Abstract
Single cell sequencing has recently been applied to many immunological studies. Flow cytometric index sorting isolates cells for single cell sequencing with protein level data linked to sequences. However, successful sequencing of index sorted samples requires careful optimization of several sort parameters, including nozzle size, flow rate, threshold rate, and yield calculations. In this study, considerations and optimization data for each of these variables are presented. Our analysis focused on index sorting, but the findings can be applied to any plate sorting protocol. Minimization of flow rates and use of the 70μm nozzle improved cell yields. Improvements in total read counts after sequencing were obtained by decreasing the threshold rate, or the number of cells processed per second. In addition, this technique provided linked protein and gene expression analysis of the cytokine interferon (IFN)γ, demonstrating that on a single cell basis IFNγ+ cells tend to express IFNG mRNA, and IFNγ− cells do not. Through rigorous optimization and quality control, we have identified parameters important to plate sorting and recommend the use of the 70μm nozzle and low flow and threshold rates for analysis of rare populations of human lymphocytes.
Keywords: Index sort, plate sort, nozzle, flow rate, threshold rate, single cell sequencing
1. Introduction
Index sorting is a powerful tool that links protein expression with single cell gene expression and sequencing analysis. Individual cells are sorted into 96- or 384- well plates with location and fluorescence intensity data saved for each cell (Osborne, 2011). This technique allows cell surface protein expression to be integrated with downstream analyses of single cells, such as T cell receptor (TCR) or B cell receptor (BCR) repertoire sequencing, and RNA sequencing, as well as additional applications (Hayashi et al., 2010). When appropriately optimized, index sorts have error rates less than 1% (Penter et al., 2018). A successful index or other plate sort requires consideration of factors that distinguish it from bulk sorting protocols. These include the dependence on nozzle size for accuracy, greater dependence on flow rate, and higher complexity of yield calculations.
Nozzle size is a key variable in both bulk and plate sorting. Nozzle sizes available on many sorters include 70, 85, 100, and 130 μm, but we focus on 70 and 100 μm nozzles because they are the most commonly used. Primary considerations for nozzle selection are cell size, rarity of the population to be sorted, and requirements for viability after sorting. The nozzle size is more crucial for index sorts with respect to cell yield. Stream stability, droplet accuracy and pressure vary with nozzle size. Specifically, the 70 μm nozzle has a more stable stream and a small drop size, which both improve accuracy of cell deposition onto the plate, as well as rapid flow (Osborne, 2011). However, the high accuracy and flow rate can limit the number of cells sorted, due to loss of cells as the plate moves. Rapid flow rates can damage cells. Each nozzle can only be used with cells 5–6 times smaller than the nozzle, so use of the 70μm nozzle is limited to cells of size less than 14 μm (Cossarizza et al., 2017). The 100 μm nozzle has a larger drop and lower sample pressure that can improve yields of delicate cells (BDBiosciences, 2012). Thus the selection of nozzle size will depend on the relative importance of these factors to the sort. The 70 μm nozzle is typically recommended for plate sorts of lymphocytes (Cossarizza et al., 2017) because the higher precision in droplet deposition and stream stability improves the likelihood that a given well in the plate contains a cell and thus nucleic acid that can be used for downstream sequencing applications.
Plate sorting has physical limitations constraining the range of possible flow rates. Accurate sorting requires each cell to be centered in the droplet, and each target cell to be in a droplet at a precise distance from non-target droplets. While bulk sorts are rate-limited by the stream and drop formation, plate sorts are further limited by the necessity to localize and center the drop over each well of a 96- or 384-well plate. This introduces additional risk of loss of cells when the drop is not centered or when cells are available to sort at a more rapid rate than the plate is able to center each well. Thus, there are more steps at which cells can be lost in plate sorts than in bulk sorts. The increased number of steps adds to the difficulty of preventing cell loss even with thorough optimization, with the result that plate sorts have substantially reduced yields in comparison to bulk sorts.
Sorting software typically provides details of relative fluorescence intensity of every parameter as well as numbers and rates of cells run through the sorter. The rate of sorting has no effect on fluorescence data but is a factor in determining the sort efficiency (percentage of cells in the sort gate that are successfully sorted) and the number of cells processed (BDBiosciences, 2012). The slow rates of plate sorts result in frequent inaccuracies of the numbers generated by the software. Consequently, optimization of yields requires manual calculations.
Each of these parameters is essential for the successful execution of a plate sort. Recognizing that resources including time, quantity of cells, and reagents are limited, substantial optimization on the part of any researcher conducting plate sorts is required. We have compiled data that we hope will direct others in the optimization and analysis of plate sorting, with a specific focus on index sorting.
2. Materials and methods
2.1. Preparation of single cell suspension
De-identified peripheral blood mononuclear cells (PBMC) were collected from the Stanford Blood Center. PBMC were isolated, frozen, and thawed as described (Higdon et al., 2016). Thawed cells were rested overnight and stimulated with a library of 15 amino acid peptides with 11 amino acid overlap for the cytomegalovirus (CMV) gene immediate early-1 (GenScript, Piscataway, NJ, USA) for 5 hours. Secretion of interferon (IFN)γ was measured using the Miltenyi IFNγ Secretion Assay kit according to the manufacturer’s protocol (Miltenyi Biotech, Bergisch Gladbach, Germany). Unstimulated cells were used as a control.
Cells were stained for 20 minutes at room temperature with fluorescently labeled antibodies for CD4 (RPA-T4), CD8 (SK1), CD3 (OKT3), CD14 (61D3), CD16 (3G8), CD19 (HIB19) from BioLegend (San Diego, CA, USA) and Abcam (Cambridge, UK), and with Zombie Aqua from BioLegend. Cells were resuspended in PBS with 0.5% bovine serum albumin and 2 mM EDTA. Cells were filtered through 35 μm nylon mesh into 5 mL round bottom tubes (Corning, Corning, NY, USA) immediately prior to sorting. Single color compensation controls were prepared using UltraComp eBeads from eBioScience (Santa Clara, CA, USA) and the above antibodies, or aliquots of cells stained with Zombie Aqua.
2.2. Set up of sorter
Experiments were carried out on a BD FACSAria III instrument equipped with BD FACSDiva V8.0 software (BD Biosciences, Franklin Lakes, NJ, USA). The instrument was equipped with Blue (488nm), Red (633nm), and Violet (405nm) lasers, and a BD Automated Cell Deposition Unit (ACDU) for plate sorting. The instrument was maintained using laser calibration with Cytometer, Setup & Tracking (CS&T) beads and drop calibration with AccuDrop beads (BD Biosciences). CS&T was run daily for both 70 and 100μm nozzles. Frequency was set to a constant value to minimize yield variation. This value was 88.0 for the 70 μm nozzle and 30.0 for the 100 μm nozzle. Amplitude was adjusted as needed to set up the droplet stream and optimize droplet break off. AccuDrop was run immediately prior to each sort to calculate the drop delay. A recirculating chiller device (Thermo Scientific, Waltham, MA, USA) was set to 5°C to maintain temperature of plates for maximal cell viability during sorting. The BD FACSAria System Family Aerosol Management Option was used to prevent formation of aerosols.
Flow rate was set to 1.0 except in experiments specifically comparing different flow rates where it was set to either 1.0 or 3.0, as specified. Cell concentration varied between 0.5 and 5×106 per mL based on desired threshold rate (range 50–800 events/second). Threshold rate is specified for each experiment.
2.3. Index sort
Cells were sorted into 96-well Hard-Shell plates (BioRad, Hercules, CA, USA) containing 12 μL of 1X One-Step RT-PCR buffer (Qiagen, Hilden, Germany) per well. ACDU positioning was calibrated using AccuDrop beads, followed by control cells of the same type as the sort population (PBMC). 200 cells were sorted per well onto aluminum sealing foil on top of 20–30 wells across the plate to test drop placement and centering. Using this method, 3 plates were tested prior to sorting to set the alignment. To check for drift in alignment, a test plate was run after every 3 plates throughout the sort, with a final test at the end. Positioning was adjusted for the plate relative to the drop using the Home Device menu and for the side stream using the side stream window. Deflection plate voltage was set at 5500 volts and 3000 volts for 70 μm and 100 μm nozzles respectively. Droplet frequencies for each nozzle were constant at 88,000 and 30,000 droplets per second. For single cell sorts the masks were set as follows: yield mask 0, purity mask 32, and phase mask 16.
Compensation was completed using single color controls and the FACSDiva compensation setup and calculation. Gates were set to identify the population of interest. The sort set up was completed for a 96-well plate, set to single cell purity (masks as described above), set to 1 cell per well of the population of interest for 88 of 96 wells. The remaining wells were used as negative controls for sequencing. Plates were prepared prior to sorting with lysis buffer in all wells as described above.
After sorting into each plate was completed, the plate was immediately covered in aluminum sealing foil (Corning) and centrifuged in an Allegra X-15R table top centrifuge (Beckman Coulter, Brea, CA, USA) at 300xg for 2 minutes to ensure that cells were in lysis buffer rather than on the side of the well. Plates were stored at −80° C immediately following centrifugation. For multi-plate experiments, sorting was paused while the plate was centrifuged and then resumed with a new plate following storage of the previous plate.
2.4. Nested PCR and sequencing of sorted single cells.
Nested PCR amplification and barcoding of single cell RNA was completed as described (Han et al., 2014). Barcoded PCR products were pooled into libraries, gel extracted using the QIAquick Gel Extraction Kit (Qiagen), and analyzed for DNA quality by Agilent Bioanalyzer (Santa Clara, CA, USA). Sequencing was completed on an Illumina MiSeq (San Diego, CA, USA) in the Stanford Functional Genomics Facility. Read counts were calculated as total count per well (IFNG) or as wells with read counts greater than bottom 10% threshold (TCR). Data processing was completed as described (Han et al., 2014).
2.5. quantitative (q)PCR of bulk sorted populations
RNA was isolated using the RNeasy Micro Kit and protocol (Qiagen), and cDNA produced using the SuperScript IV Reverse Transcriptase kit and protocol (Invitrogen, Carlsbad, CA, USA). IFNγ mRNA levels were quantitated relative to 18S ribosome and unstimulated cells (2−ΔΔCT method) using TaqMan probes (Hs00989291_m1 and Hs99999901_s1) and TaqMan Universal Master Mix on a StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
2.6. Analysis and statistics
Gating and quantification of fluorescence intensities were completed in FlowJo 10.5.0 (Tree Star, Ashland, OR, USA). Cell positions and fluorescence intensities were exported from index sort data using the FlowJo Index Sort Script (http://exchange.flowjo.com). Yield calculations and analysis of qPCR and read counts were computed in Microsoft Excel for Mac 2011 (Redmond, WA, USA). Graphs were prepared using GraphPad Prism 5 (La Jolla, CA, USA), with the exception of the read count graph in Fig. 3C, which was created using R version 3.5.1 and the R package ggplot2 (R Foundation).
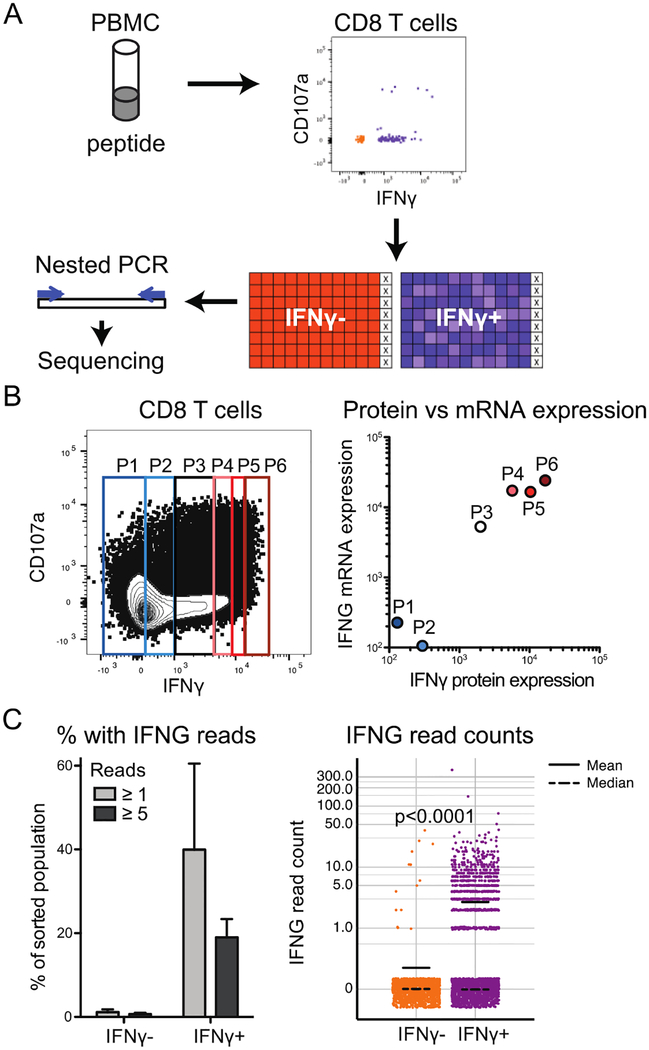
Figure 3: IFNγ protein expression predicts mRNA expression.
A) Example pipeline for use of index sorts in acquisition of sequencing data. X represents wells with lysis buffer and no cells. B) CD8 T cells were stained as indicated including capture of secreted IFNγ and sorted for the indicated levels of IFNγ. qPCR was conducted on IFNG mRNA levels in each population. IFNγ median fluorescence intensity of each population was computed. Data representative of two experiments. Correlation calculated for log-log fit on both sets of data, for R2 = 0.71. C) For plates sorted as IFNγ+ and IFNγ- as in part A (n=2 sequencing runs, 1222 cells IFNγ- and 2043 cells IFNγ+ total), the percentage of wells with ≥1 reads of IFNG or ≥5 reads of IFNG was determined (left) and read counts were quantified (right). Read counts are displayed on a log 10 scale (0 values represented as 0.1) with jitter of height 0.05 used to spread points at same value. Read counts in IFNγ+ and IFNγ- cells significantly different by one-tailed Mann-Whitney test (p<0.0001).
All statistics were computed using GraphPad Prism 5. A paired two-tailed Student’s t test was used to compare yields between samples sorted from the same individual under different conditions. Regressions were used to measure correlations between combinations of parameters including yield versus threshold rate, read count versus threshold rate, and protein quantification versus mRNA quantification. Linear or logarithmic regressions were selected as appropriate to the data scale. A one-tailed Mann-Whitney U test was used to compare read counts between IFNγ+ and IFNγ− index sorted cells.
3. Results
3.1. Calculation of yield
One of the most distinctive features of plate sorting relative to bulk sorting is the method to calculate yield. Yields are important for determining the number of cells needed in a sort and for optimizing flow rates and dilutions. Diva software calculates efficiency rates and counts of processed cells based on the number of cells sorted per second and the per second rates of error including conflict counts and electronic aborts (BDBiosciences, 2012). These numbers can be used to measure cell loss within the sorter and real time changes in yield. However, the calculations were designed for bulk sorts and efficiency rates per second are substantially less useful for plate sorts. In the studies presented here, we have therefore relied upon calculations of yield and efficiency based on starting and final numbers of cells, as described below. Similar approaches have been used to calculate yields in bulk sorts; for plate sorts, this method is also a far more reliable measure of efficiency than the software-calculated rates.
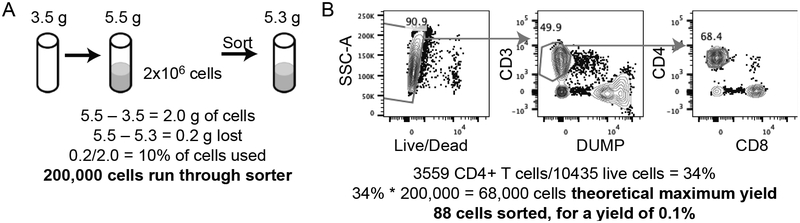
First, we determined an accurate count of live cells by hemocytometer prior to stimulation and staining. Immediately prior to sorting, we added cells and buffer to a 5 mL tube to achieve the desired concentration. This count is sufficient for yield calculations when the entire volume of cells in the tube will be used in the sort. An alternative for partially used samples is to weigh the tube containing the cells before and after sorting and calculate the difference to determine the percent of weight lost. Thus, the number of cells detected by the sorter as events can be calculated as % weight lost x total number of starting cells (Fig. 1A).
Figure 1: Example yield calculation.
A) Calculation of lymphocytes run through sorter in one healthy volunteer sample based on weight change. B) Gates and calculations of yield.
To record data on the pre-sort population of cells used in an index sort we either record a separate tube of data, or start to record the data before beginning the index sort. Both methods work on BD sorters running FACSDiva software, but may not apply to all systems. With either method, two FCS files will be produced: the index data associated with the sort itself, and recorded data of all cells run through the sorter. We then use the % in the recorded tube belonging to the index sorted population and the total number of cells recorded to calculate theoretical maximum yield. The number of cells sorted can be divided by this value to determine the % yield (Fig 1B). Recording data of the total (pre-sort) population concurrent with sorting provides an alternative method of efficiency calculation. Dividing the number of cells sorted into the plate by the number of cells recorded in that gate identifies the efficiency of the sort. In our experiments, this efficiency rate is ~12.1% +/− standard error of 1.9% at a flow rate of 1.0.
3.2. Low flow rates and 70 nozzle size maximize yield
It is critical to optimize for yield when plate sorting because yields are substantially lower for plate sorts than for bulk sorts (<5% versus ~50%), starting material may be limiting, and sorting time may be extensive. Two factors that may affect overall yield are flow rate and nozzle size.
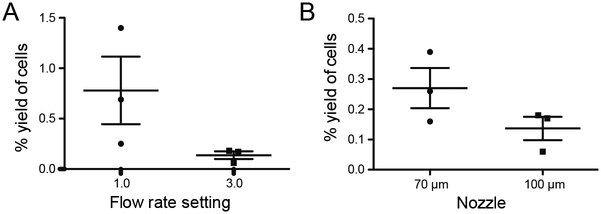
To determine optimal flow rates for our index sorts we sorted cells from identical samples at flow rate settings of 1.0 or 3.0 (Aria III sorters have a range of flow rates from 1 – 11). We compared sort yields that were calculated as described in Fig. 1. There was a trend towards improved yield when a flow rate of 1.0 was employed (Fig. 2A). We also observed improved sort efficiency, calculated as described above. Specifically, sort efficiency is 4.1 fold higher at a flow rate of 1.0 than 3.0 (3 technical replicates, n=2).
Figure 2: Sort yield depends on flow rate and nozzle size.
A) Lymphocytes from healthy volunteers (n=3) were sorted into 96-well plates at A) flow rate settings of 1 and 3 and B) at flow rate setting of 3 with indicated nozzles. Threshold rate was similar (~300 events/second) for both nozzles, but the samples were 1.5X more dilute with the 100 μm nozzle. For both A and B, yield was calculated as described above. Both trends not significant by paired Student’s t test: (A) p=0.2 (B) p=0.07.
Nozzle size may also affect yield. The 70 μm nozzle produces a finer, more directed stream relative to the 100 μm nozzle. Thus, the 100 μm nozzle is conducive to lower flow rates and in some cases improved cell survival (Osborne, 2011), but less accurate plate sorting. Direct comparison of identical samples sorted with equivalent flow rates with each nozzle demonstrates a trend towards higher index sort yield of our lymphocyte population with the 70 μm nozzle. (Fig. 2B).
3.3. Bulk and individually sorted IFNγ+ cells have higher levels of IFNG mRNA than IFNγ− cells
A variety of protocols can be employed for sequencing of single index sorted T cells, with key principles being selection of T cell isolation method, sort parameters as described, nested PCR and barcoding, and selection of sequencing platform (Fig. 3A).
An advantage of index sorting is the ability to compare expression at the transcriptional versus translational level. We have used this method to correlate expression of mRNA and secreted protein for the cytokine IFNγ (gene name: IFNG). First, to measure correlation between transcript and protein products of this gene, a bulk sort was used to isolate sextiles of CD8 T cell populations based on relative amounts of secreted IFNγ protein generated in response to stimulation with CMV peptides (Fig. 3B, left). IFNG mRNA levels in each population were then measured by qPCR. Analysis of cells sorted from two individuals found a statistically significant correlation (R2 = 0.71) between mRNA and protein levels (Fig. 3B, right). Next, cells were isolated by index sorting based on level of secreted IFNγ. Cells lacking detectable IFNγ protein also lacked IFNG mRNA based on amplification followed by next-generation sequencing (Fig. 3C). Conversely, approximately 40% of those cells expressing IFNγ protein also had detectable IFNG mRNA (Fig. 3C). The difference in IFNG reads between the two sorted populations was statistically significant (p<0.0001).
3.4. Threshold rate inversely correlates with read count
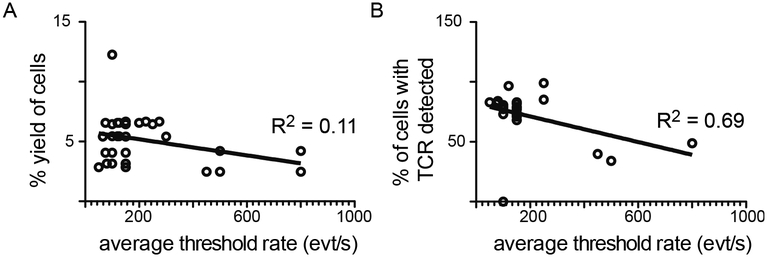
We next performed experiments to optimize threshold and flow rates. Threshold rate is the rate of cells processed by the sorter per second; flow rate is a measure of the velocity of liquid passing through the sorter (BDBiosciences, 2012). Higher flow rates are associated with increased pressure and thus decreased cell viability (Osborne, 2011). Adjustment of flow rate affects the threshold rate, and diluting cells to a lower concentration allows for finer control of threshold rate. We recorded threshold rates (cells/second) for cells sorted at flow rate of 1.0 and correlated to cell yield, calculated on the basis of loss of volume. Interestingly, there was no correlation between threshold rate and yield (R2 = 0.11, Fig. 4A). However, we also compared threshold rate to read counts obtained after nested PCR and sequencing. This comparison demonstrates that read count is inversely correlated with flow rate (R2 = 0.69, Fig 4B). In these sequencing experiments, cell deposition efficiency, or the percentage of wells containing reads, was >99% in 90% of plates
Figure 4: Decreasing threshold rate improves read counts.
For each sorted plate, average threshold rate in events (cells) per second was measured. Flow rate for all samples was 1.0. A) Yield was calculated on the basis of percent loss in volume. B) Upon sequencing, the percentage of sorted cells with TCR sequence reads detected was calculated. Threshold rate was plotted against yield (A) or % of cells with TCR (B) and a linear regression applied to the data. Slopes significantly non-zero (p<0.05). R2 displayed on graphs. Data represent sorts using 70 μm nozzle only.
4. Discussion
Index sorting is a powerful method that provides data which can be coupled to downstream analyses. Optimization of several parameters is critical for efficient sorting, especially when limiting proportions of samples are involved. We have determined that the 70 μm nozzle, flow rate of 1.0, and threshold rate under 200 events/second are optimal for sorting rare populations of human lymphocytes. In addition, we demonstrated that for the gene IFNG, protein and mRNA are co-expressed in index sorted and sequenced T cells. Most crucially, we found that measurement of read counts provides distinct information from yield calculations, and thus sequencing data are strongly recommended to interpret the success of sort optimization. We present methodology for TCR sequencing here - similar approaches can be used for BCR sequencing and other single cell protocols including RNA sequencing.
A key parameter to keep in mind regardless of experimental pipeline is that read counts are a better measure of success than yield calculations. Due to the limited accuracy of software-generated efficiency rates and processed counts in the context of index sorting, yield calculations must be made on the basis of changes in weight or volume of the cell suspension being sorted. As we have shown, these calculations are poorly correlated to threshold rate of events/second. This may be due in part to the fact that we used a limited range of low threshold rates. In addition, because of the profound difference we found in yields between threshold rates in early experiments, we switched to lower threshold rates in subsequent experiments, as reflected by fewer data points at higher threshold rates. However, the most important numbers are the read counts at the end of the sequencing process, which correlate inversely with threshold rates within the range measured (Fig. 4B). Thus, while yield calculations are useful for experimental planning, the only way to truly determine the success of an index sort is by completing the sequencing pipeline and measuring the final result. Our results also demonstrate that yields may be less than 1% of the number of cells present prior to stimulation and staining; this should be taken into consideration when planning experiments.
As our results demonstrate, yields and sort efficiency can be quite different, but both values are useful. Specifically, we typically found 2–5% yield, but 10–14% efficiency, indicating that cells are lost both during the stimulation and sample preparation (50–80%), and during the sort itself (80–90%). Thus, yields are informative for identifying how many cells are needed in the starting sample in order to have a successful sort. However, this number has limited utility for understanding at which stage cell loss occurred. For that question, adding in a measure of sort efficiency as described provides crucial details. Counting cells immediately prior to sorting, is an alternative strategy to ascertain cell loss during sorting.
In addition to the above-described parameters, plate alignment is a crucial consideration in sort optimization. Our protocol includes the best practices critical to this process, which include the following. Alignment of the droplet should be tested to deposit cells appropriately into wells across the entire plate, rather than solely into well A1. Test sorts should include the use of plates sealed with aluminum foil rather than plastic, to avoid static interference with drop deposition. In addition to using the “test sort” function, alignment must be tested using actual cells, as droplets containing cells have higher mass than empty droplets, which can impact deposition. Finally, alignment can drift over time, so it is crucial to periodically test it throughout the sort.
Several PCR protocols have been published for sequencing of TCR genes from single cells (Han et al., 2014; Picelli et al., 2014). The details vary, but certain principles are common to all. First, immediate lysis and preservation of lysate as described in the methods is crucial to maintain RNA integrity. Second, the amount of RNA present will be small enough that amplification will be required, either of a broad array of RNAs or targeted to specific genes. Thus, expression levels are semi-quantitative at best. Third, the amount of RNA in a single cell is small enough that contamination is a major concern. Recommended precautions include designating single use reagent aliquots and treating pipetmen with UV light or 10% bleach prior to completing this PCR. Fourth, barcoding will be needed at some point in the amplification or library preparation. Each of these factors requires consideration regardless of desired downstream application. When planning a single cell TCR sequencing experiment, the first step is to identify the T cell population of interest and how best to isolate it. Single cell sequencing typically has the greatest power for analysis of antigen specific populations. Approaches include peptide-major histocompatibility complex tetramer staining (Altman et al., 1996), stimulation followed by cytokine capture (Manz et al., 1995; Brosterhus et al., 1999), and staining for markers of antigen- specific activation such as CD137 and CD154 (Bacher and Scheffold, 2013). Stimulation-based approaches are most useful if the research question relates to cellular function. Cytokine capture isolates live cells and thus protects RNA integrity. Sorting protocols isolating intact RNA from fixed cells have also been published (Thomsen et al., 2016), so adaptation of intracellular staining protocols may be possible, but our studies did not directly address this approach. Similar approaches can be used for a variety of sequencing approaches. In particular, methods for BCR sequencing are quite similar, though they do require careful attention to B cell biology. For instance, antibody-secreting plasma cells have higher expression of BCR than memory or naive B cells (reviewed in Nutt et al., 2015), so feasibility of sequencing will depend on the source population.
Droplet-based technologies that allow for single cell barcoding and PCR on bulk sorted populations have recently been developed (Macosko et al., 2015; Zilionis et al., 2017; Rodda et al., 2018). These are a powerful tool for single cell protocols that can utilize much higher cell numbers than index sorting. However, the inclusion of surface expression data with these methods is currently limited to oligonucleotide-conjugated antibodies available for some 3’ RNA sequencing applications (Shahi et al., 2017). Therefore, index sorting is still generally preferable when surface expression is required. This can be crucial to analyses of differentially glycosylated proteins, post-translational modifications, and for direct comparison of transcript and protein levels.
The optimization described was used to sort limiting cell populations. However, other considerations may be more important in sorting non-limiting (>5% of total) samples. For example, sorting cells with the 70 μm nozzle and at 500 evt/s would ensure purity and reduce time between sorting and spinning down the plate, which may improve RNA integrity with a trade-off of reduced yield. Regardless of sample size, cell size remains an important concern - while the 70 μm nozzle is optimal in the absence of size concerns, additional optimization will need to be completed with the 100 μm nozzle for larger cells, or cells isolated through tissue digestion.
Another consideration when sorting limiting populations is selection of the appropriate sort purity. There are two purity options in FACSDiva compatible with plate sorts: single cell and 4-way purity. Single cell purity requires centering of the cell in the droplet, whereas 4-way purity, another mode in FACSDiva, is compatible with single cell sorting but does not require centering. Thus, changing to a less stringent purity setting can improve sort efficiency and yields, but with likely cost in purity.
It is important to note that all of our experiments were completed on BD instruments running FACSDiva software. BD makes index sorting instruments with different software and fluidics (BDBiosciences, 2014). Single cell sorting can also be carried out with 70 and 100 μm nozzles on instruments manufactured by other companies (BeckmanCoulter, 2012; Sony, 2016). The MoFlo Astrios has 7 lasers that can detect up to 51 parameters (BeckmanCoulter, 2012). While BD sorters and the MoFlo Astrios have a spatially-separated laser path (BDBiosciences, 2012; BeckmanCoulter, 2012), some Sony sorters have one path for all lasers (Sony, 2016). The single path limits the resolution of spectral overlap between fluorophores with similar emissions and therefore the complexity of the populations that can be sorted.
The difference in IFNG reads relative to IFNγ protein in index sorted data (Fig. 3C) was unexpected, and may be explained by either technical or biological factors. The primary technical factor is that the measurements were completed with two very different methods: capture of secreted protein and nested PCR amplification, both providing semi-quantitative data. In our studies, mRNA levels are dependent on multiple rounds of amplification followed by relatively shallow next generation sequencing with positive calls based on reads above a threshold of detection. It is possible that mRNA for IFNG was expressed but was below the threshold and thus undetectable with the assay used.
The primary biological factor in comparison of transcript and protein levels is the kinetics of RNA and protein synthesis and degradation. The protocol used for IFNγ capture was designed for maximal IFNγ protein, which is present after 5–6 hours of stimulation. In contrast, IFNG mRNA is maximal approximately 2 hours after stimulation (Nicolet et al., 2017). Thus, not all IFNγ+ cells will contain detectable IFNG mRNA. In combination with the technical variables discussed above, this could explain the discrepancy observed between mRNA and protein. Gene regulation should be considered for any comparison of transcript to protein. In addition to temporal differences, some genes may have low mRNA levels and long-lived protein, or high mRNA levels and rapidly degraded protein. Thus, knowledge of gene regulation at both the mRNA and protein levels can be incorporated into analyses of index sort and sequencing data order to accurately interpret the results.
In the context of sorting limited human lymphocytes into 96 well plates for PCR amplification, we recommend the 70 μm nozzle, a flow rate setting of 1.0, and dilution of cells to allow a threshold rate of 200 cells/second. Each of these features independently improved yields, as demonstrated in Fig. 2A (flow rate), Fig. 2B (nozzle size) and Fig. 4 (threshold rate). Specifically, the combination improved yields from <1 to approximately 5% of the original population. However, alternative cell populations will require separate optimization strategies for each parameter.
5. Conclusions
Overall, we have identified nozzle size, yield calculations, threshold rate, and flow rate as key factors affecting plate sort yields, efficiency and accuracy. The use of the 70 μm nozzle is strongly recommended for plate sorts of human peripheral blood lymphocytes. The yields are higher with this smaller nozzle, which also promotes more stable sorts. When further optimization is needed, we recommend optimizing the full protocol and using sequencing reads to determine the optimal sort conditions, given the ultimate importance of this readout. Researchers can use this guide to optimize core T and B cell sequencing and even extend to other cell types by testing combinations of nozzle size, flow rate and threshold rate.
Highlights.
Index sort yield calculations cannot rely on sort rates calculated on the sorter
70μM nozzle and flow rate of 1 are associated with high yield of rare sorted cells
Sort rate of less than 200 cells/second improves yield of rare sorted cells
-
IFNγ expression at protein and mRNA levels is concordant in index sorted cells
Read counts of sequencing data are improved by lower sort rates
Acknowledgments:
We thank Drs Mark Davis and Naresha Saligrama for training in nested PCR for sequencing, Drs Mrinmoy Sanyal and Lisa Blum for suggestions in troubleshooting sort alignment, the Palo Alto Veteran’s Affair (VA) Flow Cytometry Core for FACSAria III usage, and the Stanford Functional Genomics Facility for bioanalyzer and MiSeq usage.
Funding: This research was supported by the American Heart Association/Enduring Hearts [17POST33660597] and the National Institutes of Health [T32 DK007357–31, AI07290].
Abbreviations
- ACDU
Automated Cell Deposition Unit
- BCR
B cell receptor
- CMV
cytomegalovirus
- CS&T
Cytometer, Setup & Tracking
- IFN
interferon
- PBMC
peripheral blood mononuclear cells
- qPCR
quantitative PCR
- TCR
T cell receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: LEH, MAC, and JSM have no conflicts of interest to declare. CJC is employed by Coda Biotherapeutics.
References
- Altman J, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ and Davis MM, 1996, Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96. [DOI] [PubMed] [Google Scholar]
- Bacher P and Scheffold A, 2013, Flow-cytometric analysis of rare antigen- specific T cells. Cytometry Part A 83A, 692–701. [DOI] [PubMed] [Google Scholar]
- BDBiosciences. 2012. BD FACSAriaTM III User’s Guide. In: Becton, Dickinson, and Company. [Google Scholar]
- BDBiosciences. 2014. BD Influx. In. BD Biosciences. [Google Scholar]
- BeckmanCoulter. 2012. MoFlo Astrios High Speed Cell Sorter. In. Beckman Coulter. [Google Scholar]
- Brosterhus H, Brings S, Leyendeckers H, Manz RA, Miltenyi S, Radbruch A, Assenmacher M and Schmitz J, 1999, Enrichment and detection of live antigen-specific CD4+ and CD8+ T cells based on cytokine secretion. European Journal of Immunology 29, 4053–4059. [DOI] [PubMed] [Google Scholar]
- Cossarizza A, Chang H-D, Radbruch A, Akdis M, Andr I and et al. , 2017, Guidelines for the use of flow cytometry and cell sorting in immunological studies. European Journal of Immunology 47, 1584–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Glanville J, Hansmann L and Davis MM, 2014, Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nature Biotechnology 32, 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H and Agata K, 2010, Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ 52, 131–144. [DOI] [PubMed] [Google Scholar]
- Higdon LE, Lee K, Tang Q and Maltzman JS, 2016, Virtual global transplant laboratory standard operating procedures for blood collection, PBMC isolation, and storage. Transplantation Direct 2, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A and McCarroll SA, 2015, Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz R, Assenmacher M, Pfluger E, Miltenyi S and Radbruch A, 1995, Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proceedings of the National Academy of Sciences of the United States of America 92, 1921–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet BP, Guislain A and Wolkers MC, 2017, Combined Single-Cell Measurement of Cytokine mRNA and Protein Identifies T Cells with Persistent Effector Function. Journal of Immunology 198, 962–970. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Hodgkin PD, Tarlinton DM and Corcoran LM, 2015, The generation of antibody-secreting plasma cells. Nature Reviews Immunology 15, 160–171. [DOI] [PubMed] [Google Scholar]
- Osborne GW, 2011, Chapter 21 Recent advances in flow cytometric cell sorting. Methods in Cell Biology 102, 533–556. [DOI] [PubMed] [Google Scholar]
- Penter L, Dietze K, Bullinger L, Westermann J, Rahn H-P and Hansmann L, 2018, FACS single cell index sorting is highly reliable and determines immune phenotypes of clonally expanded T cells. European Journal of Immunology 48, 1248–1250. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S and Sandberg R, 2014, Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ and Cyster JG, 2018, Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P, Kim SC, Haliburton JR, Gartner ZJ and Abate AR, 2017, Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Scientific Reports 7, 44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sony. 2016. SH800S Cell Sorter. In. Sony Biotechnology Inc. [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP and Ramanathan S, 2016, Fixed single-cell transcriptomic characterization of human radial glial diversity. Nature Methods 13, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM and Mazutis L, 2017, Single-cell barcoding and sequencing using dropet microfluidics. Nature Protocols 12, 44–73. [DOI] [PubMed] [Google Scholar]