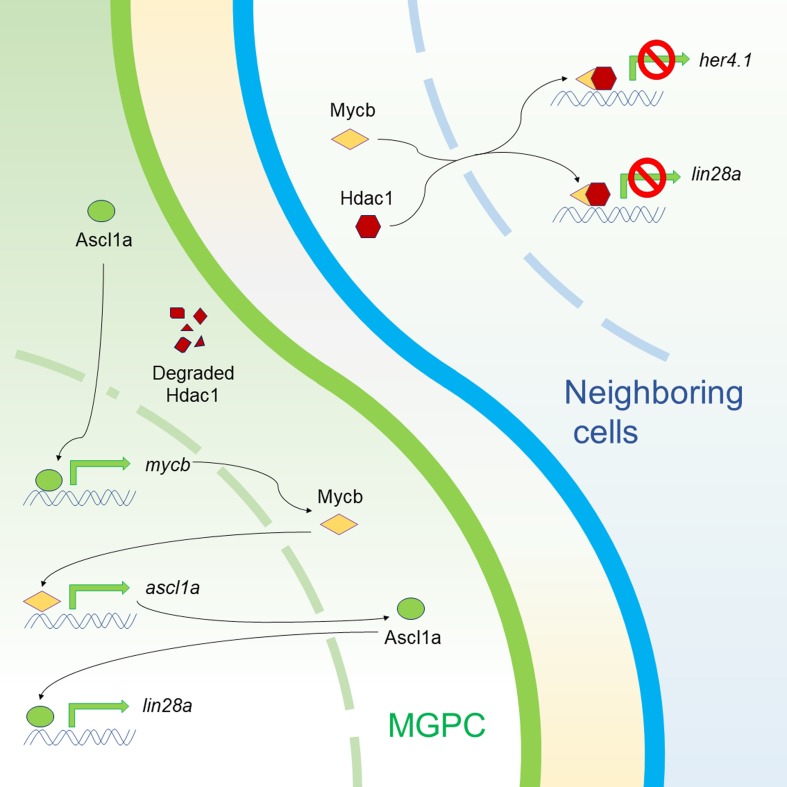
In zebrafish, the damaged retina can regenerate with the help of Muller glia–derived progenitor cells. Mitra et al. show that Mycb regulates lin28a, a facilitator of regeneration, both as an activator and repressor in selected cells. Further, Mycb in collaboration with Hdac1 represses her4.1, a negative regulator of retina regeneration.
Abstract
Cellular reprogramming leading to induction of Muller glia–derived progenitor cells (MGPCs) with stem cell characteristics is essential for zebrafish retina regeneration. Although several regeneration-specific genes are characterized, the significance of MGPC-associated Mycb induction remains unknown. Here, we show that early expression of Mycb induces expression of genes like ascl1a, a known activator of lin28a in MGPCs. Notably, mycb is simultaneously activated by Ascl1a and repressed by Insm1a in regenerating retina. Here, we unravel a dual role of Mycb in lin28a expression, both as an activator through Ascl1a in MGPCs and a repressor in combination with Hdac1 in neighboring cells. Myc inhibition reduces the number of MGPCs and abolishes normal regeneration. Myc in collaboration with Hdac1 inhibits her4.1, an effector of Delta–Notch signaling. Further, we also show the repressive role of Delta–Notch signaling on lin28a expression in post-injured retina. Our studies reveal mechanistic understanding of Myc pathway during zebrafish retina regeneration, which could pave way for therapeutic intervention during mammalian retina regeneration.
Graphical Abstract
Introduction
Compared with mammals, vertebrates such as fishes and amphibians have robust regenerative potential, which has facilitated better understanding of molecular mechanisms during tissue regeneration (Gemberling et al., 2013; Goldman, 2014; Mokalled et al., 2016; Ail and Perron, 2017; Rabinowitz et al., 2017). The zebrafish is extensively used to study regeneration of complex tissues such as retinae. Unlike mammals, zebrafish Muller glia (MG) possess remarkable ability to reprogram themselves to produce MG-derived progenitor cells (MGPCs), irrespective of the injury paradigms (Powell et al., 2016), which are capable of regenerating the damaged retina (Fausett and Goldman, 2006; Ramachandran et al., 2010b). Zebrafish retina regeneration is possible through the orchestration of various growth factors (Russell, 2003; Wan et al., 2012; Zhao et al., 2014b; Gramage et al., 2015), cytokines (Wan et al., 2014; Zhao et al., 2014b), gene transcription factors (Ramachandran et al., 2010a, 2012; Thummel et al., 2010; Nelson et al., 2012; Wan et al., 2014), epigenome modifiers (Powell et al., 2012, 2013; Mitra et al., 2018), cell cycle regulators (Ramachandran et al., 2011, 2012; Luo et al., 2012), Sonic hedgehog signaling–induced gene regulatory network (Kaur et al., 2018; Thomas et al., 2018), and differentiation factors (Munderloh et al., 2009) that are induced at the site of injury. Interestingly, mammalian MG exhibiting stem cell characteristics have been identified, which can be coaxed to grow and differentiate into retinal neurons to a limited extent (Ooto et al., 2004; Pollak et al., 2013; Ueki et al., 2015; Jorstad et al., 2017; Elsaeidi et al., 2018). Unraveling the complete cascade of gene regulatory network after zebrafish retina injury could help in deciphering the lack of efficient regeneration in mammals.
With the increasing knowledge of pluripotency-inducing factors (PIFs) in cellular reprogramming (Yu et al., 2007; Maekawa et al., 2011), studies have been undertaken to unravel the roles of naturally induced PIFs during MG reprogramming, leading to MGPC induction and retina regeneration (Ramachandran et al., 2010a; Reyes-Aguirre and Lamas, 2016; Yao et al., 2016; Gorsuch et al., 2017). However, the roles of an important PIF, Myc, during retina regeneration largely remain unknown. The c-Myc has been well characterized because of its impact on diverse biological functions. These include cellular transformation, cell cycle progression, escaping of the cell cycle arrest, inhibiting cell differentiation, and apoptosis (Amati and Land, 1994; Packham and Cleveland, 1995; Packham et al., 1996; Hoffman and Liebermann, 1998). The involvement of c-Myc in wound healing (Shi et al., 2015) and also after epithelial injury (Volckaert et al., 2013) is well documented. However, the roles of c-Myc with regards to regeneration are restricted to liver tissue of mice (Sobczak et al., 1989; Morello et al., 1990; Sanders et al., 2012) and rats (Arora et al., 2000), rat pancreas (Calvo et al., 1991), and Xenopus laevis limb (Lemaître et al., 1992) with limited knowledge about its actual mechanistic involvement.
The zebrafish has two Myc genes, namely myca and mycb. Here, we report the roles played by transcription factor Mycb in collaboration with Max, along with Ascl1a and Histone deacetylase1 (Hdac1), to regulate lin28a expression during MG reprogramming and induction of MGPCs. We show both the inductive and repressive roles played by Myc, enabling fine-tuned lin28a gene expression at the site of injury. Also, we mechanistically show the Mycb-influenced regulation of hairy enhancer of split-related 4.1 (her4.1) during injury-dependent MG reprogramming, leading to MGPC induction and differentiation that culminate in regeneration.
Results
Myc expression is associated with MGPCs in post-injured retina
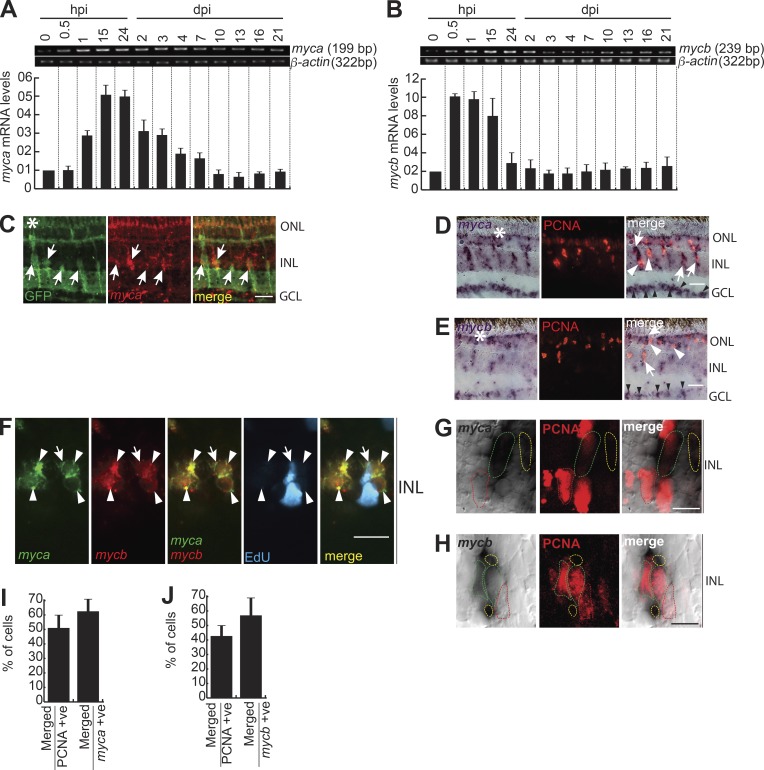
The Myca and Mycb isoforms show 80% amino acid identity (Fig. S1 A). The mycb expression was seen as early as 2 h of embryonic development, indicating its importance (Fig. S1 B). When their mRNA levels were examined after retinal injury by quantitative PCR (qPCR) and reverse transcription PCR (RT-PCR; Fig. 1, A and B), mycb showed an early expression-peak compared with myca. The mRNA in situ hybridization (ISH) of both myca and mycb exhibited a panretinal expression pattern at 12 h post injury (hpi) that became restricted to the injury site by 2 d post injury (dpi; Fig. S1, C and D). The myca expression was seen in both GFP+ and adjacent cells of 1016 tuba1a:gfp transgenic fish retina, in which MGPCs are marked with GFP upon injury (Fig. 1 C and Fig. S1 E; Fausett and Goldman, 2006). Both myca and mycb were expressed in proliferating cell nuclear antigen (PCNA)+/EdU+ MGPCs and adjacent cells at 4–6 dpi (Fig. 1, D–F; and Fig. S1, C and D). We also found specific up-regulation of myca and mycb in ganglion cell layer (GCL; Fig. 1, D and E), suggestive of their roles in optic nerve regeneration as well. In support of this, we found a strong ganglion layer–specific expression of mycb upon optic nerve lesion (Fig. S1 F). Notably, a closer evaluation of 4-dpi retina revealed that both myca and mycb are often associated with cells flanking PCNA+ MGPCs (Fig. 1, G and H). Approximately 40% of the PCNA+ cells expressed myca and mycb, whereas 60% of the myca+ and mycb+ cells had PCNA (Fig. 1, I and J). Spatial and temporal expression pattern of myca and mycb was seemingly reminiscent to previously reported genes like ascl1a, insm1a, and hb-egf (Ramachandran et al., 2010a, 2012; Wan et al., 2012). These observations suggest the existence of a myca/mycb-mediated reprogramming to induce MGPCs in damaged retina.
Figure 1.
Myc genes are rapidly induced in the injured retina. (A and B) RT-PCR (top) and qPCR (bottom) were used to assay injury-dependent myca (A), and mycb (B) gene expressions; n = 6 biological replicates. (C) FISH and IF microscopy show expression of myca mRNA and GFP in retina of 1016 tuba1a:gfp transgenic fish at 4 dpi. (D and E) ISH and IF microscopy show that myca (D) and mycb (E) mRNA is expressed in PCNA+ MGPCs and neighboring cells at 4 dpi. The white arrows indicate colabeled cells in D and E, and white arrowheads in D and E identify myca− and mycb− but PCNA+ cells near injury site. Black arrowheads indicate GCL-specific myca and mycb expression. (F) FISH microscopy shows coexpression of myca and mycb mRNA in BrdU+ cells and vicinity. The white arrow indicates EdU+, myca+, and mycb+ cells, and arrowheads mark myca+ and mycb+ cells. (G–J) A single 0.5-µm-thick Z section shows myca (G) and mycb (H) in 4-dpi retina; dotted outline in red shows PCNA+ myca−/mycb− cells, green shows colabel with PCNA and myca/mycb, and yellow indicates myca+/mycb+ but PCNA− cells; and the percentage colabeling with PCNA is quantified (I and J); n = 5 biological replicates. Bars, 10 µm; white asterisks mark the injury sites. ONL, outer nuclear layer; INL, inner nuclear layer (C–H).
Myc expression and activity in post-injured retina is essential for regeneration
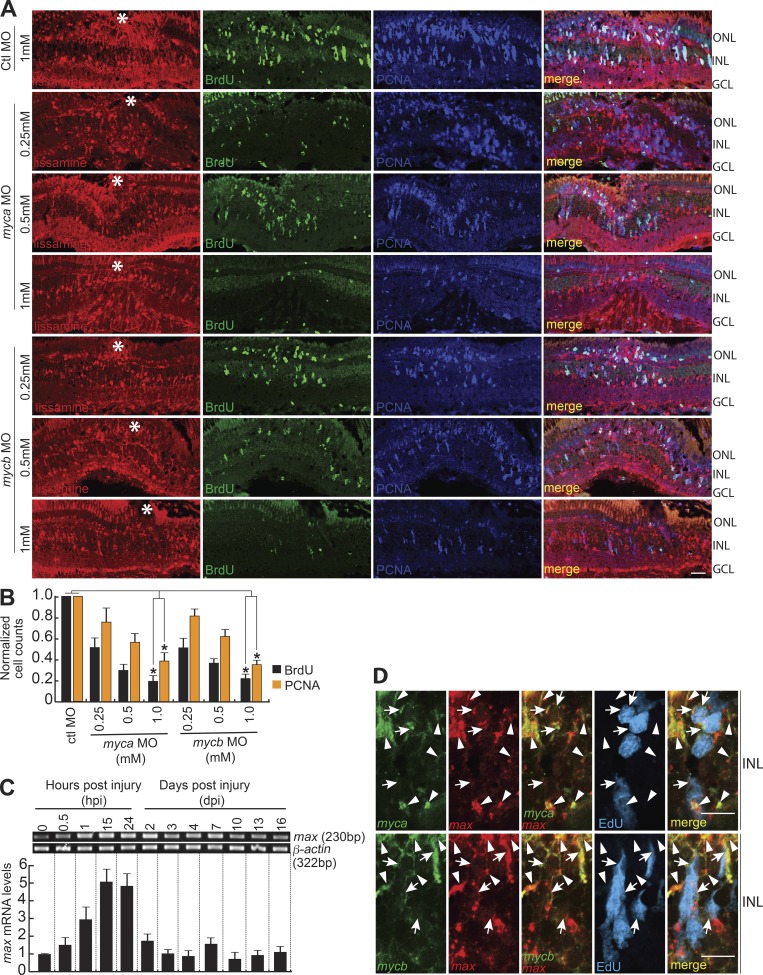
Lissamine-tagged, morpholino-modified antisense oligonucleotides (MOs) targeting myca and mycb completely blocked the translation of respective mRNA in retina (Fig. S1 G), and GFP mRNA appended with MO-binding sites, injected in embryos (Fig. S1 H). Importantly, myca/mycb knockdown using two MOs that target different regions of their mRNAs, electroporated into freshly injured retina, showed that Myca and Mycb are necessary for the generation of BrdU+/PCNA+ MGPCs (Fig. 2, A and B; and Fig. S1, I and J). It is interesting to note that myca and mycb double knockdown had an additive effect on the total number of BrdU+ cells (Fig. S2, A and B). These observations indicate the possibility of independent pathways mediated by Myca and Mycb that converge to induce MGPCs in injured retina. Notably, the decline in the proliferating population of MGPCs in myca/mycb MO-electroporated retina was not because of increased rate of cellular apoptosis revealed in a TUNEL assay (Fig. S2, C and D). Furthermore, the transfection of myca/mycb and gfp reporter mRNAs, along with respective MOs that block endogenous, but not the delivered mRNAs, could rescue the reduction in cell proliferation in the retina at 4 dpi (Fig. S3, A–C).
Figure 2.
Myc is necessary for MG dedifferentiation in the injured retina. (A) IF microscopy images of control (Ctl; 1 mM concentration) or myca/mycb-targeting lissamine-labeled MOs (0.25, 0.5, and 1 mM concentration each), electroporated into the retina of zebrafish at the time of retinal injury shows a concentration-dependent decrease in the number of MGPCs. Fish were given an intraperitoneal injection of BrdU, 3 h before euthanasia on 4 dpi. The white asterisks mark the injury sites. (B) Quantification of the number of BrdU+ and PCNA+ cells at the injury site. The data are compared with control MO. *, P < 0.001; n = 4 biological replicates. (C) RT-PCR (top) and qPCR (bottom) were used to assay injury-dependent max gene expression; n = 6 biological replicates. (D) ISH and IF microscopy show that max gene expression colabel with myca and mycb mRNA in EdU+ MGPCs and other surrounding cells at 4 dpi. White arrowheads indicate myca or mycb colabeled with max, and white arrows mark myca/mycb/max in EdU+ MGPCs. Bars, 10 µm (A and D). Error bars are SD. ONL, outer nuclear layer; INL, inner nuclear layer.
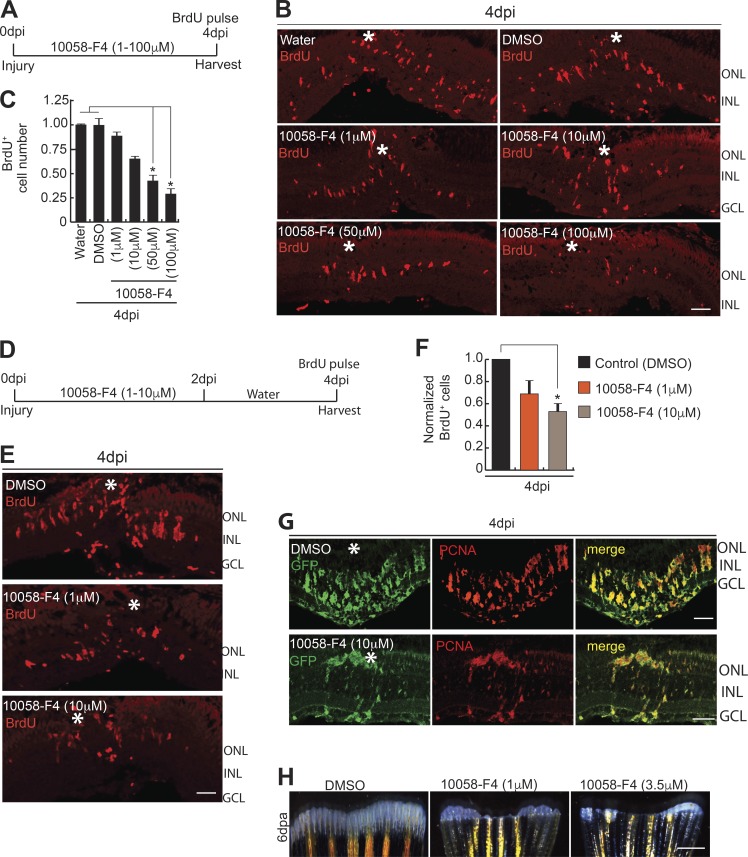
One of the basic helix-loop-helix leucine zipper family members, Max, known to interact with Myc (Yin et al., 2003; Ecevit et al., 2010), showed similar expression pattern as myca and mycb in post-injured retina (Fig. 2, C and D; and Fig. S2 E) and optic nerve lesion (Fig. S3, D–F). Max is also an obligatory partner for Myc’s gene transactivation functions (Amati and Land, 1994). Furthermore, max coexpressed with mycb in GFP+ MGPCs of 1016 tuba1a:gfp transgenic retina at 4 dpi (Fig. S3 G). The cell-sorting analysis from 1016 tuba1a:gfp transgenic retina revealed an increased expression levels of myca, mycb, and max in GFP+ MGPCs, in comparison to rest of the GFP− retinal cell types at 4 dpi (Fig. S3, H and I). These results from zebrafish retina and other reports (Yin et al., 2003; Wang et al., 2007) emphasize that Myc functions mainly in combination with Max to activate transcription and stimulate cell proliferation. This made us explore the effects of disruption of Myc–Max interaction during retina regeneration, using a pharmacological inhibitor, 10058-F4, which blocks Myc–Max interaction (Yin et al., 2003; Huang et al., 2006; Lin et al., 2007; Wang et al., 2007). This would also be an alternate way of inhibiting Myc function. We found that 10058-F4 treatments blocked up to 70% of cell proliferation in regenerating WT retina with continuous (Fig. 3, A–C) or discontinuous exposure (Fig. 3, D–F). Furthermore, we also found a drastic decline in PCNA+/GFP+ MGPCs in 1016 tuba1a:gfp retina (Fig. 3 G) at 4 dpi, according to an experimental time line (Fig. 3 A). However, there was no significant change in the rate of apoptosis in 10058-F4–treated retina, as revealed in TUNEL assay (Fig. S3, J and K). There was also no visible change in expression levels of various genes in retina at 4 dpi because of the DMSO present as solvent in the drug 10058-F4 (Fig. S3 L). These results suggest the importance of Myc–Max interaction in dedifferentiation and proliferation phases of retina regeneration. Notably, the inhibition of Myc also negatively affected fin regeneration (Fig. 3 H), suggesting that normal Myc–Max interaction may be necessary during regeneration of different tissues.
Figure 3.
Blockade of Myc–Max interaction abolishes MGPC proliferation in retina and fin regeneration. (A–C) Blockade of the Myc–Max interaction using the drug 10058-F4 treatment, as shown in timeline of experiment (A), reveals significant reduction in BrdU+ MGPCs at 4 dpi, seen by IF microscopy (B), which is quantified and normalized to Water/DMSO control (C). *, P < 0.0001; n = 5 biological replicates. (D–F) Early treatment with 10058-F4, as shown in timeline of experiment (D), also reveals a significant reduction in BrdU+ MGPCs at 4 dpi revealed by IF microscopy (E), which is quantified and normalized to DMSO control (F). *, P < 0.01; n = 5 biological replicates. (G) IF microscopy analysis of GFP and PCNA after 10058-F4 treatment shows a reduction in the number of MGPCs in 1016 tuba1a:gfp transgenic fish retina at 4 dpi. (H) Regenerating fin-blastema shows a decline in cell mass in 10058-F4–treated post-amputated fin (1 µM and 3.5 µM) at 6 d after amputation, compared with DMSO control. Error bars are SD. Bars: 10 µm (B, E, and G) and 500 µm (H). White asterisks mark the injury sites (B, E, and G). ONL, outer nuclear layer; INL, inner nuclear layer.
Myc–Ascl1a cross-talk during regeneration
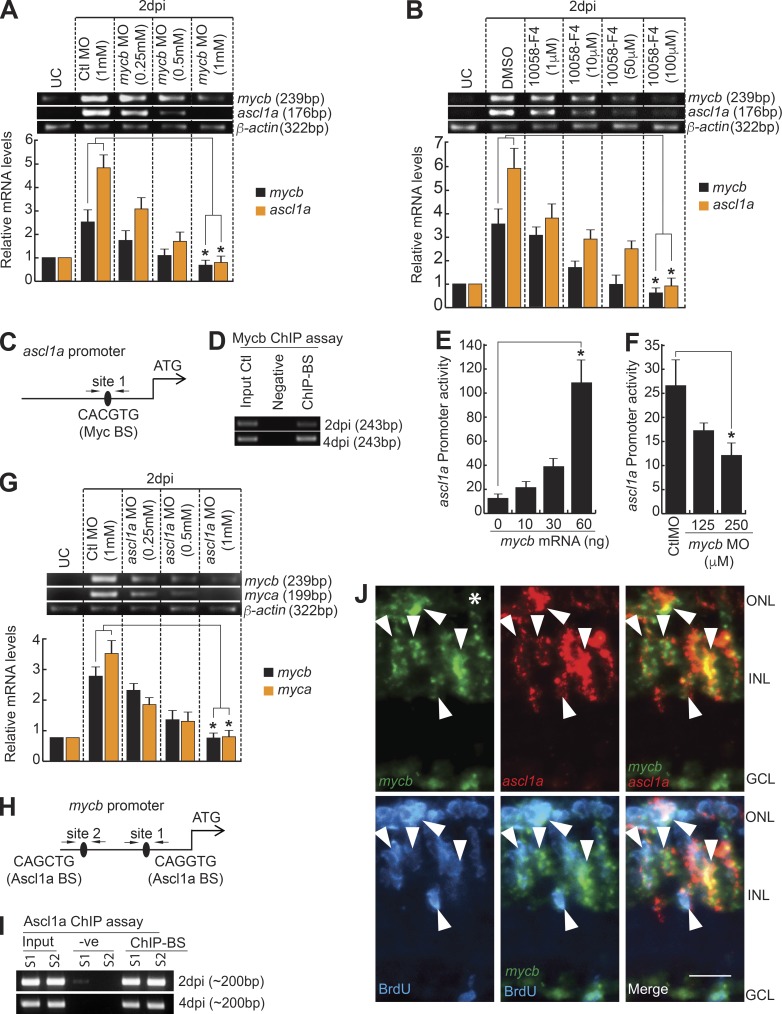
We found that myca and mycb were induced immediately after retinal injury, which is temporally ahead of previously reported ascl1a induction regimen (Ramachandran et al., 2010a). We speculated a hierarchical relationship between them. Mycb inhibition using antisense MO or the drug 10058-F4 down-regulated ascl1a and mycb (Fig. 4, A and B) and up-regulated myca and max (Fig. S4, A and B). Unlike several Myc-regulated genes (Dang, 1999; Fernandez et al., 2003; Zeller et al., 2003; Reymann and Borlak, 2008), which get affected by 10058-F4 treatment, other Myc family genes such as mycn and mycla, which showed a temporal variation in expression after retinal injury (Fig. S4 C), did not show a significant change (Fig. S4 D). Examination of ascl1a promoter revealed one putative Myc-binding site (Fig. 4 C) and chromatin immunoprecipitation (ChIP) assay using anti-Myc antibody on 2- and 4-dpi retina showed that Myc binds to this predicted site (Fig. 4 D). Zebrafish embryos coinjected with ascl1a:gfp-luciferase reporter and increasing concentrations of mycb mRNA and mycb MO separately, showed that Mycb stimulates ascl1a promoter activity (Fig. 4, E and F). This finding also suggests a developmental conservation in the regulation of these genes similar to that found in retina.
Figure 4.
Myc-Ascl1a coexpression and interdependency during regeneration. (A and B) RT-PCR (top) and qPCR (bottom) show inhibition of mycb through MO (A), and 10058-F4 (B) down-regulates ascl1a and mycb induction, relative to the control at 2 dpi. *, P < 0.0001 in A and B; n = 6 biological replicates. (C and H) Diagram of ascl1a (C) and mycb (H) promoters with putative Mycb (C)- and Ascl1a (H)-binding sites. The solid lines represent DNA sequences of the promoter. Arrows mark ChIP primers, and capital letters mark consensus sequence. (D and I) The retina ChIP assay done at 2 and 4 dpi shows that Myc bound to ascl1a promoter (D), and Ascl1a bound to mycb promoter (I). (E) Mycb overexpression up-regulates ascl1a:gfp-luciferase expression in embryos. *, P < 0.01. (F) MO-based Myc blockade inhibits ascl1a:gfp-luciferase in embryos. Promoter activity is normalized light units with internal control Renilla luciferase. *, P < 0.001. (G) RT-PCR (top) and qPCR (bottom) show MO based ascl1a knockdown down-regulates myca and mycb expression in 2 dpi retina. *, P < 0.002. (J) FISH and IF microscopy show colocalization of mycb and ascl1a expressing cells with BrdU+ MGPCs in 4-dpi retina. Arrowheads indicate colabeled mycb- and ascl1a-expressing cells. White asterisk marks the injury site. Bars, 10 µm. n = 6 biological replicates unless specified. Error bars are SD. S, site; −ve, negative, BS, binding site.
Knockdown of ascl1a resulted in a small but significant decrease in myca and mycb expression at 2 dpi (Fig. 4 G), which is similar to that seen with blockade of Wnt signaling through the drug XAV939 (Fig. S4, E and F). Blockade of Wnt signaling in regenerating retina is known to down-regulate ascl1a (Ramachandran et al., 2011). From these observations, we speculated the existence of direct and mutual regulatory relationship between Mycb and Ascl1a. Examination of mycb promoter sequence revealed two putative Ascl1a-binding sites (Fig. 4 H) and ChIP assay on 2- and 4-dpi retina confirmed that endogenous Ascl1a indeed bound onto these sites (Fig. 4 I), which is also supported by colocalization of mycb and ascl1a mRNAs (Fig. 4 J). These results strongly support the view that Ascl1a and Mycb up-regulate expression of each other during retina regeneration.
Restricted expression of mycb is through transcriptional repressor Insm1a
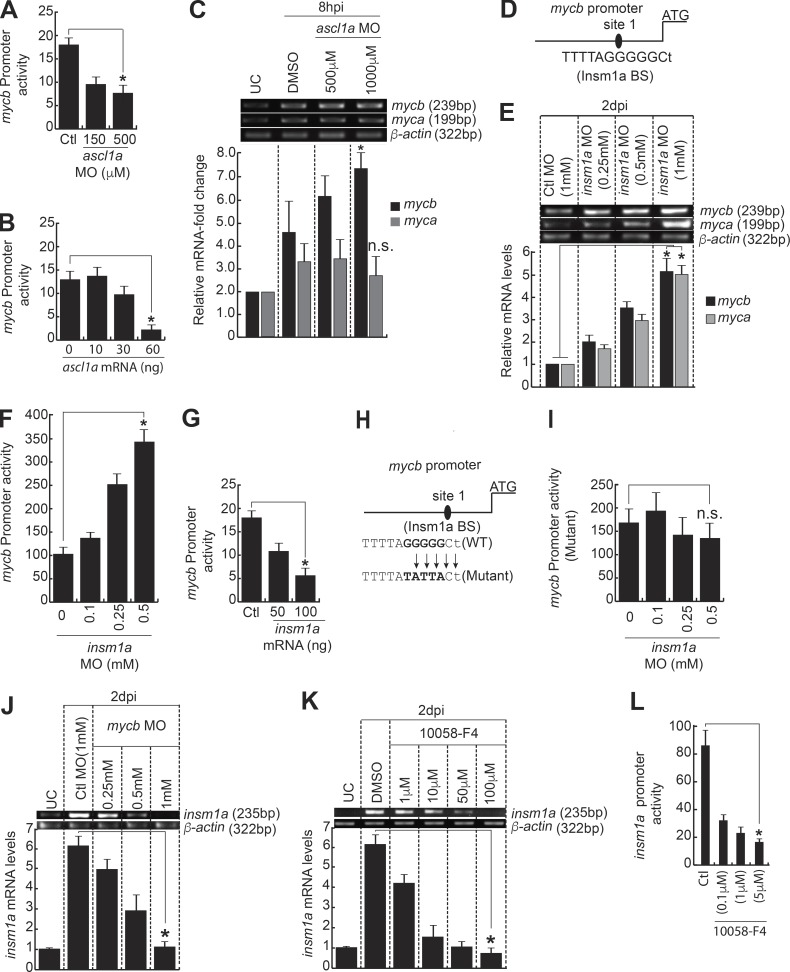
The ascl1a knockdown in zebrafish embryos coinjected with mycb:gfp-luciferase reporter with increasing concentrations of ascl1a MO showed a dose-dependent reduction in mycb promoter activity (Fig. 5 A), in agreement with retinal data, as discussed earlier (Fig. 4, G–J). However, overexpression of Ascl1a in zebrafish embryos coinjected with mycb:gfp-luciferase reporter, along with increasing concentrations of ascl1a mRNA, showed a surprising reduction in mycb promoter activity (Fig. 5 B). Interestingly, we found an increase in the mycb expression in the retina with ascl1a knockdown at an early time point of 8 hpi (Fig. 5 C), which suggests the involvement of some other transcriptional repressor molecule, regulated through Ascl1a, that affects mycb expression. We speculated that such a conundrum could be because of involvement of a potential intermediate repressor like Insm1a, which is induced by Ascl1a (Ramachandran et al., 2012).
Figure 5.
Insm1a inhibits mycb expression in regenerating retina. (A) MO-based gene knockdown of ascl1a down-regulates mycb:gfp-luciferase expression in embryos. *, P < 0.002. (B) Ascl1a overexpression inhibits mycb:gfp-luciferase expression in embryos. *, P < 0.0002. (C) MO-mediated ascl1a inhibition in retina as early as 8 h after injury causes an increase in mycb, but not myca expression. *, P < 0.009; n = 3 biological replicates. n.s., not significant. (D) Diagram of mycb promoter with putative Insm1a binding site. (E) MO-based insm1a knockdown significantly up-regulated both myca and mycb expression in injured retina at 2 dpi. *, P < 0.001. (F) The insm1a knockdown through MO up-regulates mycb:gfp-luciferase expression in zebrafish embryos by luciferase assay. *, P < 0.0001. (G) Insm1a overexpression inhibits mycb:gfp-luciferase expression in embryos. *, P < 0.001. (H) Schematic of mycb promoter with mutated Insm1a-binding site. (I) Insm1a inhibition through MO has no effect on mutated mycb:gfp-luciferase expression in zebrafish embryos by luciferase assay. n.s., not significant. (J and K) Myc inhibition through MO (J) and 10058-F4 (K) cause significant down-regulation of insm1a expression in 2-dpi retina. (L) 10058-F4–based Myc blockade inhibits insm1a:gfp-luciferase in embryos. Promoter activity is normalized light units with internal control Renilla luciferase. *, P < 0.0002. Error bars are SD. BS, binding site.
We decided to explore if Insm1a-mediated gene repression is the possible cause of reduced mycb promoter activity in zebrafish embryos with Ascl1a overexpression and increased mycb mRNA levels in 8-hpi retina with ascl1a knockdown (Fig. 5, B and C). Examination of mycb promoter revealed one putative Insm1a-binding site (Fig. 5 D). The early panretinal and the late MGPC-associated expression of insm1a are important in stringent control of several regeneration-associated genes and cell cycle exit during retina regeneration (Ramachandran et al., 2012). To evaluate this further, we examined the expression levels of myca and mycb in insm1a knockdown background in injured retina. We found a dose-dependent increase in myca and mycb mRNA levels in retina upon MO-mediated insm1a knockdown, which supported its possible inhibitory role (Fig. 5 E). We evaluated this further through coinjection of mycb:gfp-luciferase reporter in zebrafish embryos, along with insm1a MO or insm1a mRNA separately. The results showed expected up-regulation and down-regulation in mycb promoter activity, respectively (Fig. 5, F and G). Furthermore, the mutation of the single Insm1a-binding site in mycb promoter abolished the increase in promoter activity when coinjected with mycb:gfp-luciferase reporter and different concentrations of insm1a MO in zebrafish embryos (Fig. 5, H and I). These results suggest that Ascl1a-mediated induction of Insm1a could be the cause of reduction of mycb promoter activity, seen by coinjection of zebrafish embryos with mycb:gfp-luciferase reporter and increasing concentrations of ascl1a mRNA.
Similarly, the ascl1a knockdown-mediated increase in mycb mRNA seen in 8-hpi retina (Fig. 5 C) also could be because of decline in Insm1a. Earlier studies showed that induction of ascl1a is panretinal in nature, wherein both MG and non-MG cells of the retina express it at 6–8 hpi (Ramachandran et al., 2011, 2012). This kind of panretinal induction of Ascl1a, soon after injury, up-regulates the transcriptional repressor Insm1a throughout the retina. Insm1a is also necessary for repressing ascl1a and its own expression at early stages of regeneration. The Ascl1a–Insm1a regulatory loop is a prelude for initiation of MG dedifferentiation (Ramachandran et al., 2012). However, at 4 dpi, approximately only 40% of the MGPCs, which were exiting the cell cycle, expressed insm1a (Ramachandran et al., 2012). Such a stringent gene regulation in the retina would be essential for restricting the mycb expression to the vicinity of MGPCs at the site of injury from its initial panretinal induction. Furthermore, mycb knockdown and 10058-F4 treatments separately caused down-regulation of insm1a in 2-dpi retina (Fig. 5, J and K). Moreover, a decrease in insm1a promoter activity was seen in zebrafish embryos injected with insm1a:gfp-luciferase reporter and exposed to various concentrations of 10058-F4 (Fig. 5 L). These observations could be the result of decline in Ascl1a, which is an inducer of insm1a (Ramachandran et al., 2012). Collectively, these experiments reveal an efficient Myc–Ascl1a–Insm1a regulatory loop in action that contributes to the course of regeneration.
Mycb-mediated dual regulation of lin28a during regeneration
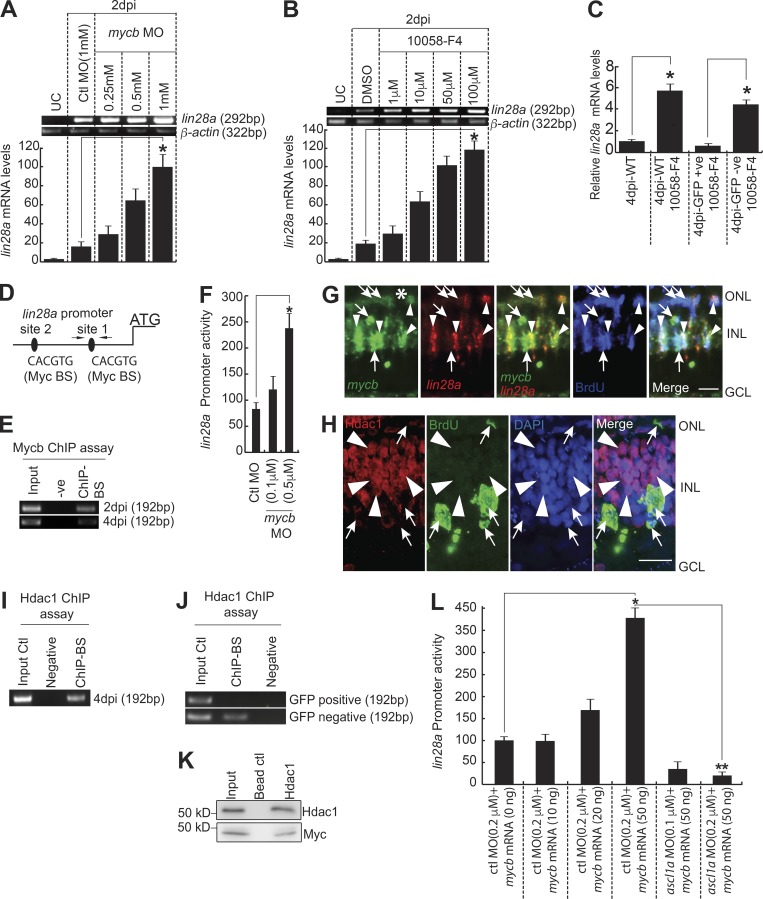
Since we found the regulation of ascl1a and insm1a through Mycb, we then probed for the expression pattern of an important regeneration-associated gene, lin28a, both in mycb knockdown and in the absence of its activity. Interestingly, absence of Myc caused a significant up-regulation of lin28a in WT retina (Fig. 6, A and B; and Fig. S4, G and H). To assess if there is any cell type bias in up-regulation of lin28a with Myc inhibition, we used 1016 tuba1a:gfp transgenic fish retina. We found a selective increase in the lin28a mRNA levels in GFP− cells compared to the GFP+ mRNA levels of 1016 tuba1a:gfp fish treated with 10058-F4 (Fig. 6 C). Examination of lin28a promoter revealed two putative Myc-binding sites (Fig. 6 D). ChIP assay done on 2- and 4-dpi retina confirmed endogenous Myc bound onto one of the two sites (Fig. 6 E). This was further confirmed by promoter activity assay in zebrafish embryos coinjected with lin28a:gfp-luciferase reporter and mycb MO (Fig. 6 F). Interestingly, lin28a mRNA showed a colocalization with only a subset of mycb+ cells, but stayed secluded in some neighboring MGPCs (Fig. 6 G). These results made us explore the possible ways in which Mycb could act as a repressor of lin28a. Myc is known to recruit bona fide transcription repressors like histone deacetylases (Hdacs) to suppress target genes (Kurland and Tansey, 2008). We speculated that similar mechanism might regulate Myc-mediated down-regulation of lin28a. To support this view further, we also saw a decline in Hdac1 expression in proliferating MGPCs pulse-labeled with BrdU at 4 dpi (Fig. 6 H). Surprisingly, we found the occupation of Hdac1 at Myc-binding sites of lin28a promoter, as revealed in a ChIP assay done in whole retina, using the same set of primers used for confirming the Myc-binding site on lin28a promoter (Fig. 6 I). Again, we did not find Hdac1 binding in Mycb-recognized DNA sequence of lin28a promoter in GFP+ MGPCs isolated from 1016 tuba1a:GFP transgenic retina (Fig. 6 J). Furthermore, to confirm the physical interaction of Myc and Hdac1, we performed a coimmunoprecipitation (co-IP) assay using Hdac1 antibody in 4-dpi retinal extract. We found that the protein complex, pulled down using Hdac1 antibody, contained Myc protein (Fig. 6 K), suggesting the existence of a physical collaboration between Hdac1 and Myc in causing repression of lin28a.
Figure 6.
Mycb-mediated regulation of lin28a in MGPCs. (A and B) RT-PCR (top) and qPCR (bottom) show Myc inhibition using antisense MO (A) or 10058-F4 (B) induce lin28a in 2-dpi retina. *, P < 0.001. UC, uninjured control. (C) qPCR analysis of lin28a mRNA from GFP+ and GFP− cells sorted from 1016 tuba1a:gfp transgenic fish retina with 1-µM 10058-F4 treatment at 4 dpi, compared with WT. *, P < 0.01. (D) Diagram of lin28a promoter with putative Mycb-binding sites. The solid lines represent DNA sequences. Arrows mark ChIP primers, and capital letters mark consensus sequence. (E) The retina ChIP assay at 2 and 4 dpi showed that Myc binds to lin28a promoter. (F) MO-based mycb knockdown up-regulates lin28a:gfp-luciferase activity. Promoter activity is normalized light units with internal control Renilla luciferase. *, P < 0.0002. (G) FISH and IF microscopy show expression of mycb and lin28a with respect to BrdU+ MGPCs in 4-dpi retina. Arrowheads indicate colabel of mycb, lin28a, and BrdU+ cells; arrows indicate mycb+ that colabel with BrdU, but are lin28a− cells. White asterisk marks the injury site. (H) IF and ISH microscopy on a single 0.5-µm thick Z section shows Hdac1 expression secludes largely from BrdU+ cells at the site of injury. Arrowheads mark hdac1+ but BrdU− cells, and arrows mark Hdac1− but BrdU+ cells. (I) ChIP assay using Hdac1 antibody reveals Hdac1 occupied Myc-binding site on lin28a promoter. (J) ChIP assay of lin28a promoter Myc-binding region, using Hdac1 antibody from GFP+ and GFP− cells from 1016 tuba1a:gfp transgenic retina. (K) Co-IP assay using Hdac1 antibody reveals Hdac1–Myc collaboration during retina regeneration. (L) MO-based ascl1a knockdown abrogates Mycb overexpression-mediated lin28a:gfp-luciferase up-regulation in embryos. *, P < 0.0002; **, P < 0.003. n = 6 biological replicates unless specified. Error bars are SD. −ve, negative; +ve, positive; BS, binding site. Bars, 10 µm (G and H). ONL, outer nuclear layer; INL, inner nuclear layer.
However, when zebrafish embryos were coinjected with lin28a:gfp-luciferase reporter and mycb mRNA, we observed a concentration-dependent up-regulation of lin28a promoter activity (Fig. 6 L). We speculated that Myc-mediated induction of Ascl1a, an activator of lin28a, could be the cause of increased lin28a promoter activity. If this is true, the effect of Ascl1a on lin28a promoter could be nullified by using ascl1a MO. We found a drastic decline in lin28a promoter activity in zebrafish embryos coinjected with a constant high mycb mRNA dose and increasing concentrations of ascl1a MO (Fig. 6 L), confirming our speculation. These results suggest that Mycb could impact the lin28a promoter indirectly as an activator in BrdU-positive MGPCs through Ascl1a and a repressor in combination with Hdac1 in BrdU-negative neighboring cells at the site of injury.
Mycb regulates her4.1 through Hdac1 in injured retina
Delta–Notch signaling–mediated regulation of cell proliferation during retina regeneration is well characterized in zebrafish. Inhibition of Delta–Notch signaling through administration of γ-secretase inhibitor N-(N-[3,5-difluorophenylacetyl]-l-alanyl)-S-phenylglycine-t-butyl ester (DAPT) caused an increase in MGPC proliferation in the injured retina (Wan et al., 2012; Conner et al., 2014). The DAPT treatment also caused an expected decline in expression levels of target genes of Delta–Notch signaling, such as her4.1, compared with control retina (Fig. S5 A). Furthermore, the induced expression of notch intracellular domain (nicd) in the retina caused panretinal expression of her4.1 (Kageyama et al., 2007; Zhou et al., 2012). The overexpression of nicd is also associated with negligible MGPC proliferation and retina regeneration (Wan et al., 2012).
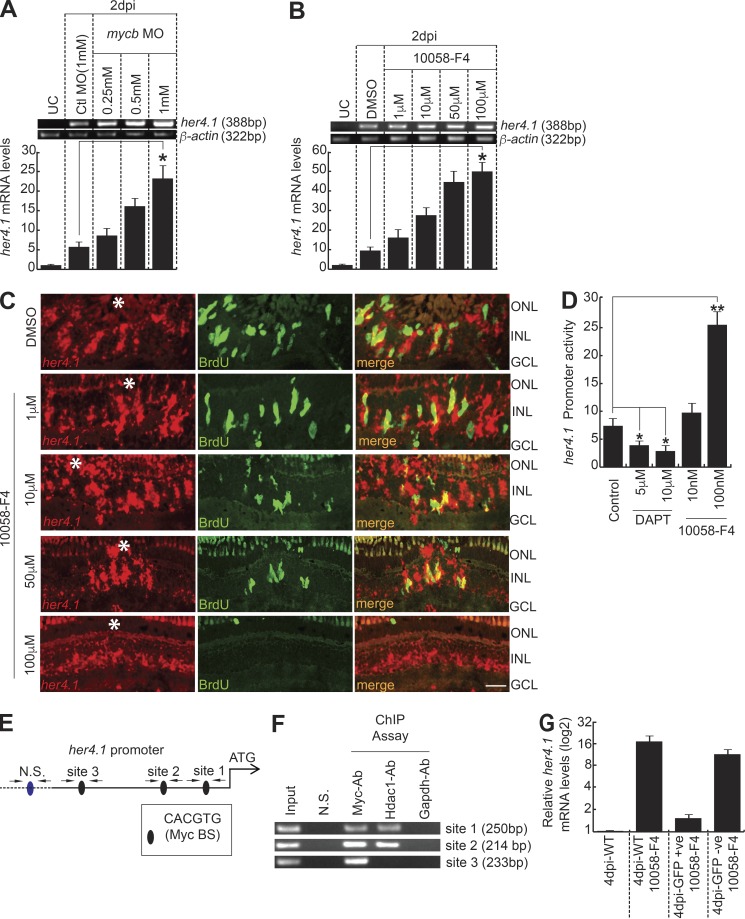
Here, we found a significant decline in MGPC proliferation because of Myc inhibition, similar to that of nicd overexpression. Based on these observations, we speculated the existence of the mechanistic involvement of Her4.1, which could cause a compromised MGPC proliferation and regeneration in the Myc-inhibited retina. In support of this hypothesis, we found a significant up-regulation of her4.1 in response to mycb knockdown and 10058-F4 treatments at 2 dpi revealed by RT-PCR and qPCR (Fig. 7, A and B). Compared with the normal injury-restricted expression of her4.1 at 4 dpi, its panretinal induction was seen with Myc inhibition, by 10058-F4 treatments in a dose-dependent manner (Fig. 7 C and Fig. S5 B). We also found similar induction of her4.1 because of indirect inhibition of mycb through XAV939-mediated blockade of Wnt signaling (Fig. S4 E and Fig. S5 C). Finally, 10058-F4–treated zebrafish embryos coinjected with her4.1:gfp-luciferase reporter caused an increase in her4.1 promoter activity, while DAPT caused the opposite (Fig. 7 D). These results suggested that the involvement of Myc–Her4.1 interplay is necessary to restrict the MGPC proliferation to the site of injury during retina regeneration.
Figure 7.
Mycb regulates her4.1 gene transcription in injured retina. (A and B) RT-PCR (top) and qPCR (bottom) show increased her4.1 induction with MO-based mycb knockdown (A) or 10058-F4 (B) relative to control MO and DMSO, respectively, in 2-dpi retina. *, P < 0.005. UC, uninjured control. (C) FISH and IF microscopy shows that 10058-F4 treatment increases her4.1 expression compared with water or DMSO-treated control in 4-dpi retina. Bar, 10 µm. White asterisks mark the injury sites. (D) Myc inhibition through 10058-F4 up-regulates her4.1:gfp-luciferase expression compared with control and DAPT-treated embryos. (E) Diagram of her4.1 promoter with putative Mycb-binding sites. The solid lines represent DNA sequences of the promoter. (F) The retina ChIP assay at 4 dpi reveals Myc and Hdac1 bound to Myc-BS on her4.1 promoter. (G) qPCR analysis of her4.1 mRNA from GFP+ and GFP− MGPCs sorted from 1016 tuba1a:gfp transgenic fish retina with 1 µM 10058-F4 treatment at 4 dpi, compared with WT. n = 3 biological replicates in all experiments. ONL, outer nuclear layer; INL, inner nuclear layer; N.S., nonspecific; BS, binding site.
We explored further to find if Myc directly regulates her4.1 through direct interactions onto its promoter sequences. A similar case was reported, wherein another member of Notch target genes, hes1, is up-regulated through sonic hedgehog signaling–dependent direct target Gli2 in retinal progenitors, which is independent of classical Delta–Notch signaling (Wall et al., 2009). In silico analysis of her4.1 regulatory sequences revealed a few putative Myc-binding sites (Fig. 7 E). Since we already demonstrated physical interaction of Mycb and Hdac1 and its occupation in lin28a regulatory sequences to cause its repression in regenerating retina (Fig. 6, I–K), we speculated that such a scenario could underlie the regulation of her4.1 as well. We performed a ChIP assay in 4-dpi retinal chromatin using antibodies against Myca/b, Hdac1, and Gapdh. Interestingly, we found that while Myc bound to three sites, Hdac1 occupied only two of these three sites tested, while Gapdh bound to none (Fig. 7 F). These results suggest the strong possibility of her4.1 being directly regulated through Myc–Hdac1 complex in the regenerating retina. We also quantified the her4.1 mRNA levels in the sorted cells from the retina of 1016 tuba1a:GFP transgenic fish treated with 10058-F4. Although there was significant decline in GFP+ MGPCs with 10058-F4 treatment (Fig. 3 G), we found a moderate increase in the her4.1 mRNA levels in these GFP+ cells, and a substantial up-regulation was seen in GFP− cells (Fig. 7 G). These observations suggest that the global increase in her4.1 levels in the 10058-F4–treated retinae could be one of the reasons for lack of normal regeneration.
Delta–Notch signaling restricts the zone of MGPCs by suppressing lin28a expression through Myc
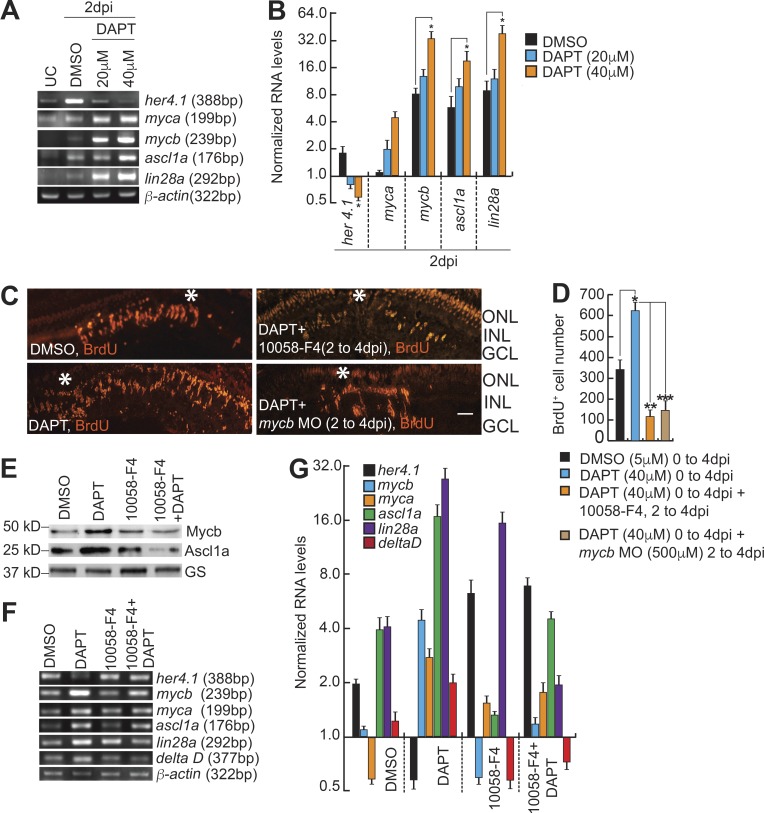
We next decided to investigate the mechanistic importance of Her4.1 up-regulation in Myc-Max–inhibited retina in detail. This could also enable us to find the reasons for reduced MGPCs in Mycb-compromised scenario. To decipher this, we first investigated the importance of Mycb and Lin28a in causing the increase in the number of MGPCs in post-injured retina with Delta–Notch signaling inhibition. The blockade of Delta–Notch signaling with DAPT treatment caused an enhancement of MGPCs, which was also accompanied by up-regulation of regeneration-associated genes like myca, mycb, ascl1a, and lin28a at 2 dpi (Fig. 8, A–C). Zebrafish embryos when treated with DAPT and injected with lin28a:gfp-luciferase reporter showed an increased lin28a promoter activity in a dose-dependent manner (Fig. S5 D).
Figure 8.
Her4.1 restricts the zone of MGPCs by suppressing lin28a expression. (A and B) RT-PCR (A) and qPCR (B) show decreased her4.1 induction and increased regeneration-associated genes’ levels with DAPT treatment relative to DMSO control in 2-dpi retina. UC, uninjured control. *, P < 0.002. (C and D) IF microscopy shows that increased MGPCs seen in DAPT-treated (40 µM) retina is blocked both by 10058-F4 (10 µM) and mycb-targeting morpholino (500 µM; C) with ∼70% and ∼85% reduction in BrdU+ cells compared with DMSO control and DAPT-treated retina, respectively (D). *, P < 0.003; **, P < 0.0001; ***, P < 0.0002. Bars, 10 µm; white asterisks mark the injury sites (C). (E–G) Western analysis of Ascl1a and Mycb show reduction in protein levels, in 10058-F4 (10 µM) + DAPT- (40 µM) blocker regimen (E); also other regeneration-associated genes seen by RT-PCR (F) and qPCR (G) compared with DMSO, 10058-F4, or DAPT-treated retinae in 2 dpi. GS, glutamine synthetase. n = 3 biological replicates in all experiments. ONL, outer nuclear layer; INL, inner nuclear layer.
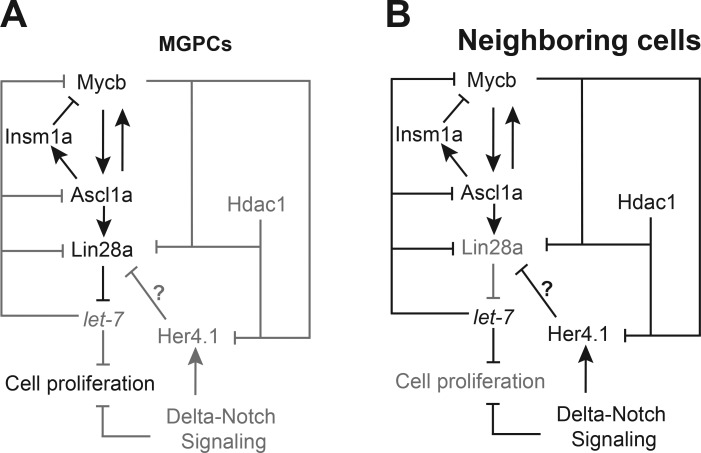
Interestingly, we found that the increased number of MGPCs, seen with DAPT treatment in 4-dpi retina could be abolished by Myc inhibition either by mycb MO or 10058-F4 exposures, in 0–4- and 2–4-dpi experimental regimes (Fig. 8, C and D; and Fig. S5, E–G). These observations suggested that the increase in the number of MGPCs seen in DAPT-treated retina was facilitated by normal Mycb-mediated gene regulations. We probed further the cause of reduction in cell proliferation through estimation of Ascl1a and Mycb protein levels in retina from double blocker experiments using DAPT and 10058-F4. We found that both Ascl1a and Mycb proteins were down-regulated in these retinae (Fig. 8 E). This prompted us to speculate that the reduction in MGPCs could be because of decline in one of the important regeneration-associated genes lin28a in the double blocker experiments as compared with DMSO control (Fig. 8, F and G). Furthermore, the reduced number of MGPCs also could be because of repression of the Notch ligand deltaD and increase in expression of her4.1 (Fig. 8, F and G). Collectively, double blocker experiments suggested that Delta–Notch signaling is active in the vicinity of MGPCs to restrict the zone of proliferation, whereas Mycb-mediated signaling is active in MGPCs to increase the cell number. Since Myc is expressed in both of these cell types, its functional absence causes significant increase in Her4.1 (Fig. 8 G), reducing the number of MGPCs. Collectively, based on these results, we could assume that the Delta–Notch signaling and Mycb-induced regenerative mechanisms are independent in some MGPCs but dependent in others. The extensive gene regulatory network and their unifying proposed mechanisms are presented as a model (Fig. 9, A and B).
Figure 9.
The gene regulatory network mediated through Myc/Hdac/Ascl1a/Lin28a/Her4.1 in MGPCs and neighboring cells. (A and B) The proposed model that depicts the mechanisms of genetic interaction of various regeneration-associated factors discussed in this study, shown separately in MGPCs (A) and neighboring cells (B) during retina regeneration. The interactions and molecules that are active are shown in black, and passive ones are in gray.
Discussion
Proto-oncogene myc, a de facto transcription activator in various developmental programs (Yan et al., 2010) and pluripotency induction (Takahashi et al., 2014), is also known to cause gene-repression in some tumors (Herkert and Eilers, 2010). Moreover, Myc’s capability of epigenetic modifications makes it one of the unique transcription factors (Amente et al., 2011; Flaisher-Grinberg et al., 2012; Kozono et al., 2015; Matkar et al., 2015). Our studies suggest that Myc genes are unique, and especially, Mycb can act as an activator or a repressor regulating dedifferentiation of MG to MGPCs and their proliferation during retina regeneration. Furthermore, our studies unravel novel Mycb-mediated signaling mechanisms and gene induction paradigms, underlying MGPCs formation.
Quantitative analysis of coexpression of both myca and mycb along with PCNA+ cells in 4-dpi retina reveals that only 40–50% of MG-derived progenitors show myca and mycb expression. The juxtaposed myca+/mycb+ cells should be the earlier MG cells that started dedifferentiating immediately after injury, which stayed restricted to injury site at 4 dpi, but some of myca+/mycb+ cells still may not enter the cell cycle. It is also important to note that up to 70% of myca/mycb expressing cells had PCNA expression. In other words, early progenitors would have myc expression, and late progenitors need not have it. Collectively, these results suggest that myca/mycb need not be present in all MGPCs, but a significant proportion of myca+/mycb+ cells show proliferation. These observations also suggest the possibility of existence of a Myc-independent cell proliferation mechanism in the retina. This could also account for the lack of complete loss of MGPCs in Myc blocked retina, either by MO or by 10058-F4.
The disparity seen in the expression of mycb with ascl1a knockdown in early and late stages of regeneration may seem contradictory. This could be because of differential expression pattern of mycb, along with ascl1a and insm1a. The immediate early panretinal expression of mycb should be initiating the induction of ascl1a, which in turn induces insm1a, a repressor of mycb, ascl1a, and insm1a itself (Ramachandran et al., 2012). This negative feedback regulation seen within the first few hours of retinal regeneration abolishes the panretinal flash expression of mycb, ascl1a, and insm1a. During these early hours of regeneration, knockdown of ascl1a causes an increase in mycb expression because of lack of insm1a induction. In this period, through another pathway, Insm1a represses a panretinal Wnt inhibitor dkk that paves the way for initiation of Wnt signaling that could restrict the expression of ascl1a (Ramachandran et al., 2011) and mycb to the injury site at a later stage. However, at 4 dpi, the insm1a is not seen in every ascl1a+ MGPC; instead, its expression is restricted to a subset of cells that are about to exit cell cycle (Ramachandran et al., 2012). At this stage, upon ascl1a knockdown, mycb expression is down-regulated due to the lack of Insm1a in those ascl1a+ cells.
We uncovered specific roles of Myc during retina regeneration. First, Mycb activates ascl1a and regulates lin28a expression, which is essential to induce multiple regeneration-associated pathways (Ramachandran et al., 2010a; Zhang et al., 2016). Second, Mycb induces transcriptional repressors like insm1a, and inhibits her4.1, essential for fine-tuned expression of ascl1a, lin28a, and mycb itself, to the active zone of regeneration. Although canonical Wnt signaling–mediated up-regulation of Lin28a is shown in mammals (Yao et al., 2016), it may also induce Myc, which probably represses Lin28a either directly via Hdac1 recruitment or through Her4/Hes, as seen in zebrafish, reducing its regenerative potential. Moreover, in zebrafish retina, Ascl1a, which is a wnt inducer and β-catenin–regulated gene (Ramachandran et al., 2011), also contributes to the Lin28a level during retina regeneration (Ramachandran et al., 2010a). Seclusion of Hdac1 from BrdU+ MGPCs at 4 dpi, support the view that the role of Mycb in these cells would be as an activator of lin28a expression through Ascl1a. The opposite will be seen in Mycb+ and Hdac1+ cells at the vicinity of cell proliferation, wherein they collaborate to cause lin28a repression. These results affirm the dual roles of Mycb as a transcriptional activator and repressor on crucial genes like lin28a.
Closer investigation of regenerating retina with compromised Notch signaling reveals that Lin28a and Mycb cause enhanced number of MGPCs at injury site, which do not sustain in the absence of Mycb. Apart from its roles in mRNA splicing (Wilbert et al., 2012), and reprogramming cellular metabolism (Shyh-Chang et al., 2013), Lin28a also mediates down-regulation of let-7 microRNA, essential for translation of several regeneration-associated genes (Ramachandran et al., 2010a), and Notch itself (Wang et al., 2010; Gökbuget et al., 2015), necessary for her4.1 induction (Gemberling et al., 2013; Zhao et al., 2014a). Interestingly, our results show that Myc collaborates with Hdac1 to cause a decline in her4.1 expression. The Her4.1 would probably suppress lin28a expression, causing a yin-yang relationship as part of restricting the zone of proliferation after focal injury. The results of double blocker experiments with DAPT and 10058-F4 in retina support this view. We find coexistence of increased her4.1 and decreased lin28a levels when both Notch signaling and Myc were blocked simultaneously. This mechanism may also underlie the possible cause of lack of MGPC induction in Her/Hes overexpressed retina (Wan et al., 2012). Moreover, the decreased Lin28a levels could cause an increase in let-7 microRNA in DAPT and 10058-F4–treated retina, which could bring down the protein levels of Myca, Mycb, and Ascl1a, as reported earlier (Ramachandran et al., 2010a), causing a reduction in the number of MGPCs.
Our data suggest that Myc plays important roles in different phases of retina regeneration (Fig. 9, A and B). First, it contributes in MG reprogramming to generate MGPCs through Ascl1a and Lin28a. Second, it restricts the zone of MGPCs through Her4.1–Lin28a axis. Finally, our studies unraveled important mechanisms by which Mycs and Hdacs mediate these effects through mutual signaling pathways, involving Ascl1a, Insm1a, Lin28a, and Her4.1, in retina regeneration. It is intriguing to speculate that GCL-specific rapid induction of mycb after optic nerve lesion also may significantly contribute to its regeneration in zebrafish. These studies suggest that Mycs and subsequent gene regulatory network are essential for retina regeneration, providing insights into signaling mechanisms that may help in understanding MG reprogramming in the injured mammalian retina, also with reference to damaged human retinae toward successful repair.
Materials and methods
Animals, fin cut, retinal injury, and drugs
Zebrafish were maintained at 26–28°C on a 14/10 h light/dark cycle. The 1016 tuba1a:gfp transgenic fish used in this study have been previously described (Fausett and Goldman, 2006). Embryos for all assays were obtained by natural breeding. The Myc-Max inhibitor, 10058-F4, and Notch-signaling blocker, DAPT, were made to a stock of 1 mM in DMSO for various experiments (all drugs were from Sigma-Aldrich). Drugs were delivered either by dipping or injected into the eye using a Hamilton syringe with a 30-G needle. Retinal injury or optic nerve lesions were performed as described previously (Fausett and Goldman, 2006; Veldman et al., 2010). Fish were anaesthetized transiently in tricaine methane sulphonate, and the right eye was gently pulled from its socket and the retina stabbed four to eight times (once or twice in each quadrant) through the sclera with a 30-G needle inserted up to the length of the bevel. Optic nerve lesions were performed similarly, except that damage was not done to retina or blood vessel while cutting the optic nerve. Both retinal injury and optic nerve lesion were performed under a dissection scope (Stemi DV4; Zeiss). All experiments were done to a minimum of three times for consistency and SD.
Primers and plasmid construction
All primers are listed in Table S1. The promoters of mycb and her4.1 were amplified from zebrafish genomic DNA using primer pairs XhoI-mycb pro-F and BamHI-mycb pro-R (∼3 kb) or XhoI-her4.1pro-F and BamHI-her4.1 pro-R (∼4 kb), respectively. The digested PCR amplicons were cloned into a pEL luciferase expression vector to create mycb:gfp-luciferase and her4.1:gfp-luciferase constructs. The ascl1a:gfp-luciferase, lin28a:gfp-luciferase, and insm1a:gfp-luciferase construct was described previously (Ramachandran et al., 2010a, 2012). The lin28a promoter site-directed mutagenesis was done as described previously (Ramachandran et al., 2010a). GFP was amplified from pEGFP-C1 plasmid with BamH1-EGFP-F and EcoR1-EGFP-R and cloned into pCS2+ vector.
Genes like ascl1a, myca, mycb, insm1a, and lin28a were cloned from cDNA amplified from zebrafish retina RNA at 4 dpi using primer pairs BamHI-ascl1a FL-F and XhoI-ascl1a FL-R (∼0.6 kb); BamHI-myca-F and XhoI-myca-R (∼1.2 kb); BamHI-mycb-F and XbaI-mycb-R (∼1.2 kb); BamHI-insm1a-F and XhoI-insm1a-R (∼1.1 kb); and BamHI-lin28a FL-F and XhoI-lin28a FL-R (∼0.6 kb). Post-digested PCR amplicons were cloned into their respective enzyme sites in pCS2+ plasmid to obtain cmv:ascl1a, cmv:myca, cmv:mycb, cmv:insm1a, and cmv:lin28a.
Total RNA isolation, RT-PCR, and qPCR analysis
Total RNA was isolated from dark-adapted zebrafish retinae of control, injured, and drug-treated/MO-electroporated group using TRIzol (Invitrogen). Combination of oligo-dT and random hexamers were used to reverse transcribe 5 µg of RNA using Superscript II reverse transcription (Invitrogen) to generate cDNA. PCR reactions used Taq or Phusion (New England Biolabs) DNA polymerase and gene-specific primers (Table S1) with previously described cycling conditions (Ramachandran et al., 2010a). qPCR was performed in triplicate with KOD SYBR qPCR mix (QKD-201; Genetix) as per manufacturer’s recommendations on a real-time PCR detection system (Eppendorf Master Cycler RealPlex4). The relative expression of mRNAs in control and injured retinae was deciphered using the ΔΔCt method and normalized to ribosomal protein l-24 or β-actin mRNA levels.
mRNA synthesis, embryo micro-injection, ChIP, and Co-IP assay
Various gene clones in pCS2+ plasmids having cDNA inserts were linearized, and capped mRNAs were synthesized using the mMESSAGE mMACHINE (Ambion) in vitro transcription system. For luciferase assay experiments, single-cell zebrafish embryos were injected with a total volume of ∼1 nl solution, containing 0.02 pg of Renilla reniformis luciferase mRNA (normalization), 5 pg of promoter:gfp-luciferase vector, and 0–6 pg of ascl1a, insm1a, or mycb mRNA. To assure consistency of results, a master mix was made for daily injections and ∼300 embryos were injected at single-cell stage. 24 h later, embryos were divided into three groups (∼70 embryos/group) and lysed for dual luciferase reporter assays (E1910; Promega).
ChIP assays to analyze endogenous Ascl1a or Mycb binding to various promoters in adult retina at 2 and 4 dpi were performed using ∼50 adult retinae after dark adaptation. Chromatin was isolated by sonication as described previously (Lindeman et al., 2009). The chromatin obtained after a brief fixing in 1% (vol/vol) formaldehyde for 10 min in room temperature and subsequent nuclear lysis were the starting material. The chromatin after sonication to make fragments of 500–800 bp in size was distributed into three equal aliquots; two were probed with an anti-zebrafish Myc and Ascl1a antibodies (described below), and the third served as a control. The antibody binding was done at 4°C with rotation. Using magnetic beads, the antibody bound chromatin were pulled down on magnetic rack. After washing, the chromatins were purified to obtain PCR-grade DNA using standard proteinase K, phenol chloroform extract before PCR analysis. Primers used for ChIP assays are described in Table S1. Co-IP was performed using the retinal lysate that were extracted using lysis buffer as per manufacturer’s recommendations and protocol reported elsewhere (Phizicky and Fields, 1995; Bonifacino et al., 2016). Co-IP was similar to ChIP in initial steps, except that the final eluted sample was run on an acrylamide gel, transferred onto a polyvinylidene fluoride membrane and probed with respective antibodies against proteins of the interaction complex obtained.
Morpholino electroporation, mRNA transfection, and knockdown-rescue
Lissamine-tagged MOs (Gene Tools) of ∼0.5 µl (0.5–1.0 mM) were injected at the time of injury using a Hamilton syringe of 10-µl volume capacity. MO delivery to cells was accomplished by electroporation as previously described (Fausett et al., 2008). An ECM 830 Electro Square Porator (BTX) was used to electroporate the retina for MO delivery. BTX830 were adjusted to deliver five consecutive 50-ms pulses at 70 V with a 950-ms interval between pulses, using BTX electrodes of 0.5-cm diameter. The control and ascl1a-targeting MOs have been previously described (Ramachandran et al., 2012). Morpholinos targeting myca, mycb and hdac1 are myca MO, 5′-AACTCGCACTCACCAGCATTTTGAC-3′; 2-myca MO, 5′-TTTAACGAATGCCGTTCCAGAATTG-3′; mycb MO, 5′-CCATACTTGAATTCAGCGGCATGGT-3′; 2-mycb MO, 5′-GAGTGCCGTAGCCGTGGTAAAAGCT-3′; insm1a MO, 5′-GCTTGACTAAAAATCCTCTGGGCAT-3′; and Ctl MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′.
Transfection mixture contained two solutions constituted in equal volumes: (1) 4–5 µg of mRNA mixed with HBSS and (2) lipofectamine messenger max reagent (LMRNA001; Invitrogen) mixed with HBSS. Both the solutions were allowed to stand at room temperature for 10 min and then mixed drop wise, followed by 30-min incubation at room temperature. The resultant solution was mixed with morpholino in equal proportion, and 0.5 µl of this mixture was used for injection in zebrafish retina, followed by electroporation as described earlier.
In vivo rescue experiments were designed for testing the specificity of myca and mycb MO antisense oligos. This was accomplished by transfection of zebrafish retina with gene-specific mRNA alongside the MO-targeting 5′ UTR region of concerned genes or control MO. For confirming the efficient mRNA transfection, GFP mRNA was also delivered by transfection in each experimental or control retina.
BrdU/EdU labeling, retina tissue preparation for mRNA ISH, immunofluorescence (IF) microscopy, TUNEL assay, and Western blotting
BrdU labeling was performed by single i.p. injection of 20 µl of BrdU (20 mM) 3 h before euthanasia and retina dissection, unless mentioned specifically. Some animals required for long-term cell tracing experiments received more BrdU injections over multiple days. Fish were given higher dose of tricaine methane sulphonate, and eyes were dissected, lens removed, fixed in 4% PFA, and sectioned as described previously (Fausett and Goldman, 2006). mRNA ISH was performed on retina sections with fluorescein or digoxigenin-labeled complementary RNA probes (FL/DIG RNA labeling kit; Roche Diagnostics; Barthel and Raymond, 2000). Fluorescence ISH was performed according to the manufacturer’s directions (T20917, B40955, and B40953; Thermo Fisher Scientific). Sense probes were used in every ISH separately as control to assess the potential of background signal. IF microscopy protocols and antibodies were previously described (Ramachandran et al., 2012). IF microscopy was performed using rabbit polyclonal antibody against human ASCL1/MASH1 (ab74065; Abcam); rat monoclonal antibody against BrdU (ab6326; Abcam); mouse monoclonal antibody against human PCNA (sc-25280; Santa Cruz); rabbit polyclonal antibody against zebrafish Myca/b (Schreiber-Agus et al., 1993; AS-55477; Anaspec); rabbit polyclonal antibody against zebrafish Hdac1 (Harrison et al., 2011; Ab41407; Abcam); mouse polyclonal antibody against GFP (ab-38689; Abcam); rabbit polyclonal antibody against GFP (ab-6556; Abcam); and rabbit polyclonal antibody against mouse glutamine synthetase (Ramachandran et al., 2010b; ab93439; Abcam) at 1:500 dilution. Before BrdU IF microscopy, retinal sections were treated with 2 N HCl at 37°C for 20 min, equilibrated with 100 mM sodium borate (pH 8.5) for 10 min twice, and then processed using standard procedures (Senut et al., 2004).
Proliferating cells were labeled by intravitreal injection of 0.5 µl of 10 mM EdU solution dissolved in DMSO. A fresh injury was made near the cornea with a Hamilton Syringe of 10-µl capacity for intravitreal injection. Eyes were enucleated after 4 h, followed by cryoprotection as described elsewhere. EdU-labeled cells were detected by treating 8-µg retinal sections with Click-iT EdU Reaction cocktail (Click-iT TM EdU Alexa Fluor 647 Imaging kit; C10340; Thermo Fisher Scientific) prepared as per manufacturer’s instructions. In brief, after the fluorescence ISH (FISH) protocol, retinal sections were fixed with 4% PFA at room temperature for 20 min, followed by permeabilization with 1% BSA in PBS with Triton X-100 (PBST) at room temperature for 10 min and blocking with 3% BSA in PBST for half an hour. After blocking, 100 µl of Click-iT reaction cocktail was overlaid with glass coverslips for half an hour, followed by washing with 1% BSA in PBST. EdU-labeled cells were detected by confocal microscopy.
BrdU-labeled MGPC lineage-tracing experiments were done in retinal sections from single-eye sections of 8-µg thickness, distributed across five slides. Individual slide was first processed for IF-based detection of specific antigen or Mrna, and then BrdU or PCNA staining was performed as mentioned above using respective antibodies (Powell et al., 2012; Ramachandran et al., 2012). The total number of BrdU+ cells and the number of colabeled BrdU+ cells that also stained with a specific ISH probe and subsequent enzymatic reaction were quantified on each slide. TUNEL assay was performed on retinal sections using In Situ Cell Death Detection Fluorescein kit (11684795910; Roche) as per manufacturer recommended protocol. Western blotting was performed using whole retina tissue from four retinae per experimental sample, lysed in Laemmli buffer, size-fractioned in 12% acrylamide gel with SDS at denaturing conditions, before transferring onto Immun-Blot polyvinylidene fluoride membrane (162-0177; Biorad Catalogue), followed by probing with specific primary antibodies, and HRP-conjugated secondary for chemiluminescence assay using Clarity Western ECL (170-5061; Biorad Catalogue).
Fluorescence and confocal microscopy and cell counting
After the staining experiments, the slides were examined with a Nikon Ni-E fluorescence microscope equipped with fluorescence optics and Nikon A1 confocal imaging system equipped with apochromat 60×/1 NA oil immersion objective lens. Imaging of bright field is done using Nikon DS-L3 camera attached onto the same microscope, as mentioned above. Cell counts were quantified by physically observing fluorescently labeled ISH, PCNA, or BrdU+ cells in retinal sections, visualized in the same microscope. We used 20× for low magnification and 40× or 60× oil objective with an NA set to 1 in almost all images. Images were from cryosections mounted on Super Frost Plus slides (Thermo Fisher Scientific), embedded with DABCO mounting medium in every retinal section discussed. The imaging was always done at room temperature. The confocal images were finally processed through deconvolution using the software NIS-Elements software and ImageJ. The final images were imported to Adobe Photoshop software (CC 2018) for conversion to 300 dpi. Every sections of the stained retina were mounted, observed, and analyzed, and at least three retinae from separate fish were used.
Fluorescence-based cell sorting
RNA and Chromatin was obtained from FACS-purified MG and MG-derived progenitors at 4 dpi, as previously described (Ramachandran et al., 2011, 2012). In brief, uninjured and injured retinae were isolated from 1016 tuba1a:gfp transgenic fish. GFP+ MGPCs from 1016 tuba1a:gfp retinae at 4 dpi were isolated by treating retinae with hyaluronidase and trypsin and then sorted on a BD FACS Aria Fusion high speed cell sorter. Approximately 40 injured retinae from 1016 tuba1a:gfp fish yielded 80,000 GFP+ and 170,000 GFP− from DMSO-treated fish (20 retinae) and 40,000 GFP+ and 220,000 GFP− (20 retinae) from 10058-F4–treated retinae.
Statistical analysis
Observed data were plotted and analyzed using standard spreadsheet software (Microsoft Excel). All data, unless specified, represent mean with SD as error bar. Data distribution was assumed to be normal, but this was not formally tested. The statistical significance by comparisons of datasets was done using a two-tailed unpaired Student’s t test for all experiments. For all other comparisons, ANOVA was performed, and subsequently, a Bonferroni–Dunn post hoc t test was done using Stat View software.
Online supplemental material
Fig. S1 shows the Myc and Max gene regulation during retina and optic nerve regeneration and development. Fig. S2 shows that knockdown of myca and mycb, separately and in combination, during regeneration decrease cell proliferation. Fig. S3 shows the rescue and expression dynamics of myc genes in retina and TUNEL assay with its blockade. Fig. S4 shows the regulation of regeneration-associated genes through Myc. Fig. S5 shows that Delta–Notch signaling and Myc show an interdependency during regeneration. Table S1 lists the DNA oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
The authors express their gratitude to Daniel Goldman (University of Michigan, Ann Arbor, MI) for sharing promoter clones of 1016 tuba, lin28a, ascl1a, and insm1a genes.
S. Mitra, M.A. Khursheed, M. Chaudhary, and A.J. Kurup acknowledge their financial support from the Indian Institute of Science Education and Research, Mohali (IISER Mohali). P. Sharma acknowledges support from Wellcome Trust/DBT India Alliance. S. Kaur acknowledges her support from the Department of Biotechnology (DBT), government of India, for Senior Research Fellowship. S. Gupta acknowledges her support from the Indian Council of Medical Research for Senior Research Fellowship. This work was supported by the Wellcome Trust/DBT India Alliance Intermediate Fellowship awarded to R. Ramachandran (IA/I/12/2/500630); R. Ramachandran also acknowledges research funding from DBT India (BT/PR9407/BRB/10/1261/2013 and BT/PR17912/MED/31/336/2016) and support from IISER Mohali.
The authors declare no competing financial interests.
Author contributions: R. Ramachandran conceived the study and designed experiments. S. Mitra, P. Sharma, and S. Kaur performed majority of experiments. M.A. Khursheed, M. Chaudhary, and S. Gupta performed a few fin cut experiments, ISH, IF microscopy, and cell counting. R. Ramachandran, S. Mitra, P. Sharma, and S. Kaur analyzed the data. A.J. Kurup performed the retinal cell sorting. R. Ramachandran wrote the manuscript with critical input from P. Sharma, S. Mitra, M. Chaudhary, and S. Gupta.
References
- Ail D., and Perron M.. 2017. Retinal Degeneration and Regeneration-Lessons From Fishes and Amphibians. Curr. Pathobiol. Rep. 5:67–78. 10.1007/s40139-017-0127-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B., and Land H.. 1994. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 4:102–108. 10.1016/0959-437X(94)90098-1 [DOI] [PubMed] [Google Scholar]
- Amente S., Lania L., and Majello B.. 2011. Epigenetic reprogramming of Myc target genes. Am. J. Cancer Res. 1:413–418. [PMC free article] [PubMed] [Google Scholar]
- Arora V., Knapp D.C., Smith B.L., Statdfield M.L., Stein D.A., Reddy M.T., Weller D.D., and Iversen P.L.. 2000. c-Myc antisense limits rat liver regeneration and indicates role for c-Myc in regulating cytochrome P-450 3A activity. J. Pharmacol. Exp. Ther. 292:921–928. [PubMed] [Google Scholar]
- Barthel L.K., and Raymond P.A.. 2000. In situ hybridization studies of retinal neurons. Methods Enzymol. 316:579–590. 10.1016/S0076-6879(00)16751-5 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Gershlick D.C., and Dell’Angelica E.C.. 2016. Immunoprecipitation. Curr. Protoc. Cell Biol. 71:7.2. [DOI] [PubMed] [Google Scholar]
- Calvo E.L., Dusetti N.J., Cadenas M.B., Dagorn J.C., and Iovanna J.L.. 1991. Changes in gene expression during pancreatic regeneration: activation of c-myc and H-ras oncogenes in the rat pancreas. Pancreas. 6:150–156. 10.1097/00006676-199103000-00004 [DOI] [PubMed] [Google Scholar]
- Conner C., Ackerman K.M., Lahne M., Hobgood J.S., and Hyde D.R.. 2014. Repressing notch signaling and expressing TNFα are sufficient to mimic retinal regeneration by inducing Müller glial proliferation to generate committed progenitor cells. J. Neurosci. 34:14403–14419. 10.1523/JNEUROSCI.0498-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1–11. 10.1128/MCB.19.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecevit O., Khan M.A., and Goss D.J.. 2010. Kinetic analysis of the interaction of b/HLH/Z transcription factors Myc, Max, and Mad with cognate DNA. Biochemistry. 49:2627–2635. 10.1021/bi901913a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaeidi F., Macpherson P., Mills E.A., Jui J., Flannery J.G., and Goldman D.. 2018. Notch Suppression Collaborates with Ascl1 and Lin28 to Unleash a Regenerative Response in Fish Retina, But Not in Mice. J. Neurosci. 38:2246–2261. 10.1523/JNEUROSCI.2126-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett B.V., and Goldman D.. 2006. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J. Neurosci. 26:6303–6313. 10.1523/JNEUROSCI.0332-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett B.V., Gumerson J.D., and Goldman D.. 2008. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J. Neurosci. 28:1109–1117. 10.1523/JNEUROSCI.4853-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P.C., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., and Amati B.. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115–1129. 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaisher-Grinberg S., Persaud S.D., Loh H.H., and Wei L.N.. 2012. Stress-induced epigenetic regulation of κ-opioid receptor gene involves transcription factor c-Myc. Proc. Natl. Acad. Sci. USA. 109:9167–9172. 10.1073/pnas.1205565109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Bailey T.J., Hyde D.R., and Poss K.D.. 2013. The zebrafish as a model for complex tissue regeneration. Trends Genet. 29:611–620. 10.1016/j.tig.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gökbuget D., Pereira J.A., Bachofner S., Marchais A., Ciaudo C., Stoffel M., Schulte J.H., and Suter U.. 2015. The Lin28/let-7 axis is critical for myelination in the peripheral nervous system. Nat. Commun. 6:8584 10.1038/ncomms9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. 2014. Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 15:431–442. 10.1038/nrn3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsuch R.A., Lahne M., Yarka C.E., Petravick M.E., Li J., and Hyde D.R.. 2017. Sox2 regulates Müller glia reprogramming and proliferation in the regenerating zebrafish retina via Lin28 and Ascl1a. Exp. Eye Res. 161:174–192. 10.1016/j.exer.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramage E., D’Cruz T., Taylor S., Thummel R., and Hitchcock P.F.. 2015. Midkine-a protein localization in the developing and adult retina of the zebrafish and its function during photoreceptor regeneration. PLoS One. 10:e0121789 10.1371/journal.pone.0121789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.R., Georgiou A.S., Spaink H.P., and Cunliffe V.T.. 2011. The epigenetic regulator Histone Deacetylase 1 promotes transcription of a core neurogenic programme in zebrafish embryos. BMC Genomics. 12:24 10.1186/1471-2164-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkert B., and Eilers M.. 2010. Transcriptional repression: the dark side of myc. Genes Cancer. 1:580–586. 10.1177/1947601910379012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B., and Liebermann D.A.. 1998. The proto-oncogene c-myc and apoptosis. Oncogene. 17:3351–3357. 10.1038/sj.onc.1202592 [DOI] [PubMed] [Google Scholar]
- Huang M.J., Cheng Y.C., Liu C.R., Lin S., and Liu H.E.. 2006. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 34:1480–1489. 10.1016/j.exphem.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Jorstad N.L., Wilken M.S., Grimes W.N., Wohl S.G., VandenBosch L.S., Yoshimatsu T., Wong R.O., Rieke F., and Reh T.A.. 2017. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature. 548:103–107. 10.1038/nature23283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., and Kobayashi T.. 2007. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 134:1243–1251. 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- Kaur S., Gupta S., Chaudhary M., Khursheed M.A., Mitra S., Kurup A.J., and Ramachandran R.. 2018. let-7 MicroRNA-Mediated Regulation of Shh Signaling and the Gene Regulatory Network Is Essential for Retina Regeneration. Cell Reports. 23:1409–1423. 10.1016/j.celrep.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozono D., Li J., Nitta M., Sampetrean O., Gonda D., Kushwaha D.S., Merzon D., Ramakrishnan V., Zhu S., Zhu K., et al. . 2015. Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc. Natl. Acad. Sci. USA. 112:E4055–E4064. 10.1073/pnas.1501967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland J.F., and Tansey W.P.. 2008. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 68:3624–3629. 10.1158/0008-5472.CAN-07-6552 [DOI] [PubMed] [Google Scholar]
- Lemaître J.M., Méchali M., and Géraudie J.. 1992. Nerve-dependent expression of c-myc protein during forelimb regeneration of Xenopus laevis froglets. Int. J. Dev. Biol. 36:483–489. [PubMed] [Google Scholar]
- Lin C.P., Liu J.D., Chow J.M., Liu C.R., and Liu H.E.. 2007. Small-molecule c-Myc inhibitor, 10058-F4, inhibits proliferation, downregulates human telomerase reverse transcriptase and enhances chemosensitivity in human hepatocellular carcinoma cells. Anticancer Drugs. 18:161–170. 10.1097/CAD.0b013e3280109424 [DOI] [PubMed] [Google Scholar]
- Lindeman L.C., Vogt-Kielland L.T., Aleström P., and Collas P.. 2009. Fish’n ChIPs: chromatin immunoprecipitation in the zebrafish embryo. Methods Mol. Biol. 567:75–86. 10.1007/978-1-60327-414-2_5 [DOI] [PubMed] [Google Scholar]
- Luo J., Uribe R.A., Hayton S., Calinescu A.A., Gross J.M., and Hitchcock P.F.. 2012. Midkine-A functions upstream of Id2a to regulate cell cycle kinetics in the developing vertebrate retina. Neural Dev. 7:33 10.1186/1749-8104-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N., and Yamanaka S.. 2011. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 474:225–229. 10.1038/nature10106 [DOI] [PubMed] [Google Scholar]
- Matkar S., Sharma P., Gao S., Gurung B., Katona B.W., Liao J., Muhammad A.B., Kong X.C., Wang L., Jin G., et al. . 2015. An Epigenetic Pathway Regulates Sensitivity of Breast Cancer Cells to HER2 Inhibition via FOXO/c-Myc Axis. Cancer Cell. 28:472–485. 10.1016/j.ccell.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., Sharma P., Kaur S., Khursheed M.A., Gupta S., Ahuja R., Kurup A.J., Chaudhary M., and Ramachandran R.. 2018. Histone Deacetylase-Mediated Müller Glia Reprogramming through Her4.1-Lin28a Axis Is Essential for Retina Regeneration in Zebrafish. iScience. 7:68–84. 10.1016/j.isci.2018.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled M.H., Patra C., Dickson A.L., Endo T., Stainier D.Y., and Poss K.D.. 2016. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science. 354:630–634. 10.1126/science.aaf2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello D., Lavenu A., and Babinet C.. 1990. Differential regulation and expression of jun, c-fos and c-myc proto-oncogenes during mouse liver regeneration and after inhibition of protein synthesis. Oncogene. 5:1511–1519. [PubMed] [Google Scholar]
- Munderloh C., Solis G.P., Bodrikov V., Jaeger F.A., Wiechers M., Málaga-Trillo E., and Stuermer C.A.. 2009. Reggies/flotillins regulate retinal axon regeneration in the zebrafish optic nerve and differentiation of hippocampal and N2a neurons. J. Neurosci. 29:6607–6615. 10.1523/JNEUROSCI.0870-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.M., Gorsuch R.A., Bailey T.J., Ackerman K.M., Kassen S.C., and Hyde D.R.. 2012. Stat3 defines three populations of Müller glia and is required for initiating maximal müller glia proliferation in the regenerating zebrafish retina. J. Comp. Neurol. 520:4294–4311. 10.1002/cne.23213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooto S., Akagi T., Kageyama R., Akita J., Mandai M., Honda Y., and Takahashi M.. 2004. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc. Natl. Acad. Sci. USA. 101:13654–13659. 10.1073/pnas.0402129101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham G., and Cleveland J.L.. 1995. c-Myc and apoptosis. Biochim. Biophys. Acta. 1242:11–28. [DOI] [PubMed] [Google Scholar]
- Packham G., Porter C.W., and Cleveland J.L.. 1996. c-Myc induces apoptosis and cell cycle progression by separable, yet overlapping, pathways. Oncogene. 13:461–469. [PubMed] [Google Scholar]
- Phizicky E.M., and Fields S.. 1995. Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 59:94–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak J., Wilken M.S., Ueki Y., Cox K.E., Sullivan J.M., Taylor R.J., Levine E.M., and Reh T.A.. 2013. ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development. 140:2619–2631. 10.1242/dev.091355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C., Elsaeidi F., and Goldman D.. 2012. Injury-dependent Müller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J. Neurosci. 32:1096–1109. 10.1523/JNEUROSCI.5603-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C., Grant A.R., Cornblath E., and Goldman D.. 2013. Analysis of DNA methylation reveals a partial reprogramming of the Müller glia genome during retina regeneration. Proc. Natl. Acad. Sci. USA. 110:19814–19819. 10.1073/pnas.1312009110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C., Cornblath E., Elsaeidi F., Wan J., and Goldman D.. 2016. Zebrafish Müller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons. Sci. Rep. 6:24851 10.1038/srep24851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J.S., Robitaille A.M., Wang Y., Ray C.A., Thummel R., Gu H., Djukovic D., Raftery D., Berndt J.D., and Moon R.T.. 2017. Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc. Natl. Acad. Sci. USA. 114:E717–E726. 10.1073/pnas.1620755114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Fausett B.V., and Goldman D.. 2010a Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 12:1101–1107. 10.1038/ncb2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Reifler A., Parent J.M., and Goldman D.. 2010b Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. J. Comp. Neurol. 518:4196–4212. 10.1002/cne.22448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Zhao X.F., and Goldman D.. 2011. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc. Natl. Acad. Sci. USA. 108:15858–15863. 10.1073/pnas.1107220108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Zhao X.F., and Goldman D.. 2012. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat. Cell Biol. 14:1013–1023. 10.1038/ncb2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Aguirre L.I., and Lamas M.. 2016. Oct4 Methylation-Mediated Silencing As an Epigenetic Barrier Preventing Müller Glia Dedifferentiation in a Murine Model of Retinal Injury. Front. Neurosci. 10:523 10.3389/fnins.2016.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann S., and Borlak J.. 2008. Transcription profiling of lung adenocarcinomas of c-myc-transgenic mice: identification of the c-myc regulatory gene network. BMC Syst. Biol. 2:46 10.1186/1752-0509-2-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. 2003. The roles of Hedgehogs and Fibroblast Growth Factors in eye development and retinal cell rescue. Vision Res. 43:899–912. 10.1016/S0042-6989(02)00416-9 [DOI] [PubMed] [Google Scholar]
- Sanders J.A., Schorl C., Patel A., Sedivy J.M., and Gruppuso P.A.. 2012. Postnatal liver growth and regeneration are independent of c-myc in a mouse model of conditional hepatic c-myc deletion. BMC Physiol. 12:1 10.1186/1472-6793-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N., Horner J., Torres R., Chiu F.C., and DePinho R.A.. 1993. Zebra fish myc family and max genes: differential expression and oncogenic activity throughout vertebrate evolution. Mol. Cell. Biol. 13:2765–2775. 10.1128/MCB.13.5.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut M.C., Gulati-Leekha A., and Goldman D.. 2004. An element in the alpha1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J. Neurosci. 24:7663–7673. 10.1523/JNEUROSCI.2281-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Shu B., Yang R., Xu Y., Xing B., Liu J., Chen L., Qi S., Liu X., Wang P., et al. . 2015. Wnt and Notch signaling pathway involved in wound healing by targeting c-Myc and Hes1 separately. Stem Cell Res. Ther. 6:120 10.1186/s13287-015-0103-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M.T., Tsanov K.M., Nguyen L., Asara J.M., Cantley L.C., and Daley G.Q.. 2013. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 155:778–792. 10.1016/j.cell.2013.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak J., Mechti N., Tournier M.F., Blanchard J.M., and Duguet M.. 1989. c-myc and c-fos gene regulation during mouse liver regeneration. Oncogene. 4:1503–1508. [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Sasaki A., Yamamoto M., Nakamura M., Sutou K., Osafune K., and Yamanaka S.. 2014. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 5:3678 10.1038/ncomms4678 [DOI] [PubMed] [Google Scholar]
- Thomas J.L., Morgan G.W., Dolinski K.M., and Thummel R.. 2018. Characterization of the pleiotropic roles of Sonic Hedgehog during retinal regeneration in adult zebrafish. Exp. Eye Res. 166:106–115. 10.1016/j.exer.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R., Enright J.M., Kassen S.C., Montgomery J.E., Bailey T.J., and Hyde D.R.. 2010. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp. Eye Res. 90:572–582. 10.1016/j.exer.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y., Wilken M.S., Cox K.E., Chipman L., Jorstad N., Sternhagen K., Simic M., Ullom K., Nakafuku M., and Reh T.A.. 2015. Transgenic expression of the proneural transcription factor Ascl1 in Müller glia stimulates retinal regeneration in young mice. Proc. Natl. Acad. Sci. USA. 112:13717–13722. 10.1073/pnas.1510595112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman M.B., Bemben M.A., and Goldman D.. 2010. Tuba1a gene expression is regulated by KLF6/7 and is necessary for CNS development and regeneration in zebrafish. Mol. Cell. Neurosci. 43:370–383. 10.1016/j.mcn.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T., Campbell A., and De Langhe S.. 2013. c-Myc regulates proliferation and Fgf10 expression in airway smooth muscle after airway epithelial injury in mouse. PLoS One. 8:e71426 10.1371/journal.pone.0071426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D.S., Mears A.J., McNeill B., Mazerolle C., Thurig S., Wang Y., Kageyama R., and Wallace V.A.. 2009. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 184:101–112. 10.1083/jcb.200805155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Ramachandran R., and Goldman D.. 2012. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev. Cell. 22:334–347. 10.1016/j.devcel.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhao X.F., Vojtek A., and Goldman D.. 2014. Retinal injury, growth factors, and cytokines converge on β-catenin and pStat3 signaling to stimulate retina regeneration. Cell Reports. 9:285–297. 10.1016/j.celrep.2014.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hammoudeh D.I., Follis A.V., Reese B.E., Lazo J.S., Metallo S.J., and Prochownik E.V.. 2007. Improved low molecular weight Myc-Max inhibitors. Mol. Cancer Ther. 6:2399–2408. 10.1158/1535-7163.MCT-07-0005 [DOI] [PubMed] [Google Scholar]
- Wang Z., Li Y., Kong D., Ahmad A., Banerjee S., and Sarkar F.H.. 2010. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 292:141–148. 10.1016/j.canlet.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert M.L., Huelga S.C., Kapeli K., Stark T.J., Liang T.Y., Chen S.X., Yan B.Y., Nathanson J.L., Hutt K.R., Lovci M.T., et al. . 2012. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell. 48:195–206. 10.1016/j.molcel.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Yu S., Lei A., Hua J., Chen F., Li L., Xie X., Yang X., Geng W., and Dou Z.. 2010. The four reprogramming factors and embryonic development in mice. Cell. Reprogram. 12:565–570. 10.1089/cell.2010.0008 [DOI] [PubMed] [Google Scholar]
- Yao K., Qiu S., Tian L., Snider W.D., Flannery J.G., Schaffer D.V., and Chen B.. 2016. Wnt Regulates Proliferation and Neurogenic Potential of Müller Glial Cells via a Lin28/let-7 miRNA-Dependent Pathway in Adult Mammalian Retinas. Cell Reports. 17:165–178. 10.1016/j.celrep.2016.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Giap C., Lazo J.S., and Prochownik E.V.. 2003. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 22:6151–6159. 10.1038/sj.onc.1206641 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. . 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318:1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zeller K.I., Jegga A.G., Aronow B.J., O’Donnell K.A., and Dang C.V.. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69 10.1186/gb-2003-4-10-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ratanasirintrawoot S., Chandrasekaran S., Wu Z., Ficarro S.B., Yu C., Ross C.A., Cacchiarelli D., Xia Q., Seligson M., et al. . 2016. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell. 19:66–80. 10.1016/j.stem.2016.05.009 [DOI] [PubMed] [Google Scholar]
- Zhao L., Borikova A.L., Ben-Yair R., Guner-Ataman B., MacRae C.A., Lee R.T., Burns C.G., and Burns C.E.. 2014a Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA. 111:1403–1408. 10.1073/pnas.1311705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.F., Wan J., Powell C., Ramachandran R., Myers M.G. Jr., and Goldman D.. 2014b Leptin and IL-6 family cytokines synergize to stimulate Müller glia reprogramming and retina regeneration. Cell Reports. 9:272–284. 10.1016/j.celrep.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Yan J., Ma Z., Zhou Y., Abbood N.N., Liu J., Su L., Jia H., and Guo A.Y.. 2012. Comparative and evolutionary analysis of the HES/HEY gene family reveal exon/intron loss and teleost specific duplication events. PLoS One. 7:e40649 10.1371/journal.pone.0040649 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.