Abstract
PIK3CA mutations are common in clinical molecular profiling, yet an effective means to target these cancers has yet to be developed. MTORC1 inhibitors are often used off-label for patients with PIK3CA mutant cancers with only limited data to support this approach. Here we describe a cohort of patients treated with cancers possessing mutations activating the PI3K signaling cascade with minimal benefit to treatment with the MTORC1 inhibitor everolimus. Previously, we demonstrated that dual PI3K/mTOR inhibition could decrease proliferation, induce differentiation, and result in a treatment response in APC and PIK3CA mutant colorectal cancer (CRC). However, reactivation of AKT was identified, indicating that the majority of the benefit may be secondary to MTORC1/2 inhibition. TAK-228, an MTORC1/2 inhibitor, was compared to dual PI3K/mTOR inhibition using BEZ235 in murine CRC spheroids. A reduction in spheroid size was observed with TAK-228 and BEZ235 (−13% and −14%, respectively) compared to an increase of >200% in control (p < 0.001). These spheroids were resistant to MTORC1 inhibition. In transgenic mice possessing Pik3ca and Apc mutations, BEZ235 and TAK-228 resulted in a median reduction in colon tumor size of 19% and 20%, respectively, with control tumors having a median increase of 18% (p = 0.02 and 0.004, respectively). This response correlated with a decrease in the phosphorylation of 4EBP1 and RPS6. MTORC1/2 inhibition is sufficient to overcome resistance to everolimus and induce a treatment response in PIK3CA mutant CRCs and deserves investigation in clinical trials and in future combination regimens.
Keywords: colon cancer, PI3K, PIK3CA, BEZ235, TAK-228, MTORC1/2, everolimus
INTRODUCTION
Precision medicine strategies hold great promise for improving the survival of patients with metastatic colorectal cancer (CRC), which is one of the leading causes of cancer-related death in the United States (1, 2). In 2009, KRAS exon 2 testing became the standard of care as a marker of resistance to the anti-epidermal growth factor receptor (EGFR) therapies, cetuximab and panitumumab (3–5). More recently, expanded KRAS, NRAS and BRAF testing have become standard, as more evidence has emerged for resistance to anti-EGFR agents in these settings (6, 7). Numerous institutions are also performing routine PIK3CA mutation testing, though the utility is not well established (1). Significant interest exists in whether cancers with PIK3CA mutations can be treated with targeted therapies, though an effective means of targeting these cancers has yet to be developed. As more patients undergo sequencing this data is being interpreted through institutional molecular tumor boards and sequencing companies are promoting the off-label use of the MTORC1 inhibitors, everolimus and temsirolimus (8).
PIK3CA mutations are present in approximately 18% of CRCs (9). These mutations occur most commonly in conjunction with APC mutations, which are present in at least 80% of CRCs. In addition, approximately 50% of PIK3CA mutant CRCs also carry KRAS, NRAS, or BRAF mutations (10, 11). This proportion of cancers with concomitant alterations likely increases significantly following exposure of these cancers to anti-EGFR antibodies. PIK3CA mutations commonly occur in 3 hotspots, including H1047R in the kinase domain and E545K and E542K in the helical domains (12). These mutations result in the expression of a constitutively active phosphoinositide 3-kinase (PI3K) activating downstream AKT and mammalian target of rapamycin (MTOR) signaling. The activation of these signaling cascades is important for cellular proliferation, protein synthesis, metabolism, inhibition of apoptosis and genomic stability among other important processes (13–15). Given the importance of the PI3K pathway in cancer, multiple therapeutic agents continue through clinical development targeting various nodes along this signaling cascade. Despite multiple investigations into the treatment of malignancies with PI3K, AKT, and MTOR inhibitors, the populations most likely to benefit from these agents largely remain to be identified, especially for CRC.
Our group has previously demonstrated that PIK3CA mutations can initiate tumorigenesis in the colon and pancreas (16–19). We have also demonstrated that PIK3CA and APC mutations are synergistic in the development and progression of CRCs (16). Recently, we investigated treatment strategies along the PI3K pathway for APC and PIK3CA mutant colon cancer using a murine model expressing a constitutively active PI3K construct, p110* and cre-mediated deletion of Apcexon14 (20). Spheroids generated from this model were resistant to the alpha isomer specific inhibitor BYL-719 and the pan-PI3K inhibitor GDC0941, yet sensitive to the dual PI3K/MTORC1/2 inhibitors BEZ235 and LY3023414 (20). These results were confirmed in vivo. Interestingly, despite inhibition of phosphorylation of AKT at 6 hours following treatment administration, prolonged therapy resulted in persistently decreased phosphorylation of ribosomal protein S6 (RPS6) and eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1), but AKT phosphorylation was restored (20). This led us to hypothesize that the majority of the benefit from these therapies was a result of MTORC1/2 inhibition alone. Here we determine if dual PI3K/MTORC1/2 inhibition is necessary for a treatment response or if MTORC1/2, or even MTORC1 inhibition alone, is sufficient in APC and PIK3CA mutant colon cancers using spheroid cultures, human isogenic cell lines and a novel murine model with colon cancers possessing loss of APC and expressing the human PIK3CAH1047R hotspot mutation.
METHODS
Precision Medicine Molecular Tumor Board Registry
Following IRB approval (UW IRB #2015–1370), retrospective analysis was performed to identify those patients with activating alterations of the PI3K/AKT/MTOR pathway for which the off-label use of everolimus was considered through the University of Wisconsin Carbone Cancer Center (UWCCC) Precision Medicine Molecular Tumor Board (8). Treatment response was evaluated per standard RECIST v1.1 response criteria (21).
Mouse husbandry and pharmacologic treatments
All animal studies were conducted under protocols approved by the Institutional Animal Care and Use Committee at the University of Wisconsin (Madison, WI) following the guidelines of the American Association for the Assessment and Accreditation of Laboratory Animal Care. Apcfl/fl mice (B6.Cg-Apctm2Rak; NCI Mouse Repository; Strain number 01XAA), Pik3caH047R mice (FVB.129S6-Gt(ROSA)26Sortm1(Pik3ca*H1047R)Egan/J; The Jackson Laboratory; Stock Number 016977) and Fc2 mice (FVB/N-Tg(Fabp1-Cre)1Jig; NCI Mouse Repository; Strain number 01XD8) were used to generate Fc1Apcfl/+Pik3caH1047R mice and these mice were genotyped as previously described (22–24). BEZ235 (LC Laboratories, Woburn, MA) and TAK-228 (MLN0128, LC Laboratories) were formulated in 0.05% AFE antifoam emulsion, 0.25% tween80, and 1.0% hydroxyethyl cellulose for in vivo treatments. BEZ235 was dosed at 30 mg/kg and TAK-228 was dosed at 1mg/kg daily by oral gavage for 14 continuous days.
Colorectal cancer cell isolation and spheroid culture
Colon cancer cells were harvested from Fc1 Apcfl/+ Pik3cap110* and Fc1 Apcfl/+ Pik3caH1047R mice and spheroid cultures generated as previously described (20). Spheroids between passage 2–9 were used for these studies. Therapeutic investigations were performed by exchanging feeding media containing the desired concentration of each agent over the spheroids entrapped in the Matrigel. BEZ235, TAK-228, and everolimus (LC Laboratories) were dissolved in DMSO to make a 10mM stock.
Histology and immunohistochemistry
Murine colon cancers were excised and fixed in 10% buffered formalin for 24 to 48 hours. Tissues were then stored in 70% ethanol until sectioned. CRCs were embedded in paraffin, and cut into 5 μm sections. Every tenth section was stained with hematoxylin and eosin (H&E). Immunohistochemistry was performed as previously described (18, 20). The primary antibodies included Ki67 (12202, 1:400, Cell Signaling Technology), phospho ERK 1/2 (Thr202/Tyr204, 4370, 1:400, Cell Signaling Technology), and phospho ribosomal protein S6 (RPS6) (Ser235/236, 4858, 1:50, Cell Signaling Technology).
Immunoblotting
Spheroid and colon tissue samples were collected and flash frozen. After 24 hours, protein was extracted and immunoblotting performed as previously described (20). The membranes were blocked with 5% non-fat dry milk for one hour and then probed with primary antibodies against phospho RPS6 (Ser235/236, 4858, Cell Signaling Technology), phospho AKT (Ser473, 4060; Thr308, 2965, Cell Signaling Technology), phospho 4EBP1 (Thr37/46, 2855, Cell Signaling Technology), total RPS6 (2217, Cell Signaling Technology), total AKT (4691, Cell Signaling Technology), or total 4EBP1 (9644, Cell Signaling Technology) in bovine serum albumin at 1:1000. β-actin (5125, Cell Signaling Technology) was used as a loading control at a ratio of 1:1000.
Spheroid hematoxylin and eosin (H&E) staining and immunofluorescence (IF)
Spheroids were plated onto glass 22mm circular coverslips and grown until mature. Coverslips were then fixed in cold 2% paraformaldehyde for 15 minutes and rinsed in PBS. H&E stains were performed as previously described (20). IF was performed as previously described (20). A conjugated antibody against Ki67 (Cell Signaling #11882 [1:50] (488 conjugate)) was used and slides were mounted with Prolong gold DAPI mounting media (Invitrogen #P36931).
Optical metabolic imaging of CRC spheroids
Optical metabolic imaging was performed using a custom-built inverted multiphoton microscope (Nikon Ti-E, Tokyo, Japan) as described previously (25, 26). Briefly, fluorescence excitation was provided by a tunable titanium:sapphire laser (Insight DS+, Spectra-Physics, Santa Clara, CA). NAD(P)H excitation was performed by tuning the laser to 750 nm, and emission was collected with a 440/40 nm filter (Chroma, Bellows Falls, VT). FAD excitation was performed at 890 nm, and emission was collected with a 550/50 nm filter (Chroma, Bellows Falls, VT). Excitation and emission light was coupled in a 40x water immersion objective (Nikon, 1.15 NA). Time-correlated single-photon counting (Becker & Hickl SPC-150, Berlin, Germany) was used to acquire intensity images over 60 seconds. Fluorescence emission was detected with a GaAsP photomultiplier tube (H7422P-40, Hamamatsu, Hamamatsu, Japan). A 4.8 μs pixel dwell time was used to acquire 256 × 256 pixel images.
MCF10A cells were treated with sodium cyanide (Complex IV inhibitor) to validate signals collected in the NAD(P)H and FAD channels. MCF10A cells were exposed to 4 mM NaCN, and NAD(P)H and FAD fluorescence intensities were recorded before and after treatment. The cyanide treatment increased NAD(P)H and decreased FAD fluorescence intensity, as expected (27), confirming the origin of the fluorescence in each channel.
Murine-derived colorectal spheroids were imaged in 35-mm glass-bottom petri dishes (MatTek Corp, Ashland, MA) two days after treatment with either BEZ235 (200 nM) or TAK-228 (200 nM). Fluorescence intensity images of NAD(P)H and FAD were collected across three fields-of-view per treatment, and three replicates were performed for each treatment.
Optical redox ratio and subpopulation analysis
NAD(P)H and FAD intensity images were created by integrating over all detected photons in each pixel. The optical redox ratio was calculated by dividing the total number of NAD(P)H photons by the total FAD photons (NAD(P)H/FAD) for each pixel. Semi-automated image segmentation with Cell Profiler software was performed on each NAD(P)H and FAD image to quantify the fluorescence in the cytoplasm of each cell, as described previously (28). All pixels within each cell cytoplasm were averaged to generate one redox ratio value per cell.
The redox ratio of individual cells was then input into a Gaussian mixture distribution model (MATLAB, version 2014a, MathWorks, Natick, Mass) (25, 29):
where g is the number of subpopulations, ϕ(y; μi, Vi) is the normal probability density function with mean μi variance Vi and πi is the mixing proportion. An Akaike information criterion (30) provided a metric to determine the goodness of fit for a given set of subpopulations (g = 1, 2, or 3), with the lowest Akaike score representing the best model. Probability density functions were normalized to ensure that the area under the curve for each treatment group was equal to 1.
Isogenic human colon cancer cells
Human SW48 and SW48 cells also carrying the PIK3CAH1047R mutation (SW48PK, Horizon Discovery, UK) were plated at 9000 cells/well in 96 well plates and allowed to adhere 48 hours. Culture medium was replaced with medium containing BEZ235 or TAK-228 at increasing concentrations. Cells were allowed to grow for 48 hours before a pre-mix WST-1 cell proliferation assay (Takara Bio USA, Mountain View, CA) was performed per manufacturer protocol.
Statistical Methods
Shifts in the mean spheroid growth rate as a function of initial diameter were determined using change-point statistical testing (31). Statistical analyses of spheroid growth and change in lumen occlusion were performed using Wilcoxon rank sum tests. P-values ≤ 0.05 were considered significant.
RESULTS
Resistance to the MTORC1 inhibitor everolimus is commonly encountered in patient’s whose cancers possess mutations that activate the PI3K pathway
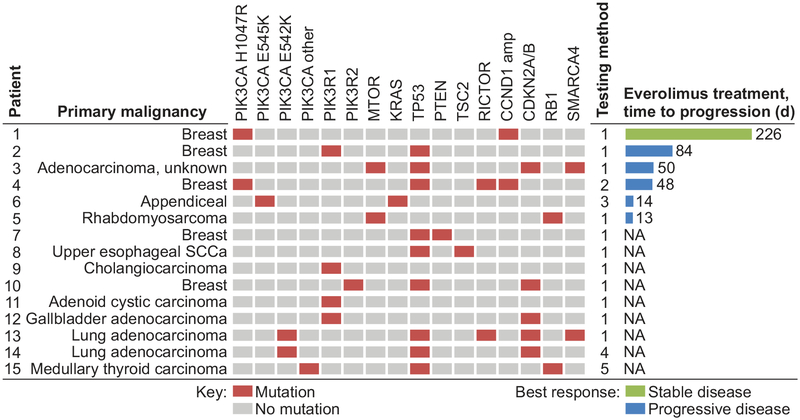
A total of 15 patients whose cancers possessed mutations known or suspected to activate PI3K/AKT/MTOR signaling were identified as part of the UWCCC Precision Medicine Molecular Tumor Board Registry (Fig. 1). These patients had treatment refractory disease across multiple cancer types with breast, lung and gastrointestinal cancers being the most common. Of these patients, 6 of them received everolimus therapy. Patient 1 in Fig. 1 received this therapy in combination with an anti-hormonal agent which resulted in stable disease for 226 days. The other 5 patients developed progressive disease as their best response, with most discontinuing therapy in less than 2 months. Though not a large enough cohort to statistically exclude the potential for benefit from MTORC1/2 inhibition, the complete lack of therapeutic activity of everolimus in this setting indicates the need to investigate potential alternative treatment strategies. This also highlights the use of these agents based on commercial sequencing recommendations with little clinical data to indicate efficacy.
Figure 1. Patients with activating alterations in phosphoinositide-3 kinase (PI3K) signaling are commonly identified and have minimal benefit from MTORC1 inhibition.
Fifteen patients with cancers possessing activating alterations within the PI3K pathway were identified in the University of Wisconsin Carbone Cancer Center Precision Medicine Molecular Tumor Board Registry. These mutations were identified in diverse cancers. Of these patients 6 of them were treated with everolimus. One patient with breast cancer received this therapy in combination with a hormonal agent resulting in stable disease for 226 days. The other five patients had progressive disease with most discontinuing therapy within 60 days secondary to clinical progression. Testing method: 1 = Foundation One; 2 = Foundation ICE; 3 = University of Wisconsin Cancer Gene Mutation Panel; 4 = Foundation T5a; 5 = NCI-MATCH. NA = not applicable.
MTORC1/2 inhibition is sufficient to induce a treatment response in Fc1 Apcfl/+ Pik3cap110* CRC spheroids
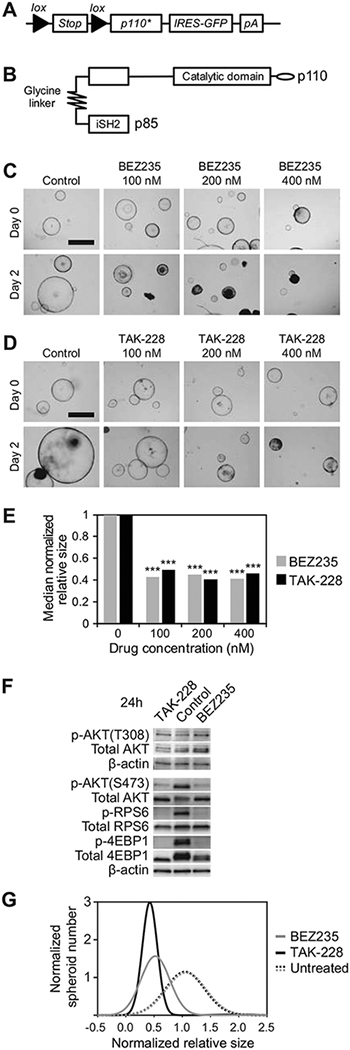
We have previously demonstrated that dual PI3K/mTOR inhibition with either BEZ235 or LY3023414 was sufficient to induce a robust therapeutic response in Fc1 Apcfl/+ Pik3cap110* CRC spheroids and in vivo (20). In tumors treated with BEZ235, a reactivation of AKT signaling as measured by increased phosphorylation of AKT was observed with prolonged treatment, potentially indicating that the upstream inhibition of PI3K was not required for its treatment effect. To evaluate if MTORC1/2 inhibition alone was sufficient to induce a response in this setting, the MTORC1/2 inhibitor, TAK-228, was used. Fc1 Apcfl/+ Pik3cap110* (Fig. 2A and 2B) CRC spheroids were allowed to mature for 48–96 hours prior to being treated with clinically relevant increasing concentrations of BEZ235 or TAK-228 in the culture media overlying the spheroids (Fig. 2C and 2D). A greater than 50% reduction in spheroid size was observed with TAK-228 treatment relative to control in all concentrations investigated (100–400nM; Fig. 2D and 2E). This reduction in spheroid size was similar to that seen in response to BEZ235 (Fig. 2C and 2E; ref. 20). Suppression of the PI3K signaling cascade was observed similarly between these agents with reductions in the phosphorylation of both RPS6 and 4EBP1 (Fig. 2F). Population distribution modeling was performed comparing the change in size of individual spheroids treated with BEZ235, TAK-228 and control (Fig. 2G). Histograms were generated following normalization to the control spheroid growth and number of spheres measured. The histogram for those spheroids treated with TAK-228 was shifted to the left and narrowed, indicating a robust treatment effect across the population of spheres compared to those treated with control.
Figure 2. MTORC1/2 induces significant responses in Fc1 Apcfl/+ Pik3cap110* CRC spheroids.
Fc1 Apcfl/+ Pik3cap110* CRC spheroids possess a lox-stop-lox sequence prior to the p110* transgene, which results in a constitutively active PI3K (A and B). These spheroids were allowed to mature for 24–96 hours prior to treatment with BEZ235, TAK-228 or control (C and D). Baseline and 48 hours post-treatment brightfield images were obtained and change in spheroid diameter measured for individual spheres. A similar median sphere size following treatment with BEZ235 and TAK-228 were observed and both significantly reduced from control (p < 0.001) (E). Reduced phosphorylation of AKT (ser473), RPS6 (Ser235/236), and 4EBP1 (Thr37/46) was observed after treatment of these spheres with BEZ235 and TAK-228 compared to control (F). Population distribution histograms demonstrating variation in change in sphere size for Fc1 Apcfl/+ Pik3cap110* CRC spheroids after 48 hours of treatment with BEZ235, TAK-228 or control (G). Size bar in panel A = 1 mm.
Fc1 Apcfl/+ Pik3caH1047R mice develop intestinal cancers similar to those encountered in Fc1 Apcfl/+ Pik3cap110* mice
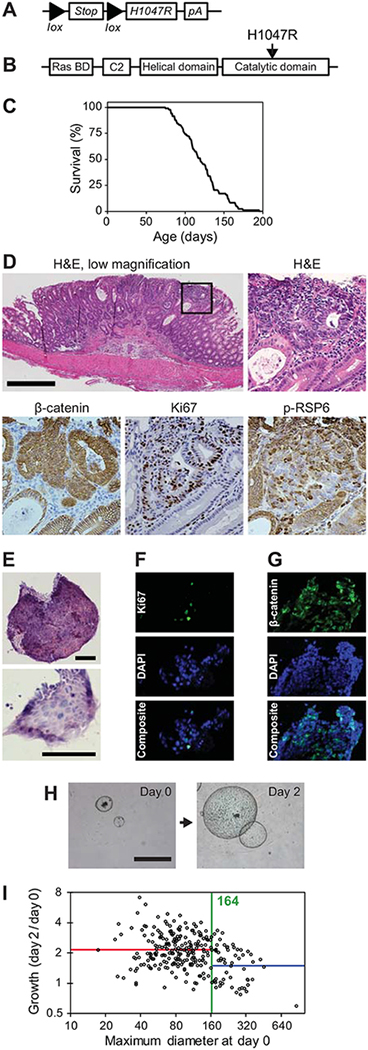
The Fc1 Apcfl/+ Pik3cap110* CRC model has activation of PI3K through the expression of a construct that has a portion of the p85 regulatory subunit linked to the p110 subunit. Recently we have demonstrated the ability of the H1047R hotspot mutation in Pik3ca to induce tumorigenesis in mice (19). The interaction between loss of APC and this hotspot mutation in Pik3ca has not yet been studied in a murine model. To explore this further we developed the Fc1 Apcfl/+ Pik3caH1047R mice (Fig. 3A and 3B). These mice live an average of 121 days (Fig. 3C). Their lifespan is limited secondary to the development of cancers within the distal small intestine and colon (Fig. 3D). These mice develop 4.9±3.2 colon tumors per mouse. A table comparing the Fc1 Apcfl/+ Pik3caH1047R mice to other similar murine models is presented in Supplementary Table S1. These colon tumors are moderately differentiated adenocarcinomas. At ~100 days of age, ~75% of these tumors are invasive adenocarcinomas (Supplementary Table S1). Of the remaining 25%, the majority are intramucosal carcinomas. As expected, these tumors demonstrate nuclear localization of CTNNB1 (β-catenin) secondary to loss of APC, phosphorylation of RPS6 and significant cellular proliferation, as measured by Ki67 nuclear staining (Fig. 3D).
Figure 3. Fc1 Apcfl/+ Pik3caH1047R mice develop invasive colon adenocarcinomas and can be utilized for translational investigations.
Fc1 Apcfl/+ Pik3caH1047R mice possess a lox-stop-lox sequence prior to the human PIK3CA H1047R hotspot mutation (A and B). These mice were allowed to age until moribund. These mice became moribund at an average age of 121 days (C). At necropsy tumors were identified within the small intestine and colon. Upon histologic sectioning, these tumors were found to be well to moderately differentiated adenocarcinomas (D, upper panels). These cancers possessed nuclear CTNNB1 (β-catenin), increased Ki67 and phosphorylation of RPS6 (D, lower panels). Spheroid cultures were able to be generated from these cancers and demonstrated a similar histology (E) and proliferating cells as measured by Ki67 (F). Nuclear CTNNB1 was also observed in the spheroids (G). The diameter of these spheres can be followed over time as a marker of proliferation (H). It was observed that larger spheroids at baseline would typically grow at a slower rate. To standardize experiments between cohorts, a standard change-point analysis was utilized (I). A change point at 164 pixels (373 μm) was identified and only those spheroids below that cut-off were utilized in further investigations. Size bars: D = 500 μm, E = 100 μm, G = 1 mm. The square panels in D are 4x enlargements of the outlined area.
Spheroid cultures were generated from Fc1 Apcfl/+ Pik3caH1047R cancers. The cancer cells in these spheroids share a similar histology (Fig. 3E) and proliferation rate (Fig. 3F) as the tumors from which they were derived. Nuclear localization of β-catenin was also confirmed in the spheroids (Fig. 3G). These spheroids can be followed longitudinally using brightfield microscopy and the change in the diameter used as a marker of cellular growth and proliferation (Fig. 3H). It was identified that the growth rate of the spheres can change as a function of spheroid size (Fig. 3I). A standard change point analysis demonstrates that spheroids less than 164 pixels (373 μm) grow at a similar rate. Spheroids whose baseline imaging size exceeded this cutoff had a reduction in growth rate (Fig. 3I). Only those spheroids whose baseline size is less than this cutoff were included in analyses for the therapeutics studies.
MTORC1/2 inhibition, but not MTORC1 inhibition alone, induces a response in Fc1 Apcfl/+ Pik3caH1047R CRC spheroids
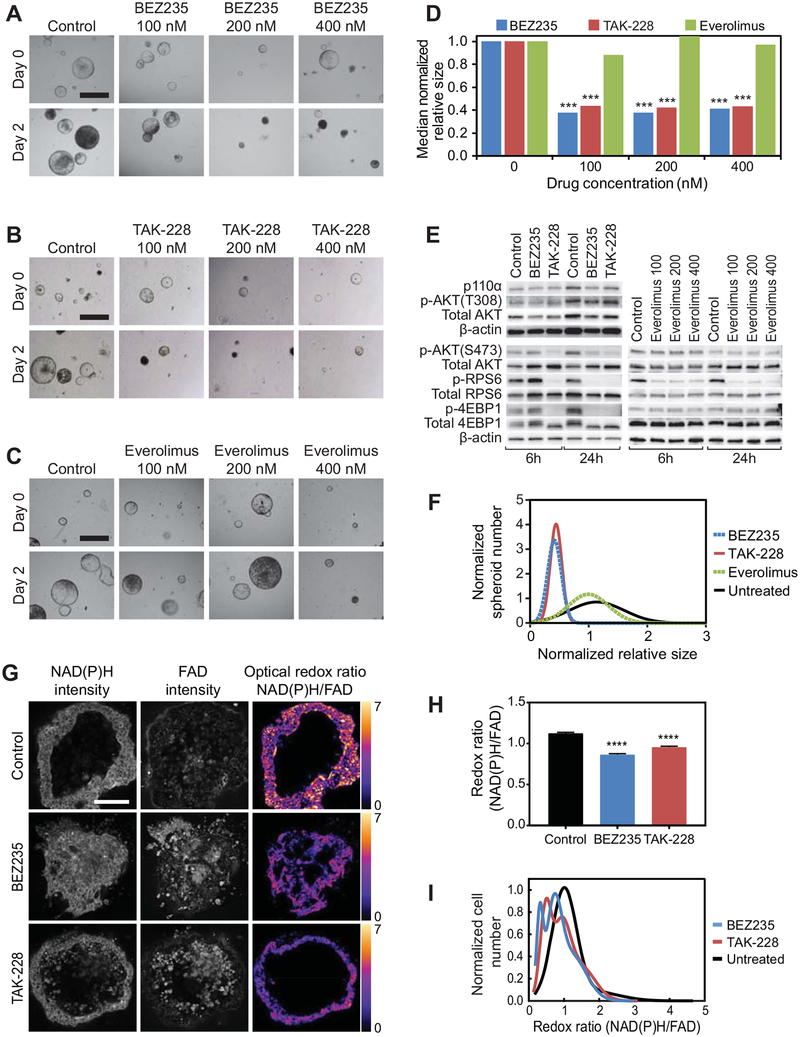
To compare the response between the dual PI3K/MTORC1/2 inhibitor, BEZ235, and the MTORC1/2 inhibitor, TAK-228, Fc1 Apcfl/+ Pik3caH1047R colon cancer spheroids were allowed to mature for 48–96 hours. Baseline 4x brightfield images were obtained and the overlaying media was exchanged with media containing increasing concentrations of BEZ235, TAK-228, everolimus or control (Fig. 4A–C). Both BEZ235 and TAK-228 resulted in a reduction of sphere size (decreasing by 13% and 14% at the 200nM concentration, respectively) with control treated spheres increasing to 229% of their size at day 0 (Fig. 4A–D). These results correlate with the ability of BEZ235 and TAK-228 to suppress the activation of the distal PI3K pathway at RPS6 and 4EBP1 (Fig. 4E).
Figure 4. Apc and Pik3caH1047R mutant CRC spheroids are sensitive to MTORC1/2, but not MTORC1 inhibition alone.
Spheroids from Fc1 Apcfl/+ Pik3caH1047R cancers were allowed to mature for 24–96 hours. Brightfield images were obtained at baseline and following treatment with BEZ235, TAK-228, everolimus or control (A-C). Similar reductions in median relative spheroid size were observed with BEZ235 and TAK-228 compared to control (p < 0.001, A, B and D). Everolimus, an MTORC1 inhibitor, did not significantly decrease spheroid growth compared to control (C and D). Decreased phosphorylation of AKT (Ser473), RPS6 (Ser235/236), and 4EBP1 (Thr37/46) was observed at 6 hours following treatment with TAK-228, but not with BEZ235 or control (E). After 24 hours, similar reductions in the phosphorylation of AKT, RPS6 and 4EBP1 were observed (E). Everolimus did not reduce the phosphorylation of 4EBP1 (D). Population distribution histograms demonstrating variation in change in sphere size for Fc1 Apcfl/+ Pik3caH1047R CRC spheroids after 48 hours of treatment with BEZ235, TAK-228, everolimus, or control (F). Optical metabolic imaging was also utilized to confirm the therapeutic response. Representative images of NAD(P)H and FAD fluorescence intensities and optical redox ratio (G). The optical redox ratio (NAD(P)H intensity/FAD intensity) is decreased in spheroids treated with TAK-228 and BEZ235 compared to untreated spheroids (G and H). Subpopulation analysis of the redox ratio calculated from fluorescence images of untreated, BEZ235-treated, and TAK-228-treated CRC spheroids (I). The area under the curve for each treatment group was normalized to equal 1. Peaks represent high frequency of cells with similar optical redox ratios. Size bar in panel A-C = 1 mm, G = 100 μm. ****p < 0.0001.
To determine if MTORC1 inhibition was sufficient to induce a treatment response in this model, Fc1 Apcfl/+ Pik3caH1047R CRC spheroids were treated with everolimus, an MTORC1 inhibitor. These cultures were treated in a similar fashion to the BEZ235 and TAK-228 experiments (Fig. 4C). Persistent spheroid growth was observed in the setting of 100–400nM everolimus and was not statistically different compared to the control treated spheres (Fig. 4D). In addition, everolimus failed to inhibit phosphorylation of 4EBP1 (Fig. 4E). Population distribution modeling demonstrates that both BEZ235 and TAK-228, but not everolimus, shift the spheroid population curve significantly to the left and narrow the distribution compared to control (Fig. 4F). These results indicate that MTORC2 inhibition in addition to MTORC1 inhibition is necessary to induce spheroid regression in this setting.
Cell-level optical metabolic imaging of spheroid cultures to identify potential resistant cell populations
To further investigate the treatment response to BEZ235 and TAK-228, optical metabolic imaging (OMI) was used to identify differences in the cell-level response of the Fc1 Apcfl/+ Pik3caH1047R colon cancer spheroids. Novel cellular-level optical imaging technologies can provide non-invasive interrogation of intact spheroid cultures to understand treatment response, heterogeneity, and mechanisms of resistance in the living, adapting spheroid. OMI exploits the intrinsic fluorescence intensity of the metabolic co-enzymes NAD(P)H and FAD to image drug response on a single-cell level across cells in a 3-dimensional sample (25–29). This single-cell analysis can dynamically quantify heterogeneous drug response over a treatment time-course, and can thus identify particularly sensitive or treatment-resistant cell sub-populations.
Forty-eight hours following treatment of Fc1 Apcfl/+ Pik3caH1047R colon cancer spheroids with BEZ235 (200nM), TAK-228 (200nM), or control, OMI was performed to image the NAD(P)H and FAD intensity of individual cells within spheroids across treatment groups (Fig. 4G). The optical redox ratio (NAD(P)H/FAD) was calculated and compared across treatment groups. A significant reduction in the redox ratio was observed in both the BEZ235 and TAK-228 treated cells compared to control (p < 0.0001, Fig. 4H). Cell-level redox ratio population density distributions demonstrate a shift of the histograms to the left for those cells treated with BEZ235 or TAK-228 (Fig. 4I). Interestingly, these histograms also detect differential sensitivity of two populations in both BEZ235 and TAK-228 treated groups, which is reflected in the two distinct redox ratio peaks for these treatment groups.
BEZ235 and TAK-228 suppress cancer cell growth in human isogenic PIK3CAH1047R mutant colon cancer cells
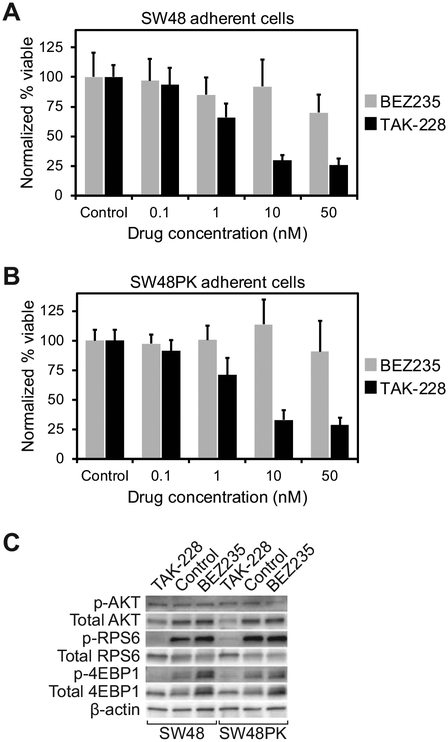
A heterozygous knock-in of the PIK3CAH1047R mutation into SW48 human colon cancer cell lines (SW48PK) was obtained. Of note, the SW48 cell line is known to possess a truncating alteration in FBXW7, which is an ubiquitin ligase that degrades CMYC, MTOR, among others (32). This would indicate that this line at baseline could be sensitive to MTOR inhibition. Increasing concentrations of BEZ235 and TAK-228 were used to treat both the SW48 and SW48PK cells and treatment response was measured using the WST-1 cellular proliferation assay. As expected, both BEZ235 and TAK-228 led to a reduction in proliferation in the SW48 cells (Supplementary Fig. S1A). A similar response was also observed in the SW48PK cells with both BEZ235 and TAK-228 (Supplementary Fig. S1B). At low concentrations (0.1–50nM), a greater treatment effect was seen with TAK-228 compared to BEZ235 (Figs. 5A and 5B). This enhanced treatment effect correlated with decreased phosphorylation of AKT, RPS6 and 4EBP1 in TAK-228 compared to BEZ235 treated cells (Fig. 5C).
Figure 5. MTORC1/2 inhibition suppresses growth and MTOR signaling in isogenic human colon cancer cells with and without the PIK3CAH1047R hotspot mutation.
SW48 human colon cancer cells were treated with increasing concentrations of BEZ235, TAK-228 or control (A). The WST-1 cell proliferation assay was utilized to measure cell viability. Similarly, SW48 cells possessing the PIK3CAH1047R hotspot mutation (SW48PK) were treated with BEZ235, TAK-228, or control with response measured with the WST-1 assay (B). Inhibition of the phosphorylation of RPS6 and 4EBP1 were greater with TAK-228 (10nM) than BEZ235 (10nM) or control after 24 hours of treatment (C).
BEZ235 and TAK-228 induce a treatment response in vivo in Apc and Pik3ca mutant colon cancers
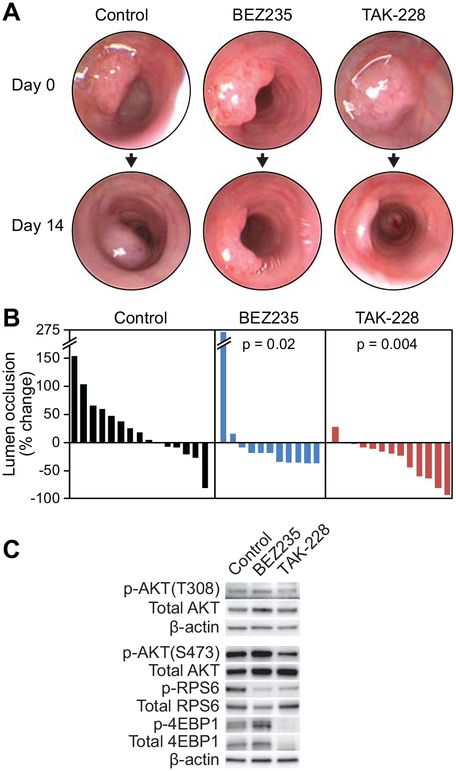
To determine the impact of MTORC1/2 inhibition on Apc and Pik3ca mutant colon cancers, Fc1 Apcfl/+ Pik3caH1047R mice were treated with BEZ235 (10 mice), TAK-228 (11 mice) or vehicle control (9 mice) for 14 days. These groups were well balanced in regards to the male-to-female ratio, baseline tumor size, and age (median age ~100 days). Murine endoscopy was utilized to determine treatment response. To interrogate the response in these mice, colonoscopies were performed on each subject at days 0 and 14 of the study (Fig. 6A). Using these images, percent lumen occlusion values for each tumor were calculated using ImageJ, as previously described (20, 24). One to two tumors were identified in the visible region of the colon per mouse. A median reduction in lumen occlusion was seen with BEZ235 and TAK-228 of 19% and 20% respectively, compared to an 18% increase in lumen occlusion of those cancers treated with control (Wilcoxon rank sum test p = 0.02 comparing BEZ235 and control, and p = 0.004 comparing TAK-228 and control, Fig. 6B). The response rate, as measured by the percent of cancers achieving at least a 30% reduction in lumen occlusion, was 45% for BEZ235 and 38% for TAK-228 (Fig. 6B). These responses correlated with a reduction in phosphorylation of RPS6 for both BEZ235 and TAK-228. Interestingly, a decrease in phosphorylation of AKT and 4EBP1 were observed with TAK-228 treatment (Fig. 6C).
Figure 6. MTORC1/2 inhibition results in a robust treatment response in Fc1 Apcfl/+ Pik3caH1047R mice.
These mice were allowed to age to ~100 days. At this time-point, the majority of these tumors are invasive cancers. Mice were treated with BEZ235, TAK-228 or control daily by oral gavage. Murine endoscopy was performed at baseline and following 14 days of treatment (A). Percent lumen occlusion was measured for each tumor and the change in percent lumen occlusion shown in a waterfall plot (B). A significant reduction in median change in lumen occlusion was observed with those tumors treated with BEZ235 (−19%) and TAK-228 (−20%) compared to control (+17%; p = 0.02 and p = 0.004, respectively) (B). A similar reduction in phosphorylation of RPS6 was seen after 14 days of treatment with BEZ235 and TAK-228, however a greater reduction in phosphorylation of AKT and 4EBP1 were observed with TAK-228 treatment (C).
DISCUSSION
PIK3CA mutations are commonly identified in patient’s cancers who undergo molecular testing. This is especially true for breast, endometrial and colorectal cancers (33). A targeted means by which to effectively treat cancers with these alterations has yet to be established clinically (14, 15). The studies to date have often had multiple limitations including the investigation of molecularly unselected patients, patients with concomitant mutations known to alter therapeutic response against this pathway (i.e. KRAS mutations), and the use of inhibitors that in our recent publications and in this manuscript have had low likelihood of therapeutic response (20).
Specifically for the treatment of CRC, almost all of the investigations using PI3K pathway inhibitors took place in phase I/II clinical trials in the late-line setting. Dual PI3K/MTOR inhibitors have demonstrated safety and some very preliminary evidence of activity (34–37). This includes 4 patients with colorectal cancer treated with BEZ235 who developed stable disease for ≥ 16 weeks and a partial response of a patient treated with LY3023414 (36–37). Many of these trials did not select specifically for PIK3CA mutant cancers and if they did, in general, utilized historical samples and did not take concomitant mutations into account. This is important as multiple investigations have demonstrated that following the use of the anti-EGFR therapies, cetuximab and panitumumab, these cancers acquire mutations that activate ERK1/2 signaling (38, 39). These mutations have been seen in KRAS, NRAS, MEK1/2 among others and are likely to result in resistance to therapies targeting the PI3K/AKT/MTOR pathway secondary to the activation of parallel signaling (38, 39).
In the above studies, we sought to answer two important questions related to the biology of cancers with PIK3CA mutations. First, we wanted to determine in a murine model if the human H1047R hotspot mutation would possess similar oncogenic properties to the p110* construct, used in recent investigations (16, 17, 20). In the p110* construct, the p110 subunit of PI3K is linked to a portion of the p85 subunit with a glycine kinker (40). Using this construct we were the first to demonstrate the ability of a constitutively active PI3K to initiate tumorigenesis through a non-canonical pathway, independent of WNT signaling (17). We then subsequently demonstrated the ability of this construct to synergize with loss of APC to increase tumor incidence and the progression of these tumors to cancer (16). To verify that the p110 construct mimics the activity of the H1047R hotspot mutation in PI3K, we first developed the Fc1 Pik3caH1047R mice (19). The Fc1 Pik3caH1047R mice developed similar cancers, also through a non-canonical mechanism, to those in Fc1 Pik3cap110* mice, but with a longer latency and a less robust activation of PI3K/AKT/MTOR signaling (19). Here we demonstrate similar synergy in tumorigenesis and progression between loss of APC and expression of a constitutively active PI3K, comparing Fc1 Apcfl/+ Pik3caH1047R mice and our previously published Fc1 Apcfl/+ Pik3cap110* mice (Fig. 2, Supplementary Table S1 and ref. 16).
Secondly, we demonstrate that MTORC1/2 inhibition is sufficient to induce a treatment response in Apc and Pik3ca mutant CRCs in spheroids, isogenic human cell lines, and in a transgenic mouse model. Whereas, the Fc1 Apcfl/+ Pik3caH1047R CRC spheroids were resistant to the MTORC1 inhibitor, everolimus. Beyond changes in the diameter of these spheroids, metabolic imaging was also utilized. In previous studies, OMI has accurately predicted early drug response in pancreatic ductal adenocarcinoma, head and neck cancer, and breast cancer (25, 26, 40). In this study, the optical redox ratio in Fc1 Apcfl/+ Pik3caH1047R colon cancer spheroids showed a significant response to BEZ235 and TAK-228 treatment (Fig. 4H) compared to untreated spheroids across all fields-of-view, consistent with changes in spheroid diameter (Fig. 4D). However, changes in tumor diameter are not sensitive to mixed populations of responsive or non-responsive cells within each tumor. Cellular-level OMI detected a heterogeneous (bi-modal) response to drug treatment in spheroids with both BEZ235 and TAK-228 treatment (Fig. 4I). These bi-modal distributions indicate a resistant (higher redox ratio population) and responsive (lower redox ratio population) tumor cell subpopulation in each treatment group (27, 41). Future studies will seek to identify the evolution of these populations over time in response to drug treatment and identify the mechanisms of their differential sensitivity.
Overall, these data presented here indicate that MTORC1/2 inhibition deserves further investigation clinically in select patient populations with PIK3CA mutations, including those without concomitant RAS or RAF alterations. BEZ235 is no longer in clinical development related to formulation issues, while agents like TAK-228 and other MTORC1/2 inhibitors are still in clinical trials. In CRC, this treatment approach should be investigated prior to the use of anti-epidermal growth factor receptor (EGFR) inhibitor therapies such as cetuximab and panitumumab. Multiple investigations have now demonstrated the selection for or acquisition of mutations in RAS/RAF/MEK signaling upon progression through anti-EGFR therapies (38, 39). This is also important for patients as PIK3CA mutations have been shown to result in a relative decreased sensitivity to anti-EGFR monoclonal antibodies (1).
Supplementary Material
Acknowledgments:
This manuscript is dedicated to Thomas R. “Tom” Freye, who inspired many as he courageously fought colon cancer and deeply believed in the power of cancer research to make a difference for patients.
Financial support: This project was supported primarily by funding from the V Foundation for Cancer Research (V Scholar Award to D. A. Deming) and Funk Out Cancer (to D. A. Deming). Additional support was provided by start-up funds from the UW Carbone Cancer Center, UW Department of Medicine, the UW School of Medicine and Public Health, and the UW Graduate School through the Wisconsin Alumni Research Foundation to D. A. Deming; and funding from P30 CA014520 (NCI Core Grant, University of Wisconsin Carbone Cancer Center), UWCCC Experimental Therapeutics (pilot award to D. A. Deming), and Wisconsin Partnership Program (to D. A. Deming). M. C. Skala was supported by a Stand Up To Cancer Innovative Research Grant, Grant Number SU2C-AACR-IRG 08–16. Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Tran NH, Cavalcante LL, Lubner SJ, Mulkerin DL, LoConte NK, Clipson L et al. Precision medicine in colorectal cancer: the molecular profile alters treatment strategies. Ther Adv Med Oncol 2015;7:252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Facts & Figures 2017. American Cancer Society: Atlanta, 2017. [Google Scholar]
- 3.Deming D, Holen K. KRAS Mutation Analysis Prior to EGFR-Directed Therapy for Metastatic Colorectal Cancer: A Review and Cost Analysis. Current Cancer Therapy Reviews 2010;6:256–261. [Google Scholar]
- 4.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091–2096. [DOI] [PubMed] [Google Scholar]
- 5.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023–1034. [DOI] [PubMed] [Google Scholar]
- 6.Tejpar S, Lenz HJ, Kohne CH, Heinemann V, Ciardiello F, Esser R et al. Effect of KRAS and NRAS mutations on treatment outcomes in patients with metastatic colorectal cancer (mCRC) treated first-line with cetuximab plus FOLFOX4: New results from the OPUS study. Journal of Clinical Oncology 2014;32:LBA444. [Google Scholar]
- 7.Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J Clin Oncol 2015;33:692–700. [DOI] [PubMed] [Google Scholar]
- 8.Burkard ME, Deming DA, Parsons B, Kenny P, Schuh M, Leal T, et al. Implementation and clinical utility of an integrated academic-community regional molecular tumor board. JCO Precision Oncology. 2017; DOI: 10.1200/PO.16.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 2009;361:2449–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Vogt PK. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 2010;9:596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang CH, Mandelker D, Gabelli SB, Amzel LM. Insights into the oncogenic effects of PIK3CA mutations from the structure of p110alpha/p85alpha. Cell Cycle 2008;7:1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francipane MG, Lagasse E. mTOR pathway in colorectal cancer: an update. Oncotarget 2014;5:49–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yap TA, Bjerke L, Clarke PA, Workman P. Drugging PI3K in cancer: refining targets and therapeutic strategies. Curr Opin Pharmacol 2015;23:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deming DA, Leystra AA, Nettekoven L, Sievers C, Miller D, Middlebrooks M et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene 2014;33:2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leystra AA, Deming DA, Zahm CD, Farhoud M, Olson TJ, Hadac JN et al. Mice expressing activated PI3K rapidly develop advanced colon cancer. Cancer Res 2012;72:2931–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne SN, Maher ME, Tran NH, Van De Hey DR, Foley TM, Yueh AE et al. PIK3CA mutations can initiate pancreatic tumorigenesis and are targetable with PI3K inhibitors. Oncogenesis 2015;4:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yueh AE, Payne SN, Leystra AA, Van De Hey DR, Foley TM, Pasch CA et al. Colon Cancer Tumorigenesis Initiated by the H1047R Mutant PI3K. PLoS One 2016;11:e0148730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foley TM, Payne SN, Pasch CA, Yueh AE, Van De Hey DR, Korkos DP et al. Dual PI3K/mTOR Inhibition in Colorectal Cancers with APC and PIK3CA Mutations. Mol Cancer Res 2017;15:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumors: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45(2):228–47. [DOI] [PubMed] [Google Scholar]
- 22.Saam JR, Gordon JI. Inducible gene knockouts in the small intestinal and colonic epithelium. J Biol Chem 1999;274:38071–38082. [DOI] [PubMed] [Google Scholar]
- 23.Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, Wong RG et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res 2011;71:2706–2717. [DOI] [PubMed] [Google Scholar]
- 24.Hung KE, Maricevich MA, Richard LG, Chen WY, Richardson MP, Kunin A et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc Natl Acad Sci U S A 2010;107:1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh AJ, Castellanos JA, Nagathihalli NS, Merchant NB, Skala MC. Optical Imaging of Drug-Induced Metabolism Changes in Murine and Human Pancreatic Cancer Organoids Reveals Heterogeneous Drug Response. Pancreas 2016;45:863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah AT, Diggins KE, Walsh AJ, Irish JM, Skala MC. In Vivo Autofluorescence Imaging of Tumor Heterogeneity in Response to Treatment. Neoplasia 2015;17:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh A, Cook RS, Rexer B, Arteaga CL, Skala MC. Optical imaging of metabolism in HER2 overexpressing breast cancer cells. Biomed Opt Express 2012;3:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh AJ, Skala MC. An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images. Multiphoton Microscopy in the Biomedical Sciences Xiv 2014;8948. [Google Scholar]
- 29.Pan W, Lin J, Le CT. Model-based cluster analysis of microarray gene-expression data. Genome Biol 2002;3:RESEARCH0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akaike H New Look at Statistical-Model Identification. Ieee Transactions on Automatic Control 1974;Ac19:716–723. [Google Scholar]
- 31.Killick R, Eckley IA. Changepoint: An R Package for Changepoint Analysis. J Stat Softw. 2014;58(3):1–19. [Google Scholar]
- 32.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 2008;321:1499–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro GI, Bell-McGuinn KM, Molina JR, Bendell J, Spicer J, Kwak EL, et al. First-in-Human Study of PF-05212384 (PKI-587), a Small-Molecule, Intravenous, Dual Inhibitor of PI3K and mTOR in Patients with Advanced Cancer. Clin Cancer Res. 2015;21:1888–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadopoulos KP, Tabernero J, Markman B, Patnaik A, Tolcher AW, Baselga J, et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of SAR245409 (XL765), a novel, orally administered PI3K/mTOR inhibitor in patients with advanced solid tumors. Clin Cancer Res. 2014;20:2445–2456. [DOI] [PubMed] [Google Scholar]
- 36.Bendell JC, Kurkjian C, Infante JR, Bauer TM, Burris HA 3rd, Greco FA, et al. A phase 1 study of the sachet formulation of the oral dual PI3K/mTOR inhibitor BEZ235 given twice daily (BID) in patients with advanced solid tumors. Invest New Drugs. 2015;33:463–471. [DOI] [PubMed] [Google Scholar]
- 37.Bendell JC, Varghese AM, Hyman DM, Bauer TM, Pant S, Callies S, et al. A first-in-human phase I study of LY3023414, an oral PI3K/mTOR dual inhibitor, in patients with advanced cancer. Clin Cancer Res. 2018;24(14):3253–62. [DOI] [PubMed] [Google Scholar]
- 38.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:827. [DOI] [PubMed] [Google Scholar]
- 39.Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med 2014;6:224ra226. [DOI] [PubMed] [Google Scholar]
- 40.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell 2009;139:573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh AJ, Skala MC. Optical metabolic imaging quantifies heterogeneous cell populations. Biomed Opt Express 2015;6:559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.