FGF2 is a cell survival factor secreted by unconventional means. Dimou et al. visualize individual FGF2 translocation events at the plasma membrane by live cell TIRF microscopy, providing insight into the kinetics and the mechanism of this process.
Abstract
FGF2 is exported from cells by an unconventional secretory mechanism. Here, we directly visualized individual FGF2 membrane translocation events at the plasma membrane using live cell TIRF microscopy. This process was dependent on both PI(4,5)P2–mediated recruitment of FGF2 at the inner leaflet and heparan sulfates capturing FGF2 at the outer plasma membrane leaflet. By simultaneous imaging of both FGF2 membrane recruitment and the appearance of FGF2 at the cell surface, we revealed the kinetics of FGF2 membrane translocation in living cells with an average duration of ∼200 ms. Furthermore, we directly demonstrated FGF2 oligomers at the inner leaflet of living cells with a FGF2 dimer being the most prominent species. We propose this dimer to represent a key intermediate in the formation of higher FGF2 oligomers that form membrane pores and put forward a kinetic model explaining the mechanism by which membrane-inserted FGF2 oligomers serve as dynamic translocation intermediates during unconventional secretion of FGF2.
Introduction
In eukaryotes, the majority of extracellular proteins is secreted through the ER/Golgi–dependent secretory pathway (Palade, 1975; Rothman, 1994; Rothman and Wieland, 1996; Schekman and Orci, 1996). However, eukaryotic cells evolved additional mechanisms to transport proteins into the extracellular space that have collectively been termed unconventional protein secretion (Dimou and Nickel, 2018). One of the most prominent examples for proteins secreted by unconventional means is FGF2 (Steringer and Nickel, 2018), a cell survival factor involved in tumor-induced angiogenesis with a broad significance for malignancies of both solid and hematological cancers (Beenken and Mohammadi, 2009; Akl et al., 2016). Following secretion from tumor cells, FGF2 exerts its biological functions by forming a ternary signaling complex with FGF high-affinity receptors and heparan sulfates on the surface of target cells. Despite exerting its biological function in the extracellular space, FGF2 lacks a signal peptide and therefore does not have access to the classical ER/Golgi-dependent secretory pathway (La Venuta et al., 2015; Brough et al., 2017). Based on biochemical reconstitution experiments and biochemical bulk measurements of FGF2 secretion from cells, the unconventional secretory mechanism of FGF2 has been shown to depend on interactions of FGF2 with ATP1A1 (Zacherl et al., 2015), Tec kinase (Ebert et al., 2010; Steringer et al., 2012), and the phosphoinositide PI(4,5)P2 (Temmerman et al., 2008; Temmerman and Nickel, 2009; Steringer et al., 2012) at the inner leaflet as well as heparan sulfates at the outer leaflet (Zehe et al., 2006). Consistently, residues in FGF2 that mediate interactions with PI(4,5)P2 (K128, R129, and K133; Temmerman et al., 2008; Steringer et al., 2017) and heparan sulfates (K133; Zehe et al., 2006; Steringer et al., 2017) as well as the residue that is phosphorylated by Tec kinase (Y81; Ebert et al., 2010; Steringer et al., 2012) have been identified. In addition, two cysteine residues (C77 and C95) on the molecular surface of FGF2 have been demonstrated to play a critical role in PI(4,5)P2-dependent formation of membrane-inserted FGF2 oligomers (Müller et al., 2015). The latter have been shown to represent dynamic intermediates of FGF2 membrane translocation (Dimou and Nickel, 2018; Steringer and Nickel, 2018). Recently, key steps of the core mechanism of FGF2 membrane translocation have been reconstituted using an inside-out membrane model system based on giant unilammelar vesicles with entirely purified components (Steringer et al., 2017). Based on the combined findings summarized above, a model of FGF2 membrane translocation has been put forward in which FGF2 oligomers are assembled in a PI(4,5)P2-dependent manner at the inner leaflet to form membrane-spanning complexes and become disassembled by membrane proximal heparan sulfates at the cell surface (La Venuta et al., 2015; Brough et al., 2017). This model provides a compelling mechanism for directional transport of FGF2 from the cytoplasm into the extracellular space (Dimou and Nickel, 2018; Steringer and Nickel, 2018). However, as opposed to the ER/Golgi-dependent secretory pathway that has been imaged by total internal reflection fluorescence (TIRF) microscopy in living cells (Schmoranzer et al., 2000), the process of unconventional secretion for any type of cargo secreted in an unconventional manner has never been observed in living cells.
In this study, we have established a novel experimental system based on high-resolution TIRF microscopy that allows for quantification of single events of both FGF2 membrane recruitment at the inner leaflet and FGF2 translocation to the outer leaflet of the plasma membrane. The observed membrane translocation events were demonstrated to depend on both the phosphoinositide PI(4,5)P2 and cell surface heparan sulfates, validating previous findings from biochemical reconstitution experiments. Furthermore, a variant form of FGF2 lacking the cis elements required for PI(4,5)P2 binding, Tec kinase, mediated tyrosine phosphorylation as well as PI(4,5)P2-dependent oligomerization, and membrane insertion was neither recruited at the inner leaflet nor detectable on cell surfaces. Intriguingly, by simultaneous imaging of membrane recruitment and translocation, we directly visualized this process in real time for the first time for any type of unconventional secretion in eukaryotes on a single event basis revealing the kinetics of FGF2 membrane recruitment and translocation. Furthermore, we quantified the oligomeric state of FGF2 at the inner plasma membrane leaflet in intact cells. While both FGF2 monomers and higher FGF2 oligomers were detectable, the predominant species was found to be a FGF2 dimer. These findings directly link to recent biochemical reconstitution and molecular dynamics studies in which a FGF2 dimer was proposed to represent a key intermediate in FGF2 oligomerization and membrane insertion (Steringer et al., 2017). Our findings establish FGF2 membrane translocation to be mediated by a two-step event of two nonoverlapping physical processes characterized by slow and fast kinetics, respectively. We propose that these steps represent (a) PI(4,5)P2-dependent oligomerization concomitant with membrane insertion of FGF2 and (b) binding of heparan sulfates to FGF2 at the outer leaflet. Indeed, the characteristics of the observed translocation events provided direct evidence for the disassembly of membrane-inserted FGF2 oligomers resulting in FGF2 translocation to the cell surface. With these data from intact cells combined with previous findings from biochemical reconstitution experiments, we are proposing a model for FGF2 membrane translocation with a central role of membrane-inserted FGF2 oligomers as dynamic intermediates of this process (La Venuta et al., 2015; Brough et al., 2017; Dimou and Nickel, 2018; Steringer and Nickel, 2018).
Results
Establishing an experimental system to visualize and quantify individual events of FGF2 membrane recruitment and secretion in living cells
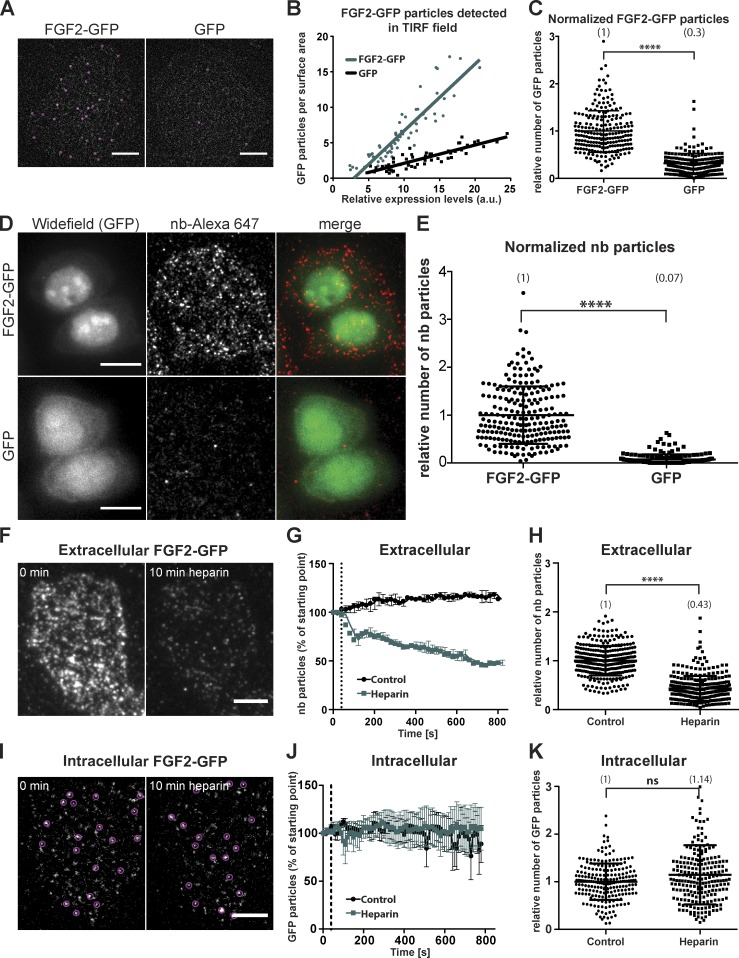
To establish an imaging system to visualize and quantify single events of FGF2 membrane recruitment and translocation to the cell surface, we studied living cells by TIRF microscopy. Stable cell lines were used that express either FGF2-GFP or GFP alone in a doxycycline-dependent manner. This allowed for titration of expression levels to establish conditions suitable for resolving recruitment of single particles of intracellular FGF2-GFP at the plasma membrane using TIRF microscopy. As shown in Fig. 1 A, individual spots of FGF2-GFP could be readily detected, suggesting association with the inner plasma membrane leaflet. By contrast, single GFP particles were only occasionally detected as defined spots in this area (Fig. 1 A and Video 1). This suggests that GFP in the cytoplasm moves too fast to be resolved as individual particles, resulting in a homogeneous background signal (Fig. 1 A and Video 1). For both FGF2-GFP and GFP, the number of individual particles per surface area was quantified and plotted as a function of relative protein expression (Fig. 1 B). This analysis revealed a linear correlation between the cellular expression level and the appearance of individual particles in the evanescent field of the TIRF setup. As expected, GFP particles were rarely observed, whereas FGF2-GFP was abundantly recruited to the plasma membrane. The data shown in Fig. 1 B were normalized by expression levels and statistically evaluated, revealing a highly significant difference between FGF2-GFP and the GFP control (Fig. 1 C). Beyond quantifying FGF2-GFP recruitment at the inner leaflet at the single-particle level, we also studied the mobility of FGF2-GFP at the plasma membrane (Fig. S1 A). Individual recruitment events could be visualized (Fig. S1 A, left panel), and single particles of FGF2-GFP were followed over time (Fig. S1 A, middle panel). Finally, the diffusional behavior of single FGF2-GFP particles was tracked, revealing different kinds of mobility (Fig. S1 A, right panels). In conclusion, a novel experimental system was established that allows for visualizing and quantifying single events of FGF2-GFP recruitment at the plasma membrane of intact cells.
Figure 1.
Establishing an experimental system to visualize and quantify individual events of FGF2 membrane recruitment at the plasma membrane and translocation to the extracellular space. (A) Single particles of FGF2-GFP were imaged in the vicinity of the plasma membrane using TIRF microscopy. Stable CHO-K1 cell lines expressing either FGF2-GFP or GFP in a doxycycline-dependent manner were used to detect GFP particles at the inner leaflet of the plasma membrane. The first frame of a time-lapse TIRF video is depicted. GFP particles were identified (circles) and quantified using the Fiji plugin TrackMate. Bar, 6 µm. (B) Single-cell analysis correlating the number of GFP particles at the inner leaflet of the plasma membrane with relative expression levels. Time-lapse TIRF videos with a total of 100 frames (80 ms/frame) were analyzed. GFP particles were quantified using the Fiji plugin TrackMate. Relative expression levels were analyzed measuring total GFP fluorescence at the first frame of each image sequence. The number of GFP particles was expressed as particles per surface area (µm2) and plotted as a function of the relative expression level of the corresponding cell. A minimum of 50 cells was analyzed for each protein. (C) Quantitative comparison of FGF2-GFP versus GFP particles at the plasma membrane in living cells. TIRF videos with a total of 300 frames (80 ms/frame) were analyzed. Raw data were analyzed using the Fiji plugin TrackMate. The number of GFP particles were normalized per surface area and relative expression levels of the corresponding cell (n > 200 cells for each condition). Mean values of each condition are given in brackets. An unpaired t test was used for statistical analysis (****, P < 0.0001). (D) CHO-K1 cells were induced with doxycycline to express FGF2-GFP or GFP for 24 h and incubated on ice for 30 min with Alexa Fluor 647–labeled anti-GFP nanobodies. Following fixation of cells, secreted FGF2-GFP bound to HSPGs on cell surfaces was imaged using both wide-field and TIRF microscopy for GFP and Alexa Fluor 647, respectively. Bar, 10 µm. (E) Quantification of single particles of FGF2-GFP and GFP at the outer leaflet of the plasma membrane using fluorescent anti-GFP nanobodies as described in D. Single particles detected per cell (n > 190) were analyzed using the Fiji plugin TrackMate. The mean values of each condition are shown in brackets with FGF2-GFP set to 1. An unpaired t test was used for statistical analysis (****, P < 0.0001). (F) Cells were induced for 24 h to express FGF2-GFP and labeled with fluorescent anti-GFP nanobodies for 30 min on ice. TIRF videos were acquired with frames of 20 s. After 40 s of image acquisition, heparin (1 mg/ml) was added. Representative images from the beginning and the end of the acquired videos are shown. Bar, 6 µm. (G) Quantification of nanobodies detected per frame for videos acquired as described in F. Three videos were analyzed per condition using the TrackMate Fiji plugin. The dotted line indicates the time point of heparin addition. The plotted data represent mean values ± SEM. (H) CHO-K1 cells induced to express FGF2-GFP with doxycycline for 24 h were treated with heparin (1 mg/ml) for 10 min or left untreated as a control. Cells were labeled with fluorescent anti-GFP nanobodies and fixed. TIRF images were acquired and analyzed using TrackMate for quantification of FGF2-GFP on cell surfaces (n > 300 cells per condition). The mean values of each condition are shown in brackets with FGF2-GFP set to 1. An unpaired t test was used for statistical analysis (****, P < 0.0001). (I) CHO-K1 cells were cultivated under conditions of low levels of FGF2-GFP expression that allow for single particle detection based on GFP fluorescence. TIRF videos were acquired with frames of 20 s, and heparin (1 mg/ml) was added 40 s after the start of data acquisition. Representative images from the beginning and the end of the acquired videos are shown, depicting GFP particles at the inner leaflet of the plasma membrane (circles) before and after addition of heparin. Bar, 6 µm. (J) Quantification of GFP particles detected per frame for videos acquired as described in I. Three videos were analyzed per condition using the TrackMate Fiji plugin. The plotted data represent mean values ± SEM. (K) CHO-K1 cells were induced to express FGF2-GFP in low levels that allow for single particle detection. TIRF videos (80 ms/frame) were acquired before and after addition of heparin (1 mg/ml) for at least 10 min. The GFP particles at the inner plasma membrane leaflet were quantified using the Fiji plugin TrackMate. For each condition, 50 frames were analyzed (n > 200 cells per condition). The mean values of each condition are shown in brackets with FGF2-GFP set to 1. An unpaired t test was used for statistical analysis. ns, not significant.
Beyond studying FGF2-GFP recruitment at the plasma membrane (Fig. 1, A–C; and Fig. S1 A), we established a procedure to quantify individual FGF2-GFP particles at the outer leaflet, i.e., following membrane translocation into the extracellular space (Fig. 1, D–K). Using Alexa Fluor 647–labeled anti-GFP nanobodies added to the extracellular medium, individual FGF2-GFP particles were detected on the cell surface of living cells (Fig. 1 D). By contrast, in cells expressing GFP alone, binding of anti-GFP nanobodies to the cell surface was almost undetectable (Fig. 1 D and Video 2). This observation was corroborated by normalization and a statistical analysis, demonstrating FGF2-GFP membrane translocation across the plasma membrane to be a highly specific process (Fig. 1 E). The extracellular localization of FGF2-GFP particles was not only defined through detection by anti-GFP nanobodies but also validated by treating cells with heparin, a heparan sulfate mimetic that elutes FGF2 from cell surface heparan sulfate proteoglycans (Florkiewicz et al., 1998; Dahl et al., 2000; Engling et al., 2002). As shown in Fig. 1 F and Video 3 as well as quantified in Fig. 1, G and H, the majority of FGF2-GFP particles detected by anti-GFP nanobodies on cell surfaces was removable by heparin treatment. By contrast, a mock treatment for the same incubation time revealed a stable association of FGF2-GFP with cell surfaces (Fig. 1 G and Fig. S1 B). As demonstrated in Fig. 1 I and quantified in Fig. 1, J and K, the total population of FGF2-GFP detected within the evanescent TIRF field based on GFP fluorescence was insensitive to heparin treatment when heparin and mock-treated conditions were compared (Fig. 1, I–K; and Fig. S1 C). This indicates that the majority of the FGF2-GFP is associated with the inner plasma membrane leaflet. In conclusion, the combined findings documented in Fig. 1 and Fig. S1 as well as in Videos 1, 2, and 3 establish a novel experimental system that allows for quantifying single events of both FGF2-GFP membrane recruitment at the inner leaflet and the appearance of secreted FGF2-GFP at the outer plasma membrane leaflet using TIRF microscopy.
FGF2-GFP membrane translocation to cell surfaces depends on membrane-proximal heparan sulfates
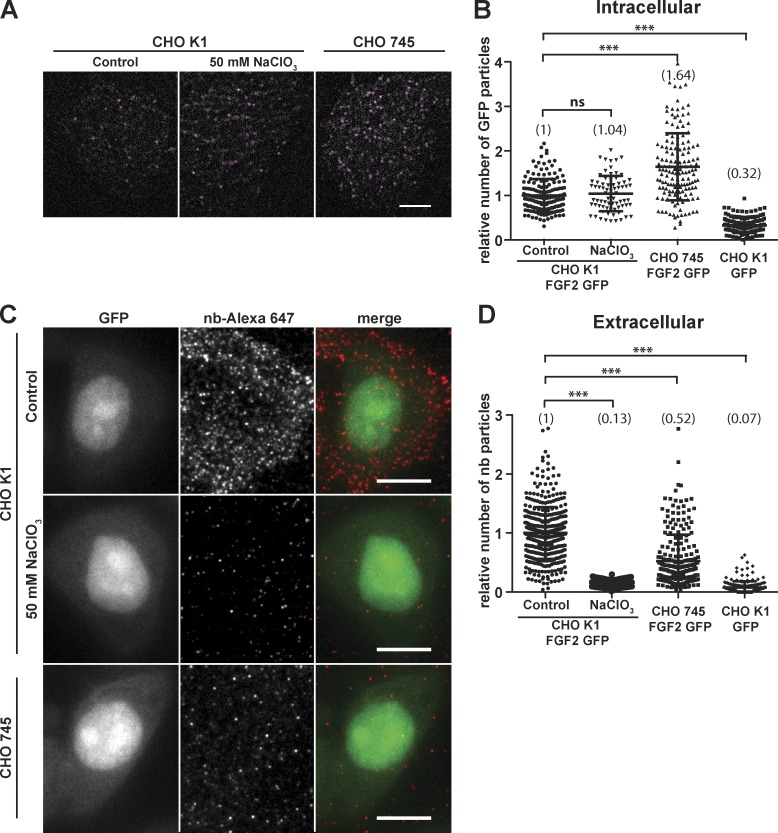
Using the experimental system described above, we tested whether FGF2-GFP membrane recruitment at the inner leaflet and translocation to the cell surface depend on heparan sulfates (Fig. 2). Three conditions were compared with (a) CHO-K1 WT cells, (b) CHO-K1 WT cells treated with NaClO3 to prevent post-translational sulfation of the carbohydrate chains of heparan sulfate proteoglycans (Safaiyan et al., 1999), and (c) CHO-745 mutant cells with a defect in xylosyltransferase I, the enzyme that catalyzes the first step of the biosynthesis of heparan sulfates as part of heparan sulfate proteoglycans (HSPGs; Esko et al., 1985). For all three conditions, both the presence of individual FGF2-GFP particles at the inner leaflet (Fig. 2, A and B, intracellular) and FGF2-GFP particles at the cell surface (Fig. 2, C and D, extracellular) were quantified. These studies revealed that HSPGs are not required for FGF2-GFP recruitment at the inner leaflet since treatment of CHO-K1 cells with NaClO3 did not affect this process (Fig. 2 B). Strikingly, in the complete absence of cell surface heparan sulfates in CHO-745 cells, the number of FGF2-GFP particles recruited to the inner plasma membrane leaflet even increased (Fig. 2 B and Video 4), corroborating HSPG-independent recruitment of FGF2 to the plasma membrane and the nature of membrane-inserted forms of FGF2 as intermediates in membrane translocation to the cell surface. By contrast to membrane recruitment, FGF2-GFP membrane translocation to the cell surface was severely impaired when functional HSPGs were missing through either NaClO3 treatment of CHO-K1 cells or the use of CHO-745 mutant cells (Fig. 2, C and D), demonstrating HSPGs to be essential for the completion of FGF2-GFP membrane translocation into the extracellular space. In conclusion, the combined findings shown in Fig. 2 and Video 4 establish that cell surface HSPGs are not required for FGF2 recruitment at the inner leaflet in intact cells but are critical for membrane translocation as the final step of unconventional secretion of FGF2.
Figure 2.
Unconventional secretion of FGF2-GFP depends on cell surface heparan sulfate proteoglycans. (A) Individual FGF2-GFP particles at the inner plasma membrane leaflet were imaged based on GFP fluorescence using TIRF microscopy as described in the legend to Fig. 1 and in Materials and methods. This analysis included CHO-K1 WT cells (control; a), CHO-K1 WT cells treated with 50 mM NaClO3 to inhibit post-translational sulfation (b), and CHO-745 mutant cells that express only the core protein without heparan sulfate chains (c). For each condition indicated, the first frame of TIRF videos is shown. Individual particles were identified using the TrackMate Fiji plugin (circles). Bar, 10 µm. (B) Quantification of FGF2-GFP particles at the inner plasma membrane leaflet under the conditions shown in A. TIRF videos of 300 frames (80 ms/frame) were analyzed using the plugin TrackMate (n > 70 cells per condition). The mean values of each condition are shown in brackets with FGF2-GFP set to 1. The statistical analysis was based on a one-way ANOVA test combined with Tukey’s post hoc test (ns, P ≥ 0.05; ***, P ≤ 0.001). (C) Individual FGF2-GFP particles on cell surfaces were imaged using fluorescent anti-GFP nanobodies. This analysis was done under the same experimental conditions shown in A. Cells were induced to express FGF2-GFP for 24 h followed by incubation with Alexa Fluor 647–labeled anti-GFP nanobodies for 30 min on ice. Cells were fixed, and images were acquired using both wide-field (GFP fluorescence) and TIRF microscopy (FGF2-GFP cell surface population). Bar, 10 µm. (D) Quantification of individual FGF2-GFP particles on cell surfaces under the conditions described in the legend to C. Data were processed using the Fiji plugin TrackMate (n > 144 cells per condition). For each condition, the mean value is shown in brackets with FGF2-GFP set to 1. The statistical analysis was based on a one-way ANOVA test combined with Tukey’s post hoc test (***, P ≤ 0.001).
FGF2-GFP membrane recruitment and translocation depend on the ability of FGF2 to interact with PI(4,5)P2
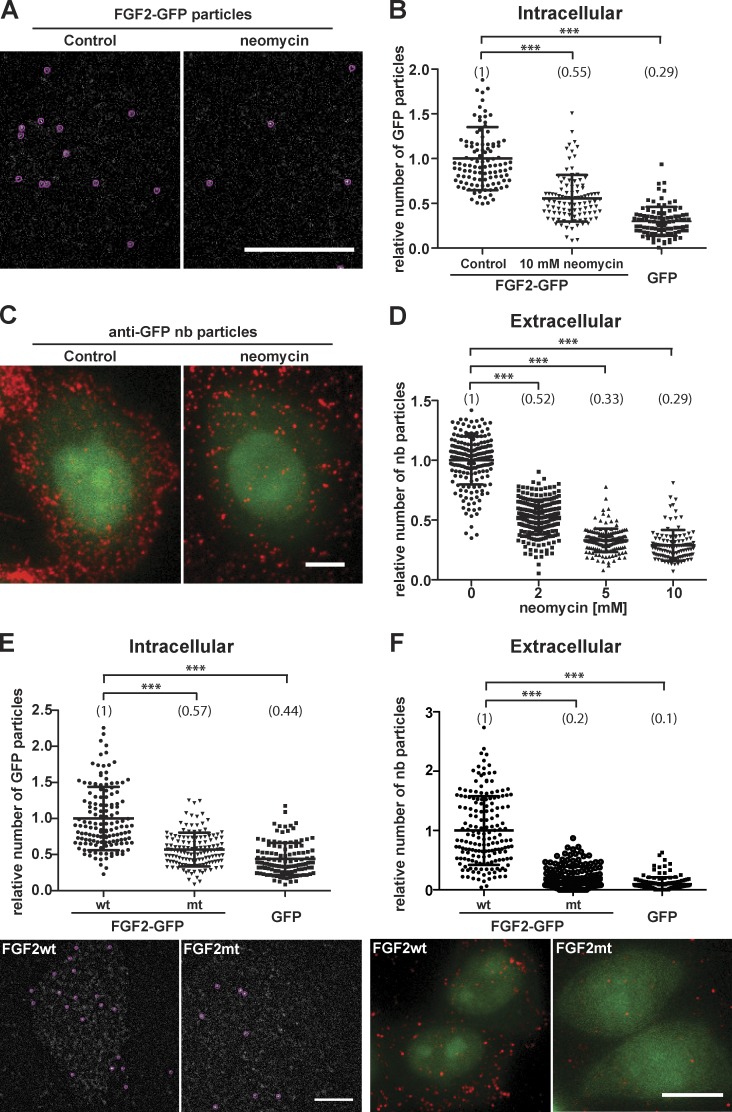
Beyond heparan sulfate proteoglycans (Fig. 2), we analyzed a role for PI(4,5)P2 in FGF2 membrane translocation (Fig. 3). First, we used the polybasic antibiotic neomycin that is known to block interactions of proteins with phosphoinositides (Arbuzova et al., 2000; Kwik et al., 2003; Temmerman et al., 2008). As shown in Fig. 3 (A and B), neomycin efficiently inhibited FGF2-GFP recruitment to the inner plasma membrane leaflet. Consistently, when we probed for FGF2-GFP particles on cell surfaces using anti-GFP nanobodies labeled with Alexa Fluor 647, we found FGF2-GFP translocation to the cell surface to be severely impaired by neomycin in a concentration-dependent manner (Fig. 3, C and D). As a second approach, we analyzed a FGF2 variant form termed FGF2-mt (FGF2-C77,95A/Y81F/KRK128,129,133QQQ) that we have previously demonstrated to be defective in both binding to PI(4,5)P2 and transport into the extracellular space using biochemical bulk secretion experiments (Temmerman et al., 2008; Temmerman and Nickel, 2009; Ebert et al., 2010; Müller et al., 2015). As shown in Fig. 3 E, membrane recruitment of FGF2-mt–GFP particles was markedly reduced when FGF2-mt–GFP was compared with the WT form of FGF2-GFP. Likewise, FGF2-mt–GFP was severely impaired in membrane translocation to the cell surface (Fig. 3 F). When compared with GFP alone as a negative control, both membrane recruitment and translocation to the cell surface of FGF2-mt–GFP were found to be close to background levels. In conclusion, the experiments shown in Figs. 2 and 3 demonstrate that individual events of membrane recruitment and translocation to the cell surface in intact cells depend on PI(4,5)P2 at the inner leaflet as well as membrane proximal heparan sulfates at the outer leaflet as demonstrated by live-cell TIRF microscopy.
Figure 3.
FGF2 membrane recruitment and translocation to cell surfaces depend on interactions of FGF2 with PI(4,5)P2 at the inner leaflet. (A) CHO-K1 cells were imaged expressing FGF2-GFP at low levels to allow for single particle detection. Where indicated, cells were treated for 3 h with 10 mM neomycin. Single FGF2-GFP particles were identified at the inner plasma membrane leaflet (red circles). The first frames of TIRF videos are shown. Bar, 6 µm. (B) Quantification of FGF2-GFP recruitment at the inner leaflet of the plasma membrane under the conditions shown in A. TIRF videos of CHO-K1 cells expressing FGF2-GFP were acquired (300 frames; 80 ms/frame). The number of GFP particles at the inner plasma membrane leaflet was quantified using the Fiji plugin TrackMate (n > 95 cells per condition). The mean value of FGF2-GFP particles for each condition is given in brackets with the control set to 1. The statistical analysis was based on a one-way ANOVA test combined with Tukey’s post hoc test (***, P ≤ 0.001). (C) Identification of single FGF2-GFP particles at the outer plasma membrane leaflet. CHO-K1 cells expressing FGF2-GFP were cultivated in the presence or absence of 5 mM neomycin as indicated. Following incubation for 24 h in the presence of doxycycline to induce FGF2-GFP expression, intact cells were stained for FGF2-GFP on cell surfaces using anti-GFP nanobodies coupled to Alexa Fluor 647. Afterward, cells were fixed and imaged using both wide-field (green channel) and TIRF microscopy (red channel), the latter identifying single FGF2-GFP particles on cell surfaces. Bar, 6 µm. (D) Quantification of single FGF2-GFP particles on cell surfaces under the conditions shown in C including a titration of neomycin at 2, 5, and 10 mM. Raw data were analyzed using the Fiji plugin TrackMate (n > 108 cells per condition). The mean value of FGF2-GFP particles on cell surfaces for each condition is given in brackets with the control set to 1. The statistical analysis was based on a one-way ANOVA test combined with Tukey’s post hoc test (****, P ≤ 0.0001). (E) CHO-K1 cells were induced to express either FGF2-GFP, FGF2mt-GFP, a secretion deficient variant form of FGF2-GFP (C77/95A, Y81F; KRK128/129/133QQQ), or GFP at levels that allow for single-particle detection at the inner plasma membrane leaflet. Time series of 300 frames (80 ms/frame) were acquired, and for each condition, single particles were identified and quantified. The first frames of TIRF videos are displayed. (Bar, 6 µm). The mean value of FGF2-GFP particles at the inner leaflet for each condition is given in brackets with the control set to 1. The statistical analysis was based on a one-way ANOVA test combined with Tukey’s post hoc test (***, P ≤ 0.001). (F) CHO-K1 cells were induced for 24 h with doxycycline to express either FGF2-GFP, FGF2mt-GFP, a secretion-deficient variant form of FGF2-GFP (C77/95A, Y81F; KRK128/129/133QQQ), or GFP. Intact cells were stained for FGF2-GFP on cell surfaces using anti-GFP nanobodies coupled to Alexa Fluor 647. Afterward, cells were fixed and imaged using both wide-field (green channel) and TIRF microscopy (red channel), the latter identifying single FGF2-GFP particles on cell surfaces (bar, 6 µm). The number of single particles on cell surfaces was analyzed using the Fiji plugin TrackMate (n > 95 cells per condition). The mean value for each condition is displayed in brackets with the WT form of FGF2-GFP set to 1. The statistical analysis was based on a one-way ANOVA test combined with Tukey’s post hoc test (***, P ≤ 0.001).
Direct visualization of individual events of FGF2 membrane translocation to the cell surface of living cells
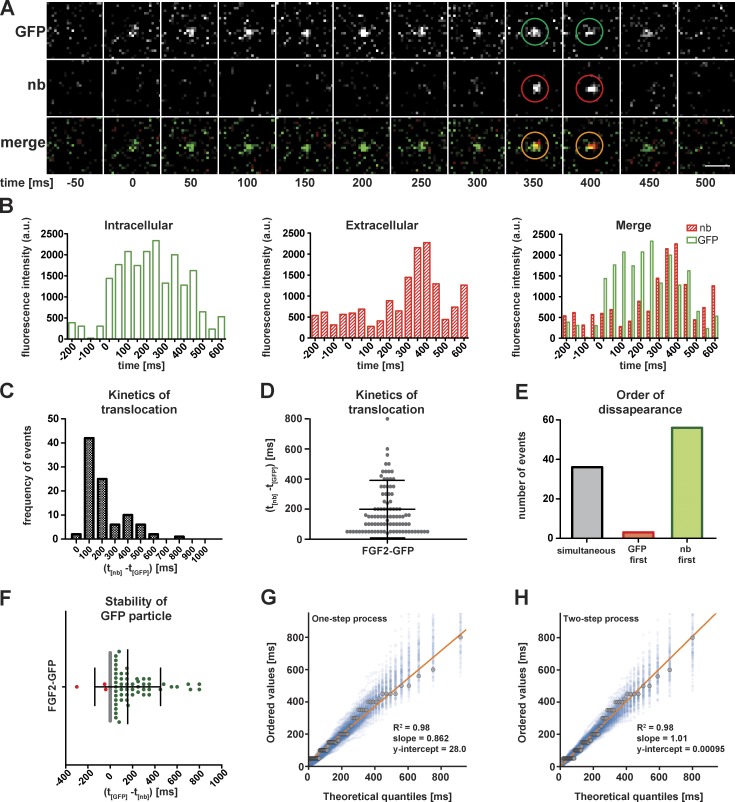
Using the experimental system described above, the key aim of this study was to visualize individual secretion events of FGF2 in intact cells. In particular, we intended to image FGF2 membrane recruitment and translocation simultaneously, capturing FGF2-GFP at the inner leaflet at specific spots and following it up in space and time until translocation into the extracellular space occurred. With time point zero being defined by the appearance of FGF2-GFP at the inner leaflet, we followed their fate over time by simultaneous imaging of both GFP fluorescence and Alexa Fluor 647 fluorescence derived from anti-GFP nanobodies in the cell culture medium, the latter capturing FGF2-GFP upon appearance at the cell surface. Thus, individual translocation events were defined as a sequence of the appearance of FGF2-GFP at a specific spot at the inner leaflet followed by anti-GFP nanobody recruitment at the outer leaflet at the very same position. Individual events of FGF2 membrane translocation were detected such as the example given in Fig. 4 A and Video 5. As shown in the bar histogram of Fig. 4 B, by plotting fluorescence intensity as a function of time, sequential steps of recruitment and translocation of FGF2-GFP could be monitored. Based on this setup, the time intervals between membrane recruitment and the appearance of FGF2-GFP at the cell surface were measured for 95 individual events of FGF2-GFP secretion from living cells. Most translocation events occurred within a time range of less than 100 to 200 ms (Fig. 4 C) with an average value of less than 200 ms (Fig. 4 D). Following translocation, nanobody fluorescence disappeared again within a relatively constant time span of ∼50 ms (Fig. 4, E and F; and Fig. S2). This indicates that, following translocation of FGF2-GFP to HSPGs on the cell surface, physical complexes of FGF2-GFP and anti-GFP nanobodies redistribute across heparan sulfate chains, resulting in intercellular spreading of FGF2-GFP (Zehe et al., 2006; Duchesne et al., 2012). Thus, following a successful membrane translocation event, nanobody fluorescence is lost at these sites as individual FGF2-GFP/anti-FGF2-GFP nanobody complexes are exchanged between cells. However, in most cases, even after the disappearance of nanobody fluorescence, sites of FGF2 membrane translocation remained positive for GFP fluorescence for extended amounts of time of up to 800 ms (Fig. 4 F and Fig. S2 A). We propose these GFP signals to be derived from oligomeric species of FGF2-GFP within the plasma membrane that serve as membrane spanning intermediates of FGF2 membrane translocation (model, Fig. 5 I). In some cases, at a specific site of the plasma membrane, even two sequential events of nanobody binding and dissociation could be observed with continuous GFP fluorescence being detected at the original site of FGF2-GFP membrane recruitment (Fig. S2 B). This observation suggests that membrane translocation of FGF2-GFP molecules into the extracellular space can occur repeatedly from one specific site in the plasma membrane, again pointing at a role for membrane-inserted FGF2-GFP oligomers as dynamic intermediates of this process. These findings are consistent with previously published biochemical reconstitution experiments (Steringer et al., 2012, 2017; Müller et al., 2015), suggesting that heparan sulfates disassemble membrane-inserted FGF2 oligomers at the outer leaflet, causing the release of FGF2-GFP units into the extracellular space.
Figure 4.
Direct visualization of individual events of FGF2 membrane translocation to the cell surface of living cells. (A) Representative example of a FGF2-GFP membrane recruitment and translocation event. TIRF imaging was performed on living cells expressing FGF2-GFP in the presence of Alexa Fluor 647–labeled anti-GFP nanobodies to detect extracellular FGF2-GFP. Successive frames (50 ms/frame) from a zoomed-in area of interest are shown. The time point of FGF2-GFP membrane recruitment was set to 0. Bar, 1 µm. (B) Bar histograms of the fluorescence intensity of the FGF2-GFP translocation event shown in A as a function of time. (C and D) A total of 95 FGF2-GFP translocation events were analyzed. The time intervals between the appearance of FGF2-GFP particles within the evanescent field and nanobody binding at the cell surface were calculated (t(nb) − t(GFP)). The distribution of observed time intervals was plotted. (E) FGF2-GFP translocation events from C and D were classified into three categories depending on the order of disappearance of GFP and Alexa Fluor 647 nanobody (nb) particles following completion of FGF2-GFP membrane translocation: (a) FGF2-GFP and Alexa Fluor 647 nanobody particles disappear simultaneously (gray), (b) GFP particles disappear before Alexa Fluor 647 nanobody particles (red), and (c) Alexa Fluor 647 nanobody particles disappear before GFP particles (green). (F) For each translocation event from C and D, the duration of GFP fluorescence remaining after nanobody disappearance in the observed position was calculated. The color code corresponds to what is shown in E. (G and H) Probability plots used to test whether one- or two-step exponential processes fit the time intervals measured between FGF2-GFP recruitment at the inner leaflet and the appearance of single FGF2-GFP molecules at the outer plasma membrane leaflet as detected by binding of anti-GFP nanobodies. Black dots correspond to measured data. The blue dots are the outcome of stochastic simulations with the hypothesized probability distribution for each case.
Figure 5.
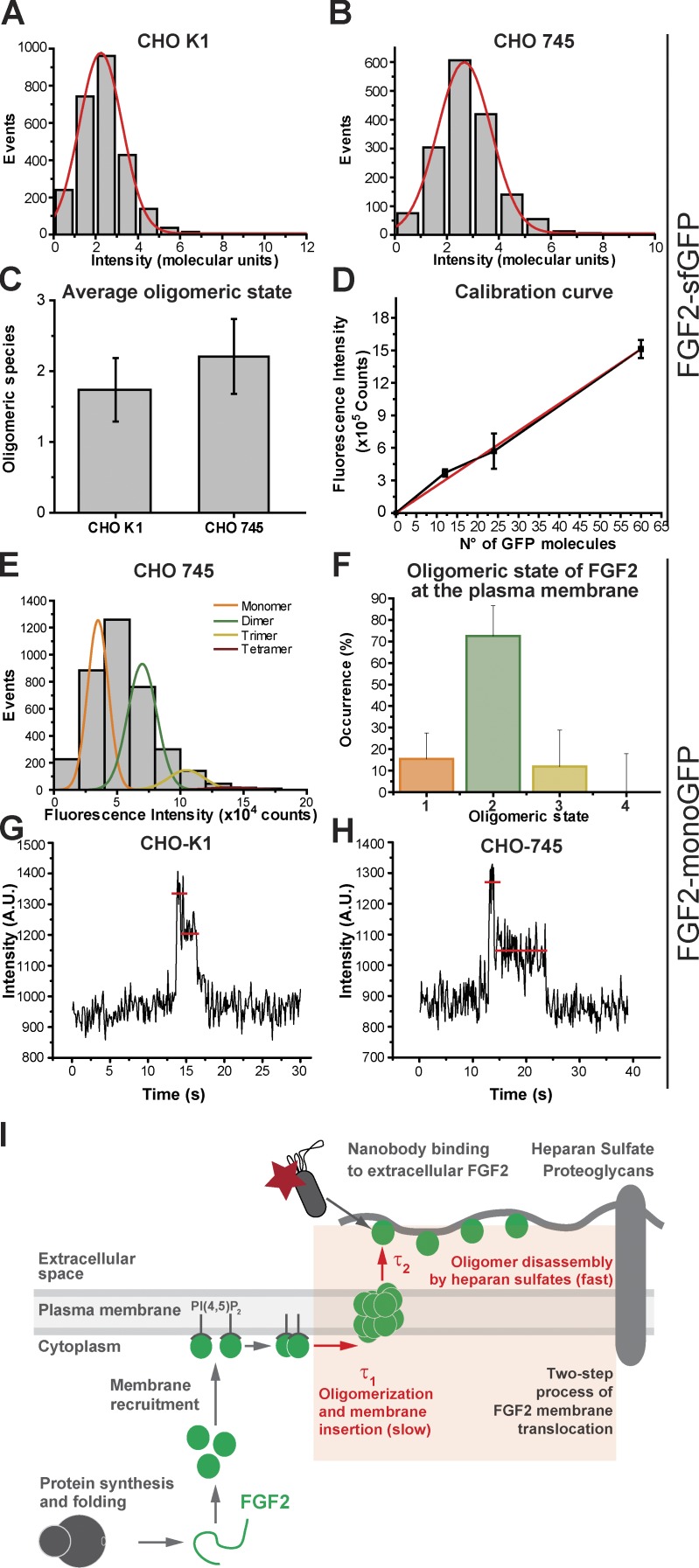
Determination of the oligomeric size distribution of FGF2-GFP by single particle brightness analysis. (A and B) Representative histograms of the fluorescence intensity distribution from individual experiments. CHO-K1 WT and CHO-745 mutant cells expressing FGF2-sfGFP at low levels were imaged to allow for single particle detection. Single FGF2-sfGFP particles localizing at the plasma membrane were identified, and their fluorescence intensity was measured and plotted. Three independent experiments were conducted with each of them including the analysis of at least 30 cells. (C) Comparison of the oligomeric state of FGF2-sfGFP at the inner plasma membrane leaflet of CHO-K1 and CHO-745 cells. The plotted data represent mean values (± SD based on three independent experiments). (D) Calibration curve of sfGFP oligomeric standards. Correlation between the mean fluorescence intensity and the sfGFP copy number for standard oligomers with different subunit numbers. Mean values and SDs from two independent replicates are shown. The red line represents the linear fitting of the data (r2 = 0.997). The slope of the curve corresponds to the theoretical fluorescence intensity value of a single sfGFP molecule. (E) Intensity distribution of FGF2-mGFP at the plasma membrane of CHO-745 cells from a representative experiment. Approximately 3,500 particles were analyzed. The resulting histograms were fitted with a sum of Gaussians to estimate the occurrence of monomers (orange), dimers (green), trimers (yellow), and tetramers (magenta). The area under each curve was used to calculate the percentage of occurrence of each oligomeric species. (F) Percentage of FGF2-mGFP monomers, dimers, trimers, and tetramers calculated after labeling correction from the averaged distributions of species from two different experiments. (G and H) Fluorescence intensity plots of two representative individual FGF2-mGFP particles in CHO-K1 and CHO-745 cells showing two sequential photobleaching steps. (I) A model proposing a two-step process of FGF2 membrane translocation with a slow component (FGF2 oligomerization and membrane insertion) and a subsequent fast component (FGF2 capturing at the outer leaflet mediated by heparan sulfates).
Estimating characteristic times of FGF2 membrane translocation events using stochastic modeling
To gain further insight into the unconventional secretory mechanism of FGF2, we measured the kinetics of FGF2 membrane translocation, i.e., the time required for FGF2-GFP to physically cross the plasma membrane. Based on the experimental design used in this study, these measurements precisely determined the time intervals between FGF2-GFP membrane recruitment at the inner leaflet and FGF2-GFP appearance at the outer leaflet based on FGF2-GFP detection by extracellular anti-GFP nanobodies. To make sure that our measurements were not limited by the time required for nanobody binding to FGF2-GFP, we first constructed a simple model governing the diffusion of nanobodies in the extracellular environment (see Materials and methods). Information was extracted on the time it takes for the nearest nanobody to reach an FGF2-GFP molecule on membrane-proximal heparan sulfates. With the nanobodies used at a final concentration of 5 nM and considering their small size with a diameter of ∼5 nm, the average time for the nearest nanobody to reach a translocation site through Brownian motion was found to be <1 ms. Taking into account the meshwork of matrix proteins on cell surfaces that may cause limitations in the accessibility of FGF2-GFP by anti-GFP nanobodies, we preincubated cells with anti-GFP nanobodies to allow for their equilibration and homogenous distribution within the protein meshwork on cell surfaces. Since the kinetics of nanobody binding to FGF2-GFP were calculated to be at least two orders of magnitude faster than the observed timescales of FGF2-GFP membrane translocation, the time interval measurements for capturing of FGF2-GFP by anti-GFP nanobodies accurately reflect the kinetics of FGF2 membrane translocation in intact cells. To analyze the considerable range of values between <100 to 800 ms (Fig. 4, C and D), we used the so-called Poisson process as a model for the distribution of waiting time values. It assumes that the waiting time between each pair of binding events follows a single-exponential probability density. Its form is defined by the mean waiting time (inverse binding rate). We also considered a more complex model in which FGF2 membrane translocation events are composed of two sequential exponential processes. Probability plots were used to test these two hypotheses against the measured waiting time data (Fig. 4, G and H). The probability plot shows the ordered values of the measured data versus theoretical quantiles predicted by the proposed stochastic model (Millar, 2011). If a sample is indeed the outcome of a given stochastic process, it will appear on the diagonal line y = x in the corresponding probability plot. In addition to the empirical data, the outcome of 200 stochastic simulations of the one- and two-step processes, respectively, was added to the probability plots (blue dots in Fig. 4, G and H). These points define envelopes that mark regions of good fit. The probability plots of Fig. 4, G and H, reveal that both one- and two-step processes seem to be good fits to the measured waiting times; however, the two-step process with a slope of 1.01 and a smaller y-intercept is a significantly better model. The average waiting time for the one-step process is 186 ms, which simply coincides with the average of the measured data. However, the two-step process divides the time interval into two distinguishable waiting times with average values of 156 ms and 30 ms. To further demonstrate the advantage of the two-step model over its one-step counterpart, we also performed the two-tailed Kolmogorov–Smirnov test for goodness of fit in each case and compared the resulting P values (note in this specific test, the higher the P value, the better the goodness of fit; Stephens, 1974). For the one-step process, the Kolmogorov–Smirnov test results in the P value of 0.0004. By contrast, the two-step process yields a significantly higher P value of 0.025, demonstrating a better fit of the experimental observations to the simulated data. Thus, the two-step process is statistically preferable to the one-step process. Our results further suggest the kinetics of FGF2 membrane translocation events to consist of a slow and a fast physical process with characteristic time intervals of 156 and 30 ms, respectively. However, due to the symmetry of the occurrence of these two events in the proposed two-step model, the sequence in which they occur cannot be determined.
Determination of the oligomeric state of FGF2-GFP at the inner plasma membrane leaflet in intact cells
Based on the observations discussed above, we further aimed at determining the oligomeric size distribution of FGF2-GFP at the inner leaflet of the plasma membrane in intact CHO-K1 (WT) and CHO-745 mutant cells (Fig. 5). As detailed in Materials and methods, two independent techniques with (a) a ratiometric analysis (Coffman and Wu, 2012) and (b) a brightness analysis (Schmidt et al., 1996) were used to correlate the fluorescent intensity of individual FGF2-GFP particles with the number of FGF2-GFP subunits they contain (Ulbrich and Isacoff, 2007; Coffman and Wu, 2012; Subburaj et al., 2015a,b). For the ratiometric approach (Fig. 5, A–D), FGF2 was fused to superfolder GFP (sfGFP), which is a fast-maturing monomeric variant of EGFP. The theoretical value of a single particle of sfGFP was calculated as the slope of a calibration curve that was obtained from defined standard oligomers of sfGFP containing 12, 24, and 60 subunits, respectively (Fig. 5 D). In Fig. 5 (A and B), the distribution of the intensity, in molecular units, of individual particles of FGF2-sfGFP is shown for CHO WT and CHO-745 cells. The molecularity of FGF2-sfGFP particles was calculated based on the ratio of fluorescence intensity of FGF2-sfGFP particles and a unit of sfGFP. The average oligomeric size of FGF2-sfGFP particles was found to be 1.75 ± 0.45 in CHO-K1 cells and 2.2 ± 0.53 in CHO-745 mutant cells (Fig. 5 C). This analysis was validated in CHO-745 cells by calculating the oligomeric state of FGF2–monomeric GFP (mGFP; containing a single-point amino acid substitution preventing GFP-induced oligomerization [mGFP = EGFP-A206K]), based on a brightness comparison to defined mGFP monomers (Subburaj et al., 2015a; Fig. 5, E and F). Fig. 5 E displays the distribution of the fluorescence of individual particles of FGF2-mGFP and its fitting to multiple Gaussians, in which each individual curve corresponds to a particle population of different stoichiometry (1-, 2-, 3-, and 4-mers). The area under each curve obtained by the fitting was used to calculate the occurrence of each species. This value was further corrected taking into account that only 70% of mGFPs are fluorescent (Fig. 5 F), as previously described (Ulbrich and Isacoff, 2007; Subburaj et al., 2015b). While FGF2-mGFP monomers and higher oligomers were detectable, this analysis revealed ∼70% of the total population of FGF2-mGFP particles at the inner plasma membrane leaflet to be represented by dimers. Since FGF2-mGFP monomers were also detectable, we propose that FGF2-mGFP gets recruited at the inner leaflet as a monomer followed by rapid dimerization. Finally, the existence of FGF2-GFP dimers at the plasma membrane was validated by step-wise photobleaching experiments (Ulbrich and Isacoff, 2007) using cells that were gently permeabilized to remove cytosolic FGF2-mGFP. While imaging individual FGF2-mGFP–positive spots, we indeed found evidence for the presence of FGF2-mGFP dimers based on a characteristic two-step bleaching behavior indicating the presence of two mGFP molecules in the same spot (Fig. 5, G and H).
A key role for PI(4,5)P2-induced FGF2 dimers as intermediates in unconventional secretion of FGF2 from intact cells has indeed been predicted by molecular dynamics simulations (Steringer et al., 2017). However, while dimers, trimers, and tetramers could be observed in intact cells in the current study, an accumulation of higher oligomers of FGF2-mGFP could be detected neither in CHO WT nor in HSPG-deficient CHO mutant cells. On the one hand, based on these findings, one may conclude that FGF2 dimers as the most abundant species may represent the translocation intermediates in intact cells. On the other hand, a rich body of evidence derived from biochemical reconstitution experiments suggests FGF2 oligomers with 8–12 subunits to represent the key intermediates of FGF2 membrane translocation (Steringer et al., 2012, 2017; Müller et al., 2015). These results were further supported by cell-based secretion experiments demonstrating higher oligomers to be required for FGF2 transport into the extracellular space (Müller et al., 2015). Yet the current study did not detect higher FGF2 oligomers with 8–12 subunits in intact cells. It is possible that limitations of our experimental setup prevent the detection of higher FGF2 oligomers that have been proposed to be highly dynamic and transient in nature. Furthermore, FGF2 secretion is a relatively infrequent process that does not lead to the massive secretion of FGF2 from cells. Therefore, oligomeric intermediates of FGF2 membrane translocation might represent rare species in intact cells that are difficult to detect. Thus, the majority of FGF2 dimers and trimers identified in this study may represent only precursors of the true translocation intermediates. In any case, beyond the reconstitution studies mentioned above, the identification of the oligomeric state of the functional intermediates of FGF2 membrane translocation in intact cells remains a key challenge for future studies on the molecular mechanism of the unconventional secretory pathway of FGF2.
Discussion
In summary, using live cell TIRF microscopy to image single events of FGF2 membrane translocation, we kinetically dissected the unconventional secretory pathway of FGF2 into two discrete steps of FGF2 membrane recruitment and translocation in intact cells (Fig. 5 I). The current study further provides a detailed kinetic analysis of FGF2 membrane translocation, revealing this process to be extremely fast with an average time interval of less than 200 ms between FGF2 membrane recruitment at the inner leaflet and FGF2 appearance in the extracellular space. To the best of our knowledge, corresponding data on other types of protein translocation across membranes such as protein import into peroxisomes and mitochondria as well as post-translational transport into the ER do not exist in the literature. When comparing FGF2 membrane translocation with cotranslational protein translocation into the ER, the latter is relatively slow since it is limited by the rate of translation with ∼6 amino acids incorporated into nascent polypeptides per second (Ingolia et al., 2011). In addition, following translocation into the ER, it takes 30–60 min for classical secretory proteins to travel along the ER/Golgi-dependent pathway until they reach the extracellular space (Matlin and Simons, 1983; Rivera et al., 2000; Boncompain et al., 2012). Thus, mediated by direct translocation across the plasma membrane, unconventional secretion of FGF2 is orders of magnitude faster than ER/Golgi-dependent protein secretion. The kinetics of FGF2 membrane translocation are actually similar to what has been reported for another type of protein translocation across membranes, nucleocytoplasmic transport. Here, based on single-molecule studies, the average time required for a protein to pass through the nuclear pore is in the range of ∼10 ms (Yang et al., 2004; Ma and Yang, 2010; Goryaynov et al., 2012). Thus, although unconventional secretion of FGF2 and protein transport across nuclear pores are mediated by mechanistically distinct processes, FGF2 membrane translocation occurs within a time range that is close to the fastest membrane translocation event reported in the currently available literature.
In conclusion, beyond the unprecedented demonstration of single events of FGF2 membrane translocation in intact cells and the kinetic analysis discussed above, our findings provide direct support for the assembly/disassembly hypothesis of FGF2 membrane translocation in living cells (La Venuta et al., 2015; Brough et al., 2017; Steringer and Nickel, 2018). The unconventional secretory pathway of FGF2 as a whole can be described in three steps with (a) PI(4,5)P2-dependent membrane recruitment, (b) PI(4,5)P2-induced oligomerization concomitant with membrane insertion, and (c) disassembly by heparan sulfates at the outer leaflet, resulting in FGF2 appearance on the cell surface (Fig. 5 I). As we used TIRF microscopy, we could specifically visualize events of FGF2 membrane recruitment at the inner leaflet of the plasma membrane. From this point on, we measured the time intervals until FGF2 appeared on the cell surface, a process referred to as FGF2 membrane translocation. Based on mathematic modeling, FGF2 translocation across the plasma membrane was found to consist of two nonoverlapping physical processes that occur after successful membrane recruitment of FGF2 at the inner leaflet. They mediate the physical passage of FGF2 across the plasma membrane, after which FGF2 becomes detectable at the outer leaflet by binding to fluorescent nanobodies. These two steps of FGF2 membrane translocation differ in terms of the length of their time intervals with a slow and a fast component. As illustrated in Fig. 5 I, we propose the slow process to represent PI(4,5)P2-dependent FGF2 oligomerization concomitant with membrane insertion as previously identified in biochemical reconstitution experiments (Steringer et al., 2012, 2017; Müller et al., 2015). We further suggest the fast process to represent disassembly of membrane-inserted FGF2 oligomers at the outer leaflet, mediated by cell surface heparan sulfates (Fig. 5 I). Thus, based on live cell imaging experiments and in combination with previously published biochemical reconstitution experiments (Steringer et al., 2017), a compelling explanation of how FGF2 can physically traverse the plasma membrane as part of its unconventional mechanism of secretion has been established.
Materials and methods
Chemicals
All chemicals used in this study were purchased from Sigma-Aldrich, unless stated otherwise.
Maintenance of cell lines
CHO K1 and 745 mutant cells were cultured using MEM-α medium supplemented with 10% FCS (Biochrom AG) and 2 mM glutamine at 37°C in the presence of 5% CO2. Human embryonic kidney EcoPack 2–293 cells (Clontech) were cultivated in collagen-coated (Collagen R; Serva Electrophoresis) plates in DMEM, supplemented with 10% FCS at 37°C in the presence of 5% CO2. Both cell lines were maintained in the presence of 100 IU/ml penicillin and 100 µg/ml streptomycin. The expression of proteins in CHO cells was induced at different concentrations (up to 1 µg/ml) of doxycycline (Clontech).
Generation of stable cell lines
To generate stable cell lines expressing various forms of FGF2-GFP fusion protein, a retroviral transduction system based on the Moloney Murine Leukemia Virus was used as described previously (Engling et al., 2002). In brief, a human embryonic kidney 293 cell line was employed for production of virus, having the pVPack-Eco packaging system integrated in its genome and stably expressing the retroviral packaging proteins (EcoPack 2–293 cells). The gene of interest was cloned into the pRevTre2 vector, containing a Tet-response element. Retrovirus production was performed using the MBS Mammalian Transfection Kit (Agilent Technologies). As target cell lines, both CHO WT and CHO-745 mutant cells were used that constitutively expressed the murine cationic amino acid transporter MCAT-1 (Albritton et al., 1989) and a Tet-On transactivator, rtTA2-M2 (Urlinger et al., 2000). This allowed for doxycycline dependent titration of protein expression (Engling et al., 2002). Cells positive for GFP fluorescence were selected in three consecutive rounds of FACS, using a FACSAria flow cytometer (BD Biosciences). Cell lines expressing FGF2-GFP (CHO-K1 and CHO-745), FGF2-mt–GFP (C77A/Y81F/C95A/K127Q/R128Q/K133Q; CHO-K1), and GFP (CHO-K1) were generated. Furthermore, CHO-K1 and -745 cell lines expressing FGF2 as a fusion protein with sfGFP or mGFP were generated and used for the determination of the oligomeric state of FGF2 at the plasma membrane. These variant forms of GFP are strictly monomeric and, therefore, artifacts from GFP-induced oligomerization of FGF2 could be excluded (Zacharias et al., 2002; Pédelacq et al., 2006).
Preparation of labeled nanobodies
GST-tagged anti-GFP nanobodies were purified using GSH-agarose. The nanobodies were eluted by incubation with 1 mg/ml 3C protease (Molox) at 6°C overnight to remove the GST tag. The resulting nanobody solution was concentrated (1 mg/ml), and the buffer was exchanged to 0.1 M NaHCO3 (pH 8.3). For labeling of nanobodies, an Alexa Fluor 647 succinimidyl ester was added at a tenfold molar excess. The mixture was incubated for 2 h at 25°C. Afterward, the solution was supplemented with 1 M Tris/HCl, pH 9.0 (10% [vol/vol]), to quench the reaction. Free dyes were removed by using desalting spin columns. The labeled nanobody preparation was stored in PBS/NaN3 at 4°C.
Preparation of standards for the analysis of the oligomeric state of FGF2-GFP variants
sfGFP standard oligomers with 12, 24, and 60 subunits, respectively, were purified as reported previously (Hsia et al., 2016) and used in the ratiometric approach for building the calibration curve (Fig. 5, A–D). Purified proteins were spread on piranha-cleaned glass slides (0.13–0.16-mm thickness; Menzel) at appropriate dilutions, and the samples were imaged immediately. In the brightness analysis approach (Fig. 5, E and F), mGFP (EGFP-A206K) obtained from a lysate of cells transfected with a vector only encoding mGFP was used for calibration. Briefly, transfected cells were harvested by trypsinization and collected by centrifugation. The sediment was resuspended in lysis buffer (RIPA) with protease inhibitors and cleared by centrifugation. Appropriate dilutions of the supernatant were spread onto piranha-cleaned glasses blocked with 10 mg/ml bovine serum albumin. After incubation for 20 min, samples were washed with PBS and imaged immediately.
Staining of FGF2-GFP on cell surfaces using fluorescently labeled anti-GFP nanobodies
CHO-K1 and CHO-745 cells were plated in eight-well Lab-Tek chambers (Thermo Fisher Scientific) and treated with 1 µg/ml doxycycline to induce protein expression. After 24 h, the medium was removed and the cells were rinsed once with Live Cell Imaging Solution (Thermo Fisher Scientific). Cells were incubated on ice with Alexa Fluor 647–labeled anti-GFP nanobodies (∼0.5 ng/ml) for 30 min. Afterward, they were rinsed three times with Live Cell Imaging Solution and fixed with 4% PFA (Electron Microscopy Sciences).
Dual-color imaging of FGF2 translocation across the plasma membrane
CHO-K1 cells were cultivated in eight-well Lab-Tek chambers (Thermo Fisher Scientific) in the absence of doxycycline to quantify total FGF2-GFP particles at low expression levels. 24 h later, the medium was removed and the cells were rinsed once with Live Cell Imaging Solution (Thermo Fisher Scientific). Cells were incubated with anti-GFP nanobodies (5 nM) in Live Cell Imaging Solution. TIRF videos were acquired at 37°C as described below.
Removal of FGF2-GFP from cell surfaces using heparin
Cells were seeded in eight-well Lab-Tek chambers in the presence of doxycycline (1 µg/ml). After 24 h, the cells were rinsed once with Live Cell Imaging Solution and then incubated for 10 min at room temperature with 500 µg/ml heparin. Afterward, FGF2-GFP on cell surfaces was stained with Alexa Fluor 647–labeled anti-GFP nanobodies followed by cell fixation with 4% PFA (Electron Microscopy Sciences).
To control FGF2-GFP particles before and after treatment with heparin, cells were seeded and cultivated in Lab-Tek chambers for 24 h in medium followed by its replacement by Live Cell Imaging Solution. Cells were treated for at least 10 min with heparin. Time-lapse videos were acquired from both heparin-treated and untreated cells.
For live cell imaging experiments, cells expressing FGF2-GFP were either cultivated in the absence of doxycycline to quantify total FGF2-GFP particles at low expression levels or induced with doxycycline to generate higher expression levels to allow for labeling of the FGF2-GFP cell surface population (incubation for 15 min with 0.5 ng/ml Alexa Fluor 647–labeled anti-GFP nanobodies at room temperature). TIRF videos were acquired with 20 s per frame. After 40 s following the start of data acquisition, heparin (1 mg/ml in Live Cell Imaging Solution) or Live Cell Imaging Solution (as control) was added.
Treatment of cells with inhibitors
To block sulfation of sugar chains of cell surface proteoglycans, cells were treated with NaClO3 for 48–72 h. Cells were seeded and cultivated in six-well plates in the presence of 50 mM NaClO3. About 12 h before live cell imaging experiments, cells were detached with trypsin and seeded in eight-well Lab-Tek chambers in medium supplemented with 50 mM NaClO3. FGF2-GFP on cell surfaces was analyzed from cells induced with doxycycline (1 µg/ml) for 24 h using Alexa Fluor 647–labeled anti-GFP nanobodies.
To inhibit the interaction of FGF2-GFP with PI(4,5)P2, neomycin was used. Cells were seeded in eight-well Lab-Tek chambers the day before the experiment. When secretion of FGF2-GFP was quantified using Alexa Fluor 647–labeled anti-GFP nanobody staining, doxycycline (1 µg/ml) and neomycin (2, 5, or 10 mM) were added 24 h before the experiment. Cell were stained, fixed, and imaged as described above. For quantification of GFP particles interacting with the inner leaflet of the plasma membrane, the cells were treated with 10 mM of neomycin for 3–4 h in the absence of doxycycline.
Optical setup
Single-particle nanoscopy analyzing membrane recruitment of FGF2-GFP to the inner plasma membrane leaflet was conducted with a custom-built microscope (Winterflood et al., 2015). Briefly, a 473-nm laser (100 mW; Laserglow Technologies) and a 643-nm laser (150 mW; Toptica Photonics) were focused onto the back-focal plane of a TIRF objective (NA, 1.49; 60×; Olympus) for highly inclined plane illumination. A quad-edge dichroic beamsplitter (405/488/561/635 nm; Semrock) was used to separate fluorescence emission from excitation light. Emission light was filtered by a quad-band bandpass filter (446/523/600/677 nm; Semrock) and focused by a 500-mm tubus lens onto the chip of a back-illuminated electron-multiplying charge-coupled device camera (Evolve; Photometrics) that was water-cooled to −85°C. Images were acquired with MicroManager (Edelstein et al., 2010). For live cell imaging, the temperature was stabilized to 35°C with a proportional-integral controller by heating the bottom plate of the encapsulated microscope.
Wide-field fluorescence and some of the TIRF imaging were performed using an Olympus IX81 xCellence CellTIRF microscope equipped with a 100× 1.4 NA oil objective lens (Olympus) and an ImagEM (C9100-13) camera (Hamamatsu). GFP fluorescence and Alexa Fluor 647 fluorescence were excited with a 100-mW/488-nm and a 140-mW/640-nm diode laser, respectively (Olympus).
Dual-color imaging was performed using a Vutara 352 microscope (Bruker Nano, Inc.) based on the single particle localization technology. A 60× 1.49 NA oil objective (Olympus) and a Flash 4.0 sCMOS camera (Hamanatsu) with a frame rate of 50 Hz were used. GFP fluorescence and Alexa Fluor 647 fluorescence were excited using the 488-nm and 639-nm laser lines, respectively. Emission signals were separated spectrally by a 640-nm long-pass dichroic, further filtered by a 640-nm long-pass emission filter (red/Alexa Fluor 647) and 640-nm short-pass emission filter (green/GFP) and passed through a custom dual-bandpass emission filter for simultaneous green/red imaging to further suppress non–Alexa Fluor 647/GFP signal. The images were recorded simultaneously, separated by spectra, and imaged on separate regions of the camera. A system calibration was performed using 100-nm TetraSpeck beads (Thermo Fisher Scientific) to experimentally record the emission point spread function of each color channel (green/GFP and red/Alexa Fluor 647) and to align the regions of the camera to a common coordinate system within the native SRX software (version 6.0). Data were recorded and exported in TIFF stacks using SRX, and further analyzed via Fiji.
For the determination of the oligomeric state of FGF2 at the plasma membrane, a modified Zeiss Axiovert 200M epifluorescence microscope was employed. The microscope was equipped with a α Plan-Fluor 100×/1.46 oil objective (Zeiss), a Laser-TIRF 3 Imaging System (Zeiss), and an electron-multiplying charge-coupled device camera (iXon 897; Andor). Samples were illuminated using a 488 laser (Ichrome MLE-LFA multi laser; Toptica) for 150 ms with a delay time between frames of 50 ms (number of frames, 100) with an intensity varying between ∼0.1 and 0.4 kW/cm2. At least 25 movies were collected per condition.
Image processing and trajectory acquisition
Image processing was performed using Fiji (Schindelin et al., 2012). Particles were identified and counted using the Fiji plugin TrackMate (Tinevez et al., 2017). Using the Log detector, the position of each detected particle was identified in each frame, and trajectories were extracted. The particles were normalized to the cell surface area (µm2). All representative images included in the manuscript were corrected for background fluorescence.
For quantification of plasma membrane–associated GFP particles, time-lapse TIRF videos were acquired. The total number of particles per frame was identified and quantified. Additionally, relative expression levels were quantified for each cell analyzed by measuring the mean intensity of GFP fluorescence at the first frame of each acquired image sequence using ImageJ. To achieve this, the frame of the cell was selected based on a bright-field image, and the mean fluorescence intensity of each cell was quantified. The number of GFP particles was initially normalized to the surface area of the corresponding cell and the expression levels, unless otherwise stated. For quantification of anti-GFP nanobody particles at the extracellular surface of fixed cells, TIRF images were acquired, and the total particles were identified and quantified using TrackMate and Fiji. Nanobody particles detected per cell were normalized to the cell surface area of the corresponding cell.
The detection of FGF2-GFP membrane translocation events at the plasma membrane was achieved by identifying colocalization of green and red particles using either the Fiji plugin ComDet (https://github.com/ekatrukha/ComDet/wiki) or by manual inspection. The detection parameters were set for both GFP and nanobody particles for detection of very dim particles with a particle size larger than 3 pixels. The maximum distance between colocalized particles was set to be 3 pixels. However, the plugin could not detect particles with high sensitivity, and therefore, the data were also analyzed manually for identification of translocation events. Fluorescence intensity of particles was counted after background correction and plotted over time. To quantify the kinetics of translocation, the time intervals between the appearance of FGF2-GFP particles within the evanescent field and nanobody binding at the cell surface were calculated (t(nb) − t(GFP)). The distribution of observed time intervals was plotted using GraphPad Prism, and a bin width of 100 ms was applied. To determine the stability of GFP particles, the time interval of the persistence of GFP fluorescence in the evanescent field following nanobody disappearance was measured for all translocation events.
Determination of the oligomeric size distribution of FGF2-GFP by single particle brightness analysis
To determine the stoichiometry of FGF2 at the plasma membrane of living cells, we performed experiments by TIRF microscopy. Images acquired were used for a stoichiometry analysis based on the fluorescence intensity of particles using an in-house algorithm implemented in Python (Python Software Foundation). Bright spots were automatically detected using an implementation of the “Difference of Gaussians” method (http://scikit-image.org/docs/dev/api/skimage.feature.html) and thresholding. Selected particles were defined by a region of interest (ROI) of a defined pixel size (2 × 2) and fitted to 2D Gaussians. Background subtraction was performed by defining a ROI around the particle’s ROI having a larger pixel size (3 × 3). Localized particles were discarded based on the distance and on the width of the 2D Gaussian to avoid overlapping ROIs or multiple particles in the same ROI. This algorithm was used to determine the brightness value for each spot. Stoichiometry counting was performed using two different, independent approaches with (a) a ratiometric approach using oligomeric standards fused to sfGFP and (b) a brightness analysis using mGFP (EGFP-A206K) as a reference. Both GFP variants contained the A206K substitution to ensure that oligomerization of FGF2 would not be a result of GFP dimerization.
In the ratiometric approach (Fig. 5, A–D), cells were expressing a fusion protein of FGF2 with sfGFP, and the fluorescence intensities of FGF2-sfGFP particles were compared with the fluorescence intensity values of standards with a known number of subunits (Coffman and Wu, 2012). The obtained brightness values for the standards (12-, 24-, or 60-mer protein constructs) and for the FGF2-sfGFP (Fig. 5, A and B) were plotted as a Gaussian distribution, which allowed for calculating the mean value of each particle population. The mean fluorescence intensity values obtained with the standards from two independent experiments were used to generate a calibration curve (Fig. 5 D). The slope of the calibration curve was calculated by linear fitting and corresponded to the fluorescence intensity value of a single sfGFP unit (Hsia et al., 2016). Finally, the fluorescence intensity values of single FGF2-sfGFP particles were converted into molecular units by dividing them by the fluorescence intensity of the single sfGFP unit. Three independent experiments were conducted, with each of them consisting of the analysis of at least 30 cells. More than 3,000 individual particles were identified and analyzed for brightness per cell type.
For the brightness analysis (Fig. 5, E and F), mGFP was employed for calibration. mGFP particles were selected by a photobleaching analysis after smoothing of the signal with a median filter. For this purpose, individual particles with a single photobleaching step were detected and used to build a histogram that was fitted to a Gaussian providing the mean intensity and SD for a single fluorophore. These values were used to calculate the theoretical values for higher oligomers (Schmidt et al., 1996). Then, brightness values of particles of FGF2-mGFP were plotted as a probability density function or as a Gaussian distribution (Fig. 5 E). Finally, the overall brightness distribution was fitted as a sum of four Gaussians imposing the theoretical mean and SD values, previously calculated for the different N-mers. The area under each curve of the multiple Gaussians was used to estimate the percentage of occurrence of each species. The single-particle analysis provides the number of labeled monomers in each analyzed particle. However, since only a fraction of FGF2-GFP fusion proteins is fluorescent, which is due to misfolding and maturation issues of mGFP (Ulbrich and Isacoff, 2007), this number does not necessarily correspond to the real stoichiometry of the particle. For example, the monomer population is overestimated since it includes higher-order oligomers containing only one fluorescent molecule of mGFP. Therefore, correction for partial labeling efficiency was performed as previously described (Subburaj et al., 2015a), considering a labeling efficiency for GFP of 70% (Ulbrich and Isacoff, 2007). The distribution of species (Fig. 5 B) corresponds to the average values obtained from two independent experiments with at least 30 cells measured per experiment. A total of 6,845 particles were identified and analyzed for their brightness.
Validation of FGF2-mGFP dimers at the plasma membrane using step-wise photobleaching experiments
Cells expressing FGF2-mGFP at levels that allow for single-particle detection were seeded on no. 1.5 glass coverslips and imaged on a custom single molecule setup at 37°C in live cell imaging buffer (phenol-red free DMEM, 25 mM Hepes, pH 7.0, 10% FBS, and 1% glutamate). Immediately before imaging, cells were treated with 0.008% saponin to remove cytosolic FGF2-mGFP and thereby reduce background fluorescence. Time series of 300 frames were acquired under TIRF illumination within 2 min of saponin addition. The image series were analyzed using ImageJ. The average intensity in a 5 × 5-pixel square centered on a bright spot was extracted and plotted.
Statistical analysis and model fitting
To estimate the typical time it takes for the nearest nanobody to reach the vicinity of FGF2-GFP on cell surfaces through Brownian motion, the mean nearest-neighbor distance between nanobodies was obtained as a function of their concentration (Chandrasekhar, 1943). A solution to the Fokker–Planck equation in a spherically symmetric geometry containing an absorbing boundary was considered as the diffusion model (Goel and Richter-Dyn, 2013). With the mean distance known, the survival probability and the first-hitting time distribution were calculated. The typical first-hitting time, i.e., the most probable time it takes for a particle to reach a target through diffusion, was numerically calculated based on these distributions. A Python code, using functions from the NumPy and SciPy packages, was developed for this purpose.
For numerical manipulation of the sampled waiting time data, implementing probability distributions, and testing proposed hypotheses, an in-house Python code was developed. Fitting proposed probability distributions to the empirical data was achieved through the well-established maximum likelihood estimation method (Millar, 2011). For the one-step process, the maximum likelihood estimation simply prescribes matching the sample average to the expected waiting time of the exponential process. But for the two-step process, it requires solving a system of nonlinear equations arising from the more complicated probability distribution function. The Newton–Krylov Jacobian-free method (Knoll and Keyes, 2004), implemented as part of the SciPy open-source software package, was used for obtaining numerical solutions that yielded expected waiting times. The probability plots used to examine the goodness of fit in the two cases (Fig. 4, G and H) were produced by the statistics module of the SciPy package and were plotted using the Matplotlib plotting library. The Kolmogorov–Smirnov test was also performed through the provided functions of the SciPy package to investigate how well a probability distribution function describes the set of our experimental observations, with the highest P value describing the best fit.
Sample sizes
The sample size of all experiments presented in this study is summarized in Table S1.
For the experiments quantifying recruitment of FGF2-GFP versus GFP at the plasma membrane of CHO-K1 cells (Fig. 1 C), a total of nine independent experiments were performed. For the cells expressing FGF2-GFP, 186 imaging fields with a total of 229 cells were analyzed. Likewise, for cells expressing GFP, 158 imaging fields with 220 cells were analyzed.
For the experiments comparing the secretion of FGF2-GFP with the GFP control in CHO-K1 cells (Fig. 1 E), four independent experiments were performed. 43 images with 217 cells were analyzed for FGF2-GFP, and 39 images with 194 cells were analyzed for GFP-expressing cells.
For the experiment shown in Fig. 1 H, in which heparin was added to the medium to elute secreted FGF2-GFP from HSPGs, six independent experiments were performed. This analysis included a total of 64 imaging fields with 320 cells for mock-treated cells and 60 imaging fields with 306 cells for heparin-treated cells.
For the experiment shown in Fig. 1 K, in which heparin was added to the medium of cells followed by quantification of FGF2-GFP recruitment at the plasma membrane, a total of six independent experiments were performed. This analysis included 62 imaging fields with 212 cells for mock-treated cells and 59 imaging fields with 222 cells for heparin-treated cells.
To compare the recruitment of FGF2 at the plasma membrane under conditions with reduced levels of sulfated HSPGs (Fig. 2 B), the following sample sizes were used: for CHO-K1 cells expressing FGF2-GFP, six independent experiments were performed, 127 images were acquired, and 159 cells were analyzed. For CHO-K1 cells expressing FGF2-GFP and treated with NaClO3, three independent experiments were performed, 57 images were acquired, and 70 cells were analyzed. For CHO-745 cells expressing FGF2-GFP, five independent experiments were performed, 93 images were acquired, and 153 cells were analyzed. Finally, the control (GFP-expressing CHO-K1 cells) was included in all six experiments, with 112 images and 150 cells analyzed.
For the experiments quantifying the secretion of FGF2 into the extracellular space under conditions characterized by reduced levels of sulfated HSPGs (Fig. 2 D), the following sample sizes were used: for CHO-K1 cells expressing FGF2-GFP, eight independent experiments were performed, with 92 images and 465 cells analyzed. For CHO-K1 cells expressing FGF2-GFP and treated with NaClO3, three independent experiments were performed, with 35 images and 144 cells analyzed. For CHO-745 cells expressing FGF2-GFP, five independent experiments were performed, with 52 images and 235 cells analyzed. Finally, the control (GFP-expressing CHO-K1 cells) was included in four independent experiments with 39 images and 194 cells analyzed.
The sample size for experiments quantifying recruitment at the plasma membrane of FGF2-GFP after treatment with neomycin (Fig. 3 B) was based on a total of four independent experiments with 91 images and 112 cells analyzed. For CHO-K1 cells expressing FGF2-GFP and treated with neomycin, 70 images with 100 cells were analyzed. Finally, for control CHO-K1 cells expressing GFP, 72 images with 95 cells were analyzed.
Experiments quantifying the secretion efficiency of FGF2-GFP in the presence of different concentrations of neomycin (Fig. 3 D) were based on a total of three independent experiments. For CHO-K1 cells expressing FGF2-GFP, 39 images with 199 cells were analyzed. For CHO-K1 cells expressing FGF2-GFP and treated with 2 mM neomycin, 56 images with 239 cells were analyzed, and for cells treated with 5 mM neomycin, 43 images with 160 cells were analyzed. Finally, for cells treated with 10 mM neomycin, 35 images with 109 cells were analyzed.
Experiments comparing plasma membrane recruitment between the WT variant form of FGF2-GFP and a secretion deficient mutant FGF2-mt–GFP; Fig. 3 E) was based on a total of four independent experiments. For CHO-K1 cells expressing FGF2wt-GFP, 99 images with 143 cells were analyzed. For CHO-K1 cells expressing FGF2mt-GFP, 75 images with 116 cells were analyzed. In addition, CHO-K1 cells expressing GFP were included as a control, with 76 images and a total of 117 cells analyzed.
The quantification of secretion efficiency (Fig. 3 F) was based on three independent experiments. For CHO-K1 cells expressing FGF2wt-GFP, 34 images with 172 cells were analyzed. For CHO-K1 cells expressing FGF2mt-GFP, 31 imaging fields with 148 cells were analyzed. In addition, CHO-K1 cells expressing GFP were included with 29 imaging fields and 148 cells analyzed.
The analysis of the kinetics of FGF2 translocation across the plasma membrane was based on eight independent experiments. A total of 60 cells were analyzed for the identification of successful FGF2-GFP translocation events.
Finally, for the analysis of the oligomeric state of FGF2 at the plasma membrane, two different approaches were used. In the ratiometric approach, three independent experiments were performed with cells expressing FGF2-sfGFP. For the CHO-K1 cell line, 82 images were acquired and a total of 3,602 particles were analyzed. For the CHO-745 cell line, 105 images were acquired and 3,296 particles were analyzed. Similarly, for the brightness analysis, 68 images were acquired and 6,845 particles were analyzed in two independent experiments in CHO-745 cells expressing FGF2-mGFP. To validate the existence of FGF2-mGFP dimers at the plasma membrane, a step-wise photobleaching approach was used. For CHO-K1 cells, four experiments were performed during which a total of five cells was analyzed with 49 particles being identified, 3 of which were found to be in the form of a dimer. For the CHO-745 cells, 10 experiments were performed (in each experiment, one field was imaged) and 17 cells were analyzed. A total of 252 particles were identified, with 23 of them in the form of a dimer.
Online supplemental materials
Fig. S1 A shows representative examples of mobility measurements of FGF2-GFP at the plasma membrane. In Fig. S1, B and C, representative images for mock-treated cells are shown that were taken at the beginning and the end of the analysis shown in Fig. 1, G and H. Fig. S2 shows representative examples of full FGF2-GFP membrane translocation events. Video 1 shows time-lapse TIRF microscopy of CHO-K1 cells expressing FGF2-GFP. Video 2 shows the secreted population of FGF2-GFP on the cell surface of CHO-K1 cells expressing FGF2-GFP by single-particle live cell imaging. Video 3 shows a time-lapse video of FGF2-GFP–expressing CHO-K1 cells treated with heparin and labeled with anti-GFP nanobodies. Video 4 shows time-lapse TIRF microscopy of FGF2-GFP under conditions where heparan sulfate biosynthesis is impaired. Video 5 shows time-lapse imaging of a representative event of FGF2-GFP membrane recruitment and translocation to the cell surface of living cells using TIRF microscopy. Table S1 lists the sample sizes and results from all experiments documented in Figs. 1, 2, 3, 4, and 5.
Supplementary Material
Acknowledgments
We thank Monika Langlotz from the Zentrum für Molekulare Biologie Heidelberg (ZMBH) Flow Cytometry Core Facility and Holger Lorenz from the ZMBH Light Microscopy Core Facility for their support in cell sorting and image acquisition/interpretation, respectively. We also thank Dr. Carl G. Ebeling (Bruker Fluorescence Microscopy) for providing support for dual color imaging experiments.
This work was supported by the Deutsche Forschungsgemeinschaft through an individual grant (Ni 423/6-1), the Collaborative Research Center (SFB/TRR186-A1), and the Deutsche Forschungsgemeinschaft Cluster of Excellence CellNetworks at Heidelberg University.
The authors declare no competing financial interests.
Author contributions: E. Dimou performed and analyzed the majority of the described experiments. E. Dimou, K. Consentino, and U. Ros performed the experiments regarding the analysis of the oligomeric state of FGF2. E. Platonova contributed to the establishment of the experimental setup. M. Sadeghi and F. Noé performed the mathematical modeling. P. Kashyap performed the step-wise photobleaching experiments. T. Katsinelos contributed to the interpretation of data and the preparation of the manuscript. K. Consentino, U. Ros, A. García-Sáez, and H. Ewers interpreted the oligomeric state of FGF2 at the plasma membrane. E. Dimou, W. Nickel, and H. Ewers contributed to the study design, interpreted all data, and wrote the manuscript with contributions from the other authors.
References
- Akl M.R., Nagpal P., Ayoub N.M., Tai B., Prabhu S.A., Capac C.M., Gliksman M., Goy A., and Suh K.S.. 2016. Molecular and clinical significance of fibroblast growth factor 2 (FGF2 /bFGF) in malignancies of solid and hematological cancers for personalized therapies. Oncotarget. 7:44735–44762. 10.18632/oncotarget.8203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albritton L.M., Tseng L., Scadden D., and Cunningham J.M.. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 57:659–666. 10.1016/0092-8674(89)90134-7 [DOI] [PubMed] [Google Scholar]
- Arbuzova A., Martushova K., Hangyás-Mihályné G., Morris A.J., Ozaki S., Prestwich G.D., and McLaughlin S.. 2000. Fluorescently labeled neomycin as a probe of phosphatidylinositol-4, 5-bisphosphate in membranes. Biochim. Biophys. Acta. 1464:35–48. 10.1016/S0005-2736(99)00243-6 [DOI] [PubMed] [Google Scholar]
- Beenken A., and Mohammadi M.. 2009. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8:235–253. 10.1038/nrd2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., and Perez F.. 2012. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 9:493–498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Brough D., Pelegrin P., and Nickel W.. 2017. An emerging case for membrane pore formation as a common mechanism for the unconventional secretion of FGF2 and IL-1β. J. Cell Sci. 130:3197–3202. 10.1242/jcs.204206 [DOI] [PubMed] [Google Scholar]
- Chandrasekhar S. 1943. Stochastic Problems in Physics and Astronomy. Rev. Mod. Phys. 15:1–89. 10.1103/RevModPhys.15.1 [DOI] [Google Scholar]
- Coffman V.C., and Wu J.Q.. 2012. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 37:499–506. 10.1016/j.tibs.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J.P., Binda A., Canfield V.A., and Levenson R.. 2000. Participation of Na,K-ATPase in FGF-2 secretion: rescue of ouabain-inhibitable FGF-2 secretion by ouabain-resistant Na,K-ATPase alpha subunits. Biochemistry. 39:14877–14883. 10.1021/bi001073y [DOI] [PubMed] [Google Scholar]
- Dimou E., and Nickel W.. 2018. Unconventional mechanisms of eukaryotic protein secretion. Curr. Biol. 28:R406–R410. 10.1016/j.cub.2017.11.074 [DOI] [PubMed] [Google Scholar]
- Duchesne L., Octeau V., Bearon R.N., Beckett A., Prior I.A., Lounis B., and Fernig D.G.. 2012. Transport of fibroblast growth factor 2 in the pericellular matrix is controlled by the spatial distribution of its binding sites in heparan sulfate. PLoS Biol. 10:e1001361 10.1371/journal.pbio.1001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A.D., Laussmann M., Wegehingel S., Kaderali L., Erfle H., Reichert J., Lechner J., Beer H.D., Pepperkok R., and Nickel W.. 2010. Tec-kinase-mediated phosphorylation of fibroblast growth factor 2 is essential for unconventional secretion. Traffic. 11:813–826. 10.1111/j.1600-0854.2010.01059.x [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14:Unit14 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engling A., Backhaus R., Stegmayer C., Zehe C., Seelenmeyer C., Kehlenbach A., Schwappach B., Wegehingel S., and Nickel W.. 2002. Biosynthetic FGF-2 is targeted to non-lipid raft microdomains following translocation to the extracellular surface of CHO cells. J. Cell Sci. 115:3619–3631. 10.1242/jcs.00036 [DOI] [PubMed] [Google Scholar]
- Esko J.D., Stewart T.E., and Taylor W.H.. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA. 82:3197–3201. 10.1073/pnas.82.10.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florkiewicz R.Z., Anchin J., and Baird A.. 1998. The inhibition of fibroblast growth factor-2 export by cardenolides implies a novel function for the catalytic subunit of Na+,K+-ATPase. J. Biol. Chem. 273:544–551. 10.1074/jbc.273.1.544 [DOI] [PubMed] [Google Scholar]
- Goel N.S., and Richter-Dyn N.. 2013. Stochastic Models in Biology. Elsevier Science, Burlington. [Google Scholar]
- Goryaynov A., Ma J., and Yang W.. 2012. Single-molecule studies of nucleocytoplasmic transport: from one dimension to three dimensions. Integr. Biol. 4:10–21. 10.1039/C1IB00041A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia Y., Bale J.B., Gonen S., Shi D., Sheffler W., Fong K.K., Nattermann U., Xu C., Huang P.S., Ravichandran R., et al. . 2016. Design of a hyperstable 60-subunit protein dodecahedron. [corrected]. Nature. 535:136–139. 10.1038/nature18010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., and Weissman J.S.. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 147:789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll D., and Keyes D.. 2004. Jacobian-free Newton-Krylov methods: a survey of approaches and applications. J. Comput. Phys. 193:357–397. 10.1016/j.jcp.2003.08.010 [DOI] [Google Scholar]
- Kwik J., Boyle S., Fooksman D., Margolis L., Sheetz M.P., and Edidin M.. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100:13964–13969. 10.1073/pnas.2336102100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Venuta G., Zeitler M., Steringer J.P., Müller H.M., and Nickel W.. 2015. The Startling Properties of Fibroblast Growth Factor 2: How to Exit Mammalian Cells without a Signal Peptide at Hand. J. Biol. Chem. 290:27015–27020. 10.1074/jbc.R115.689257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., and Yang W.. 2010. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc. Natl. Acad. Sci. USA. 107:7305–7310. 10.1073/pnas.0908269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin K.S., and Simons K.. 1983. Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell. 34:233–243. 10.1016/0092-8674(83)90154-X [DOI] [PubMed] [Google Scholar]
- Millar R. 2011. Maximum likelihood estimation and inference with examples in R, SAS, and ADMB. J. Wiley & Sons, Chichester, UK: 10.1002/9780470094846 [DOI] [Google Scholar]
- Müller H.M., Steringer J.P., Wegehingel S., Bleicken S., Münster M., Dimou E., Unger S., Weidmann G., Andreas H., García-Sáez A.J., et al. . 2015. Formation of disulfide bridges drives oligomerization, membrane pore formation, and translocation of fibroblast growth factor 2 to cell surfaces. J. Biol. Chem. 290:8925–8937. 10.1074/jbc.M114.622456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. 1975. Intracellular aspects of the process of protein synthesis. Science. 189:347–358. 10.1126/science.1096303 [DOI] [PubMed] [Google Scholar]
- Pédelacq J.D., Cabantous S., Tran T., Terwilliger T.C., and Waldo G.S.. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88. 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- Rivera V.M., Wang X., Wardwell S., Courage N.L., Volchuk A., Keenan T., Holt D.A., Gilman M., Orci L., Cerasoli F. Jr., et al. . 2000. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 287:826–830. 10.1126/science.287.5454.826 [DOI] [PubMed] [Google Scholar]
- Rothman J.E. 1994. Mechanisms of intracellular protein transport. Nature. 372:55–63. 10.1038/372055a0 [DOI] [PubMed] [Google Scholar]
- Rothman J.E., and Wieland F.T.. 1996. Protein sorting by transport vesicles. Science. 272:227–234. 10.1126/science.272.5259.227 [DOI] [PubMed] [Google Scholar]
- Safaiyan F., Kolset S.O., Prydz K., Gottfridsson E., Lindahl U., and Salmivirta M.. 1999. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J. Biol. Chem. 274:36267–36273. 10.1074/jbc.274.51.36267 [DOI] [PubMed] [Google Scholar]
- Schekman R., and Orci L.. 1996. Coat proteins and vesicle budding. Science. 271:1526–1533. 10.1126/science.271.5255.1526 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T., Schütz G., Gruber H., and Schindler H.. 1996. Local Stoichiometries Determined by Counting Individual Molecules. Anal. Chem. 68:4397–4401. 10.1021/ac960710g [DOI] [Google Scholar]
- Schmoranzer J., Goulian M., Axelrod D., and Simon S.M.. 2000. Imaging constitutive exocytosis with total internal reflection fluorescence microscopy. J. Cell Biol. 149:23–32. 10.1083/jcb.149.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. 1974. EDF Statistics for Goodness of Fit and Some Comparisons. J. Am. Stat. Assoc. 69:730–737. 10.1080/01621459.1974.10480196 [DOI] [Google Scholar]
- Steringer J.P., and Nickel W.. 2018. A direct gateway into the extracellular space: Unconventional secretion of FGF2 through self-sustained plasma membrane pores. Semin. Cell Dev. Biol. 83:3–7. 10.1016/j.semcdb.2018.02.010 [DOI] [PubMed] [Google Scholar]
- Steringer J.P., Bleicken S., Andreas H., Zacherl S., Laussmann M., Temmerman K., Contreras F.X., Bharat T.A., Lechner J., Müller H.M., et al. . 2012. Phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2)-dependent oligomerization of fibroblast growth factor 2 (FGF2) triggers the formation of a lipidic membrane pore implicated in unconventional secretion. J. Biol. Chem. 287:27659–27669. 10.1074/jbc.M112.381939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steringer J.P., Lange S., Čujová S., Šachl R., Poojari C., Lolicato F., Beutel O., Müller H.M., Unger S., Coskun Ü., et al. . 2017. Key steps in unconventional secretion of fibroblast growth factor 2 reconstituted with purified components. eLife. 6:e28985 10.7554/eLife.28985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subburaj Y., Cosentino K., Axmann M., Pedrueza-Villalmanzo E., Hermann E., Bleicken S., Spatz J., and García-Sáez A.J.. 2015a Bax monomers form dimer units in the membrane that further self-assemble into multiple oligomeric species. Nat. Commun. 6:8042 10.1038/ncomms9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subburaj Y., Ros U., Hermann E., Tong R., and García-Sáez A.J.. 2015b Toxicity of an α-pore-forming toxin depends on the assembly mechanism on the target membrane as revealed by single molecule imaging. J. Biol. Chem. 290:4856–4865. 10.1074/jbc.M114.600676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmerman K., and Nickel W.. 2009. A novel flow cytometric assay to quantify interactions between proteins and membrane lipids. J. Lipid Res. 50:1245–1254. 10.1194/jlr.D800043-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmerman K., Ebert A.D., Müller H.M., Sinning I., Tews I., and Nickel W.. 2008. A direct role for phosphatidylinositol-4,5-bisphosphate in unconventional secretion of fibroblast growth factor 2. Traffic. 9:1204–1217. 10.1111/j.1600-0854.2008.00749.x [DOI] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Ulbrich M.H., and Isacoff E.Y.. 2007. Subunit counting in membrane-bound proteins. Nat. Methods. 4:319–321. 10.1038/nmeth1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger S., Baron U., Thellmann M., Hasan M.T., Bujard H., and Hillen W.. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA. 97:7963–7968. 10.1073/pnas.130192197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterflood C.M., Platonova E., Albrecht D., and Ewers H.. 2015. Dual-color 3D superresolution microscopy by combined spectral-demixing and biplane imaging. Biophys. J. 109:3–6. 10.1016/j.bpj.2015.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Gelles J., and Musser S.M.. 2004. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 101:12887–12892. 10.1073/pnas.0403675101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., and Tsien R.Y.. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916. 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zacherl S., La Venuta G., Müller H.M., Wegehingel S., Dimou E., Sehr P., Lewis J.D., Erfle H., Pepperkok R., and Nickel W.. 2015. A direct role for ATP1A1 in unconventional secretion of fibroblast growth factor 2. J. Biol. Chem. 290:3654–3665. 10.1074/jbc.M114.590067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehe C., Engling A., Wegehingel S., Schäfer T., and Nickel W.. 2006. Cell-surface heparan sulfate proteoglycans are essential components of the unconventional export machinery of FGF-2. Proc. Natl. Acad. Sci. USA. 103:15479–15484. 10.1073/pnas.0605997103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.