Abstract
BET inhibitors (BETis), which target transcription of key oncogenic genes, are currently being evaluated in early-phase clinical trials. However, since BETis show limited single agent activity, there is increasing interest in identifying signaling pathways to enhance the efficacy of BETis. Here, we demonstrate increased MNK kinases-dependent eIF4E phosphorylation following treatment with BETis, indicating activation of a pro-survival feedback mechanism in response to BETis. BET PROTACs, which promote degradation of BET proteins, also induced eIF4E phosphorylation in cancer cells. Mechanistically, we show that the effect of BETis on MNK-eIF4E phosphorylation was mediated by p38 MAPKs. We also show that BETis suppressed RacGAP1 to induce Rac signaling-mediated eIF4E phosphorylation. Significantly, MNK inhibitors and MNK1/2 knockdown enhanced the efficacy of BETis in suppressing proliferation of cancer cells in vitro and in a syngeneic mouse model. Together, these results demonstrate a novel pro-survival feedback signaling induced by BETis, providing a mechanistic rationale for combination therapy with BET and MNK inhibitors for synergistic inhibition of cancer cells.
Keywords: BET inhibitors, Rac, MNK kinases, pancreatic cancer, thyroid cancer, syngeneic tumors
Introduction
The bromodomain (BRD) and extra-terminal domain-containing (BET) proteins, which belong to the BRD protein family, play an important role in various cellular processes (1). BET proteins, by functioning as transcriptional co-activators, can enhance cell cycle progression, limit apoptosis, and promote cell motility and metastasis in a broad range of cancers (1). BET proteins mediate their pro-tumorigenic function in part by upregulating expression of Myc and other oncogenes (2). Clinically, there is increasing interest in targeting BET proteins, and a number of potent and selective BET inhibitors (BETis) have been evaluated in early-phase clinical trials (3, 4). For example, the BET inhibitor OTX-015 (MK-8628, birabresib) has successfully undergone phase 1 clinical studies in patients with hematologic malignancies and select solid tumors (3, 4).
Although BETis have shown promising results in early-phase clinical trials, resistance to BETis has been reported in preclinical models (4). For example, cancer cells can develop resistance to BETis by activating alternative signaling pathways to upregulate MYC transcription (5–7). Recently, it was shown that increased accumulation of BET proteins also mediates resistance to BETis (8). BET proteins can be degraded through an SPOP-mediated ubiquitination (9, 10); however, cancer cells with SPOP mutation demonstrate increased levels of BET proteins and resistance to BETis (9, 10). Consequently, to overcome resistance due to increased accumulation of BET proteins, there is also increasing interest in developing inhibitors that promote degradation of BET proteins through the use of proteolysis-targeting chimeras (PROTACs) (8, 11).
Cancer cells can also develop resistance by activating the translational machinery (12). For example, breast cancer cells developing resistance to tamoxifen demonstrate evidence of selective reprogramming of mRNA translation mediated by eukaryotic initiation factor 4E (eIF4E) (13). The eIF4E activity can be regulated by MNK kinases-dependent phosphorylation, with eIF4E phosphorylation being critical for malignant transformation and promotion of tumor development in animal models (14, 15). Importantly, the MNK-eIF4E pathway has also been shown to be activated as a feedback survival mechanism in response to acute treatment with chemotherapy and targeted therapies (16, 17). However, the activation of the MNK-eIF4E pathway by BETis and the involvement of the MNK-eIF4E pathway in limiting response to BETis has not been previously evaluated.
In this study, we demonstrate that BETis and eIF4E knockdown decrease growth of cancer cells. While BETis do not repress eIF4E protein levels, we show that BETis induce MNK kinases-dependent eIF4E phosphorylation. BET PROTACs also induce MNK kinases-dependent eIF4E phosphorylation in cancer cells. Mechanistically, we show that the effect of BETis on MNK-eIF4E phosphorylation is mediated by p38 MAPKs. We also show that BETis suppress RacGAP1 to induce Rac-mediated eIF4E phosphorylation. Significantly, MNK inhibitors and MNK1/2 knockdown enhance the efficacy of BETis in suppressing proliferation of cancer cells in 3D collagen and in a syngeneic mouse model. Together, these results demonstrate increased eIF4E phosphorylation following treatment with BETis, and identify combination therapy with BET and MNK inhibitors for the treatment of cancer patients.
Materials and Methods
Cell culture –
K1, RO82-w-1, and FTC-133 cell lines were purchased from Sigma and cultured according to the manufacturer’s instructions. MDA-T22 and MDA-T85 were obtained from MD Anderson Cancer Center and were cultured in RPMI-1460 containing 10% FBS and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) as previously described (18). Human PDAC cell lines Panc1 and CD18 were obtained from ATCC. Cells were maintained in DMEM containing 10% FBS and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) (19). Mouse thyroid cancer cells TBP-3868, obtained from Dr. Sareh Parangi (MGH, Harvard Medical School) (20), were maintained in DMEM containing 10% FBS and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin). Cell lines were used within 15 passages of thawing, and continuously cultured for less than 6 months. The cells were not tested for mycoplasma. The appearance and growth characteristics of the mouse and human thyroid cancer cell lines used in this study were compared with published information to ensure their authenticity. Pancreatic cancer cells were authenticated by short tandem repeat profiling in October 2013 and also monitored by their appearance and growth characteristics to ensure their authenticity.
Chemicals –
JQ1 (2) and CGP57380 (21) were purchased from Tocris Bioscience; OTX-015 (22) and ARV-825 (11) were obtained from Selleckchem; U0126 (23) was from Cell Signaling; and SB202190 (24) and NSC23766 (25) were purchased from Calbiochem.
Embedding cells in three-dimensional type I collagen gels -
Cells were suspended in type I collagen solution (2.2 mg/mL), which was made as described previously (26), and allowed to gel for 15 minutes at 37°C. For morphological examination of cells, cell colonies in three-dimensional collagen were examined using a Zeiss Axiovert 40 CFL microscope and pictures taken with a Nikon Coolpix 4500 camera.
WST-1 proliferation assay –
Approximately 1,000 cells were embedded in 2.2 mg/mL collagen I solution and seeded in 96-well plates. After 3 days of treatment with the different inhibitors, WST-1 reagent (Sigma-Aldrich) was added into the media at a 1:100 dilution factor. Absorbance was measured according to the manufacturer’s instructions every hour for 4 hours or until maximal absorbance was reached. Coefficient of drug interaction (CDI) was calculated as CDI = AB /(A × B) (27, 28). AB, relative cell viability of the combination; A or B, relative cell viability of the single agent groups). CDI > 1 indicates an antagonistic effect; CDI = 1 indicates an additive effect; and CDI < 1 indicates a synergistic effect (with CDI < 0.6 indicates strong synergy) (27, 28).
cBioportal analysis –
Relative mRNA expression levels and survival analyses were determined by cBioPortal analysis as previously described (29). To evaluate mRNA expression levels of fibrillary collagens and eIF4E, we used the papillary thyroid carcinoma dataset (30).
Real-time PCR-based array analysis –
The Human Cytoskeleton Regulators RT2 Profiler PCR Array, which enables profiling of 84 genes involved in cytoskeleton regulation, was purchased from Qiagen and performed according to the manufacturer’s instructions and using quality controls included in the array. Expression of several representative genes from the array was validated by qRT-PCR.
Transfection –
SMARTpool On-Target siRNA targeting MNK1 and MNK2 were purchased from Dharmacon. siRNA targeting eIF4E was obtained from Ambion. All transfections with siRNA were carried out using RNAimax reagent (Life Technology) according to the manufacturer’s instructions. RacGAP1 plasmid (variant 1) was obtained from Origene. Control plasmid was obtained from Lonza. All transfection with plasmids were carried out using Lipofectamine® 3000 reagent (Life Technology) according to the manufacturer’s instructions.
qRT-PCR analysis –
Quantitative gene expression was performed with gene-specific TaqMan probes, TaqMan Universal PCR Master Mix, and the 7500 Fast Real-time PCR System from Applied Biosystems. Data were then quantified with the comparative CT method for relative gene expression.
Immunoblotting –
Whole cell extracts of cultured cells were prepared in RIPA lysis buffer supplemented with phosphatase and protease inhibitors, and separated on SDS-PAGE gel. The following antibodies and dilution factors were used: phospho-eIF4E (1:1000, Cell Signaling), total eIF4E (1:1000, Santa Cruz Biotechnology), phospho-MNK1 (1:1000, Cell Signaling), total MNK1 (1:1000, Cell Signaling), phospho-ERK1/2 (1:1000, Cell Signaling), total ERK1/2 (1:2000, Santa Cruz), BRD4 (1:1000, Abcam), Rac1 (1:2000, EMD Millipore), RacGAP1 (1:2000, Santa Cruz), and HSP90 (1:3000, Santa Cruz). Blocking agent was 5% bovine serum albumin (BSA). Secondary anti-mouse IgG (A4416) and anti-rabbit IgG (A6667) antibodies were purchased from Sigma and used at a 1:3000 dilution factor. When necessary, membrane was stripped using Restore Western Blot Stripping Buffer (Thermo Fisher Scientific).
Immunohistochemistry and immunofluorescence –
Human thyroid specimens were purchased from US Biomax and were trichrome stained or stained for eIF4E. Antigen retrieval was carried out as previously described (31, 32). Photographs for quantitative comparison were taken using FeinOptic microscope and Jenoptik ProgRes C5 camera. Cell lines (K1, MDA-T85, CD18 and Panc1) growing on glass coverslips were stained for phalloidin (Alexa Fluor 488) at a ratio of 1:500 and pictures were taken using FeinOptic microscope and Jenoptik ProgRes C5 camera.
In vivo study –
TBP-3868 cancer cells were injected subcutaneous (1.5 × 106 cells/site) into the flanks of 6–8-week-old B6129SF1/J mice (Charles River Laboratory). Mice were divided into four treatment groups: control (DMSO), JQ1 (12.5 mg/kg), CGP57380 (25 mg/kg), and a combination of JQ1 (12.5 mg/kg) and CGP57380 (25 mg/kg). Treatments were administered daily Mon-Fri for 2 weeks in a suspension containing 10% hydroxypropyl-β-cyclodextrin in double-distilled water. The mice were euthanized by CO2 inhalation and cervical dislocation, and the tumors were excised and photographed.
Statistical analysis –
Error bars represent standard deviation. All statistical analyses were done using GraphPad Instat. A p value < 0.05 was considered significant.
Study approval –
All animal work and procedures were approved by the Northwestern University Institutional Animal Care and Use Committee. In addition, all animal experiments were performed in accordance with relevant guidelines and regulations.
Results
BETis decrease growth of cancer cells in 3D collagen.
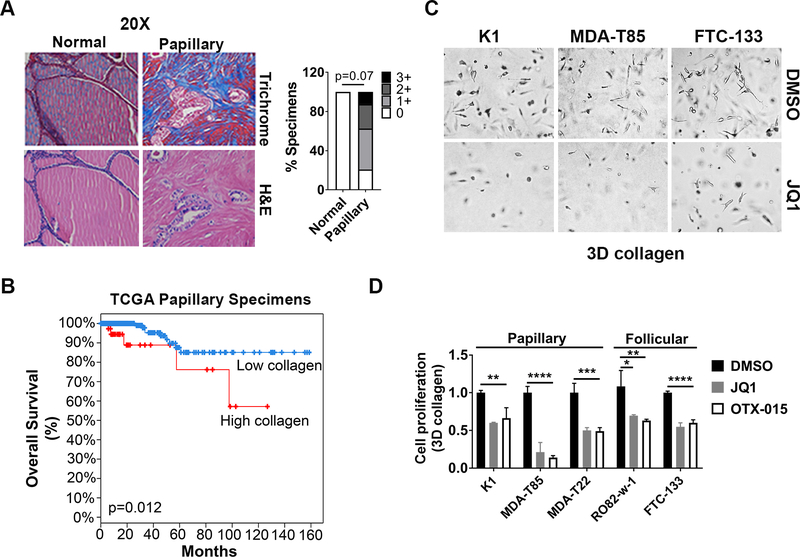
Papillary thyroid cancer (PTC), the most common subtype of thyroid cancer (33), demonstrates a collagen-rich stromal reaction (Fig. 1A). Thus, we evaluated whether BETis limit growth of thyroid cancer cells in 3D collagen. Initially, we evaluated expression of members of the fibrillar collagen family in PTC samples available in the TCGA database (29). Increased expression of fibrillar collagen was associated with worse outcome (Fig. 1B). We next evaluated the effect of BETis on the growth of PTC cells growing in 3D collagen. Treatment with the BETis JQ1 and OTX-015 significantly decreased proliferation of thyroid cancer cells in 3D collagen (Figs. 1C and 1D).
Figure 1: BET inhibitors (BETis) decrease growth of cancer cells in 3D collagen.
A. Human thyroid tissue microarray (TMA) containing 24 papillary thyroid specimens and 8 adjacent normal tissue samples was trichrome stained, and the relative staining was graded as low (0 or 1+) or high (2+ or 3+). Fisher’s exact test was used to calculate p value. B. cBioportal analysis of Kaplan-Meier survival curve of papillary thyroid cancer patients in the TCGA database with or without relative overexpression of fibrillar collagen. C and D. Thyroid cancer cells growing in 3D collagen were treated with the BETis JQ1 (1 μM) and OTX-015 (1 μM) for 72 hours. The cells were examined by phase microscopy (C), and the effect on proliferation was determined by WST-1 assay (D). *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. The results are representative of at least three independent experiments.
Increased eIF4E, which regulates growth of cancer cells in 3D collagen, is associated with worse outcome.
While transcriptional plasticity can reduce the efficacy of BETis (5–7), it is not known whether enhanced translational activity can also result in reduced efficacy of BETis. Thus, we evaluated the role of eIF4E, which regulates cap-dependent translation of specific mRNAs (12), in mediating response to BETis. Initially, we evaluated the role of eIF4E in mediating the growth of cancer cells in 3D collagen. Downregulation of eIF4E decreased growth of cancer cells in 3D collagen (Figs. 2A and 2B). Significantly, eIF4E knockdown was effective against both BRAF wild-type thyroid cancer cells and thyroid cancer cells expressing the BRAFV600E mutation, which is present in ~60% of thyroid cancers (34). Importantly, eIF4E expression is increased in cancer samples relative to normal thyroid tissue (Supplementary Fig. 1A), with increased eIF4E expression associated with decreased overall survival in these patients (Supplementary Fig. 1B) (29).
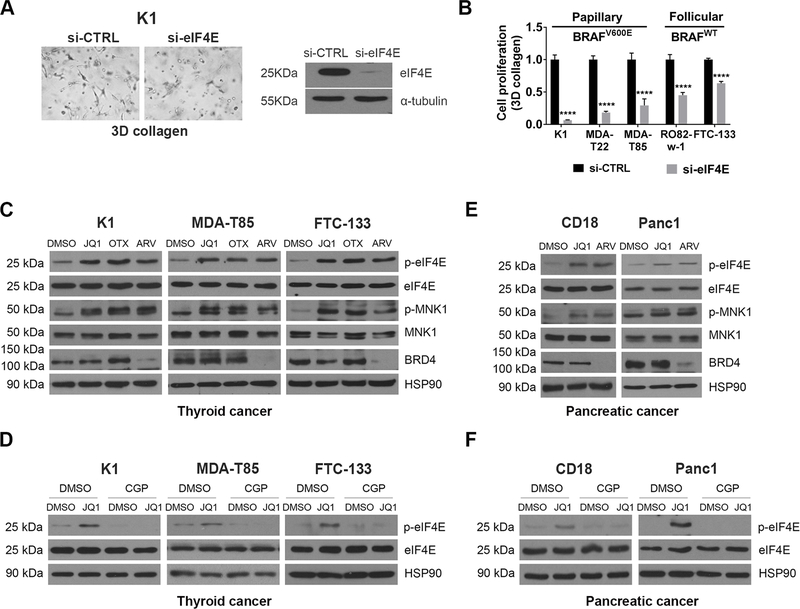
Figure 2: BETis and BET PROTACs induce eIF4E phosphorylation.
A. Thyroid cancer cells were transfected with siRNA targeting eIF4E for 48 hours, and then cultured in 3D collagen for additional 48 hours. The cells were examined by phase microscopy. Knockdown efficiency was determined by Western blotting. B. Thyroid cancer cells with or without BRAFV600E mutation were transfected with siRNA targeting eIF4E for 48 hours and then cultured in 3D collagen for additional 48 hours. The effect on proliferation was determined by WST-1 assay. ****, p<0.0001. C. Thyroid cancer cells were treated with BETis JQ1 (1 μM) and OTX-015 (OTX, 1 μM) or the BET PROTAC ARV-825 (ARV, 1 μM) for 24 hours. The effect on eIF4E and MNK1 phosphorylation and expression of eIF4E, MNK1, BRD4 and HSP90 (loading control) was determined by Western blotting. D. Thyroid cancer cells were pre-treated with CGP57380 (CGP, 10 μM) for 30 min, followed by treatment of JQ1 (1 μM) for 24 hours. The effect on eIF4E phosphorylation and expression of eIF4E and HSP90 was determined by Western blotting. E. CD18 and Panc1 pancreatic cancer cells were treated with the BET inhibitor JQ1 (1 μM) or the BET PROTAC ARV-825 (ARV, 1 μM) for 24 hours. The effect on eIF4E and MNK1 phosphorylation and expression of eIF4E, MNK1, BRD4 and HSP90 (loading control) was determined by Western blotting. F. Pancreatic cancer cells were pre-treated with CGP57380 (CGP, 10 μM) for 30 min and then treated with JQ1 (1 μM) and for 24 hours. The effect on eIF4E phosphorylation and expression of eIF4E and HSP90 was determined by Western blotting. The results are representative of at least three independent experiments.
BETis and BET PROTACs induce eIF4E phosphorylation.
Since both BET inhibitors and eIF4E knockdown decreased growth of cancer cells in 3D collagen, we evaluated the extent to which BETis repress eIF4E expression in cancer cells. While the BETis JQ1 and OTX-015 did not decrease eIF4E protein levels, we found that treatment with these inhibitors enhanced eIF4E phosphorylation (Fig. 2C). Importantly, eIF4E phosphorylation, which is regulated by the kinases MNK1 and MNK2, promotes tumor development in animal models (15). BETis also enhanced MNK1 phosphorylation in these cells (Fig. 2C). Significantly, pre-treatment with the MNK inhibitor CGP57380 blocked JQ1-induced eIF4E phosphorylation (Fig. 2D). In addition, we evaluated the effect of BET PROTACs on eIF4E phosphorylation in thyroid cancer cells. Treatment with the BET PROTAC ARV-825 decreased BRD4 protein levels (Fig. 2C). As seen with BETis, treatment with ARV-825 also induced eIF4E phosphorylation (Fig. 2C).
Since the induction of eIF4E phosphorylation by BETis and BET PROTACs in thyroid cancer cells was unexpected, we evaluated whether BETis and BET PROTACs also induce eIF4E phosphorylation in other cancer cells. As we had previously shown that BETis decrease growth of pancreatic cancer cells in 3D collagen (5, 19), we evaluated the effect of BETis and BET PROTACs on eIF4E phosphorylation in pancreatic cancer cells. Significantly, treatment with JQ1 and ARV-825 induced MNK1 and eIF4E phosphorylation in pancreatic cancer cells (Fig. 2E). As with thyroid cancer cells (Figs. 2D), treatment with the MNK inhibitor CGP57380 blocked JQ1-mediated eIF4E phosphorylation in pancreatic cancer cells (Fig. 2F).
p38 MAPKs, but not MEK-ERK, mediate BETis-induced MNK/eIF4E phosphorylation.
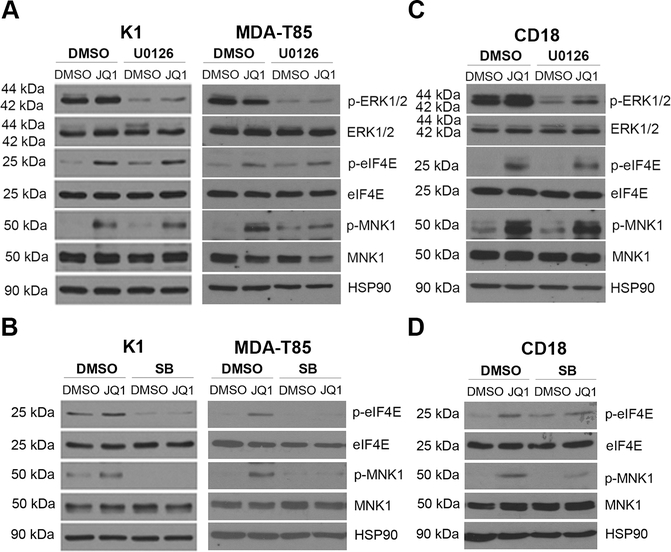
Since MNK kinases can be phosphorylated by ERK and p38 MAPKs (35), we next evaluated their roles in JQ1-induced eIF4E phosphorylation using specific MEK/ERK (U0126) and p38 MAPK (SB202190) inhibitors. Treatment with U0126 blocked ERK1/2 phosphorylation, but did not affect JQ1-induced MNK1 or eIF4E phosphorylation in cancer cells (Figs. 3A and 3C). However, treatment with the p38 MAPK inhibitor SB202190 decreased JQ1-induced MNK1 and eIF4E phosphorylation (Figs. 3B and 3D), establishing a requirement for p38 MAPK in JQ1-mediated MNK and eIF4E phosphorylation.
Figure 3: p38 MAPKs, but not MEK/ERK signaling, mediates BET inhibitor-induced eIF4E phosphorylation.
A and C. Thyroid and pancreatic cancer cells were pre-treated with the MEK1/2 inhibitor U0126 (5 μM) for 30 min and then treated with JQ1 (1 μM) and for 24 hours. The effect on ERK1/2, MNK1 and eIF4E phosphorylation and on the expression of ERK1/2, MNK1 and eIF4E expression was determined by Western blotting, using HSP90 as loading control. B and D. Thyroid and pancreatic cancer cells were pre-treated with the p38 MAPK inhibitor SB202190 (SB, 5 μM) for 30 min and then treated with JQ1 (1 μM) and for 24 hours. The effect on MNK1 and eIF4E phosphorylation and on the expression of MNK1 and eIF4E expression was determined by Western blotting, using HSP90 as loading control. The results are representative of at least three independent experiments.
BETis induce Rac-mediated cytoskeletal changes and Rac-mediated eIF4E phosphorylation.
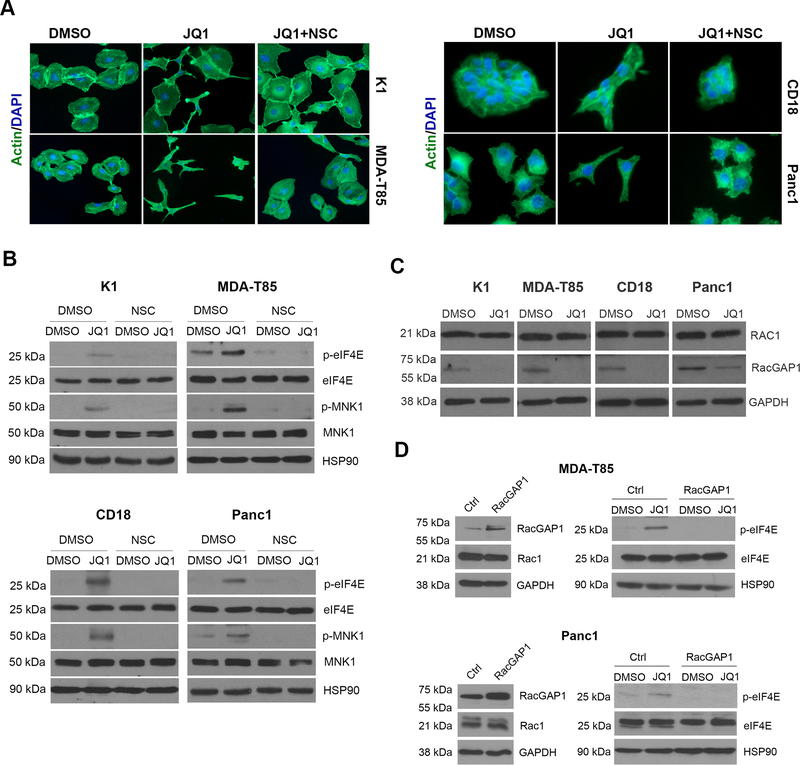
We noticed that BETis induced spindle-shaped morphology in thyroid and pancreatic cancer cells that was more clearly detected with phalloidin staining (Fig. 4A). As the change in morphology suggested increased Rac signaling, we evaluated whether blocking Rac activity could attenuate the effects of BETis on cytoskeletal changes. Pre-treatment of cancer cells with the well-established Rac inhibitor NSC23766 (36) attenuated JQ1-induced morphological and cytoskeletal changes (Fig. 4A). Importantly, the Rac inhibitor NSC23766 also blocked JQ1-induced eIF4E phosphorylation in both thyroid and pancreatic cancer cells (Fig. 4B).
Figure 4: BETis induce Rac-mediated cytoskeletal changes and Rac-mediated eIF4E phosphorylation.
A. Cancer cells growing on glass coverslips were treated with DMSO or JQ1 (1 μM) and co-treated with the Rac inhibitor NSC23766 (NSC, 50 μM) for 24 hours. The cells were then processed for phalloidin staining and the nuclei counterstained with DAPI (40x). B. Cancer cells were pre-treated with the Rac inhibitor NSC23766 (NSC, 50 μM) and then treated with JQ1 (1 μM) for 24 hours. The effect on eIF4E and MNK1 phosphorylation was determined by Western blotting. C. Cancer cells were treated with DMSO or JQ1 (1 μM) for 24 hours and the effect on Rac1 and RacGAP1 was determined by Western blotting. D. Cancer cells transfected with control (Ctrl) vector or vector expressing RacGAP1 were treated with DMSO or JQ1 (1 μM) for 24 hours. The effect on eIF4E phosphorylation and on the expression of RacGAP1, Rac1 and eIF4E expression was determined by western blotting, using GAPDH and HSP90 as loading controls. The results are representative of at least three independent experiments.
To understand the mechanism by which BETis induce Rac signaling, we evaluated the effect of JQ1 on genes known to regulate Rac signaling using an RT2PCR array (Supplementary Fig. 2). JQ1 suppressed mRNA expression of ROCK1 and ROCK2 (Supplementary Table 1 and Supplementary Fig. 2), which can counteract the effects of Rac signaling (37). While JQ1 not did not affect Rac1 protein levels (Fig. 4C), JQ1 also suppressed expression of RacGAP1 (Supplementary Table 1, Supplementary Fig. 2, and Fig. 4C), a negative regulator of Rac activity (37). As these results suggest that BETis induce relative Rac activation by suppressing negative regulators of Rac signaling, we evaluated the extent to which overexpressing RacGAP1 could attenuate the effects of JQ1. Importantly, overexpression of RacGAP1 in cancer cells blocked JQ1-induced eIF4E phosphorylation (Fig. 4D).
MNK inhibitors and MNK1/2 siRNA potentiate the effects of BETis at suppressing proliferation.
We evaluated the effect of co-treatment with BET and MNK inhibitors on the growth of cancer cells in 3D collagen. Combining the MNK inhibitor CGP57380 with the BET inhibitor JQ1 significantly suppressed the growth of thyroid and pancreatic cancer cells (Figs. 5A and 5B). The coefficient of drug interaction between BET and MNK inhibitors was 0.54 in K1 cells and 0.61 in CD18 cells (Supplementary Table 2), indicating synergistic anti-proliferative effects of combining BET and MNK inhibitors in cancer cells. Similarly, combining CGP57380 with the BET inhibitor OTX-015 or the BET PROTAC ARV-825 also significantly suppressed growth of cancer cells (Supplementary Fig. 3), and demonstrated synergistic anti-proliferative effects (Supplementary Table 2).
Figure 5: MNK inhibitors and MNK1/2 siRNA potentiate the effects of BETis at suppressing proliferation.
A and B. Cancer cells growing in 3D collagen were treated with JQ1 (1 μM) and co-treated with CGP57380 (CGP, 2 μM) for 72 hours and the effect on cell proliferation was determined using the WST-1 assay. ***, p<0.001; ****, p<0.0001 relative to the combination treatment group. C-F. Cancer cells were transfected with control siRNA (siCTRL) or combination of siRNAs against MNK1 and MNK2 (siMNK1/2) for 48 hours. The cells were then embedded in 3D collagen and treated with JQ1 (1 μM) for 48 hours. The effect on MNK1 and MNK2 knockdown was determined by Western blotting and/or by qRT-PCR. The effect on eIF4E and MNK1 phosphorylation was determined by Western blotting using HSP90 as loading control (C-D). The effect on cell proliferation was determined using the WST-1 assay (E, F). ****, p<0.0001 relative to siCTRL-transfected, DMSO-treated samples. The results are representative of three independent experiments.
We also evaluated the extent to which MNK1/2 siRNA enhanced JQ1 suppression of growth of cancer cells in 3D collagen. Initially, we evaluated the effect of MNK1/2 siRNA on JQ1-induced eIF4E phosphorylation. The MNK1/2 siRNA decreased basal and JQ1-induced eIF4E phosphorylation in cancer cells (Figs. 5C and 5D). The MNK1/2 siRNA also enhanced JQ1 suppression of growth of cancer cells in 3D collagen (Figs. 5E and 5F), and demonstrated synergistic anti-proliferative effects, evidenced by their coefficient of drug interaction of 0.66 for K1 cells and 0.75 for CD18 cells (Supplementary Table 2).
MNK inhibitors potentiate the effects of BETis at suppressing tumor growth in vivo.
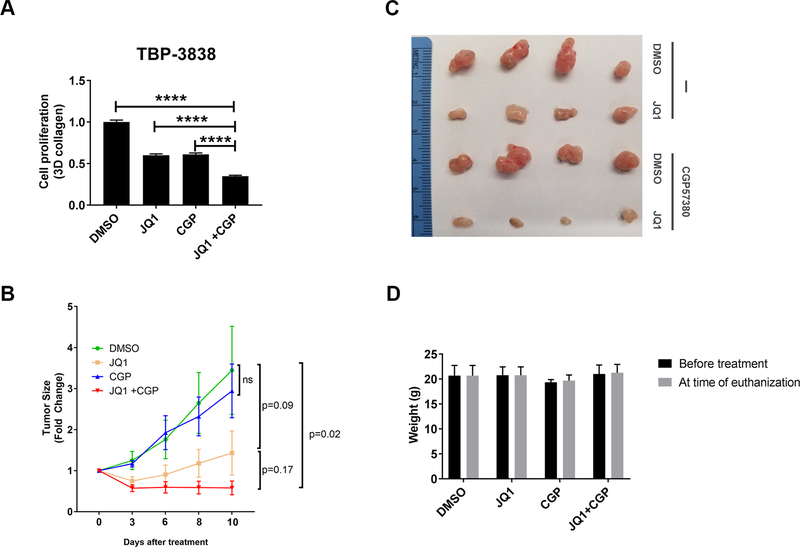
Finally, we evaluated the effect of co-treatment with BET and MNK inhibitors on tumor growth in vivo using a syngeneic model of thyroid cancer. Since human thyroid cancer cells grow poorly in vivo (18), we used the mouse papillary thyroid cancer cell line TBP-3868. This mouse thyroid cancer cell line, derived from thyroid tumors developing in a transgenic mouse model expressing B-raf(V600E) with loss of both copies of p53 in the thyroid tissue (20), has been shown to faithfully recapitulate human papillary thyroid tumors. We initially evaluated the effect of BET and MNK inhibitors on the growth of TBP-3868 cells in 3D collagen. As with human cancer cells, combining the MNK inhibitor CGP57380 with the BET inhibitor JQ1 significantly suppressed the growth of mouse TBP-3868 cells (Fig. 6A). We then injected TBP-3868 cells in the syngeneic B6129SF1/J mice, and established tumors were treated with JQ1 and/or CGP57380. Combination treatment with BET and MNK inhibitors significantly reduced tumor growth in vivo (Figs. 6B and 6C). Significantly, in contrast to JQ1 alone, combination of JQ1 and CGP resulted in tumor shrinkage relative to size of tumors at the start of treatment. Importantly, the combination treatment did not affect the weight of the treated mice (Fig. 6D). Together, these findings demonstrate crosstalk between BET proteins and the MNK-eIF4E pathway, and identify combination therapy with BET and MNK inhibitors for the treatment of cancer patients.
Figure 6: MNK inhibitors potentiate the effects of BETis at suppressing tumor growth in vivo.
A. Mouse TBP-3868 thyroid cancer cells growing in 3D collagen were treated with JQ1 (1 μM) and co-treated with CGP57380 (CGP, 2 μM) for 72 hours and the effect on cell proliferation was determined using the WST-1 assay. ****, p<0.0001 relative to the combination treatment group. B-D. Mouse 3868 thyroid cancer cells were subcutaneous injected in the flanks of syngeneic B6129SF1/J mice and allowed to form tumors for 1 week. Mice with established tumors were randomized and treated with DMSO, JQ1 (12.5 mg/kg), CGP57380 (25 mg/kg), or the combination of JQ1 (12.5 mg/kg) and CGP57380 (25 mg/kg) daily Mon-Fri for 2 weeks. Tumor growth was assessed daily, and tumor volume calculated and normalized to tumor volume at the start of treatment (B). Tumors were collected at the end of treatment and photographed (C). Mice were weighed daily and the effect of inhibitors on mouse weight at the end of treatment was compared to weight at the start of treatment (D).
Discussion
BETis have shown some efficacy in hematologic malignancies, NUT carcinomas, and in select solid tumors in clinical trials (3, 4). However, the efficacy of BETis for most solid tumors is limited by minimal single agent response or by the development of resistance to BETis (3, 4). For example, Myc-dependent pancreatic cancer and AML cells developing resistance to BETis activate alternative non-BRD transcriptional program in order to increase Myc protein levels (5, 6). In contrast, ovarian cancer cells activate compensatory pro-survival kinase network to overcome the effects of BETis (38). Also, colorectal cancer cells with intrinsic resistance to BETis have increased activation of the MEK/ERK signaling pathway (39). In this report, we show that cancer cells demonstrate increased MNK and eIF4E phosphorylation following treatment with BETis JQ1 and OTX015 as a compensatory pro-survival feedback mechanism to limit the antiproliferative effects of BETis. We also show that the BET PROTAC ARV-825 induces eIF4E phosphorylation. This is not surprising since ARV-825 consists of OTX-015 attached via a linker to pomalidomide in order to promote E3 ubiquitin ligase-mediated degradation of BRD proteins (8). Importantly, we show that blocking eIF4E phosphorylation with MNK inhibitors and MNK knockdown enhances the anti-tumorigenic effects of BETis and BET PROTACs.
We show that the p38 MAPK pathway, but not the MEK/ERK pathway, mediates BET inhibitor-induced eIF4E phosphorylation. Using the well-established Rac inhibitor NSC23766 and overexpression of RacGAP1, we also show that the Rac signaling is involved in eIF4E phosphorylation. Importantly, Rac signaling can mediate p38 MAPK activation in a number of model systems. For example, Rac mediates activation of p38 MAPKs following hyperosmotic shock of mammalian cells (40), following treatment of leukemia and breast cancer cells with all-trans-retinoic acid (41), and following treatment of leukemia cells with arsenic trioxide (42). Significantly, similar to our findings with BETis, arsenic trioxide also induces p38 MAPK-mediated eIF4E phosphorylation in leukemia cells (43). While we establish the role of Rac, p38 MAPKs and MNK kinases in regulating eIF4E phosphorylation following treatment with BETis, Rac signaling can also regulate eIF4E function by modulating the cellular localization of cytoplasmic FMR1 interacting protein 1 (CYFIP1) (44). CYFIP1 is associated with two distinct protein complexes: the wave regulator complex (WRC) to regulate actin remodeling and the eIF4E complex to inhibit translation of specific mRNAs (44). Rac activation causes redistribution of CYFIP1 from the eIF4E complex to the WRC, resulting in increased mRNA translation and actin polymerization (44). Future studies will evaluate the extent to which BETis regulate eIF4E and mRNA translation by modulating the function of CYFIP1.
In contrast to a recent study showing that JQ1 represses eIF4E in lung cancer cells (45), we have found that BETis do not repress eIF4E protein levels in thyroid and pancreatic cancer cells. This discrepancy may be due to differences in tumor types and/or duration of treatment with BETis. However, given the importance of eIF4E in cancer development and progression, the function of eIF4E is tightly regulated in cells under normal physiological conditions and in response to different signals (14, 35). eIF4E activity can be regulated by two major signaling pathways, the MAPK-MNK and the mTOR (mammalian target of rapamycin) signaling pathways (14, 35). Interestingly, both the MAPK and mTOR signaling pathways can mediate resistance to BETis. The mTOR signaling was recently shown to mediate resistance to BETis in SPOP-mutated cancer cells (10), while we demonstrate in this report the importance of the MAPK-MNK pathway in limiting the efficacy of BETis in cancer cells.
Finally, our findings suggest that MNK inhibitors should be combined with BETis and BET PROTACs for treatment of solid tumors. There are a number of BETis and BET PROTACs that are currently in preclinical development or in early phase clinical trials (4, 46). For example, the BET inhibitor OTX-015 was found to have reasonable safety profile in phase 1 clinical trials in patients with hematologic malignancies and select solid tumors (3, 4, 47, 48). Similarly, given the role of MNK kinases in mediating resistance and demonstrating synergistic activity of MNK inhibitors with other inhibitors (12, 49, 50), there is also interest in moving MNK inhibitors into clinical trials. For example, the MNK1/2 inhibitor eFT508 (Effector Therapeutics) is currently undergoing phase 1 clinical trial in patients with hematologic malignancies and solid tumors (49). Overall, our results demonstrate activation of the MNK-eIF4E pathway by BETis, and suggest that combination therapy with BET and MNK inhibitors should be pursued for the treatment of solid tumors.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by grants R01CA186885 (to H.G. Munshi) and R21CA220625 (to K. Kumar) from the NCI, a Merit award I01BX002922 (to H.G. Munshi) from the Department of Veterans Affairs, and a Translational Bridge Fellowship Award from the Lurie Cancer Center (to T.N.P).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- 1.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol. 2017;18(4):246–62. [DOI] [PubMed] [Google Scholar]
- 2.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahai V, Redig AJ, Collier KA, Eckerdt FD, Munshi HG. Targeting BET bromodomain proteins in solid tumors. Oncotarget. 2016;7(33):53997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stathis A, Bertoni F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov. 2018; 8(1)24–36. [DOI] [PubMed] [Google Scholar]
- 5.Kumar K, Raza SS, Knab LM, Chow CR, Kwok B, Bentrem DJ, et al. GLI2-dependent c-MYC upregulation mediates resistance of pancreatic cancer cells to the BET bromodomain inhibitor JQ1. Sci Rep. 2015;5:9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525(7570):543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525(7570):538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113(26):7124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23(9):1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med. 2017; 23(9):1055–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol. 2015;22(6):755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nature reviews Drug discovery. 2015;14(4):261–78. [DOI] [PubMed] [Google Scholar]
- 13.Geter PA, Ernlund AW, Bakogianni S, Alard A, Arju R, Giashuddin S, et al. Hyperactive mTOR and MNK1 phosphorylation of eIF4E confer tamoxifen resistance and estrogen independence through selective mRNA translation reprogramming. Genes Dev. 2017;31(22):2235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21(24):3232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107(32):14134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell JB, Eckerdt F, Dhruv HD, Finlay D, Peng S, Kim S, et al. Differential Response of Glioma Stem Cells to Arsenic Trioxide Therapy Is Regulated by MNK1 and mRNA Translation. Mol Cancer Res. 2018;16(1):32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Zha J, Lei M. Inhibiting ERK/Mnk/eIF4E broadly sensitizes ovarian cancer response to chemotherapy. Clin Transl Oncol. 2018;20(3):374–81. [DOI] [PubMed] [Google Scholar]
- 18.Henderson YC, Ahn SH, Ryu J, Chen Y, Williams MD, El-Naggar AK, et al. Development and characterization of six new human papillary thyroid carcinoma cell lines. J Clin Endocrinol Metab. 2015;100(2):E243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, et al. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther. 2014;13(7):1907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanden Borre P, McFadden DG, Gunda V, Sadow PM, Varmeh S, Bernasconi M, et al. The next generation of orthotopic thyroid cancer models: immunocompetent orthotopic mouse models of BRAF V600E-positive papillary and anaplastic thyroid carcinoma. Thyroid. 2014;24(4):705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21(16):5500–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henssen AG, Althoff K, Odersky A, Beckers A, Koche R, Speleman F, et al. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clin Cancer Res. 2015;22(10):2470–81. [DOI] [PubMed] [Google Scholar]
- 23.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273(29):18623–32. [DOI] [PubMed] [Google Scholar]
- 24.Glover M, Sweeny C, Davis B, O’Shaughnessy KM. A Single Amino Acid Substitution Makes WNK4 Susceptible to SB 203580 and SB 202190. Open Med Chem J. 2010;4:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpel-Massler G, Westhoff MA, Zhou S, Nonnenmacher L, Dwucet A, Kast RE, et al. Combined inhibition of HER1/EGFR and RAC1 results in a synergistic antiproliferative effect on established and primary cultured human glioblastoma cells. Mol Cancer Ther. 2013;12(9):1783–95. [DOI] [PubMed] [Google Scholar]
- 26.Shields MA, Dangi-Garimella S, Krantz SB, Bentrem DJ, Munshi HG. Pancreatic Cancer Cells Respond to Type I Collagen by Inducing Snail Expression to Promote Membrane Type 1 Matrix Metalloproteinase-dependent Collagen Invasion. J Biol Chem. 2011;286(12):10495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Ye HL, Zhang G, Yao WM, Chen XZ, Zhang FC, et al. Autophagy inhibition contributes to the synergistic interaction between EGCG and doxorubicin to kill the hepatoma Hep3B cells. PLoS One. 2014;9(1):e85771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soica C, Oprean C, Borcan F, Danciu C, Trandafirescu C, Coricovac D, et al. The synergistic biologic activity of oleanolic and ursolic acids in complex with hydroxypropyl-gamma-cyclodextrin. Molecules. 2014;19(4):4924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shields MA, Ebine K, Sahai V, Kumar K, Siddiqui K, Hwang RF, et al. Snail Cooperates with KrasG12D to Promote Pancreatic Fibrosis. Mol Cancer Res. 2013;11(9):1078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar K, DeCant BT, Grippo PJ, Hwang RF, Bentrem DJ, Ebine K, et al. BET inhibitors block pancreatic stellate cell collagen I production and attenuate fibrosis in vivo. JCI Insight. 2017;2(3):e88032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in Thyroid Cancer Incidence and Mortality in the United States, 1974–2013. JAMA. 2017;317(13):1338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. [DOI] [PubMed] [Google Scholar]
- 35.Joshi S, Platanias LC. Mnk kinase pathway: Cellular functions and biological outcomes. World journal of biological chemistry. 2014;5(3):321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon C, Cho SJ, Chang KK, Park DJ, Ryeom SW, Yoon SS. Role of Rac1 Pathway in Epithelial-to-Mesenchymal Transition and Cancer Stem-like Cell Phenotypes in Gastric Adenocarcinoma. Mol Cancer Res. 2017;15(8):1106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–42. [DOI] [PubMed] [Google Scholar]
- 38.Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S, et al. Resistance to BET Bromodomain Inhibitors Is Mediated by Kinome Reprogramming in Ovarian Cancer. Cell Rep. 2016;16(5):1273–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y, Wang L, Neitzel LR, Loganathan SN, Tang N, Qin L, et al. The MAPK Pathway Regulates Intrinsic Resistance to BET Inhibitors in Colorectal Cancer. Clin Cancer Res. 2017;23(8):2027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, et al. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5(12):1104–10. [DOI] [PubMed] [Google Scholar]
- 41.Alsayed Y, Uddin S, Mahmud N, Lekmine F, Kalvakolanu DV, Minucci S, et al. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J Biol Chem. 2001;276(6):4012–9. [DOI] [PubMed] [Google Scholar]
- 42.Verma A, Mohindru M, Deb DK, Sassano A, Kambhampati S, Ravandi F, et al. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to arsenic trioxide. J Biol Chem. 2002;277(47):44988–95. [DOI] [PubMed] [Google Scholar]
- 43.Dolniak B, Katsoulidis E, Carayol N, Altman JK, Redig AJ, Tallman MS, et al. Regulation of arsenic trioxide-induced cellular responses by Mnk1 and Mnk2. J Biol Chem. 2008;283(18):12034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bramham CR, Jensen KB, Proud CG. Tuning Specific Translation in Cancer Metastasis and Synaptic Memory: Control at the MNK-eIF4E Axis. Trends Biochem Sci. 2016;41(10):847–58. [DOI] [PubMed] [Google Scholar]
- 45.Gao Z, Yuan T, Zhou X, Ni P, Sun G, Li P, et al. Targeting BRD4 proteins suppresses the growth of NSCLC through downregulation of eIF4E expression. Cancer Biol Ther. 2018;19(5):407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abedin SM, Boddy CS, Munshi HG. BET inhibitors in the treatment of hematologic malignancies: current insights and future prospects. Onco Targets Ther. 2016;9:5943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3(4):e186–95. [DOI] [PubMed] [Google Scholar]
- 48.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3(4):e196–204. [DOI] [PubMed] [Google Scholar]
- 49.Dreas A, Mikulski M, Milik M, Fabritius CH, Brzozka K, Rzymski T. Mitogen-activated Protein Kinase (MAPK) Interacting Kinases 1 and 2 (MNK1 and MNK2) as Targets for Cancer Therapy: Recent Progress in the Development of MNK Inhibitors. Curr Med Chem. 2017;24(28):3025–53. [DOI] [PubMed] [Google Scholar]
- 50.Zhan Y, Guo J, Yang W, Goncalves C, Rzymski T, Dreas A, et al. MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma. J Clin Invest. 2017;127(11):4179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.