Abstract
The etiological agent of the neglected tropical disease African trypanosomiasis, Trypanosoma brucei, possesses an expanded and diverse repertoire of heat shock proteins, which have been implicated in cytoprotection, differentiation, as well as progression and transmission of the disease. Hsp70 plays a crucial role in proteostasis, and inhibition of its interactions with co-chaperones is emerging as a potential therapeutic target for numerous diseases. In light of genome annotations and the release of the genome sequence of the human infective subspecies, an updated and current in silico overview of the Hsp70/J-protein machinery in both T. brucei brucei and T. brucei gambiense was conducted. Functional, structural, and evolutionary analyses of the T. brucei Hsp70 and J-protein families were performed. The Hsp70 and J-proteins from humans and selected kinetoplastid parasites were used to assist in identifying proteins from T. brucei, as well as the prediction of potential Hsp70–J-protein partnerships. The Hsp70 and J-proteins were mined from numerous genome-wide proteomics studies, which included different lifecycle stages and subcellular localisations. In this study, 12 putative Hsp70 proteins and 67 putative J-proteins were identified to be encoded on the genomes of both T. brucei subspecies. Interestingly there are 6 type III J-proteins that possess tetratricopeptide repeat-containing (TPR) motifs. Overall, it is envisioned that the results of this study will provide a future context for studying the biology of the African trypanosome and evaluating Hsp70 and J-protein interactions as potential drug targets.
Electronic supplementary material
The online version of this article (10.1007/s12192-018-0950-x) contains supplementary material, which is available to authorized users.
Keywords: African trypanosomiasis, Trypanosoma brucei, Hsp70, J-protein, Hsp110
Introduction
African trypanosomiasis is a tropical disease endemic to 37 countries in sub-Saharan Africa, and those living in rural areas that depend of farming, fishing, and hunting are most at risk (WHO 2017). As the number of new cases of human African trypanosomiasis (HAT) has decreased substantially over the decades, this neglected tropical disease is targeted for eradication by 2020 (WHO 2017). The etiological agent of this disease is an extracellular blood- and tissue-borne protozoan parasite Trypanosoma brucei. T. brucei is comprised of three subspecies: Trypanosoma brucei brucei (T. b. brucei), Trypanosoma brucei gambiense (T. b. gambiense), and Trypanosoma brucei rhodesiense (T. b. rhodesiense). T. b. brucei is responsible for animal African trypanosomiasis (AAT), also known as Nagana. T. b. gambiense causes HAT and gives rise to a chronic infection and represents over 90% of reported cases in Central and Western Africa (Simarro et al. 2008), whilst T. b. rhodesiense is mainly a zoonotic disease responsible for less than 10% of reported cases of HAT in Eastern and Southern Africa and causes an acute infection, which is rapidly fatal if untreated (Brun et al. 2010). Tools for controlling the parasitic disease are limited, due to the inability to develop a vaccine, toxicity of existing drugs, and the development of parasitic resistance (Barrett and Croft 2012). The completion of the genome sequence for T. b. brucei in 2005 and the subsequent completion of other kinetoplastid genomes have facilitated transcriptome and proteome analyses. This has led to a renewed interest in the discovery of new targets and approaches for anti-trypanosomatid drugs (Wenzler et al. 2016). Amongst the potential new drug targets, heat shock proteins represent an interesting group already validated in other disease areas (Shrestha et al. 2016).
Heat shock protein 70 (Hsp70) is a prominent protein chaperone family involved in a plethora of essential cellular functions, which include but are not limited to promoting the correct protein folding of newly synthesized polypeptides and mediating protein translocation, quality control and degradation (Bukau and Horwich 1998; Agarraberes and Dice 2001; Mayer and Bukau 2005). The functional cycle of the Hsp70 chaperone system is nucleotide-dependent and regulated by co-chaperones, such as the J-protein family and nucleotide exchange factors (NEFs). Common to all J-proteins is the possession of a conserved ~ 70-amino acid region known as the J-domain (Cheetham and Caplan 1998; Craig et al. 2006; Kampinga and Craig 2010). Apart from the J-domain, members of this co-chaperone family contain a wide variety of domains, which have been used as the basis for classification of members into four classes (I–IV) (Cheetham and Caplan 1998; Botha et al. 2007). Nucleotide exchange factors (NEFs) are a group of co-chaperones that facilitate the release of the bound substrate by accelerating ADP release that essentially primes Hsp70 for the start of the next cycle (Brehmer et al. 2001; Dragovic et al. 2006). Hsp110s are one of the major eukaryotic HSPs and are divergent members of the Hsp70 family (Easton et al. 2000). Hsp110s have been shown to be potent NEFs for Hsp70 (Dragovic et al. 2006; Raviol et al. 2006), though some Hsp110 homologues have be shown to be able to bind substrate and prevent aggregation by functioning as “holdases” (Polier et al. 2008). Thus, Hsp110s possess dual roles, as chaperones and as co-chaperones of Hsp70.
The lifecycle of T. brucei is complex as these parasites must transition between two strikingly different hosts, a cold-blooded arthropod vector and a warm-blooded mammalian host. Thus, the infectious cycle results in sudden changes in growth conditions, and exposure of the parasites to a wide variety of environmental stresses (Jones et al. 2008). The Hsp70/J-protein chaperone machinery is an integral component of the heat shock response and has been found to be conserved across organisms (Boorstein et al. 1994). However, the evolution of the Hsp70 protein family has been shown to be dynamic and highly adapted to species-specific constraints (Drini et al. 2016). This has been documented by substantial variation in Hsp70 gene copy number (Daugaard et al. 2007), phylogenetically distinct subfamilies, and the evolution of atypical protein members (Hughes 1993; Boorstein et al. 1994; Gupta and Singh 1994; Kampinga and Craig 2010; Kominek et al. 2013).
A post-genomic analysis of the molecular chaperone complements in the Tritryps, T. brucei, T. cruzi, and Leishmania major (L. major), revealed an unprecedented expansion in J-protein, Hsp70 and Hsp60 complements, indicating that these protein families may play a critical role in kinetoplastid biology (Folgueira and Requena 2007). A review of the Hsp70 superfamily in the Tritryps by Louw et al. (2010a) revealed that the protein family possessed atypical Hsp70 members and features. Subsequent reviews and updated in silico analyses of Hsp70/J-protein machinery in the annotated genome sequences of intracellular kinetoplastid parasites has been conducted (Shonhai et al. 2011; Urmenyi et al. 2014; Requena et al. 2015). However, this had not been the case for the extracellular parasite, T. brucei.
Proteomic studies have compared protein expression between lifecycle stages (Gunasekera et al. 2012; Urbaniak et al. 2012; Butter et al. 2013), including the phosphoproteome (Nett et al. 2009; Urbaniak et al. 2013). The proteome of the mitochondria is available (Panigrahi et al. 2009), including the importome (Peikert et al. 2017), respiratome (Acestor et al. 2011), and mitochondrial membranes (Acestor et al. 2009). The nuclear (Goos et al. 2017), nuclear pore (DeGrasse et al. 2008) and glycosome proteomes (Colasante et al. 2006; Güther et al. 2014) have been analysed. Proteomic data is also available for the flagellum (Broadhead et al. 2006; Subota et al. 2014) and cell surface (Shimogawa et al. 2015). Considering the medical importance of these parasites, as well as the large amount of proteomic data available, an updated investigation is timely and appropriate for Hsp70 and J-proteins in trypanosomes. This study aimed to provide an updated overview of the Hsp70/J-protein chaperone machinery in T. brucei, with respect to both T. b. brucei and T. b. gambiense. The availability of the T. b. gambiense genome sequence enabled the determination and comparative analysis of the Hsp70/J-protein chaperone machinery. Other kinetoplastids included in this study were the non-parasitic Bodo saltans (Deschamps et al. 2011) and the insect-infecting Crithidia fasciculata (reviewed in Wallace 1966). The Hsp70 and J-protein families from humans and selected kinetoplastid parasites were used to assist in identifying all T. brucei Hsp70 and J-protein members, potential Hsp70–J-protein partnerships, and the inference of the cellular function of individual members and potential partnerships. African trypanosomiasis is a tropical disease that afflicts both humans and livestock. Overall, it is envisioned that the results of this study will provide a future context for studying the biology of the African trypanosome.
Materials and methods
Database mining, sequence analyses, and the determination of kinetoplastid and human orthologues
A BLASTP search using the Hsp70 proteins from T. b. brucei obtained from previous in silico studies (Folgueira and Requena 2007; Louw et al. 2010a) and human HSPA1A as queries on the TriTrypDB (version 35) database (http://tritrypdb.org/tritrypdb/; Aslett et al. 2010) was conducted in order to determine the Hsp70 superfamily encoded on the T. b. gambiense genome, as well as identify new T. b. brucei Hsp70 protein members. The e-value was set at an intermediately stringent level of e-10 for collecting as many potential Hsp70-related sequences for further analysis. Additionally, a keyword search was performed to scan the genome of T. b. gambiense for Hsp70 genes on the TriTrypDB database using the terms: “Hsp70”, “heat shock protein”, and “molecular chaperone”. The retrieved amino acid sequences from the various keyword searches were then screened for the Hsp70 domain using SMART 7 (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/; Letunic et al. 2012) and PROSITE (http://prosite.expasy.org/; Sigrist et al. 2009).
A search in the annotated T. b. brucei and T. b. gambiense genome sequences on the TriTrypDB database for proteins containing the J-domain in their amino acid sequence was conducted using the J-domain (1-77aa) from Escherichia coli (E. coli) DnaJ (EcDnaJ; NP_308042.1) as a query in a pBLAST search. The common denominator for all J-proteins is the possession of a J-domain (Cheetham and Caplan 1998), and all J-proteins are divided into the four-type classes based on their structural homology to E. coli DnaJ (Cheetham and Caplan 1998; Botha et al. 2007). The keyword search using the terms “Hsp40”, “DnaJ”, “Heat shock protein”, and “molecular chaperone” were also conducted to scan the genome of T. b. brucei and T. b. gambiense for J-protein genes on the TriTrypDB database. The retrieved amino acid sequences from the various keyword searches were screened using SMART 7 (http://smart.embl-heidelberg.de/; Letunic et al. 2012) and PROSITE (http://prosite.expasy.org/; Sigrist et al. 2009) for the presence of a J-domain.
For the identification of human and selected kinetoplastid orthologues, reciprocal BLASTP was conducted. In the first query, the putative amino acid sequences of the Hsp70 and J-proteins from both T. brucei subspecies were used as queries in a BLASTP search on the National Centre for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov), using the default parameters. The amino acid sequences of the putative orthologues were then used as second queries in BLASTP searches using default parameters on the TriTrypDB database. If the most similar orthologue in the T. brucei subspecies was exactly the Hsp70 or J-protein sequence used as the first query, the sequence of the second query was selected as an orthologue.
Phylogenetic analyses
Phylogenetic trees were constructed to analyse the phylogenetic relationship of the Hsp70 and J-protein complements in both T. brucei subspecies. The type III J-protein subfamily was omitted from the phylogenetic analysis, as the subfamily is diverse with regard to amino acid composition and protein length, with the only common feature being the J-domain. The full-length amino acid sequences for the Hsp70 superfamily and the selected J-protein subfamilies in the selected kinetoplastid parasites were obtained from the TriTrypDB database (Aslett et al. 2010), and the human and C. fasciculata protein sequences were obtained from the National Centre for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov). Partial amino sequences were omitted from the analysis. Accession numbers for the Hsp70, Hsp110, and J-protein sequences used in this study are provided in Tables 1 and 2 and in the supplementary data, Table S1 and S2. Multiple sequence alignments were performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters in MEGA 7 (Kumar et al. 2016), and are provided in the supplementary data, Fig. S1 and S4. Maximum-likelihood (ML) was utilized to find the best model of evolution and was selected by the Bayesian Information Criterion (BIC) implemented in MEGA 7. The amino acid–based Hsp70 and J-protein ML phylogeny was reconstructed using the JTT (Jones–Taylor–Thornton) model matrix (Jones et al. 1992) with gamma distribution shape parameter (G). Maximum likelihood phylogenetic trees were constructed using MEGA 7.0 (Le and Gascuel 2008). The accuracy of the reconstructed trees was assessed using a bootstrap test using 1000 replicates with a pairwise gap deletion mode. The phylogenetic trees for Hsp70/HSPA, Hsp110/HSPH, and J-proteins were unrooted.
Table 1.
The Hsp70/HSPA and Hsp110/HSPH proteins from Trypanosoma brucei with their putative T. cruzi, L. major, C. fasciculata, B. saltans, and H. sapiens orthologues
| T. brucei | T. cruzic | L. major | C. fasciculata | B. saltans | H. sapiens | |||
|---|---|---|---|---|---|---|---|---|
| Namea | Gene IDb | Gene IDb | Gene IDb | Gene IDb | Gene IDb | Gene IDb | Localisationd | Reference |
| A: Hsp70/HSPA | ||||||||
| Hsp70 | Tb927.11.11330 Tbg972.11.12660 | TcCLB.511211.170 Tc_MARK_1461 | LmjF.28.2770 | CFAC1_300044700 CFAC1_300044800 | ACI15927 | HSPA1A HSPA1B | CYT, NUC, GLYCO, FLAGELLAR, CELL SURFACE (BSF, PF) | Güther et al. 2014 |
| Subota et al. 2014 | ||||||||
| Shimogawa et al. 2015 | ||||||||
| Hsp70.4 | Tb927.7.710 Tbg972.7.640 | TcCLB.503721.39 TcCLB.511257.10 TCDM_01248 Tc_MARK_1012 | LmjF.26.1240 | CFAC1_290020800 | CUG93112 | HSPA2? | CYT, NUC, CELL SURFACE (PF) | Shimogawa et al. 2015 |
| Hsp70.c | Tb927.11.11290 Tbg972.11.12620 | TcCLB.511211.220 TCDM_07862 Tc_MARK_1466 | LmjF.28.2820 | CFAC1_300045200 | CUG86635 | HSPA6? | CYT, NUC | Goos et al. 2017 |
| GRP78A GRP78B | Tb927.11.7460 Tb927.11.7510 Tbg972.11.8650 Tbg972.11.8690 | TcCLB.506585.40 TCDM_08367 Tc_MARK_6525 Tc_MARK_6614 | LmjF.28.1200 | CFAC1_300021300 | CUE68699 CUG90530 | HSPA5 | ER | Bangs et al. 1993 |
| MtHsp70A MtHsp70B MtHsp70C | Tb927.6.3740 Tb927.6.3750 Tb927.6.3800 Tbg972.6.3510 Tbg972.6.3520 Tbg972.6.3580 | TcCLB.507029.30 Tc_MARK_1997 Tc_MARK_2001 Tc_MARK_2002 | LmjF.30.2460 LmjF.30.2470 LmjF.30.2480 LmjF.30.2490 LmjF.30.2550 | CFAC1_260048700 CFAC1_260048800 CFAC1_260049400 | ACI15928 | HSPA9 | MITO, CELL SURFACE (BSF, PF) | Panigrahi et al. 2009 |
| Niemann et al. 2013 | ||||||||
| Shimogawa et al. 2015 | ||||||||
| B: Hsp110/HSPH | ||||||||
| Hsp110 | Tb927.10.12710 Tbg972.10.15330 | TcCLB.507831.60 TCDM_05266 Tc_MARK_4982 | LmjF.18.1370 | CFAC1_140023200 | CUG91811 | HSPH1 | CYT, CELL SURFACE (PF) | Shimogawa et al. 2015 |
| Grp170 | Tb927.9.9860 Tbg972.9.5670 | TcCLB.506885.440 TcCLB.508457.20 TCDM_ 08776 Tc_MARK_4314 | LmjF.35.4710 | CFAC1_300098600 | CUG89677 | HSPH4 | ER | Field et al. 2010 |
| Hsp70.a | Tb927.9.4500 Tbg972.9.2380 | TcCLB.511585.70 TcCLB.510155.70 TCDM_06223 Tc_MARK_2135 | LmjF.01.0640 | CFAC1_050012600 | CUF98600 | – | ER, MITO | Field et al. 2010 |
| Acestor et al. 2009; | ||||||||
| Panagrahi et al. 2009 | ||||||||
| Peikert et al. 2017 | ||||||||
| Niemann et al. 2013 | ||||||||
| Hsp70.b | Tb927.7.1030 Tbg972.7.980 | TcCLB.503899.10 TCDM_01297 Tc_MARK_1045 | LmjF.26.0900 | CFAC1_290016600 | – | – | MITO | Peikert et al. 2017 |
| Niemann et al. 2013 | ||||||||
CYT cytosol, MITO- mitochondrion, NUC- nucleus, ER- endoplasmic reticulum, GYLCO- glycosomes, BSF- bloodstream form, PF- procyclic form
aThe nomenclature for the Hsp70/HSPA and Hsp110/HSPH proteins from T. b. brucei and T. b. gambiense were derived according to Folgueira and Requena (2007)
bThe gene IDs for the members of the T. b. brucei, T. b. gambiense, T. cruzi, C. fasciculata, and L. major Hsp70/HSPA and Hsp110/HSPH protein family were retrieved from the TriTrypDB database (http://tritrypdb.org/tritrypdb/; Aslett et al. 2010). The Gene IDs for the members of the B. saltans and H. sapiens Hsp70/HSPA and Hsp110/HSPH protein family were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/)
cThe gene IDs for the orthologues, identified by reciprocal BLASTP analysis, of three strains of T. cruzi are listed. T. cruzi CL Brener Esmeraldo-like (TcCLB), T. cruzi Dm28c (TCDM), and T. cruzi marinkelli strain B7 (Tc_MARK)
dSubcellular localizations for the T. brucei J-proteins were either predicted using the online prediction servers and/or determined using various proteomic datasets listed in the materials and methods
Table 2.
The J-proteins from Trypanosoma brucei with their putative T. cruzi, L. major, C. fasciculata, B. saltans, and H. sapiens orthologues
| T. bruceia | T. cruzicd | L. majorc | C. fasciculatac | B. saltansc | H. sapiensc | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type | Gene IDb | Name | Gene IDb | Gene IDb | Gene IDb | Gene IDb | Name | Localisatione | Functionf | Reference |
| I | Tb927.2.5160 Tbg972.2.3190 | J2 | TcCLB.511627.110 Tc_MARK_9726 TCDM_08977 | LmjF.27.2400 | CFAC1_210029700 | CUG85902 | DnaJA1 | CYT | Protein aggregation and refolding | Ludewig et al. 2015 |
| Terada et al. 2005 | ||||||||||
| Ahrendt and Braun 2010 | ||||||||||
| Burger et al. 2014 | ||||||||||
| Tb927.10.2290 Tbg972.10.2780 | J3 | TcCLB.511367.138 Tc_MARK_5591 TCDM_04677 | LmjF.21.0490 | CFAC1_260011100 | CUE68025 | – | CYT | Protein aggregation and refolding | Ashburner et al. 2000 | |
| Tb927.9.8410 Tbg972.9.4590 | J27 | TcCLB.510243.30 Tc_MARK_5901 TCDM_05671 | LmjF.04.0940 | CFAC1_030019200 | CUE72942 | – | MITO | Protein aggregation and folding mtDNA maintenance | Acestor et al. 2009, 2011 | |
| Panigrahi et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.11.16740 Tbg972.11.18880 | J45 | TcCLB.511531.9 Tc_MARK_2746 TCDM_02688 | LmjF.32.3300 | CFAC1_300054500 | CUG88240 | – | ER MITO | ERAD | Schlenstedt et al. 1995 | |
| Panigrahi et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.3.1430 Tbg972.3.1180 | J46 | TcCLB.509437.40 Tc_MARK_5358 TCDM_10337 | LmjF.25.1100 | CFAC1_220020000 | CUG87582 | DnaJB11 | ER | Protein folding mRNA editing | Jin et al. 2009 | |
| Tb927.9.12730 Tbg972.9.7790 | J50 | TcCLB.510743.100 Tc_MARK_4474 TCDM_00202 | LmjF.35.2980 | CFAC1_300079900 | CUG89607 | DnaJA3 | MITO | Protein aggregation and folding mtDNA maintenance | Týč et al. 2015 | |
| Acestor et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| II | Tb927.10.8540 Tbg972.10.10330 | J6 | TcCLB.506355.50 Tc_MARK_3436 TCDM_11062 | LmjF.36.6270 | CFAC1_280070900 | CUG94252 | DnaJB1 | CYT | Protein (re)folding | Freeman and Morimoto 1996 |
| Tb927.11.15130 Tbg972.11.17000 | J7 | TcCLB.509157.80 Tc_MARK_2851 TCDM_10924 | LmjF.32.1940 | CFAC1_190031300 | CUG94211 | – | MITO FLAGELLAR | ? | Acestor et al. 2009 | |
| Panigrahi et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Subota et al. 2014 | ||||||||||
| III | Tb927.11.16980 Tbg972.11.19710 | J1 | TcCLB.511537.50 Tc_MARK_2772 TCDM_02646 | LmjF.32.3030 | CFAC1_300057600 | CUE71254 | – | CYT, NUC | Protein (re)folding | Louw et al. 2010b |
| Tb927.10.5660 Tbg972.10.6880 | J5 | TcCLB.504163.100 Tc_MARK_8575 TCDM_01101 | LmjF.36.1330 | CFAC1_250022200 | CUG21319 | – | MITO | Protein folding Protein import-MITO | Panigrahi et al. 2009 | |
| Niemann et al. 2013 | ||||||||||
| Tb927.9.8410 Tbg972.9.4590 | J8 | TcCLB.510243.30 Tc_MARK_5901 TCDM_05671 | LmjF.04.0940 | CFAC1_210013600 | CUE72942 | DnaJC15? | MITO, GLYCO | Protein aggregation and folding mtDNA maintenance | Hatle et al. 2013 | |
| Acestor et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Güther et al. 2014 | ||||||||||
| Tb927.6.3850 Tbg972.6.3630 | J9 | TcCLB.511517.44 Tc_MARK_2007 TCDM_02176 | – | CFAC1_140009800 | ACI15980 | – | MITO | ? | Peikert et al. 2017 | |
| Tb927.7.6660 Tbg972.7.760 | J10 | TcCLB.510055.140 Tc_MARK_7736 TCDM_09211 | LmjF.17.0460 | CFAC1_090011100 | CUG14142 | – | MITO | Acestor et al. 2009 | ||
| Peikert et al. 2017 | ||||||||||
| Tb927.9.8160 Tbg972.9.4440 | J11 | TcCLB.510241.70 Tc_MARK_5912 TCDM_05687 | LmjF.04.0780 | CFAC1_030017200 | CUG91719 | – | MITO | Acestor et al. 2009 | ||
| Niemann et al. 2013 | ||||||||||
| Tb927.4.2970 Tbg972. 4.2970 | J12 | TcCLB.506435.50 Tc_MARK_2622 TCDM_03557 | – | CFAC1_230048800 | CUG94092 | – | MITO | ? | – | |
| Tb927.10.12530 Tbg972.10.12530 | J13 | TcCLB.509809.20 Tc_MARK_4997 | LmjF.18.1490 | CFAC1_140024700 | CUG91826 | DnaJC24? | CYT, NUC | Dipthamide synthesis | Liu et al. 2004 | |
| Tb927.5.2880 Tbg972.5.4050 | J14 | TcCLB.507063.180 | LmjF.08.0990 | CFAC1_200014300 | CUG33795 | – | CYT, NUC | ? | – | |
| Tb927.10.14730 Tbg972.10.17890 | J15 | TcCLB.503833.20 Tc_MARK_4789 TCDM_00497 | LmjF.19.0080 | CFAC1_170007200 | CUG02245 | – | CYT | ? | Nett et al. 2009 | |
| Tb927.1.1960 Tbg972.1.1130 | J16 | TcCLB.506473.20 Tc_MARK_6047 TCDM_00922 | LmjF.20.1130 | CFAC1_180019100 | CUI12704 | DnaJC2 | CYT | Translation | Otto et al. 2005 | |
| Nett et al. 2009 | ||||||||||
| Tb927.1.4300 Tbg972.1.2850 | J17 | TcCLB.506529.260 Tc_MARK_5675 TCDM_05005 | LmjF.12.1110 | CFAC1_010014800 | CUG89438 | – | CYT, NUC | ? | – | |
| Tb927.11.1010 Tbg972.11.1070 | J18 | TcCLB.506925.470 Tc_MARK_6358 TCDM_01954 | LmjF.27.0410 | CFAC1_230048800 | CUG94092 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Acestor et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.4.650 Tbg972.4.400 | J19 | TcCLB.507053.120 Tc_MARK_2469 | LmjF.34.4080 | CFAC1_290072000 | CUI15604 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.10.5040 Tbg972.10.6100 | J20 | TcCLB.510293.50 Tc_MARK_8503 TCDM_01182 | LmjF.36.0610 | CFAC1_250013400 | CUG62628 | – | MEM | Protein import | Ashburner et al. 2000 | |
| Tb927.7.540 Tbg972.7.420 | J21 | TcCLB.506287.90 Tc_MARK_965 TCDM_8503 | LmjF.26.1410 | CFAC1_290023200 | CUI15434 | – | MITO, GLYCO | Protein import | Panigrahi et al. 2009 | |
| Güther et al. 2014 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.10.6610 Tbg972.10.8080 | J22 | TcCLB.510187.330 Tc_MARK_3742 TCDM_06560 | LmjF.36.2110 | CFAC1_250032000 | CUG93498 | – | MITO, GLYCO | ? | Niemann et al. 2013 | |
| Zíková et al. 2017 | ||||||||||
| Güther et al. 2014 | ||||||||||
| Tb927.10.13830 Tbg972.10.17000 | J23 | TcCLB.507993.30 Tc_MARK_4871 | LmjF.18.0330 | CFAC1_140009800 | ACI15980 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Acestor et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.6.3120 Tbg972.6.2910 | J24 | TcCLB.511751.60 Tc_MARK_1932 | LmjF.30.1790 | CFAC1_260039300 | CUG86810 | – | CYT, NUC | ? | – | |
| Tb927.7.680 Tbg972.7.590 | J25 | TcCLB.506289.74 Tc_MARK_979 TCDM_07562 | LmjF.26.1270 | CFAC1_290021600 | CUG93121 | – | MITO | ? | Panigrahi et al. 2009 | |
| Niemann et al. 2013 | ||||||||||
| Tb927.7.6200 Tbg972.7.7170 | J26 | TcCLB.506513.30 Tc_MARK_7763 | LmjF.17.0040 | CFAC1_090006000 | CUG74938 | – | MITO, GLYCO | Protein import-MITO | Ashburner et al. 2000 | |
| Acestor et al. 2009 | ||||||||||
| Güther et al. 2014 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.7.740 Tbg972.7.670 | J28 | TcCLB.506135.40 TCDM_01252 | LmjF.26.1200 | CFAC1_290020200 | CUG93098 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.11.5710 Tbg972.11.6430 | J29 | TcCLB.506203.50 Tc_MARK_311 TCDM_00777 | LmjF.24.1080 | CFAC1_210020200 | CUF82364 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Tb927.8.1010 Tbg972.8.590 | J30 | TcCLB.508569.120 Tc_MARK_8237 TCDM_01742 | LmjF.07.0780 | CFAC1_080017400 | CUI14395 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.3.2290 Tbg972.3.2270 | J32 | TcCLB.508479.280 Tc_MARK_5111 TCDM_08321 | LmjF.25.2190 | CFAC1_230010600 | CUG43629 | – | ER | ? | Field et al. 2010 | |
| Tb927.10.9840 Tbg972.10.12010 | J33 | TcCLB.504147.60 Tc_MARK_2256 TCDM_02891 | LmjF.36.4470 | CFAC1_280050000 | CUG72683 | – | CYT | ? | – | |
| Tb927.9.10010 Tbg972.9.5780 | J34 | TcCLB.506887.90 Tc_MARK_4329 TCDM_00009 | LmjF.35.4630 | CFAC1_300097800 | CUG92668 | – | ER, MITO | Goldshmidt et al. 2008 | ||
| Acestor et al. 2009 | ||||||||||
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.7.4590 Tbg972.7.5200 | J35 | TcCLB.506605.150 Tc_MARK_7879 TCDM_05890 | LmjF.14.0110 | CFAC1_110006300 | CUF84499 | – | MITO | ? | Panigrahi et al. 2009 | |
| Tb927.3.1760 Tbg972.3.1610 | J36 | TcCLB.510091.50 Tc_MARK_5057 TCDM_03945 | LmjF.25.1690 | CFAC1_230016200 | – | DnaJC20 | MITO | FeS cluster biogenesis | Mokranjac et al. 2003 | |
| Uhrigshardt et al. 2010 | ||||||||||
| Acestor et al. 2009 | ||||||||||
| Tb927.10.12640 Tbg972.10.15230 | J37 | TcCLB.503455.10 Tc_MARK_4988 Tc_MARK_4989 TCDM_05256 | LmjF.18.1430 | – | CUG91818 | – | CYT, NUC | ? | – | |
| Tb927.6.3730 Tbg972.6.3500 | J38 | TcCLB.506941.270 | LmjF.30.2450 | CFAC1_140023800 | CUF23573 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.9.14180 Tbg972.9.8890 | J39 | TcCLB.510759.134 Tc_MARK_4558 TCDM_03405 | – | CFAC1_250032000 | – | – | MITO | ? | Niemann et al. 2013 | |
| Tb927.8.4470 Tbg972.8.4230 | J40 | TcCLB.508989.60 | LmjF.10.1050 | CFAC1_040019100 | CUG90885 | – | MITO | ? | Peikert et al. 2017 | |
| Tb927.4.3980 Tbg972.4.4110 | J41 | – | – | CFAC1_270012300 | CUG89969 | – | CYT, GOLGI | Intracellular trafficking | Ashburner et al. 2000 | |
| Tb927.10.12380 Tbg972.10.14920 | J42 | TcCLB.507625.110 Tc_MARK_5009 TCDM_09879 | LmjF.18.1650 | CFAC1_140028800 | – | – | CYT | Protein folding | Ashburner et al. 2000 | |
| Tb927.9.10950 Tbg972.9.6470 | J43 | TcCLB.508461.240 Tc_MARK_4379 TCDM_00080 | LmjF.35.4040 | CFAC1_300054500 | – | – | CYT | ? | – | |
| Tb927.8.7010 Tbg972.8.7230 | J44 | TcCLB.509911.100 Tc_MARK_1523 | LmjF.31.3100 | CFAC1_270068300 | CUE89851 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.8.1030 Tbg972.8.610 | J48 | TcCLB.508569.140 Tc_MARK_8239 TCDM_01739 | LmjF.31.3100 | – | – | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.6.2480 Tbg972.8.2240 | J49 | TcCLB.509965.229 TCDM_01497 | LmjF.30.1030 | CFAC1_260030200 | CUG89958 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Panigrahi et al. 2009 | ||||||||||
| Peikert et al. 2017 | ||||||||||
| Tb927.4.2220 Tbg972.4.2160 | J51 | TcCLB.506559.430 Tc_MARK_452 TCDM_04700 | LmjF.34.2430 | CFAC1_290053200 | CUG93032 | – | CYT | Protein folding | Ashburner et al. 2000 | |
| Tb927. 10.4900 Tbg972.10.5950 | J52 | TcCLB.504203.60 Tc_MARK_8493 TCDM_01195 | LmjF.36.0500 | CFAC1_250012000 | CUG91204 | DnaJC7 | CYT | Protein folding quality control | Brychzy et al. 2003 | |
| Tb927.7.3630 Tbg972.7.4040 | J53 | TcCLB.510407.80 Tc_MARK_8808 TCDM_06728 | LmjF.14.1330 | CFAC1_110026000 | CUG92395 | DnaJC3 | ER, MITO, GLYCO | ER protein synthesis | Yan et al. 2002 | |
| Goodman et al. 2011 | ||||||||||
| Güther et al. 2014 | ||||||||||
| Tb927.11.8420 Tbg972.11.9590 | J55 | TcCLB.506931.30 | LmjF.28.1270 | CFAC1_300022100 | CUF97845 | – | SEC | ? | – | |
| Tb927.2.3960 Tbg972.2.2180 | J56 | TcCLB.511109.90 Tc_MARK_9638 TCDM_14441 | LmjF.33.2690 | CFAC1_210037700 | – | DnaJB3? | NUC | Protein folding | Ashburner et al. 2000 | |
| Tb927.11.5920 Tbg972.11.6680 | J58 | TcCLB.508515.120 Tc_MARK_6411 TCDM_00738 | LmjF.24.1300 | CFAC1_210023600 | CUG89365 | DnaJC14 | ER, MITO, CYT | Cell surface export | Jung et al. 2016 | |
| Tb927.6.3500 Tbg972.6.3250 | J59 | TcCLB.506941.9 Tc_MARK_ 1973 TCDM_06339 TCDM_06340 | LmjF.30.2210 | CFAC1_260045500 | CUF24956 | DnaJC13 | CYT | Endosome trafficking | Girard et al. 2005 | |
| Girard and McPherson 2008 | ||||||||||
| Tb927.11.13600 Tbg972.11.15180 | J60 | TcCLB.506753.160 Tc_MARK_5565 TCDM_05717 | LmjF.08.0650 | CFAC1_050025800 | CUG87832 | – | MITO | ? | Panigrahi et al. 2009 | |
| Tb927.10.2460 Tbg972.10.3050 | J62 | TcCLB.412943.9 Tc_MARK_3379 TCDM_04012 TCDM_04013 | LmjF.34.0040 | CFAC1_290027800 | CUF08277 | – | MEM | Protein import | Ashburner et al. 2000 | |
| Tb927.11.13830 Tbg972.11.15450 | J63 | TcCLB.509161.110 Tc_MARK_2986 TCDM_09129 | LmjF.32.0590 | CFAC1_190012200 | CUI15501 | – | MITO | Acestor et al. 2009 | ||
| Tb927.4.880 Tbg972.4.680 | J65 | TcCLB.508257.160 Tc_MARK_2488 TCDM_02481 | LmjF.34.3870 | CFAC1_290069300 | CUI15574 | – | MITO | Protein folding | Ashburner et al. 2000 | |
| Tb927.10.5180 Tbg972.10.6290 | J67 | TcCLB.510297.30 TCDM_01165 | LmjF.36.0760 | CFAC1_250015000 | CUG94364 | – | MITO | Protein folding | Ashburner et al. 2000 | |
| Tb927.11.10950 Tbg972.11.12260 | J69 | TcCLB.509823.10 Tc_MARK_7219 TCDM_09821 | LmjF.36.4970 | CFAC1_280056000 | CUG89391 | – | MITO | ? | Panigrahi et al. 2009 | |
| Tb927. 8.8310 Tbg972. 8.8650 | J70 | – | – | – | – | – | GLYCO | ? | – | |
| Tb927. 9.12530 Tbg972.9.7690 | J71 | TcCLB.510741.190 Tc_MARK_4463 TCDM_00188 | LmjF.35.3090 | CFAC1_300081200 | CUG89593 | – | MITO | Protein import-MITO | Ashburner et al. 2000 | |
| Tb927.11.9060 Tbg972.11.10120 | J72 | TcCLB.507077.30 Tc_MARK_1361 TCDM_07079 | – | – | – | – | CYT | ? | – | |
| Tb927.9.1560 Tbg972.9.280 | J73 | TcCLB.510347.50 Tc_MARK_2404 TCDM_08512 | LmjF.26.2520 | CFAC1_160029000 | CUG90840 | – | NUC | Goos et al. 2017 | ||
| IV | Tb927.7.990 Tbg972.7.940 | J31 | TcCLB.506729.50 Tc_MARK_1042 TCDM_01293 | LmjF.26.0940 | CFAC1_290017000 | – | – | MITO | ? | Acestor et al. 2009 |
| Panigrahi et al. 2009 | ||||||||||
| Niemann et al. 2013 | ||||||||||
| Tb927.1.1230 Tbg972.1.570 | J47 | TcCLB.511423.170 Tc_MARK_6003 TCDM_01017 | LmjF.20.0550 | CFAC1_180012900 | CUG67522 | – | MITO | Protein folding | Panigrahi et al. 2009 | |
| Tb927.8.6310 Tbg972.8.6370 | J68 | TcCLB.503885.70 | LmjF.24.1910 | CFAC1_250049500 | CUG31915 | DnaJC19 | MITO | Protein import-MITO | Ashburner et al. 2000 | |
CYT cytosol, MITO mitochondria, NUC nucleus, ER endoplasmic reticulum, MEM plasma membrane/membrane bound, GOLGI Golgi apparatus, GYLCO glycosomes, SEC secreted
aThe nomenclature for the J-proteins of T. b. brucei and T. b. gambiense were derived from Folgueira and Requena (2007)
bGene IDs for the J-proteins from T. b. brucei, T. b. gambiense, T. cruzi, L. major and C. fasciculata were obtained from TriTrypDB database (http://tritrypdb.org/tritrypdb/; Aslett et al. 2010). The gene IDs for the J-proteins from B. saltans were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/)
cOrthologues identified from T. cruzi, L. major, C. fasciculata, and Homo sapiens were determined by reciprocal BLASTP analysis
dThe gene IDs for orthologues identified in three strains of T. cruzi are listed. T. cruzi CL Brener Esmeraldo-like (TcCLB), T. cruzi Dm28c (TCDM), and T. cruzi marinkelli strain B7 (Tc_MARK)
eSubcellular localizations for the T. b. brucei and T. b. gambiense J-proteins were either predicted using the online prediction servers and/or determined using various proteomic datasets listed in the materials and methods
fThe predicted cellular role and functions for each J-protein from T. brucei were implied from either Gene Ontology (Ashburner et al. 2000), or published literature on the identified functions/cellular roles of their identified orthologues
Protein properties, protein expression, domain mapping, and determination of the organelle distribution for the Hsp70 and J-protein complements
The molecular weight (Da) and isoelectric point (pI) of each gene was calculated using the compute pI/Mw tool from ExPASy (https://web.expasy.org/compute_pi/; Gasteiger et al. 2005). The protein expression level between lifecycle stages for each member of the Hsp70 and J-protein complement between lifecycle stages was retrieved from several proteomic datasets (Gunasekera et al. 2012; Urbaniak et al. 2012; Butter et al. 2013). Data on the phenotypic knockdown screen, using RNAi conducted by Alsford et al. (2011), for each member of the Hsp70 and J-protein complement was retrieved from the TrypsNetDB database (http://trypsnetdb.org/QueryPage.aspx; Gazestani et al. 2017). The protein domain mapping for the Hsp70 and J-protein complements was conducted using a combination of online programs that included TPRpred (http://toolkit.tuebingen.mpg.de/tprpred; Karpenahalli et al. 2007), SMART 7 (http://smart.embl-heidelberg.de/; Letunic et al. 2012), and PROSITE (http://prosite.expasy.org/; Sigrist et al. 2009).
Proteomic data from the mitochondrion (Panigrahi et al. 2009), mitochondrial importome (Peikert et al. 2017), respiratome (Acestor et al. 2011), mitochondrial membranes (outer, intermembrane space, inner, and matrix) (Acestor et al. 2009), nucleus (Goos et al. 2017), nuclear pore (DeGrasse et al. 2008), glycosomes (Colasante et al. 2006; Güther et al. 2014), flagellum (Broadhead et al. 2006; Subota et al. 2014), and cell surface (Shimogawa et al. 2015) were utilized for the determination of the organelle distribution for the T. brucei Hsp70 and J-protein complements. In the absence of experimental data, online prediction programs which included NucPred (http://www.sbc.su.se/~maccallr/nucpred/cgi-bin/single.cgi; Brameier et al. 2007), MitoPROT (http://ihg.gsf.de/ihg/mitoprot.html; Claros and Vincens 1996), MultiLoc (http://abi.inf.uni-tuebingen.de/Services/MultiLoc; Höglund et al. 2006), SignalP version 4.1 (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al. 2011), and WoLF PSORT (http://www.genscript.com/wolf-psort.html.; Horton et al. 2007) were used.
Results and discussion
Determination of the T. b. brucei and T. b. gambiense Hsp70 superfamily
A non-human infective T. brucei subspecies, T. b. brucei, is the preferred model for trypanosome research as the T. b. brucei TREU927 strain displays the full range of known T. brucei phenotypes and possesses similar biological and genetic characteristics to the human infective subspecies, T. b. rhodesiense (Gibson 2012). The T. b. rhodesiense genome has not been sequenced, but information obtained from the T. b. brucei genome can be inferred for both subspecies (Gibson 2012). However, sequencing of the T. b. gambiense genome was conducted due to this subspecies having profoundly different biological and genetic characteristics (Jackson et al. 2010) and being the most clinically relevant subspecies, as it is the etiological agent of over 90% of HAT cases (WHO 2013). Thus, an in silico analysis of the T. brucei Hsp70 superfamily, considering annotations to the T. b. brucei genome on the TriTrypDB database, available proteomics data and the release of the genome sequence for T. b. gambiense, was conducted to provide a more current and extensive overview of the Hsp70 superfamily to previous in silico analyses.
Comparative analysis of both T. b. brucei and T. b. gambiense enabled the determination of the T. b. gambiense Hsp70 superfamily and evaluated the conservation of the T. brucei Hsp70 complement. A total of 12 putative Hsp70s were identified to be encoded on both the T. b. gambiense and T. b. brucei genomes, highlighting conservation of the Hsp70 superfamily (Table 1). This number is consistent with previous in silico studies (Folgueira and Requena 2007; Louw et al. 2010a). The Hsp70 superfamily for both T. brucei subspecies were found to comprise 8 Hsp70/HSPA proteins and 4 Hsp110/HSPH proteins (Table 1). The nomenclature for the T. b. gambiense Hsp70 superfamily was adopted from the nomenclature proposed by Folgueira and Requena (2007). However, to underscore whether discussing a protein from T. b. gambiense or T. b. brucei, the abbreviations Tbg and Tbb were used in this study respectively. Thus, TbbHsp70 refers to Hsp70 from T. b. brucei. The orthologous relationships of the Hsp70 superfamily from T. b. brucei and T. b. gambiense to the selected organisms in this study are presented in Table 1.
Three T. cruzi strains (CL Brener Esmeraldo-like, Dm28c, and marinkelli strain B7) were incorporated into this study due to the discrepancy in literature on the exact number of members for the T. cruzi Hsp70 superfamily. The Hsp70 superfamily in the T. cruzi CL Brener Esmeraldo-like strain was initially reported to comprise 28 members Folgueira and Requena 2007), though more recent studies have stated 11 members encoded on the genome (Louw et al. 2010a; Shonhai et al. 2011). This in silico study identified that the T. cruzi CL Brener Esmeraldo-like strain has 13 members, the Dm28c strain has 7 members, and the marinkelli strain B7 has 12 members. The variability amongst the three strains illustrates the need for further assessment of the Hsp70 complement in T. cruzi. The number of members for the Hsp70 superfamily in the Leishmania spp. and C. fasciculata were relatively well conserved with only variability in the gene copy numbers. In comparison to the other selected organisms in this study, the Hsp70 complement in the kinetoplastid parasites is smaller than that found in Homo sapiens (H. sapiens), which both have 17 members. Gene duplication during eukaryotic evolution satisfied the demand for Hsp70 isoforms in various intracellular compartments, tissue-specific or developmental expression patterns, and functional diversity for client specificity and/or processing in the multicellular organisms (Brocchieri et al. 2008; Kabani and Martineau 2008).
T. brucei Hsp70/HSPA subfamily
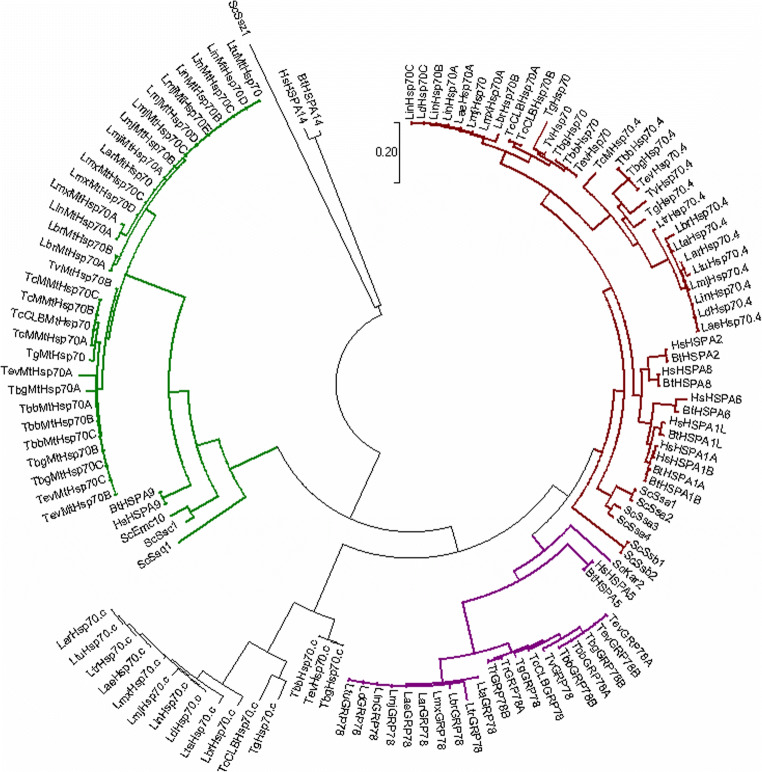
A total of 8 Hsp70/HSPA protein members were identified in this in silico study for T. b. brucei, which is consistent with previous in silico studies (Folgueira and Requena 2007; Louw et al. 2010a). Phylogenetic analysis shows that the T. brucei Hsp70/HSPA subfamily comprised 5 distinct Hsp70 groups (Hsp70, Hsp70.4, Hsp70.c, Grp78/BiP, and mtHsp70), which cluster according to protein sequence and subcellular localisation (Fig. 1). Phylogenetic analysis suggests that the T. brucei Hsp70/HSPA protein subfamily followed the same model of divergent evolution as evident amongst the other kinetoplastid parasites (Fig. 1). These five Hsp70 groups differ from each other by gene copy, protein features and domain architecture, protein expression during the lifecycle of the parasite, and the predicted or experimentally determined subcellular localisation (Table 1 and Fig. S2). These differences seem to infer that each Hsp70/HSPA protein performs a specialized cellular role(s) in the parasite. Hsp70 in both T. brucei subspecies was found to possess the canonical domain architecture of typical Hsp70s and shares high sequence identities with its orthologues in the selected organisms used in this in silico study (Fig. S2).
Fig. 1.
Phylogenetic analysis of the Hsp70 superfamily from T. brucei in relation to human and selected kinetoplastid parasites. Multiple-sequence alignment of the full-length amino acid sequences of the Hsp70/HSPA gene families in human and selected kinetoplastid parasites. The multiple sequence alignment provided in Fig. S1 was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters on the MEGA 7 software (Kumar et al. 2016). The phylogenetic tree was constructed by MEGA 7 using the Maximum-likelihood method based on the Jones–Taylor–Thornton (JTT) matrix-based model of amino acid substitution (Jones et al. 1992) with gamma distribution shape parameter (G). The alignment gaps were excluded from the analysis, and the number of amino acid sites used to construct the tree numbered 363. Bootstrap analysis was computed with 1000 replicates. Accession numbers for the T. b. brucei (Tbb), T. b. gambiense (Tbg), T. cruzi (TcCLB; CL Brener Esmeraldo), C. fasciculata (Cf), B. saltans (Bs), and L. major (Lmj) Hsp70 and Hsp110 sequences can be found in Table 1. Accession numbers for human (Hs; H. sapiens) and other kinetoplastid HSPA/Hsp70 and HSPH/Hsp110 sequences are provided in Table S1. The subcellular localisation for Hsp70s is indicated by coloured branches. Brown: cytosolic; purple: endoplasmic reticulum; and green: mitochondrion. Scale bar represents 0.2 amino acid substitutions per site (colour figure online)
Hsp70 proteins help T. brucei to adapt to changing environmental conditions, and the levels of these proteins differ during each lifecycle stage. In T. brucei, transcriptomic data often does not correlate well with protein data, and significantly larger fold changes are seen at the protein level than at the mRNA level (Urbaniak et al. 2012; Goos et al. 2017). A genome-wide comparative proteomic study between the lifecycle stages in T. brucei revealed that TbbHsp70, TbbHsp110, TbbHsp70.a, and TbbHsp70.c were downregulated in procyclic form (PF), whilst TbbmtHsp70A and TbbHsp70.4 were upregulated in PF, though with poor correlation to mRNA abundance (Urbaniak et al. 2012). In a similar study, TbbHsp70.a and TbbHsp110 were upregulated in the bloodstream from (BSF) relative to the PF, whilst TbbmtHsp70A, B, C and TbbHsp70.4 were upregulated in the PCF relative to BSF (Butter et al. 2013). The surface of T. brucei forms a vital interface with its mammalian hosts allowing it to adapt to varied environments, and cell surface proteomic analysis revealed that mtHsp70A, B, C and Hsp70 were present in both PC and BSF stages, whilst Hsp110 and Hsp70.4 were present in PF (Shimogawa et al. 2015). Protein phosphorylation plays a crucial role in the regulation of many cellular processes, and the cytosolic proteins TbbHsp70, TbbHsp70.c, and TbbHsp70.4 were determined to have at least one phosphorylation site (Nett et al. 2009; Urbaniak et al. 2013).
TbbHsp70 (Tb927.11.11330) was reported in a previous in silico study to be an unusual cytosolic Hsp70 due to the protein being shown to possess a non-canonical C-terminal RRHI motif, instead of the highly conserved C-terminal EEVD motif (Louw et al. 2010a). Following recent annotations of the T. b. brucei genome and comparison to the Hsp70 encoded on the T. b. gambiense genome (Tbg972.11.12660), the C-terminal RRHI motif has been identified as a misannotation. This is indicated to be a result of a collapse in T. b. brucei genome assembly, which caused a frameshift in the Hsp70 coding sequence, leading to changes in the C-terminal region of the protein and elimination of the EEVD motif (Droll 2013). T. b. brucei was shown to possess 5 identical copies of the TbbHsp70 gene arranged in tandem array (Glass et al. 1986), though this was collapsed into one locus following genome assembly and is constitutively co-transcribed (Lee and Van der Ploeg 1990; Huang and Van der Ploeg 1991). Duplication of the cytosolic Hsp70 gene has been shown in the other kinetoplastid parasites such as T. cruzi (Urmenyi et al. 2014), C. fasciculata (Table 1), and L. major (Requena et al. 2015; Drini et al. 2016). Amplification of HSP genes in protozoan parasites has been identified as a means of the parasites increasing chaperone activity under stressful conditions (Wiesgigl and Clos 2001).
The mRNA of Hsp70 has been shown to be regulated by a zinc finger protein, ZC3H11, where it stabilizes the mRNA after heat shock and promotes the survival of the parasite (Droll et al. 2013). HsHSPA1A/B, human orthologues of Hsp70 in both T. brucei subspecies, are major cytosolic stress-inducible Hsp70s that protect against the harmful effects of aggregates from denatured proteins during and following environmental stresses (Hartl 1996). Phenotypic knockdown, using RNAi, on the Hsp70 gene locus (Tb927.11.11330) in T. b. brucei demonstrated that it is essential to parasite survival throughout its lifecycle (Alsford et al. 2011). Based on phylogeny and orthology, it can be inferred that Hsp70 is a crucial component of the heat shock response in T. brucei, providing cytoprotection to the parasite under stressful conditions. The orthologue of Hsp70 in several Leishmania spp. has also been linked to parasite’s resistance to pentavalent antimonial treatment, as it induces Hsp70 expression which provides stress tolerance against the drug (Brochu et al. 2004; Maharjan and Madhubala 2015; Codonho et al. 2016). TbbHsp70 was detected in glycosomes with high confidence and the flagellum (Table 1) (Güther et al. 2014; Subota et al. 2014).
In this study, Hsp70.4 forms a distinct Hsp70/HSPA group found in kinetoplastid parasites, as the proteins have no mammalian orthologues (Table 1). The Hsp70.4 protein in T. brucei shares domain architecture with typical Hsp70s but possesses a divergent C-terminal EEVD motif (Fig. S2). The variation in the C-terminal EEVD motif is observed in all the kinetoplastid orthologues of Hsp70.4; DDVD in T. evansi, TDVD in T. cruzi and T. grayi, DEVD in T. vivax, TDID in B. saltans, QDVD in C. fasciculata, and EDVD in all Leishmania spp. The divergent motifs found in the kinetoplastid are proposed to be functionally equivalent to the canonical EEVD motif (Louw et al. 2010a), though the role this variation plays in the function and protein interaction with co-chaperones has not been elucidated. A previous in silico study conducted on the Hsp70 superfamily of T. cruzi noted the absence of Hsp70.4 in the T. cruzi CL Brener Esmeraldo strain genome (Louw et al. 2010a). The absence of Hsp70.4 is a result of the TcCLBHsp70.4 being on two separate loci (TcCLB.503721.39, TcCLB.511257.10) on the CL Brener Esmeraldo strain genome, both encoding for partial amino acid sequences. Additionally, a Hsp70.4 gene was found encoded on the genome sequences in the Dm28c and marinkelli strains of T. cruzi. Thus, reannotation of these loci on the T. cruzi CL Brener Esmeraldo genome is required. T. brucei Hsp70.4 is predicted to reside in the cytosol of the parasite according to its orthology and phylogeny. The Hsp70.4 orthologue in L. major has been shown through indirect immunofluorescence (IFA) staining to reside in the cytoplasm (Searle et al. 1989; Searle and Smith 1993), and to be constitutively expressed (Simpson et al. 2006), implying that the localisation and expression profile of T. brucei Hsp70.4 may be similar. TbbHsp70.4 has been shown to be non-essential as phenotypic knockdown had no detrimental effect on the survival and fitness of the parasite at any stage of its lifecycle (Alsford et al. 2011). This may indicate that the cellular functions of TbbHsp70.4 can be compensated by the other T. b. brucei cytosolic Hsp70s. However, caution should be exercised when referring to RNAi data in this study as false negatives may arise (Subramaniam et al. 2006).
Hsp70.c may represent a novel Hsp70/HSPA subfamily found only in kinetoplastids, as no clear orthologue in humans was identified (Table 1). Using various online prediction servers, the subcellular localisation was predicted to be cytosolic and nuclear, and it was found to be part of the nuclear proteome of PF T. brucei (Goos et al. 2017). Phylogenetic analysis revealed that the Hsp70.c group formed a distinct clade, as the proteins did not phylogenetically cluster with any of the other primary Hsp70/HSPA proteins (Fig. 1), which is consistent with previous phylogenetic analyses (Louw et al. 2010a; Burger et al. 2014; Requena et al. 2015). The evolutionary divergence is a result of the Hsp70 proteins possessing an atypical substrate binding domain (SBD). TbbHsp70.c (Tb927.11.11290) was shown to lack key residues that facilitate substrate recognition and were instead replaced with acidic residues (Louw et al. 2010a). The putative SBD of the Plasmodium falciparum Hsp110c was shown to be modified to handle the asparagine repeat-rich proteome of the parasite particularly during a febrile episode (Muralidharan et al. 2012). The modification of the SBD of TbbHsp70.c could be an adaptation of this particular Hsp70 to handle specific substrates in the T. b. brucei proteome during parasite differentiation. Despite these substitutions, TbbHsp70.c was still able to suppress the aggregation of the model substrates, malate dehydrogenase and rhodanese (Burger et al. 2014). Expression of TbbHsp70.c was also shown to be slightly upregulated in BSF parasites during heat shock, indicating that it could also play a potential role in parasite cytoprotection (Burger et al. 2014). Further investigation of TbbHsp70.c, and its kinetoplastid orthologues, could elucidate the cellular roles the Hsp70 fulfils in the parasites, with regard to parasite differentiation.
TbbGrp78 was the first Hsp70 isoform to be characterized from T. b. brucei, where it was shown to be a soluble luminal resident of the ER, as the C-terminal tetrapeptide MDDL maintains its subcellular localisation (Bangs et al. 1993). Both T. brucei subspecies encoded for two copies of the ER Hsp70 isoform, Grp78 (also known as BiP) (Table 1), which are 98% identical in amino acid sequence, and appear in tandem array on the genome in both subspecies. Interestingly, the duplication event of the Grp78 gene did not occur in C. fasciculata or Leishmania, as all the Leishmania spp. investigated in this study possess only one Grp78 protein (Fig. 1). It has been proposed the two Grp78 genes may be transcribed separately due to the separation of genes on chromosome XI (Louw et al. 2010a), though both Grp78A (Tb927.11.7460) and Grp78B (Tb927.11.7510) expression in T. b. brucei was shown to be upregulated at the bloodstream stage of the parasite (Bangs et al. 1993). Grp78 was further characterized by Bangs et al. (1996), where the study showed the molecular chaperone to be involved in the transport and subsequent folding of the newly synthesized variable surface glycoprotein (VSG) in the ER lumen. The upregulation and essentiality of TbbGrp78A at the bloodstream stage of the parasite may be attributed to the rapid growth of the parasites in the mammalian host and maintenance of the variable surface glycoprotein (VSG) coat of the parasite (Bangs et al. 1993).
T. brucei and several Leishmania spp. have been shown to possess a large mitochondrial Hsp70 complement, as amplification of the mtHsp70 genes is apparently a rather frequent event in kinetoplastids, with the copy numbers ranging from 2 to 5 depending on the species (Table 1). There is variability with regard to the number of mitochondrial Hsp70s in T. cruzi, as the CL Brener Esmeraldo strain was identified in this study to have one full-length gene (TcCLB.507029.30) and three partial genes encoding for mtHsp70 (TcCLB511745.10; TcCLB432677.20; TcCLB511515.40); the marinkelli strain has three genes (TcMARK_1997; TcMARK_2001; TcMARK_2002), whereas no mtHsp70 gene was found in the Dm28c strain (Table 1). The genomes of the T. cruzi marinkelli and Dm28c strains need to be further investigated to determine if the partial sequences and absence of a mtHsp70 gene respectively are sequencing errors to resolve the discrepancy in T. cruzi strains.
T. brucei possesses three mitochondrial Hsp70 homologues (mtHsp70A, mtHsp70B, mtHsp70C) (Table 1), which have been shown to appear in tandem array on the T. b. brucei chromosome VI with identical amino acid sequences (Louw et al. 2010a). TbbMtHsp70 was shown through IFA to be well distributed throughout the mitochondrion of the parasite (Klein et al. 1995) and is also an integral component of the replication and maintenance of kinetoplast DNA (kDNA) (Týč et al. 2015). The T. cruzi orthologue, TcMtHsp70, has also been implicated in mtDNA replication (Engman et al. 1989). Analysis of the T. brucei mitochondrial outer membrane proteome revealed the presence of mtHsp70A, B, and C (Niemann et al. 2013). The mammalian orthologue of the three T. brucei mitochondrial Hsp70s was identified to be HSPA9, which has been shown to facilitate the translocation and correct folding of proteins targeted for the mitochondria (Mizzen et al. 1989; Deocaris et al. 2006) (Table 1). A putative Hsp70 escort protein orthologue (TbHep1; Tb927.3.2300) was identified to be encoded on the genomes for both T. brucei subspecies, and it should be explored if TbHep1 is required to maintain solubility and functionality of the three mitochondrial Hsp70 isoforms in T. brucei.
T. brucei Hsp110/HSPH subfamily
The Hsp110/HSPH protein family in both subspecies of T. brucei was identified to comprise four members (Table 1) (Hsp110, Grp170, Hsp70.a, and Hsp70.b). All four members were shown to be considerably longer in amino acid sequence (Fig. S3), characteristic of Hsp110/HSPH protein members (Easton et al. 2000). Hsp110 is a predicted cytosolic Hsp110/HSPH protein member (Table 1; Fig. S3) that is essential throughout the lifecycle of T. b. brucei (Alsford et al. 2011). The mRNA of Hsp110 has been shown to be enriched and stabilized following heat shock in PF parasites, indicating that TbHsp110 is involved in cytoprotection and recovery following heat shock (Droll et al. 2013). Hsp110 proteins have been shown to play an important role in thermo-resistance, and the prevention of protein aggregation (Raviol et al. 2006). Hsp110 proteins are also a major component of the Hsp70 chaperone machinery, facilitating nucleotide exchange (Easton et al. 2000). Interestingly, kinetoplastid parasites only encode for one predicted cytosolic Hsp110 protein, whereas mammalian cells encode for three Hsp110 homologues (Kampinga and Craig 2010). T. brucei Hsp110 was identified in this study to be orthologous to mammalian HSPH1 (Table 1). Human HSPH1 (also known as Hsp105) has been shown to be expressed as two different isoforms, HSPH1-α and HSPH1-β (Yasuda et al. 1995). HSPH1-α is constitutively expressed but inducible to heat shock or stress, whereas HSPH1-β is strictly heat inducible (Saxena et al. 2012). HSPH-α is shown to be involved in protein biogenesis and quality control (Saxena et al. 2012). Thus, it could be suspected that Hsp110 in T. brucei forms a partnership with the predicted cytosolic Hsp70s to regulate protein biogenesis and quality control in the cytosol of the parasite. However, the cellular functions and Hsp110–Hsp70 partnerships need to be experimentally elucidated.
Grp170 and Hsp70.a are both Hsp110/HSPH protein members predicted to reside in the ER in T. brucei, as both were shown to possess N-terminal import and C-terminal ER retention signal sequences (Fig. S3). Like Grp78, mammalian Grp170 has been demonstrated to be an ER chaperone that assists in the protein folding, assembly, and transportation of secretory or transmembrane proteins (Wang et al. 2014). Grp170 in kinetoplastid parasites phylogenetically clustered with mammalian Grp170 orthologues, suggesting that the proteins may be functionally equivalent (Fig. 1). Hsp70.a is a novel ER Hsp110/HSPH protein member that is conserved in the kinetoplastid parasites (Fig. 1). TbbHsp70.a was demonstrated to be essential in parasite differentiation (Alsford et al. 2011), as the gene expression of TbbHsp70.a is upregulated during and up to 48 h post-synchronous differentiation of the parasite from the BSF to the PF life stage (Quieroz et al. 2009). Hsp70.a may be implicated in the transportation and protein folding of secretory or transmembrane proteins that could be critical for the developmental differentiation of the parasite. Suppression of the expression of TbbHsp70.a by RNAi resulted in increased accumulation of VSG in the ER and distortion of the organelle (Field et al. 2010).
In mammalian cells, the mitochondria and endoplasmic reticulum form structural and functional linkages known as mitochondria-associated ER membranes (MAMs) which are crucial to maintain cellular homeostasis (Rowland and Voeltz 2012). TbbHsp70.a, with one transmembrane domain (TMD), was also assigned to the mitochondrial membrane of PF cells with high confidence (Acestor et al. 2009). The same protein was also detected in three other mitochondrial proteomics studies (Panagrahi et al. 2009; Niemann et al. 2013; Peikert et al. 2017). Other cytosolic Hsp70s (TbbHsp70, TbbHsp70.c, and TbHsp110) were also detected in a study of the mitochondrial outer membrane proteome (Niemann et al. 2013).
Hsp70.b is a unique Hsp110/HSPH protein member, which has been shown by phylogenetic analysis to form a distinct monophyletic group (Fig. 1). Though Hsp70.b is most notably absent in B. saltans (Table 1), Hsp70.b in T. brucei is predicted to localize in the mitochondria, as the protein was shown to possess an N-terminal positively charged leader sequence (Table 1) and detected in proteomic analyses of the mitochondria (Peikert et al. 2017; Niemann et al. 2013).
The T. brucei J-protein complement
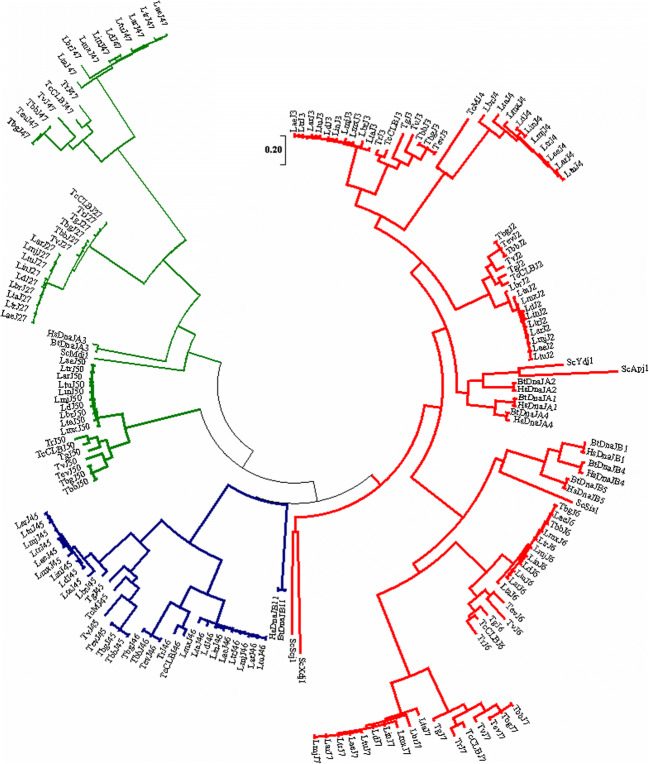
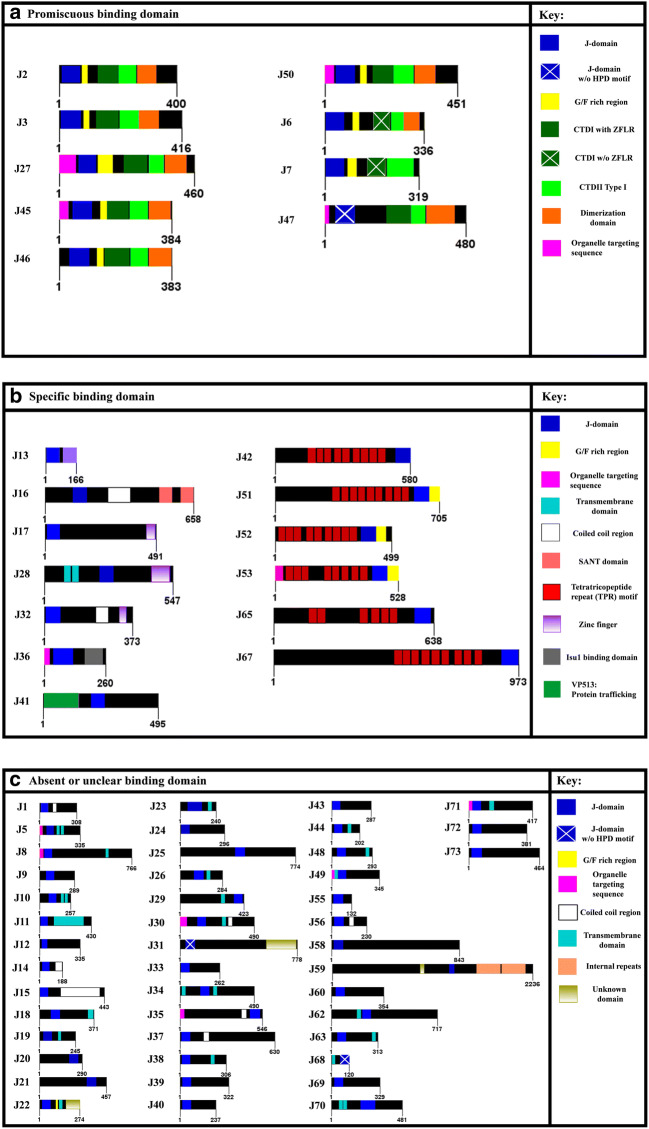
An in silico investigation identified 67 putative J-proteins encoded on the genomes of both T. brucei subspecies (Table 2). Nomenclature proposed for the T. brucei J-proteins was based on nomenclature guidelines in Folgueira and Requena (2007). For simplicity, the T. brucei J-proteins will be referred to by name, as seen in column 3 of Table 2. Phylogenetic analysis of the selected J-protein subfamilies, as illustrated in Fig. 2, shows that the J-proteins cluster based on their different classes and subcellular localisation. All identified J-protein members were further classified into the four J-protein subfamilies, I–IV, accordingly to their identified domain architecture (Table 2). The basis for classification of the J-proteins is their homology to the prokaryotic canonical J-protein, DnaJ, which is divided into an N-terminal J-domain, glycine-phenylalanine (G/F)-rich region, zinc finger-like region (ZFLR), and a C-terminal substrate-binding domain (Cheetham and Caplan 1998). Type I J-proteins possess all these canonical domains, type II J-proteins lack the ZFLR, type III J-proteins contain only the signature J-domain which can occur anywhere along the protein sequence, and type IV proteins possess a J-domain with a compromised or absent HPD motif and may also possess domain structures from the other J-protein type subfamilies (Cheetham and Caplan 1998; Botha et al. 2007). A comprehensive domain organisation of the predicted T. brucei J-proteins is illustrated in Fig. 3.
Fig. 2.
Phylogenetic analysis of selected J-protein subfamilies from T. brucei in relation to human and selected kinetoplastid parasites. Multiple-sequence alignment of the full-length amino acid sequences of the type I, II, and IV J-protein gene families in human and selected kinetoplastid parasites. The multiple-sequence alignment provided in Fig. S4 was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters on the MEGA 7 software (Kumar et al. 2016). The phylogenetic tree was constructed by MEGA 7 using the Maximum-likelihood method based on the Jones–Taylor–Thornton (JTT) matrix-based model of amino acid substitution (Jones et al. 1992) with gamma distribution shape parameter (G). The alignment gaps were excluded from the analysis, and the number of amino acid sites used to construct the tree numbered 194. Bootstrap analysis was computed with 1000 replicates. Accession numbers for the T. b. brucei (Tbb), T. b. gambiense (Tbg), T. cruzi (TcCLB; CL Brener Esmeraldo), C. fasciculata (Cf), B. saltans (Bs), and L. major (Lmj) J-protein sequences can be found in Table 2. Accession numbers for human (Hs; H. sapiens) and other kinetoplastid J-protein sequences are provided in Table S2. The subcellular localisation for J-proteins is indicated by coloured branches. Red: cytosolic; blue: endoplasmic reticulum; green: mitochondrion. Scale bar represents 0.5 amino acid substitutions per site (colour figure online)
Fig. 3.
Schematic representation of the domain architecture of the different classes of J-proteins in T. brucei. Each protein sequence for the T. brucei J-protein family is represented by an open bar with the number of amino acids indicated on either side of the protein bar. The name of the respective J-protein is indicated on the lefthand side. The various domains are highlighted by coloured blocks within the protein bar. A key is provided to give a short description of the various domains and features. The J-proteins were also categorized based on assumed client binding ability and mechanistic mode of functioning as proposed by Kampinga et al. (2009)
The total number of members for the T. b. brucei J-protein complement is larger than the previously reported 65 members by Folgueira and Requena (2007). However, 73 J-proteins were reported in T. b. brucei by Droll et al. (2013), but data mining of the available transcriptomic data revealed only 57 J-proteins indicating a discrepancy in the reported numbers for this study. A recent in silico investigation of the L. major J-protein family has also reported a larger total number of J-protein members (n = 69) encoded on the annotated genome than previously reported (n = 66) (Requena et al. 2015). However, Shonhai et al. (2011) reported fewer J-proteins (n = 61) encoded on the T. cruzi genome. This in silico study incorporated three T. cruzi strains (CL Brener Esmeraldo–like, Dm28c, and marinkelli strain B7) for in silico analysis, and investigation of the J-protein complement revealed that the T. cruzi CL Brener Esmeraldo–like strain has 66 J-protein members, Dm28c strain has 56 members, and marinkelli strain B7 has 58 members. The variability in the total J-protein member numbers highlights the need for further assessment of the T. cruzi J-protein complement. However, the J-protein complement in the kinetoplastid parasites is greatly expanded in comparison to those found in its mammalian hosts, and the significance of this expansion is yet to be elucidated.
T. brucei type I J-protein subfamily
This study identified that the type I J-protein subfamily in T. brucei comprised 6 members: J2, J3, J27, J45, J46, and J50 (Table 2). J2 was identified to be an essential type I J-protein, as knockdown via RNAi is lethal at all life stages of the parasite (Alsford et al. 2011), and it was shown to reside in the parasite cytosol (Ludewig et al. 2015). It is implicated to be an integral component of protein biogenesis, as the T. cruzi orthologue of J2 was shown to in vivo complement the yeast type I J-protein, Ydj1, and stimulate the in vitro ATPase activity of TcHsp70 (Edkins et al. 2004). The protein levels of J2 increased in response to heat stress, suggesting that the J2 protein is critical to cytoprotection in kinetoplastid parasites (Ludewig et al. 2015). The J2 orthologue in Leishmania infantum has been implicated in the differentiation process of the parasite (Tsigankov et al. 2014). J2 has demonstrated a potential partnership with Hsp70.c (Burger et al. 2014), an Hsp70 protein shown to be essential for T. brucei differentiation (Alsford et al. 2011). Thus, this Hsp70–J-protein partnership may be an integral component of parasite differentiation, and pathogenesis.
J3 is another type I J-protein predicted to localize in the cytosol of the parasite (Table 2), based on its phylogeny (Fig. 2) and domain architecture (Fig. 3). Despite the homology to J2, the two cytosolic type I J-proteins are not functionally equivalent as knockdown of J3 only resulted in loss of fitness at the PF stage of the parasite (Alsford et al. 2011), despite being shown to be expressed at all stages of the lifecycle (Aslett et al. 2010). This could suggest that the type I J-protein is required for folding of specific client proteins that are needed for PF trypanosomes. J45 and J46 are predicted to reside in the ER (Table 2), based on their phylogeny to known ER J-proteins (Fig. 2). J46 was shown to possess an N-terminal targeting sequence and similar domain architecture to its predicted human orthologue, DnaJB11 (Table 2). HsDnaJB11 is an abundant soluble ER resident type I J-protein that has been shown to co-ordinate with BiP in facilitating the folding of proteins (Jin et al. 2009; Guo and Snapp 2013).
The Hsp70/J-protein machinery was found to be indispensable for proper mitochondrial DNA maintenance and replication, as RNAi-mediated knockdown resulted in shrinkage of the highly compacted mitochondrial network, due to decreased maxicircle and minicircle copy numbers (Týč et al. 2015). J27 and J50 were predicted to reside in the mitochondrion of the parasite, based on the identification of an N-terminal targeting sequence in both J-proteins (Table 2; Fig. 3). HsDnaJA3 was identified to be the human orthologue of J50 that has been shown to co-operate with mitochondrial Hsp70s in protein translocation and folding (Iosefson et al. 2012). J50 was shown to localize in the mitochondrion of the parasite, where it was shown to form a complex with the mitochondrial Hsp70 and the nucleotide exchange factor, Mge1 (Týč et al. 2015). Analyses of the total T. brucei mitochondrial genome and the specific mitochondrial respiratory complexes in PF forms revealed the presence of a putative DnaJ protein (J27) as part of the mitochondrial respiration complex I (Panigrahi et al. 2009; Acestor et al. 2011). Proteomic analyses of three lifecycle stages showed an increase in mitochondrial protein abundance of MtHsp70A/B/C, J50, and J27 in the short stumpy and PF cells, relative to the long slender bloodstream form. A genome-wide comparative proteomic study between the lifecycle stages in T. brucei revealed that J2 and J45 were downregulated in PF form, whilst J3, J27, and J50 were upregulated in PF with an overall poor correlation to mRNA abundance (Urbaniak et al. 2012). In a similar study, J2, J45, and J46 were upregulated in the BSF relative to the PC form whilst J3, J27, and J50 were upregulated in the PCF relative to BSF (Butter et al. 2013).
T. brucei type II J-protein subfamily
Remarkably, the type II J-protein subfamily comprised only two members, J6 and J7. J7 was assigned to the mitochondrial matrix (Acestor et al. 2009) and identified in mitochondrial enriched fractions with poor signal peptide correlation (Panigrahi et al. 2009). J6 is an orthologue of Tcj6, a T. cruzi type II J-protein shown to be associated with ribosomal subunits, 80S monosomes, and smaller polysomes, and able to functionally in vivo complement a yeast mutant deficient in the orthologous gene Sis1 (Salmon et al. 2001). Tcj6 (TcCLB.506355.50) was also shown to be cytosolic, particularly concentrated around the nucleus with probable association with the ER (Salmon et al. 2001). It is speculated that J6 may perform a similar role in the cytosol of T. brucei. J7 is an essential bloodstream stage J-protein (Alsford et al. 2011). A genome-wide comparative proteomic study between the lifecycle stages in T. brucei revealed that J6 and J7 were downregulated in PF form (Urbaniak et al. 2012). In a similar study, J7 was upregulated in the BSF relative to the PC form (Butter et al. 2013).
T. brucei type III J-protein subfamily
A total of 56 members identified in the T. brucei J-protein family were found to be classified as type III J-protein members (Table 2), as these members were identified to possess a wide variety of protein domains and motifs, as illustrated in Fig. 3. The functional diversity of the type III J-protein subfamily enables the Hsp70/J-protein chaperone machinery to perform a diverse range of functions within the cell (Kaschner et al. 2015). Predicted subcellular localisation of the type III J-protein subfamily indicates that the family members localize to various organelles within the parasite (Table 2), with the majority of J proteins localized in the mitochondrion. A genome-wide comparative proteomic study between the lifecycle stages in T. brucei revealed that J20, J22, J24, J34, and J53 were downregulated in PF, whilst J1, J5, J8, J10, J11, J15, J16, J18, J21, J23, J25, J26, J28, J33, J36, J39, J51, J52, and J70 were upregulated in PF with poor correlation to mRNA abundance (Urbaniak et al. 2012). In a similar study, J1, J6, J14, J15, J20, J22, J24, J25, J34, J53, and J59 were upregulated in the BSF relative to the PC form whilst J5, J8, J10, J11, J16, J18, J21, J23, J28, J30, J32, J33, J36, J38, J39, J44, J48, J51, J52, J56, J63, J69, J70, and J73 were upregulated in the PCF relative to BSF (Butter et al. 2013). The cytosolic J15 and J16 proteins were identified as phosphoproteins with at least one phosphorylation site (Nett et al. 2009; Urbaniak et al. 2013). In a further study, a comparison of the phosphoproteins in two lifecycle stages was carried out and phosphorylation of J12, J25, J33, and J37 resulted in a 10-fold upregulation in BSF relative to PC, whilst phosphorylation of J32 resulted in a 10-fold upregulation in PC relative to BSF (Urbaniak et al. 2013). Additional J-proteins that were identified to be phosphorylated include J8, J11, J12, J14, J24, J25, J33, J34, J37, J43, J44, J51, and J59 (Urbaniak et al. 2013).
J1 was shown to be expressed in the T. b. brucei BSF stage and was unable to stimulate the ATPase activities of two different Hsp70s and did not possess independent chaperone activity, as observed for type I and II J-proteins (Louw et al. 2010b). This is not surprising as in silico analysis of the domain architecture of J1 showed the absence of a substrate-binding domain (Fig. 3). J11 was identified to be a palmitoylated protein during an analysis of palmitoylation in T. brucei (Emmer et al. 2011). J34 (also known as TbbSec63), an orthologue of ScSec63 and HsDnaJC23, is an ER membrane bound J-protein that is a component of the ER translocon, an oligomeric protein translocation pore complex that facilitates the translocation of secretory protein precursors across the ER (Engstler et al. 2007; Goldshmidt et al. 2008). RNAi-mediated knockdown of J34 was shown to be lethal (Goldshmidt et al. 2008; Alsford et al. 2011), as it affected the entry of both N-terminal ER signal peptide-containing proteins and polytopic membrane proteins (Goldshmidt et al. 2008). J34 has been implicated along with several other predicted ER chaperones to facilitate the biosynthesis and quality control of VSG proteins (Field et al. 2010). RNAi-mediated knockdown of J34 and TbbGrp78 was shown to impair protein secretion, cell viability, and presentation of variant surface glycoproteins (Field et al. 2010).
There are 6 T. brucei type III J-proteins (J42, J51, J52, J53, J65, and J67) that possess tetratricopeptide repeat-containing (TPR) motifs (Fig. 3). The tetratricopeptide repeat (TPR) is a protein–protein interaction motif that comprises a degenerate 34-amino acid sequence and has been found in many diverse proteins in all organisms (Lamb et al. 1995; D'Andrea and Regan 2003). The mammalian system is shown to possess only two TPR-containing J-proteins, DnaJC7 and DnaJC3 (Kampinga and Craig 2010). DnaJC3 (also referred to as ERdj6 or p58IPK) is a prominent type III J-protein family member in the ER, where it functions as a co-chaperone and regulator of GRP78/BiP, aiding in the refolding of misfolded proteins and thus restoring ER homeostasis (Rutkowski et al. 2007; Petrova et al. 2008). J53 was identified in this study to be the putative orthologue of DnaJC3, as it was shown to possess an N-terminal ER signal peptide (Fig. 3).
DnaJC7 (also referred to as Tpr2 or p60) is a ubiquitously expressed TPR-containing J-protein in the cytosol (Murthy et al. 1996; Ohno et al. 2014). This type III J-protein has been shown to possess two TPR domains that bind Hsp70 and Hsp90 indiscriminately, where it has been proposed to catalyze the retrograde transfer of client proteins from Hsp90 back to Hsp70 (Brychzy et al. 2003). Thus, DnaJC7 has been proposed to be a sensor of folding quality within the Hsp90 chaperoning system (Brychzy et al. 2003; Moffatt et al. 2008). Three TPR-containing J-proteins (J42, J51, and J52) have been predicted to reside in the cytosol of the parasite (Table 2). J52 was identified to be the putative orthologue of DnaJC7, but it may be speculated that the expansion of the numbers of cytosolic TPR-containing J-proteins is to offer specificity to the Hsp70/Hsp90 multichaperone heterocomplex with regard to mediating quality control of client proteins. Interestingly, only J51 has been shown to be essential to the parasite, where knockdown was shown to be lethal at the bloodstream and differentiation stages (Alsford et al. 2011). J65 and J67 are predicted to localize to the mitochondrion (Table 2). It would be worth investigating if the TPR domains of both J65 and J67 are able to act as a docking site for interaction with the mitochondrial Hsp70s, and the mitochondrial Hsp90 paralogue, TRAP-1 or HSP75.
T. brucei type IV J-protein subfamily
The T. brucei type IV J-protein subfamily comprised J31, J47, and J68, as these J-proteins were identified to possess J-domains that had abrogated HPD motifs. J68 was identified to be orthologous to HsDnaJC19 (Table 2). HsDnaJC19 is involved in the translocation of proteins into the mitochondria (Davey et al. 2006), and it can be inferred that J68 performs a similar role in the parasite. J31 was a predicted cytosolic protein, but it was assigned to the mitochondrial matrix (Acestor et al. 2009) and identified in mitochondrial enriched fractions with poor signal peptide correlation (Panigrahi et al. 2009) (Table 2). A genome-wide comparative proteomic study between the lifecycle stages in T. brucei revealed that J31 was upregulated in PF (Urbaniak et al. 2012). In a similar study, J31 and J68 were upregulated in PC form relative to the BSF (Butter et al. 2013). A comparison of the phosphoproteins in two lifecycle stages was carried out, and phosphorylation of J31 resulted in a 10-fold upregulation in BSF relative to PC (Urbaniak et al. 2013). J47 is a mitochondrial protein (Table 2) that remarkably possesses all the domains of a canonical type I J-protein. Phylogenetic analysis revealed that J47 forms a monophyletic clade with the predicted mitochondrial type I J-protein, J27 (Fig. 3). It could be assumed that J47 is a type I J-protein, with the absent HPD motif being the result of a sequencing error. However, investigation of the kinetoplastid orthologues of J47 shows that the abrogated J-domain is conserved. Investigation into the role this J-protein plays in kinetoplastid biology and its interaction with Hsp70 chaperone partners need to be elucidated.
Conclusion
This in silico study aimed to investigate the Hsp70/J-protein chaperone machinery in the T. b. brucei-annotated genome sequence, as well as to be the first to determine the Hsp70 and J-protein complements in the human infective subspecies, T. b. gambiense. These complements were comparatively analyzed in both subspecies and shown to be conserved. The T. brucei Hsp70 complement was found to comprise 12 members, with 4 belonging to the Hsp110/HSPH subfamily. This is consistent with the findings in previous in silico studies (Folgueira and Requena 2007; Louw et al. 2010a). Examination of the amino acid sequence of TbbHsp70 showed that the protein possesses a C-terminal EEVD motif, as opposed to a RRHI motif stated by Louw et al. (2010a). The misannotation is a result of a frameshift in the coding sequence after collapse of the five genes into one locus in genome assembly. Phylogenetic analysis revealed that the T. brucei Hsp70/HSPA family comprised five distinct Hsp70 groups, with multiple copies for the mitochondrial and ER Hsp70 isoforms. Hsp70.c and Hsp70.4 were both indicated to be novel cytosolic Hsp70 subgroups, as the Hsp70 proteins were only found in kinetoplastid parasites, and that the members of these Hsp70 group were found to possess atypical Hsp70 features. It is tempting to speculate that the genetic adaptation of the Hsp70 superfamily in kinetoplastid parasites is a means of coping with the environmental stresses the parasites encounter during their infectious lifecycle.
In this study, the T. brucei J-protein complement was identified to comprise 67 members. The total number of members for the T. b. brucei J-protein complement is larger than the previously reported 65 members by Folgueira and Requena (2007), with three new J-proteins (J71, J72, and J73) being identified in the annotated T. b. brucei genome sequence. Though 73 J-proteins were reported by Droll et al. (2013), there is a discrepancy to the reported numbers as data mining of the dataset revealed only 58 members. The J-protein family in T. cruzi was also identified in this study to be larger than in previously reported in silico studies (Folgueira and Requena 2007; Shonhai et al. 2011; Requena et al. 2015). However, there is discrepancy with regard to the total number of members for both Hsp70 and J-protein families in T. cruzi, as the number of members was variable in the three strains (CL Brener Esmeraldo-like, Dm28c, and marinkelli strain B7) used in this study. The T. cruzi species displays a considerable genetic and phenotypic diversity (Dvorak 1984; Tibayrenc 1998), a result of a predominantly clonal mode of evolution through large time spans (Tibayrenc et al. 1986; Tibayrenc and Ayala 2002). The population has been divided, with the use of experimental strategies, such as RAPD and multilocus isoenzyme electrophoresis (MLEE), into seven distinct T. cruzi lineages (Marcili et al. 2009; Zingales et al. 2012). The three T. cruzi CL Brener Esmeraldo-like, Dm28c, and marinkelli strains used in this study are from the TcV, TcI, and TcVII lineages, respectively (Marcili et al. 2009; Zingales et al. 2012; Grisard et al. 2014). Thus, the discrepancy observed regarding numbers of members for the Hsp70 and J-proteins is a result of the genetic diversity displayed by the various T. cruzi lineages, highlighting the need for further assessment of these complements in T. cruzi.
Members of each of the J-protein subfamilies (I–IV) were identified in both T. brucei subspecies, though the majority of the J-protein family were found to comprise type III J-protein members. Despite this overwhelming number of J-proteins in T. brucei, very few of these have been biochemically characterized to date. RNAi interference of several of the J-proteins in T. b. brucei were shown to be lethal at one or more stages of the parasite lifecycle, highlighting that the proteins may perform roles unique to the biology of the parasite. Comparative analysis of the T. brucei J-proteins in relation to the selected organisms of this study was conducted to infer cellular function, and potential Hsp70-J-protein partnerships. Obviously, many of the inferences stated in this study will need to be confirmed experimentally. However, it has become increasingly evident that the Hsp70/J-protein machinery is essential to the survival, pathogenicity, and differentiation of the parasite. However, the molecular details of the Hsp70/J-protein chaperone interactions and pathways need to be further elucidated, as some of these pathways may represent a novel means of chemotherapeutic intervention for African trypanosomiasis.
Electronic supplementary material
Alignment of the Hsp70 superfamily from T. brucei in relation to human and other selected kinetoplastids. Multiple sequence alignment of the full-length amino acid sequences was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters in the MEGA7 software (Kumar et al. 2016). Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers for the J-protein amino acid sequences used in this study are provided in Table 1 and Table S1. (DOCX 220 kb)
Schematic representation of the domain architecture of the HSPA/Hsp70 proteins in T. brucei. Each protein sequence for the T. brucei HSPA/Hsp70 family is represented by an open bar with the number of amino acids indicated on either side of the protein bar with the various protein domains and other associated features that were identified using Prosite (Sigrist et al. 2009) and SMART (Letunic et al. 2012). These domains and associated features include the N-terminal nucleotide binding domain (NBD; blue), substrate binding domain (SBD I; light green), C-terminal region (SBD II; yellow) and targeting signal peptides (S; pink). The molecular weight (MW), and isoelectric point (pI) for each T. brucei Hsp70 protein was calculated using the compute pI/Mw tool from ExPASy (https://web.expasy.org/compute_pi/; Gasteiger et al. 2005). Data on the phenotypic knockdown screen, using RNAi conducted by Alsford et al. (2011), for HSPA/Hsp70 protein member is provided: ALL-All lifecycle stages; BSF- Bloodstream; DIFF- Differentiation; NE- Non-essential; ND-Not determined. (PNG 273 kb)
Schematic representation of the domain architecture of the HSPH/Hsp110 proteins in T. brucei. Each protein sequence for the T. brucei HSPH/Hsp110 family is represented by an open bar with the number of amino acids indicated on either side of the protein bar with the various protein domains and other associated features that were identified using Prosite (Sigrist et al. 2009) and SMART (Letunic et al. 2012). These domains and associated features include the N-terminal nucleotide binding domain (NBD; blue), substrate binding domain (SBD I; dark green), C-terminal region (SBD II; yellow) and targeting signal peptides (S; pink). The molecular weight (MW), and isoelectric point (pI) for each T. brucei Hsp110 protein was calculated using the compute pI/Mw tool from ExPASy (https://web.expasy.org/compute_pi/; Gasteiger et al. 2005). Data on the phenotypic knockdown screen, using RNAi conducted by Alsford et al. (2011), for HSPH/Hsp110 protein member is provided: ALL-All lifecycle stages; BSF- Bloodstream; DIFF- Differentiation. (PNG 199 kb)
Alignment of the Type I, II and IV J-protein subfamilies from T. brucei in relation to human and other selected kinetoplastids. Multiple sequence alignment of the full-length amino acid sequences was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters in the MEGA7 software (Kumar et al. 2016). Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers for the J-protein amino acid sequences used in this study are provided in Table 2 and Table S2. (DOCX 104 kb)
Accession numbers for HSPA/Hsp70 and HSPH/Hsp110 amino acid sequences used in this study. (DOCX 27 kb)
Accession numbers for J-protein amino acid sequences used in this study. (DOCX 23 kb)
Funding information
This work was funded by a grant from the National Research Foundation (NRF), grant number 87663. S.J.B. is the recipient of an NRF Doctoral Innovation Scholarship. M.J. is the recipient of an NRF DAAD Fellowship.
References
- Acestor N, Panigrahi AK, Ogata Y, Anupama A, Stuart KD. Protein composition of Trypanosoma brucei mitochondrial membranes. Proteomics. 2009;9:5497–5508. doi: 10.1002/pmic.200900354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acestor N, Zíková A, Dalley RA, Anupama A, Panigrahi AK, Stuart KD. Trypanosoma brucei mitochondrial respiratome: composition and organization in procyclic form. Mol Cell Proteomics MCP. 2011;10:M110.006908. doi: 10.1074/mcp.M110.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarraberes FA, Dice JF. A molecular chaperon complex at the lysosomal membrane is required for protein translocation. J Cell Biol. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- Ahrendt E, Braun JE. Channel triage: emerging insights into the processing and quality control of hERG potassium channels by DnaJA proteins 1, 2 and 4. Channels (Austin) 2010;4(5):335–336. doi: 10.4161/chan.4.5.13090. [DOI] [PubMed] [Google Scholar]
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21(6):915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(Database issue):D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD, Uyetake L, Brickman MJ, Balber AE, Boothroyd JC. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J Cell Sci. 1993;105(4):1101–1113. doi: 10.1242/jcs.105.4.1101. [DOI] [PubMed] [Google Scholar]
- Bangs JD, Brouch EM, Ransom DM, Roggy JL. A soluble secretory reporter system in Trypanosoma brucei. Studies on endoplasmic reticulum targeting. J Biol Chem. 1996;271(31):18387–18393. doi: 10.1074/jbc.271.31.18387. [DOI] [PubMed] [Google Scholar]
- Barrett MP, Croft SL. Management of trypanosomiasis and leishmaniasis. Br Med Bull. 2012;104:175–196. doi: 10.1093/bmb/lds031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38(1):1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Botha M, Pesce ER, Blatch GL. The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: regulating chaperone power in the parasite and the host. Int J Biochem Cell Biol. 2007;39:1781–1803. doi: 10.1016/j.biocel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Brameier M, Krings A, MacCallum RM. NucPred—predicting nuclear localization of proteins. Bioinformatics. 2007;23:1159–1160. doi: 10.1093/bioinformatics/btm066. [DOI] [PubMed] [Google Scholar]
- Brehmer D, Rüdiger S, Gässler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8:427–432. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- Brocchieri L, de Macario EC, Macario AJL. hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol. 2008;8:19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu C, Haimeur A, Ouellette M. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite Leishmania. Cell Stress Chaperones. 2004;9(3):294–303. doi: 10.1379/CSC-15R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Blum J, Chappius F, Burri C. Human African trypanosomiasis. Lancet. 2010;375(9709):148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WMJ. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22(14):3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92(3):351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Burger A, Ludewig ML, Boshoff A. Investigating the chaperone properties of a novel heat shock protein, Hsp70.c, from Trypanosoma brucei. J Parasitol Res. 2014;2014:172582. doi: 10.1155/2014/172582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butter F, Bucerius F, Michel M, Cicova Z, Mann M, Janzen CJ. Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite’s host adaptation machinery. Mol Cell Proteomics MCP. 2013;12:172–179. doi: 10.1074/mcp.M112.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3(1):28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Codonho BS, Costa S, Peloso E de F, Joazeiro PP, Gadelha FR, Giorgio S. HSP70 of Leishmania amazonensis alters resistance to different stresses and mitochondrial bioenergetics. Mem Inst Oswaldo Cruz. 2016;111(7):460–468. doi: 10.1590/0074-02760160087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante C, Ellis M, Ruppert T, Voncken F (2006) Comparative proteomics of glycosomes from bloodstream form and procyclic culture formTrypanosoma brucei brucei. PROTEOMICS 6(11):3275-3293 [DOI] [PubMed]
- Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28(12):655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581(19):3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Davey KM, Parboosingh JS, McLeod DR, Chan A, Casey R, Ferreira P, Snyder FF, Bridge PJ, Bernier FP. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet. 2006;43(5):385–393. doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrasse JA, Chait BT, Field MC, Rout MP. High-yield isolation and subcellular proteomic characterization of nuclear and subnuclear structures from trypanosomes. Methods Mol Biol Clifton NJ. 2008;463:77–92. doi: 10.1007/978-1-59745-406-3_6. [DOI] [PubMed] [Google Scholar]
- Deocaris CC, Kaul SC, Wadhwa R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones. 2006;11(2):116–128. doi: 10.1379/CSC-144R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P, Lara E, Marande W, López-García P, Ekelund F, Moreira D. Phylogenomic analysis of kinetoplastids supports that trypanosomatids arose from within bodonids. Mol Biol Evol. 2011;28:53–58. doi: 10.1093/molbev/msq289. [DOI] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drini S, Criscuolo A, Lechat P, Imamura H, Skalický T, Rachidi N, Lukeš J, Dujardin JC, Späth GF. Species- and strain-specific adaptation of the HSP70 super family in pathogenic Trypanosomatids. Genome Biol Evol. 2016;8(6):1980–1995. doi: 10.1093/gbe/evw140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droll D, Minia I, Fadda A, Singh A, Stewart M, Queiroz R, Clayton C. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013;9(4):e1003286. doi: 10.1371/journal.ppat.1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak JA. The natural heterogeneity of Trypanosoma cruzi: biological and medical implications. J Cell Biochem. 1984;24(4):357–371. doi: 10.1002/jcb.240240406. [DOI] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR. The Hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5(4):276–290. doi: 10.1379/1466-1268(2000)005<0276:THAGSP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edkins AL, Ludewig MH, Blatch GL. A Trypanosoma cruzi heat shock protein 40 is able to stimulate the adenosine triphosphate hydrolysis activity of heat shock protein 70 and can substitute for a yeast heat shock protein 40. Int J Biochem Cell Biol. 2004;36(8):1585–1598. doi: 10.1016/j.biocel.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Emmer BT, Nakayasu ES, Souther C, Choi H, Sobreira TJP, Epting CL, Nesvizhskii AI, Almeida IC, Engman DM (2011) Global analysis of protein palmitoylation in African trypanosomes. Eukaryot Cell 10(3):455-463 [DOI] [PMC free article] [PubMed]
- Engman DM, Kirchhoff LV, Donelson JE. Molecular cloning of mtp70, a mitochondrial member of the hsp70 family. Mol Cell Biol. 1989;9(11):5163–5168. doi: 10.1128/mcb.9.11.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstler M, Pfohl T, Herminghaus S, Boshart M, Wiegertjes G, Heddergott N, Overath P. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131(3):505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- Field MC, Sergeenko T, Wang Y-N, Böhm S, Carrington M. Chaperone requirements for biosynthesis of the trypanosome variant surface glycoprotein. PLoS One. 2010;5(1):e8468. doi: 10.1371/journal.pone.0008468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueira C, Requena JM. A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol Rev. 2007;31(4):359–377. doi: 10.1111/j.1574-6976.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15(12):2969–2979. doi: 10.1002/j.1460-2075.1996.tb00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. New York: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gazestani VH, Yip CW, Nikpour N, Berghuis N, Salavati R. TrypsNetDB: an integrated framework for the functional characterization of trypanosomatid proteins. PLoS Negl Trop Dis. 2017;11(2):e0005368. doi: 10.1371/journal.pntd.0005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. The origins of the trypanosome genome strains Trypanosoma brucei brucei TREU 927, T. b. gambiense DAL 972, T. vivax Y486 and T. congolense IL3000. Parasit Vectors. 2012;5:71. doi: 10.1186/1756-3305-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard M, McPherson PS. RME-8 regulates trafficking of the epidermal growth factor receptor. FEBS Lett. 2008;582(6):961–966. doi: 10.1016/j.febslet.2008.02.042. [DOI] [PubMed] [Google Scholar]
- Girard M, Poupon V, Blondeau F, McPherson PS. The DnaJ-domain protein RME-8 functions in endosomal trafficking. J Biol Chem. 2005;280:40135–40143. doi: 10.1074/jbc.M505036200. [DOI] [PubMed] [Google Scholar]
- Glass DJ, Polvere RI, Van der Ploeg LH. Conserved sequences and transcription of the hsp70 gene family in Trypanosoma brucei. Mol Cell Biol. 1986;6(12):4657–4666. doi: 10.1128/mcb.6.12.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt H, Sheiner L, Butikofer P, Roditi I, Uliel S, Gunzel M, et al. Role of protein translocation pathways across the endoplasmic reticulum in Trypanosoma brucei. J Biol Chem. 2008;283:32085–32098. doi: 10.1074/jbc.M801499200. [DOI] [PubMed] [Google Scholar]
- Goodman AG, Tanner BC, Chang ST, Esteban M, Katze MG. Virus infection rapidly activates the P58(IPK) pathway, delaying peak kinase activation to enhance viral replication. Virology. 2011;417(1):27–36. doi: 10.1016/j.virol.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goos C, Dejung M, Janzen CJ, Butter F, Kramer S. The nuclear proteome of Trypanosoma brucei. PLoS One. 2017;12:e0181884. doi: 10.1371/journal.pone.0181884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisard EC, Teixeira SMR, de Almeida LGP, et al. Trypanosoma cruzi clone Dm28c draft genome sequence. genome announcements. Genome Announc. 2014;2(1):e01114–e01113. doi: 10.1128/genomeA.01114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera K, Wüthrich D, Braga-Lagache S, Heller M, Ochsenreiter T. Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics. 2012;13:556. doi: 10.1186/1471-2164-13-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Snapp EL. ERdj3 regulates BiP occupancy in living cells. J Cell Sci. 2013;126(6):1429–1439. doi: 10.1242/jcs.118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Singh B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol. 1994;4:1104–1114. doi: 10.1016/S0960-9822(00)00249-9. [DOI] [PubMed] [Google Scholar]
- Güther MLS, Urbaniak MD, Tavendale A, Prescott A, Ferguson MAJ. High-confidence glycosome proteome for procyclic form Trypanosoma brucei by epitope-tag organelle enrichment and SILAC proteomics. J Proteome Res. 2014;13:2796–2806. doi: 10.1021/pr401209w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381(6583):571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hatle KM, Gummadidala P, Navasa N, Bernardo E, Dodge J, Silverstrim B, Fortner K, Burg E, Suratt BT, Hammer J, Radermacher M, Taatjes DJ, Thornton T, Anguita J, Rincon M. MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations. Mol Cell Biol. 2013;33(11):2302–2314. doi: 10.1128/MCB.00189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglund A, Dönnes P, Blum T, Adolph HW, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, van der Ploeg LH. Maturation of polycistronic pre-mRNA in Trypanosoma brucei: analysis of trans splicing and poly(A) addition at nascent RNA transcripts from the hsp70 locus. Mol Cell Biol. 1991;11(6):3180–3190. doi: 10.1128/mcb.11.6.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Nonlinear relationships among evolutionary rates identify regions of functional divergence in heat-shock protein-70 genes. Mol Biol Evol. 1993;10:243–255. doi: 10.1093/oxfordjournals.molbev.a039997. [DOI] [PubMed] [Google Scholar]
- Iosefson O, Sharon S, Goloubinoff P, Azem A. Reactivation of protein aggregates by mortalin and Tid1-the human mitochondrial Hsp70 chaperone system. Cell Stress Chaperones. 2012;17:57–66. doi: 10.1007/s12192-011-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, Chukualim B, Capewell P, MacLeod A, Melville SE, Gibson W, Barry JD, Berriman M, Hertz-Fowler C. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLoS Negl Trop Dis. 2010;4(4):e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zhuang M, Hendershot LM. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009;48:41–49. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kim J, Roh SH, Jun I, Sampson RD, Gee HY, Choi JY, Lee MG. The HSP70 co-chaperone DNAJC14 targets misfolded pendrin for unconventional protein secretion. Nat Commun. 2016;7:11386. doi: 10.1038/ncomms11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Martineau CN. Multiple hsp70 isoforms in the eukaryotic cytosol: mere redundancy or functional specificity? Curr Genomics. 2008;9(5):338–248. doi: 10.2174/138920208785133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanquay RM, Bruford EA, et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14(1):105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschner LA, Sharma R, Shrestha OK, Meyer AE, Craig EA. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim Biophys Acta. 2015;1853(5):1035–1045. doi: 10.1016/j.bbamcr.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KG, Olson CL, Engman DM. Mitochondrial heat shock protein 70 is distributed throughout the mitochondrion in a dyskinetoplastic mutant of Trypanosoma brucei. Mol Biochem Parasitol. 1995;70:207–209. doi: 10.1016/0166-6851(95)00013-Q. [DOI] [PubMed] [Google Scholar]
- Kominek J, Marszalek J, Neuveglise C, Craig EA, Williams BL. The complex evolutionary dynamics of Hsp70s: a genomic and functional perspective. Genome Biol Evol. 2013;5:2460–2477. doi: 10.1093/gbe/evt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1879. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20(7):257–259. doi: 10.1016/S0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshield G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lee MG, Van der Ploeg LH. Transcription of the heat shock 70 locus in Trypanosoma brucei. Mol Biochem Parasitol. 1990;41(2):221–231. doi: 10.1016/0166-6851(90)90185-O. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24(21):9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw CA, Ludewig ML, Mayer J, Blatch GL. The Hsp70 chaperones of the Tritryps are characterized by unusual features and novel members. Parasitol Int. 2010;59(4):497–505. doi: 10.1016/j.parint.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Louw CA, Ludewig MH, Blatch GL. Overproduction, purification and characterisation of Tbj1, a novel type III Hsp40 from Trypanosoma brucei, the African sleeping sickness parasite. Protein Expr Purif. 2010;69(2):168–177. doi: 10.1016/j.pep.2009.09.023. [DOI] [PubMed] [Google Scholar]
- Ludewig MH, Boshoff A, Horn D, Blatch GL. Trypanosoma brucei J protein 2 is a stress inducible and essential Hsp40. Int J Biochem Cell Biol. 2015;60:93–98. doi: 10.1016/j.biocel.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Maharjan M, Madhubala R. Heat shock protein 70 (HSP70) expression in antimony susceptible/resistant clinical isolates of Leishmania donovani. Nepal J Biotechnol. 2015;3(1):22–28. doi: 10.3126/njb.v3i1.14225. [DOI] [Google Scholar]
- Marcili A, Valente VC, Valente SA, Junqueira AC, da Silva FM, Pinto AY, et al. Trypanosoma cruzi in Brazilian Amazonia: lineages TCI and TCIIa in wild primates, Rhodnius spp. and in humans with Chagas disease associated with oral transmission. Int J Parasitol. 2009;39(5):615–623. doi: 10.1016/j.ijpara.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen LA, Chang C, Garrels JI, Welch WJ. Identification, characterization, and purification of two mammalian stress proteins present in mitochondria, grp 75, a member of the hsp 70 family and hsp 58, a homolog of the bacterial groEL protein. J Biol Chem. 1989;264:20664–20675. doi: 10.1016/S0021-9258(19)47115-9. [DOI] [PubMed] [Google Scholar]
- Moffatt NS, Bruinsma E, Uhl C, Obermann WM, Toft D. Role of the cochaperone Tpr2 in Hsp90 chaperoning. Biochemistry. 2008;47:8203–8213. doi: 10.1021/bi800770g. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003;22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Oksman A, Pal P, Lindquist S, Goldberg DE. Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeatrich parasite proteome during malarial fevers. Nat Commun. 2012;3:1310. doi: 10.1038/ncomms2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy AE, Sohal SK, Carrington M, Bishop RP, Allsopp BA. Identification and characterization of two novel tetratricopeptide repeat-containing genes. DNA Cell Biol. 1996;15:727–735. doi: 10.1089/dna.1996.15.727. [DOI] [PubMed] [Google Scholar]
- Nett IRE, Martin DMA, Miranda-Saavedra D, Lamont D, Barber JD, Mehlert A, Ferguson MAJ. The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol Cell Proteomics MCP. 2009;8:1527–1538. doi: 10.1074/mcp.M800556-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann M, Wiese S, Mani J, Chanfon A, Jackson C, Meisinger C, Warscheid B, Schneider A. Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol Cell Proteomics MCP. 2013;12:515–528. doi: 10.1074/mcp.M112.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Kanayama T, Moore R, Ray M, Negishi M. The roles of co-chaperone CCRP/DNAJC7 in Cyp2b10 gene activation and steatosis development in mouse livers. PLoS One. 2014;9(12):e115663. doi: 10.1371/journal.pone.0115663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H, Conz C, Maier P, Wölfle T, Suzuki CK, Jenö P, et al. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci U S A. 2005;102(29):10064–10069. doi: 10.1073/pnas.0504400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ogata Y, Zíková A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peikert CD, Mani J, Morgenstern M, Käser S, Knapp B, Wenger C, Harsman A, Oeljeklaus S, Schneider A, Warscheid B. Charting organellar importomes by quantitative mass spectrometry. Nat Commun. 2017;8:15272. doi: 10.1038/ncomms15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Quieroz R, Benz C, Fellenberg K, Hoheisel J, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics. 2009;10:495. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena JM, Montalvo AM, Fraga J. Molecular chaperones of Leishmania: central players in many stress-related and -unrelated physiological processes. Biomed Res Int. 2015;2015:301326. doi: 10.1155/2015/301326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18(9):3681–3691. doi: 10.1091/mbc.e07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D, Montero-Lomeli M, Goldenberg S. A DnaJ-like protein homologous to the yeast cochaperone Sis1 (TcJ6p) is involved in initiation of translation in Trypanosoma cruzi. J Biol Chem. 2001;276:43970–43979. doi: 10.1074/jbc.M102427200. [DOI] [PubMed] [Google Scholar]
- Saxena A, Banasavadi-Siddegowda YK, Fan Y, Bhattacharya S, Roy G, Giovannucci DR, Frizzell RA, Wang X. Human heat shock protein 105/110 kDa (Hsp105/110) regulates biogenesis and quality control of misfolded cystic fibrosis transmembrane conductance regulator at multiple levels. J Biol Chem. 2012;287:19158–19170. doi: 10.1074/jbc.M111.297580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/ Kar2p via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle S, Smith DF. Leishmania major: characterisation and expression of a cytoplasmic stress-related protein. Exp Parasitol. 1993;77(1):43–52. doi: 10.1006/expr.1993.1059. [DOI] [PubMed] [Google Scholar]
- Searle S, Campos AJ, Coulson RM, Spithill TW, Smith DF. A family of heat shock protein 70-related genes are expressed in the promastigotes of Leishmania major. Nucleic Acids Res. 1989;17(13):5081–5095. doi: 10.1093/nar/17.13.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa MM, Saada EA, Vashisht AA, Barshop WD, Wohlschlegel JA, Hill KL. Cell surface proteomics provides insight into stage-specific remodeling of the host-parasite interface in Trypanosoma brucei. Mol Cell Proteomics MCP. 2015;14:1977–1988. doi: 10.1074/mcp.M114.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonhai A, Maier AG, Przyborski JM, Blatch GL. Intracellular protozoan parasites of humans: the role of molecular chaperones in development and pathogenesis. Protein Pept Lett. 2011;18:143–157. doi: 10.2174/092986611794475002. [DOI] [PubMed] [Google Scholar]
- Shrestha L, Bolaender A, Patel HJ, Taldone, T (2016) Heat shock protein (HSP) drug discovery and development: targeting heat shock proteins in disease. Curr Top Med Chem 16(25):2753-2764 [DOI] [PMC free article] [PubMed]
- Sigrist CJ, Cerutti L, De Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2009;38:161–166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro PP, Jannin J, Cattand P. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 2008;5:174–180. doi: 10.1371/journal.pmed.0050055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, Stevens JR, Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Subota I, Julkowska D, Vincensini L, Reeg N, Buisson J, Blisnick T, Huet D, Perrot S, Santi-Rocca J, Duchateau M, Hourdel V, Rousselle J-C, Cayet N, Namane A, Chamot-Rooke J, Bastin P. Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localization and dynamics. Mol Cell Proteomics MCP. 2014;13:1769–1786. doi: 10.1074/mcp.M113.033357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam C, Veazey P, Redmond S, Hayes-Sinclair J, Chambers E, Carrington M, Gull K, Matthews K, Horn D, Field MC. Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryot Cell. 2006;5:1539–1549. doi: 10.1128/EC.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Yomogida K, Imai T, Kiyonari H, Takeda N, Kadomatsu T, Yano M, Aizawa S, Mori M. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO J. 2005;24(3):611–622. doi: 10.1038/sj.emboj.7600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M. Beyond strain typing and molecular epidemiology: integrated genetic epidemiology of infectious disease. Parasitol Today. 1998;14:323–329. doi: 10.1016/S0169-4758(98)01286-1. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/S1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Ward P, Moya A, Ayala FJ. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc Natl Acad Sci U S A. 1986;83:1335–1339. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigankov P, Gherardini PF, Helmer-Citterich M, Späth GF, Myler PJ, Zilberstein D. Regulation dynamics of Leishmania differentiation: deconvoluting signals and identifying phosphorylation trends. Mol Cell Proteomics. 2014;13(7):1787–1799. doi: 10.1074/mcp.M114.037705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Týč J, Klingbeil MM, Lukeš J. Mitochondrial heat shock protein machinery Hsp70/Hsp40 is indispensable for proper mitochondrial DNA maintenance and replication. MBio. 2015;6(1):e02425–e02414. doi: 10.1128/mBio.02425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, Rouault TA. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet. 2010;19(19):3816–3834. doi: 10.1093/hmg/ddq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Guther MLS, Ferguson MAJ. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One. 2012;7:e36619. doi: 10.1371/journal.pone.0036619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Martin DMA, Ferguson MAJ. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res. 2013;12:2233–2244. doi: 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urmenyi TP, Silva R, Rondinelli E. The heat shock proteins of Trypanosoma cruzi. In: Santos A, Branquinha M, d’Avila-Levy C, Kneipp L, Sodré C, editors. Proteins and proteomics of Leishmania and Trypanosoma. Subcell Biochem, vol. Dordrecht: Springer; 2014. p. 74. [DOI] [PubMed] [Google Scholar]
- Wallace FG. The trypanosomatid parasites of insects and arachnids. Exp Parasitol. 1966;18:124–193. doi: 10.1016/0014-4894(66)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Pezeshki AM, Yu X, Guo C, Subjeck JR, Wang XY. The endoplasmic reticulum chaperone GRP170: from immunobiology to cancer therapeutics. Front Oncol. 2014;4:377. doi: 10.3389/fonc.2014.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzler Y, Schumann Burkard G, Schmidt RS, Mäser P, Bergner A, Roditi I, Brun R (2016) A new approach to chemotherapy: drug-induced differentiation kills African trypanosomes. Sci Rep 6(1) [DOI] [PMC free article] [PubMed]
- Wiesgigl M, Clos J. The heat shock protein 90 of Leishmania donovani. Med Microbiol Immunol. 2001;190(1–2):27–31. doi: 10.1007/s004300100074. [DOI] [PubMed] [Google Scholar]
- World Health Organization & WHO Expert Committee on the Control and Surveillance of Human African Trypanosomiasis (2013: Geneva, Switzerland). (2013). Control and surveillance of human African trypanosomiasis: report of a WHO expert committee. World Health Organization. http://www.who.int/iris/handle/10665/95732. Accessed 28 September 2018
- World Health Organization . Report of the second WHO stakeholders meeting on rhodesiense human African trypanosomiasis, Geneva, 26–28 April 2017. Geneva: World Health Organization; 2017. [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99(25):15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Nakai A, Hatayama T, Nagata K. Cloning and expression of murine high molecular mass heat shock proteins, HSP105. J Biol Chem. 1995;270(50):29718–29723. doi: 10.1074/jbc.270.50.29718. [DOI] [PubMed] [Google Scholar]
- Zíková A, Verner Z, Nenarokova A, Michels PAM, Lukeš J. A paradigm shift: the mitoproteomes of procyclic and bloodstream Trypanosoma brucei are comparably complex. PLoS Pathog. 2017;13:e1006679. doi: 10.1371/journal.ppat.1006679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, et al. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12(2):240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the Hsp70 superfamily from T. brucei in relation to human and other selected kinetoplastids. Multiple sequence alignment of the full-length amino acid sequences was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters in the MEGA7 software (Kumar et al. 2016). Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers for the J-protein amino acid sequences used in this study are provided in Table 1 and Table S1. (DOCX 220 kb)
Schematic representation of the domain architecture of the HSPA/Hsp70 proteins in T. brucei. Each protein sequence for the T. brucei HSPA/Hsp70 family is represented by an open bar with the number of amino acids indicated on either side of the protein bar with the various protein domains and other associated features that were identified using Prosite (Sigrist et al. 2009) and SMART (Letunic et al. 2012). These domains and associated features include the N-terminal nucleotide binding domain (NBD; blue), substrate binding domain (SBD I; light green), C-terminal region (SBD II; yellow) and targeting signal peptides (S; pink). The molecular weight (MW), and isoelectric point (pI) for each T. brucei Hsp70 protein was calculated using the compute pI/Mw tool from ExPASy (https://web.expasy.org/compute_pi/; Gasteiger et al. 2005). Data on the phenotypic knockdown screen, using RNAi conducted by Alsford et al. (2011), for HSPA/Hsp70 protein member is provided: ALL-All lifecycle stages; BSF- Bloodstream; DIFF- Differentiation; NE- Non-essential; ND-Not determined. (PNG 273 kb)
Schematic representation of the domain architecture of the HSPH/Hsp110 proteins in T. brucei. Each protein sequence for the T. brucei HSPH/Hsp110 family is represented by an open bar with the number of amino acids indicated on either side of the protein bar with the various protein domains and other associated features that were identified using Prosite (Sigrist et al. 2009) and SMART (Letunic et al. 2012). These domains and associated features include the N-terminal nucleotide binding domain (NBD; blue), substrate binding domain (SBD I; dark green), C-terminal region (SBD II; yellow) and targeting signal peptides (S; pink). The molecular weight (MW), and isoelectric point (pI) for each T. brucei Hsp110 protein was calculated using the compute pI/Mw tool from ExPASy (https://web.expasy.org/compute_pi/; Gasteiger et al. 2005). Data on the phenotypic knockdown screen, using RNAi conducted by Alsford et al. (2011), for HSPH/Hsp110 protein member is provided: ALL-All lifecycle stages; BSF- Bloodstream; DIFF- Differentiation. (PNG 199 kb)
Alignment of the Type I, II and IV J-protein subfamilies from T. brucei in relation to human and other selected kinetoplastids. Multiple sequence alignment of the full-length amino acid sequences was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters in the MEGA7 software (Kumar et al. 2016). Degree of amino acid conservation is symbolized by the following: (*) all fully conserved residues; (:) one of the residues is fully conserved and (.) residues are weakly conserved. Accession numbers for the J-protein amino acid sequences used in this study are provided in Table 2 and Table S2. (DOCX 104 kb)
Accession numbers for HSPA/Hsp70 and HSPH/Hsp110 amino acid sequences used in this study. (DOCX 27 kb)
Accession numbers for J-protein amino acid sequences used in this study. (DOCX 23 kb)