Turan and Grosche et al. show that herpes simplex virus type 1 (HSV-1) infection leads to autophagic degradation of nuclear lamins in immature dendritic cells, facilitating HSV-1 nuclear egress and the formation of infectious progeny virus. In mature dendritic cells, autophagy is inhibited due to elevated KIF1B and KIF2A protein levels.
Abstract
Dendritic cells (DCs) are crucial for the induction of potent antiviral immune responses. In contrast to immature DCs (iDCs), mature DCs (mDCs) are not permissive for infection with herpes simplex virus type 1 (HSV-1). Here, we demonstrate that HSV-1 infection of iDCs and mDCs induces autophagy, which promotes the degradation of lamin A/C, B1, and B2 in iDCs only. This in turn facilitates the nuclear egress of progeny viral capsids and thus the formation of new infectious particles. In contrast, lamin protein levels remain stable in HSV-1–infected mDCs due to an inefficient autophagic flux. Elevated protein levels of KIF1B and KIF2A in mDCs inhibited lamin degradation, likely by hampering autophagosome–lysosome fusion. Therefore, in mDCs, fewer progeny capsids were released from the nuclei into the cytosol, and fewer infectious virions were assembled. We hypothesize that inhibition of autophagic lamin degradation in mDCs represents a very powerful cellular counterstrike to inhibit the production of progeny virus and thus viral spread.
Introduction
As professional antigen-presenting cells, dendritic cells (DCs) are crucial players in the induction of effective antiviral immune responses. Immature DCs (iDCs) are present in the vast majority of peripheral tissues, where they encounter and take up antigen (Mellman and Steinman, 2001). As a consequence, DCs mature and migrate along a chemokine gradient toward draining lymph nodes to enter paracortical T cell zones to activate and prime naive antigen-specific T lymphocytes (Banchereau and Steinman, 1998; Palucka and Banchereau, 1999). For major histocompatability complex (MHC) class II presentation, endocytosed antigens are targeted to lysosomes via receptor-mediated endocytosis (Geuze, 1998). In lysosomes, antigens are partially degraded to generate specific peptides for MHC class II presentation (Watts, 2001). Macroautophagy (henceforth autophagy) is an additional route by which cytoplasmic and nuclear antigens (e.g., upon viral infection) can be provided to MHC class II molecules (Dengjel et al., 2005; Crotzer and Blum, 2009).
Autophagy is a conserved cellular degradation pathway to digest intracellular components such as proteins or whole organelles (e.g., mitochondria and peroxisomes) via the lysosomal machinery (Takeshige et al., 1992). Up-regulation of autophagy, mainly due to starvation or related stress, therefore provides a source of amino acids from degraded proteins for the synthesis of new proteins (Takeshige et al., 1992). Mechanistic target of rapamycin (mTOR) is a key regulator of autophagy and plays an important role in cell survival (Wu et al., 2009; Yu et al., 2010). Phosphorylated and thus activated mTOR inhibits autophagy by controlling UNC-51-like kinase 1 (ULK1) ubiquitination (Nazio et al., 2013). The activated ULK1/2 kinase complex, including focal adhesion kinase family interacting protein of 200 kD (FIP200), and subsequent activation of the beclin-1–Vps34–AMBRA1 complex are necessary to initiate phagophore formation (Bodemann et al., 2011). Among others, p62 marks cytoplasmic cargo for degradation by autophagy. p62 recognizes polyubiquitinated proteins that are too large to be degraded by the proteasome and delivers them to the autophagy pathway, where its cargo and p62 itself become degraded (Bjørkøy et al., 2006). During autophagophore maturation, microtubule-associated protein light chain 3 (LC3) I is proteolytically cleaved and attached to phosphatidylethanolamine to form LC3-II. This lipidated form is then inserted into the autophagosomal membrane. Conversion of LC3B-I to LC3B-II indicates the presence of mature autophagosomes and therefore autophagy induction (Kabeya et al., 2000, 2004). Finally, lysosomes fuse with autophagosomes, and the resulting autophagolysosomes are degraded (e.g., by hydrolysis). Interestingly, autophagy is induced not only upon starvation or cellular stress but also in fibroblasts and neurons upon infection with herpes simplex virus type 1 (HSV-1; McFarlane et al., 2011; reviewed in O’Connell and Liang, 2016).
HSV-1 represents the prototype of the α-herpesvirus family and is characterized by a fast lytic replication cycle. Common to all α-herpesviruses, HSV-1 establishes latency in sensory neurons and ganglia after primary infection (Whitley and Roizman, 2001; Rechenchoski et al., 2017). Replication of HSV-1 occurs in the nucleus, where the DNA is packaged into viral capsids that subsequently traverse the inner and outer nuclear membrane to leave the nucleus for secondary envelopment in the cytoplasm. During this process, the nuclear lamina constitutes a main barrier for the nuclear egress of viral capsids (Mou et al., 2008). The nuclear lamina mainly consists of lamin proteins that belong to the group of type V intermediate filament proteins. These lamin proteins are grouped into type A and B, namely lamin A/C, lamin B1, and lamin B2 (Dechat et al., 2010). Therefore, viruses whose capsids are assembled in the nucleus have evolved mechanisms to disassemble and cross this lamin meshwork for nuclear egress. Viral and cellular proteins, in particular protein kinases, which induce destabilization of the nuclear lamina, are important to mediate nuclear egress of HSV-1. Previous studies revealed that in Hep2 cells, HSV-1 induces the phosphorylation of lamin A/C by the viral protein kinase US3, leading to a disrupted organization of nuclear lamina (Mou et al., 2007). As a similar mechanism was shown for nuclear egress of human cytomegalovirus (Milbradt et al., 2010), it was hypothesized that this mode of lamina reorganization is a conserved mechanism for the nuclear egress for herpesviruses.
Interestingly, earlier studies revealed that HSV-1–infected iDCs released infectious progeny virions into the culture medium (Mikloska et al., 2001), while mature DCs (mDCs) produced very few progeny virions (Kruse et al., 2000). Since HSV-1 proteins are expressed in both iDCs and mDCs upon infection (Goldwich et al., 2011), we hypothesized that nuclear egress or a subsequent step of virus assembly might be hampered in mDCs. In the present study, we demonstrate that the autophagic degradation of lamin A/C, B1, and B2 facilitates the nuclear egress of newly assembled HSV-1 capsids in iDCs. Furthermore, we provide evidence that the elevated expression of two kinesin family members, KIF1B and KIF2A, contribute to the inefficient autophagy in mDCs, resulting in hampered lamin degradation and thus reduced nuclear egress of HSV-1 capsids.
Results
HSV-1 capsids are trapped in the nucleus of mDCs, but not iDCs
Previous studies demonstrated that human iDCs are permissive for an HSV-1 infection as infectious progeny virions are released into the cell culture supernatant (Mikloska et al., 2001). Noteworthy, and in sharp contrast to iDCs, mDCs are not permissive for an HSV-1 infection, and infectious virus particles are barely detectable in the supernatant of infected mDCs (Kruse et al., 2000; Goldwich et al., 2011). This observation is even more striking, since viral proteins of all three gene expression phases are present in HSV-1–infected mDCs (Goldwich et al., 2011). Thus, regarding the generation of infectious progeny virus, there is a fundamental difference between iDCs and mDCs.
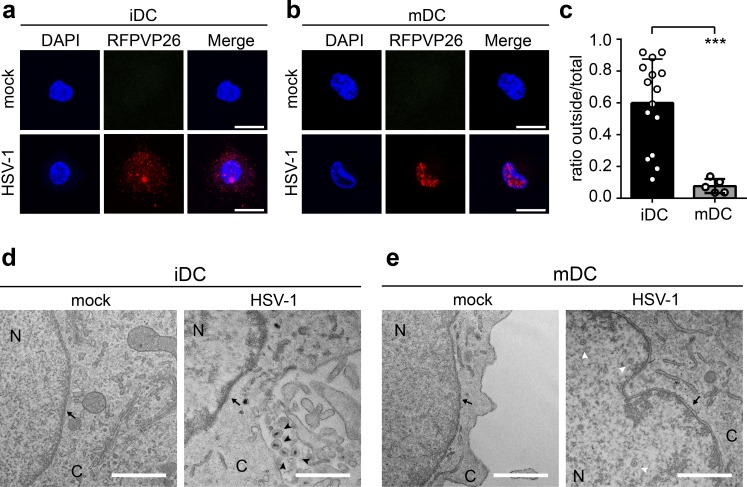
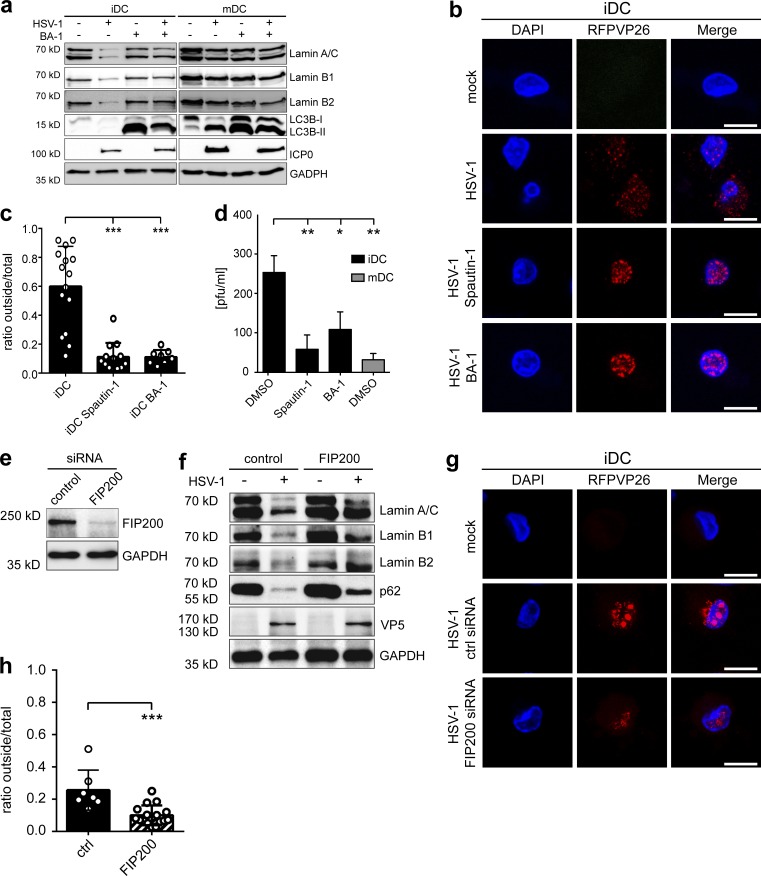
To further investigate the underlying mechanisms, we analyzed the intracellular capsid localization in HSV-1–infected human monocyte–derived iDCs and mDCs. For this purpose, iDCs or mDCs were infected with an HSV-1 reporter virus, which expresses the VP26 capsid surface protein with an RFP fusion tag (HSV1-RFPVP26) to investigate the intracellular distribution of HSV-1 capsids via fluorescence microscopy (Fig. 1, a–c). While ∼60% of all capsids were detected in the cytoplasm of HSV-1–infected iDCs at 24 h postinfection (24 hpi; Fig. 1, a and c), only ∼10% were localized in the cytoplasm of infected mDCs (Fig. 1, b and c), indicating that nuclear capsid egress might have been impaired. In addition, HSV-1 WT–infected iDCs and mDCs were analyzed using electron microscopy. In line with our previous data, newly formed enveloped capsids were detected in the cytoplasm of HSV-1–infected iDCs (Fig. 1 d, black arrowheads), while viral capsids were exclusively observed within the nucleus in infected mDCs (Fig. 1 e, white arrowhead). According to these data, we hypothesize that in contrast to iDCs, HSV-1 capsids are not able to efficiently pass the nuclear lamina in mDCs.
Figure 1.
Viral capsids are retained in the nucleus of mDCs, but not iDCs. (a and b) iDCs (a) and mDCs (b) were mock- or HSV1-RFPVP26–infected and cultured on poly-L-lysine–coated coverslips for 24 h. Nuclei were stained using DAPI (blue). Scale bars, 10 µm. (c) Quantification of intranuclear and cytoplasmic capsids in iDCs (black bar, n = 15) versus mDCs (gray bar, n = 5) based on RFPVP26 signals. Values are shown as ratios of integral intensities of cytoplasmic (“outside”) versus total capsid signals. Error bars indicate SD. Significant changes were analyzed using the Mann–Whitney U test and are indicated by asterisks (***, P < 0.001). (d and e) Transmission electron microscopy images of mock- or HSV-1 WT–infected iDCs (d) and mDCs (e). Original magnification 15,000. Scale bars, 1 µm. Black arrows, nuclear membrane; white arrowheads, capsids; black arrowheads, enveloped capsids. C, cytoplasm; N, nucleus.
Nuclear lamin proteins are degraded in HSV-1–infected iDCs, but not in mDCs
The nuclear lamina represents the main barrier for the nuclear egress of DNA-filled HSV-1 capsids (Mou et al., 2008). Hence, disruption of the nuclear lamina promotes access of viral capsids to the inner nuclear membrane and subsequently egress into the cytoplasm (Leach and Roller, 2010).
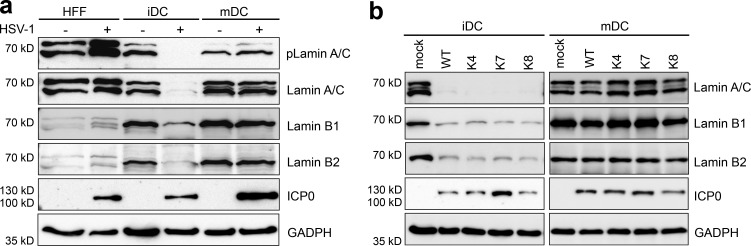
To investigate whether HSV-1 differentially regulates lamin turnover in iDCs and mDCs, both cell types were analyzed upon HSV-1 infection for 24 h by immunoblotting (Fig. 2 a). While lamin A/C was prominently expressed in HSV-1–infected mDCs compared with their uninfected counterparts, it was almost absent in HSV-1–infected iDCs at late stages of infection (i.e., 24 hpi; Fig. 2 a, second panel). HSV-1 infection is known to induce a phosphorylation of lamin A/C in fibroblastic or epithelial cells, which loosens the lamin network and facilitates the egress of viral capsids from the nucleus (Leach and Roller, 2010). However, and in sharp contrast to high levels of lamin A/C phosphorylation in HSV-1–infected human foreskin fibroblasts (HFFs), lamin A/C was not phosphorylated in iDCs or mDCs during the course of infection (Figs. 2 a and S2).
Figure 2.
Nuclear lamin protein levels are down-regulated in HSV-1–infected iDCs, but not mDCs. (a) HFF, iDCs, and mDCs (2 × 106) were mock- or HSV1-RFPVP26–infected (MOI of 2) and harvested 24 hpi. Cells were directly lysed and subjected to immunoblot analyses using specific antibodies raised against phospho-lamin A/C (pLamin A/C), lamin A/C, lamin B1, lamin B2, or ICP0. GAPDH was used to verify equal loading. (b) iDCs and mDCs were mock- or HSV-1–infected or infected with HSV-1 clinical isolates (K4, K7, and K8) and harvested 24 hpi. Samples were analyzed for protein expression of lamin A/C, lamin B1, lamin B2, and ICP0. GAPDH was used as a loading control. Experiments were performed at least three times with cells from different healthy donors and representative data are shown.
Moreover, the protein levels of lamin B1 and lamin B2 were also dramatically reduced in infected iDCs but were not affected in infected mDCs (Fig. 2 a, third and fourth panels, respectively). To analyze whether this phenotype was a general response to an HSV-1 infection, we tested three clinical HSV-1 isolates from different patients (Fig. 2 b, K4, K7, and K8). Confirming our data using the “WT” strain, reduced levels of lamin A/C, lamin B1, and lamin B2 were observed in iDCs, but not in mDCs, upon infection with either of these HSV-1 isolates. These data support the hypothesis that the nuclear lamin proteins might prevent nuclear egress of HSV-1 capsids in mDCs and could therefore explain the lack of newly produced virions in the supernatant of HSV-1–infected mDCs (Kruse et al., 2000; Mikloska et al., 2001).
HSV-1 infection induces autophagy in both iDCs and mDCs
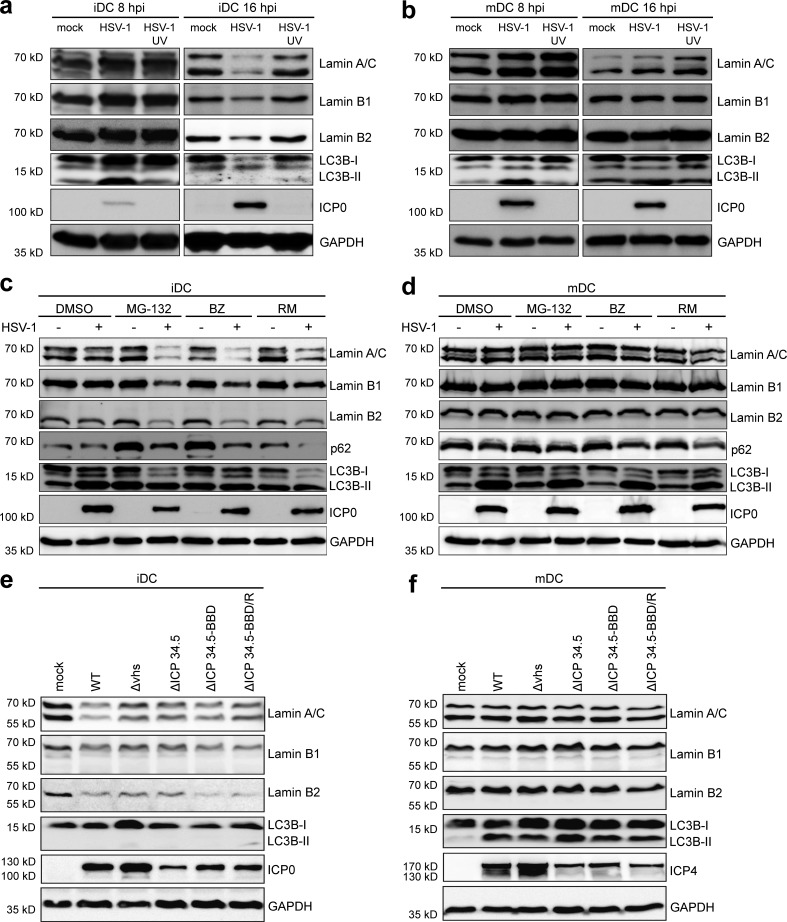
Previous studies reported that lamin B1 is degraded via autophagy in primary human fetal lung fibroblasts upon senescence induced by an oncogenic RAS protein (Dou et al., 2015). Therefore, iDCs and mDCs were infected with HSV-1 or UV-inactivated HSV-1 or mock treated and analyzed for LC3B-I and LC3B-II expression levels to monitor autophagy (Kabeya et al., 2000, 2004) using immunoblotting at early (8 hpi) and late (16 hpi) infection phases (Fig. 3, a and b).
Figure 3.
Induction of autophagy leads to an accelerated down-regulation of nuclear lamin protein levels in HSV-1–infected iDCs but not mDCs. (a and b) iDCs (a) and mDCs (b) were mock-, HSV-1–, or HSV-1 UV–infected and harvested 8 or 16 hpi. Samples were analyzed using immunoblotting for detection of lamin A/C, lamin B1, lamin B2, LC3B, or ICP0 levels. GAPDH was used as internal loading control. (c and d) iDCs (c) and mDCs (d) were mock- or HSV-1–infected and treated with different compounds known to induce autophagy (MG-132; BZ, bortezomib; RM, rapamycin) or DMSO as negative control. Cells were harvested 8 hpi, directly lysed, and subjected to immunoblot analyses using specific antibodies against lamin A/C, lamin B1, lamin B2, p62, LC3B, or ICP0. GAPDH was used to verify equal loading. (e and f) iDCs (e) and mDCs (f) were mock-, HSV-1 WT–, HSV-1 Δvhs–, HSV-1 ΔICP34.5–, HSV-1 ΔICP34.5-BBD–, or HSV-1 ΔICP34.5-BBD/R–infected and harvested 16 hpi. Expression levels of lamin A/C, lamin B1, lamin B2, LC3B, or ICP0/ICP4 were analyzed using immunoblot analyses. GAPDH was used as internal loading control. Experiments were performed at least three times with cells from different healthy donors.
Early during infection (i.e., at 8 hpi), lamin degradation was absent, while LC3B-II levels were already increased, indicating an induction of autophagy in both iDCs and mDCs (Fig. 3, a and b, respectively). In contrast, later, at 16 hpi, lamin, LC3B-I, and LCB-II protein levels were severely reduced in HSV-1–infected iDCs (Fig. 3 a). The decrease in LC3B levels at later time points might indicate autophagic flux, as autophagolysosomes are ultimately degraded (Mizushima and Yoshimori, 2007). Interestingly, and in sharp contrast to HSV-1–infected iDCs, lamin and LC3B-II levels remained stably high in HSV-1–infected mDCs until 16 hpi (Fig. 3 b). Furthermore, UV-inactivated HSV-1 did neither affect lamin expression levels nor LC3B-II signals in iDCs and mDCs, compared with the respective mock controls at 8 and 16 hpi (Fig. 3, a and b). These data indicate that the initiation of HSV-1 gene expression is necessary for autophagy induction.
To investigate whether lamin degradation occurred via autophagy, we infected iDCs and mDCs with HSV-1 for 8 h, when the lamin levels still remained unaltered upon infection, and furthermore treated these cells with different compounds known to induce autophagy. In particular, autophagy is activated by the proteasomal inhibitors MG-132 and bortezomib due to amino acid starvation (Klionsky and Emr, 2000) or rapamycin via its inhibition of mTOR phosphorylation. Accordingly, there was an increase in lamin A/C, lamin B1, and lamin B2 degradation in HSV-1–infected iDCs in the presence of these inducers of autophagy (Fig. 3 c), while the protein levels were unaffected in mDCs (Fig. 3 d). Notably, while the p62 protein levels were reduced in iDCs, p62 was barely affected in HSV-1–infected mDCs, suggesting inefficient autophagic degradation in mDCs.
To analyze the role of HSV-1 vhs, a viral encoded mRNA-specific RNase that immediately triggers a shutoff of cellular protein synthesis (Smiley, 2004), and HSV-1 ICP34.5, which can inhibit autophagy (Orvedahl et al., 2007), iDCs (Fig. 3 e) or mDCs (Fig. 3 f) were infected with the indicated HSV-1 strains for 16 h. The protein levels of lamin A/C, lamin B1, lamin B2 and LC3B-II were significantly reduced in iDCs infected with HSV-1 WT, Δvhs, ΔICP34.5, or ΔICP34.5-BBD (beclin-binding domain) as well as the rescue virus ΔICP34.5-BBD/R. In contrast, lamin protein levels and LC3 lipidation remained stable in mDCs upon infection with any HSV-1 strain or mutant. These results suggest that the differential regulation of lamin protein levels in iDCs versus mDCs did not depend on vhs or ICP34.5. Moreover, the lamin A/C protein levels were not simply reduced by transcriptional silencing, which was observed in HSV-1–infected iDCs or mDCs (Fig. S3). In particular, the protein levels of lamin A/C were also reduced upon infection with HSV-1 Δvhs in iDCs, but not mDCs, although their mRNA levels had not been changed in comparison to mock-treated cells.
Nuclear lamin proteins are cargos of autophagosomes
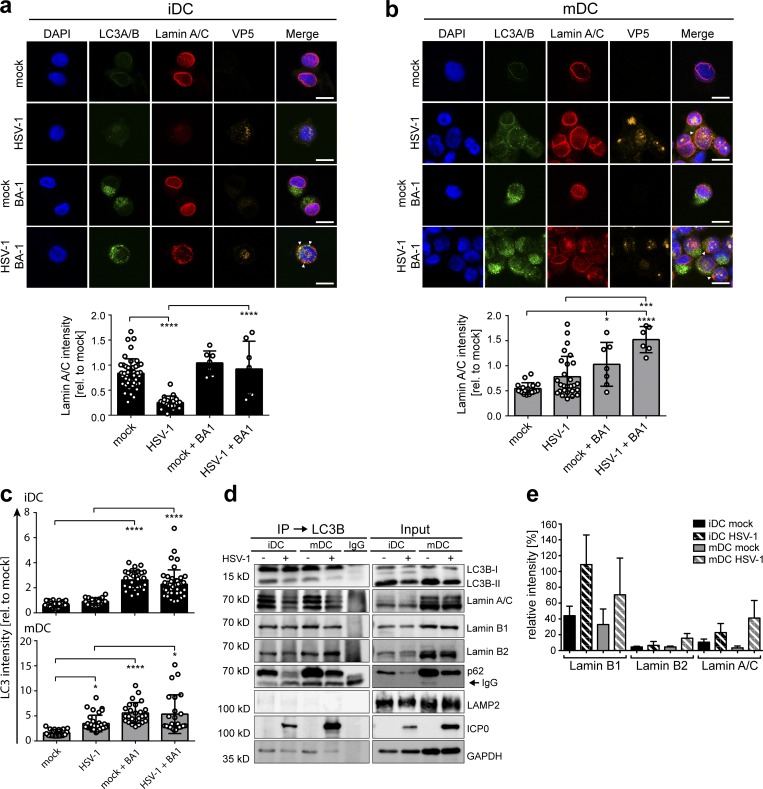
To determine whether lamins were degraded via autophagy in iDCs, we analyzed their subcellular localization during an HSV-1 infection. Therefore, iDCs and mDCs were mock- or HSV-1–infected in the presence or absence of bafilomycin A1 (BA-1), which inhibits the V-ATPase–dependent acidification of the endocytic pathway and autophagosome–lysosome fusion (Mauvezin and Neufeld, 2015). After 16 h of infection, the cells were fixed, labeled for LC3A/B, lamin A/C, and the HSV-1 major capsid protein VP5 (Fig. 4, a and b), and analyzed by fluorescence microscopy. Upon HSV-1 infection, lamin A/C protein levels were reduced to ∼30% of uninfected iDCs (Fig. 4 a). Interestingly, BA-1 treatment prevented the HSV-1–mediated loss of lamin A/C in iDCs and instead resulted in an ∼2.5-fold accumulation of LC3A/B (Fig. 4 c). In contrast, lamin levels were not altered in HSV-1–infected mDCs (Fig. 4 b), but the levels of LC3A/B increased significantly (by two- to fourfold) upon HSV-1 infection or BA-1 treatment when compared with uninfected, untreated cells (Fig. 4 c). Remarkably, lamin A/C (Fig. 4, a and b, arrowheads), lamin B1 (Fig. S3, a and b), and lamin B2 (Fig. S3, c and d) were no longer exclusively targeted to the nuclear envelope in iDCs and mDCs but also to autophagosomal organelles containing LC3A/B. These data strongly suggest that the lamins had been targeted to autophagosomes upon HSV-1 infection in iDCs as well as in mDCs.
Figure 4.
Nuclear lamins colocalize with autophagosomes in iDCs and mDCs. (a and b) iDCs (a) and mDCs (b) were mock- or HSV-1–infected and treated with or without BA-1. After 24 h of incubation, cells were analyzed via immunofluorescence microscopy for expression of LC3A/B (green), lamin A/C (red), and VP5 (orange). Nuclei were stained using DAPI (blue). Scale bars, 10 µm. Arrowheads display colocalization of LC3A/B and lamin A/C. Lamin A/C immunofluorescence signals were quantified as mean intensities. Values are shown relative to mock signal (i.e., without BA-1; iDC mock, n = 47; iDC HSV-1, n = 25; iDC mock + BA1, n = 7; iDC HSV-1+BA1, n = 7; mDC mock, n = 17; mDC HSV-1, n = 27; mDC mock + BA1, n = 7; mDC HSV-1 + BA1, n = 6). (c) Mean signal intensities of LC3A/B (a and b; and Fig. S4) were quantified in iDCs (top, black bars) and mDCs (bottom, gray bars). Values are shown relative to mock signal of the respective experiment (iDC mock, n = 28; iDC HSV-1, n = 19; iDC mock + BA1, n = 30; iDC HSV-1 + BA1, n = 42; mDC mock, n = 20; mDC HSV-1, n = 34; mDC mock + BA1, n = 25; mDC HSV-1 + BA1, n = 24). Error bars indicate SD. Significant changes were analyzed using a one-way ANOVA and Bonferroni multiple comparison post hoc tests and are indicated by asterisks (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001). (d and e) iDCs and mDCs (8 × 106) were mock- or HSV-1–infected and treated with BA-1. Cells were lysed 16 hpi and used for LC3B-immunoprecipitation. Samples were analyzed using immunoblot for expression of LC3B, lamin A/C, lamin B1, lamin B2, p62, LAMP2, ICP0, or GAPDH (d) and mass spectrometry–based proteomic label-free comparison of lamin A/C, lamin B1, and lamin B2 (e). Experiments were performed three times with cells from different healthy donors.
In a next step, immunoprecipitation for LC3B was performed to analyze whether lamin proteins are cargos of autophagosomes. Thus, iDCs or mDCs were mock-treated or infected with HSV-1 in the presence of BA-1 to prevent autophagosomal and lysosomal degradation. Cells were harvested 16 hpi and subjected to immunoblotting of total extracts as well as LC3B immunoprecipitation. As a positive control for efficient precipitation, LC3B and p62 protein levels were monitored using immunoblotting (Fig. 4 d, first and fifth panels). Since the lysosomal marker LAMP2 is not expected to be present in autophagosomes upon BA-1 treatment, it was therefore used as a negative control (Fig. 4 d, sixth panel). Indeed, LAMP2 was not detected in the LC3B-specific immunoprecipitates, while LC3B and also p62 had been both precipitated (Fig. 4 d). Interestingly, lamin A/C, lamin B1, and lamin B2 were detected in the LC3B precipitates of HSV-1– and mock-infected iDCs and mDC (Fig. 4 d, second, third, and fourth panels, respectively). ICP0 was used as an indicator for HSV-1 infection, and GAPDH was used as an internal loading control (Fig. 4 d, seventh and eighth panels, respectively). Furthermore, a quantitative mass spectrometry–based label-free analysis confirmed that the lamin proteins had been coprecipitated with LC3B at basal levels and that an infection of iDCs as well as mDCs with HSV-1 even increased the ratio of lamin to p62 at 16 hpi (Fig. 4 e and Table S2).
Degradation of the nuclear lamina via autophagy facilitates HSV-1 nuclear egress
Next, we asked whether HSV-1–induced degradation of lamins could be prevented by inhibiting autophagosomal degradation. Hence, iDCs and mDCs were mock-treated or HSV-1–infected in the presence or absence of BA-1, harvested at 16 hpi, and subjected to immunoblot analyses (Fig. 5 a). Protein levels of lamin A/C, lamin B1, and lamin B2 were clearly reduced in HSV-1–infected iDCs compared with the mock control, while treatment with BA-1 prevented their reduction (Fig. 5 a, first, second, and third panels). As expected, the lamin protein levels remained stable in HSV-1–infected mDCs. These observations were confirmed using a different autophagy inhibitor (i.e., spautin-1; see Fig. S5). Notably, while untreated HSV-1–infected iDCs showed reduced LC3B levels due to autophagic flux, HSV-1–infected mDCs showed an accumulation of LC3B-II.
Figure 5.
Inhibition of autophagy-mediated lamin degradation prevents nuclear egress of newly built capsids. (a–d) iDCs or mDCs were mock- or HSV-1–infected and treated with BA-1 1 h before infection. Cells were harvested 16 hpi, and samples were subjected to immunoblot analyses using antibodies specific for lamin A/C, lamin B1, lamin B2, LC3B, and ICP0 (a). GAPDH was used as a loading control. iDCs and mDCs were mock or HSV1-RFPVP26 infected and treated with spautin-1, BA-1, or DMSO as negative control (b and c). Cells were analyzed using confocal microscopy (b). Nuclei were stained using DAPI (blue). Scale bars, 10 µm. Quantification of intranuclear and cytoplasmic capsids based on RFPVP26 signals (c). Values are shown as ratios of cytoplasmic (“outside”) versus total capsid signals based on integral intensities (iDC, n = 15, iDC spautin-1, n = 13; iDC BA-1, n = 8). Error bars indicate SD. Significant changes were analyzed using the Mann–Whitney U test. Viral titers of the supernatants of iDCs (black bars) and mDCs (gray bar) were determined using a plaque assay (d). (e–h) IDCs (6 × 106) were electroporated with control or FIP200 siRNA. After 48 h, DCs were used for infection experiments. Cells were subjected to immunoblot analyses of FIP200 and GAPDH as loading control (e). DCs were mock- or HSV-1–infected and analyzed for protein levels of lamin A/C, lamin B1, lamin B2, and p62 using immunoblotting 20 hpi (f). VP5 was used as infection control and GAPDH to verify equal loading. DCs were mock- or HSV1-RFPVP26–infected and analyzed using confocal microscopy at 20 hpi (g and h). Nuclei were stained using DAPI (blue). Scale bars, 10 µm. Quantification of intranuclear and cytoplasmic capsids based on RFPVP26 signals (h). Values are shown as ratios of cytoplasmic (“outside”) versus total capsid signals based on integral intensities (control, n = 8; FIP200, n = 16). Error bars indicate SD. Significant changes were analyzed using the Mann–Whitney U test. Significant changes are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Experiments were performed three times with cells from different healthy donors.
In a following step, we investigated the subcellular capsid distribution in respect to autophagosomal degradation. Therefore, iDCs and mDCs were infected with HSV1-RFPVP26 for 16 h in the presence or absence of the autophagy inhibitors BA-1 or spautin-1 and analyzed by fluorescence microscopy. At this time point, the HSV-1 capsids were localized in a 40:60% ratio in both the nucleoplasm and the cytoplasm of control iDCs, but the nuclear fraction increased to 90% upon treatment with spautin-1 or BA-1 (Fig. 5, b and c). These results suggest that the degree of autophagosomal lamin degradation correlated with the efficacy of nuclear egress of HSV-1 capsids.
We also determined the amount of infectious virus released from HSV-1–infected DCs treated with or without autophagy inhibitors within 16 h (Fig. 5 d). Supernatants of uninfected iDCs were used as negative control. HSV-1–infected iDCs released approximately eightfold more progeny viruses into the supernatant when compared with HSV-1–infected mDCs, consistent with earlier reports (Kruse et al., 2000; Mikloska et al., 2001). Moreover, HSV-1–infected iDCs treated with autophagy inhibitors released significantly less progeny virus into the cell culture supernatants when compared with the DMSO-treated control (Fig. 5 d). These results show that inhibiting autophagy, and therefore preventing lamin degradation, led to reduced viral titers in the supernatant of iDCs and thus hampered the generation of infectious viral particles.
We furthermore blocked autophagy by siRNA-mediated silencing of FIP200 expression, which is required for the initiation of autophagy (Nazio et al., 2013). Thus, iDCs were electroporated and successful knockdown was verified via immunoblot analysis after 48 h (Fig. 5 e). Afterwards, cells were mock-treated or HSV-1–infected, and protein levels of lamins and p62 were evaluated at 20 hpi (Fig. 5 f). Protein levels of p62 were enriched in FIP200-silenced cells when compared with control iDCs, which indicates an inhibition of autophagic flux. Consistent with our previous data, lamin A/C, B1, B2, and p62 levels were significantly reduced in HSV-1–infected iDCs treated with a scramble siRNA when compared with the mock control. Notably, specific inhibition of autophagy by silencing FIP200 expression strongly hampered the degradation of lamins upon infection. Finally, using fluorescence microscopy, the subcellular capsid distribution was analyzed in control versus FIP200-silenced iDCs after infection with HSV1-RFPVP26 for 20 h (Fig. 5, g and h). While ∼30% of HSV1-RFPVP26 capsids were localized in the cytoplasm in control iDCs, only 10% of cytoplasmic capsids were found upon FIP200 silencing.
Taken together, our data suggest that lamin was degraded in iDCs upon HSV-1 infection by autophagy and that this weakening of the lamin barrier at the inner nuclear envelope facilitated nuclear egress of HSV-1 capsids.
Infection of mDCs with HSV-1 leads to autophagosome accumulation
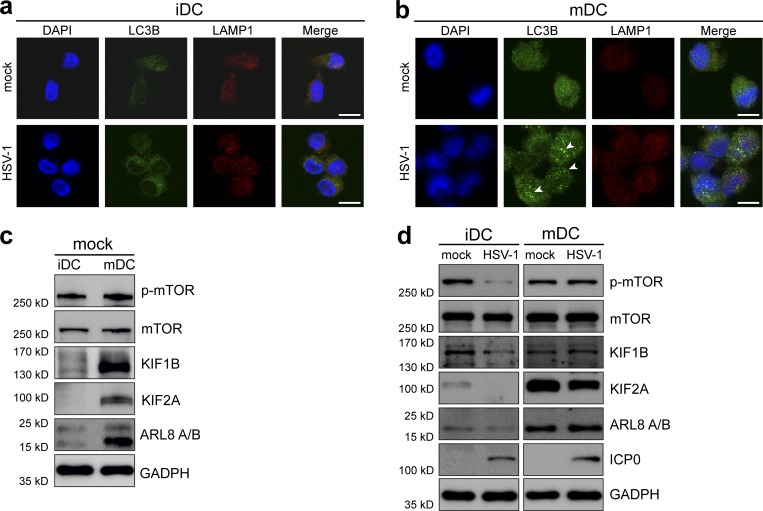
Since our results indicated that the autophagic flux might be hampered in mDCs, we investigated whether autophagosomes and lysosomes were able to fuse and to induce degradation of autophagolysosomes. Therefore, iDCs and mDCs were mock- or HSV-1–infected, and immunofluorescence analyses were performed at 8 hpi. DCs were labeled with anti-LC3B, a marker for autophagosomes, and anti-LAMP1, a specific lysosomal protein. HSV-1– and mock-infected iDCs showed comparable LC3B and LAMP1 signal intensities (Fig. 6 a). Remarkably, LC3B levels were considerably higher in mDCs and concentrated in certain cytoplasmic regions upon HSV-1 infection (Fig. 6 b, arrowheads). LAMP1 expression levels were comparable between iDCs and mDCs (Fig. 6, a and b). These results indicate that LC3B accumulates in HSV-infected mDCs when compared with infected iDCs. Thus, these data suggest that the fusion of autophagosomes with lysosomes might be hampered in mDCs.
Figure 6.
Up-regulation of KIF1B and KIF2A during DC maturation inhibits fusion of autophagosomes with lysosomes in mDCs. (a and b) iDCs (a) or mDCs (b) were mock- or HSV-1–infected. Cells were subjected to confocal immunofluorescence microscopy at 8 hpi. DCs were labeled with antibodies specific for LC3B (green) and LAMP1 (red). Nuclei were stained with DAPI (blue). Arrowheads mark autophagosome accumulations. Scale bars, 10 µm. (c) iDCs and mDCs (3 × 106) were subjected to immunoblot analyses. Protein levels of phospho-mTOR, mTOR, KIF1B, KIF2A, and ARL8 A/B were investigated. GAPDH was used as internal loading control. The experiment was performed at least four times with cells from different healthy donors. (d) iDCs and mDCs (3 × 106) were mock- or HSV-1–infected and analyzed using immunoblot at 16 hpi. Levels of phospho-mTOR, mTOR, KIF1B, KIF2A, and ARL8 A/B were detected with the respective antibodies. Infection of DCs was confirmed using ICP0. GAPDH was used to verify equal loading. Experiments were performed at least three times with cells from different healthy donors.
In this context, the kinesin family members KIF1B and KIF2A represent interesting factors contributing to autophagosome–lysosome fusion. KIF1B and KIF2A were described to induce mTOR activity, which in turn inhibits autophagy (Poüs and Codogno, 2011). Furthermore, the presence of KIF1B, KIF2A, and Arf-like GTPase (ARL)8 A/B leads to a peripheral localization of lysosomes (Hofmann and Munro, 2006; Poüs and Codogno, 2011), thus inhibiting autophagosome–lysosome fusion (Korolchuk and Rubinsztein, 2011). Interestingly, immunoblots showed that the protein levels of phosphorylated mTOR (p-mTOR; Fig. 6 c, first panel), KIF1B (Fig. 6 c, third panel), KIF2A (Fig. 6 c, fourth panel), and ARL8 A/B (Fig. 6 c, fifth panel) were elevated in mDCs when compared with iDCs (Fig. 6 c, third and fourth panels, respectively). These data are consistent with earlier reports indicating that elevated levels of KIF1B, KIF2A, and ARL8 A/B are associated with an increased phosphorylation of mTOR (Santama et al., 1998; Hofmann and Munro, 2006; Poüs and Codogno, 2011).
Next, we analyzed the expression of these proteins during an HSV-1 infection (Fig. 6 d). In mock-infected iDCs, KIF1B and KIF2A were only slightly expressed and further decreased upon HSV-1 infection (Fig. 6 d, third and fourth panels, respectively). Interestingly, this was accompanied by a decrease of p-mTOR levels during an HSV-1 infection of iDCs (Fig. 6 d, first panel). In contrast, the weak expression of ARL8 A/B remained unaltered in mock- and HSV-1–infected iDCs (Fig. 6 d, fifth panel). mDCs expressed overall higher levels of KIF1B, KIF2A, and ARL8 A/B compared with their immature counterparts (Fig. 6 d, third, fourth, and fifth panels, respectively). Similarly to iDCs, a correlation between KIF1B, KIF2A, as well as ARL8 A/B, expression levels, and phosphorylated mTOR could be observed in infected mDCs. In contrast to a reduction of p-mTOR in HSV-1–infected iDCs, significant phosphorylation levels of mTOR remained stable upon HSV-1 infection of mDCs accompanied by stable expression of KIF1B, KIF2A, and ARL8 A/B (Fig. 6 d, first, third, fourth, and fifth panels). Taken together, these data indicate that high protein levels of KIF1B, KIF2A, and ARL8 A/B lead to an activation of mTOR via its phosphorylation and thus to an inhibition of autophagy in mDCs.
Kinesin family members KIF1B and KIF2A are responsible for inhibition of autophagic degradation of lamins in mDCs
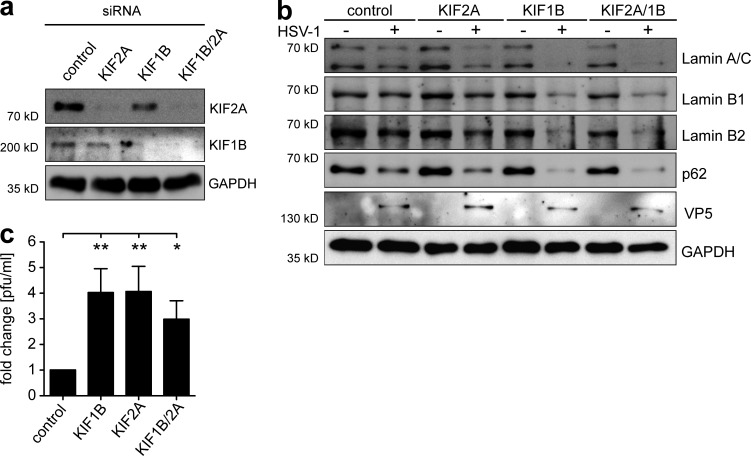
Having demonstrated the elevated protein expression of KIF1B and KIF2A in mDCs, we next examined whether this was responsible for the inefficient autophagic flux and therefore for the failure of lamin degradation upon HSV-1 infection. To investigate this, iDCs were electroporated with siRNAs targeting KIF1B and/or KIF2A followed by the maturation of the cells. Successful silencing of KIF1B and/or KIF2A was monitored at 2 d after electroporation (Fig. 7 a). Subsequently, cells were mock- or HSV-1–infected and subjected to immunoblot analyses to monitor lamin protein levels at 24 hpi (Fig. 7 b). Supporting our hypothesis, lamin degradation was observed in HSV-1–infected mDCs, when the expression of KIF2A, KIF1B, or both had been ablated (Fig. 7 b, first, second, and third panels). Knockdown of KIF1B alone or the combined knockdown of KIF1B/KIF2A enhanced the reduction of lamin and p62 expression in HSV-1–infected mDCs, when compared with KIF2A-silenced mDCs (Fig. 7 b, first and fourth panels). These results clearly indicate that KIF1B and KIF2A both contribute to the inhibition of HSV-1–induced lamin degradation in mDCs, and that silencing the expression of at least one kinesin was sufficient to promote autophagic degradation of lamins.
Figure 7.
RNAi-mediated silencing of KIF1B and KIF2A facilitates lamin degradation and nuclear egress of progeny HSV-1 capsids. IDCs (3 × 106) were electroporated with control siRNA, KIF1B siRNA, KIF2A siRNA or both, KIF1B and KIF2A siRNA. Afterward, maturation was induced via addition of the cytokine cocktail. After 48 h, matured DCs were used for further experiments. (a) Cells were subjected to immunoblot analyses of KIF1B and KIF2A protein levels. GAPDH was used as loading control. (b) DCs were mock- or HSV-1–infected and analyzed for lamin A/C, lamin B1, lamin B2, and p62 levels using immunoblotting 20 hpi. VP5 was used as infection control and GAPDH to verify equal loading. (c) Viral titers of the supernatants were determined using a plaque assay. Error bars indicate SD. Significant changes are indicated by asterisks (*, P < 0.05; **, P < 0.01). The experiment was performed four times with cells from different healthy donors.
Finally, a plaque assay was performed to determine the HSV-1 titers in the supernatants of HSV-1–infected mDCs that were pretreated with the respective siRNAs. HSV-1–infected mDCs treated with KIF1B, KIF2A or both siRNAs released more infectious HSV-1 particles into the supernatants than mDCs treated with scrambled siRNA (Fig. 7 c). Taken together these data indicate that siRNA-mediated knockdown of KIF1B and/or KIF2A leads to an enhanced lamin degradation in HSV-1–infected mDCs and subsequently to increased viral loads in the respective cell culture supernatants.
Combining our data, we conclude that autophagy-mediated lamin degradation facilitates nuclear egress of HSV-1 capsids and thus the release of progeny virus into the cell culture supernatant.
Discussion
As potent antigen-presenting cells, DCs play an important role in limiting the propagation of pathogens by the induction of potent (e.g., antiviral) immune responses (Banchereau and Steinman, 1998). In this regard, HSV-1 evolved a plethora of immune evasion strategies to overcome the host’s immune system and to establish successful replication (Pollara et al., 2004; Cunningham et al., 2010). In the present study, we report that the HSV-1 infection of iDCs resulted in autophagic degradation of the nuclear lamina proteins lamin A/C, B1, and B2, facilitating nuclear egress of newly formed viral capsids. In contrast, HSV-1 replication was significantly impaired in mDCs, and we proved that elevated KIF1B and KIF2A levels in mDCs were responsible for the inhibition of autophagy and thus for impaired nuclear egress of HSV-1 capsids. These observations provide an explanation why HSV-1–infected mDCs mainly release noninfectious L-particles, since L-particles lack the viral capsid and DNA and are therefore not infectious (McLauchlan and Rixon, 1992). Nevertheless, they are also able to interfere with DC biology, such as via the down-modulation of the CD83 molecule on the surface of mDCs and thus inhibition of antiviral immune responses (Heilingloh et al., 2015).
In the present study, we show that in HSV-1–infected iDCs, nuclear lamins were degraded by autophagy to facilitate nuclear egress of newly assembled capsids. Given previous investigations of the nuclear lamina by Moir et al., this finding was rather unexpected. Using light microscopy, the authors showed discontinuities in the lamina of proliferating epidermal cells, which might be large enough to allow capsids access to the inner nuclear membrane (Moir et al., 2000). Moreover, a disruption of nuclear lamins is not essential for nuclear egress of HSV-1 capsids in Vero cells (Vu et al., 2016). Apart from this, phosphorylation of lamin A/C as well as lamin B is crucial to disorganize the nuclear lamina and to facilitate the egress of HSV-1 capsids through the nuclear membrane in several cell types (Park and Baines, 2006; Wu et al., 2016). As a similar mechanism contributes to nuclear egress of progeny viral capsids of the human cytomegalovirus, it might be conserved throughout the herpesvirus family and for all cell types (Marschall et al., 2005; Milbradt et al., 2009, 2010). However, our data show that lamin A/C phosphorylation was absent in iDCs and mDCs, in contrast to HFFs (Figs. 2 and S2), and other mechanisms, including autophagy, might be important for the egress of progeny viral capsids in this nonproliferating cell type.
However, only little is known about the degradation of nuclear proteins via autophagy. Dou et al. demonstrated that the nuclear lamina protein lamin B1 is degraded in an autophagy-dependent manner in primary human fetal lung fibroblasts upon oncogenic insult (Dou et al., 2015). In particular, LC3 can directly interact with lamin B1, but not with lamin A/C or lamin B2, exclusively in the nucleoplasm, and the export of lamin B1 from the nucleus to the cytoplasm results in its lysosomal degradation. In contrast, HSV-1 infection of iDCs does not only lead to the autophagic degradation of lamin B1, but also of lamin A/C and lamin B2 (Figs. 3 and 4). HSV-1 inhibits autophagy in fibroblasts and primary neurons (Tallóczy et al., 2002) via the viral late phase proteins ICP34.5 and Us11 (Orvedahl et al., 2007; Lussignol et al., 2013). However, our data clearly indicate that in monocyte-derived DCs ICP34.5 does not prevent autophagy initiation, as it does also not in bone marrow–derived DCs (Gobeil and Leib, 2012). Moreover, ICP34.5 is not involved in the differential regulation of lamin degradation by autophagy in iDCs versus mDCs, since infection with an ICP34.5-deletion mutant mirrored the findings using HSV-1 WT (Fig. 3, e and f). Furthermore, HSV-1 induces a specific type of autophagy, namely nuclear envelope-derived autophagy (NEDA) in several cell types (Radtke et al., 2013). NEDA uses, in contrast to macroautophagy, the nuclear envelope as a source for the wrapping membranes to enclose the substrates to be degraded by autophagy. Since NEDA is differentially regulated compared with macroautophagy, the authors hypothesized that NEDA is a cellular stress response, which is triggered late during HSV-1 infection when the classical macroautophagy has already been inhibited (Radtke et al., 2013).
Autophagy is important for the maintenance of cellular homeostasis as well as during episodes of cellular stress (Crotzer and Blum, 2009). Furthermore, autophagy is particularly important when fighting against bacterial as well as viral infections (Deretic et al., 2013). Thus, the induction of autophagy upon HSV-1 infection is most likely a response of the infected cell to combat this infection. HSV-1 in turn utilizes autophagy to facilitate the nuclear egress of progeny capsids and therefore viral maturation and spread. In addition, autophagy is identified as a major route for antigen processing and loading onto MHC class I and II complexes for antigen presentation to CD8- and CD4-positive T cells (Zhou et al., 2005; Schmid et al., 2007; Budida et al., 2017). Therefore, the high autophagic activity of iDCs is necessary to facilitate effective antigen presentation. However, upon DC maturation, autophagy is no longer as essential and could consequently be down-regulated.
Our data show that autophagy induction as well as autophagosome maturation occurs in mDCs but, due to the elevated protein levels of KIF1B and KIF2A, autophagosome–lysosome fusion is impaired. Earlier studies demonstrated that a protein complex consisting of KIF1B, KIF2A, and ARL8 A/B leads to a peripheral localization of lysosomes and therefore reduces the likelihood of autophagosomes meeting lysosomes (Santama et al., 1998; Hofmann and Munro, 2006; Poüs and Codogno, 2011). The subcellular localization of lysosomes and autophagosomes is therefore a key determinant of the rate of autophagosome–lysosome fusion. While depletion of KIF1B and KIF2A increases autophagosome–lysosome fusion, overexpression of these kinesins results in peripheral localization of lysosomes and thereby reduces autophagosome–lysosome fusion (Korolchuk and Rubinsztein, 2011). Nevertheless, the peripheral localization might not be the only mechanism to inhibit autophagy in mDCs. The elevated levels of these proteins in mDCs induce mTOR phosphorylation, which in turn also inhibits the autophagic pathway. Furthermore, the autophagy pathway is enhanced through inhibition of mTOR by knockdown of KIF1B and KIF2A or both.
In conclusion, our data show that the nuclear egress of HSV-1 capsids in iDCs is dependent on autophagic degradation of nuclear lamins and that HSV-1 depends on autophagy for efficient virus assembly in iDCs. In contrast, an up-regulation of KIF1B and KIF2A protein expression suppresses autophagy in mDCs and impairs the nuclear egress of newly assembled viral capsids. This cellular mechanism represents a novel powerful counterstrike to inhibit viral spread in the host cells and the host organism.
Materials and methods
Generation of DCs
Monocyte-derived DCs were generated from peripheral blood mononuclear cells from different healthy donors as described earlier (Heilingloh et al., 2014). In brief, peripheral blood mononuclear cells were isolated using a Lymphoprep gradient (Nycomed Pharma AS), and subsequently, monocytes were separated using plastic adherence. Monocytes were differentiated to iDCs by the addition of 800 U/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; Milteny) and 250 U/ml IL-4 (Milteny). On day 5, iDCs were used for further experiments or subsequently matured by adding 10 ng/ml TNF-α (Beromun), 1 µg/ml prostin E2 (PGE2; Pfizer), 200 U/ml IL-1β (CellGenix), 40 U/ml GM-CSF, 1,000 U/ml IL-6 (CellGenix), and 250 U/ml IL-4 to the medium. The phenotypic maturation status was verified using flow cytometry.
Approvals and legal requirements
For the generation of monocyte-derived DCs from leukapheresis products of healthy donors, a positive vote from the local ethics committee has been obtained (reference number 4556). The present study was performed in accordance with the Declaration of Helsinki and recommendations of the ethics committee of the Friedrich-Alexander Universität Erlangen-Nürnberg, with written informed consent from all donors.
Generation of pHSV1(17+)Lox-mRFPVP26
We generated pHSV1(17+)Lox-EGFPVP26 (HSV1-GFPVP26) and pHSV1(17+)Lox-mRFPVP26 (HSV1-RFPVP26) from the bacterial artificial chromosome (BAC) pHSV1(17+)Lox by replacing the first seven N-terminal amino acids of VP26 by EGFP or mRFP as reported before for pHSV1(17+)Lox-CherryVP26 (Sandbaumhüter et al., 2013). The sequences encoding the fluorescent proteins were amplified from pEP-mRFP-in and pEP-GFP-in, and the PCR fragments were transformed together with the BAC pHSV1(17+)Lox into Escherichia coli GS1783 for homologous recombination (Tischer et al., 2006; Nagel et al., 2008). BAC modifications were analyzed by digests with seven restriction enzymes and in-house sequencing of the BAC sequences around the insertion. BAC DNA was purified from overnight E. coli cultures using the Nucleobond BAC100 kit (Machery and Nagel) and resuspended in 50 µl 10 mM Tris-HCl, pH 8, and 50 µg/ml RNase A. Approximately 25 µg of DNA was digested with 35 U restriction enzymes (AscI, BamHI, EcoRI, EcoRV, HindIII, NotI, or XhoI; Fermentas) for 3.5 h, and loaded on a 0.6% (wt/vol) agarose gel in 0.5× TBE buffer (0.44 M Tris-HCl, 0.44 M boric acid, and 10 mM EDTA). Fragments were separated by gel electrophoresis at 66 mA for 17 h (Fig. S1 a). We calculated the expected restriction fragment sizes based on the published sequence of HSV1(17+) (GenBank accession number NC_001806). 10 µg BAC DNA was transfected in a 60-mm dish with subconfluent Vero cells using the MBS mammalian transfection kit. Cells and medium were collected after development of cytopathic effects. Samples were freeze-thawed three times and used to amplify the viral stocks.
Characterization of HSV1(17+)Lox-mRFPVP26
Extracellular viral particles secreted into the extracellular media were harvested by centrifugation. 6 × 107 or 1.3 × 107 plaque-forming units (PFUs) per lane of HSV1(17+)Lox or HSV1(17+)Lox-mRFPVP26, respectively, were loaded onto a 6–18% gradient gel and transferred (48 mM Tris, 380 mM glycine, 20% [vol/vol] methanol, and no SDS) onto nitrocellulose membranes (Pall Corp.). After blocking in 5% low-fat milk in PBS with 0.1% (vol/vol) Tween 20 membranes were probed with a mouse monoclonal antibody against VP5 (H1.4; Meridian Life Science) and rabbit polyclonal antibodies against gC (R47; Eisenberg et al., 1987) and VP26 (amino acids 95–112; Desai et al., 1998) followed by incubation with secondary antibodies coupled to alkaline phosphatase (Dianova). Membranes were transferred into TSM buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 5 mM MgCl2) for 20 min and stained with 0.2 mM nitroblue tetrazolium chloride and 0.8 mM 5-bromo-4-chloro-indolyl-3-phsophate in TSM buffer. For documentation, membranes were scanned with a ScanJet 6300 scanner (Fig. S1 b; Hewlett Packard).
Virus strains, virus preparation, and virus titration
In the present study, the HSV-1(17+) cytomegalovirus-EGFP/UL43 strain expressing EGFP under the control of the cytomegalovirus promoter, inserted into the UL43 gene, was used as the WT strain (HSV-1; BioVex). The following HSV-1 deletion strains were used in this study: HSV-1 strain 17 Δvhs (HSV-1-GFPΔKan-UL41(vhs); provided by Martin Messerle, Hannover Medical School, Hannover, Germany), HSV-1 strain 17 ΔICP34.5 (17termA, premature termination in the ICP34.5 gene; Bolovan et al., 1994), HSV-1 ICP34.5Δ68–87 (in-frame deletion of amino acids 68–87 of ICP34.5 = ΔICP34.5-BBD, beclin-binding domain; Orvedahl et al., 2007), and HSV-1 ICP34.5Δ68-87R (marker rescue virus = ΔICP34.5_BBD/R; Orvedahl et al., 2007; provided by David Leib, Geisel School of Medicine at Dartmouth, Hanover, NH). HSV-1 clinical isolates derived from patients (K4, K7, and K8, kindly provided by Manfred Marschall, Institute for Clinical and Molecular Virology, Erlangen, Germany) were used for further infection experiments. Preparation of virus stocks was performed using a modified protocol described by Sodeik et al. (1997). Briefly, subconfluent BHK-21 cells (used for HSV-1 WT, HSV-1 Δvhs, and HSV1-RFPVP26) or Vero cells (used for ICP34.5 mutant strains) were infected with RPMI 1640 supplemented with 20 mM Hepes (5 ml/175 cm2 flask) containing a low multiplicity of infection (MOI; 0.01). After 1–2 h, 25 ml D10 medium was added and cells were cultivated for 3–4 d, until complete cytopathic effect was observed. Medium was harvested, cell debris was removed via centrifugation at 2,575 g and 4°C for 10 min, and virus containing supernatant was centrifuged at 39,742 g at 4°C for 2 h. Virus pellets were overlaid with a small volume of MNT buffer (30 mM MES, 100 mM NaCl, and 20 mM Tris) at 4°C overnight. Afterwards, virus pellets were resuspended, aliquoted, and stored at −80°C until further use. For UV inactivation (HSV-1 UV), virions were exposed six times to 0.12 J/cm2 in a Vilber Luormat and mixed gently after each irradiation cycle.
Plaque assay
Vero cells, used for viral titration, were cultured in D10 medium (DMEM; Lonza) supplemented with 10% FCS (Merck), 2 mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin. Titration of cell culture supernatants was performed using Vero cells at 100% of confluency. Cells were washed with RMPI 1640 (Lonza) supplemented with 0.1% BSA (Sigma-Aldrich) and 20 mM Hepes (Lonza) before 200 µl supernatant from cell culture was added. After incubation on a rocking platform for 1 h at room temperature, the inoculum was removed, and 1 ml D10 medium containing 10 µg/ml human IgG (Sigma-Aldrich) was added to each well. Cells were cultured in an incubator for 3 d until visible plaques had formed. Media were discarded, and the cells were fixed with 9% formaldehyde in PBS for 10 min. Afterwards, the formaldehyde solution was removed and 1 ml crystal violet solution (5% crystal violet in ethanol, 1:50 dilution in H2O) was added and incubated for 10 min. Subsequently, wells were washed with water and air-dried. Finally, plaques were counted, and the viral titer was calculated and indicated in PFUs per milliliter.
Single-step growth kinetics and plaque assays
Subconfluent Vero cells were inoculated with 5 PFUs per cell and supernatants were harvested at 9, 12, 18, and 24 hpi, respectively. Extracellular virus titers were determined by plaque assays as described previously (Döhner et al., 2002). Briefly, Vero cells were cultured to just confluence in six-well dishes and incubated for 1 h at room temperature on a rocking platform with 10-fold serial dilutions of the different virus suspensions in CO2-independent medium (Life Technologies Gibco) with 0.1% (wt/vol) cell culture grade bovine serum albumin (PAA Laboratories). The inoculum was removed, and 2 ml/well growth medium containing 20 µg/ml pooled human IgG (Sigma-Aldrich) was added. The cells were incubated for 3 d, fixed in absolute methanol, and stained with 0.1% (wt/vol) crystal violet and 2% (vol/vol) ethanol in H2O (Fig. S1 c).
Infection procedure
DCs (2 × 106) were infected in a total volume of 300 µl RPMI 1640 medium supplemented with 20 mM Hepes at an MOI of 2. Infection was performed for 1 h at 37°C at 300 rpm in a shaking heating block (Eppendorf). Subsequently, cells were transferred to RPMI 1640 medium containing 1% autologous serum, 10 mM Hepes, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 40 U/ml GM-CSF, and 250 U/ml IL-4, adjusted to a final concentration of 106 cells/ml.
Where indicated, cells were treated with 10 µM MG-132, 2 µM bortezomib, or 10 nM rapamycin (all from Enzo Life Sciences) at 1 hpi. To inhibit autophagy and lysosomal degradation, cells were incubated with 10 µM spautin-1 or 1 µM BA-1 (all from Sigma-Aldrich) 1 h before infection.
Immunofluorescence microscopy
To analyze the subcellular localization of viral capsids, iDCs or mDCs were infected with HSV1-RFPVP26 and allowed to adhere on poly-L-lysine (Sigma-Aldrich)–coated glass coverslips. Cells were fixed using 4% paraformaldehyde in PBS for 20 min. For nuclear staining and mounting, Roti-Mount FluorCare DAPI (Carl Roth) was used. Microscopic images were acquired using an LSM780 confocal microscope equipped with a Plan-Apochromat 100×/1.40 oil M27 objective and ZEN 2011 SP3 acquisition software (black edition; Zeiss). As imaging medium, we used Immersol 518 F (Zeiss). For image processing and contrast adjustments, CorelDRAW X6 software (Corel) was used. For quantification of capsid distribution in one confocal section, we manually segmented the nucleus and calculated the proportion of RFPVP26 signal that is located outside the nucleus (cytoplasm) versus total intensities for individual cells using the Fiji software (Schindelin et al., 2012) and a custom-written macro (see ZIP file in Online supplemental material).
To label lamin proteins LC3B and LAMP1, the cells were fixed and permeabilized using 100% ice-cold methanol. Blocking was performed using 1% BSA in PBS. Antibodies and fluorochromes used for immunofluorescence are listed in Table S1. Confocal microscopy was performed as described above. To quantify the immunofluorescence signal intensities of lamin and LC3A/B in one confocal section, we manually defined cell regions of individual cells using Fiji software (Schindelin et al., 2012) and a custom-written macro (see ZIP file in Online supplemental material).
Transmission electron microscopy
Cells were grown in poly-L-lysine–coated 35 mm glass bottom dishes (ibidi) and fixed with glutaraldehyde (2.5% in 100 mM phosphate buffer) for 2 h at the indicated time points post infection. Cells were then contrasted en bloc with 1% osmium tetroxide, 1% thiocarbohydrazide, 1.5% potassium ferrocyanide, and 0.5% uranyl acetate before they were dehydrated in an ascending ethanol series (50%, 70%, 90%, and 3× 100%, 10 min each) and stepwise (EPON/EtOH 1:1 for 2 h, EPON overnight) flat-embedded in EPON resin (Poly/Bed 812; Polysciences Europe). Subsequently, EPON-embedded samples were polymerized for 60°C at 24 h. After removal of the glass slide with 40% hydrofluoric acid, the EPON disk containing the cells of interest was removed from the dish, mounted on EPON resin blocks, and processed for ultramicrotomy. Serial sections were cut using a Leica EM UC7 ultramicrotome set to a thickness of 50 nm and routinely poststained with 1% uranyl acetate and 3.8 mM lead citrate. Sections were investigated using a LaB6-equipped JEOL 1400Plus transmission electron microscope. Digital images were acquired using a 4,096 × 4,096-pixel CMOS camera (TemCam-F416; TVIPS) and image postprocessing of the resulting 16-bit TIFF files was performed using Adobe Photoshop CS6.
Cell lysis and immunoblotting
Cells were harvested and solubilized in 2× Rotiload (Carl Roth) supplemented with 1 mM MgCl2 and 12.5 U/ml benzonase (Sigma-Aldrich). Samples were incubated at 37°C and with constant shaking at 300 rpm in a heating block for 15 min and then denaturated at 95°C for 10 min. Proteins were separated using SDS-PAGE and subsequently transferred onto nitrocellulose membranes. After blocking with 5% (wt/vol) dry milk or 1× RotiBlock (Carl Roth) the membranes were incubated with the primary antibodies at 4°C overnight. After incubation with the appropriate secondary HRP-labeled antibodies, detection was performed using ECL Prime Western blotting Detection Reagent (GE Healthcare). Antibodies used for immunoblot analyses are listed in Table S1.
RNA isolation, cDNA synthesis, and quantitative real-time PCR
For RNA isolation, DCs were harvested, washed once with cold PBS, and homogenized using the QIAshredder kit (QIAGEN) according to the manufacturer’s instructions. Total RNA was subsequently isolated using the RNeasy MiniKit (QIAGEN) according to the manufacturer’s recommendations. Genomic DNA was removed via on-column DNA-digestion using the RNase-free DNase set (QIAGEN). Finally, cDNA was transcribed with the First Strand cDNA synthesis kit (Thermo Fisher Scientific) as specified by the manual. Quantitative real-time PCR was performed using Sso Advanced SYBR Green Supermix (BioRad) and lamin A/C (forward, 5′-CAAGAAGGAGGGTGACCTGA-3′; reverse, 5′-GCATCTCATCCTGAAGTTGCTT-3′)– or S14 (forward, 5′-GGCAGACCGAGATGAATCCTCA-3′; reverse, 5′-CAGGTCCAGGGGTCTTGGTCC-3′)-specific primers. The following quantitative real-time PCR cycling profile was performed: 95°C, 3 min; (95°C, 10 s; 63°C, 30 s; 72°C, 30 s) 40 cycles; 65–95°C, 0.5°C/s using the Touch Thermal Cycler CFX96 TM Real Time System (BioRad). Data analysis was performed using CFX Manager 3.0 software (BioRad).
Immunoprecipitation of LC3B
For immunoprecipitation of autophagosomes, 8 × 106 iDCs or mDCs were treated with 1 µM BA-1 1 h before infection. Afterwards, samples were mock treated or infected with HSV-1 (MOI of 2) and cultured in DC medium supplemented with 40 U/ml GM-CSF, 250 U/ml IL-4, and 1 µM BA-1. Cells were harvested 18 hpi and lysed directly in 500 µl lysis buffer (10% glycerol, 2 mM EDTA, pH 8, 137 mM NaCl, 10 mM sodium phosphate, pH 7.2, 0.5% NP-40, 2 mM PMSF, 20 mM sodium fluoride, and 2 mM Na-orthovanadate). After incubation at 4°C in an overhead rotator for 1 h, lysates were precleared via the addition of 25 µl protein A magnetic beads to each sample. Following incubation at 4°C in an overhead rotator for 1 h, 30 µl each lysate was mixed with 4× Rotiload (input control) and denatured at 95°C for 10 min. For immunoprecipitation, 1 µg of the LC3B-specific antibody (Table S1) was added to each sample. As negative control, pure lysis buffer was incubated with antibody in parallel. Samples were rotated at 4°C overnight. Next, 70 µl protein A magnetic beads was added to each sample followed by an incubation at 4°C in an overhead rotator for 5 h. Immunoprecipitates were washed five times with PBS supplemented with 0.1% Tween. Finally, magnetic beads were resuspended in 60 µl of 2× Rotiload and incubated for 15 min at 95°C. Samples were analyzed by immunoblot analysis or subjected to quantitative label-free mass spectrometry.
Mass spectrometry analysis
For comparative proteomic analysis, protein extracts as well as immunoprecipitates were processed by a modified filter-aided sample preparation method (Wiśniewski et al., 2009) as previously described by Lamm et al. (2017). Briefly, protein samples were loaded on a 10-kD-cutoff filter (Vivacon 500; Sartorius) and washed with 8M urea supplemented with 50 mM TEAB (wash buffer) to discard remaining detergents and to denature proteins for enhanced accessibility during protease digestion. Proteins were reduced with 25 mM DTT in wash buffer followed by alkylation with 25 mM 2-chloroacedtamide (Sigma-Aldrich) in wash buffer in darkness for 30 min. Upon each buffer change or washing step, samples were centrifuged until dry. Protein samples were then incubated with 50 µl of 6 M urea supplemented with 50 mM TEAB and 0.5 µg Lys-C (Wako Chemicals) at a shaking incubator at 37°C for 3 h. Subsequently, 250 µl of 50 mM TEAB was added adjusting a final concentration of 1 M urea for tryptic digestion using 1 µg trypsin (sequencing-grade modified trypsin; Promega) at 37°C overnight. After protein digestion, obtained tryptic peptides were collected in the flow through after centrifugation. The resulting peptide solution was acidified to a final concentration of 0.1% TFA for subsequent desalting using C18 stage tips. Subsequently, peptide samples were vacuum concentrated and resuspended in 5% formic acid. All peptide samples were separated by reverse phase chromatography with a linear increase of acetonitrile on a nano flow Ultimated 3000 HPLC (Dionex) with a flow rate of 200 nl/min. Separated peptides were ionized by an EASY-Spray ion source (Thermo Fisher Scientific) with 2.0 kV and 275°C of the transfer capillary. All samples were analyzed by an Orbitrap Fusion tribrid (Thermo Scientific) working in a positive polarity mode. Detailed mass spectrometry scan settings were previously described by Kraner et al. (2017). For label-free quantification, raw file analysis was performed with PEAKS Studio 8.0 (Bioinformatics Solutions; Zhang et al., 2012) against the combined human and HHV1 uniprot.org database (March 2017, 71,014 entries). Oxidation of methionine was set as dynamic modification and carbamidomethylation of cysteines as static modification. Only identified proteins with a false discovery rate of <1% were used for quantitative analysis.
Silencing of KIF1B and KIF2A expression
iDCs (3 × 106) were electroporated with 75 pmol siRNA targeting KIF1B (Santa Cruz Biotechnology) and/or KIF2A (Santa Cruz Biotechnology) using the P3 Primary Cell 4D-Nucleofector X Kit (pulse: EH-100) and the 4D-Nucleofector Device (Lonza) according to the manufacturer’s instructions. Control siRNA (fluorescein conjugate)-A (Santa Cruz Biotechnology) was used as negative control. After electroporation, the nucleovette was immediately flushed with 500 µl warm RPMI 1640, and cells were transferred into a 24-well tissue culture plate. RPMI 1640 supplemented with 1% autologous serum, 10 mM Hepes, 2 mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, 40 U/ml GM-CSF, and 250 U/ml IL-4 was added adjusting a final concentration of 106 cells/ml. After 4 h, maturation was induced by the addition of 10 ng/ml TNF-α, 1 mg/ml PGE2, 200 U/ml IL-1β, and 1000 U/ml IL-6. 2 d later, cells were used for further experiments.
Silencing of FIP200 expression
iDCs (6–8 × 106) were electroporated with 75 pmol (750 nM) siRNA targeting FIP200 (Santa Cruz Biotechnology) or control siRNA (fluorescein conjugate)-A (Santa Cruz Biotechnology) according to a method described previously (Gerer et al., 2017). In brief, siRNA was transferred to a 4-mm electroporation cuvette. iDCs were washed once with electroporation medium (OptiMEM without phenol red; Life Technologies Gibco), dissolved in electroporation medium adjusted to a final concentration of 6 × 106 cells/100 µl, and added to the cuvette. The cuvette was electroporated in a Gene Pulser Xcell (BioRad) with the following settings: square-wave pulse, 500 V, 1 ms. After electroporation, cells were transferred into RPMI 1640 supplemented with 1% autologous serum, 10 mM Hepes, 2 mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, 40 U/ml GM-CSF, and 250 U/ml IL-4 adjusted to a final concentration of 106 cells/ml. Cells were used for further experiments after an incubation of 48 h.
Statistics
Statistical analyses were performed using GraphPad Prism 6 (GraphPad Prism Software). For statistical analyses, we used a Mann–Whitney U test for pairwise nonparametric tests between infected iDCs and mDCs or the infected, untreated control group and the infected condition upon treatment. For multiple datasets, we used a one-way ANOVA and Bonferroni multiple comparison post hoc tests to determine the variance in the experimental results obtained. Significance was accepted at P < 0.05. Error bars are shown as mean ± SD or as mean ± SEM.
Online supplemental material
Fig. S1 shows the molecular and functional characterization of HSV1-RFPVP26, which was used for infection experiments. Fig. S2 shows the phosphorylation status of lamin A/C during HSV-1 WT infection kinetics of iDCs, mDCs, and HFF cells. Fig. S3 shows transcriptional and translational regulation of lamin A/C in HSV-1 WT– versus Δvhs–infected iDCs and mDCs. Fig. S4 shows microscopic data of lamin B1 and lamin B2 immunofluorescence staining as well as their quantification in iDCs and mDCs. Fig. S5 shows lamin A/C, B1, and B2 protein levels upon HSV-1 infection of iDCs and mDCs with or without inhibition of autophagy using spautin-1. Table S1 summarizes all antibodies for immunoblotting, immunofluorescence, or immunoprecipitation experiments used in this study. Table S2 shows raw data obtained from mass spectrometric analyses of anti-LC3B immunoprecipitates. Our custom-written macro for the quantification of HSV1-RFPVP26 signals located inside versus outside the nucleus is available in the Data S1 ZIP file and designated as MacroCapsidLocalization. The custom-written macro for the quantification of lamin and LC3B immunofluorescence signal intensities is also available in the Data S1 ZIP file and designated as MacroLaminLC3B.
Supplementary Material
Acknowledgments
We thank Delia Cosgrove for proofreading the manuscript and Professor Manfred Marschall and Dr. Jens Milbradt (Institute of Clinical and Molecular Virology, Erlangen, Germany) for discussion. We thank Professor David Leib for providing us with the HSV-1 ICP34.5 deletion strains. We thank the Imaging Center Essen for help with light and electron microscopy and the Optical Imaging Centre Erlangen (Erlangen, Germany; Dr. Benjamin Schmid) for help with the quantification of the immunofluorescence data.
This work was supported by Deutsche Forschungsgemeinschaft projects STE432/11-1 (A. Steinkasserer), GZ:KR4476/2-1 (A. Krawczyk), CRC900 (TP2; B. Sodeik), and EXC62 REBIRTH (B. Sodeik).
The authors declare no competing financial interests.
Author contributions: C. Silke Heilingloh and A. Steinkasserer designed the study and the experiments. A. Turan, L. Grosche, A. Krawczyk, P. Mühl-Zürbes, C. Draßner, A. Düthorn, S. Voortmann, H. Jastrow, M. Kummer, M. Kraner, J. Dörrie, N. Schaft, K. Döhner, and C. Silke Heilingloh performed experiments and analyzed and interpreted the data; C. Silke Heilingloh, A. Turan, L. Grosche, M. Hasenberg, and M. Kraner wrote the paper. K. Döhner and B. Sodeik generated and provided essential reagents. A. Krawczyk, P. Mühl-Zürbes, A. Düthorn, M. Hasenberg, H. Jastrow, M. Kummer, K. Döhner, B. Sodeik, and A. Steinkasserer critically revised the manuscript. All authors approved the final version of the manuscript.
References
- Banchereau J., and Steinman R.M.. 1998. Dendritic cells and the control of immunity. Nature. 392:245–252. 10.1038/32588 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., and Johansen T.. 2006. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2:138–139. 10.4161/auto.2.2.2405 [DOI] [PubMed] [Google Scholar]
- Bodemann B.O., Orvedahl A., Cheng T., Ram R.R., Ou Y.H., Formstecher E., Maiti M., Hazelett C.C., Wauson E.M., Balakireva M., et al. 2011. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 144:253–267. 10.1016/j.cell.2010.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolovan C.A., Sawtell N.M., and Thompson R.L.. 1994. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J. Virol. 68:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budida R., Stankov M.V., Döhner K., Buch A., Panayotova-Dimitrova D., Tappe K.A., Pohlmann A., Sodeik B., and Behrens G.M.N.. 2017. Herpes simplex virus 1 interferes with autophagy of murine dendritic cells and impairs their ability to stimulate CD8+ T lymphocytes. Eur. J. Immunol. 47:1819–1834. 10.1002/eji.201646908 [DOI] [PubMed] [Google Scholar]
- Crotzer V.L., and Blum J.S.. 2009. Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 182:3335–3341. 10.4049/jimmunol.0803458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham A.L., Donaghy H., Harman A.N., Kim M., and Turville S.G.. 2010. Manipulation of dendritic cell function by viruses. Curr. Opin. Microbiol. 13:524–529. 10.1016/j.mib.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Dechat T., Adam S.A., Taimen P., Shimi T., and Goldman R.D.. 2010. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2:a000547 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Müller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., et al. 2005. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 102:7922–7927. 10.1073/pnas.0501190102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., and Akira S.. 2013. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13:722–737. 10.1038/nri3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P., DeLuca N.A., and Person S.. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 247:115–124. 10.1006/viro.1998.9230 [DOI] [PubMed] [Google Scholar]
- Döhner K., Wolfstein A., Prank U., Echeverri C., Dujardin D., Vallee R., and Sodeik B.. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell. 13:2795–2809. 10.1091/mbc.01-07-0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Xu C., Donahue G., Shimi T., Pan J.A., Zhu J., Ivanov A., Capell B.C., Drake A.M., Shah P.P., et al. 2015. Autophagy mediates degradation of nuclear lamina. Nature. 527:105–109. 10.1038/nature15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg R.J., Ponce de Leon M., Friedman H.M., Fries L.F., Frank M.M., Hastings J.C., and Cohen G.H.. 1987. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb. Pathog. 3:423–435. 10.1016/0882-4010(87)90012-X [DOI] [PubMed] [Google Scholar]
- Gerer K.F., Hoyer S., Dörrie J., and Schaft N.. 2017. Electroporation of mRNA as Universal Technology Platform to Transfect a Variety of Primary Cells with Antigens and Functional Proteins. Methods Mol. Biol. 1499:165–178. 10.1007/978-1-4939-6481-9_10 [DOI] [PubMed] [Google Scholar]
- Geuze H.J. 1998. The role of endosomes and lysosomes in MHC class II functioning. Immunol. Today. 19:282–287. 10.1016/S0167-5699(98)01269-9 [DOI] [PubMed] [Google Scholar]
- Gobeil P.A.M., and Leib D.A.. 2012. Herpes simplex virus γ34.5 interferes with autophagosome maturation and antigen presentation in dendritic cells. MBio. 3:e00267–e12. 10.1128/mBio.00267-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwich A., Prechtel A.T., Mühl-Zürbes P., Pangratz N.M., Stössel H., Romani N., Steinkasserer A., and Kummer M.. 2011. Herpes simplex virus type I (HSV-1) replicates in mature dendritic cells but can only be transferred in a cell-cell contact-dependent manner. J. Leukoc. Biol. 89:973–979. 10.1189/jlb.0310180 [DOI] [PubMed] [Google Scholar]
- Heilingloh C.S., Mühl-Zürbes P., Steinkasserer A., and Kummer M.. 2014. Herpes simplex virus type 1 ICP0 induces CD83 degradation in mature dendritic cells independent of its E3 ubiquitin ligase function. J. Gen. Virol. 95:1366–1375. 10.1099/vir.0.062810-0 [DOI] [PubMed] [Google Scholar]
- Heilingloh C.S., Kummer M., Mühl-Zürbes P., Drassner C., Daniel C., Klewer M., and Steinkasserer A.. 2015. L Particles Transmit Viral Proteins from Herpes Simplex Virus 1-Infected Mature Dendritic Cells to Uninfected Bystander Cells, Inducing CD83 Downmodulation. J. Virol. 89:11046–11055. 10.1128/JVI.01517-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., and Yoshimori T.. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812. 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., and Emr S.D.. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721. 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., and Rubinsztein D.C.. 2011. Regulation of autophagy by lysosomal positioning. Autophagy. 7:927–928. 10.4161/auto.7.8.15862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraner M.E., Müller C., and Sonnewald U.. 2017. Comparative proteomic profiling of the choline transporter-like1 (CHER1) mutant provides insights into plasmodesmata composition of fully developed Arabidopsis thaliana leaves. Plant J. 92:696–709. 10.1111/tpj.13702 [DOI] [PubMed] [Google Scholar]
- Kruse M., Rosorius O., Krätzer F., Stelz G., Kuhnt C., Schuler G., Hauber J., and Steinkasserer A.. 2000. Mature dendritic cells infected with herpes simplex virus type 1 exhibit inhibited T-cell stimulatory capacity. J. Virol. 74:7127–7136. 10.1128/JVI.74.15.7127-7136.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm C.E., Kraner M.E., Hofmann J., Börnke F., Mock H.P., and Sonnewald U.. 2017. Hop/Sti1 - A Two-Faced Cochaperone Involved in Pattern Recognition Receptor Maturation and Viral Infection. Front. Plant Sci. 8:1754 10.3389/fpls.2017.01754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach N.R., and Roller R.J.. 2010. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology. 406:127–137. 10.1016/j.virol.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussignol M., Queval C., Bernet-Camard M.F., Cotte-Laffitte J., Beau I., Codogno P., and Esclatine A.. 2013. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 87:859–871. 10.1128/JVI.01158-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., and Stamminger T.. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357–33367. 10.1074/jbc.M502672200 [DOI] [PubMed] [Google Scholar]
- Mauvezin C., and Neufeld T.P.. 2015. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 11:1437–1438. 10.1080/15548627.2015.1066957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane S., Aitken J., Sutherland J.S., Nicholl M.J., Preston V.G., and Preston C.M.. 2011. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J. Virol. 85:4212–4221. 10.1128/JVI.02435-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., and Rixon F.J.. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269–276. 10.1099/0022-1317-73-2-269 [DOI] [PubMed] [Google Scholar]
- Mellman I., and Steinman R.M.. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell. 106:255–258. 10.1016/S0092-8674(01)00449-4 [DOI] [PubMed] [Google Scholar]
- Mikloska Z., Bosnjak L., and Cunningham A.L.. 2001. Immature monocyte-derived dendritic cells are productively infected with herpes simplex virus type 1. J. Virol. 75:5958–5964. 10.1128/JVI.75.13.5958-5964.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbradt J., Auerochs S., Sticht H., and Marschall M.. 2009. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90:579–590. 10.1099/vir.0.005231-0 [DOI] [PubMed] [Google Scholar]
- Milbradt J., Webel R., Auerochs S., Sticht H., and Marschall M.. 2010. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285:13979–13989. 10.1074/jbc.M109.063628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., and Yoshimori T.. 2007. How to interpret LC3 immunoblotting. Autophagy. 3:542–545. 10.4161/auto.4600 [DOI] [PubMed] [Google Scholar]
- Moir R.D., Yoon M., Khuon S., and Goldman R.D.. 2000. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151:1155–1168. 10.1083/jcb.151.6.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F., Forest T., and Baines J.D.. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81:6459–6470. 10.1128/JVI.00380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F., Wills E.G., Park R., and Baines J.D.. 2008. Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane. J. Virol. 82:8094–8104. 10.1128/JVI.00874-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel C.H., Döhner K., Fathollahy M., Strive T., Borst E.M., Messerle M., and Sodeik B.. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J. Virol. 82:3109–3124. 10.1128/JVI.02124-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M., Gretzmeier C., Dengjel J., Piacentini M., Fimia G.M., and Cecconi F.. 2013. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15:406–416. 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- O’Connell D., and Liang C.. 2016. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy. 12:451–459. 10.1080/15548627.2016.1139262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., and Levine B.. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 1:23–35. 10.1016/j.chom.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Palucka K., and Banchereau J.. 1999. Dendritic cells: a link between innate and adaptive immunity. J. Clin. Immunol. 19:12–25. 10.1023/A:1020558317162 [DOI] [PubMed] [Google Scholar]
- Park R., and Baines J.D.. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494–504. 10.1128/JVI.80.1.494-504.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollara G., Jones M., Handley M.E., Rajpopat M., Kwan A., Coffin R.S., Foster G., Chain B., and Katz D.R.. 2004. Herpes simplex virus type-1-induced activation of myeloid dendritic cells: the roles of virus cell interaction and paracrine type I IFN secretion. J. Immunol. 173:4108–4119. 10.4049/jimmunol.173.6.4108 [DOI] [PubMed] [Google Scholar]
- Poüs C., and Codogno P.. 2011. Lysosome positioning coordinates mTORC1 activity and autophagy. Nat. Cell Biol. 13:342–344. 10.1038/ncb0411-342 [DOI] [PubMed] [Google Scholar]
- Radtke K., English L., Rondeau C., Leib D., Lippé R., and Desjardins M.. 2013. Inhibition of the host translation shutoff response by herpes simplex virus 1 triggers nuclear envelope-derived autophagy. J. Virol. 87:3990–3997. 10.1128/JVI.02974-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechenchoski D.Z., Faccin-Galhardi L.C., Linhares R.E.C., and Nozawa C.. 2017. Herpesvirus: an underestimated virus. Folia Microbiol. (Praha). 62:151–156. 10.1007/s12223-016-0482-7 [DOI] [PubMed] [Google Scholar]
- Sandbaumhüter M., Döhner K., Schipke J., Binz A., Pohlmann A., Sodeik B., and Bauerfeind R.. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell. Microbiol. 15:248–269. 10.1111/cmi.12075 [DOI] [PubMed] [Google Scholar]
- Santama N., Krijnse-Locker J., Griffiths G., Noda Y., Hirokawa N., and Dotti C.G.. 1998. KIF2beta, a new kinesin superfamily protein in non-neuronal cells, is associated with lysosomes and may be implicated in their centrifugal translocation. EMBO J. 17:5855–5867. 10.1093/emboj/17.20.5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., and Münz C.. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92. 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J.R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063–1068. 10.1128/JVI.78.3.1063-1068.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B., Ebersold M.W., and Helenius A.. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021. 10.1083/jcb.136.5.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., and Ohsumi Y.. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301–311. 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z., Jiang W., Virgin H.W. IV, Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., and Levine B.. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 99:190–195. 10.1073/pnas.012485299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer B.K., von Einem J., Kaufer B., and Osterrieder N.. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- Vu A., Poyzer C., and Roller R.. 2016. Extragenic Suppression of a Mutation in Herpes Simplex Virus 1 UL34 That Affects Lamina Disruption and Nuclear Egress. J. Virol. 90:10738–10751. 10.1128/JVI.01544-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts C. 2001. Antigen processing in the endocytic compartment. Curr. Opin. Immunol. 13:26–31. 10.1016/S0952-7915(00)00177-1 [DOI] [PubMed] [Google Scholar]
- Whitley R.J., and Roizman B.. 2001. Herpes simplex virus infections. Lancet. 357:1513–1518. 10.1016/S0140-6736(00)04638-9 [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Wu S., Pan S., Zhang L., Baines J., Roller R., Ames J., Yang M., Wang J., Chen D., Liu Y., et al. 2016. Herpes Simplex Virus 1 Induces Phosphorylation and Reorganization of Lamin A/C through the γ134.5 Protein That Facilitates Nuclear Egress. J. Virol. 90:10414–10422. 10.1128/JVI.01392-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.T., Tan H.L., Huang Q., Ong C.N., and Shen H.M.. 2009. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy. 5:824–834. 10.4161/auto.9099 [DOI] [PubMed] [Google Scholar]
- Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465:942–946. 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G.A., and Ma B.. 2012. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics. 11:010587 10.1074/mcp.M111.010587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Li P., Lin Y., Lott J.M., Hislop A.D., Canaday D.H., Brutkiewicz R.R., and Blum J.S.. 2005. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 22:571–581. 10.1016/j.immuni.2005.03.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.