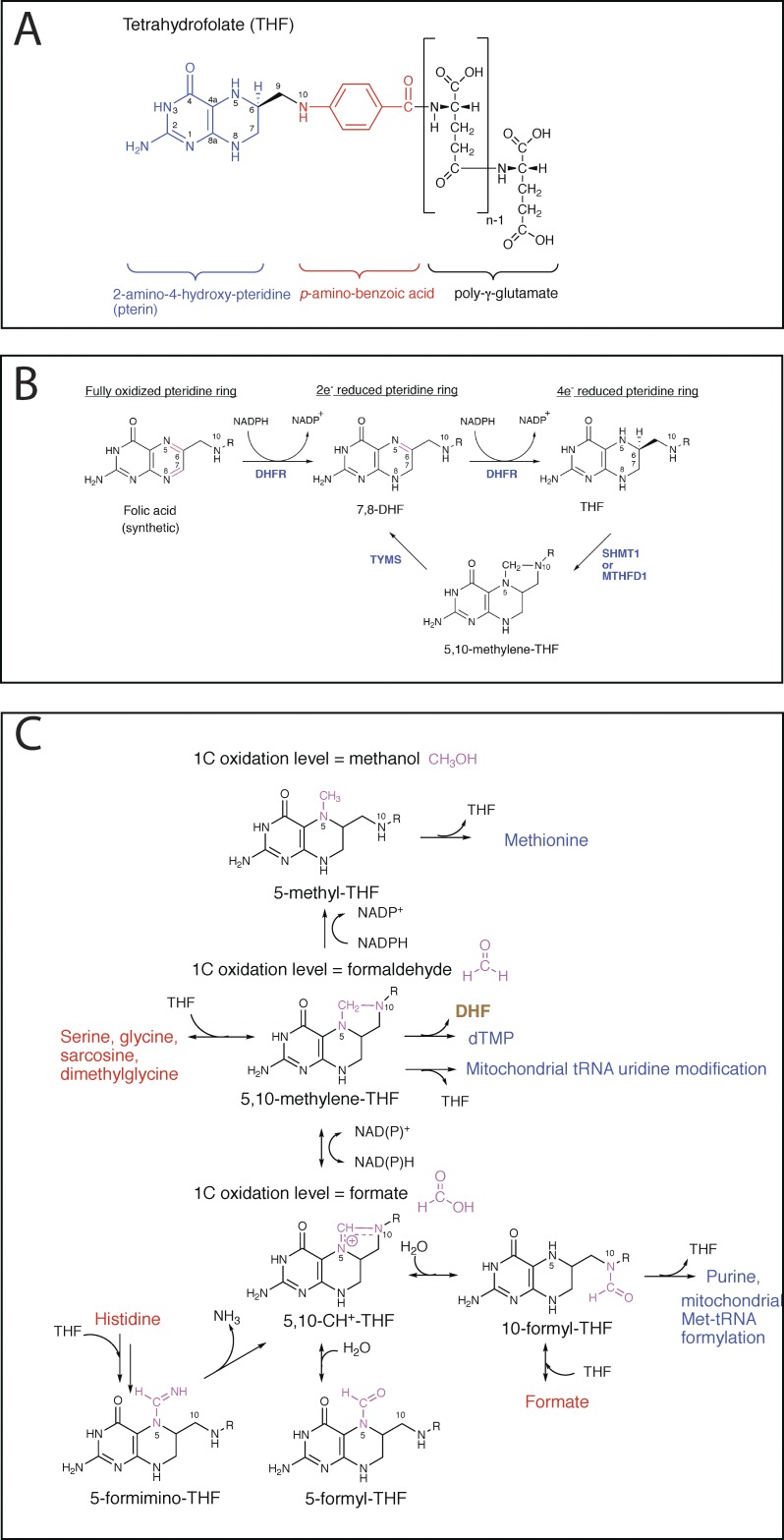
By convention, “folates” is a generic term, referring to a large family of compounds consisting of a 2-amino-4-hydroxy-pteridine ring, linked by a methylene (CH2) group to a p-aminobenzoyl moiety, which is in turn linked through amide bond to the α-amino group of a monoglutamate or poly-γ-glutamate (Fig. 1 A; Blakley, 1987). One-carbon (1C) units can be attached to N5, N10, or both. On the other hand, the name “folic acid” is reserved for the synthetic form with the fully oxidized pteridine ring and no 1C substitution (Fig. 1 B).
Figure 1.
Folate chemistry. (A) Chemical structure of THF. (B) Redox reactions for the pteridine ring of folates. (C) The 1C unit is derived from donors such as serine, exists in three different oxidation states, and is used for various biochemical processes.
The number of distinct molecular species within the folate family could be as many as 150, due to the combinatorial effect of varying 1C and pteridine oxidation states and varying polyglutamate chain lengths (Krumdieck et al., 1983). Nevertheless, typically <50 species are detectable in natural animal and plant sources (Gregory, 1989), with the pteridine ring predominantly in the most reduced, tetrahydro state (Fig. 1 B), N5 and N10 either unsubstituted or substituted with one of six different 1C units (Fig. 1 C), and the polyglutamate chain length being predominantly five to eight (Lin et al., 1993).
Folates as 1C carriers or electron carriers
In most folate-mediated reactions, tetrahydrofolate (THF) serves as a 1C carrier, which obtains the 1C unit from a donor (e.g., serine) and then transfers it to an acceptor, typically a biosynthetic intermediate. The attached 1C units exist in three different oxidation states that are equivalent to methanol, formaldehyde, and formate (Fig. 1 C) and can be interconverted by cellular enzymes using nicotinamide adenine dinucleotide phosphate (NADP) or nicotinamide adenine dinucleotide (NAD) as a cosubstrate.
Although the pteridine ring in folates is capable of undergoing redox reactions, this capability is rarely used. A notable exception occurs in the thymidylate synthase–catalyzed reaction (Fig. 1 B), in which the folate pteridine ring undergoes a two-electron oxidation. To sustain THF levels, therefore, DHFR is required to act downstream of thymidylate synthase to recycle DHF back to THF.
Chemical lability of reduced folates
Naturally occurring, reduced folates are chemically labile. In fact, the first identified folate, folic acid, was likely an artifact due to the lengthy isolation procedures, during which natural folates were largely decomposed by reacting with molecular oxygen, and a minor component among the oxidized products was folic acid (Krumdieck et al., 1983). The chemical lability is an important aspect of folates, not only presenting a technical challenge for their chemical analysis, but also reflecting their tendency to break down in vivo. Antioxidants, ascorbate in particular, are effective in minimizing folate decomposition and have proved useful in various analytical methods for folates (Quinlivan et al., 2006).
Different classes of folates, defined by the pteridine oxidation state and the 1C identity, display markedly different stabilities and follow distinct degradation pathways. We describe the pathways for each class, roughly in the order of increased stability.
THF
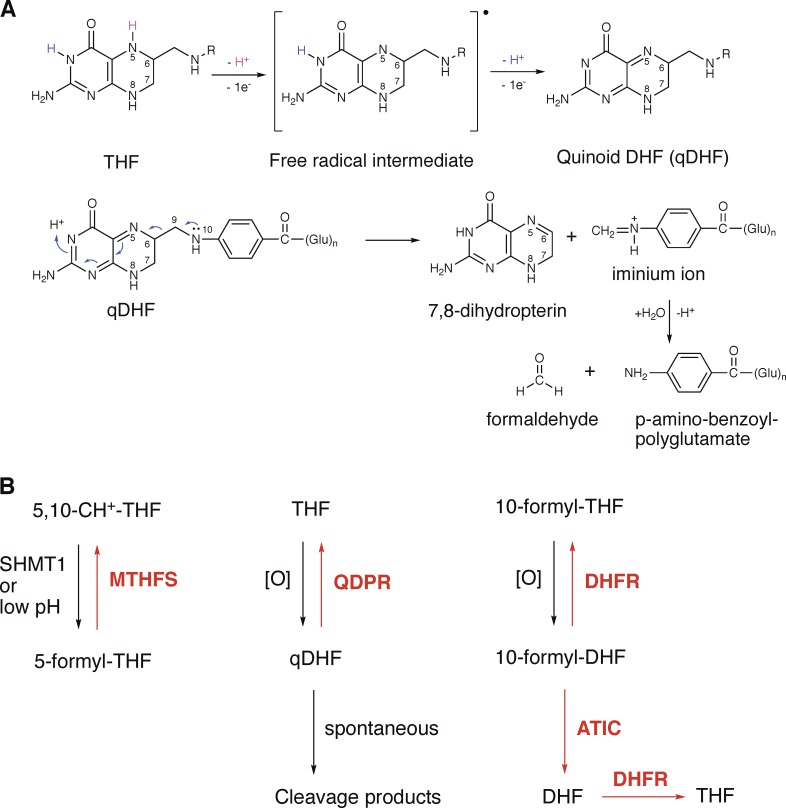
In the absence of antioxidants, the half-life of THF in aqueous solutions at 37°C is typically <30 min (Pollock and Kaufman, 1978). The pathway of THF decomposition is mediated by quinoid DHF (qDHF), generated via a free radical mechanism (Fig. 2 A). qDHF is unstable and readily cleaves into 7,8-dihydropterin, formaldehyde, and p-aminobenzoylglutamate (pABG; Reed and Archer, 1980; Burgos-Barragan et al., 2017). Formaldehyde is a potent cross-linker for proteins and nucleic acids (Burgos-Barragan et al., 2017).
Figure 2.
Folate decomposition and damage repair. (A) Chemical pathway of THF decomposition. (B) Cellular enzymatic systems for folate damage repair.
10-Formyl-THF
In the absence of antioxidants, 10-formyl-THF is readily oxidized to 10-formyl-DHF (half-life, typically <30 min at 37°C), and on prolonged incubation, is further oxidized to 10-formyl-folic acid (Baggott et al., 1995; Baggott, 2000).
5,10-CH2-THF and 5,10-CH+-THF
There appear to be few reports on the stability of 5,10-CH2-THF and 5,10-CH+-THF, presumably because 5,10-CH2-THF is in equilibrium with THF and formaldehyde (Kallen, 1971), and 5,10-CH+-THF would hydrolyze to 10-formyl-THF and, to a much lesser degree, 5-formyl-THF at neutral pHs (Stover and Schirch, 1993).
DHF
DHF is much more stable than THF, with a half-life of 4.5 h at 30°C, pH 7.0 (Reed and Archer, 1980). DHF can undergo either pteridine ring oxidation to folic acid or C9-N10 cleavage to pABG and 6-formyl-dihydropterin or dihydroxanthopterin (Chippel and Scrimgeour, 1970; Reed and Archer, 1980). The partition into these three pathways seems to depend on the pH and the type of buffer (Chippel and Scrimgeour, 1970; Reed and Archer, 1980).
5-Methyl-THF
5-Methyl-THF can be readily oxidized to 5-methyl-5,6-DHF. In the presence of thiols or ascorbate, 5-methyl-5,6-DHF readily reverts to 5-methyl-THF. Conversely, when subject to more oxidative conditions, 5-methyl-5,6-DHF either rearranges to a 5-methyl-dihydro-pyrazino-s-trazine compound or cleaves into an unidentified pterin and pABG (Gregory, 1989).
Folic acid and 5-formyl-THF
These two species are stable to oxidation, except under harsh conditions such as in the presence of KMnO4 (Maruyama et al., 1978).
Paradoxical stability of folates in vivo
Given the intrinsic tendency of reduced folates to decompose, it is surprising that folates were found to be extremely slowly turned over in human whole body or in cultured mammalian cells (Krumdieck et al., 1978; von der Porten et al., 1992; Lawrence et al., 2014). For example, using a 2H-folic acid tracer, folate breakdown rates in adult humans were estimated to be ∼0.5% per day (half-life, ∼19 d; von der Porten et al., 1992). This is in stark contrast to the situation in plants, where folate breakdown is very rapid, at a rate of ∼10% per day (Orsomando et al., 2006).
A series of excellent reviews (Linster et al., 2013; Lerma-Ortiz et al., 2016; Sun et al., 2017) emphasized two important facets of metabolism that were largely overlooked in classic textbooks of biochemistry. First, enzymes are not always exquisitely specific and efficient, and many of them also catalyze unwanted side reactions. Moreover, many metabolites are chemically labile and reactive, and thus prone to damage. To counter such metabolite damage, cells have often evolved dedicated metabolite repair enzymes. In an example related to folate metabolism, 5-formyl-THF, a dead-end metabolite formed from 5,10-CH+-THF by a side reaction of serine hydroxymethyltransferases (SHMT1/2; Stover and Schirch, 1992) or spontaneously at mildly acidic pHs (Baggott, 2000), is salvaged by methenyl-THF synthetase (MTHFS) back to 5,10-CH+-THF in eukaryotes and most prokaryotes (Fig. 2 B; Linster et al., 2013).
We recently found that cellular THF decomposition can be prevented by an enzyme, quinoid dihydropteridine reductase (QDPR); specifically, the qDHF intermediate of THF decomposition can be reduced by QDPR back to THF at the expense of two reducing equivalents from NADH (Fig. 2 B; Zheng et al., 2018). Moreover, the oxidation product of 10-formyl-THF, 10-formyl-DHF, can be reduced back to 10-formyl-THF by DHFR, or to THF by tandem actions of ATIC and DHFR (Fig. 2 B; Zheng et al., 2018). Thus, the oxidative damage to the two least stable folates, THF and 10-formyl-THF, can be enzymatically repaired in mammalian cells.
It is likely that other factors also contribute to folate stability in vivo. For example, folates can be stabilized through protein binding (Wittwer and Wagner, 1981; Cook and Wagner, 1982; Min et al., 1988; Jones and Nixon, 2002). In addition, the reduced intracellular milieu maintained by glutathione- and thioredoxin-mediated systems may play an indispensable role as well.
Folate pathways: Identified and unidentified components
Among the three processes we discuss in this section, folate uptake and catabolism typically occur on a much slower timescale than most 1C transfer reactions, consistent with the cell’s differential needs for folate and 1C. Folate is needed only in catalytic amounts to function as a cofactor, whereas 1C must be rapidly transferred via heavy-traffic pathways to support demanding processes such as nucleotide synthesis.
Folate uptake and intracellular accumulation
Folate uptake at the cell surface
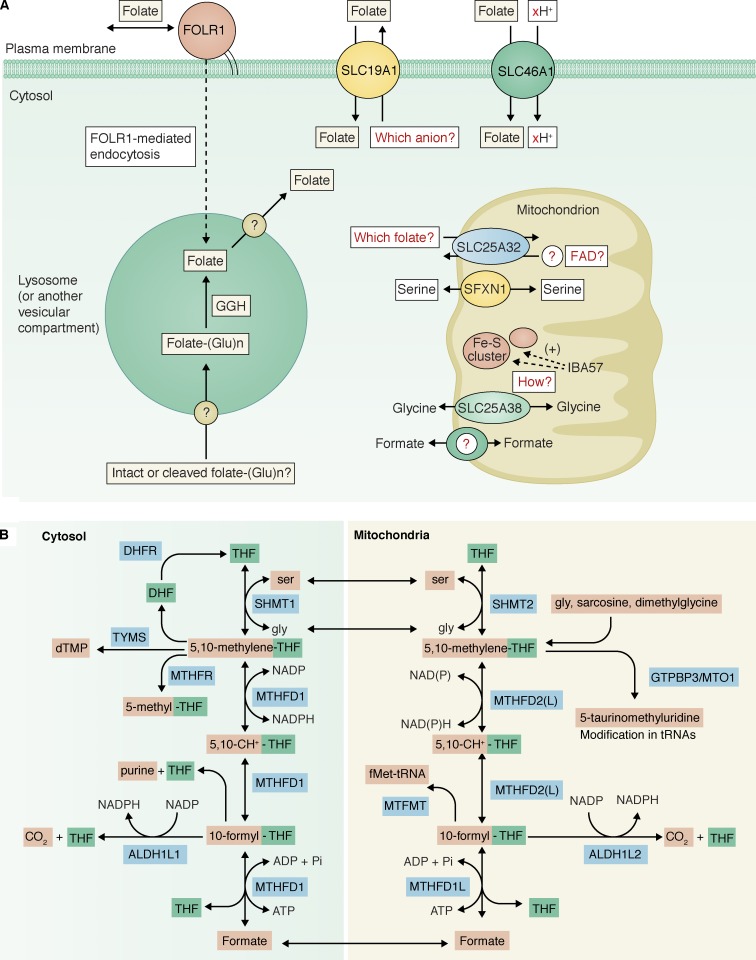
At the plasma membrane, two transmembrane carriers (SLC19A1 and SLC46A1) and three glycosylphosphatidylinositol-anchored receptors (FOLR1–3) can mediate folate uptake (Fig. 3 A; Zhao et al., 2011). The substrate of these transporters and receptors is folate monoglutamate, consistent with the fact that the circulating folate is predominantly 5-methyl-THF monoglutamate (Ratanasthien et al., 1974). SLC19A1, also known as the reduced folate carrier, is a widely expressed antiporter that exchanges a reduced folate for another anion substrate. Candidates for the counter substrate of SLC19A1, including thiamine phosphates and 5-aminoimidazole-4-carboxamide ribonucleotide monophosphate (ZMP or AICAR), have been suggested (Visentin et al., 2012). ZMP is an intermediate in purine biosynthesis and is known to accumulate in folate deficiency, in both bacteria (Stetten and Fox, 1945; Bochner and Ames, 1982) and mammal cells (Herbert et al., 1964; McGeer et al., 1965). Therefore, using ZMP as a counter substrate for SLC19A1 would represent an elegant feedback mechanism for regulating intracellular folate homeostasis.
Figure 3.
Folate transport and metabolic compartmentation. (A) Knowledge gaps in cellular folate metabolism. (B) Parallel cytosolic and mitochondrial folate pathways.
SLC46A1, also known as the proton-coupled folate transporter, is most active at acidic extracellular pHs. Consistent with its low pH optimum, SLC46A1 catalyzes symport of protons with the folate substrate, notably in the acidic, upper region of the small intestine, where it is highly expressed. Loss-of-function mutations in SLC46A1 are the only known cause of the rare disease hereditary folate malabsorption (Qiu et al., 2006; Zhao et al., 2007).
The folate receptors, strongly expressed in certain epithelial tissues and some gynecological cancers, mediate folate uptake through an endocytic pathway, but the exact mechanism is a matter of debate (Paulos et al., 2004; Chadda et al., 2007). Whatever the mechanism, to support cellular folate-dependent functions, the internalized folate monoglutamate must be effluxed from an internal vesicular compartment into the cytosol (Fig. 3 A). It has been suggested that SLC46A1 could mediate this efflux (Zhao et al., 2009), yet the same authors also pointed out that FOLR1 can function normally in some cells that lack SLC46A1 expression, indicating that there is at least one alternative transporter (Zhao et al., 2011).
Folate uptake into the mitochondria
At the mitochondrial inner membrane, SLC25A32, a member of the mitochondrial carrier family that consists of ∼50 proteins, imports reduced folate monoglutamates into the mitochondrial matrix (Fig. 3 A). Surprisingly, however, human SLC25A32 cDNA was able to complement the growth defects of a yeast mutant lacking a homologous gene, FLX1, that encodes a putative mitochondrial flavin adenine dinucleotide (FAD) transporter (Spaan et al., 2005). Recently, compound heterozygous mutations in SLC25A32 were found in a patient with riboflavin-responsive exercise intolerance, further supporting a link between SLC25A32 and FAD metabolism (Schiff et al., 2016). Thus, these studies have raised the question of whether mammalian SLC25A32 is a carrier for folate, FAD, or both.
The virtual absence of mitochondrial folates in SLC25A32 mutant Chinese hamster ovary (CHO) cells strongly argues that folate is a bona fide substrate for SLC25A32 (Titus and Moran, 2000; McCarthy et al., 2004). In contrast, FLX1-deficient yeast still has sizable mitochondrial FAD (Tzagoloff et al., 1996). Moreover, mammalian mitochondria (Barile et al., 2000; Calvo et al., 2016), and possibly yeast mitochondria as well (Bafunno et al., 2004), can synthesize FAD from riboflavin using riboflavin kinase and FAD synthetase, so it is not apparent why FAD needs to be imported from the cytosol. Given that the transport mechanism of SLC25A32 remains to be further delineated (Lawrence et al., 2011), it is possible that SLC25A32 is an antiporter that couples folate import with FAD export (Fig. 3 A). Consistent with this possibility, yeast FLX1 has been suggested to be a mitochondrial FAD export carrier (Bafunno et al., 2004).
Cytosolic and mitochondrial folate retention by polyglutamation
Folate polyglutamation, catalyzed by folylpolyglutamate synthetase (FPGS), plays an indispensable role for intracellular folate accumulation. In cells lacking FPGS, folate monoglutamates that have been taken up will readily efflux out of the cell, resulting in only ∼10% of normal folate accumulation (Osborne et al., 1993). Thus, polyglutamation adds multiple negative charges to the imported folates and thereby dramatically enhances their intracellular accumulation. Moreover, polyglutamation markedly increases the affinity of most folate-dependent enzymes for their folate substrate (Matthews et al., 1987).
DHF, THF, and 10-formyl-THF are efficient substrates for FPGS, whereas 5-methyl-THF, 5-formyl-THF, and folic acid are poor substrates (Cichowicz and Shane, 1987). Because 5-methyl-THF is the major circulating folate, its conversion by methionine synthase (MTR) to THF is thought to be the first metabolic step in cells that precedes polyglutamation (Cook et al., 1987).
FPGS is expressed in most tissues as at least two isoforms that result from alternative splicing and alternative translational start sites, one of which targets to the mitochondrial matrix (Lawrence et al., 2014). This enables the establishment of distinct, nonmiscible cytosolic and mitochondrial folate pools that operate in two parallel pathways, described next.
Parallel cytosolic and mitochondrial 1C pathways
Parallel 1C pathways exist in the cytosol and in mitochondria, respectively, each composed of a core set of serine hydroxymethyltransferase, CH2-THF dehydrogenase/CH+-THF cyclohydrolase, and 10-formyl-THF synthetase activities (Fig. 3 B; Shin et al., 2014). These two pathways are connected by small metabolites that can readily traverse the mitochondrial inner membrane, such as serine, glycine, and formate. The consensus for the interrelationship of the two pathways is that the mitochondrial 1C pathway produces formate, which in turn is used in the cytosolic pathway for purine, dTMP, and methionine biosynthesis, or excreted out of the cell if in excess (Christensen and MacKenzie, 2006; Tibbetts and Appling, 2010; Meiser et al., 2016; Ducker and Rabinowitz, 2017).
Recently, SFXN1 has been identified as a mitochondrial serine transporter (Kory et al., 2018). SLC25A38 has been suggested to be a mitochondrial glycine transporter (Fernández-Murray et al., 2016; Lunetti et al., 2016). However, the genes encoding mitochondrial transporters for formate, sarcosine, and dimethylglycine remain unknown (Fig. 3 A).
Another well-recognized role for the mitochondrial 1C pathway is glycine synthesis (Titus and Moran, 2000; Christensen and MacKenzie, 2006; Tibbetts and Appling, 2010; Jain et al., 2012; Ducker and Rabinowitz, 2017). A long-standing question is why the cytosolic SHMT1 cannot produce sufficient glycine. To generate glycine continuously via SHMT1, THF has to be continuously regenerated. For the cytosolic 1C pathway operating in the direction of glycine synthesis, THF regeneration is impeded by the unfavorable NADP/NADPH and ADP/ATP ratios in the cytosol (Veech et al., 1979; Christensen and MacKenzie, 2006). Our recent work showed that overexpression of ALDH1L1 can overcome this thermodynamic barrier by bypassing the unfavorable 10-formyl-THF synthetase step, thereby allowing continuous THF regeneration and glycine synthesis in the cytosol (Zheng et al., 2018).
The mitochondrial 1C pathway also supports mitochondrion-specific functions. It is well known that mitochondrial 10-formyl-THF is used for N-formylation of the initiator methionine in mitochondrial methionyl-tRNAMet. Loss-of-function mutations in the human formyltransferase gene, MTFMT, have been causally linked to Leigh syndrome, a mitochondrial disease (Tucker et al., 2011). Unexpectedly, a recent study showed that SHMT2-knockout HCT116 cells contained normal levels of formylmethionine in mitochondrially translated peptides (Morscher et al., 2018). In contrast, another recent study found severe depletion of mitochondrial formyl-methionyl-tRNAMet in SHMT2-knockout Jurkat cells (Minton et al., 2018). It remains to be clarified whether these discrepancies can be accounted for by the differences in the cell line context, and if so, what the context-specific modulators are.
Recent research is also beginning to reveal a surprising role for mitochondrial 5,10-CH2-THF in mitochondrial protein translation. These studies, first performed on prokaryotic systems, highlight the synergy between genetic, structural, and biochemical approaches. It was first established, through genetics, that two bacterial proteins, MnmE and MnmG, are involved in modifying uridine at the wobble position of certain bacterial transfer RNAs (tRNAs) to 5-methyluridine derivatives (Elseviers et al., 1984; Hagervall et al., 1987), yet the biochemical understanding was lagging behind.
A breakthrough came when x-ray crystallography revealed that MnmE contains a folate-binding fold strikingly resembling that in several known folate-binding enzymes (Scrima et al., 2005; Scrutton and Leys, 2005). This revelation enabled the long-awaited in vitro reconstitution experiments, in which various THF derivatives were tested (together with other known cosubstrates) for the ability to stimulate the reaction of the MnmE/MnmG complex with tRNAs (Moukadiri et al., 2009; Armengod et al., 2012). Indeed, 5,10-CH2-THF was found to be active and was suggested to be an essential cofactor for MnmE/MnmG.
The human orthologues of MnmE and MnmG, GTPBP3 and MTO1, have been predicted to be responsible for the occurrence of 5-taurinomethyluridine at the wobble position in certain mitochondrial tRNAs (Suzuki et al., 2011; Suzuki and Suzuki, 2014). To date, successful in vitro reconstitution for this putative activity has not been published. However, compelling genetic evidence has been provided by a recent study: knocking out either MTO1 or SHMT2 (which supplies mitochondrial 5,10-CH2-THF) in HCT116 cells resulted in no detectable levels of 5-taurinomethyluridine in mitochondrial tRNAs (Morscher et al., 2018). Consistent with the crucial role of this wobble uridine modification in stabilizing codon–anticodon pairing, these knockout cells were defective in mitochondrial protein translation (Morscher et al., 2018). Thus, the mitochondrial folate pathway provides the 5,10-CH2-THF substrate for GTPBP3/MTO1-mediated mitochondrial tRNA modification in human cells, an important finding with relevance to the mitochondrial disorders myoclonic epilepsy with ragged red fibers and mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes, which are caused by defective 5-taurinomethyluridine modification (Suzuki et al., 2011; Suzuki and Suzuki, 2014).
Our understanding of mitochondrial folate metabolism is not yet complete, however. Like MnmE, another bacterial protein YgfZ contains a folate-binding fold in its crystal structure, and its folate-binding activity was confirmed by biophysical experiments (Teplyakov et al., 2004). YgfZ homologues (named IBA57 for the human gene) occur in all domains of life and, through a poorly understood mechanism, are essential for the assembly or maintenance of certain iron-sulfur proteins in bacteria (Waller et al., 2010, 2012) and in yeast (Gelling et al., 2008) and human (Sheftel et al., 2012) mitochondria (Fig. 3 A). At least in bacteria, the function of YgfZ in iron-sulfur cluster metabolism is folate dependent (Waller et al., 2010, 2012). Further investigation into IBA57 is much needed to fully decipher the mitochondrial respiration defects caused by impaired mitochondrial folate metabolism.
Folate catabolism
Folate catabolism is extremely slow under normal conditions, but can be greatly accelerated with methotrexate treatment (Zheng et al., 2018). The bulk of folate catabolism in vivo is accounted for by the appearance of p-acetamidobenzoylglutamate (pAcABG) and pterins, products of folate C9-N10 bond cleavage, in urine (Murphy et al., 1976; Krumdieck et al., 1978). Whereas the C9-N10 bond cleavage is probably spontaneous, reflecting folate intrinsic instability, formation of pAcABG from the direct cleavage product pABG requires an enzyme, the NAT1 acetyltransferase (Minchin, 1995; Wakefield et al., 2007).
The cellular pathways for folate catabolism are poorly understood. Because plasma membrane efflux is prohibited for polyglutamated folates, pABG, or pAcABG, conversion of folate polyglutamate into folate monoglutamate, or deglutamation, is required for cellular clearance of folate catabolites. The only known enzyme capable of folate deglutamation is γ-glutamyl-hydrolase (GGH), which is either contained within lysosomes or secreted into the extracellular space (Hugonnet, 2012). Consistent with the role of GGH in folate deglutamation, GGH up-regulation was associated with a shift in intracellular folate distribution to shorter polyglutamate species in a hepatoma cell line (Yao et al., 1995). To access GGH, polyglutamated folates, pABG, or pAcABG have to be imported into the lysosome. A lysosomal carrier specific for polyglutamated rather than monoglutamated methotrexate was reported in the 1990s (Barrueco et al., 1992; Sirotnak and Tolner, 1999), but many questions remain unanswered, such as its gene identity and whether it preferentially imports cleaved folates (Fig. 3 A). The acidic pH (∼4.5) of lysosomal lumen is expected to precipitate some spontaneous reactions of folates, such as the conversion of 5,10-CH+-THF to 5-formyl-THF (Baggott, 2000). Finally, it remains to be determined whether the monoglutamate product of GGH is directly secreted via lysosomal exocytosis (Settembre et al., 2013) or first released into the cytosol via a lysosomal carrier.
Approaches to identify missing components
Historically, many components involved in folate metabolism were discovered by biochemical purification to chase down the protein responsible for an activity of interest (Tan et al., 1977; Huennekens, 1996) and by forward genetic screens followed by cDNA complementation (Dixon et al., 1994; Titus and Moran, 2000). The use of radioactive folic acid tracer in rats also greatly accelerated the discoveries of folate transporters/receptors and folate-binding enzymes for which folate was not an expected substrate but later proved to be an integral component of their catalysis (Wagner, 1982; Garcia et al., 2016). More recently, searching for paralogs in the then just-completed human genome database allowed identification of missing genes for known activities (Prasannan et al., 2003; Bolusani et al., 2011).
It is possible that saturation has not been reached in traditional forward genetic screens where chemical-induced mutagenesis could have target bias. Moreover, in some cases, the causal gene remains elusive. For example, glycine auxotrophic mutants of CHO cells discovered in the 1970s (Chasin et al., 1974; McBurney and Whitmore, 1974) fall into four complementation groups: GLYA (=SHMT2), GLYB (=SLC25A32), AUXB1 (=FPGS), and GLYC/AUXB2. Although the growth defect of the GLYC mutant was complemented by human DHFR2 (also named DHFRL1) cDNA (Anderson et al., 2011), the DHFR2 gene does not exist in the rodent from which the CHO line was derived. Thus, the GLYC/AUXB2 gene remains to be identified, for example by RNA or whole-exome sequencing.
The radioactive folic acid tracer approach had been limited to folate binders that are highly abundant and show high affinity (dissociation constant = submicromolar or lower) toward its folate ligand (Min et al., 1988). The same limitations would be encountered if a folate affinity column (Wittwer and Wagner, 1980) were used to pull down potential binders (Burdine and Kodadek, 2004). Although protein abundance is less an issue with the advent of highly sensitive mass spectrometry instruments, the requirement for tight binding remains a hurdle. Thus, new proteomic approaches such as thermal proteome profiling (Martinez Molina et al., 2013; Franken et al., 2015) and limited proteolysis–small molecule mapping (Piazza et al., 2018), developed to capture both strong and weak protein–ligand interactions in cells, can be powerfully complementary.
If an activity is known to localize to an intracellular organelle (e.g., lysosomes or mitochondria), then the search for the gene can be dramatically narrowed down to dozens or fewer candidates by referring to published inventories of genes encoding proteins for that organelle (Chapel et al., 2013; Pagliarini and Rutter, 2013; Calvo et al., 2016). A caveat is that these inventories have incomplete coverage and finite false discovery rates (Pagliarini and Rutter, 2013), and moreover, for the lysosome, it might be difficult to distinguish resident proteins from cargo proteins delivered there for degradation (Chapel et al., 2013). Nevertheless, recent success stories, such as the discoveries of the mitochondrial pyruvate carrier (Bricker et al., 2012; Herzig et al., 2012) and the mitochondrial calcium uniporter (Baughman et al., 2011), highlight the excellent potential for this approach.
The above organelle proteomics–based approach correlates an activity with a list of proteins on the basis of subcellular colocalization. Yet there are other ways to infer correlations. For example, genes with unknown functions can be linked to genes with well-defined functions on the basis of coevolution, coexpression, protein–protein interactions, suppression and synthetic interactions, and for microbial orthologues of a candidate human gene, gene clustering and gene fusion (https://string-db.org/ and https://thebiogrid.org/; Pagliarini and Rutter, 2013; Niehaus et al., 2015). To further understand the function of a gene of interest, one may zero in on short stretches of protein-encoding or promoter sequences and correlate the protein or DNA sequences with well-defined protein kinase phosphorylation or transcription factor binding motifs, as detailed in previous reviews (Frame and Cohen, 2001; Manning and Cantley, 2007) and exemplified by some recent studies (Ben-Sahra et al., 2013, 2016; Ye et al., 2014; DeNicola et al., 2015; Lee et al., 2017). Ultimately, the hypotheses thus generated need to be tested using genetic, biochemical, and pharmacological approaches.
Interaction of the folate cycle with the methionine cycle
ATP, NAD, and NADP are universal cofactors in metabolism. Therefore, folate pathways are connected to almost every other metabolic pathway through shared ATP, NAD, and NADP pools. Although folate pathways can contribute to the production of NADPH and ATP, their fluxes are also directly affected by compartment-specific ATP/ADP, NAD/NADH, and NADPH/NADP ratios. Recently it has been proposed that NADPH produced by ALDH1L2 in the mitochondrial 1C pathway is important to limit mitochondrial ROS (Piskounova et al., 2015; Yang and Vousden, 2016). Here, we further discuss the methionine cycle, which is linked to folate cycle via a more “specialized” metabolite, 5-methyl-THF (Fig. 4).
Figure 4.
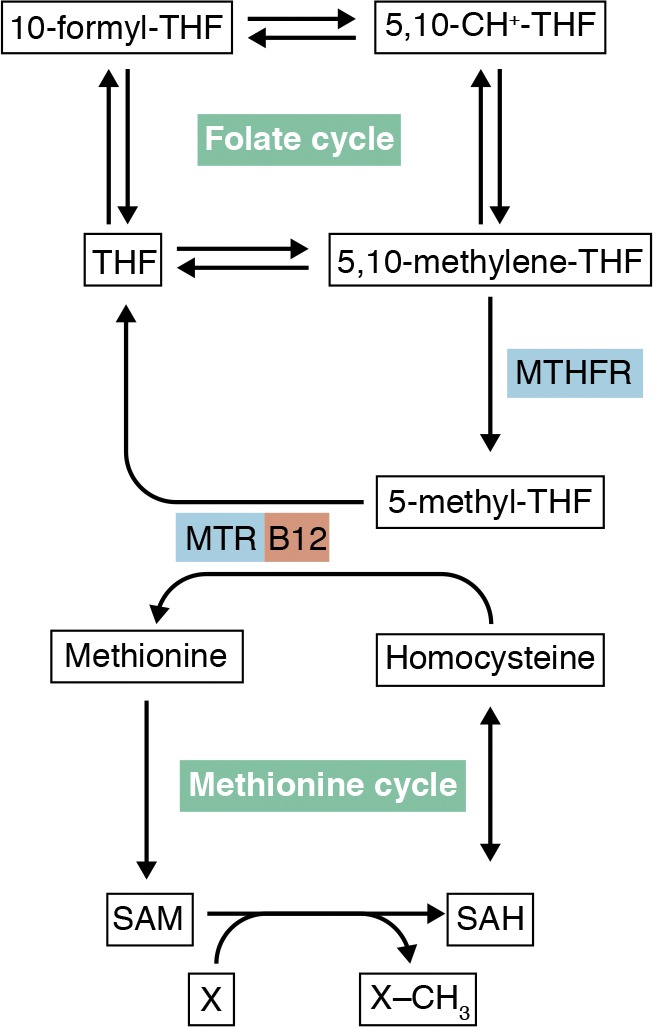
The folate cycle is linked to the methionine cycle via MTHFR and vitamin B12–using MTR.
The link between folate and methionine cycles has long been recognized, since a deficiency in either folate or vitamin B12 (an essential cofactor for MTR) produces megaloblastic anemias that are hematologically indistinguishable (Shane and Stokstad, 1985). The megaloblastic anemia in both cases is ascribed to defective DNA synthesis and can be reversed by large doses of folates. Thus, vitamin B12 deficiency causes functional folate deficiency, and this is best explained by the methyl trap model (Shane and Stokstad, 1985). Methylenetetrahydrofolate reductase (MTHFR)–catalyzed conversion of 5,10-CH2-THF to 5-methyl-THF is irreversible in cells, due to the large negative standard free-energy change and high cytosolic NADPH/NADP ratios (Matthews and Drummond, 1990). In addition, methionine synthase is the only enzyme that can remove the methyl group from 5-methyl-THF. Thus, a lack of vitamin B12 and thus MTR activity would irreversibly trap folates into 5-methyl-THF, at the expense of 5,10-CH2-THF and 10-formyl-THF, both of which are crucial to DNA synthesis.
The original methyl trap model has been expanded to include two subsequent developments. First, MTHFR is allosterically inhibited by S-adenosyl-methionine (SAM) with an inhibition constant of 2.8 µM (Shane and Stokstad, 1985). Thus, large doses of methionine can increase intracellular SAM, block the methyl trap, and thereby ameliorate both folate and B12-deficient megaloblastic anemia. Second, as discussed above, the MTR-catalyzed conversion of 5-methyl-THF to THF is likely the first folate metabolic step in cells that precedes polyglutamation, so vitamin B12 deficiency would also compromise intracellular folate polyglutamation and retention. This explains the clinically observed, diminished total cellular folate content in vitamin B12 deficiency.
It is currently unresolved to what extent the folate cycle contributes to cellular methylations in normal physiology and diseases such as cancers. MTHFR-knockout mice are viable, and some clinical symptoms of human MTHFR deficiency may be ascribed to hyperhomocysteinemia rather than alterations in methylation (Schwahn et al., 2003). Many cultured cell lines are unable to proliferate when the medium methionine is replaced with homocysteine (Borrego et al., 2016; Lien et al., 2017), and indeed stable isotope labeling and mass spectrometry analysis indicated that for many cell lines, the contribution of serine and folate pathways to the active methyl group is insignificant compared with medium methionine (Pike et al., 2010; Shyh-Chang et al., 2013; Ducker et al., 2016; Maddocks et al., 2016). Understanding the mechanistic bases for such methionine dependency in cancer cell lines should be an important future direction, especially in light of the recent discoveries that many oncogenic mutations directly impact DNA and histone methyltransferases and demethylases (Garraway and Lander, 2013).
Functions of folate metabolism and disease connections
The biochemical outputs of folate metabolism include nucleotide synthesis, serine-glycine interconversion, mitochondrial tRNA modification, and methyl group biogenesis, which in turn supports cellular functions such as proliferation, mitochondrial respiration, and epigenetic regulation. Abnormal folate metabolism has been causally linked with a myriad of diseases. However, for each disease, it is often unclear which biochemical processes and cellular functions are culprits.
The earliest known folate-related disease is probably megaloblastic anemia. Systemic folate deficiency, whether dietary, alcohol or drug induced, or due to germline mutations in DHFR or SLC46A1 (Qiu et al., 2006; Banka et al., 2011; Cario et al., 2011), invariably results in megaloblastic anemia. The molecular basis for megaloblastic anemia has been ascribed to deranged DNA synthesis, which results in chromosome breakage, and continued cell growth without division during red blood cell production (Shane and Stokstad, 1985). In some other cell types, folate deficiency can cause uracil misincorporation into DNA, leading to chromosome breakage (Blount et al., 1997). It has been proposed that such breaks could contribute to the increased risk of cancer (Blount et al., 1997).
Cerebral folate deficiency, caused by folate receptor autoantibodies or germline mutations in FOLR1 or SLC46A1, is manifested in neurological impairments and shows some resemblance to severe MTHFR deficiency (Watkins and Rosenblatt, 2014). Thus, folate metabolism plays a crucial, yet poorly defined, role in the brain.
Insufficient folate intake around and during pregnancy has been associated with increased risk of neural tube defects, which is the basis for the mandatory folate fortification of foods in the US since 1998 (Momb and Appling, 2014). Strong evidence for the role of the mitochondrial folate pathway in neural tube development has been provided by a MTHFD1L-knockout mouse model, which presents neural tube defects with 100% penetrance while on a folate-proficient diet (Momb et al., 2013). In addition, mice deficient in the AMT or GLDC gene, encoding a subunit of the glycine cleavage system, also developed neural tube defects with partial penetrance (Narisawa et al., 2012; Pai et al., 2015). It was further shown with the MTHFD1L or GLDC-knockout mice that supplementation with formate rescued the neural tube defects (Momb et al., 2013; Pai et al., 2015). In a recent study, further deletion of MTHFR in GLDC-null mouse embryos resulted in significant protection from neural tube defects, suggesting that folate and formate-supported nucleotide synthesis, rather than MTHFR-mediated methyl group biogenesis, is crucial for neural tube development (Leung et al., 2017).
Conversely, excess folate intake may enhance proliferation of malignant tumors (Strickland et al., 2013). Moreover, genes encoding enzymes in serine biosynthesis and the mitochondrial folate pathway are often amplified or overexpressed in cancers (Locasale et al., 2011; Possemato et al., 2011; Nilsson et al., 2014). It has been suggested that metabolic activities under oncogenic control in cancers can be categorized based on whether they are transforming, enabling, or neutral (Vander Heiden and DeBerardinis, 2017). As attested by the successes of methotrexate and other antifolates in treating cancers, it is likely that serine biosynthesis and mitochondrial folate metabolism belong to the first two categories, and therefore are potential therapeutic targets (Mullarky et al., 2016; Pacold et al., 2016; Ducker et al., 2017). On the other hand, restriction of dietary serine and glycine has been proposed to treat cancers that lack up-regulation of de novo serine biosynthesis (Maddocks et al., 2017).
Future perspectives
Throughout this review, we have highlighted areas that await further development and discussed how to explore them using existing technologies. Yet it is hard to predict where upcoming technologies will lead us. From a biochemical perspective, we envision a future where the entire human proteome is structurally solved (Petsko, 2014) and the folate binding fold (Scrutton and Leys, 2005) might be revealed in many more proteins, thus pushing forward a new level of understanding of folate metabolism in human health and disease.
Acknowledgments
This work was supported by a Department of Defense Breast Cancer Research Program postdoctoral fellowship (W81XWH-13-1-0251 to Y. Zheng), a National Cancer Institute grant (R35 CA197588 to L.C. Cantley), a Lustgarten Foundation grant (388207 to L.C. Cantley), and a gift from the Mindy and Jon Gray family (L.C. Cantley).
L.C. Cantley is a founder and member of the Science Advisory Board of Agios Pharmaceuticals and Petra Pharmaceuticals, companies that are developing drugs to treat cancer. Petra Pharmaceuticals provides funds to support the laboratory of L.C. Cantley. Y. Zheng declares no competing financial interests.
Author Contributions: Y. Zheng and L.C. Cantley conceived and designed the study; Y. Zheng wrote the paper; and L.C. Cantley reviewed the paper.
References
- Anderson D.D., Quintero C.M., and Stover P.J.. 2011. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc. Natl. Acad. Sci. USA. 108:15163–15168. 10.1073/pnas.1103623108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengod M.E., Moukadiri I., Prado S., Ruiz-Partida R., Benítez-Páez A., Villarroya M., Lomas R., Garzón M.J., Martínez-Zamora A., Meseguer S., and Navarro-González C.. 2012. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie. 94:1510–1520. 10.1016/j.biochi.2012.02.019 [DOI] [PubMed] [Google Scholar]
- Bafunno V., Giancaspero T.A., Brizio C., Bufano D., Passarella S., Boles E., and Barile M.. 2004. Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 279:95–102. 10.1074/jbc.M308230200 [DOI] [PubMed] [Google Scholar]
- Baggott J.E. 2000. Hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate at pH 2.5 to 4.5. Biochemistry. 39:14647–14653. 10.1021/bi001362m [DOI] [PubMed] [Google Scholar]
- Baggott J.E., Johanning G.L., Branham K.E., Prince C.W., Morgan S.L., Eto I., and Vaughn W.H.. 1995. Cofactor role for 10-formyldihydrofolic acid. Biochem. J. 308:1031–1036. 10.1042/bj3081031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banka S., Blom H.J., Walter J., Aziz M., Urquhart J., Clouthier C.M., Rice G.I., de Brouwer A.P.M., Hilton E., Vassallo G., et al. 2011. Identification and characterization of an inborn error of metabolism caused by dihydrofolate reductase deficiency. Am. J. Hum. Genet. 88:216–225. 10.1016/j.ajhg.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile M., Brizio C., Valenti D., De Virgilio C., and Passarella S.. 2000. The riboflavin/FAD cycle in rat liver mitochondria. Eur. J. Biochem. 267:4888–4900. 10.1046/j.1432-1327.2000.01552.x [DOI] [PubMed] [Google Scholar]
- Barrueco J.R., O’Leary D.F., and Sirotnak F.M.. 1992. Facilitated transport of methotrexate polyglutamates into lysosomes derived from S180 cells. Further characterization and evidence for a simple mobile carrier system with broad specificity for homo- or heteropeptides bearing a C-terminal glutamyl moiety. J. Biol. Chem. 267:19986–19991. [PubMed] [Google Scholar]
- Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L., et al. 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 476:341–345. 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I., Howell J.J., Asara J.M., and Manning B.D.. 2013. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 339:1323–1328. 10.1126/science.1228792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I., Hoxhaj G., Ricoult S.J.H., Asara J.M., and Manning B.D.. 2016. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 351:728–733. 10.1126/science.aad0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakley R.L. 1987. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature and symbols for folic acid and related compounds. Recommendations 1986. Eur. J. Biochem. 168:251–253. 10.1111/j.1432-1033.1987.tb13413.x [DOI] [PubMed] [Google Scholar]
- Blount B.C., Mack M.M., Wehr C.M., MacGregor J.T., Hiatt R.A., Wang G., Wickramasinghe S.N., Everson R.B., and Ames B.N.. 1997. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. USA. 94:3290–3295. 10.1073/pnas.94.7.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B.R., and Ames B.N.. 1982. ZTP (5-amino 4-imidazole carboxamide riboside 5′-triphosphate): a proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell. 29:929–937. 10.1016/0092-8674(82)90455-X [DOI] [PubMed] [Google Scholar]
- Bolusani S., Young B.A., Cole N.A., Tibbetts A.S., Momb J., Bryant J.D., Solmonson A., and Appling D.R.. 2011. Mammalian MTHFD2L encodes a mitochondrial methylenetetrahydrofolate dehydrogenase isozyme expressed in adult tissues. J. Biol. Chem. 286:5166–5174. 10.1074/jbc.M110.196840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego S.L., Fahrmann J., Datta R., Stringari C., Grapov D., Zeller M., Chen Y., Wang P., Baldi P., Gratton E., et al. 2016. Metabolic changes associated with methionine stress sensitivity in MDA-MB-468 breast cancer cells. Cancer Metab. 4:9 10.1186/s40170-016-0148-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.-C., Cox J.E., Cardon C.M., Van Vranken J.G., Dephoure N., et al. 2012. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 337:96–100. 10.1126/science.1218099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine L., and Kodadek T.. 2004. Target identification in chemical genetics: the (often) missing link. Chem. Biol. 11:593–597. 10.1016/j.chembiol.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Burgos-Barragan G., Wit N., Meiser J., Dingler F.A., Pietzke M., Mulderrig L., Pontel L.B., Rosado I.V., Brewer T.F., Cordell R.L., et al. 2017. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature. 548:549–554. 10.1038/nature23481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Clauser K.R., and Mootha V.K.. 2016. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario H., Smith D.E.C., Blom H., Blau N., Bode H., Holzmann K., Pannicke U., Hopfner K.-P., Rump E.-M., Ayric Z., et al. 2011. Dihydrofolate reductase deficiency due to a homozygous DHFR mutation causes megaloblastic anemia and cerebral folate deficiency leading to severe neurologic disease. Am. J. Hum. Genet. 88:226–231. 10.1016/j.ajhg.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K.J. 2003a A short history of nutritional science: part 3 (1912-1944). J. Nutr. 133:3023–3032. 10.1093/jn/133.10.3023 [DOI] [PubMed] [Google Scholar]
- Carpenter K.J. 2003b A short history of nutritional science: part 4 (1945-1985). J. Nutr. 133:3331–3342. 10.1093/jn/133.11.3331 [DOI] [PubMed] [Google Scholar]
- Chadda R., Howes M.T., Plowman S.J., Hancock J.F., Parton R.G., and Mayor S.. 2007. Cholesterol-sensitive Cdc42 activation regulates actin polymerization for endocytosis via the GEEC pathway. Traffic. 8:702–717. 10.1111/j.1600-0854.2007.00565.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapel A., Kieffer-Jaquinod S., Sagné C., Verdon Q., Ivaldi C., Mellal M., Thirion J., Jadot M., Bruley C., Garin J., et al. 2013. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell. Proteomics. 12:1572–1588. 10.1074/mcp.M112.021980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L.A., Feldman A., Konstam M., and Urlaub G.. 1974. Reversion of a Chinese hamster cell auxotrophic mutant. Proc. Natl. Acad. Sci. USA. 71:718–722. 10.1073/pnas.71.3.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippel D., and Scrimgeour K.G.. 1970. Oxidative degradation of dihydrofolate and tetrahydrofolate. Can. J. Biochem. 48:999–1009. 10.1139/o70-156 [DOI] [PubMed] [Google Scholar]
- Christensen K.E., and MacKenzie R.E.. 2006. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. BioEssays. 28:595–605. 10.1002/bies.20420 [DOI] [PubMed] [Google Scholar]
- Cichowicz D.J., and Shane B.. 1987. Mammalian folylpoly-gamma-glutamate synthetase. 2. Substrate specificity and kinetic properties. Biochemistry. 26:513–521. 10.1021/bi00376a025 [DOI] [PubMed] [Google Scholar]
- Cook R.J., and Wagner C.. 1982. Purification and partial characterization of rat liver folate binding protein: cytosol I. Biochemistry. 21:4427–4434. 10.1021/bi00261a036 [DOI] [PubMed] [Google Scholar]
- Cook J.D., Cichowicz D.J., George S., Lawler A., and Shane B.. 1987. Mammalian folylpoly-gamma-glutamate synthetase. 4. In vitro and in vivo metabolism of folates and analogues and regulation of folate homeostasis. Biochemistry. 26:530–539. 10.1021/bi00376a027 [DOI] [PubMed] [Google Scholar]
- DeNicola G.M., Chen P.-H., Mullarky E., Sudderth J.A., Hu Z., Wu D., Tang H., Xie Y., Asara J.M., Huffman K.E., et al. 2015. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47:1475–1481. 10.1038/ng.3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon K.H., Lanpher B.C., Chiu J., Kelley K., and Cowan K.H.. 1994. A novel cDNA restores reduced folate carrier activity and methotrexate sensitivity to transport deficient cells. J. Biol. Chem. 269:17–20. 10.1111/hsc.12375 [DOI] [PubMed] [Google Scholar]
- Ducker G.S., and Rabinowitz J.D.. 2017. One-Carbon Metabolism in Health and Disease. Cell Metab. 25:27–42. 10.1016/j.cmet.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker G.S., Chen L., Morscher R.J., Ghergurovich J.M., Esposito M., Teng X., Kang Y., and Rabinowitz J.D.. 2016. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 23:1140–1153. 10.1016/j.cmet.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker G.S., Ghergurovich J.M., Mainolfi N., Suri V., Jeong S.K., Hsin-Jung Li S., Friedman A., Manfredi M.G., Gitai Z., Kim H., and Rabinowitz J.D.. 2017. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA. 114:11404–11409. 10.1073/pnas.1706617114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elseviers D., Petrullo L.A., and Gallagher P.J.. 1984. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 12:3521–3534. 10.1093/nar/12.8.3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber S., and Diamond L.K.. 1948. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 238:787–793. 10.1056/NEJM194806032382301 [DOI] [PubMed] [Google Scholar]
- Farber S., Cutler E.C., Hawkins J.W., Harrison J.H., Peirce E.C. II, and Lenz G.G.. 1947. The Action of Pteroylglutamic Conjugates on Man. Science. 106:619–621. 10.1126/science.106.2764.619 [DOI] [PubMed] [Google Scholar]
- Fernández-Murray J.P., Prykhozhij S.V., Dufay J.N., Steele S.L., Gaston D., Nasrallah G.K., Coombs A.J., Liwski R.S., Fernandez C.V., Berman J.N., and McMaster C.R.. 2016. Glycine and Folate Ameliorate Models of Congenital Sideroblastic Anemia. PLoS Genet. 12:e1005783 10.1371/journal.pgen.1005783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame S., and Cohen P.. 2001. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359:1–16. 10.1042/bj3590001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken H., Mathieson T., Childs D., Sweetman G.M.A., Werner T., Tögel I., Doce C., Gade S., Bantscheff M., Drewes G., et al. 2015. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 10:1567–1593. 10.1038/nprot.2015.101 [DOI] [PubMed] [Google Scholar]
- Gabaldón T., and Koonin E.V.. 2013. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 14:360–366. 10.1038/nrg3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia B.A., Luka Z., Loukachevitch L.V., Bhanu N.V., and Wagner C.. 2016. Folate deficiency affects histone methylation. Med. Hypotheses. 88:63–67. 10.1016/j.mehy.2015.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway L.A., and Lander E.S.. 2013. Lessons from the cancer genome. Cell. 153:17–37. 10.1016/j.cell.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Gelling C., Dawes I.W., Richhardt N., Lill R., and Mühlenhoff U.. 2008. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol. Cell. Biol. 28:1851–1861. 10.1128/MCB.01963-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory J.F., III 1989. Chemical and Nutritional Aspects of Folate Research: Analytical Procedures, Methods of Folate Synthesis, Stability, and Bioavailability of Dietary Folates. In Advances in Food and Nutrition Research. Elsevier, New York, pp. 1–101. [DOI] [PubMed] [Google Scholar]
- Hagervall T.G., Edmonds C.G., McCloskey J.A., and Björk G.R.. 1987. Transfer RNA(5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities. J. Biol. Chem. 262:8488–8495. [PubMed] [Google Scholar]
- Hanson A.D., and Gregory J.F. III. 2011. Folate biosynthesis, turnover, and transport in plants. Annu. Rev. Plant Biol. 62:105–125. 10.1146/annurev-arplant-042110-103819 [DOI] [PubMed] [Google Scholar]
- Herbert V., Streiff R.R., Sullivan L.W., and McGeer P.L.. 1964. Deranged purine metabolism manifested by aminoimidazolecarboxamide excretion in megaloblastic anaemias, haemolytic anaemia, and liver disease. Lancet. 2:45–46. 10.1016/S0140-6736(64)90042-X [DOI] [PubMed] [Google Scholar]
- Herzig S., Raemy E., Montessuit S., Veuthey J.-L., Zamboni N., Westermann B., Kunji E.R.S., and Martinou J.-C.. 2012. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 337:93–96. 10.1126/science.1218530 [DOI] [PubMed] [Google Scholar]
- Horlbeck M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A., Chen Y., Fields A.P., Park C.Y., Corn J.E., Kampmann M., and Weissman J.S.. 2016. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5:e19760 10.7554/eLife.19760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huennekens F.M. 1994. The methotrexate story: a paradigm for development of cancer chemotherapeutic agents. Adv. Enzyme Regul. 34:397–419. 10.1016/0065-2571(94)90025-6 [DOI] [PubMed] [Google Scholar]
- Huennekens F.M. 1996. In search of dihydrofolate reductase. Protein Sci. 5:1201–1208. 10.1002/pro.5560050626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J.-E. 2012. γ-Glutamyl hydrolase. In Handbook of proteolytic enzymes. Rawlings N.D., and Salveson G., editors. Elsevier Ltd., Amsterdam. [Google Scholar]
- Hutchings B.L., Bohonos N., and Peterson W.H.. 1941. Growth factors for bacteria. XIII. Purification and properties of an eluate factor required by certain lactic acid bacteria. J. Biol. Chem. 141:521–528. [Google Scholar]
- Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., and Mootha V.K.. 2012. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 336:1040–1044. 10.1126/science.1218595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.L., and Nixon P.F.. 2002. Tetrahydrofolates are greatly stabilized by binding to bovine milk folate-binding protein. J. Nutr. 132:2690–2694. 10.1093/jn/132.9.2690 [DOI] [PubMed] [Google Scholar]
- Kallen R.G. 1971. [183] Tetrahydrofolic acid and formaldehyde. Methods Enzymol. 18:705–716. 10.1016/S0076-6879(71)18140-2 [DOI] [Google Scholar]
- Kory N., Wyant G.A., Prakash G., Uit de Bos J., Bottanelli F., Pacold M.E., Chan S.H., Lewis C.A., Wang T., Keys H.R., et al. 2018. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science. 362:eaat9528 10.1126/science.aat9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumdieck C.L., Fukushima K., Fukushima T., Shiota T., and Butterworth C.E. Jr. 1978. A long-term study of the excretion of folate and pterins in a human subject after ingestion of 14C folic acid, with observations on the effect of diphenylhydantoin administration. Am. J. Clin. Nutr. 31:88–93. 10.1093/ajcn/31.1.88 [DOI] [PubMed] [Google Scholar]
- Krumdieck C.L., Tamura T., and Eto I.. 1983. Synthesis and analysis of the pteroylpolyglutamates. Vitam. Horm. 40:45–104. 10.1016/S0083-6729(08)60432-X [DOI] [PubMed] [Google Scholar]
- Lawrence S.A., Hackett J.C., and Moran R.G.. 2011. Tetrahydrofolate recognition by the mitochondrial folate transporter. J. Biol. Chem. 286:31480–31489. 10.1074/jbc.M111.272187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S.A., Titus S.A., Ferguson J., Heineman A.L., Taylor S.M., and Moran R.G.. 2014. Mammalian mitochondrial and cytosolic folylpolyglutamate synthetase maintain the subcellular compartmentalization of folates. J. Biol. Chem. 289:29386–29396. 10.1074/jbc.M114.593244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Zheng Y., Cho S., Jang C., England C., Dempsey J.M., Yu Y., Liu X., He L., Cavaliere P.M., et al. 2017. Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell. 171:1545–1558.e18. 10.1016/j.cell.2017.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma-Ortiz C., Jeffryes J.G., Cooper A.J.L., Niehaus T.D., Thamm A.M.K., Frelin O., Aunins T., Fiehn O., de Crécy-Lagard V., Henry C.S., and Hanson A.D.. 2016. ‘Nothing of chemistry disappears in biology’: the Top 30 damage-prone endogenous metabolites. Biochem. Soc. Trans. 44:961–971. 10.1042/BST20160073 [DOI] [PubMed] [Google Scholar]
- Leung K.-Y., Pai Y.J., Chen Q., Santos C., Calvani E., Sudiwala S., Savery D., Ralser M., Gross S.S., Copp A.J., and Greene N.D.E.. 2017. Partitioning of One-Carbon Units in Folate and Methionine Metabolism Is Essential for Neural Tube Closure. Cell Reports. 21:1795–1808. 10.1016/j.celrep.2017.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien E.C., Ghisolfi L., Geck R.C., Asara J.M., and Toker A.. 2017. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci. Signal. 10:eaao6604 10.1126/scisignal.aao6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.F., Huang R.F., and Shane B.. 1993. Regulation of folate and one-carbon metabolism in mammalian cells. III. Role of mitochondrial folylpoly-gamma-glutamate synthetase. J. Biol. Chem. 268:21674–21679. [PubMed] [Google Scholar]
- Linster C.L., Van Schaftingen E., and Hanson A.D.. 2013. Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 9:72–80. 10.1038/nchembio.1141 [DOI] [PubMed] [Google Scholar]
- Locasale J.W. 2013. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 13:572–583. 10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale J.W., Grassian A.R., Melman T., Lyssiotis C.A., Mattaini K.R., Bass A.J., Heffron G., Metallo C.M., Muranen T., Sharfi H., et al. 2011. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43:869–874. 10.1038/ng.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetti P., Damiano F., De Benedetto G., Siculella L., Pennetta A., Muto L., Paradies E., Marobbio C.M.T., Dolce V., and Capobianco L.. 2016. Characterization of Human and Yeast Mitochondrial Glycine Carriers with Implications for Heme Biosynthesis and Anemia. J. Biol. Chem. 291:19746–19759. 10.1074/jbc.M116.736876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D.K., Labuschagne C.F., Adams P.D., and Vousden K.H.. 2016. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell. 61:210–221. 10.1016/j.molcel.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D.K., Athineos D., Cheung E.C., Lee P., Zhang T., van den Broek N.J.F., Mackay G.M., Labuschagne C.F., Gay D., Kruiswijk F., et al. 2017. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 544:372–376. 10.1038/nature22056 [DOI] [PubMed] [Google Scholar]
- Manning B.D., and Cantley L.C.. 2007. AKT/PKB signaling: navigating downstream. Cell. 129:1261–1274. 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., and Nordlund P.. 2013. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 341:84–87. 10.1126/science.1233606 [DOI] [PubMed] [Google Scholar]
- Maruyama T., Shiota T., and Krumdieck C.L.. 1978. The oxidative cleavage of folates. A critical study. Anal. Biochem. 84:277–295. 10.1016/0003-2697(78)90511-0 [DOI] [PubMed] [Google Scholar]
- Matthews R.G., and Drummond J.T.. 1990. Providing one-carbon units for biological methylations: mechanistic studies on serine hydroxymethyltransferase, methylenetetrahydrofolate reductase, and methyltetrahydrofolate-homocysteine methyltransferase. Chem. Rev. 90:1275–1290. 10.1021/cr00105a010 [DOI] [Google Scholar]
- Matthews R.G., Ghose C., Green J.M., Matthews K.D., and Dunlap R.B.. 1987. Folylpolyglutamates as substrates and inhibitors of folate-dependent enzymes. Adv. Enzyme Regul. 26:157–171. 10.1016/0065-2571(87)90012-4 [DOI] [PubMed] [Google Scholar]
- McBurney M.W., and Whitmore G.F.. 1974. Isolation and biochemical characterization of folate deficient mutants of Chinese hamster cells. Cell. 2:173–182. 10.1016/0092-8674(74)90091-9 [DOI] [PubMed] [Google Scholar]
- McCarthy E.A., Titus S.A., Taylor S.M., Jackson-Cook C., and Moran R.G.. 2004. A mutation inactivating the mitochondrial inner membrane folate transporter creates a glycine requirement for survival of chinese hamster cells. J. Biol. Chem. 279:33829–33836. 10.1074/jbc.M403677200 [DOI] [PubMed] [Google Scholar]
- McGeer P.L., Sen N.P., and Grant D.A.. 1965. Excretion of 4(5)-amino-5(4)-imidazolecarboxamide and formimino-l-glutamic acid in folic acid and vitamin B12 deficient rats. Can. J. Biochem. 43:1367–1374. 10.1139/o65-152 [DOI] [PubMed] [Google Scholar]
- Meiser J., Tumanov S., Maddocks O., Labuschagne C.F., Athineos D., Van Den Broek N., Mackay G.M., Gottlieb E., Blyth K., Vousden K., et al. 2016. Serine one-carbon catabolism with formate overflow. Sci. Adv. 2:e1601273 10.1126/sciadv.1601273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H., Shane B., and Stokstad E.L.. 1988. Identification of 10-formyltetrahydrofolate dehydrogenase-hydrolase as a major folate binding protein in liver cytosol. Biochim. Biophys. Acta. 967:348–353. 10.1016/0304-4165(88)90097-9 [DOI] [PubMed] [Google Scholar]
- Minchin R.F. 1995. Acetylation of p-aminobenzoylglutamate, a folic acid catabolite, by recombinant human arylamine N-acetyltransferase and U937 cells. Biochem. J. 307:1–3. 10.1042/bj3070001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton D.R., Nam M., McLaughlin D.J., Shin J., Bayraktar E.C., Alvarez S.W., Sviderskiy V.O., Papagiannakopoulos T., Sabatini D.M., Birsoy K., and Possemato R.. 2018. Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs. Mol. Cell. 69:610–621.e5. 10.1016/j.molcel.2018.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell H.K., Snell E.E., and Williams R.J.. 1941. The concentration of “folic acid.”. J. Am. Chem. Soc. 63:2284 10.1021/ja01853a512 [DOI] [Google Scholar]
- Momb J., and Appling D.R.. 2014. Mitochondrial one-carbon metabolism and neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 100:576–583. 10.1002/bdra.23268 [DOI] [PubMed] [Google Scholar]
- Momb J., Lewandowski J.P., Bryant J.D., Fitch R., Surman D.R., Vokes S.A., and Appling D.R.. 2013. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc. Natl. Acad. Sci. USA. 110:549–554. 10.1073/pnas.1211199110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morscher R.J., Ducker G.S., Li S.H.-J., Mayer J.A., Gitai Z., Sperl W., and Rabinowitz J.D.. 2018. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 554:128–132. 10.1038/nature25460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukadiri I., Prado S., Piera J., Velázquez-Campoy A., Björk G.R., and Armengod M.E.. 2009. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 37:7177–7193. 10.1093/nar/gkp762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarky E., Lucki N.C., Beheshti Zavareh R., Anglin J.L., Gomes A.P., Nicolay B.N., Wong J.C.Y., Christen S., Takahashi H., Singh P.K., et al. 2016. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc. Natl. Acad. Sci. USA. 113:1778–1783. 10.1073/pnas.1521548113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M., Keating M., Boyle P., Weir D.G., and Scott J.M.. 1976. The elucidation of the mechanism of folate catabolism in the rat. Biochem. Biophys. Res. Commun. 71:1017–1024. 10.1016/0006-291X(76)90756-7 [DOI] [PubMed] [Google Scholar]
- Narisawa A., Komatsuzaki S., Kikuchi A., Niihori T., Aoki Y., Fujiwara K., Tanemura M., Hata A., Suzuki Y., Relton C.L., et al. 2012. Mutations in genes encoding the glycine cleavage system predispose to neural tube defects in mice and humans. Hum. Mol. Genet. 21:1496–1503. 10.1093/hmg/ddr585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus T.D., Thamm A.M., de Crécy-Lagard V., and Hanson A.D.. 2015. Proteins of unknown biochemical function: A persistent problem and a roadmap to help overcome it. Plant Physiol. 169:1436–1442. 10.1104/pp.15.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., Huang J., Asplund A., and Mootha V.K.. 2014. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5:3128 10.1038/ncomms4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsomando G., Bozzo G.G., de la Garza R.D., Basset G.J., Quinlivan E.P., Naponelli V., Rébeillé F., Ravanel S., Gregory J.F. III, and Hanson A.D.. 2006. Evidence for folate-salvage reactions in plants. Plant J. 46:426–435. 10.1111/j.1365-313X.2006.02685.x [DOI] [PubMed] [Google Scholar]
- Osborne C.B., Lowe K.E., and Shane B.. 1993. Regulation of folate and one-carbon metabolism in mammalian cells. I. Folate metabolism in Chinese hamster ovary cells expressing Escherichia coli or human folylpoly-gamma-glutamate synthetase activity. J. Biol. Chem. 268:21657–21664. [PubMed] [Google Scholar]
- Pacold M.E., Brimacombe K.R., Chan S.H., Rohde J.M., Lewis C.A., Swier L.J.Y.M., Possemato R., Chen W.W., Sullivan L.B., Fiske B.P., et al. 2016. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 12:452–458. 10.1038/nchembio.2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini D.J., and Rutter J.. 2013. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 27:2615–2627. 10.1101/gad.229724.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai Y.J., Leung K.-Y., Savery D., Hutchin T., Prunty H., Heales S., Brosnan M.E., Brosnan J.T., Copp A.J., and Greene N.D.E.. 2015. Glycine decarboxylase deficiency causes neural tube defects and features of non-ketotic hyperglycinemia in mice. Nat. Commun. 6:6388 10.1038/ncomms7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulos C.M., Reddy J.A., Leamon C.P., Turk M.J., and Low P.S.. 2004. Ligand binding and kinetics of folate receptor recycling in vivo: impact on receptor-mediated drug delivery. Mol. Pharmacol. 66:1406–1414. 10.1124/mol.104.003723 [DOI] [PubMed] [Google Scholar]
- Petsko G.A. 2014. Crystallography without crystals. Chem. Eng. News. 92:42–43. [Google Scholar]
- Piazza I., Kochanowski K., Cappelletti V., Fuhrer T., Noor E., Sauer U., and Picotti P.. 2018. A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell. 172:358–372.e23. 10.1016/j.cell.2017.12.006 [DOI] [PubMed] [Google Scholar]
- Pike S.T., Rajendra R., Artzt K., and Appling D.R.. 2010. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J. Biol. Chem. 285:4612–4620. 10.1074/jbc.M109.079855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E., Agathocleous M., Murphy M.M., Hu Z., Huddlestun S.E., Zhao Z., Leitch A.M., Johnson T.M., DeBerardinis R.J., and Morrison S.J.. 2015. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 527:186–191. 10.1038/nature15726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R.J., and Kaufman S.. 1978. Dihydropteridine reductase may function in tetrahydrofolate metabolism. J. Neurochem. 31:115–123. 10.1111/j.1471-4159.1978.tb12439.x [DOI] [PubMed] [Google Scholar]
- Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.-K., Jang H.G., Jha A.K., et al. 2011. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 476:346–350. 10.1038/nature10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasannan P., Pike S., Peng K., Shane B., and Appling D.R.. 2003. Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls. J. Biol. Chem. 278:43178–43187. 10.1074/jbc.M304319200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A., Jansen M., Sakaris A., Min S.H., Chattopadhyay S., Tsai E., Sandoval C., Zhao R., Akabas M.H., and Goldman I.D.. 2006. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 127:917–928. 10.1016/j.cell.2006.09.041 [DOI] [PubMed] [Google Scholar]
- Quinlivan E.P., Hanson A.D., and Gregory J.F.. 2006. The analysis of folate and its metabolic precursors in biological samples. Anal. Biochem. 348:163–184. 10.1016/j.ab.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Ratanasthien K., Blair J.A., Leeming R.J., Cooke W.T., and Melikian V.. 1974. Folates in human serum. J. Clin. Pathol. 27:875–879. 10.1136/jcp.27.11.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.S., and Archer M.C.. 1980. Oxidation of tetrahydrofolic acid by air. J. Agric. Food Chem. 28:801–805. 10.1021/jf60230a044 [DOI] [Google Scholar]
- Schiff M., Veauville-Merllié A., Su C.H., Tzagoloff A., Rak M., Ogier de Baulny H., Boutron A., Smedts-Walters H., Romero N.B., Rigal O., et al. 2016. SLC25A32 Mutations and Riboflavin-Responsive Exercise Intolerance. N. Engl. J. Med. 374:795–797. 10.1056/NEJMc1513610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R.T. 1989. The discovery of gene amplification in mammalian cells: to be in the right place at the right time. BioEssays. 11:69–73. 10.1002/bies.950110208 [DOI] [PubMed] [Google Scholar]
- Schwahn B.C., Chen Z., Laryea M.D., Wendel U., Lussier-Cacan S., Genest J. Jr., Mar M.H., Zeisel S.H., Castro C., Garrow T., and Rozen R.. 2003. Homocysteine-betaine interactions in a murine model of 5,10-methylenetetrahydrofolate reductase deficiency. FASEB J. 17:512–514. 10.1096/fj.02-0456fje [DOI] [PubMed] [Google Scholar]
- Scotti M., Stella L., Shearer E.J., and Stover P.J.. 2013. Modeling cellular compartmentation in one-carbon metabolism. Wiley Interdiscip. Rev. Syst. Biol. Med. 5:343–365. 10.1002/wsbm.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A., Vetter I.R., Armengod M.E., and Wittinghofer A.. 2005. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J. 24:23–33. 10.1038/sj.emboj.7600507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N.S., and Leys D.. 2005. Crystal structure of DMGO provides a prototype for a new tetrahydrofolate-binding fold. Biochem. Soc. Trans. 33:776–779. 10.1042/BST0330776 [DOI] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D.L., and Ballabio A.. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14:283–296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., and Zhang F.. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16:299–311. 10.1038/nrg3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane B., and Stokstad E.L.. 1985. Vitamin B12-folate interrelationships. Annu. Rev. Nutr. 5:115–141. 10.1146/annurev.nu.05.070185.000555 [DOI] [PubMed] [Google Scholar]
- Sheftel A.D., Wilbrecht C., Stehling O., Niggemeyer B., Elsässer H.-P., Mühlenhoff U., and Lill R.. 2012. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell. 23:1157–1166. 10.1091/mbc.e11-09-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M., Bryant J.D., Momb J., and Appling D.R.. 2014. Mitochondrial MTHFD2L is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase expressed in both adult and embryonic tissues. J. Biol. Chem. 289:15507–15517. 10.1074/jbc.M114.555573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J.J., Zhu H., et al. 2013. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 339:222–226. 10.1126/science.1226603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotnak F.M., and Tolner B.. 1999. Carrier-mediated membrane transport of folates in mammalian cells. Annu. Rev. Nutr. 19:91–122. 10.1146/annurev.nutr.19.1.91 [DOI] [PubMed] [Google Scholar]
- Spaan A.N., Ijlst L., van Roermund C.W.T., Wijburg F.A., Wanders R.J.A., and Waterham H.R.. 2005. Identification of the human mitochondrial FAD transporter and its potential role in multiple acyl-CoA dehydrogenase deficiency. Mol. Genet. Metab. 86:441–447. 10.1016/j.ymgme.2005.07.014 [DOI] [PubMed] [Google Scholar]
- Stetten M.R., and Fox C.L. Jr. 1945. An amine ed formed by bacteria during sulfonamide bacteriostasis. J. Biol. Chem. 161:333–349. [PubMed] [Google Scholar]
- Stokstad E.L., and Jukes T.H.. 1987. Sulfonamides and folic acid antagonists: a historical review. J. Nutr. 117:1335–1341. 10.1093/jn/117.7.1335 [DOI] [PubMed] [Google Scholar]
- Stover P., and Schirch V.. 1992. Enzymatic mechanism for the hydrolysis of 5,10-methenyltetrahydropteroylglutamate to 5-formyltetrahydropteroylglutamate by serine hydroxymethyltransferase. Biochemistry. 31:2155–2164. 10.1021/bi00122a037 [DOI] [PubMed] [Google Scholar]
- Stover P., and Schirch V.. 1993. The metabolic role of leucovorin. Trends Biochem. Sci. 18:102–106. 10.1016/0968-0004(93)90162-G [DOI] [PubMed] [Google Scholar]
- Strickland K.C., Krupenko N.I., and Krupenko S.A.. 2013. Molecular mechanisms underlying the potentially adverse effects of folate. Clin. Chem. Lab. Med. 51:607–616. 10.1515/cclm-2012-0561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Jeffryes J.G., Henry C.S., Bruner S.D., and Hanson A.D.. 2017. Metabolite damage and repair in metabolic engineering design. Metab. Eng. 44:150–159. 10.1016/j.ymben.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Suzuki T., and Suzuki T.. 2014. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 42:7346–7357. 10.1093/nar/gku390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Nagao A., and Suzuki T.. 2011. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45:299–329. 10.1146/annurev-genet-110410-132531 [DOI] [PubMed] [Google Scholar]
- Tan L.U., Drury E.J., and MacKenzie R.E.. 1977. Methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. A multifunctional protein from porcine liver. J. Biol. Chem. 252:1117–1122. [PubMed] [Google Scholar]
- Teplyakov A., Obmolova G., Sarikaya E., Pullalarevu S., Krajewski W., Galkin A., Howard A.J., Herzberg O., and Gilliland G.L.. 2004. Crystal structure of the YgfZ protein from Escherichia coli suggests a folate-dependent regulatory role in one-carbon metabolism. J. Bacteriol. 186:7134–7140. 10.1128/JB.186.21.7134-7140.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts A.S., and Appling D.R.. 2010. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 30:57–81. 10.1146/annurev.nutr.012809.104810 [DOI] [PubMed] [Google Scholar]
- Titus S.A., and Moran R.G.. 2000. Retrovirally mediated complementation of the glyB phenotype. Cloning of a human gene encoding the carrier for entry of folates into mitochondria. J. Biol. Chem. 275:36811–36817. 10.1074/jbc.M005163200 [DOI] [PubMed] [Google Scholar]
- Tucker E.J., Hershman S.G., Köhrer C., Belcher-Timme C.A., Patel J., Goldberger O.A., Christodoulou J., Silberstein J.M., McKenzie M., Ryan M.T., et al. 2011. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 14:428–434. 10.1016/j.cmet.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Jang J., Glerum D.M., and Wu M.. 1996. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J. Biol. Chem. 271:7392–7397. 10.1074/jbc.271.13.7392 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., and DeBerardinis R.J.. 2017. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 168:657–669. 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech R.L., Lawson J.W., Cornell N.W., and Krebs H.A.. 1979. Cytosolic phosphorylation potential. J. Biol. Chem. 254:6538–6547. [PubMed] [Google Scholar]
- Visentin M., Zhao R., and Goldman I.D.. 2012. Augmentation of reduced folate carrier-mediated folate/antifolate transport through an antiport mechanism with 5-aminoimidazole-4-carboxamide riboside monophosphate. Mol. Pharmacol. 82:209–216. 10.1124/mol.112.078642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Porten A.E., Gregory J.F. III, Toth J.P., Cerda J.J., Curry S.H., and Bailey L.B.. 1992. In vivo folate kinetics during chronic supplementation of human subjects with deuterium-labeled folic acid. J. Nutr. 122:1293–1299. 10.1093/jn/122.6.1293 [DOI] [PubMed] [Google Scholar]
- Wagner C. 1982. Cellular folate binding proteins; function and significance. Annu. Rev. Nutr. 2:229–248. 10.1146/annurev.nu.02.070182.001305 [DOI] [PubMed] [Google Scholar]
- Wakefield L., Cornish V., Long H., Griffiths W.J., and Sim E.. 2007. Deletion of a xenobiotic metabolizing gene in mice affects folate metabolism. Biochem. Biophys. Res. Commun. 364:556–560. 10.1016/j.bbrc.2007.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller J.C., Alvarez S., Naponelli V., Lara-Nuñez A., Blaby I.K., Da Silva V., Ziemak M.J., Vickers T.J., Beverley S.M., Edison A.S., et al. 2010. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc. Natl. Acad. Sci. USA. 107:10412–10417. 10.1073/pnas.0911586107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller J.C., Ellens K.W., Hasnain G., Alvarez S., Rocca J.R., and Hanson A.D.. 2012. Evidence that the folate-dependent proteins YgfZ and MnmEG have opposing effects on growth and on activity of the iron-sulfur enzyme MiaB. J. Bacteriol. 194:362–367. 10.1128/JB.06226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J.J., Sabatini D.M., and Lander E.S.. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 343:80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins D., and Rosenblatt D.S.. 2014. Inherited Disorders of Folate and Cobalamin Transport and Metabolism. In The Online Metabolic and Molecular Bases of Inherited Disease. Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., Gibson K.M., and Mitchell G., editors. The McGraw-Hill Companies, New York. [Google Scholar]
- Werkheiser W. 1961. Specific binding of 4-amino folic acid analogues by folic acid reductase. J. Biol. Chem. 236:888 10.1002/1097-0142(195105)4:33.0.CO;2-X/pdf [DOI] [Google Scholar]
- Wittwer A.J., and Wagner C.. 1980. Identification of folate binding protein of mitochondria as dimethylglycine dehydrogenase. Proc. Natl. Acad. Sci. USA. 77:4484–4488. 10.1073/pnas.77.8.4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer A.J., and Wagner C.. 1981. Identification of the folate-binding proteins of rat liver mitochondria as dimethylglycine dehydrogenase and sarcosine dehydrogenase. Purification and folate-binding characteristics. J. Biol. Chem. 256:4102–4108. [PubMed] [Google Scholar]
- Yang M., and Vousden K.H.. 2016. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer. 16:650–662. 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- Yao R., Rhee M.S., and Galivan J.. 1995. Effects of gamma-glutamyl hydrolase on folyl and antifolylpolyglutamates in cultured H35 hepatoma cells. Mol. Pharmacol. 48:505–511. 10.1002/(ISSN)2052-1707 [DOI] [PubMed] [Google Scholar]
- Ye J., Fan J., Venneti S., Wan Y.-W., Pawel B.R., Zhang J., Finley L.W.S., Lu C., Lindsten T., Cross J.R., et al. 2014. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4:1406–1417. 10.1158/2159-8290.CD-14-0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Min S.H., Qiu A., Sakaris A., Goldberg G.L., Sandoval C., Malatack J.J., Rosenblatt D.S., and Goldman I.D.. 2007. The spectrum of mutations in the PCFT gene, coding for an intestinal folate transporter, that are the basis for hereditary folate malabsorption. Blood. 110:1147–1152. 10.1182/blood-2007-02-077099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Min S.H., Wang Y., Campanella E., Low P.S., and Goldman I.D.. 2009. A role for the proton-coupled folate transporter (PCFT-SLC46A1) in folate receptor-mediated endocytosis. J. Biol. Chem. 284:4267–4274. 10.1074/jbc.M807665200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Diop-Bove N., Visentin M., and Goldman I.D.. 2011. Mechanisms of membrane transport of folates into cells and across epithelia. Annu. Rev. Nutr. 31:177–201. 10.1146/annurev-nutr-072610-145133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Lin T.-Y., Lee G., Paddock M.N., Momb J., Cheng Z., Li Q., Fei D.L., Stein B.D., Ramsamooj S., et al. 2018. Mitochondrial One-Carbon Pathway Supports Cytosolic Folate Integrity in Cancer Cells. Cell. 175:1546–1560.e17. 10.1016/j.cell.2018.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]