Abstract
The molecular characteristics of CNG (cyclic nucleotide-gated) channels in auditory/vestibular hair cells are largely unknown, unlike those of CNG mediating sensory transduction in vision and olfaction. In the present study we report the full-length sequence for three CNGA3 variants in a hair cell preparation from the trout saccule with high identity to CNGA3 in olfactory receptor neurons/cone photoreceptors. A custom antibody targeting the N-terminal sequence immunolocalized CNGA3 to the stereocilia and subcuticular plate region of saccular hair cells. The cytoplasmic C-terminus of CNGA3 was found by yeast two-hybrid analysis to bind the C-terminus of EMILIN1 (elastin microfibril interface-located protein 1) in both the vestibular hair cell model and rat organ of Corti. Specific binding between CNGA3 and EMILIN1 was confirmed with surface plasmon resonance analysis, predicting dependence on Ca2+ with Kd = 1.6× 10−6 M for trout hair cell proteins and Kd = 2.7× 10 −7 M for organ of Corti proteins at 68 μM Ca2+ . Pull-down assays indicated that the binding to organ of Corti CNGA3 was attributable to the EMILIN1 intracellular sequence that follows a predicted transmembrane domain in the C-terminus. Saccular hair cells also express the transcript for PDE6C (phosphodiesterase 6C), which in cone photoreceptors regulates the degradation of cGMP used to gate CNGA3 in phototransduction. Taken together, the evidence supports the existence in saccular hair cells of a molecular pathway linking CNGA3, its binding partner EMILIN1 (and β1 integrin) and cGMP-specific PDE6C, which is potentially replicated in cochlear outer hair cells, given stereociliary immunolocalizations of CNGA3, EMILIN1 and PDE6C.
Keywords: cyclic nucleotide-gated channel α3 (CNGA3), elastin microfibril interface-located protein 1 (EMILIN1), inner ear hair cell, phosphodiesterase 6C (PDE6C), surface plasmon resonance (SPR), pull-down assay
INTRODUCTION
CNG (cyclic nucleotide-gated) channels underlie sensory transduction in vision and olfaction [1,2] and additionally have a role in gustatory signal transduction [3,4]. In mammals, at least six genes encode CNG subunits [5]. Native CNG channels are tetrameric proteins [6] composed of at least two structurally related subunit types, α (CNGA1, CNGA2, CNGA3 and CNGA4) and β (CNGB1 and CNGB3). Each subunit contains six putative transmembrane domains, cytoplasmic N- and C-termini, a CNBD (cyclic nucleotide-binding domain) and a conserved pore region [1]. Although heterologous expression of α-subunits alone (except CNGA4) forms functional homomeric channels, the co-assembly of appropriate α- and β-subunits creates heteromeric CNG channels with properties that more closely resemble native CNG channels [7,8]. CNG channels are highly permeable to Ca2+ , and Ca2+ entry provides a negative feedback signal in its association with calmodulin that assists in adaptation of CNG channels in photoreceptors [9,10] and olfactory sensory neurons [10].
The existence of a cAMP-gated CNG channels in cochlear hair cells, moderately selective for monovalent cations and, unusually, specific in activation by cAMP and not cGMP, has been reported [11]. We determined that mRNA for the olfactory subunits CNGA2, CNGA4 and CNGB1b is expressed in cochlear hair cells/organ of Corti, with subunits for CNGA2 and CNGA4 immunolocalized to hair cell stereocilia [12]. The inclusion of CNGA4, in particular, would contribute to cAMP sensitivity. Additionally, evidence was obtained for transcript expression of the cGMP-sensitive CNGA3 mRNA in a subdissected organ of Corti fraction [13]. The nature of this CNGA3 subunit, whether identical to cone, olfactory or gustatory CNGA3 sequence, was not determined.
We now provide evidence that CNGA2 and three variants of CNGA3 are expressed in another hair cell model, a vestibular hair cell layer isolated from the trout saccule. PPI (protein–protein interaction) protocols demonstrate binding of one CNGA3 variant (CNGA3-3) to EMILIN1 (elastin microfibril interface-located protein 1) in the teleost hair cell, a finding that was replicated with CNGA3 protein expressed in rat organ of Corti and found to be attributable to the C-terminal sequence of EMILIN1, which is predicted to be intracellular. Preliminary reports on CNG function have been published [14–17].
EXPERIMENTAL
Isolation of a model hair cell preparation from the trout saccule
The model hair cell preparation from the trout saccule was isolated as described previously [18], corresponding to 1.5×106 saccular hair cells, which are free of intact supporting cells.
RT (reverse transcription)–PCR, primers and cloning
Isolated hair-cell layers were homogenized in 4 M guanidine thiocyanate containing 1% Sarkosyl. Total RNA was extracted with phenol/chloroform and isopropanol precipitation [19]. DNAse was added to remove residual genomic DNA, and the total RNA was immediately reverse-transcribed using a Superscript First-Strand cDNA Synthesis System (Invitrogen) and oligo-dT primers, random hexamer primers or SMART RACE (rapid amplification of cDNA ends) primers (SMART RACE cDNA amplification kit; BD Biosciences Clontech) to yield cDNA.
PCRs were carried out on trout saccular hair cell cDNA using Advantage 2 DNA polymerase (BD Biosciences) with denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 68 °C for 2 min with a total of 40 cycles. Negative controls were derived from the elimination of the reverse-transcription step or omission of cDNA with substitution of sterilized water (water blanks) in the PCR reaction. The PCR products were analysed with 1.5% agarose gel electrophoresis (Nusieve 3:1 agarose, Lonza).
Degenerate primers (see Supplementary Table S1 and Figure S1 at http://www.BiochemJ.org/bj/443/bj4430463add.htm) were designed that cross the nucleotide sequences for the olfactory CNG subunits expressed in catfish (GenBank® accession number M-83111) and zebrafish (GenBank® accession number XM-001337717). The PCR products were inserted into the pGEM-T Easy vector (Promega) and JM109 competent bacteria were transformed by heat shock (42 °C). A total of 60 clones of the 220-bp and 364-bp PCR products were sequenced, indicating expression of CNGA2 and CNGA3 transcripts in the saccular hair cell preparation.
CNGA3 sequence in saccular hair cells was subsequently extended with a series of degenerate oligonucleotide primers based upon CNGA3 expressed in Danio rerio and Oncorhynchus mykiss (trout) pineal photoreceptors, designed with OLIGO2 (National Biosciences) and Accelrys Gene 2.5 software (Supplementary Table S1 and Figure S1) The C-termini were obtained with 3′-RACE (SMART RACE, BD Biosciences), with the specific upstream primer 5′-GATGAGGAAATCGCTAATGCAG-3′ and 3′-RACE downstream primer 5′-AAGCAGTGGTATCAACGCAGAGT-3′. The products were cloned and sequenced, yielding 1941-bp, 1986-bp and 2136-bp CNGA3 variants. PDE6C (phosphodiesterase 6C) cDNA was amplified from trout saccular cDNA with degenerate primers (Supplementary Table S1) and the PCR products were sequenced.
Yeast two-hybrid analysis: trout saccular hair cell CNGA3-3
Bait construction
Yeast two-hybrid screening for the interacting proteins of trout hair cell CNGA3 variant 3 (CNGA3-3) was performed using the Matchmaker library construction and screening kit (Clontech). The trout CNGA3-3 sequence containing a C-terminal cytoplasmic loop (amino acids 632–712) was used as bait from PCR that was carried out on trout hair cell cDNA, with upstream primer 5′-GCCGAATTCGCTACGCTGATGAAGGACAAC-3′ and downstream primer 5′-TGCGGATCCTTACTTGTCGGCCACCACCTCAGA-3′ (restriction sites are underlined). The corresponding amplification product was gel-purified and restriction-digested with EcoRI and BamHI enzymes and ligated into a similarly digested pGBKT7 bait vector to produce a fusion construct with the GAL4 DNA-binding domain. The fusion product was confirmed to be transcriptionally inactive in the appropriate nutrient-deficient medium.
Prey library construction
A trout hair cell cDNA library was constructed from the isolated hair cell layer in the vector pGADT7-Rec using a homologous recombination technique in the host strain AH109 from equal proportions of oligo(dT) and random hexamers directing RT of hair cell mRNA (Matchmaker library construction and screening kit; Clontech) [20]. In brief, 5 μg of total RNA was reverse-transcribed in two reactions, one containing oligo(dT) and another containing random hexamer. An equal proportion of both of the cDNAs was used to construct the prey library.
Mating and screening
For screening of interacting proteins, cells containing bait constructs in strain Y187 were mixed with the cells containing prey constructs in AH109. These cells were allowed to mate in 2× YPDA (yeast peptone dextrose adenine) medium overnight at 30 °C. After incubation, the cells were plated on the selection medium SD/ – Ade/ – His/ – Leu/ – Trp. After 5–7 days, the larger colonies that appeared on the plates were screened for prey insert by PCR. The purified PCR products were sequenced and prey transcripts were identified using BLAST (National Center for Biotechnology Information).
Co-transformation
Co-transformation was performed to confirm the interaction of CNGA3-3 with EMILIN1. Approximately 500 ng of each vector (mixture of bait plasmid constructs and prey plasmid constructs) were employed to co-transform the freshly prepared AH109 competent yeast strain using the standard protocol (Clontech yeast two-hybrid system). The transformants were plated on quadruple dropout, nutrient-deficient, selection medium (SD/ – Ade/ – His/ – Leu/ – Trp) for 3–5 days. Transformants were also plated on a less stringent deficient medium (SD/ – Leu/ – Trp) and colonies picked from the SD/ – Leu/ – Trp medium were streaked on the higher stringency medium (SD/ – Ade/ – His/ – Leu/ – Trp). Negative controls included the combination of bait or prey with empty vector selected under the same conditions as above. To confirm further the binding interaction of trout CNGA3-3 with EMILIN1, we performed co-transformation with reversal of bait and prey, i.e. CNGA3-3 as prey and EMILIN1 as bait, with corresponding negative controls in the selection medium SD/ – Ade/ – His/ – Leu/ – Trp.
Yeast two-hybrid co-transformation analysis: rat organ of Corti CNGA3 and EMILIN1
The cochlear subdissected fractions used in RT–PCR have been morphologically documented previously [21]. The cytoplasmic C-terminus of rat CNGA3 was PCR amplified from rat brain cDNA using specific primers (upper primer, 5′-GCCCCATGGCAGATTCTGATGAAGGACAACCTAATCG-3′; lower primer, 5′-TGCGGATCCTTAGTCAGCTTTTGAAGCATCCTCGGAATTC-3′, nucleotides 1873–2133; GenBank® accession number AJ272428; underlining indicates restriction sites). This PCR product was sequence-verified and cloned into the bait vector pGBKT7 in a +1 frame. The C-terminus of EMILIN1 in rat organ of Corti was targeted in RT–PCR using specific primers (upstream primer, 5′-GCCGAATTCTATGACCCA-GAGACAGGTGTA-3′; downstream primer, 5′-TGCGGATCC-TTACTCGTAAAGCAGGGCTCCGCT-3′, nucleotides 3169-–3499; NM_001106710) and cloned into the prey vector pGADT7. For the yeast two-hybrid assay, AH109 yeast was co-transformed with ~ 500 ng of each vector using the standard protocol (Clontech) and the transformants were plated on nutrient-deficient selection medium (SD/ – Ade/ – His/ – Leu/ – Trp) for 3–5 days. Again, transformants were also plated on a less-stringent deficient medium (SD/ – Leu/ – Trp) and colonies were picked and streaked on higher stringency medium (SD/ – Ade/ – His/ – Leu/ – Trp). Negative controls consisted of either the bait or the prey combined with the opposite empty vector. To further confirm the binding interaction of rat CNGA3 with EMILIN1, we performed the reverse with rat CNGA3-C as prey and EMILIN1 as bait in selection medium SD/ – Ade/ – His/ – Leu/ – Trp. Negative controls comprised empty bait or prey vectors paired with counterpart vectors containing CNGA3-C or EMILIN1, respectively.
Expression and purification of hexahistidine fusion proteins
Trout hair cell CNGA3-3 and EMILIN1 cDNA as well as rat organ of Corti CNGA3 and EMILIN1 cDNA were prepared for insertion into pRSET vectors (Invitrogen). Restriction sites BamHI and EcoRI (underlined) were included in PCR primers for the C-termini of trout hair cell CNGA3-3 with upstream primer (5′-GCCGGATCCGCTACGCTGATGAAGGACAAC-3′) and downstream primer (5′-TGCGAATTCTTACTTGTCGGCC-ACCACCTCAGA-3′) (GenBank® accession number HQ542179) and trout EMILIN1 upstream primer (5′-GCCGGATCCTAT-GACCCGCGGACCGGCATC-3′) and downstream primer (5′-TGCGAATTCTTACATATGTTCATACAGCAGCAT-3′) applied to trout saccular hair cell cDNA. Likewise, the C-terminus of rat organ of Corti CNGA3 cDNA was amplified with upstream (5′-GCCGGATCCCAGATTCTGATGAAGGACAACCTAATCG-3′) and downstream (5′-TGCCCATGGTTAGTCAGCTTTTGAAGCATCCTCGGAATTC-3′) primers (GenBank® accession number AJ272428) (BamHI and NcoI restriction sites) and rat EMILIN1 cDNA amplified with upstream (5′-GCCGGATCCTATGACCCAGAGACAGGTGTA-3′) and downstream (5′-TGCGAATTCTTACTCGTAAAGCAGGGCTCCGCT-3′) primers (GenBank® accession number NM_001106710) containing the BamHI and EcoRI restriction sites. PCR products were digested with restriction enzymes and cloned into similarly digested pRSET bacterial expression vectors (Invitrogen). The plasmids were prepared in Escherichia coli (One Shot® TOP10F′ cells, Invitrogen). Selected clones were sequence-verified before expression studies.
Expression vector pRSETs containing the desired sequences were used to transform E. coli BL21 (DE3) cells. Clones were selected, and expression of fusion proteins was induced with 1 mM IPTG (isopropyl β-d-thiogalactopyranoside) in LB (Luria–Bertani) culture medium. The cultures were incubated for 4 h at 37 °C, and cells were harvested, lysed in 10 mM phosphate buffer with 8 M urea (pH 8.0), centrifuged at 20818 g for 25 min at 22°C and analysed using SDS/PAGE (4–12% gels), with protein bands visualized using Coomassie Blue staining. Fusion proteins were purified via Ni2+-nitrilotriacetic acid affinity spin columns (Qiagen) washed with 10 mM phosphate and 8 M urea, pH 6.3, and eluted with 10 mM phosphate and 8 M urea, pH 4.5. Urea was removed by dialysis against PBST containing 1× protease inhibitors (Sigma–Aldrich), and protein concentration was determined with the Qubit fluorescent system (Invitrogen).
SPR (surface plasmon resonance) analysis
Real-time measurement of the interaction at 22 °C between CNGA3 and EMILIN1 expressed in trout saccular hair cells and rat organ of Corti was studied by SPR as previously described [20,22] using a Biacore 3000 biosensor system (Biacore). With this technique, binding of the mobile molecule (analyte) to the immobilized molecule (ligand) can be detected by a change in the refractive index of the film. pRSET-A fusion proteins were made for the C-termini of CNGA3 (CNGA3-3 for trout) and EMILIN1. Fusion proteins were affinity-purified as described above. Affinity-purified EMILIN-1 fusion polypeptide (ligand) was immobilized on a CM5 research grade sensor chip by amine coupling [22]. Binding analysis was carried out by injecting the affinity-purified CNGA3 fusion peptide (analyte) into the flow cells (ligand and reference cell), and the interaction (response units, RU) between analyte and ligand was recorded. Kinetic sensorgram curves were evaluated with BIAevaluation 3.0 software [22]. Ca2+ levels in experimental solutions were determined with inductively coupled plasma MS [20].
Western blot analysis
Purity of the fusion proteins was ascertained by SDS/PAGE [4–12% NuPAGE gel followed by overnight blocking with 5% (w/v) non-fat dried skimmed milk powder at 4 °C] with Coomassie Blue staining. Blots were incubated with anti-Xpress monoclonal antibody (Invitrogen, 1:5000 dilution) for 3 h at room temperature (22 °C), washed with PBS containing 0.1% Tween 20, incubated with horseradish peroxidase-conjugated goat anti-(mouse IgG) secondary antibody (1:10 000 dilution) for 1 h at room temperature, and proteins were detected using Western Lightning chemiluminescence (PerkinElmer Life Sciences).
Pull-down assays
The C-terminus of trout hair cell CNGA3-3 was amplified with primers containing restriction sites as designated for pRSET. The upstream primer for rat CNGA3 was the same as for pRSET and the downstream primer was modified to include a NotI restriction site (underlined): 5′-TGCGCGGCCGCTTAGTCAGCTTTTGAAGCATCCTCGGAATTC-3′. The corresponding restriction-digested PCR products were ligated into a similarly restriction-digested pGEX-6P-1 vector with a GST (glutathione transferase) fusion tag (Amersham Biosciences) and used to transform E. coli BL21 (DE3) cells to express the fusion protein induced by IPTG. Bacterial cell lysates were either used directly in pull-down assays, or the fusion products were affinity-purified using glutathione–Sepharose beads (GE Healthcare). GST conjugates were mixed with hexahistidine-tagged trout or rat EMILIN1 in binding buffer (PBST buffer, pH 7.4,1 × protease inhibitor mixture) for 3 h at room temperature. Glutathione–Sepharose beads (30 μl of 50% slurry) were added to the reaction mixture for 1 h at room temperature, followed by centrifugation for 2 min at 1000 g. The beads were washed six times in the binding buffer, incubated at 70 °C for 5 min in gel-loading buffer (30 μl) and centrifuged. The supernatant was electrophoresed in a 4–12% NuPAGE gel (Invitrogen), electroblotted on to a nylon membrane, and EMILIN1 was immunodetected using an anti-Xpress antibody.
Pull-down assays were used to determine the PPIs between CNGA3–C-terminus and a ‘before transmembrane’ (a) domain, amino acids 784–849 (rat EMILIN1, GenBank® accession number NP_001100180.1), a ‘transmembrane domain’ (b), amino acids 846–876 and an ‘after transmembrane domain’ (c), amino acids 874–900 (see Figure 6G). The three portions of EMILIN1 were amplified with primers containing restriction sites (BamHI and EcoRI; underlined) as designated for GST. Primers used for PCR (upstream and downstream) were: EMLIN1 (a), 5′-GCCGGATCCTATGACCCAGAGACAGGT-3′, 5′-TGCGAATTCTTACGGGCTAGGCTGACTTTC-3′; EMILIN1 (b)5′-GccGGATCCCCTAGCCCGGGCGCTCTG-3′, 5′-TGCGAATTCTTACAGCTGCCCCATGACCAG-3′, and EMILIN1 (c) 5′-GCCGGATCCGGGCAGCTGGCACACTCA-3′, 5′-TGCGAATTCTTACACCTGTTCAAGGTCTGT-3′.
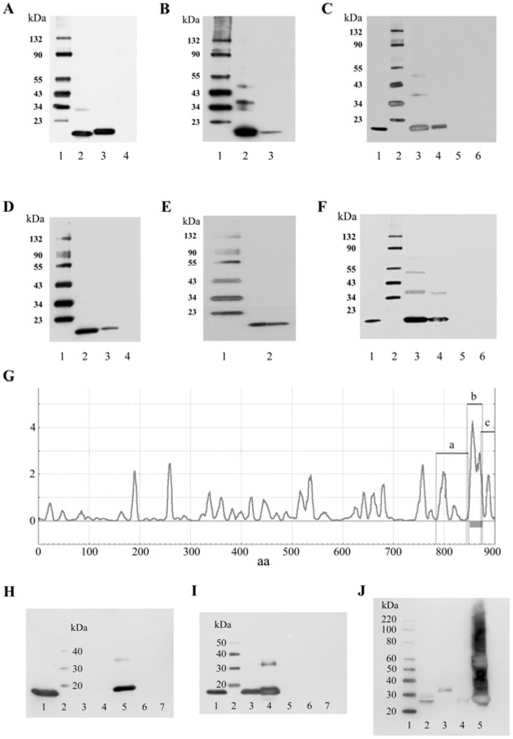
Figure 6. GST pull-down assays of binding between CNGA3 and EMILIN1 for trout saccular hair cell and rat organ of Corti proteins.
(A) Western blot of trout hair cell CNGA3-3 (TA3-C) and EMILIN1 (TEM) C-terminus fusion proteins expressed in pRSET-A and detected with anti-Xpress monoclonal antibody (Invitrogen). Lane 1, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 2, affinity-purified TA3-C fusion protein at 13 kDa. Lane 3, affinity-purified TEM fusion protein at 16 kDa. Lane 4, pRSET-A bacterial lysate as negative control. (B) Western blot of rat EMILIN1 (REM) and trout EMILIN1 detected with an anti-EMILIN1 mouse monoclonal antibody (Abnova) which has cross-reactivity for rat and trout sequence. Lane 1, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 2, affinity-purified REM fusion protein at 16 kDa. Lane 3, affinity-purified TEM fusion protein at 16 kDa. (C) GST pull-down assay of binding between TA3-C and TEM-like protein. Lane 1, purified TEM fusion product used in the pull-down assay, detected with anti-Xpress monoclonal antibody. Lane 2, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 3, binding of TEM fusion protein (~300 ng) with GST–TA3-C cleared lysate, pulled down with glutathione–Sepharose, and detected with anti-Xpress antibody. Lane 4, binding of TEM fusion protein (~100 ng) with GST–TA3-C lysate. Lane 5, TEM (~300 ng) incubated with GST bacterial lysate as a negative control. Lane 6, TEM fusion protein mixed with Sepharose beads under the same conditions as above, representing another negative control. (D) Western blot of the C-termini of rat CNGA3 (RA3-C) and EMILIN1 (REM) fusion proteins in pRSET-A detected with anti-Xpress antibody. Lane 1, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 2, affinity-purified rat organ of Corti RA3-C fusion protein at 14 kDa. Lane 3, affinity-purified REM fusion protein at 16 kDa. Lane 4, pRSET-A bacterial lysate as negative control. (E) Western blot of CNGA3 C-terminus fusion protein (RA3-C) in pRSET-A. Lane 1, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 2, affinity-purified rat organ of Corti CNGA3 (RA3-C) fusion protein at 14 kDa, detected with anti-CNGA3 rabbit polyclonal antibody (Alomone). (F) GST pull-down assay of binding between RA3-C and REM. Lane 1, purified REM fusion protein used in the pull-down assay, detected with anti-Xpress antibody. Lane 2, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 3, binding of REM fusion protein (~300 ng) with GST–RA3-C cleared lysate, pulled down with glutathione–Sepharose, and detected with anti-Xpress antibody. Lane 4, binding of REM fusion protein (~100 ng) with GST–RA3-C lysate. Lane 5, REM (~300 ng) incubated with GST bacterial lysate as a negative control. Lane 6, REM fusion protein mixed with Sepharose beads under the same conditions as above, representing another negative control. (G) Transmembrane helix preference for rat EMILIN1 (GenBank® accession number NP_001100180.1) with predicted transmembrane domain b (amino acids 846–876), before-transmembrane segment a (amino acids 784–849) and after-transmembrane segment c (amino acids 873–900). (H) Pull-down assay for segments of the C-terminus of EMILIN1 in GST and CNGA3. Lane 1, CNGA3 C-terminus detected with anti-Xpress, 14 kDa. Lane 2, protein standards (LC5603, Invitrogen) with molecular masses indicated to the left-hand side in kDa. Lane 3, region before-transmembrane domain (a) plus CNGA3 detected with anti-Xpress. Lane 4, transmembrane domain (b) plus CNGA3. Lane 5, region after-transmembrane domain (c) plus CNGA3. Lane 6, CNGA3 plus GST lysate negative control. Lane 7, CNGA3 plus GST beads, negative control. (I) Lane 1, CNGA3 C-terminus detected with anti-Xpress, 14 kDa. Lane 2, protein standards with molecular masses indicated to the left-hand side in kDa, Invitrogen. Lane 3, pull-down of 100 ng purified CNGA3 with EMILIN1 c. Lane 4, pull-down of 200 ng purified CNGA3 with EMILIN1 c. Lane 5, same as lane 4, but with no beads. Lane 6, 200 ng purified CNGA3 plus GST lysate, i.e. negative control. Lane 7, 200 ng purified CNGA3 plus GST beads, negative control. (J) Western blot for EMILIN1 C-terminal segments detected with anti-GST. Lane 1, protein standards with molecular masses indicated to the left-hand side in kDa. Lane 2, 50 ng of purified EMILIN1 c. Lane 3, 50 ng of purified EMILIN1 a. Lane 4, 50 ng of purified EMILIN1 b. Lane 5, GST lysate (contains the GST vector).
Immunohistochemical analyses
A custom antibody was designed to target the trout CNGA3 N-terminus sequence that is conserved in the cone/olfactory CNGA3 sequence (see Figure 3, trout hair cell CNGA3-1–3, amino acids 22–40). The peptide CKKKTPNDNLDSIENGTPRSNSE was synthesized and used as a trout hair cell-specific CNGA3 antigen to raise a polyclonal chick IgY antibody (Covance). Immunolocalization was carried out according to previously published procedures [23]. In brief, the intact trout saccule was fixed in 4% (w/v) paraformaldehyde and 0.1% glutaraldehyde in phosphate buffer at 4 °C for 2–3 h, washed in PBS, taken through a graded ethanol series into xylene and paraffin-embedded. Sections (4 μm) were used for immunolocalization of CNGA3 with primary antibody at 1:100 000 dilution, biotinylated goat anti-chick IgY-B secondary antibody (SC-2430, Santa Cruz Biotechnology), and DAB (diaminobenzidine) as chromogen (BioGenex).
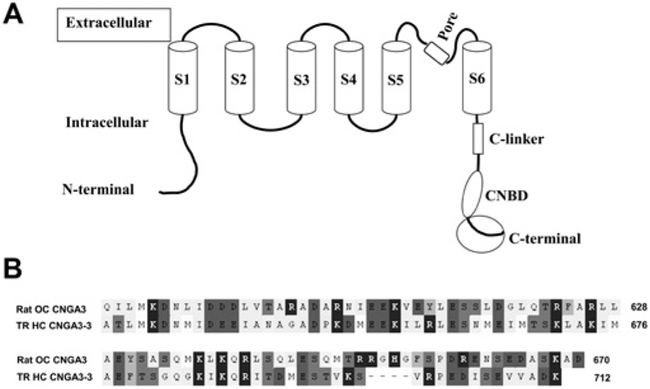
Figure 3. CNGA3 C-termini in rat and trout.
(A) Schematic depiction of CNGA3 with S1–S6 transmembrane regions, connecting intracellular and extracellular sequence, the C-linker, CNBD and the cytoplasmic C-terminal sequence (encircled). S4 represents the voltage sensor. (B) Alignment (with OMIGA 2.0) of the cytoplasmic C-terminal sequences of rat organ of Corti CNGA3 (amino acid numbering according to GenBank® accession number AJ272428) and trout saccular hair cell CNGA3-3 (HQ542179) used as bait in yeast two-hybrid protocols and in pull-down and SPR analyses.
CNGA3 in the rat organ of Corti was immunolocalized with previously described protocols [12] using a rabbit polyclonal anti-rat CNGA3 primary antibody (amino acids 594–611, # APC-060, Alomone) at 1:75 dilution, and a chick anti-rabbit IgG-B secondary antibody, mouse/human adsorbed (sc-2986, Santa Cruz Biotechnology), detected with DAB. EMILIN1 was immunolocalized in rat organ of Corti with a mouse monoclonal antibody (H00011117-M01, clone 4A3, Abnova; 1:1000 dilution) targeting full-length human EMILIN1 with cross-reactivity to rat EMILIN1 (GenBank® accession number EDM02967) with 86% amino acid identity. The secondary antibody was biotinylated affinity-purified horse anti-mouse IgG, rat-adsorbed (Vector). Immunofluorescence was detected with chick anti-mouse-Alexa Fluor® 488 secondary antibody (A21200, Molecular Probes). PDE6C was immunolocalized in rat organ of Corti with mouse polyclonal antibody (H00005146-A01, Abnova; 1:1000 dilution) targeting the human cone PDE6C sequence EINQVAVEKYLEENPQFAKEYFDRKLRVEVLGEIFKNSQVPVQSSMSFSELTQVEESALCLELLWTVQEEGGTPEQGVHRALQRLAHLLQADRCSMFL, which is 69% identical, 78% positive to rat cone PDE6C sequence. The secondary antibody was either horse anti-mouse IgG for DAB (Santa Cruz Biotechnology) or chick anti-mouse-Alexa Fluor® 488 antibody for fluorescence (Molecular Probes). Light images were obtained with a Leitz Diaplan microscope, photographed with an Olympus OM-4T camera, and the negatives were digitized. Immunofluorescence was examined with an Olympus BX60 microscope with a fluorescein (470–490 nm) excitation filter (500–800 nm emission), photographed with a PM 20 camera system, and the negatives were digitized. For negative controls, the primary antibodies were either pre-absorbed with their respective antigens or omitted.
Confocal microscopy
B629SF2/J wild-type mice (5–6 weeks old) were anaesthetized with 55% urethane, 0.9 g/kg of body weight, administered intraperitoneally, transcardially perfused with fixative [4% (w/v) paraformaldehyde and 0.1% glutaraldehyde] and decapitated. Experimental procedures reported in the present study were performed according to the guidelines issued by the National Institutes of Health, and approved by the Animal Investigation Committee of Wayne State University. All efforts were made to minimize both the suffering of animals and the number of animals used. The temporal bones were rapidly harvested, and the cochleas were isolated and perfused with fixative [4% (w/v) paraformaldehyde and 0.1% glutaraldehyde] at 4 °C via the round window, exiting via a small hole at the helicotrema. The labyrinthine tissues were isolated, washed with PBS three times for 10 min, pretreated in 0.3% Triton X-100 (T-8787, Sigma) in PBS for 5–10 min, incubated in 0.1% sodium borohydride for 30 min, blocked in normal sera (corresponding to the species used to raise the secondary antibodies) for 30 min at room temperature and incubated with primary antibody at 4 °C overnight, followed by incubation in secondary antibody at room temperature for 30 min. The primary antibody was rabbit polyclonal anti-rat CNGA3 amino acids 594–611 (APC-060; Alomone), 1:75 dilution. The secondary antibody was chick anti-rabbit-Alexa Fluor® 488 (Molecular Probes), 1:2000 dilution. F-actin (filamentous actin) was detected with rhodamine-phalloidin (Molecular Probes), 1:100 dilution. Z-stack 1 μm images of cochlea surface preparations were captured with a Zeiss Meta LSM 510 microscope with 543 nm excitation and 565–615 nm emission (red) and 488 nm excitation and 500–550 nm emission (green).
RESULTS
CNG subtypes CNGA2 and CNGA3 are expressed in the trout hair cell layer
Degenerate primers that have cross-reactivity for the nucleotide sequence of olfactory CNG subunits were applied to trout saccular hair cell cDNA and the products were cloned. The first set of primers (CNGA2/3-1, Supplementary Table S1) applied in PCR targeted the CNGA2 and CNGA3 sequence (deletion of 220 bp, Supplementary Figure S2 at http://www.BiochemJ.org/bj/443/bj4430463add.htm). The second set of primers (CNGA2/3-2), which would elicit amplification of either CNGA2 or CNGA3 (Supplementary Table S1), directed amplification of PCR products with a deletion of 364 bp (Supplementary Figure S2). Analysis of 60 clones of the 220-bp and 364-bp PCR products indicated expression of CNGA2 and three variants of CNGA3 in the saccular hair cell preparation (Figure 1, line 7, amino acids 1–120 and line 3, amino acids 429–548, respectively) with α subunit distribution (relative cloning frequency) for CNGA2/CNGA3-1/CNGA3-2/CNGA3-3 of 2:8:4:4.
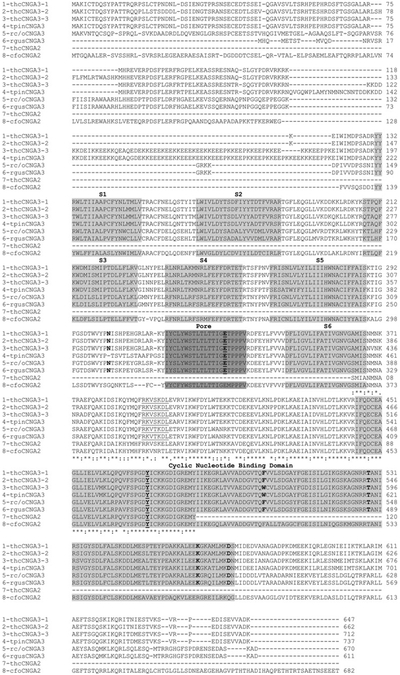
Figure 1. Clustal sequence alignment of the three trout saccular hair cell CNGA3 subtypes.
Line 1, trout hair cell (thc) CNGA3-1 (GenBank® accession number HQ542177). Line 2, trout hair cell CNGA3-2 (GenBank® accession number HQ542178). Line 3, trout hair cell CNGA3-3 (GenBank® accession number HQ542179). Line 4, trout pineal photoreceptor (tpin) CNGA3 (GenBank® accession number AF393839). Line 5, rat cone/olfactory (rc/o) CNGA3 (GenBank® accession number EDL992404 for olfactory protein sequence). Line 6, rat gustatory (rgus) CNGA3 (GenBank® accession number NM_053495). Line 7, trout hair cell CNGA2. Line 8, catfish olfactory (cfo) CNGA2 (GenBank® accession number P55934 for protein sequence). The six transmembrane domains, S1–S6 (light grey), the pore region between S5 and S6 (dark grey) and the CNBD (light grey) are highlighted. The Clustal designations for identity and homology across CNGA3 and CNGA2 sequences are indicated starting at residue 429 and finishing at residue 547 (CNGA3-3). Also in bold and/or underlined are amino acid residues believed to have specific functions in higher vertebrates and which are conserved in the trout hair cell CNGA3 sequence, with numbering according to that of CNGA3-3: N-glycosylation site, Asn367 putatively affecting pore function; predicted PKA site, RKVSKDL in C-linker, amino acids 455–461; Cys513, affecting coupling of cGMP to the pore; Tyr536 within the CNBD, a putative phosphorylation site conserved across CNG subunits in higher vertebrates; Trp566, Thr593, Lys629 and Asp637 within the CNBD represent amino acids thought to be important in specifying cGMP sensitivity in CNGA1 and CNGA3 channels.
Three full-length CNGA3 variants with cone/olfactory CNGA3 sequence signatures
Degenerate primers and their relative positions utilized to obtained full-length sequence for three variants of CNGA3 ion channel from trout saccular hair cell layer are indicated in Supplementary Table S1 and Figure S1, with respective PCR products on agarose gels illustrated in Supplementary Figure S2. The overlapping PCR products were cloned, yielding sequence for three variants of CNGA3 expressed in the hair cell layer of the trout saccule, CNGA3-1, CNGA3-2 and CNGA3-3, 1946-bp, 1986-bp and 2186-bp in length respectively (GenBank® accession numbers HQ542177, HQ542178 and HQ542179). The deduced amino acid sequences for trout saccular hair cell CNGA3-1 (647 amino acids), CNGA3-2 (662 amino acids) and CNGA3-3 (712 amino acids) were aligned and compared with CNGA3 sequence from trout pineal gland, rat olfactory/cone photoreceptor and rat taste buds (CNGgust), and further compared with trout hair cell CNGA2 and catfish CNGA2 (Figure 1). Overall, CNGA3-1, CNGA3-2 and CNGA3-3 exhibited 86 %, 82% and 95% amino acid identity respectively, to trout pineal photoreceptor CNGA3, 67 %, 68% and 58% identity to rat olfactory CNGA3, and 73 %, 89% and 62% identity to rat gustatory CNGA3.
Evidence of expression of the protein corresponding to CNGA3 sequence has been obtained in trout saccular hair cells. A custom antibody was designed to target the N-terminal sequence (amino acids 22–40) in trout saccular hair cell CNGA3-1 and CNGA3-3 (100% identity for both variants) and CNGA3-2 (68% identity) that was 100% identical with the sequence reported for a trout cGMP-gated channel (GenBank® accession number AF-393839), termed trout pineal CNGA3 (Figure 1). This N-terminal region displays identity (42%) to the CNGA3 sequence found in the olfactory/cone photoreceptor CNGA3 sequence of higher vertebrates, which is not replicated in gustatory CNGA3 (Figure 1). CNGA3 with this custom antibody was immunolocalized to the subcuticular plate region and stereociliary array of saccular hair cells in the trout saccule (Figure 2A), with negative controls devoid of immunoreactivity (Figure 2B).
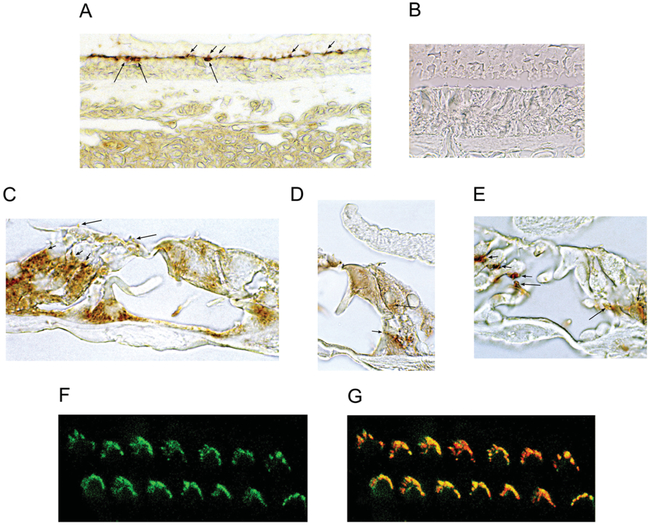
Figure 2. Immunoreactivity for CNGA3 in the trout saccule and rat and mouse organ of Corti.
(A) A custom antibody (Covance) was developed in chick against an N-terminal epitope of trout saccular hair cell CNGA3. This primary antibody was used at 1:100 000 dilution, coupled with a biotinylated goat anti-chick IgY secondary antibody, and detected with DAB. CNGA3 immunoreactivity was observed in saccular hair cell stereociliary arrays (short arrows) and in corresponding subcuticular plate sites (long arrows). (B) Negative control showing hair cell sensory epithelium in trout saccule with no primary antibody present, but secondary antibody included. (C) In the organ of Corti of the adult rat, immunoreactivity for CNGA3 was detected in efferent endings at the base of outer hair cells (short arrows) and in outer hair stereocilia (long arrows). (D) Beneath the inner hair cell of the ratorgan of Corti, CNGA3 immunoreactivity was observed in efferent nerve fibres above the habenula perforata crossing the inner pillar cell to enter the tunnel of Corti (short arrows) before crossing through the tunnel to end on outer hair cells and on Deiters’ cells. CNGA3 immunoreactivity was also detected at the base of the inner hair cell (long arrow). (E) In mouse organ of Corti, CNGA3 immunoreactivity was found in efferent tunnel crossing fibres (long arrows) and in efferent nerve endings at the base of outer hair cells (short arrows). (F) In Z-stack confocal microscopy, CNGA3 immunofluorescence (green) was associated with outer hair cell stereociliary arrays and in (G) overlapping phalloidin (red) labelling of F-actin in a 1 μm optical section (overlap in yellow) of mouse cochlea.
Trout saccular hair cell CNGA3-3 binds to EMILIN1 in yeast two-hybrid analysis
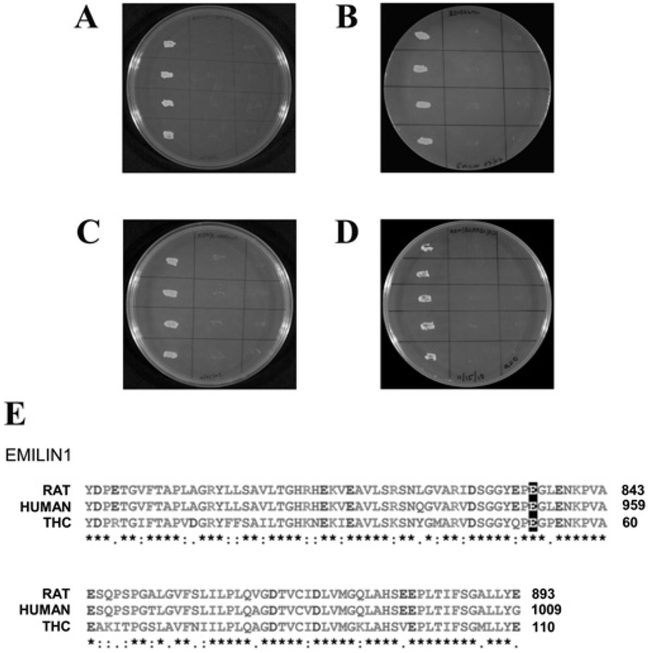
Yeast two-hybrid mating protocols were used to identify protein-binding partners for the C-terminus of trout saccular hair cell CNGA3-3 (amino acids 632–712) (Figure 1, line 3 and Figure 3). The corresponding nucleotide sequence was inserted into pGBKT7 (Clontech) in bait construction and tested in mating protocols with a trout saccular hair-cell cDNA library in prey construct pGADKT7-Rec [20]. The transformation efficiency, calculated on the basis of the number of colonies which appeared on the SD/ – Leu plates after 5 days, was 8.4× 105 per 3 μg of vector. Overall, 127 prey clones were sequenced and EMILIN1 was identified as a protein-binding partner for CNGA3-3 C-terminus bait in quadruple drop-out media. Binding was confirmed in yeast two-hybrid co-transformation protocols (Figure 4A), switching bait and prey (Figure 4B). The amino acid sequence ascertained as binding to CNGA3-3 was the C-terminus of EMILIN1, with 75% and 76% identity with EMILIN1 of rat and human respectively (Figure 4E) and 81% identity with Danio rerio EMILIN1 (GenBank® accession number NP_001025378, not shown).
Figure 4. Interaction of trout hair cell CNGA3-3 C-terminus with EMILIN1.
(A) First lane, binding of trout CNGA3-3 as bait to EMILIN1-like protein as prey, determined on quadruple drop-out medium with yeast two-hybrid co-transformation protocols. Second lane, negative control with CNGA3-3 as bait and empty vector as prey. Third lane, negative control with empty vector as bait and EMILIN1 as prey. (B) Reversal of bait and prey. First lane, EMILIN1 as bait and CNGA3–3 as prey. Second lane, negative control with EMILIN1 as bait and empty vector as prey. Third lane, negative control with empty vector as bait and CNGA3–3 as prey. (C) The C-terminus of rat CNGA3 as bait and rat EMILIN1 as prey in selection medium SD/–Trp/–Leu/–His/–Ade. First lane, interaction between bait and prey fusion peptides. Second lane, negative control with rat CNGA3 as bait and empty vector as prey. Third lane, negative control with empty vector as bait and EMILIN1 as prey. (D) Reversal of bait and prey. First lane, rat EMILIN1 as bait and CNGA3-C as prey. Second lane, negative control with EMILIN1 as bait and empty vector as prey. Third lane, negative control, with empty vector as bait and CNGA3-C as prey. (E) Clustal alignment of EMILIN1 in rat, human and trout saccular hair cells (THC). Alignment illustrating that trout EMILIN1 prey sequence is 74% identical with rat and human EMILIN1 (GenBank® accession numbers NP_001100180 and NP_008977 respectively). The highlighted glutamate residues (E) correspond to Glu993 in human EMILIN1, required for binding of EMILIN1 to integrin α4β1.
CNGA3 expressed in rat organ of Corti binds to EMILIN1 in yeast two-hybrid co-transformation analysis
Given that CNGA3 C-terminus mRNA had been previously identified in a rat microdissected organ of Corti subfraction [13], we explored the possibility that the CNGA3 C-terminus would also bind the C-terminus of EMILIN1 expressed in rat organ of Corti. The C-terminus of rat organ of Corti CNGA3, identical with that for rat cone/olfactory/gustatory sequence (Figure 1, line 5, amino acids 584–670), was found as bait in yeast two-hybrid co-transformation protocols to bind rat organ of Corti EMILIN1 as prey (Figure 4C) in high stringency selection media. Binding also occurred for the reverse, with CNGA3 as prey and EMILIN1 (Figure 4D) as bait. No growth was found in negative controls employing empty vectors in quadruple drop-out medium (Figures 4C and 4D).
SPR analysis of PPIs between trout saccular hair cell CNGA3-3 and EMILIN1
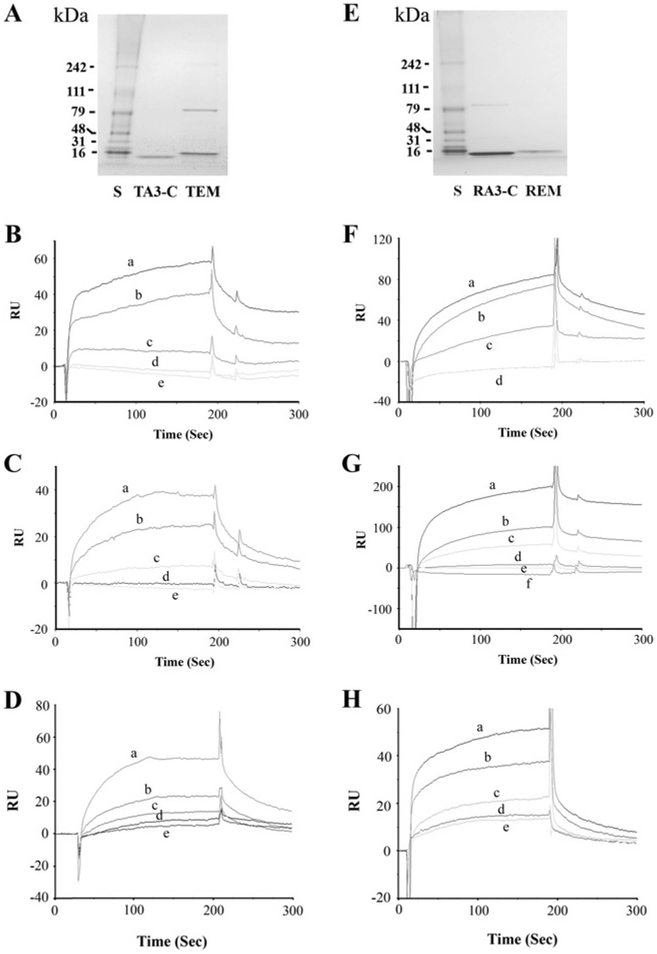
The affinity-purified C-termini of trout hair cell CNGA3-3 (TA3-C) and the affinity-purified C-terminus of trout EMILIN1 (TEM) (Figure 5A) were found to bind in the presence of Ca2+ (Figures 5B and 5C). The specific binding of trout hair cell CNGA3-3 (ligand) to the C-terminus of EMILIN1 (analyte) was enhanced by Ca2+ ; elevated at 26.5 μM Ca2+ relative to binding with 1 mM EGTA, with further increases in binding observed up to 68 μM Ca2+ (Figure 5B). No binding was detected for negative controls HBS-N plus 1 mM EGTA or HBS-N alone. In separate experiments, we obtained similar results when the ligand and analyte were reversed (EMILIN1 as ligand and CNGA3-3 as analyte). Binding was again indicated between EMILIN1 and CNGA3-C at 26.5 μM Ca2+, which was further promoted by increases in Ca2+ up to 68 μM (Figure 5C). Binding for EMILIN1 as ligand and CNGA3-3 (0–160 nM) as analyte, in a kinetic series at 68 μM Ca2+ (Figure 5D), yielded a Kd of 1.6×10−6 M. The results were normalized by subtracting the SPR response (RU) for buffer alone and averaged from a single experiment performed in triplicate. All of the experiments were performed multiple times, and samples for different Ca2+ concentrations were analysed randomly. There was no binding to the reference cell surface, which was blocked by ethanolamine. Thus these results indicate calcium-dependent binding of the C-terminus of trout saccular hair cell CNGA3-3 to the C-terminus of EMILIN1.
Figure 5. SPR analysis of binding interaction between CNGA3 and EMILIN1.
(A) Coomassie Blue detection of affinity-purified fusion proteins. S, pre-stained protein standards with molecular masses indicated to the left (kDa) on an SDS/PAGE gel (Bench-Mark, Invitrogen). TA3-C, affinity-purified C-terminus of trout hair cell CNGA3-3 fusion protein at 13 kDa. TEM, C-terminus of trout EMILIN1 fusion protein at 16 kDa. (B) Binding of TA3-C as ligand to TEM as analyte (100 nM), enhanced by Ca2+ . Response units (RU) are indicated for binding at 68 μMCa2+ (a), 26.5 μM Ca2+ (b), and 1 mM EGTA (c), compared with the response for 10 mM Hepes, pH 7.4, and 150 mM NaCl (HBS-N) plus 1mM EGTA (d) or HBS-N buffer alone (e). (C) Binding of TEM as ligand to TA3-C as analyte (100 nM) increases with elevation of Ca2+ . Response units are indicated for binding at 68 μM Ca2+ (a), 26.5 μM Ca2+ (b) and 1 mM EGTA (c), compared with the response for HBS-N plus 1mM EGTA (d) or HBS-N buffer alone (e). (D) Kinetics of binding between TEM as ligand and TA3-C as analyte, at 68 μM Ca2+ and 10 (e), 20 (d), 40 (c), 80 (b) or 160 (a) nM analyte. Kd = 1.6×10 −6 M. (E) Affinity-purified rat fusion proteins stained with Coomassie Blue. S, pre-stained protein standards with molecular masses indicated to the left (kDa) on an SDS/PAGE gel. RA3-C, affinity purified C-terminus of CNGA3 fusion protein at 14 kDa. REM, C-terminus of EMILIN1 fusion protein at 16 kDa. (F) Ca2+ -dependence of the interaction of REM as ligand and RA3-C as analyte (100 nM). Response units are indicated for binding at 68 μM Ca2+ (a), 26.5 μM Ca2+ (b) and 1 mM EGTA (c), compared with the response for HBS-N plus 1 mM EGTA (d) or overlapping plot for HBS-N buffer alone (d). (G) Ca2+ -dependence of the response for the reverse interaction of rat RA3-C as ligand with REM as analyte (100 nM). Response units (RU) are indicated for binding with REM at 166 μM Ca2+ (a), 68 μM Ca2+ (b), 26.5 μM Ca2+ (c) and 1 mM EGTA (d), compared with the response for HBS-N plus 1 mM EGTA (e) or HBS-N buffer alone (f). (H) Kinetics of binding between REM as ligand and RA3-C as analyte, at 68 μM Ca2+ and 10 (e), 20 (d), 40 (c), 80 (b) and 160 (a) nM analyte. Kd = 2.7×10−7 M.
Rat CNGA3 directly binds EMILIN1 by SPR analysis
The affinity-purified C-termini of CNGA3 and EMILIN1 (RA3-C and REM, Figure 5E) were found to bind in the presence of Ca2+ by SPR analysis. Binding increased for rat organ of Corti EMILIN1 as ligand and CNGA3 as analyte as Ca2+ was elevated (68 μM Ca2+ compared with 26.5 μM Ca2+, Figure 5F). No response was observed in the absence of Ca2+ (plus EGTA) for HBS-N. SPR showed the similar results when the ligand and analyte were reversed (rat CNGA3 as ligand and EMILIN1 as analyte) with binding increasing with elevation of Ca2+ up to 166 μM (Figure 5G). A series of kinetic experiment responses for rat EMILIN1 as ligand and CNGA3 (0–160 nM) as analyte in the presence of 68 μM Ca2+ (Figure 5H) yielded a Kd interaction value of 2.7 × 10 −7 M.
GST pull-down assay of binding between trout CNGA3-3 and EMILIN1
The PPI between trout saccular hair cell CNGA3-3 and EMILIN1 observed with yeast two-hybrid analysis was also confirmed by pull-down assay. Western blots indicated the predicted molecular masses for trout hair cell CNGA3 and EMILIN1 C-terminus purified fusion proteins of ~ 13 and ~ 16 kDa (TA3-C and TEM) respectively, detected with an anti-Xpress antibody (Figure 6A). A gene-specific mouse monoclonal antibody (Abnova) which targeted the EMILIN1 sequence of both rat and fish also labelled the EMILIN1 fusion protein (Figure 6B). Binding between trout hair cell EMILIN1 and trout hair cell CNGA3-3 was demonstrated by pull-down assay (Figure 6C) for 300 ng and 100 ng of EMILIN1 (TEM). Negative controls included trout EMILIN1 incubated with either GST–bacterial lysate or with Sepharose beads under the same conditions as for the experimental samples.
GST pull-down assay of binding between rat CNGA3 and EMILIN1
Binding between rat CNGA3 and EMILIN1 observed with yeast two-hybrid analysis was further confirmed by pull-down assay. Western blots indicated the predicted molecular masses for rat organ of Corti CNGA3 and EMILIN1 purified fusion proteins of ~ 14 and ~ 16 kDa (RA3-C and REM) respectively, detected with either anti-Xpress antibody (Figure 6D) or the gene-specific antibodies anti-CNGA3 rabbit polyclonal antibody (Alomone) (Figure 6E) and anti-EMILIN1 mouse monoclonal antibody (Abnova) (Figure 6B). Binding between rat CNGA3 and EMILIN1 was demonstrated by pull-down assay for 300 ng and 100 ng of EMILIN1 fusion protein (REM, Figure 6F). Negative controls included rat EMILIN1 incubated with either GST–bacterial lysate or unconjugated Sepharose beads under similar conditions as for the experimental samples (Figure 6F).
To define further the molecular requirements for PPIs between rat EMILIN1 and CNGA3, we considered the possibility that EMILIN1 might possess an intracellular carboxy domain. Transmembrane domain analysis by secondary structure software (http://split.pmfst.hr/split/4/) and the DAS transmembrane prediction server (www.sbc.su.se/~miklos/DAS/) indicated a transmembrane domain for rat EMILIN1 (Genbank® accession number NP_001100180, amino acids 846–876) (Figure 6G) and for mouse EMILIN1 (GenBank® accession number NP_598679, amino acids 964–993) (not shown). We divided the rat EMILIN1 C-terminal sequence into three portions in accordance with the software predictions: before-transmembrane (a), amino acids 784–849; transmembrane (b), amino acids 846–876; and after-transmembrane (c), amino acids 873–900 (Figure 6G). In a direct comparison in pull-down assays, the CNGA3 C-terminus was found to bind to the after-transmembrane segment (Figure 6H, lane 5), but not to bind to either the before-transmembrane (Figure 6H, lane 3) or the transmembrane segments (Figure 6H, lane 4), consistent with intracellular–intracellular peptide binding between CNGA3 and EMILIN1.
Immunolocalization of CNGA3 and EMILIN1 in mammalian organ of Corti
In an overview of the organ of Corti of the adult rat and mouse, CNGA3 immunoreactivity was associated with efferent nerve fibres crossing the habenula and passing across the tunnel to end on outer hair cells/Deiters’ cells (Figures 2C and 2E). Immunoreactivity was observed at neural sites at the base of the cochlear inner hair cell (Figure 2D) and weakly associated with the cochlear hair cell stereocilia as detected with the chromogen DAB (Figure 2C), the latter consistent with previously determined expression of CNGA3 mRNA rat microdissected organ of Corti subfraction [13]. CNGA3 immunofluorescence (green) was localized with confocal microscopy to outer hair cell stereociliary arrays in mouse organ of Corti (Figures 2F and 2G), overlapping (yellow) fluorescence (red) for phalloidin targeting F-actin (Figure 2G).
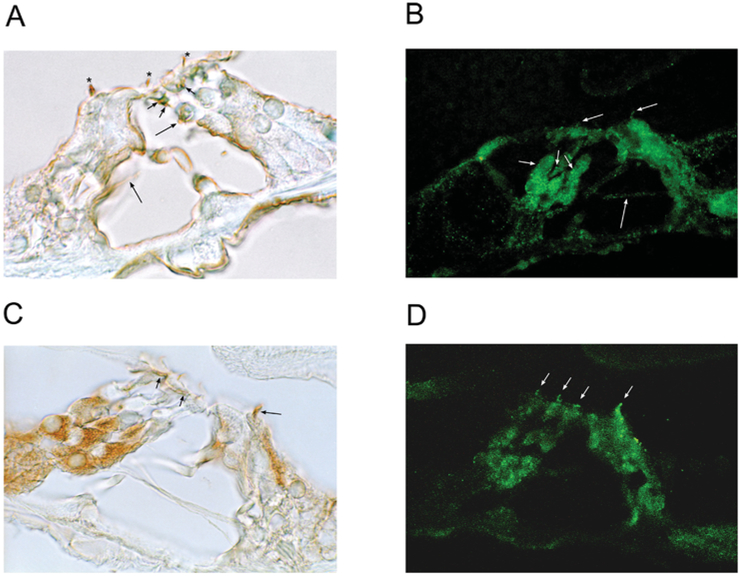
Immunoreactivity for EMILIN1 in the rat organ of Corti, whether detected with DAB (Figure 7A) or immunofluorescence (green, Figure 7B), was found in tunnel-crossing efferent nerve fibres and at sites of contact at the base of the outer hair cells and Deiters’ cells. EMILIN1 immunoreactivity was found in nerves at the level of the subcuticular plate region of outer hair cells (Figure 7A). Immunoreactivity for EMILIN1 was also associated with the hair cell stereociliary arrays, extending along the lengthwise axis of the stereocilia (Figure 7B).
Figure 7. Immunolocalization of EMILIN1 and PDE6C in rat organ of Corti.
(A) EMILIN1 was immunolocalized with DAB to nerve fibres (efferents) crossing the tunnel of Corti ending at the base of outer hair cells (long arrows) in rat organ of Corti. Small nerve fibres at subcuticular plate sites of outer hair cells were immunoreactive (short arrows) and stereociliary arrays of both inner and outer hair cells exhibited immunoreactivity for EMILIN1 (asterisks). (B) With fluorescence detection, EMILIN1 immunoreactivity was again found in olivocochlear axonal efferent fibres crossing the tunnel (long arrow), making contact with the base of the outer hair cells and Deiters’ cells (short arrows). Immunoreactivity was clearly associated with stereocilia of both inner and outer hair cells (mid-length arrows). (C) PDE6C immunoreactivity was associated with Deiters’ cells; however, this was not due to overlapping efferent/afferent contacts, since neither the tunnel crossing efferents nor the type II afferents were immunoreactive. PDE6C immunoreactivity was found at subcuticular sites on outer hair cells (short arrows) and was strongly present on stereocilia of the inner hair cell (mid-length arrow). (D) PDE6C immunofluorescence was associated with stereocilia of both inner and outer hair cells (arrows).
PDE6C in trout and rat hair cell models
PDE6C is the primary regulator of cytoplasmic cGMP concentration in rod and cone photoreceptors gating the CNGA3 channel. Given that the CNGA3-3 sequence expressed in trout saccular hair cells is a cone/olfactory subtype, we wondered if PDE6C would also be expressed in the teleost saccular hair cell model. Two sets of degenerate primers targeting Danio rerio PDE6C sequence (Supplementary Table S1), cGMP-specific, cone, alpha prime (GenBank® accession number NM_200871) directed amplification of trout saccular hair cell PDE6C cDNA (Figure 8 and Supplementary Figure S2). In the rat organ of Corti, immunoreactivity for PDE6C was associated with the subcuticular plate region of OHC and again (as for EMILIN1) distributed along the long axis of the cochlear hair cell stereocilia (Figures 7C and 7D).
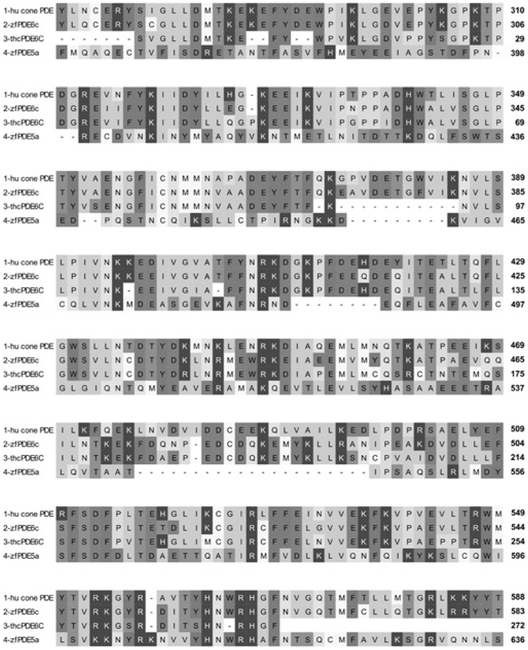
Figure 8. PDE6C expression in the trout saccular hair cell preparation.
OMIGA 2 alignment of PDE6C amino acid sequences. Line 1, human cone photoreceptor (hu cone) PDE6C (GenBank® accession number CAA64079). Line 2, zebrafish (zf) PDE6C (GenBank® accession number NP_957165). Line 3, trout saccular hair cell (thc) PDE6C deduced from RT–PCR. Line 4, zebrafish (zf) PDE5a (GenBank® accession number NP_001116732).
DISCUSSION
CNG channels have been implicated in sensory transduction of vision, olfaction and taste, with further roles determined in Ca2+ signalling. A CNG channel is considered to represent one candidate for the inner ear mechanosensory transduction channel, on the basis of electrophysiological considerations [24]. We previously obtained evidence that the olfactory CNG subunits CNGA2 and CNGA4 are expressed in hair cells of the organ of Corti, with the subunit channel protein immunolocalized to the hair cell stereociliary arrays [12,13]. CNGα subunit expression has now been examined in a vestibular model hair cell preparation isolated from the trout saccule. This preparation, owing to its large hair cell population free of intact supporting cells (and blood vessels) and consequent high representation of hair cell mRNA, has singularly provided the full-length sequences for ion channels and receptors expressed in hair cells [23,25–28]. In the present study, cloning of amplification products from RT–PCR has provided evidence of mRNA expression of CNGA2 and CNGA3 subunits in the trout saccular hair cells with a relative distribution frequency for CNGA2/CNGA3-1/CNGA3-2/CNGA3-3 of 2:8:4:4, consistent with the hypothesis that olfactory/cone photoreceptor-type CNG subunits are expressed in vestibular hair cells as well as in the organ of Corti [12,13]. We have obtained full-length coding sequence for the three variants of CNGA3 expressed in trout saccular hair cells and demonstrate PPIs for CNGA3 with EMILIN1 expressed in saccular hair cells and organ of Corti, using yeast two-hybrid, pull-down and SPR analysis.
CNGA3 expression in the trout saccular hair cell layer
CNGA3 is a cGMP-gated channel known to have a role in sensory transduction of cone photoreceptors [2,29], taste receptor cells (gustation) [3,4] and a subset of olfactory primary sensory neurons in the main olfactory epithelium [30,31]. The trout saccular hair cell CNGA3 variants were found to have transmembrane domains, pore and CNBD largely similar to CNGA3 sequences found in higher vertebrates conserved across evolution.
Although the overall features of sensory and non-sensory versions of CNGA3 are recognizable as specific to CNGA3 channels, there are also differences in the amino acid sequences that differentiate CNGA3 channels in different tissues and differentiate between hair cell CNGA3 variants. For example, the N-terminus is a source of variation. None of the rat CNGA3 sequences are identical for the first 65 amino acids. Within this region, saccular hair cell CNGA3-3 amino acid sequence was 24% identical with rat cone/olfactory sequence and 28% identical with bovine renal and human testes sequence, compared with identity to CNGA3-1 and CNGA3-2 variants (98% and 80 %, respectively). None of the trout saccular hair cell CNGA3 variants were characterized by the N-terminus found in CNGA3 used in taste sensory transduction (amino acids 27–42).
There is a degree of conservation for the next region within the N-terminus, amino acids 66–88, between bovine renal, human testes and rat cone/olfactory sequence which was replicated in CNGA3-2, but not in other hair cell variants. This region is followed by human CNGA3 exon 5 (amino acids 131–147, GenBank® accession number AAC 17440) specifically found in human retina CNGA3 [32], with identity to trout pineal CNGA3 sequence (amino acids 119–136). This sequence is not found in the rat olfactory CNGA3 sequence (GenBank® accession number EDL99244) nor in any of the saccular hair cell CNGA3 variants, a feature which by itself would point to more of an olfactory-like sequence rather than a cone-like sequence for trout saccular hair cell CNGA3. Another variation in the N-terminus of CNGA3 sequences appears next with alternating lysine and glutamate residues appearing in bovine renal and trout pineal sequences replicated in hair cell variant CNGA3–3 (amino acids 119–184). Overall, the trout saccular hair cell CNGA3 variants appeared to reflect both olfactory and photoreceptor-like features on the basis of the N-terminus sequence.
Channel domains in CNGA3 variants
The fourth transmembrane helix, ‘S4’ of CNGA3 channels, was conserved in the trout hair cell CNGA3-1, CNGA3-2 and CNGA3-3 subunits, although with some variation between lysine and arginine residues as the third positively charged residue in the motif (+)xx(+)xx(+)xx(+)xx [33]. The possibility that one of the trout CNGA3 variants could act as a CNG3B subunit appears negated in part due to the fact that the β-subunit, although exhibiting the basic S4 motif [34], has only three (+)xx repeats [33], whereas all three of the trout CNGA3 variants have four repeats.
CNGs in inner ear hair cells may influence Ca2+ signalling, modulating both exocytosis of the hair-cell transmitter [25,28,35] and adaptation of mechanosensory transduction [36]. The S5 and S6 domains and pore-flanking domains, influencing Ca2+ permeation properties of CNGs [37], are significantly conserved across the three CNGA3 variants having identity with cone, olfactory and gustatory CNGA3 sequences. The sequence in the actual pore region of trout hair cell CNGA3 sub-types is identical with other CNGA3 subunits.
Within the C-linker region, the three amino acids Ile448, Asp490 and Asp503 (CNGA3-3) are recognized as major determinants of gating in CNGA3 channels, pointing to a cone photoreceptor-like CNGA3 profile as opposed to an olfactory CNGA2 signature [38]. The C-linker peptide also contains a potential PKA-targeted sequence, RKVSKDL, conserved across vertebrate CNGA3 sequences and found at amino acids 455–461 in trout saccular hair cell CNGA3-3. The implication would be that CNGA3 channel function may be regulated by PKA phosphorylation via AC (adenylate cyclase) activity, which in trout saccular hair cells is attributable to AC9, AC7 or AC5/6 [23]. The CNBD region itself for the CNGA3 variants contained tryptophan, threonine, lysine and aspartate residues (amino acids 566, 593, 629 and 637 for CNGA3-3), which are required to enhance sensitivity to cGMP activation of cGMP-preferring photoreceptor channels CNGA1 and CNGA3 [39,40].
Exon structure of trout saccular hair cell CNGA3 splice variants
Although the genomic sequence for trout is unavailable, the trout saccular hair cell CNGA3 transcript variants may be compared with the genomic sequence for another teleost, zebrafish, for which exon organization of CNGA3 is known (chromosome 6 genomic scaffolds Zv9, NW_001879150.3). We suggest that the variation between the saccular hair cell CNGA3 variants is due to alternative splicing, and, additionally for two variants, omission of one of seven exons expressed in zebrafish CNGA3. The N-terminus is covered by exons 1–5. Only CNGA3-3 has exon 4. CNGA3-2 has an extended version of exon 3, different in sequence to variants 1 and 3, all representing presumed splice variants of exon 3 in zebrafish. The extension for exon 3 in CNGA3-2 accounts for amino acids 66–88, which is found in higher vertebrates (see above). Membrane-spanning region S1 to the end of the sequence is covered by the middle of exon 5 (S1) and all of exons 6 (S2) and 7 (S3 to end). There was close to 100% identity between variants from the middle to the end of exon 5. Nucleotides for exon 6 were 100% identical for variants 1 and 2 and 90% identical for variant 3. The differences between variants 1 and 2 with variant 3 continued in exon 7, with variant 3 being 91% identical with the two others (themselves close to 100% identity), indicating the likelihood of alternative splicing for variant 3 compared with variants 1 and 2.
PPI between CNGA3 and EMILIN1
In the present study, we provide evidence that the cytoplasmic C-terminus of CNGA3 binds the C-terminal region of EMILIN1 for both trout saccular hair cell proteins and rat organ of Corti proteins. The similarity of results for trout and rat was not surprising given that the respective sequences have regions of conservation across evolution: the trout saccular hair cell CNGA3 C-terminal sequence is overall 42% identical (81% positive) with rat sequence (EDL99244). The C-terminus of EMILIN1 in trout hair cells is 75% identical with rat sequence (NP_001100180). Specific binding between CNGA3-3 and EMILIN1 was confirmed with pull-down assays and SPR analysis, predicting dependence on Ca2+. In the presence of 68 μM Ca2+, binding between CNGA3-3 and EMILIN1 was characterized by a Kd = 1.6×10 −6 M for trout and Kd = 2.7×10−7 M for rat proteins. Immunolocalizations carried out on the respective tissues indicated sites of protein expression consistent with the observed PPIs. CNGA3 was immunolocalized to the subcuticular plate region and stereocilia of trout saccular hair cells and also co-localized with phalloidin in the stereocilia of the cochlear outer hair cells. EMILIN1 was likewise immunolocalized to stereocilia of hair cells, specifically both inner and outer hair cells of the rat organ of Corti.
EMILIN1 is a secreted extracellular matrix multidomain glycoprotein, a member of the elastic fibre system [41,42] that includes a gC1q-like globular domain close to its C-terminus. The question that arises is how EMILIN1, an extracellular matrix protein, can bind to the intracellular C-terminus of CNGA3 in situ. PPIs may mediate intracellular regulatory mechanisms for protein trafficking, but little is known concerning interactions for CNGA3 inserted into the plasma membrane. Several lines of evidence point to the possibility that EMILIN1 might have an intracellular presence. Analysis of C1q domain proteins indicated a final C-terminus stretch of amino acids not encompassed by the C1q domain for EMILIN1 [43]. In the present study, software analysing membrane protein secondary structure predicted a transmembrane segment for the rat and mouse EMILIN1 sequence 54 amino acids from the end of the C-terminus. Pull-down assays indicated that, of the three segments of EMILIN1 prey sequence, the peptide following the transmembrane region in the prey sequence was responsible for the PPI between EMILIN1 and CNGA3, with no binding detected for the other two segments before the transmembrane domain and the transmembrane domain itself. These results are consistent with an intracellular PPI, although not representing definitive proof of such an interaction in stereocilia of intact hair cells in vivo, which would require additional experiments beyond the scope of the present study.
The gC1q domain of EMILIN1 is considered to be a ligand for β1 integrin, with a monoclonal antibody to β1 integrin blocking EMILIN1 gC1q-mediated cell attachment [44]. Integrins and other cell-surface receptors regulate many aspects of cytoskeleton dynamics in different cell types [45]. In the cochlea, integrins play a number of different roles during development [46] and stereociliary cytoskeletal deficits have been described in inner ear hair cells in mice deficient in integrin α8β1 [47]. We have obtained evidence that the transcript for β1 integrin, which combines with all of the α-subunits, is expressed in the model hair cell preparation from the trout saccule (D. Selvakumar, M. J. Drescher and D. G. Drescher, unpublished work). Consequently, a complex may exist between β1-integrin, EMILIN1 and CNGA3 in saccular hair cells.
Prior to the present study, no information has been presented on molecular function or localization of EMILIN1 protein in the organ of Corti of the adult mammal. EMILIN2 has been suggested to have a structural role in the cochlear basilar membrane [48]. During development, levels of EMILIN2 mRNA in the mouse cochlea increased from P0 to a maximum level at P14, before falling to 18% of the maximum by P37. Levels of EMILIN1 mRNA increased from P0 to a maximum at P4, beginning to descend by P8 to reach 38% of the maximum level by P14, a level which was maintained at P37 [48], consistent with relative constancy of expression in the adult.
PDE6C
Another potential key component in a molecular pathway for CNGA3 is PDE6. PDE6A and PDE6C are the primary regulators of cytoplasmic cGMP concentration in rod and cone photoreceptors respectively. PDE6A in rod photoreceptors actually binds to a splice variant of cGMP-gated CNGB1, GARP2 (glutamic acid-rich protein-2) [49]. Precise regulation of cGMP level is essential for normal operation of the visual transduction cascade. Persistent imbalance in cGMP metabolism disrupts the visual signalling pathway and leads to cell death and retinal degeneration (e.g. retinitis pigmentosa [50]). In the present study, we demonstrated expression of PDE6C in the trout saccular hair cell model, which is consistent with a cone photoreceptor-like association of CNGA3 and PDE in teleost saccular hair cells. PDE6C was immunolocalized to cochlear hair cell stereocilia, a site also consistent with CNGA3 immunolocalization.
There exist several connections between PDE6 and ear-related pathologies. A PDE6A mutation is reported to produce retinitis pigmentosa in a canine model [51]. Retinitis pigmentosa is one part of Usher syndrome [52], which affects both vision and hearing [53]. Human PDE6C and PDE6A (GenBank® accession numbers CAA64079 and NP_000431 respectively) are related, 64% identical, 81% positive. The rod PDE6A catalytic subunit can restore cone function in a knockout of PDE6C catalytic subunit [54]. PDE6C is located on human chromosome 10 (10q24) similarly to cadherin 23 (10q22.1) and protocadherin 15 CD3 (10q21.1), proteins affected in Usher syndrome.
PDE inhibitors produce clinically relevant effects in the inner ear, both on hearing as well as on vestibular function. Sildenafil (Viagra®) is a selective inhibitor of PDE5, with one exception: it is cross-reactive for PDE6 [55]. Documented side effects for sildenafil are alterations in visual function [56] and hearing loss [57–60]. A case report [57] prompted the FDA to search the AERS (Adverse Event Reporting System) for postmarketing reports of hearing impairment for all drugs in the class of PDE5 inhibitors. The search resulted in 29 reports of sudden hearing loss, both with and without accompanying vestibular symptoms (tinnitus, vertigo or dizziness), in strong temporal relationship to dosing with Viagra (sildenafil), Cialis (tadalafil) or Levitra (vardenadil).
In addition to PDE6C, another enzyme required for cGMP degradation in the regulation of CNGA3 channels, guanylate kinase 1, has been found to bind protocadherin 15 CD3 in yeast two-hybrid mating protocols (N.A. Ramakrishnan, M. J. Drescher and D. G. Drescher, unpublished work). Protocadherin 15 CD3 itself is localized to stereociliary tip links and to the subcuticular plate region of cochlear hair cells [61,62]. Taken together, the evidence supports the possibility of a molecular pathway in vestibular and cochlear hair cells coupling CNGA3 to EMILIN1 (and β1 integrin) and to enzymes required for cGMP degradation, PDE6C and guanylate kinase 1.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr Stanley Terlecky for the use of the SPR facility and Biacore 3000 instrument.
FUNDING
This work was supported by the National Institutes of Health [grant numbers DC004076 (to M.J.D.) and DC000156 (to D.G.D.)].
Abbreviations used:
- AC
adenylate cyclase
- CNBD
cyclic nucleotide-binding domain
- CNG
cyclic nucleotide-gated
- DAB
diaminobenzidine
- EMILIN1
elastin microfibril interface-located protein 1
- F-actin
filamentous actin
- GST
glutathione transferase
- IPTG
isopropyl β-D-thiogalactopyranoside
- PDE6C
phosphodiesterase 6C
- PPI
protein–protein interaction
- RACE
rapid amplification of cDNA ends
- RT
reverse transcription
- SPR
surface plasmon resonance
Footnotes
REFERENCES
- 1.Zagotta WN and Siegelbaum SA (1996) Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci 19, 235–263 [DOI] [PubMed] [Google Scholar]
- 2.Biel M and Michalakis S (2009) Cyclic nucleotide-gated channels. Handb. Exp. Pharmacol 191, 111–136 [DOI] [PubMed] [Google Scholar]
- 3.Misaka T, Kusakabe Y, Emori Y, Gonoi T, Arai S and Abe K (1997) Taste buds have a cyclic nucleotide-activated channel, CNGgust. J. Biol. Chem 272, 22623–22629 [DOI] [PubMed] [Google Scholar]
- 4.Lee HM, Park YS, Kim W and Park CS (2001) Electrophysiological characteristics of rat gustatory cyclic nucleotide-gated channel expressed in Xenopus oocytes. J. Neurophysiol 85, 2335–2349 [DOI] [PubMed] [Google Scholar]
- 5.Bradley J, Frings S, Yau KW and Reed R (2001) Nomenclature for ion channel subunits. Science 294, 2095–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu DT, Tibbs GR and Siegelbaum SA (1996) Subunit stoichiometry of cyclic nucleotide-gated channels and effects of subunit order on channel function. Neuron 16, 983–990 [DOI] [PubMed] [Google Scholar]
- 7.Chen TY, Peng YW, Dhallan RS, Ahamed B, Reed RR and Yau KW (1993) A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature 362, 764–767 [DOI] [PubMed] [Google Scholar]
- 8.Liman ER and Buck LB (1994) A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron 13, 611–621 [DOI] [PubMed] [Google Scholar]
- 9.Fain GL, Matthews HR, Cornwall MC and Koutalos Y (2001) Adaptation in vertebrate photoreceptors. Physiol. Rev 81, 117–151 [DOI] [PubMed] [Google Scholar]
- 10.Trudeau MC and Zagotta WN (2003) Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J. Biol. Chem 278, 18705–18708 [DOI] [PubMed] [Google Scholar]
- 11.Kolesnikov SS, Rebrik TI, Zhainazarov AB, Tavartkiladze GA and Kalamkarov GR (1991) A cyclic-AMP-gated conductance in cochlear hair cells. FEBS Lett 290, 167–170 [DOI] [PubMed] [Google Scholar]
- 12.Drescher MJ, Barretto RL, Chaturvedi D, Beisel KW, Hatfield JS, Khan KM and Drescher DG (2002) Expression of subunits for the cAMP-sensitive, “olfactory” cyclic nucleotide-gated ion channel in the cochlea: implications for signal transduction. Mol. Brain Res 98, 1–14 [DOI] [PubMed] [Google Scholar]
- 13.Drescher MJ, Chaturvedi D and Drescher DG (2001) Cyclic nucleotide gated ion channel subunit expression in the rat cochlea. Assoc. Res. Otolaryngol. Abstr 24, 40–41 [Google Scholar]
- 14.Selvakumar D, Drescher MJ and Drescher DG (2008) Molecular cloning of cyclic nucleotide-gated channel subunits expressed in teleost saccular hair cells. Assoc. Res. Otolaryngol. Abstr 31, 88 (30) [Google Scholar]
- 15.Selvakumar D, Drescher M and Drescher D (2010) Cyclic-nucleotide-gated (CNG) ion channels in saccular hair cells. Assoc. Res. Otolaryngol. Abs 33, 28 [Google Scholar]
- 16.Selvakumar D, Drescher MJ, Khan K and Drescher DG (2011) The CNGA3 channel subunit binds to the intracellular C1q domain of EMILIN1. Assoc. Res. Otolaryngol. Abs 34, 572 [Google Scholar]
- 17.Dowdall J, Drescher M and Drescher D (2010) Expression of phosphodiesterases in hair cells modulating cAMP and cGMP signaling. Assoc. Res. Otolaryngol. Abs 33, 591 [Google Scholar]
- 18.Drescher DG, Khan KM, Arden RL, Drescher MJ, Kerr TP and Hatfield JS (1989) Protein associated with the sensory cell layer of the rainbow trout saccular macula. Brain Res 485, 225–235 [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF and Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn, pp. 7.18–7.29. Cold Spring Harbor Laboratory Press, Cold Spring Harbor [Google Scholar]
- 20.Ramakrishnan NA, Drescher MJ, Barretto RL, Beisel KW, Hatfield JS and Drescher DG (2009) Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip-link protein protocadherin 15 CD3. J. Biol. Chem 284, 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh CK, Drescher MJ, Hatfield JS and Drescher DG (1999) Selective expression of serotonin receptor transcripts in the mammalian cochlea and its subdivisions. Mol. Brain Res 70, 135–140 [DOI] [PubMed] [Google Scholar]
- 22.Drescher DG, Ramakrishnan NA and Drescher MJ (2010) Surface plasmon resonance (SPR) analysis of binding interactions of proteins in inner-ear sensory epithelia. Methods Mol. Biol 493, 323–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drescher MJ, Cho WJ, Folbe A, Kewson DT, Abu-Hamdan M, Oh CK, Selvakumar D, Ramakrishnan NA, Hatfield JS, Khan KM et al. (2010) An adenylyl cyclase signaling pathway predicts direct dopaminergic input to vestibular hair cells. Neuroscience 171, 1054–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricci AJ, Kachar B, Gale J and Van Netten SM (2006) Mechano-electrical transduction: new insights into old ideas. J. Membr. Biol 209, 71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakrishnan NA, Swanson GS, Green GE, Lakhani RS, Ahsan SF, Pasha R, Drescher MJ and Drescher DG (2002) L-type, α1D subunit of the voltage-gated Ca2+ channel in the saccular hair-cell epithelium of the rainbow trout. Mol. Brain Res 109, 69–83 [DOI] [PubMed] [Google Scholar]
- 26.Cho WJ, Drescher MJ, Hatfield JS, Bessert DA, Skoff RP and Drescher DG (2003) Hyperpolarization-activated, cyclic AMP-gated, HCN1-like cation channel: the primary, full-length HCN isoform expressed in a saccular hair-cell layer. Neuroscience 118, 525–534 [DOI] [PubMed] [Google Scholar]
- 27.Drescher DG, Ramakrishnan NA, Drescher MJ, Chun W, Wang X, Myers S, Green GE, Karpenko A and Nguyen T (2004) Cloning and characterization of nicotinic acetylcholine receptor α9 subunits expressed by saccular hair cells of the rainbow trout (Oncorhynchus mykiss). Neuroscience 127, 737–752 [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan NA, Sheikhali SA, Drescher MJ, Khan KM, Hatfield JS and Drescher DG (2006) Molecular identification of an N-type Ca2+ channel in saccular hair cells. Neuroscience 139, 1417–1434 [DOI] [PubMed] [Google Scholar]
- 29.Yu WP, Grunwald ME and Kau KW (1996) Molecular cloning, functional expression and chromosomal localization of a human homolog of the cyclic nucleotide-gated ion channel of retinal cone photoreceptors. FEBS Lett 393, 211–215 [DOI] [PubMed] [Google Scholar]
- 30.Meyer MR, Angele A, Kremmer E, Kaupp UB and Müller FA (2000) cGMP-signaling pathway in a subset of olfactory sensory neurons. Proc. Natl. Acad. Sci. U.S.A 97, 10595–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leinders-Zufall T, Cockerham RE, Michalakis S, Biel M, Garbers DL, Reed RR, Zufall F and Munger SD (2007) Contribution of the receptor guanylyl cyclase GC-D to chemosensory function in the olfactory epithelium. Proc. Natl. Acad. Sci. U.S.A 104, 14507–14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassar SC, Chen J, Zhang D and Gopalakrishnan M (2004) Tissue specific expression of alternative splice forms of human cyclic nucleotide gated channel subunit CNGA3. Mol. Vision 10, 808–813 [PubMed] [Google Scholar]
- 33.Paillart C, Zhang K, Rebrik TI, Baehr W and Korenbrot JI (2006) Cloning and molecular characterization of cGMP-gated ion channels from rod and cone photoreceptors of striped bass (M. saxatilis) retina. Vis. Neurosci 23, 99–113 [DOI] [PubMed] [Google Scholar]
- 34.Faillace MP, Bernabeu RO and Korenbrot JI (2004) Cellular processing of cone photoreceptor cyclic GMP-gated ion channels: a role for the S4 structural motif. J. Biol. Chem 279, 22643–53 [DOI] [PubMed] [Google Scholar]
- 35.Drescher MJ and Drescher DG (1992) Glutamate, of the endogenous primary α-amino acids, is specifically released from hair cells by elevated extracellular potassium. J. Neurochem 58, 93–98 [DOI] [PubMed] [Google Scholar]
- 36.Ricci AJ and Fettiplace R (1997) The effects of calcium buffering and cyclic AMP on mechano-electrical transduction in turtle auditory hair cells. J. Physiol 501, 111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert R, Eismann E, Ludwig J, Baumann A and Kaupp UB (1999) Molecular determinants of a Ca2+ -binding site in the pore of cyclic nucleotide-gated channels: S5/S6 segments control affinity of intrapore glutamates. EMBO J. 18, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zong X, Zucker H, Hofmann F and Biel M (1998) Three amino acids in the C-linker are major determinants of gating in cyclic nucleotide-gated channels. EMBO J. 17, 353–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott SP, Cummings J, Joe JC and Tanaka JC (2000) Mutating three residues in the bovine rod cyclic nucleotide-activated channel can switch a nucleotide from inactive to active. Biophys. J. 78, 2321–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetreault ML, Henry D, Horrigan DM, Matthews G and Zimmerman AL (2006) Characterization of a novel cyclic nucleotide-gated channel from zebrafish brain. Biochem. Biophys. Res. Commun 348, 441–449 [DOI] [PubMed] [Google Scholar]
- 41.Doliana R, Mongiat M, Bucciotti F, Giacomello E, Deutzmann R, Volpin D, Bressan GM and Colombatti A (1999) EMILIN, a component of the elastic fiber and a new member of the C1q/tumor necrosis factor superfamily of proteins. J. Biol. Chem 24, 16773–16781 [DOI] [PubMed] [Google Scholar]
- 42.Colombatti A, Doliana R, Bot S, Canton A, Mongiat M, Mungiguerra G, Paron-Cilli S and Spessotto P (2000) The EMILIN protein family. Matrix Biol 19, 289–301 [DOI] [PubMed] [Google Scholar]
- 43.Hu Y-L, Pan X-M, Xiang L-X and Shao J-Z (2010) Characterization of C1Q in teleosts: insight into the molecular and functional evolution of C1Q family and classical pathway. J. Biol. Chem 285, 28777–28786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spessotto P, Cervi M, Mucignat MT, Mungiguerra G, Sartoretto I, Doliana R and Colombatti A (2003) β1 integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. J. Biol. Chem 278, 6160–6167 [DOI] [PubMed] [Google Scholar]
- 45.Hall A and Nobes CD (2000) Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc., B 355, 965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyama K, Ozeki M, Hamajima Y and Lin J (2005) Expression of the integrin genes in the developing cochlea of rats. Heart Res 201, 21–26 [DOI] [PubMed] [Google Scholar]
- 47.Littlewood-Evans A and Müller U (2000) Stereocilia defects in the sensory hair cells of the inner ear in mice deficient in integrin α8β1. Nat. Genet 24, 424–428 [DOI] [PubMed] [Google Scholar]
- 48.Amma LL, Goodyear R, Faris JS, Jones I, Ng L, Richardson G and Forrest D (2003) An emilin family extracellular matrix protein identified in the cochlear basilar membrane. Mol. Cell. Neurosci 23, 460–472 [DOI] [PubMed] [Google Scholar]
- 49.Pentia DC, Hosier S and Cote RH (2006) The glutamic acid-rich protein-2 (GARP2) is a high affinity rod photoreceptor phosphodiesterase (PDE6)-binding protein that modulates its catalytic properties. J. Biol. Chem 231, 5500–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cote RH (2004) Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int. J. Impot. Res 16 (Suppl. 1), S28–S33 [DOI] [PubMed] [Google Scholar]
- 51.Tuntivanich N, Pittler SJ, Fischer AJ, Omar G, Kiupel M, Weber A, Yao S, Steibel JP, Khan NW and Petersen-Jones SM (2009) Characterization of a canine model of autosomal recessive retinitis pigmentosa due to a PDE6A mutation. Invest. Ophthalmol. Vis. Sci 50, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremer H, van Wijk E, Märker T, Wolfrum U and Roepman R (2006) Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum. Mol. Genet. Spec 15 (Suppl. 2), R262–R270 [DOI] [PubMed] [Google Scholar]
- 53.Friedman TB, Schultz JM, Ahmed ZM, Tsilou ET and Brewer CC (2011) Usher syndrome: hearing loss with vision loss. Adv. Otorhinolaryngol 70, 56–65 [DOI] [PubMed] [Google Scholar]
- 54.Kolandaivelu S, Chang B and Ramamurthy V (2011) Rod phosphodiesterase-6 (PDE6) catalytic subunits restores cone function in a mouse model lacking PDE6 catalytic subunit. J. Biol. Chem 286, 33252–33259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bischoff E (2004) Potency, selectivity, and consequences of nonselectivity of PDE inhibition. Int. J. Potency Res 16, S11–S14 [DOI] [PubMed] [Google Scholar]
- 56.Laties A and Zrenner E (2002) Viagra (sildenafil citrate) and ophthalmology. Prog. Retin. Eye Res 21, 485–506 [DOI] [PubMed] [Google Scholar]
- 57.Mukherjee B and Shivakumar T (2007) A case of sensorineural deafness following ingestion of sildenafil. J. Laryngol. Otol 121, 395–7 [DOI] [PubMed] [Google Scholar]
- 58.Hong BN, Yi TH, Kim SY and Kang TH (2008) High dosage sildenafil induces hearing impairment in mice. Biol. Pharm. Bull 31, 1981–1984 [DOI] [PubMed] [Google Scholar]
- 59.Okuyucu S (2009) Effect of phosphodiesterase-5 inhibitor on hearing. J. Laryngol. Otol 12, 1–5 [DOI] [PubMed] [Google Scholar]
- 60.Khan AS, Sheikh Z, Khan S, Dwivedi R and Benjamin E (2011) Viagra deafness–sensorineural hearing loss and phosphodiesterase-5 inhibitors. Laryngoscope 121, 1049–54 [DOI] [PubMed] [Google Scholar]
- 61.Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess M, Lilley KS, Wilcox ER, Riazuddin S et al. (2006) The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci 26, 7022–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Müller U and Kachar B (2007) Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.