Abstract
The intestinal microbiota actively converts dietary flavanols into phenolic acids, some of which are bioavailable in vivo and may promote resilience to select neurological disorders by interfering with key pathologic mechanisms. Since every person harbors a unique set of gut bacteria, we investigated the influence of the gut microbiota’s interpersonal heterogeneity on the production and bioavailability of flavonoid metabolites that may interfere with the misfolding of alpha (α)-synuclein, a process that plays a central role in Parkinson’s disease and other α-synucleinopathies. We generated two experimental groups of humanized gnotobiotic mice with compositionally diverse gut bacteria and orally treated the mice with a flavanol-rich preparation (FRP). The two gnotobiotic mouse groups exhibited distinct differences in the generation and bioavailability of FRP-derived microbial phenolic acid metabolites that have bioactivity towards interfering with α-synuclein misfolding or inflammation. We also demonstrated that these bioactive phenolic acids are effective in modulating the development and progression of motor dysfunction in a Drosophila model of α-synucleinopathy. Lastly, through in vitro bacterial fermentation studies, we identified select bacteria that are capable of supporting the generation of these bioavailable and bioactive phenolic acids. Outcomes from our studies provide a better understanding of how interpersonal heterogeneity in the gut microbiota differentially modulates the efficacy of dietary flavanols to protect against select pathologic mechanisms. Collectively, our findings provide the basis for future developments of probiotic, prebiotic, or synbiotic approaches for modulating the onset and/or progression of α-synucleinopathies and other neurological disorders involving protein misfolding and/or inflammation.
Keywords: microbiome, humanized gnotobiotic mice, polyphenol metabolism, phenolic acids, α-synucleinopathy, Drosophila
1. INTRODUCTION
Dietary polyphenols, in particular compounds belonging to the flavanol subclass, hold great potential as gut-brain therapeutics due to their ability to promote resilience against a variety of neurological disorders by contemporaneously modulating multiple pathogenic mechanisms underlying these disorders [1–9]. The majority of dietary polyphenols are extensively metabolized during gastrointestinal (GI) absorption and/or post-absorptive xenobiotic metabolism, converting them into polyphenolic metabolites [10–11]. In addition, dietary polyphenols are extensively metabolized by the GI microbiota into phenolic acids [12]. Thus, orally consumed polyphenols, including flavanols, are typically bioavailable in phenolic metabolite forms that, ultimately, are responsible for supporting biological activities in vivo.
Accumulating evidence demonstrates that select microbiota fermentation of phenolic acid metabolites may mechanistically contribute to neurological resilience. For example, we demonstrated that phenolic acid metabolites generated from microbial fermentation of dietary flavanols may promote neurological resilience by interfering with protein misfolding and overly active inflammatory processes, key pathogenic mechanisms underlying diverse neurological disorders [1. 13]. In particular, we have shown that select brain-penetrating phenolic acid metabolites, such as 3-hydroxybenzoic acid (3-HBA), and 3-(3-hydroxyphenyl)propionic acid (3-HPPA) potently inhibit the misfolding of beta-amyloid (Aβ) peptides into soluble neurotoxic Aβ aggregates, which play a key role in Alzheimer’s disease (AD) [13]. We have also shown that dihydrocaffeic acid (DHCA), a phenolic acid metabolite found in the systemic circulation, significantly reduces lipopolysaccharide-mediated induction of inflammatory responses in cultured primary peripheral blood cells [1].
Every person harbors a unique set of roughly 100 different bacterial species constituting their gut microbiota [14–15]. Interpersonal differences in the metabolic potential of each individual’s gut microbiota might differentially affect the bioavailability of phenolic acid metabolites from dietary polyphenols, resulting in divergent efficacy in the promotion of neurological resilience. We recently demonstrated that dietary supplementation with a standardized flavanol-rich preparation (FRP) is effective at promoting resilience against stress-induced depression phenotypes (J. Wang and G.M. Pasinetti, unpublished observation). Based on this, the present investigation used two experimental groups of humanized gnotobiotic mice with compositionally diverse gut bacteria derived from heterogeneous human GI microbiota to investigate their effects on the generation and bioavailability of FRP-derived phenolic acids. Based on our finding that some of the FRP-derived phenolic acid metabolites specifically accumulate in the brain, we conducted in vitro studies to investigate the effects of brain-penetrating phenolic acid metabolites on inhibiting the misfolding of the alpha (α)-synuclein protein, which plays a key role in α-synucleinopathies such as Parkinson’s disease (PD), dementia with Lewy bodies and multiple system atrophy [16]. In parallel in vivo studies, we also investigated the effects of brain-penetrating phenolic acid metabolites in modulating the development and progression of motor dysfunction in a Drosophila model of α-synucleinopathy. Lastly, using in vitro bacteria fermentation studies, we characterized select bacteria species from the two experimental groups of humanized gnotobiotic mice that support the generation of bioavailable, bioactive phenolic acids. Outcomes from our studies will provide a better understanding of how interpersonal heterogeneity in the GI microbiota may differentially modulate dietary flavanol-mediated promotion of resilience to neurological disorders, with particular implications to α-synucleinopathies and other neurological disorders involving inflammation and protein misfolding.
2. MATERIALS AND METHODS
2.1. Chemicals, solvents, peptides and proteins
Chemicals were obtained from Sigma-Aldrich (St Louis, MO, USA) and, unless otherwise stated, were of the highest purity available. Solvents were high-performance liquid chromatography (HPLC) grade and were obtained from Fisher Scientific. Water was double-distilled and deionized using a Milli-Q system (Millipore Corp., Bedford, MA). Monomeric α-synuclein peptide was purchased from rPeptide (Watkinsville, GA). Glutathione S-transferase (GST) was purchased from Sigma-Aldrich.
2.2. Human gut microbiota culture collections
Arrayed culture collections were generated by high throughput isolation of unique species from two healthy human donors as previously described [17–18]. Bacterial isolates were identified at the species level using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker Biotyper) and 16S rDNA amplicon Sanger sequencing. Isolates from each microbiota were grown for 48 hours to stationary phase in separate wells of a deep 96-well plate and stored in 15% glycerol at −80°C as a pooled cocktail for administration to mice. Bacteria compositions for each of the two arrayed culture collections isolated from different healthy humans are presented in Table I.
Table I: Bacteria compositions of human microbiota #HuA and #HuB from two healthy donors.
Shown are bacteria species in microbiota #HuA and #HuB; bacteria species were identified using MALDI-TOF mass spectrometry (Bruker Biotyper) and 16S rDNA amplicon Sanger sequencing. Four bacteria species, Bacteroides ovatus, Bacteroides thetaiotanomicron, Bacteroides uniformis and Eggerthella lenta, are common in both microbiotas. The majority of the bacteria species (6 out of 10 of microbiota #HuA and 16 of 20 of micriobiota#HuB) are unique among the two microbiota.
| HuA | HuB |
|---|---|
| Anaerotruncus colihominis | Alistipes onderdonkii |
| Baceroides ovatus | Bacteroides caccae |
| Bacteroides thetaiotaomicron | Bacteroides fagilis |
| Bacteroides uniformis | Bacteroides ovatus |
| Closteridium innocuum | Bacteroides salyersiae |
| Clostridium scindens | Bacteroides stercoris |
| Eggerthella lenta | Bacteroides thetaiotaomicron |
| Enterococcus faecalis | Bacteroides uniformis |
| Lactobacillus paracasei | Bacteroides vulgatus |
| Ruminococcus gnavus | Bifidobacterium adolescentis |
| Bifidobacterium bifidum | |
| Bifidobacterium longum | |
| Collinsella aerofaciens | |
| Eggerthella lenta | |
| Enterococcus durans | |
| Enterococcus faecium | |
| Excherichia coli | |
| Flavonifractor plautii | |
| Odoribacter | |
| Parabacteroides johnsonii | |
2.3. Generation of humanized gnotobiotic mice
Germ-free C57Bl/6J mice were bred in flexible vinyl isolators. Male, 5-week old germ-free mice were inoculated with pooled stationary phase cultures of microbiota HuA (n=6) or HuB (n=7) via oral gavage administered in a volume of 0.3-ml in a biosafety cabinet. Gnotobiotic mice were maintained in filter-topped cages. All animals were maintained on a 12:12-h light/dark cycle with lights on at 07:00 in a temperature-controlled (20 ± 2°C) gnotobiotic vivarium with ad libitum access to sterile food and water, including polyphenol-free diet and FRP-infused drinking water. For the duration of the experiment, changes of cages, food, and water were conducted using aseptic techniques in a biosafety cabinet. All procedures were approved by the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai.
2.4. Flavanol-rich preparation (FRP)
The FRP used in this study was a flavanol-rich cocoa preparation, which is commercially available as CocoaVia® online through cocoavia.com (lot #144532). We have independently analyzed and archived the CocoaVia® preparation in compliance with National Center for Complementary and Integrative Health (NCCIH) Product Integrity guidelines. Polyphenol contents in this FRP were analyzed using HPLC/MS. Results confirmed that FRP polyphenols are confined to the flavanol subclass: catechin (C), epicatechin (Ec), proanthocyanidin dimers (P2 dimers), and gallic acid (GA) (Supplementary Figure S1).
2.5. Gnotobiotic FRP treatment and tissue collection
The overall experimental design is schematically depicted in Fig. 2A. Animals were first gavaged with the heterogeneous microbiota populations HuA or HuB, and were then maintained for three days on an autoclave-sterilized standard polysaccharide-rich diet (LabDiet 5K67). Three days after microbiota transplantation, mice were transferred to an irradiated germ-free polyphenol-free rodent diet (Modified AIN-93G purified rodent diet with corn oil replacing soybean oil; Dyet#101591, Dyets Inc, PA) for the duration of the study. After 15 days on the polyphenol-free diet, FRP treatment was initiated by exchanging the regular drinking water with FRP-infused drinking water that was sterilized by filtration through a 0.2 μM filter (Sigma-Aldrich). The FRP drinking water contained 176 mg FRP flavanol per L of drinking water, calculated to provide each mouse with 40 mg FRP flavanol / kg BW / day, a dose we observed to significantly promote neurological resilience to the development of depression-type behaviors in a repeated social defeat stress mouse model (J. Wang and G.M. Pasinetti, unpublished observation). To simulate long-term flavanol consumption, animals were maintained on the FRP drinking water solution for 14 days. All FRP drinking water solutions were routinely replaced with freshly made FRP drinking water every 4–5 days. Animals were housed 3–4 per cage. For a given cage, we monitored the amount of liquid (water or FRP drinking water) in a fresh water bottle provided and the amount of liquid remaining when we replaced the water bottle with a fresh water bottle. We then estimated the animals’ liquid consumption by first dividing water consumed by the number of days (3–4 days) the water bottle was available to the cage, and then further divided the quotient by the number of animals in the cage. We also monitored the animals’ body weight over the course of the study. On FRP treatment day 15, individual animals were given 40 mg/kg BW FRP as a single 0.3-ml bolus via oral gavage and were then anesthetized 6 hours later by an intraperitoneal injection of a ketamine (100 mg/kg) and xylazine (10 mg/kg) mixture. Thereafter we collected circulating blood into heparinized collection tubes via cardiac puncture. Animals were then transcardially perfused with ice-cold saline to remove residual circulating blood, followed by the collection of perfused brain specimens and cecum contents (cecum specimens). Perfused brain specimens and cecum specimens were homogenized in 0.2% formic acid (1:5 w/v) to help stabilize phenolic compounds. Blood specimens were centrifuged and the separated plasma layer was collected and adjusted to a final formic acid content of 0.2%. All tissue homogenates and plasma specimens were stored at −80°C.
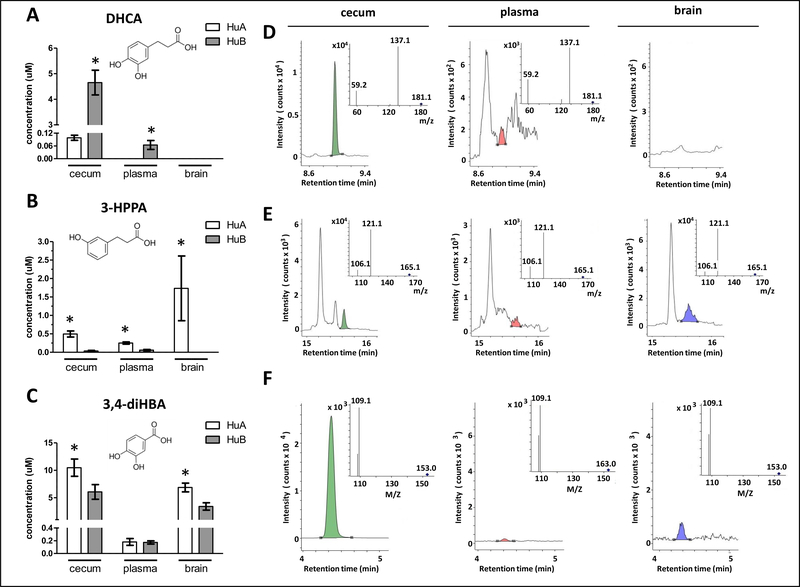
Fig. 2. Distinct differences in the generation and biological availability of select FRP-derived microbial phenolic acid metabolites.
FRP-derived microbial phenolic acid metabolites in cecum, plasma and perfused brain specimens from HuA (n=6) and HuB (n=7) mice were monitored by UPLC-QQQ/MS, 6 hours after the last FRP flavanol treatment on day 11. (A, B, C) Concentration of DHCA (A), 3-HPPA (B), and 3,4-diHBA (C) in cecum, plasma and brain specimens from the two groups of gnotobiotic mice. Bar graphs represent group phenolic acid concentration in mean ± SEM values. Statistics for each phenolic acids: one-way ANOVA < 0.05; * p < 0.05, post-hoc group comparisons between HuA and HuB mice. Insets: molecular structures of DHCA, 3-HPPA, and 3,4-diHBA. (D, E, F) Representative mass spectra of DHCA (D), 3-HPPA (E), and 3,4-diHBA (F) detected in cecum (green peaks), plasma (red peaks) and brain (blue peaks). Insets: corresponding MS/MS spectra of target compounds identified in mouse tissues.
2.6. Extraction and quantitative analysis of phenolic acids from tissue specimens
Targeted analytes were FRP flavanols catechin (C) and epicatechin (EC), and 17 phenolic acids known to be generated by the microbial metabolism of flavanols [13]. Concentrations of individual analytes in tissue, and bacterial broth specimens were analyzed by UPLC-QQQ/MS (Supplemental Fig. 2). Contents of C, EC and phenolic acids from cecum, plasma and brain specimens were analyzed by UPLC-QQQ/MS with the addition of trans-cinnamic acid (d7) as an internal standard (IS). Quantitation was achieved with calibration curves established using the analyte-to-IS peak area of quantifier ions.
Frozen plasma specimens were thawed on ice and then conditioned to room temperature. Trans-cinnamic acid (d7) (2 μg/mL), was diluted in 0.4 M NaH2PO4 buffer (pH 5.0) to a final concentration of 100 ng/mL. A 100 μL aliquot of thawed plasma was incubated with 300 μL of NaH2PO4 buffer containing IS and 50 μL NaH2PO4 buffer containing 2000 U of β-glucuronidase (with sulfatase contamination) [13] at 37°C for 45 minutes after purging with nitrogen. The enzymatic reaction was stopped by adding ethyl acetate (500 μL). Frozen cecum specimens were thawed on ice and then conditioned to room temperature for analysis. An aliquot (0.5 mL) of cecum homogenate was mixed with 300 μL of NaH2PO4 buffer containing the IS and similarly extracted with ethyl acetate (500 μL). Individual plasma (or cecum) mixtures were vortexed vigorously for 1 min, followed by centrifugation at 3000 xg for 5 min using a micro-centrifuge. The upper organic phase (~450 μL) was transferred to a 1-dram glass vial. After two more extractions with ethyl acetate (500 μL), the pooled supernatant was mixed with 10 μL of 2% ascorbic acid and dried under a gentle stream of nitrogen. The residue was reconstituted in 100 μL of 45% methanol containing 0.1% formic acid and centrifuged at 16,000 xg for 10 min. For each sample extract, 5 μL was injected into an UPLC-QQQ/MS system for analysis under dynamic multiple reaction monitoring (dMRM) mode.
Frozen brain homogenate specimens were thawed on ice and then conditioned to room temperature. Thawed brain homogenates (1 mL) were spiked with 5 μL of t-cinnamic acid (d7) (2 μg/mL) and then incubated with 100 μL of NaH2PO4 buffer containing 2000 U of β-glucuronidase (with sulfatase contamination) 13 at 37°C for 45 minutes after purging with nitrogen. Termination of the enzymatic reaction and protein precipitation were achieved by adding 1 mL of 4 % HCl in methanol. The mixture was allowed to stand still for 5 min in ice-cold water for complete protein denaturation and precipitation. After centrifuging at 16000 xg for 10 min, the supernatant (~800 μL) was transferred to a glass vial containing 20 μL of 2% ascorbic acid. The residue was re-mixed with 0.5 mL of acidified methanol, vigorously vortexed and then sonicated for 3 min in ice-cold water. After centrifuging and transferring the supernatant (~500 μL) to the glass vial, the residue was re-extracted once again with acidified methanol as aforementioned. The pooled supernatant was then dried under a gentle stream of nitrogen and reconstituted in 100 μL of 60% methanol containing 0.1% formic acid and centrifuged at 16,000 xg for 10 min. For each sample extract, 5 μL was injected into an UPLC-QQQ/MS system for analysis under dynamic multiple reaction monitoring (dMRM) mode.
The analyses were carried out on an Agilent 1290 Infinity II UPLC system interfaced with an Agilent 6470 Triple Quadrupole Mass Spectrometer with an electrospray ionization (ESI) source (Agilent Technology, Palo Alto, CA, USA) and a Waters Acquity UPLC BEH C8 column (2.1 × 150 mm, 1.7 μm) (Milford, Massachusetts, USA) using a method developed previously at Rutgers University. Briefly, a binary mobile phase system consisting of 0.2% ascorbic acid (AA) in water (phase A) and 0.1% AA in acetonitrile (phase B) at a flow rate of 0.3 mL/min was used. Thermostats of the column and the autosampler were set at 40°C and 4°C, respectively. The liquid chromatography gradient started at 96% phase A and 4% phase B, held for 1.5 min before increasing % phase B to 12% in 12.5 min, to 90% in 1 min and held for another 2 min, and then returned to initial conditions in 1 min. The column was equilibrated for another 5 min before the next injection. Mass spectral data acquisition was achieved under negative polarity (ESI-) and dMRM mode. Identification of phenolic acid metabolites, (+)-C and (−)-EC was achieved by comparing their parent-product ion pair transitions and retention times with those of the authentic standards. Detailed MRM transitions for individual analytes are shown in Supplemental Fig. 2. Quantitation was achieved with calibration curves established using the analyte-to-IS peak area of quantifier ions. All tested compounds were analyzed and archived in compliance with NCCIH Product Integrity guidelines.
2.7. Bacterial fermentation studies
Bacterial strains Bacteroides ovatus ATCC8483, Eggerthella lenta ATCC25559, and Escherichia coli MG1655 were obtained from ATCC (Manassas, VA) and cultured in nutrient-rich anaerobic broth under anaerobic conditions at 37°C for 48 hours to reach stationary phase. Each bacterial culture was then diluted 1:50 into 10 mL of fresh bacterial broth with or without 50 μM each of (+)-catechin [(+)-C] and (−)-epicatechin [(−)-EC], and then cultured at 37°C under anaerobic conditions. Samples were recovered at 3 and 24 hours and were immediately centrifuged at 4,000 × g for 5 minutes. The clarified bacteria broth fractions (supernatant) were recovered, acidified with formic acid to a final 0.2%, and stored at −80°C.
2.8. Extraction and quantitative analysis of phenolic acid metabolites from bacteria broth
Banked bacteria broth specimens were thawed on ice and conditioned to room temperature before processing. After vortexing, an aliquot (500 μL) of bacterial broth were acidified with 100 μL of 4 M HCl solution and spiked with 50 μL of t-cinnamic acid (d7) (2 μg/mL). The mixture was then extracted with ethyl acetate (500 μL), followed by vigorous vortexing for 1 min, and centrifugation at 3000 xg for 5 min. The upper organic phase (~450 μL) was transferred to a 1-dram glass vial. After two more extractions with ethyl acetate (500 μL), the combined supernatants were mixed with 10 μL of 2% ascorbic acid and dried under a gentle stream of nitrogen. The residue was reconstituted in 1000 μL of 45% methanol containing 0.1% formic acid and centrifuged at 16,000 xg for 10 min. For each sample extract, 5 μL was injected into an UPLC-QQQ-MS system for analysis using the same analytical method as described for analyzing animal tissues. All tested compounds were analyzed and archived in compliance with NCCIH Product Integrity guidelines.
2.9. Photoinduced cross-linking of unmodified proteins (PICUP) assay
α-synuclein PICUP assays were conducted essentially as previously described for PICUP assessments of AD-type Aβ peptides [19]. In a volume of 18 μL, monomeric α-synuclein (35 μM) was dissolved in 10 mM phosphate, pH 7.4 in the presence of either vehicle or individual phenolic acids. Two doses of phenolic acids were tested: a low dose (LD, equal molar concentration of phenolic acids relative to α-synuclein) or at a high dose (HD, 4-fold higher molar concentration of phenolic acid relative to α-synuclein). Following, 1 μL of 1 mM tris(2,2′-bipyridyl)dichlororuthenium(II) and 1 μL of 20 mM ammonium persulfate was added. The mixture was irradiated for 1 second with visible light, and then quenched with 10 μL of Tricine sample buffer (Invitrogen) containing 5% (v/v) β-mercaptoethanol. Samples were analyzed by gel electrophoresis followed by silver staining. Parallel studies using glutathione S-transferase (GST, 50 μM) controls for potential effects of phenolic acids on the cross-linking chemistry were conducted.
2.10. Thioflavin T spectroscopic (ThT) assay
The α-synuclein ThT assay was conducted essentially as previously described for ThT assessments of AD-type Aβ peptides [19]. In a final reaction volume of 200 μL, monomeric α-synuclein (70 μM) was dissolved in an aggregation buffer (100 mM phosphate buffer, pH 7.4) in the presence of either vehicle or individual phenolic acids at LD or HD. The mixture was incubated at 37°C for up to 94 h. ThT fluorescence was determined in triplicates at intervals using a Hitachi F-4500 fluorometer. Excitation and emission wavelengths were 450 and 482 nm, respectively.
2.11. Electron microscopy (EM) assay
The α-synuclein EM assay was conducted essentially as previously described for ThT assessments of AD-type Aβ peptides [19] in the presence of vehicle or individual test phenolic acids at LD or HD. After 94 hours of incubation at 37°C, samples were examined using a JEOL CX100 transmission electron microscopy.
2.12. Fly stocks, culture and fly crossing
Flies were maintained on a cornmeal–molasses–yeast medium and at room temperature (22 °C) with 60–65% humidity. The following Drosophila lines were obtained from the Bloomington Stock Center: elavC155-GAL4, UAS-alpha-synucleinA53T.
Female flies carrying the driver elavC155-GAL4 were crossed to males carrying the UAS-alpha-synucleinA53T. The first generation (F1) female offspring expressing A53T alpha-synuclein served as our PD fly model. In parallel, female flies carrying the driver elavC155-GAL4 were crossed to males carrying the UAS-LacZ, and the resultant F1 served as control
2.13. Negative Geotaxis Behaviour Assay (Climbing Assay)
Ten age-matched female flies of the same genotype were placed in a vial marked with a line drawn horizontally 8 cm above the surface. The flies were gently tapped to the bottom surface and given 10 s to demonstrate climbing activity as a negative geotaxic response. After 10 s, the number of flies that successfully climbed above the 8-cm line was recorded. This assay was repeated 10 times and the averaged data were represented as percentages where the number of flies above the 8-cm mark was divided by the total number of flies tested within each group. For each negative geotaxis assay, a minimum of 10 groups (100 flies) per genotype was tested [20].
2.14. Statistics
An unpaired two-samples Mann–Whitney U test comparison was used for single-variable analysis among two groups. One-way factorial ANOVA was used for single-variable analysis across multiple groups and two-way ANOVA was used to two-variable analysis across multiple groups, followed by Bonferonni post hoc analysis to compare select pairs. Significance was set at P<0.05 to define group differences.
3. RESULTS
3.1. Gnotobiotic mice with compositionally diverse human microbiotas generate unique phenolic acid profiles in the cecum after orally consuming FRP flavanols.
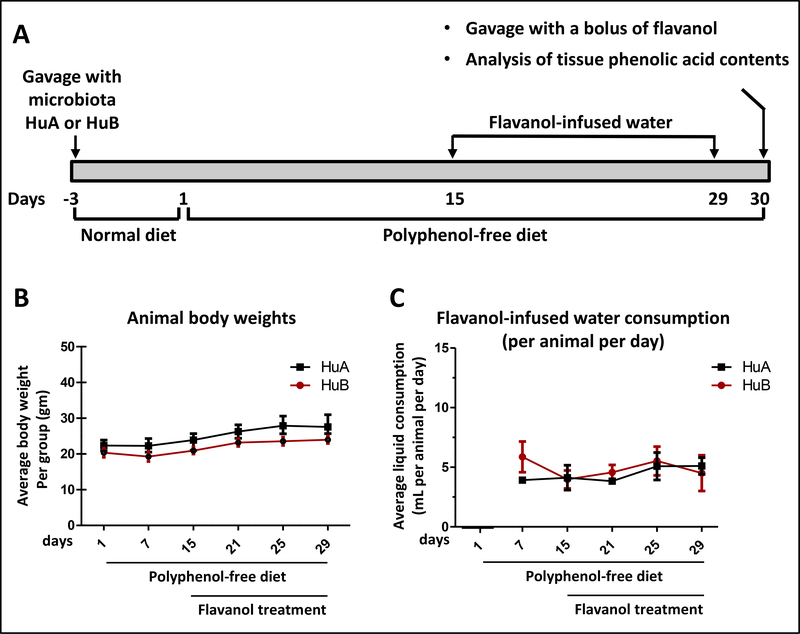
Human microbiota arrayed culture collections (HuA and HuB) comprised, respectively, of 10 and 20 bacterial species, were generated from two healthy human donors (Table I). Each isolate was identified with matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker Biotyper) and 16S rRNA amplicon Sanger sequencing. We then colonized germ-free C57Bl/6J mice with the HuA (n=6) or the HuB (n=7) microbiota culture collection by oral gavage. Humanized gnotobiotic mice carrying HuA or HuB microbiota were treated with FRP flavanols for 15 days to simulate long term administration (Fig. 1A). We found no detectable difference in body weight between the two groups of gnotobiotic mice during the course of the experiment (Fig. 1B). We also observed no detectable differences in the amount of flavanol-infused drinking water consumed by the two groups of gnotobiotic mouse groups (Fig. 1C). On day 15, animals were gavaged with a single 40 mg FRP/kg BW bolus as this dose promotes resilience to depression phenotypes induced by psychological stress (J. Wang and G.M. Pasinetti, unpublished observation). Tissue specimens (cecum, plasma and perfused brain) were collected 6 hours later for the analysis of the predominant flavanols, (+)-C and (−)-EC, and 17 targeted phenolic acids that are known to be generated by microbiota metabolism of flavanols [13] (Supplementary Fig. S2).
Fig. 1. Body weight and liquid intake of HuA and HuB mice.
(A) Schematics of the overall experimental design. (B, C) Body weight (B) and liquid intake (C) of HuA and HuB mice. In (B,C), points represent the mean (±SE) per mouse group with an n value of 6–7 per group. There was no detectable difference in body weight, as well as in the consumptions of water or FRP-infused drinking water between the two groups of gnotobiotic mice during the course of the study.
We found 15 out of the 17 target microbial phenolic acid metabolites (CA, DHCA, FA, GA, HA, 3-HBA, 3,4-diHBA, 4-HCA, 3-HPAA, 3,4-diHPAA, 3-HPPA, 4-HPPA, 3,4-diHPVA, HVA, and VA) detectable at μM to sub-μM concentration levels in the cecal compartment across the two gnotobiotic mouse groups (Table II), suggesting that both microbiotas, HuA and HuB, can generate a diverse array of phenolic acids from FRP flavanols. Between the two mouse groups, we observed that 6 FRP-derived microbial phenolic acid metabolites (GA, 3-HBA, 3,4-diHBA, 4-HCA, 3-HPAA, and 3-HPPA) were detected in significantly higher concentrations in cecum specimens from HuA than HuB mice. In contrast, 5 phenolic acid metabolites (CA, DHCA, FA, HA, and 3,4-diHPVA) were detected in significantly higher concentrations in cecum specimens from HuB mice (Table II). Lastly, no detectable difference in cecal concentration between the two groups was observed for the remaining 4 phenolic acid metabolites (3,4-diHPAA, 4-HPPA, HVA, and VA) (Table II). Collectively, these results demonstrate that interpersonal gut microbiota heterogeneity may significantly affect the in vivo generation of FRP-derived phenolic acid metabolites.
Table II: Contents of phenolic compounds in cecum specimens from gnotobiotic mouse modes HuA and HuB.
We survey the concentration of phenolic acid metabolites in cecum specimens obtained from HuA and HuB mice, 6 hours after the last treatment of mice with FRP flavanols, on day 11. UPLC-QQQ/MS was used to monitor the concentration of microbial-generated phenolic acid metabolites in cecum specimens from gnotobiotic mice with HuA (n=6) or HuB (n=7) following FRP flavanol treatment. Analytes included C and EC (FRP flavanols), and 17 FRP flavanol-derived microbial phenolic acid metabolites (CA, DHCA, FA, GA, HA, 3-HBA, 3,4-diHBA, 4-HCA, 3-HPPA, 3-HPAA, 4-HPPA, 3,4-diHPAA, 3-HPVA, 4-HPVA, 3,4-diHPVA, VA, and HVA). Concentrations of individual phenolic compounds are shown as group mean ± SD values. For each of the phenolic compounds, p-value is calculated by unpaired Mann-Whitney U test comparisons between the two gnotobiotic mouse groups. P-values highlighted in red letters denote significant difference (p < 0.05) between the two groups. Abbreviations: (+)-C, (+)-catechin; (-)-EC, (-)-epicatechin; CA, caffeic acid; DHCA, dihydrocaffeic acid; FA, ferulic acid; GA, gallic acid, HA, hippuric acid; 3-HBA, 3-benzoic acid; 3-HBA, 3-hydroxybenzoic acid; 3,4-diHBA, 3,4-dihydroxybenzoic acid; 4-HCA, 4-hydroxycinnamic acid; 3-HPPA, 3-(3-hydroxyphenyl)propionic acid; 3-HPAA, 3-hydroxyphenylacetic acid; 3,4-diHPPA, 3-(4-hydroxyphenol)propionic acid; 3,4-diHPAA, 3,4-dihydroxyphenylacetic acid; 3-HPVA, 5-(3-Hydroxyphenyl)valeric acid; 4-HPVA, 5-(4-hydroxyphenyl)valeric acid, 3,4-diHPVA, 5-(3,4-dihydroxyphenyl)valeric acid; VA, vanillic acid; HVA, homovanillic acid.
| Cecum concentration (μM) | |||
|---|---|---|---|
| Phenolic Compounds | HuA | HuB | p-value |
| (+)-C | 5.7± 1.2 | 2.9± 1.7 | 0.0082 |
| (-)-Ec | 24.6± 3.7 | 10.2± 6.5 | 0.0012 |
| CA | 0.1± 0.01 | 0.2± 0.04 | 0.0012 |
| DHCA | 0.1± 0.03 | 4.7± 1.3 | 0.0012 |
| FA | 0.034± 0.02 | 0.1± 0.033 | 0.0012 |
| GA | 2.5± 0.3 | 1.4± 0.6 | 0.0082 |
| HA | 0± 0 | 0.08± 0.04 | 0.0021 |
| 3-HBA | 0.072± 0.07 | 0± 0 | 0.0015 |
| 3,4-diHBA | 11.7± 2.8 | 5.6± 3.2 | 0.0047 |
| 4-HCA | 0.09± 0.01 | 0.06± 0.007 | 0.0012 |
| 3-HPAA | 18± 7.3 | 0.08± 0.5 | 0.0012 |
| 3,4-diHPAA | 1.7± 0.2 | 1.3± 0.5 | 0.1807 |
| 3-HPPA | 0.5± 0.2 | 0± 0 | 0.0032 |
| 4-HPPA | 0.6± 0.3 | 0.3± 0.1 | 0.0734 |
| 3-HPVA | 0± 0 | 0± 0 | ----- |
| 4-HPVA | 0± 0 | 0± 0 | ----- |
| 3,4-diHPVA | 0.03± 0.004 | 0.09± 0.4 | 0.0012 |
| HVA | 0.2± 0.1 | 0.2± 0.1 | 0.7308 |
| VA | 0.8± 0.2 | 0.5± 0.1 | 0.0513 |
3.2. Gnotobiotic mice with compositionally diverse human microbiotas exhibit distinct differences in the bioavailability of FRP-derived microbial phenolic acid metabolites.
Among the 15 FRP-derived microbial phenolic acid metabolites identified in cecum specimens (Table II), 12 of the phenolic acid metabolites (DHCA, FA, GA, HA, 3,4-diHBA, 4-HCA. 3-HPAA, 3,4-diHPAA, 3-HPPA, 4-HPPA, HVA, and VA) were detectable in μM to sub-μM concentrations in plasma specimens from HuA and/or HuB gnotobiotic mice (Table III, left panel). Between the two experimental groups of humanized gnotobiotic mice, we found a significantly higher concentration of DHCA in plasma from mice with HuB and a significantly higher concentration of 3-HPPA in plasma from mice with HuA (Table III, left panel). For the remaining 10 FRP-derived microbial phenolic acid metabolites, no significant difference in plasma concentration was observed between HuA and HuB (Table III, left panel).
Table III: Contents of FRP-derived microbial phenolic acid metabolites in plasma and perfused brain specimens from HuA and HuB gnotobiotic mice.
We survey the concentration of phenolic acid metabolites in plasma and perfused brain specimens obtained from the HuA and HuB mice, 6 hours after the last treatment of mice with FRP flavanols on day 11. UPLC-QQQ/MS was used to monitor the concentration of 17 FRP-derived microbial phenolic acid metabolites (CA, DHCA, FA, GA, HA, 3-HBA, 3,4-diHBA, 4-HCA, 3-HPPA, 3-HPAA, 4-HPPA, 3,4-diHPAA, 3-HPVA, 4-HPVA, 3,4-diHPVA, VA, and HVA) in plasma (left panel) and perfused brain specimens (right panel) from HuA (n=6) and HuB (n=7) mice. Concentrations of individual FRP-derived microbial phenolic acid metabolites are shown as group mean ± SD values. For each of the phenolic acid metabolites, p-value is calculated by unpaired Mann–Whitney U test comparisons between the two gnotobiotic mouse groups; p-values highlighted in red letters denote significant difference between the two groups.
| Plasma concentration (μM) | Brain concentration (μM) | |||||
|---|---|---|---|---|---|---|
| Phenolic Compounds | HuA | HuB | p-value | HuA | HuB | p-value |
| CA | 0 ± 0 | 0 ± 0 | ----- | 0 ± 0 | 0 ± 0 | ----- |
| DHCA | 0 ± 0 | 0.65 ± 0.42 | 0.0057 | 0 ± 0 | 0 ± 0 | ----- |
| FA | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.4452 | 0 ± 0 | 0 ± 0 | ----- |
| GA | 0.32 ± 0.04 | 0.35 ± 0.08 | 0.9452 | 10.4 ± 8.2 | 3.7 ± 1.3 | 0.1014 |
| HA | 0.3 ± 0.2 | 0.2 ± 0.2 | 1 | 0.09 ± 0.08 | 0.06 ± 0.05 | 0.5338 |
| 3-HBA | 0 ± 0 | 0 ± 0 | ----- | 0 ± 0 | 0 ± 0 | ----- |
| 3,4-diHBA | 0.2 ± 0.2 | 0.2 ± 0.1 | 0.366 | 7.5 ± 1.4 | 3.5 ± 1.3 | 0.0043 |
| 4-HCA | 0.1 ± 0.02 | 0.1 ± 0.02 | 0.2343 | 0 ± 0 | 0 ± 0 | ----- |
| 3-HPAA | 0.7 ± 0.2 | 0.6 ± 0.1 | 0.9452 | 0 ± 0 | 0 ± 0 | ----- |
| 3,4-diHPAA | 0.5 ± 0.29 | 0.26 ± 0.12 | 0.2343 | 0 ± 0 | 0 ± 0 | ----- |
| 3-HPPA | 0.25 ± 0.08 | 0.06 ± 0.05 | 0.0012 | 1.7 ± 2 | 0 ± 0 | 0.0021 |
| 4-HPPA | 0.063 ± 0.07 | 0.057 ± 0.07 | 1 | 0 ± 0 | 0 ± 0 | ----- |
| 3-HPVA | 0 ± 0 | 0 ± 0 | ----- | 0.09 ± 0.1 | 0.12 ± 0.1 | 0.4452 |
| 4-HPVA | 0 ± 0 | 0 ± 0 | ----- | 0 ± 0 | 0.05 ± 0 | ----- |
| 3,4-diHPVA | 0 ± 0 | 0 ± 0 | ----- | 0 ± 0 | 0 ± 0 | ----- |
| HVA | 0.2 ± 0.06 | 0.2 ± 0.09 | 0.8357 | 0 ± 0 | 0 ± 0 | ----- |
| VA | 0.14 ± 0.06 | 0.2 ± 0.06 | 0.6282 | 0 ± 0 | 0 ± 0 | ----- |
Five FRP-derived microbial phenolic acid metabolites (GA, HA, 3,4-diHBA, 3-HPPA, and 3-HPVA) (Table III, right panel) were detected at μM to sub-μM concentration levels in perfused brain specimens from the two gnotobiotic mouse groups. Brain concentrations for 2 of these phenolic acids (3,4-diHBA and 3-HPPA) were significantly higher in HuA mice compared to HuB mice (Table III, right panel). For the remaining 3 phenolic acid metabolites detected in brain specimens (GA, HA, and 3-HPVA), no significant difference in brain concentration was observed between the HuA and HuB mice (Table III, right panel).
The relative concentrations of the microbial-derived phenolic acid metabolites from FRP in cecum, plasma, and brain specimens for the two gnotobiotic mouse groups are summarized in Fig. 2. Significantly higher concentrations of DHCA are found accumulated in the cecum and plasma specimens of HuB compared to HuA mice (Fig. 2A, D). Consistent with our previous observation in rats in which DHCA does not cross the blood-brain barrier [13], DHCA was not detected in brain specimens from either HuA or HuB gnotobiotic mice. In comparison to HuB mice, HuA mice generated and accumulated significantly higher concentrations of 3-HPPA and 3,4-diHBA in the colon, coincidental with significantly higher concentrations of 3-HPAA in plasma and the brain (Fig. 2B, E) as well as significantly higher concentrations of 3,4-diHBA in the brain (Fig. 2C, F).
Collectively, these results demonstrate that interpersonal heterogeneity in human gut microbiotas can drive significant differences in the bioavailability of FRP-derived microbial phenolic acid metabolites in gnotobiotic mice.
3.3. Select brain-accumulating phenolic acid metabolites interfere with the assembly of monomeric α-synuclein peptide into neurotoxic α-synuclein aggregates, in vitro.
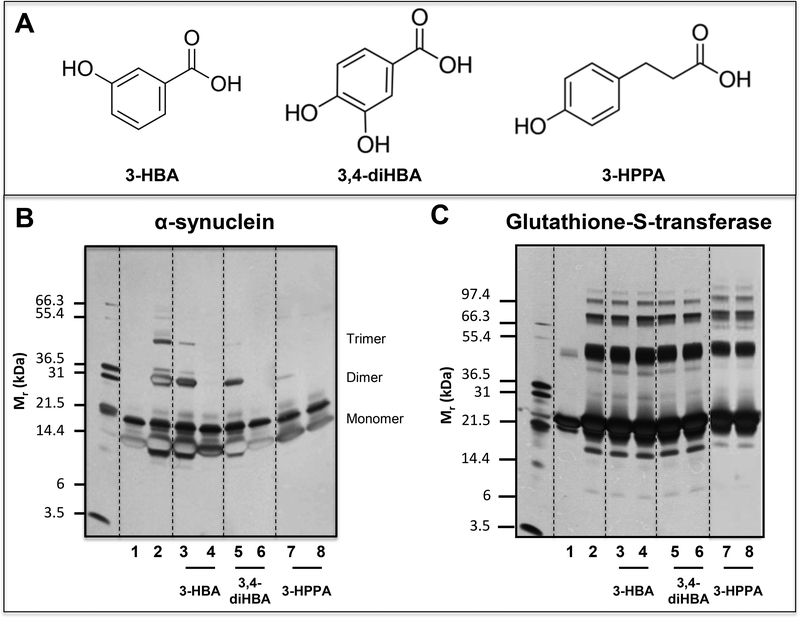
A diverse group of neurodegenerative diseases, including AD and α-synucleinopathies, all share a common key pathophysiologic feature involving misfolding of disease-specific proteins into neurotoxic aggregates and deposition of these aggregates in the brain [21–22]. We previously observed that 3-HBA and 3-HPPA, two brain-accumulating phenolic acids derived from microbial fermentation of flavanols, are effective in interfering with the assembly of Aβ peptides into misfolded neurotoxic aggregates, which play central roles in AD neuropathology [13]. Similar processes, including abnormal interactions and self-assembly of proteins into structurally organized aggregates, are involved in the misfolding of diverse disease-specific proteins, including the misfolding of α-synuclein in α-synucleinopathies and the misfolding of tau in tauopathies [19, 23–28]. We therefore investigated whether 3-HPPA and 3,4-diHBA, two FRP-derived brain-accumulating microbial phenolic acid metabolites (Fig. 2B,C), may interfere with α-synuclein misfolding. In addition, based on our previous observation that 3-HBA, another flavanol-derived brain-accumulating microbial phenolic acid metabolite with structural similarity to 3,4-diHBA, is effective in inhibiting Aβ misfolding [13], we also assessed whether 3-HBA could similarly interfere with the misfolding of α-synuclein. The structures of the three targeted compounds are shown in Fig. 3A.
Fig. 3. Select brain accumulating, FRP-derived microbial phenolic acid metabolites potently interfere with protein-protein interactions among α-synuclein peptides.
Using the PICUP protocol, we assess effects of three structurally closely related phenolic acid metabolites, 3-HBA, 3,4-diHBA, and 3-HPPA on the initial protein-protein interactions necessary for the assembly of monomeric α-synuclein into multimeric forms. PICUP assays were conducted in the presence of vehicle or individual phenolic acids at a low dose (LD, equal molar concentration of phenolic acid relative to α-synuclein) or at a high dose (HD, 4-fold higher molar concentration of phenolic acid relative to α-synuclein). Monomeric and cross-linked oligomeric α-synuclein forms were resolved by SDS-PAGE and visualized by silver staining. We confirmed that the addition of phenolic acids did not lead to observable change in the pH of the reaction mixture. (A) Molecular structures of the 3-HBA, 3,4-diHBA, and 3,4-diHBA. (B) Effects of individual phenolic acids on the oligomerization of monomeric α-synuclein peptides. (C) Control PICUP studies using glutathione-S-transferase confirming that the addition of 3-HBA, 3,4-diHBA or 3-HPPA at both and LD or the HD did not interfere with the PICUP cross-linking mechanism. In (B, C) Lane 1, peptide (α-synuclein or glutathionne-S-transferase) without cross linking; lane 2, peptide cross-linked in the presence of vehicle; lane 3, peptide cross-linked in the presence of a LD of 3-HBA; lane 4, peptide cross-linked in the presence of a HD of 3-HBA; lane 5, peptide cross-linked in the presence of a low dose of 3,4-diHBA; lane 6, peptide cross-linked in the presence of a HD of 3,4-diHBA; lane 7, peptide cross-linked in the presence of a LD of 3-HPPA; lane 8, peptide cross-linked in the presence of a HD of 3-HPPA. Each of the gel shown is representative of three independent experiments.
Using the photoinduced cross-linking of unmodified protein (PICUP) assay, we monitored the effects of the individual brain-available phenolic acids on initial protein-protein interactions that are necessary for the assembly of α-synuclein into higher-ordered misfolded forms. Two doses were tested: a low dose (LD) and a high dose (HD), corresponding, respectively, to equal molar concentrations or 4-fold higher concentration of phenolic acid relative to α-synuclein. 3-HBA, 4-diHBA, and 3-HPPA inhibited the formation of α-synuclein dimers and trimers (Fig. 3B, lanes 4, 6, 8) at HD. At LD, all three compounds showed a dose-dependent reduction in anti-aggregation activity. Comparative efficacy anti-aggregation activity at LD are: 3-HPPA > 3,4-diHBA > 3-HBA (Fig. 3B, lanes 3, 5, 7). As we have done in the past [19], in a parallel control study using glutathione-S-transferase (GST), we confirmed that none of the phenolic acids interfered with the PICUP cross-linking mechanism (Fig. 3C).
3.4. Select brain-accumulating phenolic acid metabolites interfere with the assembly of monomeric α-synuclein peptide into α-synuclein protofibrils, in vitro.
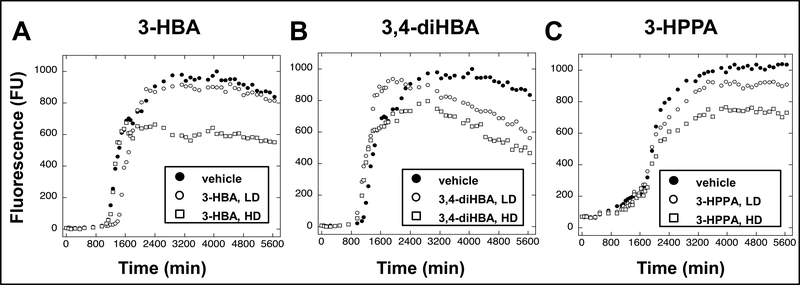
Using the thioflavin T (ThT) spectroscopic assay, we assessed for the effects of 3-HBA, 3,4-diHBA, and 3-HPPA on the assembly of α-synuclein peptides into α-synuclein fibrils, in vitro. We observed that 3-HBA, 3,4-diHBA and 3-HPPA attenuated α-synuclein protofibrils formation at the HD (Fig. 4A, B, C). The LD of 3,4-diHBA and 3-HPPA also attenuated α-synuclein fibril formation, but to a lesser extent compared to the HD (Fig. 4B, C), while the LD of 3-HBA did not reduce α-synuclein protofibril formation (Fig. 4A).
Fig. 4. Select brain-accumulating microbial-generated phenolic acid metabolites potently interfere with α-synuclein protofibril formation.
Spontaneous assembly of monomeric α-synuclein into protofibrils was monitored using the ThT assay. ThT binds to beta-sheet contents α-synuclein protofils and increasing ThT flourescence (FU, in arbitrary fluorescence units) reflects the presence of higher contents of α-synuclein protofibrils. We note that ThT fluorescence is not a direct measure of fibril content. However, since β-sheet formation correlates with fibril formation, ThT fluorescence is a useful surrogate marker [32]. (A, B, C) Monomeric α-synuclein was incubated for 0–94 hours at 37°C in the presence of vehicle (○), or individual phenolic acids at a low dose (LD, equal molar concentration of phenolic acid relative to α-synuclein) (●), or a high dose (HD, 4-fold higher molar concentration of phenolic acid relative to α-synuclein) (■). Phenolic acids tested were 3-HBA (A), 3,4-diHBA (B) or 3-HPPA (C).
3.5. Effects of brain-accumulating phenolic acid metabolites on the morphologies of α-synuclein assemblies, in vitro.
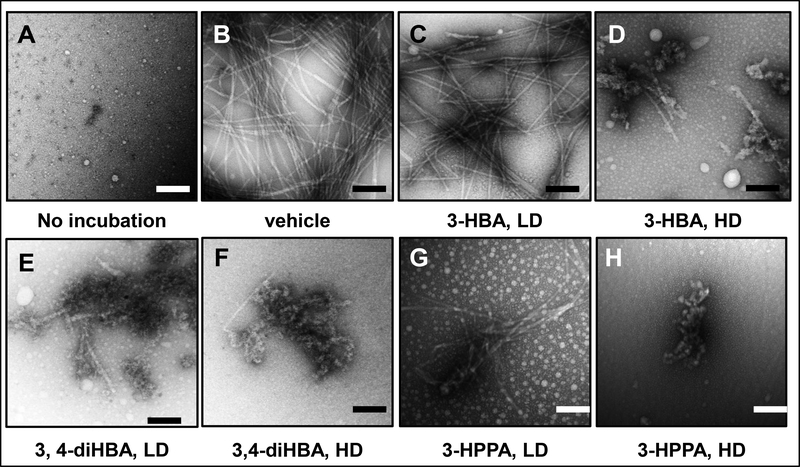
Using electronic microscopy, we confirmed that monomeric α-synuclein peptides were spontaneously assembled into classical PD-type non-branched α-synuclein fibrils (Fig. 5B). We observed that α-synuclein fibril formation was completely abolished by 3,4-diHBA and 3-HPPA at the HD (Fig. 5F, H) and severely attenuated by 3,4-diHBA and 3-HPPA at the LD (Fig. 5E, G). At the HD, 3-HBA also completely abolished α-synuclein fibril formation (Fig. 5D) whereas α-synuclein fibril formation was partially inhibited by a LD of 3-HBA (Fig. 5C).
Fig. 5. Effects of brain-accumulating microbial-generated phenolic acid metabolites on α-synuclein protofibril morphology.
Monomeric α-synuclein (70 μM) was incubated in the presence of vehicle or individual brain-accumulating microbial-generated phenolic acid metabolites at 37°C for 94 hours in 20 mM Tris, pH 7.4, 100 mM NaCl. EM was used to determine the morphologies of the resultant α-synuclein assemblies. (A, B) monomeric α-synuclein (A) and α-synuclein incubated in the presence of vehicle (B). (C, D, E, F, G, H) α-synuclein incubated in the presence of individual phenolic acids at a low dose (LD, equal molar concentration of phenolic acid to α-synuclein) or a high dose (HD, 4-fold higher molar concentration of phenolic acid to α-synuclein): LD or HD 3-HBA (C, D), LD or HD 3,4-diHBA (E, F), LD or HD 3-HPPA (G, H). Scale bars indicate 100 nm
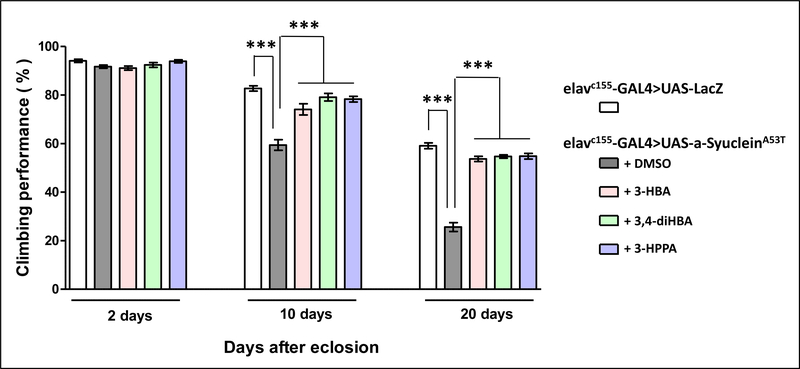
3.6. Effects of brain-accumulating phenolic acid metabolites in modulating α-synucleinopathy, in vivo, in a Drosophila model of PD.
Autosomal dominant missense G209A mutation in the α-synuclein gene resulting in the expression of the mutant Ala53Thr (A53T) α-synuclein protein is linked to familial PD [27]24. In Drosophila, neuronal expression of the A53T mutant human α-synuclein protein has been shown to recapitulate key pathologic features of PD, including adult-onset loss of dopaminergic neurons and locomotor dysfunction 25. Using this A53T mutant α-synuclein Drosophila model of PD, we investigated for effects of 3-HBA, 3,4-diHBA and 3-HPPA on modulating PD pathologic phenotypes, in vivo, by monitoring locomotive functions using a negative geotaxis behavior assay (climbing assay) in adult flies at 2, 10, and 20 days after eclosion (DAE) [20]. As shown previously, neuronal expression of A53T mutant α-synuclein caused locomotive dysfunction in adult flies, as evidenced by impaired climbing of mutant α-synuclein expressing flies compared to control (LacZ expressing) flies at both 10 and 20 DAE (Fig. 6). In contrast, we demonstrated that in comparison to vehicle (DMSO)-treated mutant flies, treatment with 3-HBA, 3,4-diHBA and 3-HPPA significantly improved the climbing performance of mutant α-synuclein expressing flies (Fig. 6). Thus, not only do 3-HBA, 3,4-diHBA, and 3-HPPA reduce the assembly of α-synuclein into neurotoxic aggregates in vitro (Fig. 3, 4, 5), but these data from A53T mutant α-synuclein flies suggests that 3-HBA, 3,4-diHBA, and 3-HPPA also reduce mutant α-synuclein-mediated neurotoxicity in vivo.
Fig. 6. Brain-accumulating phenolic acid metabolites beneficially modulates mutant α-synuclein mediated locomotive activity decline in a Drosophila model of α-synucleinopathy.
An established Drosophila PD model resulting from transgenic neuronal expression of the A53T mutant human α-synuclein protein 25 was used in these studies. We monitored locomotive functions in control flies (expressing LacZ) mutant flies using a negative geotaxic behavior assay (climbing assay) 26. Shown are climbing performance (expressed as percentage of flies that successfully performed the climbing test) of adult flies at the ages of 2, 10 and 20 DAE. Within each age group, bar graphs represent mean (±SEM) of control flies, vehicle (DMSO) treated A53T mutant flies, or A53T mutant flies treated with 3-HBA, 3-HPPA, or 3,4-diHBA (1 μM); n=10 flies per control, vehicle-treated or phenolic-acid treated group. Significance level was established by one-way ANOVA (P ≤ 0.0001); *** Bonferroni post-hoc test P ≤ 0.0001.
3.7. Select bacteria are capable of supporting the generation of brain-accumulating, bioactive phenolic acid metabolites, in vitro.
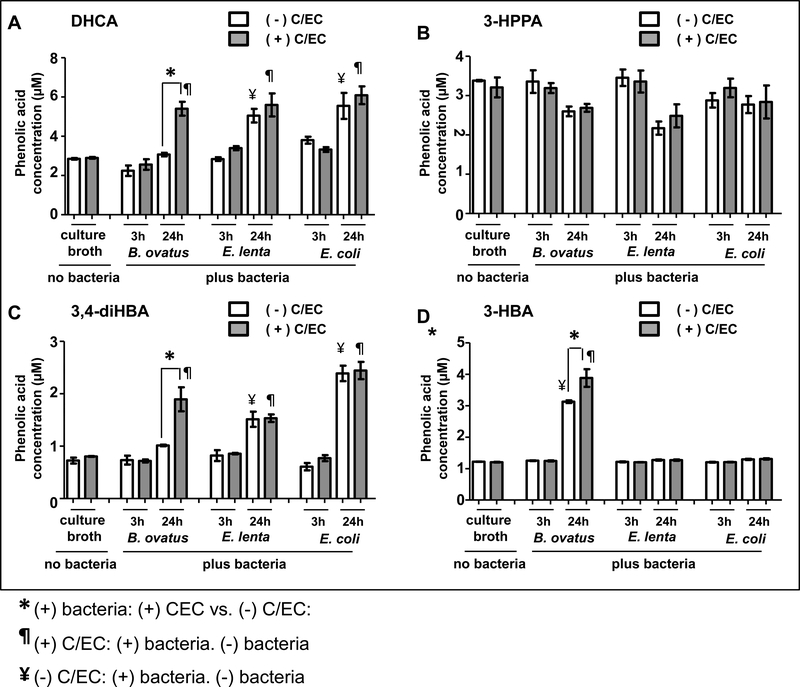
To determine the potential of individual bacteria to generate biologically available, bioactive phenolic acid metabolites, we selected three species, representing three different phyla found in the human gut microbiota, for in vitro fermentation studies. Specifically, we choose Bacteroides ovatus (B. ovatus), as one of the most common representatives of the Bacterodetes phylum, Eggerthella lenta (E. lenta) as a representative of the Firmicutes phylum that was previously identified to metabolize (+)-catechin (C) and (−)-epicatechin (EC) [29–30], and Escherichia coli (E. coli) as a facultative anaerobe and a common species in the gut from the phylum Proteobacterium. Fermented broth concentrations of 4 bioavailable, bioactive phenolic metabolites (DHCA, 3,4-diHBA, 3-HBA and 3-HPPA) were monitored. DHCA was detected in plasma (Table III) and has been shown to exert potent anti-inflammatory activities [1]. Moreover, we observed the accumulation of 3-HPPA (Table III), 3,4-diHBA (Table III) and 3-HBA [13] in the brain and demonstrated these brain-targeting phenolic metabolites are bioactive in inhibiting misfolding of α-synuclein (Figs. 3, 4, 5) and/or Aβ proteins [13], in vitro, as well as in modulating mutant α-synuclein-mediated PD phenotypes, in vivo (Fig. 6).
B. ovatus alone was able to convert C/EC into DHCA, 3,4-diHBA, and 3-HBA. In particular, we observed significantly higher concentration of DHCA (Fig. 7A), 3,4-diHBA (Fig. 7C) and 3-HBA (Fig. 7D) in the culture broth when B. ovatus was inoculated in the presence versus the absence of C/EC. In contrast, comparisons of B. ovatus in the presence or absence of C/EC resulted in no observable difference in the culture broth content of 3-HPPA (Fig. 7B). Moreover, we observed neither E. lenta nor E. coli was able to convert C/EC to DHCA, 3-HPPA, 3,4-diHBA or 3-HBA. Comparing E. lenta or E. coli fermentation in the presence versus absence of C/EC, we found no observable difference in the culture broth content of DHCA (Fig. 7A), 3-HPPA (Fig. 7B), 3,4-diHBA (Fig. 7C) or 3-HBA (Fig. 7D).
Fig. 7. Differential efficacy of individual bacteria to support the generation of biologically available, bioactive phenolic acid metabolites, in vitro.
Using in vitro bacteria fermentation protocols, we assess for effects of individual bacterium from microbiome HuA and HuB to support the generation of biologically available, bioactive phenolic acid metabolites. Individual bacteria (B. ovatus, E. lenta, and E. coli) were cultured in rich anaerobic broth under anaerobic conditions at 37 °C, in the absence or in the presence of C/EC for 3 or 24 h. Phenolic acid concentrations in culture broth samples (n=3 repetitions per treatment group) were assessed by UPLC-QQQ/MS. (A, B, C, D) Bar graphs represents mean ± SD the concentration of DHCA (A), 3-HPPA (B), 3,4-diHBA (C), and 3-HBA (D) in i) cultured broth containing bacteria and C/EC vs. cultured broth containing bacteria but no C/EC, ii) cultured broth containing C/EC and bacteria vs. cultured broth containing C/EC but no bacteria, and iii) cultured broth containing bacteria and no C/EC vs. cultured broth containing no bacteria and no C/EC. Statistics: one-way ANOVA<0.001 in (A, C,D); one-way ANOVA<0.018 in (B). Bonferonni post hoc analysis to compare select pairs: *, P<0.05, (+) bacteria: (−) C/EC vs. (+) C/EC; ¥, p<0.05, (−) C/EC: (−) bacteria vs. (+) bacteria; ¶, p<0.05, (+) C/EC: (−) bacteria vs. (+) bacteria.
Interestingly, we observed that B. ovatus, E. lenta, and E. coli were also able to generate DHCA, 3-HPPA, 3,4-diHBA, and 3-HBA through a C/EC-independent process. Compared to a culture broth with no bacteria or C/EC, broth fermented with E. lenta and E. coli in the absence of C/EC resulted in significantly higher concentrations of DHCA (Fig. 7A) and 3,4-diHBA (Fig. 7C), while fermentation with B. ovatus in the absence of C/EC resulted in significantly higher concentration of 3-HBA (Fig. 7D). The rich anaerobic culture broth endogenously contains many phenolic acids (Supplementary Fig. S3A) that conceivably could serve as precursor(s) to the C/EC-independent derivatization of DHCA, 3-HPPA, 3,4-diHBA, and 3-HBA by B. ovatus, E. lenta or E. coli. For example, the concentration of 3-HPPA in the culture broth is 52 μM (Supplementary Fig. 3A). Based on a proposed C/EC metabolic pathway shown in Supplementary Fig. S3B for the derivatization of DHCA, 3,4-diHBA, 3-HBA, and 3-HPPA from C/EC or other phenolic acids, it is possible that the high concentration of 3-HPPA in the culture broth could support C/EC-independent generation of 3-HBA by B. ovatus.
4. DISCUSSION
The ultimate goal of this study was to identify groups of microbes that can enhance the generation of beneficial phenolic acids, such as those identified in this study, that impact inflammation and protein misfolding and these could subsequently be used as therapies for neurological diseases. We are specifically interested in the culturable fraction of GI microbiota that can serve as consistent and reproducible sources of bacteria for experimental investigations and therapeutic development. Using the clonal array technique, we have previously established that the majority of the genus-level microbiota organisms are captured in our culture libraries [17], allowing us to be confident in the diversity and representative microbiota populations we have named HuA and HuB.
Using two groups of humanized gnotobiotic mice with distinct culturable GI microbiota compositions, we demonstrated the feasibility that interpersonal heterogeneity in gut microbiota differentially affects the generation and bioavailability of microbiota-derived phenolic acid metabolites from dietary flavanols. In particular, mice with HuA and HuB GI microbiota exhibited distinct differences in the generation and bioavailability of plasma-circulating DHCA and brain-accumulating 3-HPPA and 3,4-diHBA (Fig. 2). We have previously shown that DHCA is capable of modulating inflammatory responses [1]. We have also shown that 3-HPPA and 3-HBA, derived from GI microbiota fermentation of a grape seed polyphenol extract, are effective in interfering with the misfolding of AD-related Aβ peptides [13]. New evidence from our current study demonstrates the in vitro efficacy of 3-HPPA, 3-HBA, and 3,4-diHBA to inhibit the misfolding of α-synuclein (Fig. 3, 4, 5). Using transgenic flies with neuronal expression of A53T mutant human α-synuclein, we demonstrated, for the first time, the efficacy of 3-HPPA, 3-HBA and 3,4-diHBA to modulate A53T mutant human α-synuclein neurotoxicity, in vivo. We note the protective effects of 3-HPPA, 3-HBA, and 3,4-diHBA in vivo may reflect the efficacy of these phenolic acid metabolites to interfere with α-synuclein misfolding. Ongoing studies are investigating whether additional biological activities, such as antioxidant effects, may contribute to the efficacy of these brain-accumulating phenolic acids in modulating α-synuclein-mediated phenotypes, in vivo.
Inflammation and α-synuclein misfolding are both key pathological mechanisms underlying α-synucleinopathies such as Parkinson’s disease, dementia with Lewy bodies and multiple system atrophy [21, 23. 31]. Thus, interpersonal heterogeneity in gut microbiota may modulate resilience to α-synucleinopathy-associated disorders by FRP and other flavanol-rich preparations by differentially affecting the generation and bioavailability of DHCA, 3-HBA, 3,4-diHBA, 3-HPPA, and perhaps other uncharacterized bioactive FRP-derived microbial phenolic acid metabolites. In addition to α-synucleinopathies, both inflammation and protein misfolding are key pathologic mechanisms underlying multiple neurological disorders such as AD, Huntington’s disease, amyotrophic lateral sclerosis, progressive supranuclear palsy, cortical basal degeneration, Pick’s disease, and familial frontotemporal dementias with Parkinsonism linked to chromosome 17 17. The misfolding of diverse disease-specific proteins all involve similar processes 19; therefore, compounds that are effective in inhibiting α-synuclein aggregation might also be effective in inhibiting the misfolding of other proteins. Consistent with this, we observed that 3-HBA is effective at inhibiting Aβ misfolding [13] and is also effective in inhibiting α-synuclein misfolding (Fig. 3, 4, 5). Thus, results from our present study demonstrate distinct differences in the bioavailability of DHCA (effective in modulating inflammation) as well as 3-HPPA, 3-HBA and 3,4-diHBA (effective in inhibiting protein misfolding), which link interpersonal differences in gut microbiota with resilience against several neurological disorders.
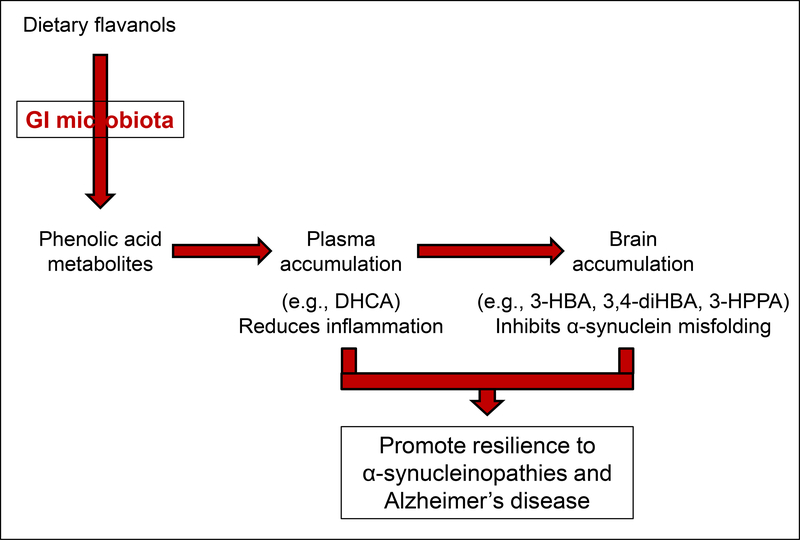
Collectively, our study demonstrates the proof-of-concept that interpersonal heterogeneity of the gut microbiota may differentially affect the generation, and thereby the bioavailability, of microbial-generated phenolic acid metabolites that have been derived from dietary flavanols, which ultimately may lead to interpersonal differences in the efficacy of dietary flavanols in modulating mechanisms underlying neurological resilience (Fig. 8). We note a limitation of our investigation is the small sample size as we only investigated two microbiota compositions. Future investigations involving larger sample sizes of microbiota will be necessary to confirm that interpersonal heterogeneity in gut microbiota may lead to interpersonal differences in the efficacy of dietary flavanols towards modulating neurological resilience.
Fig. 8. GI microbiome, through interaction with dietary flavanols may modulate resilience of host to neurological disorders through diet-gut microbiome interactions.
The schematics showed that orally consumed flavanols are converted by GI microbiota into phenolic acid metabolites, some of which are biologically available in the plasma and/or the brain. Highlighted are effects of plasma-accumulating DHCA in the modulating inflammation and brain-accumulating 3-HBA, 3,4-diHBA, and 3-HPPA in inhibiting with protein folding, and that both anti-inflammation and protein-misfolding activities of these biologically available, bioactive phenolic acid metabolites would promote resilience for a diverse neurological disorders for which inflammation and misfolding are significant pathological processes. The implication of this overview is that interpersonal differences in GI microbiota would affect generation and thereby bioavailability of bioactive phenolic acid metabolites, would differentially affect the efficacy of dietary flavanols on the modulation of neurological resilience of diverse neurological disorders involving inflammation and/or protein misfolding.
To maximize availability, repeatability, and comparisons with existing knowledge of these species in the context of other polyphenols and metabolism studies, we chose to specifically study widely-used strains of these species for their individual metabolic potential. Using in vitro fermentation with three bacteria species from ATCC that are commonly found in the gut microbiota, B. ovatus, E. lenta and E. coli, we began to characterize individual GI bacterial species that are supportive to the generation of select bioavailable and bioactive phenolic acid metabolites from dietary flavanols. E. lenta is known for its capability to cleave the C-ring of flavanols while both B. ovatus and E. coli are known for their capacity to catalyze O-deglycosylation of flavonols [29–30]. In our present investigation, we observed that among the three bacterial strains investigated, only B. ovatus was effective in supporting metabolic conversion of C/EC to DHCA, 3,4-diHBA, and 3-HBA (Fig. 7A, C, D). Interestingly, we also observed that B. ovatus, E. lenta, and E. coli were capable of generating DHCA, 3,4-diHBA and/or 3-HBA through metabolic processes independent of C/EC (Fig. 7A, C, D). As a number of phenolic acids are endogenously present in the culture broth (Supplementary Fig. 3), we hypothesized that some of these phenolic acids may also act as precursors for the generation of DHCA, 3,4-diHBA, and 3-HBA by B. ovatus, E. lenta, and E. coli. In the future, further studies will be conducted to clarify the metabolic processes underlying the generation of specific bioavailable, bioactive phenolic acid metabolites, and to identify individual (or combinations) of bacteria that optimize the generation of these phenolic acids. These future studies also will facilitate the development of probiotic, prebiotic and/or synbiotic approaches for promoting neurological resilience.
Supplementary Material
ACKNOWLEDGEMENTS:
This study was supported by Grant Number P50 AT008661–01 from the National Center for Complementary and Integrative Health (NCCIH) and the National Institutes of Health Office of Dietary Supplements (ODS) and, in part, by the Altschul Foundation. GMP holds a Senior VA Career Scientist Award. We acknowledge that the contents of this study do not represent the views of the NCCIH, the ODS, the NIH, the U.S. Department of Veterans Affairs, or the United States Government.
ABBREVIATIONS:
- 3-HBA
3-hydroxybenzoic acid
- 3-HPPA
3-(3-hydroxyphenyl)propionic acid
- 3-HPAA
3-hydroxyphenylacetic acid
- 3-HPVA
5-(3-Hydroxyphenyl)valeric acid
- 4-HCA
4-hydroxycinnamic acid
- 4-HPVA
5-(4-hydroxyphenyl)valeric acid
- 3,4-diHBA
3,4-dihydroxybenzoic acid
- 3,4-diHPPA
3-(4-hydroxyphenol)propionic acid
- 3,4-diHPAA
3,4-dihydroxyphenylacetic acid
- 3,4-diHPVA
5-(3,4-dihydroxyphenyl)valeric acid
- AA
ascorbic acid
- α-synuclein
alpha-synuclein
- B, ovatus
Bacteroides ovatus
- C
(+)-catechin
- CA
caffeic acid
- DHCA
dihydrocaffeic acid
- dMEM
dynamic multiple reaction monitoring
- E. coli
Escherichia coli
- E. lenta
Eggerthella lenta
- EC
(−)-epicatechin
- EM
electron microscopy
- ESI
electrospray ionization
- FA
ferulic acid
- FRP
flavanol-rich preparation
- GA
gallic acid
- GI
gastrointestinal
- GST
Glutathione S-transferase
- HA
hippuric acid
- HD
high dose
- HVA
homovanillic acid
- IS
internal standard
- LD
low dose
- NCCIH
National Center for Complementary and Integrative Health
- ODS
National Institutes of Health Office of Dietary Supplements
- P2 dimers
proanthocyanidin dimers
- PICUP
Photoinduced cross-linking of unmodified proteins
- ThT assay
Thioflavin T spectroscopic assay
- VA
vanillic acid
Footnotes
CONFLICT OF INTEREST:
The authors declare that they have no conflict of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wang J, Hodes GE, Zhang H, Zhang S, Zhao W, Golden SA, Bi W, et al. Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice. Nat Commun. 2018; 9(1):477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng T, Wang W, Li Q, Han X, Xing J, Qi C, Lan X, Wan J, Potts A, Guan F, Wang J. Cerebroprotection of flavanol (−)-epicatechin after traumatic brain injury via Nff2-dependent and –independent pathways. Free Radic Biol Med. 2016; 9:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubner L, Wang J, Ho L, Pasinetti GM. Recommendations for development of new standardized forms of Cocoa breeds and cocoa extract processing for the prevention of Alzheimer’s disease: role of cocoa in promotion of cognitive resilience and healthy brain agina. J Alzheimers Dis. 2015; 48(4):879–889. [DOI] [PubMed] [Google Scholar]
- 4.Stringer TP, Guerrieri D, Vivar C, van Praag H. Plant-derived flavanol (−)epicatechin mitigates anxiety in association with elevated hippocampal monoamine and BDNF levels, but does not influence pattern separation in mice. Transl Psychiatry. 2015; 5: e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nehlig A The neuroprotective effects of cocoa flavanol and its influence on cognitive performance. Br J Clin Pharmacol. 2013; 75(3):705–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox CJ, Choudhry F, Peacey E, Perkinton MS, Richardson JC, Howlett DR, Lichtenthaler SD, et al. Dietary (−)-epicatechin as a potent inhibitor of βγ-secretase amyloid precursor protein processing. Neurobiol Aging. 2015; 36(1):178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Ferruzzi MG, Ho L, Blount J, Janle EM, Gong B, Pan Y, et al. Brain-targeted proanthocyanidin metabolites for Alzheimer’s disease treatment. J Neurosci. 2012; 32(15):5144–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Santa-Maria I, Ho L, Ksiezak-Reding H, Ono K, Teplow DB, Pasinetti GM. Grape derived polyphenols attenuate tau neuropathology in a mouse model of Alzheimer’s disease. J Alzheimers Dis. 2010; 22(2):653–661. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Ho L, Zhao W, Ono K, Rosensweig C, Chen L, Humala N, et al. Grape-derived polyphenolics prevent Abeta oligomerization and attenuate cognitive deterioration in a mouse model of Alzheimer’s disease. J Neurosci. 2008; 28(25):6388–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hribar U, Ulrih NP. The metabolism of anthocyanins Curr Drug Metab 2014; 15:3–13. [DOI] [PubMed] [Google Scholar]
- 11.Hu FB, Rimm E, Smith_Warner SA, Feskanich D, Stampfer MJ, Ascherio A, et al. Reproducibility and validity of dietary patterns assessed with a food-frequency questionnaire. Am J Clin Nutr 1999; 69:243–249. [DOI] [PubMed] [Google Scholar]
- 12.van Duynhoven J, Vanghan EE, Jacons DM, Kemperman RA, van Velzen EJ, Gross G, Roger LC, et al. Metabolic fate of polyphenols in the human superorganism. Proc Natl Acad Sci U.S.A. 2011; 108(Supplement 1):4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Ho L, Faith J, Ono K, Janle EM, Lachcik PJ, Cooper BR, et al. (2015) Role of intestinal microbiota in the generation of polyphenol-derived phenolic acid mediated attenuation of Alzheimer’s disease β-amyloid oligomerization. Mol Nutr Food Res. 59(6):1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, et al. The long-term stability of the human gut microbiota. Science 2013; 341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spillantini MG, Goedert M. The alpha-synucleinopathies: Parkinson’s disease, dementia with Lewy bodies multiple system atrophy. Ann NY Acad Sc. 2000; 920:16–27. [DOI] [PubMed] [Google Scholar]
- 17.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U.S.A. 2011; 108(15):6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014; 6(220):220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono K, Condron MM, Ho L, Wang J, Zhao W, Pasinetti GM, Teplow DB. Effects of grape seed-derived polyphenols on amyloid beta-protein self-assembly and cytotoxicity. J Biol Chem. 2008; 283(47):32176–32187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali YO, Escala W, Ruan K, Zhai RG. Assayng locomotor, learning, and memory deficits in Drosophila models of neurodegeneration. J Vis Exp. 2011; 49:e2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho L, Pasinetti GM. Polyphenolic compounds for treating neurodegenerative disorders involving protein misfolding. Expert Rev Proteomics. 2010; 7(4):579–589. [DOI] [PubMed] [Google Scholar]
- 22.Shamsi TN, Athar T, Parveen R, Fatima S. A review on protein misfolding, aggregation and strategies to prevent related ailments. Int J Biol Macromol. 2017; 105(Pt 1):993–1000. [DOI] [PubMed] [Google Scholar]
- 23.Shigemitsu Y, Hiroaki H. Common molecular pathogenesis of disease-related intrinsically disordered proteins revealed by NMR analysis. J Biochem. 2018; 163(1):11–18. [DOI] [PubMed] [Google Scholar]
- 24.Biza K, Nastou KC, Tsiolaki PL, Mastrokalou CV, Hamodrakas SJ, Iconomidou VA. The amyloid interactome: exploring protein aggregation. PLos One 2017; 12(3):e0173163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolshette NB, Thakur KK, Bidkar AP, Trandafir C, Kumar P, Gogoi R. Protein folding and misfolding in the neurodegenerative disorders: a review. Rev Neurol. 2014; 170(3):151–161. [DOI] [PubMed] [Google Scholar]
- 26.Breyldo L, Wu JW, Uversky VN. α-synuclein misfolding and Parkinson’s disease. Biochim Biophys Acta. 2012; 1822(2):261–285. [DOI] [PubMed] [Google Scholar]
- 27.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Eplen JT, Schols L, Riess O. Ala30Pro mutation in the gene Encoding α-synuclein in Parkinson’s disease. Nat. Genet 1998; 18: 106–108. [DOI] [PubMed] [Google Scholar]
- 28.Feany MB, Bender WW. A Drosophila modle of Parkinson’s disease. Nature. 2000. 404(6776):394–398. [DOI] [PubMed] [Google Scholar]
- 29.Takagaski A, Nanjo F. Bioconversion of (−)-epicatechin, (+)-epicatechin, (−)-catechin, and (+)-catechin by (−)epigallocatechin-metabolizing bacteria. Biol Pharm Bull. 2015; 38(5):789–794. [DOI] [PubMed] [Google Scholar]
- 30.Braune A, Blaut M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes. 2016; 7(3):216–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lema Tome CM, Tyson T, Rey NL, Grathwohl S, Britschgi M, Brundin P. Inflammation and a-synuclein’s prion-like behavior in Parkinson’s disease – is there a link? Mol Neurobiol. 2013; 47(2):561–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine H Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999; 309:274–284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.