Abstract
The habenulo-interpeduncular system (HIPS) is now recognized as a critical circuit modulating aversion, reward, and social behavior. There is evidence that dysfunction of this circuit leads to psychiatric disorders. Because psychiatric diseases may originate in developmental abnormalities, it is crucial to investigate the developmental mechanisms controlling the formation of the HIPS. Thus far, this issue has been the focus of limited studies. Here, we explored the developmental processes underlying the formation of the medial habenula (MHb) and its unique output, the interpeduncular nucleus (IPN), in mice independently of their gender. We report that the Otx2 homeobox gene is essential for the proper development of both structures. We show that MHb and IPN neurons require Otx2 at different developmental stages and, in both cases, Otx2 deletion leads to disruption of HIPS subcircuits. Finally, we show that Otx2+ neurons tend to be preferentially interconnected. This study reveals that synaptically connected components of the HIPS, despite radically different developmental strategies, share high sensitivity to Otx2 expression.
SIGNIFICANCE STATEMENT Brain reward circuits are highly complex and still poorly understood. In particular, it is important to understand how these circuits form as many psychiatric diseases may arise from their abnormal development. This work shows that Otx2, a critical evolutionary conserved gene implicated in brain development and a predisposing factor for psychiatric diseases, is required for the formation of the habenulo-interpeduncular system (HIPS), an important component of the reward circuit. Otx2 deletion affects multiple processes such as proliferation and migration of HIPS neurons. Furthermore, neurons expressing Otx2 are preferentially interconnected. Therefore, Otx2 expression may represent a code that specifies the connectivity of functional subunits of the HIPS. Importantly, the Otx2 conditional knock-out animals used in this study might represent a new genetic model of psychiatric diseases.
Keywords: circuits formation, genetic models, homeoprotein, neuronal migration, psychiatric diseases, reward circuits
Introduction
Mounting evidence suggests that mood disorders and drug addiction are associated with abnormalities of brain reward circuits. The “neurodevelopmental hypothesis,” which was formulated 50 years ago and continuously receives support, states that these disorders take root during development. However, few studies have thus far focused on the development of reward circuits. An important component of these circuits, the habenulo-interpeduncular system (HIPS), composed of the medial habenula (MHb) connected to the interpeduncular nucleus (IPN) via the fasciculus retroflexus, represents a major connection between the limbic system in the forebrain and the monoaminergic systems in the midbrain and hindbrain. The HIPS is thought to be a regulator of mood and reward (for review, see McLaughlin et al., 2017), although major controversies remain (Yamaguchi et al., 2013; Hsu et al., 2016). The circuitry between MHb and IPN is classically subdivided into two parts. The dorsal part of the MHb (dMHb), which expresses Tac1 and contains substance P, targets the lateral interpeduncular nucleus (Lat-IPN), whereas the ventral part of the MHb (vMHb), which expresses Chat and contains acetylcholine, targets the medial IPN (Contestabile et al., 1987). These anatomically and genetically distinct parts may support different functions (Yamaguchi et al., 2013).
Although dysregulation of HIPS development may lead to mood disorders such as anxiety, not much is known about this issue in mammals. It has been shown that early patterning of the epithalamus, giving rise to the habenula, critically depends on signals expressed by the zona limitans intrathalamica (ZLI) such as Shh (Chatterjee et al., 2014) and that the transcription factor Brn3a is crucial for differentiation and maintenance of habenular neurons (Quina et al., 2009; Serrano-Saiz et al., 2018). The homeodomain transcription factor Otx2 is expressed in the epithalamus. This expression is conserved from fish to humans (Simeone et al., 1992; Mori et al., 1994; Larsen et al., 2010), suggesting an evolutionary conserved function. In particular, Otx2 expression in the habenula has been reported (Courtois et al., 2003; Mallika et al., 2015), although little is known about its dynamics during development. Regarding IPN development, the origin of the different IPN subparts has been studied in the chick (Lorente-Cánovas et al., 2012). IPN neurons originate from isthmic and rhombomere 1 (r1) regions. They can be divided into three main groups: the Nkx6.1+ prodromal domain, the Pax7+ rostral IPN (rIPN) and the caudal IPN (cIPN), largely Otx2+ with one subpopulation Pax7+/Otx2−. To form the IPN, IPN neurons migrate long distances through specific routes. Little is known about molecular pathways that guide them. One study reported that Shh deletion affected rIPN but not cIPN formation (Moreno-Bravo et al., 2014). Another study reported that the knock-out of the IPN-restricted Goosecoid like (Gscl) homeobox gene led to the disappearance of some molecular markers while the IPN was still forming. Interestingly, Gscl mutants were shown to exhibit abnormal sleep patterns (Funato et al., 2010).
Considering that abnormalities of HIPS development may lead to mood disorders, it is expected that genes controlling HIPS development may be recognized as susceptibility genes linked to these diseases. One particularly interesting candidate for mood disorders is Otx2. It is a susceptibility gene for bipolar disorders (Sabunciyan et al., 2007; Jukic et al., 2015) and is already known to play important functions in reward circuits. In mice, Otx2 downregulation in the ventral tegmental area (VTA) is induced by early stress and is tightly linked to the risk of developing depressive and anxiety-like disorders (Peña et al., 2017). Beyond the reward system, Otx2 is essential for the development of brain, cerebellum, pineal gland, and eye (for review, see Beby and Lamonerie, 2013). The role of Otx2 specifically in HIPS development has yet to be investigated and the list of genes known to be involved in this development is very short. In this study, we therefore investigated whether Otx2 is important for the formation of both the habenula and the IPN. We report crucial and multiple functions of this developmental gene at multiple steps of HIPS development.
Materials and Methods
Mice and tamoxifen injections.
All mice used in this study were maintained in the animal facilities of the Institut de Biologie Valrose at Université de Nice Sophia Antipolis, Nice, France. To visualize Otx2 expression pattern we used the reporter mouse line, in which Otx2 protein is fused to the fluorescence protein GFP (Otx2-GFP/+; Fossat et al., 2007). Otx2CreERT2 and Otx2flox/flox mice were generated as described previously (Fossat et al., 2006). The Dbx1Cre mouse line (Bielle et al., 2005) was obtained from A. Pierani (Institut Jacques Monod, Paris, France). Cre lines were mated with reporter mouse lines ROSA26YFP (Srinivas et al., 2001) and TAUGFP-NLSlacz (obtained from The Jackson Laboratory and developed by S. Arber; Hippenmeyer et al., 2005). All mouse lines were maintained in the 129/Sv background. Embryonic day 0.5 (E0.5) corresponded to the day that a vaginal plug was detected. Tamoxifen (Sigma-Aldrich) diluted in sunflower oil at 10 mg/ml was injected in pregnant females at 5 μl/g bodyweight. The care and handling of the animals prior or during the experimental procedures followed European Union rules and were approved by the French Animal Care and local ethic Committee (CIEPAL NCE/2015-202).
Immunohistochemistry and in situ hybridization.
Preparation of the brains for vibratome and cryostat sections were performed as described previously (Ruiz-Reig et al., 2018). Immuhistochemical staining was performed in a vibratome (80 μm) and cryostat (12–16 μm) sections using the following primary antibodies: chicken anti-GFP (Aves Laboratories, 1:2000), sheep anti-GFP (AbD Serotec, 1:200), mouse anti-Brn3a (Merck/Millipore, 1:250), rabbit anti-Er81 (generous gift frp, Silvia Arber, 1:1000), rabbit anti-Ki67 (Abcam, 1:100), goat anti-Otx (R&D Systems, 1:500), mouse anti-Pax7 (DSHB, 1:200), rabbit anti-Gscl (generous gift from Naomi Galili and Clayton A. Buck, University of Pennsylvania, 1:200; Galili et al., 1998), mouse anti-Nkx6.1 (DSHB, 1:100), mouse anti-GATA3 (Santa Cruz Biotechnology, 1:50), mouse anti-EN1 (DSHB, 1:100), rabbit anti-Pax6 (Merck/Millipore, 1:500), mouse IgM anti-RC2 (DSHB, 1:500), rabbit anti-ER (Thermo Fisher Scientific, 1:200), rabbit anti-GABA (Sigma-Aldrich, 1:1000), guinea pig anti-VGlut1 (Synaptic Systems, 1:500), rabbit anti-substance P (Millipore, 1:500), goat anti-DCC (R&D Systems, 1:400), and goat anti-ChAT (Millipore, 1:500). Alexa Fluor secondary antibodies (Jackson ImmunoResearch, 1:500) were also used. Prior incubation of adult brain sections with mouse antibodies, a blocking step of 2 h with mouse Fab fragments was performed. Primary antibodies against Brn3a, En1, Gata3, and Pax7 required prior antigen retrieval treatment (citrate buffer, pH 6, for 10 min at 95°C). Notes on goat anti-Otx (R&D Systems): these antibodies are known to recognize to some levels other homologous proteins to Otx2, such as Otx1, Crx, and maybe Otp (suggested by the expression pattern in the hypothalamus). For that reason, we used the Otx2-GFP line to monitor Otx2 expression with more precision. Nevertheless, these antibodies are useful in many cases because their affinity to Otx2 largely exceeds their affinity to other molecules. In the habenula and the IPN of Ox2 cKO animals, the absence of anti-Otx2 immunoreactivity suggested that these cells do not express other homologs or at an undetectable level, at least in the absence of Otx2.
In situ hybridization was performed in cryosections (12–16 μm) as described previously (Martinez-Lopez et al., 2015). The following probes were used: Gbx2 (Wassarman et al., 1997), Shh (Echelard et al., 1993), Dbx1 (Lu et al., 1994), and Netrin1 (Serafini et al., 1996). Tac1 and Dcc probes were prepared by PCR based on the Allen Atlas website with the following primers: Tac1 forward: CCCCTGAACGCACTATCTATTC, Tac1 reverse: TAATACGACTCACTATAGGGAGACAGGAAACATGCTGCTAGGATA, Dcc forward: ATGGTGACCAAGAACAGAAGGT, Dcc reverse: TAATACGACTCACTATAGGGAGAAATCACTGCTACAATCACCACG.
EdU, BrdU injections, and TUNEL staining.
To label dividing cells, pregnant females were subjected to intraperitoneal injection of EdU and BrdU (50 mg/kg body weight). Embryos were collected at different embryonic stages and processed for Edu staining (Click-iT EdU Imaging Kits; Invitrogen). For BrdU staining, vibratome sections were pretreated with HCl 2 n during 30 min followed by a 10 min incubation with borate buffer and immunodetection using primary antibody mouse anti-BrdU (Invitrogen, 1:50). To detect cell death, we used TUNEL staining (In situ Cell Death Detection Kit, TMR red; Roche) in vibratome sections. Positive controls, performed by pretreating sections with DNAse1 (1500 U/ml) for 10 min at room temperature, validated the efficiency of the TUNEL assay.
Stereotaxic injections.
Before surgical procedures, animals were anesthetized by intraperitoneal injection of an anesthetic combination (ketamine 87 mg/ml, xylazine 13 mg/ml) at a dosage of 1 ml/kg. After verification of deep anesthesia by pinching the tail, mice were placed on a stereotaxic apparatus (Stoelting). The bregma coordinates used for unilateral injections were as follows: MHb, anterior/posterior (AP) −1.6, medial/lateral (ML) +0.25, dorso/ventral (DV) −2.75. The incisor bar was leveled with the interaural line. One hole was drilled and injections of AAVs expressing EYFP upon Cre recombination were performed at a speed of 100 nl/min with a 500 nl total volume (pAAV-Ef1a-DIO ChETA-EYFP; Penn Core). The cannula was left in place for 5 min to infuse before removal and suture of the skin.
Imaging, experimental design, and statistical analysis.
Images were captured with a laser scanning confocal microscope (Zeiss 780) and with a stereomicroscope coupled to a digital camera (Zeiss axioplan2). Images were processed using ZEN and ImageJ software and figures were prepared using the Adobe Photoshop and Illustrator CS5 software package. A minimum of three animals and three slices per animal were used for all the analyses and quantifications (see Table 1 for details). All quantifications are presented as the mean ± SE. For convenience when presenting statistics, groups of data from Otx2flox/flox, Otx2CreERT/+, and Otx2CreERT/flox were named a–c, respectively. The lines Dbx1Cre;Otx2flox/+ and Dbx1Cre;Otx2flox/flox were named d and e, respectively. When comparing two groups of data between mutants and controls using Student's t test, unequal variance was first rejected by performing the F test when p > 0.05. All statistical comparisons failed to show unequal variance. Unpaired, two-tailed t test assuming equal variance was then used to compare mean data from two groups. The size of samples (n), degree of freedom, and p-values are indicated for each data comparison (Table 1). The exact p-values are indicated when p > 0.01. Minimal statistical significance was fixed at p < 0.05. p-values are represented on the graphs as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
Table 1.
Summary of datasets and statistics
| Figure | Datasets | Parameter tested | t test p-value | T value | df | Significance |
|---|---|---|---|---|---|---|
| 1A–H | E11.5, E13.5, E15.5, E18.5 (each stage n = 3) | GFP/Ki67 and GFP/Brn3a IFs | NA | NA | NA | NA |
| 1I | E18.5 (n = 3) | GFP/Brn3a/Er81 IFs | NA | NA | NA | NA |
| 1J | E18.5 (n = 3) | Tac1 ISH | NA | NA | NA | NA |
| 1K–O | P0 (each injection stage n = 3) | Labeling EdU | NA | NA | NA | NA |
| 2A–E′′ | E13.5 (each genotype n = 3) | YFP/Otx2/DAPI IF, Shh ISH and Gbx2 ISH | NA | NA | NA | NA |
| 2F–F′′, graph G | E18.5 (each genotype n = 4) | Mean Brn3a+ area | [a] vs [c] p < 0.001; [b] vs [c] p < 0.01; [a] vs [b] p = 0.64 | 6.28; 3.97; 0.49 | 6; 6; 6 | ***; **; ns |
| Mean Er81+ area | [a] vs [c] p < 0.01; [b] vs [c] p = 0.048; [a] vs [b] p = 0.66 | 4.55; 2.48; 0.46 | 6; 6; 6 | **; ns; ns | ||
| Mean Brn3a+ Er81− area | [a] vs [c] p < 0.001; [b] vs [c] p < 0.001; [a] vs [b] p = 0.58 | 9.71; 7.75; 0.58 | 6; 6; 6 | ***; ***; ns | ||
| 2F–F′′, graph H | E18.5 ([a] and [b] n = 3) ([c] n = 4) | Mean Brn3a+ cells | [a] vs [c] p < 0.01; [b] vs [c] p < 0.01; [a] vs [b] p = 0.72 | 6.13; 5.41; 0.39 | 5; 5; 4 | **; **; ns |
| Mean Er81+ cells | [a] vs [c] p = 0.028; [b] vs [c] p = 0.042; [a] vs [b] p = 0.64 | 3.06; 2.72; 0.50 | 5; 5; 4 | *; *; ns | ||
| Mean Brn3a+ Er81− cells | [a] vs [c] p < 0.01; [b] vs [c] p < 0.01; [a] vs [b] p = 0.25 | 6.52; 5.69; 1.33 | 5; 5; 4 | **; **; ns | ||
| 2I–I′′, graph K | E18.5 (each genotype n = 3) | Mean EdU+ (E13.5 injection) cells in MHb | [a] vs [c] p = 0.84; [b] vs [c] p = 0.36; [a] vs [b] p = 0.62 | 0.22; 1.02; 0.54 | 4: 4; 4 | ns; ns; ns |
| 2J–J′′, graph L | E18.5 ([a] n = 4) ([b] and [c] n = 3) | Mean EdU+ (E15.5 injection) cells in MHb | [a] vs [c] p = 0.033; [b] vs [c] p = 0.011; [a] vs [b] p = 0.62 | 2.92; 4.45; 0.53 | 5; 4; 5 | *; *; ns |
| 3A–C′ | All stages (each genotype n = 3), controls (n = 1) | TUNEL staining | NA | NA | NA | NA |
| 3D–E′4, graph F | E13.5 ([a]n = 3) ([c] n = 4) E15.5 ([a] n = 3) ([c] n = 5) | Mean MHb VZ length | E13.5 [a] vs [c] p = 0.28; E15.5 [a] vs [c] p < 0.001 | 1.21; 7.91 | 5; 6 | ns; *** |
| 3D–E′4, graph G | E13.5 ([a] n = 3) ([c] n = 4) E15.5 ([a] n = 3) ([c] n = 5) | Mean Ki67+ cells | E13.5 [a] vs [c] p = 0.022; E15.5 [a] vs [c] p = 0.019 | 3.29; 3.20 | 5; 6 | *; * |
| 3D–E′4, graph H | E13.5 ([a] n = 3) ([c] n = 4) E15.5 ([a] n = 3) ([c] n = 5) | Mean BrdU+ EdU− cells | E13.5 [a] vs [c] p = 0.45; E15.5 [a] vs [c] p < 0.01 | 0.81; 4.27 | 5; 6 | ns; ** |
| 4A | E15.5 (n = 3) | Dbx1 ISH | NA | NA | NA | NA |
| 4B–D | E18.5 (n = 3) | YFP/Brn3a/Pax6 and Otx2/Brn3a/Pax6 Ifs | NA | NA | NA | NA |
| 4F–H, graph H | E18.5 (n = 3) | Mean Brn3a+; ER81+; Brn3a+ Er81− cells | [d] vs [e] p < 0.001; [d] vs [e] p = 0.038; [d] vs [e] p < 0.01 | 10.36; 3.05; 6.01 | 4; 4; 4 | ***; *; ** |
| 4I–N | E18.5 (each genotype n = 3) | EdU/Brn3a IF and Nurr1 IF/Tac1 ISH | NA | NA | NA | NA |
| 5B,C | P10 (n = 3) | GFP/Pax7 and GFP/Gscl IFs | NA | NA | NA | NA |
| 5D–Dd4 | E11.5 (n = 3) | Labeling GFP and several markers IFs | NA | NA | NA | NA |
| 5E,F | P0 (n = 3) | Labeling EdU/GFP IF | NA | NA | NA | NA |
| 5G,H | E13.5 E18.5 (n = 5) E15.5 (n = 6) | GFP IF | NA | NA | NA | NA |
| 5J,M | Adult (n = 2) | GFP/GABA IF | NA | NA | NA | NA |
| 6A–D | E13.5, E15.5 (each genotype n = 3) | ER/RC2 IF | NA | NA | NA | NA |
| 6E–L | E18.5 (each genotype n = 3) | Several markers IF | NA | NA | NA | NA |
| 7A–F | E13.5 (each genotype n = 3) | DCC ISH, Netrin1 ISH and ER/DCC IF | NA | NA | NA | NA |
| 8A–D | P29 (n = 3) | Several markers IF | NA | NA | NA | NA |
| 8F,G | P60 (n = 3) | eYFP/SP IF | NA | NA | NA | NA |
| 8H,I | P21 (each genotype n = 3) | ChAT/SP IF | NA | NA | NA | NA |
The lines of the table follow from top to bottom the number of the main figures by ascending order. All data shown in the manuscript are presented and the details of the statistical analyzes performed are indicated when applicable. T value, df, and significance are shown in respective order of the t test p-values.
*p < 0.05;
**p < 0.01;
***p < 0.001.
[a], Otx2flox/flox; [b], Otx2CreERT2/+; [c], Otx2CreERT2/flox; [d], Dbx1Cre;Otx2flox/+; [e], Dbx1Cre;Otx2flox/flox; df, degree of freedom; n, number of animals used; NA, not applicable; ns, not significant; IF, immunofluorescence; ISH, in situ hybridization.
Results
Otx2 is expressed throughout the development of the habenula
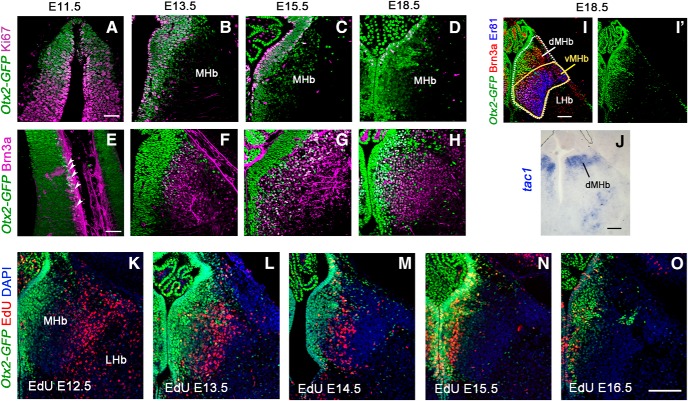
We undertook the precise description of Otx2 expression both temporally and topographically in the habenula (Fig. 1). Because antibodies against Otx2 recognize to some extent other homologous proteins such as Otx1, we used an Otx2-GFP knock-in mouse that expresses a version of Otx2 fused to GFP to strictly monitor Otx2 expression (Fossat et al., 2007). To determine whether Otx2 is expressed in neural progenitors or in postmitotic neurons, we compared Otx2-GFP expression with Ki67 staining, a cell proliferation marker, and with Brn3a staining, a well known postmitotic marker of medial habenular neurons (Quina et al., 2009). In the epithalamus region, the progenitor domain was doubly labeled with Otx2 and Ki67 at each stage analyzed starting from E11.5, indicating sustained Otx2 expression during neurogenesis (Fig. 1A–D). Only rare cells were Brn3a+ at E11.5, corresponding to cells exiting the proliferative zone (Fig. 1E, arrowheads). From E13.5 on, Otx2 was coexpressed with Brn3a in immature postmitotic neurons once they had moved outside of the proliferative domain. Otx2 expression decreased as cells migrated away from the midline with a medial-high, lateral-low expression gradient (Fig. 1F–H). At E18.5, development of the habenula appeared completed and the medial and lateral habenula (MHb and LHb, respectively) could be anatomically recognized. Otx2 expression was restricted to the MHb, whereas Brn3a was expressed in all neurons of the MHb and in some sparse neurons of the LHb (Quina et al., 2009). We used another marker, Er81 (also known as Etv1), to visualize the two main subparts of the habenula; the vMHb (Er81+) and dMHb (Er81−) that correspond to cholinergic and substance P-ergic subregions, respectively (Fig. 1I,J). Interestingly, Otx2 expression was not restricted to one of these subparts but exhibited a mediolateral gradient in the MHb, with highly expressing neurons in the medial part (Otx2High) and neurons presenting low to absent expression (Otx2Low) in the lateral part (Fig. 1I′). Proportionally, Otx2High neurons contributed to a large part of the dMHb and to a small part of the vMHb (Fig. 1I,I′). To understand how neurogenesis occurs and populates the different parts of the habenula, we analyzed animals at P0 that had been injected with EdU at various stages of development. The medial habenula started to be clearly labeled at E12.5 and this labeling followed a wave from lateral to medial parts with a peak of density for the most medial part at E15.5 (Fig. 1K–O). As described previously (Angevine, 1970), habenula neurogenesis occurs via a centrifugal mode, with the early-born neurons settling outside and the later-born neurons occupying a ventricular position. Therefore, the latest-born habenular neurons are those that strongly maintain Otx2 expression, whereas the earliest-born neurons found in the lateral part are characterized by low or absent expression of Otx2 (Fig. 1K–O).
Figure 1.
Temporal expression of Otx2 and neurons birth-dating in the medial habenula. A–H, Coronal sections of Otx2-GFP mouse embryos at different stages of development showing the expression of Otx2 (GFP+) in progenitor cells labeled with Ki67 in the ventricular zone of the Hb (A–D) and the expression of Otx2 (GFP+) in medial habenular neurons labeled with Brn3a (E–H). White arrowheads in E show few postmitotic neurons double labeled with Otx2 (GFP) and Brn3a at E11.5. I, I′, Coronal section of E18.5 Otx2-GFP mouse embryo immunolabeled for Brn3a to mark all MHb neurons and for Er81 to mark the ventral MHb. GFP colocalized with both markers in the medial part of the MHb. J, In situ hybridization for Tac1 showing the dorsal part of the MHb. K, O, Immunofluorescence of GFP (Otx2) and EdU labeling in coronal sections at the level of the MHb in P0 Otx2-GFP brains when EdU was injected at different embryonic stages. Scale bars: A–H (shown in A and E), 50 μm; I–O (shown in I, J, and O), 100 μm.
Otx2 deletion leads to neurogenesis defects in the Hb
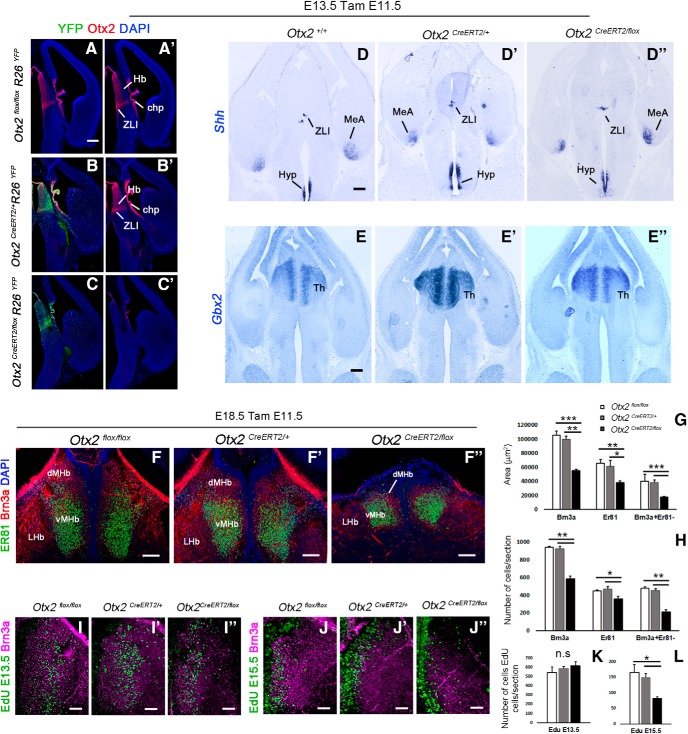
We next tested whether Otx2 is required for the formation of the Hb. According to our birth-dating analysis, MHb neurons start to appear from ∼E12.5 for the most lateral part and are all born before E17.5 (Fig. 1K–O). Because they all derive from Otx2+ progenitors, we took advantage of the Otx2CreERT2/+ and Otx2flox/flox mouse lines (Fossat et al., 2006) to inactivate Otx2 from E11.5 by an intraperitoneal injection of tamoxifen (TamE11.5). We then compared Otx2CreERT2/flox(TamE11.5) conditional mutants with Otx2flox/flox(TamE11.5) control littermates or with Otx2CreERT2/+(TamE11.5) embryos (from Otx2+/+ females) as additional controls at E18.5 (Fig. 2).
Figure 2.
Conditional deletion of Otx2 produces habenular hypoplasia. A–C′, Coronal sections of E13.5 control and mutant embryos injected with tamoxifen at E11.5 showing the expression of the reporter YFP and Otx2 in the diencephalon. In situ hybridization for Shh (D–D′′) and Gbx2 (E–E′′) in coronal section of E13.5 mutant and control mice injected with tamoxifen at E11.5. F–F′′, Immunostaining for Er81 (Etv1) and Brn3a in coronal sections at the level of the habenula in E18.5 embryos that have been injected with tamoxifen at E11.5 stage. G, Graph representing the average area in square micrometers of the MHb per coronal section at E18.5. The MHb areas are significantly reduced in mutant mice (black column, Otx2CreERT2/flox) compared with control mice (white and gray columns, Otx2flox/flox and Otx2CreERT2/+ respectively). H, Graph representing the average numbers of Brn3a+ cells, Er81+ cells, and Brn3a+Er81− cells per coronal section in the MHb at E18.5. The number of Brn3a+ cells in the MHb is reduced in mutant mice compared with control mice. However, the reduction of the number of Er81+ cells in the ventral MHb is less severe compared with the reduction of the Brn3a+ Er81− cells in the dorsal MHb. I–L, EdU labeling combined with Brn3a immunostaining in coronal sections showing Edu+ cells in the MHb in control and mutant E18.5 embryos when the EdU was injected in pregnant females at E13.5 (F–H) and E15.5 (I–K). K, L, Graph representing the average number of EdU cells in the MHb per coronal section in control and mutant E18.5 embryos. Note that the number of Edu cells does not change significantly when the EdU was injected at E13.5. However, there were fewer EdU+ cells in mutant mice compared with controls when EdU was injected at E15.5. n.s., Nonsignificant. p-values are represented on the graphs as follows: *p < 0.05; **p < 0.01; ***p < 0.001. However, there were fewer EdU+ cells in mutant mice compared with controls when EdU was injected at E15.5. MeA, Medial amygdala; Hyp, hypothalamus; Th, thalamus. Scale bars, A–E′′ (shown in A, D, and E), 100 μm; F–F′′, 100 μm; I–J′′, 50 μm.
Otx2 deletion induced at E11.5 does not affect early patterning of the diencephalon
Because early Otx2 ablation has been shown previously to affect forebrain patterning, we first verified that Otx2 ablation at E11.5 did not affect early steps of diencephalon development, which would prevent analysis of a direct role of Otx2 in Hb formation. It was shown in zebrafish, for example, that in hypomorphic mutants for Otx1 and Otx2 expression, the ZLI was not induced properly (Scholpp et al., 2007), leading to the disappearance of the thalamus. We therefore analyzed the expression of molecular markers of the ZLI and thalamic region. At E13.5, 2 d after tamoxifen injection, Otx2 was already largely absent from the forebrain of mutant mice, indicating high recombination efficiency (Fig. 2C,C′), whereas both controls showed unaltered Otx2 expression (Fig. 2A–B′). High recombination efficiency in the progenitors expressing Otx2 at the time of injection (Fig. 1) was also demonstrated using a Rosa26::flox-stop-flox-YFP(R26YFP) reporter line of Cre activity. Indeed, a large number of cells generated from these progenitors and labeled with YFP was found in the dorsal diencephalic region (Fig. 2B,C). The ZLI, marked by Shh, was properly induced at its correct location in all mice (Fig. 2D–D′′). The thalamic region, labeled with Gbx2 transcripts, was present and displayed a normal size in all mice (Fig. 2E–E′′). These data rule out an indirect effect of Otx2 deletion at E11.5 that would alter early patterning of the diencephalon.
Otx2 deletion specifically affects the pool of late born habenular neurons
We then investigated whether Otx2 deletion could affect subsequent development of the epithalamus region and particularly the formation of the MHb. To visualize the different subparts of the habenula in Otx2CreERT2/flox(tamE11.5) conditional mutants, we used the three markers Brn3a, Er81, and Otx2. We observed a large reduction in MHb area (44 ± 5.6% and 47 ± 3.8% reduction relative to Otx2CreERT2/+ and Otx2flox/flox controls, respectively) and number of habenular Brn3a+ cells (37 ± 3.1% and 40 ± 2.6% reduction relative to Otx2CreERT2/+ and Otx2flox/flox controls, respectively; Fig. 2F–H, Table 1). Moreover, counts of Brn3a+ and Er81+ MHb neurons revealed a reduction that was more pronounced in the Er81−/Brn3a+ (dMHb) population than in the Er81+/Brn3a+ (vMHb) population (Fig. 2G,H, Table 1). Therefore, Otx2 deletion preferentially affected Er81−/Brn3a+ neurons that compose the dMHb. These data were compatible with the fact that the Otx2High region was the one preferentially reduced because it proportionally contributes more to the dMHb (Fig. 1I). To further test this possibility and in the absence of specific molecular markers of this subpopulation except Otx2, we used the fact that Otx2High neurons are born later than Otx2Low neurons (Fig. 1K–O). In control animals, as expected, E13.5 and E15.5 EdU injections preferentially labeled Otx2low and Otx2high cells, respectively (Fig. 2I–L, Table 1). In Otx2CreERT2/flox(tamE11.5) mice, no difference could be statistically detected in early born neurons compared with any of the two controls (Fig. 2K, Table 1). By contrast, a very large decrease was seen in the late-born pool normally populating the Otx2High region (Fig. 2L, Table 1). These data demonstrate that Otx2 deletion preferentially affects late-born MHb neurons, which would normally maintain strong expression of Otx2.
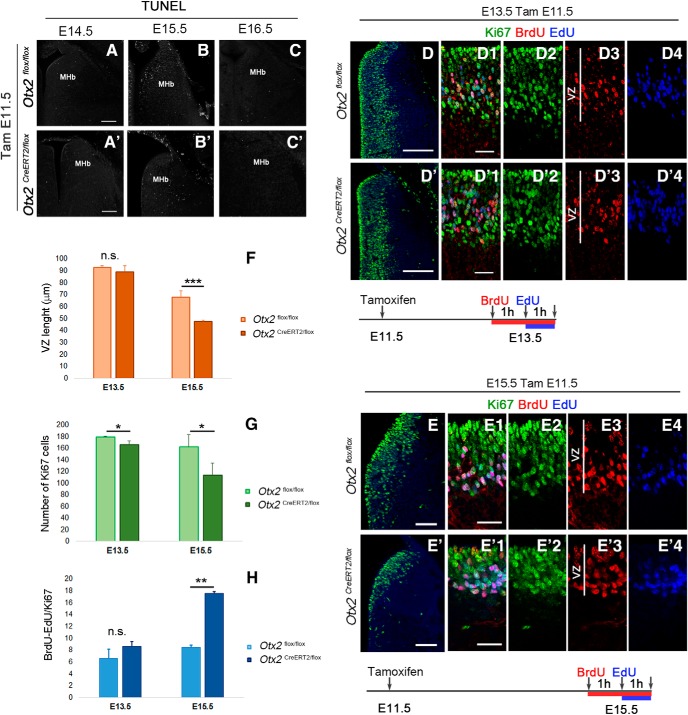
Otx2 deletion affects the proliferation of Hb progenitors
To understand the mechanisms behind the decreased number of late-born neurons, we investigated two possibilities. This decrease may result either from the loss of Otx2 in the progenitors affecting progenitor proliferation and generation of late-born neurons or from increased cell death of progenitor or postmitotic cells. In the absence of evidence of cell death (Fig. 3A–C′), we were left with the abnormal progenitor proliferation hypothesis. One immediate observation was that the thickness of the ventricular zone (VZ) appeared reduced in mutants at E15.5 but was not affected at E13.5 (Fig. 3D–E′, 4F, Table 1). Measures of the number of proliferative progenitors labeled with anti-Ki67 revealed a small and large decrease at E13.5 and E15.5, respectively (Fig. 3G, Table 1). We performed EdU/BrdU experiments to measure the number of cells exiting the S phase as a function of time and in proportion to the total number of Ki67 cells (Fig. 3H, Table 1). Cells singly labeled with BrdU were considered as the cells exiting the S phase within 1 h. Interestingly, the number of cells exiting the S phase was largely increased in the mutant (207 ± 12.6% of control), showing that the S phase is shorter in the absence of Otx2. It therefore appears that Otx2 deletion leads to a premature reduction of the pool of epithalamic progenitors that may be explained by an alteration of their cell cycle.
Figure 3.
Loss of Otx2 alters habenular progenitor cell cycle. A–C′, Tunnel staining to detect cell death in coronal sections at the level of the MHb at different embryonic stages in control (A–C) and Otx2 mutant embryos (A′–C′) injected with tamoxifen at E11.5. D–E′, Immunofluorescence for Ki67 labeling progenitor cells in the habenular region in coronal section at E13.5 (D, D′) and E15.5 (E, E′) of control (D, E) and mutant (D′, E′) embryos tamoxifen injected at E11.5. D1–E′4, High magnification of the VZ showing immunolabeling of Ki67 and BrdU cells and EdU staining in control and mutant mice at E13.5 (D1–D′4) and E15.5 (E1–E′4) when BrdU was injected 2 h and EdU 1 h before pregnant females were killed. Graphs representing the length in micrometers of the VZ (F), the average number of Ki67 cells in coronal section per area (G), and the average number of BrdU+ EdU− cells relative to the area (H) in control and mutant E13.5 and E15.5 embryos. p-values are represented on the graphs as follows: *p < 0.05; **p < 0.01; ***p < 0.001. n.s., Nonsignificant. Scale bars, A–C′ (shown in A, and A′, 100 μm; D–E′, 100 μm; D1–E′4 (shown in D1, D′1, E1, and E′1), 25 μm.
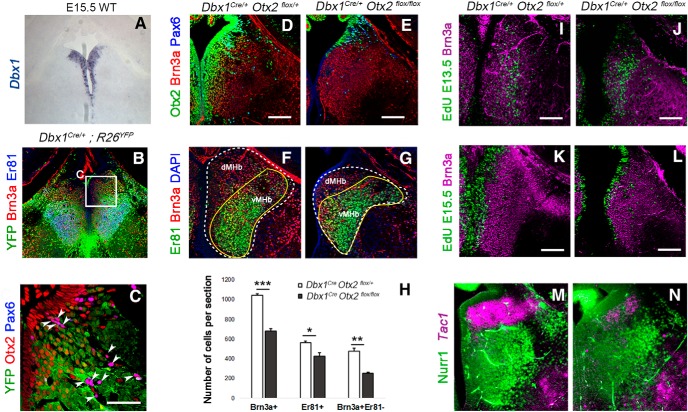
Reduction of the pool of late-born neurons is not due to a potential secondary effect of Otx2 deletion in tissues other than the habenula
To confirm that the phenotype we observed is due to a deletion of Otx2 in habenular progenitors and not to an indirect effect of the concomitant deletion of Otx2 in adjacent tissues such as choroid plexus and pineal gland, we restricted the deletion of Otx2 to the MHb using a Dbx1::Cre driver line. Dbx1 is expressed in epithalamic VZ (Vue et al., 2007). We confirmed this by following the Dbx1 cell lineage using the R26YFP reporter line. At E18.5, in Dbx1::Cre;R26YFP, all habenular Brn3a+ neurons were YFP+ (Fig. 4B,C). Only rare cells that were Brn3a− were also YFP− in the MHb region. These cells are yet to be defined, but their strong Pax6 expression makes them clearly distinct from other cell populations in the MHb (Fig. 4C, white arrowheads). In Dbx1::Cre;Otx2flox/flox mutants, Otx2 expression was largely absent from the habenula region at E18.5 compared with control mice (Fig. 4D,E), confirming that Dbx1::Cre line is an efficient tool to delete Otx2 in habenular progenitors. Otx2 deletion in Dbx1::Cre;Otx2flox/flox led to a significant reduction in Brn3a+ cells (Fig. 4F–H, Table 1). This effect was again more pronounced in the Er81−/Brn3a+ than in the Er81+/Brn3a+ population, suggesting that, as in Otx2CreERT2/flox mice, late-born neurons were preferentially affected (Fig. 4H, Table 1). We confirmed this by showing that the number of neurons marked with EdU injections at E15.5 was strongly decreased, whereas neurons labeled at E13.5 seemed unaffected (Fig. 4I–L). These results also showed that Otx2 deletion does not specifically affect the classically described substance P-ergic/tac1-expressing or cholinergic/Er81-expressing neurons of the dMHb and vMHb, respectively (see also Introduction section). Instead, it affects the late-born neurons independently of their neurotransmitter content. Consistent with this idea, only a partial reduction of Er81+ neurons (Fig. 4F,G) and Tac1-expressing neurons (Fig. 4M,N) was observed in Otx2 cKO.
Figure 4.
Specific deletion of Otx2 in Dbx1 domain. A, Dbx1 expression pattern in the ventricular zone of the MHb at E15.5. B, C, Coronal section of a E18.5 Dbx1Cre control embryo revealing Dbx1 lineage in the habenula. Immunofluorescence for YFP, Brn3a, and Er81 (Etv1) (B). High magnification of a similar area as indicated in B showing Otx2/Pax6+ cells that are negative for YFP and therefore not generated from Dbx1+ progenitors (C). D, E, Immunostaining in coronal sections for Otx2, Brn3a, and Pax6 in control (D) and Otx2 conditional mutant embryos (E) at E18.5. F, G, Immunolabeling for Er81 and Brn3a in coronal sections of control and mutant E18.5 embryos. H, Graph representing the average number of Brn3a+ cells (Brn3a+), Er81+ cells (Er81+), and Brn3a+ Er81− (Brn3a+ Er81−) cells per coronal section in the MHb of control and Otx2 mutant E18.5 embryos. I–L, EdU staining and Brn3a immunolabeling in coronal sections of the MHb in E18.5 control and Otx2 mutant embryos when the EdU was injected at E13.5 (I, J) and E15.5 (K, L). M, N, In situ hybridization using Tac1 RNA probe in combination with immunostaining of Nurr1, a marker of the vMHb, in coronal sections of control (M) and Otx2 cKO animals (N) at E18.5. p-values are represented on the graphs as follows: *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars, A, B, 175 microns, C, 50 microns (shown in C); D–L 100 microns, M, N, 75 microns (shown in D, E, I–L).
In conclusion, Otx2 seems to control the development of the MHb by sustaining the expansion of the progenitor pool, thus affecting the total number of derived neurons. This effect is due to cell-autonomous Otx2 activity in Hb progenitors.
Otx2 is also expressed in the unique output of the Hb, the IPN
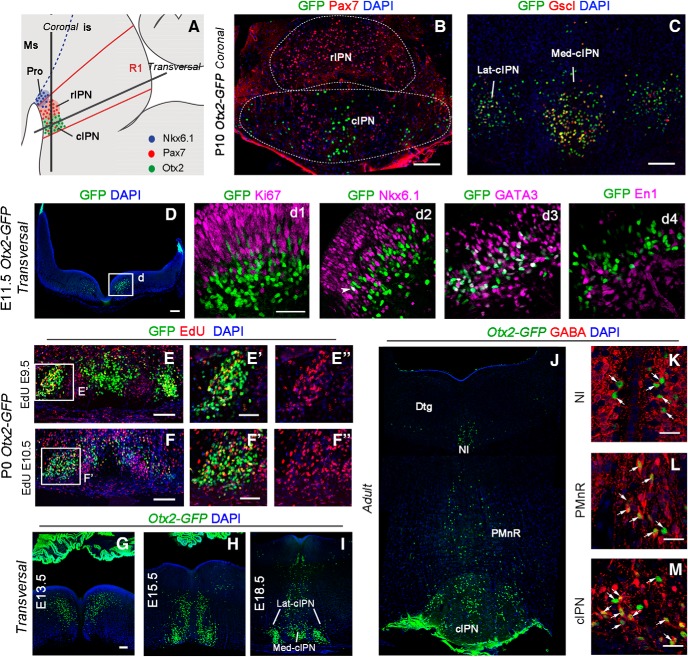
We then studied Otx2 expression in the main target of the medial habenula, the IPN, using the Otx2-GFP knock-in line. The IPN, which is located in the basal plate of rhombomere 1 (r1), is composed of several subnuclei (Lorente-Cánovas et al., 2012; Fig. 5A). We confirmed that Otx2+ neurons only populated the cIPN (Fig. 5A–C). The cIPN could be further divided into lateral (Lat-cIPN) and medial subnuclei (Med-cIPN) (Fig. 5B,C). Otx2+ neurons composed the Lat-cIPN and the central-most part of the Med-cIPN, whereas a Pax7+/Otx2− population was located between them (Fig. 5B). The Otx2 population could be further characterized by the specific expression of the Gscl protein (Fig. 5C). We then investigated how the cIPN forms and looked for Otx2+ cells at earlier stages. Cell cohorts expressing Otx2 in basal r1 could already be detected at E11.5 (Fig. 5D). These cells were found in the outer ventricular zone, in between Ki67+ cells, or in the mantle layer at the level of the Nkx6.1+ progenitor domain (Fig. 5Dd1,Dd2). Some rare cells starting to accumulate Otx2-GFP in the progenitor zone were also weakly Nkx6.1+; however, none coexpressed Ki67 (Fig. 5Dd1,Dd2). These results suggest that Otx2 expression is induced in cells generated from Nkx6.1+ progenitors soon after they become postmitotic and start to downregulate Nkx6.1 expression. To further narrow down the origin of Otx2-expressing cells, we explored additional markers and found that the Otx2+ population expressed Gata3 and GABA, but not En1, and therefore could be defined as the equivalent of a subset of spinal cord V2b GABAergic neurons (Karunaratne et al., 2002) (Fig. 5Dd1,Dd2,M). To characterize the timing of neurogenesis of Otx2+ IPN neurons, we injected EdU at several stages of development and analyzed animals at P0. Otx2+ IPN cells were only marked when EdU was injected at E9.5 and E10.5, but not later (Fig. 5E–F′). We concluded that Otx2+ IPN neurons were generated between E9.5 and E10.5. These cells then started to migrate in three successive steps (Fig. 5G–I). At E13.5 (Fig. 5G), cells were seen as a stream going tangentially toward the midline (Step 1) without crossing it and turning down to migrate radially toward the ventral r1 (Step 2). At E15.5 (Fig. 5H), many cells had already reached the ventral r1 and started to aggregate (Step 3) as bilateral nuclei, whereas others were still on the migration path. At E18.5 (Fig. 5I), some sparse Otx2+ cells could still be seen in the dorsal region and close to the midline in the migration path. In the ventral region, Otx2+ cells were also compacting in the midline, thus contributing to Lat-cIPN and Med-cIPN (Fig. 5I). This organization was maintained until adulthood (Fig. 5J). In addition, this analysis revealed additional expression of Otx2 in r1, in GABAergic cells of the median and paramedian raphe (MnR and PMnR, respectively), in the nucleus incertus, and around the dorsal tegmental nucleus (Fig. 5K–M).
Figure 5.
Otx2 expression pattern in the IPN. A, Schematic illustration of a sagittal section of IPN subnuclei in the ventral r1 in embryonic brain. B, Coronal section of P10 Otx2-GFP mouse brain at the level of the IPN. The rostral IPN is characterized by high density of Pax7 neurons. Otx2 neurons (GFP) in the cIPN form two subnuclei, the medial and the lateral one, with some Pax7 neurons in between. C, Coronal section of the IPN showing colocalization between Otx2 and Gscl in the cIPN. D, Transversal section of E11.5 Otx2-GFP embryo at the level of the r1 showing Otx2 neurons in the basal plate. Dd1–Dd4, High magnification of the box area indicated in D showing potential Otx2 neurons (GFP+) in r1 that also contain Ki67 (Dd1), Nkx6.1 (Dd2), GATA3 (Dd3), and En1 (Dd4) at embryonic stage E11.5. E, F, High magnification of the IPN area on P0 Otx2-GFP embryo showing colocalization between Otx2 (GFP+) and Edu injected at E9.5 (E) and E10.5 (F). E′–F′′, High magnification of the boxed area. G–M, Immunolabeling in transversal section of the r1 in Otx2-GFP embryos showing the distribution of Otx2 neurons (GFP+) in the basal plate at different embryonic (G–I) and adult stages (J–M). K–M, Colocalization between Otx2 and GABA in the nucleus incertus, PMnR, and cIPN. Ms, Mesencephalon; is, isthmus; Pro, prodromal division of the IPN; Dtgm dorsal tegmental nucleus; NI, nucleus incertus. Scale bars, B–D and G–I, 100 μm; Dd1–Dd4 (shown in Dd1) and E′–F′′ (shown in E′ and F′), 50 μm; K–M, 25 μm.
These results show that IPN neurons are generated from Nkx6.1+ progenitors, start to express Otx2 soon after they become postmitotic, and migrate extensively through tangential and then radial migration paths to finally concentrate in the Lat-cIPN and Med-cIPN.
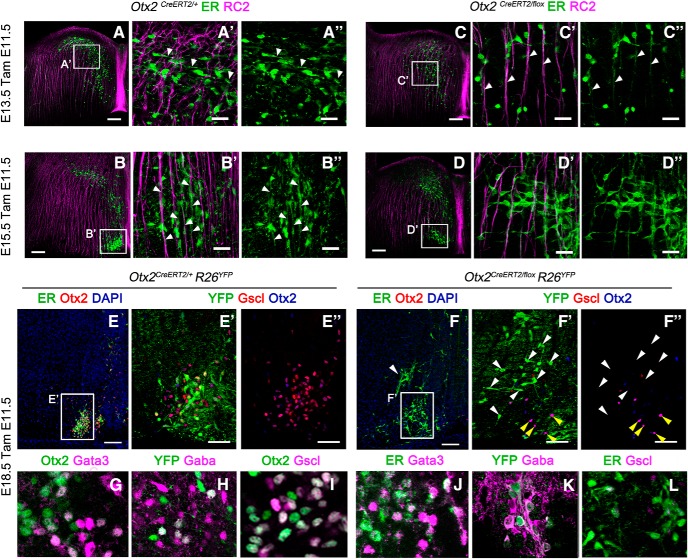
Otx2 deletion leads to migration defects of IPN neurons
We then tested whether Otx2 expression is also required for the development of the IPN. Because Otx2 is not expressed at the progenitor stage (Fig. 5Dd1), we could test whether Otx2 plays a postmitotic role in the migration and formation of the caudal IPN. From our EdU birth-dating experiments, we concluded that the cell population fated to express Otx2 and form the caudal IPN is already born by E11.5 (Fig. 5E–F′′). We therefore took advantage of our Otx2CreERT2/flox mouse line to inactivate Otx2 at E11.5, when IPN neurons are already born. To follow the recombined cells in Otx2CreERT2/flox(tamE11.5) mutants, we used two complementary tools: anti-ER antibodies to recognize Cre-ERT2 protein produced from the Otx2CreERT2 allele and the R26RYFP Cre reporter line to trace recombined cells. As described above, IPN neurons reach the ventral r1 through three successive steps. At E13.5, in control Otx2CreERT2/+(TamE11.5); R26RYFP animals, the leading processes of Otx2 cells labeled with anti-ER antibodies were projecting toward the midline and perpendicular to radial glia cells, indicating that Otx2+ cells were migrating tangentially (Fig. 6A–A′′, white arrowheads, phase 1). At E15.5, Otx2+ cells changed their direction with their leading processes aiming ventrally following the radial glia (Fig. 6B–B′′, white arrowheads, phase 2). At E18.5, Otx2+ cells had already reached the cIPN (Fig. 6E–E′′, phase 3). We then analyzed cell migration in Otx2CreERT2/flox(tamE11.5) mutants. At E13.5, Otx2 KO cells labeled with anti-ER antibodies and showing no detectable Otx2 precociously migrated ventrally along the radial glia, skipping the first step of tangential migration toward the midline (Fig. 6C–C′′, white arrowheads, phase 1′). At E15.5, some of these cells had reached the ventral part but in a more lateral position (Fig. 6D–D′′, phase 2′). Finally, at E18.5, sparse mutant cells were found more dorsally and laterally compared with control cells (Fig. 6F–F′′, white arrowheads, phase 3′). Interestingly, the rare non-recombined cells that had retained Otx2 and were not YFP-immunoreactive were found at their correct location (Fig. 6F′,F′′, yellow arrowheads), pointing to a cell-autonomous function of Otx2.
Figure 6.
Migrational defects in IPN neurons due to Otx2 deletion is not a consequence of fate switch. A–D, Immunofluorescence to visualize Otx2 neurons (ER) and radial glia cells (RC2) in transversal section of r1 at the levels of the caudal IPN in control (A, B) and mutant (C, D) embryos at the E13.5 and E15.5 stages injected with tamoxifen at E11.5. A′–D′′, High magnification of the area indicated in A–D. The white arrowheads indicate the orientation of the Otx2+ cells leading process with respect to the radial glia. E, F, Immunofluorescence for CreERT2 (ER) and Otx2 in transversal sections of E18.5 control and mutant embryos showing the disposition of Otx2+ cells when the migration is completed. The white arrowhead in F indicates the aberrant position of ER+ cells that have lost the expression of Otx2.(E′–F′′), High magnification of the area indicated in E and F showing cells labeled with YFP, Gscl, and Otx2 in the caudal IPN. Note that the YFP+ cells are the one that lose the expression of both Otx2 and Gscl and are mislocalized (white arrowheads), whereas the Otx2+ cells that are not recombined migrate properly (yellow arrowheads), indicating a cell-autonomous defect. G–L, Immunofluorescence in transversal sections of E18.5 control and mutant embryos showing the expression of the main V2b interneurons markers in Otx2+ cells. Scale bars, A–F, 100 μm, E′–F′, 50 μm; A′–D′′, G–L, 25 μm.
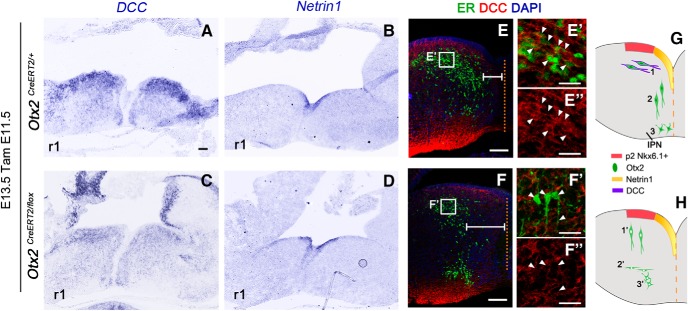
These results demonstrate a role of Otx2 at the postmitotic level in the HIPS. Cells that normally form the caudal IPN do not migrate properly in the absence of Otx2. Impaired motility seems unlikely because these cells are still able to migrate radially. The major defect seems to be an aberrant or absent tangential migration toward the floor plate that could be the consequence of abnormal expression of guidance molecule receptors in these cells. Alternatively, this phenotype could be due to a change of cell identity that translates into an alternative migration behavior. However, these cells kept their expression of Gata3 (Fig. 6G,J) and their GABAergic nature (Fig. 6H,K). This implies that, despite the absence of Otx2, these cells most likely retain their identity as V2b interneurons. However, one marker, Gscl, was found to be absent (Fig. 6E–F′,I,L), demonstrating that Otx2-ablated cells were molecularly affected. Because Gscl has no known role in migration, we sought for other cues that might explain the migration defect of Otx2-ablated cells. Few guidance molecules secreted by hindbrain floor plate have been shown to attract neurons toward it. The most documented mechanism is based on Dcc/Netrins interplay (Bloch-Gallego et al., 1999; Kim et al., 2015). We hypothesized that IPN cells may express Dcc at E13.5 and be attracted to the known source of Netrin in the floor plate. Expression of Dcc was indeed found at particularly high levels in Otx2+ cells at the most dorsal location in r1 (Fig. 7A,E–E′′). To test whether deletion of Otx2 affected Dcc/Netrin guidance, we analyzed Netrin 1 and Dcc mRNA expression by in situ hybridization. Although Netrin 1 expression was unaltered in the absence of Otx2 (Fig. 7B,D), Dcc expression was decreased in the dorsally located IPN cells that had lost Otx2 (Fig. 7A,C,E–E′′,F–F′′, white arrowheads). This could explain why these cells were no longer attracted toward the floor plate and precociously started to migrate radially. Such lack of attraction was clearly visible by measuring the distance between the Otx2 cells and the midline, which was always wider in mutant mice compared with control (Fig. 7E,F, white lines). Therefore, in the absence of Otx2, IPN cells retained their V2b characteristics but failed to migrate tangentially, most likely due to a defect in the Dcc/Netrin signaling mechanism (Fig. 7G,H).
Figure 7.
Aberrant expression of DCC expression in Otx2 mutant mice. A–D, In situ hybridization for Netrin1 (A, B) and DCC (C, D) in transversal sections of r1 in E13.5 control and Otx2 mutant embryos tamoxifen injected at E11.5. E, F, Immunofluorescence for ER and DCC in transversal section of control and mutant E13.5 embryos showing the disposition of ER+ cells with respect the midline (dashed orange line) indicated with the white lines. E′–F′′, High magnification of the area indicated in E and F showing decreased DCC staining in mutant ER+ cells located in the dorsal area (F′, F′′, white arrowheads) compared with control ER+ cells (E′, E′′, white arrowheads). G, H, Schematic illustration of our hypothesized mechanism for IPN precursor migration. This illustration shows the different migration phases in control and mutant embryos of IPN precursors. In control conditions, Otx2+ cells are generated from Nkx6.1 domain (progenitor domain 2, red area) and, due to DCC expression (violet contour), migrate tangentially toward the dorsal midline highly expressing Netrin1 (G1). After that, Otx2 neurons downregulate DCC expression, which may allow them to not stay trapped at this level and start to migrate radially to more ventral position (G2) to finally form the IPN (G3). In mutant mice, in the absence of Otx2 and due to a weaker expression of DCC, IPN neurons are no longer attracted to midline position. Instead, they start a radial migration precociously (H1′) and therefore they settle down earlier (H2′), resulting in an aberrant disposition of IPN neurons in more dorsal and lateral position compared with control mice (H3′). Scale bars: A–F, 100 μm; E′–F′′, 25 μm.
Otx2 developmental requirement is shared by interconnected subcircuits with specific outputs
Next, we investigated whether shared Otx2 expression by a subset of habenular neurons and IPN neurons could endow them with specific connectivity properties. Because specific parts of the system retain high expression of Otx2 and require Otx2 activity to develop properly, we tested whether these specific parts formed interconnected subcircuits that would consequently share HIPS subfunctions.
Otx2+ subcircuit between the Hb and the IPN
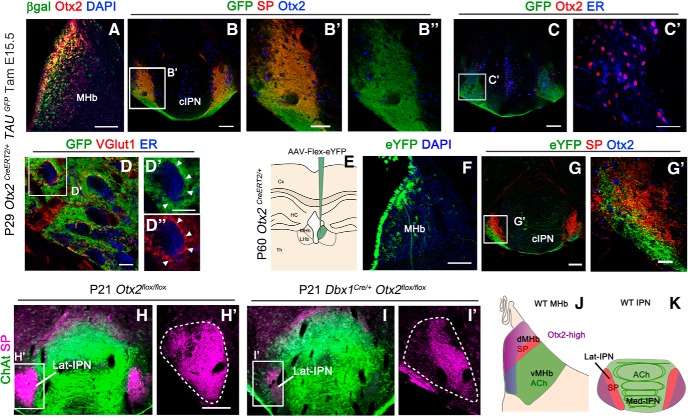
The classical description of the connectivity between the habenula and the IPN, as described in the Introduction, is divided between substance P-ergic fibers from the dMHb connecting the Lat-IPN and cholinergic vMHb neurons that connect to the Med-IPN. Otx2 expression does not follow this division, but instead follows neuronal birth date, with late-born Otx2high neurons being closer to the midline. We therefore tested whether Otx2high neurons have specific targets in the IPN. To clearly visualize axon terminals from Otx2 neurons, we used Otx2CreERT2/+; TauGFP−lacz/+ mice, which express myristylated GFP upon Cre-mediated recombination (Hippenmeyer et al., 2005). We injected tamoxifen at E15.5 to label Otx2High cells and analyzed the brain at postnatal stages (Fig. 8A). In the IPN, GFP+ fibers mainly innervated the Lat-cIPN and were largely doubly labeled with SP (Fig. 8B–B′′). To visualize potential synaptic connections between Otx2High habenular and Otx2+ IPN neurons, we first labeled the cytoplasm of Otx2+ neurons with anti-ER (Fig. 8C,C′). We then doubly labeled GFP+ habenular fibers for GFP and VGlut1, a synaptic marker of habenular neurons that are all glutamatergic. This also allowed us to exclude potential confounds with endings from Otx2+ IPN neurons that are GABAergic. Interestingly, doubly labeled GFP+/V-Glut1+ terminals were found decorating the cell surface of IPN cells marked with anti-ER antibodies (Fig. 8D–D′′). Because we could not formally rule out that the glutamatergic fibers in the IPN were exclusively from habenular neurons, we took advantage of the late expression of Otx2 in the HIPS that is maintained in the adult brain to specifically label these cells. We stereotaxically injected an inducible AAV construct that expresses eYFP upon Cre recombination (AAV-DIO-EYFP) in the habenula of Otx2CreERT2/+ adult mice (Fig. 8E). Two weeks after transduction, mice were tamoxifen injected for 2 consecutive days and brains were analyzed 2 weeks later (Fig. 8F–G′). In all analyzed brains, only a subset of cells belonging to the Otx2High population expressed eYFP+, probably due to their stronger expression of CreERT2 and thus to their higher recombination efficiency. These cells were concentrated in the medial-most part of the dMHb, with rare cells found in the vMHb (Fig. 8F). Again, the fibers coming from these neurons were mainly found in the Lat-cIPN. Interestingly, these fibers formed a bundle that had only partial overlap with the more laterodorsally located SP+ fibers (Fig. 8G–G′). Therefore, the medial-most part of Otx2High neurons is specifically connected to the lateral-most region of the Lat-cIPN.
Figure 8.
Otx2 expression in interconnected subcircuits. A, Coronal section of P29 control mice showing the expression of β-gal+ cells when tamoxifen was injected at E15.5 and Otx2 expression in the MHb. B, Immunofluorescence for GFP, substance P (SP) and Otx2 in the caudal IPN in coronal section. B′, B′′, High magnification of the area indicated in B showing colocalization between GFP and SP fibers in the Otx2+ region at the level of the lateral and caudal IPN. C, Immunofluorescence for GFP, Otx2, and ER in the caudal IPN in coronal section. C′, High magnification of the area indicated in C showing that all the Otx2 neurons are labeled with ER. D, Immunofluorescence for GFP, VGlut1, and ER in the laterocaudal IPN. D′, High magnification of the area indicated in D showing GFP terminals labeled with VGlut1 in contact with Otx2 cells labeled with anti-ER antibodies. E, Schematic illustration of an EYFP-expressing associated adenovirus that has been stereotaxically injected at the level of the MHb. F, Coronal section of the MHb showing neurons labeled with eGFP after virus injection. G, Coronal section of the IPN showing EYFP fibers from the MHb and SP fibers in the caudal and lateral IPN, where Otx2 IPN neurons are localized. G′, High magnification of the area indicated in G showing that EYFP and SP fibers correspond to two different populations. H–I′, Immunostaining of ChAT and SP on coronal sections at the level of the IPN of control (H, H′) and Otx2 cKO mutant (I,I′) at P21. H′ and I′ correspond to a high magnification of the white frame in H and I, respectively. J, K, Schematic illustration of a coronal sections at the level of the habenula and IPN showing the different populations of habenular neurons (J) and their respective field of projections in the IPN (K) with the same color code. Cx, Cerebral cortex; HC, hippocampus; Th, thalamus. Scale bars: A–C, F, G, 100 μm; B′, C′, 50 μm; D, D′, 10 μm; G′, 25 μm.
Otx2 is required for connecting the Otx2+ Hb-IPN subcircuit
Finally, because Otx2 deletion impairs the generation of Otx2High neurons (see above), we tested whether this could lead to connectivity defects between the MHb and the Lat-cIPN. Because Otx2 deletion of in Otx2CreERT2/flox(tamE11.5) mice is lethal at birth, we used Dbx1::Cre;Otx2flox/flox mutants that are viable and fertile to study dMHb and vMHb projections clearly identifiable only at postnatal stages. Cholinergic innervations from the vMHb were still present without obvious defects in Otx2 cKO (Fig. 8H,I). By contrast, SP fibers were largely reduced, especially in the lateral-most region that corresponds to the terminal field of innervation from Otx2High neurons of the MHb (Fig. 8H–I′).
Together, these results show an Otx2-linked chronotopic organization of habenular projections with late-born Otx2High habenular neurons preferentially targeting the lateral region of the cIPN, where they establish synaptic contacts with Otx2 GABAergic neurons (Fig. 8J,K). Furthermore, genetic ablation of Otx2High habenular neurons in Otx2cKO leads to subcircuit disruption between these neurons and the Lat-cIPN.
Discussion
Here, we have described for the first time the role of Otx2 in HIPS development. We reveal the complex pattern of Otx2 expression and demonstrate that, in the absence of Otx2, the habenular progenitors have an altered cell cycle, giving rise to fewer late-born neurons, whereas early-born populations are unaffected. In the IPN, Otx2 acts at the postmitotic level, playing a new role in neuron migration. We show that Otx2 functionally contributes to the building of specific subcircuits of the HIPS. Habenular late-born neurons, highly expressing and depending on Otx2, target IPN neurons also expressing and depending on Otx2. Otx2 expression therefore marks specific HIPS subcircuits that intersect with the classical dichotomy of substance-P-ergic dMHb contacting the Lat-IPN and cholinergic vMHb contacting the Mid-IPN. The expression of Otx2 may therefore be a better marker of conserved and important subfunctions of the HIPS.
Otx2 and habenular neurogenesis
Otx2 deletion affects the cell cycle of habenular progenitors, leading to reduced number of late-born neurons. We excluded that these progenitors die or exhibit slower proliferation because, if this were the case, then it should have caused a reduced number or delayed generation of early-born neurons as well, which we did not observe. We thus hypothesized that these progenitors display an equivalent rate of proliferation but differentiate earlier, resulting in a decreased number of neurons. We found a shorter S phase in mutant progenitors. In medulloblastoma, Otx2 was shown to control the G1–S and G2–M transitions (Bunt et al., 2012; El Nagar et al., 2018). The G1–S transition is particularly important for neuron precursors because it is the time when the decision to proliferate or to undergo their last division is made. In the cortex, a shorter S-phase correlates with lower proliferative potential (Arai et al., 2011). It is thus likely that, in the absence of Otx2, habenular progenitors have a decreased proliferative potential, leading to precocious depletion of the progenitor pool and thus to fewer neurons.
Our results cannot be explained by other known mechanisms affecting habenular size. First, we did not detect any patterning defect that could lead to smaller habenula. Second, we did not detect any obvious habenular asymmetry unlike previous results (Concha and Wilson, 2001) or any symmetry alteration in Otx2 mutants. We also tested the role of Otx2 in postmitotic cells by ablating Otx2 when all neurons are born (F. D'Autréaux and N. Ruiz-Reig, personal communication). Contrary to what happens when Brn3a is deleted (Muzyka et al., 2018), we did not detect any effects on identity or survival of habenular neurons. Therefore, Otx2 deletion affects habenular precursor proliferation rather than maintenance of habenular neuron identity and survival.
Otx2 roles in migration
Otx2 expression, which is initiated at the postmitotic stage in IPN neurons, is required for their tangential migration. This is the first report of an Otx2 role in tangential migration and one of the rare studies exploring migration of the numerous hindbrain cells. We ruled out cell fate switch to explain altered migration behavior and showed a Netrin/Dcc interplay defect in Otx2 mutants. Netrin-1 is expressed in a timely manner in the floor plate to attract IPN neurons. Otx2+ IPN neurons express high and low levels of Dcc during tangential and radial migration, respectively. In the mutant, newborn neurons displayed weak Dcc expression that may explain their lack of attraction and their precocious radial migration. Otx2 has known roles in migration. In the mouse embryo, the proximo-distal axis conversion to definitive anteroposterior axis requires Otx2. It controls the migration of distal visceral endoderm cells by repressing Wnt signaling and promoting β-catenin degradation (Kimura-Yoshida et al., 2005). Canonical Wnt signaling is known to affect migration but not preferentially tangential migration. We thus do not think that a defect of this pathway can explain the phenotype of mutant IPN cells. Cre-mediated deletion of Shh using the En1 promoter was shown to affect the migration of the rIPN neurons, sparing the Otx2+ cIPN (Moreno-Bravo et al., 2014). We found that Otx2 cIPN neurons do not express En1. Furthermore, high En1 expression seems to occur at a more rostral location than the region generating the cIPN (F. D'Autréaux, personal communication). This may explain why the caudal IPN is not affected in this study.
Otx2 functions are highly context dependent
The search for a general rule of Otx2 functions has left many scientists with more questions than answers. Studies in the developing mouse brain have revealed highly context-dependent (Buecker et al., 2014; Kaur et al., 2015) and time-dependent (Fossat et al., 2006) functions of Otx2. We found here that Otx2 plays radically different functions depending on which stage and cell type is considered. At the progenitor stage, Otx2 controls the proliferative status of epithalamus cells. In the IPN, Otx2 acts postmitotically to control the migration of IPN precursors. We could not directly test for migration defect of Otx2high habenular neurons in the absence of Otx2 because they are not generated. However, Otx2low habenular neurons seemed to conserve their centrifugal mode of migration. In fact, if newborn neurons are simply laterally pushed away by younger ones, then they may marginally rely on migration cues to form the habenula, which would explain why Otx2 is not required for such “passive” migration. A recent study (Kaur et al., 2015) showed a role for OTX2 in self-renewal and migration of neural precursors. In this study, OTX2 overexpression in a cell context yielded the same effect on proliferation as OTX2 knock-down in another one. Furthermore, genes affected in the two cell contexts were nonoverlapping. Developmental genes can thus be recruited at several time points, even in the same tissue, to play nonoverlapping functions with nonoverlapping partners or gene targets. This suggests that analysis of gene targets and partners may not be sufficient to reach a conclusion on the common mechanisms that underlie the multiple functions of Otx2.
Otx2 functions from an evolutionary perspective
A good way of considering the multiple roles of Otx2 is from an evolutionary point of view. The link between Otx2 and the visual system has been largely described (for review, see Beby and Lamonerie, 2013). In a more general way, Otx2 appears as a marker of sensory-related circuits (Harden et al., 2006; Sen et al., 2013; Steventon et al., 2016). Here, we show that Otx2 is critical for both habenula and IPN development. Otx2 is evolutionary conserved and its expression pattern and functions are equally highly conserved (Sen et al., 2013). It is therefore tempting to try to predict from its expression pattern potentially conserved and important brain circuits. Investigating Otx2 expression outside of the visual pathway, we found that it strikingly marked numerous regions implicated in emotional processing such as the VTA, the lateral septum, the periaqueductal gray, the laterodorsal and dorsal tegmentum, the nucleus incertus, the medial raphe, and the HIPS. These subcircuits, as well as Otx2 itself, may thus perform conserved and important functions in the limbic system. Actually, this Otx2+ circuit inside the limbic system might still be related to the visual pathway. In zebrafish, the MHb is a target of visual inputs through a thalamic structure not precisely defined (Zhang et al., 2017). This structure has been recently described as the perihabenular nucleus in mammals (Fernandez et al., 2018). It would be interesting to verify whether it expresses Otx2 as well. In zebrafish, the habenula is asymmetric with predominant dMHb and small vMHb homologs on the left side. Interestingly, this is the side preferentially targeted by the visual pathway. Because Otx2 mostly affects the dMHb, one can also predict that the functions associated with this part of the habenula will be preferentially affected. However, the exact functions of the dMHb remain elusive and Otx2 deletion restricted to the habenula may help to solve these issues.
Conclusions
We found that Otx2 expression delineates a specific subcircuit of the HIPS and is also required for this circuit to form. The HIPS is a modulator of aversion and reward processing and is involved in brain disorders such as anxiety, depression, and addiction. It is thus reasonable to hypothesize that abnormal expression of Otx2 in the HIPS may lead to psychiatric diseases. There is now evidence that some mood disorders may take root during development. Therefore, to find the causes of these disorders, we need to focus on the developmental mechanisms shaping the structures involved. This study paves the way to investigate specific subfunctions of the HIPS from a developmental perspective.
Footnotes
This work was supported by the University of Nice (CSI funding) and the National Center for Scientific Research (CNRS). N.R.-R. is funded by a postdoctoral fellowship from the City of Nice, France (Aide Individuelle aux Chercheurs, 2016). We would like to particularly thank Alizée Costantini for preliminary data, Aurélie Biancardini for mouse husbandry, Renaud Rebillard and Mouna Sekoni Khemiri for genotyping. We thank Alexandra Pierani for the Dbx1Cre line; Naomi Galili, Jonathan Epstein, and Clayton A. Buck for the Gscl antibodies; Silvia Arber for the Er81 antibodies; Paola Bovolenta and Michele Studer for ISH probes; and the iBV PRISM platform, especially Magali Mondin and Simon Lachambre.
The authors declare no competing financial interests.
N. Ruiz-Reig′s present address: Institute of Neuroscience of the Universite Catholique de Louvain, 1200 Brussels, Belgium.
References
- Angevine JB., Jr (1970) Time of neuron origin in the diencephalon of the mouse. an autoradiographic study. J Comp Neurol 139:129–187. 10.1002/cne.901390202 [DOI] [PubMed] [Google Scholar]
- Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, Huttner WB (2011) Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun 2:154. 10.1038/ncomms1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beby F, Lamonerie T (2013) The homeobox gene Otx2 in development and disease. Exp Eye Res 111:9–16. 10.1016/j.exer.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Bielle F, Griveau A, Narboux-Nême N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A (2005) Multiple origins of cajal-retzius cells at the borders of the developing pallium. Nat Neurosci 8:1002–1012. 10.1038/nn1511 [DOI] [PubMed] [Google Scholar]
- Bloch-Gallego E, Ezan F, Tessier-Lavigne M, Sotelo C (1999) Floor plate and netrin-1 are involved in the migration and survival of inferior olivary neurons. J Neurosci 19:4407–4420. 10.1523/JNEUROSCI.19-11-04407.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J (2014) Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14:838–853. 10.1016/j.stem.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, Kool M (2012) OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer 131:E21–E32. 10.1002/ijc.26474 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Guo Q, Weber S, Scholpp S, Li JY (2014) Pax6 regulates the formation of the habenular nuclei by controlling the temporospatial expression of shh in the diencephalon in vertebrates. BMC Biol 12:13. 10.1186/1741-7007-12-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Wilson SW (2001) Asymmetry in the epithalamus of vertebrates. J Anat 199:63–84. 10.1046/j.1469-7580.2001.19910063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contestabile A, Villani L, Fasolo A, Franzoni MF, Gribaudo L, Oktedalen O, Fonnum F (1987) Topography of cholinergic and substance P pathways in the habenulo-interpeduncular system of the rat. an immunocytochemical and microchemical approach. Neuroscience 21:253–270. 10.1016/0306-4522(87)90337-X [DOI] [PubMed] [Google Scholar]
- Courtois V, Chatelain G, Han ZY, Le Novère N, Brun G, Lamonerie T (2003) New Otx2 mRNA isoforms expressed in the mouse brain. J Neurochem 84:840–853. 10.1046/j.1471-4159.2003.01583.x [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP (1993) Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75:1417–1430. 10.1016/0092-8674(93)90627-3 [DOI] [PubMed] [Google Scholar]
- El Nagar S, Chakroun A, Le Greneur C, Figarella-Branger D, Di Meglio T, Lamonerie T, Billon N (2018) Otx2 promotes granule cell precursor proliferation and shh-dependent medulloblastoma maintenance in vivo. Oncogenesis 7:60. 10.1038/s41389-018-0070-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DC, Fogerson PM, Lazzerini Ospri L, Thomsen MB, Layne RM, Severin D, Zhan J, Singer JH, Kirkwood A, Zhao H, Berson DM, Hattar S (2018) Light affects mood and learning through distinct retina-brain pathways. Cell 175:71–84.e18. 10.1016/j.cell.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Chatelain G, Brun G, Lamonerie T (2006) Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout. EMBO Rep 7:824–830. 10.1038/sj.embor.7400751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Le Greneur C, Béby F, Vincent S, Godement P, Chatelain G, Lamonerie T (2007) A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors. BMC Dev Biol 7:122. 10.1186/1471-213X-7-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato H, Sato M, Sinton CM, Gautron L, Williams SC, Skach A, Elmquist JK, Skoultchi AI, Yanagisawa M (2010) Loss of goosecoid-like and DiGeorge syndrome critical region 14 in interpeduncular nucleus results in altered regulation of rapid eye movement sleep. Proc Natl Acad Sci U S A 107:18155–18160. 10.1073/pnas.1012764107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Epstein JA, Leconte I, Nayak S, Buck CA (1998) Gscl, a gene within the minimal DiGeorge critical region, is expressed in primordial germ cells and the developing pons. Dev Dyn 212:86–93. [DOI] [PubMed] [Google Scholar]
- Harden MV, Newton LA, Lloyd RC, Whitlock KE (2006) Olfactory imprinting is correlated with changes in gene expression in the olfactory epithelia of the zebrafish. J Neurobiol 66:1452–1466. 10.1002/neu.20328 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S (2005) A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 3:e159. 10.1371/journal.pbio.0030159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Morton G, Guy EG, Wang SD, Turner EE (2016) Dorsal medial habenula regulation of mood-related behaviors and primary reinforcement by tachykinin-expressing habenula neurons. eNeuro 3: ENEURO.0109-16.2016. 10.1523/ENEURO.0109-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic MM, Carrillo-Roa T, Bar M, Becker G, Jovanovic VM, Zega K, Binder EB, Brodski C (2015) Abnormal development of monoaminergic neurons is implicated in mood fluctuations and bipolar disorder. Neuropsychopharmacology 40:839–848. 10.1038/npp.2014.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunaratne A, Hargrave M, Poh A, Yamada T (2002) GATA proteins identify a novel ventral interneuron subclass in the developing chick spinal cord. Dev Biol 249:30–43. 10.1006/dbio.2002.0754 [DOI] [PubMed] [Google Scholar]
- Kaur R, Aiken C, Morrison LC, Rao R, Del Bigio MR, Rampalli S, Werbowetski-Ogilvie T (2015) OTX2 exhibits cell-context-dependent effects on cellular and molecular properties of human embryonic neural precursors and medulloblastoma cells. Dis Model Mech 8:1295–1309. 10.1242/dmm.020594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Fontelonga T, Roesener AP, Lee H, Gurung S, Mendonca PRF, Mastick GS (2015) Motor neuron cell bodies are actively positioned by Slit/Robo repulsion and Netrin/DCC attraction. Dev Biol 399:68–79. 10.1016/j.ydbio.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Nakano H, Okamura D, Nakao K, Yonemura S, Belo JA, Aizawa S, Matsui Y, Matsuo I (2005) Canonical wnt signaling and its antagonist regulate anterior-posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev Cell 9:639–650. 10.1016/j.devcel.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Larsen KB, Lutterodt MC, Møllgård K, Møller M (2010) Expression of the homeobox genes OTX2 and OTX1 in the early developing human brain. J Histochem Cytochem 58:669–678. 10.1369/jhc.2010.955757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente-Cánovas B, Marín F, Corral-San-Miguel R, Hidalgo-Sánchez M, Ferrán JL, Puelles L, Aroca P (2012) Multiple origins, migratory paths and molecular profiles of cells populating the avian interpeduncular nucleus. Dev Biol 361:12–26. 10.1016/j.ydbio.2011.09.032 [DOI] [PubMed] [Google Scholar]
- Lu S, Wise TL, Ruddle FH (1994) Mouse homeobox gene dbx: sequence, gene structure and expression pattern during mid-gestation. Mech Dev 47:187–195. 10.1016/0925-4773(94)90089-2 [DOI] [PubMed] [Google Scholar]
- Mallika C, Guo Q, Li JY (2015) Gbx2 is essential for maintaining thalamic neuron identity and repressing habenular characters in the developing thalamus. Dev Biol 407:26–39. 10.1016/j.ydbio.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez JE, Moreno-Bravo JA, Madrigal MP, Martinez S, Puelles E (2015) Red nucleus and rubrospinal tract disorganization in the absence of Pou4f1. Front Neuroanat 9:8. 10.3389/fnana.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin I, Dani JA, De Biasi M (2017) The medial habenula and interpeduncular nucleus circuitry is critical in addiction, anxiety, and mood regulation. J Neurochem 142:130–143. 10.1111/jnc.14008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bravo JA, Perez-Balaguer A, Martinez-Lopez JE, Aroca P, Puelles L, Martinez S, Puelles E (2014) Role of shh in the development of molecularly characterized tegmental nuclei in mouse rhombomere 1. Brain Struct Funct 219:777–792. 10.1007/s00429-013-0534-6 [DOI] [PubMed] [Google Scholar]
- Mori H, Miyazaki Y, Morita T, Nitta H, Mishina M (1994) Different spatio-temporal expressions of three otx homeoprotein transcripts during zebrafish embryogenesis. Brain Res Mol Brain Res 27:221–231. 10.1016/0169-328X(94)90004-3 [DOI] [PubMed] [Google Scholar]
- Muzyka VV, Brooks M, Badea TC (2018) Postnatal developmental dynamics of cell type specification genes in Brn3a/Pou4f1 retinal ganglion cells. Neural Dev 13:15. 10.1186/s13064-018-0110-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña CJ, Kronman HG, Walker DM, Cates HM, Bagot RC, Purushothaman I, Issler O, Loh YE, Leong T, Kiraly DD, Goodman E, Neve RL, Shen L, Nestler EJ (2017) Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 356:1185–1188. 10.1126/science.aan4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, Turner EE (2009) Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. J Neurosci 29:14309–14322. 10.1523/JNEUROSCI.2430-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Reig N, Andres B, Lamonerie T, Theil T, Fairén A, Studer M (2018) The caudo-ventral pallium is a novel pallial domain expressing Gdf10 and generating Ebf3-positive neurons of the medial amygdala. Brain Struct Funct 223:3279–3295. 10.1007/s00429-018-1687-0 [DOI] [PubMed] [Google Scholar]
- Sabunciyan S, Yolken R, Ragan CM, Potash JB, Nimgaonkar VL, Dickerson F, Llenos IC, Weis S (2007) Polymorphisms in the homeobox gene OTX2 may be a risk factor for bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 144B:1083–1086. 10.1002/ajmg.b.30523 [DOI] [PubMed] [Google Scholar]
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C (2007) Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development 134:3167–3176. 10.1242/dev.001461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Reichert H, VijayRaghavan K (2013) Conserved roles of ems/Emx and otd/Otx genes in olfactory and visual system development in Drosophila and mouse. Open Biol 3:120177. 10.1098/rsob.120177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M (1996) Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87:1001–1014. 10.1016/S0092-8674(00)81795-X [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Leyva-Díaz E, De La Cruz E, Hobert O (2018) BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr Biol 28:2813–2823.e2. 10.1016/j.cub.2018.06.045 [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E (1992) Nested expression domains of four homeobox genes in developing rostral brain. Nature 358:687–690. 10.1038/358687a0 [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1:4. 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A (2016) Directional cell movements downstream of Gbx2 and Otx2 control the assembly of sensory placodes. Biol Open 5:1620–1624. 10.1242/bio.020966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, Nakagawa Y (2007) Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol 505:73–91. 10.1002/cne.21467 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Lewandoski M, Campbell K, Joyner AL, Rubenstein JL, Martinez S, Martin GR (1997) Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 124:2923–2934. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Danjo T, Pastan I, Hikida T, Nakanishi S (2013) Distinct roles of segregated transmission of the septo-habenular pathway in anxiety and fear. Neuron 78:537–544. 10.1016/j.neuron.2013.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BB, Yao YY, Zhang HF, Kawakami K, Du JL (2017) Left habenula mediates light-preference behavior in zebrafish via an asymmetrical visual pathway. Neuron 93:914–928.e4. 10.1016/j.neuron.2017.01.011 [DOI] [PubMed] [Google Scholar]