Abstract
The present study was carried out with the objective of development of species-specific loop-mediated isothermal amplification (LAMP) assay for identification of tissue of cattle origin. The cattle-specific LAMP primer set was designed by targeting mitochondrial D-loop gene. The conditions for LAMP reaction for amplification of template DNA from cattle using designed cattle-specific primer set were optimized for the components of mixture and temperature of reaction. Amplified products were analysed using SYBR Green I dye and by agarose gel electrophoresis. The developed species-specific LAMP assay was evaluated for its specificity, sensitivity and validated in laboratory on samples from known, coded, binary meat admixture with other than cattle at relative percentage of 20%, 10%, 5% and 1%, Phire tissue direct PCR master mix treated tissues of cattle and on species-specific polymerase chain reaction assay positive samples. The developed LAMP assay using self-designed primer set was highly specific, amplifying the DNA template exclusively from cattle tissue under the optimized LAMP reaction conditions. The sensitivity assay using serially diluted DNA templates revealed lowest level of detection as 0.01 ng of absolute DNA from target species. Laboratory validation substantiated the accuracy of assay in known/unknown (coded) samples and up to the 1% level of admixture in binary meat sample. DNA present in supernatant of Phire Animal tissue kit treated samples were also amplified successfully eliminating the extra step of extraction of genomic DNA. The developed assays exhibited comparable results with previously established species-specific PCR assay taken as gold standards. Thus, it was concluded that developed species-specific loop-mediated isothermal amplification assay was effective in identification of tissue of cattle origin.
Keywords: Species-specific, Loop-mediated isothermal amplification (LAMP), Cattle, Sensitivity and laboratory validation
Introduction
Meat and meat products are important parts of the human food chain. Their quality and origin are important issues to consumers, government agencies and retailers. In recent year species authentication has been gaining sensible importance throughout the globe. The fraudulent substitution is more common in economically poor countries with high population, owing to increasing demand of meat and meat products and their high cost (Spink and Moyer 2011). Although, in most of the cases of species substitution of meat, the motivation may be profit making, the public health vulnerability also coexists in view of prevalence of bovine spongiform encephalopathy (BSE) and food allergies. The religious taboos and individual preference (vegetarian) determining choices not to intake a particular type of meat or meat sources also make the issue of meat authentication vital (Ortea et al. 2012).
Several reliable and powerful detection methods for meat species identification have been developed in the past and amplification of mitochondrial DNA sequences using various techniques is still considered as the most reliable tool. PCR based DNA amplification techniques used for species identification are PCR-RFLP (Doosti et al. 2014), PCR- RAPD (Mohindra et al. 2007), simple or multiplex PCR (Hou et al. 2015), RT-PCR (Fajardo et al. 2008a) and species-specific PCR (Kumar et al. 2012). With the advancement in research, it was observed that the PCR-based assays were not suitable for on point species identification due to requirement of sophisticated equipment’s and laboratory conditions. Most of the nucleic acid amplification tests (NAT) are percluded as routine diagnostic tool in low resource setting laboratories because of their non suitability for point-of-care test (PoCT) or portable detection systems as required in applications like food investigation, operation in fully equipped laboratories with good infrastructure as well as highly trained employees and post amplification processing.
Loop-mediated isothermal amplification (LAMP) is an established nucleotide amplification method which combines simplicity as well as high specificity. LAMP assay is relatively fast because the method amplifies DNA in isothermal conditions eliminating the need for thermal cycling (Cho et al. 2014). It employs auto cycling strand displacement DNA synthesis using Bacillus stearothermophilus (Bst) DNA polymerase and a set of 4–6 primers that bind to unique sites on the target sequence, making them highly specific (Notomi et al. 2000). Additionally, gel electrophoresis to visualize the amplified DNA is not required (Tomlinson and Boonham 2008) and LAMP amplified product can be visualized by naked eye using DNA binding dyes (SYBR Green I) and colorimetric indicator dye such as Calcein (Parida et al. 2008) and HNB (Goto et al. 2009). There are very few reports on use of LAMP assay for species identification, particularly for tissue of cattle origin. Some of the noted attempts made for identification of pork using LAMP assays are by Abdullahi et al. (2017), Ran et al. (2016), Yang et al. (2015), Kanchanaphum et al. (2014) and Cho et al. (2014). In the present study, we attempted to identify cattle tissues and also evaluated its specificity from closely related species such as buffalo, sheep and goat. The developed assay was also evaluated for its sensitivity and validated on samples of diverse origin to ascertain their utility.
Materials and methods
Collection of meat/tissue samples
Meat/tissue samples were collected from Cattle, Buffalo, Sheep, Goat and Pig. Fresh meat samples of common food animals sheep, goat and pig were collected from experimental abattoir of Livestock Products Technology Division, Indian Veterinary Research Institute, India; buffalo meat samples were collected from municipal abattoirs of Bareilly (UP) and cattle tissue samples were collected from post-mortem hall of Indian Veterinary Research Institute, India. The samples were collected under aseptic conditions in a pre-sterilized plastic container maintained at chilling temperature and transported to the laboratory under refrigeration condition to be stored frozen at − 20 °C until DNA isolation.
Chemical, reagents and plastic-wares
The chemicals and reagents used in the study were of molecular biology grade with high purity and were procured from QIAGEN, SIGMA—ALDRICH, NEW ENGLAND BIOLAB, THERMO-FISHER and SRL, etc. Double distilled sterilized water was used for preparation of various buffer, whereas reagents used for DNA work were prepared in nuclease-free molecular biology grade water. The plastic wares were purchased from TARSONS and other reputed firms, whereas glass-wares were procured from RIVERA.
Scientific equipments, commercial kits and computer-aided softwares
The various scientific equipments used in the present investigations were as follows: Thermocycler (Agilent gradient thermocycler, USA), Temperature-controlled dry bath (Genei, India), Water bath (Perfit, India) Electrophoresis assembly (Genei, India), Micro-centrifuge (Remi, India), Micropipettes (Thermo Scientific, USA), Gel documentation system (AlphaImager, USA), Table top centrifuge (Biofuge, USA), Nanodrop spectrophotometer (Thermo-fisher, USA), Cyclo mixture (Remi, India), Ice-flacking machine (Icematic, USA), Spinix vortex machine (Tarson, India), Electronic balance (Sartorius, Germany), Refrigerator and Deep freezer (Blue star, India), etc. DNeasy blood and tissue kit (Qiagen, Germany), Phire tissue direct PCR master mix (Thermo Scientific, USA) and available softwares (DNA Star, Primer Explorer V4, Primer BLAST, Gene Tool etc) were also used.
Extraction of genomic DNA from tissue (muscle)
High quality genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Germany) as per the manufacturer’s instructions. All the centrifugation steps were performed at room temperature (15–25 °C). The genomic DNAs from muscle of cattle, buffalo, sheep, goat and pork were extracted. The concentration and purity of the isolated genomic DNA was checked using UV spectrophotometric method. Spectrophotometric readings were taken at OD260 and OD280 using Nanodrop against triple distilled water. Horizontal submarine agarose gel electrophoresis was used to check the quality of genomic DNA using 0.8% (w/v) agarose gel. Electrophoresis was performed at 110V for 45–50 min. After electrophoresis, the gel was visualized under UV transilluminator and documented by gel documentation system.
The Phire tissue direct PCR master mix (Thermo Scientific) was used to eliminate the additional steps of DNA isolation its quality, purity and concentration check. Direct tissue sample from various species were subjected to protocol as per manufacture’s instruction. An approximately 0.5 mm in diameter from animal tissue was taken using sampling tool and placed into the PCR reaction tube (200 µL of volume) already containing 20 µl dilution buffer. 0.5 µL of DNA release additive was added to the mixture and the tube were placed into the preheated heat block (98 °C) for 2 min. 2 µL of supernatant was used as a DNA template for a 25 µL of LAMP reaction mixture. The supernatant supposed to be rich with genomic DNA was used for LAMP reaction and validation of developed assay.
Designing of species-specific (LAMP) primers
The DNA sequences of mitochondrial D-loop gene of different species viz., cattle (EF524135.1), buffalo (DQ364182.1), goat (AM282549.1), sheep (AM279285.1) and pig (GQ141902.1) were retrieved from the National Centre for Biotechnology Information (NCBI) database for designing of LAMP primer set. For designing of cattle LAMP primers set mitochondrial gene D-loop sequence was targeted. Species-specific LAMP primers were designed through homology comparisons of the mitochondrial gene regions from these species using Megalign (DNA- STAR) (Table 1). The species-specific LAMP primers set was designed from conserved sequences of different regions/genes of mitochondrial genome using available softwares (Primer Explorer V4 Eiken Chemical Co., Ltd and DNA STAR).
Table 1.
Details of species-specific primers set used for identification of tissue of cattle origin
| Species | Primer code | Type | Nucleotide sequence 5′–3′ | Product length |
|---|---|---|---|---|
| Cattle | SaP-BOVSARU/F3 | Forward outer | CAAATCCATCCTCAACAACAT | 206 |
| SaP-BOVSARU/B3 | Backward outer | GAATGGACCACTTTAGATGAG | ||
| SaP-BOVSARU/F4 | Forward inner | GATCCTCTGCTTAGCGGGTT-GCCACTAGATCACGAGCTTAA | ||
| SaP-BOVSARU/B4 | Backward inner | ACCGTGGGGGTCGCTATTTAATGGCCCTGAAGAAAGAAC | ||
| SaP-BOVSARU/L5 | Loop forward | TCACGCGGCATGGTA | ||
| SaP-BOVSARU/L6 | Loop backward | ATGAATTTTACCAGGCATCTG |
Optimization of LAMP reaction and analysis of LAMP products
The reaction mixture components and temperature for LAMP reaction using cattle specific LAMP primers set were optimized. The thermocycler was used as heating block to achieve the isothermal amplification. The amplification reactions were carried out using gradient heat blocks in the range of 52–70 °C for amplification and reaction mixture component as per reported literature to determine the optimum temperature of amplification for LAMP primers. Once the optimal amplification temperature was determined for LAMP primers set based on analysis of LAMP products, further refinements were made by optimizing the components of LAMP reaction mixture viz: concentration of dNTP, MgSO4, ratio of outer and inner primer used, BstDNA polymerase and betaine.
The reaction mixture was prepared in a final volume of 25 µl in a 200-µl PCR tube. All the ingredients were taken using filter tips to avoid any cross-contamination. The PCR tube containing reaction mixture was flash spun on a table-top micro-centrifuge to get the reactants at the bottom. Every-time negative control (without template DNA) was kept to make sure the LAMP system was devoid of contamination.
A successful LAMP amplification was visualized by the addition of 1 µl of 1:10 diluted 10,000 X SYBR Green I dye (Sigma–Aldrich) to the tubes post amplification. The instant change in colour was visible with naked eye under day light. Additionally, horizontal submarine agarose gel electrophoresis was also used for analysis of LAMP products using 1.2% (w/v) agarose gel and performed at 110 V for 45–60 min.
Determination of specificity and sensitivity of developed species-specific LAMP Assay
DNA templates extracted from muscle tissues of different species viz: cattle, buffalo, sheep, goat and pig were subjected to cross reactivity test with cattle-specific LAMP assay. The species-specific amplification pattern was confirmed by both changes in colour of amplified LAMP product after addition of SYBR green I dye as well as by ladder-like electrophoretic pattern of amplified product by gel electrophoresis in only target species, without any cross reaction with other. The developed LAMP assay using cattle-specific LAMP primer set was evaluated for their sensitivity in serially diluted concentration of DNA template of cattle prepared by tenfold serial dilutions. The amplification patterns were observed for each diluted DNA sample to determine the lowest amount of absolute DNA template required for detectable amplification. The degree of intensity of colour of amplified product after addition of SYBR green I as well as electrophoretic pattern observable during gel electrophoresis was used for analysis of LAMP amplification.
Laboratory validation of the developed LAMP assays for detection of species origin of meat in known/coded samples, meat admixture, phire tissue direct PCR master mix treated tissue samples
DNA templates extracted from tissues collected from ten different cattle were analysed by developed species-specific LAMP assay to determine its utility for identification of species origin of tissue. The coded samples from different animal species including cattle were also analysed to validate the developed LAMP assay. The developed species-specific LAMP assay was also evaluated for their ability to amplify the DNA extracted from binary meat admixture of cattle and other species at relative percentage of 20%, 10%, 5% and 1%. Cattle tissue was subjected to Phire tissue direct PCR master mix treatment and species specific LAMP assay was validated by using supernatant from treated tissue sample as a source of DNA template.
Comparison of developed species-specific LAMP assay with species-specific PCR assay taken as gold standard
A total of ten DNA samples from cattle species were analysed using previously developed species-specific PCR assay by Kumar et al. (2012). The same DNA samples were also subjected to developed species-specific LAMP assay. The results of identification of specie origin of tissue by both the assay were compared.
Results
The concentration of genomic DNA from meat tissues ranged between 53.4 and 62.4 ng/µl and 37.4–51.3 ng/µl from meat admixture. The DNA pooled from raw meat and meat admixture sample was genomic DNA, which also included the mitochondrial DNA; hence it was used to amplify the mitochondrial sequences very well.
Based on the instant colour change of LAMP product after addition of SYBR green I dye, visualized by naked eye under day light and electrophoretic pattern of amplified LAMP product the temperature for LAMP reaction was optimized as extension at 64 °C for 45 min, followed by denaturation at 80 °C for 10 min. The optimized quantity and concentration of various components of LAMP reaction was as follows (Table 2):
Table 2.
Quantity and concentration of various components used in LAMP master mix
| Component | Concentration | Quantity (µl) |
|---|---|---|
| Thermopol buffer | 10 × | 2.5 |
| d-NTP mix | 10 mM | 1 |
| MgSO4 | 100 mM | 0.75 |
| Forward outer primer | (25 pM/µl) | 0.5 |
| Backward outer primer | (25 pM/µl) | 0.5 |
| Forward inner primer | (25 pM/µl) | 3 |
| Backward inner primer | (25 pM/µl) | 3 |
| Loop forward | (25 pM/µl) | 1 |
| Loop backward | (25 pM/µl) | 1 |
| Betaine | (5 M) | 3 |
| Bst DNA Polymerase | (6 Unit/µl) | 1 |
| Nuclease-free water | 5.75 | |
| DNA templet | (50 ng/µl) | 2 |
| Total | 25 | |
The result of LAMP reaction using cattle-specific LAMP primers set is presented in the form of instant colour change of LAMP amplified product after addition of SYBR Green dye I, visualized by naked eye under day light and electrophoretic ladder-like band pattern of amplified LAMP products in Fig. 1a, b. It was evident from the result that cattle LAMP primers set exclusively amplified the template DNA of cattle origin. Unchanged colour tubes and non-appearance of electrophoretic pattern on gel electrophoresis of products from these tubes, containing DNA template other than cattle, substantiated non amplification of their DNA template with used primer set. The electrophoretic pattern of amplified PCR product using cattle-specific outer LAMP primer set revealed amplification of cattle DNA to amplicon size of 206 bp and also a detection limit of 10 ng absolute DNA content for this PCR assay (Fig 2a, b).
Fig. 1.
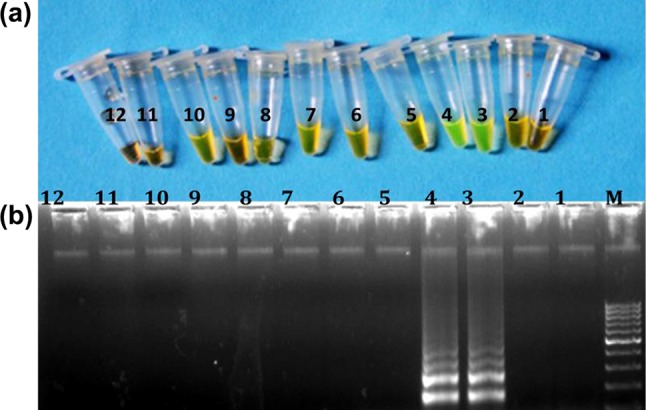
Specificity of the cattle-specific LAMP primer set. a Naked eye visualization of LAMP amplified product after addition of SYBER Green I dye under day light. b Electrophoretic pattern of LAMP amplified product. Lane M: 100 bp ladder (fermentas) 1–2: negative control, 3–4: cattle, 5–6: buffalo, 7–8: sheep, 9–10: goat and 11–12: pig
Fig. 2.
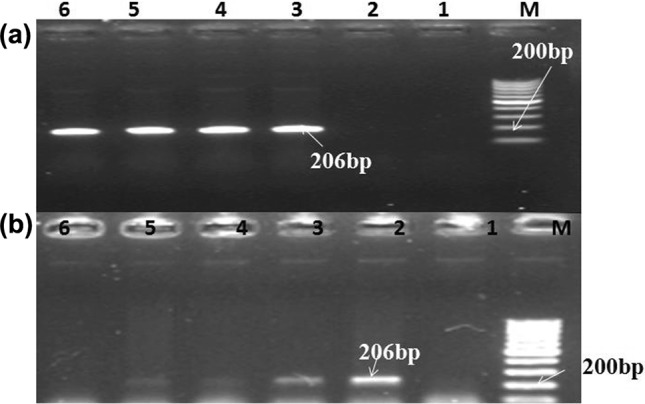
PCR verification and detection of cattle species using LAMP outer primers: a electrophoretic pattern of amplified products (206 bp) from PCR using SaP-BOVSARU/F3 and SaPBOV- SARU/B3 primer set. Lane M: 100 bp ladder (fermentas) 1–2: negative control, 3–6: cattle. b Detection limit of PCR using SaP-BOVSARU/F3 and SaPBOV- SARU/B3 primerset. Lane M : 100 bp ladder (fermentas) 1: negative control, 2: 100 ng, 3: 10 ng, 4: 1 ng, 5: 0.1ngand 6: 0.01 ng
The sensitivity result of cattle-specific LAMP assay using cattle-specific LAMP primer set is presented in the form of instant color change of LAMP amplified products after addition of SYBR Green I dye, visualized by naked eye under day light and electrophoretic ladder-like band pattern of LAMP amplified products in Fig. 3a, b. The results revealed that developed assay was able to the amplify DNA template from cattle up to the lowest absolute DNA content of 0.01 ng. There was no detectable amplification of template DNA below the concentration 0.01 ng.
Fig. 3.
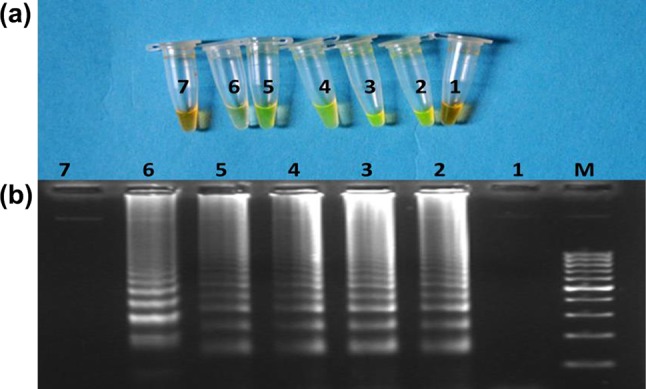
Sensitivity of the cattle-specific LAMP assay. a Naked eye visualization of LAMP amplified product after addition of SYBER Green I dye under day light. b Electrophoretic pattern of LAMP amplified product. Lane M:100 bp ladder fermentas, 1: negative control, 2: 100 ng, 3: 10 ng, 4:1 ng. 5: 0.1 ng, 6: 0.01 ng and 7: 0.001 ng
The results of LAMP reactions carried out on DNA templates from ten individuals of cattle and coded DNA samples from cattle/other species using species-specific LAMP assay revealed that developed species-specific LAMP amplification-based assay was useful in identifying the cattle origin of meat. The outcomes of LAMP reaction carried out on template DNA from meat admixture are represented in Fig. 4a, b. The change in colour of LAMP products after addition of SYBR Green I dye, visualized by naked eye under day light as well as ladder-like electrophoretic pattern of LAMP products on gel electrophoresis was observable up-to lowest concentration examined, i.e. 1% of cattle species in meat admixture. The results of LAMP reactions carried out with supernatant from Phire tissue direct PCR master mix treated tissues of cattle showed that designed species-specific LAMP primer set was capable of amplifying the DNA template present in supernatant. Comparison of developed species-specific LAMP assay with species-specific PCR assay taken as gold standard revealed the complete match, indicative of acceptability of developed assay, as better alternate for identification of species origin of tissue of cattle (Fig 5).
Fig. 4.
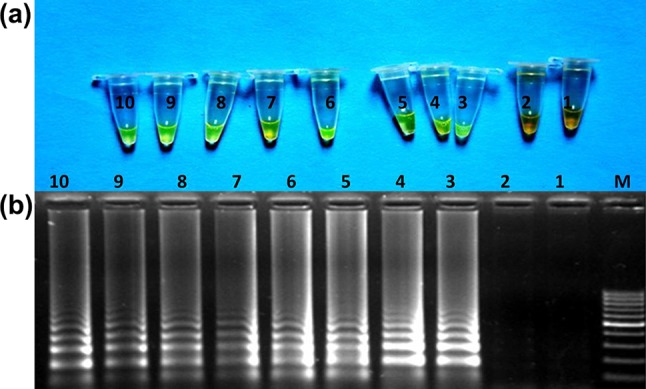
Laboratory validation of the LAMP assay on DNA samples from meat admixtures of cattle and other species using cattle-specific primer set. Naked eye visualization of LAMP amplified product after addition of SYBER Green I dye under day light. Electrophoretic pattern of LAMP amplified product. M :100 bp ladder (Fermentas) 1: negative control, 2: other species, 3–4: 20% cattle, 5–6: 10% cattle, 7–8: 5% cattle and 9–10: 1% cattle
Fig. 5.
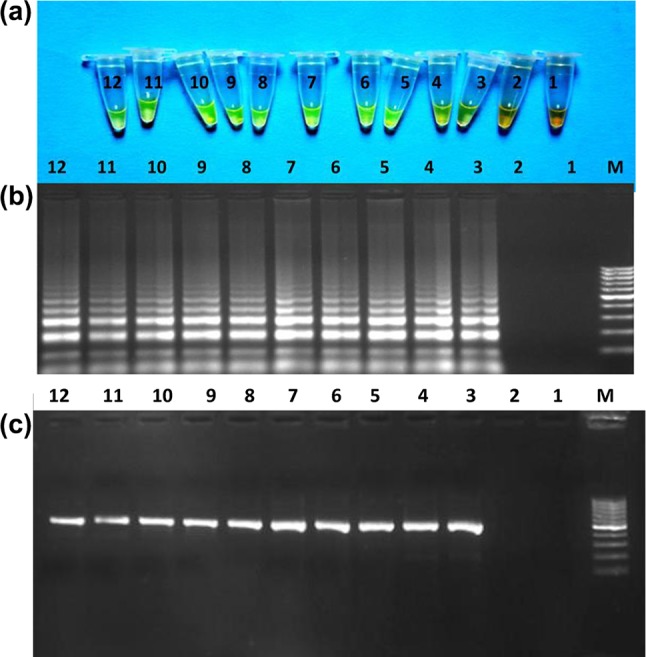
Comparison of developed species-specific LAMP assay with species-specific PCR assay taken as gold standard. a Naked eye visualization of LAMP amplified product after addition of SYBER Green I dye under daylight. b Electrophoretic pattern of LAMP amplified product. c Electrophoretic pattern of amplified products (477 bp) from PCR using cattle-specific LPT V/Cyt.B 03CF&13 CR primer set. Lane M: 100 bp ladder fermentas, 1–2: negative control and 3–12: samples from ten different cattle
Discussion
A notable high yield of genomic DNA was observed from meat samples using DNeasy® Tissue Kit (QIAGEN), which was sufficient to carry out LAMP reaction (Abdulmawjood et al. 2014). The OD260:280 of 1.9 to 2.1 reflected the absence of RNA contamination and others (Ghatak et al. 2013). The clear band without smearing in electrophoretic pattern of extracted DNA from meat/meat admixture samples was indicative of good quality DNA (Zhang et al. 2007). Inter species variations were also observed in concentration and quality of extracted DNA, but adequate enough from all the tissue samples for LAMP reaction. The amount and quality of extracted DNA from meat are reported to be affected by the species, breed, tissue, feeding condition and processing procedure (Li et al. 2006).
Isothermal amplification of template DNA using cattle-specific LAMP primer set in range of 58–68 °C with optimum at 64 °C was in accordance with finding of Ranjbar and Davoud (2015). Heat inactivation of activity of Bst DNA polymerase has been reported above 70 °C temperature, while at low temperatures binding ability gets affected adversely. Fernández-Soto et al. (2014) reported 100% activity of Bst DNA polymerase in the range of 60 °C–65 °C which reaches just 20% beyond 70 °C. Zhou et al. (2016) also found that Bst DNA polymerase dosage ranging from 6.0 to 8.0 U showed good results in LAMP reaction. Bst DNA polymerase is a Mg+ 2 dependent enzyme, which utilizes magnesium as a chelate with nucleotidyl di- or tri-phosphates or the NTP substrate and the metal cofactor serves as a mediator of phosphoryl or nucleotidyl transfer (Cowan 2002). Kil et al. (2015) has also found the optimum amplification of LAMP assay at concentration of 3–4 mM of MgSO4. Deoxynucleoside Triphosphate (dNTP) is source of nucleotides for amplification of target template DNA. Jeong et al. (2015) cited that the amplified LAMP products were obtained using a concentration of dNTPs ranging from 0.4 to 1.4 mM, but no reaction products were obtained at 0.2 mM. Betaine (N,N,N-trimethyl glycine) is routinely used in LAMP amplification to lower the melting temperature, increase the specificity and facilitate the separation of DNA templates (En et al. 2008). Previous studies revealed that 0.8 M betaine resulted in elevated sensitivity and increased the effectiveness of the LAMP assay (George et al. 2011 and; Nagdev et al. 2011). In the initial steps of the LAMP reaction, all four primers were used, but later during the cycling reaction of LAMP amplification, only the inner primers were used for strand displacement DNA synthesis. Liu et al. (2010) also reported that in LAMP reactions the concentrations of inner primer ratio are often higher (4–10 times) than outer primer. DNA-binding dyes, i.e, SYBR Green I, possess specific molecular structures that allow them to bind selectively to double-stranded DNA (dsDNA). SYBR Green I dye is widely used for analysis of LAMP amplified product which emits a weak fluorescence signal in the presence of single-stranded DNA (ssDNA), but emits strongly upon binding to double-stranded DNA (dsDNA). SYBR Green I dye was added to LAMP products at the end point of LAMP reaction due to its inhibitory effect on DNA polymerization at higher concentrations (Fishbach et al. 2015). All the positive LAMP reactions produced a ladder-like electrophoretic band pattern characteristic of the LAMP amplification and indicate that stem-loop DNA with inverted repeats of the target sequence was produced (Notomi et al. 2000).
DNA-based methods have raised interest as a useful tool in the identification of species in animal products and other food materials, due to the unique features of DNA, such as its structure remains conserved within all tissues of an individual (Girish et al. 2004; Saini et al. 2007). A number of researchers working for species discrimination in meat have targeted the mitochondrial DNA sequences because of its unique properties such as maternal inheritance (Rojas et al. 2011), multiple numbers of copies (approximately 1000 copies) than nuclear DNA (Koh et al. 2011), relatively high tolerance for environmental stresses such as heat, salt and pressure, 10-times higher mutation rate than in nuclear genes, rare possibility of recombination and high efficiency of amplification than nuclear markers (Rastogi et al. 2007). They act as naturally amplified source of genetic variation (Girish et al. 2004; Fajardo et al. 2006) and due to their stability attributes, simplicity, abundance in multiple copies per cells, additional protection given by the specialized mitochondrial membranes and short sequence (Ghovvati et al. 2009), increase the probability of positive result even in highly fragmented DNA (Mane et al. 2011). Specificity of LAMP amplification is higher than PCR assay because in LAMP reaction six primers are used as compared to two in PCR, which bind eight distinct regions on target DNA template. The cattle-specific LAMP primer set based on D loop gene sequence was found to be highly specific for cattle DNA template. The high specificity of cattle LAMP primer set in developed LAMP assays was due to well designing of LAMP primers and targeting of the variable region of mit. gene D loop. The observed specificity of the cattle-specific LAMP primer set without any cross-reactivity in other species was crucial of the fact that the set of six primers with eight binding site hybridized correctly and exclusively to their target gene sequence before DNA amplification occurred. The species-specific LAMP approach has been used earlier also for the identification of cattle tissue targeting diverse mitochondrial genes (Zahradnik et al. 2015; Li et al. 2017; Ma et al. 2016).
The intensity of colour of LAMP products after addition of SYBR Green I dye, visualized by naked eye under day light as well as electrophoretic band pattern of LAMP products on gel electrophoresis, diminished gradually with decrease in concentration of template DNA. LAMP assay is highly sensitive than conventional PCR, attributed to the fact that four primers bind to six distinct regions within a fairly small segment of a genome for amplification, with concentration higher than that used in traditional PCR methods. In addition to that LAMP reactions are also less prone to the presence of irrelevant DNA than conventional PCR (Notomi et al. 2000). Cho et al. (2014) designed LAMP assays targeting species-specific mitochondrial DNA to identify and discriminate eight meat species viz: cattle, pig, horse, goat, sheep, chicken, duck and turkey with limits of detection (LODs) from 10 pg/µL to 100 fg/µL levels in raw and cooked meat.A relatively lower limit of detection in present study might be due variation in DNA isolation, selection of tissues and its condition.Wang et al. (2014) also found that the sensitivity of the LAMP method was 10- to 1000-fold higher than conventional PCR or multiplex PCR.
Earlier researchers have also reported successful use of species-specific LAMP assay from mitochondrial DNA sequences for identification of cattle, pig, horse, goat, sheep, chicken, duck, and turkey (Cho et al. 2014), beef (Li et al. 2016) and pork (Lee et al. 2016) in admixed meat products .Successful LAMP reaction established the utility of supernatant from Phire tissue direct PCR master mix treated tissue of cattle as a source of DNA while using developed species-specific LAMP assay. LAMP amplified product after addition of SYBR Green I dye and electrophoretic band pattern of LAMP amplified product from direct LAMP assay were almost similar in results obtained with genomic DNA extracted from DNeasy® Tissue Kit (QIAGEN). Genomic DNA isolation from tissue/muscle using DNeasy® Tissue Kit (QIAGEN) was time-consuming process characterized by a lengthy incubation period with proteinase K to digest muscle/tissue followed by organic solvent extraction and centrifugation steps. Direct LAMP assay without prior DNA purification successfully amplified the DNA template present in supernatant, attributed to the fact that LAMP assay has higher amplification efficiency and Bst DNA polymerases used in assay are more tolerant to inhibitors present in supernatant. Lee et al. (2016) also applied direct real time LAMP assay to various processed meat products for identification of species origin of meat by using same protocol as Phire tissue direct PCR master mix.
The cost-effectiveness advantage of the LAMP assay could further strengthen its use as a tool for authentication of species origin of meat in developing countries where resources are limited. In the present study LAMP assay based on species-specific LAMP primers set was 100% similar to PCR assay based on species-specific primers. Ma et al. (2016), also reported similar match score for species-specific real-time LAMP assay for beef and mutton in comparison to real-time PCR.
Thus it was concluded that developed species- specific LAMP assay using species-specific primer set based on mt. D loop DNA sequence is quite effective in the identification of tissue of cattle origin and could identify the species origin of meat with even absolute DNA content of 0.01 ng. The assay is also very effective to identify the source of meat in unknown (coded) samples and various combinations of binary admixtures of meat samples.
Acknowledgements
The authors gratefully acknowledge Indian council of Agricultural research-Indian Veterinary Research Institute (IVRI) Izatnagar, India for providing necessary facilities and financial support to accomplish this research.
Author contribution
The authors’ contributions are as follows: Sarita Kumari: carried out the majority of the research work, DNA isolation and species-specific LAMP assay; RRK: responsible for the conception of the project, designed the study and planned the experiments, manuscript drafting. SKM and DK coordinated and carried out data interpretation, Preeti Rana: DNA isolation from diverse samples, field samples, Dhananjay Kumar and Jyoti Jawla: validation of findings by repeating the experiments. All authors read and approved this manuscript.
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical statement
The authors declare their commitment for scientific integrity and research ethics of reported research work and assure the absence of scientific fraud and misrepresentations, inappropriate manipulation of graphics files, redundant publications, and plagiarism in the manuscript.
Contributor Information
Sarita Kumari, Email: dr.sarita.bhamu@gmail.com.
Rajiv Ranjan Kumar, Phone: +915812311711, Phone: +919897299102, Email: dr_rajivranjan@yahoo.com.
Sanjod Kumar Mendiratta, Email: mendiratta_65@yahoo.co.in.
Deepak Kumar, Email: deep_biotek@yahoo.com.
Preeti Rana, Email: preetirana321@gmail.com.
Dhananjay Kumar, Email: ivridhananjay@gmail.com.
Jyoti Jawla, Email: jawlajyoti@gmail.com.
References
- Abdullahi UF, Igwenagu E, Aliyu S, Mu’azu A, Naim R, Wan-Taib WR. A rapid and sensitive Loop-mediated isothermal amplification assay for detection of pork DNA based on porcine tRNAlys and ATPase 8 genes. Int Food Res J. 2017;24(4):1357–1361. [Google Scholar]
- Abdulmawjood A, Grabowski N, Fohler S, Kittler S, Nagengast H, Klein G. Development of loop-mediated isothermal amplification (LAMP) assay for rapid and sensitive identification of ostrich meat. PLoS One. 2014;9(6):1007–1017. doi: 10.1371/journal.pone.0100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho AR, Dong HJ, Cho S. Meat species identification using loop-mediated isothermal amplification assay targeting species-specific mitochondrial DNA. Korean J Food Sci Anim Resour. 2014;34(6):799–807. doi: 10.5851/kosfa.2014.34.6.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan JA. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals. 2002;15(3):225–235. doi: 10.1023/A:1016022730880. [DOI] [PubMed] [Google Scholar]
- Doosti A, Dehkordi PG, Rahimi E. Molecular assay to fraud identification of meat products. J Food Sci Technol. 2014;51(1):148–152. doi: 10.1007/s13197-011-0456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- En FX, Wei X, Jian L, Qin C. Loop-mediated isothermal amplification establishment for detection of pseudorabies virus. J Virol Methods. 2008;151(1):35–39. doi: 10.1016/j.jviromet.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo V, González I, López-Calleja I, Martín I, Hernández PE, García T. PCR-RFLP authentication of meats from red deer (Cervuselaphus), fallow deer (Damadama), roe deer (Capreoluscapreolus), cattle (Bostaurus), sheep (Ovisaries), and goat (Capra hircus) J Agric Food Chem. 2006;54(4):1144–1150. doi: 10.1021/jf051766r. [DOI] [PubMed] [Google Scholar]
- Fajardo V, Gonzalez I, Martyn I, Rojas M, Hernandez PE, Garcya T. Real-time PCR for detection and quantification of red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) in meat mixtures. Meat Sci. 2008;79(2):289–298. doi: 10.1016/j.meatsci.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Fernández Soto P, Mvoulouga PO, Akue JP, Abán JL, Santiago BV, Sánchez MC. Development of a highly sensitive loop-mediated isothermal amplification (LAMP) method for the detection of Loa loa. PloS One. 2014;9(4):946–964. doi: 10.1371/journal.pone.0094664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach J, Xander NC, Frohme M, Glökler JF. Method summary. Biotechniques. 2015;58(4):189–194. doi: 10.2144/000114275. [DOI] [PubMed] [Google Scholar]
- George G, Mony P, Kenneth J. Comparison of the efficacies of loop-mediated isothermal amplification, fluorescence smear microscopy and culture for the diagnosis of tuberculosis. PloS One. 2011;6(6):210–217. doi: 10.1371/journal.pone.0021007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatak S, Muthukumaran RB, Nachimuthu SK. A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. J Biomol Tech JBT. 2013;24(4):224–231. doi: 10.7171/jbt.13-2404-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghovvati S, Nassiri MR, Mirhoseini SZ, Moussavi AH, Javadmanesh A. Fraud identification in industrial meat products by multiplex PCR assay. Food Control. 2009;20(8):696–699. doi: 10.1016/j.foodcont.2008.09.002. [DOI] [Google Scholar]
- Girish PS, Anjaneyulu ASR, Viswas KN, Anand M, Rajkumar N, Shivakumar BM. Sequence analysis of mitochondrial 12S rRNA gene can identify meat species. Meat Sci. 2004;66(3):551–556. doi: 10.1016/S0309-1740(03)00158-X. [DOI] [PubMed] [Google Scholar]
- Goto M, Honda E, Ogura A, Nomoto A, Hanaki KI. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxynaphthol blue. Biotechniques. 2009;46(3):167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- Hou B, Meng X, Zhang L, Guo J, Li S, Jin H. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci. 2015;101:90–94. doi: 10.1016/j.meatsci.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Jeong J, Cho SY, Lee WH, Lee K, Ju HJ. Development of a rapid detection method for Potato virus X by reverse transcription loop-mediated isothermal amplification. Plant Pathol J. 2015;31(3):219–225. doi: 10.5423/PPJ.OA.03.2015.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanaphum P, Maneenin S, Chaiyana W. Analysis of pork meat using loop mediated isothermal amplification (LAMP) to confirm halal status. Int J Biosci. 2014;4:62–68. [Google Scholar]
- Kil EJ, Kim S, Lee YJ, Kang EH, Lee M, Cho SH. Advanced loop-mediated isothermal amplification method for sensitive and specific detection of Tomato chlorosis virus using a uracil DNA glycosylase to control carry-over contamination. J Virol Methods. 2015;213:68–74. doi: 10.1016/j.jviromet.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Koh BRD, Kim JY, Na HM, Park SD, Kim YH. Development of species-specific multiplex PCR assays of mitochondrial 12S rRNA and 16S rRNA for the identification of animal species. Korean J Vet Serv. 2011;34(4):417–428. doi: 10.7853/kjvs.2011.34.4.417. [DOI] [Google Scholar]
- Kumar A, Kumar RR, Sharma BD, Mendiratta SK, Sharma D, Gokulakrishnan P. Species specific polymerase chain reaction (PCR) assay for identification of pig (Sus domesticus) meat. Afr J Biotech. 2012;11(89):15590–15595. doi: 10.5897/AJB12.1753. [DOI] [Google Scholar]
- Lee SY, Kim MJ, Hong Y, Kim HY. Development of a rapid on-site detection method for pork in processed meat products using real-time loop-mediated isothermal amplification. Food Control. 2016;66:53–61. doi: 10.1016/j.foodcont.2016.01.041. [DOI] [Google Scholar]
- Li YJ, Fan JY. Rapid visual identification of bovine meat by loop mediated isothermal amplification combined with immunochromatographic strip. BioChip J. 2017;11(1):8–13. doi: 10.1007/s13206-016-1102-y. [DOI] [Google Scholar]
- Li B, Bai S, Xu Y, Zhang W, Ma J. Identification of sika deer and red deer using partial cytochrome b and 12 s ribosomal RNA genes. J For Res. 2006;17(2):160–162. doi: 10.1007/s11676-006-0038-9. [DOI] [Google Scholar]
- Liu J, Huang C, Wu Z, Zhang X, Wang Y. Detection of tomato aspermy virus infecting chrysanthemums by LAMP. Sci Agric Sin. 2010;43(6):1288–1294. [Google Scholar]
- Ma B, Dai M, Fang J, Wu Y, Zhang M. Visual loop-mediated isothermal amplification (LAMP) method for identification bovine and ovine gene in animal foodstuff. Am J Food Technol. 2016;11:193–203. doi: 10.3923/ajft.2016.240.252. [DOI] [Google Scholar]
- Mane BG, Mendiratta SK, Tiwari AK, Sharma BD, Bhilegaokar KN, Anjaneyulu ASR. Detection of pork in admixed meat and meat products by species-specific PCR technique. Indian J Anim Sci. 2011;8:11–16. [Google Scholar]
- Mohindra V, Khare P, Lal KK, Punia P, Singh RK, Barman AS. Molecular discrimination of five Mahseer species from Indian peninsula using RAPD analysis. Acta Zool Sinica. 2007;53(4):725–732. [Google Scholar]
- Nagdev KJ, Kashyap RS, Parida MM, Kapgate RC, Purohit HJ, Taori GM. Loop-mediated isothermal amplification for rapid and reliable diagnosis of tuberculous meningitis. J Clin Microbiol. 2011;49(5):1861–1865. doi: 10.1128/JCM.00824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):63–69. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortea I, Pascoal A, Cañas B, Gallardo JM, Barros-Velázquez J, Calo-Mata P. Food authentication of commercially-relevant shrimp and prawn species: From classical methods to Foodomics. Electrophoresis. 2012;33(15):2201–2211. doi: 10.1002/elps.201100576. [DOI] [PubMed] [Google Scholar]
- Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18(6):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran G, Ren L, Han X, Liu X, Li Z, Pang D. Development of a rapid method for the visible detection of pork DNA in halal products by loop-mediated isothermal amplification. Food Anal Methods. 2016;9(3):565–570. doi: 10.1007/s12161-015-0246-z. [DOI] [Google Scholar]
- Ranjbar R, Afshar D. Development of a loop-mediated isothermal amplification assay for rapid detection of Yersinia enterocolitica via targeting a conserved locus. Iran J Microbiol. 2015;7(4):185–190. [PMC free article] [PubMed] [Google Scholar]
- Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, Shouche YS. Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007;76(4):666–674. doi: 10.1016/j.meatsci.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Rojas M, González I, Pavon MA, Pegels N, Hernández PE, García T. Mitochondrial and nuclear markers for the authentication of partridge meat and the specific identification of red-legged partridge meat products by polymerase chain reaction. Poult Sci. 2011;90(1):211–222. doi: 10.3382/ps.2010-00895. [DOI] [PubMed] [Google Scholar]
- Saini M, Das DK, Dhara A, Swarup D, Yadav MP, Gupta PK. Characterisation of peacock (Pavocristatus) mitochondrial 12S rRNA sequence and its use in differentiation from closely related poultry species. Br Poult Sci. 2007;48(2):162–166. doi: 10.1080/00071660701285897. [DOI] [PubMed] [Google Scholar]
- Spink J, Moyer DC. Backgrounder: defining the public health threat of food fraud. Minneap Min Natl Cent Food Prot Def. 2011;76(9):157–162. doi: 10.1111/j.1750-3841.2011.02417.x. [DOI] [PubMed] [Google Scholar]
- Tomlinson J, Boonham N. Potential of LAMP for detection of plant pathogens. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour. 2008;3(66):1–7. [Google Scholar]
- Wang X, Seo DJ, Lee MH, Choi C. Comparison of conventional PCR, multiplex PCR, and loop-mediated isothermal amplification assays for rapid detection of Arcobacter species. J Clin Microbiol. 2014;52(2):557–563. doi: 10.1128/JCM.02883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Fu S, Peng X, Li LE, Song T, Li L. Identification of pork in meat products using real-time loop-mediated isothermal amplification. Biotechnol Biotechnol Equip. 2014;28(5):882–888. doi: 10.1080/13102818.2014.963789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradnik C, Martzy R, Mach RL, Krska R, Farnleitner AH, Brunner K. Loop-mediated isothermal amplification (LAMP) for the detection of horse meat in meat and processed meat products. Food Anal Methods. 2015;8(6):1576–1581. doi: 10.1007/s12161-014-0072-8. [DOI] [Google Scholar]
- Zhang CL, Fowler MR, Scott NW, Lawson G, Slater A. A TaqMan real-time PCR system for the identification and quantification of bovine DNA in meats, milks and cheeses. Food Control. 2007;18(9):1149–1158. doi: 10.1016/j.foodcont.2006.07.018. [DOI] [Google Scholar]
- Zhou D, Wang C, Li Z, Chen Y, Gao S, Guo J. Detection of bar transgenic sugarcane with a rapid and visual loop-mediated isothermal amplification assay. Front Plant Sci. 2016;7:279–284. doi: 10.3389/fpls.2016.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]


