Abstract
Background:
In mice, bacteria from the mouth can translocate to the pancreas and impact pancreatic cancer progression. In humans, oral bacteria associated with periodontal disease have been linked to pancreatic cancer risk. It is not known if DNA bacterial profiles in the pancreas and duodenum are similar within individuals.
Methods:
Tissue samples were obtained from 50 subjects with pancreatic cancer or other conditions requiring foregut surgery at the Rhode Island Hospital (RIH), and from thirty-four organs obtained from the National Disease Research Interchange. 16S rRNA gene sequencing was performed on 189 tissue samples (pancreatic duct, duodenum, pancreas), 57 swabs (bile duct, jejunum, stomach), and 12 stool samples.
Results:
Pancreatic tissue samples from both sources (RIH and NDRI) had diverse bacterial DNA, including taxa typically identified in the oral cavity. Bacterial DNA across different sites in the pancreas and duodenum were highly subject-specific in both cancer and non-cancer subjects. Presence of genus Lactobacillus was significantly higher in non-cancer subjects compared with cancer subjects and the relative abundance of Fusobacterium spp., previously associated with colorectal cancer, was higher in cancer subjects compared to non-cancer subjects.
Conclusions:
Bacterial DNA profiles in the pancreas were similar to those in the duodenum tissue of the same subjects, regardless of disease state, suggesting that bacteria may be migrating from the gut into the pancreas. Whether bacteria play a causal role in human pancreatic cancer needs to be further examined.
Impact:
Identifying bacterial taxa that differ in cancer patients can provide new leads on etiologically relevant bacteria.
Keywords: Microbiome, pancreatic cancer, duodenum, bacteria, 16S rRNA gene sequencing, bacterial dissemination
Introduction
In 2018, an estimated 55,440 individuals will be diagnosed with pancreatic cancer in the US, and only 8% of these individuals are expected to survive the next five years [1]. Given this high fatality rate, and the silent progression of early disease, identifying risk factors for the prevention and early detection of pancreatic cancer is critical to reducing its mortality. To date, known risk factors for pancreatic cancer, including smoking, obesity, diabetes, heavy alcohol consumption, family history and markers of genetic susceptibility, cannot, even collectively, be used for early detection and risk stratification of pancreatic cancer in the general population [2].
Studies have suggested a link between bacteria and pancreatic cancer risk [3], highlighting the need to more critically explore the underlying factors that affect the microbiome of the oral cavity and upper digestive tract in both cancer patients and cancer-free individuals. The current research on oral bacteria and pancreatic cancer risk stems from a number of observational studies that reported a higher risk of pancreatic cancer among individuals with periodontitis, when compared to those without periodontitis [3, 4]. Periodontitis, an inflammatory disease of the gums, is largely driven by keystone pathogens and pathobionts [5]. Two large prospective cohort studies have reported positive associations between periodontal disease pathogens and subsequent pancreatic cancer risk [6, 7]; in these two studies, detection of elevated antibodies to Porphyromonas gingivalis, measured in blood collected prior to cancer diagnosis, was associated with a two-fold higher risk of pancreatic cancer [6], and presence (vs absence) of P. gingivalis in saliva collected prior to cancer diagnosis was associated with a 60% increase in risk of pancreatic cancer [7]. Aggregatibacter actinomycetemcomitans, another periodontal pathogen, was also associated with pancreatic cancer risk in the prospective study using saliva [7].
Few investigations to date have attempted to detect bacteria in pancreatic tissue. Earlier studies reported the presence of bacteria in pancreatic ducts of subjects with chronic pancreatitis or bile duct obstruction [8–10]. Other studies have investigated the presence of specific bacterial DNA in the pancreatic tissue of pancreatic cancer subjects, namely species of Helicobacter [11] and Fusobacterium [12]. The most comprehensive molecular microbiome studies to date reported the presence of a diverse bacterial populations in fluids collected from the bile duct, pancreas and jejunum of subjects undergoing pancreaticoduodenectomy [13], and in pancreatic cyst fluid removed endoscopically from pancreatic cysts [14]. In mice, bacteria have been shown to translocate from the mouth to the pancreas, and germ-free mice have reduced progression of pancreatic ductal adenocarcinoma [15].
Metagenomics studies on DNA isolated from tissue samples from cancer subjects have been conducted for lung [16], colorectal [17], esophageal [18], stomach [19], and breast cancer [20]. These studies demonstrate that 16S rRNA gene sequencing can be effectively conducted on fresh tissue samples where the ratio of bacterial to human DNA is much lower than at other human sites (e.g., stool or oral cavity)[21]. Moreover, these studies have shown that bacterial profiles at different organ sites are often unique [16, 20] and that changes may be associated with cancer [18, 19]. In two recent studies, bacterial DNA was measured in tumor tissue samples obtained from patients with pancreatic ductal adenocarcinoma (PDAC) using 16S rRNA gene sequencing [15, 22]; however, comparison of microbiota in pancreas and different gastrointestinal tissue was not conducted in these patients.
To date, no study to our knowledge has characterized the overall microbiome in pancreatic and normal surrounding tissue samples, a critical step to understand whether and how bacteria may play a role in carcinogenesis. In an effort to address the specific question of whether the pancreas has its own microbiome, we recruited subjects from the Rhode Island Hospital (Providence, RI) with planned foregut surgery to obtain tissue samples for 16S rRNA gene microbiome analysis. In addition, for comparison to controls, we obtained pancreatic and duodenum tissue from National Disease Research Interchange (NDRI) from non-cancer subjects.
Materials and Methods
Study population and sample collection
Seventy-seven subjects, enrolled between January 2014 and March 2016, were included in this study. Subjects were eligible if identified as candidates for surgery of the foregut by Dr. Charpentier (the lead surgeon at the RIH) and included those with pancreatic cancer, pancreatic cystic neoplasms, pancreatitis, bile duct or small bowel diseases. All recruited subjects were between 31–86 years old (Table 1). Participants were asked to complete a self-administered questionnaire to provide data on demographic and behavioral factors, and included a question on past use of antibiotics; this variable was included in the statistical analysis to control for changes that may have occurred due to antibiotic use in recent past. Questions on family history of cancer, use of other over-the-counter medications were also included on the questionnaire. Stool collection kits with ethanol as a fixative (95% (wt/wt) ethanol) were provided prior to surgery [23]; participants were asked to return the samples using a pre-paid box.
Table 1.
Distribution of demographic, lifestyle and health conditions variables among patients with diseases of the foregut, primarily pancreatic diseases, and deceased controls.
| RIH Subjects (n=77) | NDRI Subjects (n=34) | ||
|---|---|---|---|
| Characteristic | Mean (SD) | Characteristic | Mean (SD) |
| Age | 63 ± 13 | Age | 68 ± 15 |
| BMI | 27 ± 6 | BMI | 29 ± 6.5 |
| N (%) | N (%) | ||
| Sex | Sex | ||
| Male | 38 (49) | Male | 21 (62) |
| Female | 39 (51) | Female | 13 (38) |
| Race | Race | ||
| Caucasian | 72 (93.5) | Caucasian | 30 (88) |
| Black | 2 (2.6) | Black | 2 (6) |
| Other | 2 (2.6) | Other | 2 (6) |
| Smoking status | Smoking status | ||
| Ever smoker | 44 (58) | Ever smoker | 23 (68) |
| Chemotherapy | Cause of Death | ||
| Never | 52 (76.5) | Heart failure | 17 (50) |
| Prior to past 6 months | 7 (10.3) | Cardiopulmonary arrest | 5 (15) |
| In past 6 months | 9 (13.2) | Cerebrovascular accident | 1 (3) |
| Respiratory arrest | 2 (6) | ||
| Antibiotic use | Abdominal aortic aneurysm | 1 (3) | |
| Never | 13 (18.1) | Intracerebral hemorrhage | 1 (3) |
| Prior to past 6 months | 32 (44.2) | Liver cirrhosis | 1 (3) |
| In past 6 months | 21 (29.2) | Overdose | 1 (3) |
| Missing | 6 (8.3) | Parkinson’s disease | 1 (3) |
| Pneumonia | 1 (3) | ||
| Stent prior to surgery (yes) | 19 | Pulmonary embolism | 1 (3) |
| Pre-OP EUS | 20 | Pulmonary fibrosis | 1 (3) |
| Surgery for: | |||
| Pancreatic cancer | 51 (66.2) | ||
| Chronic pancreatitis or pancreatic cysts | 18 (23.4) | ||
| Other | 8 (10.4) | ||
A protocol was established for processing tissue samples collected during surgery to reduce contamination. A technician from the Pathology Department was informed in advance of the surgery date and time, and was paged as soon as the specimens had been obtained. Surgical tissue samples were frozen within one hour of the surgery time, as well as tissues swab samples from the stomach, jejunum, and bile duct that were collected using DNA-free forensic sterile swabs whenever possible. During surgery, the surgeon also recorded (on a surgery form for the study) if the patient had received prior pre-OP endoscopic ultrasound (EUS), had previously had their gallbladder removed, or had received prior placement of a stent (for treatment of symptoms); all subjects received a single dose of perioperative antibiotics immediately prior skin incision at the time of the operation. Tissue samples (pancreatic tumor tissue, pancreatic cysts, normal pancreatic tissue, pancreatic ducts and duodenums) were prepared by a Rhode Island Hospital pathologist; cancerous and non-cancerous tissues were identified, separated and labeled. All samples were de-identified and stored at −80°C until processing.
Upon review of pathology records, ICD10 codes were assigned to each subject; 39 subjects had pancreatic cancer (ICD10 codes C25.0-C25.9; the majority of cases were adenocarcinomas, only 2 subjects had neuroendocrine tumors of the pancreas), 12 subjects had periampullary cancer (ICD10 codes C24.0-C24.1), 18 subjects had other pancreatic conditions (ICD10 codes K86.0-K86.3), and the remaining 8 had other gastrointestinal conditions. The study was approved by Lifespan’s Research Protection Office for recruitment at RIH, as well as the Institutional Review Boards for Human Subjects Research at Brown University, Tufts University and the Forsyth Institute.
In addition, we obtained pancreatic specimens without known conditions of pancreatic diseases from the National Disease Research Interchange (NDRI) to serve as control samples in the absence of available healthy pancreatic tissue in non-cancer subjects. Snap-frozen ‘control’ whole-pancreas and duodenum (~ 5cm) human specimens from 34 deceased donors were obtained from NDRI with an average post-mortem recovery time of 13 hours. Control pancreas (head and tail), pancreatic ducts and duodenums were dissected under sterile conditions, and stored at −80°C until processing. To remove additional contamination, we removed a thin tissue layer around each sample prior to extracting DNA. Details for DNA extraction and sequencing procedures are provided in the Supplementary Methods.
16S rRNA amplicon Illumina sequencing
The 16S rRNA gene dataset consists of Illumina MiSeq sequences targeting the V3–V4 hypervariable regions. The DNA target sequencing was performed by the Forsyth Institute Sequencing Core. To evaluate effect of running samples on MiSeq runs at different times, we included bacterial mock community samples on each run and then compared their relative abundances across the MiSeq runs; the results for the mock communities were consistent across run, demonstrating minor fluctuations (Supplementary Figure 1).
The MiSeq reporter analysis was used to discard low quality sequences and to generate FASTQ files containing only filtered quality sequences, subsequently the overlapping paired-end reads were stitched together and further processed using used a multi-stage BLASTN-base search taxonomy read assignment pipeline that maximizes species level classification [24].
Taxonomic assignment pipeline of 16S rRNA amplicon sequencing data
Sequences were BLASTN-searched against a combined set of 16S rRNA reference sequences that consist of the HOMD (version 14.5)[25], Greengenes Gold [26], and the NCBI 16S rRNA reference sequence set. All assigned reads were subject to several down-stream bioinformatics analyses, including alpha and beta diversity assessments, provided in the QIIME (Quantitative Insights Into Microbial Ecology [27]) software package version 1.9.1.
Statistical analysis
Samples with < 500 total read counts were excluded from all analysis. In addition, only OTUs with a minimal read count of 100 sequences (across all samples) were included in the analyses. For QIIME analyses, we normalized the number of sequences in the different MiSeq runs by rarefying each library to 500 reads to account for differences in sequencing depth across runs (increasing rarefaction cutpoint to higher read number did not result in changes in alpha-diversity results or OTU numbers in samples; 500 reads was used as the cutpoint to reduce number of samples lost from the analysis). Range of sequencing counts for the different sample types are provided in Supplementary Figure 2. Across samples, OTU relative abundance was computed as the ratio of an OTU’s absolute abundance to the total number of reads for that sample.
To create relative abundance plots, we restricted bacterial taxa (at genus-level) present at >2% relative abundance and with >35% prevalence in both NDRI and RIH samples (this was done to simplify comparison between the RIH and NDRI samples). Jaccard Index was used for paired comparison of proportion of shared microbiota taxa present at >2% relative abundance in tissue/swab samples within subjects.
To examine the variation in the microbial profile across the different habitats/sites (Supplemental Table 1) among the NDRI and RIH subjects, we calculated the distance/dissimilarity between samples using the Bray-Curtis and Sorensen indices [28]. Computed distances were subsequently used to generate principal coordinate analysis (PCoA) plots to visualize the arrangement of the samples in the ordination space. PERMANOVA (available in QIIME) was used to test whether the distances are more similar within a group of samples than that from other groups of samples.
To identify demographic and clinical correlates of pancreatic microbial composition, we fit a series of zero-inflated beta regression models to examine associations between genus-level relative abundances and demographic (i.e., age, gender, race, BMI) and clinical (i.e., health status, chemotherapy, antibiotics use prior to surgery, anxiety medications, presence of stent prior to surgery, whether pre-operative endoscopic ultrasound [pre-OP EUS] was conducted prior to surgery, tumor surgery classification by International Code of Disease [ICD10 code]). In the results, we refer to relative mean abundance among non-zero observations (μ) merely as relative mean abundance. More details are provided in the Supplemental Methods.
We explored which factors obtained in the questionnaires and medical files in RIH subjects were associated with bacterial communities in the pancreatic tissue samples. The most influential factors were sequencing run, presence of stent, and chemotherapy prior to surgery (only 5 patients with available tissue/swab samples had chemotherapy in the past 6 months); each of these factors was significantly associated with a large number of genera tested in marginal models. Given that the mock bacterial communities were similar across runs (see Supplemental Materials), it is possible that “run” was associated with certain genera due to differences in number of samples per sequencing run. To adjust for potential confounding, we considered this covariate in the final models comparing cancer to non-cancer subjects and the different ICD-codes among the RIH subjects. Similarly, age, BMI and sex were adjusted for as these features were shared between the studies and were found to explain variation in the relative abundance of some of the genera. Smoking was not found to explain variation in relative abundance in our data.
Results
The present analysis included a total of 246 pancreatic tissue and swab samples collected from 82 subjects (50 subjects from RIH providing 133 samples [57 swabs, 76 tissue] and 34 subjects from NDRI providing 113 tissue samples; Supplemental Table 1). In addition, 12 RIH subjects provided stool samples. There were no significant differences in the distribution of age, gender, BMI, race, and smoking status between RIH and NDRI subjects (Table 1). The Illumina-based sequencing of V3–V4 hypervariable regions of the bacterial 16S rRNA gene resulted in a total of 19,498,743 high quality sequences (with a median sequence length of 427 nucleotides).
Taxonomy
Over 99% of the reads from RIH pancreatic samples were attributed to 5 bacterial phyla (45.9% Proteobacteria, 35.6% Firmicutes, 9.5 % Bacteroidetes, 4.3% Fusobacteria, and 3.9% Actinobacteria). The remaining low abundance phylotypes (0.6% of the total) belonged to six bacterial phyla (Synergistetes, TM7, Deinococcus-Thermus, Verrucomicrobia, Spirochaetes, and Tenericutes). 99.6% of the reads observed among the NDRI pancreatic samples belonged to the same five bacterial phyla as observed in RIH subjects. The phylum Tenericutes (Bacteria) was present only in RIH samples, and the phylum Euryarchaeota (Archaea) was present only in NDRI samples, but both of these phyla were uncommon.
While the microbial communities in the pancreatic tissues were dominated by the phyla Firmicutes and Proteobacteria, substantial inter-individual variability was observed. In RIH samples, Proteobacteria relative abundance ranged from 2 to 99%, and similarly, Firmicutes relative abundance ranged from 0.6 to 84%. Large inter-individual variability was also observed in the NDRI samples.
Within and between sample diversity analysis
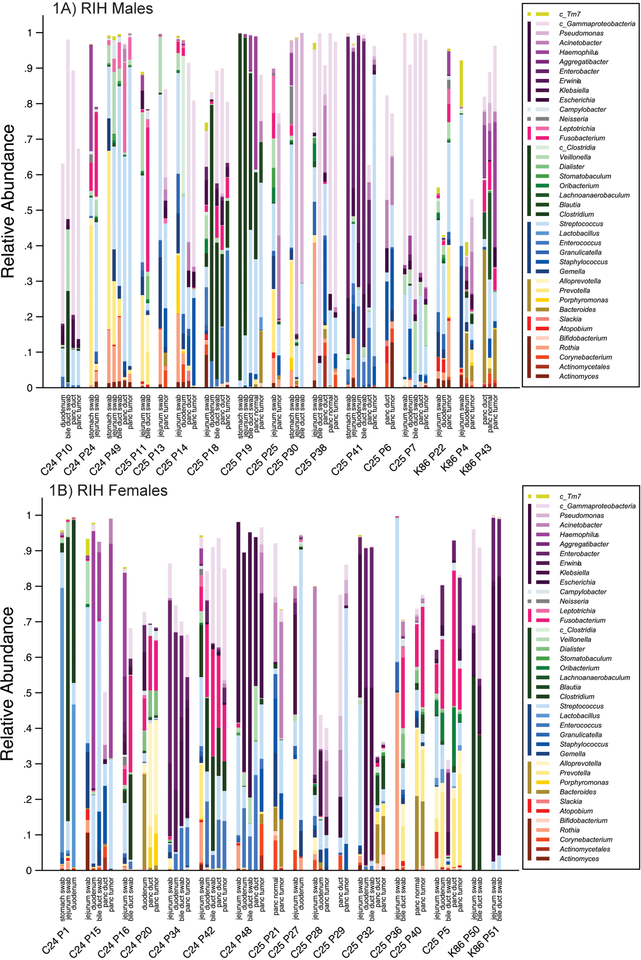
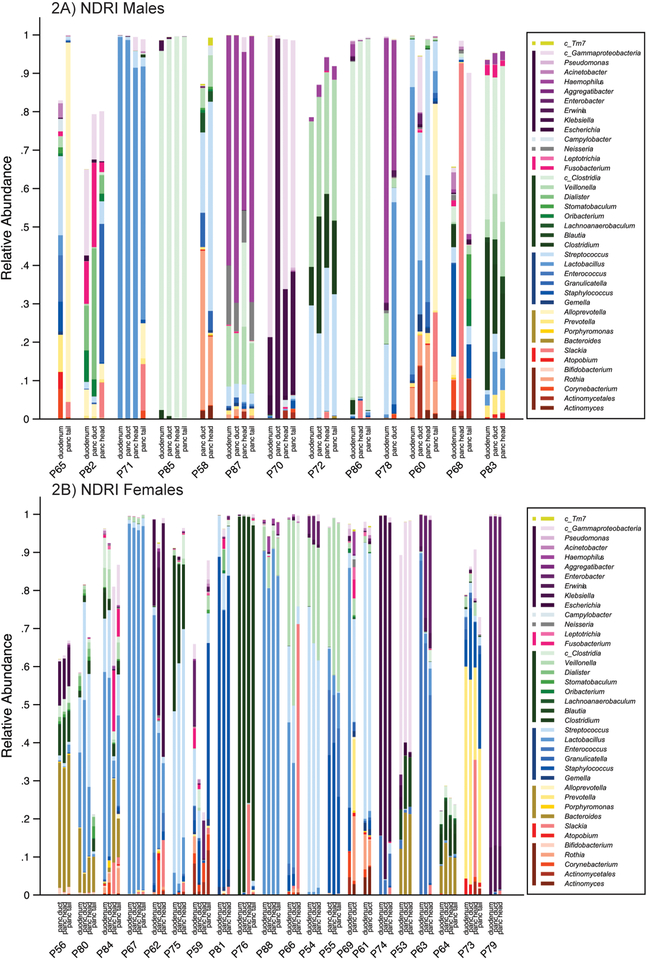
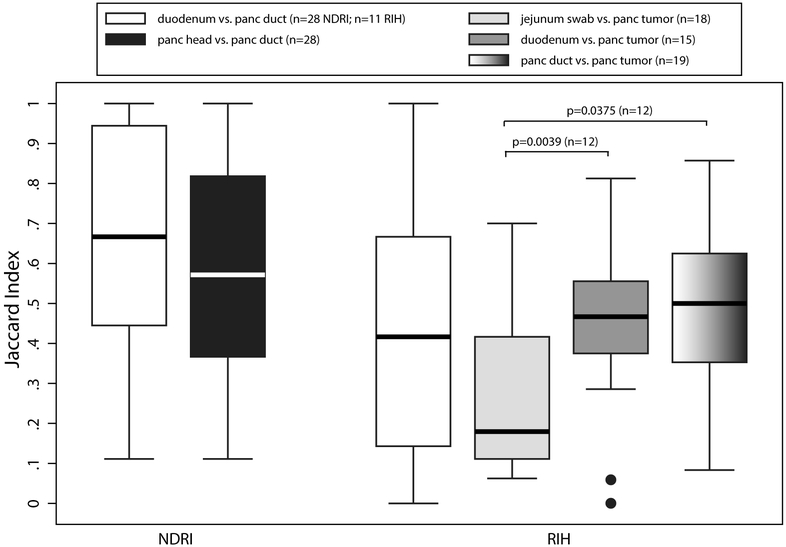
Mean relative abundance for bacterial taxa (mostly at the genus-level) in the pancreatic tissue samples (duct, head, tail, normal and tumor), duodenum tissue samples, and jejunum, bile duct and stomach swabs are presented for each subject with more than one available sample in the RIH Figure 1 and NDRI in Figure 2 (males and females are presented separately for ease of comparison – no major differences were observed by sex). Three striking patterns emerge: 1) bacterial profiles in the pancreas are subject-specific rather than site-specific, 2) bacterial profiles in duodenum tissue are remarkably similar to those in pancreatic tissue in the same subjects, 3) concordance of paired comparisons of bacterial profiles in cancer subjects (RIH) are slightly lower across tissue type or site than those for non-cancer subjects (NDRI) (Figure 3). Subjects from RIH with only one sample available (n=5) demonstrate similar bacterial profiles as those with multiple samples. Bacterial taxa commonly recognized as oral bacteria, including Fusobacterium spp., Prevotella spp., Dialister spp., Veillonella spp., and Haemophilus spp. were identified in many of the tissue samples, both cancer and non-cancer subjects (Figure 1). Other oral bacterial taxa, including Parvimonas micra, Selenomonas noxia, Capnocytophaga spp., Peptostreptococcus spp. and Solobacterium moorei were also identified in tissue samples but were less common (present in 20%−35% of all samples).
Figure 1.
(A) RIH Males with ICD code (C25, C24 or K86); (B) RIH Females with ICD code (C25, C24, or K86). Distribution of bacteria relative abundance by genus level in all the studied body habitats based on read taxa attribution using V3–V4 hypervariable region of 16S rRNA genes. All names are at genera level except for those with c_ which denotes class for multigenera taxa (within that class). Colored bars next to legend reflect taxa at class level: TM7 (lime); Gammaproteobacteria (purple); Epsilonproteobacteria (light grey); Betaproteobacteria (dark grey); Fusobacteriia (pink); Clostridia (green); Bacilli (blue); Bacteroides (Gold); Coriobacteriia (red); Actinobacteria (marron).
Figure 2.
(A) NDRI Males; (B) NDRI Females. Distribution of bacteria relative abundance by genus level in all the studied body habitats based on read taxa attribution using V3–V4 hypervariable region of 16S rRNA genes. All names are at genera level except for those with c_ which denotes class for multigenera taxa (within that class). Colored bars next to legend reflect taxa at class level: TM7 (lime); Gammaproteobacteria (purple); Epsilonproteobacteria (light grey); Betaproteobacteria (dark grey); Fusobacteriia (pink); Clostridia (green); Bacilli (blue); Bacteroides (Gold); Coriobacteriia (red); Actinobacteria (marron).
Figure 3.
Jaccard Index (proportion of shared genera) for paired comparison of tissue samples in NDRI and RIH subjects.
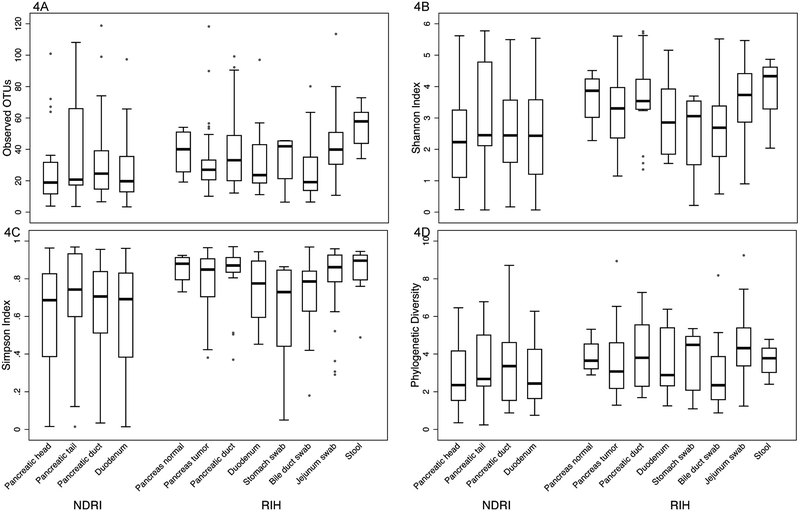
With the exception of the stool and the jejunum, all the bacterial communities were characterized as habitats with low bacterial richness including the pancreatic sites, duodenums and the bile ducts (Figure 4A). Among RIH subjects, the microbial communities of the stool samples were represented by higher richness than the microbial communities in the tumors of the pancreas (p=0.007), duodenums (p=0.013) and bile duct swabs (p=0.017). Likewise, the stool bacterial communities had higher richness than the NDRI pancreatic heads (p=0.012), pancreatic ducts (p=0.020) and duodenums (p=0.005). The microbial communities in the jejunum swabs showed more richness than the communities in the RIH pancreatic head (p=0.014) and duodenums (p=0.028). In general, the bacterial communities in the pancreas of RIH subjects had slightly higher richness when compared to those from the pancreas of the NDRI matching sample types. Similar results were observed using additional alpha diversity measures of the bacterial communities (Figure 4B–D). As expected, the stool samples were the most diverse with a Shannon index ≥ 4 (Figure 4B). As the number of phyla represented in high abundance in these samples was relatively low (~5), we observed relatively low levels of phylogenetic distances across all samples (Figure 4D).
Figure 4.
Comparative alpha diversity analyses of bacterial communities in anatomical sites (based on a simulated data set subsampled from the input OTU table). Alpha diversity metrics: (A) Richness, (B) Shannon diversity index, (C) Simpson index, and (D) Phylogenetic diversity.
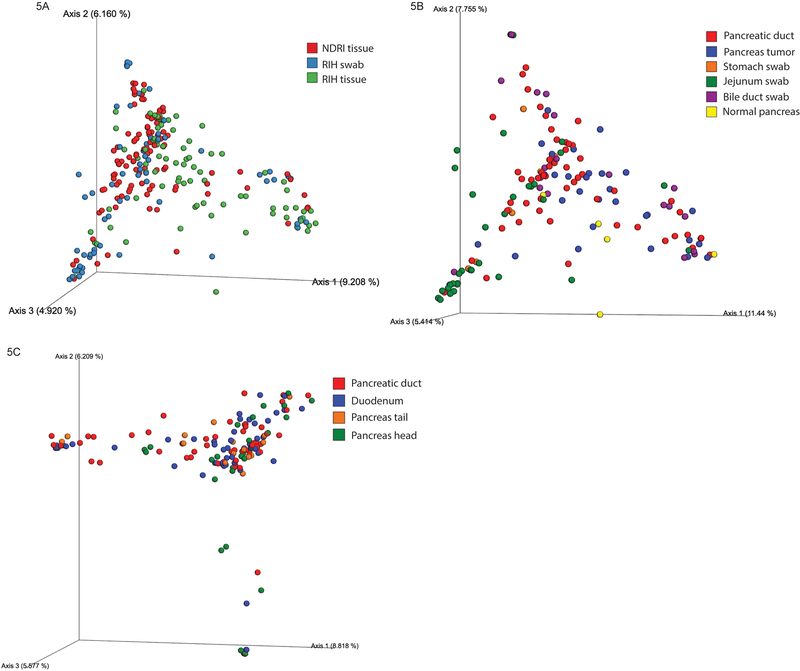
The ordination beta-diversity analysis revealed that the majority of samples belonged to a single cluster, without any visually apparent groupings by the nature of the sample, health status or anatomical site (Figure 5A–C). However, the PERMANOVA tests revealed statistically significant differences between NDRI and RIH samples (p<0.001), and for the swab samples obtained from the bile duct, jejunum and stomach (compared to pancreas tissue samples). Differences between sites within the pancreas (i.e., head, tail, duct), and compared to the duodenum (for NDRI and RIH, separately), were not statistically significant (after accounting for multiple comparisons). The principal component analyses of both Bray-Curtis and Sorensen distances between all samples (tissues and swabs) showed that both RIH and NDRI samples clustered mostly by subject.
Figure 5.
PCoA plots showing the relatedness of microbial communities among samples from RIH subjects and NDRI donors using the Bray-Curtis dissimilarity index. Individual datasets are colored according to their (A) RIH and NDRI sample type, (B) RIH anatomical site, and (C) NDRI anatomical site.
Associations of host factors with microbial communities
Using multiple regression analyses, we examined presence or absence, and relative mean abundance of bacterial taxa (at the genus and species level) among present (non-zero) observations using all tissue and swab samples comparing RIH subjects to NDRI subjects. Table 2 presents the bacterial taxa (at the genus level) that were present in at least 20% of all tissue and swab samples; each model includes both a zero-inflated component (testing for differences in presence/absence of bacterial taxa) and a relative mean abundance comparison. Lactobacillus taxa were present in almost all non-cancer tissue samples (estimated proportion of presence [P1]=0.98), but were much less likely to be present in cancer tissue samples (P1=0.58, p<0.0001), and mean relative abundance was higher in non-cancer subjects (μ=0.06 vs μ=0.02 in RIH subjects, p<0.0001; Table 2). In contrast, a number of bacterial taxa, including Porphyromonas, were present in higher mean relative abundance in cancer subjects than non-cancer subjects (Table 2, and species level data presented in Supplemental Table 2). Oral bacteria Fusobacterium spp. and Prevotella spp. had higher mean relative abundance in cancer subjects than non-cancer subjects (p-values <0.0001 according to Wald tests for μ). Although these two bacteria do not appear in Table 2 because they were not significant according to the joint permutation (based test for prevalence and mean relative abundance at the genus-level), a number of Fusobacterium species, e.g., Fusobacterium nucleatum _subsp._vincentii, were much more prevalent in RIH samples and are significant in the species-level models (Supplemental Table 2).
Table 2.
Results from multivariable zero-inflated beta regression models comparing bacteria presence/absence and relative abundance in tissue and swab samples from NDRI and RIH subjects*
| Genus | Total Read Counts | Non-zero Samples | Estimated Mean Relative Abundance (μ)** | Estimated Proportion of Presence (P1) | Global Perm Test* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RIH | NDRI | Wald p-value | RIH | NDRI | Wald p-value | p-value | p-adjusted^ | AIC difference | |||
| Lactobacillus | 1075844 | 154 | 0.0209 | 0.0640 | < 0.0001 | 0.5270 | 0.9846 | < 0.0001 | < 0.002 | < 0.02 | 35.22 |
| Pseudomonas | 137618 | 90 | 0.0077 | 0.0026 | < 0.0001 | 0.5523 | 0.2480 | 0.0001 | < 0.002 | < 0.02 | 13.55 |
| Parvimonas | 45705 | 73 | 0.0091 | 0.0048 | 0.0157 | 0.5251 | 0.1813 | < 0.0001 | < 0.002 | < 0.02 | 10.41 |
| Acinetobacter | 13915 | 152 | 0.0083 | 0.0040 | < 0.0001 | 0.7606 | 0.5069 | 0.0008 | < 0.002 | < 0.02 | 10.12 |
| Ralstonia | 532 | 61 | 0.0001 | 0.0002 | < 0.0001 | 0.1687 | 0.4098 | 0.0010 | < 0.002 | < 0.02 | 8.24 |
| Kluyvera | 36883 | 54 | 0.0097 | 0.0045 | 0.0353 | 0.2536 | 0.0361 | < 0.0001 | < 0.002 | < 0.02 | 6.94 |
| Bilophila | 102226 | 60 | 0.0070 | 0.0013 | < 0.0001 | 0.2000 | 0.0197 | 0.0001 | < 0.002 | < 0.02 | 2.95 |
| Gemella | 76895 | 133 | 0.0193 | 0.0064 | < 0.0001 | 0.7648 | 0.6134 | 0.0249 | 0.004 | 0.03 | 5.21 |
| Slackia | 34060 | 82 | 0.0084 | 0.0266 | < 0.0001 | 0.2400 | 0.3958 | 0.0223 | 0.008 | 0.05 | 4.72 |
| Lachnoanaerobaculum | 19696 | 92 | 0.0037 | 0.0029 | 0.2536 | 0.6163 | 0.2944 | 0.0008 | 0.012 | 0.07 | 0.80 |
| Solobacterium | 16204 | 67 | 0.0069 | 0.0019 | < 0.0001 | 0.5132 | 0.2351 | 0.0041 | 0.014 | 0.07 | 0.02 |
| Blautia | 156957 | 92 | 0.0030 | 0.0027 | 0.4899 | 0.3081 | 0.4915 | 0.0193 | 0.020 | 0.09 | 1.62 |
| Porphyromonas | 20741 | 86 | 0.0046 | 0.0028 | 0.0192 | 0.3639 | 0.5223 | 0.0465 | 0.022 | 0.10 | 0.86 |
| Anaerococcus | 49115 | 63 | 0.0053 | 0.0069 | 0.2285 | 0.1708 | 0.3281 | 0.0164 | 0.026 | 0.11 | 0.56 |
| Selenomonas | 2407 | 73 | 0.0002 | 0.0002 | 0.1128 | 0.4335 | 0.2425 | 0.0205 | 0.038 | 0.15 | 0.86 |
| Staphylococcus | 303413 | 196 | 0.0105 | 0.0175 | 0.0001 | 0.8357 | 0.9146 | 0.1077 | 0.042 | 0.15 | 5.99 |
| Megasphaera | 21221 | 69 | 0.0028 | 0.0017 | 0.0266 | 0.4267 | 0.2150 | 0.0134 | 0.046 | 0.15 | −1.45 |
| Actinomyces | 34042 | 153 | 0.0064 | 0.0034 | < 0.0001 | 0.8013 | 0.7065 | 0.1642 | 0.052 | 0.15 | −1.62 |
| Prevotella | 222237 | 179 | 0.0240 | 0.0125 | < 0.0001 | 0.8790 | 0.9046 | 0.5502 | 0.052 | 0.15 | −1.70 |
| Bifidobacterium | 12181 | 88 | 0.0021 | 0.0016 | 0.1358 | 0.3098 | 0.1140 | 0.0073 | 0.052 | 0.15 | −2.33 |
| Abiotrophia | 1086 | 43 | 0.0008 | 0.0001 | < 0.0001 | 0.1502 | 0.0666 | 0.0421 | 0.054 | 0.15 | −1.42 |
| Rothia | 227122 | 173 | 0.0274 | 0.0166 | 0.0022 | 0.9458 | 0.8861 | 0.0537 | 0.080 | 0.22 | −0.24 |
All models are adjusted for age, sex, BMI and log library size. Only bacteria (at genus-level) associated with source of samples at p<0.10 before correcting for multiple comparisons are shown. Permutation testing accounts for within subject correlation via random intercept.
Among non-zero samples.
Adjusted for multiple testing
| Table 2 Full OTU (in same order) |
|---|
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Lactobacillaceae;g__Lactobacillus |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;g__Pseudomonas |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Peptostreptococcaceae_[XIII];g__Parvimonas |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Acinetobacter |
| k__Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Burkholderiaceae;g__Ralstonia |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__Kluyvera |
| k__Bacteria;p__Proteobacteria;c__Deltaproteobacteria;o__Desulfovibrionales;f__Desulfovibrionaceae;g__Bilophila |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Gemellaceae;g__Gemella |
| k__Bacteria;p__Actinobacteria;c__Coriobacteriia;o__Eggerthellales;f__Eggerthellaceae;g__Slackia |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Lachnospiraceae;g__Lachnoanaerobaculum |
| k__Bacteria;p__Firmicutes;c__Erysipelotrichi;o__Erysipelotrichales;f__Erysipelotrichaceae;g__Solobacterium |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Lachnospiraceae;g__Blautia |
| k__Bacteria;p__Bacteroidetes;c__Bacteroides;o__Bacteroidales;f__Porphyromonadaceae;g__Porphyromonas |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Peptostreptococcaceae_[XIII];g__Anaerococcus |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Veillonellaceae;g__Selenomonas |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Staphylococcaceae;g__Staphylococcus |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Aerococcaceae;g__Abiotrophia |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Veillonellaceae;g__Megasphaera |
| k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Actinomycetaceae;g__Actinomyces |
| k__Bacteria;p__Bacteroidetes;c__Bacteroides;o__Bacteroidales;f__Prevotellaceae;g__Prevotella |
| k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Bifidobacteriales;f__Bifidobacteriaceae;g__Bifidobacterium |
| k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Micrococcaceae;g__Rothia |
Table 3 presents the bacterial taxa (at genus level) for which statistically significant associations remained after multiple comparison correction (at p<0.10) when comparing bacterial taxa in tumor tissue (RIH) by ICD code to those identified in normal pancreatic head tissue from NDRI subjects (labeled as “controls” in Table 3; given that the bacterial profiles were highly similar by subject, we included only pancreatic head tissue for this analysis). In the marginal models (prior to adjusting for other covariates), a total of 16 bacterial genera were identified as being significantly associated with disease status prior to correction for multiple comparisons (Supplemental Table 3); a number of these taxa have representative strains in the Human Oral Microbiome Database (http://www.homd.org) (e.g., Fusobacterium, Capnocytophaga, Prevotella, Porphyromonas, Parvimonas, Selenomonas, and Haemophilus). Mean relative abundances for some of these taxa (namely, Capnocytophaga, Prevotella, Selenomonas) were higher in samples coming from subjects diagnosed with pancreatic cancer (ICD C25) compared to NDRI samples. The model with Porphyromonas had the strongest association overall (p=4.5 × 10−7); the relative mean abundance for periampullary cancer tissue samples was substantially higher than that of NDRI samples (p=5.8 × 10−19), as were the IPMNs (K86.2) samples (p=3.6 × 10−7). The associations with Porphyromonas remained elevated in multiple regression models (Table 3).
Table 3.
Results from multivariable zero-inflated beta regression models comparing bacteria presence/absence and relative abundance across subject disease ICD codes*
| Genus | Estimated Mean Relative Abundance (μ)** | Estimated Proportion of Presence (P1) | Likelihood Ratio Tests | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total read counts | Non-zero samples | Control (N=29) | C24 (N=7) |
C25 (N=16) |
K86.2 (N=6) |
Control (N=29) |
C24 (N=7) |
C25 (N=16) |
K86.2 (N=6) |
p-value | AIC difference | p-adjusted^ | |
| Simonsiella | 231 | 15 | 0.0001 | 0.0001 | 0.0018 | 0.0112 | 0.1640 | 0.5559 | 0.5140 | 0.1574 | <0.0001 | 35.41 | <0.0001 |
| Helicobacter | 1475 | 7 | 0.4670 | <0.0001 | 0.0001 | 0.0001 | 0.0079 | 0.6287 | 0.0005 | 0.0039 | <0.0001 | 35.38 | <0.0001 |
| Porphyromonas | 7008 | 15 | 0.0005 | 0.0512 | 0.0022 | 0.0087 | 0.3886 | 0.1261 | 0.0351 | 0.3175 | <0.0001 | 29.01 | <0.0001 |
| Capnocytophaga | 316 | 12 | <0.0001 | <0.0001 | 0.0001 | 0.0017 | 0.1375 | 0.2673 | 0.0865 | 0.3766 | <0.0001 | 28.87 | <0.0001 |
| Ralstonia | 153 | 15 | 0.0008 | 0.0061 | 0.0452 | 0.0010 | 0.2955 | 0.0008 | 0.6080 | 0.6144 | <0.0001 | 17.80 | 0.0026 |
| Bilophila | 10685 | 13 | 0.0002 | 0.0663 | 0.0139 | 0.0002 | 0.0696 | 0.0547 | 0.0099 | 0.1653 | <0.0001 | 17.76 | 0.0027 |
| Pseudomonas | 64128 | 31 | 0.0131 | 0.0231 | 0.0469 | 0.5237 | 0.2139 | 0.8512 | 0.6192 | 0.6193 | 0.0001 | 15.35 | 0.0076 |
| Acinetobacter | 7916 | 43 | 0.0192 | 0.0615 | 0.1637 | 0.1208 | 0.4407 | 1.0000 | 0.7870 | 0.7825 | 0.0002 | 13.81 | 0.0147 |
| Gemella | 7769 | 24 | 0.0028 | 0.0113 | 0.0113 | 0.0071 | 0.2778 | 0.5284 | 0.8466 | 1.0000 | 0.0006 | 11.83 | 0.0343 |
| Enterococcus | 28254 | 29 | 0.0230 | 0.0176 | 0.0067 | 0.0173 | 0.4277 | 1.0000 | <0.0001 | 0.4857 | 0.0007 | 11.35 | 0.0419 |
| Propionibacterium | 19 | 10 | <0.0001 | <0.0001 | 0.0001 | <0.0001 | 0.1589 | 0.1454 | 0.0105 | 0.0597 | 0.0011 | 10.29 | 0.0655 |
| Peptoclostridium | 4137 | 14 | 0.0031 | 0.0191 | 0.1199 | 0.0088 | 0.2598 | 0.0003 | 0.2297 | 0.5380 | 0.0017 | 9.19 | 0.1034 |
| Solobacterium | 1402 | 10 | 0.0001 | 0.0002 | 0.0006 | 0.0003 | 0.2789 | 0.8483 | 0.1583 | 0.1326 | 0.0038 | 7.20 | 0.2340 |
| Salmonella | 37 | 7 | 0.0003 | 0.6282 | 0.0001 | 0.0015 | 0.0336 | 0.0159 | 0.0080 | 0.1074 | 0.0089 | 5.09 | 0.5455 |
| Lactobacillus | 251585 | 40 | 0.1343 | 0.0717 | 0.1408 | 0.0891 | 0.9565 | 0.6933 | 0.4354 | 1.0000 | 0.0146 | 3.85 | 0.8897 |
| Enterobacter | 64118 | 30 | 0.0374 | 0.0323 | 0.0303 | 0.0140 | 0.4467 | 0.6746 | <0.0001 | 0.2106 | 0.0177 | 3.35 | 1.00 |
| Lactococcus | 3592 | 17 | 0.0018 | 0.0818 | 0.8557 | 0.0009 | 0.2025 | 0.0684 | 0.0855 | 0.2273 | 0.0194 | 3.12 | 1.00 |
| Clostridium | 100517 | 29 | 0.0643 | 0.0459 | 0.0416 | 0.0697 | 0.4600 | 0.7216 | 0.0470 | 0.5243 | 0.0267 | 2.27 | 1.00 |
| Bacteroides | 153955 | 34 | 0.0186 | 0.0171 | 0.0818 | 0.0470 | 0.3719 | 0.8799 | 0.7322 | 0.7174 | 0.0381 | 1.33 | 1.00 |
| Raoultella | 31688 | 9 | 0.0593 | 0.1574 | 0.0041 | 0.0081 | 0.0010 | 0.0040 | <0.0001 | <0.0001 | 0.0523 | 0.47 | 1.00 |
All models are adjusted for age, sex, BMI and sequencing run. Only bacteria (at genus-level) associated with ICD code (overall) at p≤0.05 prior to correcting for multiple comparisons are shown. Due to missing BMI on two individuals, numbers for the fully-adjusted models were based on 58 tissue samples. Marginal models with all samples are shown in Supplemental Table 3.
Among non-zero samples.
Adjusted for multiple testing.
| Table 3 Full OTU (in same order) |
|---|
| k__Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Neisseriales;f__Neisseriaceae;g__Simonsiella |
| k__Bacteria;p__Proteobacteria;c__Epsilonproteobacteria;o__Campylobacterales;f__Helicobacteraceae;g__Helicobacter |
| k__Bacteria;p__Bacteroidetes;c__Bacteroides;o__Bacteroidales;f__Porphyromonadaceae;g__Porphyromonas |
| k__Bacteria;p__Bacteroidetes;c__Flavobacteria;o__Flavobacteriales;f__Flavobacteriaceae;g__Capnocytophaga |
| k__Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Burkholderiaceae;g__Ralstonia |
| k__Bacteria;p__Proteobacteria;c__Deltaproteobacteria;o__Desulfovibrionales;f__Desulfovibrionaceae;g__Bilophila |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;g__Pseudomonas |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Acinetobacter |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Gemellaceae;g__Gemella |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Enterococcaceae;g__Enterococcus |
| k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Propionibacteriaceae;g__Propionibacterium |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Peptostreptococcaceae;g__Peptoclostridium |
| k__Bacteria;p__Firmicutes;c__Erysipelotrichi;o__Erysipelotrichales;f__Erysipelotrichaceae;g__Solobacterium |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Enterobacteriaceae;g__Salmonella |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Lactobacillaceae;g__Lactobacillus |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__Enterobacter |
| k__Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Streptococcaceae;g__Lactococcus |
| k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Clostridiaceae;g__Clostridium |
| k__Bacteria;p__Bacteroidetes;c__Bacteroides;o__Bacteroidales;f__Bacteroidaceae;g__Bacteroides |
| k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Enterobacteriaceae;g__Raoultella |
The multivariable regression models for the pancreatic tissue samples identified bacterial taxa (at the genus-level) that had not been significant in the marginal regression models, including Simonsiella, Helicobacter, and Bilophia (Table 3 vs Supplemental Table 3). Helicobacter was commonly identified in periampullary pancreatic tumors (C24) but at very low levels; in contrast, Helicobacter was infrequently identified in the NDRI samples, but was a dominant genus when present (relative mean abundance 47%; Table 3).
We further examined the RIH pancreatic tumor tissue samples without the NDRI samples given the difference in source of tissue and to account for clinical factors such as prior chemotherapy. Porphyromonas were also strongly associated with ICD code in both marginal (Supplemental Table 4) and multiple regression models suggesting clinical covariates were not confounding the main findings for these bacteria.
To test whether the associations would be similar using pancreatic duct tissue samples (vs tumor tissue), we repeated the analysis using RIH and NDRI samples obtained from the pancreatic ducts. The associations for Porphyromonas remained detectable and statistically significant in these analyses (p=1.53 × 10−11).
Using tissue samples obtained from the duodenum, we compared relative abundance of bacterial taxa in NDRI and RIH subjects to examine whether any bacteria from the pancreatic tissue analyses were also noticeably different in the duodenum samples. Of the significant associations noted in the pancreatic tissues, Selenomonas was also elevated in the duodenum tissue of pancreatic cancer subjects compared to duodenum tissue from NDRI subjects (p=3.9 × 10−12). A weak association was also observed for Gemella for the duodenum samples, consistent with an overall elevated mean relative abundance in the RIH samples compared to the NDRI samples (Table 2); other associations were either not significant or not consistent in direction of differences.
We only had one pancreatic duct stent to examine microbial community; the bacterial taxa from this stent were characterized as the members of the genera Klebsiella and Enterobacter.
Discussion
Using pancreatic and duodenum tissue samples from subjects with pancreatic cysts or pancreatic cancer, and comparing them to pancreatic tissue samples obtained from donors who died of non-cancer causes, we were able to demonstrate that pancreatic tissue contains a number of different bacterial taxa, including taxa that are known to inhabit the oral cavity. Our findings provide evidence that the pancreas is not a sterile organ and that there is substantial between-person variability in relative abundance of bacterial taxa at the genera level in the pancreas, but we also observed marked within-person stability across site (Figures 1 and 2); bacterial composition at different sites in the pancreas (i.e., duct, head and tail) as well as the duodenum were highly similar in the same individuals. Finally, we noted lower presence and relative abundance of Lactobacillus in cancer subjects compared to non-cancer subjects, and a significant increase in the mean relative abundance of periodontal-related pathogens in the tissue of pancreatic subjects when compared to non-cancer subjects.
Dissemination of oral bacteria to different parts of the body has been well-reported, and oral bacteria have been linked to a number of chronic diseases, including cardiovascular diseases [29, 30]. Fusobacterium nucleatum has been associated with colon cancer in a number of cross-sectional studies [31, 32]. Mouse models of colorectal cancer provide some support for a causal link [17, 33], demonstrating how this bacterium has the ability to initiate recruitment of tumor-infiltrating immune cells. Moreover, a recent study demonstrated similar microbiome profiles in primary colon cancer tumors and liver metastases from the same individuals (resected at a later time point), especially for Fusobacterium positive tumors [17], suggesting stability in the microbiome as the tumor progresses and metastasizes. Given the findings from this study, where multiple tissue specimens were examined in the same subjects, it may also be plausible that each individual has a unique microbiome profile that exists in different gastrointestinal tissue and that certain profiles increase cancer susceptibility by impacting the immune environment to allow for tumor promotion and growth. Bacterial taxa found in this study were highly consistent with those reported in a microbiome study on colon cancer; enriched bacterial taxa associated with Fusobacterium nucleatum positive tumors were similar to those we identified in this study (e.g. Bacteroides, Prevotella, Selenomonas, and Leptotrichia)[17].
Presence of Lactobacillus spp. was significantly reduced in both periampullary and PDAC cancers compared to non-cancer patients (including those with pancreatic cysts). Certain strains of this bacterium have been identified as playing a key role in mediating anti-inflammatory pathways in calorie-restricted mice [34]. Further research on the role of these bacteria in pancreatic cancer should be conducted.
Previous studies have reported associations between periodontal disease pathogens and pancreatic cancer risk, especially Porphyromonas gingivalis [6, 7]. Periodontal disease is an inflammatory disease of the gums that can, in advanced conditions of periodontitis, result in systemic inflammation. In this study, we observed significantly higher mean relative abundance levels (at the genus-level) for two bacterial taxa previously associated with periodontitis in pancreatic tissue, including Porphyromonas and Selenomonas [35–38]; however, only Porphyromonas remained statistically significant after adjusting for age, sex, BMI and library size. Porphyromonas was also elevated in the pancreatic duct tissue of periampullary pancreatic cancers, but no statistically significant associations were noted for the other oral bacterial taxa. Whether Porphyromonas play a role in pancreatic carcinogenesis will need to be further examined in other studies and confirmed in animal models. Proposed mechanisms for carcinogenesis include the ability of certain bacteria to induce a pro-inflammatory response in the tumor microenvironment [33]; inhibit the immune response targeted at eliminating tumor cells [39]; and modulate key cellular pathways associated with cell division [40].
A similar study using swab specimens from the pancreas, bile and jejunum, was conducted on subjects with pancreatic cancer undergoing pancreaticoduodenectomy [13]. In that study, many bacterial taxa were present in fluids obtained from the pancreatic ducts and the common bile duct, including Prevotella, Haemophilus, Aggregatibacter, and Fusobacterium [13]. Consistent with our findings, microbial communities in the pancreas, bile and jejunum fluids were similar within individuals [13]. Mean relative abundance for the bacterial genus Klebsiella was high in the samples from pancreatic cancer subjects in that study [13]; in our study, we found Klebsiella to be one of two taxa on a swab taken from the stent itself. Placement of stent prior to surgery may impact the type of bacteria present in the pancreas, as observed in our study. In a separate study, metagenomics was conducted on freshly frozen duodenum samples from 5 normal and 5 obese individuals; Streptococcus (30–32%) and Actinomyces (12–17%) were the most common bacterial taxa identified in those samples, and relatively higher counts of Gemella were also identified in all 10 subjects [41]. Porphyromonas were not identified in the duodenal samples [41].
In a recent study examining tumor resistance to the drug gemcitabine, bacteria were found in tumor tissues of 65 PDAC patients (out of 113), and 51.7% of bacterial taxa belonged to the class Gammaproteobacteria [22], which is highly consistent with our findings (Figure 1). Similar to our study, there was large inter-individual variability in relative abundance of bacteria in each tumor, but in contrast to our study, only 3 out of 20 organ donors were found to be positive for bacteria, and no normal tissue samples were included from the same patients [22]. In addition, a high number of reads for Porphyromonas was found in one (out of 65) pancreatic cancer tissue specimens (mean relative abundance of 0.123; Supplementary Material [22]). In our study, read counts for Porphyromonas spp. were also extremely high in two RIH subject. In a separate study, 408 genera of bacteria were identified in pancreatic cyst fluids obtained from patients through endoscopy [14]; many of the taxa found in pancreatic cysts were similar to those in tissue from our study, including the presence of Fusobacterium. Furthermore, Porphyromonas was present in 33% of fluid samples and relative abundances for those taxa were similar to those in our study (non-zero cysts mean relative abundance: 0.00178, range 0.0001–0.004) [14].
In a recent study, Bifidobacterium spp. was found to increase in abundance in the feces of mice with Kras mutations (genetically modified to increase pancreatic cancer) as disease progressed, compared to wildtype mice [15]. Furthermore, gut repopulation of the germ-free (Kras) mouse with Bifidobacterium pseudolongum increased T-cell infiltration and tumor growth [15]. Similarly, we also noted a higher prevalence for the genus Bifidobacterium in cancer subjects compared to non-cancer subjects (Table 2).
Several studies have looked at the involvement of bacteria in biliary and pancreatic diseases and have observed a high number of bacterial taxa present in the calcified pancreatic duct epithelium and in pancreatic abscess [8, 42–45]. Anaerobic bacterial taxa have been found at a variable rate in pancreatitis; the results depend on the process for bacterial identification [8, 42, 43]. Previous studies have also reported the presence of bacteria in bile [46, 47]. In a study of 6 subjects with gallstones, 16S rRNA gene sequencing identified high relative abundances of Escherichia, Klebsiella and Pyramidobacter in the bile, and the bacterial profile of the bile was very similar to the duodenum in the same subjects [47]. Pyramidobacter species was originally isolated from the oral cavity [48] and was also found in our study samples, but at low levels (<20% of all samples).
Several bacterial taxa we observed with elevated relative mean abundance in RIH samples have been previously identified in immunocompromised patients and are largely believed to be opportunistic pathogens, including Acinetobacter [49] and Kluyvera [50]. The genus Gemella, which was found at higher relative abundance in pancreatic cancer subjects when compared to NDRI samples, has been previously associated with a number of infections, including endocarditis, soft-tissue abscesses, empyema, bloodstream infection, and bone infections [51–54]. Because our analysis was based on a cross-sectional study design, we expected to identify bacteria that were present as a result of opportunistic nosocomial infections given that the majority of RIH subjects were likely immunocompromised from their cancer. However, our results show that even normal pancreatic tissue harbors a microbial community.
The strength of this study was the collection of specimens specifically for the purpose of microbiome analysis, with precautions made to reduce contamination during collection and processing of samples. Moreover, multiple types of samples were collected on each patient at RIH, including obtaining tissue or swabs from multiple sites, to allow for inter vs. intra-individual differences at different sites. Finally, the multivariable regression analyses was conducted to adjust for potential confounding by known pancreatic cancer risk factors, including BMI and smoking, as well as other factors that may cause bias, including pre-OP EUS and prior chemotherapy.
The major limitation of this analysis was the small number of subjects with pancreatic cysts and pancreatic cancer; despite recruiting 77 subjects, not all subjects had tissue resections during surgery (as more advanced pancreatic cancer patients are often not operable). We did not have sufficient power to examine in great detail the differences in bacterial composition between different pancreatic cancer subtypes, including IPMNs; however, we were the first to include ICD 24 tumors and to explore differences with ICD 25 tumors. Moreover, cancer versus non-cancer comparisons of bacterial presence/absence and relative abundances were based on subjects spread across two different data sources (i.e., RIH and NDRI). Differences in microbiota between these two sources may have been due to differences in collection methods and collection times; DNA was extracted from frozen tissue using the same protocol and methods, but tissue samples were either collected during surgery (RIH) or from organs that were rapidly frozen after death (NDRI samples had a mean time of 13 hours to processing of samples). Consequently, it is possible that the identified genera (and overall differences in bacterial taxonomy) merely reflect study-specific differences, rather than real cancer-specific differences.
In this culture-independent study, we detected many bacterial taxa in pancreatic tissue from cancer subjects as well as non-cancer subjects. Furthermore, the bacterial profiles in the pancreas were more similar within individuals across different sites of the pancreas (i.e., head, tail, ducts) and duodenum than between individuals at each site. Bacterial taxa known to inhabit the oral cavity were common in the pancreas microbiome and several periodontal pathogens were also identified in pancreatic tissue samples. Further research is needed to address if and how bacteria may be related to pancreatic carcinogenesis or disease progression.
Supplementary Material
Acknowledgement
We thank the participants who graciously enrolled in this study. We thank Ms. Priyanka Joshi for her tremendous help with recruitment of subjects at the RIH, and Dr. Ross Taliano for assisting with the preparation of the tissue specimens in the Pathology Department at the RIH. We also thank Drs. Murray Resnick, Kara Lombardo, Alexis Kokaras, Emily Walsh and Ms. Laura Gantt and Naisi Zhao for their help with this project.
Financial support: The research reported in this publication was supported by the NIH/National Cancer Institute grants R01 CA166150 and P30 CA168524.
Footnotes
Availability of data and material
The datasets analyzed for this manuscript will be available through the NCBI under the BioProject accession number: PRJNA421501. R codes generated to analyze these data will be available upon request.
Potential competing interests: None.
References:
- 1.ACS. Cancer Facts & Figures 2018. In. Atlanta: American Cancer Society, Inc; 2018. [Google Scholar]
- 2.Klein AP, Lindstrom S, Mendelsohn JB, et al. An absolute risk model to identify individuals at elevated risk for pancreatic cancer in the general population. PLoS One 2013;8(9):e72311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis 2013;34(10):2193–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaud DS, Fu Z, Shi J, et al. Periodontal Disease, Tooth Loss, and Cancer Risk. Epidemiol Rev 2017;39(1):49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajishengallis G Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol 2014;35(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaud DS, Izard J, Wilhelm-Benartzi CS, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013;62(12):1764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67(1):120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swidsinski A, Schlien P, Pernthaler A, et al. Bacterial biofilm within diseased pancreatic and biliary tracts. Gut 2005;54(3):388–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider J, Schenk P, Obermeier A, et al. Microbial colonization of pancreatic duct stents: a prospective analysis. Pancreas 2015;44(5):786–90. [DOI] [PubMed] [Google Scholar]
- 10.Scheithauer BK, Wos-Oxley ML, Ferslev B, et al. Characterization of the complex bacterial communities colonizing biliary stents reveals a host-dependent diversity. ISME J 2009;3(7):797–807. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson HO, Stenram U, Ihse I, et al. Helicobacter species ribosomal DNA in the pancreas, stomach and duodenum of pancreatic cancer patients. World J Gastroenterol 2006;12(19):3038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitsuhashi K, Nosho K, Sukawa Y, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015;6(9):7209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers MB, Aveson V, Firek B, et al. Disturbances of the Perioperative Microbiome Across Multiple Body Sites in Patients Undergoing Pancreaticoduodenectomy. Pancreas 2017;46(2):260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Fuhler GM, Bn N, et al. Pancreatic cyst fluid harbors a unique microbiome. Microbiome 2017;5(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pushalkar S, Hundeyin M, Daley D, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8(4):403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G, Gail MH, Consonni D, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol 2016;17(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358(6369):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott DRF, Walker AW, O’Donovan M, et al. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol Hepatol 2017;2(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Zhou J, Xin Y, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol 2016;28(3):261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hieken TJ, Chen J, Hoskin TL, et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci Rep 2016;6:30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segata N, Haake SK, Mannon P, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012;13(6):R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357(6356):1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzosa EA, Morgan XC, Segata N, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A 2014;111(22):E2329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Hebshi NN, Nasher AT, Idris AM, et al. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol 2015;7:28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol 2010;192(19):5002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6(3):610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecological Monographs 1957;27:325–349. [Google Scholar]
- 29.LaMonte MJ, Genco RJ, Hovey KM, et al. History of Periodontitis Diagnosis and Edentulism as Predictors of Cardiovascular Disease, Stroke, and Mortality in Postmenopausal Women. J Am Heart Assoc 2017;6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson FC 3rd, Yumoto H, Takahashi Y, et al. Innate immune signaling and Porphyromonas gingivalis-accelerated atherosclerosis. J Dent Res 2006;85(2):106–21. [DOI] [PubMed] [Google Scholar]
- 31.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22(2):292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22(2):299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14(2):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan F, Zhang L, Li M, et al. Predominant gut Lactobacillus murinus strain mediates anti-inflammaging effects in calorie-restricted mice. Microbiome 2018;6(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faveri M, Figueiredo LC, Duarte PM, et al. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol 2009;36(9):739–49. [DOI] [PubMed] [Google Scholar]
- 36.Stingu CS, Schaumann R, Jentsch H, et al. Association of periodontitis with increased colonization by Prevotella nigrescens. J Investig Clin Dent 2013;4(1):20–5. [DOI] [PubMed] [Google Scholar]
- 37.Liu B, Faller LL, Klitgord N, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 2012;7(6):e37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goncalves LF, Fermiano D, Feres M, et al. Levels of Selenomonas species in generalized aggressive periodontitis. J Periodontal Res 2012;47(6):711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015;42(2):344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelakis E, Armougom F, Carriere F, et al. A Metagenomic Investigation of the Duodenal Microbiota Reveals Links with Obesity. PLoS One 2015;10(9):e0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid SW, Uhl W, Friess H, et al. The role of infection in acute pancreatitis. Gut 1999;45(2):311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brook I, Frazier EH. Microbiological analysis of pancreatic abscess. Clin Infect Dis 1996;22(2):384–5. [DOI] [PubMed] [Google Scholar]
- 44.Hill MC, Dach JL, Barkin J, et al. The role of percutaneous aspiration in the diagnosis of pancreatic abscess. AJR Am J Roentgenol 1983;141(5):1035–8. [DOI] [PubMed] [Google Scholar]
- 45.Tsui NC, Zhao E, Li Z, et al. Microbiological findings in secondary infection of severe acute pancreatitis: a retrospective clinical study. Pancreas 2009;38(5):499–502. [DOI] [PubMed] [Google Scholar]
- 46.Wu T, Zhang Z, Liu B, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics 2013;14:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye F, Shen H, Li Z, et al. Influence of the Biliary System on Biliary Bacteria Revealed by Bacterial Communities of the Human Biliary and Upper Digestive Tracts. PLoS One 2016;11(3):e0150519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downes J, Vartoukian SR, Dewhirst FE, et al. Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int J Syst Evol Microbiol 2009;59(Pt 5):972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 1996;9(2):148–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarria JC, Vidal AM, Kimbrough RC 3rd. Infections caused by Kluyvera species in humans. Clin Infect Dis 2001;33(7):E69–74. [DOI] [PubMed] [Google Scholar]
- 51.Mosquera JD, Zabalza M, Lantero M, et al. Endocarditis due to Gemella haemolysans in a patient with hemochromatosis. Clin Microbiol Infect 2000;6(10):566–8. [DOI] [PubMed] [Google Scholar]
- 52.La Scola B, Raoult D. Molecular identification of Gemella species from three patients with endocarditis. J Clin Microbiol 1998;36(4):866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Lechuz JM, Cuevas-Lobato O, Hernangomez S, et al. Extra-abdominal infections due to Gemella species. Int J Infect Dis 2002;6(1):78–82. [DOI] [PubMed] [Google Scholar]
- 54.Fangous MS, Hemon F, Graf P, et al. Bone infections caused by Gemella haemolysans. Med Mal Infect 2016;46(8):449–452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.