Kinesin-8 motor proteins control chromosome alignment in a variety of species, but the specific biochemical activity responsible is unclear. Edzuka and Goshima find that Drosophila kinesin-8 (Klp67A) exhibits both microtubule plus end–stabilizing and –destabilizing activities in vitro. In cells, Klp67A, and likely human kinesin-8 (KIF18A) as well, stabilize the kinetochore–microtubule attachment during mitosis.
Abstract
Kinesin-8 is required for proper chromosome alignment in a variety of animal and yeast cell types. However, it is unclear how this motor protein family controls chromosome alignment, as multiple biochemical activities, including inconsistent ones between studies, have been identified. Here, we find that Drosophila kinesin-8 (Klp67A) possesses both microtubule (MT) plus end–stabilizing and –destabilizing activity, in addition to kinesin-8's commonly observed MT plus end–directed motility and tubulin-binding activity in vitro. We further show that Klp67A is required for stable kinetochore–MT attachment during prometaphase in S2 cells. In the absence of Klp67A, abnormally long MTs interact in an “end-on” fashion with kinetochores at normal frequency. However, the interaction is unstable, and MTs frequently become detached. This phenotype is rescued by ectopic expression of the MT plus end–stabilizing factor CLASP, but not by artificial shortening of MTs. We show that human kinesin-8 (KIF18A) is also important to ensure proper MT attachment. Overall, these results suggest that the MT-stabilizing activity of kinesin-8 is critical for stable kinetochore–MT attachment.
Introduction
Equal segregation of sister chromatids into daughter cells relies on proper attachment of microtubules (MTs) to a specialized site on the chromosome, the kinetochore. Kinetochores consist of dozens of proteins, including those that bind to DNA or MTs, and many of them form subcomplexes for normal function (Musacchio and Desai, 2017). The Ndc80 complex is localized to the kinetochore during mitosis and functions as the major MT attachment site: “end-on” attachment of MTs to kinetochores absolutely depends on this conserved protein complex (Cheeseman et al., 2006; Powers et al., 2009; Musacchio and Desai, 2017). In yeast and animals, the Dam1 and Ska complexes, respectively, support MT binding of the Ndc80 complex (Tien et al., 2010; Schmidt et al., 2012). However, these complexes might not be the sole critical factors for MT attachment, as other MT-associated proteins, such as motor proteins, are also enriched at the kinetochore (Musacchio and Desai, 2017).
Besides attachment, kinetochores regulate the dynamics of the associated MTs. A major regulator is cytoplasmic linker-associated protein (CLASP), which promotes persistent growth of kinetochore MTs (Maiato et al., 2003, 2005). In its absence, MTs continuously shrink, and spindles collapse (Maiato et al., 2005). In vitro, CLASP retards MT growth and acts as a potent inhibitor of MT “catastrophe” and as an inducer of “rescue” (Al-Bassam et al., 2010; Moriwaki and Goshima, 2016; Yu et al., 2016). Another key regulator of kinetochore MT dynamics is the kinesin-8 motor protein.
Kinesin-8 is a widely conserved kinesin subfamily. Its motor domain lies at the N terminus, followed by coiled-coil and tail regions. The mitotic functions of kinesin-8 have been well described for budding yeast Kip3 (Cottingham and Hoyt, 1997; Straight et al., 1998; Tytell and Sorger, 2006; Wargacki et al., 2010), fission yeast Klp5/Klp6 (Garcia et al., 2001; West et al., 2002), Drosophila melanogaster Klp67A (Goshima and Vale, 2003; Gandhi et al., 2004; Savoian et al., 2004; Savoian and Glover, 2010), and mammalian KIF18A (Mayr et al., 2007; Stumpff et al., 2008) and KIF18B (McHugh et al., 2018). Kinesin-8 is generally enriched at the outer region of the mitotic kinetochore, where plus ends of kinetochore MTs are present, and its depletion affects spindle length and chromosome alignment. In human KIF18A RNAi, the amplitude of chromosome oscillation in the abnormally elongated spindle is dramatically elevated, such that chromosome congression cannot be achieved. In the absence of budding yeast Kip3, kinetochores are unclustered in the spindle, indicating chromosome alignment defects. Fission yeast klp5/6 mutant also exhibits chromosome misalignment associated with Mad2-dependent mitotic delay. Overall, the loss of kinesin-8 consistently perturbs chromosome alignment in a variety of cell types.
Despite the conserved phenotype and localization associated with kinesin-8, its biochemical activity toward MTs is inconsistent between reports. The best-studied budding yeast Kip3 has plus end–directed, processive motility and also has strong MT-depolymerizing activity; it can depolymerize MTs stabilized by nonhydrolyzable GTP (GMPCPP) and promote catastrophe (growth-to-shrinkage transition) in dynamic MTs (Gupta et al., 2006; Varga et al., 2006). The C-terminal tail has MT- and tubulin-binding activities, which allow this motor to cross-link and slide antiparallel MTs (Su et al., 2011, 2013). However, MT depolymerization activity has not been detected for fission yeast proteins Klp5/Klp6 and MT nucleation activity has been reported instead (Erent et al., 2012). Humans have two mitotic kinesin-8s, KIF18A and KIF18B, and kinetochore function has been observed for KIF18A. KIF18A, like Kip3, exhibits processive motility toward plus ends, and accumulates at plus ends on its own (Mayr et al., 2007; Du et al., 2010). The tail region of KIF18A has MT and tubulin affinity, which is similar to Kip3 (Mayr et al., 2011; Weaver et al., 2011). However, its impact on MT dynamics has been controversial. In one study, KIF18A was concluded to have MT-depolymerizing activity, based on its depolymerization activity toward stabilized MTs (Mayr et al., 2007). In another study, however, this activity was reported to be undetectable, and instead, it dampened MT dynamicity; KIF18A suppressed both growth and shrinkage of MTs (Du et al., 2010). Although the former activity is more consistent with Kip3, the latter activity appears to be more congruous with the cellular phenotype associated with KIF18A (Stumpff et al., 2008).
In the present study, we investigated Klp67A, the sole mitotic kinesin-8 in Drosophila. In addition to conserved MT-based motility, we identified both MT-stabilizing and -destabilizing activities in a single experimental condition. Functional analysis in the S2 cell line indicated that, with these two activities, kinesin-8Klp67A not only regulates MT length, but also stabilizes kinetochore–MT attachment.
Results
Kinesin-8Klp67A shows plus end–directed motility and tubulin-binding activity in vitro
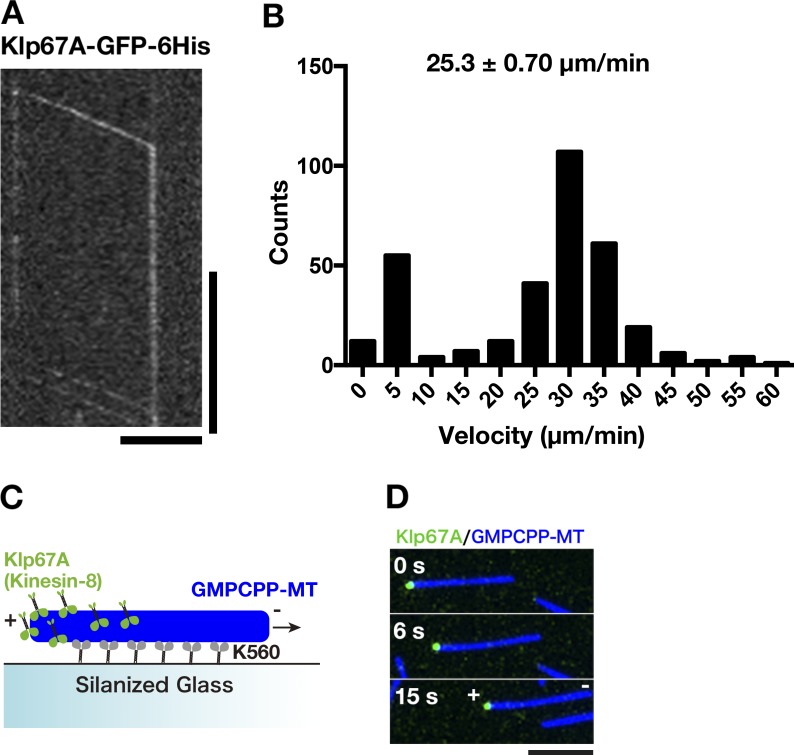
Generally observed biochemical activities among mitotic kinesin-8 motors are processive motility (Gupta et al., 2006; Varga et al., 2006; Stumpff et al., 2011; McHugh et al., 2018) and tubulin binding at the nonmotor region (Mayr et al., 2011; Su et al., 2011; Weaver et al., 2011). To determine the biochemical activity of Drosophila kinesin-8Klp67A, we purified recombinant GFP-tagged full-length protein (Fig. S1 A). First, we performed single motor motility assays and found that kinesin-8Klp67A-GFP is a processive motor with mean velocity 25 ± 0.7 µm/min (± SEM; Fig. 1, A and B; and Video 1). The GFP signal quickly accumulated at one end of MTs: therefore, to visualize how each motor accumulates at the MT end, we photo-bleached the preaccumulated kinesin-8Klp67A-GFP signals at the end, followed by observing unbleached motors. In 32 out of 33 cases, we observed that the motile motor on the MT lattice reached and stayed at the end of the MT polymers (Fig. 1 A andVideo 1). Thus, kinesin-8Klp67A-GFP processively moved to the end and resided there for a certain period; this behavior is identical to that observed for Kip3 (Varga et al., 2009). Next, we determined the directionality of motility by localizing kinesin-8Klp67A-GFP on MTs that underwent gliding by the plus end–directed kinesin-1 motor (Fig. 1 C). Kinesin-8Klp67A-GFP was enriched at the trailing end of MTs, indicating that kinesin-8Klp67A is a plus end–directed motor, as seen with other kinesin-8s (Fig. 1 D).
Figure 1.
Kinesin-8Klp67A is a processive plus end–directed motor. (A) Kymograph showing processive motility of kinesin-8Klp67A-GFP (100 pM) along MTs and accumulation at the MT end. Kinesin-8Klp67A-GFP enriched at the MT end was first photo-bleached, followed by observation of unbleached kinesin-8Klp67A-GFP. Horizontal bar, 5 µm; vertical bar, 20 s. (B) Plot of kinesin-8Klp67A-GFP run velocity, calculated based on kymographs (n = 331). Immobile GFP signals were not counted (the “0” column represents velocity between 0 and 5 µm/min). (C and D) Gliding of stabilized MTs (blue) by the plus end–directed human kinesin-1 motor (K560 construct; nonfluorescent). Rightward motility of MTs indicate that the left end corresponds to MT plus end. Kinesin-8Klp67A-GFP (green; 4 nM) accumulated specifically at the plus end.
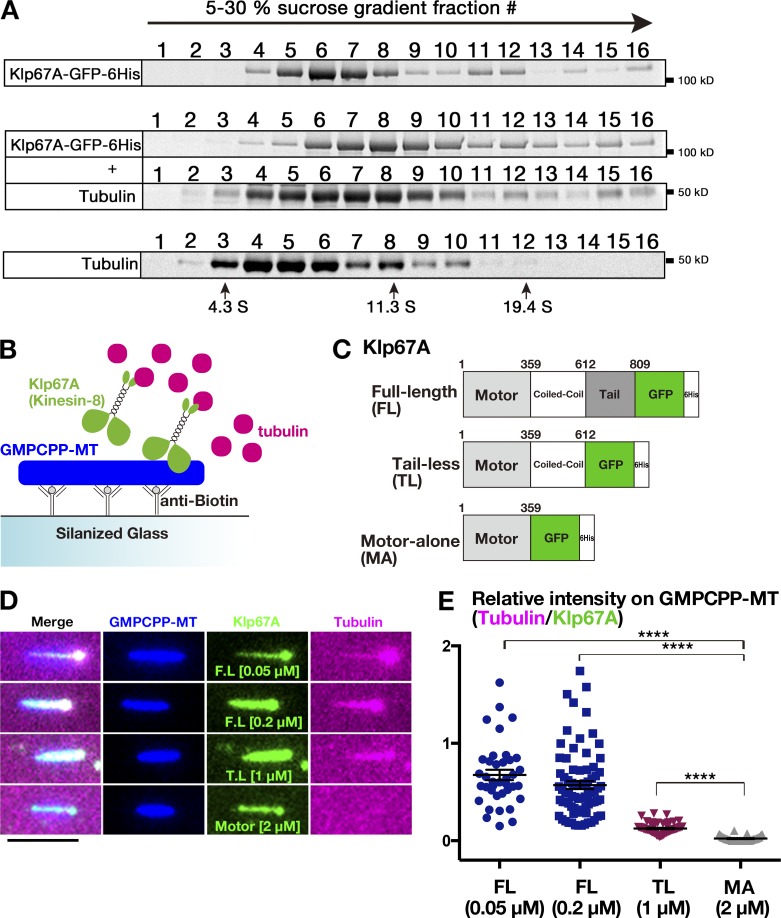
To test if kinesin-8Klp67 binds to tubulin, like Kip3 and KIF18A, we performed sucrose gradient centrifugation of recombinant kinesin-8Klp67A-GFP in the presence and absence of tubulin. Kinesin-8Klp67A-GFP was co-fractionated with tubulin as a larger complex, indicating that kinesin-8Klp67A directly binds to tubulin (Fig. 2 A). Next, to verify this finding and further identify the region responsible for tubulin binding, we performed a tubulin recruitment assay, in which kinesin-8Klp67A (full length and truncations) was localized along the stabilized MT seed, and fluorescently labeled free tubulin was added and observed (Fig. 2, B and C; and Fig S1 B). Consistent with the sucrose gradient centrifugation analysis, tubulin was efficiently recruited to the MT seed by kinesin-8Klp67A full length (Fig. 2, D and E). C-terminally truncated “tail-less” construct (1–612 aa), which was shown to rescue the spindle length and chromosome alignment phenotypes (Savoian and Glover, 2010), also recruited tubulin onto MT seeds, albeit less efficiently than full length. However, when nonmotor regions were entirely eliminated, tubulin was hardly recruited. These results indicate that kinesin-8Klp67A binds to tubulin at the nonmotor region and suggest that the binding is important for kinesin-8Klp67A function.
Figure 2.
Tubulin-binding activity of kinesin-8Klp67A. (A) Co-fractionation of kinesin-8Klp67A-GFP and tubulin after sucrose gradient centrifugation. Each fraction was subjected to SDS-PAGE, followed by staining with Sypro Ruby. (B–D) Tubulin recruitment by kinesin-8Klp67A. Tubulin (magenta; 10 µM) and kinesin-8Klp67A (green; full-length, tail-less [1–612 aa], motor-alone [1–359 aa]), which bound to GMPCPP-stabilized MTs (blue), were mixed, and tubulin localization along MTs was investigated. Bar, 5 µm. (E) Quantification of tubulin intensity on the MT seed. Each dot represents a value obtained from a single MT and error bars represent SEM. FL versus MA: P = 3.8 ×10−14, and TL versus MA: P = 3.8 ×10−10 by Games-Howell test. n = 38 (50 nM), 77 (200 nM; full length), 33 (motor alone), and 58 (tail-less).
Kinesin-8Klp67A has both MT-destabilizing and -stabilizing activities in vitro
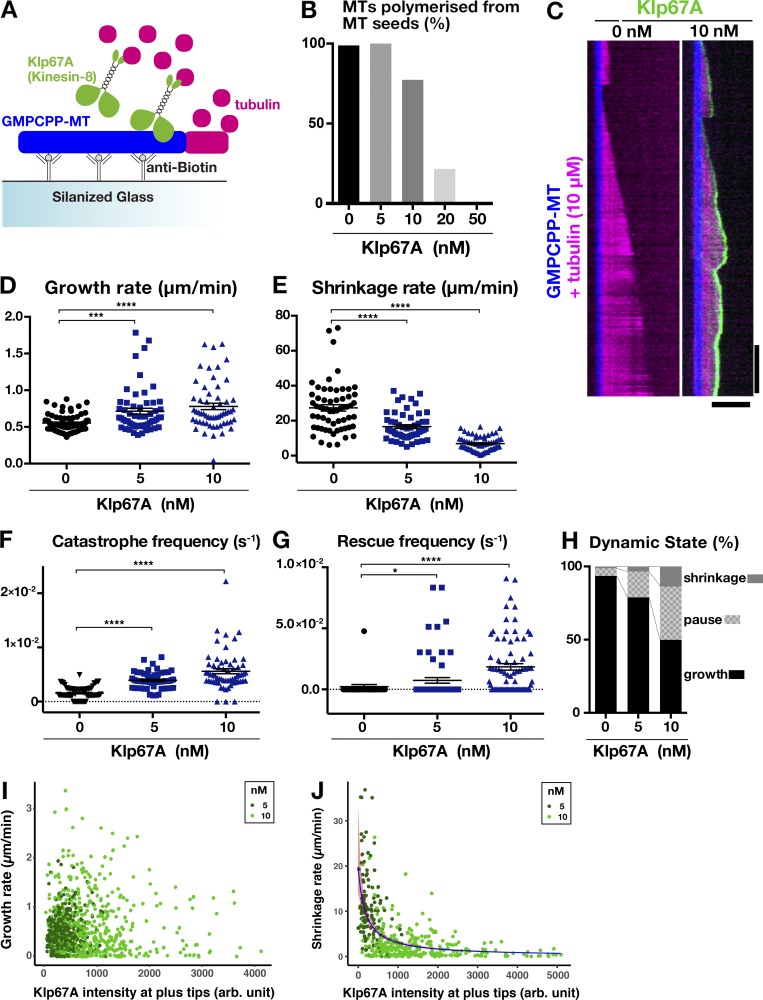
We next determined the effect of kinesin-8Klp67A on MT polymerization dynamics in an in vitro assay (Fig. 3 A). When we mixed 5, 10, 20, or 50 nM kinesin-8Klp67A with GMPCPP-stabilized MT seeds and free tubulin (10 µM), dynamic MTs from the seeds were rarely observed or nonexistent at 20 or 50 nM, respectively (Fig. 3 B). Furthermore, at high concentrations, the excess force exerted by kinesin-8Klp67A glided and bundled MT seeds. In the subsequent experiment, we used 5 or 10 nM kinesin-8Klp67A.
Figure 3.
Kinesin-8Klp67A regulates MT plus end dynamics in vitro. (A) Schematic presentation of the in vitro MT polymerization assay. (B) Inhibitory effect of kinesin-8Klp67A on MT polymerization from the seed (n = 84, 74, 84, 74, and 94; from left to right). (C) MT dynamics are represented by kymographs. MTs stabilized with GMPCPP are colored blue, dynamic MTs are magenta, and kinesin-8Klp67A-GFP (0 or 10 nM) is green. Horizontal bar, 5 µm; vertical bar, 120 s. (D–H) Parameters of MT plus end dynamics. Experiments were performed twice, and the combined data are displayed (the change in rate/frequency by kinesin-8Klp67A was reproduced). n = 41 + 33 (0 nM), 25 + 31 (5 nM), and 20 + 41 (10 nM). Each dot represents a value obtained from a single MT, and error bars represent SEM. Note that rescue was rarely observed in the absence of kinesin-8Klp67A. *, P < 0.03; ***, P < 0.002; ****, P < 0.0001 or by Games-Howell (D–F) or Steel Dwass (G) tests. (I and J) Correlation between the amount of kinesin-8Klp67A-GFP at the tip and growth (I; n = 397 [5 nM] and 494 [10 nM]) or shrinkage (J; n = 116 [5 nM] and 293 [10 nM]) rate. Negative correlation was found for shrinkage rate (P < 2 × 10−16, likelihood ratio test).
First, we could not observe any MT depolymerization activity of kinesin-8Klp67A toward GMPCPP-stabilized MTs, which differs from that for yeast Kip3 (Gupta et al., 2006; Varga et al., 2006), human KIF18A reported by (Mayr et al., 2007; Locke et al., 2017), or the bona fide MT-depolymerizing Drosophila kinesin-13Klp10A (Rogers et al., 2004) that was used as a positive control in our experiments (Fig. S1 C). However, we could not exclude the possibility that kinesin-8Klp67A has a weak seed depolymerization activity that was undetectable with this motor concentration.
Next, we quantified the dynamics parameters in the presence of 5 or 10 nM kinesin-8Klp67A (Fig. 3, C–G). As expected from the spindle-lengthening phenotype, kinesin-8Klp67A elevated the catastrophe frequency of the MTs. Interestingly, kinesin-8Klp67A also increased the rescue frequency and slowed down shrinkage under the same assay conditions, which resulted in more frequent MT pausing (Fig. 3 H). Thus, we observed both MT-stabilizing and -destabilizing effects reported for kinesin-8KIF18A by Du et al. (2010), and Locke et al. (2017) and Mayr et al. (2007), respectively. The growth rate was increased in a slight but statistically significant manner, which is consistent with a recent report concerning human kinesin-8KIF18B (McHugh et al., 2018).
Finally, we analyzed the correlation between the amount of kinesin-8Klp67A-GFP at the plus end and the MT growth/shrinkage rate. We measured and plotted GFP intensity at the tip and the velocity of the MTs (Fig. 3, I and J). GFP intensity at the shrinking tip was, on average, slightly higher than that at the growing tip (Fig. S1 D). Whether this small difference is of physiological relevance is unclear. Interestingly, there was a strong anti-correlation between GFP intensity and shrinking velocity (P < 2 × 10−16, gamma regression, likelihood-ratio tests; Fig. 3 J), whereas no clear correlation was identified for growth (P = 0.99; Fig. 3 I). The data are consistent with the model that kinesin-8Klp67A accumulated at the tip induces catastrophe, but simultaneously prohibits drastic shrinkage. On the other hand, the mechanism by which kinesin-8Klp67A increases the MT growth rate is unclear.
Kinesin-8Klp67A depletion causes instability of kinetochore–MT attachment
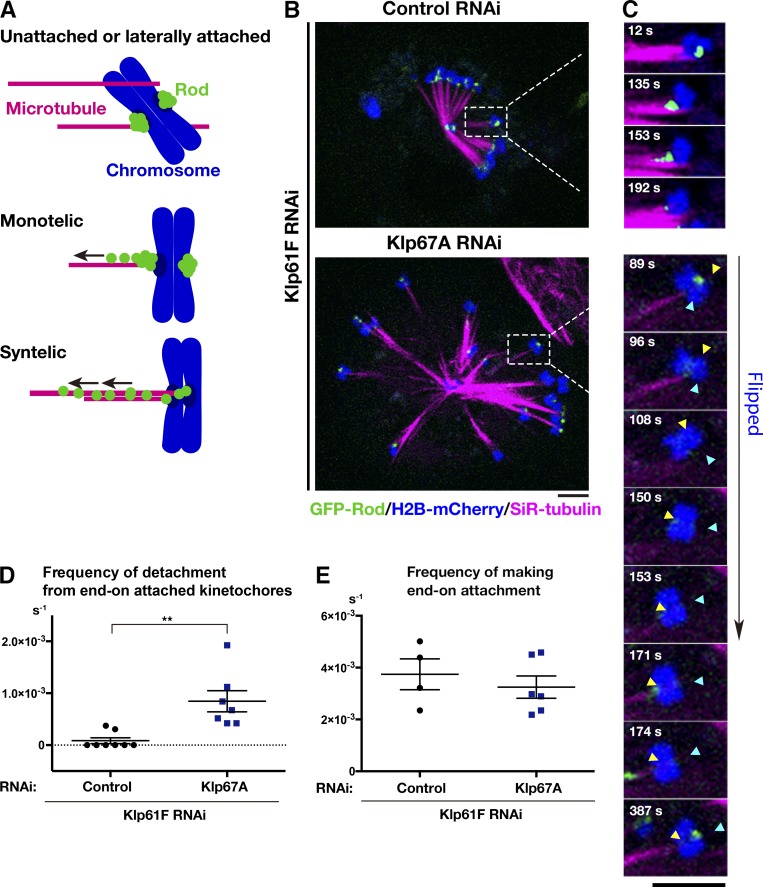
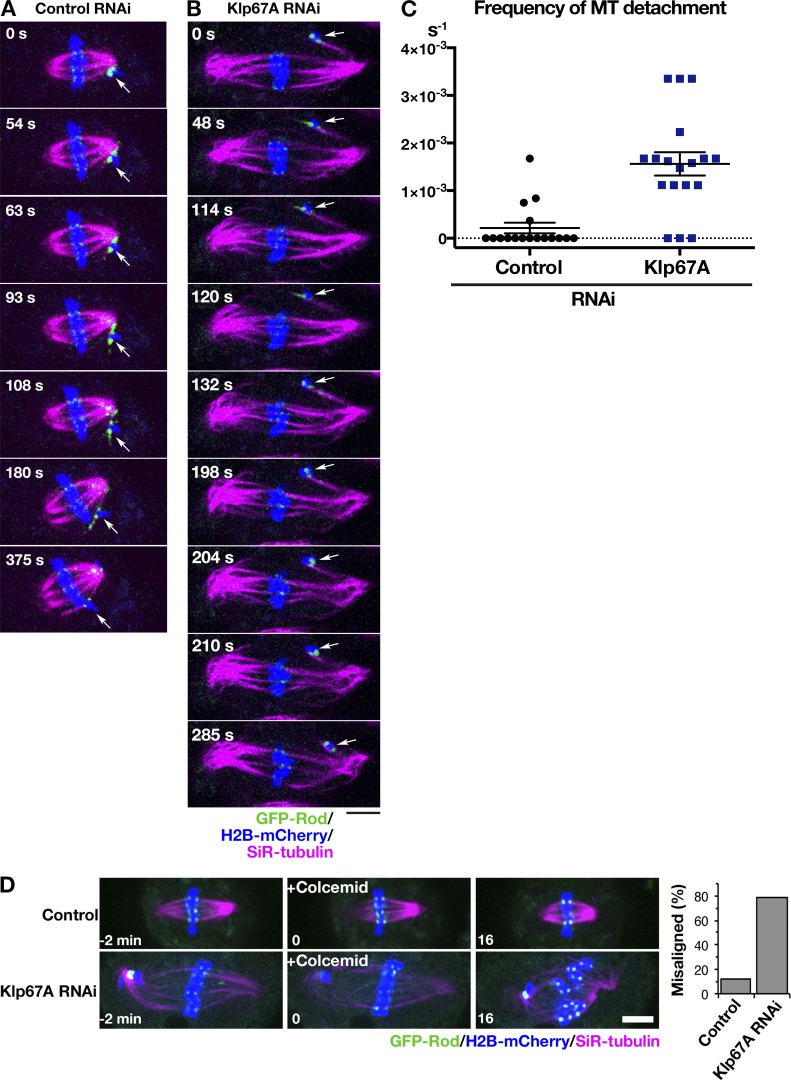
To explore the function of kinesin-8Klp67A in the spindle, we observed its RNAi phenotype in living S2 cells (RNAi knockdown was confirmed by immunoblotting; Fig. S2 A). In addition to chromosomes and MTs, we traced GFP-Rod: GFP-Rod accumulates at unattached or laterally attached kinetochores and, once MTs are attached in an end-on fashion, it is transported away from the kinetochore along the MTs (Basto et al., 2004; Głuszek et al., 2015). Therefore, GFP-Rod exhibits “streaming” upon MT end-on attachment, while residual proteins are still visible at the kinetochore; thus, it serves as an ideal marker for kinetochore dynamics, as well as for its attachment status (Fig. 4 A). To precisely monitor and evaluate the dynamics of individual chromosome and GFP-Rod signals in the uniformly shaped spindle, we induced monopolar spindles by depleting Klp61F, the kinesin-5 motor protein required for spindle bipolarization. MTs were stained with the SiR-tubulin dye. In control cells that were singly depleted of kinesin-5Klp61F, we observed that the majority of sister kinetochores were attached to MTs from the pole (“syntelic” attachment), with weak GFP-Rod signals detected (Fig. 4 B and Video 2). The syntelically attached kinetochores were static and scarcely changed position during the observation time. In some instances, we observed “monotelic” attachment at the beginning, where one of the sister kinetochores was not associated with MTs and, therefore, a strong GFP-Rod signal was detected (Fig. 4 C, 12 s). However, they were converted into syntelic attachments during imaging, which was characterized by GFP-Rod streaming along the newly formed kinetochore MTs (Fig. 4 C, top, 153 s). Once syntelic attachment was established, MTs were rarely dissociated from kinetochores (Fig. 4 D, control).
Figure 4.
Kinesin-8Klp67A is required for stable kinetochore–MT attachment: monopolar spindle. (A) Diagram of GFP-Rod and MT attachment mode. (B) Monopolar spindles formed after RNAi of kinesin-5Klp61F (top) or double kinesin-5Klp61F/kinesin-8Klp67A (bottom). Blue, chromosome (H2B-mCherry); green, GFP-Rod; and magenta, MT (SiR-tubulin). (C) Monotelic-to-syntelic conversion in the control monopolar spindle (top); upon new MT association, GFP-Rod stream was observed along the kinetochore MT (153 s), and subsequently, the GFP signal diminished at the kinetochore (192 s). Chromosome flipping (bottom) was observed in the absence of kinesin-8Klp67A (89–171 s; sister kinetochores are indicated by yellow and blue arrows). Monotelic-to-syntelic conversion was also observed for this chromosome (96 s). (D) Increased frequency (event no. per chromosome per second) of MT detachment in the absence of kinesin-8Klp67A (**, P < 0.009; Welch’s t test). Events were counted when MTs terminated end-on attachment. RNAi and imaging were performed four times, data were quantitatively analyzed twice, and the two datasets were combined. Each dot in the graph represents mean frequency for a cell. A total of 86 chromosomes in eight cells (control) and 56 chromosomes in eight cells (kinesin-8Klp67A RNAi) were analyzed. (E) Frequency (event no. per chromosome per second) of newly acquired end-on attachment that was indicated by GFP-Rod stream along kinetochore MTs. The attachment number was divided by the total time the chromosomes spent in a monotelic or unattached state. Each dot in the graph represents mean frequency for a cell. A total of 18 chromosomes in four cells (control) and 18 chromosomes in six cells (kinesin-8Klp67A RNAi) were analyzed. Error bars indicate SEM. Bars, 5 µm.
When kinesin-5Klp61F and kinesin-8Klp67A were codepleted, monopolar spindles with much longer MTs were assembled (Fig. 4 B). In addition, kinetochore dynamics were dramatically different in these cells (Video 2). Some kinetochores were visibly motile, and monotelic attachment was more frequently observed for those chromosomes. Most of the unattached kinetochores acquired end-on attachment during the imaging period, as indicated by GFP-Rod streaming (e.g., Fig. 4 C, 171–174 s), although the associations were often transient. Quantification indicated detachment of MTs from the kinetochore was significantly more frequently observed in the absence of kinesin-8Klp67A, whereas end-on attachment event was detected at a frequency similar to that in control cells (Fig. 4, D and E). Interestingly, MT detachment was usually associated with chromosome flipping, where a sister kinetochore originally distal from the pole was flipped to face the pole (e.g., Fig. 4 C, 89–153 s).
To verify that the observed phenotype was not an artifact of SiR-tubulin staining, we observed a cell line that expressed GFP-Rod, H2B-mCherry, and EB1-SNAP (fluorescent SiR-SNAP was added to the medium), in which EB1 served as a marker of kinetochore MTs (Fig. S2 B). In addition, we simply observed GFP-Rod and H2B-mCherry without MT markers (Fig. S2 D). In the absence of kinesin-8Klp67A, chromosome flipping was 19-fold more frequently observed than in control cells (n = 15 and 27), confirming the role of kinesin-8Klp67A in stable kinetochore–MT attachment in the monopolar spindle (Fig. S2 C).
We next observed GFP-Rod behavior in the bipolar spindle with and without kinesin-8Klp67A (Fig. 5). As previously reported, abnormally elongated spindles with unaligned chromosomes were observed in the absence of kinesin-8Klp67A. Since MTs were crowded in the spindle, it was impossible to observe MT attachment status for most kinetochores. Nevertheless, when we focused on completely unaligned chromosomes that were remote from the main body of the spindle, we observed a phenotype similar to that observed in the monopolar assay. In the control cell displayed in Fig. 5 A and Video 3, a chromosome (arrow) was not immediately captured by MTs and remained near the pole; it had strong GFP-Rod signals. However, MTs were generated independent of centrosomes and bound to the kinetochore (63 s). Once those MTs were formed, the flow of GFP-Rod was visible, concomitant with the decrease of the kinetochore signal intensity (93–180 s). Thus, a bi-oriented chromosome with “amphitelic” attachment was finally observed, and it was translocated toward the spindle equator (375 s). In contrast, MT attachment was unstable, and MT detachment was observed in kinesin-8Klp67A–depleted cells. In the case displayed in Fig. 5 B and Video 3, a misaligned chromosome (arrow) achieved amphitelic attachment at 114 s, as evident by GFP-Rod stream along chromosome-bound MTs, but then such a configuration was disrupted at 198 s, and the chromosome was flipped (198–285 s). MT detachment frequency was quantified in Fig. 5 C.
Figure 5.
Kinesin-8Klp67A is required for stable kinetochore–MT attachment: bipolar spindle. (A and B) Time-lapse imaging of spindle MTs (magenta; SiR-tubulin staining), chromosomes (blue; H2B-mCherry), and GFP-Rod (green) after control or kinesin-8Klp67A RNAi. The behavior of an initially unaligned chromosome (arrows) was dramatically different. (C) Increased frequency (event no. per cell per second) of MT detachment in the absence of kinesin-8Klp67A (P < 0.001; Welch’s t test). Events were counted when MTs terminated end-on attachment. RNAi and imaging were performed three times, data were quantitatively analyzed twice, and the two datasets were combined. Error bars represent SEM. (D) Sensitivity to MT depolymerization. One or more chromosomes at the metaphase plate became unaligned upon colcemid treatment at much higher frequency in the absence of kinesin-8Klp67A. Bars, 5 µm.
Finally, we treated the metaphase cells with a MT-destabilizing drug and preferentially depolymerized nonkinetochore MTs. In control cells, kinetochore MTs kept the chromosomes aligned at the metaphase plate in 88% of the cases (n = 25) for ≥ 15 min, as expected (Goshima et al., 2008). In contrast, at least one chromosome that had been located at the metaphase plate was misaligned in 78% of the cells in the absence of kinesin-8Klp67A (Fig. 5 D).
From these results, we concluded that the kinetochore–MT association becomes unstable in the absence of kinesin-8Klp67A.
Artificial MT destabilization does not rescue the kinesin-8Klp67A–depleted phenotype
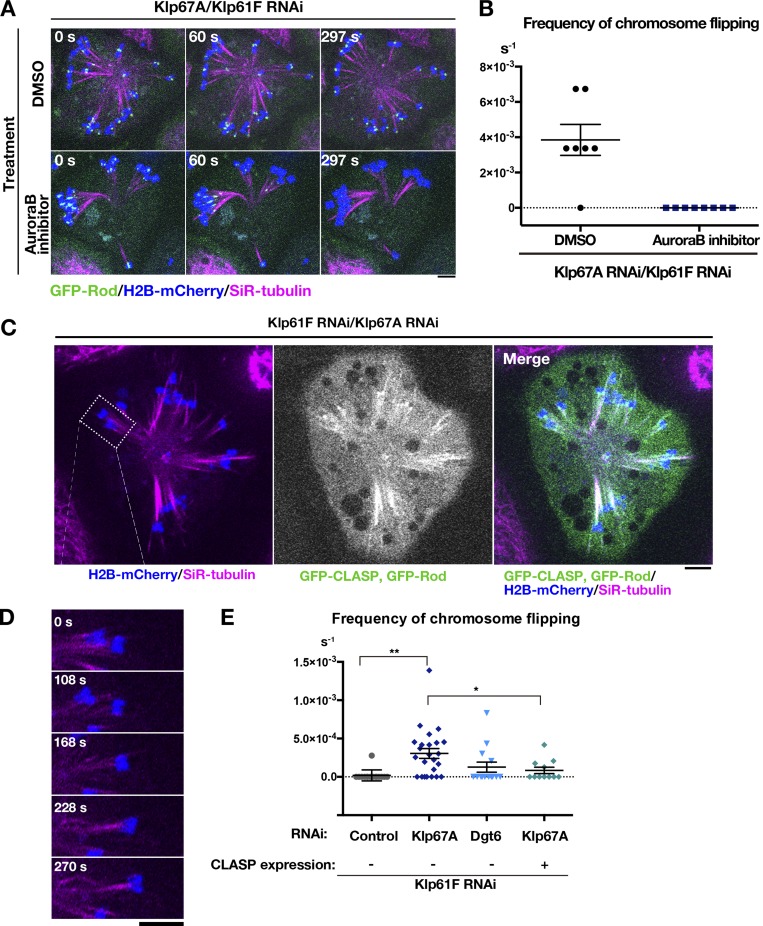
Attachment instability might be the consequence of longer MTs in the absence of kinesin-8Klp67A. To test this possibility, we depleted kinesin-5Klp61F and Dgt6, an essential subunit of the MT amplifier augmin, to generate monopolar spindles with long MTs; MTs are elongated in this condition due to the reduction of MT nucleation sites within the spindle (Goshima et al., 2008). AugminDgt6/kinesin-5Klp61F RNAi-treated cells indeed exhibited much longer and pendulous MTs, similar to those observed after kinesin-8Klp67A depletion (Fig. S3 A and Video 4). Monotelic and syntelic attachments were both observed, similar to those seen in kinesin-8Klp67A–depleted cells. However, once attached, MTs were more persistent, and chromosome flipping was rarely observed (Fig. 6 E).
Figure 6.
MT attachment stability was restored by inhibiting Aurora B kinase or CLASPMast/Orbit overexpression. (A) Monopolar spindles were induced after double kinesin-5Klp61F/kinesin-8Klp67A RNAi and treated with 20 µM Binucleine-2, an inhibitor of Drosophila Aurora B kinase, or control DMSO. (B) Frequency of chromosome flipping (per cell, per second; ± SEM) in Binucleine-2–treated cells (n = 7 [control DMSO treated] and 8 cells [Binucleine-2 treated]). (C) GFP-CLASPMast/Orbit overexpression in the monopolar spindle depleted of kinesin-5Klp61F and kinesin-8Klp67A. (D) A representative chromosome (inset in C). Chromosomes were static when GFP-CLASPMast/Orbit was overexpressed. (E) Frequency of chromosome flipping (per chromosome, per second; ± SEM). Expression of GFP-CLASPMast/Orbit significantly reduced the frequency of flipped chromosomes (*, P < 0.034; **, P < 0.0013; Games-Howell test). RNAi (and overexpression) were performed two or more times, and combined data are presented. Each dot in the graph represents mean frequency for a cell. A total of 168 chromosomes in 15 cells (control), 364 chromosomes in 24 cells (kinesin-8Klp67A RNAi), 128 chromosomes in 13 cells (augminDgt6 RNAi), and 128 chromosomes in 11 cells (kinesin-8Klp67A RNAi and GFP-CLASPMast/Orbit overexpression) were analyzed. Bars, 5 µm.
To further exclude the possibility that abnormally elongated MTs due to reduced catastrophe are the major cause of MT attachment instability, we exposed a low dosage of colcemid to kinesin-8Klp67A RNAi-treated cells and shortened the bipolar spindle lengths to the control levels (Fig. S3, B and C). Unaligned chromosomes were still frequently observed and mitosis was significantly delayed (Fig. S3, B and D). These results suggested that regulation of MT length alone cannot explain the attachment instability of kinesin-8Klp67A–depleted cells.
Aurora B kinase inhibition or CLASPMast/Orbit overexpression rescues attachment instability caused by kinesin-8Klp67A depletion
In mammalian cells, inhibition of Aurora B kinase stabilizes syntelic attachment, at least partly due to dephosphorylation of Ndc80: Ndc80 is a critical kinetochore component for MT binding, and its MT binding affinity is decreased by Aurora B phosphorylation (Cheeseman et al., 2002; DeLuca et al., 2006; Lampson and Grishchuk, 2017; Musacchio and Desai, 2017). We added the inhibitor of Drosophila Aurora B, Binucleine-2 (Smurnyy et al., 2010), to S2 cells depleted of kinesin-8Klp67A/kinesin-5Klp61F, and observed that most kinetochores stably attached MTs in a syntelic manner, as indicated by a decrease in GFP-Rod signals (Fig. 6, A and B; and Video 5). The result indicates that kinetochores retain an ability to bind stably to MTs in the absence of kinesin-8Klp67A when Aurora B activity is low. Our interpretation is that the MT-binding potential of the dephosphorylated form of Ndc80 complexes is preserved in the absence of kinesin-8Klp67A.
In the absence of kinesin-8Klp67A, persistent poleward motility of chromosomes was observed, which would involve kinetochore MT shrinkage (Video 2). We hypothesized that the MT stabilization activity of kinesin-8Klp67A, namely slowing down MT shrinkage and inducing rescue/pausing, is critical for MT attachment stability. If that were the case, we reasoned that MT stabilization by other means might partially suppress the MT detachment phenotype. To this end, we expressed Drosophila CLASP (also called Mast or Orbit) in kinesin-8Klp67A/kinesin-5Klp61F RNAi cells and observed the consequent chromosome dynamics. Since we attached GFP to CLASPMast/Orbit to identify cells overexpressing CLASPMast/Orbit, GFP-Rod signals could not be used to evaluate MT–kinetochore attachment status. Nevertheless, chromosome flipping frequency was significantly reduced in GFP-CLASPMast/Orbit overexpressing cells, supporting our hypothesis (Fig. 6, C–E; and Video 6). However, spindle length, chromosome alignment, and mitotic duration were not restored by GFP-CLASPMast/Orbit overexpression (Fig. S4). The suppression was thus specific to MT attachment stability.
GFP-Mad2 accumulation after KIF18A RNAi in HeLa cells
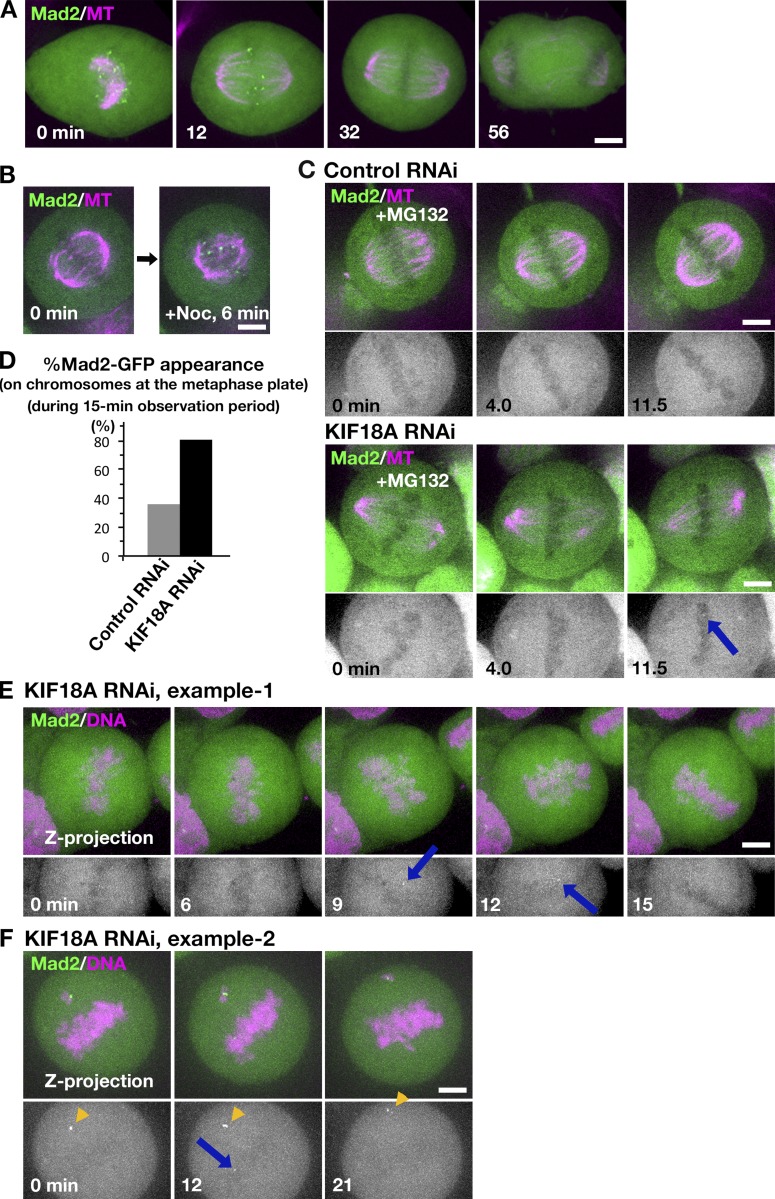
Several studies have characterized loss-of-function phenotypes for human kinesin-8, KIF18A (Mayr et al., 2007; Stumpff et al., 2008, 2012; Janssen et al., 2018; Kim and Stumpff, 2018). The components of the spindle assembly checkpoint have been also analyzed in a few studies using fixed cells (Mayr et al., 2007; Janssen et al., 2018; Kim and Stumpff, 2018). To confirm the phenotype and also possibly gain new insights into KIF18A function, we performed our own RNAi analysis using living HeLa cells expressing GFP-Mad2; Mad2 is a major component of spindle assembly checkpoint and is rapidly recruited to the kinetochores that are not properly attached to MTs (Joglekar, 2016). For example, upon laser cutting of kinetochore MTs, Mad2 signals appeared within a few minutes (Dick and Gerlich, 2013).
We first confirmed that punctate signals of GFP-Mad2 were rarely detected on aligned chromosomes, but were often observed in prometaphase, during which the majority of the kinetochores were unattached to MTs (Fig. 7 A). They were also detectable when spindle MTs at metaphase were depolymerized with nocodazole (Fig. 7 B).
Figure 7.
GFP-Mad2 dynamics in the absence of KIF18A in HeLa cells. (A) GFP-Mad2 localization during mitosis in the control HeLa cell. Images were acquired from 11 z sections (separated by 2 µm) and are displayed after maximum projection. (B) Accumulation of GFP-Mad2 on kinetochores upon MT depolymerization by nocodazole. Image was acquired every 30 s at a single focal plane. (C) Metaphase-arrested cells were imaged every 30 s at a single focal plane after control or KIF18A RNAi. Arrows indicate the appearance of a punctate GFP-Mad2 signal at the metaphase plate. (D) Frequency of the cells in which GFP-Mad2 appeared on chromosomes at the metaphase plate during the 15-min observation period. n = 47 for each. (E and F) GFP-Mad2 appearance at the metaphase plate in KIF18 RNAi-treated cells (arrows). As a reference, brighter GFP-Mad2 signals on persistently unaligned chromosomes are shown in F (arrowheads). Maximum projection images are shown. Bars, 5 µm.
Second, we observed GFP-Mad2 every 4 s in the monopolar spindle assembled by inhibiting kinesin-5 in the presence or absence of KIF18A, which was an assay analogous to those performed in S2 cells. However, in HeLa, GFP-Mad2 was constantly observed and chromosomes were dynamic even in the control cells: we could not identify a defect in the KIF18A RNAi-treated cells by our eyes (n = 41 cells; Video 7).
We then observed GFP-Mad2 dynamics in the bipolar spindle in the absence of KIF18A in two conditions. First, we treated cells with MG132 to arrest them at metaphase and performed imaging of a single focal plane of multiple mitotic cells every 30 s for 15 min (Fig. 7 C). We analyzed a total of 47 metaphase cells for control and KIF18A-depeleted cells. In this artificially arrested condition, unaligned chromosomes with strong punctate Mad2 signals were observed in control and KIF18A RNAi samples at similar frequencies (40 and 36%). In contrast, Mad2 signals also appeared on aligned chromosomes, often transiently, in 81% of the cells missing KIF18A, while only 36% of the control cells displayed such behavior (Fig. 7, C and D, arrow indicates the GFP-Mad2 signal). The results suggested that, in the absence of KIF18A, a subpopulation of seemingly aligned chromosomes had an improper attachment. Moreover, the transient signal appearance suggested that Mad2-negative kinetochores (i.e., properly attached kinetochores) occasionally altered their attachment status and became Mad2 positive. Alternatively, the Mad2 appearance/disappearance might simply reflect the kinetochore motility in and out of focal plane of the microscope.
To further test if Mad2-negative chromosomes could become Mad2 positive in the absence of KIF18A, we acquired z-stack images of normally cycling cells every 3 min for 30 min after RNAi (0.5 µm separation, >30 sections, by which the entire chromosome sets were covered at the beginning of imaging). Interestingly, in 70% cells (n = 23), GFP-Mad2 signals appeared on chromosomes that had not displayed any signals in previous time frames (Fig. 7, E and F, blue arrows). The signal intensity was generally weaker than that observed on persistently unaligned chromosomes (Fig. 7 F, yellow arrowheads), supporting the idea that the GFP-Mad2 emerged on those chromosomes during metaphase instead of persisting from early prometaphase. From these results, we concluded that once-disappeared GFP-Mad2 could reemerge in the absence of KIF18A. Thus, KIF18A may be required to maintain proper attachment status between MTs and kinetochores.
Discussion
Biochemical activity of kinesin-8Klp67A
We observed the processive plus end–directed motility, catastrophe-induction, and rescue/pausing activities of kinesin-8Klp67A toward MTs. This combination of activities has not been observed for another five Drosophila MT plus end–regulating proteins we have characterized so far using an identical assay (Li et al., 2012; Moriwaki and Goshima, 2016). Motility and catastrophe induction can be deduced from the amino acid sequences of Klp67A’s motor domain; the motor has been shown to be critical in a previous study using a rigor mutant (Savoian and Glover, 2010). In contrast, how rescue/pausing activity and also mild growth-accelerating activity are executed remains unclear; it might involve the usage of the tubulin-binding region next to the motor domain to increase the local concentration of tubulin.
Whether human KIF18A is a depolymerase (Mayr et al., 2007; Locke et al., 2017) or an MT dynamics suppressor (Du et al., 2010) has been debated. However, those studies used distinct assay and buffer conditions in vitro; it is possible that KIF18A can execute both activities in cells, namely, being endowed with a similar set of activities to Drosophila Klp67A. Budding yeast Kip3 is an established MT depolymerase, but rescue and pausing frequencies are also reduced in the mutant in vivo (Gupta et al., 2006), for which the tubulin-binding region is involved (Su et al., 2011). Our study suggests that the multiple activities observed here for kinesin-8Klp67A in a single experimental condition are widely conserved among kinetochore-localized kinesin-8s.
Kinesin-8Klp67A is required for stable kinetochore–MT attachment
Previous studies using RNAi or mutants of kinesin-8Klp67A consistently reveal its role in spindle length regulation (Goshima and Vale, 2003, 2005; Savoian et al., 2004; Gatt et al., 2005; Goshima et al., 2005; Buster et al., 2007; Wang et al., 2010). This was confirmed in this study and most likely is attributed to the catastrophe-inducing function. However, experiments involving colcemid treatment indicated that MT elongation with reduced catastrophe is not the causal factor leading to chromosome misalignment. GFP-Rod imaging further uncovered a specific role of kinesin-8Klp67A at the kinetochore–MT interface: it ensures persistent MT attachment to the kinetochore.
Why are MTs frequently detached in the absence of kinesin-8Klp67A? We observed that overexpression of the MT rescue/pausing factor CLASP largely rescued the flipping behavior of chromosomes. Since flipping is associated with the detachment event, we interpret that CLASP overexpression reduced MT detachment rates. The data therefore suggest that kinetochores are prone to detach from MTs during rapid and persistent depolymerization. On the other hand, in vitro study using Ndc80-decorated beads and depolymerizing MTs indicated robust load-bearing attachment of the beads during depolymerization (Powers et al., 2009). However, this observation might be reconciled with our findings, as Aurora B kinase could dampen Ndc80’s MT binding ability in cells. Consistent with this notion, we observed stable MT association when Aurora B kinase was inhibited. We propose that kinesin-8Klp67A constitutes an additional layer of the MT attachment interface.
Functional similarity and difference between Drosophila and human kinesin-8s
It was previously reported that KIF18A depletion increases the amplitude of chromosome oscillation, where MT dynamics regulation, rather than attachment per se, is defective (Stumpff et al., 2008). This behavior is consistent with the in vitro MT stabilization activity (Du et al., 2010). However, a more recent study suggested that kinetochore–MT interaction is also perturbed in this condition (Kim and Stumpff, 2018). The study attributes the phenotype partly to defects in KIF18A-dependent PP1 delocalization (Kim and Stumpff, 2018); however, those investigations also showed the presence of a PP1-independent function of KIF18A for chromosome alignment. It remains to be determined if PP1 mislocalization contributes to chromosome misalignment in the absence of kinesin-8Klp67A, which does not possess the PP1-binding motif (De Wever et al., 2014).
While we were revising the manuscript, a new report on human KIF18A function was published, in which KIF18A depletion phenotype was investigated in a haploid cell line as well as HeLa cells (Janssen et al., 2018). In this report, MTs and Mad1 (binding partner of Mad2) were immuno-stained in the metaphase-arrested cell in the absence of KIF18A, and three interesting conclusions have been drawn: (1) Mad1 is detected on multiple, but not all, kinetochores in the absence of KIF18A, which was consistent with a previous report using Mad2 (Mayr et al., 2007); (2) similar numbers of MTs are associated with each kinetochore, regardless of the presence or absence of Mad1 accumulation (i.e., MT–kinetochore attachment is not disrupted in the absence of KIF18A); (3) nevertheless, tension is not sufficiently applied to Mad1-positive kinetochores. Our data obtained in live cells confirmed the first point. We detected even fewer numbers of Mad2 signals than Janssen et al. (2018); this might be the due to the difference in knockdown efficiency or methodology (live cell imaging vs. immuno-staining after preextraction of the cell). In contrast, neither study has revealed the mechanism by which tension is reduced, despite the apparently similar numbers of MTs attached in an end-on fashion to the kinetochore. However, it would be reasonable to assume that kinetochore–MT attachment is somewhat skewed in the absence of KIF18A. Our data suggests that this could occur even when attachment has been once established. In this regard, it is intriguing that we could not identify an apparent defect in the monopolar spindle. KIF18A may be particularly critical in the bipolar spindle, in which it might fine tune the attachment mode for the kinetochore to generate sufficient tension.
Our data and those in Janssen et al. (2018) both agreed that MT attachment does not require KIF18A, which is consistent with the conclusion drawn from the analysis using Drosophila Klp67A. However, in the case of Drosophila, the loss of Klp67A caused MT detachment from the kinetochore, whereas the KIF18A depletion phenotype was milder. This was possibly due to the presence of other regulators of kinetochore–MT attachment in mammals, such as Ska and SKAP/astrin complexes, which are missing in Drosophila (Schmidt et al., 2012; Kern et al., 2017). Nevertheless, proper attachment does require KIF18A function, as reflected by the appearance of Mad1/Mad2 signals and the loss of tension in the absence of KIF18A (Janssen et al., 2018). Thus, KIF18A ensures the attachment mode for the kinetochore to generate sufficient tension. Several studies suggest an involvement of KIF18A in carcinogenesis (Zhang et al., 2010; Hitti et al., 2016). It would be intriguing to investigate how KIF18A contributes to chromosome segregation in various cancer cell models.
Materials and methods
RNAi and cell line selection
S2 cell culture and RNAi were performed as previously described (Goshima et al., 2007; Bettencourt-Dias and Goshima, 2009; Ito and Goshima, 2015). In brief, Schneider’s medium (Gibco) supplemented with 10% serum was used for cell culture. Cell lines were selected with hygromycin or puromycin following plasmid transformation with the TransIT-Insect reagent (TaKaRa). Plasmids used in this study are listed in Table S1, whereas dsRNA sequences used here are available at Goshima and Vale (2005) and Goshima et al. (2007). For RNAi experiments, cells were treated with dsRNAs for 3–4 d and then plated on concanavalin A–coated glass-bottom dishes for microscopy. HeLa cells stably expressing GFP-Mad2 (a gift of T. Kiyomitsu; Nagoya University, Nagoya, Japan) were cultured in DMEM with 10% serum. RNAi was conducted with RNAiMax (Invitrogen) and 25 nM siRNA as previously described (UAAAUUACCCGAACAAGAATT; Tanenbaum et al., 2009). Imaging was started at 24 h.
Microscopy
S2 MTs were stained with 15 nM SiR-tubulin, whereas EB1-SNAP was visualized with 15–30 nM SiR-SNAP. Live S2 imaging was performed with a spinning-disc confocal microscope (Nikon Ti; 100× 1.45 NA or 60× 1.40 NA lens; EM charge-coupled device camera ImagEM [Hamamatsu]; CSU-X1 [Yokogawa]). A total internal reflection fluorescence microscope was used in the in vitro MT dynamics experiment (Nikon Ti; 100× 1.49 NA lens; EM charge-coupled device camera Evolve [Roper]). 488-/561-/640-nm excitation lasers were associated with both microscopes. Microscopes were controlled by micromanager and images were processed with ImageJ (National Institutes of Health). For the colcemid treatment experiment in Fig. 5 D, S2 cells expressing GFP-Rod and H2B-mCherry were stained with 15 nM SiR-tubulin and treated with 25 µM MG132 (≥ 30 min). Cells were imaged every 1 min for 20 min, and 5 µg/ml colcemid was supplied at 2 min. In Fig. S3 (B–D), cells were treated with 60 ng/ml colcemid for 2 h; images were acquired every 2 min. All imaging was performed at ∼25°C. HeLa cells expressing GFP-Mad2 by CMV promoter were stained with 50 nM SiR-tubulin. Metaphase arrest, MT depolymerization, and monopolar spindle formation were induced by 25 µM MG132, 100 nM nocodazole, and 5 µM STLC (a kinesin-5 inhibitor), respectively (≥1 h).
Protein purification
S2 tubulin was purified by using a previously described method, using GST-tagged TOG1 domain (Saccharomyces cerevisiae Stu2, 1–306 aa; Widlund et al., 2012; Moriwaki and Goshima, 2016). Klp67A-GFP-6His was expressed in Escherichia coli SoluBL21 cells (250 ml culture in l-rich medium, 0.1–0.5 mM IPTG at 18°C for 16–20 h). Cells were resuspended in Lysis Buffer (50 mM MOPS-NaOH, pH 7.2, 250 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 0.5 mM PMSF, peptide cocktail [1 µg/ml leupeptin, pepstatin, chymostatin, and aprotinin], 2 mM 2-mercaptoethanol, and 0.1 mM ATP), sonicated with a homogenizer (450DA; Branson), bound to Ni-NTA (4°C, 60–90 min), washed with Wash Buffer (Lysis Buffer supplemented with 20 mM imidazole and 0.2% Tween), followed by elution 5–8 times with Elution Buffer (MRB80 [80 mM Pipes-KOH, pH 6.8, 1 mM EGTA, and 4 mM MgCl2], 100 mM KCl, 250 mM imidazole, 2 mM 2-mercaptoethanol, and 1 mM ATP). The eluate was subjected to sucrose gradient sedimentation. Gel filtration chromatography was not used because the procedure caused protein loss due to unknown reasons. A 2.5–40% sucrose gradient was made with 2 ml buffer (MRB80, 100 mM KCl, 0.1 mM ATP, 1 mM DTT, and sucrose) in a 2.2-ml, 11- × 35-mm ultracentrifugation tube (347357; Beckman Coulter). Protein solution (200 µl) was applied and centrifuged with a TLS-55 rotor (214,000 g, 4 h, 4°C), and 16 fractions were collected. Fractions containing Klp67A-GFP-6His were identified with SDS-PAGE and Coomassie staining, followed by flash freezing. The tail-less Klp67A (1–612 aa)–GFP-6His and motor-alone Klp67A (1–359 aa)–GFP-6His were purified in a manner identical to full-length Klp67A-GFP-6His, except that the gradient sedimentation step was omitted, and the buffer was exchanged instead to MRB80 containing 100 mM KCl, 20% sucrose, 0.1 mM ATP, and 1 mM DTT, using the desalting column PD MiniTrap G-25 (GE Healthcare). The solution was flash frozen with liquid nitrogen and stored at −80°C. 6His-Klp10A was purified with Ni-NTA, as previously described (Moriwaki and Goshima, 2016).
Sucrose gradient sedimentation of the Klp67A–tubulin complex
Purified Klp67A-GFP-6His (∼2 µM) solution was dialyzed with Tube-O-DIALYZE, Micro, and 8K MWCO (TaKaRa) in MRB80 containing 75 mM KCl, 5% sucrose, 0.1 mM ATP, and 1 mM DTT for 6 h at 4°C. The dialyzed Klp67A was mixed with 4 µM pig tubulin, 1 mM ATP, and 1 mM GTP and incubated for 10 min at room temperature. The mixed solution was loaded into sucrose gradient buffer (MRB80, 75 mM KCl, 1 mM ATP, 1 mM GTP, 1 mM DTT, and sucrose) to make 5–30% sucrose gradient in a 2.2-ml, 11- × 35-mm ultracentrifugation tube and centrifuged with TLS-55 rotor (214,000 g, 5.5 h, 4°C). The sedimented solution was divided into 16 fractions, and the peak fractions were identified with SDS-PAGE and Sypro Ruby Staining.
In vitro MT polymerization assay
The in vitro MT polymerization assay was performed essentially following a method previously described (Li et al., 2012; Moriwaki and Goshima, 2016). A silanized coverslip was coated with anti-biotin (1–5% in 1× MRB80; Invitrogen), and the nonspecific surface was blocked with Pluronic F127 (1% in 1× MRB80; Invitrogen). Biotinylated MT seeds (50–100 µM tubulin mix containing 10% biotinylated pig tubulin and 10% Alexa Fluor 647–labeled pig tubulin with 1 mM GMPCPP) were specifically attached to the functionalized surface by biotinylated tubulin–anti-biotin links. After the chamber was washed with 1× MRB80, MT growth was initiated by flowing 10 µM tubulin (containing 80% S2 tubulin and 20% Alexa Fluor 568–labeled pig tubulin) and Klp67A-GFP into the assay buffer (1× MRB80, 75 mM KCl, 1 mM GTP, 1 mM ATP, 0.5 mg/ml κ-casein and 0.1% methylcellulose, and 5.5% sucrose) and an oxygen scavenger system (50 mM glucose, 400 µg/ml glucose oxidase, 200 µg/ml catalase, and 4 mM DTT). The samples were sealed with candle wax. During experiments, the samples were maintained at ∼25°C, and images were collected every 3 s for 15 min using total internal reflection fluorescence microscopy. MT grew from both ends, but only the plus end dynamics were analyzed. MT depolymerization assay was conducted in an identical condition, except that no tubulin was included. MT gliding assay was performed following a previous report (Miki et al., 2015) with slight modification to the buffer. In brief, the flow chamber was washed with 1× MRB80, and purified kinesin-1 motor (K560-6His) was flowed into the chamber. After washing with MRB80 containing 0.5 mg/ml κ-casein, the motility buffer (1× MRB80, 75 mM KCl, GMPCPP-stabilized MTs with Alexa Fluor 647 labels, 1 mM ATP, 0.5 mg/ml κ-casein and 0.1% methylcellulose, and 5% sucrose), with an oxygen scavenger system (50 mM glucose, 400 mg/ml glucose oxidase, 200 mg/ml catalase and 4 mM DTT) and 4 nM Klp67A-GFP, was flowed into the chamber. The single kinesin motility assay was conducted following other publications (Naito and Goshima, 2015) with slight modification to the buffer. A silanized coverslip was coated with anti-biotin antibody, and a solution containing 1% pluronic acid was loaded into the chamber. After washing once with 1× MRB80, GMPCPP-stabilized MTs labeled with Alexa Fluor 647 and biotin were loaded. After a 1× MRB80 wash, Klp67A-GFP was loaded into the chamber with a buffer identical to that used for the in vitro MT polymerization assay. GFP was bleached for 3 min with a 20-mW, 488-nm laser (maximum power), followed by imaging unbleached GFP with 40% power.
Data analysis
MT attachment instability was determined by counting the detachment events over time (monotelic to unattachment/lateral interaction or syntelic to monotelic conversion), whereas MT attachment frequency was obtained for monotelically attached chromosomes. MT plus end dynamics in vitro were analyzed based on kymographs, following others (Moriwaki and Goshima, 2016): catastrophe frequency was determined by dividing the number of shrinkage events by the sum of growth and pause times, whereas the transition from shrinkage to pause or growth was considered a rescue event, and the rescue frequency (for shrinkage time) was calculated. When MTs did not grow or shrink more than two pixels (0.32 µm) for five or more frames (15 s), this period was defined as a pause. Intensity of fluorescent tubulin on GMPCPP-MT was measured with the line tool associated with Fiji/ImageJ (images acquired at 5 min of time lapse were used). The obtained value was divided by the fluorescent intensity of GMPCPP-MT. To investigate the correlation between Klp67A-GFP intensity at the tip and the growth/shrinkage rate, kymographs were analyzed with the segmented line tool associated with Fiji/ImageJ. At each time point, the maximum GFP intensity within five pixels (0.8 µm) around the tip was obtained, and after background intensity subtraction, the mean GFP intensity during the growth (or shrinkage) was calculated. The plot was analyzed with general linear regression, where GFP intensity and growth/shrinkage rate were used as explanatory and response variables, respectively, and gamma distribution was assumed. Reciprocal function was used as link function. To make the response variable exclusively positive values, 0 µm/min growth rate was approximated to 10−9 µm/min. The approximate curve was drawn with 95% Wald confidence interval. P values were obtained with likelihood-ratio test. Image analysis was performed with Jython scripts associated with Fiji/ImageJ. Further statistical analysis and data visualizations were performed on R or Prism.
Online supplemental material
Fig. S1 shows purification of kinesin-8Klp67A-GFP. Fig. S2 shows MT attachment instability after kinesin-8Klp67A RNAi. Fig. S3 shows no rescue of chromosome misalignment after artificial MT shortening in the absence of kinesin-8Klp67A. Fig. S4 shows no rescue of chromosome misalignment after CLASPMast/Orbit overexpression in the absence of kinesin-8Klp67A. Video 1 shows processive motility and plus end accumulation of kinesin-8Klp67A. Video 2 shows MT attachment stability in the monopolar spindle in the presence or absence of kinesin-8Klp67A. Video 3 shows MT attachment stability in the bipolar spindle in the presence or absence of kinesin-8Klp67A. Video 4 shows MT attachment stability in the absence of augminDgt6. Video 5 shows stable syntelic attachment in the monopolar spindle after Aurora B inhibition in the absence of kinesin-8Klp67A. Video 6 shows the rescue of MT attachment stability by CLASPMast/Orbit overexpression in the absence of kinesin-8Klp67A. Video 7 shows GFP-Mad2 dynamics in the monopolar spindle after KIF18A RNAi in HeLa cells. Table S1 shows plasmids used in this study.
Supplementary Material
Acknowledgments
We thank Kosuke Ariga for helping data analysis, Tomoko Nishiyama for technical support, Tomomi Kiyomitsu for valuable comments on the manuscript, and Elsa Tungadi for proofreading.
This work was funded by Japan Society for the Promotion of Science (JSPS) KAKENHI (15KT0077 and 17H01431) and Laura and Arthur Colwin Endowed Summer Research Fellowship Fund (2015) of the Marine Biological Laboratory to G. Goshima. T. Edzuka is a recipient of a JSPS pre-doctoral fellowship (16J02807).
The authors declare no competing financial interests.
Author contributions: T. Edzuka and G. Goshima conceived and designed the research project. T. Edzuka performed most of the experiments and analyzed the data. G. Goshima performed some experiments, analyzed the data, and wrote the paper.
References
- Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S.C., and Chang F.. 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell. 19:245–258. 10.1016/j.devcel.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., and Karess R.. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14:56–61. 10.1016/j.cub.2003.12.025 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., and Goshima G.. 2009. RNAi in Drosophila S2 cells as a tool for studying cell cycle progression. Methods Mol. Biol. 545:39–62. 10.1007/978-1-60327-993-2_3 [DOI] [PubMed] [Google Scholar]
- Buster D.W., Zhang D., and Sharp D.J.. 2007. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol. Biol. Cell. 18:3094–3104. 10.1091/mbc.e06-11-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R. III, Chan C.S., Drubin D.G., and Barnes G.. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cottingham F.R., and Hoyt M.A.. 1997. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138:1041–1053. 10.1083/jcb.138.5.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- De Wever V., Nasa I., Chamousset D., Lloyd D., Nimick M., Xu H., Trinkle-Mulcahy L., and Moorhead G.B.. 2014. The human mitotic kinesin KIF18A binds protein phosphatase 1 (PP1) through a highly conserved docking motif. Biochem. Biophys. Res. Commun. 453:432–437. 10.1016/j.bbrc.2014.09.105 [DOI] [PubMed] [Google Scholar]
- Dick A.E., and Gerlich D.W.. 2013. Kinetic framework of spindle assembly checkpoint signalling. Nat. Cell Biol. 15:1370–1377. 10.1038/ncb2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., English C.A., and Ohi R.. 2010. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol. 20:374–380. 10.1016/j.cub.2009.12.049 [DOI] [PubMed] [Google Scholar]
- Erent M., Drummond D.R., and Cross R.A.. 2012. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PLoS One. 7:e30738 10.1371/journal.pone.0030738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R., Bonaccorsi S., Wentworth D., Doxsey S., Gatti M., and Pereira A.. 2004. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell. 15:121–131. 10.1091/mbc.e03-05-0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Vardy L., Koonrugsa N., and Toda T.. 2001. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 20:3389–3401. 10.1093/emboj/20.13.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt M.K., Savoian M.S., Riparbelli M.G., Massarelli C., Callaini G., and Glover D.M.. 2005. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J. Cell Sci. 118:2671–2682. 10.1242/jcs.02410 [DOI] [PubMed] [Google Scholar]
- Głuszek A.A., Cullen C.F., Li W., Battaglia R.A., Radford S.J., Costa M.F., McKim K.S., Goshima G., and Ohkura H.. 2015. The microtubule catastrophe promoter Sentin delays stable kinetochore-microtubule attachment in oocytes. J. Cell Biol. 211:1113–1120. 10.1083/jcb.201507006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907. 10.1091/mbc.e05-02-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., and Vale R.D.. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988. 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., and Vale R.D.. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429. 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L. Jr., Carvalho P., Roof D.M., and Pellman D.. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923. 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Hitti E., Bakheet T., Al-Souhibani N., Moghrabi W., Al-Yahya S., Al-Ghamdi M., Al-Saif M., Shoukri M.M., Lánczky A., Grépin R., et al. . 2016. Systematic Analysis of AU-Rich Element Expression in Cancer Reveals Common Functional Clusters Regulated by Key RNA-Binding Proteins. Cancer Res. 76:4068–4080. 10.1158/0008-5472.CAN-15-3110 [DOI] [PubMed] [Google Scholar]
- Ito A., and Goshima G.. 2015. Microcephaly protein Asp focuses the minus ends of spindle microtubules at the pole and within the spindle. J. Cell Biol. 211:999–1009. 10.1083/jcb.201507001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen L.M.E., Averink T.V., Blomen V.A., Brummelkamp T.R., Medema R.H., and Raaijmakers J.A.. 2018. Loss of Kif18A Results in Spindle Assembly Checkpoint Activation at Microtubule-Attached Kinetochores. Curr. Biol. 28:2685–2696.e4. 10.1016/j.cub.2018.06.026 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P. 2016. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology (Basel). 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D.M., Monda J.K., Su K.C., Wilson-Kubalek E.M., and Cheeseman I.M.. 2017. Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. eLife. 6:e26866 10.7554/eLife.26866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., and Stumpff J.K.. 2018. Kif18A promotes Hec1 dephosphorylation to coordinate chromosome alignment with kinetochore microtubule attachment. bioRxiv. doi: 10.1101/304147 (Preprint posted April 18, 2018). [Google Scholar]
- Lampson M.A., and Grishchuk E.L.. 2017. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology (Basel). 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K., and Goshima G.. 2012. Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 199:849–862. 10.1083/jcb.201206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J., Joseph A.P., Peña A., Möckel M.M., Mayer T.U., Topf M., and Moores C.A.. 2017. Structural basis of human kinesin-8 function and inhibition. Proc. Natl. Acad. Sci. USA. 114:E9539–E9548. 10.1073/pnas.1712169114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Fairley E.A., Rieder C.L., Swedlow J.R., Sunkel C.E., and Earnshaw W.C.. 2003. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 113:891–904. 10.1016/S0092-8674(03)00465-3 [DOI] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A., and Rieder C.L.. 2005. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7:42–47. 10.1038/ncb1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M.I., Hümmer S., Bormann J., Grüner T., Adio S., Woehlke G., and Mayer T.U.. 2007. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17:488–498. 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- Mayr M.I., Storch M., Howard J., and Mayer T.U.. 2011. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PLoS One. 6:e27471 10.1371/journal.pone.0027471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh T., Gluszek A.A., and Welburn J.P.I.. 2018. Microtubule end tethering of a processive kinesin-8 motor Kif18b is required for spindle positioning. J. Cell Biol. 217:2403–2416. 10.1083/jcb.201705209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Nishina M., and Goshima G.. 2015. RNAi screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in Physcomitrella patens. Plant Cell Physiol. 56:737–749. 10.1093/pcp/pcv002 [DOI] [PubMed] [Google Scholar]
- Moriwaki T., and Goshima G.. 2016. Five factors can reconstitute all three phases of microtubule polymerization dynamics. J. Cell Biol. 215:357–368. 10.1083/jcb.201604118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., and Desai A.. 2017. A Molecular View of Kinetochore Assembly and Function. Biology (Basel). 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito H., and Goshima G.. 2015. NACK kinesin is required for metaphase chromosome alignment and cytokinesis in the moss Physcomitrella patens. Cell Struct. Funct. 40:31–41. 10.1247/csf.14016 [DOI] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., and Asbury C.L.. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875. 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., and Sharp D.J.. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. 10.1038/nature02256 [DOI] [PubMed] [Google Scholar]
- Savoian M.S., and Glover D.M.. 2010. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J. Cell Sci. 123:767–776. 10.1242/jcs.055905 [DOI] [PubMed] [Google Scholar]
- Savoian M.S., Gatt M.K., Riparbelli M.G., Callaini G., and Glover D.M.. 2004. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 117:3669–3677. 10.1242/jcs.01213 [DOI] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. . 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smurnyy Y., Toms A.V., Hickson G.R., Eck M.J., and Eggert U.S.. 2010. Binucleine 2, an isoform-specific inhibitor of Drosophila Aurora B kinase, provides insights into the mechanism of cytokinesis. ACS Chem. Biol. 5:1015–1020. 10.1021/cb1001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., and Murray A.W.. 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143:687–694. 10.1083/jcb.143.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., and Wordeman L.. 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 14:252–262. 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Du Y., English C.A., Maliga Z., Wagenbach M., Asbury C.L., Wordeman L., and Ohi R.. 2011. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol. Cell. 43:764–775. 10.1016/j.molcel.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Wagenbach M., Franck A., Asbury C.L., and Wordeman L.. 2012. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell. 22:1017–1029. 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Qiu W., Gupta M.L. Jr., Pereira-Leal J.B., Reck-Peterson S.L., and Pellman D.. 2011. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell. 43:751–763. 10.1016/j.molcel.2011.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Arellano-Santoyo H., Portran D., Gaillard J., Vantard M., Thery M., and Pellman D.. 2013. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 15:948–957. 10.1038/ncb2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Janssen A., Geers E.F., Alvarez-Fernández M., and Medema R.H.. 2009. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19:1703–1711. 10.1016/j.cub.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., and Davis T.N.. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723. 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell J.D., and Sorger P.K.. 2006. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172:861–874. 10.1083/jcb.200509101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., and Howard J.. 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962. 10.1038/ncb1462 [DOI] [PubMed] [Google Scholar]
- Varga V., Leduc C., Bormuth V., Diez S., and Howard J.. 2009. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 138:1174–1183. 10.1016/j.cell.2009.07.032 [DOI] [PubMed] [Google Scholar]
- Wang H., Brust-Mascher I., Cheerambathur D., and Scholey J.M.. 2010. Coupling between microtubule sliding, plus-end growth and spindle length revealed by kinesin-8 depletion. Cytoskeleton (Hoboken). 67:715–728. 10.1002/cm.20482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargacki M.M., Tay J.C., Muller E.G., Asbury C.L., and Davis T.N.. 2010. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle. 9:2581–2588. 10.4161/cc.9.13.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L.N., Ems-McClung S.C., Stout J.R., LeBlanc C., Shaw S.L., Gardner M.K., and Walczak C.E.. 2011. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr. Biol. 21:1500–1506. 10.1016/j.cub.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom T., and McIntosh J.R.. 2002. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 115:931–940. [DOI] [PubMed] [Google Scholar]
- Widlund P.O., Podolski M., Reber S., Alper J., Storch M., Hyman A.A., Howard J., and Drechsel D.N.. 2012. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell. 23:4393–4401. 10.1091/mbc.e12-06-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Signorile L., Basu S., Ottema S., Lebbink J.H.G., Leslie K., Smal I., Dekkers D., Demmers J., and Galjart N.. 2016. Isolation of functional tubulin dimers and of tubulin-associated proteins from mammalian cells. Curr. Biol. 26:1728–1736. 10.1016/j.cub.2016.04.069 [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhu C., Chen H., Li L., Guo L., Jiang W., and Lu S.H.. 2010. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 31:1676–1684. 10.1093/carcin/bgq134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.