Rosen et al. identify a role for the centrosomal protein Bsg25D/Ninein in nuclear positioning and microtubule organization in Drosophila muscle fibers. Genetic, cell biological, and atomic force microscopy analyses demonstrate that complex interactions between Bsg25D and the microtubule-associated protein Ensconsin govern myonuclear positioning in Drosophila.
Abstract
Skeletal muscle consists of multinucleated cells in which the myonuclei are evenly spaced throughout the cell. In Drosophila, this pattern is established in embryonic myotubes, where myonuclei move via microtubules (MTs) and the MT-associated protein Ensconsin (Ens)/MAP7, to achieve their distribution. Ens regulates multiple aspects of MT biology, but little is known about how Ens itself is regulated. We find that Ens physically interacts and colocalizes with Bsg25D, the Drosophila homologue of the centrosomal protein Ninein. Bsg25D loss enhances myonuclear positioning defects in embryos sensitized by partial Ens loss. Bsg25D overexpression causes severe positioning defects in immature myotubes and fully differentiated myofibers, where it forms ectopic MT organizing centers, disrupts perinuclear MT arrays, reduces muscle stiffness, and decreases larval crawling velocity. These studies define a novel relationship between Ens and Bsg25D. At endogenous levels, Bsg25D positively regulates Ens activity during myonuclear positioning, but excess Bsg25D disrupts Ens localization and MT organization, with disastrous consequences for myonuclear positioning and muscle function.
Introduction
In many cell types, nuclei occupy specific subcellular localizations that are functionally important (Gundersen and Worman, 2013). In mammalian muscle, nuclei are positioned at the muscle cell periphery in a manner that maximizes internuclear distance. Myonuclear mispositioning is a feature of certain muscle diseases (Romero, 2010; Folker and Baylies, 2013), and model organisms that are mutant for genes required to position myonuclei exhibit decreased muscle function (Zhang et al., 2010; Elhanany-Tamir et al., 2012; Folker et al., 2012; Metzger et al., 2012; Schulman et al., 2014). These findings argue that correct nuclear positioning is essential for muscle function.
Skeletal muscle development and structure are highly conserved between the fruit fly Drosophila melanogaster and humans. In both humans and Drosophila, muscle cells are multinucleated and formed from the iterative fusion of muscle precursor cells, each of which contributes a nucleus to the growing myotube. After fusion, myonuclei in each Drosophila myotube are present as a single cluster. Subsequently, the myonuclei undergo coordinated movements that ultimately leave them evenly distributed along the length of the muscle cell. The first step of nuclear positioning occurs at stage 14 (10–11 h after egg laying [AEL]), when the myonuclei separate into two clearly defined groups along the myotube’s long axis. Then, during stages 15 (11–13 h AEL) and 16 (13–16 h AEL), the two clusters of myonuclei migrate away from the myotube’s center toward opposite muscle poles. During stage 17 (16–24 h AEL), the last stage of embryonic development, myonuclei spread out from the two clusters and fill in the myofiber evenly, such that the distance between myonuclei is maximized (Metzger et al., 2012). Finally, this even spacing is maintained, likely by active mechanisms, during the lifetime of the larval myofibers (Elhanany-Tamir et al., 2012; Manhart et al., 2018).
The regulation of myonuclear positioning is poorly understood. A key player in the process is Ensconsin (Ens)/MAP7, a microtubule (MT)-associated protein (MAP). Ens promotes Kinesin-based MT transport by relieving Kinesin from its autoinhibited conformation (Barlan et al., 2013) or by recruiting Kinesin to MTs (Sung et al., 2008). ens loss-of-function mutants exhibit a complete block in myonuclear separation and pole-ward cluster migration through stage 16; at this stage, control myonuclei reside in two clusters near opposite myotube poles, while ens mutant myonuclei are present as a single cluster (Metzger et al., 2012). Loss of Kinesin heavy chain (Khc) impairs myonuclear movement (Metzger et al., 2012), as do mutations in the genes encoding the MT minus end–directed motor protein Dynein heavy chain and the motor protein adaptor Sunday driver (Folker et al., 2012, 2014; Schulman et al., 2014). Taken together, these findings demonstrate the centrality of MTs and associated proteins to myonuclear positioning. Interestingly, ens is the only mutant isolated to date where myonuclear movement appears to be completely blocked. While maternal products may partially ameliorate the phenotypes of Khc, Dhc, and syd zygotic mutants, the uniqueness of the ens phenotype raises the possibility that Ens plays additional roles in myonuclear movement beyond its regulation of MT-based transport. Indeed, in other cell types, Ens has been shown to be a MT polymerizing factor (Gallaud et al., 2014). How Ens promotes nuclear movement in muscle, and what other proteins regulate its critical activity, are outstanding questions in the field.
MT networks, which are essential for myonuclear positioning, undergo dramatic changes during muscle development. In mammalian cell culture, following myoblast fusion, centrosomes are lost and centrosomal proteins relocalize to alternative MT organizing centers (MTOCs) in the acentrosomal myotube, most prominently the myonuclear envelopes (Tassin et al., 1985). In Drosophila, MTs similarly reorganize during muscle development; starting in the embryonic myotube, most MTs run parallel to the cell’s long axis, but by the third-instar stage of larval development, immunofluorescent staining for Tubulin clearly reveals that MT arrays also extend from myonuclear envelopes (Elhanany-Tamir et al., 2012; Metzger et al., 2012; Manhart et al., 2018). Only two genes, pavarotti and RacGAP50C, have been implicated in the regulation of muscle MT reorganization in the embryo (Guerin and Kramer, 2009). Much more needs to be learned about how this transition occurs.
Proteins that regulate MT networks in other cell types are strong candidates for being factors in muscle MT organization and MT-dependent myonuclear positioning. One such factor is the MT minus end–binding protein Ninein, encoded by the gene NIN. Pioneering cell culture experiments showed that Ninein localizes to centrosomes (Bouckson-Castaing et al., 1996) and noncentrosomal MTOCs (Mogensen et al., 2000) and that Ninein anchors MTs to MTOCs and promotes MT nucleation by docking γ-Tubulin (Abal et al., 2002; Casenghi et al., 2005; Delgehyr et al., 2005; Shinohara et al., 2013). More recently, cell culture studies and RNAi-based in vivo approaches have affirmed critical roles for Ninein in regulating MT organization in numerous cell types. Surprisingly, recently generated null NIN mutants in invertebrates are relatively normal: Caenorhabditis elegans mutants for the NIN orthologue noca-1 are sterile with defects in the noncentrosomal MT arrays of the germline and epidermis but have grossly normal morphology and develop to adulthood (Wang et al., 2015). Mutants for the Drosophila NIN orthologue Bsg25D are viable and fertile (Kowanda et al., 2016; Zheng et al., 2016). Loss of Ninein/NOCA-1 in C. elegans is partially compensated for by the MT minus end protein Patronin (Wang et al., 2015); no redundant factors have been identified in Drosophila. Since Bsg25D/Ninein is conserved from invertebrates to humans, Bsg25D may have important functions not readily detectable by single mutant analysis. How does Bsg25D function, and what proteins does it interact with in muscle?
In this work, we find that Bsg25D acts with Ens to regulate myonuclear positioning. Muscle-specific Bsg25D overexpression caused myonuclear positioning defects in embryonic myotubes, and overexpression of Bsg25D and ens reciprocally affected the speed and direction of myonuclear movement. In mature myofibers, overexpressed Bsg25D caused nuclear positioning phenotypes, MT defects, and reduced muscle stiffness and function. We conclude that Bsg25D, under wild-type conditions, binds to Ens and promotes its activity in myonuclear positioning, whereas when Bsg25D is expressed at high levels, it alters Ens localization and function, phenocopying ens mutants, and ultimately disrupts muscle MT networks and muscle function. Our results shed further light on the function of Ens in the process of nuclear movement and find a novel in vivo role for Bsg25D/Ninein, a poorly understood centrosomal protein, in myonuclear positioning.
Results
Bsg25D and Ens interact in vitro and colocalize in embryonic myotubes
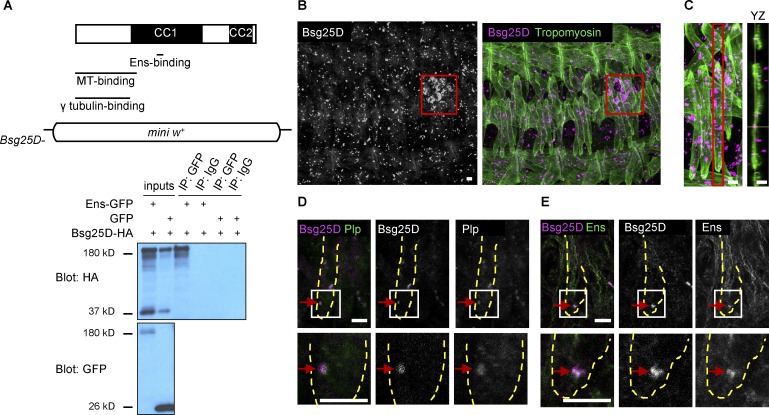
We have previously shown that myonuclei in ens mutant embryos fail to separate into two clusters and migrate toward muscle poles (Metzger et al., 2012). To better understand the mechanisms underlying myonuclear positioning, we performed a yeast two-hybrid screen to identify proteins that interact with Ens. In addition to clones of Khc, which we have previously shown to genetically and physically interact with Ens (Metzger et al., 2012), we recovered numerous clones corresponding to Bsg25D, the Drosophila homologue of vertebrate Ninein (Kowanda et al., 2016; Zheng et al., 2016). The sequence common to all recovered Bsg25D clones (nucleotides 1463–1,577/amino acids 487–526, using isoform Bsg25D-PB as a reference) falls entirely within the gene’s fourth exon, which is present in all Bsg25D alternatively spliced transcripts; thus, the Ens-binding region is present in all Bsg25D isoforms (Fig. 1 A). This region is found in the first of two spans of Bsg25D that contain numerous coiled coil domains. The Ens-binding region is nonoverlapping with the region of Bsg25D thought to be necessary for γ-Tubulin binding, based on alignment between Bsg25D and mouse Ninein, where the latter region was experimentally defined (Delgehyr et al., 2005). The Ens-binding region also does not overlap with the N-terminal Bsg25D region (amino acids 1–353) that has been shown to bind MTs in vitro (Kowanda et al., 2016). The Ens–Bsg25D interaction was validated in S2 cells, where overexpressed Ens and full-length Bsg25D coimmunoprecipitated (Fig. 1 A).
Figure 1.
Bsg25D and Ens physically interact and colocalize in myotubes in vivo. (A) Top: Map of Bsg25D indicating coiled–coil (CC) domain–rich regions and regions that bind Ens (this work), MTs (Kowanda et al., 2016), and γ-Tubulin (predicted from alignment to mouse Ninein; Delgehyr et al., 2005). In Bsg25D−, the entire coding region has been removed and replaced by a mini-white (mini w+) reporter cassette. Bottom: Coimmunoprecipitation of overexpressed Ens-GFP and Bsg25D-HA from S2 cells. The largest Bsg25D-HA band is the main protein species, and smaller bands likely represent cleavage products. IP, immunoprecipitation. (B) Bsg25D expression in the stage 16 embryo. Left and right panels are the same image with and without Tropomyosin, a muscle marker. Left: Bsg25D, gray. Right: Bsg25D, magenta; Tropomyosin, green. Red box indicates primordial germ cells. (C) Left: Extended focus projection of one stage 16 hemisegment. Right: Orthogonal (YZ) section of indicated (red box) lateral transverse myotube. Crosshairs identify a Bsg25D+ punctum inside myotube. Bsg25D, magenta; Tropomyosin, green. (D) Immunofluorescent staining of stage 16 myotube for Bsg25D and Plp. Image is a single slice from a Z-stack. Bsg25D, magenta; Plp, green; colocalization, white (see red arrows). (E) Immunofluorescent staining of stage 16 myotube for Bsg25D and Ens. Image is a single slice from a Z-stack. In the merged image, Bsg25D, magenta; Ens, green; colocalization, white (highlighted by arrows). In D and E, images in bottom row are higher magnification views of boxed regions, and dashed lines outline lateral transverse muscles. In all images, scale bars = 5 µm.
Next, we examined Bsg25D expression in vivo. In situ hybridization performed by the Berkeley Drosophila Genome Project showed Bsg25D transcripts in embryonic muscle (Tomancak et al., 2002, 2007; Hammonds et al., 2013). By immunofluorescent antibody staining using an existing antibody against Bsg25D (Iampietro et al., 2014), we found that Bsg25D protein was present in discrete puncta in numerous cell types in the embryo. (The Bsg25D signal was absent from Bsg25D null mutants [Fig. S1 A].) This punctate expression pattern was expected, as Bsg25D is known to localize to centrosomes in mononucleated cells (Kowanda et al., 2016; Zheng et al., 2016). Notably, it was most strongly expressed in primordial germ cells, as has been previously observed (Zheng et al., 2016; Fig. 1 B, red outlined box). Combined immunofluorescent labeling of Bsg25D and the muscle marker Tropomyosin showed that from stage 14 to stage 16, the stages during which myonuclei split into two clusters and migrate toward muscle poles, Bsg25D was broadly but weakly expressed in the cytoplasm and strongly enriched in cytoplasmic puncta of myotubes (Fig. 1 B). Analysis of orthogonal slices of Z-stacks confirmed the presence of Bsg25D puncta inside myotubes (Fig. 1 C). At stage 16, there were between zero and eight Bsg25D puncta per myotube (mode = 1, mean = 2.1). Puncta were excluded from the nucleus but otherwise had no conspicuous localization. Single puncta were also visible in some unfused myoblasts. We used genetic reporters and antibody staining to determine whether the Bsg25D puncta in myotubes reside in various organelles and subcellular structures. We did not observe Bsg25D colocalization with markers of Golgi apparatus, ER, or endosomes, though we did observe that 95% of Bsg25D puncta were in proximity to Arl8 (n = 60 myotube puncta), a protein that localizes to lysosomes (Bagshaw et al., 2006; Hofmann and Munro, 2006; Fig. S1 B). Counter to our expectations (Bouckson-Castaing et al., 1996; Mogensen et al., 2000; Kowanda et al., 2016; Zheng et al., 2016), we also failed to detect colocalization between Bsg25D puncta and the MTOC component γ-Tubulin or the MT component α-Tubulin (Fig. S1 B). Interestingly, however, all myotube Bsg25D puncta (n = 59) colocalized with the MTOC component Pericentrin-like protein (Plp), the sole Drosophila Pericentrin orthologue (Figs 1 D and S1 C). This finding is consistent with physical interactions observed between their mammalian orthologues in cell culture (Chen et al., 2014).
Since we found that Bsg25D and Ens physically interact in vitro, we performed immunofluorescent antibody staining to determine whether they colocalize in embryonic myotubes. While the bulk of Ens protein localized to MTs, as expected for a MAP (Fig. S1 D), there was also Ens protein present in Bsg25D puncta; thus, Ens and Bsg25D colocalize in cytoplasmic puncta in embryonic myotubes (Fig. 1 E). Outside of these puncta, the broad, diffuse cytoplasmic Bsg25D signal was difficult to resolve, so it is possible that Bsg25D and Ens are both present in other parts of the cell as well. Since Bsg25D and Ens bind in vitro and colocalize in myotubes, we hypothesized that Bsg25D functions with ens during myonuclear positioning.
Loss of Bsg25D enhances ens myonuclear positioning defects
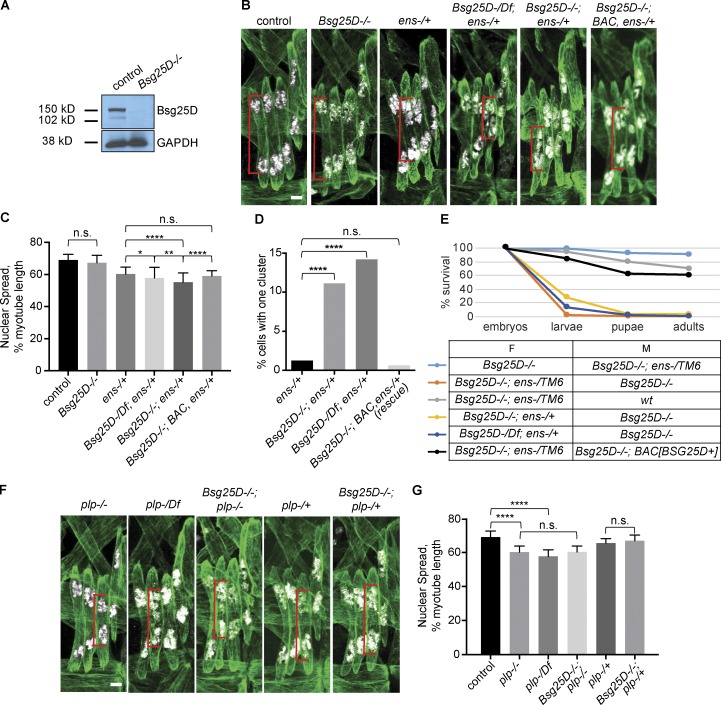
To determine whether Bsg25D functions in myonuclear positioning, we generated a null Bsg25D mutant. We used an accelerated homologous recombination approach (Baena-Lopez et al., 2013) to remove the entire Bsg25D locus, including all predicted alternatively spliced transcripts (Fig. 1 A and Materials and methods). Western blotting showed Bsg25D protein was absent from larval lysates (Fig. 2 A and Materials and methods).
Figure 2.
Bsg25Dgenetically interacts with ens, but not plp. (A) Western blot showing loss of Bsg25D protein in Bsg25D−/− larval lysates. (B) Extended-focus projections of representative stage 16 hemisegments from indicated genotypes. (C) Bar graph showing mean nuclear spread and SD. For each genotype, n (number of hemisegments) is control, n = 39; Bsg25D−/−, n = 61; ens−/+, n = 46; Bsg25D-/Df;ens−/+, n = 43; Bsg25D;ens−/+, n = 71; Bsg25D−/−;BAC, ens−/+, n = 44. P values were calculated by Student’s t test. (D) Graph showing the percentage of myotubes that have a single nuclear cluster. P values were calculated by Fisher’s exact test from contingency tables comparing ens-/+ to each other genotype. n values (number of myotubes for each genotype) are ens−/+, 239; Bsg25D−/−;ens−/+, 296; Bsg25D-/Df;ens−/+, 176; Bsg25D−/−;BAC, ens−/+ (rescue), 160. (E) Viability graph showing survival during development for individuals derived from indicated crosses. The following number of individuals were included for each genotype: Bsg25D−/−×Bsg25D−/−;ens−/TM6, 140; Bsg25D−/−;ens-/TM6×Bsg25D−/−, 153; Bsg25D−/−;ens-/TM6×wt, 140; Bsg25D−/−;ens−/+×Bsg25D−/−, 113; Bsg25D-/Df;ens−/+×Bsg25D−/−, 148; Bsg25D−/−;ens-/TM6×Bsg25D−/−;BAC[Bsg25D+], 144. F, female; M, male. (F) Extended-focus projections of representative stage 16 hemisegments from indicated genotypes. (G) Bar graph showing mean nuclear spread and SD. For each genotype, number of hemisegments is plp−/−, n = 47; plp-/Df, n = 24; Bsg25D−/−; plp−/−, n = 39; plp-/+, n = 34; Bsg25D−/−;plp-/+, n = 29. P values were calculated by Student’s t test. The control data in G are the same as in C; for representative image, see first panel in B. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. In all confocal images, red brackets indicate sample nuclear spread measurements. Tropomyosin, green; nuclei, white. Scale bars = 5 µm.
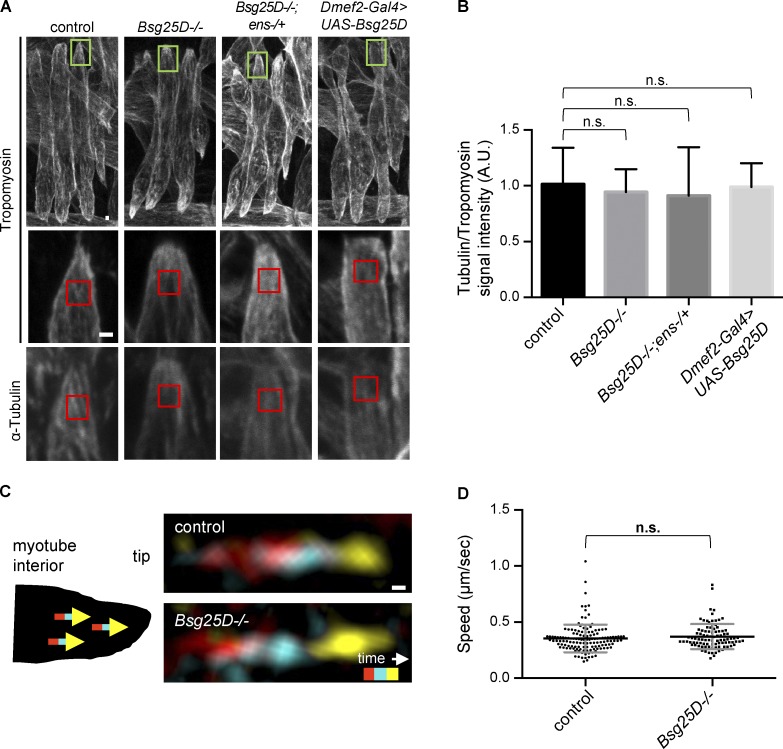
In agreement with recent reports (Kowanda et al., 2016; Zheng et al., 2016), we found that Bsg25D homozygous null mutants (Bsg25D−/−) lacking maternal and zygotic Bsg25D were viable and fertile, with a modest decrease in the frequency of survival to adulthood relative to wild-type controls (70% vs. 96%; Fig. S2 A). A standard larval motility assay revealed moderately impaired muscle function (Fig. S2 B). Using a transgenic reporter that labels myonuclei in a specific set of muscles (i.e., the lateral transverse muscles; Richardson et al., 2007), we found that myonuclear positioning in stage 16 Bsg25D−/− myotubes was normal (Fig. 2 B). Thus, Bsg25D is not essential for viability, fertility, or myonuclear positioning.
We next considered the possibility that Bsg25D plays a role in myonuclear positioning but that the pathway it affects is robust enough to withstand the loss of Bsg25D if the other components are present at wild-type levels. To investigate this possibility, we generated double mutants in which Bsg25D and candidate interactor genes were targeted. We found that ens−/+ embryos lacking all maternal and zygotic Bsg25D had significantly more impaired myonuclear positioning than ens−/+ embryos containing the normal complement of Bsg25D (Fig. 2, B–D): at stage 16, Bsg25D−/−;ens−/+ myonuclear clusters traversed less of the distance toward the muscle poles. Further, in some myotubes, myonuclei are present as one cluster, resembling those found in ens−/− homozygotes (Metzger et al., 2012). Enhancement of myonuclear positioning defects was also observed when one copy of the null Bsg25D allele was replaced by a deficiency removing the gene (Fig. 2, B–D), which argues that the enhancement effect was specific to the Bsg25D region. Moreover, we rescued the Bsg25D−/−;ens−/+ enhanced phenotype by reintroducing Bsg25D+ from the large genomic Bacterial Artificial Chromosome (BAC) insertion CH321-49G22 (Fig. 2, B–D). In addition, Bsg25D−/−;ens−/− double mutants showed the ens−/− phenotype, which is the most severe clustering phenotype (Fig. S3 A). We conclude that endogenous Bsg25D and ens likely affect a common pathway that moves myonuclei in embryonic myotubes.
Bsg25D and ens genetically interact in nonmuscle cells as well, as embryonic lethality occurred when Bsg25D and Ens were jointly removed in a specific genetic combination. Nearly 100% of embryos derived from Bsg25D−/−;ens−/TM6 mothers and Bsg25D−/− fathers failed to hatch (Fig. 2 E). However, embryos derived from the reciprocal cross survived well (Fig. 2 E). Bsg25D−/−;ens−/TM6 females crossed with wild-type males laid eggs that hatched, demonstrating that those females are fertile. We also observed widespread death when the third chromosome balancer TM6 was replaced by a wild-type chromosome, indicating that the genetic interaction was not due to mutations on the balancer chromosome. The phenotype also persisted when one of the Bsg25D alleles was replaced with a deficiency completely uncovering Bsg25D. Furthermore, restoring Bsg25D+ through a BAC transgene achieved nearly complete rescue (Fig. 2 E, compare black and salmon-colored lines in graph). We conclude that Bsg25D and ens are jointly required to support normal embryonic development. Since genotypes with even the strongest myonuclear positioning defects survive embryonic development (Metzger et al., 2012), the observed lethality must be due to issues in nonmuscle cells.
To gain further insights to the role of Bsg25D, we tested for genetic interactions between Bsg25D and other genes involved in myonuclear positioning or MT biology. We did not detect interactions between Bsg25D and the genes encoding Khc and Dhc (which are essential for myonuclear positioning) or patronin and the γ-Tubulin homologue γTub23C (which interact with Ninein/noca-1 in C. elegans [Wang et al., 2015]; Fig. S3, B and C; and data not shown). We also tested pericentrin-like protein (plp) mutants. Interestingly, plp−/− mutants had myonuclear positioning defects; nuclear clusters usually split correctly but failed to move the normal distance toward the muscle poles by stage 16 (Fig. 2, F and G). The same phenotype was observed when one allele of plp was replaced by a deficiency uncovering the gene (Fig. 2, F and G). However, loss of Bsg25D did not further enhance myonuclear positioning defects in either plp homozygotes or heterozygotes (Fig. 2, F and G); thus, although we observed colocalization between the two proteins in myotubes, we did not uncover a role for Bsg25D–Plp interactions in myonuclear positioning. Taken together, these results reveal dosage-sensitive genetic interactions only between Bsg25D and ens; these are not observed with other genes involved in MT biology or myonuclear positioning.
Muscle-specific overexpression of Bsg25D causes defects in embryonic myonuclear positioning by sequestering Ens
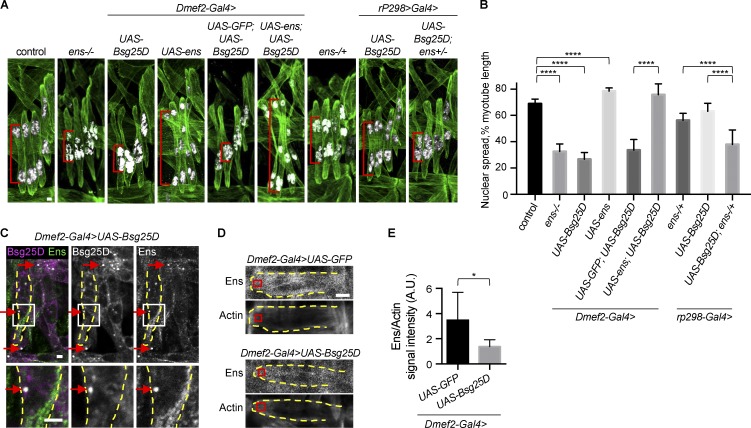
Having demonstrated that Bsg25D functions in myonuclear positioning, we next performed gain-of-function analysis with the Gal4/upstream activating sequence (UAS) system (Brand and Perrimon, 1993) using the muscle-specific Dmef2 promoter to probe the activity of Bsg25D in muscle. Strikingly, in stage 16 myotubes overexpressing Bsg25D, myonuclei were present in a single cluster near the myotube center; in contrast, control myotubes at this stage had two nuclear clusters near opposite muscle poles (Fig. 3 A). This suggests that overexpressed Bsg25D blocked myonuclear cluster separation (stage 14) and movement toward the muscle poles (stages 15 and 16).
Figure 3.
Overexpressed Bsg25D causes nuclear positioning defects in embryonic myotubes by sequestering endogenous Ens. (A) Extended-focus projections of representative stage 16 hemisegments from indicated genotypes. Green, Tropomyosin; white, nuclei. Red brackets indicate sample nuclear spread measurements. (B) Bar graph showing mean nuclear spread and SD. For each genotype, the number of hemisegments is as follows: control, n = 22; Dmef2-Gal4>UAS-Bsg25D, n = 34; Dmef2-Gal4>UAS-ens, n = 47; Dmef2-Gal4>UAS-GFP:UAS-Bsg25D, n = 25; Dmef2-Gal4>UAS-ens;UAS-Bsg25D, n = 42; ens−/+, n = 37; rP298-Gal4>UAS-Bsg25D, n = 33; rP298-Gal4>UAS-Bsg25D;ens−/+, n = 37. Control data and the representative image are the same as in Fig. 2. ****, P < 0.0001. (C) Immunofluorescent antibody staining for Bsg25D and Ens in an embryo overexpressing Bsg25D. Arrows show examples of colocalization. Dashed yellow lines outline a lateral transverse myotube. In the merged image: magenta, Bsg25D; green, Ens; white, colocalization. Images in the bottom row are higher magnification views of boxed regions. (D) Representative images of stage 16 VL1 myotubes from indicated genotypes. Each image is a single slice from a confocal stack. Red boxes show where the signal intensity was quantified. (E) Graph showing Ens intensity, normalized to actin intensity. Number of myotubes is six for both genotypes, mean ± SD. *, P < 0.05. Scale bars = 2 µm (A and C) and 5 µm (D).
That Bsg25D overexpression phenocopied loss of ens (Fig. 3 A; Metzger et al., 2012) led us to hypothesize that overexpressed Bsg25D inhibits or sequesters endogenous Ens in myotubes. In support of this hypothesis, we found that Ens overexpression, but not overexpression of a control protein (GFP), could rescue Bsg25D-induced nuclear positioning defects. In fact, embryos overexpressing Bsg25D and Ens had even greater nuclear spread than control embryos (Fig. 3, A and B). This finding led us to examine the effects of overexpressing Ens in otherwise wild-type embryos. We found that overexpressed Ens caused novel myonuclear positioning defects; nuclear spread was increased and nuclei were often present as individuals rather than as part of clusters (Fig. 3, A and B). In sum, Ens overexpression and Bsg25D overexpression confer reciprocal phenotypes, and Ens overexpression is epistatic to Bsg25D overexpression.
As further support of our hypothesis that overexpressed Bsg25D inhibits or sequesters endogenous Ens, we detected genetic interactions between overexpressed Bsg25D and endogenous ens. Both loss of one allele of ens and overexpression of Bsg25D with rP298-Gal4, a muscle-specific driver that produces a lower level of gene expression than Dmef2-Gal4, conferred reduced poleward movement of nuclear clusters (Fig. 3, A and B). However, in ens−/+ embryos expressing rP298>Bsg25D, a much stronger nuclear positioning phenotype occurred (Fig. 3, A and B). This implies that Bsg25D affects the same pathway as loss of ens.
Finally, in support of our hypothesis, we found that overexpressed Bsg25D alters Ens localization. Like endogenous Bsg25D, overexpressed Bsg25D was present both broadly in the cytoplasm and in bright cytoplasmic puncta (Fig. 3 C). Ens normally localizes to MTs (Fig. S1 D), but in Bsg25D-overexpressing myotubes, Ens was strongly enriched at sites of Bsg25D accumulation in the cytoplasm (Fig. 3 C). Outside of Bsg25D+ puncta, Ens signal was lower in myotubes overexpressing Bsg25D than in controls overexpressing GFP (Fig. 3, D and E). This argues that overexpression of Bsg25D reduces the amount of Ens on myotubes. Based on our genetic and imaging analyses, we conclude that overexpressed Bsg25D exerts its effect on myonuclear positioning by regulating Ens activity, at least in part by mislocalizing Ens.
Bsg25D and Ens levels regulate the dynamics of myonuclear positioning
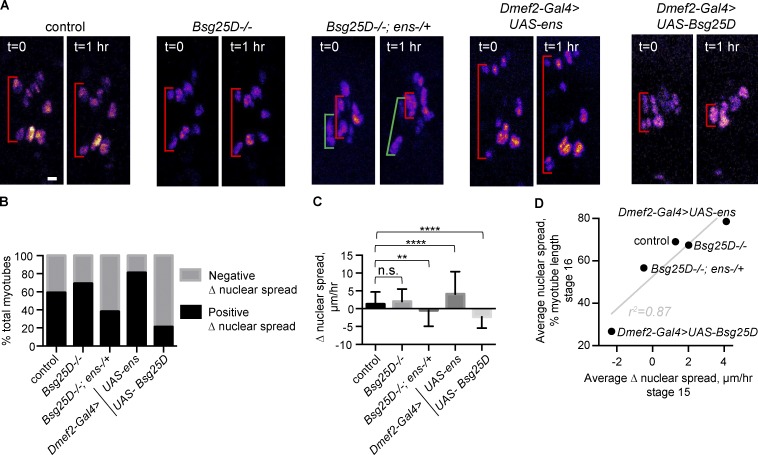
We hypothesized that modulating Bsg25D and Ens levels affects nuclear positioning at stage 16 by disrupting the normal dynamics of myonuclear movement during stage 15, when nuclear clusters normally move toward muscle poles. To address this hypothesis, we performed time-lapse microscopy during stage 15, as done previously (Folker et al., 2012; Videos 1, 2, 3, 4, and 5). We measured the distance from the dorsal-most nucleus to the ventral-most nucleus (nuclear spread) at the beginning and end of the time-lapse series. We divided the difference between the final and initial nuclear spreads for each cell by the time elapsed and defined this quotient as Δ nuclear spread (Fig. 4 A).
Figure 4.
Dynamics of myonuclear cluster separation are affected by levels of Bsg25D and Ens. (A) First and last frames from time-lapse series of myonuclear movement during stage 15 in indicated genotypes. Red brackets show change in nuclear spread over time. In Bsg25D−/−;ens−/+ images, green and red brackets show adjacent myotubes undergoing opposite Δ nuclear spreads. All images are extended focus projections of Z-stacks. Nuclei are multicolored. Scale bar = 5 µm. (B) Graph showing percentages of myotubes undergoing positive versus negative Δ nuclear spread during stage 15. (C) Mean and SD of Δ nuclear spread during stage 15. For each genotype in B and C, the number of myotubes is as follows: control, n = 112; Bsg25D−/−, n = 72; Bsg25D−/−;ens−/+, n = 86; Dmef2-Gal4>UAS-ens, n = 85; Dmef2-Gal4>UAS-Bsg25D, n = 62. P values were calculated by Student’s t test. **, P < 0.01; ****, P < 0.0001. (D) Correlation between stage 15 mean Δ nuclear spread and stage 16 mean nuclear spread in fixed samples. Linear regression line is in gray; r2 is the coefficient of determination.
We observed that only 59% of control myotubes exhibited positive Δ nuclear spread over a 1-h period during stage 15. This was initially surprising, because we have previously seen that from stages 14 to 16, nuclear clusters move toward myotube poles (Folker et al., 2012; Metzger et al., 2012). Our interpretation is that nuclear clusters move toward the muscle poles unevenly, with periods where the nuclear spread increases and periods where it decreases. Nuclear clusters all eventually arrive near the muscle poles because the net positive (poleward) movement is greater than negative (toward the center of the muscle) movement; however, during relatively short time-lapse recordings, the nuclear spread in individual cells can decrease quite dramatically.
The percentage of myotubes exhibiting positive versus negative Δ nuclear spread varied with genotype (Fig. 4 B). Embryos with muscle-specific Ens overexpression contained the highest percentage of myotubes with positive Δ nuclear spread (81%) and embryos with muscle-specific Bsg25D overexpression the lowest (21%). In Bsg25D-overexpressing embryos at stage 15, myonuclei were either present in a single cluster or two slightly separated clusters. Negative Δ nuclear spread was due to loosely packed single clusters compacting or barely separated clusters coming back together (for example, Fig. 4 A). Bsg25D-overexpressing cells exhibiting positive Δ nuclear spread had single nuclear clusters that appeared to separate slightly without dividing into two distinct clusters.
We detected a novel behavior in myonuclei from Bsg25D−/−;ens−/+ embryos. In some cells, two distinct nuclear clusters migrated a considerable distance toward each other and reformed one cluster. In this genotype, there was high variability in dynamic cluster behavior among myotubes; the SD of Δ nuclear spread was greater in Bsg25D−/−;ens−/+ cells than in controls (4.47 µm/h vs. 3.42 µm/h, P = 0.008; Fig. 4 A).
For each genotype, we averaged Δ nuclear spread for each cell, including both negative and positive values (Fig. 4 C). We found no statistically significant difference between control and Bsg25D−/− mutants. Control, Bsg25D−/−, and Ens-overexpressing cells had a positive average Δ nuclear spread. Bsg25D−/−;ens−/+ and Bsg25D-overexpressing cells had a negative average Δ nuclear spread. Across genotypes, there was a strong correlation (r = 0.94) between average Δ nuclear spread at stage 15 and average nuclear spread in fixed embryos at stage 16 (Fig. 4 D). Thus, the rate of nuclear spreading at stage 15 predicts and likely determines the degree of nuclear spread observed at stage 16.
Bsg25D overexpression causes defects in myonuclear positioning in fully differentiated larval muscle that are rescued by addition of Ens
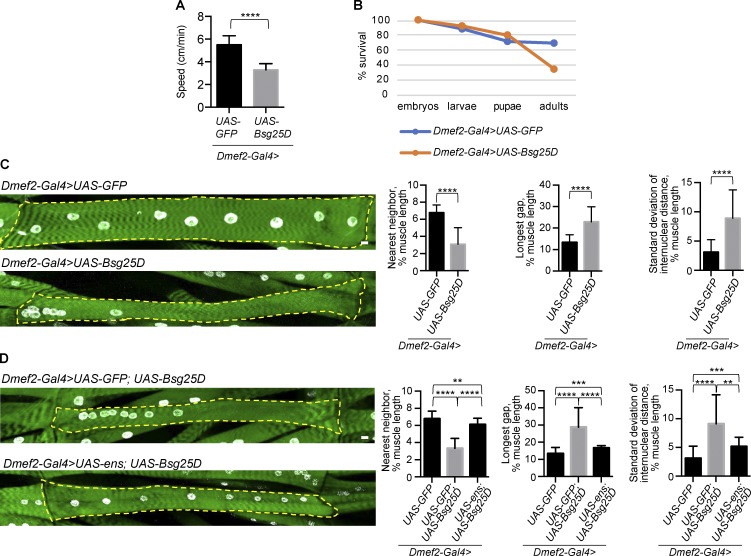
Having characterized the effect of Bsg25D overexpression on embryonic myotubes, we next investigated Bsg25D overexpression in mature myofibers. Myofibers are established in the embryo at stage 17 and continue to grow and function during larval stages. We focused on the third-instar larval stage, a time point when the muscles are amenable to functional analysis and antibody staining (Folker et al., 2012). Using the Dmef2-Gal4 driver, which becomes active in embryonic muscle and persists through the larval stage, we overexpressed either Bsg25D or GFP (as a control) and measured larval motility as a readout of muscle function. Control larvae outperformed Bsg25D-overexpressing larvae (Fig. 5 A), indicating that Bsg25D overexpression perturbs muscle function. Animals with muscle-specific overexpression of Bsg25D exhibited decreased survival to adulthood, with peak lethality occurring during the pupal stage (Fig. 5 B). Moreover, adult survivors were unable to fly, indicating that overexpressed Bsg25D also perturbs muscle function in adults (data not shown).
Figure 5.
Bsg25D overexpression induces nuclear positioning defects in larval myofibers that can be rescued by Ens. (A) Larval motility in larvae overexpressing either GFP (control) or Bsg25D in muscle. For Dmef2-Gal4>UAS-GFP, n = 30 larvae. For Dmef2-Gal4>UAS-Bsg25D, n = 12 larvae. P values were calculated by Student’s t test. (B) Viability graph showing survival during development. n = 100 individuals for each genotype. (C and D) Representative extended focus projections of stained myofibers and nuclear positioning analyses. Green, phalloidin; white, nuclei. Dashed yellow line outlines individual myofibers. Scale bars = 10 µm. Graphs depict mean and SD for three methods for quantifying myonuclear positioning (see Materials and methods). For each genotype, the same images were analyzed with each method. For Dmef2-Gal4>UAS-GFP, n = 40 myofibers. For Dmef2-Gal4>UAS-Bsg25D, n = 45. For Dmef2-Gal4>UAS-GFP;UAS-Bsg25D, n = 23; Dmef2-Gal4>UAS-ens;UAS-Bsg25D, n = 19. Dmef2-Gal4>UAS-GFP data are the same in C and D. P values were calculated by Student’s t test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
After assessing muscle function in Bsg25D-overexpressing larvae, we examined muscle structure. In control larvae with muscle-specific GFP expression, myonuclei were evenly spaced throughout muscle fibers, as expected (Fig. 5 C). In larvae that had developed with sustained muscle-specific Bsg25D overexpression, however, myonuclei were frequently clustered together (Fig. 5 C). This phenotype was highly penetrant and robustly quantifiable by three different analyses. First, the average distance between each nucleus and its nearest neighbor was decreased. Second, the average longest stretch of myofiber devoid of myonuclei was increased. Third, the average SD of the distances between sequential nuclei (SD of internuclear distance) within a myofiber was increased, indicating irregular, uneven positioning of nuclei (Fig. 5 C). Unlike stage 16 myotubes, where Bsg25D-induced myonuclear positioning defects were nearly uniform (i.e., one cluster of myonuclei near the myotube’s center), larval muscle fibers overexpressing Bsg25D exhibited nuclear clusters that varied in both nuclear number and position within the fiber (Figs. 5 C and S4 A). The total number of nuclei present per larval muscle was the same in Bsg25D-overexpressing and control larvae (Fig. S4 B). As with other genotypes affecting myonuclear positioning (Elhanany-Tamir et al., 2012), Bsg25D overexpression conferred a decrease in nuclear size (Fig. S4 C).
Since Ens overexpression restored nuclear spread in Bsg25D-overexpressing embryonic myotubes (Fig. 3, A and B), we hypothesized that “rescued” myotubes would develop into myofibers with normal myonuclear positioning. Indeed, when we overexpressed Ens along with Bsg25D starting in embryonic myotubes, the resulting larval myofibers had nearly normal nuclear positioning; in contrast, control myofibers expressing GFP with Bsg25D had severe nuclear positioning defects (Fig. 5 D). These two genotypes had the same number of myonuclei per myofiber (Fig. S4 D). From these experiments, we conclude that sustained Ens overexpression rescues nuclear positioning defects in Bsg25D-overexpressing mature muscles.
Overexpressed Bsg25D disrupts MT organization in myofibers
Since Bsg25D and Ens have been shown to regulate MT organization in other contexts (Mogensen et al., 2000; Delgehyr et al., 2005; Gallaud et al., 2014; Srivatsa et al., 2015; Kowanda et al., 2016; Zheng et al., 2016), we hypothesized that Bsg25D–Ens interactions control myonuclear positioning by regulating the muscle MT network. However, we did not observe a difference in the density of the MT network in embryonic myotubes among control, Bsg25D−/−, Bsg25D−/−;ens−/+, and Bsg25D-overexpressing genotypes (Fig. 6, A and B). Using time-lapse microscopy to follow genetically encoded EB1-YFP, a fluorescently tagged MT plus end–binding protein that tracks the growing ends of MTs, we observed that EB1-YFP “comets” moved with the same speed in control and Bsg25D−/− myotubes (control: 0.36 ± 0.12 µm/s, Bsg25D−/−: 0.37 ± 0.11 µm/s; Videos 6 and 7 and Fig. 6, C and D), indicating that MT polymerization rates were unaffected by complete loss of Bsg25D.
Figure 6.
MT density and dynamics are normal in Bsg25D−/− embryonic myotubes. (A) Representative extended-focus projections of Tropomyosin and α-Tubulin signal in stage 16 lateral transverse myotubes. Green boxes in the top row indicate muscle tips magnified in the second and third rows. Red boxes represent regions where signal intensity was quantified. Scale bar = 1 µm. (B) Bar graph showing the mean and SD of the ratio of Tubulin to Tropomyosin signal for indicated genotypes. The number of embryos for each genotype is as follows: control, n = 9; Bsg25D−/−, n = 14; Bsg25D−/−;ens-/+, n = 11; Dmef2-Gal4>UAS-Bsg25D, n = 14. P values were calculated by Student’s t test. The entire experiment was performed twice with similar results; images and graph are from one replicate. (C) Representative EB1-YFP comets for indicated genotypes. Schematic shows that comets run toward myotube tips. Images were generated by merging three frames from a time-lapse series, pseudocolored red (t1 = 0 s), cyan (t2 = 1.6 s), and yellow (t3 = 3.2 s). Scale bar = 0.1 µm. (D) Scatter plot showing mean speed ± SD (micrometers per second) of EB1-YFP comets. The number of EB1-YFP comets for each genotype is as follows: control, n = 140; Bsg25D−/−, n = 94. P value was calculated by Student’s t test.
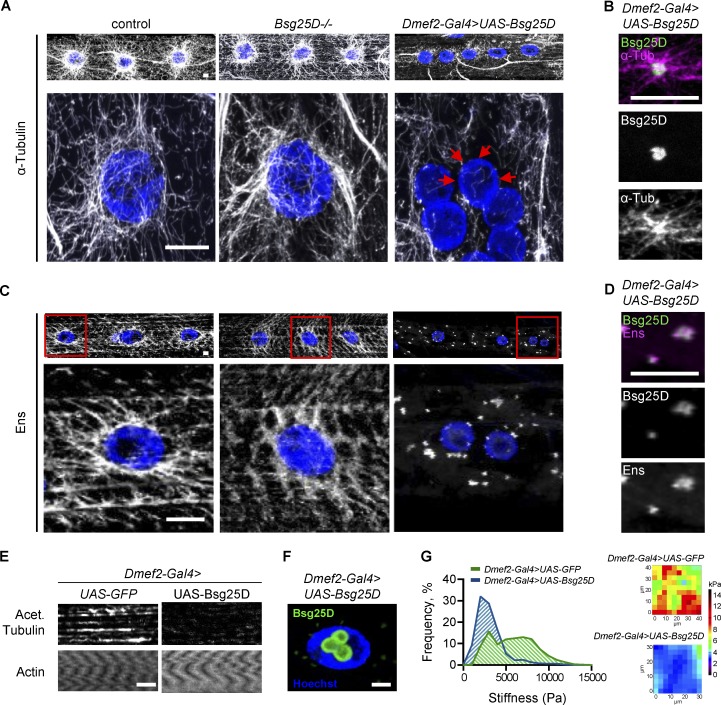
Next, we examined the effect of Bsg25D levels on the mature MT organization of larval myofibers. Larval myofibers are advantageous for genetic analysis of MT organization because they are larger, more regularly shaped, and possess a more clearly organized MT network than myotubes. In control muscle fibers, as previously observed (Elhanany-Tamir et al., 2012; Metzger et al., 2012), there were two spatially defined populations of MTs: superficial MTs that emanate from the nuclear envelope of each nucleus, and deeper MTs oriented parallel to the long axis of the fiber. While Bsg25D null mutants had normal myonuclear positioning (Fig. S4, E and F) and MT organization (Fig. 7 A), Bsg25D-overexpressing muscle fibers had severe defects in MT organization (Fig. 7 A). Longitudinal MTs were present, but nuclear MT arrays were completely absent. Instead, MTs appeared to grow from Bsg25D-positive ectopic MTOCs in the cytoplasm, as determined by the colocalization of Bsg25D with the centers of star-shaped MT asters (Fig. 7 B). Overexpressed Bsg25D also localized inside myonuclei (Fig. 7 F), as has been seen with overexpressed human Ninein in cell culture. This cell culture localization was linked to posttranslational SUMO modification of Ninein (Cheng et al., 2006), and we observed colocalization of the sole Drosophila SUMO protein, SUMO-2, and overexpressed Bsg25D in myonuclei, but not in Bsg25D-positive cytoplasmic puncta (Fig. S5). This is consistent with overexpressed nuclear Bsg25D being sumoylated and raises the possibility that sumoylation is necessary for overexpressed Bsg25D to localize to myonuclei.
Figure 7.
Bsg25D overexpression perturbs normal MT organization and forms Ens-positive MTOCs in larval myofibers. (A) Top: α-Tubulin signal in extended-focus projections of larval myofibers. Bottom: Higher magnification images of nuclear MT arrays. Gray, α-Tubulin; blue, nuclei. Red arrows show myonuclear envelope without MTs. (B) Extended-focus projections of Bsg25D and α-Tubulin signal in a cytoplasmic Bsg25D punctum in Dmef2-Gal4>UAS-Bsg25D larval myofiber. Bsg25D, green; α-Tubulin, magenta; colocalization, white. (C) Top: Ens signal in extended-focus projections of larval myofibers of indicated genotypes. Bottom: Higher magnification images of nuclei delineated by red boxes. Gray, Ens; blue, nuclei. (D) Extended-focus projections of Bsg25D and Ens signal in a cytoplasmic Bsg25D punctum in Dmef2-Gal4>UAS-Bsg25D larval myofiber. Bsg25D, green; Ens, magenta; colocalization, white. (E) Staining of larval myofibers with phalloidin (labeling Actin) and an antibody against acetylated Tubulin. Images are single slices from Z-stacks, immediately below myonuclei. Staining for actin confirms the presence of intact myofibers. (F) Single slice through a Dmef2-Gal4>UAS-Bsg25D myofiber nucleus. Blue, Hoechst; green, Bsg25D. In all images, scale bars = 10 µm. In B, D, and F, Bsg25D localization is visualized by Eos, which fluorescently tags the N-terminus of overexpressed Bsg25D. (G) Left: Frequency histogram of all stiffness measurements from AFM for indicated genotypes. For Dmef2-Gal4>UAS-GFP, n = 21 myofibers. For Dmef2-Gal4>UAS-Bsg25D, n = 16 myofibers. In each genotype, multiple measurements were taken per myofiber; see materials and methods. P value was calculated by Student’s t test: ****, P < 0.0001. Right: Examples of an AFM force map for each genotype.
Since we have shown that Bsg25D overexpression affects Ens localization in embryonic myotubes (Fig. 3 C), we next determined how it affects Ens localization in mature myofibers. Immunofluorescent staining in control myofibers showed Ens broadly expressed with increased signal around myonuclei, consistent with Ens (a MAP) being present on MTs (Fig. 7 C). This localization was not affected by loss of Bsg25D (Fig. 7 C). However, in myofibers overexpressing Bsg25D, Ens was lost from nuclear peripheries and instead colocalized with Bsg25D puncta in the cytoplasm (Fig. 7, C and D). However, Ens did not label the MTs emanating from Bsg25D puncta (Fig. 7, B vs. D). Thus, when Bsg25D is overexpressed, Ens becomes mislocalized from MTs to cytoplasmic Bsg25D puncta.
To further investigate the Bsg25D-induced disruptions to the MT network in larval myofibers, we performed immunofluorescent antibody staining to detect MTs bearing an acetylation modification associated with MT stability. Control muscles exhibited acetylated MTs emanating from the nuclear envelope, as well as in the deeper longitudinal tracts (Fig. 7 E). In Bsg25D-overexpressing myofibers, the antibody against acetylated MTs only weakly labeled longitudinal MTs (Fig. 7 E). This indicates the longitudinal MTs in Bsg25D-overexpressing muscles were hypoacetylated relative to controls. (There was no acetylated tubulin signal around myonuclei, because there were no MTs there.) It was recently shown that MT acetylation in neuronal branches correlates with the subcellular localization of Ens (Tymanskyj et al., 2017), raising the possibility that overexpressed Bsg25D causes MT hypoacetylation by sequestering Ens away from MTs. Taken together, Bsg25D overexpression causes defects in MT organization and posttranslational acetylation; the latter may be indicative of a loss of MT stability.
We next investigated whether Bsg25D overexpression affected the mechanical properties of myofibers by performing atomic force microscopy (AFM) to measure stiffness in control (GFP-overexpressing) and Bsg25D-overexpressing myofibers. We found a significant decrease in the stiffness of Bsg25D-overexpressing myofibers (Dmef2-Gal4>UAS-GFP mean ± SD = 5,895 ± 2,646 Pa, Dmef2-Gal4>UAS-Bsg25D mean ± SD = 3,167 ± 1,735 Pa, P < 0.0001; Fig. 7 G), perhaps due to the changes in MT organization that we documented, although the role of MTs in regulating myofiber stiffness appears to be complex and variable between systems (Collinsworth et al., 2002; Nishimura et al., 2006; Kerr et al., 2015). Alternatively, other cytoskeletal elements such as actin or myosin, which have been shown to be critical for generating stiffness in skeletal muscle (Collinsworth et al., 2002), may have been disorganized or reduced in these muscles. Together, this series of experiments shows that overexpressed Bsg25D causes defects in the intracellular organization and mechanical properties of larval myofibers.
Discussion
Bsg25D functions in myonuclear positioning
The Bsg25D null mutants we generated, like those in two recent reports, are viable and fertile (Kowanda et al., 2016; Zheng et al., 2016). Zheng et al. (2016) did not detect any mutant abnormalities, whereas Kowanda et al. (2016) observed a decrease in embryonic hatching and partially penetrant mitotic defects. Our null Bsg25D mutant has a lower rate of embryonic death than the mutant generated by Kowanda et al. (2016). The differences between the Bsg25D mutant phenotypes generated by different groups likely reflect the sensitivity of Bsg25D mutant phenotypes to genetic background. Because our null Bsg25D mutant did not have obvious defects, our analysis of Bsg25D required other approaches: generation of double mutants and gain-of-function analysis. Through these approaches, we uncovered a role for Bsg25D in myonuclear positioning, where it works with Ens. To our knowledge, Ninein null mice have not been reported; it will be interesting to observe what aspects of Ninein function are conserved in mammals.
Bsg25D and Ens regulate the dynamics of myonuclear movement
Our live imaging of stage 15 embryos yielded unexpected insights about the dynamics of myonuclear movement. We initially thought that nuclear clusters separated at stage 14 and then migrated toward their respective poles in a straightforward fashion. However, in all genotypes tested, we observed both myotubes where the nuclear spread increased during our window of observation and myotubes where nuclear spread decreased. These observations argue for a more complex model of myonuclear positioning in which the forces driving nuclear clusters toward muscle poles (for example, Bsg25D and Ens) are opposed by unknown forces driving clusters toward each other. Thus, the relative levels of these opposing forces during stages 14–16 determine myonuclear spread.
Overexpression of Bsg25D affects multiple aspects of muscle organization and function
Gain-of-function analysis revealed that overexpressed Bsg25D has potent activity in embryonic myotubes and mature myofibers. In myotubes it perturbs myonuclear positioning; in myofibers it perturbs myonuclear positioning, MT organization, muscle stiffness, and muscle function. The severity of these phenotypes suggests it is critical for muscle cells to limit the amount of Bsg25D protein present. In overexpression experiments in the ovary, Bsg25D localization and activity is regulated by its 3′ UTR (Kowanda et al., 2016); however, we found that Bsg25D overexpression constructs with or without 3′ UTR confer similar phenotypes in myotubes (data not shown), suggesting that the 3′ UTR is not critical to Bsg25D regulation in muscle. Developing muscle presumably employs other mechanisms to limit the levels of Bsg25D.
Unlike in myofibers, Bsg25D overexpression in embryonic myotubes confers no detectable MT defects. Consistent with these observations, overexpressed Bsg25D did not colocalize with α-Tubulin or γ-Tubulin (data not shown). It is unclear how myotubes resist the MT-altering effects of overexpressed Bsg25D that we observed in myofibers and others have observed in other systems (Stillwell et al., 2004; Kowanda et al., 2016; Zheng et al., 2016); perhaps myotubes lack an essential cofactor necessary for Bsg25D to induce ectopic MTOCs. In general, our data support that MTs in developing myotubes have different behaviors from MTs in myofibers and in other tissues.
While overexpressed Bsg25D does not perturb MTs in embryonic myotubes, does endogenous Bsg25D regulate MT organization? We failed to detect differences in MT density and polymerization rate between control and Bsg25D−/− myotubes; this is in contrast to the C. elegans larval epidermis, where Bsg25D/Noca-1 mutants were found to have lower-than-normal MT polymerization rates by an EB1-based assay analogous to ours (Wang et al., 2015). There are two caveats to our conclusion that Bsg25D does not function in MT regulation in Drosophila myotubes. First, we could not draw conclusions about the number of polymerizing MTs in each genotype because the EB1-YFP signal was weaker in mutant cells than control cells, making the number of traceable EB1-YFP comets an unreliable proxy for the number of polymerizing MTs. Second, it remains possible that MT phenotypes in Bsg25D−/− myotubes are masked by a redundantly acting factor. Though our negative data do not definitively rule out a role for endogenous Bsg25D in regulating MTs in muscle, the absence of clear MT phenotypes in Bsg25D−/− myotubes and myofibers leads us to favor the conclusion that Bsg25D functions in myonuclear positioning without playing a major role in MT regulation.
Ens and Bsg25D exhibit complex interactions
Our work reveals that Bsg25D and Ens work together in myonuclear positioning. It is not clear exactly where in the myotube they interact to promote myonuclear movement. In immunostained embryos, the most conspicuous Bsg25D signal in myotubes is in cytoplasmic puncta, which also stain positive for Ens. However, there is also a broad, low-level Bsg25D signal throughout the cytoplasm that our microscopy techniques cannot further resolve. Given that Ens coats MTs, it is plausible that Bsg25D interacts with Ens broadly on those structures. Indeed, studies have shown Bsg25D/Ninein interacting with the MT network as a cargo (Dammermann and Merdes, 2002; Moss et al., 2007), and a Bsg25D fragment has been shown to directly bind MTs in vitro (Kowanda et al., 2016).
In both myotubes and myofibers, co-overexpression of Ens relieves the myonuclear clustering caused by overexpression of Bsg25D. The mechanisms by which Bsg25D and Ens function likely differ between these two developmental stages. In embryonic myotubes, Bsg25D overexpression phenocopies the ens mutant phenotype, so inhibition of Ens by high levels of Bsg25D is sufficient to explain the nuclear positioning defects. However, the nuclear positioning phenotypes in mature larval myofibers caused by long-term overexpression of Bsg25D are more severe than those caused by loss of Ens; thus, inhibition of Ens is not a sufficient explanation for the Bsg25D-induced defects. We propose that Bsg25D causes nuclear positioning defects in larval myofibers through the combined effect of multiple mechanisms, including inhibition of Ens, inhibition of other proteins involved in myonuclear positioning such as Dynein, and disruption of MT organization. Co-overexpressed Ens then suppresses at least some of these Bsg25D activities, restoring normal nuclear positioning.
The finding that embryos with reduced maternal/zygotic ens and a total absence of Bsg25D fail to survive to larval stages demonstrates that critical Bsg25D–Ens interactions occur outside of the muscle as well. Evidence from the literature raises the possibility that they interact in the female germline, as groups have separately reported overlapping localization patterns for Bsg25D and Ens proteins, with both found at the anterior pole of the developing oocyte during midoogenesis (Sung et al., 2008; Kowanda et al., 2016). Moreover, misexpression of Bsg25D and loss of function of ens cause common phenotypes: reductions in Dynein and Gurken at the posterior pole and anterior-dorsal corner of the oocyte, respectively, and resulting embryos lacking dorsal appendages (Sung et al., 2008; Kowanda et al., 2016). Thus, it may be the case that forced expression of Bsg25D binds and disrupts endogenous Ens in the female germline as it does in muscle.
Centrosomal proteins participate in myonuclear movement
We find that in addition to Bsg25D, a second centrosomal protein, Plp (the Drosophila Pericentrin homologue), is involved in myonuclear positioning. The function of centrosomal proteins in muscle, which lacks centrosomes, has been unclear. Our work suggests that a general function of centrosomal proteins in muscle is to position myonuclei. This hypothesis is supported by the recent finding that the centrosomal protein PCM-1 is required for myonuclear positioning in C2C12 myotubes (Espigat-Georger et al., 2016). These findings underscore the conservation of mechanisms regulating nuclear movement and muscle organization. It is intriguing that although Bsg25D and Plp colocalize in myotubes, and each participates in myonuclear positioning, loss of Bsg25D does not enhance myonuclear positioning defects in plp mutants. It may be the case that the two proteins ultimately contribute to myonuclear positioning via separate pathways.
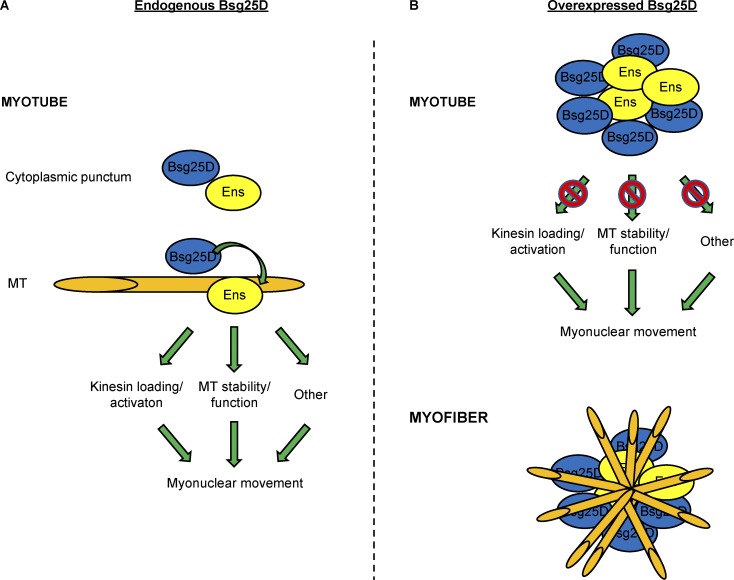
Model
We propose that endogenous Bsg25D positively regulates Ens in a manner dependent on their physical interaction (Fig. 8). The critical interaction could either be at Bsg25D–Ens cytoplasmic puncta or on Ens-coated MTs. Bsg25D could conceivably promote Ens activity by affecting its protein levels, subcellular localization, access to binding partners, or posttranslational modification, or by other means. In our model, embryos with the full complement of ens have enough Ens activity to withstand loss of the activator Bsg25D without showing diminished myonuclear movement. However, in embryos where Ens activity is limited by loss of one allele of ens, loss of Bsg25D further reduces Ens activity and perturbs myonuclear movement.
Figure 8.
Model of Bsg25D–Ens interactions. (A) Model of the Bsg25D–Ens interaction in wild-type myotubes. (B) Model of the Bsg25D–Ens interaction when Bsg25D is overexpressed. See Discussion for model details. Bsg25D, blue; Ens, yellow; MTs, orange.
When Bsg25D is present at high concentrations in myotubes, it confers severe nuclear positioning phenotypes through its interactions with Ens (Fig. 8 B). Overexpressed Bsg25D forms puncta that recruit endogenous Ens away from its normal localization on MTs, leading to a block in Ens-dependent myonuclear movement toward the muscle poles. In myofibers, these Bsg25D- and Ens-positive puncta serve as ectopic MTOCs. MTs are lost from the nuclear envelope, possibly because necessary factors are recruited to the ectopic MTOCs. Nuclei are mispositioned, in part due to altered MT organization, and muscle function is decreased.
Conclusion
We have identified Drosophila Bsg25D as a novel Ens interactor and shown that it functions with Ens in myonuclear positioning. We have found that overexpressed Bsg25D blocks myonuclear movements in embryonic myotubes and that sustained Bsg25D overexpression disrupts both myonuclear positioning and MT organization in mature larval myofibers; nuclear positioning defects in embryos and larvae are rescued by cooverexpression of Ens. Thus, Bsg25D–Ens interactions are critical for myonuclear positioning and muscle development. The human homologues of the two proteins have important functions that are illustrated by their association with disease; loss of Ninein causes Seckel syndrome (Dauber et al., 2012), while high levels of MAP7 are associated with types of colon cancer and leukemia (Blum et al., 2008; Fu et al., 2016). The use of model systems to investigate how gain and loss of Ninein/Bsg25D and Ens/MAP7 affect cells in vivo will lend insights to human health.
Materials and methods
Drosophila genetics
Standard Drosophila genetics were performed at 25°C. The following stocks were used: apterousME-NLS::dsRed (Richardson et al., 2007), ensswo (Metzger et al., 2012), Df-ensΔ3277 (Sung et al., 2008), khc8 (Brendza et al., 1999), dhc4–19 (Gepner et al., 1996), plp5 (Martinez-Campos et al., 2004), Dmef2-Gal4 (Ranganayakulu et al., 1998), rP298-Gal4 (Menon and Chia, 2001), ubiquitin-Gal4[3xP3-GFP] (Baena-Lopez et al., 2013), UAS-Ens-HA (Metzger et al., 2012), UAS-2xEGFP (Halfon et al., 2002), UAS-EB1-YFP (Bulgakova et al., 2013), and patroninΔ2 (gift from M. Gonzalez-Gaitan, University of Geneva, Geneva, Switzerland). w1118 (3605), yw (1495), UAS-Rab7.GFP (42706), UASp-GFP.Golgi (30902), UAS-GFP.KDEL (9899), Df(2L)BSC693 (26545), Df(3L)BSC441 (24945), hs-FLP, hs-SceI (25679), γ-tub23CA6-2 (5728), and γ-tub23CA14-9 (7041) were obtained from the Bloomington Drosophila Stock Center. P3-83 containing BAC clone CH321-49G22, referred to in the text as BAC[Bsg25D+], was obtained from Genetivision. The following lines were developed for this work (details below): Bsg25Dnull, UAS-eos::Bsg25D, and UAS-ens::EGFP. ensswo and Df-ensΔ3277 homozygotes have the same nuclear positioning phenotype at stage 16 (data not shown). The Df-ensΔ3277 allele was used for genetic interaction experiments with Bsg25D overexpression (the three rightmost panels in Fig. 3 A). All other experiments with ens mutants used the ensswo allele.
Generation of mutant and transgenic Drosophila stocks
Bsg25Dnull (referred to in this work as Bsg25D−/−) flies were generated by accelerated homologous recombination without CRISPR/Cas9 (Baena-Lopez et al., 2013). Homology arms 5′ and 3′ of Bsg25D were PCR amplified and sequentially cloned into pTVCherry (gift from J.P. Vincent, Francis Crick Institute, London, UK). The following primers, containing the indicated restriction enzyme sites, were used: 5p homology arm: 5p forward NheI, 5′-GATCGCTAGCAGCCATTCGCCTCGCTCTTC-3′; 5p reverse KpnI, 5′-GATCGGTACCGCTGCCGTTCCAATCGATCG-3′; 3p homology arm: 3p forward BglII, 5′-GATCAG-ATCTGACAAACCTGAGGAGGGAGT-3′; 3p reverse AvrII, 5′-GATCCCTAGGCGTAGAAAGGGTAGAAATAT-3′.
The pTVCherry-Bsg25D donor cassette was then randomly integrated in w1118 flies by p-element transformation (Genetic Services, Inc.). Potential transgenic flies were screened for by mini-white expression, and a third chromosome transgenic line was obtained. Following the published accelerated homologous recombination cross scheme (Baena-Lopez et al., 2013), 14 putative mutant lines were recovered. One was validated as Bsg25Dnull by PCR using primers designed to amplify genomic regions specific to either wild-type or mutant chromosomes, followed by sequencing. The Bsg25D−/− line used in this study had the mini w+ cassette inserted between bp334 in the Bsg25D 5′ UTR and bp5919 at the beginning of the Bsg25D 3′ UTR based on the published FLYBASE Bsg25D genomic sequence, leading to the removal of all protein coding sequences and, as a result, all protein isoforms (Fig. 2 A). The following PCR primers were used for mutant verification: wild-type-specific Bsg25D: 1537 forward, 5′-ACAATACGGACGAGGACCAG-3′; 3907 reverse, 5′-TTCCTTTGCAGCCTTGAGTT-3′; mutant-specific Bsg25D pair 1: 7224 forward, 5′-GAGTTGCTGTTGCTGCAGAG-3′; 11081 reverse, 5′-TTGACCTCAGCGTCGTAGTG-3′; mutant-specific Bsg25D pair 2: 7326 forward, 5′-TGAAGATACCCCACCAAACC-3′; 10289 reverse, 5′-GCGCACTCAGCAAAACATTA-3′. Mutant flies were confirmed by Western blotting for Bsg25D multiple times with biological replicates.
UAS-eos::Bsg25D (referred to in this paper as UAS-Bsg25D) flies were generated as follows: Eos was amplified from pME-eos (gift from D. Raible, University of Washington, Seattle, WA) using the following forward and reverse primers containing restriction sites for NotI and XhoI, respectively: F_NotI_eos_NotI, 5′-CACCGCGGCCGCATGAGTGCGATTAAGCCAGACATG-3′; R_XhoI_eos_no_stop_XhoI, 5′-CTCGAGTCGTCTGGCATTGTCAGGCAATCC-3′. The PCR product was cloned into pUAST. Next, Bsg25D was amplified from expressed sequence tag clone LD21844 (Drosophila Genome Resource Center [DGRC]), corresponding to transcript Bsg25D-PB, and cloned into pUAST-eos in frame with the N-terminal Eos. The following primers, both containing KpnI restriction sites, were used for PCR: F-CACC-KpnI-Bsg25D, 5′-CACCGGTACCATGGAGGTATCCGCCGATCCGTAC-3′; R-KpnI-stop-Bsg25D, 5′-GGTACCCTAAGGCATGCCAGGCAGTCCACC-3′. pUAS-eos::Bsg25D was randomly integrated in w1118 flies by p-element transformation (Genetic Services, Inc.). Potential transgenic flies were selected by mini-white expression. The eos::Bsg25D line was used for all fly studies on overexpressed Bsg25D.
UAS-ens::EGFP flies were generated as follows. Full-length ens (LD09626; DGRC) was PCR-amplified using the following primers containing EcoRI and BamHI restriction sites, respectively: 5′-GAATTCATGGCGAGTCTTGGGGGCCAACAC-3′ and 5′-GGATCCCAGCAGCGATATATCTTTATTTTCGTG-3′. The PCR product was cloned into pUAST (DGRC) previously modified to contain a C-terminal EGFP tag. pUAS-Ens::EGFP was randomly integrated in w1118 flies by p-element transformation (Best Gene, Inc.), and potential transgenic flies were screened for by mini-white expression.
Protein sequence analysis
Bsg25D protein sequence analysis was performed by InterProScan, which identifies domains by scanning protein sequences against predictive models provided by several different databases (Jones et al., 2014). Alignment of Bsg25D to mouse Ninein was done using the Blastp algorithm (National Center for Biotechnology Information).
Viability assays
Viability assays were done at 25°C essentially as previously described (Schulman et al., 2014). For comparison of GFP- and Bsg25D-overexpressing flies, embryos were bleached and selected at stage 16. For other viability assays, embryos were selected without bleaching under halocarbon oil at stage 5. In all cases, a minimum of 100 embryos were selected over multiple days. The number of hatched and unhatched embryos was counted, and all first-instar larvae were transferred to a vial with fresh food. The number of pupae and adults that eclosed was assessed. Viability is represented as a percentage of the initial number of embryos.
Fluorescent antibody staining
Embryos were prepared for staining as previously described (Richardson et al., 2007). Larvae were dissected and flat mounted in HL3.1 dissecting buffer and fixed for 20 min in formalin. Embryos and larvae were incubated in primary antibody overnight at 4°C or for 1 h at room temperature at the following concentrations: rat anti-Tropomyosin (1:500; Abcam), rabbit anti-DsRed (1:400; Clontech), chicken anti-GFP (1:500; Abcam), mouse anti-α-Tubulin (1:500; Sigma), mouse anti-acetylated α-Tubulin (1:50; Santa Cruz Biotechnology), mouse anti-γ Tubulin clone GTU-88 (1:500; Sigma), guinea pig anti-Bsg25D (Iampietro et al., 2014; 1:400; gift from E. Lecuyer, Montreal Clinical Research Institute [IRCM], Montreal, Canada), rat anti-Ens (1:100; gift from P. Rorth, University of Copenhagen, Copenhagen, Denmark), rabbit anti-Plp (gift from J. Raff, University of Oxford, Oxford, UK), rabbit anti-Arl8 (1:100; Developmental Studies Hybridoma Bank), and mouse anti-SUMO-2 8A2 (1:50; Developmental Studies Hybridoma Bank). Overexpressed eos::Bsg25D was detected either by Bsg25D antibody staining or by Eos signal. For both embryo and larva staining, Alexa Fluor 488–, Alexa Fluor 555–, and Alexa Fluor 647–conjugated secondary antibodies (Invitrogen) were applied 1:400 for 1 h at room temperature. Hoechst and Alexa Fluor-conjugated phalloidin (either Alexa Fluor 488 or Alexa Fluor 555; Life Technologies) were added with the secondary antibody at 1:400 and 1:200, respectively. Samples were mounted in ProLong Gold antifade reagent (Invitrogen).
Fixed sample imaging and analysis
Z-stacks of fixed samples were acquired using a Leica SP5 laser-scanning confocal microscope equipped with the LAS AF software using a 63× 1.4 NA HCX PL Apochromat oil objective and processed in Volocity (Perkin Elmer) or ImageJ (National Institutes of Health). For analysis of nuclear positioning at stage 16, the Line function in ImageJ was used to measure the distances from the dorsal-most nucleus to the dorsal myotube pole, the ventral-most nucleus to the ventral myotube pole, and the total myotube length. “Nuclear spread” was calculated by subtracting the first two values from the third and expressing the difference as a percentage of total myotube length. For statistical purposes, the mean of the four lateral transverse muscles in a hemisegment was considered one sample. Nuclear positioning data were collected from a maximum of four hemisegments (abdominal hemisegments 2–5) per embryo. Embryos were fixed over the course of multiple days and then pooled for staining and analysis. Myonuclear positioning in the ventral longitudinal (VL) 4 muscle of third-instar larvae was also assessed using the Line function in ImageJ. “Nearest neighbor” was defined as the distance between a nucleus and the nearest neighboring nucleus. “Largest gap” refers to the longest stretch of muscle fiber void of myonuclei, and internuclear distance refers to the distance between consecutive myonuclei along the longitudinal muscle axis. All three terms are presented as percentages of muscle length. Statistical analysis of nuclear positioning in larval muscles employed weighted averages to account for different numbers of myofibers analyzed for each larva. n refers to the total number of myofibers. MT density was calculated by measuring the intensity of Tubulin and Tropomyosin near the tips of the muscles, essentially as published (Folker et al., 2012). For statistics, the average of Tubulin/Tropomyosin ratios from multiple cells in the same embryo was considered one sample. Student’s t tests were performed using Prism software or Excel. Analysis of Ens and Phalloidin signal intensities in embryonic VL myotubes were performed in essentially the same fashion from single slices of confocal stacks.
Time-lapse imaging and analysis
Embryos to be subjected to time-lapse imaging of myonuclear movements were prepared as reported previously (Folker et al., 2012). Embryos were bleached for 4 min, rinsed and mounted in halocarbon oil on a custom-made slide with an air-permeable membrane, and covered with a glass coverslip. Time-lapse series of myonuclear movements were acquired on a Zeiss LSM 700 equipped with the ZEN software and using a Plan-Apochromatic 20×/0.8 M27 objective with 1.7× optical zoom. Stacks were acquired every 3 min for 1 h at 25°C. The line function in ImageJ was used to measure distance between clusters. To calculate Δ nuclear spread, the distance from the dorsal-most nucleus to the ventral-most nucleus in a cell at the beginning of the time-lapse series was subtracted from the same parameter at the end of the series. The difference was then divided by time elapsed to yield Δ nuclear spread, measured in micrometers per hour. Student’s t tests and correlation analysis were performed using Prism software. F-tests were performed in Excel.
Embryos to be used for EB1-YFP analysis were prepared and mounted as above. Time-lapse series of EB1-YFP comets in the tips of stage 16 lateral transverse muscles were acquired on a Leica TCS SP8 confocal microscope at 25°C using the HCX PL APO 63×/1.30 glycerol lens with 40× optical zoom using the Leica proprietary software. Comets were recorded from 19 cells (control) and 26 cells (Bsg25D−/−). Z-stacks consisted of ∼10 slices with a step size of 400 nm; stacks were acquired without intervals (i.e., nonstop) at a frame rate of ∼23 frames per second for ∼90 s. Time-lapse datasets were deconvolved by a blind deconvolution algorithm in Autoquant X3 software (Media Cybernetics). EB1-YFP comets were manually tracked in three dimensions using Imaris (Bitplane). For each EB1-YFP comet, the displacement distance between its position in the first and last frames of its run was exported and divided by the time elapsed to determine EB1-YFP comet speed. EB1-YFP speed was compared between groups by Student’s t test.
Cell culture, coimmunoprecipitation, and Western blotting
For Drosophila S2 cell expression studies, pUAS-3xHA::Bsg25D was generated as follows. Full-length Bsg25D (LD21844; DGRC) was PCR amplified using the following primers: 5′-CACCATGGAGGTATCCGCGATCCGTAC-3′ and 5′-CTAAGGCATGCCAGGCAGTCCACC-3′. The amplicon was TOPO cloned into pENTR (Life Technologies) and then Gateway cloned using LR Clonase (Life Technologies) into pTHW (DGRC), which contains a UAS followed by an N-terminal 3xHA tag. The generation of pUAS-Ens::EGFP is described above (in Generation of mutant and transgenic Drosophila stocks), as the same plasmid was used for transforming flies and driving expression in S2 cells.
Experiments in S2 cells were performed to validate a physical interaction between Ens and Bsg25D detected in a yeast two-hybrid screen previously described (Metzger et al., 2012). S2 cells were cultured in Schneider’s media plus 10% FBS using standard procedures. Protein expression was achieved by transfecting a plasmid expressing Gal4 from a constitutively active promoter (Ubiquitin-Gal4) along with pUAS-3xHA::Bsg25D and pUAS-Ens-EGFP or pUAS-3xHA::Bsg25D and pUAS-2xEGFP. Transfections used Effectene (QIAGEN). Cells were lysed 2 d after transfection in lysis buffer (150 mM NaCl, 2 mM MgOAc, 20 mM Tris, pH 7.5, 5% glycerol, 0.5% NP-40, 2 mM DTT, and one Complete Mini tab [Roche]). Following centrifugation, lysate supernatant was collected and protein concentration was determined by Bradford assay (Bio-Rad). 400 µg lysate was incubated with Protein G-Agarose beads overnight at 4°C using rabbit anti-GFP (Torrey Pines Biolab) or rabbit anti-IgG (Santa Cruz Biotechnology) for mock immunoprecipitation controls. Following incubation, beads were washed four times with lysis buffer and boiled in 4× Laemmli buffer. Proteins released from beads and 40 µg (10%) inputs were then subjected to SDS-PAGE, transferred overnight to polyvinylidene difluoride membrane, and blotted with rat anti-HA (Roche), secondary stained at 1:5,000 with anti-rat HRP (Jackson ImmunoResearch), and detected with chemiluminescence (Denville Scientific). Membranes were then stripped according to standard practices and reblotted with mouse anti-GFP antibody (Roche) and HRP-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch) and then developed. Coimmunoprecipitation between 3xHA::Bsg25D and Ens-GFP was demonstrated with multiple biological replicates.
To compare Bsg25D protein expression in control and Bsg25D−/− genotypes, w1118 and Bsg25D−/− third-instar larvae were dissected in HL3.1 relaxing buffer. The intestines of the larvae were removed, and the resultant pelts were lysed in lysis buffer (recipe above). Following Bradford determination of protein concentration (Bio-Rad), 80 µg lysate per genotype was loaded on a 7.5% SDS-PAGE protein gel. The gel was transferred to nitrocellulose membrane overnight, blocked with 5% milk (for Bsg25D) or 5% BSA (for GAPDH) in TBS-Tween, and blotted overnight with guinea pig anti-Bsg25D antibody (1:1,000; gift from E. Lecuyer) or mouse anti-GAPDH primary antibody (1:10,000; Abcam). HRP-conjugated secondary antibodies against guinea pig (1:5,000; Jackson ImmunoResearch) or mouse (1:5,000; Jackson ImmunoResearch) were applied before development by chemiluminescence (Kindle Biosciences).
AFM
Larvae were dissected in HL3.1 dissecting buffer and fixed in formalin for 10 min before microscopy. Bright-field images of Drosophila larvae, for determination of location of stiffness measurements, were acquired using an inverted microscope (Axio Observer Z1; Zeiss) as the AFM base (LD Plan-Neofluar 20×/0.4 objective). An Asylum Research MFP-3D-BIO Atomic Force Microscope was used to collect force maps from the Drosophila larvae tissue. A CP CONT-PS-C (NanoAndMore.com) probe with a 6.1-µm polystyrene bead was used for all measurements. The Asylum Research GetReal calibration method was used for the determination of the spring constant (0.2 N/m). Each force map sampled a 20 × 20-µm to 40 × 40-µm region, depending on muscle width, in a 10 × 10-grid under fluid conditions (PBS). The trigger point was set to 50 nN with an approach velocity of 10 µm/s. The force-indentation curves were fit to the Hertz model for spherical tips using Asylum Research Software to determine the Young’s modulus, with an assumed Poisson’s ratio value of 0.45 for the sample (Chen et al., 1996). For each genotype, at least three larvae were used; from each larva, at least 400 individual measurements, corresponding to four different muscles, were taken. Force maps of stiffness along with individual stiffness values for each measured point were then exported from Asylum Research Software for analysis. For statistics, the average of all the measurements from a single muscle was considered one sample.
Larval motility assays
Laval motility assays were performed essentially as previously described, either by using a charge-coupled device camera and Ethovision software (Noldus) to record third-instar larvae crawling and automatically calculate crawling speed (Metzger et al., 2012) or by placing larvae at the center of an apple juice plate surrounded by zones defined by concentric circles and observing which zone they reached in 30 s of crawling at 25°C (Fernandes and Schock, 2014). A minimum of 47 larvae per genotype were used. In all cases, data were acquired over multiple days. P values were calculated by Student’s t test.
Online supplemental material
Fig. S1 shows the subcellular localization of Bsg25D puncta in developing myotubes. Fig. S2 shows Bsg25D−/− viability and larval motility data. Fig.S3 shows myonuclear positioning in Bsg25D−/−;ens−/− myotubes and the lack of genetic interactions between Bsg25D and Khc or Dhc. Fig. S4 shows the effects of Bsg25D overexpression on myonuclear clusters. Fig. S5 shows that overexpressed Bsg25D colocalizes with SUMO-2 in myonuclei. Videos show nuclear movements in control (Video 1), Bsg25D−/− (Video 2), Bsg25D−/−;ens−/+ (Video 3), Bsg25D overexpression (Video 4), and Ens overexpression (Video 5). Videos 6 and 7 show control and Bsg25D−/− EB1-YFP comets, respectively.
Supplementary Material
Acknowledgments
We thank Eric Lecuyer, Jordan Raff, Pernille Rorth, and the Developmental Studies Hybridoma Bank for antibodies; J.P. Vincent, David Raible, and the DGRC, supported by National Institutes of Health grant 2P40OD010949-10A1, for plasmids; and Marcos Gonzalez-Gaitan, Pernille Rorth, J.P. Vincent, and the Bloomington Stock Center for flies. We also thank Cyrille Alexandre for providing technical advice on the generation of the Bsg25Dnull mutant; members of the Baylies Lab for helpful discussion; and Jennifer Zallen, Kari Lenhart, and Jodi Schottenfeld-Roames for critical reading of the manuscript.
This work was supported by Portuguese Science and Technology Foundation grant SFRH/BD/52041/2012 (to M. Azevedo), National Institutes of Health, National Institute of General Medical Sciences grant GM078318 and National Institute of Arthritis and Musculoskeletal and Skin Diseases grant AR108981 (to M.K. Baylies), and National Cancer Institute Core Grant P30 CA 008748 (to Memorial Sloan Kettering Cancer Center). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interests.
Author contributions: J.N. Rosen and M.K. Baylies designed the study. J.N. Rosen and M. Azevedo performed the experiments with the help of D.B. Soffar (Western analysis, fly stocks), V.P. Boyko (EB1 tracking experiments), M.B. Brendel (AFM microscopy), and V.K. Schulman (initial training in assessing nuclear position). J.N. Rosen, M. Azevedo, and M.K. Baylies analyzed the data. J.N. Rosen and M.K. Baylies wrote the paper, and all authors participated in generating the final version of the paper.
References
- Abal M., Piel M., Bouckson-Castaing V., Mogensen M., Sibarita J.B., and Bornens M.. 2002. Microtubule release from the centrosome in migrating cells. J. Cell Biol. 159:731–737. 10.1083/jcb.200207076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez L.A., Alexandre C., Mitchell A., Pasakarnis L., and Vincent J.-P.. 2013. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development. 140:4818–4825. 10.1242/dev.100933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw R.D., Callahan J.W., and Mahuran D.J.. 2006. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344:1186–1191. 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- Barlan K., Lu W., and Gelfand V.I.. 2013. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 23:317–322. 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum C., Graham A., Yousefzadeh M., Shrout J., Benjamin K., Krishna M., Hoda R., Hoda R., Cole D.J., Garrett-Mayer E., et al. 2008. The expression ratio of Map7/B2M is prognostic for survival in patients with stage II colon cancer. Int. J. Oncol. 33:579–584. 10.3892/ijo_00000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckson-Castaing V., Moudjou M., Ferguson D.J., Mucklow S., Belkaid Y., Milon G., and Crocker P.R.. 1996. Molecular Characterisation of Ninein, a New Coiled-Coil Protein of the Centrosome. J. Cell Sci. 109:179–190. [DOI] [PubMed] [Google Scholar]
- Brand A.H., and Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. 10.1101/lm.1331809 [DOI] [PubMed] [Google Scholar]
- Brendza K.M., Rose D.J., Gilbert S.P., and Saxton W.M.. 1999. Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J. Biol. Chem. 274:31506–31514. 10.1074/jbc.274.44.31506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova N.A., Grigoriev I., Yap A.S., Akhmanova A., and Brown N.H.. 2013. Dynamic microtubules produce an asymmetric E-cadherin-Bazooka complex to maintain segment boundaries. J. Cell Biol. 201:887–901. 10.1083/jcb.201211159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casenghi M., Barr F.A., and Nigg E.A.. 2005. Phosphorylation of Nlp by Plk1 negatively regulates its dynein-dynactin-dependent targeting to the centrosome. J. Cell Sci. 118:5101–5108. 10.1242/jcs.02622 [DOI] [PubMed] [Google Scholar]
- Chen C.T., Hehnly H., Yu Q., Farkas D., Zheng G., Redick S.D., Hung H.F., Samtani R., Jurczyk A., Akbarian S., et al. 2014. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr. Biol. 24:2327–2334. 10.1016/j.cub.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.J., Novakofski J., Jenkins W.K., and O’Brien W.D.. 1996. Young’s Modulus Measurements of Soft Tissues with Application to Elasticity Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 43:191–194. 10.1109/58.484478 [DOI] [Google Scholar]
- Cheng T.S., Chang L.K., Howng S.L., Lu P.J., Lee C.I., and Hong Y.R.. 2006. SUMO-1 modification of centrosomal protein hNinein promotes hNinein nuclear localization. Life Sci. 78:1114–1120. 10.1016/j.lfs.2005.06.021 [DOI] [PubMed] [Google Scholar]
- Collinsworth A.M., Zhang S., Kraus W.E., and Truskey G.A.. 2002. Apparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiation. Am. J. Physiol. Cell Physiol. 283:C1219–C1227. 10.1152/ajpcell.00502.2001 [DOI] [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber A., Lafranchi S.H., Maliga Z., Lui J.C., Moon J.E., McDeed C., Henke K., Zonana J., Kingman G.A., Pers T.H., et al. 2012. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. J. Clin. Endocrinol. Metab. 97:E2140–E2151. 10.1210/jc.2012-2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., and Bornens M.. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565–1575. 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Elhanany-Tamir H., Yu Y.V., Shnayder M., Jain A., Welte M., and Volk T.. 2012. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 198:833–846. 10.1083/jcb.201204102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espigat-Georger A., Dyachuk V., Chemin C., Emorine L., and Merdes A.. 2016. Nuclear alignment in myotubes requires centrosome proteins recruited by nesprin-1. J. Cell Sci. 129:4227–4237. 10.1242/jcs.191767 [DOI] [PubMed] [Google Scholar]
- Fernandes I., and Schock F.. 2014. The nebulin repeat protein Lasp regulates I-band architecture and filament spacing in myofibrils. J. Cell Biol. 206:559–572. 10.1083/jcb.201401094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., and Baylies M.K.. 2013. Nuclear positioning in muscle development and disease. Front. Physiol. 4:363 10.3389/fphys.2013.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., Schulman V.K., and Baylies M.K.. 2012. Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development. 139:3827–3837. 10.1242/dev.079178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., Schulman V.K., and Baylies M.K.. 2014. Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development. 141:355–366. 10.1242/dev.095612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Fu H., Zhou L., Xu K., Pang Y., Hu K., Wang J., Tian L., Liu Y., Wang J., et al. 2016. High expression of MAP7 predicts adverse prognosis in young patients with cytogenetically normal acute myeloid leukemia. Sci. Rep. 6:34546 10.1038/srep34546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud E., Caous R., Pascal A., Bazile F., Gagné J.P., Huet S., Poirier G.G., Chrétien D., Richard-Parpaillon L., and Giet R.. 2014. Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells. J. Cell Biol. 204:1111–1121. 10.1083/jcb.201311094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepner J., Li M., Ludmann S., Kortas C., Boylan K., Iyadurai S.J., McGrail M., and Hays T.S.. 1996. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 142:865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin C.M., and Kramer S.G.. 2009. RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension. Development. 136:1411–1421. 10.1242/dev.031823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G.G., and Worman J.H.. 2013. Nuclear positioning. Cell. 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M.S., Gisselbrecht S., Lu J., Estrada B., Keshishian H., and Michelson A.M.. 2002. New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis. 34:135–138. 10.1002/gene.10136 [DOI] [PubMed] [Google Scholar]
- Hammonds A.S., Bristow C.A., Fisher W.W., Weiszmann R., Wu S., Hartenstein V., Kellis M., Yu B., Frise E., and Celniker S.E.. 2013. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14:R140 10.1186/gb-2013-14-12-r140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Iampietro C., Bergalet J., Wang X., Cody N.A., Chin A., Lefebvre F.A., Douziech M., Krause H.M., and Lécuyer E.. 2014. Developmentally regulated elimination of damaged nuclei involves a Chk2-dependent mechanism of mRNA nuclear retention. Dev. Cell. 29:468–481. 10.1016/j.devcel.2014.03.025 [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30:1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J.P., Robison P., Shi G., Bogush A.I., Kempema A.M., Hexum J.K., Becerra N., Harki D.A., Martin S.S., Raiteri R., et al. 2015. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 6:8526 10.1038/ncomms9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanda M., Bergalet J., Wieczorek M., Brouhard G., Lécuyer É., and Lasko P.. 2016. Loss of function of the Drosophila Ninein-related centrosomal protein Bsg25D causes mitotic defects and impairs embryonic development. Biol. Open. 5:1040–1051. 10.1242/bio.019638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhart A., Windner S., Baylies M., and Mogilner A.. 2018. Mechanical positioning of multiple nuclei in muscle cells. PLOS Comput. Biol. 14:e1006208 10.1371/journal.pcbi.1006208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., and Raff J.W.. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S.D., and Chia W.. 2001. Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev. Cell. 1:691–703. 10.1016/S1534-5807(01)00075-2 [DOI] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E.S., Richardson B.E., Gomes E.R., and Baylies M.K.. 2012. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 484:120–124. 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen M., Malik M.A., Piel M., Bouckson-Castaing V., and Bornens M.. 2000. Microtubule Minus-End Anchorage at Centrosomal and Non-Centrosomal Sites: The Role of Ninein. J. Cell Sci. 113:3013–3023. [DOI] [PubMed] [Google Scholar]
- Moss D.K., Bellett G., Carter J.M., Liovic M., Keynton J., Prescott A.R., Lane E.B., and Mogensen M.M.. 2007. Ninein is released from the centrosome and moves bi-directionally along microtubules. J. Cell Sci. 120:3064–3074. 10.1242/jcs.010322 [DOI] [PubMed] [Google Scholar]
- Nishimura S., Nagai S., Katoh M., Yamashita H., Saeki Y., Okada J., Hisada T., Nagai R., and Sugiura S.. 2006. Microtubules modulate the stiffness of cardiomyocytes against shear stress. Circ. Res. 98:81–87. 10.1161/01.RES.0000197785.51819.e8 [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G., Elliott D.A., Harvey R.P., and Olson E.N.. 1998. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development. 125:3037–3048. [DOI] [PubMed] [Google Scholar]
- Richardson B.E., Beckett K., Nowak S.J., and Baylies M.K.. 2007. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 134:4357–4367. 10.1242/dev.010678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero N.B. 2010. Centronuclear myopathies: a widening concept. Neuromuscul. Disord. 20:223–228. 10.1016/j.nmd.2010.01.014 [DOI] [PubMed] [Google Scholar]
- Schulman V.K., Folker E.S., Rosen J.N., and Baylies M.K.. 2014. Syd/JIP3 and JNK signaling are required for myonuclear positioning and muscle function. PLoS Genet. 10:e1004880 10.1371/journal.pgen.1004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H., Sakayori N., Takahashi M., and Osumi N.. 2013. Ninein is essential for the maintenance of the cortical progenitor character by anchoring the centrosome to microtubules. Biol. Open. 2:739–749. 10.1242/bio.20135231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsa S., Parthasarathy S., Molnár Z., and Tarabykin V.. 2015. Sip1 downstream Effector ninein controls neocortical axonal growth, ipsilateral branching, and microtubule growth and stability. Neuron. 85:998–1012. 10.1016/j.neuron.2015.01.018 [DOI] [PubMed] [Google Scholar]
- Stillwell E.E., Zhou J., and Joshi H.C.. 2004. Human ninein is a centrosomal autoantigen recognized by CREST patient sera and plays a regulatory role in microtubule nucleation. Cell Cycle. 3:923–930. 10.4161/cc.3.7.947 [DOI] [PubMed] [Google Scholar]
- Sung H.H., Telley I.A., Papadaki P., Ephrussi A., Surrey T., and Rørth P.. 2008. Drosophila ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Dev. Cell. 15:866–876. 10.1016/j.devcel.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Tassin A.M., Maro B., and Bornens M.. 1985. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100:35–46. 10.1083/jcb.100.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S.E., Richards S., et al. 2002. Systematic Determination of Patterns of Gene Expression during Drosophila Embryogenesis. Genome Biol. 3:research0088.1 10.1186/gb-2002-3-12-research0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Berman B.P., Beaton A., Weiszmann R., Kwan E., Hartenstein V., Celniker S.E., and Rubin G.M.. 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8:R145 10.1186/gb-2007-8-7-r145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymanskyj S.R., Yang B., Falnikar A., Lepore A.C., and Ma L.. 2017. MAP7 Regulates Axon Collateral Branch Development in Dorsal Root Ganglion Neurons. J. Neurosci. 37:1648–1661. 10.1523/JNEUROSCI.3260-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wu D., Quintin S., Green R.A., Cheerambathur D.K., Ochoa S.D., Desai A., and Oegema K.. 2015. NOCA-1 functions with γ-tubulin and in parallel to Patronin to assemble non-centrosomal microtubule arrays in C. elegans. eLife. 4:e08649 10.7554/eLife.08649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Felder A., Liu Y., Guo L.T., Lange S., Dalton N.D., Gu Y., Peterson K.L., Mizisin A.P., Shelton G.D., et al. 2010. Nesprin 1 is critical for nuclear positioning and anchorage. Hum. Mol. Genet. 19:329–341. 10.1093/hmg/ddp499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Mennella V., Marks S., Wildonger J., Elnagdi E., Agard D., and Megraw T.L.. 2016. The Seckel syndrome and centrosomal protein Ninein localizes asymmetrically to stem cell centrosomes but is not required for normal development, behavior, or DNA damage response in Drosophila. Mol. Biol. Cell. 27:1740–1752. 10.1091/mbc.e15-09-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.