Abstract
The type III secretion system (T3SS) plays an important role in the pathogenesis of Pseudomonas aeruginosa. Expression of the T3SS is controlled under a complicate regulatory network. In this study, we demonstrate that NrtR (PA4916) is involved in the T3SS expression and pathogenesis of P. aeruginosa in a mouse acute pneumonia model. Overexpression of the T3SS central activator ExsA or exogenous supplementation of cAMP restored the expression of T3SS in the ΔnrtR mutant, suggesting that NrtR might regulate T3SS through the cAMP-Vfr signaling pathway. Further experiments demonstrated that the decrease of cAMP content is not due to the expression change of adenylate cyclases or phosphodiesterase in the ΔnrtR mutant. As it has been shown that nadD2 is upregulated in the ΔnrtR mutant, we overexpressed nadD2 in wild type PAK, which reduced the intracellular cAMP level and the expression of the T3SS genes. Meanwhile, deletion of nadD2 in the ΔnrtR mutant restored the expression and secretion of the T3SS. Co-immunoprecipitation assay revealed an interaction between NadD2 and the catalytic domain of the adenylate cyclase CyaB. Further in vitro assay indicated that NadD2 repressed the enzymatic activity of CyaB. Therefore, we have identified a novel regulatory mechanism of T3SS in P. aeruginosa.
Keywords: Pseudomonas aeruginosa, NrtR, cAMP, CyaB, type III secretion system
Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium that can cause both acute and chronic infections in individuals with compromised immunity such as cancer patients and those with cystic fibrosis (Crowe et al., 1982; Sherertz and Sarubbi, 1983; Lyczak et al., 2002; Sousa and Pereira, 2014).
The type III secretion system (T3SS) is an important virulence factor of P. aeruginosa, through which effector proteins are directly injected into the cytosols of eukaryotic host cells, inhibiting host defense by inducing cell death in polymorphonuclear phagocytes, macrophages, and epithelial cells (Dacheux et al., 1999; Hauser and Engel, 1999; Kaufman et al., 2000). Expression of the T3SS confers an increased virulence in P. aeruginosa and is associated with poor clinical outcomes (El-Solh et al., 2012), whereas strains with defective T3SS display attenuated virulence in mouse acute infection models (Smith et al., 2004). To date, four effector proteins have been identified and well characterized in P. aeruginosa, i.e., ExoS, ExoT, ExoU, and ExoY (Hauser, 2009). However, majority of P. aeruginosa isolates do not encode all of the four effectors (Feltman et al., 2001). For example, strain PAK expresses ExoS, ExoT, and ExoY, while strain PA14 expresses ExoU, ExoT, and ExoY.
In P. aeruginosa, the T3SS is induced in response to a variety of environmental conditions, such as direct contact with host cell, calcium depletion and the presence of serum (Kim et al., 2005; Hayes et al., 2010). The expression of T3SS is activated by ExsA, an AraC-type DNA binding protein, which recognizes and binds to two adjacent highly conserved consensus sequences in the promoter region of the T3SS genes (Hovey and Frank, 1995; Brutinel et al., 2008). The ExsA activity and transcriptional regulation on T3SS are intimately coupled to secretion by a partner-switching model involving three other proteins: ExsC, ExsD, and ExsE (Brutinel and Yahr, 2008). Under non-inducing condition, the secretable repressor ExsE is kept inside bacterial cytosol and binds to ExsC, and ExsD binds to and inactivates ExsA. Whereas under inducing environment, ExsE is secreted by the T3SS machinery, which releases ExsC to sequester its low affinity partner ExsD, resulting in free ExsA that activates the transcription of whole T3SS gene cluster (McCaw et al., 2002; Dasgupta et al., 2004; Rietsch et al., 2005; Urbanowski et al., 2005, 2007; Thibault et al., 2009; Diaz et al., 2011).
The virulence factor regulator Vfr is a cAMP-dependent transcriptional regulator. It was originally identified as an activator of extracellular protease and exotoxin A expression. Now it is appreciated as a global regulator of virulence gene expression, including T3SS and pili biosynthesis genes (West et al., 1994; Wolfgang et al., 2003; Marsden et al., 2016). Intracellular cAMP is generated by adenylate cyclases CyaA and CyaB and hydrolyzed by the phosphodiesterase CpdA in P. aeruginosa (Wolfgang et al., 2003; Fuchs et al., 2010). Besides, other regulators and proteins are also known to affect expression of the T3SS, such as the rhl quorum sensing system (Hogardt et al., 2004), stationary-phase sigma factor RpoS (Hogardt et al., 2004), transcriptional activator PsrA (Shen et al., 2006), global regulator MexT (Jin et al., 2011), alginate biosynthesis protein MucA (Wu et al., 2004), RNA-binding proteins RsmA and Crc (Mulcahy et al., 2006; Dong et al., 2013), small proteins PtrA, PtrB, and PtrC (Ha et al., 2004; Wu and Jin, 2005; Jin et al., 2011), tryptophan synthase TrpA (Shen et al., 2008), tryptophan dioxygenase KynA (Shen et al., 2008), pseudouridinase enzyme TruA (Ahn et al., 2004), nitrite reductase NirS (Van Alst et al., 2009), magnesium transporter MgtE (Anderson et al., 2010), two-component system AlgZR (Intile et al., 2014), DNA binding protein Fis (Deng et al., 2017), multiple virulence regulator SuhB and RNA helicase DeaD (Li et al., 2013; Intile et al., 2015). However, the molecular mechanism by which most of these proteins regulate T3SS is not fully elucidated yet. All of these suggest that expression of T3SS is controlled under a complicate regulatory network. Therefore, we aimed to identify novel regulators of T3SS and their regulatory mechanisms.
In the present study, we identified that NrtR (PA4916) is required for the expression of T3SS in P. aeruginosa. In vivo studies suggest that NrtR plays an important role in the pathogenesis of P. aeruginosa. Further studies demonstrated that NrtR affects expression of the T3SS through cAMP/Vfr signaling system that lies upstream of the ExsA. We demonstrated that NadD2 is involved in the NrtR mediated regulation of the T3SS by inhibiting the CyaB enzymatic activity and subsequent reducing intracellular cAMP level.
Materials and Methods
Ethics Statement
All animal studies complied with National and Nankai University guidelines regarding the use of animals in research. All animal experiment protocols have been approved by the institutional animal care and use committee of the College of Life Sciences of Nankai University (permit number NK-04-2012).
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Both E. coli and P. aeruginosa were grown in Luria-Bertani broth (LB) medium (Deng et al., 2017) or on LB agar (Deng et al., 2017) plates at 37°C. Whenever needed, antibiotics were used at following concentrations (μg/ml): for E. coli, ampicillin 100, tetracycline 10, spectinomycin 50, streptomycin 25, gentamicin 10; for P. aeruginosa, carbenicillin 150, tetracycline 50, spectinomycin 200, streptomycin 200, gentamicin 100. When needed, 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) or 50 mM cAMP were added to culture medium. Primers used to make various constructs and in RT-qPCR are listed in Table 2.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F- ϕ 80dlacZΔM15 endA1 recA1 hsdR17(rK- mK+) supE44 thi-1 relA1 Δ(lacZYA-argF)U169 gyrA96 deoR | TransGen |
| S17-1 | RP4-2 Tc::Mu Km::Tn7 Tpr Smr Pro Res- Mod+ | Dr. Ramphal |
| HB101 | Source for wild type lacP1 promoter sequence | TransGen |
| P. aeruginosa strains | ||
| PA14 | Wild type P. aeruginosa strain | Liberati et al., 2006 |
| PAK | Wild type P. aeruginosa strain | David Bradley |
| exsA::Tn | PA14 with exsA disrupted by insertion of Tn | Liberati et al., 2006 |
| PA4336::Tn | PA14 with PA4336 inserted with Tn | Liberati et al., 2006 |
| PA4916::Tn | PA14 with PA4916 inserted with Tn | Liberati et al., 2006 |
| PA0020::Tn | PA14 with PA0020 inserted with Tn | Liberati et al., 2006 |
| PA4753::Tn | PA14 with PA4753 inserted with Tn | Liberati et al., 2006 |
| exsA::Ω | PAK with exsA disrupted by insertion of cassette Ω; Spr, Smr | Li et al., 2013 |
| ΔnadD2 | PAK with nadD2 deleted | This study |
| ΔnadD2-nrtR | PAK with nadD2-nrtR operon deleted | This study |
| ΔPA4916 | PAK with PA4916 deleted | This study |
| ΔPA4916/att7::PA4916 | ΔPA4916 with PA4916 inserted into the chromosome with mini-Tn7 insertion | This study |
| Δvfr | PAK with vfr gene deleted | This study |
| ΔnrtRΔvfr | PAK with both nrtR and vfr gene deleted | This study |
| Plasmids | ||
| pDN19 | Shuttle vector between E. coli and P. aeruginosa; Tcr | Li et al., 2013 |
| pMMB67EH | Shuttle vector between E. coli and P. aeruginosa; Ampr | Li et al., 2016 |
| pE1553 | Promoterless pUCP20; Ampr | Li et al., 2016 |
| pE1553a | cyaA-flag with own promoter in pE1553; Ampr | This study |
| pE1553b | cyaB-flag with own promoter in pE1553; Ampr | This study |
| pE1553-cpdA | cpdA-flag with own promoter in pE1553; Ampr | This study |
| pMMB67-cyaA | cyaA-flag cloned into pMMB67EH driven by tac promoter; Ampr | This study |
| pMMB67-cyaB | cyaB-flag cloned into pMMB67EH driven by tac promoter; Ampr | This study |
| pMMB67-nadD2 | nadD2 gene of PAK on pMMB67EH driven by tac promoter; Ampr | This study |
| pMMB67-4918-20 | PA4918-20 gene of PAK on pMMB67EH driven by tac promoter; Ampr | This study |
| pMMB67EH-cyaB217-463-His | CyaB217-463-His cloned into pMMB67EH driven by tac promoter; Ampr | This study |
| pMMB67EH-cyaA-His | CyaA-His cloned into pMMB67EH driven by tac promoter; Ampr | This study |
| pUCP24 | Shuttle vector between E. coli and P. aeruginosa; Gmr | Shi et al., 2015 |
| pUCP24-nadD2-Flag | nadD2-Flag cloned into pUCP24; Gmr | This study |
| exsA | exsA gene of PAK on pDN19 driven by lac promoter; Tcr | Li et al., 2013 |
| lacP1 | LacP1 promoter of E. coli fused to promoterless lacZ on pDN19lacZΩ; Spr, Smr, Tcr | This study |
| pUC18T-mini-Tn7T-Gm | mini-Tn7 base vector insertion into chromosome attTn7 site, Gmr | Li et al., 2013 |
| pUC18T-mini-Tn7T-PexsC-exsCEBA-Flag-ExsD | exsCEBAD gene with exsA-Flag tagged on pUC18T-mini-Tn7T driven by exsC promoter, Gmr | This study |
| pTNS3 | Helper plasmid encoding Tn7 site-specific transposition pathway; Ampr | Deng et al., 2017 |
| pEX18Tc | Gene knockout vector; Tcr | Deng et al., 2017 |
| pZF01 | PA4916 gene deletion on pEX18Tc; Tcr | This study |
| pEX18Tc-ΔnadD2 | nadD2 gene deletion on pEX18Tc; Tcr | This study |
| pEX18Tc-ΔnadD2-nrtR | nadD2-nrtR operon deletion on pEX18Tc; Tcr | This study |
| pEX18Tc-Δvfr | vfr gene deletion on pEX18Tc; Tcr | This study |
| pEX18Tc-ΔcyaA | cyaA gene deletion on pEX18Tc; Tcr | This study |
| pEX18Tc-ΔcyaB | cyaB gene deletion on pEX18Tc; Tcr | This study |
| pZF02 | PA4916 gene on pUC18T-mini-Tn7T-Tc; Tcr | This study |
| pET16b | Expression vector, Kanr | Novagen |
| pET16b-nadD2 | nadD2 gene of PAK cloned into pET16b | This study |
| pET28a | Expression vector, Kanr | Novagen |
| pET28a-cyaB217-463 | cyaB gene encoding amino acid positions 217–463 cloned into pET28a | This study |
Table 2.
Primers used in this study.
| Primer | Sequence 5′–3′b | Source |
|---|---|---|
| PA4916UFa | CTCGGAATTCTTCCAGACGAAGAAGTCGTAG | This study |
| PA4916UR | CTGCTCTAGACGTCACTCCTCTCTTCAGCCC | This study |
| PA4916DF | GAGCTCTAGACCTGCCGCGCTTGCTAGACG | This study |
| PA4916DR | GCCCAAGCTTAAATCATCGAGTCGCTGGTCCCC | This study |
| nadD2UF | GGAATTCGAAGACCTCCACCTCCAGTGTCG | This study |
| nadD2UR | GCTCTAGAAGAGAGGAGTGACGATGAGTTCAG | This study |
| nadD2DF | GCTCTAGAAGAAAATACCTTCCACTGCG | This study |
| nadD2DR | CCCAAGCTTGAGCAGGTTCTGCACAATGC | This study |
| cyaAUF | GGAATTCCGGCATCCGTTGTTCCGCGCGGAGATCCAG | This study |
| cyaAUR | GCTCTAGAGGGCGTCCGGGCACAGGCAAGGCCAGGCG | This study |
| cyaADF | GCTCTAGACCCAGCGCCGCACCGCGCGGGGCTCGAC | This study |
| cyaADR | CCCAAGCTTCGCCGGCGAAGGCAAGGTCTCGATCCTC | This study |
| cyaBUF | GGAATTCGGAAAGTCAGGTCGGACGCTTCCGCGATG | This study |
| cyaBUR | GCTCTAGAGCGCTGGAGAGGATCCCTGTGTATTTTCG | This study |
| cyaBDF | GCTCTAGAGTTCGTCGAACGCCGCCGGCAGTTCGTCGCGCC | This study |
| cyaBDR | CCCAAGCTTCCGCTCGGCTGGGCCGCGCGGCGCTGGC | This study |
| vfrUF | CTCGGAATTCGTAGCAGATGTCGTAGATGTTG | This study |
| vfrUR | CTGGGGTACCCGAGTCCCGAAAGAATAAAG | This study |
| vfrDF | GAGGGGTACCTGGTGCATGTGAAAGGAAAGAC | This study |
| vfrDR | GCCCAAGCTTGCGACCAGCCTGCACGAG | This study |
| PA4916PF | GCGAGCTCTCCTTGCTGCCCAGGCGCAGC | This study |
| PA4916PR | AACTGCAGTCACTTGCCGAAGGCGTGGCGGTGG | This study |
| PA4916F | AACTGCAGGGCGGTCTGAAGAGAGGAGTGACG | This study |
| PA4916R | CCCAAGCTTTGCCGCACCCGTTTGTCAGG | This study |
| cyaAFown | GGGGTACCGCAGCGCATCCTCGCCAGCGGCGAG | This study |
| cyaAFtac | GGGGTACCCTGGCCTTGCCTGTGCCCGGACGCCC | This study |
| cyaAR | CCCAAGCTTTCACTTGTCGTCATCGTCCTTGTAGTCTTGTTCCAGCAGCGCCTGGTTCAGCGCCG | This study |
| cyaBFown | GGGGTACCTCGCCGAGTTCTACCCCTACTACCTGCAG | This study |
| cyaBFtac | GGGGTACCATACACAGGGATCCTCTCCAGCGCATG | This study |
| cyaBR | CCCAAGCTTTCACTTGTCGTCATCGTCCTTGTAGTCGAGGATGACCTTGTCGCGCAGGCGTTCGG | This study |
| nadD2OF | GGAATTCCAAGCACTTGTACTACAAAATTTCGCAG | This study |
| nadD2ORHis | CCCAAGCTTTCAATGATGATGATGATGATG GCCCGCCTGGCGTTCGCCGCCATAGCAGTG | This study |
| nadD2ORFlag | CCCAAGCTTTCACTTGTCGTCATCGTCCTTGTAGTC GCCCGCCTGGCGTTCGCCGCCATAGCAGTG | This study |
| PA4918-20OF | GCTCTAGACAAGCGGAGGCTTCCATGAATCGCCCCAGC | This study |
| PA4918-20OR | CCCAAGCTTTCAGGGCGCCTTCGGCAGTTCGCGCTTGTG | This study |
| cpdAFown | GGGGTACCCGCAGGCCTCGCGCCGGGTCGCGCTGAGCG | This study |
| cpdAR | CCCAAGCTTTCACTTGTCGTCATCGTCCTTGTAGTCGTATCCGGCGGTGTCGTAGTCCACTTC | This study |
| pET16b-nadD2F | TTTTCTCCATGGGCCGTGATGAAATAAGTTCCCGGATTCGCCGA | This study |
| pET16b-nadD2R | CCGCTCGAGGCCCGCCTGGCGTTCGCCGCCATAGCAG | This study |
| pET28a-cyaB217-463F | CATGCCATGGGCAAGAGCGTGCGCCTGGAAACCCAGC | This study |
| pET28a-cyaB217-463R | CCGCTCGAGGAGGATGACCTTGTCGCGCAGGCGTTCGG | This study |
| pMMB67EH-cyaB217-463F | GGGGTACCAAGAGCGTGCGCCTGGAAACCCAGC | This study |
| pMMB67EH-cyaB217-463R | CCCAAGCTTTCAATGATGATGATGATGATG GAGGATGACCTTGTCGCGCAGGCGTTCGG | This study |
| pMMB67EH-cyaA-hisR | CCCAAGCTTTCAATGATGATGATGATGATG TTGTTCCAGCAGCGCCTGGTTCAGCGCCG | This study |
| cyaB-SDF | AATTCATACACAGGGATCCTCTCCAGCGCATGGGTAC | This study |
| cyaB-SDR | CCATGCGCTGGAGAGGATCCCTGTGTATG | This study |
| lacP1F | GAATTCGCCCAATACGCAAACCGC | This study |
| lacP1R | GGATCCTCAGGCGAAAGGGGGATGTGCTG | This study |
| pTn7L | ATTAGCTTACGACGCTACACCC | This study |
| pglmS-up | CTGTGCGACTGCTGGAGCTGA | This study |
| qPCR primer | ||
| qcyaAF | CTTCAAGGAGCAGGTATTC | This study |
| qcyaAR | TTCGAGATGGCGATAGAC | This study |
| qcyaBF | GACCTGCTCAACAACTACC | This study |
| qcyaBR | GACGAACTTGTCGATGGT | This study |
| qcpdAF | GCGGATCGACCTGATTCTC | This study |
| qcpdAR | CTGCGGAAGCGTGTGTAG | This study |
| qPA4918F | GTCATCGAATACCTGAGG | This study |
| qPA4918R | GTTCTTCACGCAGTAGTC | This study |
| qnadD2F | GGTGTATTGCGAACCGATC | This study |
| qnadD2R | GCTTCCAGCAGGTCGATG | This study |
| qexsAF | CACGTCGGATAATCCTGATT | This study |
| qexsAR | TAGCGGAGAGGCATGAATA | This study |
| qprpLF | TATCGTATTTCGCCGACTCCC | This study |
| qprpLR | GCGAGTTGCCGTTGTTCAG | This study |
| qtoxAF | CGAGATGGGCGACGAGTTG | This study |
| qtoxAR | TGATGACCGTGGGCTTGATGT | This study |
| qrpsLF | CAAGCGCATGGTCGACAAGAG | This study |
| qrpsLR | ACCTTACGCAGTGCCGAGTTC | This study |
aF, forward; R, reverse; U, upstream of specific gene; D, downstream of specific gene; P, promoter of specific gene; own, native promoter of specific gene; tac, tac promoter; O, for over-expression plasmid construction; SD, Shine-Dalgarno sequence. bThe underlines are the sites of restriction enzymes.
For the deletion of the nrtR gene, a 1,058 bp fragment immediately upstream of the nrtR start codon and a 1,308 bp fragment downstream of the nrtR stop codon were PCR amplified, digested with XbaI-EcoRI and HindIII-XbaI, respectively. The two fragments were then ligated into pEX18Tc that was digested with EcoRI and HindIII, resulting in pZF01. Similar manipulation was used to construct the vfr, cyaA, cyaB, nadD2 and nadD2-nrtR deletion plasmids (detailed descriptions in Supplementary Text).
For the nrtR complementation plasmid, a 735 bp nrtR-containing fragment and a 465 bp fragment containing the promoter of nadD2-nrtR operon was amplified by PCR using PAK genomic DNA as template (primers shown in Table 2). The 735 bp and 465 bp PCR products were digested with PstI/HindIII and PstI/SacI, respectively, and then ligated into the vector pUC18T-mini-Tn7T digested with HindIII and SacI, resulting in pZF02. The plasmid was introduced into the ΔnrtR mutant by electroporation, along with the helper plasmid pTNS3 (Choi and Schweizer, 2006). Insertion of the nrtR gene into the chromosome was confirmed by PCR with primers PTn7L and PglmS-up (primers shown in Table 2).
For overexpression of nadD2, a 643 bp nadD2-containing fragment with its putative Shine-Dalgarno (SD) sequence was PCR amplified using PAK genomic DNA as the template (primers shown in Table 2). The PCR product was digested with HindIII and EcoRI, and then ligated into a shuttle vector pMMB67EH which was digested with the same restriction enzymes, resulting in pMMB67-nadD2. pMMB67-cyaA, pMMB67-cyaB, pMMB67-4918-20 (encoding PA4918-4920), pMMB67EH-cyaA-His and pUCP24-nadD2-Flag were constructed with the similar manipulation. For the translational fusion of CyaB-Flag, cyaB with c-terminal Flag-tag and its upstream 500 bp region was PCR amplified using PAK genomic DNA as template (primers shown in Table 2), digested with HindIII and KpnI, and then ligated into a promoterless pUCP20 (Li et al., 2016), resulting in pE1553b. Similar manipulation was used to construct the CyaA and CpdA translational fusion plasmids pE1553a and pE1553-cpdA. For expression of catalytic domain of CyaB, open reading frame from amino acids 217 to 463, which corresponds to the catalytic domain of CyaB (Fulcher et al., 2010), was PCR amplified and digested with KpnI-HindIII. This digested fragment, together with the annealed native SD region of cyaB (primers shown in Table 2), was cloned into the pMMB67EH, resulting in pMMB67EH-cyaB217-463-His. The lacP1 reporter was created as described by Fulcher et al. (2010), except that the lacP1 promoter was cloned into the vector pDN19lacZΩ.
Western Blot Assay
Overnight bacterial cultures were diluted 50-fold into fresh LB with or without 5 mM EGTA. The P. aeruginosa strains were then cultured to an OD600 of 1.0 in a shaking incubator. Then the bacterial pellet and supernatant were separated by centrifugation at 13,000 ×g for 2 min. Supernatant and pellet samples from equivalent number of bacterial cells were mixed with SDS-PAGE loading buffer, boiled for 10 min at 99°C, separated on 12% SDS-PAGE (15% SDS-PAGE for ExsA-Flag), transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore), and probed with a rabbit polyclonal antibody against ExoS, the RNA polymerase beta subunit (RNAP, Abcam) or a mouse monoclonal antibody against Flag (Sigma). Signals were detected with the ECL-plus kit (Millipore).
Cytotoxicity Assay
Bacterial cytotoxicity was determined by measuring detachment of mammalian cells after P. aeruginosa infection. 1.4 × 105 HeLa cells (Deng et al., 2017) were seeded into each well of a 24-well plate and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Deng et al., 2017) containing 10% fetal bovine serum (FBS) (Deng et al., 2017), penicillin (100 μg/ml) and streptomycin (50 μg/ml) at 37°C with 5% CO2 the night before infection. Three hours before infection, cells were washed twice with phosphate-buffered saline (PBS) (Deng et al., 2017) and incubated in DMEM with 10% FBS. Log phase bacteria were used to infect HeLa cells at a multiplicity of infection (MOI) of 50. 50 mM cAMP was added into DMEM medium at the start of infection as indicated. Three hours post infection, the culture medium was removed from each well, and cells remaining attached were washed twice with PBS and stained with 500 μl 0.1% crystal violet in 10% ethanol for 15 min at 37°C. After discarding the staining solution, each well was washed twice with 1 ml distilled water and dried in air. A 200 μl volume of 95% ethanol was added into each well and incubated at room temperature for 30 min with gentle shaking. The dissolved crystal violet was subjected to measurement of absorbance at a wavelength of 590 nm.
Murine Acute Pneumonia Model
Bacterial overnight culture was inoculated into fresh LB medium with 50-fold dilution and grown to an OD600 of 1.0. The bacteria were collected by centrifugation and adjusted to 1 × 109 CFU/ml in PBS. The exact number of bacteria for each inoculum was further determined by serial dilution and plating. Six- to eight-week-old female BALB/c mice were anesthetized with 100 μl chloral hydrate (7.5%) by intraperitoneal injection, and then intranasally inoculated with 10 μl of bacterial suspension in each nostril, giving a total infection bacterial number of approximately 2 × 107 per mouse. After 12 h, the mice were sacrificed and the lungs were dissected and homogenized in 1% protease peptone. The bacterial loads were determined by serial dilution and plating. The experimental results were analyzed with the GraphPad Prism software.
Histology
Twelve hours post infection with P. aeruginosa strains or sterile PBS, mouse lungs were removed and fixed with 10% paraformaldehyde. Fixed tissues were dehydrated in grades of ethanol, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Images were taken with a 20× objective lens.
Total RNA Isolation and RT-qPCR
Bacterial overnight culture was inoculated into fresh LB medium with 50-fold dilution and grown to an OD600 of 1.0 under T3SS inducing and non-inducing conditions. Total RNA was isolated using an RNA prep Pure cell/Bacteria Kit (Tiangen Biotech). cDNA was synthesized with a PrimeScript Reverse Transcriptase and random primers (Takara). The cDNA was mixed with indicated primers (shown in Table 2) and SYBR premix Ex Taq II (Takara). The 30S ribosomal protein encoding gene rpsL was used as an internal control.
cAMP Assay
Intracellular cAMP concentration was measured as previously described (Fulcher et al., 2010). Overnight bacteria were subcultured with 50-fold dilution into LB and grown to an OD600 of 1.0. 1.5 ml of the bacteria were harvested by centrifugation at 13,000 ×g for 2 min at 4°C and washed twice with cold 0.9 M NaCl. Pellets were resuspended in 100 μl of 0.1 M HCl and incubated on ice for 10 min with occasional vortex to lyse bacteria. Cellular debris were removed by centrifugation at 13,000 ×g for 5 min at 4°C and the supernatant was used to measure intracellular cAMP using an enzyme-linked immunosorbant assay (ELISA kit, Cayman Chemical) following the manufacturer’s protocol for sample acetylation. For protein concentration determination, duplicate bacterial pellets were resuspended in 100 μl PBS and lysed by three freeze/thaw cycles followed by centrifugation at 13,000 ×g for 5 min at 4°C. The protein concentration of the supernatant was measured by the BCA protein assay (Beyotime Biotechnology). Assay values for cAMP levels were converted to intracellular concentrations per mg of protein.
Co-immunoprecipitation Assay
The co-immunoprecipitation assay was performed as previously described with minor modifications (Shi et al., 2015). ΔnadD2 containing pUCP24-nadD2-Flag and pMMB67EH-cyaB217-463-His, pMMB67EH-cyaA-His or the empty vector pMMB67EH were grown overnight and diluted 50-fold into fresh LB medium. When the OD600 reached 0.6, 1 mM IPTG was added to induce the expression of CyaB217-463-His or CyaA-His at 16°C for 18 h. Bacteria were harvested by centrifugation at 5,000 g for 10 min, resuspended in lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 3 mM β-mercaptoethanol, 20 mM imidazole, 0.5% NP-40, pH 8.0) and lysed by sonication. Supernatants were collected by centrifugation and incubated with Ni-NTA agarose beads for 1 h at 4°C. The beads were washed five times with the lysis buffer and boiled in the SDS-PAGE loading buffer. Samples were separated by 15% SDS-PAGE (CyaB217–463-His) or 12% SDS-PAGE (CyaA-His) and probed with an anti-Flag (Sigma) or anti-His antibody (Millipore).
Expression and Purification of CyaB217-463 or NadD2 Protein
The full-length nadD2 or catalytic domain of the cyaB (amino acid positions 217–463) was PCR amplified from PAK chromosomal DNA with primers shown in Table 2 and cloned into pET16b or pET28a, resulting in pET16b-nadD2 or pET28a-cyaB217-463, respectively. Overnight culture of the E. coli strain BL21 (DE3) carrying pET16b-nadD2 or pET28a-cyaB217-463 was subcultured with 50-fold dilution into 500 ml fresh LB medium at 37°C. When the OD600 reached 0.6, 1 mM IPTG was added to induce the protein expression at 16°C for 16 h. The bacteria were collected by centrifugation at 4°C, 5,000 ×g, for 20 min and resuspended in the lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 3 mM β-mercaptoethanol, 10 mM imidazole, 0.5% NP-40, pH 8.0), followed by sonication. The bacterial lysate was centrifuged at 15,000 rpm for 20 min at 4°C and the supernatant was applied to a Ni-NTA column (Qiagen). After the Ni-NTA column was washed four times with the lysis buffer containing 20 mM imidazole, the protein was eluted with 400 mM imidazole prepared in lysis buffer, followed by dialysis against enzymatic reaction buffer (100 mM NaCl, 20 mM Tris-HCl, 10 mM MgCl2). The purified protein was examined by SDS-PAGE, and quantified by BCA protein assay (Beyotime Biotechnology). Expression and purification of the LasR protein was described in a previous report (Fan et al., 2018).
Adenylate Cyclase Activity Assays
The adenylyl cyclase activity assays were performed as previously described with minor modifications (Topal et al., 2012). Briefly, the reaction was performed in a 50 μl volume of 100 mM NaCl, 20 mM Tris-HCl (pH 7.0), 10 mM MgCl2, 5 mM ATP, and 2.0–2.25 μg purified CyaB protein, with or without 4.5–5.5 μg NadD2 or LasR protein. Samples were incubated at 30°C for 30 min, and then heated at 95°C for 4 min. The reaction mixtures were centrifuged at 15,000 rpm for 1 min, followed by filtration with a 0.22 μm filter, and then the cAMP concentrations were determined using an ELISA kit (Cayman Chemical) according to the manufacturers’ instructions.
Other Methods
The nrtR gene knock out was generated by homologous recombination as described previously (Hoang et al., 1998). β-galactosidase activity assay was conducted to determine the lacP1 promoter transcriptional activity as described before (Wu et al., 2004). The measurement of ATP concentration was carried out following the manufacturer’s instruction (Beyotime Biotechnology). DNA manipulations were performed according to Molecular Cloning (Sambrook et al., 1989).
Statistical Analysis
GraphPad Prism software was used to perform the statistical analyses. Results were analyzed by Mann–Whitney test or the Student’s t-test (two-tailed).
Results
The PA4916 Mutant Is Defective in T3SS-Dependent Cytotoxicity
In our previous screen for T3SS related genes, we found 23 genes that affect T3SS (Li et al., 2013). Among them, PA0020, PA3202, PA4336, PA4630, PA4916, and PA4753 encoded products are annotated as hypothetical proteins1 (Winsor et al., 2016) with unknown biological functions. To confirm their relationships with T3SS as well as exclude strain specific phenotype, mutants with Transposon (Tn) insertions in the PA0020, PA4336, PA4916, and PA4753 from the PA14 Non-Redundant Transposon Insertion Mutant Set (PA14NR Set) were selected for further tests (Liberati et al., 2006), however, mutant of the PA3202 or PA4630 is not available in the PA14NR Set. As T3SS plays a major role in cytotoxicity (Hauser et al., 1998), we infected HeLa cells with those mutants. Detached cells due to cytotoxicity were washed away, and the remaining cells were observed and quantified by crystal violet staining. Similar to wild type PA14, mutant strains of PA0020, PA4336 and PA4753 detached most of the HeLa cell within 3 h, whereas the ΔPA4916 mutant caused minimal detachment (Figure 1A), indicating a defective cytotoxicity.
FIGURE 1.
Cytotoxicity of indicated strains and the role of PA4916 in the expression and secretion of ExoS. (A,B) HeLa cells were infected with indicated strains at a MOI of 50. Three hours post infection, cells attached to the 24-well plate were washed with PBS and stained with crystal violet. The cell associated crystal violet was dissolved in ethanol and quantified by measuring OD590. HeLa cells with no bacterial infection served as a control. (C) Bacteria were cultured to an OD600 of 1.0 in LB with or without 5 mM EGTA. Proteins in supernatants and pellets from equivalent bacterial cells were loaded onto SDS-PAGE gels and probed with an antibody against ExoS or RNA polymerase beta subunit. S, supernatant; P, pellet.
To further confirm the role of PA4916, the whole open reading frame of PA4916 was deleted from the PAK chromosome via DNA recombination, resulting in ΔPA4916. As shown in Figure 1B, this mutant also displayed a reduced cytotoxicity, which was restored nearly to that of wild type by complementation with an intact PA4916 gene.
PA4916 Is Required for ExoS Expression and Involved in Pathogenesis of P. aeruginosa
To verify whether the reduced cytotoxicity is due to a defective T3SS, wild type PAK and the ΔPA4916 mutant were grown under T3SS inducing and non-inducing conditions (in the presence and absence of 5 mM EGTA), and the expression and secretion of ExoS were examined by Western blot. Under T3SS inducing condition, the expression and secretion of ExoS were highly induced in the wild type PAK, however, faint ExoS was observed in the pellet and not detected in the supernatant of ΔPA4916. Complementation with an intact PA4916 gene restored the expression and secretion of ExoS in the ΔPA4916 mutant background (Figure 1C).
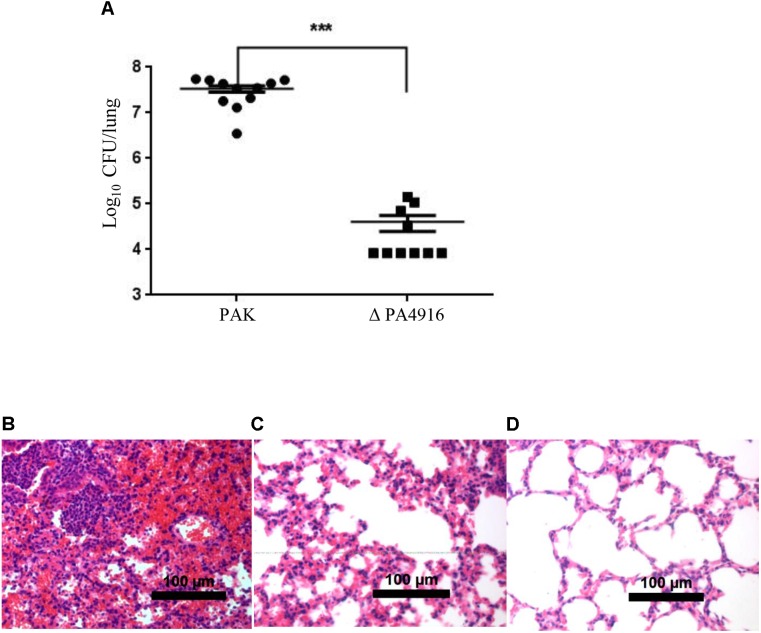
The T3SS plays an important role in acute infections (Sadikot et al., 2005). The functional connection between PA4916 and the T3SS promoted us to examine its role in the pathogenesis of a mouse acute pneumonia model. Six- to eight-week-old female BALB/c mice were infected intranasally with 2 × 107 CFU of wild type PAK or the ΔPA4916 mutant. Twelve hours post infection, lungs were isolated and homogenized. Bacterial loads were determined by serial dilution and plating. Compared to the wild type PAK strain, the number of ΔPA4916 mutant isolated from the lungs was significantly lower (Figure 2A), suggesting a defective virulence. Reduced growth rate in the ΔPA4916 mutant might lead to a reduced virulence; however, the deletion mutant ΔPA4916 showed a rate of growth indistinguishable from that of wild type PAK strain when cultured in LB medium (Supplementary Figure S1).
FIGURE 2.
Role of PA4916 in the mouse acute pneumonia model. (A) Bacteria were grown to an OD600 of 1.0. Female BALB/c mice (6- to 8-week-old) were inoculated intranasally with 2 × 107 CFU of wild-type PAK or its isogenic ΔPA4916 mutant. After 12 h, the mice were sacrificed, and the lungs were dissected and homogenized. The bacteria load in each lung was determined by serial dilution and plating. ∗∗∗P < 0.001, by Mann–Whitney test. (B–D) Pathology sections of lungs infected with indicated strains. Mice were infected with 2 × 107 CFU of wild type PAK (B), ΔPA4916 mutant (C) or sterile PBS (D). Lungs from infected mice were removed, fixed, sectioned, and stained with hematoxylin and eosin. Images were taken with a 20× objective lens.
To further validate the role of PA4916 in the bacterial virulence, the lungs infected by wild type or ΔPA4916 mutant were observed following histological section and staining. Lungs from mice infected with PAK for 12 h had significant neutrophil infiltration, edema and tissue damage (Figure 2B). Most of the airways in the lungs of these mice were completely occluded with neutrophil and pyocyte infiltration. In contrast, infections with the ΔPA4916 mutant showed significantly reduced inflammatory characteristics (Figure 2C), with substantially fewer neutrophils present in the alveolar spaces, compared to the infections with PAK, though the inflammation was more intense compared to the PBS instilled control (Figure 2D). The extent of inflammation caused by the ΔPA4916 mutant correlated with the ability of this strain to colonize the lungs of infected mice. A recent study has also shown that PA4916 plasposon mutagenesis abrogated virulence of a robust mucoid P. aeruginosa cystic fibrosis airway isolate and named PA4916 as NrtR (Okon et al., 2017). So we further explored the regulation mechanism of PA4916 on the T3SS and referred PA4916 as NrtR hereafter.
Plasmid-Expressed exsA Restores the T3SS in ΔnrtR Mutant
ExsA is a central regulator of T3SS (Hovey and Frank, 1995). To investigate whether NrtR regulates T3SS through ExsA, total RNA was isolated and the mRNA levels of exsA were compared between PAK and the ΔnrtR mutant. As shown in Figure 3A, under both T3SS inducing and non-inducing conditions, the exsA mRNA level was significantly decreased in the ΔnrtR mutant, which was restored by complementation with a nrtR gene. To further confirm the role of exsA in the NrtR mediated regulation of T3SS, a plasmid carrying an exsA gene driven by a lac promoter (Totten and Lory, 1990; Li et al., 2013) was introduced into the ΔnrtR or an exsA::Ω mutant. Plasmid-expressed ExsA restored the expression of ExoS in both the exsA::Ω and ΔnrtR mutants (Figure 3B). Although the expression of ExoS was similar in the exsA::Ω/exsA and ΔnrtR/exsA mutants, the ExsA amount in the ΔnrtR/exsA mutant was much less than that in the exsA::Ω/exsA mutant (Figure 3C). Considering the fact that lac promoter is controlled by catabolite repression and T3SS is regulated by the cAMP-Vfr signaling pathway, the decreased ExsA amount may indicate a reduced cAMP level in the ΔnrtR mutant strain.
FIGURE 3.
Plasmid mediated expression of exsA restored T3SS in the ΔnrtR mutant. (A) The relative exsA mRNA levels in PAK, the ΔnrtR mutant and ΔnrtR/att7:: nrtR strain. Total RNA was isolated under T3SS inducing and non-inducing conditions and the exsA mRNA levels were determined by real-time PCR using rpsL as the internal control. ∗P < 0.05, ∗∗P < 0.01, by Student’s t-test. (B) Bacteria harboring an exsA-Flag driven by a lac promoter or the empty vector pDN19 were grown to an OD600 of 1.0 in LB with or without EGTA. Proteins from equivalent number of bacterial cells of indicated strains were separated on SDS-PAGE and probed with an anti-ExoS antibody or an anti-RNA polymerase beta subunit antibody. Expression levels of the ExsA-Flag were determined with an anti-FLAG antibody (C).
The cAMP-Vfr Signaling Pathway Is Involved in NrtR Mediated Regulation of T3SS
To investigate if the cAMP-Vfr signaling pathway is involved in the NrtR mediated T3SS regulation, we initially compared the cAMP contents between PAK and ΔnrtR mutant with a transcriptional fusion of the lacP1 promoter and a lacZ gene (lacP1-lacZ), whose expression has been shown to correlate to intracellular cAMP levels (Fulcher et al., 2010). A Δvfr mutant was included as a control of ΔnrtR mutant (Fulcher et al., 2010). As expected, the LacZ levels in the ΔnrtR mutant under both T3SS inducing and non-inducing conditions were lower than those of the PAK strain, which were restored by complementation with a nrtR gene (Figure 4A). This result was further confirmed by direct measurement of intracellular cAMP levels with a cAMP ELISA detection kit (Figure 4B). A previous study has shown that exogenous addition of 50 mM cAMP restored the phenotypes of an adenylate cyclase mutant of P. aeruginosa (Fulcher et al., 2010). Therefore, we constructed a c-terminus Flag-tagged ExsA driven by its native promoter and examined the effect of exogenous addition of 50 mM cAMP on the T3SS in the ΔnrtR mutant. As shown in Figure 4C,D, exogenous addition of cAMP restored the exsA expression levels in the ΔnrtR mutant at both transcriptional and protein levels, but not in the Δvfr mutant. Furthermore, the expression and secretion of ExoS and cytotoxicity of ΔnrtR were restored to wild type level by exogenous addition of 50 mM cAMP (Figure 4E,F). As expected, exogenous addition of 50 mM cAMP did not affect the expression and secretion of ExoS and cytotoxicity of the Δvfr mutant (Figure 4E,F). In addition, ΔnrtRΔvfr double mutant strain was constructed. The exsA transcriptional level, as well as the expression and secretion of ExoS of ΔnrtRΔvfr were compared with ΔnrtR and Δvfr mutant. The results showed that, like Δvfr mutant, exogenous cAMP addition did not affect exsA transcriptional level, as well as the expression and secretion of T3SS in ΔnrtRΔvfr double mutant strain (Figure 4C,E). These results demonstrate that NrtR regulates T3SS through the cAMP-Vfr signaling pathway in P. aeruginosa, likely by altering the intracellular cAMP level. Furthermore, twitching motility and the expression levels of toxA and prpL which were demonstrated to be affected by cAMP were examined in the ΔnrtR mutant (Wolfgang et al., 2003). As expected, the expression levels of both toxA and prpL were decreased significantly in ΔnrtR mutant (Supplementary Figure S2). However, the twitching motility of the ΔnrtR mutant showed no detectable change compared to the wild type PAK strain (Supplementary Figure S2).
FIGURE 4.
Decreased cAMP contributed to the T3SS defect in the ΔnrtR mutant. (A,B) cAMP level was decreased in ΔnrtR mutant and Δvfr mutant. β-galactosidase assay was used to examine the transcriptional activity of lacP1 promoter fused to a lacZ gene in indicated strains under T3SS inducing and non-inducing conditions (A). (B) Intracellular cAMP levels were measured using an ELISA kit. Error bars represent standard deviations. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. (C–F) Exogenous addition of cAMP recovers the expression of ExsA, ExoS and cytotoxicity of the ΔnrtR mutant, while not of or ΔnrtRΔvfr mutant. (C) Relative mRNA levels of exsA in indicated strains with or without cAMP addition at the beginning of subculture in the presence or absence of 5 mM EGTA, with rpsL as an internal control. ns, not significant, ∗P < 0.05, ∗∗∗P < 0.001 by Student’s t-test. (D) Indicated strains containing an exsA-Flag driven by its native promoter were grown at 37°C with or without 5 mM EGTA and 50 mM cAMP as indicated. Protein samples from equal number of bacteria were separated by SDS-PAGE and probed with an anti-Flag antibody or an anti-RNA polymerase beta subunit antibody. (E) Expression of ExoS in indicated strains were grown with or without 5 mM EGTA and 50 mM cAMP. The protein levels were detected with an antibody against ExoS or RNA polymerase beta subunit. S, supernatant; P, pellet. (F) Cytotoxicity of indicated strains in the presence or absence of 50 mM cAMP. HeLa cells were infected with indicated strains at a MOI of 50. 50 mM final concentration of cAMP was added to DMEM medium as indicated. Three hours post infection, cells attached to the 24-well plate were washed with PBS and stained with crystal violet. The cell associated crystal violet was dissolved in ethanol and quantified by measuring OD590. HeLa cells with no bacterial infection (blank and blank+50 mM cAMP) served as a control. ∗∗P < 0.01, by Student’s t-test.
Decreased Intracellular cAMP Is Not Caused by Altered Expression of CyaA or CyaB in the ΔnrtR Mutant
In P. aeruginosa, cAMP is synthesized by the adenylate cyclases CyaA and CyaB (Wolfgang et al., 2003; Marsden et al., 2016). To explore if the reduced cAMP level is caused by decreased expression of the adenylate cyclases in ΔnrtR mutant, we determined the expression levels of the two genes by RT-qPCR. As shown in Figure 5A, the mRNA level of cyaA was lower in the ΔnrtR mutant than those in both wild type PAK and the complemented strain, while the cyaB was similar among these three strains. To confirm this observation, C-terminal Flag-tagged CyaA or CyaB driven by their respective native promoters, were transformed into PAK and the ΔnrtR mutant, and their protein expression levels were examined by Western blot assay. Consistent with the RT-qPCR result, similar level of CyaB-Flag, while slightly lower level of CyaA-Flag protein were observed in the ΔnrtR mutant compared to that in PAK (Figure 5B). To further understand if the observed slight reduction of the adenylate cyclase is the cause of the decreased cAMP in ΔnrtR mutant, the functional C-terminal Flag tagged CyaA or CyaB was driven by a tac promoter and transformed into the ΔnrtR mutant. Similar levels of expression and secretion of ExoS were observed by Western blot assay between ΔnrtR/pMMB67EH and ΔnrtR/pMMB67-cyaA or ΔnrtR/pMMB67-cyaB (Figure 5C,D). These results suggest that the reduced cAMP level in the ΔnrtR strain is not due to alteration of adenylate cyclases expression.
FIGURE 5.
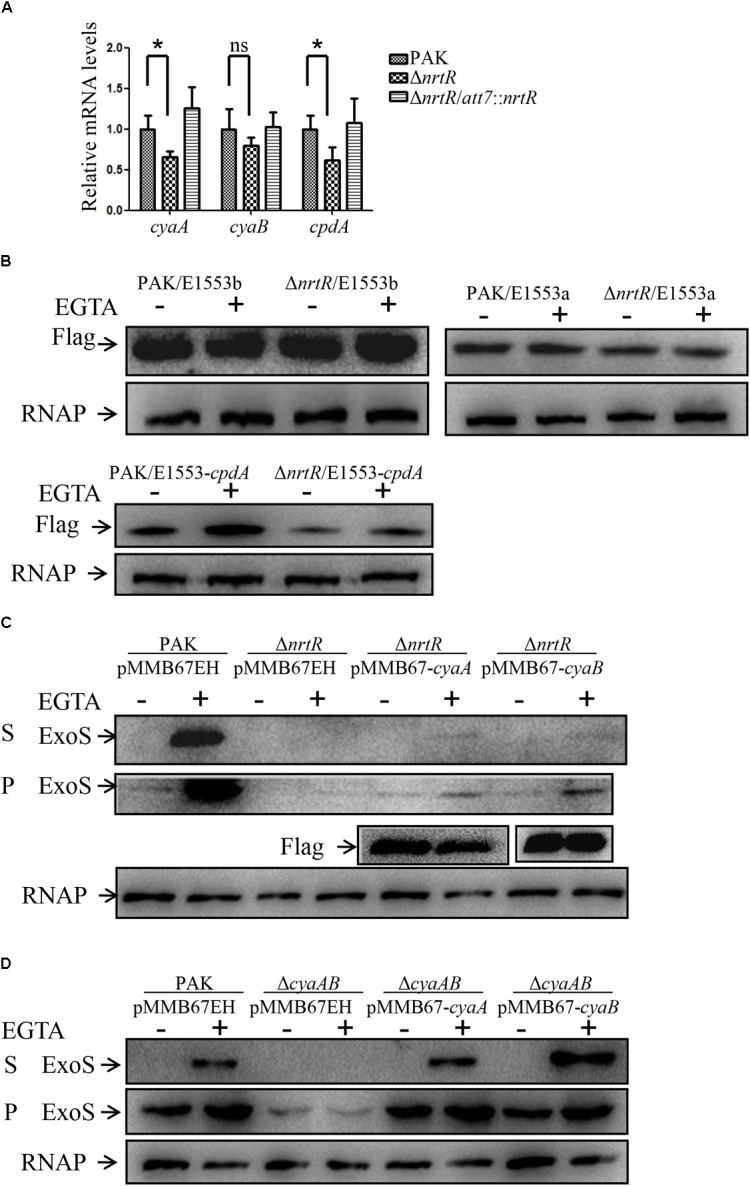
The decrease of cAMP is not caused by changed expression of CyaA, CyaB or CpdA. (A) Relative mRNA levels of cyaA, cyaB, and cpdA. Total RNA of indicated strains was isolated and the mRNA levels of these genes were determined by real time PCR with rpsL serving as an internal control. ns, not significant, ∗p < 0.05 by Student’s t-test. (B) Indicated strains containing a cyaA-Flag (cyaB-Flag or cpdA-Flag) driven by its native promoter were grown at 37°C with or without 5 mM EGTA until OD600 of 1.0. Samples from equal number of bacteria were separated by SDS-PAGE and probed with an anti-Flag antibody or an anti-RNA polymerase beta subunit antibody. (C,D) Indicated strains containing a cyaA-Flag or cyaB-Flag driven by a tac promoter or the empty vector pMMB67EH were grown to an OD600 of 1.0 in LB with 1 mM IPTG with or without 5 mM EGTA. Proteins from equivalent bacterial cells of indicated strains were separated by SDS-PAGE and probed with an anti-ExoS antibody, an anti-Flag antibody or an anti-RNA polymerase beta subunit antibody. S, supernatant; P, pellet.
Decreased Intracellular cAMP Is Not Caused by Altered Expression of CpdA in the ΔnrtR Mutant
Since we did not get evidence for an effect of altered expression of adenylate cyclases on the T3SS in the ΔnrtR mutant, we next wanted to investigate if the degradation of cAMP was affected in the ΔnrtR mutant. To date, CpdA is the only known phosphodiesterase degrading cAMP in P. aeruginosa (Fuchs et al., 2010). One possibility for the observed decrease in cAMP levels and subsequent T3SS in the ΔnrtR mutant could be that CpdA expression is upregulated in the ΔnrtR mutant. This would in turn result in an increased degradation of cAMP and a decrease in T3SS. To test this possibility, total RNA was isolated and mRNA levels of cpdA were compared among wild type PAK, ΔnrtR mutant and ΔnrtR complemented strain by RT-qPCR. As shown in Figure 5A, the mRNA level of cpdA was lower, rather than higher, in the ΔnrtR mutant than those in both wild type PAK and complemented strain. To confirm this observation, C-terminal Flag-tagged CpdA driven by its native promoter, was transformed into PAK and the ΔnrtR mutant, and their protein expression levels were examined by Western blot assay. Consistent with the RT-qPCR result, lower CpdA-Flag protein level were observed in the ΔnrtR mutant than that in PAK (Figure 5B). These results thus suggest that decreased cAMP and subsequent T3SS does not occur through increased CpdA-mediated cAMP degradation.
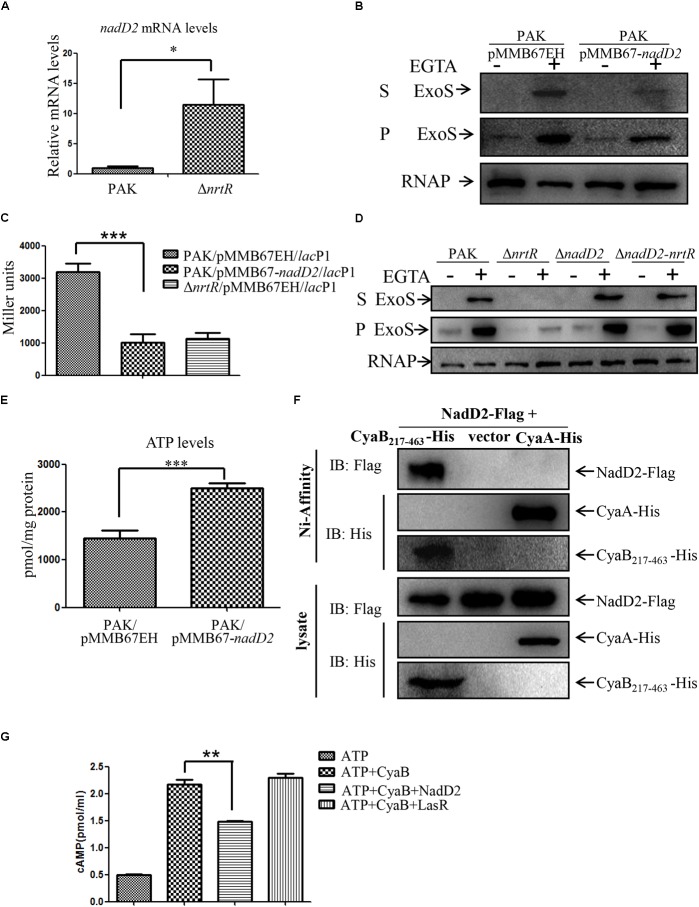
Increased NadD2 Level Contributes to the Decreased cAMP and T3SS in the ΔnrtR Mutant
NrtR encodes a transcriptional regulator which binds to the DNA intergenic region between the nadD2- nrtR and PA4918-4920 operons to repress their expression (Okon et al., 2017). RT-qPCR assay showed that the mRNA levels of nadD2 and PA4918 in the ΔnrtR mutant were much higher than those in PAK (Figure 6A and Supplementary Figure S3A). Thus, we examined whether NadD2 or PA4918-4920 operon was involved in the NrtR mediated regulation of T3SS. Overexpression of nadD2 in wild type PAK reduced the expression and secretion of the ExoS, as well as the intracellular cAMP level (Figure 6B,C). However, overexpression of PA4918-4920 operon in PAK showed no inhibitory effect on the expression and secretion of T3SS (Supplementary Figure S3B). In addition, deletion of nadD2 in wild type PAK did not affect the expression and secretion of ExoS, whereas deletion of nadD2 restored the expression and secretion of ExoS in the ΔnrtR mutant (Figure 6D). These results indicate that the increased NadD2 level might be responsible for the decreased cAMP and T3SS in the ΔnrtR mutant. NadD2 is an ATP consuming enzyme (Okon et al., 2017). Adenylate cyclases catalyzed the synthesis of cAMP from ATP. Reduced ATP availability due to increased NadD2 level might result in the decreased cAMP level in PAK/pMMB67-nadD2. However, the PAK/pMMB67-nadD2 strain displayed a higher ATP level than the strain with an empty vector (PAK/pMMB67EH) (Figure 6E).
FIGURE 6.
Increased expression of NadD2 contributes to the decreased intracellular cAMP and expression of T3SS in the ΔnrtR mutant. (A) Relative mRNA levels of nadD2. Total RNA of indicated strains was isolated and mRNA levels of nadD2 were determined by real time PCR with rpsL serving as an internal control. Data represents the mean ± standard deviation. ∗p < 0.05 by Student’s t-test. (B) PAK containing nadD2 driven by a tac promoter or the empty vector pMMB67EH were grown to an OD600 of 1.0 in LB containing 1 mM IPTG with or without 5 mM EGTA. Proteins from equivalent bacterial cells of indicated strains were separated by SDS-PAGE and probed with an antibody against ExoS or RNA polymerase beta subunit. S, supernatant; P, pellet. (C) The cAMP levels were decreased in the PAK/pMMB67-nadD2 strain. β-Galactosidase assay was used to examine the LacZ level driven by the lacP1 promoter in indicated strains with 1 mM IPTG induction. Error bars represent standard deviations. ∗∗∗P < 0.001 by Student’s t-test. (D) Indicated bacteria were cultured to an OD600 of 1.0 in LB with or without 5 mM EGTA. Proteins in supernatants and pellets from equivalent bacterial cells were loaded onto SDS-PAGE gels and probed with an antibody against ExoS or RNA polymerase beta subunit. S, supernatant; P, pellet. (E) ATP levels in indicated strains. ∗∗∗P < 0.001 by Student’s t-test. (F) Interaction between NadD2 and CyaB217-463 or CyaA. ΔnadD2 carrying pUCP24-nadD2-Flag with pMMB67EH-cyaB217-463-His, pMMB67EH-cyaA-His or pMMB67EH were grown to an OD600 of 0.6 and incubated with 1 mM IPTG for 18 h at 16°C. Bacteria were lysed and subjected to chromatography with Ni-NTA beads. His-tagged CyaB217–463 or CyaA and FLAG-tagged NadD2 were detected by Western blot assay. (G) Inhibition of CyaB adenylyl cyclase activity by NadD2. 2.0–2.25 μg CyaB was incubated with 5 mM ATP. 4.5–5.5 μg NadD2 or LasR was added as indicated. After 30 min at 30°C, the cAMP levels were measured using an ELISA kit. Error bars represent standard deviations. ∗∗P < 0.01 by Student’s t-test.
Since the decreased cAMP level is not caused by the changed expression of the adenylate cyclases in the ΔnrtR mutant, it is possible that their enzymatic activities might be affected by NadD2. To test this possibility, a CyaA-His or a CyaB217-463-His fusion protein (the catalytic domain of CyaB without the transmembrane region), was constructed and overexpressed in ΔnadD2 carrying a NadD2-Flag fusion protein. The His-Tagged CyaA and CyaB217-463 were purified with Ni-affinity chromatography. As shown in Figure 6F, NadD2-Flag was co-purified with the CyaB217-463-His, but not with CyaA-His.
Next, we examined whether NadD2 directly represses the adenylyl cyclase activity of CyaB. CyaB217-463 and NadD2 were expressed in E. coli and purified (Supplementary Figure S4A). The purified NadD2 was not contaminated by ATPase or phosphodiesterase (Supplementary Figures S4B,C). The purified catalytic domain of CyaB (CyaB217-463) was incubated with ATP with or without NadD2 and the cAMP level was determined with a cAMP ELISA kit (Cayman Chemical). An unrelated protein LasR was used as a negative control. As shown in Figure 6G, the cAMP level was reduced by the presence of NadD2 but not LasR, indicating a repression of the adenylyl cyclase activity by the NadD2. These results suggest that NadD2 might suppress the enzymatic activity of CyaB.
Discussion
In the present study, we identified that NrtR is required for the T3SS and involved in pathogenesis of P. aeruginosa in a murine acute pneumonia model. Further experimental results demonstrated that NrtR regulates expression of T3SS through the cAMP/Vfr signaling system. NadD2, which is in the same operon of nrtR and repressed by NrtR, is involved in the NrtR mediated regulation of the T3SS by inhibition of adenylyl cyclase activity of CyaB in P. aeruginosa.
In our previous study, we identified PA0020, PA4336, PA4916 and PA4753 as T3SS related genes by screening Tn insertion mutant library of PAK with an ELISA assay (Li et al., 2013). While in this study, only the nrtR mutant in PA14 background displayed a significant change in T3SS related cytotoxicity. The previous screening by ELISA was performed with EGTA as the T3SS inducing condition, whereas in the cytotoxicity assay, contact with host cell is the inducing signal. The different results in the two tests indicate that PA0020, PA4336 and PA4753 might play different roles in bacterial response to the two signals. In addition, the differences of the Tn insertion sites in each of the genes might cause different effects on the gene function, thus leading to different phenotypes. Further studies are required to fully understand the functions of those genes.
NrtR encodes a putative ADP-ribose pyrophosphatase with a Nudix hydrolase domain. Nudix protein specifically hydrolyzes varieties of substrates with a common structure of a Nucleoside diphosphates linked to moiety, X, such as (d) NTPs, coenzymes and capped RNAs (O’Handley et al., 1996; Cartwright et al., 2000; Song et al., 2013). It has been reported that some of these proteins may play important regulatory roles in response to stress, invasion to host cell and in pathogenesis (Edelstein et al., 2005; Urick et al., 2005; Liu et al., 2012). A recent study reported that nrtR of P. aeruginosa PA14 encodes a transcriptional repressor, which has no ADP-ribose pyrophosphatase activity and can bind to the DNA intergenic region between nadD2-nrtR and PA4918-4920 operons to repress their expression (Okon et al., 2017). Consistent with this, our study demonstrated that the transcriptional levels of nadD2 and PA4918 increased 11- and 46-fold, respectively, in the ΔnrtR mutant (Figure 6A and Supplementary Figure S3A). Overexpression of NadD2 repressed the expression of T3SS in PAK strain, whereas overexpression of PA4918-4920 in PAK did not show any inhibitory effect on the expression of T3SS (Supplementary Figure S3B), thus NrtR positively controls the expression of T3SS in P. aeruginosa through repression of nadD2 specifically. nadD2 deletion restored the expression and secretion of T3SS in ΔnrtR mutant, but had no influence on the expression and secretion of T3SS in wild type PAK strain, indicating that the altered expression level of NadD2 is not sufficient to repress the expression of T3SS in wild type PAK strain.
NadD2, encoding a nicotinate mononucleotide adenylyltransferase, is located downstream of the transcriptional repressor NrtR and upstream of the PA4918-4920 (pncA-pncB1-nadE) operon. The interaction between NadD2 and other proteins were predicted using the STRING database2), a pre-computed database to predict both physical and functional interactions. The high confidence score (>0.7) exist between NadD2 and proteins encoded by its neighboring genes on the chromosome. It has been demonstrated that pncA, pncB1, and nadE encode the nicotinamidase, nicotinate phosporibosyltransferase and Nad synthase, respectively (Okon et al., 2017). Therefore, NrtR negatively regulates the salvage pathway I of the NAD biosynthesis. In addition, the co-immunoprecipitation assay in the present study suggests that NadD2 interacts with CyaB, but not with CyaA, indicating that NadD2 influences the cAMP production through inhibition of the adenylyl cyclase activity of CyaB. However, the inability of functional CyaA to complement the expression of T3SS in ΔnrtR mutant suggests that NrtR may also influence the adenylyl cyclase activity of CyaA indirectly.
cAMP, as an important second messenger, has been shown to regulate the T3SS, exotoxin A, protease IV and type IV pili biosynthesis (Wolfgang et al., 2003). As the cAMP was decreased in the ΔnrtR mutant, we also tested the twitching motility and the expression levels of toxA and prpL in the ΔnrtR mutant. As expected, the expression levels of both toxA and prpL were decreased significantly in ΔnrtR mutant. However, the twitching motility of the ΔnrtR mutant showed no detectable change compared to the wild type PAK strain (Supplementary Figure S2). It might be possible that the T3SS is more sensitive to changes in the cAMP level than twitching motility. The generation of cAMP in P. aeruginosa relies on CyaA and CyaB, while its degradation relies on phosphodiesterase CpdA (Fuchs et al., 2010). Expression of CpdA is lower in the ΔnrtR mutant, which may be caused by decreased levels of intracellular cAMP, as the cpdA can be directly activated by Vfr in response to intracellular cAMP as a feedback loop (Fuchs et al., 2010).
The intracellular cAMP levels are modulated by calcium concentration (Inclan et al., 2011), and the EGTA-induced calcium depletion has been shown to increase intracellular cAMP levels in P. aeruginosa (Wolfgang et al., 2003). Consistent with these results, our study demonstrated that intracellular cAMP levels were increased in both wild type PAK and ΔnrtR mutant under EGTA inducing condition, even in the Δvfr mutant. Considering that EGTA addition did not affect the expression of adenylate cyclases in both PAK and ΔnrtR mutant, this increase suggests that EGTA might increase the activity of adenylate cyclases. The fact that the activation of lacP1 was Vfr-dependent in P. aeruginosa might result in no obvious increase of reporter activity in Δvfr mutant under EGTA inducing condition (Fulcher et al., 2010). In contrast to the almost eliminated reporter activity, the Δvfr mutant displayed an approximately 50% reduction in intracellular cAMP compared to the PAK strain, which is consistent with previous report (Fulcher et al., 2010).
The T3SS of P. aeruginosa can be induced by EGTA addition (calcium depletion) (Wolfgang et al., 2003). Previous studies reported that most secretion apparatus component and effector genes were regulated by calcium depletion except for the ExsA, whose expression was relatively unaffected (Wolfgang et al., 2003). Inconsistent with this finding, in our study, the mRNA levels of exsA showed a significant increase under EGTA inducing condition. This may be caused by the different experimental methods used in these two studies (transcriptomic analysis vs. RT-qPCR). Furthermore, the ETGA-dependent increase of ExsA protein levels was demonstrated previously (Intile et al., 2014; Ince et al., 2015). However, in contrast to the previous reports and our transcriptional levels of exsA, in this study, the protein levels of ExsA were not affected by EGTA addition both in PAK and the ΔnrtR mutant. In the previous studies, P. aeruginosa was grown in Trypticase soy broth, while we cultured bacteria in L-broth medium. The difference between the transcriptional levels and protein levels of exsA might be due to the sensitivity of RT-qPCR or some unknown post-transcriptional regulatory mechanism.
Recently, Okon et al. (2017) demonstrated that NrtR regulates nicotinamide adenine dinucleotide (NAD) biosynthesis and is involved in the virulence of a P. aeruginosa clinical isolate. Our study revealed novel functions of NrtR and NadD2 in the cAMP biosynthesis in P. aeruginosa.
Author Contributions
YJ, WW, and SJ conceived and designed the experiments, and wrote the paper. YJ, MZ, FZ, QP, YW, QZ, and CL performed the experiments. YJ, WW, FB, ZC, and SJ analyzed the data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by National Science Foundation of China (31600110 and 31670130), Science and Technology Committee of Tianjin (17JCQNJC09200, 15JCZDJC33000, and 15JCYBJC53900), Program of International S&T Cooperation (2015DFG32500), and the State Key Laboratory of Medicinal Chemical Biology (2017005).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00085/full#supplementary-material
Growth curve of PAK and the ΔPA4916 mutant.
Role of NrtR in twitching motility or expression of prpL and toxA in P. aeruginosa. (A) Twitching motilities of the indicated strains were examined on 1% LB agar with a Δvfr mutant serving as a control. The twitching zones were visualized with 0.1% crystal violet staining. (B) Relative mRNA levels of prpL and toxA. Total RNA of indicated strains was isolated and mRNA levels of prpL and toxA were determined by real time PCR with rpsL serving as an internal control. Data represents the mean ± standard deviation. ∗∗p < 0.01, ∗∗∗p < 0.001 by Student’s t-test.
PA4918 is not involved in the regulation of T3SS. (A) Relative mRNA levels of PA4918. Total RNA of indicated strains was isolated and mRNA levels of PA4918 were determined by real time PCR with rpsL serving as an internal control. Data represents the mean ± standard deviation. ∗∗∗p < 0.001 by Student’s t-test. (B) PAK containing the PA4918-20 operon driven by a tac promoter or the empty vector pMMB67EH were grown to an OD600 of 1.0 in LB with 1 mM IPTG with or without 5 mM EGTA. Proteins from equivalent bacterial cells of indicated strains were separated by SDS-PAGE and probed with an anti-ExoS antibody or an anti-RNA polymerase beta subunit antibody. S, supernatant; P, pellet.
Coomassie blue staining of purified CyaB217-463 and NadD2. (B) Purified NadD2 was not contaminated by ATPase. Equal amount ATP was incubated with or without 5 μg NadD2. After 30 min at 30°C, the ATP levels were measured using an ATP detection kit. Error bars represent standard deviations. ns, not significant, by Student’s t-test. (C) Purified NadD2 was not contaminated by phosphodiesterase. Equal amount cAMP was incubated with or without 5 μg NadD2. After 30 min at 30°C, the cAMP levels were measured using an ELISA kit. Error bars represent standard deviations. ns, not significant, by Student’s t-test.
References
- Ahn K.-S., Ha U., Jia J., Wu D., Jin S. (2004). The truA gene of Pseudomonas aeruginosa is required for the expression of type III secretory genes. Microbiology 150 539–547. 10.1099/mic.0.26652-0 [DOI] [PubMed] [Google Scholar]
- Anderson G. G., Yahr T. L., Lovewell R. R., O’Toole G. A. (2010). The Pseudomonas aeruginosa magnesium transporter MgtE inhibits transcription of the type III secretion system. Infect. Immun. 78 1239–1249. 10.1128/iai.00865-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel E., Vakulskas C., Brady K., Yahr T. (2008). Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 68 657–671. 10.1111/j.1365-2958.2008.06179.x [DOI] [PubMed] [Google Scholar]
- Brutinel E., Yahr T. (2008). Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 11 128–133. 10.1016/j.mib.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright J. L., Gasmi L., Spiller D. G., McLennan A. G. (2000). The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its derivatives. J. Biol. Chem. 275 32925–32930. 10.1074/jbc.M005015200 [DOI] [PubMed] [Google Scholar]
- Choi K. H., Schweizer H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1 153–161. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- Crowe K. E., Bass J. A., Young V. M., Straus D. C. (1982). Antibody response to Pseudomonas aeruginosa exoproducts in cancer patients. J. Clin. Microbiol. 15 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux D., Attree I., Schneider C., Toussaint B. (1999). Cell death of human polymorphonuclear neutrophils induced by a Pseudomonas aeruginosa cystic fibrosis isolate requires a functional type III secretion system. Infect. Immun. 67 6164–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N., Lykken G., Wolfgang M., Yahr T. (2004). A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 53 297–308. 10.1111/j.1365-2958.2004.04128.x [DOI] [PubMed] [Google Scholar]
- Deng X., Li M., Pan X., Zheng R., Liu C., Chen F., et al. (2017). Fis regulates type III secretion system by influencing the transcription of exsA in Pseudomonas aeruginosa strain PA14. Front. Microbiol. 8:669. 10.3389/fmicb.2017.00669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M. R., King J. M., Yahr T. L. (2011). Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front. Microbiol. 2:89. 10.3389/fmicb.2011.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. H., Zhang X. F., Zhang L. H. (2013). The global regulator Crc plays a multifaceted role in modulation of type III secretion system in Pseudomonas aeruginosa. Microbiologyopen 2 161–172. 10.1002/mbo3.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein P. H., Hu B., Shinzato T., Edelstein M. A. C., Xu W., Bessman M. J. (2005). Legionella pneumophila NudA is a nudix hydrolase and virulence factor. Infect. Immun. 73 6567–6576. 10.1128/iai.73.10.6567-6576.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Solh A. A., Hattemer A., Hauser A. R., Alhajhusain A., Vora H. (2012). Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit. Care Med. 40 1157–1163. 10.1097/CCM.0b013e3182377906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Xu C., Pan X., Dong Y., Ren H., Jin Y., et al. (2018). Mechanisms of RsaL mediated tolerance to ciprofloxacin and carbenicillin in Pseudomonas aeruginosa. Curr. Genet. 10.1007/s00294-018-0863-3 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Feltman H., Schulert G., Khan S., Jain M., Peterson L., Hauser A. R. (2001). Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147(Pt 10), 2659–2669. 10.1099/00221287-147-10-2659 [DOI] [PubMed] [Google Scholar]
- Fuchs E. L., Brutinel E. D., Klem E. R., Fehr A. R., Yahr T. L., Wolfgang M. C. (2010). In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J. Bacteriol. 192 2779–2790. 10.1128/jb.00168-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N. B., Holliday P. M., Klem E., Cann M. J., Wolfgang M. C. (2010). The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 76 889–904. 10.1111/j.1365-2958.2010.07135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha U., Kim J., Badrane H., Jia J., Baker H., Wu D., et al. (2004). An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol. Microbiol. 54 307–320. 10.1111/j.1365-2958.2004.04282.x [DOI] [PubMed] [Google Scholar]
- Hauser A. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7 654–665. 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R., Engel J. N. (1999). Pseudomonas aeruginosa induces type-III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 67 5530–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R., Kang P. J., Engel J. N. (1998). PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27 807–818. 10.1046/j.1365-2958.1998.00727.x [DOI] [PubMed] [Google Scholar]
- Hayes C. S., Aoki S. K., Low D. A. (2010). Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44 71–90. 10.1146/annurev.genet.42.110807.091449 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweize R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Hogardt M., Roeder M., Schreff A. M., Eberl L., Heesemann J. (2004). Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology 150 843–851. 10.1099/mic.0.26703-0 [DOI] [PubMed] [Google Scholar]
- Hovey A., Frank D. (1995). Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177 4427–4436. 10.1128/jb.177.15.4427-4436.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince D., Sutterwala F. S., Yahr T. L. (2015). Secretion of flagellar proteins by the Pseudomonas aeruginosa type III secretion-injectisome system. J. Bacteriol. 197 2003–2011. 10.1128/jb.00030-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inclan Y. F., Huseby M. J., Engel J. N. (2011). FimL regulates cAMP synthesis in Pseudomonas aeruginosa. PLoS One 6:e15867. 10.1371/journal.pone.0015867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intile P. J., Balzer G. J., Wolfgang M. C., Yahr T. L. (2015). The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 197 2664–2674. 10.1128/jb.00231-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intile P. J., Diaz M. R., Urbanowski M. L., Wolfgang M. C., Yahr T. L. (2014). The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 196 357–366. 10.1128/jb.01199-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Yang H., Qiao M., Jin S. (2011). MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J. Bacteriol. 193 399–410. 10.1128/jb.01079-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman M. R., Jia J., Zeng L., Ha U., Chow M., Jin S. (2000). Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of ExoS. Microbiology 146 2531–2541. 10.1099/00221287-146-10-2531 [DOI] [PubMed] [Google Scholar]
- Kim J., Ahn K., Min S., Jia J., Ha U., Wu D., et al. (2005). Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151 3575–3587. 10.1099/mic.0.28277-0 [DOI] [PubMed] [Google Scholar]
- Li K., Xu C., Jin Y., Sun Z., Liu C., Shi J., et al. (2013). SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio 4:e0419-13. 10.1128/mBio.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Long Y., Liu Y., Liu Y., Chen R., Shi J., et al. (2016). HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced persister cells. Front. Cell Infect. Microbiol. 6:125. 10.3389/fcimb.2016.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati N. T., Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., et al. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U.S.A. 103 2833–2838. 10.1073/pnas.0511100103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Semino-Mora C., Dubois A. (2012). Mechanism of H. pylori intracellular entry: an in vitro study. Front. Cell Infect. Microbiol. 2:13. 10.3389/fcimb.2012.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak J. B., Cannon C. L., Pier G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15 194–222. 10.1128/cmr.15.2.194-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden A. E., Intile P. J., Schulmeyer K. H., Simmons-Patterson E. R., Urbanowski M. L., Wolfgang M. C., et al. (2016). Vfr directly activates exsa transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 198 1442–1450. 10.1128/jb.00049-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaw M., Lykken G., Singh P., Yahr T. (2002). ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 46 1123–1133. 10.1046/j.1365-2958.2002.03228.x [DOI] [PubMed] [Google Scholar]
- Mulcahy H., O’Callaghan J., O’Grady E. P., Adams C., O’Gara F. (2006). The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 74 3012–3015. 10.1128/iai.74.5.3012-3015.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Handley S. F., Frick D. N., Bullions L. C., Mildvan A. S., Bessman M. J. (1996). Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins: cloning, purification, and characterization of the enzyme. J. Biol. Chem. 271 24649–24654. 10.1074/jbc.271.40.24649 [DOI] [PubMed] [Google Scholar]
- Okon E., Dethlefsen S., Pelnikevich A., Barneveld A. V., Munder A., Tummler B. (2017). Key role of an ADP - ribose - dependent transcriptional regulator of NAD metabolism for fitness and virulence of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 307 83–94. 10.1016/j.ijmm.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Rietsch A., Vallet-Gely I., Dove S. L., Mekalanos J. J. (2005). ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102 8006–8011. 10.1073/pnas.0503005102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W., Prince A. S. (2005). Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171 1209–1223. 10.1164/rccm.200408-1044SO [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis J. (1989). Molecular Cloning: a Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shen D.-K., Filopon D., Chaker H., Boullanger S., Derouazi M., Polack B., et al. (2008). High-cell-density regulation of the Pseudomonas aeruginosa type III secretion system: implications for tryptophan catabolites. Microbiology 154 2195–2208. 10.1099/mic.0.2007/013680-0 [DOI] [PubMed] [Google Scholar]
- Shen D. K., Filopon D., Kuhn L., Polack B., Toussaint B. (2006). PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect. Immun. 74 1121–1129. 10.1128/iai.74.2.1121-1129.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherertz R. J., Sarubbi F. A. (1983). A three-year study of nosocomial infections associated with Pseudomonas aeruginosa. J. Clin. Microbiol. 18 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Jin Y., Bian T., Li K., Sun Z., Cheng Z., et al. (2015). SuhB is a novel ribosome associated protein that regulates expression of MexXY by modulating ribosome stalling in Pseudomonas aeruginosa. Mol. Microbiol. 98 370–383. 10.1111/mmi.13126 [DOI] [PubMed] [Google Scholar]
- Smith R. S., Wolfgang M. C., Lory S. (2004). An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72 1677–1684. 10.1128/IAI.72.3.1677-1684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.-G., Bail S., Kiledjian M. (2013). Multiple nudix family proteins possess mRNA decapping activity. RNA 19 390–399. 10.1261/rna.037309.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A. M., Pereira M. O. (2014). Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs-a review. Pathogens 3 680–703. 10.3390/pathogens3030680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault J., Faudry E., Ebel C., Attree I., Elsen S. (2009). Anti-activator ExsD Forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J. Biol. Chem. 284 15762–15770. 10.1074/jbc.M109.003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal H., Fulcher N. B., Bitterman J., Salazar E., Buck J., Levin L. R., et al. (2012). Crystal structure and regulation mechanisms of the CyaB adenylyl cyclase from the human pathogen Pseudomonas aeruginosa. J. Mol. Biol. 416 271–286. 10.1016/j.jmb.2011.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totten P. A., Lory S. (1990). Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172 7188–7199. 10.1128/jb.172.12.7188-7199.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski M. L., Brutinel E. D., Yahr T. L. (2007). Translocation of ExsE into chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect. Immun. 75 4432–4439. 10.1128/iai.00664-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski M. L., Lykken G. L., Yahr T. L. (2005). A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 102 9930–9935. 10.1073/pnas.0504405102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urick T., I-Chang C., Arena E., Xu W., Bessman M. J., Ruffolo C. G. (2005). The pnhA Gene of Pasteurella multocida encodes a dinucleoside oligophosphate pyrophosphatase member of the nudix hydrolase superfamily. J. Bacteriol. 187 5809–5817. 10.1128/jb.187.16.5809-5817.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alst N. E., Wellington M., Clark V. L., Haidaris C. G., Iglewski B. H. (2009). Nitrite reductase NirS Is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa. Infect. Immun. 77 4446–4454. 10.1128/iai.00822-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. E., Sample A. K., Runyen-Janecky L. J. (1994). The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176 7532–7542. 10.1128/jb.176.24.7532-7542.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., Brinkman F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44 D646–D653. 10.1093/nar/gkv1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M. C., Lee V. T., Gilmore M. E., Lory S. (2003). Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4 253–263. 10.1016/S1534-5807(03)00019-4 [DOI] [PubMed] [Google Scholar]
- Wu W., Badrane H., Arora S., Baker H. V., Jin S. (2004). MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J. Bacteriol. 186 7575–7585. 10.1128/jb.186.22.7575-7585.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Jin S. (2005). PtrB of Pseudomonas aeruginosa suppresses the type III secretion system under the stress of DNA damage. J. Bacteriol. 187 6058–6068. 10.1128/jb.187.17.6058-6068.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curve of PAK and the ΔPA4916 mutant.
Role of NrtR in twitching motility or expression of prpL and toxA in P. aeruginosa. (A) Twitching motilities of the indicated strains were examined on 1% LB agar with a Δvfr mutant serving as a control. The twitching zones were visualized with 0.1% crystal violet staining. (B) Relative mRNA levels of prpL and toxA. Total RNA of indicated strains was isolated and mRNA levels of prpL and toxA were determined by real time PCR with rpsL serving as an internal control. Data represents the mean ± standard deviation. ∗∗p < 0.01, ∗∗∗p < 0.001 by Student’s t-test.
PA4918 is not involved in the regulation of T3SS. (A) Relative mRNA levels of PA4918. Total RNA of indicated strains was isolated and mRNA levels of PA4918 were determined by real time PCR with rpsL serving as an internal control. Data represents the mean ± standard deviation. ∗∗∗p < 0.001 by Student’s t-test. (B) PAK containing the PA4918-20 operon driven by a tac promoter or the empty vector pMMB67EH were grown to an OD600 of 1.0 in LB with 1 mM IPTG with or without 5 mM EGTA. Proteins from equivalent bacterial cells of indicated strains were separated by SDS-PAGE and probed with an anti-ExoS antibody or an anti-RNA polymerase beta subunit antibody. S, supernatant; P, pellet.
Coomassie blue staining of purified CyaB217-463 and NadD2. (B) Purified NadD2 was not contaminated by ATPase. Equal amount ATP was incubated with or without 5 μg NadD2. After 30 min at 30°C, the ATP levels were measured using an ATP detection kit. Error bars represent standard deviations. ns, not significant, by Student’s t-test. (C) Purified NadD2 was not contaminated by phosphodiesterase. Equal amount cAMP was incubated with or without 5 μg NadD2. After 30 min at 30°C, the cAMP levels were measured using an ELISA kit. Error bars represent standard deviations. ns, not significant, by Student’s t-test.