The TPX2/Aurora A heterodimeric kinase canonically orchestrates mitotic events. Byrum et al. identify two new roles for this complex in regulating DNA double-stranded break repair and the protection of DNA forks during replication stress.
Abstract
53BP1 is a chromatin-associated protein that regulates the DNA damage response. In this study, we identify the TPX2/Aurora A heterodimer, nominally considered a mitotic kinase complex, as a novel binding partner of 53BP1. We find that TPX2/Aurora A plays a previously unrecognized role in DNA damage repair and replication fork stability by counteracting 53BP1 function. Loss of TPX2 or Aurora A compromises DNA end resection, BRCA1 and Rad51 recruitment, and homologous recombination. Furthermore, loss of TPX2 or Aurora A causes deprotection of stalled replication forks upon replication stress induction. This fork protection pathway counteracts MRE11 nuclease activity but functions in parallel to BRCA1. Strikingly, concurrent loss of 53BP1 rescues not only BRCA1/Rad51 recruitment but also the fork instability induced upon TPX2 loss. Our work suggests the presence of a feedback mechanism by which 53BP1 is regulated by a novel binding partner and uncovers a unique role for 53BP1 in replication fork stability.
Introduction
DNA double-stranded breaks (DSBs) pose a substantial threat to genome integrity. Two primary mechanisms, nonhomologous end joining and homologous recombination (HR), repair DSBs (Chapman et al., 2012). HR is promoted by the tumor suppressor BRCA1, which recruits CtIP and Rad51, facilitating end resection and strand invasion of the sister chromatid. Conversely, 53BP1 promotes nonhomologous end joining through its downstream effectors RIF1, PTIP, and the REV7-shieldin complex, which block DNA end resection and the recruitment of HR proteins (Callen et al., 2013; Feng et al., 2013; Noordermeer et al., 2018).
Recent studies using single-molecule DNA fiber assays show that another function of BRCA1 is to protect stalled replication forks from degradation by MRE11 (Schlacher et al., 2012). Loss of PTIP prevents MRE11 accumulation at stalled forks and rescues nascent DNA shortening in BRCA1-deficient cells, conferring chemoresistance despite sustaining defects in HR. This suggests that protection of the replication fork is a key mechanism by which BRCA-deficient cancers survive (Ray Chaudhuri et al., 2016). Interestingly, unlike PTIP, loss of 53BP1 does not rescue shortened DNA tracks in BRCA1-deficient cells (Ray Chaudhuri et al., 2016). However, 53BP1 is enriched at stalled replication forks and forms nuclear bodies in G1 that sequester chromosomal lesions caused by replication stress during the previous cell cycle (Lukas et al., 2011; Dungrawala et al., 2015). Thus, although 53BP1 is primed to function in response to replication stress, its role in this context is still unclear.
Here, through a proteomic interaction screen, we identify TPX2 as a direct 53BP1 interactor, which in turn recruits the Aurora A kinase. TPX2 and Aurora A canonically play critical roles in orchestrating mitotic spindle events (Neumayer et al., 2014). Our work uncovers two novel nonmitotic functions of the TPX2/Aurora A heterodimer in regulating HR and replication fork stability and implicates a feedback mechanism modulating 53BP1 function.
Results and discussion
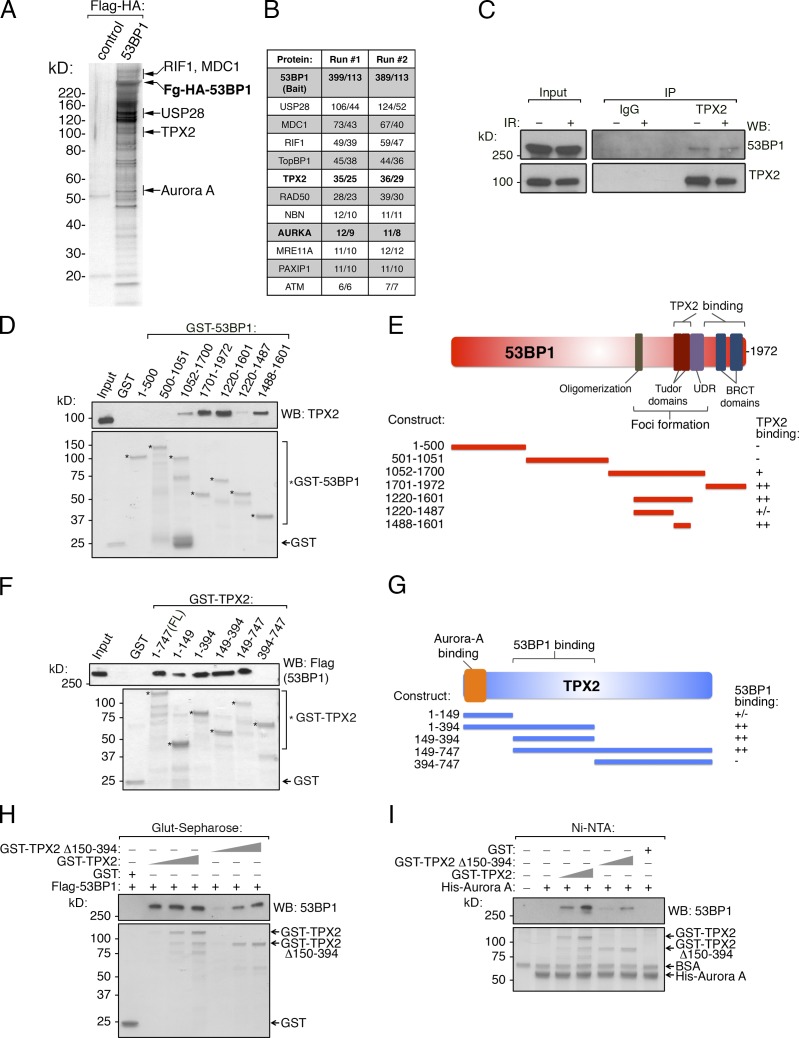
To gain a better understanding of 53BP1 function, we stably expressed and purified 53BP1 from HeLa-S cells by tandem affinity purification (TAP), as previously described (Nakatani and Ogryzko, 2003). Mass spectrometry (MS) analysis identified known 53BP1 interactors, including RIF1, MDC1, and USP28 (Fig. 1, A and B; and Table S1; Zimmermann and de Lange, 2014). By MS, we also copurified TPX2 and its kinase partner Aurora A, a heterodimer involved in mitotic progression and spindle assembly (Fig. 1, A and B; Neumayer et al., 2014). Immunoprecipitation of endogenous TPX2 confirmed its association with 53BP1 with or without γ-irradiation (Fig. 1 C). Next, we demonstrated that recombinant 53BP1 and TPX2 physically interact by in vitro pull-down assays (Figs. 1, D–H; and S1 A). Deletion analysis of 53BP1 revealed that the TPX2-binding region overlaps with the Tudor and BRCT domains, which are critical for 53BP1 localization to chromatin (Fig. 1, D and E). We did not observe TPX2 recruitment to sites of DNA damage induced by γ-irradiation or laser microirradiation (Fig. S1 B), suggesting that the interaction may occur primarily off the chromatin template. Furthermore, TPX2 residues 150–394 were important for the interaction with 53BP1 (Fig. 1, F–H). We next assessed the interaction between recombinant 53BP1 and Aurora A and found that, unlike TPX2, 53BP1 does not directly bind to Aurora A in vitro (Fig. 1 I). However, upon the addition of recombinant TPX2, 53BP1 bound immobilized His-Aurora A. This interaction was significantly reduced when TPX2-Δ150-394 was substituted for WT TPX2 (Fig. 1 I), suggesting that TPX2 mediates the interaction between 53BP1 and Aurora A using this region. Using phosphorylation of histone H3 serine 10 as a measure of Aurora A catalytic activity, we found that the TPX2-Δ150-394 53BP1-interaction mutant (hereafter referred to as TPX2-BP1-IM) activated Aurora A similarly to WT TPX2 in vitro, suggesting that this key mitotic function is maintained in this mutant protein (Fig. S1 C; Crosio et al., 2002).
Figure 1.
Identification of TPX2/Aurora A as 53BP1 effectors. (A) Flag-HA-53BP1 and associated proteins were isolated from HeLa-S nuclear extract using sequential Flag and HA immunoaffinity purification. The final eluted material was analyzed by SDS-PAGE and silver staining. (B) The 53BP1 complex from A was analyzed by MS/MS twice, and the total/unique peptide numbers for each protein are shown for each run. (C) Nuclear extracts from HeLa-S cells were immunoprecipitated with control IgG or TPX2 antibody and Western blotted as shown. (D) GST, or the indicated GST-53BP1 fragments, were immobilized on glutathione-Sepharose, and binding with full-length recombinant His-TPX2 was tested. Bound material was analyzed by Western blot or Coomassie Blue staining. (E) Schematic of 53BP1 and summary of TPX2 binding data. (F) GST, GST-TPX2, or the indicated GST-TPX2 fragments were immobilized on glutathione-Sepharose and assessed for binding to full-length Flag-53BP1, as in D. (G) Schematic of TPX2 and summary of 53BP1 binding data. (H) GST, and increasing equimolar amounts of GST-TPX2 or GST- TPX2-Δ150-394, were tested for 53BP1 binding as done in F. (I) Recombinant His-Aurora A was immobilized on Ni-NTA and incubated with GST, GST-TPX2, or GST- TPX2-Δ150-394. Bound material was washed, subsequently incubated with Flag-53BP1, washed again, and analyzed for binding by Western blot or Coomassie Blue staining.
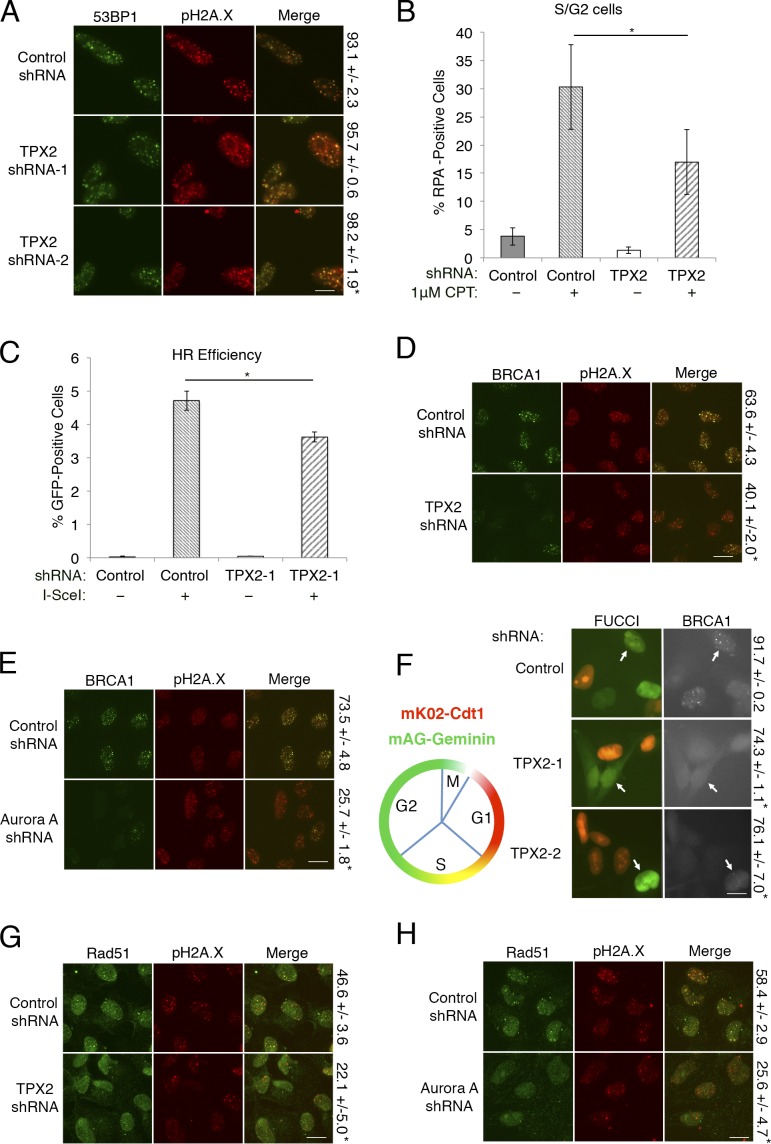
Given that the TPX2 binding region on 53BP1 includes the Tudor domains, we next asked whether TPX2 interferes with 53BP1 localization to DSBs. We depleted TPX2 from U2OS cells using shRNAs and analyzed 53BP1 irradiation-induced foci by immunofluorescence (Figs. 2 A and S1 D). 53BP1 foci were largely unaffected by loss of TPX2. Similarly, TPX2 loss did not have a significant effect on Rif1 foci formation, suggesting that TPX2 does not contribute to the recruitment of 53BP1 or its effectors (Figs. 2 A and S1 E).
Figure 2.
TPX2 and Aurora A regulate BRCA1 recruitment and function. (A) U2OS cells expressing the indicated shRNAs were irradiated (5 Gy) and analyzed for 53BP1 and pH2A.X foci formation. Percentage of cells with ≥10 53BP1 foci ± SD is shown on the right. Scale bar, 10 µm. Foci were quantitated from three biological replicates with n ≥ 200 cells per experiment. Statistics: Student’s two-tailed t test; *, P < 0.05. (B) U2OS cells expressing the indicated shRNAs were treated with camptothecin (CPT) as shown, and chromatin-bound RPA was quantified by flow cytometry. Bar graph indicates the percentage of S/G2 cells that are RPA positive, as determined by 7-AAD staining from four biological replicates. Error bars represent ± SD. Statistics: Student’s two-tailed t test; *, P < 0.05. (C) Quantitation of HR efficiency of DR-GFP U2OS cells treated with the indicated shRNA and with or without the Flag-ISce expression vector. Cells were analyzed for GFP positivity via flow cytometry in three biological replicates as described in Materials and methods. Error bars indicate ± SD. Statistics: Student’s two-tailed t test; *, P < 0.05. (D and E) U2OS cells expressing the indicated shRNAs were irradiated (5 Gy) and analyzed for BRCA1 and p.H2A.X foci formation. Percentage of cells with ≥10 BRCA1 foci ± SD is shown on the right. Scale bar, 10 µm. Foci were quantitated from three biological replicates with n ≥ 250 cells per experiment. Statistics: Student’s two-tailed t test; *, P < 0.05. (F) Schematic of FUCCI cell cycle indicator expression pattern of mK02-Cdt (red; G1) and mAG-Geminin (green; S/G2/M; left). U2OS-FUCCI cells were treated with the indicated shRNAs, and S/G2-phase cells (yellow or green cells; indicated by white arrows) were analyzed for BRCA1 foci formation (right). Percentage of S/G2 cells with ≥10 BRCA1 foci ± SD is shown on the right. Scale bar, 10 µm. Foci were quantitated from three biological replicates with n ≥ 250 cells per experiment. Statistics: Student’s two-tailed t test; *, P < 0.05. (G and H) U2OS cells expressing the indicated shRNAs were irradiated (5 Gy) and analyzed for Rad51 foci. Percentage of cells with four or greater Rad51 foci ± SD is shown on the right. Scale bar, 10 µm. Foci were quantitated from three biological replicates with n ≥ 250 cells per experiment. Statistics: Student’s two-tailed t test; *, P < 0.05.
Since 53BP1 protects DNA ends from resection and subsequent repair by HR, we used flow cytometry to measure chromatin-bound RPA levels in S/G2 cells as a proxy for DNA end resection. Strikingly, we saw a considerable decrease in RPA binding in response to TPX2 loss after damage with camptothecin (Figs. 2 B and S1 F). To functionally assess HR, we used the DR-GFP reporter system and found that loss of TPX2 reduces HR efficiency (Fig. 2 C). Seeing that BRCA1 facilitates DNA end resection and HR, we used immunofluorescence to visualize BRCA1 foci following depletion of TPX2 or Aurora A. Remarkably, knockdown of either protein resulted in a significant decrease in BRCA1 irradiation-induced foci (Figs. 2, D and E; and S1 D). While TPX2- or Aurora A–depleted cells did exhibit a reduction in S/G2 phase compared with control cells, it was not sufficient to explain the magnitude of BRCA1 foci loss (Fig. S1 G). Additionally, using a U2OS-FUCCI cell cycle indicator cell line (Sakaue-Sawano et al., 2008), we found that TPX2 or Aurora A depletion decreased BRCA1 foci formation in S/G2 cells, further suggesting that cell cycle defects alone could not explain this phenotype (Figs. 2 F and S1 H). The loss of BRCA1 foci in TPX2-depleted cells could also be rescued by reexpression of TPX2 (Fig. S1, I and J). Furthermore, BRCA1 promotes HR by initiating Rad51 recruitment, and consistently, knockdown of TPX2 or Aurora A resulted in decreased Rad51 foci (Fig. 2, G and H; Prakash et al., 2015). Together, our data suggest that TPX2 regulates BRCA1 recruitment to sites of DSBs as well as its role in repair by HR.
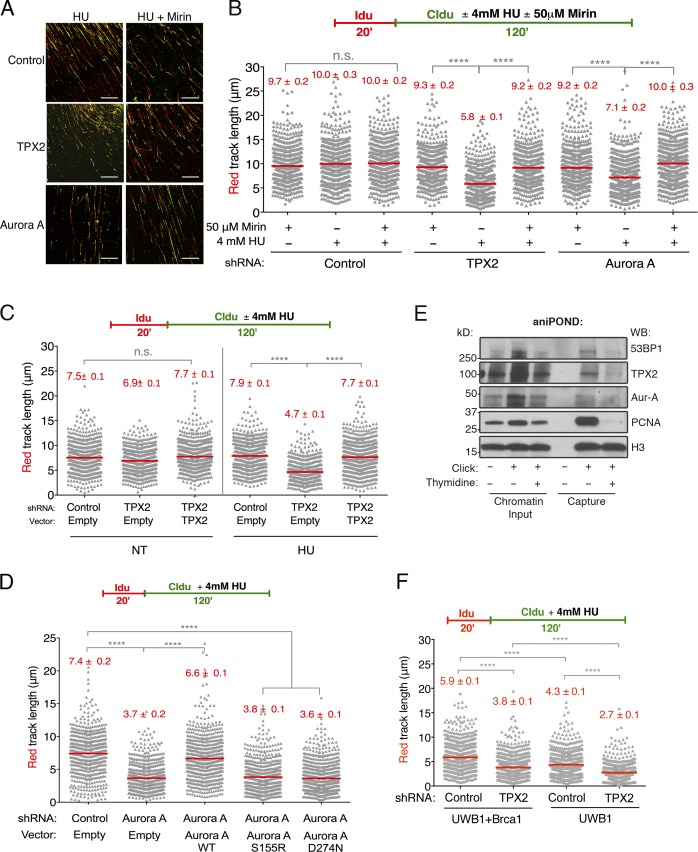
In addition to its function in end resection, BRCA1 has been shown to protect stalled replication forks from degradation by MRE11 upon replication stress induction (Schlacher et al., 2012). Therefore, we investigated whether TPX2 and Aurora A are also needed to carry out this additional function via genome-wide single-molecule DNA fiber analysis. TPX2- or Aurora A–depleted U2OS cells were labeled with the first thymidine analogue 5-iodo-2′-deoxyuridine (IdU; red) followed by treatment with hydroxyurea (HU) and concomitant labeling with the second thymidine analogue, 5-chloro-2′-deoxyuridine (CldU; green). Shortening of the IdU (red) tract was measured as a readout of degradation, as described (Lemaçon et al., 2017). Control, TPX2-depleted, and Aurora A–depleted cells produced nearly identical IdU-labeled DNA track lengths in the absence of HU, indicating that replication fork speed was unaffected (Fig. S2 A). Notably, when treated with HU, the IdU tracks of both the TPX2-depleted and Aurora A–depleted cells were significantly shorter compared with control cells (Fig. 3, A and B). Furthermore, this track-shortening phenotype was rescued upon the addition of Mirin, an MRE11-specific inhibitor. Track shortening due to TPX2 depletion could also be rescued by exogenous TPX2 expression (Fig. 3 C). Strikingly, replication fork stability was not rescued by a catalytically inactive Aurora A (D274N; Bayliss et al., 2003), nor was it rescued by an Aurora A mutant that does not interact with TPX2 (S155R; Bibby et al., 2009; Figs. 3 D and S2, B and C). These data suggest that Aurora A kinase activity as well as its interaction with TPX2 are critical for its role in replication fork stability.
Figure 3.
TPX2 and Aurora A promote replication fork stability. (A) Representative DNA fiber images of control, TPX2-depleted, and Aurora A–depleted U2OS cells treated with HU (4 mM) for 120 min. IdU, red; CldU, green. Scale bar, 50 µm. (B) Schematic of single-molecule DNA fiber track analysis (top) and size distribution of IdU track length in TPX2- and Aurora A–depleted cells in the presence and absence of HU or Mirin (bottom). Mirin (50 µM) was added concomitantly with HU treatment, as indicated. Out of two biological replicates; n ≥ 300 tracks were scored for each dataset. Bars represent median. Statistics: Mann–Whitney; ****, P < 0.0001; n.s., not significant. (C) Labeling and HU treatment schematic (top) and size distribution of IdU track length in U2OS cells expressing the indicated combination of shRNA and TPX2 expression vector as performed in B (bottom). (D) Size distribution of IdU track length in U2OS cells expressing the indicated combination of shRNA and Aurora A expression vector as performed in B. (E) U2OS cells were pulse labeled with 10 µM EdU and subjected to a 1-h thymidine chase (10 µM). Proteins bound to nascent DNA were purified by aniPOND as described in Materials and methods and analyzed by Western blot. (F) Size distribution of IdU track length in BRCA1-deficient (UWB1) and -proficient (UWB1 + BRCA1) cells in the presence of HU. Cells were infected with control or TPX2 shRNA before IdU and CldU labeling, as indicated. Fiber analysis was performed as in B.
Like BRCA1, TPX2 and Aurora A appear to play a critical role in preventing extensive MRE11-mediated degradation of stalled replication intermediates. Consistent with this, we found that TPX2 and Aurora A are present at nascent DNA using the accelerated native isolation of proteins on nascent DNA (aniPOND) assay (Fig. 3 E; Leung et al., 2013). Importantly, a thymidine chase reduced the abundance of these proteins on nascent DNA, suggesting that they are associated with the replication fork.
As TPX2/Aurora A loss impairs DNA-end resection and BRCA1 foci formation and also compromises replication fork stability, we next asked whether TPX2 and BRCA1 function in a common fork protection pathway. We depleted TPX2 from both BRCA1-deficient UWB1 cells and from UWB1 cells complemented with BRCA1 (UWB1 + BRCA1) and performed DNA fiber analysis (Figs. 3 F and S2, D and E). As expected, in the presence of HU, BRCA1-deficient UWB1 cells produced significantly shorter IdU track lengths in comparison to the UWB1 + BRCA1 cells. Comparably shortened tracks were also observed upon HU treatment in TPX2-depleted UWB1 + BRCA1 cells (Fig. 3 F). However, TPX2-depleted UWB1 cells produced even shorter IdU tracks than cells deficient for either BRCA1 or TPX2 alone, suggesting that TPX2 and BRCA1 function in separate pathways to protect stalled forks. These phenotypes were also observed in BRCA-deficient HCC 1937 cells, confirming that the observed effect is not cell type specific (Fig. S2, F and G).
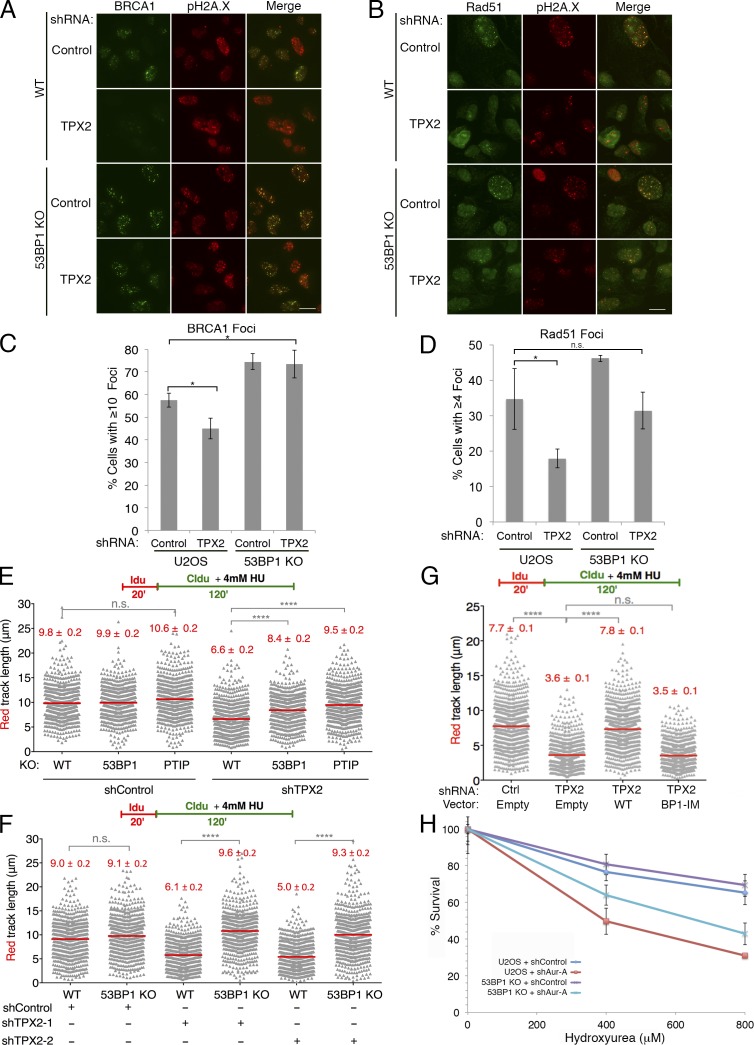
Due to the biochemical interaction between 53BP1 and TPX2, we next investigated how 53BP1 may impact TPX2/Aurora A function in DSB repair and replication fork stability. Strikingly, we found that 53BP1 deficiency prevented the loss of BRCA1 and Rad51 foci formation upon TPX2 knockdown (Figs. 4, A–D; and S2 H). In addition, our aniPOND analysis demonstrated that, like TPX2 and Aurora A, 53BP1 localizes to replication forks (Fig. 3 E). Thus, we tested whether concurrent loss of 53BP1 may also prevent stalled replication fork degradation due to TPX2 depletion. Indeed, knockout of 53BP1 partially rescued IdU track shortening in TPX2-depleted U2OS cells, while a complete rescue was observed in mouse embryonic fibroblasts (MEFs; Figs. 4, E and F; and S2, H–J). The interaction between TPX2 and 53BP1 was apparently important for the function of TPX2 in replication fork stability, as TPX2-BP1-IM failed to rescue TPX2 fork degradation (Figs. 4 G and S3, A and B). Importantly, this mutant form of TPX2 localized to mitotic spindles similar to WT TPX2 (Fig. S3 C). This supports the notion that its functions in mitosis are preserved, consistent with its ability to activate Aurora A kinase (Fig. S1 C). Furthermore, using aniPOND, we found that TPX2 depletion caused a moderate increase in 53BP1 binding to newly replicated DNA, and this effect appeared augmented when cells were treated with HU to stall replication forks (Fig. S3 D). Together, our data suggest that TPX2/Aurora A promote DSB repair and replication fork stability by negatively regulating 53BP1.
Figure 4.
TPX2/Aurora A negatively regulate 53BP1 in the DNA damage response. (A) WT U2OS or 53BP1 knockout cells expressing the indicated shRNAs were irradiated (5 Gy) and analyzed for BRCA1 and pH2A.X foci formation. (B) U2OS cells treated as in A were also analyzed by Rad51 and pH2A.X foci. Scale bars, 10 µm. (C) Quantitation of BRCA1 foci from A. Foci were quantitated from three biological replicates with n ≥ 200 cells per experiment. Error bars indicate ±SD. Statistics: Student’s two-tailed t test; *, P < 0.05. (D) Quantitation of Rad51 foci from B. Foci were quantitated from three technical replicates with n ≥ 200 cells per experiment. Error bars indicate ±SD. Statistics: Student’s two-tailed t test; *, P < 0.05. (E) Size distribution of IdU track length in U2OS cells. WT, 53BP1 KO, or PTIP KO cells were treated with control or TPX2 shRNA before IdU and CldU labeling, as indicated. Out of two biological replicates; n ≥ 300 tracks scored for each dataset. Bars represent median. Statistics: Mann–Whitney; ****, P < 0.0001; n.s., not significant. (F) Size distribution of IdU track length in MEFs. WT or 53BP1−/− cells were treated with control or TPX2 shRNA before labeling and HU treatment as indicated. Fiber analysis was performed as in E. (G) Size distribution of IdU track length in U2OS cells expressing the indicated combination of shRNA and TPX2 expression vector. Out of three biological replicates; n ≥ 300 tracks scored for each dataset. Bars represent median. Statistics: Mann–Whitney; ****, P < 0.0001; n.s., not significant. (H) U2OS WT or 53BP1-knockout cells were treated with the indicated shRNAs and subsequently treated with HU, as described in Materials and methods. Cell viability after 48 h of drug exposure was measured by MTS assay. Graph is a representative of three biological replicate experiments. Error bars indicate ±SD with n = 5 technical replicates.
In contrast to TPX2 depletion, loss of 53BP1 failed to prevent IdU track shortening in BRCA1-depleted U2OS cells, consistent with previous reports (Fig. S3, E–G; Ray Chaudhuri et al., 2016). This further substantiates our finding that TPX2 and BRCA1 protect stalled replication forks via separate pathways. As a positive control for these experiments, we also depleted TPX2 and BRCA1 from PTIP-knockout U2OS cells, as PTIP is required for MRE11 localization at stalled replication forks. As expected, loss of PTIP restored IdU track length in both BRCA1- and TPX2-depleted U2OS cells (Figs. 4 E, S2 H, and S3, E–G).
Given our evidence that TPX2/Aurora A are important for replication fork stability, we tested whether loss of Aurora A increased sensitivity to HU. Indeed, U2OS cells deficient for Aurora A were significantly more sensitive to HU (Figs. 4 H and S3 H). Strikingly, loss of 53BP1 significantly reduced HU sensitivity upon Aurora A depletion (Fig. 4 H). Thus, our data demonstrate that the functions of TPX2/Aurora A in maintaining replication fork stability depend on 53BP1.
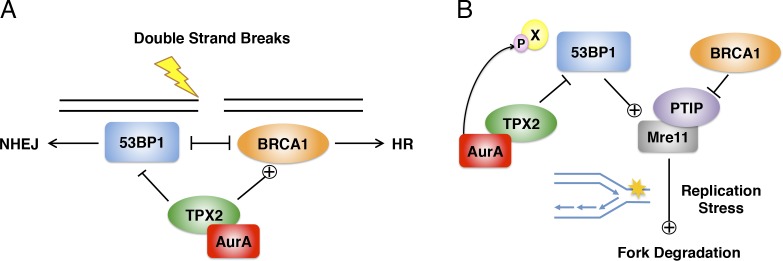
This work uncovers a previously unappreciated function of TPX2 and Aurora A in promoting DNA end resection at DSBs and replication fork stability by negatively regulating 53BP1 function. We propose a model in which TPX2/Aurora A regulate DSB repair and replication fork stability via two distinct mechanisms that are both dependent on 53BP1 (Fig. 5). TPX2/Aurora A appear to regulate 53BP1 function during DSB repair, preventing BRCA1 antagonism and thus allowing for DNA end resection and HR to take place (Fig. 5 A). This is reminiscent of the TIRR protein, which similarly antagonizes 53BP1 by binding to its Tudor domains (Drané et al., 2017; Dai et al., 2018). In the context of replication stress, TPX2/Aurora A promote the protection of stalled replication intermediates from extensive degradation by MRE11, again by inhibiting 53BP1, through a mechanism dependent on Aurora A kinase activity (Fig. 5 B). Therefore, our model suggests a novel role for 53BP1 in counteracting replication fork stability, but in a manner that is distinct from BRCA1. Finally, our model provides a rationale for targeting Aurora A kinase for improved chemosensitization, particularly in BRCA1-deficient tumors.
Figure 5.
TPX2/Aurora A regulate DSB repair and replication fork stability via negative regulation of 53BP1. (A) Model of TPX2/Aurora A regulation of 53BP1 in response to double-strand breaks. (B) Model of TPX2/Aurora A function in response to replication stress. See text for details.
Materials and methods
Cell culture
U2OS, HeLa, HeLa-S, 293T cells, and MEFs were cultured in DMEM (Invitrogen), supplemented with 10% FBS (Atlanta Biologicals) and 100 U/ml penicillin-streptomycin (Gibco) at 37°C and 5% CO2. BRCA1 mutant ovarian cancer cell line UWB1.289 (UWB1) and its complemented derivative expressing WT BRCA1 (UWB1 + BRCA1; DelloRusso et al., 2007) cells were grown in 50% RPMI medium, 50% mammary epithelial cell growth medium bullet kit (Lonza CC-3150) completed with 3% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. Human triple-negative breast cancer cells HCC 1937 (Tassone et al., 2003, 2005) were cultured in RPMI complemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. Sf9 cells were grown in suspension in Sf-900 II serum-free medium (Gibco), supplemented with 100 U/ml penicillin-streptomycin (Gibco) at 27°C.
Plasmids
53BP1 and TPX2 cDNAs were isolated from total human RNA and cloned into pOZ-Flag-HA, pENTR-3C, or pENTR4. I-SceI cDNA (a kind gift from Maria Jasin, Sloan Kettering Institute, New York, NY) was subcloned into pENTR-3C by PCR. For mammalian cell expression, cDNAs were subcloned into pHAGE-CMV-Flag, pMSCV-Flag-HA, or untagged pMSCV as needed by Gateway recombination (Sowa et al., 2009). For expression in insect cells, 53BP1 cDNA was subcloned into pDEST-BB-Flag by Gateway recombination. For bacterial expression, cDNAs were subcloned by PCR into pGEX-4T1 or pET-28a-Flag. All constructs derived by PCR, including deletions and point mutations, were confirmed by Sanger sequencing.
Transfection, virus production, and transduction
Retrovirus (pMSCV vectors) and lentivirus (pHAGE or pLKO.1 vectors) were produced by cotransfection of 293T with the appropriate packaging vectors (retrovirus: pVSV-G and pGag-Pol; lentivirus: pHDM-VSV-G, pHDM-tat1b, pHDM-Hg-PM2, and pRC-CMV-RaII; Mulligan et al., 2008). Viral supernatant was collected after 48–72 h and sterile filtered with a 0.45-µm filter. The indicated cell lines were then incubated with the viral supernatant and 4 µg/ml polybrene. For knockdown of TPX2, Aurora A, and BRCA1, cells were infected with the lentiviral shRNA vector 60–64 h before analysis by immunofluorescence, flow cytometry, or fiber assay. Lentiviral shRNA vectors were obtained from Sigma (control shRNA: SHC002) and GE Dharmacon (clone Id: TPX2-1, TRCN0000074533, 5′-TTAAAGGAAGTAACTACATGG-3′ (antisense); TPX2-2, TRCN0000074536, 5′-AAGATTAGCCTTTCTCAAAGG-3′ (antisense); TPX2-1 (mouse), TRCN0000120812, 5′-AAATAACTACAAGAAGTCTGG-3′ (antisense); TPX2-2 (mouse), TRCN0000120813, 5′-AATTCTAGGATCAAGTTCCCG-3′ (antisense); Aurora A-1, TRCN0000000655, 5′-TATAAGTAGCACAATTCTCGT-3′ (antisense); Aurora A-2, TRCN0000000656, 5′-TTCGAATGACAGTAAGACAGG-3′ (antisense); and BRCA1, TRCN0000009823, 5′-ATTCATGCCAGAGGTCTTATA-3′ (antisense). TPX2-1 and Aurora A-1 shRNAs were used for experiments in which only one knockdown is shown. For TPX2 and Aurora A rescue experiments, cells were infected with the pMSCV-TPX2 retroviral vector and selected with blasticidin (10 µg/ml) for 72 h before shRNA infection.
Antibodies
All antibodies used in this study are listed in Table S2.
Purification of TAP-53BP1 complex and MS/MS analysis
Purification of 53BP1 was performed using an established tandem immunoaffinity method (Nakatani and Ogryzko, 2003). Flag-HA-53BP1 was stably expressed after transduction of pOZ-Flag-HA-53BP1 retrovirus into HeLa-S cells. Cells were harvested and resuspended in a 5× volume of hypotonic buffer (10 mM Tris, pH 7.3, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM PMSF, 10 mM β-mercaptoethanol, and protease inhibitors; Pierce). Cells were pelleted by spinning at 2,500 rpm, resuspended in 1× pellet volume of hypotonic buffer, and homogenized using a Dounce tissue grinder (Wheaton). Nuclear material was pelleted, resuspended in 0.5× pellet volume of low-salt buffer (20 mM Tris, pH 7.3, 20 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.2 mM PMSF, 10 mM β-mercaptoethanol, and protease inhibitors), and Dounced again. 0.5× pellet volume of high-salt buffer (20 mM Tris, pH 7.3, 1.2M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.2 mM PMSF, 10 mM β-mercaptoethanol, and protease inhibitors) was slowly added to nuclear extract, which was subsequently stirred for 30–45 min. Extract was spun down at 14,000 rpm for 30 min, and the soluble material was dialyzed in BC100 buffer (20 mM Tris, pH 7.3, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.2 mM PMSF, and 1 mM β-mercaptoethanol).
53BP1 complex was purified from the nuclear extract using anti-Flag (M2) resin (Sigma), followed by purification using anti-HA (F-7) resin (Santa Cruz) in TAP buffer (50 mM Tris-HCl, pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT, and protease inhibitors). For both Flag and HA purifications, nuclear extract was rotated with resin for 4 h, washed extensively with TAP buffer, and eluted with 0.4 mg/ml Flag or HA peptide (Sigma). After elution, the complex was TCA precipitated, and associated proteins were identified by liquid chromatography-MS/MS using an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific) and Sequest software (Sowa et al., 2009).
Endogenous immunoprecipitation
HeLa-S cells were mock treated or irradiated (5 Gy) using an XCELL 50 x-ray (Kubtec), and nuclear extract was prepared following a 1-h recovery period at 37°C as described above (Nakatani and Ogryzko, 2003). The nuclear extract was precleared with protein A/G-agarose (Santa Cruz) and then incubated overnight at 4°C with 3 µg antibody. The extract was then incubated with fresh protein A/G-agarose for 1 h at 4°C, centrifuged, and washed five times with TAP buffer. Bound material was eluted using Laemmli buffer and analyzed by Western blotting.
Recombinant protein purification
Recombinant GST-tagged TPX2 proteins were purified from Rosetta (DE3) using an ÄKTA-pure FPLC (GE Healthcare). Cells were resuspended in GST-lysis buffer (50 mM Tris-HCl, pH 7.3, 250 mM NaCl, 0.05% Triton X-100, 3 mM β-ME 20, and protease inhibitors) and lysed by sonication. After centrifugation and filtration, the extract was loaded onto a GSTrap HP column (GE Healthcare). After washing with 20 column volumes of lysis buffer, the protein was eluted using lysis buffer containing 10 mM glutathione. The eluted proteins were dialyzed into TAP buffer.
Flag-53BP1 was purified from Sf9 cells infected with pDEST-BB-Flag-53BP1 recombinant baculovirus. Cells were resuspended in lysis buffer (20 mM Tris, pH 7.3, 150 mM NaCl, 8% glycerol, 0.2% NP-40, 0.1% Triton X-100, 2 mM βME, and protease inhibitors) and sonicated to complete lysis. After centrifugation, the cell extract was incubated with anti-Flag (M2) beads (Sigma) for 3 h. The bound material was eluted using lysis buffer containing 0.4 mg/ml Flag peptide (Sigma) and dialyzed into TAP wash buffer.
GST-tagged protein pull-down assays
All binding assays were performed in TAP buffer with 1% BSA in a total volume of 100 µl, using 10 µl glutathione-Sepharose beads (GE Healthcare) per reaction. The beads were preblocked with 3% BSA in TAP buffer. Each reaction contained ∼5 µg GST-tagged protein and ∼250 ng Flag-53BP1. Binding reactions were incubated at 4°C for 1 h with rotation. Beads were then washed extensively in TAP buffer, and a final wash was performed with 1× PBS. Bound material was eluted with SDS loading buffer, analyzed by SDS-PAGE, and stained with Coomassie Blue or Western blotted as indicated.
Aurora A kinase assay
His-Flag-Aurora A was incubated with GST, GST-TPX2, or GST-TPX2-BP1-IM in TAP buffer for 30 min at 4°C with rotation. Recombinant human histone H3.1 (1 µg; NEB) and ATP (10 mM final concentration) were subsequently added to each reaction for a total volume of 30 µl. Kinase reactions were incubated at room temperature for 30 min and stopped with the addition of 30 µl Laemmli buffer. Phospho-H3 (S10) levels were measured by Western blot.
AniPOND
aniPOND assays were performed as previously described with some modifications (Leung et al., 2013; Wiest and Tomkinson, 2017). Cells were pulse labeled with 10 µM EdU in DMEM for 15 min, washed with PBS, and then incubated in medium with 10 µM thymidine for 1 h or with 4 mM HU for 2 h. Medium was removed, and cells were immediately lysed and harvested with nuclei extraction buffer (20 mM Hepes, pH 7.2, 40 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% NP-40). Nuclei pellets were washed with PBS and then resuspended in click reaction mix (in order of addition: 25 mM biotin picolyl azide [Click Chemistry Tools], 10 mM (+)-sodium l-ascorbate, and 2 mM CuSO4) and rotated at 4°C for 1 h. Samples were then washed with PBS, and pellets were resuspended in 500 µl Buffer B1 (25 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 8.0, 1% NP-40, and protease inhibitors). Samples were rotated for 30 min at 4°C, spun down at maximum speed for 10 min at 4°C, resuspended again in 500 µl Buffer B1, and rotated for an additional 30 min at 4°C. Nuclei were again pelleted and resuspended in 500 µl Buffer B1. Samples were then sonicated using a Model 50 Sonic Dismembrator (Thermo Fisher Scientific) 12 × 10 s at 20 amplitude to solubilize DNA-bound proteins. Samples were spun down at maximum speed for 10 min, and the supernatant was collected. 500 µl Buffer B2 (150 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 8.0, 0.5% NP-40, and protease inhibitors) was added to the supernatant to bring the total sample size to ∼1 ml. Samples were rotated overnight (16–20 h) with streptavidin-coated beads (Thermo Fisher Scientific). A chromatin input sample was collected immediately before streptavidin capture. Beads were washed extensively with Buffer B2 and boiled in Laemmli buffer to elute bound material. A small amount of material was then analyzed for H3 via Western blotting, and densitometry of the bands was performed using ImageJ (National Institutes of Health). Loading adjustments were made based on these measurements, and the proteins of interest were then run on an SDS-PAGE gel and analyzed by Western blotting.
CRISPR/Cas9-mediated knockouts
U2OS knockout cells were created using CRISPR/Cas9 genome editing at the Genome Engineering and iPSC Center at Washington University School of Medicine (St. Louis, MO). All clones used for this study were confirmed by deep sequencing. The 53BP1 knockout clone was further confirmed by Western blot analysis. The guide RNA sequences used to generate the knockout cell line were as follows: 53BP1, 5′-GATACAGCTCAACACAGACA-3′; and PTIP, 5′-ATTTCTTATTGAGGGTTAGC-3′.
Immunofluorescence microscopy
U2OS cells were seeded onto microscope coverslips and irradiated using an XCELL 50 x-ray (Kubtec). Cells were incubated for 1 or 6 h at 37°C, as indicated, for recovery. The cells were extracted with 1× PBS containing 0.2% Triton X-100 and protease inhibitors (Pierce) for 10 min on ice and fixed with 3.2% PFA in 1× PBS. The cells were then washed extensively with IF Wash Buffer (1× PBS, 0.5% NP-40, and 0.02% NaN3) and blocked with IF Blocking Buffer (IF Wash Buffer plus 10% FBS) for at least 30 min. Cells were incubated with primary antibodies diluted in IF Blocking Buffer for 1 h to overnight at 4°C. Primary antibodies used were as follows: mouse anti-53BP1 (1:1,000; BD Biosciences; 612522), rabbit anti-pH2AX (1:1,000; Abcam; 26350), rabbit anti-Rif1 (1:1,000; Bethyl; A300-569A), mouse anti-BRCA1 (1:1,000; Santa Cruz; 6954), rabbit anti-Rad51 (1:1,000; Santa Cruz; 8349), and mouse anti-HA (1:1,000; BioLegend; 901501). Cells were stained with secondary antibodies (conjugated with 1:1,000 diluted Alexa Fluor 488, 594, or Pacific Blue; Invitrogen) and Hoechst 33342 (Sigma) and mounted using Prolong Gold mounting medium (Invitrogen). Epifluorescence microscopy was performed on an Olympus fluorescence microscope (BX-53) using an ApoN 60×/1.49 NA, UPlanS-Apo 60×/1.35, or an UPlanS-Apo 100×/1.4 oil-immersion lens with immersion oil from Millipore (104699). Images were obtained at room temperature using an Olympus XM10 monochrome camera and cellSens Dimension software. Raw images were exported into Adobe Photoshop, and for any adjustments in image contrast or brightness, the levels function was applied. For foci quantitation, at least 200 cells were analyzed in triplicate, and a Student’s two-tailed t test was used to determine statistical significance.
For laser microirradiation experiments, cells were cultured for 24 h before irradiation with 1 µM BrdU (Sigma; cat #B9285). UVA laser (50 mW) striping was conducted using an inverted microscope (Eclipse Ti; Nikon) with a Palm MicroBeam laser microdissection workstation. After irradiation, cells were incubated at 37°C for 15 min, and then processed for immunofluorescence as stated above. Primary antibodies used were rabbit anti-TPX2 (1:500, Bethyl; A300-429A) and mouse anti–Aurora A (1:200; Abcam; 13824).
Flow cytometry
All flow cytometry experiments were performed on a BD FACSCalibur three-color flow cytometer and analyzed using FlowJo software. To measure chromatin-bound RPA, U2OS cells were treated with 1 µM camptothecin for 1 h before collection. Cells were washed with PBS + 2% FBS and fixed for 15 min in BD Cytofix/Cytoperm (BD Biosciences) at room temperature. Cells were then washed with Perm/Wash buffer (BD Biosciences) and stained with rat anti-RPA (1:200; Cell Signaling, 2208S). Cells were washed and stained with Alexa Fluor 488 goat anti-rat secondary (1:1,000; Invitrogen, A11006). Cells were again washed with Perm/Wash buffer and resuspended in PBS + 2% FBS containing 7-AAD for analysis (Forment et al., 2012). For quantification of HR, U2OS-DR-GFP cells (a gift from Maria Jasin) were infected with lentiviral shRNAs and subsequently infected 24 h later with pHAGE-Flag-ISce-I. 72 h after ISceI infection, cells were collected and resuspended in PBS, and GFP+ cells were measured via flow cytometry. Both RPA and DR-GFP assays were performed in triplicate, and 30,000 events were measured per experiment. To obtain cell cycle profiles of TPX2- and Aurora A–depleted cells, cells were collected 64 h after shRNA infection and fixed in 70% ethanol at −20°C overnight. Cells were washed in PBS with 1% BSA, treated with 100 µg/ml RNase, and stained with 50 µg/ml propidium iodide (Sigma). 10,000 events were measured per experiment. A Student’s two-tailed t test was used to determine statistical significance for all flow cytometry experiments.
DNA fiber analysis
DNA fiber analysis was performed as described previously (Quinet et al., 2017). Briefly, asynchronously growing cells were labeled with two thymidine analogues: 20 µM IdU (Sigma) followed by 200 µM CldU (Sigma) for the indicated times according to the labeling scheme used. For untreated conditions, we labeled first with IdU for 20 min, followed by three washes with 1× PBS, and then CldU for an additional 20 min. For the experiments with HU, cells were labeled first with IdU for 20 min, followed by three washes with 1× PBS, and then CldU plus 4 mM HU for an additional 2 h. After labeling, the cells were harvested and resuspended in PBS at 150,000 cells/ml. Cells were lysed and spread on positively coated slides and stained according to the protocol previously described (Prakash et al., 2015). Images of the fibers were sequentially acquired (for double label) with LAS AF software using TCS SP5 confocal microscope (Leica). A 63×/1.4 oil-immersion objective was used. A minimum of 300 tracks were measured for each sample using ImageJ, and for the DNA track lengths, the pixel length values were converted into micrometers using the scale bars created by the microscope. Data were plotted on a scatter dot graph plot. DNA fiber experiments were done in biological duplicate unless stated otherwise in the figure legend. Statistical significance was assessed using unpaired nonparametric Mann–Whitney compared ranks t test. Differences in fiber length ≤15% are not considered significant. Additional information on the minimal number of tracts that should be measured for a reliable estimation of changes in fork speed within a given sample has been previously described (Técher et al., 2013; Thangavel et al., 2015).
MTS survival assay
Cells were treated with either control or Aurora A lentiviral shRNAs for 48 h and then seeded in 96-well plates at a concentration of 5,000 cells per well. The following day, cells were treated with the indicated dose of HU and incubated at 37°C and 5% CO2 for an additional 48 h. At that time, the medium was replaced with fresh medium containing the CellTiter MTS reagent (Promega). Cell viability was assessed by measuring the absorbance of each well at 490 nM using an Epoch Microplate Spectrophotometer (BioTek). All MTS assays were performed in quintuplicate.
Online supplemental material
Table S1 shows the total and unique peptides for each identified protein in the MS analysis of Flag-HA-53BP1. Table S2 lists all antibodies used in this study. Fig. S1 shows supporting data for Figs. 1 and 2, which demonstrate that TPX2/Aurora A impair BRCA1 foci formation and DNA end resection in S/G2 but do not localize to DSBs. Fig. S2 contains supporting data for Figs. 3 and 4, showing that loss of TPX2/Aurora A results in replication fork deprotection but does not affect replication fork speed. Fig. S3 shows additional data for Fig. 4, demonstrating that TPX2/Aurora A protects stalled replication forks in a manner that is independent of its mitotic functions, but through a direct interaction with 53BP1.
Supplementary Material
Acknowledgments
We wish to thank B. Sleckman (Weill Cornell Medical School, New York, NY), G. Oltz (Washington University in St. Louis, St. Louis, MO), and P.T. Stukenberg (University of Virginia, Charlottesville, VA) for their advice.
A.K. Byrum is supported by a Cell and Molecular Biology Training grant (5T32GM007067-40). This work was supported by the National Institutes of Health (R01 CA118487 to Y. Shi, R01 GM108648 to A. Vindigni, and R01 CA193318 to N. Mosammaparast), Boston Children’s Hospital (to Y. Shi), the Siteman Cancer Center (to N. Mosammaparast), the Children’s Discovery Institute of St. Louis Children’s Hospital (MC-II-2015-453 to N. Mosammaparast), and the Department of Defense Breast Cancer Research Program Breakthrough Award (BC151728 to A. Vindigni).
The authors declare no competing financial interests.
Author contributions: A.K. Byrum: conceptualization, investigation, methodology, visualization, and writing (original draft and editing); D. Carvajal-Maldonado: investigation, methodology, visualization, and writing (review and editing); M.C. Mudge: resources; D. Valle-Garcia: investigation; M.C. Majid: resources; R. Patel: resources and investigation; M.E. Sowa: investigation and methodology; S.P. Gygi: methodology, resources, and supervision; J.W. Harper: methodology, resources, and supervision; Y. Shi: conceptualization, resources, writing (review and editing), and supervision; A. Vindigni: conceptualization, investigation, methodology, visualization, writing (original draft and editing), project administration, funding acquisition, and supervision; N. Mosammaparast: conceptualization, investigation, methodology, visualization, writing (original draft and editing), project administration, funding acquisition, and supervision.
References
- Bayliss R., Sardon T., Vernos I., and Conti E.. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862. 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bibby R.A., Tang C., Faisal A., Drosopoulos K., Lubbe S., Houlston R., Bayliss R., and Linardopoulos S.. 2009. A cancer-associated aurora A mutant is mislocalized and misregulated due to loss of interaction with TPX2. J. Biol. Chem. 284:33177–33184. 10.1074/jbc.M109.032722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.T., Faryabi R.B., Polato F., Santos M., Starnes L.M., et al. . 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 153:1266–1280. 10.1016/j.cell.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R., and Boulton S.J.. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 47:497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G.M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C.D., and Sassone-Corsi P.. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874–885. 10.1128/MCB.22.3.874-885.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Zhang A., Shan S., Gong Z., and Zhou Z.. 2018. Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR. Nat. Commun. 9:2123 10.1038/s41467-018-04557-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelloRusso C., Welcsh P.L., Wang W., Garcia R.L., King M.C., and Swisher E.M.. 2007. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res. 5:35–45. 10.1158/1541-7786.MCR-06-0234 [DOI] [PubMed] [Google Scholar]
- Drané P., Brault M.E., Cui G., Meghani K., Chaubey S., Detappe A., Parnandi N., He Y., Zheng X.F., Botuyan M.V., et al. . 2017. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature. 543:211–216. 10.1038/nature21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H., Rose K.L., Bhat K.P., Mohni K.N., Glick G.G., Couch F.B., and Cortez D.. 2015. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol. Cell. 59:998–1010. 10.1016/j.molcel.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Fong K.W., Wang J., Wang W., and Chen J.. 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288:11135–11143. 10.1074/jbc.M113.457440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment J.V., Walker R.V., and Jackson S.P.. 2012. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 81:922–928. 10.1002/cyto.a.22155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., and Vindigni A.. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8:860 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K.H., Abou El Hassan M., and Bremner R.. 2013. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques. 55:204–206. 10.2144/000114089 [DOI] [PubMed] [Google Scholar]
- Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøfte M., Chan K.L., Hickson I.D., Bartek J., and Lukas J.. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253. 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- Mulligan P., Westbrook T.F., Ottinger M., Pavlova N., Chang B., Macia E., Shi Y.J., Barretina J., Liu J., Howley P.M., et al. . 2008. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol. Cell. 32:718–726. 10.1016/j.molcel.2008.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y., and Ogryzko V.. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430–444. 10.1016/S0076-6879(03)70037-8 [DOI] [PubMed] [Google Scholar]
- Neumayer G., Belzil C., Gruss O.J., and Nguyen M.D.. 2014. TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. 71:3027–3047. 10.1007/s00018-014-1582-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer S.M., Adam S., Setiaputra D., Barazas M., Pettitt S.J., Ling A.K., Olivieri M., Álvarez-Quilón A., Moatti N., Zimmermann M., et al. . 2018. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 560:117–121. 10.1038/s41586-018-0340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Zhang Y., Feng W., and Jasin M.. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7:a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet A., Carvajal-Maldonado D., Lemacon D., and Vindigni A.. 2017. DNA Fiber Analysis: Mind the Gap! Methods Enzymol. 591:55–82. 10.1016/bs.mie.2017.03.019 [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., et al. . 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 535:382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. . 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 132:487–498. 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., and Harper J.W.. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell. 138:389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone P., Tagliaferri P., Perricelli A., Blotta S., Quaresima B., Martelli M.L., Goel A., Barbieri V., Costanzo F., Boland C.R., and Venuta S.. 2003. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br. J. Cancer. 88:1285–1291. 10.1038/sj.bjc.6600859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone P., Blotta S., Palmieri C., Masciari S., Quaresima B., Montagna M., D’Andrea E., Eramo O.P., Migale L., Costanzo F., et al. . 2005. Differential sensitivity of BRCA1-mutated HCC1937 human breast cancer cells to microtubule-interfering agents. Int. J. Oncol. 26:1257–1263. [PubMed] [Google Scholar]
- Técher H., Koundrioukoff S., Azar D., Wilhelm T., Carignon S., Brison O., Debatisse M., and Le Tallec B.. 2013. Replication dynamics: biases and robustness of DNA fiber analysis. J. Mol. Biol. 425:4845–4855. 10.1016/j.jmb.2013.03.040 [DOI] [PubMed] [Google Scholar]
- Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A., et al. . 2015. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 208:545–562. 10.1083/jcb.201406100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest N.E., and Tomkinson A.E.. 2017. Optimization of Native and Formaldehyde iPOND Techniques for Use in Suspension Cells. Methods Enzymol. 591:1–32. 10.1016/bs.mie.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Zimmermann M., and de Lange T.. 2014. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.