Abstract
Background:
Janus kinases (JAK1–3, TYK2) mediate cytokine signals in the regulation of hematopoiesis and immunity. JAK2 clinical mutations cause myeloproliferative neoplasms and leukemia and the mutations strongly concentrate in the regulatory pseudokinase domain, JAK homology 2, JH2. Current clinical JAK inhibitors target the tyrosine kinase domain and lack mutation- and pathway-selectivity.
Objective:
To characterize mechanisms and differences for pathogenic and cytokineinduced JAK2 activation to enable design of novel selective JAK inhibitors.
Methods:
Systematic analysis of JAK2 activation requirements using structure-guided mutagenesis, cell signaling assays, microscopy, and biochemical analysis.
Results:
Distinct structural requirements identified for activation of different pathogenic mutations. Specifically, the predominant JAK2 mutation V617F is the most sensitive to structural perturbations in multiple JH2 elements (C helix (αC), SH2-JH2 linker and ATPbinding site). In contrast, activation of K539L is resistant to most perturbations. Normal cytokine signaling shows distinct differences in activation requirements: JH2 ATP-binding site mutations have only a minor effect on signaling, while JH2 αC mutations reduce homomeric (JAK2-JAK2) EPO signaling, and almost completely abrogate heteromeric (JAK2-JAK1) IFNγ signaling, potentially by disrupting a dimerization interface on JH2.
Conclusions:
These results suggest that therapeutic approaches targeting the JH2 ATPbinding site and αC could be effective in inhibiting most pathogenic mutations. JH2 ATPsite targeting have potential for reduced side-effects by retaining EPO and IFNγ functions. Simultaneously, however, we identify the JH2 αC interface as a potential target for pathway-selective JAK inhibitors in diseases with unmutated JAK2, thus providing new insights for the development of novel pharmacological interventions.
Keywords: Janus kinase, JAK2 V617F, cytokine signaling, myeloproliferative neoplasm, kinase activation, drug design
Introduction
Janus kinases (JAKs) are non-receptor tyrosine kinases critically involved in cellular signaling, regulating the immune system, development, differentiation, and growth 1. Signaling through JAKs are numerous proinflammatory cytokines, including interleukins (IL-)2, IL-3, IL-4, IL-6, IL-9, IL-12, IL-13, IL-15, IL-23, and granulocyte-macrophage colony stimulating factor (GM-CSF), making JAK inhibition a tempting drug target for the treatment of inflammatory diseases 2. Similarly, aberrant signaling caused by activating gain-of-function (GOF) mutations in JAKs underlie multiple neoplastic diseases, including myeloproliferative neoplasms 3. Indeed, the recent advent of JAK inhibitors for the treatment of both of these disease groups has made understanding the mechanisms of JAK-STAT signaling highly relevant to the clinical immunologist 4.
JAKs associate with type I and type II cytokine receptors and mediate cytokine signals from activated receptors to signal transducers and activators of transcription (STATs), which upon phosphorylation by JAKs, move to the nucleus to activate transcription. The four JAKs in mammals (JAKs 1–3, TYK2) signal at homodimeric (JAK2) or heterodimeric/oligomeric (all JAKs) receptors and consist of four domains (N-to-C): a 4.1-band, ezrin, radexin, moiesin (FERM) domain, a Src homology 2 (SH2)-like domain, a pseudokinase domain (JAK homology 2, JH2), and a protein tyrosine kinase domain (JH1). FERM-SH2 mediate association to cytokine receptors 5. JH2 serves a dual role; it inhibits the tyrosine kinase activity of JH1 in the basal state, and is required for full activation upon cytokine stimulation 6–9.
JH2 is a mutational hotspot for clinical JAK mutations. The somatic JAK2 V617F mutation in exon 14, for example, causes ligand-independent JAK2 activation and underlies >95% of polycythemia vera and >50% of essential thrombocythemia and primary myelofibrosis cases 10. Other JAK2 GOF mutations are located in JH2 in exon 12 (residues 506–547, including K539L), exon 16 (including R683S/G), and some in JH1 3. JH2 GOF mutations in other JAKs cause leukemias and loss-of-function mutations cause immune deficiencies 3, highlighting the dual regulatory role of JH2 6,7,11. Recent structural information of the JH2JH1 interaction explains the inhibitory function of JH2 9,12. In this interaction, the C helix (αC) side of JH2 binds to JH1 in a front-to-back orientation (Figure 1 A) leading to conformational restriction of JH1 and inhibition of kinase activity 12.
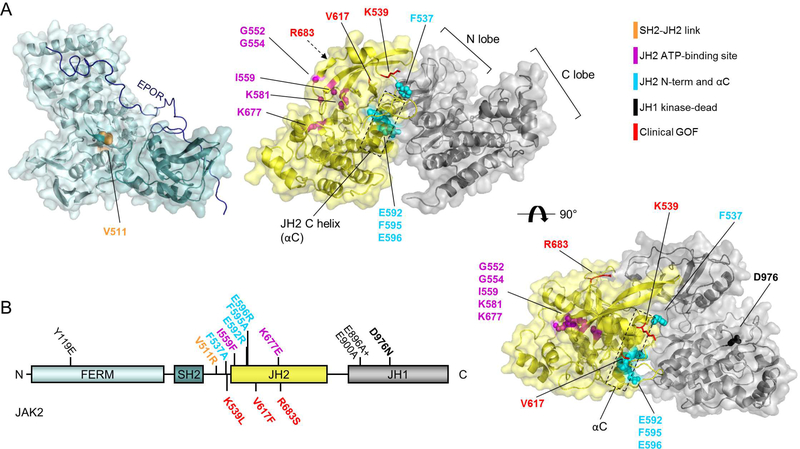
Figure 1: JAK2 domain structure.
A: Structures of JAK2 FERM-SH2 (left, PDB: 4Z32) with model of EPOR JAK2-binding peptide shown in dark blue (modelled based on Interferon λ 1 receptor (IFNLR1) peptide bound to JAK1 FERM-SH2, PDB: 5L04), and JAK2 JH2-JH1 inhibitory interaction 12. Right: JAK2 JH2-JH1 top view. B: Domain structure of JAK2. See also Table 1.
While multiple GOF mutations lie in the JH2-JH1 interface, disruption of the JH2-JH1 interaction alone does not fully explain the high activation potential of all GOF mutations—including JAK2 V617F or JAK2 K539L. We speculate that these mutations utilize the known, but molecularly incompletely characterized, stimulatory function of JH2 to activate JAK2.
Current clinical JAK inhibitors used to treat diseases caused by JAK2 GOF mutations target JH1 and thus do not distinguish between mutated and wild-type (WT) JAK2 and are unable to eradicate the disease. Furthermore, they frequently lead to anemia caused by suppression of normal erythropoietin (EPO) signaling due to inhibition of JAK2 WT functions 13. In contrast, in inflammatory diseases, in which usually no JAK mutations are present (with rare exceptions, see e.g., ref 14) current inhibitors are effective in approximately half of the patients, but also affect unwanted cytokine functions and show side-effects such as reactivation of viral infections and anemia 4. Thus, there is a clinical need for more effective and selective JAK inhibitors able to discriminate between pathogenic and cytokine-induced signaling and/or discriminate between different types of JAK-mediated signaling pathways.
However, a potential paradigm shift in JAK inhibition is emerging, as molecular characterization of JH2 is suggesting an alternative approach and implies JH2 to be a valid target for novel modulators of JAK activity 15. JH2 harbors the majority of human pathogenic JAK mutations, and we recently identified the JAK JH2 ATP-binding site as a potential drug target by demonstrating that activation by the pathogenic JAK2 JH2 GOF mutations K539L, V617F, and R683S is reliant on the stabilizing effect of ATP binding on JH2 16. Furthermore an ATP-competitive compound targeting TYK2 JH2 has been demonstrated to efficiently and specifically inhibit cytokine signaling 17.
Here, we provide a systematic analysis of the molecular basis for different JH2-targeting intervention strategies. We identify distinct differences in activation mechanisms between clinical JAK2 GOF mutations in terms of reliance on specific activating JH2 molecular interfaces and JAK2-mediated receptor dimerization. Analysis of cytokine-induced signaling shows differences in JH2 interface requirements between homodimeric (EPO) and heterodimeric (Interferon γ, IFNγ) JAK2 activation. These results provide novel insights into pathogenic and cytokine induced JAK2 activation mechanisms that have implications for development of mutant- and potentially pathway-preferring inhibitors.
Materials and Methods
See Supplementary material for full details of Materials and Methods. Briefly, for immunoblotting and luciferase reporter assays, JAK2-deficient γ2A human fibrosarcoma cells 18 were transfected with the designated combination of human JAK2-HA, human HAEPOR (both in pCIneo), and human STAT5-HA (in pXM) using FuGENE HD (Promega) for 24–48 h. For reporter assays, a Firefly luciferase reporter plasmids for STAT5 (Spi-Luc 1) or STAT1 (IRF-GAS 19) were added along with a constitutively expressing Renilla luciferase plasmid. Cytokine stimulation was done in starvation medium without FBS for 30 min (for immunoblotting) or 5 h (for reporter assays) unless otherwise specified, with recombinant human EPO (Roche), or IFNγ (Peprotech). For immunoblotting, cells were washed with PBS, lysed in Triton X-100 lysis buffer, and complete lysates run on lab-made SDS-PAGE gels. Immunoblots were blocked with bovine serum albumin and incubated with primary antibodies: HA Tag (Aviva Systems Biology), phospho-JAK2 (Millipore), phospho-STAT5 (Cell Signaling), phospho-STAT1 (Cell Signaling), STAT1 (BD Biosciences), or actin (Millipore), and a mixture of goat-anti-rabbit and goat-anti-mouse DyLight secondary antibodies (both Thermo Fisher Scientific). Blots were read using an Odyssey CLx (LI-COR), and immunoblot signals quantified using Image Studio software (LI-COR) by manually assigning bands (See Supplementary Material and Figure S1). Reporter assays were detected using the DualGlo reporter assay kit (Promega) according to manufacturer’s instructions and normalized to readings from wells of unstimulated cells transfected with JAK2-HA WT.
For qPCR analysis, γ2A cells were transfected for 28 h, starved for 16 h, stimulated for 2 h with 10 U/ml EPO or 10 ng/ml IFNγ, and RNA extracted using TRI Reagent (Molecular Research Center) according to manufacturer’s instructions. IRF1 gene expression was measured from reverse-transcribed total RNA using specific primers (5’GCATGAGACCCTGGCTAGAG-3’ and 5’-CTCCGGAACAAACAGGCATC-3’) and normalized to the expression of TATA-box binding protein (TBP).
For in vitro kinase assays, recombinant JAK2 JH2-JH1 (residues 513–1132–6×His) WT, I559F, and E592R proteins were expressed in High Five insect cells (Thermo Fisher Scientific) using the Bac-to-Bac expression system (Invitrogen) according to manufacturer’s instructions. Cells were lysed by freeze-thawing, clarified by centrifugation, and recombinant proteins purified using Ni-NTA agarose (Qiagen) followed by sizeexclusion chromatography in a HiLoad 16/600 Superdex 75 pg column (GE Healthcare). Protein concentrations were measured by Bradford assay (Bio-Rad) and enzymatic activity determined with Lance Ultra kinase assay (PerkinElmer) under conditions recommended by the manufacturer. Kinase reactions were performed in triplicate and results shown are representative from 2–3 individual experiments.
For microscopy, cells were seeded on 35 mm glass bottom dish (MatTek), transfected with JAK2-YFP fusion constructs (in pEGFP) or EPOR-YFP/EPOR-CFP (in pBOF 20) overnight and starved for 8 h. Cells were fixed with 4% paraformaldehyde and 0.1% glutaraldehyde for 15 minutes at room temperature, washed, and kept in PBS at 4 °C before imaging on a Zeiss LSM 780 laser scanning confocal microscope using a Plan Apochromat 63×/1.4 oil immersion objective. FRET was monitored by acceptor photobleaching 21 and FRET efficiency was calculated from manually segmented cell membrane areas.
Results
Suppressing mutations reveal differences in activation mechanisms of GOF mutations
Studies on JAK2 activation mechanisms have identified several mutations capable of suppressing activation by pathogenic JAK2 GOF mutations (see Table 1, Figure 1, Figure 2). These mutations, termed here ‘suppressing mutations’, localize in JH2 αC (F595A), in the C-terminus of the SH2-JH2 linker (F537A) and in the JH2 ATP-binding site (see Table 1). Recently Leroy et al. identified an additional residue in the outer face of JH2 αC (JAK2 E596) as an important link in the activation mechanism of V617F, but not of K539L, R683G, or of T875N 22. Notably, these suppressing mutations are functionally distinct from mutations that completely destabilize JH2 structure (e.g. JAK2 F739R refs 16,23 or deletion of JH2 αG 8), which mimic JH2 deletion resulting in increased basal activation and irresponsiveness to cytokines.
Table 1:
Used JAK2 mutations and their presumed mode of action or experimental 530 rationale. See also Figure 1.
| Mutation | Substructure | Rationale / mode of action | Reference |
|---|---|---|---|
| Y119E | FERM F1 | Mimics Y119 phosphorylation. Previously reported to induce dissociation of JAK2 from receptor. | 29,32 |
| V511R | SH2 | Designed to disrupt SH2-JH2 linker from FERMSH2. | - |
| F537A | SH2-JH2 link | F537 proposed to stack with F595 in JAK2 JH2 WT. Known to inhibit V617F. | 26 |
| K539L | SH2-JH2 link | Activating by unknown mechanism. Causes PV. | 38 |
| G552A + G554A | JH2 β1: Glyrich loop | Designed to remove flexible glycines usually needed for ATP binding. | 16 |
| I559F | JH2 β2 | Designed to sterically inhibit ATP binding. Verified to inhibit ATP binding 16. | 16 |
| K581A | JH2 β3 | Removes conserved β3 lysine. | 16,39 |
| E592R | JH2 αC | Outer face of JH2 αC | 12 |
| F595A | JH2 αC | Inner face of JH2 αC. Known to inhibit V617F and others by potentially destabilizing JH2 and making space for F617 (ref 12). | 12,23–25 |
| E596R | JH2 αC | Outer face of JH2 αC. Known to inhibit V617F and others. Mechanism unknown. | 22 |
| V617F | JH2 β4-β5 loop | Activating, potentially by disturbing SH2-JH2 linker. Causes MPNs. | 40–43 |
| K677E | JH2 β6-β7 loop | Designed to inhibit ATP binding electrostatically. Verified to inhibit ATP binding. | 16 |
| R683S | JH2 β7-β8 loop | Activating, probably by breaking R683-D873 interaction over inhibitory JH2-JH1 interface. Causes ALL. | 44,45 |
| T875N | JH1 β2-β3 loop | Activating, mechanism probably similar to R683S. Causes AMKL. | 46 |
| L884P | JH1 β3-αC loop | Activating by unknown mechanism. Homologous to JAK3 L857P found in ALL. | 47 |
| E896A + E900A | JH1 αC | Outer face of JH1 αC. | - |
| D976N | JH1 β6-β7 loop | D in HRD. Mutation is catalytically inactive (i.e., kinase dead). | - |
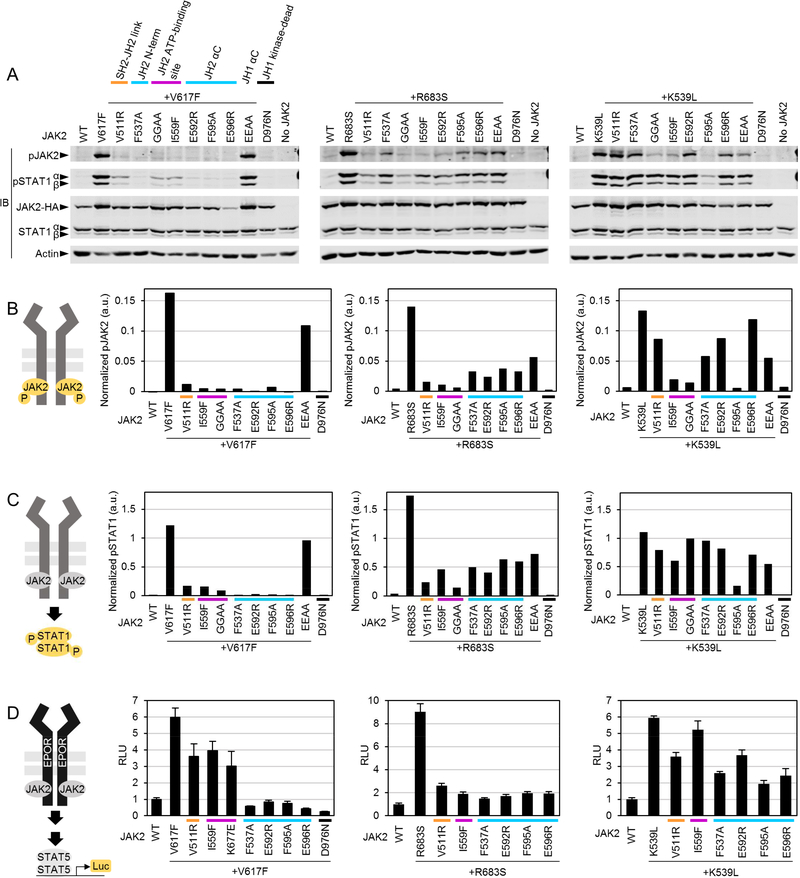
Figure 2: Suppressing JAK2 mutations reveal distinct activation mechanisms for different JAK2 gain-of-function (GOF) mutations.
A: Representative immunoblots of whole-cell lysates from JAK2-deficient γ2A cells transiently transfected with full-length JAK2-HA mutants as indicated. pJAK2, JAK2 activation loop phosphorylation JAK2(Y1007/1008); pSTAT1, STAT1(Y701) phosphorylation. GGAA, G552A+G554A. EEAA, E896A+E900A. Experiment was repeated twice with similar results. B and C: Quantification of immunoblots shown in A. a.u., arbitrary units. D: STAT5 reporter assay in the presence of transfected EPOR-HA. Averages and standard deviations from triplicate wells are shown as fold induction relative to unstimulated JAK2 WT. RLU, relative luminescence units. All reporter experiments were repeated twice with similar results.
The activation mechanisms and requirements of regulatory interfaces for different GOF mutations and cytokine-induced JAK2 activation have not been systemically analyzed. We thus set out to compare ligand-independent (pathogenic) and normal ligand-dependent JAK2 activation using suppressing mutations in JAK2-deficient γ2A fibroblast cells. We focused on the three previously identified regulatory regions in JH2: the ATP-binding site (JAK2 mutations I559F, G552A+G554A, or K677E) 16, the outer face of JH2 including αC (F595A, E596R) 12,22–25, and the C-terminus of the SH2-JH2 linker (F537A) 26. Additionally, we tested a novel JH2 αC outer face mutation, E592R, in order to analyze the involvement of the N-terminus of the JH2 αC. We further hypothesized that JH2 functions as a structural linker between FERM-SH2 and JH1 and, when structurally sound, is able to position JH1 for trans-autophosphorylation. To test this, we aimed to break up the putative interaction between FERM-SH2 and JH2 by introducing V511R to disrupt the short β sheet between the SH2-JH2 linker and the FERM F1-F2 loop (Figure 1). We also included JH1 αC outer face mutations (E896A+E900A) analogous to the JH2 mutations E592R/E596R to test the function of JH1 αC as a potential interaction interface.
We analyzed activation by three different pathogenic GOF JAK2 mutations predicted to have differing activation mechanisms (Figure 2): V617F (exon 14), which has been suggested to alter the conformation of the SH2-JH2 linker and thus indirectly affect the inhibitory JH2-JH1 interaction 12,26; R683S (exon 16), which is predicted to activate primarily by breaking the inhibitory JH2-JH1 interaction 9,12; and K539L (exon 12), which lies at the N terminus of JH2, and thus might also affect the SH2-JH2 linker, but whose activation mechanism has not been studied in detail.
In accordance with previous reports 16,22,24,25 we found that ligand-independent JAK2 JH1 activation loop (Y1007-Y1008, pJAK2) hyperphosphorylation caused by V617F is suppressed by JH2 ATP pocket and αC mutations (Figure 2 A and B, first panel). V617Finduced pJAK2 is also suppressed by V511R, suggesting that the activation mechanism of V617F requires correct linking of JH2 to SH2. However, JAK2V617F activation is not sensitive to perturbation of JH1 αC (Figure 2 A and B, first panel). Downstream pSTAT1 analysis correlated with pJAK2 levels. Effects of suppressing mutations on STAT5 activation were analyzed in reporter assays with EPOR-HA coexpression (Figure 2 D), where the inhibition profile correlated with pJAK2 and pSTAT1 analysis with strongest inhibition with αC mutations and F537A.
Activation by R683S was sensitive to all suppressing mutations in pJAK2 and pSTAT1 analysis as well as in STAT5 transcriptional activation, and the ATP-binding site mutants showed slightly more suppression than mutations in αC. Interestingly, K539L was clearly the most resistant to suppression, and only the αC mutation F595A strongly suppressed K539L in pJAK2, pSTAT1, and STAT5 activation. JH2 ATP-binding site mutations affected mainly JAK2 phosphorylation. These data suggest a distinct activation mechanism for K539L over V617F and R683S.
Taken together, these results indicate that interactions involving JH2 are critical for hyperactivation of all JAK2 GOF mutants, but that the specific JH2-mediated interactions differ between the GOF mutations.
The effect of suppressing mutations on cytokine activation
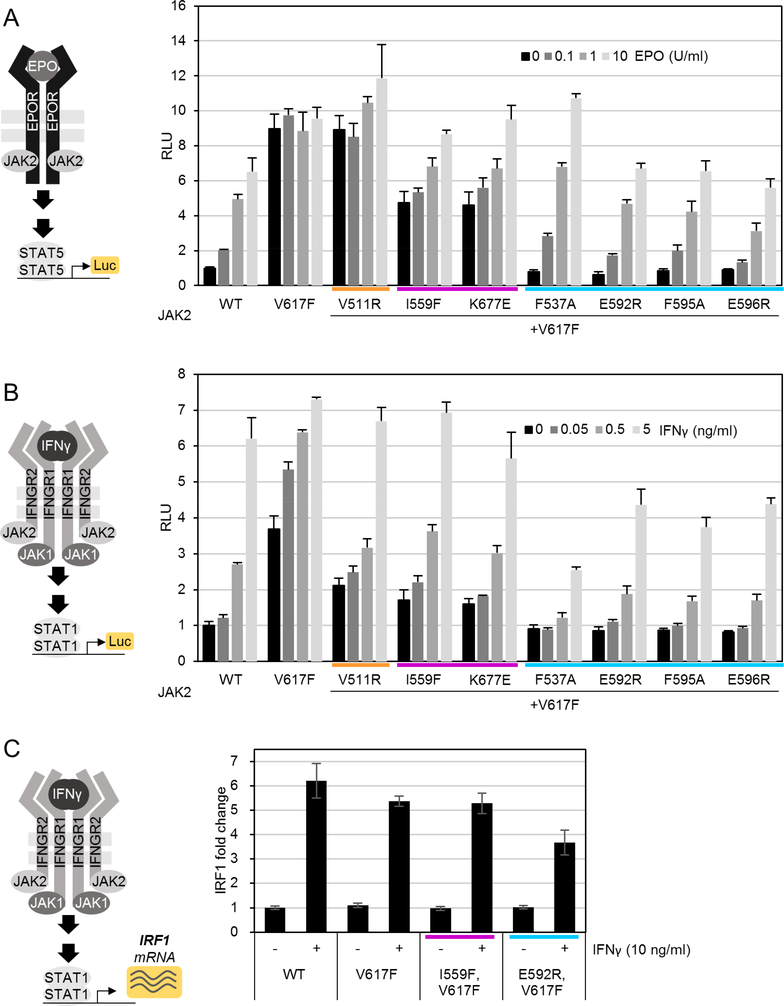
Cytokine stimulation titrations with JAK2-HA V617F+suppressor double-mutant constructs showed that even strong suppression of basal V617F-induced activity did not inhibit cytokine-induced STAT5 transcriptional activation for EPO or STAT1 activation for IFNγ (Figure 3 A and B, respectively). Rather, the most potent suppressor mutations (F537A and all αC mutations) restored EPO sensitivity to be indistinguishable from JAK2 WT (Figure 3 A). For IFNγ, cytokine sensitivity was also restored, which was further corroborated with qPCR of induction of expression of the IFNγ-responsive gene Interferon regulatory factor 1 (IRF1). Interestingly, however, IFNγ-induced STAT1 activation with αC and F537A mutations with V617F were lower than with JAK2 WT (Figure 3 B and C) suggesting potential specific involvement of these regions in IFNγ signaling.
Figure 3: Suppression of V617F activation by secondary mutations restores cytokine sensitivity.
A: STAT5 reporter assay in the presence of transfected EPOR-HA. B: IFNγ/STAT1 reporter. A and B as described for Figure 2. C: quantitative PCR (qPCR) of IFNγ-induced interferon regulatory factor 1 (IRF1). Averages and standard deviations from two biological replicates each done in technical triplicates in qPCR are shown. Mutations are color-coded by type as in Figure 2.
Previous work has suggested that suppressing mutations do not inhibit cytokine-induced signaling in a JAK2 WT background 16,22,24, but detailed analysis of sensitivity to different modes of JAK2-mediated signaling (homo- vs. heterodimeric) has been lacking. We thus analyzed cytokine-induced JAK2 activation using the mutation panel in a JAK2 WT background. Immunoblot analysis of JAK2-mediated STAT5 phosphorylation on homodimeric EPO receptor showed that, despite lower basal signaling activity, EPOinduced signaling was preserved in suppressing mutations (Figure 4 A), and JH2 ATPbinding site mutations were virtually identical to JAK2 WT in their response to EPO. F595A and E592R in αC and F537A, however, showed diminished EPO-induced STAT5 phosphorylation (Figure 4 A). Reporter assays showed similar results, albeit with differences even more pronounced (Figure 4 C).
Figure 4: Analysis of suppressing mutations in JAK2 WT background.
A and B: Quantifications from immunoblots, see also Figure S2 A and B. C: STAT5 reporter assay in the presence of transfected EPOR-HA. D: IFNγ/STAT1 reporter assay. C and D as described for Figure 2. E: qPCR of IRF1 expression as described for Figure 3. Wild-type sample (WT) same as in Figure 3 C. Mutations are color-coded by type as in Figure 2. Experiments were repeated twice with similar results.
Strikingly, the same JH2 αC and SH2-JH2 linker mutations practically abolished heteromeric JAK2-JAK1-mediated STAT1 phosphorylation upon IFNγ stimulation (Figure 4 B), while JH2 ATP-site mutations were again indistinguishable from JAK2 WT. In accordance with pSTAT1-immunoblot data, IFNγ-induced STAT1 transcription activity was almost completely abrogated for all αC mutations (including E596R) and F537A, whereas JH2 ATP-site mutations and V511R showed no significant decrease (Figure 4 D). IRF1qPCR further corroborated these results with E592R and I559F (Figure 4 E).
We also measured STAT1 activation with longer IFNγ stimulation times to estimate rule out simply delayed signaling kinetics 27, and found no activation of STAT1 with E592R or F537A mutants even at long time-scales (Figure S2 D).
Characterization of JAK2 GOF activation mechanisms by suppressor mutations
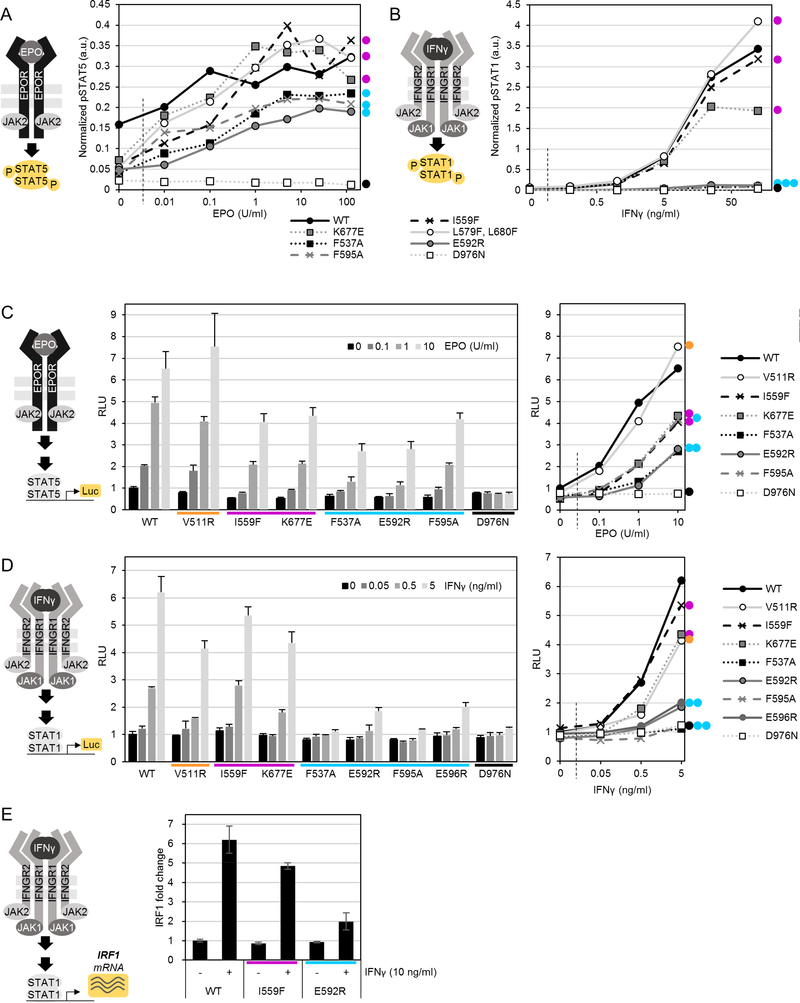
JAK2 V617F hyperactivation relies on the interaction with cytokine receptors but the underlying mechanisms are still elusive 28–30. Disruption of FERM and receptor binding of JAKs to receptors is a potential mechanism of suppression 28, and it has been suggested that some JAK3 JH2 mutations (including a JH2 ATP-binding site mutation) could affect subcellular localization of JAK3 31. We assessed subcellular localization of our suppressing mutations by imaging JAK2-YFP fusion proteins, but found no effect for either mutation on subcellular localization, either with or without added EPOR-HA (Figure 5 A). In contrast, JAK2-YFP with Y119E, known to cause dissociation from receptors 29,32, showed exclusively cytoplasmic JAK2 (Figure 5 A).
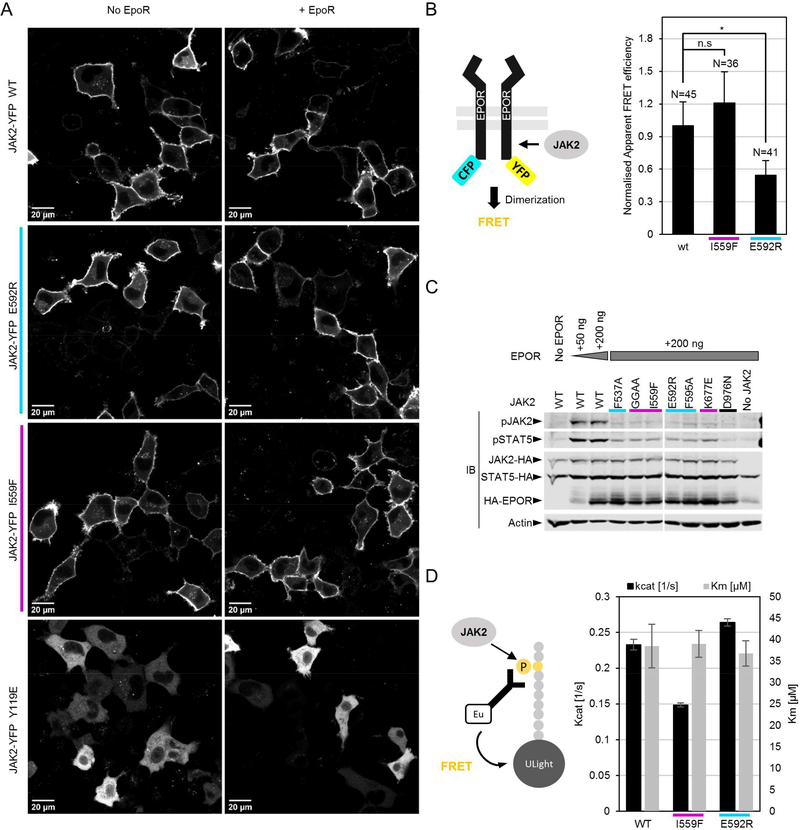
Figure 5: Suppressing mutations localize correctly to the membrane, but differentially affect dimerization of JAK2-EPOR and kinase activity of recombinant JAK2 JH2-JH1.
A: Representative confocal microscopy micrographs of fixed γ2A cells expressing the indicated JAK2-YFP mutations. B: Analysis of basal JAK2-EPOR dimerization. Normalized apparent FRET efficiency calculated from manually segmented cell membranes as detailed in Materials and Methods. Number of individual cells analyzed for each condition is indicated. Significance assessed by Student’s t test (unpaired). n.s. = not significant; *p < 0.05. C: Immunoblot analysis of whole-cell lysate from γ2A cells transiently transfected with the JAK2-HA constructs and EPOR-HA as shown. D: Kinase assay with purified recombinant JAK2 JH2-JH1. Shown are averages and standard deviation from triplicate measurements.
We speculated that mutation-induced JAK2-receptor dimerization is part of the activation mechanism of JAK2 GOF mutations. We thus analyzed whether suppressing mutations directly affect the propensity of JAK2 to dimerize on receptors. Using a FRET-based EPOR-CFP/YFP receptor dimerization assay in the JAK2-deficient γ2A fibroblast cell line (lacking endogenous EPOR expression) along with our JAK2-HA mutant constructs showed that E592R significantly reduces basal JAK2-EPOR dimerization (Figure 5 B). I559F, in contrast, showed a slight increase in dimerization, but this was within experimental noise and not significant.
To directly assess whether decreased dimerization propensity also translates to decreased receptor-mediated JAK2 activation, we assessed basal activation of otherwise wild-type JAK2. EPOR-HA overexpression (known to induce ligand-independent activation 16) alongside expression of JAK2 mutants showed that all suppressing mutations, irrespective of their mode of action, suppress EPOR-induced JAK2 activation (Figure 5 C), suggesting that other mechanisms beyond lowering of dimerization propensity (as shown for E592R (Figure 5 B), and potentially also true for other αC mutations) are likely at play to explain the mode of action of suppressing JH2 ATP-binding site mutations. We thus measured whether mutating the JH2 ATP-binding site or αC directly affects the enzymatic activity of JH1. Indeed, kinase assays with recombinant JAK2 JH2-JH1 fragments in vitro showed unchanged Km, ATP values for both JH2 mutations I559F and E592R, but lowered kinase reaction catalysis rates (kcat) for I559F (Figure 5 D).
Taken together, these data suggest that the mechanisms of suppression of JH2 ATPbinding site and αC mutations are different. Inhibiting ATP binding to JH2 directly lowers catalytic activity of JH1, potentially through lowering the stability of the JH2 αC 16, and thus strengthening the JH2-JH1 interaction 12. Altering the outer face charge of αC directly (e.g., with E592R), on the other hand, inhibits the propensity of JAK2 to dimerize and thereby hinders JH1 activity by suppressing trans-autophosphorylation (Figure 5 B, D).
Discussion
The molecular mechanisms of JAK activation by cytokine or mutation have long been elusive and here we have performed a systematic analysis of JAK2 activation mechanisms using structure-guided mutagenesis. Our results shed light on not only the mechanism of cytokine-independent JAK2 activation, but also identify a previously unknown interface on JH2 involved in JAK2-mediated receptor dimerization and needed especially for heteromeric JAK signaling.
Our results enable grouping of activating JAK2 mutations based on their requirements for distinct structural elements and thus activation mechanisms. Both V617F and R683S were sensitive to mutations affecting the JH2 ATP binding site and αC, albeit their suppressing effects showed differences, i.e., αC mutations completely abrogated V617F but not R683S, while JH2 ATP-binding site mutations showed similar suppression of both. The effect of suppressing mutations were more pronounced at pJAK2 and pSTAT1 than on STAT5 reporter assays which may reflect technical differences (e.g., stability of luciferase), or be indicative of signal amplification in the JAK-STAT pathway. Previously, we have shown that a suppressing JH2 ATP-binding site mutation reverts the increased hematocrit in a mouse V617F MPN model 16.
The resistance of K539L compared to V617F and R683S to suppressing mutations is interesting, since K539 and V617 reside near each other in the JH2 structure (Figure 1), and suggests a distinct activation mechanism for K539L. R683S likely activates through breaking the autoinhibitory interface resulting in increased conformational freedom and activation of JH1. This freed JH1 does, however, still rely on correct JH2-mediated positioning for ligand-independent activation, as well as JAK2-mediated receptor dimerization (see V511R and αC mutations, respectively in Figure 2). In contrast, K539L is unlikely to simply interfere with the autoinhibitory interaction, and our inhibitory profile analysis is consistent with a more direct activation mechanism of K539L, potentially involving direct activation of JH1, e.g., through interaction with K857 on JH1 22. Consistent with previous reports, K539L is effectively inhibited only by F595A, which is known primarily for participating in stabilizing interactions in JH2 αC in the context of V617F hyperactivation 24,25, but which has been suggested to alter the stability of the JH2 αC also more broadly.
For V617F, our results show complete inhibition by αC mutations including E592R, which our FRET-data indicate to interfere with JAK2-mediated receptor dimerization (Figure 5 B). We thus hypothesize that V617F activates JAK2 mostly by enhancing the propensity of JAK2 to dimerize on a receptor. This is in line with previous reports with recombinant JAK2 and TYK2 JH2-JH1 fragments, which showed only modest activation of kinase activity with the JAK2 V617F or analogous TYK2 V678F mutation in an isolated in vitro setting, which does not include JAK-mediated receptor dimerization effects 9,33. Our mutagenesis data furthermore suggests, that the dimerization interface directly includes the JH2 αC, with E592 (and probably E596) involved.
Our analysis of suppressing mutations in an otherwise wild-type background shows that, contrary to previous reports 16,22,24, suppressing mutations do affect JAK2 WT activity by lowering both basal (Figure 5 C), as well as ligand-dependent activation (Figure 4). Interestingly, quantitative comparison of potency of individual suppressing mutations to inhibit activation by V617F and cytokine reveals that these two correlate clearly (Figure S3). The correlating suppression of ligand-dependent and -independent JAK2 activation suggests that the same JH2 interface (αC and C-terminus of SH2-JH2 linker) is used in both settings. Furthermore, imaging data of JAK2-YFP shows unaltered subcellular distribution of JAK2 carrying suppressing mutations (Figure 5 A) ruling out direct destabilization of JAK2/FERM as an explanation for suppression.
Our results also reveal that JH2 αC suppressing mutations most likely inhibit JAK2 activation by suppressing JAK2 dimerization (Figure 5 B), while JH2 ATP-binding site mutations exert their suppressing effects by directly affecting tyrosine kinase activity of JAK2 JH1 (Figure 5 D), potentially through partial destabilization of JH2 αC 12,16. We thus conclude, that V617F and R683S most likely activate JAK2 by increasing its propensity for dimerization, and that this is counteracted by suppressing mutations in the JH2 αC (E592R, E596R, F595A) and SH2-JH2 linker (F537A). For the case of R683S, which lies directly in the JH2-JH1 autoinhibitory interface (Figure 1 and refs 9,12), we speculate that weakening of the autoinhibitory interaction releases JH2, which relieves autoinhibition on JH1, as well as exposing the dimerization interface on JH2.
The molecular details of heteromeric JAK activation have remained largely unknown. Our analysis of cytokine-stimulation of JAK2 carrying suppressing mutations strikingly suggest that the same interface needed for activation by V617F or R683S by dimerization of (receptor-bound) JAK2, is crucial especially for heteromeric activation of JAK2. This finding refines earlier work that showed a critical role for JH2 in JAK activation 7,11. Previous studies have shown in several cytokine receptor systems that catalytic activity of both JAKs is not required for heteromeric JAK activation 20,34. For instance, in IFNγ signaling STAT1 does not require enzymatically functional JAK1 35, but does require the presence of JAK1 JH2 36. Pathway-specific JAK substructures have also been implicated in JAK2 JH1 for EPO signaling 37. Our results refine these findings by identifying the Cterminus of the SH2-JH2 linker and JH2 αC as critical for heteromeric JAK activation. However, whether the JH2 interface identified here participates in JAK2-JAK2 or JAK2JAK1 dimers/multimers on IFNGR remains a topic for future research.
Currently available JAK inhibitors show beneficial clinical responses, but there is a clear need for more effective, optimally disease-selective drugs with less side-effects. The key question for this goal is to understand the differential mechanisms defining pathogenic and different cytokine-induced activation modes. Our results presented here provide insights into these questions and identify specific regions in JH2 that are differentially required for JAK2 activation in different contexts. These findings pave the way for the design of novel, potentially mutant and/or pathway-selective pharmacological compounds.
Supplementary Material
Figure S1: Validation of immunoblotting quantification method. A: Control immunoblot from whole-cell lysate of γ2A cells transiently transfected with JAK2-HA WT or JAK2-HA V617F as indicated. Lysates were run at different dilutions in lysis buffer to gauge linearity of immunoblotting signal. For detection, immunoblot is cut into three pieces (indicated with dashed lines) and the pieces double stained with Anti-pJAK2 + Anti-HA or Anti-pSTAT1 + STAT1, or single-stained with Anti-Actin. B: Example of quantification procedure using ImageStudio software (LICOR Biosciences). Shown is an example area from the immunoblot shown in (A) and indicated by a white dotted line. Bold blue boxes are the manually assigned areas of interest, each encompassing a single band (note, e.g., that for STAT1, only STAT1α is quantified). The light blue boxes on top and below each band are used to calculate the local background from the lane. From the background, the median signal intensity is used as a background value, which is deducted from the total signal from the area of interest. C: pJAK2 and total JAK2-HA quantifications showing the approximate linearity of the signal over the measured intensity range. Last panel shows the ratio of pJAK2 and JAK2-HA signal intensities, which is used as a measure of phosphorylation status (“Normalized pJAK2”). The point of deviation from linearity on the normalized pJAK2 panel shows the limits of pJAK2+JAK2-HA quantifiability (down to pJAK2 signal intensities of ~50), thus limiting the direct comparison of pJAK2 values to samples with relatively strong pJAK2 signals. D: pSTAT1 and total STAT1 quantifications as delineated above for (C). Note the exceptional linearity of STAT1 signals (and the ensuing stability of the normalized pSTAT1 value in the last panel), thus rendering pSTAT1 better suited for comparison of samples over a wide range of pSTAT1 signal intensities. E: Quantification of Actin signals showing poor linearity over the tested range.Thus Actin was not used for normalization in quantification of immunoblot data.
Figure S2: Characterization of effects of suppressing mutation on cytokine signaling in JAK2 WT background. Related to Figure 4. A and B: Immunoblots related to Figure 4 A and B, respectively. Immunoblots of whole-cell lysates from γ2A cells transiently transfected with full-length JAK2-HA (and mutants thereof), STAT5-HA, and EPOR-HA (A) or full-length JAK2-HA mutants only (B), and stimulated with the indicated amount of cytokine for 30 min. Quantification of immunoblots was done as shown in Figure S1. C: Related to Figure 4 E, qPCR of IRF1 from RNA extracted from γ2A cells transiently transfected with JAK2-HA mutants and stimulated with EPO for 2 h showing the specificity of IRF1 induction. D: Immunoblots of whole-cell lysates from γ2A cells transiently transfected with full-length JAK2-HA mutants and stimulated with 1 μg/ml IFNγ for the indicated times before lysis. JAK2-HA E592R and F537A show no induction of pSTAT1 even at longer stimulation times.
Figure S3: JAK2 activation by V617F relies on the same interfaces to activate as JAK2 activation by cytokine. Suppression calculated as STAT5 or STAT1 reporter activity relative to basal activity of JAK2 V617F (y-axes) and stimulated wild-type JAK2 (10 U/ml EPO or 5 ng/ml IFNγ; x-axes). Data for figure is from the same experiments as for Figures 2 and 3.
Acknowledgements
We thank Juha Saarikettu (PhD) for expert assistance with qPCR experiments and Krista 346 Lehtinen and Merja Lehtinen for excellent technical assistance. Tampere Imaging Facility (TIF) and Tampere facility of Protein Services (PS) are acknowledged for their service.
Funding sources:
Dr. Hammarén, Dr. George Abraham, Dr. Virtanen, and Dr. Silvennoinen report grants from Academy of Finland, grants from Sigrid Juselius Foundation, grants from Jane and Aatos Erkko Foundation, grants from Finnish Cancer Foundation, grants from Prostate Cancer Fondation, grants from Tampere University Hospital District competitive research funding, during the conduct of the study; Dr. Hubbard reports grant NIH R01AI101256; In addition, Dr. Hammarén reports personal fees from Pfizer Oy, Finland, outside the submitted work; and Dr. Silvennonen has a patent US patent no. 8,841,078, AU 2011214254, CAN 2789186, EPO 11741946.5 issued. Ms. Peussa has nothing to disclose.
Abbreviations
- JAK
Janus kinase
- GOF
gain-of-function
- MPN
myeloproliferative neoplasm
- JH
JAK homology
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (1).Yamaoka K, Saharinen P, Pesu M, Holt V 3rd, Silvennoinen O, O’Shea JJ The Janus kinases (Jaks). Genome Biol 2004;5(12):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Schwartz DM, Bonelli M, Gadina M, O’Shea JJ. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nature Reviews Rheumatology 2016;12(1):25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hammarén HM, Virtanen AT, Raivola J, Silvennoinen O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 2018;S1043–4666(18):30127–3. [DOI] [PubMed] [Google Scholar]
- (4).Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nature Reviews Drug Discovery 2017;16(12):843–862. [DOI] [PubMed] [Google Scholar]
- (5).Ferrao R, Lupardus PJ. The Janus Kinase (JAK) FERM and SH2 Domains: Bringing Specificity to JAK–Receptor Interactions. Frontiers in Endocrinology 2017;8(18):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol 2000;20(10):3387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yeh TC, Dondi E, Uzé G, Pellegrini S. A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-α signaling. Proc Natl Acad Sci U S A 2000;97(16):89916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Saharinen P, Vihinen M, Silvennoinen O. Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol Biol Cell 2003;14(4):14481459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lupardus PJ, Ultsch M, Wallweber H, Bir Kohli P, Johnson AR, Eigenbrot C. Structure of the pseudokinase–kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci U S A 2014. May 19;111(22):80258030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Skoda RC, Duek A, Grisouard J. Pathogenesis of myeloproliferative neoplasms. Exp Hematol 2015;43(8):599–608. [DOI] [PubMed] [Google Scholar]
- (11).Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem 2002;277(49):47954–63. [DOI] [PubMed] [Google Scholar]
- (12).Shan Y, Gnanasambandan K, Ungureanu D, Kim ET, Hammaren H, Yamashita K, et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat Struct Mol Biol 2014. 06/11;21:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sonbol MB, Firwana B, Zarzour A, Morad M, Rana V, Tiu RV. Comprehensive review of JAK inhibitors in myeloproliferative neoplasms. Ther Adv Hematol 2013;4(1):15–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Del Bel KL, Ragotte RJ, Saferali A, Lee S, Vercauteren SM, Mostafavi SA, et al. JAK1 gain-of-function causes an autosomal dominant immune dysregulatory and hypereosinophilic syndrome. J Allergy Clin Immunol 2017. June;139(6):2016–2020.e5. [DOI] [PubMed] [Google Scholar]
- (15).Leroy E, Constantinescu SN. Rethinking JAK2 inhibition: Towards novel strategies of more specific and versatile Janus kinase inhibition. Leukemia 2017. January 25;31(5):10231038. [DOI] [PubMed] [Google Scholar]
- (16).Hammaren HM, Ungureanu D, Grisouard J, Skoda RC, Hubbard SR, Silvennoinen O. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc Natl Acad Sci U S A 2015. March 30;112(15):4642–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tokarski JS, Zupa-Fernandez A, Tredup JA, Pike K, Chang C, Xie D, et al. Tyrosine Kinase 2-Mediated Signal Transduction in T Lymphocytes Is Blocked by Pharmacological Stabilization of its Pseudokinase Domain. J Biol Chem 2015. March 11;290(17):11061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kohlhuber F, Rogers NC, Watling D, Feng J, Guschin D, Briscoe J, et al. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol 1997. February;17(2):695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFN gamma, and is likely to autoregulate the p91 gene. EMBO J 1994. January 1;13(1):158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Haan C, Rolvering C, Raulf F, Kapp M, Drückes P, Thoma G, et al. Jak1 Has a Dominant Role over Jak3 in Signal Transduction through [gamma] c-Containing Cytokine Receptors. Chem Biol 2011;18(3):314–323. [DOI] [PubMed] [Google Scholar]
- (21).Bastiaens P, Majoul IV, Verveer PJ, Söling H, Jovin TM. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J 1996;15(16):4246–4253. [PMC free article] [PubMed] [Google Scholar]
- (22).Leroy E, Dusa A, Colau D, Motamedi A, Cahu X, Mouton C, et al. Uncoupling JAK2 V617F activation from cytokine-induced signalling by modulation of JH2 alphaC helix. Biochem J 2016. June 1;473(11):1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, Hubbard SR. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol 2012;19:754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dusa A, Mouton C, Pecquet C, Herman M, Constantinescu SN. JAK2 V617F Constitutive Activation Requires JH2 Residue F595: A Pseudokinase Domain Target for Specific Inhibitors. PLoS One 2010;5(6):207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gnanasambandan K, Magis A, Sayeski PP. The constitutive activation of Jak2-V617F is mediated by a π stacking mechanism involving phenylalanines 595 and 617. Biochemistry 2010;49(46):9972–9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Toms AV, Deshpande A, McNally R, Jeong Y, Rogers JM, Kim CU, et al. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol 2013;20(10):1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Keil E, Finkenstadt D, Wufka C, Trilling M, Liebfried P, Strobl B, et al. Important scaffold function of the Janus kinase 2 uncovered by a novel mouse model harboring a Jak2 activation-loop mutation. Blood 2014. January 23;123(4):520–529. [DOI] [PubMed] [Google Scholar]
- (28).Wernig G, Gonneville JR, Crowley BJ, Rodrigues MS, Reddy MM, Hudon HE, et al. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim protooncogenes. Blood 2008;111(7):3751–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yao H, Ma Y, Hong Z, Zhao L, Monaghan SA, Hu MC, et al. Activating JAK2 mutants reveal cytokine receptor coupling differences that impact outcomes in myeloproliferative neoplasm. Leukemia 2017. January 6;31(10):2122–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lu X, Huang LJS, Lodish HF. Dimerization by a cytokine receptor is necessary for constitutive activation of JAK2V617F. J Biol Chem 2008;283(9):5258–5266. [DOI] [PubMed] [Google Scholar]
- (31).Hofmann SR, Lam AQ, Frank S, Zhou YJ, Ramos HL, Kanno Y, et al. Jak3independent trafficking of the common gamma chain receptor subunit: chaperone function of Jaks revisited. Mol Cell Biol 2004. June;24(11):5039–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Funakoshi-Tago M, Pelletier S, Moritake H, Parganas E, Ihle JN. Jak2 FERM domain interaction with the erythropoietin receptor regulates Jak2 kinase activity. Mol Cell Biol 2008;28(5):1792–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sanz A, Ungureanu D, Pekkala T, Ruijtenbeek R, Touw IP, Hilhorst R, et al. Analysis of Jak2 catalytic function by peptide microarrays: The role of the JH2 domain and V617F mutation. PLoS One 2011;6(4):e18522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li Z, Gakovic M, Ragimbeau J, Eloranta M, Rönnblom L, Michel F, et al. Two Rare Disease-Associated Tyk2 Variants Are Catalytically Impaired but Signaling Competent. J Immunol 2013;190(5):2335–2344. [DOI] [PubMed] [Google Scholar]
- (35).Briscoe J, Rogers N, Witthuhn B, Watling D, Harpur A, Wilks A, et al. Kinase-negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. EMBO J 1996;15(4):799–809. [PMC free article] [PubMed] [Google Scholar]
- (36).Eletto D, Burns SO, Angulo I, Plagnol V, Gilmour KC, Henriquez F, et al. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat Commun 2016. Dec 23;7:13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Haan C, Kroy DC, Wuller S, Sommer U, Nocker T, Rolvering C, et al. An unusual insertion in Jak2 is crucial for kinase activity and differentially affects cytokine responses. J Immunol 2009. March 1;182(5):2969–2977. [DOI] [PubMed] [Google Scholar]
- (38).Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med 2007;356(5):459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol 2011;18(9):971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005;365(9464):1054–1061. [DOI] [PubMed] [Google Scholar]
- (41).James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434(7037):1144–1148. [DOI] [PubMed] [Google Scholar]
- (42).Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-offunction mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005;352(17):1779–1790. [DOI] [PubMed] [Google Scholar]
- (43).Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJP, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005;7(4):387–397. [DOI] [PubMed] [Google Scholar]
- (44).Bercovich D, Ganmore I, Scott LM, Wainreb G, Birger Y, Elimelech A, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down’s syndrome. Lancet 2008;372(9648):1484–1492. [DOI] [PubMed] [Google Scholar]
- (45).Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2009. June 9;106(23):9414–9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mercher T, Wernig G, Moore SA, Levine RL, Gu TL, Frohling S, et al. JAK2T875N is a novel activating mutation that results in myeloproliferative disease with features of megakaryoblastic leukemia in a murine bone marrow transplantation model. Blood 2006. October 15;108(8):2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Losdyck E, Hornakova T, Springuel L, Degryse S, Gielen O, Cools J, et al. Distinct Acute Lymphoblastic Leukemia (ALL)-associated Janus Kinase 3 (JAK3) Mutants Exhibit Different Cytokine-Receptor Requirements and JAK Inhibitor Specificities. J Biol Chem 2015. November 27;290(48):29022–29034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Validation of immunoblotting quantification method. A: Control immunoblot from whole-cell lysate of γ2A cells transiently transfected with JAK2-HA WT or JAK2-HA V617F as indicated. Lysates were run at different dilutions in lysis buffer to gauge linearity of immunoblotting signal. For detection, immunoblot is cut into three pieces (indicated with dashed lines) and the pieces double stained with Anti-pJAK2 + Anti-HA or Anti-pSTAT1 + STAT1, or single-stained with Anti-Actin. B: Example of quantification procedure using ImageStudio software (LICOR Biosciences). Shown is an example area from the immunoblot shown in (A) and indicated by a white dotted line. Bold blue boxes are the manually assigned areas of interest, each encompassing a single band (note, e.g., that for STAT1, only STAT1α is quantified). The light blue boxes on top and below each band are used to calculate the local background from the lane. From the background, the median signal intensity is used as a background value, which is deducted from the total signal from the area of interest. C: pJAK2 and total JAK2-HA quantifications showing the approximate linearity of the signal over the measured intensity range. Last panel shows the ratio of pJAK2 and JAK2-HA signal intensities, which is used as a measure of phosphorylation status (“Normalized pJAK2”). The point of deviation from linearity on the normalized pJAK2 panel shows the limits of pJAK2+JAK2-HA quantifiability (down to pJAK2 signal intensities of ~50), thus limiting the direct comparison of pJAK2 values to samples with relatively strong pJAK2 signals. D: pSTAT1 and total STAT1 quantifications as delineated above for (C). Note the exceptional linearity of STAT1 signals (and the ensuing stability of the normalized pSTAT1 value in the last panel), thus rendering pSTAT1 better suited for comparison of samples over a wide range of pSTAT1 signal intensities. E: Quantification of Actin signals showing poor linearity over the tested range.Thus Actin was not used for normalization in quantification of immunoblot data.
Figure S2: Characterization of effects of suppressing mutation on cytokine signaling in JAK2 WT background. Related to Figure 4. A and B: Immunoblots related to Figure 4 A and B, respectively. Immunoblots of whole-cell lysates from γ2A cells transiently transfected with full-length JAK2-HA (and mutants thereof), STAT5-HA, and EPOR-HA (A) or full-length JAK2-HA mutants only (B), and stimulated with the indicated amount of cytokine for 30 min. Quantification of immunoblots was done as shown in Figure S1. C: Related to Figure 4 E, qPCR of IRF1 from RNA extracted from γ2A cells transiently transfected with JAK2-HA mutants and stimulated with EPO for 2 h showing the specificity of IRF1 induction. D: Immunoblots of whole-cell lysates from γ2A cells transiently transfected with full-length JAK2-HA mutants and stimulated with 1 μg/ml IFNγ for the indicated times before lysis. JAK2-HA E592R and F537A show no induction of pSTAT1 even at longer stimulation times.
Figure S3: JAK2 activation by V617F relies on the same interfaces to activate as JAK2 activation by cytokine. Suppression calculated as STAT5 or STAT1 reporter activity relative to basal activity of JAK2 V617F (y-axes) and stimulated wild-type JAK2 (10 U/ml EPO or 5 ng/ml IFNγ; x-axes). Data for figure is from the same experiments as for Figures 2 and 3.