Abstract
The aim of this study is to identify, clone and express a Mycobacterium avium subsp. paratuberculosis specific immunogenic antigen candidate, in order to develop better reagents for diagnosis and vaccines for the protection of the host. Therefore, MAP2191 gene (a member of MAPmce5 operon) from MAP, was isolated and characterized by Bioinformatics tools and in vitro experiments. Then, a novel Mce-whole protein encoded by MAP2191 gene was amplified and sub-cloned into E. coli. We tried to express the Mce/whole protein in different condition along with a positive expression control (pET28a-Mce/truncated plasmid that we know express well), to ensure that nothing is wrong regarding culture/induction condition. The level of the recombinant protein expression was analyzed by means of SDS-PAGE and Western blotting. Western blot analysis toward full-length MAP2191 protein and its truncation only demonstrated Mce/truncated protein. The concurrence of in-silico prediction of primary structure of MAP2191 protein results along with experimental results confirmed that expression of Mce/whole protein was affected by the hydrophobicity nature of this protein. Our data support the hypothesis that the presence of hydrophobic regions in protein structure can influence the level of recombinant protein expression. This stresses the importance of gene selection and the protein sequence checking of the hydrophobic content in any protein purification project in order to achieve a large amount of desirable proteins.
Key Words: Johne's disease, Control, mce gene, MAP2191, Hydrophobicity
INTRODUCTION
Johne’s disease (JD) or paratuberculosis is a chronic infection of the intestinal tract of animals, which is caused by Mycobacterium avium subsp. paratuberculosis (MAP) [1, 2]. JD is primarily a disease of ruminants (cattle, sheep, goats, buffaloes etc.), but can affect non-ruminants, especially wildlife [3, 4]. High to very high prevalence of JD has been frequently reported from dairy farms world-wide [5, 6]. JD is recognized as an important public health pathogen duo to the presence of live MAP bacilli in animal milk (both raw and pasteurized) and other dairy products [1,2,7]. There are potential association of this multi-host pathogen with human diseases like Crohn’s disease and several other chronic inflammatory syndromes [8, 9]. Thus, current trends in diagnosis and disease control of JD are searching for identification and characterization specific immunogenic antigen candidates of MAP [10, 11].
Mammalian cell entry (Mce) protein was first recognized in Mycobacterium tuberculosis (MTB) and was shown have a possible role in the virulence of MTB, which allows non-pathogenic Escherichia coli to enter into non-phagocytic cells [13-15]. Although Mce genes were originally identified in MTB, homologous of these genes are widely distributed throughout the Mycobacterium genus [16-18]. Mce proteins were characterized as invasion‐like proteins localized at the cell surface of the mycobacteria [19]. Eight homologous regions of mce gene families have predicted to be located in the outer membrane of all the MAP isolates [20]. In MAP K-10, mce5 operon included a cluster of six homologous genes (MAP2189-MAP2194) that predicted to encode proteins involved in lipid uptake [21, 22]. These mce5 genes play important roles in MAP invasion, survival and virulence [22]. MAP2191 gene is a member MAPmce5 operon, having potential antigenic nature and immunogenic T- and B-cell epitopes specific for MAP based on our in-silico analyses. This prompted us to perform an antigen discovery study with the objective of characterizing full length of MAP2191 gene both at genome and protein level using cloning, gene expression techniques and bioinformatic analysis. In this study, we tried to express the Mce/whole protein and discovered some interesting trends that might be appreciated by future investigators in their recombinant protein purification studies.
MATERIALS AND METHODS
Morphological and molecular characterization of MAP: MAP reference strain ‘S 5’ Indian Bison Type (provided by Central Institute for Research on Goats (CIRG), Makhdoom, Mathura, Uttar Prades in India), was grown on the slant of Herold’s egg yolk agar (HEY) medium as per Singh et al., [23]. Molecular confirmation of the strain was done by IS900 [24], IS1311 PCR [25] and IS1311 PCR-REA [26]. All primers sequences are listed in Table 1.
Table 1.
Primers used for characterization of Mycobacterium avium subsp. paratuberculosis
| Target | Primer | Primer Sequence | Product size (bp) |
|---|---|---|---|
| IS 900 | P90 | 5’ GAAGGGTGTTCGGGGCCGTCGCTTAGG 3’ | 413 |
| P91 | 5’ GGCGTTGAGGTCGATCGCCCACGTGAC 3’ | ||
| IS 1311 | M56 | 5’ GCGTGAGGCTCTGTGGTGAA 3’ | 608 |
| M119 | 5’ ATGACGACCGCTTGGGAGAC 3’ | ||
| MAP2191 | F | 5’ACTGGATCCCTGAAATACCGTGGCGCAAAC 3’- BamH1 | 1065 |
| R | 5’CTAAAGCTTTCATCGCGAACCGCCCGGGATG 3’– HindIII |
In -silico analyses of MAP 2191 gene and protein identification: In-silico analysis of the MAP2191 protein sequence was carried out using the basic local alignment search (BLASTp) analysis and SOPMA web site. Mce/whole protein coding sequence (MAP2191 full length protein referred by this short name) was subjected to BLASTp analysis at the NCBI GenBank site to confirm the MAP sequence identity. The antigenicity and hydrophobicity analysis of the Mce/whole protein molecule were also done using the CLC Genomics Workbench 7.5.1 program (CLC bioQIAGEN, Germany) software.
Preparation of full-length version of the MAP2191 gene by PCR: Full length of the MAP2191 gene, namely mce/whole gene successfully amplified with primers were designed to have BamHI (forward primer) and HindIII (reverse primer) restriction enzyme sites as depicted in Table 1. PCR master mix (2X) (Cat no. #K0171, Thermo Scientific) containing all components of the reaction mixture (dNTPs, Taq DNA polymerase, 10X PCR buffer, MgCl2 and loading dye) were used. Positive control (containing DNA from the reference strain as template) and negative control (containing nuclease free water) are also prepared along with the test DNA samples. The PCR products was confirmed by nucleic acid sequencing.
Cloning of mce -whole gene in TA-cloning vector: The confirmed and purified gene segment of respective mce/whole gene was cloned in pTZ57R/T cloning vector using ligase enzyme (Fermentas, USA), as per manufacturer’s instructions. The ligation product (pTZ57R/T plasmids carrying mce/whole gene) was successfully transformed with heat shock at 42◦C for 90 Sec into E.coli XL10-Gold ultra-competent cell (Stratagene, Agilent).White transformed E.coli colonies were selected and further screened by colony PCR with mce/whole primers for the selection of recombinant clones.Final confirmations of selected plasmids were done using restriction digestion followed by DNA sequencing.
Sub-cloning of mce -whole gene in expression vector: Expression vector was prepared by ligation of BamH1/HindIII digested, de-phosphorylated (by the fast alkaline phosphatase enzyme, Cat. No: #EF0651, Fermentas, USA), and gel purified pET28a and BamH1/HindIII digested and gel purified mce/whole gene fragment (insert) by using ligase enzyme (Fermentas, USA). Specific product was cut from the gel and the gel cut was purified using agarose gel DNA purification Kit (Thermo scientific, USA). Colonies were confirmed for the presence of the insert by colony PCR followed by double digestion. One of the confirmed pET28a-mce/whole vectors was sequenced using T7 universal primers for further confirmation.
Expression and purification of recombinant Mce-whole protein: Sequences-confirmed pET28a-mce/whole vector was used to transform E. coli Rosetta (Novagene, WI, USA) and E. coli BL-21 (DE3, Novagene, WI, USA) competent cells. The expression of the transformed cloned was induced by adding different concentrations of isopropyl-β-D thiogalactopyranoside (IPTG) (0.1-1.5 mM) at four different temperatures (37°C, 30°C, 23°C and 16°C) and different growth time intervals (2, 4, 8, 12, 16, 24, 30 hours). Each collected samples were further analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) to enquired expected size of the differently expressed Mce-whole protein in the different IPTG induced time point samples versus the un-induced sample. Sonicated extracts (crude, supernatant and pellets) of IPTG induced recombinant E. coli strains containing pET28a-mce/whole plasmid was also separated by SDS-PAGE and analyzed for the presence of Mce/whole gene by western blotting. To test the protein expression and purification efficiency, Mce/truncated protein co-purified along with Mce/whole protein in our experiment as a positive expression control.
RESULTS
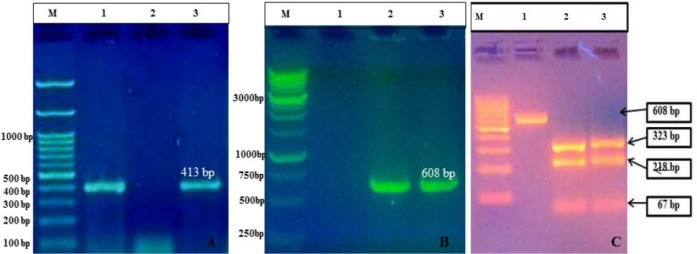
MAP reference strain (S5) colonies on HEY slant supplemented with Mycobactin J. were small, translucent, straw color, having nipple over raised part and loosely attached to the surface of the medium. Bacterial cells from MAP colonies were strongly acid-fast on Ziehl-Neelsen staining. The presence of the specific PCR products was considered as positive for MAP with IS900 PCR (Fig. 1A), IS1311 PCR (Fig. 1B) and IS1311 PCR-REA (Fig. 1C).
Figure 1.
Molecular Characterization of MAP ‘S5’ DNA: (A) IS900 PCR: lane M- 100 base Marker (#SM0243, Fermentas), lanes 1 and 3: IS900 PCR products; (B) IS1311 PCR: lane M: 1KB Marker (#MBT51, Hi-Media), lanes2 and 3: IS1311 PCR products; (C) IS1311-REA genotyping:lane M- 100 base Marker (#SM0243, Fermentas), Lane 1: IS1311 PCR product,lane 2: Digested PCR product of IS1311 from Positive control, lane 3: Digested PCR product of IS1311 from sub-cultured colonies.
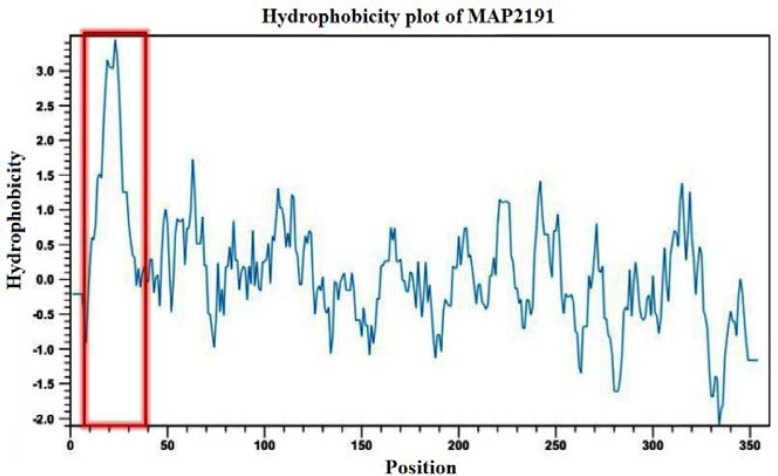
According to alignment sequence comparison analysis, MAP2189-MAP2194 cluster organization is conserved among MAP K-10 and MAP S 5 strain sequences and most epitopes are shared. The homology analysis also showed the MAP2191 protein was highly specific as unique to MAP. The MAP2191 gene is 1065bp and has 354 amino acids with an estimated molecular weight of 37.510 kDa, an isoelectric point of 4.9, a net charge of -3.8 at pH 7.0 and good water solubility. Web-based analysis of the full length of MAP2191 protein using SOPMA web site revealed this protein displayed 41.53% α-helix content, 11.58% β-turn content, 16.95% extended strand and 29.94% random coil structures. CLC Genomics Workbench software analysis predicted the presence of a highly hydrophobic loop in N-terminal amino acid sequence of the full length MAP2191 protein from residues 10 to 32 (Fig. 2). In addition, the C-terminal portion of MAP2191 protein was more hydrophilic than N-terminal portion and had a high antigenic index as shown in-silico analysis results.
Figure 2.
Hydrophobicity analysis of MAP2191 protein. The presence of hydrophobic N-terminal loop is shown in a red box.
The mce/whole gene was successfully amplified from MAP ‘S 5’ DNA by PCR. A band of approximately 1083bp (1065 mce gene and extra 18bp used to bring the coding sequence in-frame and for restriction enzyme sites) was considerate as positive for the whole gene. PCR product’s sequencing showed 100% homology of mce/whole nucleotide sequence with MAP2191 gene sequences present in the complete genome sequence of MAP K10 available in the GenBank (Accession number: NC_002944.2).
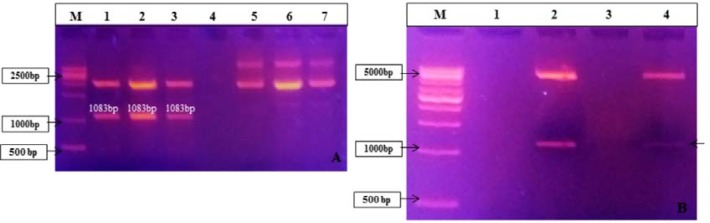
The present of mce-whole gene segment (insert) in the TA cloning vector and in the pET28a expression vector was confirmed using specific colony PCR and restriction analysis (Figs. 3). Gene sequencing also confirmed the positive mce-whole-pTZ57R/T and mce-whole-pET28a clones.
Figure 3.
(A) Restriction digestion of confirmed colony PCR positive pZ57R/T mce-whole clones: lane M: 1kb DNA ladder (#SM0313, Fermentas), lanes 1, 2 and 3: Positive clones of PTZ57R/T mce-whole plasmid digested with BamHI and HindIII enzymes; Lanes 5, 6 and 7: Plasmid isolated of positive PTZ57R/T mce-whole clones. (B) Restriction Digestion of confirmed colony PCR positive pET28a mce-whole clones; lane M: 1kb DNA ladder (#SM0313, Fermentas),lanes 2 and 4: Positive clones of pET28a mce-whole plasmid.
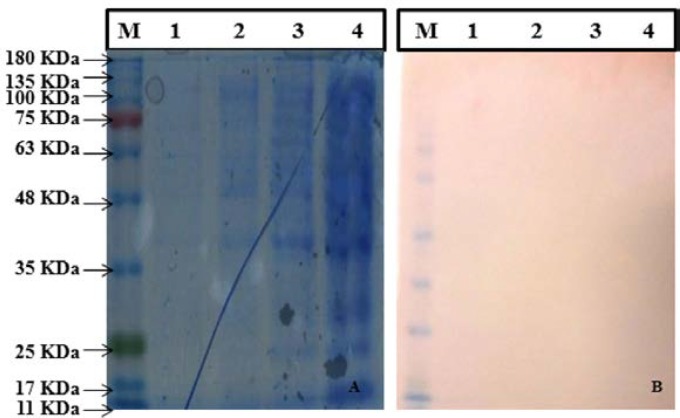
SDS-PAGE results indicated that there was no obvious expression of Mce/whole protein in all fractions collected from different IPTG concentration and different time point samples.Although, no band at the same size of Mce/whole protein was detected in running the crude, supernatant and pellets of sonicated cells on an SDS-PAGE gel (Fig. 4A). No such band was obtained with sonicated extracts of IPTG-induced E. coli strains harboring pET28a-mce plasmid by Anti-His-Tag monoclonal antibody (Fig. 4B).
Figure 4.
(A) Analysis of Mce-whole Protein expressed by SDS PAGE. (Left to Right), lane M: Protein molecular weight marker (SL7012, CinnaGen), lane 1: pET28a.Mce.whole Elution 1, lane 2: pET28a.Mce.whole Supernatant, lane 3: pET28a.Mce.whole Pellet, lane 4. pET28a.Mce.whole Crude; (B) Western blot analysis (from left to right) lane M: Protein molecular weight marker (SL7012, CinnaGen), lane 1: pET28a.Mce.whole Elution 1, lane 2: p1ET28a.Mce.whole Supernatant, lane 3: pET28a.Mce.whole Pellet, lane 4. pET28a.Mce.whole Crude.
DISCUSSION
Johne’s disease (JD) in animals presents as a chronic disease characterized by enteritis, weakness, diarrhea, emaciation and death [27]. Diagnosis, control and laboratory confirmation of MAP is always challenging [1, 4]. Mammalian cell entry (mce) genes are known to be virulence factors involved in mycobacterial entry and survival within macrophages and can facilitate host invasiveness [12, 28, 29]. In the present study the MAP2191 gene (a member of MAPmce5 operon) from MAP, was isolated and characterized by Bioinformatics tools and in vitro experiments. Bioinformatics analysis results revealed the protein’s amino acids 10-32 formed a hydrophobic loop at the N-terminal end of full-length MAP2191 protein and the presence of such hydrophobic region can block expression of the Mce/whole protein.
Although a lot of protein expression systems have been developed today, but achieving a pure protein is still a problematic duo to the presence of labor-intensive cloning, expression and purification steps [30]. In our study, expression of full-length MAP2191 protein was performed similar to the cloning and expression strategies used to produce recombinant Mce/truncated protein, but no expression of Mce/whole protein was observed in induced E. coli cell containing pET28a-mce/whole plasmid on SDS-PAGE at different IPTG-concentration and temperatures. The level of expression of the Mce/whole maybe being too less to be detectable on gel, so we have also gone for protein specific antibody where we could say there is no complete expression. Western blot analysis toward full-length MAP2191 protein and its truncation only demonstrated Mce/truncated protein, which was similar to that predicted for Mce/whole protein.
Two usual concepts that will result in very low or no yield of protein during protein purification studies are protein toxicity and codon bias [31]. We were sure that our expression vector is correct and there was no mutation in the sequence based on sequencing results. If Mce/whole protein happened to be toxic for E. coli, we should see that transformed E. coli strain grow slower than the simple vector transformed strain in liquid culture [44]. In the case of protein proteolytic degradation problem, we tried to express the full-length protein in another E. coli strain like Rosetta expression cell in soluble condition with the pET28a vector system. Since, anti-His-Tag monoclonal antibody could not detect any Mce/whole protein from either Bl-21 or Rosetta strains and the results were the same when we used of Rosetta strain as an alternative host.We checked for our specific protein in cell lysate, supernatant and pellet also, because some protein is insoluble, which will present in pellet, but still didn’t get expression in any of the fractions. Therefore, in-silico prediction of primary structure of MAP2191 protein results along with experimental results confirmed that expression of Mce/whole protein was affected by the hydrophobicity nature of this protein. Our results were exactly in agreement with those of Cho et al., (2007), who described cloning and expression of C- and N-terminal portion of PepA protein separately and recorded only C-terminal end was successfully expressed due to the presence of hydrophilic eras [32]. In their study also N-terminal portion of PepA protein was predicted to be more hydrophobic than the C-terminal portion based on in-silico analysis. Lu et al., (2006), also reported a short sequence of 22 amino acids, the most hydrophilic region of Mce1A protein, is the only functional domain of this protein [14]. Strych et al., (2001), also reported that the presence of N-terminal hydrophobic region may interfere with Alr protein dimerization, folding or ligand binding [33].
In summary, our study results showed that the presence of hydrophobic structure, especially in first initialing amino acids in the protein’s chain can influence the protein expression system. Our data provide a valuable guide for future investigators with their own protein expression project to scan the protein sequence for the hydrophobic region in the initiating steps of protein purification to have an idea about final expected protein yield and to make more informed choices.
Acknowledgements:
The financial support of this research by the Animal Health Division at Central Institute for Research on Goats (CIRG), Makhdoom, Mathura, India, and Shiraz University, Iran, are gratefully acknowledged.
Conflict of Interest:
The authors declare that they have no competing interests.
References
- 1.Chaubey KK, Singh SV, Bhatia AK. ‘Indigenous’ and ‘Ethanol Vortex’ ELISA kits for diagnosis of Mycobacterium avium subsp paratuberculosis infection in cattle: Is there a ‘globally relevant kit’ in the ‘Reverse Ice-burg’ environment? Indian J Exp Biol. 2018;56:279–286. [Google Scholar]
- 2.Machado G, Kanankege K, Schumann V, Wells S, Perez A, Alvarez J. Identifying individual animal factors associated with Mycobacterium avium subsp paratuberculosis (MAP) milk ELISA positivity in dairy cattle in the Midwest region of the United States. BMC Vet Res. 2018;14:28. doi: 10.1186/s12917-018-1354-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathnaiah G, Zinniel DK, Bannantine JP, Stabel JR, Grohn YT, Collins MT, Barletta RG. Pathogenesis, Molecular genetics and genomics of Mycobacterium avium subsp paratuberculosis the etiologic agent of Johne's disease. Front Vet Sci . 2017;4:187. doi: 10.3389/fvets.2017.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahzad , Singh SV, Gupta S, Chaubey KK, Singh M, Dayal D, Dhama K, Hemati Z, Mansi S. Comparison of antibody response and direct microscopy between male and female goats in response to infection of Mycobacterium avium subspecies paratuberculosis in goatherds endemic for Johne’s disease. JSM Microbiol . 2017;5:1035. [Google Scholar]
- 5.Rossi G, Grohn YT, Schukken YH, Smith RL. The effect of Mycobacterium avium ssp Paratuberculosis infection on clinical mastitis occurrence in dairy cows. J Dairy Sci. 2017;100:7446–7454. doi: 10.3168/jds.2017-12721. [DOI] [PubMed] [Google Scholar]
- 6.Hussain T, Zhao D, Shah SZA, Wang J, Yue R, Liao Y, Sabir N, Yang L, Zhou X. MicroRNA27a-3 pregulates antimicrobial responses of murine macrophages infected by Mycobacterium avium subspecies paratuberculosis by targeting interleukin-10 and TGF-β-activated protein kinase 1 binding protein 2. Front Immunol. 2018;8:1915. doi: 10.3389/fimmu.2017.01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh M, Singh SV, Gupta S, Chaubey KK, Stephan BJ, Sohal JS, Dutta M. 'Nano-immunetest' for the detection of live Mycobacterium avium subspecies paratuberculosis bacilli in the milk samples using magnetic nano-particles and chromogen. Vet Res Commun. 2018;42:183–194. doi: 10.1007/s11259-018-9721-5. [DOI] [PubMed] [Google Scholar]
- 8.Gupta S, Singh SV, Gururaj K, Chaubey KK, Singh M, Lahiri B, Agarwal P, Kumar A, Misri J, Hemati Z, Bhatia AK. Comparison of IS900 PCR with ‘Taqman probe PCR’ and ‘SYBR Green Real time PCR’ assays in patients suffering with thyroid disorder and sero-positive for Mycobacterium avium subspecies paratuberculosis. Indian J Biotechnol . 2017;228 [Google Scholar]
- 9.Kuenstner JT, Naser S, Chamberlin W, Borody T, Graham DY, McNees A, Hermon-Taylor J, Hermon-Taylor A, Dow CT, Thayer W, Biesecker J, Collins MT, Sechi LA, Singh SV, Zhang P, Shafran I, Weg S, Telega G, Rothstein R, Oken H, Schimpff S, Bach H, Bull T, Grant I, Ellingson J, Dahmen H, Lipton J, Gupta S, Chaubey K, Singh M, Agarwal P, Kumar A, Misri J, Sohal J, Dhama K, Hemati Z, Davis W, Hier M, Aitken J, Pierce E, Parrish N, Goldberg N, Kali M, Bendre S, Agrawal G, Baldassano R, Linn P, Sweeney RW, Fecteau M, Hofstaedter C, Potula R, Timofeeva O, Geier S, John K, Zayanni N, Malaty HM, Kahlenborn C, Kravitz A, Bulfon A, Daskalopoulos G, Mitchell H, Neilan B, Timms V, Cossu D, Mameli G, Angermeier P, Jelic T, Goethe R, Juste RA, Kuenstner L. The Consensus from the Mycobacterium avium ssp paratuberculosis (MAP) Conference 2017. Front Public Health . 2017;5:208. doi: 10.3389/fpubh.2017.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dheenadhayalan V, Shin KS, Chang CF, Chang CD, Wang SJ, McDonough S, McDonough P, Stehman S, Shin S, Torres A, Chang YF. Cloning and characterization of the genes coding for antigen 85A, 85B and 85C of Mycobacterium avium subsp paratuberculosis. DNA Seq. 2002;13:287–294. doi: 10.1080/1042517021000019269. [DOI] [PubMed] [Google Scholar]
- 11.Okuni JB, Kateete DP, Okee M, Nanteza A, Joloba M, Ojok L. Application of antibodies to recombinant heat shock protein 70 in immune-histochemical diagnosis of Mycobacterium avium subspecies paratuberculosis in tissues of naturally infected cattle. Ir Vet J. 2017;270:10. doi: 10.1186/s13620-017-0088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohn WW, vander-Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem. 2017;283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arruda S, Bomfim G, Knights R, Huimo-Byron T, Riley LW. Cloning of a M tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 14.Lu S, Tager LA, Chitale S, Riley LW. A cell-penetrating peptide derived from mammalian cell uptake protein of Mycobacterium tuberculosis. Anal Biochem . 2006;353:7–14. doi: 10.1016/j.ab.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 15.Takenami I, de Oliveira CC, Lima FR, Soares J, Machado A Jr, Riley LW, Arruda S. ImmunoglobulinG response to mammalian cell entry1A (Mce1A) protein as biomarker of active tuberculosis. Tuberculosis (Edinb) 2016;100:82–88. doi: 10.1016/j.tube.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Haile Y, Caugant DA, Bjune G, Wiker HG. Mycobacterium tuberculosis mammalian cell entry operon mce homologs in Mycobacterium other than tuberculosis MOTT. FEMS Immunol Med Microbiol. 2002;332:125–132. doi: 10.1111/j.1574-695X.2002.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Fernandez J, Papavinasasundaram K, Galán B, Sassetti CM, García JL. Molecular and functional analysis of the mce4 operon in Mycobacterium smegmatis. Environ Microbiol . 2017;19:3689–699. doi: 10.1111/1462-2920.13869. [DOI] [PubMed] [Google Scholar]
- 18.Lima FR, Takenami I, Cavalcanti MA, Riley LW, Arruda S. ELISA-based assay of immunoglobulinG antibodies against mammalian cell entry1A (Mce1A) protein: a novel diagnostic approach for leprosy. Mem Inst Oswaldo Cruz. 2017;112:844–849. doi: 10.1590/0074-02760160549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Li J, Li B, Wang J, Liu CH. Mycobacterium tuberculosis Mce3C promotes mycobacteria entry into macrophages through activation of β2 integrin-mediated signalling pathway. Cell Microbiol . 2017;12:800. doi: 10.1111/cmi.12800. [DOI] [PubMed] [Google Scholar]
- 20.Cangelosi GA, Do JS, Freeman R, Bennett JG, Semret M, Behr MA. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob Agents Chemother. 2006;502:461–468. doi: 10.1128/AAC.50.2.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander DC, Turenne CY, Behr MA. Insertion and deletion events that define the pathogen Mycobacterium avium subsp paratuberculosis. J Bacteriol. 2009;913:1018–1025. doi: 10.1128/JB.01340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semret M, Alexander DC, Turenne CY, de-Haas P, Overduin P, van Soolingen D, Cousins D, Behr MA. Genomic polymorphisms for Mycobacterium avium subsp paratuberculosis diagnostics. J Clin Microbiol . 2005;43:3704–3712. doi: 10.1128/JCM.43.8.3704-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh N, Singh SV, Gupta VK, Sharma VD, Sharma RK, Katoch VM. Isolation and identification of Mycobacterium paratuberculosis from naturally infected goatherds in India. Indian J Vet Pathol. 1996;20:104–108. [Google Scholar]
- 24.Millar D, Ford J, Sanderson J, Withey S, Tizard M, Doran T, Hermon-Taylor J. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows’ milk in England and Wales. Appl Environ Microbiol . 1996;62:3446–3452. doi: 10.1128/aem.62.9.3446-3452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sevilla I, Singh SV, Garrido JM, Aduriz G, Rodriguez S, Geijo MV, Cousins D, Behr MA. Molecular typing of Mycobacterium avium subspecies paratuberculosis strains from different hosts and regions. Rev Sci Techn. 2005;24:1061–1066. [PubMed] [Google Scholar]
- 26.Whittington RJ, Marsh IB, Whitlock RH. Typing of IS1311 polymorphisms confirms that bison (Bison bison) with paratuberculosis in Montana are infected with strain of Mycobacterium avium subspecies paratuberculosis distinct from that occurring in cattle and other domestic livestock. Mole Cell Probes. 2001;15:139–145. doi: 10.1006/mcpr.2001.0346. [DOI] [PubMed] [Google Scholar]
- 27.Mukartal SY, Rathannama D, Narayanswamy HD, Gupta S, Chaubey KK, Singh M, Hemati Z, Nishanth C, Pachoori A, Dhama K, Singh SV. Assessment of Ovine Johne’s disease in the Mandya sheep breed in South India using multiple diagnostic tests and bio-typing of Mycobacterium avium subspecies paratuberculosis infection. Cogent Food Agric. 2017;3 [Google Scholar]
- 28.Rodriguez DC, Ocampo M, Varela Y, Curtidor H, Patarroyo MA, Patarroyo ME. Mce4F Mycobacterium tuberculosis protein peptides can inhibit invasion of human cell lines. Pathog Dis. 2015;73:1–12. doi: 10.1093/femspd/ftu020. [DOI] [PubMed] [Google Scholar]
- 29.Castellanos E, Aranaz A, de Juan L, Dominguez L, Linedale R, Bull TJ. A 16 kb naturally occurring genomic deletion including mce and PPE genes in Mycobacterium avium subspecies paratuberculosis isolates from goats with Johne's disease. Vet Microbiol . 2012;159:60–68. doi: 10.1016/j.vetmic.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Sekar SC, Goswami PP, Deb R. Expression and purification of a gene encoding a 97 kDa PE protein of Mycobacterium avium subsp paratuberculosis 3. Biotech. 2016;6:198. doi: 10.1007/s13205-016-0506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol . 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho D, Shin SJ, Talaat AM, Collins MT. Cloning, expression, purification and sero-diagnostic evaluation of fourteen Mycobacterium paratuberculosis proteins. Protein Expr Purif . 2007;53:411–420. doi: 10.1016/j.pep.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Strych U, Penland RL, Jimenez M, Krause KL, Benedik MJ. Characterization of the alanineracemases from two mycobacteria. FEMS Microbiol Lett . 2001;196:93–98. doi: 10.1111/j.1574-6968.2001.tb10547.x. [DOI] [PubMed] [Google Scholar]