Detyrosinated microtubules constitute a minor subpopulation of microtubules in epithelial cells. Lysosomes are specifically enriched on detyrosinated microtubules through a kinesin-1–dependent mechanism. This spatial constraining of lysosomes to a subset of microtubules enables them to efficiently encounter and fuse with autophagosomes to initiate autophagy.
Abstract
Microtubule post-translational modifications impart functional diversity to microtubules by affecting their dynamics, organization, and interaction with proteins. Using super-resolution microscopy, we show that only a small subpopulation of microtubules are detyrosinated in epithelial cells, while acetylated and tyrosinated microtubules comprise the majority of all microtubules. Surprisingly, lysosomes are enriched by approximately threefold on detyrosinated microtubules. Further, their motility on detyrosinated microtubules is impaired, showing shorter runs and more frequent and longer pauses. Lysosome enrichment is mediated through a kinesin-1–dependent mechanism, since knocking down this motor abolishes enrichment. Finally, correlative live-cell and super-resolution microscopy showed that lysosomes interact with autophagosomes on detyrosinated microtubules. Removal of detyrosinated microtubules or knockdown of kinesin-1 leads to a decrease in the percentage of autolysosomes, a fusion intermediate of autophagosomes and lysosomes. Taken together, our data reveal a new role of detyrosinated microtubules as hubs that spatially concentrate lysosomes on a small subset of microtubules and facilitate their interaction and fusion with autophagosomes to initiate autophagy.
Introduction
Lysosomes are dynamic organelles that move bidirectionally along microtubules (Bálint et al., 2013; Verdeny-Vilanova et al., 2017); interact and fuse with compartments from the endocytic, secretory, and autophagy pathways; and degrade proteins and other macromolecules that they receive from these pathways (Luzio et al., 2007). In addition to their classic role in degradation, recent discoveries have dramatically expanded the functional significance of lysosomes as nutrient sensors, docking sites for active mTORC1, and regulators of transcription and metabolic homeostasis (Sancak et al., 2008, 2010; Zoncu et al., 2011; Lim and Zoncu, 2016). These new discoveries have placed lysosomes as central organelles controlling cell growth and survival (Lim and Zoncu, 2016). It has also emerged that lysosome positioning within the cytoplasm is essential for regulating its many functions (Pu et al., 2016). Retrograde motility of lysosomes is mediated by the dynein motor, whereas several kinesin motors belonging to different kinesin families have been implicated in anterograde lysosome motility (Pu et al., 2016). In particular, KIF5B, a member of the kinesin-1 family, KIF3A, a member of the kinesin-2 family, and KIF1A/KIF1Bβ, members of the kinesin-3 family, have been shown to be responsible for the peripheral movement and distribution of lysosomes (Matsushita et al., 2004; Brown et al., 2005; Loubéry et al., 2008; Granger et al., 2014; Bentley et al., 2015; Guardia et al., 2016). Several of the adapter and accessory proteins involved in linking these motors to the lysosome membrane have also been identified, including Rab7, RILP, and the BORC complex (Pu et al., 2016). The stop-and-go motility of lysosomes mediated by the presence of these multiple motors has also been extensively studied using live-cell imaging (Jordens et al., 2001; Bálint et al., 2013; Verdeny-Vilanova et al., 2017). Therefore, a lot is known about the machinery involved in moving lysosomes along microtubules and mediating their fusion with other compartments. However, the microtubule cytoskeleton has a complex three-dimensional architecture, connecting the perinuclear region to the cell periphery. Given the high density of microtubules and the diversity of the pathways that a single lysosome can follow in traveling from one part of the cell to another, how lysosome positioning can be regulated and how lysosomes can efficiently encounter and fuse with compartments of the endocytic and the autophagy pathways are less clear.
Microtubules assemble from heterodimers of α- and β-tubulin and carry out a host of crucial functions ranging from intracellular trafficking to chromosome segregation during mitosis. Tubulins within microtubules are subject to a large number of post-translational modifications (PTMs; Magiera and Janke, 2014; Song and Brady, 2015). Most α-tubulin contains a terminal tyrosine residue, which can be removed by a newly discovered cytosolic carboxypeptidase (Aillaud et al., 2017; Nieuwenhuis et al., 2017) leading to detyrosination. Acetylation, on the other hand, happens on Lys40 of α-tubulin in the microtubule lumen and is the only PTM that is not on the microtubule surface (Magiera and Janke, 2014). Akin to the histone code, microtubule PTMs are thought to generate microtubule diversity and hence regulate the various functions of microtubules (Song and Brady, 2015). Recent work has highlighted the differential motility of motor proteins such as dynein and kinesin on microtubules containing different PTMs (Dunn et al., 2008; Cai et al., 2009; Sirajuddin et al., 2014). In addition, the role of specific microtubule PTMs in a myriad of biological processes, ranging from regulation of retrograde trafficking in neurons to guidance of chromosomes by kinesin motors, is starting to be identified (Friedman et al., 2010; Barisic et al., 2015; Herms et al., 2015; Nirschl et al., 2016; Tas et al., 2017).
Here, we aimed to determine whether microtubules with specific PTMs can play a role in mediating efficient lysosome–autophagosome encounters and fusion along the complex cytoskeletal network. Using a combination of super-resolution microscopy and live-cell imaging, we show that, instead of equally partitioning among all microtubules, at any given time, lysosomes are specifically enriched on a small subset of detyrosinated microtubules. Autophagosomes are also enriched on the same small subset of microtubules, albeit to a lesser extent. This enrichment spatially constrains lysosomes, autophagosomes, and potentially other endocytic compartments to a minority of microtubule tracks, along which they can encounter and fuse with each other. Indeed, autolysosomes, which are fusion intermediates of autophagosomes with lysosomes, were similarly enriched on detyrosinated microtubules, and heterotypic interactions among lysosomes and autophagosomes happened much more frequently along detyrosinated microtubules. Kinesin-1 knockdown abolished lysosome enrichment on detyrosinated microtubules, suggesting that this enrichment is mediated through a mechanism involving this motor protein. Finally, loss of enrichment either through removal of detyrosinated microtubules or knockdown of kinesin-1 led to decreased fusion between lysosomes and autophagosomes. Our results highlight a new role of detyrosinated microtubules in mediating efficient autophagy.
Results
Detyrosinated microtubules form a small subset of all microtubules in epithelial cells
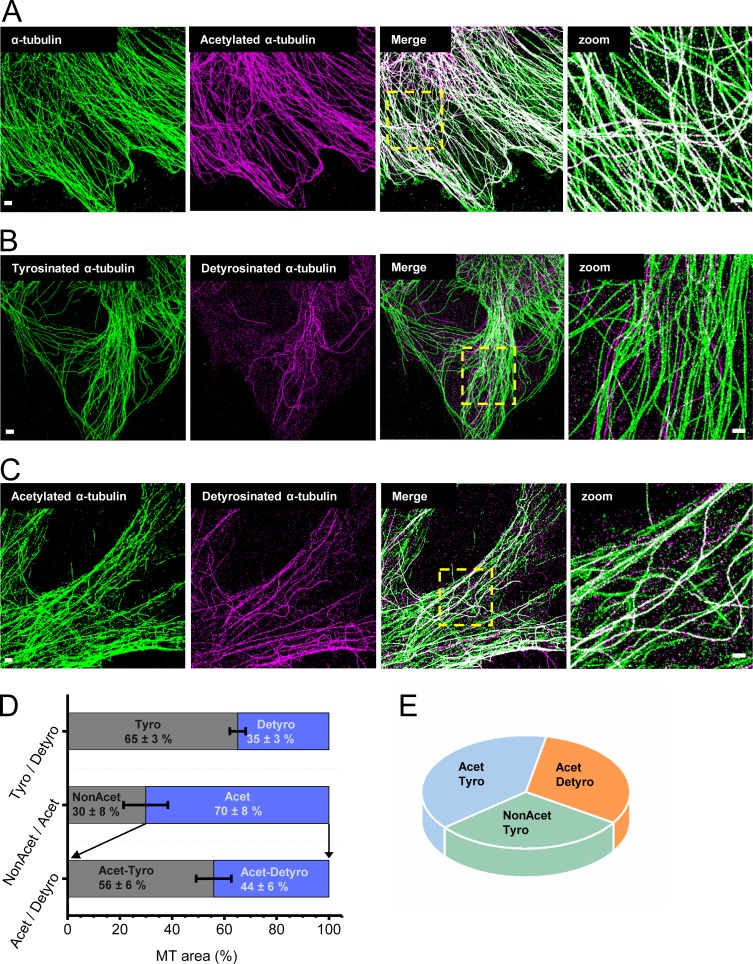
To quantify the percentage of microtubules containing different PTMs in epithelial cells, we performed sequential multi-color super-resolution microscopy (Tam et al., 2014b; Fig. 1). We labeled total α-tubulin as well as detyrosinated, tyrosinated, and acetylated α-tubulin, using antibodies that recognize these specific PTMs; performed multi-color super-resolution microscopy; and quantified the percentage of post-translationally modified microtubule subsets (Fig. 1 and Fig. S1 A). Multi-color super-resolution images of total α-tubulin, together with acetylated α-tubulin, revealed that a majority of microtubules were acetylated (70 ± 8% acetylated, n = 10 cells; Fig. 1, A and D; and Fig. S1 B). Tyrosinated and detyrosinated microtubules formed distinct subsets that excluded one another, as expected (Fig. 1 B), and detyrosinated microtubules constituted a small subpopulation of all microtubules (35 ± 3%, n = 10 cells; Fig. 1, B and D; and Fig. S1 C; Verdeny-Vilanova et al., 2017). Interestingly, there was more cell-to-cell variability in the proportion of acetylated microtubules (Fig. S1 B), while the proportion of detyrosinated microtubules was very similar among different cells (Fig. S1 C). Finally, there was a large overlap between acetylated and detyrosinated microtubules (Fig. 1 C). In fact, almost all detyrosinated microtubules were also acetylated, whereas only a subpopulation (44 ± 6%, n = 10 cells) of acetylated microtubules overlapped with detyrosinated microtubules, suggesting that the rest of the acetylated microtubule subset (56 ± 6%, n = 6) was tyrosinated (Fig. 1, C and D; and Fig. S1 D). These results were consistent, whether detyrosinated microtubules were labeled first, followed by acetylated microtubules or vice versa, suggesting that the two antibodies did not interfere with each other’s epitope recognition and binding (Fig. S1, E and F). Further, the percentage of acetylated–detyrosinated and acetylated–tyrosinated microtubules was similar for a range of antibody concentrations and reached saturation (Fig. S1, E and F). Based on these results, we concluded that microtubules contained different combinatorial degrees of PTMs (Fig. 1 E) that may differentially affect microtubule function, with detyrosinated microtubules overall constituting a minority subpopulation (∼35%).
Figure 1.
Detyrosinated microtubules form a small subset of all microtubules. (A–C) Two-color super-resolution images of total α-tubulin (green), acetylated α-tubulin (magenta), and overlay (A); tyrosinated α-tubulin (green), detyrosinated α-tubulin (magenta), and overlay (B); and acetylated α-tubulin (green), detyrosinated α-tubulin (magenta), and overlay (C). (D) Bar plots show the proportion of detyrosinated α-tubulin, acetylated α-tubulin, and double-modified α-tubulin containing both acetylation and detyrosination marks quantified as the percentage of total microtubule (MT) area corresponding to these PTMs. The bars indicate the median, and the error bar is the SD, analyzed for 10 different cells in each case. (E) Pie diagram showing the major subpopulations of modified microtubules that could be identified from the super-resolution images. Bars are 2 µm (left) and 500 nm (Zoom).
Lysosomes and their fusion intermediates are specifically enriched on detyrosinated microtubules
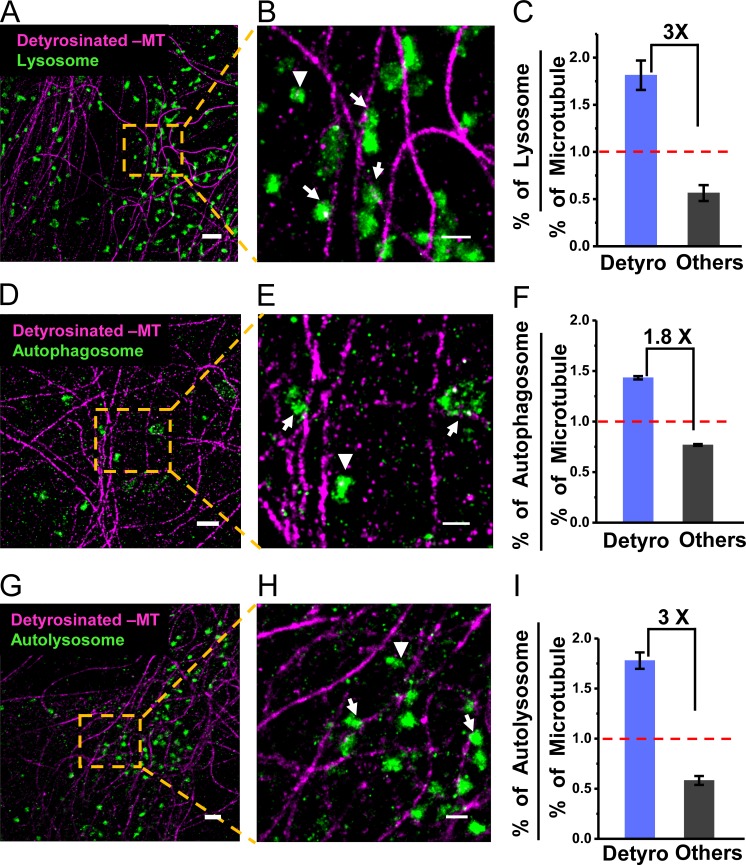
To determine the distribution of membrane-bound compartments along microtubules containing different PTMs, we carried out multi-color, super-resolution imaging of lysosomes together with detyrosinated microtubules (Fig. 2, A and B; Verdeny-Vilanova et al., 2017). Lysosomes were labeled by expressing LAMP2-mCherry and subsequently immunostaining mCherry using super-resolution–compatible antibodies (Materials and methods). We manually scored the association of lysosomes to detyrosinated microtubules based on their spatial proximity to the microtubule in the super-resolution images. We scored any lysosome whose edge was closer than 50 nm to the nearest microtubule as being associated with that microtubule (Fig. S2, A and B). The 50-nm threshold was selected based on the localization error in super-resolution images (∼12 nm) and the potential size of a motor cross-bridge between the microtubule and organelle (motor proteins are ∼70–100 nm long when fully extended, but are typically partially bent or folded when bound to organelles and microtubules; Hirokawa et al., 1989). This analysis showed that 63 ± 5% of lysosomes (818 lysosomes out of 1,316, n = 3 experiments) were associated with detyrosinated microtubules (Fig. 2 C). Given that detyrosinated microtubules constitute only ∼35% of the total microtubule population, these results show that lysosomes are enriched by approximately threefold on detyrosinated microtubules compared with microtubules with other PTMs (∼1.8 lysosome per detyrosinated microtubule versus ∼0.6 lysosome per microtubule with other PTMs). Three-color super-resolution imaging of lysosomes, together with detyrosinated and tyrosinated microtubules, further confirmed that lysosomes were specifically enriched on detyrosinated microtubules and depleted from tyrosinated microtubules (325/953 lysosomes or 34 ± 7%, n = 3 experiments; Fig. S2 C). A similar enrichment was not found on acetylated microtubules, as 71 ± 5% of lysosomes were associated to acetylated microtubules (1,459/2,067 lysosomes, n = 3 experiments; Fig. S2 D), similar to the total percentage of acetylated microtubules in the cells.
Figure 2.
Lysosomes, autophagosomes, and autolysosomes are enriched on detyrosinated microtubules. Two-color super-resolution images showing lysosomes (green; A and B), autophagosomes (green; D and E), and autolysosomes (green; G and H) together with detyrosinated microtubules (magenta). Arrows and arrowheads indicate examples of lysosomal, autophagosomal, or autolysosomal compartments, associated and not associated with detyrosinated microtubules, respectively. Quantification of the percentage of lysosomes (C), autophagosomes (F), and autolysosomes (I) associated with detyrosinated or other microtubule subpopulations normalized to the percentage of the respective microtubule subpopulations. The bars and error bars represent the median and SD, respectively, for three different cells analyzed in each case. The total number of lysosomes analyzed, n = 1,316 (on Detyro = 818, others = 498); for autophagosomes, n = 210 (on Detyro = 105, others = 105); and for autolysosomes, n = 302 (on Detyro = 185, others = 117). Bars, 1 µm for A, D, and G, and 500 nm for B, E, and H.
We next aimed to determine the subcellular distribution of autophagosomes and autolysosomes on microtubules with different PTMs. In cells overexpressing the autophagosome marker LC3B, or lysosome marker LAMP2, most autophagosomes were already fused with lysosomes (Fig. S2, E and F). To specifically label the pure population of autophagosomes before their fusion with lysosomes, we used the tandem fluorescent-tagged LC3B plasmid, where LC3B is tagged to both monomeric RFP and EGFP (Kimura et al., 2007). Lysosomal acidification quenches EGFP fluorescence but not RFP; thus, the EGFP fluorescence reports only autophagosomes before fusion, while RFP fluorescence reports both autophagosomes and autolysosomes. The very low number of EGFP-positive puncta (i.e., autophagosomes) under basal conditions was increased by approximately twofold after nutrient starvation (Materials and methods), indicating an increased biogenesis of autophagosomes in starved cells (Fig. S2 G). Nutrient starvation did not affect the percentage of detyrosinated microtubules (34 ± 6%, n = 4 cells). Multi-color super-resolution imaging (Fig. 2, D and E) and quantitative analysis showed that there was an approximately twofold enrichment of autophagosomes on detyrosinated microtubules (50 ± 0.6%, 105/210 autophagosomes, n = 3 experiments; Fig. 2 F). Autolysosomes found in LC3B-overexpressing cells were also highly enriched on detyrosinated microtubules (61 ± 3%, 185/302 autolysosomes, n = 3 experiments; Fig. 2, G–I). Finally, mitochondria were not enriched on detyrosinated microtubules (Fig. S2 H), suggesting that the enrichment was specific to autophagy and lysosomal compartments. Taken together, these results show that the various compartments of the autophagy and degradation pathways are specifically associated with a small subpopulation of detyrosinated microtubules.
Lysosome motility is impaired on detyrosinated microtubules
Given the previous in vitro results showing that the motility of single motors depends on the microtubule PTMs (Sirajuddin et al., 2014), we next aimed to determine whether the motility of membranous compartments, whose transport is typically driven by more than one motor type, was different depending on the PTM of the microtubules on which they moved. To this end, we performed correlative live-cell and super-resolution microscopy of lysosomes together with detyrosinated microtubules (Bálint et al., 2013; Tam et al., 2014a; Verdeny-Vilanova et al., 2017; Fig. 3, A–D; Fig. S3 A; and Video 1).
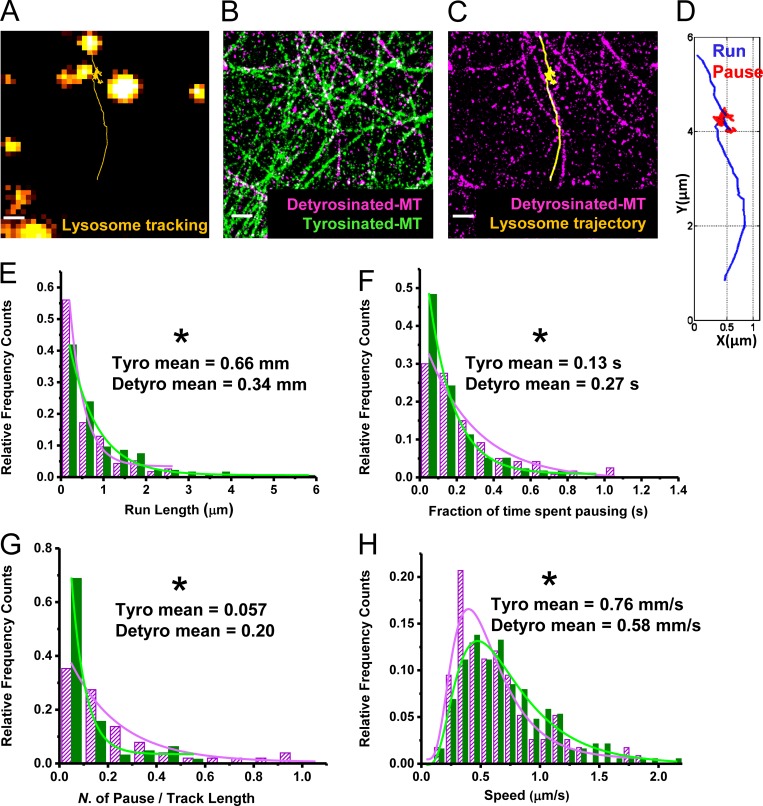
Figure 3.
Lysosomes show hindered motility on detyrosinated microtubules. (A) Trajectory (yellow) shows the motility of a lysosome (orange). (B) Super-resolution image of the same region showing detyrosinated α-tubulin (magenta) and tyrosinated α-tubulin (green). (C) Lysosome trajectory from A overlaid with the super-resolution image of detyrosinated α-tubulin from B. (D) Lysosome trajectory color coded to indicate the processive runs (blue) and the nonprocessive pauses (red). (E–H) Comparison of run length (E), fraction of time spent pausing (F), number of pauses per total track length (G), and speed (H) for lysosomes moving on detyrosinated (magenta) and tyrosinated (green) microtubules. Total number of tracks analyzed, n = 64 for tyrosinated and n = 51 for detyrosinated, from five different cells. The values indicate the mean and the SD. The asterisk indicates the statistical significance of the difference in the motility parameter of the tyrosinated versus the detyrosinated populations, determined by two-tailed two-sample t test (*, P < 0.05). Bars, 500 nm.
We split the single-particle trajectories of lysosomes into two categories using a previously developed analysis algorithm: (i) active, processive motion and (ii) nonprocessive motion, referred to as pauses (Verdeny-Vilanova et al., 2017; see Materials and methods and Fig. 3 D). We then calculated the speed of the active phases as well as the run length (defined as the distance moved during the active phase in between two pauses; see Materials and methods). Lysosome mobility was impaired on detyrosinated microtubules compared with tyrosinated microtubules, resulting in shorter run lengths and longer as well as more frequent pauses (Fig. 3, E–G). This impairment was observed for lysosomes moving in both the antero- and retrograde directions (Fig. S3 B). Furthermore, lysosomes moved with slightly slower speed during the active phase on detyrosinated microtubules (Fig. 3 H). Besides this mobile population of lysosomes, a large population of immobile lysosomes was also present and enriched on detyrosinated microtubules, whereas both mobile and immobile lysosomes were depleted from tyrosinated microtubules (Fig. S3 C), in line with the immunofluorescence results in fixed cells. Finally, similar hindered motility, with shorter run length, longer pause time, and more frequent pausing, was also observed for autolysosomes (Fig. S3 D).
Lysosome–autophagosome interactions and fusion happen on detyrosinated microtubules
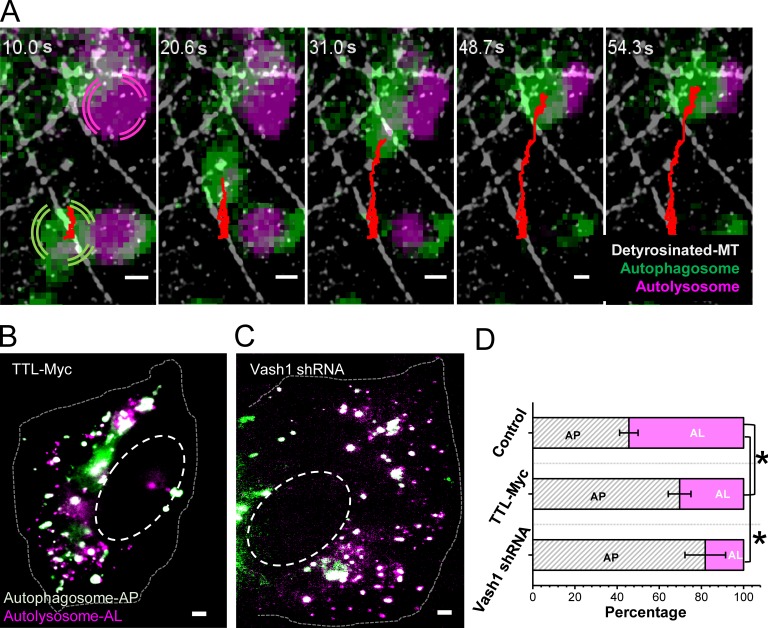
Taken together, our results from multi-color super-resolution and correlative imaging show that compartments of the degradative and autophagy pathways are specifically enriched on a subset of microtubules on which they are less mobile. We hypothesized that the hindered motility may be due to encounters, interactions, and fusion events between autophagosomes and lysosomes, which may happen more frequently on detyrosinated microtubules due to the enrichment of these vesicles on this small microtubule subset. To test this hypothesis, we performed multi-color live-cell imaging of lysosomes together with autophagosomes, followed by super-resolution imaging of detyrosinated microtubules (Fig. 4 A, Fig. S4 A, and Video 2). We observed heterotypic interactions between lysosomes and autophagosomes. Aligning the trajectories of the interacting compartments with the super-resolution images showed that these interactions indeed coincided with detyrosinated microtubules (Fig. 4 A, Fig. S4 A, and Video 2). Overall, we scored 78.8% heterotypic interactions on detyrosinated microtubules compared with other microtubule subtypes.
Figure 4.
Heterotypic interactions between autophagosomes and lysosomes happen on detyrosinated microtubules. (A) Snapshots from a time-lapse video where an autophagosome (green) moves toward and interacts with an autolysosome (magenta) on detyrosinated microtubules (white) in cells expressing the tandem fluorescent-tagged LC3B plasmid. The autophagosome trajectory is shown in red. Bars, 500 nm. (B–D) Wide-field images showing autophagosomes (white) and autolysosomes (magenta) in starved cells transiently overexpressing tyrosine ligase enzyme TTL (B) or starved cells transiently expressing Vasohibin 1 shRNA (C). Bar, 2 µm. (D) Quantification of the percentage of autophagosomes (AP) and autolysosomes (AL) in control starved cells (Fig. S2 G) and for the conditions corresponding to B and C. The bars indicate the median, and the error bar is the SD analyzed for 10 different cells in each case. The asterisk indicates the statistical significance for a two-tailed two-sample t test (*, P < 0.05).
To more specifically test the impact of microtubule detyrosination on autophagosome–lysosome fusion, we genetically modulated detyrosination levels by either overexpressing tubulin tyrosine ligase (TTL) or knocking down the detyrosinating enzyme Vasohibin 1. Both TTL overexpression and Vasohibin 1 knockdown almost completely abolished detyrosinated microtubules in cells positively transfected with the TTL plasmid or the Vasohibin 1 shRNA (Fig. S4, B–F). Acetylated microtubules, on the other hand, were not affected (Fig. S4, G–K). We next quantified the percentage of autolysosomes in starved cells after TTL overexpression or Vasohibin 1 knockdown. In both cases, there was a substantial decrease in the percentage of autolysosomes compared with control starved cells (Fig. 4, B–D; and Fig. S2 G), suggesting a defect in autophagosome–lysosome fusion. Similar results were obtained when detyrosination was removed by treatment with the drug parthenolide, a sesquiterpene lactone that leads to loss of detyrosinated microtubules (Fig. S4, L–P). Overall, our data support an important role of detyrosinated microtubules in autophagosome–lysosome fusion. While we typically observe autophagosomes and lysosomes moving along detyrosinated microtubules right before their interaction and fusion, we cannot fully rule out a role of tyrosinated microtubules in moving these compartments toward detyrosinated microtubules on which they may dock and fuse. In support of such a mechanism, in our correlative imaging experiments, we observed some lysosome-switching events onto detyrosinated microtubules from other microtubule subtypes.
Kinesi-1 is responsible for lysosome enrichment on detyrosinated microtubules
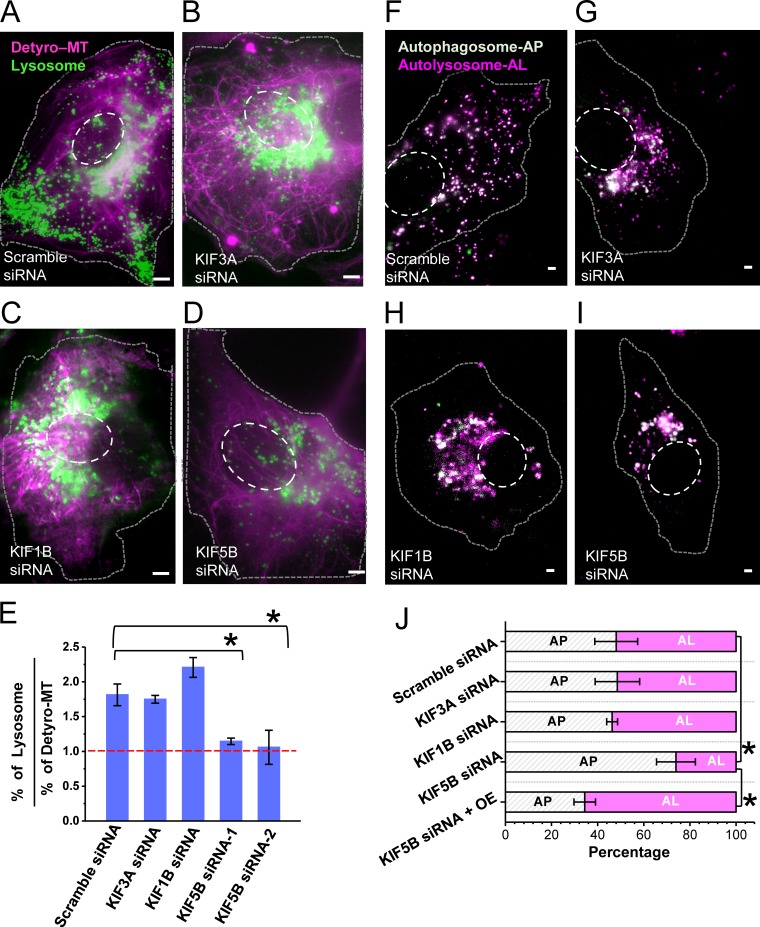
To determine the role of motor proteins in moving lysosomes on differentially post-translationally modified microtubules, we knocked down the three major kinesin motors (KIF5B or kinesin-1; KIF1B, belonging to the kinesin-3 family; and KIF3A, belonging to the kinesin-2 family of motor proteins). These motors have previously been shown to transport lysosomes (Brown et al., 2005; Cardoso et al., 2009; Rosa-Ferreira and Munro, 2011; Guardia et al., 2016; Farías et al., 2017). Knockdown of all three kinesin motors led to a perinuclear clustering of lysosomes in a subpopulation of cells (Fig. 5, A–D; and Fig. S5, A and B; 48% cells with perinuclear lysosome clustering), which was not observed in control cells transfected with scrambled siRNA (Fig. 5 A). These results are consistent with these kinesin motors’ role in anterogradely transporting lysosomes. The fact that not all cells had perinuclear clustering of lysosomes after knockdown could be due to partial siRNA transfection and knockdown efficiency. We further immunostained cells transfected with scrambled, KIF5B, KIF1B, or KIF3A siRNA using antibodies against these motors. Cells that showed a clustered lysosome phenotype also had a lower amount of the respective motors compared with cells that did not show such a phenotype or control cells transfected with scrambled siRNA (Fig. S5, B–E), further suggesting that knockdown was effective only in a subpopulation of cells. We then determined the association of lysosomes to detyrosinated microtubules, focusing on cells that showed a phenotype of lysosome clustering in the perinuclear region. Knockdown of KIF5B or KIF3A did not change the percentage of detyrosinated microtubules (Fig. S5 H). Further, knockdown of KIF3A did not alter the enrichment of lysosomes on detyrosinated microtubules (Fig. 5 E). KIF1B knockdown slightly increased this enrichment (Fig. 5 E), consistent with previous reports that this motor may be responsible for moving subcellular compartments on the tyrosinated microtubule (Guardia et al., 2016). On the other hand, KIF5B knockdown almost completely abolished this enrichment (Fig. 5 E). Similar results were obtained using two independent siRNA pools for knocking down KIF5B (Table S1 and Fig. 5 E), ruling out that the results may be due to off-target effects of siRNA. These results suggest that KIF5B is the motor responsible for enriching lysosomes on detyrosinated microtubules. Consistent with this interpretation, KIF5B knockdown led to an increased association of lysosomes with tyrosinated microtubules (Fig. S5, I and J). Finally, knockdown of KIF5B, but not of KIF1B or KIF3A, led to a decrease in the percentage of autolysosomes (Fig. 5, F–J), suggesting impaired fusion between autophagosomes and autolysosomes. KIF5B overexpression after KIF5B knockdown rescued the percentage of cells with perinuclear clustered lysosomes (this percentage decreased from 48% to 7%) and restored the percentage of autolysosomes to control levels (Fig. 5 J and Fig. S5 K), once again ruling out off-target siRNA effects. Overall, these results suggest that KIF5B helps enrich and constrain lysosomes on detyrosinated microtubules, and this enrichment facilitates their fusion with autophagosomes.
Figure 5.
KIF5B knockdown abolishes the enrichment of lysosomes on detyrosinated microtubules and impacts lysosome–autophagosome fusion. (A–D) Conventional epifluorescence images of cells stably expressing LAMP-2 mCherry, transiently transfected with scrambled siRNA (A), KIF3A siRNA (B), KIF1B siRNA (C), and KIF5B siRNA (D), followed by immunofluorescence for detyrosinated microtubules (magenta). Lysosomes (green) in scrambled siRNA–transfected cells are distributed throughout the cell including the perinuclear region and periphery, whereas lysosomes in cells transfected with KIF3A, KIF1B, and KIF5B siRNA cluster at the perinuclear region with very few lysosomes at the cell periphery. (E) Percentage of lysosomes associated with detyrosinated microtubules normalized to the overall percentage of detyrosinated microtubules, quantified from super-resolution images. The bars indicate the median, and the error bar is the SD for three different cells analyzed in each case. The total number of lysosomes analyzed for scrambled siRNA, n = 355 (on Detyro = 223, others = 132); KIF3A siRNA, n = 332 (on Detyro = 201, others = 131); KIF1B siRNA, n = 396 (on Detyro = 291, others = 105); and KIF5B siRNA, n = 312 (on Detyro = 124, others = 188). Bar, 2 µm. (F–I) Conventional epifluorescence images of cells transiently expressing LC3B-GFP-RFP, cotransfected with scrambled siRNA (F), KIF3A siRNA (G), KIF1B siRNA (H), and KIF5B siRNA (I). (J) Quantification of the percentage of autophagosomes and autolysosomes for 10 different cells in each case. The asterisk indicates the statistical significance for a two-tailed two-sample t test (*, P < 0.05). OE, overexpressing KIF5B.
Discussion
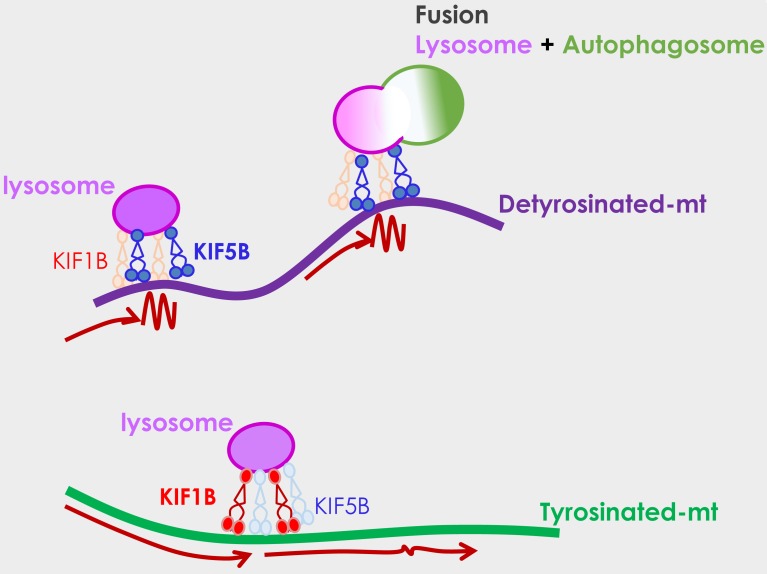
Here, using a combination of live-cell imaging and super-resolution microscopy, we show a new role of detyrosinated microtubules in epithelial cells as hubs for spatially concentrating lysosomes on a small subset of microtubules through a mechanism mediated by the motor protein kinesin-1, therefore enhancing their encounter, interaction, and fusion with autophagosomes (Fig. 6). Indeed, autophagosomes as well as autolysosomes were also enriched on detyrosinated microtubules, and removal of detyrosination or knockdown of kinesin-1 (but not kinesin-2 or kinesin-3) led to a decrease in the percentage of autolysosomes.
Figure 6.
Proposed model. Lysosomes (magenta) and autophagosomes (green) are enriched on a small subset of microtubules that are detyrosinated, where they can encounter each other, interact, and undergo fusion. KIF5B preferentially biases the localization of lysosomes to detyrosinated microtubules, where their mobility is hindered (wiggly line compared with long arrows). KIF1B, on the other hand, is more involved in lysosome motility along tyrosinated microtubules. The potential preference of KIF5B for detyrosinated microtubules and/or its hindered motility on detyrosinated microtubules may help concentrate lysosomes on this small population of microtubules, enabling their efficient fusion with autophagosomes to initiate autophagy.
Previous work showed that newly forming autophagosomes in response to nutrient deprivation are located on dynamic, hyperacetylated microtubules, whereas mature autophagosomes are located on acetylated microtubules that are more stable (Geeraert et al., 2010). Although the role of detyrosinated microtubules was not examined in this study, the differences in the microtubule localization of immature and mature autophagosomes may be explained by the presence of detyrosination on the more stable population of acetylated microtubules. Stable microtubules are resistant to drugs such as nocodazole, but not to other drugs such as vinblastine. Treatment with vinblastine (but not nocodazole) was shown to impair lysosome–autophagosome fusion (Xie et al., 2010). This impairment in fusion was attributed to disruption of acetylated microtubules by vinblastine, although the role of other stable microtubule populations including detyrosinated microtubules was not explored. These results are consistent with our data, since detyrosinated microtubules are likely also sensitive to vinblastine.
Our results showed that knockdown of KIF5B, but not of KIF1B or KIF3A, led to a loss of lysosome enrichment on detyrosinated microtubules, suggesting a role of KIF5B in this enrichment. On the other hand, KIF1B knockdown led to a slight increase in lysosome enrichment on detyrosinated microtubules, and this motor may play a role in moving lysosomes on tyrosinated microtubules and away from detyrosinated microtubules (Guardia et al., 2016). Further, the motility of lysosomes and autolysosomes was hindered along detyrosinated microtubules with shorter run lengths and more frequent pauses. These results are consistent with in vitro studies that showed that KIF5B has shorter run lengths on detyrosinated microtubules (Sirajuddin et al., 2014). Hence, the mechanism of this impaired motility may be explained by a direct influence of the detyrosination state of the microtubule on KIF5B’s single-molecule properties, although further work is needed to dissect the exact mechanisms involved. The enrichment of lysosomes on detyrosinated microtubules may be a result of this hindered motility of kinesin-1, which may have a tethering effect once lysosomes become localized on detyrosinated microtubules. Indeed, besides the mobile lysosomal population, a large population of immobile lysosomes was also localized on detyrosinated microtubules. In addition, kinesin-1 may interact with other microtubule-associated proteins that may be enriched on detyrosinated microtubules, leading to preferential recruitment to and/or hindered mobility and tethering on detyrosinated microtubules. In Drosophila melanogaster cells, ensconsin was shown to be an essential cofactor of kinesin-1 (Barlan et al., 2013). It would be interesting in the future to determine whether this microtubule-associated protein is enriched on detyrosinated microtubules.
Recently, the multi-subunit complex BORC was identified to couple two different kinesins, KIF5B (kinesin-1 family) and KIF1A/KIF1Bβ (kinesin-3 family) to the lysosomal membrane, regulating the anterograde motility of lysosomes to the cell periphery (Pu et al., 2015; Guardia et al., 2016). Interestingly, in HeLa cells, KIF5B was shown to move lysosomes on more centrally located acetylated microtubules, whereas KIF1A moved lysosomes on more peripheral tyrosinated microtubules (Guardia et al., 2016). Our results are consistent with this work but highlight a more important role of detyrosinated microtubules in lysosome positioning and mobility in epithelial cells. While HeLa cells lack detyrosinated microtubules, microtubule detyrosination plays an important role in many biological processes in several cell types, including neurons (Barisic et al., 2015; Herms et al., 2015; Nirschl et al., 2016). Our results add to this growing list of literature, highlighting a new role of this important PTM in mediating lysosome–autophagosome interactions to facilitate efficient autophagy.
Materials and methods
Cell culture and treatments
A stable cell line for GFP-tubulin and mCherry-tubulin was derived from African green monkey (Cercopithecus aethiops) kidney epithelial cells (BS-C-1; CCL-26; American Type Culture Collection). Cell culture consisted of complete growth medium (Eagle’s minimum essential medium with Earle’s salts and nonessential amino acids, plus 10% [vol/vol] FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 500 µg/ml Geneticin [G418 Sulfate; Thermo Fisher Scientific], and penicillin-streptomycin) at 37°C and 5% carbon dioxide.
A double stably transfected cell line for GFP-tubulin and LAMP2-mCherry (Lgp120NL-mCherry) was derived from the BS-C-1 GFP-tubulin cell line following pHyg-LGP120NL-mCherry plasmid transfection and clone selection. Cell culture medium included an additional 100 µg/ml Hygromycin B (10687010; Invitrogen).
A double stably transfected cell line for GFP-tubulin and LC3B-mCherry (mCherry-hLC3B-pcDNA3.1) was derived from the BS-C-1 GFP-tubulin cell line following pHyg-hLC3B-mCherry plasmid transfection and clone selection. Cell culture medium included an additional 100 µg/ml Hygromycin B (10687010; Invitrogen). mCherry-hLC3B-pcDNA3.1 was a gift from D. Rubinsztein, Cambridge Institute for Medical Research, Cambridge, UK (Jahreiss et al., 2008; plasmid 40827; Addgene).
BS-C-1 wild-type or stable cell lines for mCherry tubulin derived from BS-C-1 were transiently transfected with plasmid ptfLC3, a gift from T. Yoshimori, Osaka University, Osaka, Japan (Kimura et al., 2007; plasmid 21074; Addgene).
Cells were plated on fiduciary markers (Carboxyl Fluorescent Yellow microspheres 260 nm or Carboxyl Fluorescent Nile Red microspheres 240 nm; Spherotech) and fibronectin (20 µg/ml)–coated eight-well Lab-Tek 1 coverglass chambers (Nunc). Cell culture media and additives were purchased from Gibco (Life Technologies).
A complete list of siRNAs used to knock down motor proteins in this work is shown in Table S1.
Drug treatments and starvation
Cells were treated with 10 µM nocodazole (Sigma-Aldrich) in complete growth medium for 2 h at 37°C, to depolymerize the microtubules completely, and subsequently recovered with 10 µM parthenolide (Sigma-Aldrich) in complete growth medium for 16 h at 37°C, to suppress the formation of detyrosinated microtubules in the recovered cells.
Cells transfected with ptfLC3 were incubated with Hanks’ Balanced Salt Solution (Life Technologies) for 2 h at 37°C to induce a starvation condition and generate additional pure autophagosomes.
Immunostaining
Cells were fixed and immunostained as described in Bálint et al. (2013). Briefly, fixation was done with 3% (vol/vol) paraformaldehyde and 0.1% glutaraldehyde in PBS. A 0.1% NaBH4 solution in PBS was used to quench background fluorescence. Cells were blocked with 3% (wt/vol) BSA and 0.2% Triton X-100 (vol/vol; Thermo Fisher Scientific) in PBS and incubated with the appropriate dilution of primary and secondary antibodies in the same blocking buffer. Cells were rinsed with washing buffer (0.2% BSA and 0.05% Triton X-100; Thermo Fisher Scientific) between antibody incubations. A complete list of antibodies used in this work is provided in Table S2. The secondary antibodies used were AffiniPure donkey anti-rabbit IgG (H+L, 711-005-152; Jackson ImmunoResearch) at a dilution of 1:100, AffiniPure donkey anti-rat IgG (H+L, 712-005-150; Jackson ImmunoResearch) at a dilution of 1:100, and AffiniPure donkey anti-chicken IgY (IgG; H+L, 703-005-155; Jackson ImmunoResearch,) at a dilution of 1:100. For stochastic optical reconstruction microscopy (STORM), the secondary antibodies were labeled with an Alexa Fluor 405–Alexa Fluor A647 or Alexa Fluor 405–Cyb activator/reporter dye pair combination at concentrations of 0.12–0.15 mg/ml (Bates et al., 2007).
Live-cell imaging
Live-cell imaging was performed with a custom-built, wide-field fluorescence microscope as described before (Bálint et al., 2013). Dual-color live-cell imaging was performed by using two laser sources: a 488-nm laser line for exciting GFP-tubulin or GFP-LC3B, and a 560-nm fiber laser for exciting mCherry-tubulin, cy3b, LAMP2-mCherry, or mCherry-LC3B. The emitted fluorescence was split by a quad band set (TRF89902-ET-405/488/561/647 Laser Quad Band Set for total internal reflection fluorescence applications; Chroma Technology) for the two-color tracking experiments or by two separate emission filters (ET525/50 and ET605/52; Chroma Technology) for the rest of the experiments. The exposure time in the dual-color live-cell imaging experiments was 100 ms.
STORM imaging
STORM was acquired with the same custom-built microscope or an Oxford Nanoimaging system. For the custom-built setup, laser light at 647 nm from an argon-krypton laser (Spectrum IC70; Coherent) was used for exciting Alexa Fluor 647 (Invitrogen), and a 405-nm solid-state laser (Cube; Coherent) was used for reactivating the Alexa Fluor 647 (Invitrogen) via an activator dye (Alexa Fluor 405). The emitted light from Alexa Fluor 647 (Invitrogen) was collected by the 100× objective, filtered by an emission filter (ET705/72m; Chroma), and imaged onto the EM-CCD camera at 20 ms per frame. For multi-color STORM, the emitted light from Alexa Fluor 647 and Cy3b was filtered by a quad band set (TRF89902-ET-405/488/561/647 Laser Quad Band Set for total internal reflection fluorescence applications; Chroma Technology).
Some of the images were acquired on the Nanoimager-S microscope (Oxford Nanoimaging) with the following configuration: 405-, 488-, 561-, and 640-nm lasers, 498–551- and 576–620-nm band-pass filters in channel 1, and 665–705-nm band-pass filters in channel 2, 100× 1.4 NA objective (Olympus), and a Hamamatsu Flash 4 V3 sCMOS camera. Localization microscopy images were acquired with 16-ms exposure for 30,000 frames with 405-nm activation and then processed using the NimOS localization software (Oxford Nanoimaging).
Data analysis
Image registration
Channel registration between the live-cell videos of lysosomes or autophagosomes (mCherry emission) excited by a 560-nm wavelength laser line, acquired using the quad band filter set and the multicolor super-resolution images of modified and unmodified microtubules (Cy3b emission and Alexa Fluor 647 emission), excited with two different alternating laser lines at wavelengths of 560 and 647 nm, and acquired with the same quad band filter set, was done as previously reported (Verdeny-Vilanova et al., 2017). Briefly, to account for sample drift between the live-cell and the super-resolution imaging, we calculated a rigid shift in x, y, and z, using fiduciary markers on the glass surface of the samples and visible both in the live-cell video and the super-resolution image. The rigid shift was applied to the super-resolution raw localizations. The alignment precision (∼10 nm) was calculated as the root mean square difference in the aligned position of the fiduciary markers present in the sample in x, y, as previously described (Verdeny-Vilanova et al., 2017).
Single-particle tracking
Lysosome and autophagosome positions were tracked by a semiautomated, custom-written, particle-tracking software. To determine the x and y positions, trajectories were analyzed by performing a 2D Gaussian fit to the object point spread function. Sample drift during acquisition was calculated by tracking the x, y, and z positions of the fiduciary markers adsorbed onto the glass surface and subsequently subtracted from the trajectories.
A custom-written MATLAB script was used to determine active and passive phases from the 2D trajectory information, as previously described (Verdeny-Vilanova et al., 2017). We performed a moving window analysis (four-point segments) along the trajectory data points. We then calculated the ratio between the total displacement between the initial and final points of the segment and the sum of displacements between the points within the segment. Because the segments overlap, each point within a trajectory will appear in multiple segments. Thus, for every data point, the ratios were averaged over all segments containing this specific point. This ratio estimates the linearity of each segment, and hence, values close to 1 correspond to active phases. A threshold ratio of 0.8 was chosen to distinguish between active and passive phases. Additionally, we used an angle criterion (successive displacement vectors showing angles less than 90° are categorized as passive) to further filter the initial categorization. The chosen parameters were optimized by visual inspection and comparison of a select number of trajectories (n = 10) to a published method based on a Hidden Markov Model analysis (Monnier et al., 2015). After this analysis, we considered the segments >12 data points and calculated the power-law exponent from the mean square displacement (MSD) to further confirm the previous categorization. The exponents were calculated by fitting the MSD curves to a power law: [MS(Δt)] = α × log(Δt) + C. Active transport was separated from passive transport based on α > 1.5. In all cases, the determined power-law exponent matched our initial categorization, confirming that this analysis was effective. Next, we considered the segments of trajectories of <12 data points, for which MSD could not be calculated with high confidence. The segments initially categorized as passive and containing fewer than five data points (500 ms) were considered to be inconclusive and were merged with the segment before and after them. The segments initially categorized as active with a total displacement <160 nm (one pixel) were categorized as passive and the rest as active. The displacement (160 nm) was chosen based on the fact that the vesicles on average move at speeds of ∼0.4 µm/s and, in five frames (500 ms), the expected displacement is ∼200 nm. Additional parameters, such as run length, processivity, and average speed, were also computed using the same custom-written program after the active/passive categorization of the trajectories.
STORM data analysis
STORM images were analyzed and rendered as previously described, using custom-written software (Insight3, provided by B. Huang, University of California, San Francisco, San Francisco, CA; Huang et al., 2008). Spectral cross-talk in two-color experiments was subtracted as previously described (Dani et al., 2010).
The microtubule and lysosome positions were extracted from the super-resolution raw localizations and fitting by a Gaussian distribution function. A threshold at 20% of the localizations was taken to be the outer edge of the lysosome or microtubules, and the distance between the lysosome and microtubule was calculated as the difference between their respective outer edge positions. When this distance was <50 nm, we considered lysosomes to be in contact with the microtubule.
The percentage of microtubule area for acetylated versus nonacetylated tubulin was calculated using ImageJ (National Institutes of Health) by estimating the total pixel area from a binary image of microtubules generated from a sequential, two-color super-resolution image acquired with the exact same imaging conditions with the same fluorophore as previously described (Tam et al., 2014b). The area percentage for nonacetylated microtubules was calculated by subtracting the area fraction of acetylated tubulin obtained from the binary image from that of total α-tubulin. For calculating the detyrosinated and tyrosinated microtubule area, we considered the sum of the area fraction of detyrosinated and tyrosinated as the total α-tubulin area and subtracted the fraction of detyrosinated area from the total to get the tyrosinated microtubule area percentage and vice versa. From the acetylated and detyrosinated sequential images, we approximated that all of the detyrosinated images were acetylated microtubules forming the acetylated–detyrosinated fraction. The acetylated microtubules did not completely overlap with the detyrosinated ones; thus, the nonoverlapping subset formed the acetylated–tyrosinated fraction. We obtained the detyrosinated fraction from the binary images and subtracted it from the acetylated fraction to get the acetylated–tyrosinated microtubule percentage. We performed similar analyses with raw localizations of the sequential STORM images for different microtubules subsets, and the results were consistent with the ImageJ analysis.
Statistical analysis
Data were tested for normality to meet the criteria for the use of a statistical two-sample t test. When data were not normally distributed, they were log-transformed to make them so, and the two-sample t test was performed on the log-transformed dataset. P < 0.05 was taken to indicate statistical significance.
Online supplemental material
Fig. S1 shows the method used for quantification of the proportion of different post-translationally modified microtubules and the variability among the cells (n = 10) for each case. Fig. S2 shows the method and the criterion used for quantifying the proportion of lysosomes and other compartments associated to or not associated with a given post-translationally modified microtubule. In addition, the proportion of lysosomes on acetylated microtubules and the proportion of mitochondria on detyrosinated microtubules are shown. Finally, the distribution of autophagosomes, lysosomes, and autolysosomes on the cell lines used in these experiments (LAMP2- and LC3B-overexpressing cells) is shown. Fig. S3 shows the advantage of correlative live-cell and STORM imaging in comparison to conventional imaging in terms of achieving high spatial and temporal resolution. Also, the quantification of the percentage of mobile and immobile lysosomes associated with detyrosinated microtubules and the motility analysis for anterograde and retrograde transport of lysosomes and autolysosomes on tyrosinated versus detyrosinated microtubules are shown. Fig. S4 shows an example cell with autophagosomes and autolysosomes making heterotypic contacts on detyrosinated microtubules. In addition, the impact of nocodazole and parthenolide treatment on detyrosinated and acetylated microtubules is shown. Fig. S5 shows the immunofluorescence staining of KIF3A and KIF5B in cells transfected with scrambled siRNA, KIF3A siRNA, or KIF5B siRNA, together with the quantification of the immunofluorescence intensity of KIF3A and KIF5B in control and knockdown cells. Also, the quantification of the proportion of detyrosinated microtubules in control and knockdown cells is shown. Video 1 shows the correlative live-cell and STORM imaging of lysosome moving on detyrosinated microtubules associated with Fig. 4 (A–D). Video 2 shows the correlative live-cell and STORM imaging of autophagosome–lysosome interactions on detyrosinated microtubules associated with Fig. 5 A. Table S1 lists siRNAs used in this study and Table S2 lists antibodies used in this study.
Supplementary Material
Acknowledgments
We thank Michael S. Marks (University of Pennsylvania, Philadelphia, PA) for critical reading of the manuscript, comments, and suggestions. We thank David Rubinsztein for the mCherry-hLC3B-pcDNA3.1; Tamotsu Yoshimori for the ptfLC3 plasmid; Ben Prosser (University of Pennsylvania) for the Vasohibin-1 shRNA and TTL overexpression plasmid; and Bo Huang for the Insight3 program.
M. Lakadamyali acknowledges funding from the Perelman School of Medicine, University of Pennsylvania, and European Research Council grant 337191-MOTORS.
The authors declare no competing financial interests.
Author contributions: N. Mohan carried out the experiments, performed the data analysis, and wrote the software. E.M. Sorokina provided reagents, prepared samples, and carried out the knockdown and transfection experiments. I. Vilanova Verdeny wrote the software. A. Sandoval Alvarez provided the cell lines. M. Lakadamyali and N. Mohan designed the experiments. M. Lakadamyali supervised the research. M. Lakadamyali and N. Mohan wrote the manuscript. All authors provided feedback on the manuscript.
References
- Aillaud C., Bosc C., Peris L., Bosson A., Heemeryck P., Van Dijk J., Le Friec J., Boulan B., Vossier F., Sanman L.E., et al. . 2017. Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science. 358:1448–1453. 10.1126/science.aao4165 [DOI] [PubMed] [Google Scholar]
- Bálint Š., Verdeny Vilanova I., Sandoval Álvarez Á., and Lakadamyali M.. 2013. Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc. Natl. Acad. Sci. USA. 110:3375–3380. 10.1073/pnas.1219206110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Silva e Sousa R., Tripathy S.K., Magiera M.M., Zaytsev A.V., Pereira A.L., Janke C., Grishchuk E.L., and Maiato H.. 2015. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science. 348:799–803. 10.1126/science.aaa5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K., Lu W., and Gelfand V.I.. 2013. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 23:317–322. 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M., Huang B., Dempsey G.T., and Zhuang X.. 2007. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 317:1749–1753. 10.1126/science.1146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M., Decker H., Luisi J., and Banker G.. 2015. A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. J. Cell Biol. 208:273–281. 10.1083/jcb.201408056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.L., Maier K.C., Stauber T., Ginkel L.M., Wordeman L., Vernos I., and Schroer T.A.. 2005. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 6:1114–1124. 10.1111/j.1600-0854.2005.00347.x [DOI] [PubMed] [Google Scholar]
- Cai D., McEwen D.P., Martens J.R., Meyhofer E., and Verhey K.J.. 2009. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol. 7:e1000216 10.1371/journal.pbio.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C.M., Groth-Pedersen L., Høyer-Hansen M., Kirkegaard T., Corcelle E., Andersen J.S., Jäättelä M., and Nylandsted J.. 2009. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One. 4:e4424 10.1371/journal.pone.0004424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A., Huang B., Bergan J., Dulac C., and Zhuang X.. 2010. Superresolution imaging of chemical synapses in the brain. Neuron. 68:843–856. 10.1016/j.neuron.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S., Morrison E.E., Liverpool T.B., Molina-París C., Cross R.A., Alonso M.C., and Peckham M.. 2008. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J. Cell Sci. 121:1085–1095. 10.1242/jcs.026492 [DOI] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Webster B.M., Mastronarde D.N., Verhey K.J., and Voeltz G.K.. 2010. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190:363–375. 10.1083/jcb.200911024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeraert C., Ratier A., Pfisterer S.G., Perdiz D., Cantaloube I., Rouault A., Pattingre S., Proikas-Cezanne T., Codogno P., and Poüs C.. 2010. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 285:24184–24194. 10.1074/jbc.M109.091553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger E., McNee G., Allan V., and Woodman P.. 2014. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 31:20–29. 10.1016/j.semcdb.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia C.M., Farías G.G., Jia R., Pu J., and Bonifacino J.S.. 2016. BORC Functions Upstream of Kinesins 1 and 3 to Coordinate Regional Movement of Lysosomes along Different Microtubule Tracks. Cell Reports. 17:1950–1961. 10.1016/j.celrep.2016.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A., Bosch M., Reddy B.J., Schieber N.L., Fajardo A., Rupérez C., Fernández-Vidal A., Ferguson C., Rentero C., Tebar F., et al. . 2015. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 6:7176 10.1038/ncomms8176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Pfister K.K., Yorifuji H., Wagner M.C., Brady S.T., and Bloom G.S.. 1989. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 56:867–878. 10.1016/0092-8674(89)90691-0 [DOI] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M., and Zhuang X.. 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 319:810–813. 10.1126/science.1153529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreiss L., Menzies F.M., and Rubinsztein D.C.. 2008. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 9:574–587. 10.1111/j.1600-0854.2008.00701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Lim C.Y., and Zoncu R.. 2016. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214:653–664. 10.1083/jcb.201607005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubéry S., Wilhelm C., Hurbain I., Neveu S., Louvard D., and Coudrier E.. 2008. Different microtubule motors move early and late endocytic compartments. Traffic. 9:492–509. 10.1111/j.1600-0854.2008.00704.x [DOI] [PubMed] [Google Scholar]
- Luzio J.P., Pryor P.R., and Bright N.A.. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- Magiera M.M., and Janke C.. 2014. Post-translational modifications of tubulin. Curr. Biol. 24:R351–R354. 10.1016/j.cub.2014.03.032 [DOI] [PubMed] [Google Scholar]
- Matsushita M., Tanaka S., Nakamura N., Inoue H., and Kanazawa H.. 2004. A novel kinesin-like protein, KIF1Bbeta3 is involved in the movement of lysosomes to the cell periphery in non-neuronal cells. Traffic. 5:140–151. 10.1111/j.1600-0854.2003.00165.x [DOI] [PubMed] [Google Scholar]
- Monnier N., Barry Z., Park H.Y., Su K.C., Katz Z., English B.P., Dey A., Pan K., Cheeseman I.M., Singer R.H., and Bathe M.. 2015. Inferring transient particle transport dynamics in live cells. Nat. Methods. 12:838–840. 10.1038/nmeth.3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis J., Adamopoulos A., Bleijerveld O.B., Mazouzi A., Stickel E., Celie P., Altelaar M., Knipscheer P., Perrakis A., Blomen V.A., and Brummelkamp T.R.. 2017. Vasohibins encode tubulin detyrosinating activity. Science. 358:1453–1456. 10.1126/science.aao5676 [DOI] [PubMed] [Google Scholar]
- Nirschl J.J., Magiera M.M., Lazarus J.E., Janke C., and Holzbaur E.L.. 2016. α-Tubulin Tyrosination and CLIP-170 Phosphorylation Regulate the Initiation of Dynein-Driven Transport in Neurons. Cell Reports. 14:2637–2652. 10.1016/j.celrep.2016.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Guardia C.M., Keren-Kaplan T., and Bonifacino J.S.. 2016. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129:4329–4339. 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M., Rice L.M., and Vale R.D.. 2014. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16:335–344. 10.1038/ncb2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., and Brady S.T.. 2015. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 25:125–136. 10.1016/j.tcb.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J., Cordier G.A., Bálint Š., Sandoval Álvarez Á., Borbely J.S., and Lakadamyali M.. 2014a A microfluidic platform for correlative live-cell and super-resolution microscopy. PLoS One. 9:e115512 10.1371/journal.pone.0115512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J., Cordier G.A., Borbely J.S., Sandoval Álvarez A., and Lakadamyali M.. 2014b Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS One. 9:e101772 10.1371/journal.pone.0101772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas R.P., Chazeau A., Cloin B.M.C., Lambers M.L.A., Hoogenraad C.C., and Kapitein L.C.. 2017. Differentiation between Oppositely Oriented Microtubules Controls Polarized Neuronal Transport. Neuron. 96:1264–1271.e5. 10.1016/j.neuron.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdeny-Vilanova I., Wehnekamp F., Mohan N., Sandoval Álvarez Á., Borbely J.S., Otterstrom J.J., Lamb D.C., and Lakadamyali M.. 2017. 3D motion of vesicles along microtubules helps them to circumvent obstacles in cells. J. Cell Sci. 130:1904–1916. 10.1242/jcs.201178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Nguyen S., McKeehan W.L., and Liu L.. 2010. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 11:89 10.1186/1471-2121-11-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., and Sabatini D.M.. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12:21–35. 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.