Cantor previews work from the Mosammaparast laboratory showing that the mitotic regulators TPX2 and Aurora A protect DNA forks during replication stress by counteracting 53BP1 function.
Abstract
In this issue, Byrum et al. (2019. J. Cell Biol. https://doi.org/10.1083/jcb.201803003) surprisingly identify an interaction between 53BP1 and the mitotic regulators TPX2 and Aurora A that lead them to novel mechanistic insights about DNA double-stranded break repair regulation and a new fork protection pathway during replication stress.
The breast cancer suppressor protein BRCA1 is a well-known suppressor of the DNA repair protein 53BP1 and associated nonhomologous end joining (NHEJ) pathway. NHEJ indiscriminately sticks ends of DNA together. If not properly controlled during S phase, NHEJ contributes to gross genome instability, such as the aberrant fusion of chromosomes. In addition to being destructive to the genome, NHEJ is also a barrier to genome preservation via homologous recombination (HR). HR requires the suppression of NHEJ (1). A key determinant regulating the choice between HR and NHEJ pathways is the structure of the broken DNA end. NHEJ factors load on blunt DNA ends, whereas the recombination protein RAD51 loads when one end of a double stranded DNA fiber is removed or peeled back by a process called DNA end resection. The exposed DNA 3′tail-bound RAD51 fiber initiates a genome search to identify a suitable match to guide the accurate repair of the broken DNA. BRCA1 suppresses NHEJ by promoting DNA end resection that enables subsequent RAD51 loading in a process mediated by BRCA2 that also displaces the single-stranded DNA binding protein RPA (1). In this issue, Byrum et al. show that the TPX2/Aurora A kinase heterodimer complex that orchestrates mitotic spindle events (2) has unexpected functions that maintain genome integrity: the TPX2/Aurora A complex counteracts 53BP1 chromatin accumulation to ensure genome stability.
Starting with a mass spectrometry analysis of 53BP1 binding partners, Byrum et al. (3) identified an interaction between 53BP1 and TPX2 as well as its kinase partner Aurora A. After validating that 53BP1 binds to TPX2 in vitro, which then together interacts with Aurora A, the authors set out to analyze the function of this interaction in DNA double strand break repair. Byrum et al. (3) observed that TPX2/Aurora A promotes BRCA1 localization to breaks and that depletion of either TPX2 or Aurora A resulted in a significant decrease in both BRCA1 and RAD51 DNA damage-induced focal nuclear accumulation that was not due to cell cycle changes. Moreover, end resection as assessed by chromatin-bound RPA was significantly reduced in cells deficient for TPX2. As found in BRCA1-deficient cells, TPX2- or Aurora A–deficient cells displayed a shortening (degradation) of replication tracts when replication is stalled with hydroxyurea (HU). Consistent with loss of nascent DNA due to unregulated nuclease activity, subsequent inactivation of the nuclease MRE11 restored fork lengths. Thus, Byrum et al. (3) concluded that, similar to BRCA1 (4), the TPX2–53BP1 interaction protects newly replicated DNA from MRE11 nuclease degradation. This fork protection function is likely accomplished by the presence of TPX2 and Aurora A in replisomes, as determined by aniPOND (accelerated native isolation of proteins on nascent DNA [5]). Demonstrating the functional significance of fork protection, Byrum et al. (3) observed that cells deficient for TPX2/Aurora A are sensitive to HU and loss of 53BP1 significantly reduced HU sensitivity.
Despite these common functions, Byrum et al. (3) teased the contribution of TPX2/Aurora A apart from the BRCA1 pathway. Specifically, they showed that TPX2 and BRCA1 exist in distinct pathways that converge on 53BP1. The finding of distinct pathways explains why TPX2 depletion reduces BRCA1 and RAD51 foci, but not if 53BP1 is eliminated first. By suppressing 53BP1, TPX2/Aurora A facilitate BRCA1-dependent DNA end resection, RAD51 loading, and HR. Exemplifying separate pathways, unlike BRCA1, TPX2 directly binds 53BP1 and is not recruited to sites of DNA damage despite the TPX2–53BP1 interaction being stable in the presence of DNA damage. Given that TPX2 is not at sites of DNA damage, it follows that the localization of 53BP1 to chromatin or sites of DNA damage is unaffected by TPX2/Aurora A loss. Counterintuitively, Byrum et al. found that the region of 53BP1 binding to TPX2 also mediated 53BP1 localization to chromatin (3); however, evidence suggests there are additional TPX2 interaction domains on 53BP1.
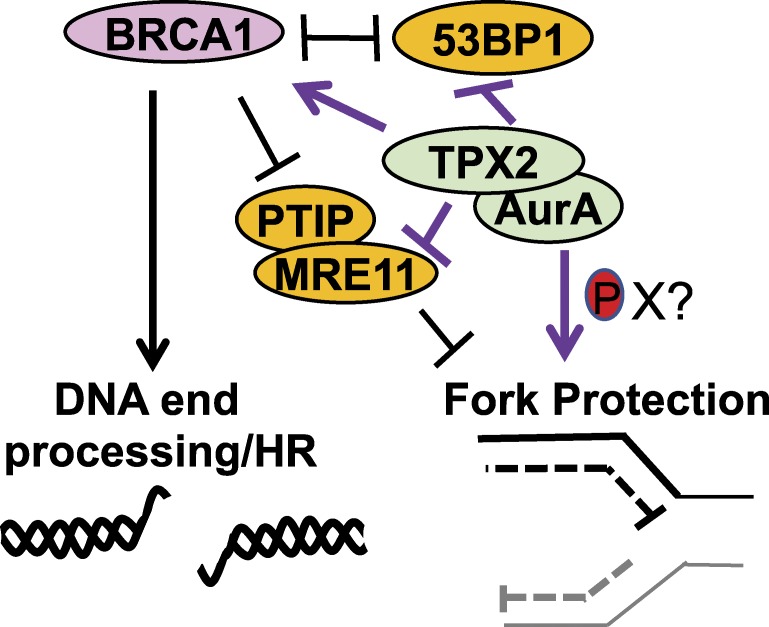
With fork protection, TPX2 and BRCA also play distinct roles. Most notably, unlike BRCA1, TPX2 suppresses 53BP1 at stalled replication forks as part of its fork protection function. As such, Byrum et al. observed that fork degradation due to TPX2 loss was rescued by the concurrent loss of 53BP1 (3). Specifically, knockout of 53BP1 partially and completely rescued track lengths in TPX2-depleted U2OS cells and in mouse embryonic fibroblasts, respectively. In direct contrast, fork degradation due to BRCA1 loss was not rescued by concurrent loss of 53BP1, as found here and published previously (6). However, fork degradation due to BRCA1 loss was rescued when PTIP, a factor required for MRE11 localization at stalled replication forks (6), was eliminated and MRE11 was restricted from accessing stalled forks. Also consistent with distinct pathways of fork protection, combined loss of TPX2 and BRCA1 generated an additive shortening of fork lengths (Fig. 1).
Figure 1.
TPX2/Aurora A promotes BRCA1-dependent double strand break repair and replication fork protection by counteracting 53BP1 and fork degradation by MRE11 in a pathway distinct from BRCA1.
In addition to directing break and fork activities in a parallel pathway to BRCA1, this TPX2 function is separate from its mitotic kinase activities. Notably, Byrum et al. provide data indicating that Aurora A catalytic activity and mitotic functions do not require the TPX2–53BP1 interaction (3). The 53BP1 interaction–defective mutant of TPX2 localized to mitotic spindles similar to wild-type TPX2. However, kinase activity was proposed to mediate TPX2 function in genome preservation, because the catalytically inactive Aurora A mutant did not rescue fork protection. Moreover, the Aurora A mutant disrupted for TPX2 binding failed to restore fork protection. Thus, Aurora A kinase activity may be directed to appropriate substrates through the TPX2–53BP1 complex.
Key questions remain: What are the Aurora A kinase substrates at the fork? Does the kinase also target HR factors? It is challenging to speculate, as the Aurora catalytic activity or TPX2 binding mutants were not evaluated for rescue of HR. While also not addressed, the parallel functions of TPX2/Aurora A and BRCA1 in both HR and fork protection could lead to a synthetic lethal relationship. Why is it important for a mitotic kinase to regulate HR and replication fork stability? One likely possibility is that modulating 53BP1 function is critical for cell fitness and therefore requires coordination through feedback with other cellular events. In particular, communication could be essential to ensure that DNA replication and its processing intermediates are suitable for the complex transactions that will take place in mitosis. Most importantly, these findings raise the promise of targeting the Aurora A kinase for improved chemo-sensitization of cancer cells, especially those with HR or fork protection defects.
Acknowledgments
This work was supported by National Institutes of Health grant R01 CA176166-01A1.
The author declares no competing financial interests.
References
- 1.Ceccaldi R., et al. . 2016. Trends Cell Biol. 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumayer G., et al. . 2014. Cell. Mol. Life Sci. 71:3027–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrum A., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201803003. [DOI] [Google Scholar]
- 4.Schlacher K., et al. . 2011. Cell. 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung K.H., et al. . 2013. Biotechniques. 55:204–206. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri A.R., et al. . 2016. Nature. 535:382–387.27443740 [Google Scholar]