Abstract
We have identified a member of the growth arrest and DNA damage (Gadd45) protein family, Gadd45γ, which is known to be critically involved in DNA repair, as a key player in the regulation of immediate early gene (IEG) expression underlying the consolidation of associative fear memory in adult male C57BL/6 mice. Gadd45γ temporally influences learning-induced IEG expression in the prelimbic prefrontal cortex (PLPFC) through its interaction with DNA double-strand break (DSB)-mediated changes in DNA methylation. Our findings suggest a two-hit model of experience-dependent IEG activity and learning that comprises (1) a first wave of IEG expression governed by DSBs and followed by a rapid increase in DNA methylation, and (2) a second wave of IEG expression associated with the recruitment of Gadd45γ and active DNA demethylation at the same site, which is necessary for memory consolidation.
SIGNIFICANCE STATEMENT How does the pattern of immediate early gene transcription in the brain relate to the storage and accession of information, and what controls these patterns? This paper explores how Gadd45γ, a gene that is known to be involved with DNA modification and repair, regulates the temporal coding of IEGs underlying associative learning and memory. We reveal that, during fear learning, Gadd45γ serves to act as a coordinator of IEG expression and subsequent memory consolidation by directing temporally specific changes in active DNA demethylation at the promoter of plasticity-related IEGs.
Keywords: DNA double-strand break, DNA methylation, DNA repair, immediate early gene, memory consolidation, neuroepigenetic
Introduction
Memory consolidation requires learning-induced gene expression, protein synthesis, and changes in synaptic plasticity (Izquierdo and McGaugh, 2000; Kandel, 2001). For decades, models of cued and contextual fear conditioning have been used to advance the understanding of the cellular, molecular, and circuit-level mechanisms of learning and memory. Although the amygdala and hippocampus have historically been the primary focus of studies on fear-related learning, it is becoming evident that the prefrontal cortex (PFC) is also involved in the initial encoding of fear memories, with the dorsomedial or prelimbic region of the PFC playing a particularly important role (Giustino and Maren, 2015; Dejean et al., 2016; Widagdo et al., 2016; Klavir et al., 2017; Rizzo et al., 2017; Santos et al., 2017). With respect to genomic responses to learning, several members of a particularly unique class of inducible genes known as the immediate early genes are rapidly and transiently expressed in response to neural activity, and are thought to be important for information processing in the brain because of the tight temporal relationship between their expression and learning. Specifically, activity-regulated cytoskeleton-associated protein (Arc), fos proto-oncogene (c-Fos), and neuronal PAS domain protein 4 (Npas4) have all been shown to be critically involved in memory formation (Minatohara et al., 2016; Sun and Lin, 2016; Gallo et al., 2018). However, a detailed understanding of the molecular mechanisms by which these IEGs are regulated during learning and the formation of memory remains to be developed.
It has become increasingly evident that there is an important epigenetic layer of regulatory control over gene expression and memory, which includes DNA modification (Baker-Andresen et al., 2013). DNA methylation and active DNA demethylation are directly involved in gene expression underlying memory formation (Li et al., 2014), and several recent studies have extended these findings to include IEGs. For example, RNA-directed changes in DNA methylation regulate c-Fos expression by directing DNA methyltransferase (DNMT) activity to the c-Fos promoter (Baker-Andresen et al., 2013). DNA hydroxymethylation regulates both c-Fos and Arc expression in the hippocampus (Li et al., 2014), and chronic drug exposure increases DNA methylation at the c-Fos and Arc promoters, which has a profound effect on learning and memory (Miller et al., 2010; Baker-Andresen et al., 2013; Li et al., 2014). Several active DNA demethylation pathways have been proposed, and each has been shown to be involved in regulating gene expression related to plasticity and memory (Li et al., 2013, 2014; Kaas et al., 2013; Rudenko et al., 2013; Ratnu et al., 2014). Perhaps most direct pathway involves independent members of the Gadd45 family of DNA repair proteins (α, β, and γ), where each has been shown to form a complex with DNA repair enzymes to guide the removal of 5-mC by either base or nucleotide excision repair (Barreto et al., 2007; Rai et al., 2008; Ma et al., 2009).
Early reports indicated that in sensory and motor neurons, Gadd45α is induced by nerve injury (Befort et al., 2003), whereas Gadd45β was found to be activity-dependent and involved in neurogenesis (Ma et al., 2009). With respect to experience-dependent plasticity in the adult brain, Gadd45β has also been shown to influence memory formation, although reports differ with regards to whether its knockdown leads to enhancement (Sultan et al., 2012) or impairment of contextual fear memory (Leach et al., 2012). Although fear-related learning led to a significant increase in Gadd45γ mRNA expression in the hippocampus and amygdala (Sultan et al., 2012), and the expression of Gadd45β, Gadd45γ, and Arc and a relationship with DNA methylation in a mouse model of depression has been reported (Grassi et al., 2017), whether any members of the Gadd45 family of proteins contribute to memory formation through direct effects on IEG expression has yet to be determined.
Materials and Methods
Mice.
Nine- to 11-week-old C57BL/6J (Animal Resource Centre) male mice were housed four per cage, maintained on a 12 h light/dark schedule, and allowed ad libitum access to food and water. All testing was conducted during the light phase in red-light-illuminated testing rooms following protocols approved by the Animal Ethics Committee of the University of Queensland.
Culture and usage of primary mouse cortical neurons.
Primary mouse cortical neuron cultures were prepared and maintained as previously described (Lin et al., 2011). To knock down expression of genes of interest, cultured neurons at 2–3 d in vitro were exposed to lentivirus, followed by replacement of the culture medium. To investigate activity-dependent gene regulation, cultured neurons were depolarized by the addition of 20 mm KCl to the culture medium after 7–10 d. Cells were collected for molecular analysis immediately at the end of the KCl exposure time.
Culture of Neuro2A, HT-22, and HEK293T cells.
Neuro2A (N2A) cells were maintained in medium consisting of 50% DMEM, high glucose (Invitrogen), and 50% Opti-MEM (Invitrogen) supplemented with 5% fetal bovine serum and 1% Pen/Strep. HT-22 and HEK293T cells were maintained in DMEM, high glucose (Invitrogen) supplemented with 10% fetal bovine serum and 1% Pen/Strep.
RNA isolation and reverse transcription.
To extract RNA, cells were lysed with NucleoZOL (Macherey-Nagel) directly in the culture dish and RNA was isolated following the manufacturer's instructions. Reverse transcription was performed using the QuantiTect kit (Qiagen) using the provided RT Primer Mix and following the manufacturer's instructions.
Quantitative PCR for gene expression.
Quantitative PCRs were prepared in duplicate, in a 10 μl reaction volume, using 2× SYBR master mix (Qiagen), 500 μm of each primer, and 1 μl per reaction of a cDNA sample (the cDNA dilution factor varied according to target abundance). Reactions were run on the Rotor-Gene Q platform and results were analyzed using the δδcT method, normalized to the reference gene PGK (phosphoglycerate kinase). qPCR primers used including: qGadd45α F: CCGAAAGGATGGACACGGTG; qGadd45α R: TTATCGGGGTCTACGTTGAGC; qGadd45β F: CACCCTGATCCAGTCGTTCT; qGadd45β R: TTGCCTCTGCTCTCTTCACA; qGadd45γ F: GGGAAAGCACTGCACGAACT; qGadd45γ R: AGCACGCAAAAGGTCACATTG; qCyr61 F: CTGCGCTAAACAACTCAACGA; qCyr61 R: GCAGATCCCTTTCAGAGCGG; qc-Fos F: GAACGGAATAAGATGGCTGC; qc-Fos R: TTGATCTGTCTCCGCTTGG; qNpas4 F: CTGCATCTACACTCGCAAGG; qNpas4 R: GCCACAATGTCTTCAAGCTCT; qArc F: AAGTGCCGAGCTGAGATGC; qArc R: CGACCTGTGCAACCCTTTC; qPGK F: TGCACGCTTCAAAAGCGCACG; qPGK R: AAGTCCACCCTCATCACGACCC. Data were analyzed using Student's t test or one-way ANOVA where appropriate.
DNA extraction.
PLPFC tissue from naive and trained mice was dissociated by dounce homogenization in 500 μl PBS. Preparation of DNA was performed using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer's instructions.
Protein extract and Western blot.
Total protein was extracted using NP40 lysis buffer (ThermoFisher) following the manufacturer's protocol. Protein concentration was determined using the Qubit protein assay (Invitrogen). For Western blot, samples were prepared on ice (to final volume of 20 μl), and then vortexed and denatured for 10 min at 90°C. PAGE was performed and proteins transferred onto nitrocellulose membrane (Bio-Rad). The membrane was blocked using Li-Cor blocking buffer for 1 h at room temperature, washed with Tris-buffered saline containing 1% Tween 20 (TBS-T) three times for 5 min each, then incubated with different antibodies (Gadd45α: Cell Signaling Technology, catalog #4632; Gadd45β: Abcam, catalog #ab105060; Gadd45γ: Abcam, catalog #ab105060; Beta-actin: Cell Signaling Technology, catalog #3700) diluted in Li-Cor blocking buffer overnight at 4°C. Membranes were then washed three times with TBS-T, incubated for 1 h with anti-mouse secondary antibody (1:15,000; Li-Cor) and anti-rabbit secondary antibody (1:15000; Li-Cor) diluted in blocking buffer, and washed three times with TBST for 10–20 min. Optical density readings of the membrane were taken using the Li-Cor FX system.
Methylated DNA immunoprecipitation.
Methylated DNA immunoprecipitation (MeDIP) was performed using the Active Motif kit (catalog #55009). In brief, DNA was fragmented with the Covaris M220, and then heat denatured. Following this, fragments containing at least one 5-mC were captured with a 5-mC antibody bound to protein G Dynabeads (Invitrogen). The beads were then washed and DNA fragments with 5-mC were eluted for quantitative PCR.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed using the Invitrogen ChIP kit, with a modified protocol. Lysate from cells or tissue was fixed with 1% formaldehyde and cross-linked cell lysates were sheared in 1% SDS lysis buffer using the Covaris M220, generating chromatin fragments with an average length of 300 bp. Various antibodies (Gadd45α: Cell Signaling Technology, catalog #4632; Gadd45β: Abcam, catalog #ab105060; Gadd45γ: Santa Cruz Biotechnology, catalog #sc-393261; γH2a.X: Abcam, catalog #ab2893; topoisomerase 2 beta (Topo IIβ): Santa Cruz Biotechnology, catalog #sc-365071; DNMT3a: Active Motif, catalog #39206; rabbit IgG: Millipore, catalog #12-370; mouse IgG: Millipore, catalog #12-371) were used to perform immunoprecipitation; additionally, each antibody was paired with an equivalent quantity of IgG as a negative control for validation purposes. Antibodies were incubated with the sheared lysates overnight at 4°C with gentle agitation, and protein-DNA-antibody complexes were then precipitated with Protein G Dynabeads for 1 h at 4°C, followed by three washes in low-salt buffer, and three washes in high-salt buffer. The precipitated protein-DNA complexes were eluted from the captured antibody with 1% SDS and 0.1 m NaHCO3, then incubated overnight at 65°C in 200 mm NaCl to reverse formaldehyde cross-linkages. Samples were then digested with Proteinase K, and the DNA extracted with phenol-chloroform and purified by ethanol precipitation. DNA was used for quantitative PCR using primers specific to the proximal promoter regions of IEGs (Chip-Cyr61 F: GGTCAAGTGGAGAAGGGTGA; Chip-Cyr61 R: GCACCTCGA GAGAAGGACAC; Chip-Fos F: GAAAGCCTGGGGCGTAGAGT; Chip-Fos R: CCTCAGCTGGCGCCTTTAT; Chip-Npas4 F: GATCGTGGGAGAGGTTCAAA; Chip-Npas4 R: TCACAACTGGGGGTCTTTTC; Chip-Arc F: CCTCAGCTGCCTTTGGTTAG; Chip-Arc R: GAAAAAGCCTTGCCTGAGTG).
Quantitative PCR for ChIP and MeDIP samples.
Quantitative PCRs were prepared in duplicate, using 2× SYBR master mix (Qiagen), 500 μm of each primer, and 1 μl of template DNA purified as described previously after immunoprecipitation. Reactions were run on the Rotor-Gene Q platform and results were analyzed using the δδcT method normalized to input samples.
Lentivirus construction and packaging.
Lentivirus was packaged according to our previously published protocol (Lin et al., 2011). Briefly, HEK293T cells were grown to 70–80% confluence in triple-layer flasks. Cells were transfected with the plasmids pMDG, pRSV-rev, pMDLg/pRRE, and the transfer vector (gene-specific shRNA cloned into FG12) using Lipofectamine 3000 reagent according to the manufacturer's instructions. Transfected cells were cultured for 48 h, then the culture media was collected, clarified, filtered, and lentivirus particles were concentrated by ultracentrifugation. Concentrated viral pellets were resuspended in PBS and snap-frozen. Five lentiviruses were produced: one carrying a control shRNA with no specificity to any known mouse transcript, and one targeting each of Gadd45α, Gadd45β, Gadd45γ and Cyr61 (Gadd45α shRNA: CCACATTCATCACAATGGA; Gadd45β shRNA: ACGAACTGTCATACAGATT; Gadd45γ shRNA: AGATCCATT TCACGTTGAT; Cyr61 shRNA: TCCAGCCCAACTGTAAACA; Universal control shRNA: GCGCGATAGCGCTAATAAT).
Titration of virus.
Four × 105 293T cells/well of a 6 well plate were plated the day before titration of virus. On the following day, the number of cells in each well were estimated by counting. Then, four dilutions of each virus were prepared, corresponding to 0.5, 1, 2, and 5 μl per well, and added to the HEK cells which were then incubated for 2–3 d. The percentage of cells expressing e-GFP was used to calculate the virus titer by using the following formula: (% GFP-positive cells) × (μl of virus added to well)/(number of cells in well before infection) = infectious units (IU)/ml × 103 = IU/ml. The titers of shRNA lentivirus for Gadd45α,Gadd45β, Gadd45γ, Cyr61, and control are ∼1.5 × 108 IU/ml, 1.3 × 108 IU/ml, 1 × 108 IU/ml, 2 × 108 IU/ml, and 1.4 × 108 IU/ml, separately.
Stereotaxic surgery.
Double cannulae (Plastics One) were implanted in the anterior–posterior plane, along the midline, into the PLPFC. The cannula placements were centered at +1.80 in the anterior–posterior plane, and −2.20 mm in the dorsal-ventral plane, relative to bregma. Following surgery, animals were single-housed and given at least 1 week to recover. After the recovery period, mice were infused with lentivirus through both cannulae; 0.5 μl was infused through each cannula on 2 consecutive days, for a total delivered volume of 2.0 μl per animal. Following virus infusion, a 1 week waiting period was observed to ensure stable expression of the viral construct. For antibody injection, saline or Gadd45γ antibody was premixed with 0.1% Triton ×100 and 1 μl was infused into each side 3 h after cued fear conditioning. Etoposide (Sigma-Aldrich, catalog #E1383) was dissolved in 1% DMSO at two different concentrations (1 ng or 0.1 μg/μl) and mice were bilaterally infused with 0.5 μl of etoposide or 1% DMSO as control.
DsiRNA design and injection.
Three separate dicer-substrate siRNAs (DsiRNAs) against Cyr61 were designed using an online design tool provided by Integrated DNA Technologies (https://sg.idtdna.com/site/order/designtool/index/DSIRNA_CUSTOM). DsiRNAs were tested in HT-22 cells and compared with a universal negative control that does not recognize any sequences in human, mouse, or rat transcriptomes. After determining which DsiRNA-Cyr61 had the maximum knockdown efficiency, we reordered DsiRNA with 2′-O-methylated modified nucleotides to provide further stability and avoid triggering an innate immune response which can be harmful to the animal (DsiRNA-Cyr61 sense: mGmArUmGrUmUrUrUrCrCrAmArGmArAmUrGmUrCrArUrGrAmUG; DsiRNA-Cyr61 antisense: rCrAmUrCrArUrGrAmCrAmUrUmCrUrUrGrGrArArArArCmArUmCmUmC; DsiRNA-cont sense: mCmGrUmUrAmArUrCrGrCrGmUrAmUrAmArUmArCrGrCrGrUmAT; DsiRNA-cont antisense: rArUmArCrGrCrGrUmArUmUrAmUrArCrGrCrGr ArUrUrAmArCmGmAmC). Two concentrations of DsiRNA-Cyr61 were tested (100 nm/μl or 10 nm/μl) by bilateral infusion of 1 μl into the PLPFC.
Fear conditioning and recall tests.
Mice underwent cued fear conditioning in operant chambers (Coulbourn Instruments), which have two transparent walls and two stainless steel walls, with a square shape and steel grid floor. The training context was scented using a dilute solution of lemon essence. The training protocol consisted of three pairings of a 120 s, 80 dB, pure tone stimulus (CS) with a 1 s, 0.7 mA, foot shock (US), separated by an empty 10 s trace interval. The pretrial interval, intertrial interval, and post-trial interval were all 120 s long. Movement was captured by cameras in the boxes and processed with the software FreezeFrame 4 to determine the percentage-based freezing score, with “freezing” defined as 1 s or more of immobility. The context-only control mice were placed in the same context for 16 min (the same duration as the training protocol) without the cue sound or the shock. Twenty-four hours after training, mice were returned to the context for recall tests. Following a 2 min acclimation period (to reduce context generalization), freezing was assessed during two 120 s CS presentations (with 120 s intertrial interval). Memory was inferred by the observing percentage of time spent freezing during the recall tests.
Open-field test.
Following completion of other behavioral testing, mice were tested in open fields to check for off-target behavioral effects that could cause a change in freezing score unrelated to memory (specifically, increased generalized anxiety or reduced spontaneous locomotion). Open-field tests were conducted in a sound-attenuated room dimly illuminated with white lighting (60 ± 3 lux). Mice were placed into the center of a white plastic open field (30 × 30 × 30 cm) and recorded with an overhead camera for 20 min. Videos were analyzed using Noldus EthoVision 11 to determine the distance traveled, and the number of entries into and cumulative time spent in the center of the arena (defined as a 15 × 15 cm square concentric with the base of the arena).
Statistical analyses.
All statistics were performed using Prism7. Following analysis of descriptive statistics, two-tailed unpaired Student's t test was used for direct comparisons between fear and contextual control groups at each time point. One-way or two-way ANOVA was chosen for multiple comparisons when whichever was appropriate. All multiple-comparison tests were adjusted using a Sidak correction. Error bars represent SEM. Significant differences were accepted at p < 0.05.
Results
Learning-induced Gadd45γ expression in the PLPFC is required for the formation of cued fear memory
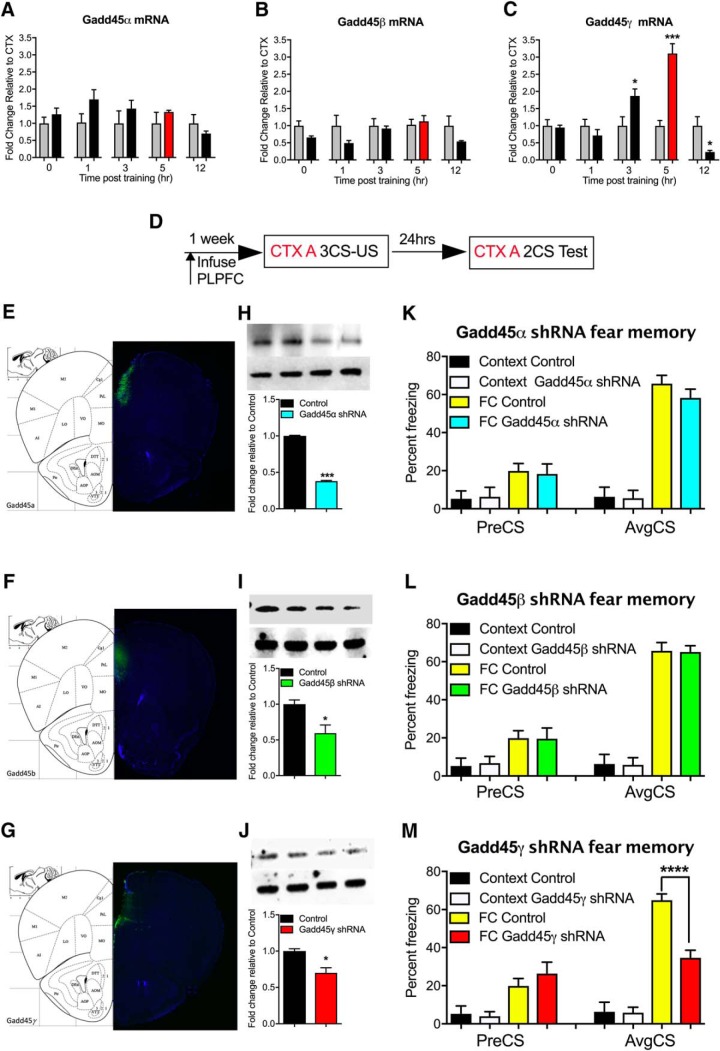
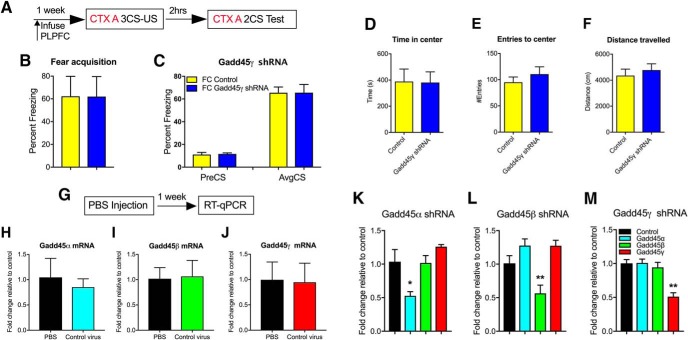
To establish which members of the Gadd45 family are associated with learning, we first examined Gadd45α, Gadd45β, and Gadd45γ mRNA expression in stimulated cortical neurons, in vitro, and in the PLPFC in response to cued fear learning in adult mice. In vitro, neural activation led to a significant increase in Gadd45β and Gadd45γ, but not Gadd45α, mRNA transcripts (data not shown). However, contrary to previous findings in the hippocampus and amygdala (Leach et al., 2012; Sultan et al., 2012), we found that only Gadd45γ mRNA exhibited a significant increase in the PLPFC in response to cued fear conditioning (Fig. 1A–C; t(6) = 2.63, CTX 3 h vs FC 3 h, p = 0.0392, t(6) = 6.46, CTX 5 h vs FC 5 h, p = 0.007, and t(6) = 3.87, CTX 12 h vs FC 12 h, p = 0.029, unpaired t test). Given that the level of mRNA expression does not necessarily reflect the functional relevance of the transcript (Nainar et al., 2016) and considering previous findings of a role for Gadd45β in regulating contextual fear memory, we next used established protocols (Lin et al., 2011) to design and test shRNAs against all three members of the Gadd45 family. In independent experiments, each shRNA was infused into the PLPFC ∼1 week before behavioral training (Fig. 1D–G). A decrease in the protein level of all three Gadd45 family members was confirmed by Western blot (Fig. 1H–J; t(2) = 67.92, Control vs Gadd45α shRNA, p = 0.0002; t(2) = 4.42 Control vs Gadd45β shRNA, p = 0.0474; t(2) = 5.48 Control vs Gadd45γ shRNA, p = 0.0317, unpaired t test). There was no effect of knockdown of any member of the Gadd45 family on the acquisition of freezing behavior during cued fear learning (data not shown). Knockdown of Gadd45α and Gadd45β had no effect on memory retention. In contrast, Gadd45γ shRNA-treated mice exhibited a significant impairment in fear memory when tested 24 h later (Fig. 1K–M; F(3,34) = 42.15, ****p < 0.0001; Sidak post hoc analysis: FC Control vs FC Gadd45γ shRNA, ****p < 0.0001, one-way ANOVA), with no effect on short-term memory when tested 2 h post-training, further implying a selective role for Gadd45γ in the PLPFC in the consolidation phase of cued fear memory formation (Fig. 2A–C). There was no effect of Gadd45γ shRNA on locomotor or anxiety-like behavior in the open-field test (Fig. 2D–F). Given the connection between Gadd45β and inflammatory signaling (Jarome et al., 2015), we also performed a PBS-injection control experiment, which revealed no nonspecific effect of lentivirus infusion on Gadd45 family member mRNA expression (Fig. 2G–J). Considering the evolutionarily conserved nature of the three Gadd45 family members, as well as their sequence similarity, we next determined whether there was an influence of individual gene knockdown on the mRNA expression of the other Gadd45 family members. Additionally, we confirmed that each shRNA is specific to the Gadd45 family member it was designed against, and does not influence expression of the other two genes, further confirming the high degree of specificity of our shRNA design (Fig. 2K–M; F(3,8) = 7.53, p = 0.01; Sidak post hoc analysis: Gadd45α shRNA vs control, p = 0.032; F(3,8) = 10.11, p = 0.004; Sidak post hoc analysis: Gadd45β shRNA vs control, p = 0.042; F(3,8) = 15.19, p = 0.001; Sidak post hoc analysis: Gadd45γ shRNA vs control, p = 0.001, one-way ANOVA). Together, these data demonstrate a selective and critical role for Gadd45γ, but not Gadd45α or Gadd45β, within the PLPFC in the regulation of cued fear memory.
Figure 1.
Gadd45γ expression is activity-dependent and necessary for cued fear learning in the PLPFC. A–C, Fold-change for fear conditioned animals which is calculated relative to each group's own context control at that time point. A, Following cued fear conditioning there was no significant change in Gadd45α or (B) Gadd45β mRNA levels. C, There was a significant increase in Gadd45γ 3 and 5 h post-conditioning and decrease at 12 h time point. D, Time course of behavioral training and shRNA infusion. E–G, Representative images show the location of infused lentivirus expressing shRNA knockdown effect at ∼55, 50, and 40% for (H) Gadd45α, (I) Gadd45β, and (J) Gadd45γ, separately, at the protein level. There were no significant differences in percentage freezing during the fear test for animals infused with either Gadd45α shRNA (K) or Gadd45β shRNA lenti-virus (L) compared with the control, which has no specificity to any known mouse transcript. M, There was a significant decrease in the percentage freezing for animals infused with Gadd45γ shRNA compared with control at test (N = 8–14 per group). FC, Fear conditioned; CTX, context exposure; CS, conditioned stimulus (tone); US, unconditioned stimulus (shock); PreCS, contextual freezing before first CS; AvgCS, average freezing at test. Error bars represent SEM. * p < 0.05, *** p < 0.001, **** p < 0.0001.
Figure 2.
Control experiments showing no nonspecific effect of shRNA lentivirus injection on behavior and gene expression. A, Time course of behavioral training. Gadd45γ knockdown had no effect on fear acquisition (B) and short memory when tested 2 h post-training (C; N = 8 per group). Open-field test results show that there is no effect of on generalized anxiety as measured by time spent in the center of the open field (D) or the number of entries to the center zone (E); likewise, there is no effect on locomotion (F; N = 7–8 per group). G, Time course of behavioral training. Compared with PBS injection, lenti-viral transduction itself had on effect on mRNA expression of (H) Gadd45α, (I) Gadd45β, and (J) Gadd45γ (N = 4 per group). K, Additionally, Gadd45α shRNA had no effect on other two family members. L, Gadd45β shRNA had no effect on other two family members and (M) Gadd45γ shRNA had no effect on other two family members (N = 3 per group for K–M). PreCS, Contextual freezing before first CS; AvgCS, average freezing at test. Error bars represent SEM. * p < 0.05, ** p < 0.01.
Cued fear learning leads to two peaks of IEG expression in the PLPFC
We next used a candidate gene approach to examine whether Gadd45γ influences IEG expression and the formation of fear memory. The IEGs Arc, c-Fos, and Npas4 represented prime choices because of their well known role in regulating fear-related learning and memory (Tischmeyer and Grimm, 1999; Ploski et al., 2008, 2011; Katche et al., 2010). We also included a newly discovered IEG, Cyr61, as it has recently been shown to be highly expressed in the brain and to be induced by neural activity (Albrecht et al., 2000; Sakuma et al., 2015). An analysis of the temporal pattern of learning-induced IEG mRNA expression revealed that Arc, c-Fos, Npas4, and Cyr61 exhibited two significant peaks of expression in the PLPFC in response to cued fear conditioning (Fig. 3A–D; t(8) = 3.49, CTX 0 h vs FC 0 h, p = 0.008 and t(8) = 2.44, CTX 5 h vs FC 5 h, p = 0.040 for Arc; t(8) = 2.78, CTX 0 h vs FC 0 h, p = 0.024 and t(8) = 2.28, CTX 5 h vs FC 5 h, p = 0.048 for c-Fos; t(8) = 3.66, CTX 0 h vs FC 0 h, p = 0.006 and t(8) = 2.82, CTX 5 h vs FC 5 h, p = 0.023 for Npas4; t(8) = 3.26, CTX 0 h vs FC 0 h, p = 0.012 and t = 3.92, CTX 5 h vs FC 5 h, p = 0.002 for Cyr61, unpaired t test), which is concomitant with a similar pattern of RNA Polymerase II (RNA pol II) occupancy within the promoter region of all four IEGs (Fig. 2E–H; t(6) = 3.36, CTX 0 h vs FC 0 h, p = 0.015 and t(6) = 2.53, CTX 5 h vs FC 5 h, p = 0.035 for Arc; t(6) = 4.65, CTX 0 h vs FC 0 h, p = 0.004 and t(6) = 2.92, CTX 5 h vs FC 5 h, p = 0.019 for c-Fos; t(6) = 3.64, CTX 0 h vs FC 0 h, p = 0.011 and t(6) = 2.50, CTX 5 h vs FC 5 h, p = 0.037 for Npas4; t(6) = 3.44, CTX 0 h vs FC 0 h, p = 0.014 and t(6) = 2.36 CTX 5 h vs FC 5 h, p = 0.046 for Cyr61, unpaired t test). This is reminiscent of earlier observations by Igaz et al., 2002 in which two waves of transcription, one occurring immediately after training and one occurring 3–6 h later, were shown to be required for the formation of hippocampal-dependent fear memory (Igaz et al., 2002), as well as a variety of other reports in which double IEG peaks have been observed in the context of learning (Katche et al., 2010; Lefer et al., 2012). However, in those studies, the molecular mechanisms underlying multiple waves of IEG expression were not determined.
Figure 3.
Fear learning leads to a distinct pattern of IEG expression in the PLPFC regulated by DSBs. All panels show fold-change for fear conditioned animals which is calculated relative to each group's own context control at that time point. A, There was a significant increase in mRNA expression 0 and 5 h post-fear conditioning for Arc, (B) c-Fos, (C) Npas4, and (D) Cyr61. E, The IEG mRNA induction was associated with a significant increase in RNA Pol II occupancy 0 and 5 h as well post-fear conditioning for Arc, (F) c-Fos, (G) Npas4, and (H) Cyr61. I, There was also a significant increase in γH2A.X occupancy immediately following cued fear conditioning for Arc, (J) c-Fos, (K) Npas4, and (L) Cyr61. M, We also observed a significant increase in Topo IIβ binding immediately following cued fear conditioning for Arc, (N) c-Fos, (O) Npas4, and (P) Cyr61. N = 4–6 per group for all panels. FC, Fear conditioned; CTX, context exposure. Error bars represent SEM. * p < 0.05, ** p < 0.01.
Learning-induced IEG expression is regulated by DSBs and a time-dependent increase in DNA methylation
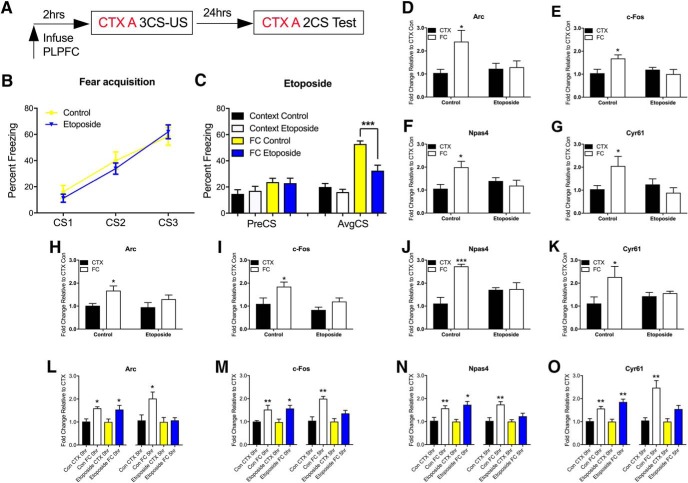
Emerging evidence suggests that DNA double-strand breaks (DSBs) and related DNA repair pathways may also be required for the rapid induction of IEG expression (Madabhushi et al., 2015; Su et al., 2015). These processes interact with dynamic changes in DNA methylation (Pastink et al., 2001; O'Hagan et al., 2008; Wossidlo et al., 2010; Niehrs and Schäfer, 2012; Morano et al., 2014). In line with this, Gadd45β has been shown to be recruited to genomic loci in response to stimuli that generate DSBs (i.e., radiation; Xiao et al., 2000; Smith et al., 2011). Based on these observations, we questioned whether members of the Gadd45 family would mediate the interaction between DSBs, DNA methylation, and DNA repair to influence IEG expression and memory formation. To first determine whether these IEGs are subject to DSBs in the adult brain, we probed their proximal promoters for evidence of learning-induced DSBs following fear conditioning. γH2A.X has been shown to be an excellent marker for DSBs (Kuo and Yang, 2008), and Topo IIβ is known to generate DSBs and is involved in the recruitment of DNA repair enzymes (Wossidlo et al., 2010; Calderwood, 2016). Indeed, previous work has shown a critical role for Topo IIβ in the regulation of IEG expression (Madabhushi et al., 2015). ChIP assay revealed a significant increase in both γH2A.X (Fig. 3I–L; t(8) = 4.07, CTX 0 h vs FC 0 h p = 0.003 for Arc; t(8) = 3.52, CTX 0 h vs FC 0 h, p = 0.008 for c-Fos; t(8) = 2.31, CTX 0 h vs FC 0 h, p = 0.049 for Npas4; t(8) = 3.81, CTX 0 h vs FC 0 h, p = 0.005 for Cyr61, unpaired t test) and Topo IIβ (Fig. 3M–P; t(8) = 3.68, CTX 0 h vs FC 0 h, p = 0.006 for Arc; t(8) = 2.39, CTX 0 h vs FC 0 h, p = 0.043 for c-Fos; t(8) = 3.51, CTX 0 h vs FC 0 h, p = 0.008; t(8) = 2.67 CTX 0 h vs FC 0 h, p = 0.028, unpaired t test) binding within the promoter of all four IEGs immediately following fear conditioning, corresponding to the first peak of IEG expression, but not at the second.
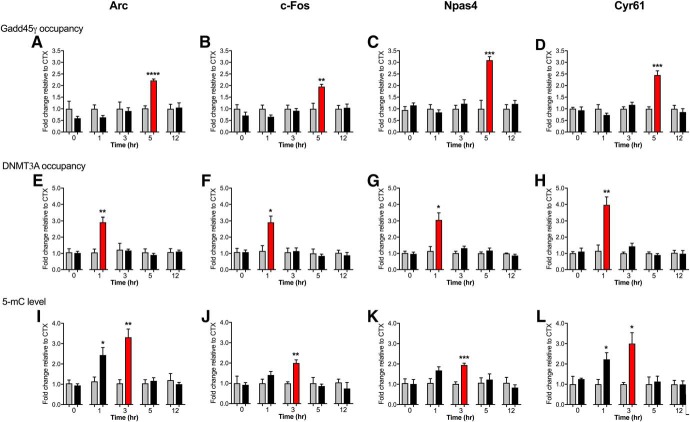
Given that Topo IIβ is associated with the induction and rapid repair of DSBs, which occurs within the first hour post-training, we questioned the underlying mechanism of the second peak of IEG expression. To test the hypothesis that the formation of DSBs leads to the recruitment of Gadd45γ, we performed ChIP to determine the time-dependent recruitment of Gadd45γ to the promoter of each IEG in response to fear learning. There was a significant increase in Gadd45γ binding at the 5 h time point for all IEGs (Fig. 4A–D; t(8) = 10.17, CTX 5 h vs FC 5 h, ****p < 0.0001 for Arc; t(8) = 4.24, CTX 5 h vs FC 5 h, p = 0.003 for c-Fos; t(8) = 6.02, CTX 5 h vs FC 5 h, p = 0.0003 for Npas4; t(8) = 5.99, CTX 5 h vs FC 5 h, p = 0.0003 for Cyr61, unpaired t test). Given that DSBs are repaired before the second wave of transcription, we considered whether there are other mechanisms that serve to maintain the promoter region of these IEGs in a poised state following the repair of the originating DSB, in order for the second wave of transcription to proceed. DNA methylation has been shown to increase at sites of DSBs (Morano et al., 2014) and changes in DNA methylation are associated with memory formation (Day and Sweatt, 2010). We therefore assessed the DNA methylation state at these loci. DNMT3A ChIP and MeDIP revealed a significant increase in DNMT3A occupancy at the site of DSBs in all loci after the first peak of IEG expression (Fig. 4E–H; t(8) = 4.5, CTX 1 h vs FC 1 h, p = 0.002 for Arc; t(8) = 3.29, CTX 1 h vs FC 1 h, p = 0.011 for c-Fos; t(8) = 3.32, CTX 1 h vs FC 1 h, p = 0.011 for Npas4; t(8) = 4.17, CTX 1 h vs FC 1 h, p = 0.003 for Cyr61), which was followed by an increase in 5-mC up to 3 h post-fear conditioning (Fig. 4I–L; t(8) = 2.65, CTX 1 h vs FC 1 h, p = 0.029 and t(8) = 4.29, CTX 3 h vs FC 3 h, p = 0.003 for Arc; t(8) = 4.88, CTX 3 h vs FC 3 h, p = 0.001 for c-Fos; t(8) = 6.7, CTX 3 h vs FC 3 h, p = 0.0002 for Npas4; t(8) = 2.74, CTX 1 h vs FC 1 h, p = 0.026 and t(8) = 2.97, CTX 3 h vs FC 3 h, p = 0.018 for Cyr61, unpaired t test). The data imply a general increase in learning-related DNA methylation that follows the repair of learning-induced DSBs.
Figure 4.
Fear conditioning associated DSBs are followed by time-dependent increases in DNA methylation, and later by Gadd45γ recruitment. There was a significant increase in Gadd45γ occupancy at 5 h for (A) Arc, (B) c-Fos, (C) Npas4, and (D) Cyr61. E, A significant increase in DNMT3A occupancy was observed 1 h after fear conditioning for Arc, (F) c-Fos, (G) Npas4, and (H) Cyr61. I, A significant increase in 5-mC levels as measured by methylated DNA immunoprecipitation was found post-fear conditioning for Arc, (J) c-Fos, (K) Npas4, and (L) Cyr61. N = 6 per group for all panels. FC, Fear conditioned; CTX, context exposure. Error bars represent SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Gadd45γ regulates learning-induced IEG expression in a temporally specific manner through an interaction with DNA methylation
Bearing in mind that Gadd45γ binding occurred specifically at the 5 h time point, which matches the second wave of IEG expression, we next asked whether Gadd45γ is involved in the temporal regulation of IEG expression. Gadd45γ knockdown significantly reduced the occupancy of Gadd45γ (Fig. 5A–D; F(1,16) = 6.38, p = 0.022; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.007 for Arc; F(1,16) = 15.77, p = 0.001; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, ****p < 0.0001 for c-Fos; F(1,16) = 7.54, p = 0.014; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.002 for Npas4; F(1,16) = 8.38, p = 0.011; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.002 for Cyr61, two-way ANOVA) and blocked IEG expression at the second peak only (Fig. 5E–H; F(1,15) = 6.57, p = 0.021; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.045 for Arc; F(1,15) = 6.15, p = 0.025; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.029 for c-Fos; F(1,15) = 5.67, p = 0.031; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.017 for Npas4; F(1,15) = 5.64, p = 0.031; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.008 for Cyr61, two-way ANOVA). In addition, 5-mC levels declined sharply at the time point at which Gadd45γ was bound, whereas Gadd45γ knockdown led to persistently high levels of 5-mC (Fig. 5I–L; F(1,14) = 4.78, p = 0.046; Sidak post hoc analysis: Gadd45γ shRNA CTX 5 h vs Gadd45γ shRNA FC 5 h, p = 0.013 for Arc; F(1,14) = 5.47, p = 0.035; Sidak post hoc analysis: Gadd45γ shRNA CTX 5 h vs Gadd45γ shRNA FC 5 h, p = 0.011 for c-Fos; F(1,14) = 5.01, p = 0.042; Sidak post hoc analysis: Gadd45γ shRNA CTX 5 h vs Gadd45γ shRNA FC 5 h, p = 0.017 for Npas4; F(1,14) = 9.82, p = 0.007; Sidak post hoc analysis: Gadd45γ shRNA CTX 5 h vs Gadd45γ shRNA FC 5 h, p = 0.0003 for Cyr61, two-way ANOVA). Together, these data suggest that although DSBs are required for the initial activation of the IEG expression, it is the second peak of IEG expression during the consolidation phase of memory that is critically regulated by Gadd45γ-mediated changes in DNA methylation.
Figure 5.
Gadd45γ regulates learning-induced IEG expression in a temporally specific manner through interaction with DNA methylation. Presence of Gadd45γ shRNA led to significantly less Gadd45γ occupancy at (A) Arc, (B) c-Fos, (C) Npas4, and (D) Cyr61. Gadd45γ knockdown also led to a significant decrease in mRNA expression at 5 h for (E) Arc, (F) c-Fos, (G) Npas4, and (H) Cyr61, without any influence on mRNA expression of Arc, c-Fos, Npas4, and Cyr61 at 0 h time point (E–H). Gadd45γ shRNA infusion result in remaining high level of promoter DNA methylation at 5 h for (I) Arc, (J) c-Fos, (K) Npas4, and (L) Cyr61. N = 4–6 per group for all panels. FC, Fear conditioned; CTX, context exposure. Error bars represent SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Gadd45γ occupancy is necessary for the second wave of IEG expression and the consolidation of cued fear memory
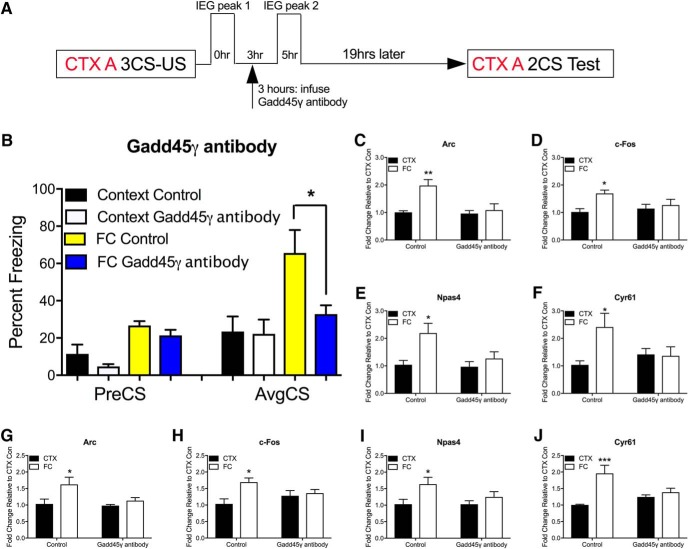
To further reveal the mechanism by which Gadd45γ regulates the formation of fear memory, we manipulated Gadd45γ activity immediately before the second wave of IEG expression, without disturbing the first peak. There are no commercially available pharmacological Gadd45γ antagonists so we turned to an antibody decoy approach. This is a rapid and transient method to block Gadd45γ activity, which permits an examination of the precise temporal role of Gadd45γ during the consolidation phase of fear memory. A similar approach has previously been used to examine the role of BDNF in memory consolidation. The biological activity of Gadd45γ was temporally neutralized by infusing an anti-Gadd45γ antibody into the PLPFC region (1 μg/side) before the second peak of IEG expression. Injection of the Gadd45γ antibody 3 h post-fear training (Fig. 6A) significantly impaired fear memory (Fig. 6B; F(4,21) = 6.02, p = 0.002; Sidak post hoc analysis: FC control vs FC Gadd45γ antibody, p = 0.036, one-way ANOVA). Importantly, Gadd45γ occupancy at each IEG promoter was significantly reduced 5 h after fear training (Fig. 6C–F; F(1,12) = 10.4, p = 0.007, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.003 for Arc; F(1,12) = 6.48, p = 0.026, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.021 for c-Fos; F(1,12) = 8.47, p = 0.013, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.013 for Npas4; F(1,12) = 10.4, p = 0.049, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.023 for Cyr61, two-way ANOVA), and this effect was accompanied by a reduction in the second peak of IEG mRNA expression (Fig. 6G–J; F(1,12) = 6.87, p = 0.022, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.024 for Arc; F(1,12) = 7.11, p = 0.021, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.011 for c-Fos; F(1,12) = 6.77, p = 0.023, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.039 for Npas4; F(1,12) = 15.1, p = 0.002, Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.0009 for Cyr61, two-way ANOVA). Together, these data demonstrate the temporal requirement of Gadd45γ binding for the second peak of IEG expression, and suggest that this is critically important for memory consolidation.
Figure 6.
Gadd45γ is necessary for cued fear learning in a temporally specific manner. A, Time course of behavioral training and anti-Gadd45γ antibody infusion. B, There was a significant decrease in the percentage freezing for animals infused with Gadd45γ-blocking antibody compared with control at test (N = 7–8 per group). Following antibody injection, there was a significant reduction in Gadd45γ occupancy at the promoter of (C) Arc, (D) c-Fos, (E) Npas4, and (F) Cyr61. The induction of IEG mRNA expression was blocked at 5 h post-training compared with control (G) Arc, (H) c-Fos, (I) Npas4, and (J) Cyr61. N = 4 per group for C–J. FC, Fear conditioned; CTX, context exposure; CS, conditioned stimulus (tone); US, unconditioned stimulus (shock); PreCS, contextual freezing before first CS; AvgCS, average freezing at test. Error bars represent SEM. * p < 0.05, ** p < 0.01, *** p < 0.001.
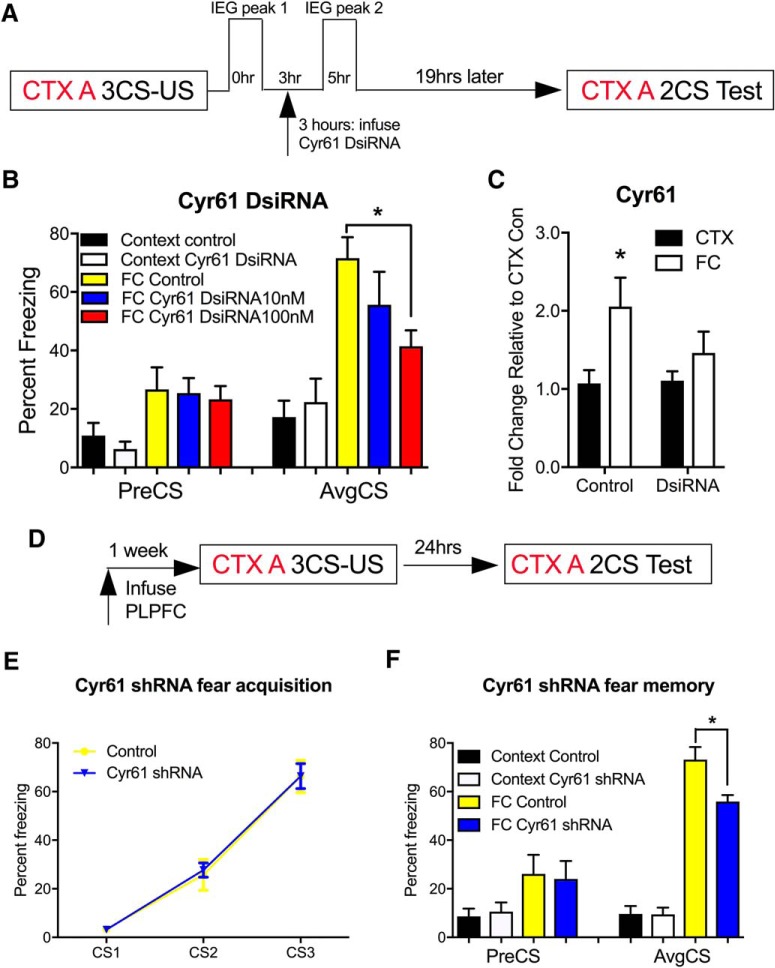
Temporally controlled knockdown of Cyr61 impairs the formation of fear memory
With the exception of Cyr61, each of the candidate IEG have previously been shown to influence the formation of fear memory (Albrecht et al., 2000; Igaz et al., 2002). To extend these findings to include the novel IEG, Cyr61, we designed and validated a DsiRNA directed against Cyr61. We infused two different concentrations of the Cyr61 DsiRNA to determine the most efficient knockdown of Cyr61 in the PLPFC following the same behavioral timeline as the Gadd45γ antibody experiment (Fig. 7A). Cyr61 DsiRNA (100 nm) administered after the first peak of Cyr61 expression led to a significant impairment in fear memory, although notably to a lesser degree than Gadd45γ knockdown (Fig. 7B; F(4,27) = 6.17 p = 0.001; Sidak post hoc analysis: FC control vs FC DsiRNA 100 nm, p = 0.035, one-way ANOVA) by significantly inhibited Cyr61 mRNA expression (Fig. 7C; F(1,16) = 7.09, p = 0.017; Sidak post hoc analysis: CTX control vs FC control, p = 0.027, two-way ANOVA). In a separate experiment, we used a lentiviral packaged Cyr61 shRNA. Knockdown of Cyr61 led to a similar impairment in fear memory, which strongly indicates a functional role for this novel, brain-enriched IEG in fear-related learning and memory (Figs. 1D, 7D–F; F(3,24) = 27.37, ****p < 0.0001; Sidak post hoc analysis: FC Control vs FC Cyr61 shRNA, p = 0.002, two-way ANOVA).
Figure 7.
Cyr61 mRNA expression within PLPFC is necessary for cued fear learning. A, Time course of behavioral training and Cyr61 DsiRNA infusion. B, PLPFC infusion of Cyr61 DsiRNA (100 nm) administered 3 h after fear training blocks fear memory consolidation (N = 7–8 per group). C, Cyr61 DsiRNA (100 nm) also significantly reduced Cyr61 mRNA expression at 5 h post-fear training (n = 5 per group). D, Time course of behavioral training and shRNA infusion. E, There were no significant differences between the Cyr61 shRNA and control groups during fear acquisition. F, There was a significant decrease in the percentage freezing for animals infused with Cyr61 shRNA compared with control at test (N = 7–8 per group). FC, Fear conditioned; CTX, context exposure. Error bars represent SEM. * p < 0.05.
Inhibition of DSB repair also impairs the formation of fear memory
Etoposide is a well established inhibitor of Topo IIβ that traps the enzyme at DSB sites and prevents their repair (Madabhushi et al., 2015). If DSBs associated with the first peak of IEG expression are repaired before the second peak of IEG expression, and this is necessary for establishing the poised state of IEG promoters, then administration of etoposide into the PLPFC should inhibit the second peak and impair fear memory. We first performed a dose–response experiment, which suggested that a high dose of etoposide (0.1 μg) but not a low dose (1 ng) administered into the PLPFC 2 h before training impaired the consolidation of fear memory resulting in a memory defect 24 h after training (data not shown). This finding was subsequently replicated in a larger cohort, confirming that etoposide infusion had no effect on fear acquisition, but significantly impaired fear memory when tested 24 h later (Fig. 8A–C; F(3,34) = 24.17, ****p < 0.001; Sidak post hoc analysis: FC control vs FC etoposide, p = 0.001, two-way ANOVA). In separate cohorts of mice, we also found that etoposide treatment significantly inhibited the accumulation of 5-mC at 3 h post-training (Fig. 8D–G; F(1,12) = 5.16, p = 0.042; Sidak post hoc analysis: control CTX 3 h vs control FC 3 h, p = 0.019 for Arc; F(1,12) = 6.69, p = 0.024; Sidak post hoc analysis: control CTX 3 h vs control FC 3 h, p = 0.031 for c-Fos; F(1,12) = 7.30, p = 0.019; Sidak post hoc analysis: control CTX 3 h vs control FC 3 h, p = 0.017 for Npas4; F(1,12) = 6.11, p = 0.029; Sidak post hoc analysis: control CTX 3 h vs control FC 3 h, p = 0.048 for Cyr61; two-way ANOVA), as well as Gadd45γ binding at the 5 h time point (Fig. 8H–K; F(1,12) = 8.3, p = 0.014; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.041 for Arc; F(1,12) = 8.43, p = 0.013; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.034 for c-Fos; F(1,12) = 16.04 p = 0.002; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.0002 for Npas4; F(1,12) = 4.98 p = 0.045; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.03 for Cyr61, two-way ANOVA). This then led to a reduction in the second peak of IEG expression, with no effect on the first peak of IEG expression (Fig. 8L–O; F(1,12) = 5.39, p = 0.038; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.019 for Arc; F(1,12) = 6.21, p = 0.0004; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.0006 for c-Fos; F(1,12) = 14.67, p = 0.002; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.003 for Npas4; F(1,12) = 25.4, p = 0.0003; Sidak post hoc analysis: control CTX 5 h vs control FC 5 h, p = 0.0005 for Cyr61, two-way ANOVA).
Figure 8.
Etoposide infusion into the PLPFC inhibits fear memory formation by blocking repair of DSBs. A, Time course of behavioral training and etoposide infusion. B, There were no significant differences between etoposide (0.1 μg/side) and control groups in percentage freezing during fear acquisition. C, There was a significant decrease in the percentage freezing for animals infused with etoposide compared with control at test (N = 9–12 per group). D, Etoposide infusion blocked the temporal accumulation of 5-mC within the promoter region of Arc, (E) c-Fos, (F) Npas4, and (G) Cyr61. H, Etoposide infusion also inhibited Gadd45γ occupancy at promoter of Arc at the 5 h time point, (I) c-Fos, (J) Npas4, and (K) Cyr61. Etoposide led to a significant decrease in mRNA at 5 h post-training (L) Arc, (M) c-Fos, (N) Npas4, and (O) Cyr61. Etoposide infusion had no effect on Arc, c-Fos, Npas4, and Cyr61 mRNA expression at the 0 h time point (L–O). N = 4 per group for D–O. FC, Fear conditioned; CTX, context exposure. Error bars represent SEM. * p < 0.05, ** p < 0.01, *** p < 0.001.
Discussion
In this study, we have made three important discoveries. First, we found that the expression of four plasticity-related IEGs peak twice in response to cued fear learning, with an increase in the presence of the DNA damage marker γH2A.X and the recruitment of Topo IIβ to IEG promoters corresponding to the first peak of IEG expression, whereas an increase in DNA methylation and a time-dependent recruitment of Gadd45γ is associated with the second peak of IEG expression. Second, we have shown that inhibition of Topo IIβ activity before the first peak of IEG expression impairs the consolidation of fear memory, as does the disruption of Gadd45γ binding before the second peak, strongly suggesting that these molecular events are both necessary for memory consolidation. Third, we have demonstrated that DsiRNA-mediated knockdown of a novel brain-enriched IEG, Cyr61, before the second peak of its expression impairs the consolidation of cued fear memory, a finding which extends the understanding of learning-induced IEGs in the PLPFC to include the epigenetic regulation of Cyr61 in the consolidation of associative fear memory.
Together, our results indicate that there is a tight temporal relationship between learning-induced DSBs, DNA repair, and DNA (de)methylation in the regulation of learning-induced IEG expression, and we highlight Gadd45γ as a central regulator of the temporal coding of IEG transcription that is required for fear memory consolidation. To our knowledge this is the first demonstration of a causal influence of Gadd45γ on the consolidation of cued fear memory, and the first model synthesizing the functional interaction of DSBs, DNA methylation, and DNA repair-mediated DNA demethylation in a learning context. In addition, these data add mechanistically to the literature describing memory consolidation as a process which is not simply linear and proportional to the passage of time, but is instead continually and dynamically stabilized and destabilized across time.
Whereas previous studies observed that global Gadd45β knock-out affected contextual but not cued fear conditioning (Leach et al., 2012), we have found that Gadd45γ, but not Gadd45α or Gadd45β, is required in the PLPFC for cued fear conditioning. This suggests that Gadd45γ may function in a region-specific, and thus task-specific, manner, an idea which is supported by the observation of Gadd45γ recruitment to the promoter of all of the IEGs at the same 5 h time point, with similarly reduced IEG mRNA expression at this time point following Gadd45γ knockdown. Selective temporal control over IEG expression is also supported by the observation that knockdown of Gadd45γ, which binds to a variety of IEGs, impairs fear conditioning by a considerably larger margin than by knockdown of a single IEG, as in the case of Cyr61. We find this particularly interesting because it has been shown that Gadd45β binds more slowly to genes encoding neurotrophic factors (Ma et al., 2009). Thus, in addition to specificity for different brain regions and potential cell types, members of the Gadd45 family may also be targeting different classes of genes that have different temporal expression patterns.
Perhaps even more intriguing is the fact that all four IEGs we investigated are a hot spot for DNA breakage, and that this is related to dynamic DNA methylation states in all four cases. The time course analysis revealed that DSBs occur in the promoter regions of Arc, c-Fos, Npas4, and Cyr61 in response to fear learning, which is followed by a time-dependent increase in DNA methylation. Increased DNA methylation following DSBs has been suggested to occur in only a few cells after aberrant repair (Morano et al., 2014). Our data suggest that this phenomenon may be more widespread than previously thought, and in the brain may be used to control the rapid induction and temporal nature of learning-induced IEG expression (O'Hagan et al., 2008; Wossidlo et al., 2010; Morano et al., 2014). Furthermore, recent work has shown that both 5-formylcytosine, and 5-mC within CA dinucleotides, can epigenetically prime gene activity throughout the brain (Song et al., 2013; Stroud et al., 2017). In this study, we did not determine whether a reduction in 5-mC corresponding to the recruitment of Gadd45γ reflects removal of 5-mC by DNA repair mechanisms, or further conversion of 5-mC to these downstream derivatives. Future studies will investigate whether Gadd45γ interacts with other factors to participate in the conversion of 5-mC to 5-formylcytosine, and whether there are metaplastic changes in IEG activation following the consolidation of fear memory. Whether DSBs and their recognition by Gadd45γ is required for the regulation of all IEGs that are induced by cued fear learning is not yet known. Furthermore, we did not identify any common motifs within the promoters of the four IEGs included in this investigation, which would have suggested a common target for the process of activity-induced DNA break and repair. Future studies will expand on this analysis to include a genome-wide profile of DSBs and Gadd45γ occupancy to determine whether this represents a general principle of temporal control over learning-induced gene expression, or if it is restricted to a subset of rapidly induced IEGs.
Gadd45γ is recruited to the promoter of each IEG only at the 5 h time point, which corresponds to the second wave of IEG expression. This observation is made more intriguing by the long-held position that there are two time periods during which protein synthesis inhibitors impair consolidation, with observations of these two critical windows being immediately following learning and 3–6 h later (Grecksch and Matthies, 1980; Freeman et al., 1995; Chew et al., 1996). The dual observation that these genes are subject to both DSBs and the later recruitment of Gadd45γ is not to be ignored as it suggests a two-tier process whereby DSBs are needed to activate IEGs at the first time point, and activity-dependent DNA demethylation and repair guided by Gadd45γ is critical for this pattern of IEG expression to trigger processes required for long-term memory. As described by Igaz et al. (2002), two peaks of learning-induced gene expression fit within the lingering consolidation hypothesis, which states that there are processes which occur across time and result in continued destabilization and re-stabilization of memory traces, contrasted to the simpler model describing a linear relationship between memory strength and time (Dudai and Eisenberg, 2004). This distinction is theoretically important as the latter assumes certain time periods, such as 24 h post-learning, could be used as a control where manipulations should have no effect. In fact, Katche et al. (2010) have shown that the manipulation of c-Fos 24 h after initial training impairs long-term memory storage. Here, we add mechanistically to this model by demonstrating that a DNA modification switch, which is activated by DSBs and regulated by DNA demethylation and repair, is intimately involved in this process; we also extend these findings to include the neural plasticity-related IEGs Arc, c-Fos, Npas4, and Cyr61.
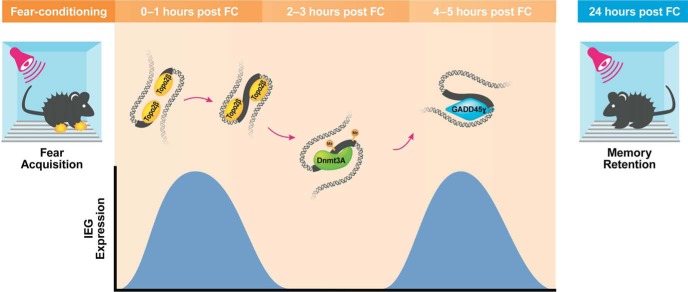
The model would make further predictions that DNA methylation and repair may not only be critical for the initial learning-induced induction and encoding of information at the level of gene expression, but also indicate a retrieval-induced recapitulation of transcriptional activity. Supporting this idea is the recent observation that retrieval-induced Tet3 gene expression is necessary for retrieval and reconsolidation (Liu et al., 2018), as well as selective retrieval impairment by infusion of DNA methyltransferase inhibitors into the PLPFC (Han et al., 2010), although further work is required to establish this. It also remains to be determined whether the phenomenon of DSB followed by DNA methylation and the recruitment of Gadd45γ generalizes to other classes of genes, or whether other mechanisms of DNA modification also follow this time course. Regardless of these open questions, our data imply a two-hit model of IEG expression whereby the initial induction is dependent on learning-induced DSBs and increased DNA methylation, and the second wave depends on Gadd45γ-mediated DNA demethylation and repair (Fig. 9), both of which represent critical aspects of the memory consolidation process.
Figure 9.
Fear learning leads to a rapid induction in IEG expression in the PLPFC that is mediated by Topo2β associated DSBs within their promoter region, and which is followed by an increase in DNMT3a-mediated DNA methylation at the same locus. The increased deposition of 5-mC results in the recruitment of the DNA repair associated protein Gadd45γ, which is then necessary for the induction of a second peak of IEG expression and the consolidation of fear memory.
Footnotes
This work was supported by Grants from the NIH (5R01MH105398-TWB) and the NHMRC (APP1033127-TWB), and by postgraduate scholarships from the University of Queensland and the ANZ Trustees Queensland for Medical Research and University of Queensland development fellowship to X.L., from NSERC and the University of Queensland to P.R.M., and from the Westpac Bicentennial Foundation and the University of Queensland to L.J.L. We thank Rowan Tweedale for helpful editing of the paper.
The authors declare no competing financial interests.
*X.L. and P.R.M. contributed equally to this work.
References
- Albrecht C, von der Kammer H, Mayhaus M, Klaudiny J, Schweizer M, Nitsch RM (2000) Muscarinic acetylcholine receptors induce the expression of the immediate early growth regulatory gene CYR61. J Biol Chem 275:28929–28936. 10.1074/jbc.M003053200 [DOI] [PubMed] [Google Scholar]
- Baker-Andresen D, Ratnu VS, Bredy TW (2013) Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci 36:3–13. 10.1016/j.tins.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, Döderlein G, Maltry N, Wu W, Lyko F, Niehrs C (2007) Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445:671–675. 10.1038/nature05515 [DOI] [PubMed] [Google Scholar]
- Befort K, Karchewski L, Lanoue C, Woolf CJ (2003) Selective up-regulation of the growth arrest DNA damage-inducible gene Gadd45 alpha in sensory and motor neurons after peripheral nerve injury. Eur J Neurosci 18:911–922. 10.1046/j.1460-9568.2003.02827.x [DOI] [PubMed] [Google Scholar]
- Calderwood SK. (2016) A critical role for topoisomerase IIb and DNA double strand breaks in transcription. Transcription 7:75–83. 10.1080/21541264.2016.1181142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SJ, Vicario DS, Nottebohm F (1996) Quantal duration of auditory memories. Science 274:1909–1914. 10.1126/science.274.5294.1909 [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD (2010) DNA methylation and memory formation. Nat Neurosci 13:1319–1323. 10.1038/nn.2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean C, Courtin J, Karalis N, Chaudun F, Wurtz H, Bienvenu TC, Herry C (2016) Prefrontal neuronal assemblies temporally control fear behaviour. Nature 535:420–424. 10.1038/nature18630 [DOI] [PubMed] [Google Scholar]
- Dudai Y, Eisenberg M (2004) Rites of passage of the engram: reconsolidation and the lingering consolidation hypothesis. Neuron 44:93–100. 10.1016/j.neuron.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Freeman FM, Rose SP, Scholey AB (1995) Two time windows of anisomycin-induced amnesia for passive avoidance training in the day-old chick. Neurobiol Learn Mem 63:291–295. 10.1006/nlme.1995.1034 [DOI] [PubMed] [Google Scholar]
- Gallo FT, Katche C, Morici JF, Medina JH, Weisstaub NV (2018) Immediate early genes, memory and psychiatric disorders: focus on c-fos, Egr1 and arc. Front Behav Neurosci 12:79. 10.3389/fnbeh.2018.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustino TF, Maren S (2015) The role of the medial prefrontal cortex in the conditioning and extinction of fear. Front Behav Neurosci 9:298. 10.3389/fnbeh.2015.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi D, Franz H, Vezzali R, Bovio P, Heidrich S, Dehghanian F, Lagunas N, Belzung C, Krieglstein K, Vogel T (2017) Neuronal activity, TGFβ-signaling and unpredictable chronic stress modulate transcription of Gadd45 family members and DNA methylation in the hippocampus. Cereb Cortex 27:4166–4181. 10.1093/cercor/bhx095 [DOI] [PubMed] [Google Scholar]
- Grecksch G, Matthies H (1980) Two sensitive periods for the amnesic effect of anisomycin. Pharmacol Biochem Behav 12:663–665. 10.1016/0091-3057(80)90145-8 [DOI] [PubMed] [Google Scholar]
- Han J, Li Y, Wang D, Wei C, Yang X, Sui N (2010) Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur J Pharmacol 642:93–98. 10.1016/j.ejphar.2010.05.050 [DOI] [PubMed] [Google Scholar]
- Igaz LM, Vianna MR, Medina JH, Izquierdo I (2002) Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci 22:6781–6789. 10.1523/JNEUROSCI.22-15-06781.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo I, McGaugh JL (2000) Behavioural pharmacology and its contribution to the molecular basis of memory consolidation. Behav Pharmacol 11:517–534. 10.1097/00008877-200011000-00001 [DOI] [PubMed] [Google Scholar]
- Jarome TJ, Butler AA, Nichols JN, Pacheco NL, Lubin FD (2015) NF-κB mediates Gadd45β expression and DNA demethylation in the hippocampus during fear memory formation. Front Mol Neurosci 8:54. 10.3389/fnmol.2015.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD (2013) TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79:1086–1093. 10.1016/j.neuron.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. (2001) The molecular biology of memory storage: a dialogue between genes and synapses. Science 294:1030–1038. 10.1126/science.1067020 [DOI] [PubMed] [Google Scholar]
- Katche C, Bekinschtein P, Slipczuk L, Goldin A, Izquierdo IA, Cammarota M, Medina JH (2010) Delayed wave of c-Fos expression in the dorsal hippocampus involved specifically in persistence of long-term memory storage. Proc Natl Acad Sci U S A 107:349–354. 10.1073/pnas.0912931107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klavir O, Prigge M, Sarel A, Paz R, Yizhar O (2017) Manipulating fear associations via optogenetic modulation of amygdala inputs to prefrontal cortex. Nat Neurosci 20:836–844. 10.1038/nn.4523 [DOI] [PubMed] [Google Scholar]
- Kuo LJ, Yang LX (2008) Gamma-H2AX: a novel biomarker for DNA double-strand breaks. In Vivo 22:305–309. [PubMed] [Google Scholar]
- Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, Gould TJ (2012) Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Mem 19:319–324. 10.1101/lm.024984.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefer D, Perisse E, Hourcade B, Sandoz J, Devaud JM (2012) Two waves of transcription are required for long-term memory in the honeybee. Learn Mem 20:29–33. 10.1101/lm.026906.112 [DOI] [PubMed] [Google Scholar]
- Li X, Wei W, Ratnu VS, Bredy TW (2013) On the potential role of active DNA demethylation in establishing epigenetic states associated with neural plasticity and memory. Neurobiol Learn Mem 105:125–132. 10.1016/j.nlm.2013.06.009 [DOI] [PubMed] [Google Scholar]
- Li X, Wei W, Zhao QY, Widagdo J, Baker-Andresen D, Flavell CR, D'Alessio A, Zhang Y, Bredy TW (2014) Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci U S A 111:7120–7125. 10.1073/pnas.1318906111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, Bredy TW (2011) The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci 14:1115–1117. 10.1038/nn.2891 [DOI] [PubMed] [Google Scholar]
- Liu C, Sun X, Wang Z, Le Q, Liu P, Jiang C, Wang F, Ma L (2018) Retrieval-induced upregulation of Tet3 in pyramidal neurons of the dorsal hippocampus mediates cocaine-associated memory reconsolidation. Int J Neuropsychopharmacol 21:255–266. 10.1093/ijnp/pyx099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H (2009) DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle 8:1526–1531. 10.4161/cc.8.10.8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH (2015) Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161:1592–1605. 10.1016/j.cell.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD (2010) Cortical DNA methylation maintains remote memory. Nat Neurosci 13:664–666. 10.1038/nn.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minatohara K, Akiyoshi M, Okuno H (2016) Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front Mol Neurosci 8:78. 10.3389/fnmol.2015.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano A, Angrisano T, Russo G, Landi R, Pezone A, Bartollino S, Zuchegna C, Babbio F, Bonapace IM, Allen B, Muller MT, Chiariotti L, Gottesman ME, Porcellini A, Avvedimento EV (2014) Targeted DNA methylation by homology-directed repair in mammalian cells. transcription reshapes methylation on the repaired gene. Nucleic Acids Res 42:804–821. 10.1093/nar/gkt920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainar S, Marshall PR, Tyler CR, Spitale RC, Bredy TW (2016) Evolving insights into RNA modifications and their functional diversity in the brain. Nat Neurosci 19:1292–1298. 10.1038/nn.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Schäfer A (2012) Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol 22:220–227. 10.1016/j.tcb.2012.01.002 [DOI] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genetics 4:e1000155. 10.1371/journal.pgen.1000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastink A, Eeken JC, Lohman PH (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 480–481:37–50. 10.1016/S0027-5107(01)00167-1 [DOI] [PubMed] [Google Scholar]
- Ploski JE, Pierre VJ, Smucny J, Park K, Monsey MS, Overeem KA, Schafe GE (2008) The activity-regulated cytoskeletal-associated protein (Arc/Arg3.1) is required for memory consolidation of Pavlovian fear conditioning in the lateral amygdala. J Neurosci 28:12383–12395. 10.1523/JNEUROSCI.1662-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE (2011) The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PloS One 6:e23760. 10.1371/journal.pone.0023760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell 135:1201–1212. 10.1016/j.cell.2008.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnu VS, Wei W, Bredy TW (2014) Activation-induced cytidine deaminase regulates activity-dependent BDNF expression in post-mitotic cortical neurons. Eur J Neurosci 40:3032–3039. 10.1111/ejn.12678 [DOI] [PubMed] [Google Scholar]
- Rizzo V, Touzani K, Raveendra BL, Swarnkar S, Lora J, Kadakkuzha BM, Liu XA, Zhang C, Betel D, Stackman RW, Puthanveettil SV (2017) Encoding of contextual fear memory requires de novo proteins in the prelimbic cortex. Biol Psychiatry Cogn Neurosci Neuroimaging 2:158–169. 10.1016/j.bpsc.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH (2013) Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79:1109–1122. 10.1016/j.neuron.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma K, Komatsu H, Maruyama M, Imaichi S, Habata Y, Mori M (2015) Temporal and spatial transcriptional fingerprints by antipsychotic or propsychotic drugs in mouse brain. PLoS One 10:e0118510. 10.1371/journal.pone.0118510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos TB, Kramer-Soares JC, Favaro VM, Oliveira MGM (2017) Involvement of the prelimbic cortex in contextual fear conditioning with temporal and spatial discontinuity. Neurobiol Learn Mem 144:1–10. 10.1016/j.nlm.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Smith IF, Shuai J, Parker I (2011) Active generation and propagation of Ca2+ signals within tunneling membrane nanotubes. Biophys J 100:L37–L39. 10.1016/j.bpj.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C (2013) Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153:678–691. 10.1016/j.cell.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Su SC, Hrvatin S, Greben AW, Renthal W, Boxer LD, Nagy MA, Hochbaum DR, Kinde B, Gabel HW, Greenberg ME (2017) Early-life gene expression in neurons modulates lasting epigenetic states. Cell 171:1151–1164.e16. 10.1016/j.cell.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Ming GL, Song H (2015) DNA damage and repair regulate neuronal gene expression. Cell Res 25:993–994. 10.1038/cr.2015.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD (2012) Genetic deletion of gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci 32:17059–17066. 10.1523/JNEUROSCI.1747-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lin Y (2016) Npas4: linking neuronal activity to memory. Trends Neurosci 39:264–275. 10.1016/j.tins.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischmeyer W, Grimm R (1999) Activation of immediate early genes and memory formation. Cell Mol Life Sci 55:564–574. 10.1007/s000180050315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V, Bredy TW (2016) Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J Neurosci 36:6771–6777. 10.1523/JNEUROSCI.4053-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, Schöler H, Walter J (2010) Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J 29:1877–1888. 10.1038/emboj.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Chicas A, Olivier M, Taya Y, Tyagi S, Kramer FR, Bargonetti J (2000) A DNA damage signal is required for p53 to activate gadd45. Cancer Res 60:1711–1719. [PubMed] [Google Scholar]