A variety of human rheumatologic disease-causing mutations have recently been identified. Some of these mutations are found in viral nucleic acid-sensing proteins, but whether viruses can influence the onset or progression of these human diseases is less well understood. One such autoinflammatory disease, called STING-associated vasculopathy with onset in infancy (SAVI), affects children and leads to severe lung disease. We generated mice with a SAVI-associated STING mutation and infected them with γHV68, a common DNA virus that is related to human Epstein-Barr virus. Mice with the human disease-causing STING mutation were more vulnerable to infection than wild-type littermate control animals. Furthermore, the STING mutant mice developed lung fibrosis similar to that of patients with SAVI. These findings reveal that a human STING mutation creates severe immunodeficiency, leading to virus-induced lung disease in mice.
KEYWORDS: MHV68, STING, adaptive immunity, autoimmunity, gammaherpesvirus, innate immunity, pulmonary fibrosis, vasculopathy
ABSTRACT
We previously generated STING N153S knock-in mice that have a human disease-associated gain-of-function mutation in STING. Patients with this mutation (STING N154S in humans) develop STING-associated vasculopathy with onset in infancy (SAVI), a severe pediatric autoinflammatory disease characterized by pulmonary fibrosis. Since this mutation promotes the upregulation of antiviral type I interferon-stimulated genes (ISGs), we hypothesized that STING N153S knock-in mice may develop more severe autoinflammatory disease in response to a virus challenge. To test this hypothesis, we infected heterozygous STING N153S mice with murine gammaherpesvirus 68 (γHV68). STING N153S mice were highly vulnerable to infection and developed pulmonary fibrosis after infection. In addition to impairing CD8+ T cell responses and humoral immunity, STING N153S also promoted the replication of γHV68 in cultured macrophages. In further support of a combined innate and adaptive immunodeficiency, γHV68 infection was more severe in Rag1–/– STING N153S mice than in Rag1–/– littermate mice, which completely lack adaptive immunity. Thus, a gain-of-function STING mutation creates a combined innate and adaptive immunodeficiency that leads to virus-induced pulmonary fibrosis.
IMPORTANCE A variety of human rheumatologic disease-causing mutations have recently been identified. Some of these mutations are found in viral nucleic acid-sensing proteins, but whether viruses can influence the onset or progression of these human diseases is less well understood. One such autoinflammatory disease, called STING-associated vasculopathy with onset in infancy (SAVI), affects children and leads to severe lung disease. We generated mice with a SAVI-associated STING mutation and infected them with γHV68, a common DNA virus that is related to human Epstein-Barr virus. Mice with the human disease-causing STING mutation were more vulnerable to infection than wild-type littermate control animals. Furthermore, the STING mutant mice developed lung fibrosis similar to that of patients with SAVI. These findings reveal that a human STING mutation creates severe immunodeficiency, leading to virus-induced lung disease in mice.
INTRODUCTION
STING is a cytosolic DNA sensor that signals upon the binding of cyclic GMP-AMP (cGAMP), a cyclic dinucleotide second messenger produced by cGAMP synthase (cGAS) in response to viral or aberrant host DNA (1, 2). STING can also be activated by bacterial cyclic dinucleotides, a phenomenon that can impact the mobilization of hematopoietic progenitor cells (3). Activation of the cGAS-STING signaling pathway leads to upregulation of antiviral type I interferon (IFN)-stimulated genes (ISGs) and proinflammatory cytokines (1, 4, 5). Patients with autosomal dominant gain-of-function STING mutations have a pediatric autoinflammatory syndrome known as STING-associated vasculopathy with onset in infancy (SAVI) (6). The peripheral blood mononuclear cells of SAVI patients exhibit a type I IFN gene expression signature, and the clinical disease phenotype includes skin rash, vasculopathy, and pulmonary fibrosis. Cell culture studies using transfected cells, peripheral blood mononuclear cells, and fibroblasts have suggested that SAVI-associated STING mutants are constitutively active (6).
One of the most common SAVI-associated mutations is STING N154S (N153S in mice) (6). We previously generated heterozygous STING N153S mice and found that older adult mice develop spontaneous autoinflammation with IgM autoantibodies, lymphopenia, and perivascular inflammation in the lung that occurs independently of IFN regulatory factor 3 (IRF3) (7). However, unlike patients with SAVI, our STING N153S mice did not develop pulmonary fibrosis (7). Despite having T cell and NK cell cytopenia, SAVI patients are not regarded as having an immunodeficiency. Nevertheless, SAVI patients are naturally exposed to viruses and other pathogens, and they have been reported to suffer from secondary infections (e.g., recurrent pneumonias) (6). In contrast, our STING N153S mice, which also have T and NK cell cytopenia (7), are housed in an enhanced, specific-pathogen-free animal facility. Since STING is a sensor of viral DNA, we reasoned that viruses may be required to trigger certain rheumatologic manifestations of the human disease, including pulmonary fibrosis. Furthermore, we hypothesized that the STING N153S gain-of-function mutation may create resistance to viral infection, since this STING mutant is thought to activate the antiviral type I IFN response. Indeed, type I and type III IFN-mediated immunity can limit virus infection and persistence in mice, independently of adaptive immunity (8, 9).
Here we show that intranasal inoculation with murine gammaherpesvirus 68 (γHV68) leads to uncontrolled viral replication, pneumonia, and pulmonary fibrosis in heterozygous STING N153S mice, but not in wild-type (WT) littermate control animals or in mice deficient in STING signaling. Pulmonary fibrosis was prevented with the antiviral drug cidofovir, demonstrating that viral replication was required for this lung pathology. Enhanced viral replication in cultured macrophages, as well as viral pathogenesis studies with STING N153S mice lacking type I IFN signaling (Ifnar1–/– STING N153S mice) or adaptive immunity (Rag1–/– STING N153S mice), demonstrated a combined innate and adaptive immunodeficiency. Thus, an autosomal dominant gain-of-function mutation in STING causes susceptibility to γHV68 infection and pulmonary fibrosis in mice.
RESULTS
STING N153S mice are highly vulnerable to viral infections.
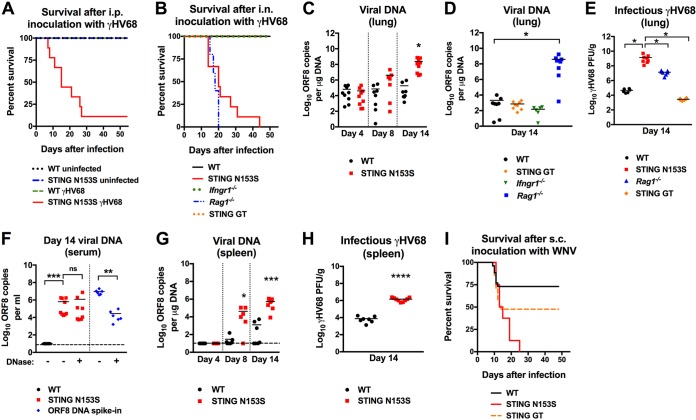
Autosomal dominant mutations in STING, including STING N154S (N153S in mice), promote constitutive signaling to activate TBK1 and upregulate antiviral ISGs (6). Therefore, we hypothesized that heterozygous mice with the STING N153S mutation may be resistant to viral infections due to upregulation of ISGs. Unexpectedly, we discovered that heterozygous STING N153S mice were more vulnerable than WT littermate control animals to infection with murine gammaherpesvirus 68 (γHV68), a double-stranded DNA virus known to activate the cGAS-STING pathway (10, 11) (Fig. 1A and B). Enhanced vulnerability to infection was observed after intraperitoneal (i.p.) inoculation of older adult mice with 1 × 106 PFU of γHV68 (89% mortality in STING N153S mice versus 0% mortality in WT mice [P < 0.0001] [Fig. 1A]). To examine the effects of age and route of inoculation, we also performed intranasal (i.n.) inoculations of 7- to 8-week-old STING N153S mice with 2 × 105 PFU of γHV68. Intranasal inoculation also resulted in 100% lethality of STING N153S mice, in contrast to 0% mortality in WT littermate control mice (Fig. 1B). To determine the relative severity of immunodeficiency of STING N153S mice using an established model of γHV68 pathogenesis, we intranasally inoculated Ifngr1−/− mice, which lack the type II IFN receptor known to be important for γHV68 control (12), as well as STING goldenticket (GT) mice, which are deficient in STING signaling (13). In further support of the conclusion that STING N153S mice are severely immunodeficient, Ifngr1−/− and STING GT mice were much less vulnerable to γHV68 than STING N153S mice, since the former exhibited no lethality within 60 days after infection (Fig. 1B).
FIG 1.
STING N153S mice are highly vulnerable to viral infections. (A) Kaplan-Meier curve showing 60-day mortality of 25- to 30 week-old STING N153S mice and WT littermates following intraperitoneal inoculation with 106 PFU of γHV68 or PBS (uninfected). Shown are results for 7 to 13 mice per genotype pooled from 3 independent experiments. (B) Kaplan-Meier curve showing mortality rates of 7- to-8-week-old STING N153S, WT littermate, Ifngr1–/–, Rag1–/–, and STING goldenticket (GT) mice that were inoculated intranasally with 2 × 105 PFU of γHV68. Survival was assessed for 60 days after infection. Shown are results for 8 (WT), 9 (STING N153S), 16 (Ifngr1–/–), and 5 (Rag1–/–) mice pooled from at least 2 independent experiments. (C) Viral burdens in the lung on days 4, 8, and 14 after intranasal inoculation of STING N153S mice and WT littermates with 2 × 105 PFU of γHV68. Shown are results for 7 to 9 mice per genotype at each time point from 3 independent experiments. (D) Viral burdens in WT, STING GT, Ifngr1–/–, and Rag1–/– mouse lungs at day 14 after intranasal inoculation with 2 × 105 PFU of γHV68. Shown are results for 8 mice per genotype pooled from at least 2 independent experiments. (E) PFU of γHV68 in the lungs of WT, STING N153S, Rag1–/–, and STING GT animals 14 days after infection with γHV68. Shown are results for 6 to 8 mice per genotype. Data were pooled from at least 2 independent experiments. (F) γHV68 viral burdens in the serum on day 14 after intranasal inoculation of STING N153S mice and WT littermates with 2 × 105 PFU of γHV68. Shown are results for 8 to 9 mice per group, pooled from two independent experiments. The dashed line denotes the limit of sensitivity of the assay. (G) Viral burdens in the spleen on days 4, 8, and 14 after intranasal inoculation of STING N153S mice and WT littermates with 2 × 105 PFU of γHV68. Shown are results for 7 to 9 animals per genotype, pooled from at least 2 independent experiments. The dashed line denotes the limit of sensitivity of the assay. (H) PFU of γHV68 in the spleens of WT and STING N153S mice 14 days after infection with γHV68. Shown are results for 7 to 9 mice per genotype, pooled from at least 2 independent experiments. (I) Kaplan-Meier curve showing mortality rates of 7-to-8 week-old STING N153S, WT littermate, and STING GT mice after subcutaneous inoculation with 102 FFU of WNV. Shown are results for 14 (WT), 8 (STING N153S), and 21 (STING GT) mice. Kaplan-Meier curves (A, B, and I) were analyzed by the log rank test. The data in other panels were analyzed by the unpaired t test (C and H), Kruskal-Wallis test (D), one-way ANOVA (E), or Mann-Whitney test (F and G). All data represent the means ± standard errors of the means of results from at least 2 independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
To begin to define mechanisms of γHV68 pathogenesis in STING N153S mice, we measured viral burdens in the lungs, serum, and spleen after intranasal inoculation (Fig. 1C to H). Since STING signaling activates cell-intrinsic antiviral ISGs that are most important during early infection (5), we reasoned that the gain-of-function STING N153S mutation would preferentially restrict viral replication at early time points. Unexpectedly, we observed no difference in viral burdens in the lungs on day 4 after intranasal inoculation of 7-week-old STING N153S and WT littermate control animals (3.8 × 104 copies/μg in STING N153S mice versus 3.3 × 104 copies/μg in WT mice [P > 0.9]) (Fig. 1C). However, by day 8, we began to see a trend toward higher viral DNA levels in the lungs of STING N153S mice (4 × 106 copies/μg in STING N153S mice versus 5 × 104 copies/μg in WT mice [P > 0.1]), and by day 14, we observed a ∼10,000-fold increase in the amount of viral DNA in the lungs of STING N153S mice over that in WT littermate control animals (Fig. 1C). Previous work has demonstrated that antigen-specific CD8+ T cells control the viral burden during γHV68 infection (14, 15). Additionally, STING N153S mice are known to have T and NK cell cytopenia (7), which may contribute to immunodeficiency. Thus, we reasoned that adaptive immunity may be impaired in the STING N153S mice. In accord with prior studies, we detected high levels of γHV68 DNA on day 14 in the lungs in Rag1−/− mice. In contrast, viral burdens in the lungs of STING GT and Ifngr1−/− mice were more comparable to the levels of viral DNA in WT animals (Fig. 1D).
We directly compared levels of infectious γHV68 in the lungs of these mice by plaque assays. Levels of infectious γHV68 were ∼3,000-fold greater in the lungs of STING N153S animals than in WT littermate lungs, ∼100-fold greater than in Rag1–/– lungs, and ∼10,000-fold greater than in STING GT lungs (Fig. 1E). In the serum, γHV68 DNA was still detectable in STING N153S mice but not in WT mice (Fig. 1F). γHV68 DNA in the serum was likely virion-associated DNA, since it was resistant to DNase treatment (Fig. 1F). The spleens of STING N153S mice also had higher levels of γHV68 DNA and infectious virus than WT control spleens on days 8 and 14 after infection (Fig. 1G and H). These results demonstrate that STING N153S causes severe immunodeficiency and failure to control lytic-phase viral replication.
Since STING also promotes antiviral immunity against RNA viruses, including West Nile virus (WNV) (16), which is phylogenetically unrelated to γHV68, we also tested the susceptibility of STING N153S mice to infection with WNV. Subcutaneous (s.c.) inoculation with a pathogenic strain of WNV caused 100% mortality in STING N153S mice, in contrast to ∼50% mortality in STING GT mice and only ∼25% mortality in WT littermate control animals (Fig. 1I). Thus, a single missense mutation in just one STING allele is sufficient to create vulnerability to lethal infection with unrelated viruses.
STING N153S mice develop viral pneumonia and pulmonary fibrosis after γHV68 infection.
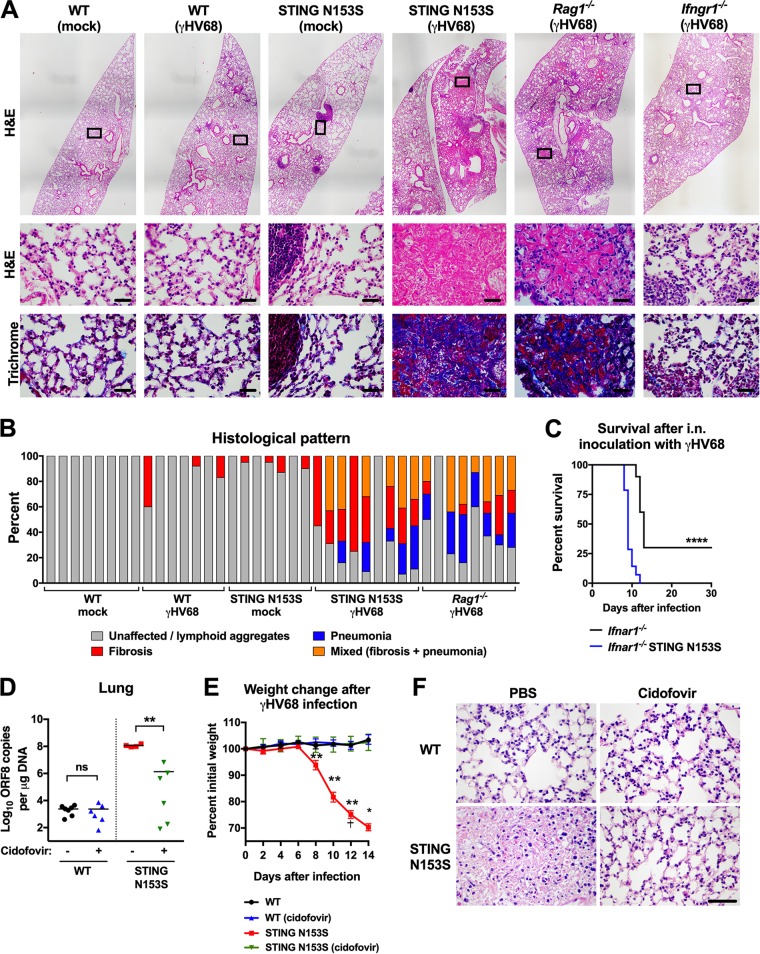
Patients with SAVI have been reported to have secondary infections and recurrent pneumonias (6), but whether viral infections may contribute to an autoimmune disease phenotype in humans is not known. To examine virus-induced lung disease in mice, we performed histological examinations of the lungs of STING N153S mice and WT littermate control animals on day 14 after infection with γHV68 (Fig. 2A and B). In agreement with our viral burden findings (Fig. 1C to E), the STING N153S mice developed severe viral pneumonia and pulmonary fibrosis, producing a phenotype similar to that of Rag1−/− mice (Fig. 2A and B). In contrast, WT littermate control and Ifngr1−/− mice had only mild lung disease at day 14 (Fig. 2A and B). To determine the effects of type I IFN on lung disease, we crossed the STING N153S mice to animals that lack the type I IFN receptor (Ifnar1−/− mice) and intranasally inoculated Ifnar1−/− STING N153S and Ifnar1−/− littermate control mice with γHV68. Ifnar1−/− STING N153S mice were more vulnerable to γHV68 infection than Ifnar1−/− littermates (100% lethality in STING N153S mice versus 70% lethality in Ifnar1−/− littermates [P < 0.0001]) (Fig. 2C). Thus, STING N153S causes vulnerability to infection that extends beyond effects on type I IFN signaling. Unfortunately, since the Ifnar1−/− STING N153S mice succumbed to infection at a much earlier time point than their Ifnar1−/− littermates, it was not possible to determine the contribution of type I IFN to pulmonary fibrosis.
FIG 2.
STING N153S mice develop viral pneumonia and pulmonary fibrosis after γHV68 infection. Seven- to 8-week-old mice were intranasally inoculated with 2 × 105 PFU of γHV68 or PBS (mock) and were euthanized at day 14 after infection. Lungs were fixed in 4% PFA, sectioned, and stained with hematoxylin and eosin (H&E) or Gomori's trichrome stain, followed by histological analysis. (A) Representative lung sections from mock-infected and γHV68-infected WT, STING N153S, Rag1–/–, and Ifngr1–/– mice. Sections were stained with H&E (top and center) or trichrome (bottom). Bar, 20 μm. (B) Percentages of ×20 fields of view with the indicated histological patterns (unaffected or small lymphoid aggregates, fibrosis, pneumonia, or mixed). Shown are results for 7 to 9 mice per genotype, pooled from two independent experiments. (C) Kaplan-Meier curves showing 30-day mortality of 11- to 15-week-old Ifnar1−/− STING N153S mice and Ifnar1−/− littermates following intranasal inoculation with 2 × 105 PFU of γHV68. Shown are results for 10 to 14 mice per genotype, pooled from three independent experiments. The curves were analyzed by the log rank test. (D to F) Fourteen days after intranasal inoculation with 2 × 105 PFU of γHV68. Animals received 25 mg/kg of cidofovir or an equivalent volume of vehicle only (PBS) via intraperitoneal injections on days 4, 5, 8, and 11 after infection. (D and E) γHV68 burdens in the lungs of WT or STING N153S animals receiving PBS or cidofovir treatment (D) and percentages of change from starting weight for γHV68-infected WT and STING N153S animals (E). Shown are results for 4 to 7 mice per condition and genotype, pooled from two independent experiments. Data were analyzed by the Mann-Whitney test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. The dagger indicates that this animal was removed at this time point after meeting euthanasia criteria. (F) Representative ×40 H&E images of sections from WT and STING N153S animals receiving PBS or cidofovir treatment. Bar, 20 μm.
To determine whether lytic infection with γHV68 is required for pulmonary fibrosis in STING N153S mice, we treated 7- to 8-week-old WT and STING N153S animals with cidofovir beginning 4 days after infection. Cidofovir treatment reduced viral burdens in the lungs of STING N153S mice but not in those of WT mice (1.14 × 108 copies/μg versus 1.34 × 106 copies/μg [P < 0.01]), which corresponded with the prevention of infection-related weight loss and lung pathology at day 14 after infection (Fig. 2D to F). Thus, lytic infection is required for γHV68-induced lung disease and lethality in STING N153S mice.
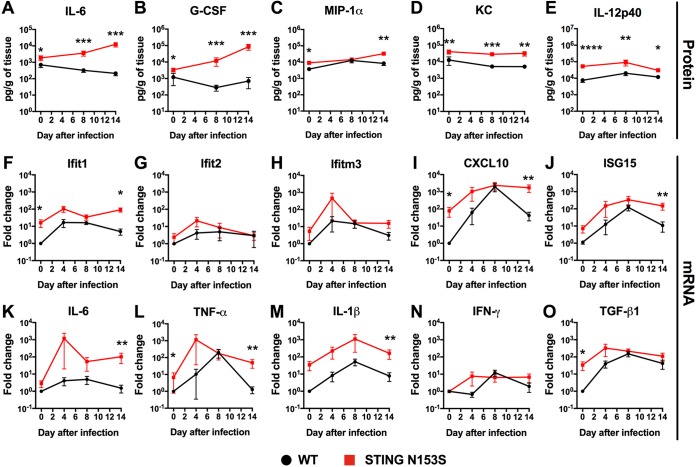
We reasoned that enhanced production of chemokines and proinflammatory cytokines may contribute to virus-related immunopathology. To examine whether the STING N153S mutation impacts cytokine production during infection, we performed multiplex cytokine analysis of the lungs on days 0, 8, and 14 after infection with γHV68 (Fig. 3A to E; Table 1). Whereas levels of interleukin 6 (IL-6) and granulocyte colony-stimulating factor (G-CSF) were elevated at late time points after infection, when viral burdens were ∼10,000-fold higher in STING N153S mice than in WT littermate control animals (Fig. 3A and B), macrophage inflammatory protein 1α (MIP-1α), keratinocyte chemoattractant (KC), and IL-12p40 were upregulated even prior to infection in the lungs of STING N153S mice (Fig. 3C to E). These findings suggest that the gain-of-function STING N153S mutation increases the production of macrophage-derived chemokines in the lungs of uninfected mice. However, the majority of cytokines and chemokines exhibited very similar expression patterns in WT and STING N153S animals at all time points during infection (Table 1). Quantitative real-time reverse transcription-PCR (qRT-PCR) analysis of lung tissue revealed ∼10- to ∼100-fold upregulation of certain ISGs (IFN-induced protein with tetratricopeptide repeats 1 [Ifit1], CXC motif chemokine ligand 10 [CXCL10], and ISG15), ∼10-fold upregulation of transforming growth factor β (TGF-β) at baseline, and ∼10-fold upregulation of proinflammatory cytokines (IL-6, tumor necrosis factor alpha [TNF-α], and IL-1β) at late time points after infection, a finding that might be explained by high viral burdens on day 14 (Fig. 3F to O). Elevated baseline levels of some ISGs and cytokines likely contribute to elevation at late time points as well.
FIG 3.
Cytokine and ISG expression levels in WT and STING N153S mouse lung homogenates prior to and during infection with γHV68. (A to E) WT and STING N153S mice were inoculated intranasally with 2 × 105 PFU of γHV68. Animals were euthanized at day 8 or 14 after infection; mock-infected animals were euthanized on day 0. Cytokine and chemokine levels were assessed by a Luminex assay of lung homogenates. Data represent the means ± standard errors of the means of results for 7 to 9 mice per group at each time point, pooled from 2 independent experiments. (F to O) STING N153S and WT littermate control mice were inoculated intranasally with 2 × 105 PFU of γHV68. Animals were euthanized at day 4, 8, or 14 after infection; mock-infected animals were euthanized on day 0. ISG (F to J) and cytokine (K to O) gene expression levels in lung homogenates were assessed by qRT-PCR and are reported as the fold change relative to expression levels for the WT. Results from 6 mice per group at each time point were pooled from 2 independent experiments. All data represent the means ± standard errors of the means from 2 independent experiments. Results were analyzed by the Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TABLE 1.
Cytokine levels in WT and STING N153S lung homogenates prior to and during infection with γHV68a
| Cytokineb | LODc (pg/g) | Level (pg/g of tissue) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mock infection |
Day 8 after infection |
Day 14 after infection |
|||||||||||
| WT |
STING N153S |
WT |
STING N153S |
WT |
STING N153S |
||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| IL-1α | 2.25 | 2,950 | 1,873 | 4,741 | 2,892 | 892 | 353 | 5,757 | 7,886 | 858 | 249 | 2,343 | 1,304 |
| IL-1β | 1.85 | 1,937 | 1,339 | 2,508 | 1,225 | 417 | 244 | 2,459 | 3,319 | 592 | 502 | 1,079 | 675 |
| IL-2 | 0.88 | 1,858 | 2,040 | 1,472 | 675 | 398 | 160 | 337 | 123 | 325 | 190 | 159 | 328 |
| IL-3 | 0.45 | 46 | 190 | 67 | 142 | 9 | 16 | 16 | 28 | 111 | 201 | 11 | 22 |
| IL-4 | 0.32 | 123 | 304 | 137 | 245 | 160 | 92 | 115 | 97 | 45 | 93 | 11 | 5 |
| IL-5 | 0.59 | 960 | 778 | 812 | 297 | 262 | 142 | 279 | 141 | 287 | 176 | 155 | 94 |
| IL-6 | 0.71 | 699 | 626 | 1,865 | 1,860 | 325 | 231 | 3,608 | 3,249 | 212 | 155 | 12,467 | 10,306 |
| IL-9 | 2.36 | 1,958 | 2,263 | 1,016 | 1,839 | 617 | 255 | 462 | 394 | 398 | 900 | 62 | 32 |
| IL-10 | 3.57 | 1,789 | 1,541 | 3,011 | 1,521 | 3,438 | 2,240 | 2,995 | 1,423 | 1,394 | 1,057 | 1,943 | 855 |
| IL-12(p40) | 8.81 | 7,539 | 5,501 | 52,699 | 24,545 | 20,226 | 16,461 | 90,693 | 93,701 | 11,844 | 7,296 | 30,459 | 19,296 |
| IL-12(p70) | 4.28 | 1,711 | 1,739 | 1,613 | 1,096 | 1,106 | 456 | 1,260 | 681 | 766 | 712 | 783 | 509 |
| IL-13 | 10.96 | 10,266 | 10,463 | 8,335 | 6,455 | 672 | 713 | 652 | 666 | 3,939 | 6,600 | 1,220 | 1,743 |
| IL-17A | 0.49 | 450 | 368 | 281 | 135 | 93 | 26 | 158 | 120 | 140 | 208 | 52 | 62 |
| Eotaxin | 22.92 | 304,216 | 642,378 | 703,751 | 1,263,328 | 931,207 | 834,322 | 499,161 | 843,408 | 34,380 | 13,778 | 10,237 | 3,474 |
| G-CSF | 3.97 | 1,211 | 2,381 | 3,390 | 2,737 | 281 | 312 | 11,874 | 16,821 | 698 | 1,212 | 85,054 | 86,806 |
| GM-CSF | 21.6 | 2,281 | 1,172 | 2,370 | 1,111 | 22 | 0 | 843 | 1,030 | 1,343 | 1,436 | 1,867 | 1,202 |
| IFN-γ | 0.92 | 2,809 | 1,887 | 1,921 | 948 | 513 | 329 | 302 | 226 | 878 | 280 | 554 | 541 |
| KC | 1.15 | 12,785 | 19,214 | 40,670 | 34,909 | 5,162 | 3,204 | 28,292 | 12,174 | 5,083 | 2,533 | 32,282 | 26,956 |
| MCP-1 | 43.09 | 36,610 | 43,423 | 50,221 | 25,564 | 42,790 | 31,808 | 33,136 | 48,194 | 19,537 | 11,692 | 256 | 132 |
| MIP-1α | 0.43 | 3,756 | 2,050 | 9,068 | 5,282 | 12,726 | 10,362 | 14,477 | 8,434 | 8,418 | 5,705 | 33,277 | 13,085 |
| MIP-1β | 2.61 | 6,274 | 7,863 | 5,688 | 6,137 | 5,252 | 2,057 | 3,028 | 424 | 2,001 | 525 | 1,699 | 987 |
| RANTES | 1.62 | 82,621 | 67,099 | 164,089 | 120,460 | 530,764 | 601,268 | 24,005 | 56,366 | 142,988 | 69,619 | 794 | 912 |
| TNF-α | 5.05 | 23,864 | 14,393 | 21,234 | 6,908 | 1,934 | 608 | 2,036 | 678 | 7,094 | 10,641 | 8,753 | 7,290 |
WT and STING N153S mice were inoculated intranasally with 2 × 105 PFU of γHV68. Animals were euthanized at day 8 or 14 after infection; mock-infected animals were euthanized on day 0. Cytokine and chemokine levels were assessed by Luminex assays of lung homogenates. Results for selected cytokines are displayed graphically in Fig. 3. Data represent results for 7 to 9 mice per group at each time point, pooled from 2 independent experiments.
MCP-1, monocyte chemoattractant protein 1.
LOD, limit of detection.
Impaired antigen-specific CD8+ T cell responses in STING N153S mice infected with γHV68.
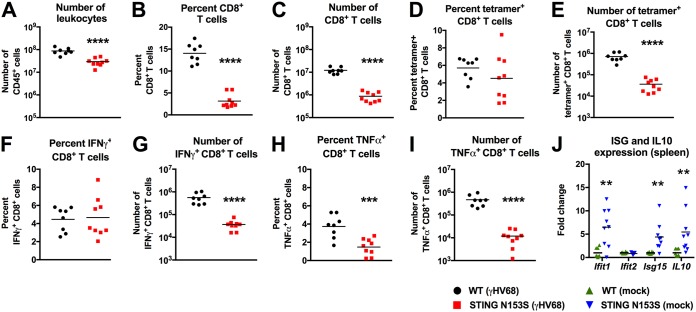
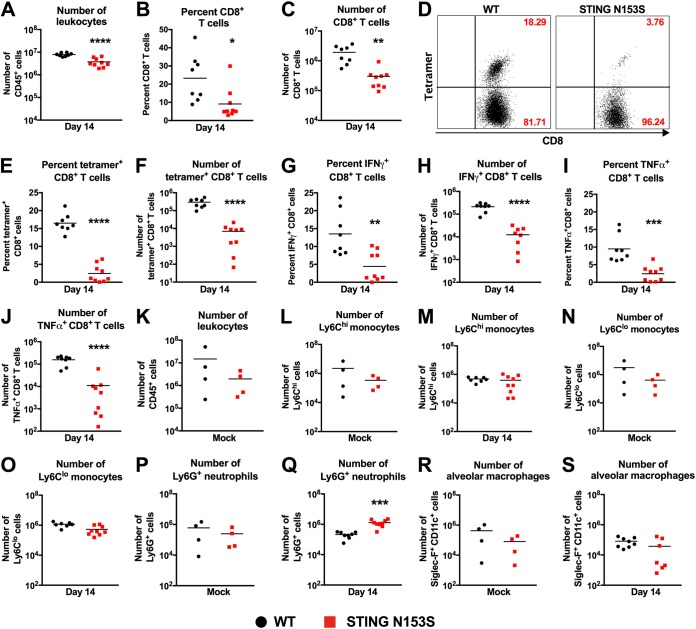
Since STING signaling influences adaptive immunity (2, 17, 18), we assessed adaptive immune responses in WT and STING N153S mice on day 14 after infection with γHV68. We reported previously that STING N153S mice have a ∼7-fold decrease in the number of CD8+ and CD4+ T cells (7), much like SAVI patients (6), but we did not characterize the ability of STING N153S T cells to launch antigen-specific responses. Furthermore, since depletion of CD8+ T cells protects against pulmonary fibrosis in γHV68-infected Ifngr1−/− mice (19), we reasoned that it would be important to characterize antigen-specific CD8+ T cell responses in the STING N153S mice. On day 14 after intranasal inoculation with γHV68, flow cytometric analysis of splenocytes stained with the γHV68 ORF6 tetramer revealed similar percentages of antigen-specific CD8+ T cells in the spleens of WT and STING N153S mice, but diminished total numbers in the latter, suggesting that STING N153S CD8+ T cells can be primed in the spleen (Fig. 4A to E). Similarly, we found diminished numbers of IFN-γ- and TNF-α-positive CD8+ T cells in STING N153S mice after restimulation with an ORF6 immunodominant peptide (Fig. 4F to I). Since ISGs and IL-10 can inhibit T cell responses (20, 21), we measured the expression levels of ISGs and IL-10 in uninfected spleens. Indeed, we found 4- to 6-fold upregulation of Ifit1, ISG15, and IL-10 prior to infection (Fig. 4J). Examination of T cells in the lung revealed that an even larger deficit in antigen-specific CD8+ T cell numbers was observed with tetramer staining and peptide restimulation (Fig. 5A to J), suggesting that impairment of antigen-specific CD8+ T cell responses may contribute to STING N153S-related immunodeficiency.
FIG 4.
CD8+ T cell responses and ISG expression in the spleens of WT and STING N153S mice after γHV68 infection. Mice were inoculated intranasally with 2 × 105 PFU of γHV68 and were sacrificed 14 days after infection. Spleens were harvested and leukocytes isolated for flow cytometric analysis. (A) Total number of CD45+ cells. (B and C) Frequency (B) and number (C) of CD8+ cells in the CD45+ population. (D and E) Frequency (D) and number (E) of ORF6 tetramer-positive CD8+ cells in the CD8+ population. (F to I) Splenocytes were restimulated with the immunodominant ORF6-derived peptide for 4 h. Cells were intracellularly stained and were analyzed for cytokine production via flow cytometry to calculate the frequency (F and H) and numbers (G and I) of restimulated CD8+ T cells producing IFN-γ (F and G) and TNF-α (H and I). Data represent the mean results for 8 to 9 mice per group, pooled from 2 independent experiments. Results for panels A to I were analyzed by an unpaired t test. (J) Splenocytes of uninfected WT and STING N153S mice were analyzed for expression of ISGs by qRT-PCR. Data represent the mean results for 8 to 9 mice per group, pooled from 2 independent experiments. Results were analyzed by the Mann-Whitney test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
FIG 5.
Impaired antigen-specific CD8+ T cell responses in the lungs of STING N153S mice infected with γHV68. (A to J) Mice were inoculated intranasally with 2 × 105 PFU of γHV68 and were euthanized 14 days after infection. Lungs were harvested and leukocytes isolated for flow cytometric analysis. (A) Total numbers of CD45+ cells in the lungs of infected WT and STING N153S animals. (B and C) Frequency (B) and number (C) of CD8+ cells in the CD45+ population. (D) Representative FACS plots of ORF6 tetramer-positive and -negative CD8+ T cells in the lungs of WT (left) and STING N153S (right) mice. Numbers indicate the percentage of events in each gate. (E and F) Frequency (E) and number (F) of ORF6 tetramer-positive CD8+ cells in the CD8+ population. (G to J) Lung leukocytes were restimulated with the immunodominant ORF6-derived peptide for 4 h. Cells were intracellularly stained and were analyzed for cytokine production via flow cytometry to calculate the frequency (G and I) and numbers (H and J) of restimulated CD8+ T cells producing IFN-γ (G and H) and TNF-α (I and J). Data represent the means for 8 or 9 mice per group, pooled from two independent experiments. (K to S) Mice were inoculated intranasally with 2 × 105 PFU of γHV68 or PBS (mock) and were euthanized 14 days after infection. (K) Total numbers of CD45+ cells in the lungs of mock-infected WT and STING N153S animals. (L to S) Total numbers of monocytes (L to O), neutrophils (P and Q), and alveolar macrophages (R and S) in the lungs of WT and STING N153S animals. Data represent the means for 4 to 9 mice per group, pooled from at least two independent experiments. Results were analyzed by an unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Examination of total lung leukocytes and myeloid cells by flow cytometry revealed no difference in the total numbers of leukocytes or monocytes in the lungs of WT and STING N153S mice, whether we looked before or after infection (Fig. 5K to O). There was a small increase relative to WT littermate control animals in the number of neutrophils in the lungs of STING N153S mice on day 14 after infection (Fig. 5P and Q). Macrophages serve as targets and reservoirs of γHV68 (22, 23), but there was no difference in the number of alveolar macrophages in the lungs between WT and STING N153S mice (Fig. 5R and S).
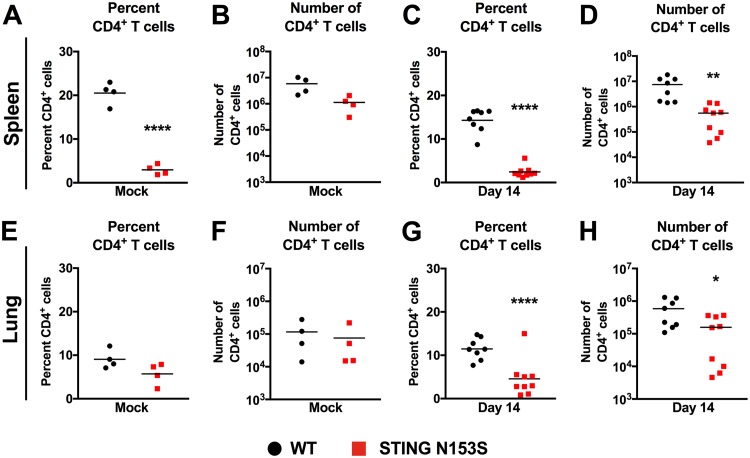
Since CD4+ T cells also restrict γHV68 infection (24), we examined the percentages and numbers of CD4+ T cells before and after infection in both the spleen and the lung (Fig. 6A to H). In the spleen, we observed diminished numbers of STING N153S CD4+ T cells after infection with γHV68 (Fig. 6A to D). As with the spleen, WT and STING N153S lungs contained similar numbers of CD4+ T cells prior to infection (Fig. 6E and F), but STING N153S lungs exhibited diminished numbers of CD4+ T cells 14 days after infection (Fig. 6G and H).
FIG 6.
CD4+ T cells in the spleens and lungs of STING N153S mice. Mice were inoculated intranasally with 2 × 105 PFU of γHV68 or with PBS (mock) and were euthanized 14 days after infection. Splenocytes and pulmonary leukocytes were isolated for flow cytometric analysis. (A and B) Frequency (A) and number (B) of CD4+ cells in the spleens of mock-infected WT and STING N153S animals. (C and D) Frequency (C) and number (D) of CD4+ cells in the spleens of γHV68-infected WT and STING N153S animals. (E and F) Frequency (E) and number (F) of CD4+ cells in the lungs of mock-infected WT and STING N153S animals. (G and H) Frequency (G) and number (H) of CD4+ cells in the lungs of γHV68-infected WT and STING N153S animals. Data represent the means of results for 4 to 9 mice per group, pooled from at least two independent experiments. Results were analyzed by an unpaired t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
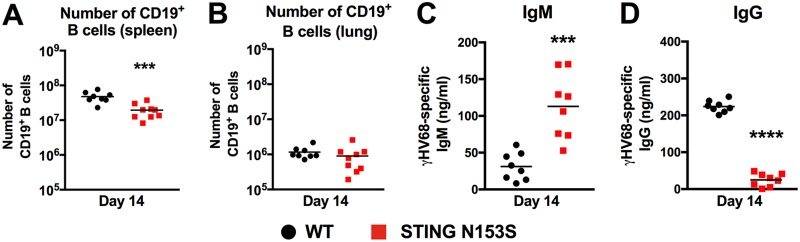
Humoral immunity limits γHV68 infection in the context of T cell cytopenia (25). Therefore, we reasoned that impaired antibody responses may also contribute to immunodeficiency in the T cell cytopenic STING N153S mice. On day 14 after infection, we observed a small decrease in the number of B cells in the spleen (Fig. 7A) and no effect on the number of B cells in the lung (Fig. 7B). However, evaluation of γHV68-specific antibody responses by enzyme-linked immunosorbent assay (ELISA) revealed that the STING N153S mice had ∼2-fold more γHV68-specific IgM and a 10-fold reduction in the level of γHV68-specific IgG 2 weeks after infection (Fig. 7C and D), despite much greater viral burdens in the STING N153S mice (Fig. 1). Low levels of virus-specific IgG are likely related to an impairment of class switching. Thus, in addition to disruptive effects on T cells, STING N153S also interferes with the virus-specific IgG response.
FIG 7.
B cell numbers and γHV68-specific IgM and IgG levels in WT and STING N153S mice. Mice were inoculated intranasally with 2 × 105 PFU of γHV68 and were euthanized 14 days after infection. Splenocytes and pulmonary leukocytes were isolated for flow cytometric analysis. (A and B) Total numbers of CD19+ cells in the spleens (A) and lungs (B) of STING N153S and WT mice 14 days after infection with γHV68. (C and D) Virus-specific IgM (C) and IgG (D) in the sera of STING N153S and WT mice 14 days after infection with γHV68. Data represent the means of results for 8 to 9 mice per group, pooled from two independent experiments. Results were analyzed by an unpaired t test. ***, P < 0.001; ****, P < 0.0001.
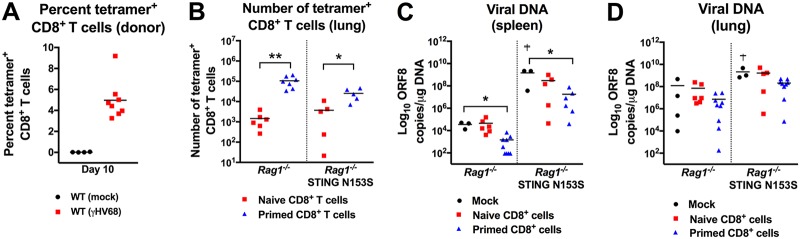
To determine whether WT CD8+ T cells might be sufficient to control γHV68 infection in STING N153S recipient mice, we crossed the STING N153S mice to Rag1−/− animals, which lack adaptive immunity. Next, we adoptively transferred equal numbers of naïve or primed antigen-specific WT CD8+ T cells into Rag1−/− and Rag1−/− STING N153S recipient mice. We confirmed the presence of γHV68-specific CD8+ T cells in the spleens of WT donor mice (Fig. 8A). Cells were pooled from multiple donors, and equal numbers of antigen-specific cells were transferred. The transfer of primed WT CD8+ T cells resulted in greater numbers of antigen-specific CD8+ T cells in the lungs of Rag1−/− mice and Rag1−/− STING N153S recipient mice than did the transfer of naïve WT CD8+ T cells (Fig. 8B). Furthermore, transfer of primed, but not naïve, CD8+ T cells resulted in diminished viral burdens in the spleens (Fig. 8C), and a trend toward diminished viral burdens in the lungs, of Rag1−/− and Rag1−/− STING N153S recipients (Fig. 8D). This effect of primed antigen-specific CD8+ T cells on viral burden occurred despite a survivor bias, since 2 out of 5 phosphate-buffered saline (PBS)-injected (mock) Rag1−/− STING N153S recipients died prior to day 14. However, despite the transfer of primed CD8+ T cells, the viral burdens in the spleens of Rag1−/− STING N153S recipients of WT CD8+ T cells were ∼10,000-fold higher than in those of Rag1−/− recipients (1.8 × 107 genome copies/μg in Rag1−/− STING N153S mice versus 1.5 × 103 genome copies/μg in Rag1−/− mice).
FIG 8.
Adoptive transfer of naïve and primed WT CD8+ T cells into Rag1−/− and Rag1−/− STING N153S recipient mice. (A to D) Adoptive transfer of PBS (mock), naïve WT CD8+ T cells, or primed WT CD8+ T cells. Splenocytes were enriched for CD8+ T cells via negative selection, and 5 × 105 CD8+ T cells were retro-orbitally transferred into intranasally infected Rag1–/– or Rag1–/– STING N153S recipients 1 day after inoculation with γHV68. Tissues were harvested 14 days after infection and viral burdens measured by qPCR. (A) Frequency of ORF6 tetramer-positive cells in the CD8+ population of WT donor mouse spleens on day 10 after infection with γHV68. (B) Numbers of ORF6 tetramer-positive CD8+ cells in the lungs of Rag1−/− STING N153S and Rag1−/− mice that received naïve or primed WT CD8+ T cells. Five to six mice per group were used. Data were analyzed by an unpaired t test. (C and D) Viral burdens measured by qPCR in the spleens (C) and lungs (D) of Rag1−/− STING N153S and Rag1−/− mice that received PBS, naïve WT CD8+ T cells, or primed WT CD8+ T cells. Daggers indicate survivor bias. Three to nine mice per group were used. Data were analyzed by the Kruskal-Wallis test. All data represent means pooled from at least 2 independent experiments. *, P < 0.05; **, P < 0.01.
STING N153S causes innate and adaptive immunodeficiency.
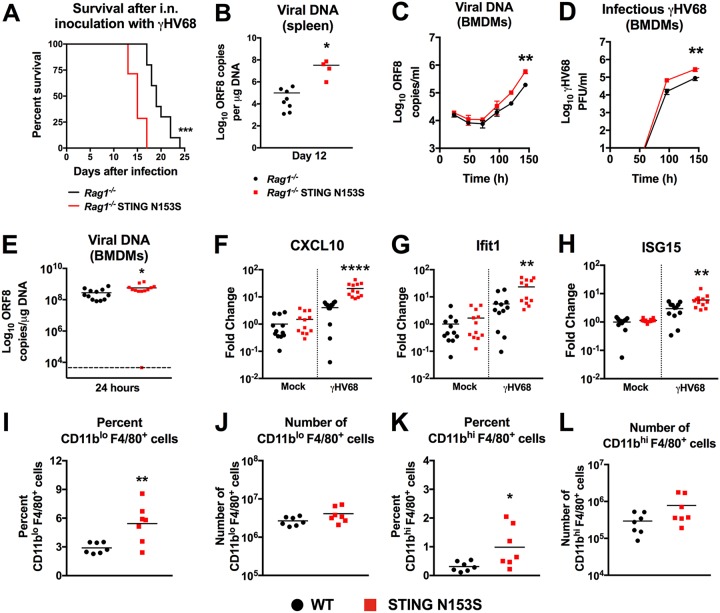
Without any adoptive transfer of CD8+ T cells, the Rag1−/− STING N153S mice were more vulnerable to lethal infection with γHV68 than Rag1−/− littermate control animals (Fig. 9A). Since Rag1−/− mice lack adaptive immunity, we reasoned that STING N153S may also cause an innate immunodeficiency. In further support of this hypothesis, we found higher levels of viral DNA in the spleens of Rag1−/− STING N153S mice than in those of their Rag1−/− littermates (Fig. 9B). Multistep viral growth curve analysis in WT and STING N153S bone marrow-derived macrophages (BMDMs) revealed a ∼5-fold increase in γHV68 replication in STING N153S BMDMs (Fig. 9C and D). Infection at a high multiplicity of infection (MOI) facilitates γHV68-mediated induction of proinflammatory cytokines (26, 27). STING N153S BMDMs contained ∼2-fold higher levels of viral DNA 24 h after infection at a high MOI (Fig. 9E). Infected STING N153S BMDMs also exhibited higher expression of ISGs than WT BMDMs, indicating that STING N153S macrophages retain the ability to activate antiviral ISGs (Fig. 9F to H). Enhanced replication of γHV68 in macrophages may contribute to the high expression of cytokines and ISGs in STING N153S mice. Furthermore, STING N153S causes expansion of myeloid cells in the spleen (7), and so we reasoned that there may be more cellular targets for infection in the STING N153S mice than in WT mice. However, flow cytometric analysis of splenic myeloid cells revealed no difference in the numbers of macrophages in WT and STING N153S spleens prior to infection (Fig. 9I to L), suggesting that differences in target cell numbers are unlikely to explain enhanced viral replication in Rag1−/− STING N153S mice. Thus, a single amino acid change on one STING allele is sufficient to cause severe innate and adaptive immunodeficiency in mice.
FIG 9.
Enhanced replication of γHV68 in STING N153S bone marrow-derived macrophages (BMDMs). (A) Kaplan-Meier curves showing 30-day mortality of 17- to 20-week-old Rag1−/− STING N153S mice and Rag1−/− littermates following intranasal inoculation with 2 × 105 PFU of γHV68. Shown are results for 7 to 10 mice per genotype, pooled from 2 independent experiments. The curves were analyzed by a log rank test. (B) γHV68 viral DNA levels in the spleens of Rag1−/− STING N153S mice and Rag1−/− littermates on day 12 after infection. (C) Multistep growth curve analysis of viral DNA levels in WT and STING N153S BMDMs after infection with γHV68 at an MOI of 0.05. (D) Multistep growth curve analysis of γHV68 in WT and STING N153S BMDMs after infection at an MOI of 0.05. Viral titers were measured by plaque assay with 2 wells per time point in 3 independent experiments. Data in panels C and D were analyzed by 2-way ANOVA. (E) Genome copies of ORF8 from BMDMs 24 h after infection with γHV68 at an MOI of 10. (F to H) ISG expression in WT and STING N153S BMDMs 24 h after infection with γHV68. Data in panels E to H were analyzed by the Mann-Whitney test. (I to L) Flow cytometric analysis and quantitation of total numbers of CD11blo F4/80+ and CD11bhi F4/80+ macrophages in the spleens of uninfected WT and STING N153S mice. All data represent the means for 6 to 12 biological replicates from at least 2 independent experiments. Data in panels B and I to L were analyzed by an unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
We found that a dominant gain-of-function mutation in a single STING allele creates a combination of severe immunodeficiency and immunopathology. We initially anticipated that the STING N153S mutation would enhance cell-intrinsic antiviral immunity by upregulating ISGs, thereby protecting mice against viruses. However, we found that this human disease-causing mutation in STING unexpectedly created a combined innate and adaptive immunodeficiency. Thus, aside from an established role in autoinflammation, our study of γHV68 pathogenesis in mice reveals that a dominant STING mutation impairs T cell responses, humoral immunity, and cell-intrinsic innate immunity.
How STING N153S disrupts T and B cell immunity remains to be fully elucidated. In addition to impairment of CD8+ T cell responses, we found that CD4+ T cell deficiency in the STING N153S mice coincides with impaired production of γHV68-specific IgG. A deficiency in the number of CD4+ T cells likely limits class switching and subsequent production of antiviral IgG, which controls γHV68 infection in the context of T cell cytopenia (25). Thus, a failure of humoral immunity may explain, at least in part, why adoptive transfer of primed CD8+ T cells into Rag1−/− STING N153S recipients was not sufficient to prevent severe infection.
STING agonists can trigger lymphocyte activation (28), proliferation defects (29), and apoptosis of certain lymphocyte subsets, including T cells (28). Our previously published experiments using mixed bone marrow chimeras demonstrated that T cell cytopenia in the STING N153S mice may result from a defect occurring at the early stages of T cell development, prior to the migration of progenitors to the thymus (7). Thus, the constitutively active STING N153S mutant may induce apoptosis, aberrant trafficking, or other dysregulation of lymphoid progenitor cells. Furthermore, high expression of the anti-inflammatory cytokine IL-10 in the spleen may blunt T cell responses (20, 21). Similarly, marked elevated expression of TGF-β in the lungs of STING N153S mice may simultaneously promote fibrosis and inhibit T cell responses (30–32). High expression of inhibitory cytokines may result from enhanced signaling of the STING N153S mutant in certain subsets of B cells, T cells, or macrophages (33, 34). Thus, in addition to causing T cell cytopenia, STING N153S may inhibit adaptive immune responses via cell-extrinsic effects in the spleen and the lung.
Type I IFN and ISGs protect against viral infections (2, 35, 36), but high expression of ISGs in the lungs of STING N153S mice did not protect against γHV68. In addition to disruption of adaptive immunity in STING N153S mice, our viral pathogenesis studies using Rag1−/− and Rag1−/− STING N153S animals revealed that STING N153S also causes an innate immunodeficiency. In mice, the innate immune defects resulting from STING N153S may include reduced numbers of innate immune cells (e.g., NK cells). However, NK cells are thought to be dispensable for antiviral immunity against γHV68 (37), although it remains plausible that the antiviral effects of NK cells might become relevant in the context of T cell cytopenia. Additionally, the STING N153S mutation led to enhanced replication of γHV68 in cultured BMDMs, demonstrating a deleterious effect of STING N153S on cell-intrinsic innate immunity, despite the fact that STING N153S macrophages efficiently upregulate type I IFN-stimulated genes. Type I IFN-independent effects of STING include the upregulation of endoplasmic reticulum (ER) stress-associated autophagy (38), as well as the activation of NF-κB (39), which can interact with TpI2/AP1 to promote lytic replication of γHV68 (40). Autophagy-related genes also promote lytic replication and reactivation of γHV68 from latency (41, 42). Thus, in addition to impacting type I IFN signaling, STING N153S may have other effects that promote γHV68 replication independently of type I IFN. Understanding how the STING gain-of-function mutation may dysregulate STING-related autophagy and apoptosis will be a topic of future study.
Viruses are known to trigger autoimmunity in genetically susceptible humans (43–48), so we hypothesized that a DNA virus may influence the lung disease phenotype in STING N153S mice. However, despite strong epidemiological associations between viruses and autoimmune diseases (43–48), there are few experimental demonstrations of pathogens exacerbating autoimmune disease in the context of a human mutation. Although many prior studies have examined the impact of γHV68 in immunodeficient knockout mice, including Ifngr1–/– mice (49–51), our report is the initial description of γHV68-induced lung disease in mice with a human genetic defect. Like a prior study of norovirus infection combined with a Crohn’s disease susceptibility allele (52), our characterization of γHV68-induced pulmonary fibrosis in STING N153S mice reveals how a human disease-related phenotype can be uncovered when a virus and a susceptibility gene are combined. Viral infections in humans with STING gain-of-function mutations may similarly have the ability to influence lung disease phenotypes.
STING N153S enhances the production of TGF-β and IL-6 in the lung even prior to infection, and these cytokines can promote pulmonary fibrosis (53–55). However, treatment with cidofovir prevented pneumonia and pulmonary fibrosis, indicating that lytic infection was required for lung pathology in the STING N153S mice. Prior work indicates that herpesvirus infections can induce immunopathology in humans, and an antiviral targeting another cytomegalovirus is currently undergoing a clinical trial for the treatment of idiopathic pulmonary fibrosis. Our animal model proves that a virus can induce lung disease in mice with a human disease-associated mutation and that virus-induced immunopathology in STING N153S mice can be prevented with an antiviral drug.
SAVI patients with gain-of-function STING mutations and T cell cytopenia are not thought to be immunodeficient, although some of these patients have had pneumonias, otitis media, and secondary infections, which led to the death of 2 out of 6 patients in the initial description of the disease (6). However, the etiology of these infections in a very limited number of SAVI patients is unclear, especially since these patients are treated with immunomodulatory and immunosuppressive medications that can increase the risk of infections (56). Thus, we must underscore that our studies with mice only suggest what may be possible in humans, and therefore, we cannot draw a conclusion at this point as to whether viruses may act as a trigger for immunopathology in SAVI patients.
In conclusion, our work demonstrates an unexpected role for a dominant STING mutation in causing a combined innate and adaptive immunodeficiency. STING is expressed in many cell types, including nonhematopoietic cells, and the STING N153S mutation is likely to cause a variety of cell-type-specific effects that create this combination of immunological abnormalities. Therefore, future studies using cell-type-specific STING N153S knock-in mice will help to more precisely define how a dominant gain-of-function STING mutant simultaneously triggers autoinflammation and immunodeficiency in mice.
MATERIALS AND METHODS
Design.
The goal of our study was to use viruses to interrogate innate and adaptive immune responses in STING N153S mice. Power analysis was conducted for Institutional Animal Care and Use Committee-approved in vivo studies in order to determine the number of animals needed per experimental group. At least two independent experiments were conducted to replicate findings. WT littermate control animals were used in all studies of STING N153S mice, and no outliers were excluded from analyses. The number of replicates for each experiment is given in the figure legends.
Study approvals.
Animal protocols were approved by the Institutional Animal Care and Use Committees at the Washington University School of Medicine (assurance no. A-3381-01).
Mice.
Mice were housed in specific-pathogen-free mouse facilities at the Washington University School of Medicine. STING N153S mice were generated on a C57BL/6 background using CRISPR/Cas9 as described previously (7). In all experiments using STING N153S mice, WT littermates were used as controls. Ifngr1−/− mice were obtained from The Jackson Laboratory (stock no. 003288; B6.129S7-Ifngr1tm1Agt/J), as were Rag1−/− mice (stock no. 002216; B6.129S7-Rag1tm1Mom/J) and STING goldenticket mice (TMEM173gt/gt) (stock no. 017537; C57BL/6J-Tmem173gt/J). Ifnar1−/− mice (57) were backcrossed onto a C67BL/6 background. Age-matched WT controls on congenic backgrounds were purchased from The Jackson Laboratory as controls for strains other than the STING N153S mouse line.
Viruses.
The γHV68 strain WUMS (ATCC VR1465) was originally isolated from the organs of a bank vole in Czechoslovakia in 1976 (58). The γHV68 strain was passaged and viral titers measured by infectious plaque assay in 3T12 cells (ATCC CCL-164). The West Nile virus (WNV) strain (3000.0259) was isolated in New York in 2000 and was passaged twice in C6/36 Aedes albopictus cells (59), followed by measurement of viral titers by focus-forming assay as described previously (60).
Viral pathogenesis studies.
For γHV68 survival analysis, mice were inoculated either intraperitoneally, with one dose of 1 × 106 PFU of γHV68 delivered in 200 μl of PBS, or intranasally, with two doses of 1 × 105 PFU of γHV68 for a total dose of 2 × 105 PFU as described previously (61). For WNV survival analysis, mice were inoculated subcutaneously via footpad injection with 100 focus-forming units (FFU) diluted in 50 μl of PBS.
Viral burden analysis.
Lungs and spleens were weighed and homogenized in 500 μl (lungs) or 200 μl (spleens) of PBS, using zirconia beads and a MagNA lyser (Roche Life Sciences). Total DNA was isolated from the homogenate using the DNeasy Blood and Tissue kit (catalog no. 69504; Qiagen). Genome copies of γHV68 were quantified using AmpliTaq Gold DNA polymerase (catalog no. N8080241; Thermo Fisher) and PrimeTime (Integrated DNA Technologies) quantitative PCR (qPCR) assays for ORF8 as described previously (62). Standard curves for qPCR assays were used for absolute quantification of genome copy numbers. Infectious γHV68 was measured using plaque assays as described previously (26, 63).
DNase treatment of γHV68-infected serum.
Sera from infected animals were incubated with 5 U of DNase I (Thermo Fisher, USA) for 1 h at 37°C and were processed for DNA extraction with the DNeasy Blood and Tissue kit (Qiagen) as per the manufacturer’s protocol. For control samples, γHV68 DNA was spiked into uninfected sera of WT mice. DNA was extracted from spike serum after DNase or mock treatment and γHV68 DNA levels measured by qPCR.
Tissue collection and histological analysis.
Infected and uninfected mice were anesthetized with tribromoethanol (Avertin) at specified time points, and blood was collected via terminal exsanguination. Animals were perfused with 20 ml of PBS and tissues harvested for histological or molecular analysis. Tissues for cytokine and RNA analysis were immediately placed on dry ice and stored at −80°C. For histological analysis, lungs were immediately fixed in 4% paraformaldehyde (PFA) at room temperature. After 48 h, tissues were washed, resuspended in 70% ethanol, and embedded in paraffin. Tissues were sectioned and were stained with hematoxylin and eosin or Gomori trichrome, followed by image acquisition using a Nikon Eclipse E400 microscope and NIS Elements software. The histological pattern of each ×20 field for the entire trichrome-stained slide was documented by a blinded histologist and was reported as a percentage of total fields, with at least 10 fields analyzed per section.
Cidofovir treatment of γHV68-infected mice.
Cidofovir (catalog no. S1516; Selleck Chemicals) was diluted in PBS to a concentration of 6.25 mg/ml and was administered by intraperitoneal injection at a dose of 2 5 mg/kg of body weight. Mice received cidofovir on days 4, 5, 8, and 11 after infection (∼100 μl per mouse), and control animals received an equivalent volume of PBS. Individual mice were weighed every 2 days after infection, and the most recent weight was used to calculate each cidofovir dose. Mice were euthanized 14 days after infection and lungs harvested for viral burden analysis and histology.
Studies of BMDMs.
Bone marrow-derived macrophages (BMDMs) were generated from bone marrow cells flushed from the femurs and tibias of gender- and age-matched donor mice. Then 2.5 × 106 cells were incubated at 37°C in 10-cm cell culture plates in complete medium containing Dulbecco’s modified Eagle medium (DMEM) (catalog no. D6429; Sigma), 10% fetal bovine serum (FBS) (catalog no. SH30070-03; HyClone), 1% penicillin and streptomycin (catalog no. 15140; Gibco), 1% sodium pyruvate (catalog no. 11360; Gibco), and 1% l-glutamate (catalog no. 35050; Gibco), supplemented with 40 ng/ml of macrophage colony-stimulating factor (M-CSF) (catalog no. 300-25; PeproTech), for 6 days. On day 7, BMDMs were replated and thereafter were cultured in complete medium with 20 ng/ml M-CSF. Twenty-four hours after replating in 12-well plates, BMDMs were infected with γHV68 at an MOI of 10. Cells were harvested 24 h after infection for analysis by a TaqMan qRT-PCR assay.
Viral growth curve analysis.
Mouse embryonic fibroblasts (MEFs) were generated from 14-day-old WT, STING N153S, and STING GT fetuses and were cultured at 37°C under 5% CO2 in DMEM containing 10% FBS, 1% HEPES buffer, and 1% penicillin-streptomycin solution (50 U/ml). For γHV68 growth curves, 3 × 105 MEFs per well were seeded into 6-well plates and were infected at an MOI of 0.05. Supernatants were collected at 24, 48, 72, 96, and 120 h after infection and were then analyzed by plaque assay as described previously (26, 63). For γHV68 growth curves in BMDMs, 1 × 105 cells were seeded into 24-well plates. After 12 h, cells were infected with γHV68 at an MOI of 0.05 for 1 h at 37°C. Supernatants were collected at 24, 48, 96, 144, and 196 h after infection, and infectious virus was quantified by plaque assay.
Virus-specific antibody ELISAs.
γHV68-specific IgG and IgM ELISAs were performed as described previously (64). Briefly, γHV68 was diluted in PBS and was used to coat Nunc MaxiSorp flat-bottom 96-well plates (Thermo Fisher, USA) for 12 h. For standard curves, plates were coated with serially diluted mouse IgG (Sigma-Aldrich, USA) or IgM (EMD Millipore) in duplicate. Wells were blocked with 1% bovine serum albumin in PBS for 1 h at room temperature and were washed three times with wash buffer (PBS–0.1% Tween) before the addition of samples. Appropriate dilutions of serum samples from infected mice were made in ELISA diluent and were plated in wells coated with virus. Plates were incubated for 2 h at room temperature. Wells were washed three times, and bound antibody was detected with horseradish peroxidase-conjugated IgG and IgM (Sigma-Aldrich, USA) antibodies. Plates were incubated for 2 h at room temperature, washed three times, and incubated with 50 μl of 1× TMB ELISA substrate (Thermo Fisher, USA). Reactions were stopped after 10 min with 25 μl of 2 N H2SO4 (Sigma-Aldrich, USA) per well. Plates were analyzed on a BioTek Synergy 2 Multi Detection microplate reader (BioTek) using Gen5 software (BioTek).
Gene expression analysis.
Total RNAs from lung homogenates, spleen homogenates, and BMDMs were isolated using the RNeasy kit (Qiagen) per the manufacturer’s protocol. The TaqMan RNA-to-Ct 1-Step kit (Applied Biosystems) was used to measure mRNA expression. Primer and probe assays were obtained from Integrated DNA Technologies. ΔΔCT values were calculated and then normalized to the values for the mock-infected WT samples.
Multiplex cytokine analysis.
Lungs were harvested and homogenized in 500 μl of PBS. Cytokine and chemokine levels were measured on the Luminex platform using the Bio-Plex Pro Mouse Cytokine Group I Panel 23-plex assay kit (Bio-Rad). Cytokines and chemokines tested included IL-1α, IL-1β, IL-2, IL-3 IL-4, IL-5, IL-6, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-17, eotaxin, G-CSF, granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, MIP-Iα, MIP-1β, RANTES (CCL5), and TNF.
Measurement of antigen-specific CD8+ T cell responses.
Spleens and lungs were harvested 14 days after infection following cardiac perfusions with 20 ml of PBS. Spleens were crushed and filtered through a 70-μm cell strainer to generate single-cell suspensions. Lungs were digested using a MACS lung dissociation kit (catalog no. 130095927; Miltenyi Biotech) and were enriched for leukocytes using a Percoll gradient. Tetramers that bind to T cell receptors specific to the γHV68 ORF6-derived peptide AGPHNDMEI (65) were generated by the Andrew and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs (CHiiPs) for staining of antigen-specific T cells. Intracellular IFN-γ and TNF-α staining was performed after ex vivo stimulation with the same ORF6-derived peptide for 4 h in the presence of brefeldin A (Sigma). Cells were stained with the following antibodies and were processed by multicolor flow cytometry on a BD FACSCanto or LSRFortessa X-20 system: CD4 (catalog no. 100437; BioLegend), CD8α (catalog no. 100734; BioLegend), CD8β (catalog no. 126609; BioLegend), CD19 (catalog no. 115528; BioLegend), CD45 (catalog no. 103139; BioLegend), IFN-γ (catalog no. 51731182; eBioscience), and TNF-α (catalog no. 506306; BioLegend).
Adoptive transfer of CD8+ T cells.
For primed CD8+ T cell transfers, spleens were harvested from WT mice 10 days after intranasal inoculation with 2 × 105 PFU of γHV68. For naïve CD8+ T cell transfer, spleens were isolated from naïve WT mice. Single-cell suspensions were obtained after the disruption of tissue through a 70-μm filter. After erythrocyte lysis in ammonium-chloride-potassium (ACK) buffer, splenocytes were washed in RPMI medium containing 10% fetal bovine serum and 0.1% 2-mercaptoethanol. CD8+ T cells were enriched by negative selection with magnetic beads from Miltenyi Biotec according to the manufacturer’s protocol (CD8α+ T cell isolation kit, mouse; catalog no. 130-104-075). Purity was assessed by flow cytometry. One day after intranasal inoculation with γHV68, 5 × 105 CD8+ T cells were transferred to Rag1–/– STING N153S or Rag1–/– animals via intravenous injection. Lungs were harvested from recipient animals for viral burden assessment 14 days after infection (13 days after the transfer of CD8+ T cells).
Statistical analysis.
Unless otherwise specified, all data were analyzed using GraphPad Prism software as specified in the figure legends. Flow cytometry data were analyzed using Cytobank or FlowJo, v10.4.1.
ACKNOWLEDGMENTS
We thank the Immunomonitoring Laboratory of the Andrew M. and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs at Washington University for assistance with the generation of the γHV68 ORF6 tetramer. We also acknowledge the Washington University Pulmonary Morphology Core for assistance with tissue processing and staining.
H.I. is supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (MI-F-2018-712). D.J.P. is supported by the Washington University Chancellors Graduate Fellowship Program and the Initiative to Maximize Student Development. The Miner laboratory is supported by grants from the NIH (K08AR070918) and the Rheumatology Research Foundation. The Baldridge laboratory is supported by grants from the NIH (K22AI127846).
B.G.B., H.I., T.L.A., C.A.M., D.J.P., and A.M.S. performed experiments and analyzed data. B.G.B., H.I., and T.L.A. wrote portions of the initial manuscript and edited subsequent versions of the manuscript. M.T.B. guided experiments and edited the manuscript. J.J.M. conceived the project, performed experiments, wrote the initial complete version of the manuscript, analyzed data, and edited subsequent versions of the manuscript.
We declare no competing interests.
REFERENCES
- 1.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi H, Kobayashi CI, Nakamura-Ishizu A, Karigane D, Haeno H, Yamamoto KN, Sato T, Ohteki T, Hayakawa Y, Barber GN, Kurokawa M, Suda T, Takubo K. 2015. Bacterial c-di-GMP affects hematopoietic stem/progenitors and their niches through STING. Cell Rep 11:71–84. doi: 10.1016/j.celrep.2015.02.066. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signaling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee C-CR, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R. 2014. Activated STING in a vascular and pulmonary syndrome. N Engl J Med 371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner JD, Irizarry-Caro RA, Bennion BG, Ai TL, Smith AM, Miner CA, Sakai T, Gonugunta VK, Wu J, Platt DJ, Yan N, Miner JJ. 2017. STING-associated vasculopathy develops independently of IRF3 in mice. J Exp Med 214:3279–3292. doi: 10.1084/jem.20171351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 2015. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nice TJ, Osborne LC, Tomov VT, Artis D, Wherry EJ, Virgin HW. 2016. Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog 12:e1005684. doi: 10.1371/journal.ppat.1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang K, Wang J, Wu M, Li M, Wang Y, Huang X. 2015. Mesenchymal stem cells detect and defend against gammaherpesvirus infection via the cGAS-STING pathway. Sci Rep 5:7820. doi: 10.1038/srep07820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schock SN, Chandra NV, Sun Y, Irie T, Kitagawa Y, Gotoh B, Coscoy L, Winoto A. 2017. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ 24:615–625. doi: 10.1038/cdd.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutia BM, Clarke CJ, Allen DJ, Nash AA. 1997. Pathological changes in the spleens of gamma interferon receptor-deficient mice infected with murine gammaherpesvirus: a role for CD8 T cells. J Virol 71:4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehtisham S, Sunil-Chandra NP, Nash AA. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J Virol 67:5247–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gredmark-Russ S, Cheung EJ, Isaacson MK, Ploegh HL, Grotenbreg GM. 2008. The CD8 T-cell response against murine gammaherpesvirus 68 is directed toward a broad repertoire of epitopes from both early and late antigens. J Virol 82:12205–12212. doi: 10.1128/JVI.01463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You F, Wang P, Yang L, Yang G, Zhao YO, Qian F, Walker W, Sutton R, Montgomery R, Lin R, Iwasaki A, Fikrig E. 2013. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat Immunol 14:1237–1246. doi: 10.1038/ni.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ, Metchette K, Dubensky TW Jr, Gajewski TF. 2015. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep 11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, Villunger A, Radtke F, Ablasser A. 2017. Signalling strength determines proapoptotic functions of STING. Nat Commun 8:427. doi: 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Flaherty BM, Matar CG, Wakeman BS, Garcia A, Wilke CA, Courtney CL, Moore BB, Speck SH. 2015. CD8+ T cell response to gammaherpesvirus infection mediates inflammation and fibrosis in interferon gamma receptor-deficient mice. PLoS One 10:e0135719. doi: 10.1371/journal.pone.0135719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorentino DF, Zlotnik A, Vieira P, Mosmann TR, Howard M, Moore KW, O’Garra A. 1991. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J Immunol 146:3444–3451. [PubMed] [Google Scholar]
- 21.Taga K, Tosato G. 1992. IL-10 inhibits human T cell proliferation and IL-2 production. J Immunol 148:1143–1148. [PubMed] [Google Scholar]
- 22.Weck KE, Kim SS, Virgin HI, Speck SH. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J Virol 73:3273–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan CSE, Lawler C, Stevenson PG. 2017. CD8+ T cell evasion mandates CD4+ T cell control of chronic gamma-herpesvirus infection. PLoS Pathog 13:e1006311. doi: 10.1371/journal.ppat.1006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuller KA, Flano E. 2009. CD4 T cells mediate killing during persistent gammaherpesvirus 68 infection. J Virol 83:4700–4703. doi: 10.1128/JVI.02240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim IJ, Flano E, Woodland DL, Blackman MA. 2002. Antibody-mediated control of persistent gamma-herpesvirus infection. J Immunol 168:3958–3964. doi: 10.4049/jimmunol.168.8.3958. [DOI] [PubMed] [Google Scholar]
- 26.Cieniewicz B, Dong Q, Li G, Forrest JC, Mounce BC, Tarakanova VL, van der Velden A, Krug LT. 2015. Murine gammaherpesvirus 68 pathogenesis is independent of caspase-1 and caspase-11 in mice and impairs interleukin-1β production upon extrinsic stimulation in culture. J Virol 89:6562–6574. doi: 10.1128/JVI.00658-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, O’Hare P, Adler H, Sun R, Ploegh HL, Damania B, Upton JW, Fitzgerald KA, Paludan SR. 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin B, Ilyukha V, Sorokin M, Buzdin A, Vannier E, Poltorak A. 2017. Activation of STING in T cells induces type I IFN responses and cell death. J Immunol 199:397–402. doi: 10.4049/jimmunol.1601999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerboni S, Jeremiah N, Gentili M, Gehrmann U, Conrad C, Stolzenberg M-C, Picard C, Neven B, Fischer A, Amigorena S, Rieux-Laucat F, Manel N. 2017. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J Exp Med 214:1769–1785. doi: 10.1084/jem.20161674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Border WA, Noble NA. 1994. Transforming growth factor beta in tissue fibrosis. N Engl J Med 331:1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 31.Naik PN, Horowitz JC, Moore TA, Wilke CA, Toews GB, Moore BB. 2012. Pulmonary fibrosis induced by gamma-herpesvirus in aged mice is associated with increased fibroblast responsiveness to transforming growth factor-beta. J Gerontol A Biol Sci Med Sci 67:714–725. doi: 10.1093/gerona/glr211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, Du Bois RM, Borok Z. 2005. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 166:1321–1332. doi: 10.1016/S0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosser EC, Mauri C. 2015. Regulatory B cells: origin, phenotype, and function. Immunity 42:607–612. doi: 10.1016/j.immuni.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Saraiva M, O'Garra A. 2010. The regulation of IL-10 production by immune cells. Nat Rev Immunol 10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Sastre A, Biron CA. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 36.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usherwood EJ, Meadows SK, Crist SG, Bellfy SC, Sentman CL. 2005. Control of murine gammaherpesvirus infection is independent of NK cells. Eur J Immunol 35:2956–2961. doi: 10.1002/eji.200526245. [DOI] [PubMed] [Google Scholar]
- 38.Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G, Green DR, Blander JM. 2017. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 171:809–823.e13. doi: 10.1016/j.cell.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abe T, Barber GN. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol 88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Feng J, Chen S, Peng L, He WW, Qi J, Deng H, Sun R. 2010. Tpl2/AP-1 enhances murine gammaherpesvirus 68 lytic replication. J Virol 84:1881–1890. doi: 10.1128/JVI.01856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S, Buck MD, Desai C, Zhang X, Loginicheva E, Martinez J, Freeman ML, Saitoh T, Akira S, Guan JL, He YW, Blackman MA, Handley SA, Levine B, Green DR, Reese TA, Artyomov MN, Virgin HW. 2016. Autophagy genes enhance murine gammaherpesvirus 68 reactivation from latency by preventing virus-induced systemic inflammation. Cell Host Microbe 19:91–101. doi: 10.1016/j.chom.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou XC, Dong SH, Liu ZS, Liu S, Zhang CC, Liang XZ. 2018. Regulation of gammaherpesvirus lytic replication by endoplasmic reticulum stress-induced transcription factors ATF4 and CHOP. J Biol Chem 293:2801–2814. doi: 10.1074/jbc.M117.813675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon BJ, Heyward WL, Templin DW, Clement D, Lanier AP. 1989. Hepatitis B-associated polyarteritis nodosa in Alaskan Eskimos: clinical and epidemiologic features and long-term follow-up. Hepatology 9:97–101. doi: 10.1002/hep.1840090116. [DOI] [PubMed] [Google Scholar]
- 44.Revenga Arranz F, Díaz Díaz R, Iglesias Díez L, Cassis Herce B, Sánchez Gómez F, Fuertes Ortiz A. 1995. Cryoglobulinemic vasculitis associated with hepatitis C virus infection. A report of eight cases. Acta Derm Venereol 75:234–236. [DOI] [PubMed] [Google Scholar]
- 45.James JA, Neas BR, Moser KL, Hall T, Bruner GR, Sestak AL, Harley JB. 2001. Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum 44:1122–1126. doi:. [DOI] [PubMed] [Google Scholar]
- 46.Kelly BG, Lok SS, Hasleton PS, Egan JJ, Stewart JP. 2002. A rearranged form of Epstein-Barr virus DNA is associated with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 166:510–513. doi: 10.1164/rccm.2103058. [DOI] [PubMed] [Google Scholar]
- 47.Tang YW, Johnson JE, Browning PJ, Cruz-Gervis RA, Davis A, Graham BS, Brigham KL, Oates JA Jr, Loyd JE, Stecenko AA. 2003. Herpesvirus DNA is consistently detected in lungs of patients with idiopathic pulmonary fibrosis. J Clin Microbiol 41:2633–2640. doi: 10.1128/JCM.41.6.2633-2640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClain MT, Poole BD, Bruner BF, Kaufman KM, Harley JB, James JA. 2006. An altered immune response to Epstein-Barr nuclear antigen 1 in pediatric systemic lupus erythematosus. Arthritis Rheum 54:360–368. doi: 10.1002/art.21682. [DOI] [PubMed] [Google Scholar]
- 49.Ebrahimi B, Dutia BM, Brownstein DG, Nash AA. 2001. Murine gammaherpesvirus-68 infection causes multi-organ fibrosis and alters leukocyte trafficking in interferon-gamma receptor knockout mice. Am J Pathol 158:2117–2125. doi: 10.1016/S0002-9440(10)64683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mora AL, Woods CR, Garcia A, Xu J, Rojas M, Speck SH, Roman J, Brigham KL, Stecenko AA. 2005. Lung infection with gamma-herpesvirus induces progressive pulmonary fibrosis in Th2-biased mice. Am J Physiol Lung Cell Mol Physiol 289:L711–L721. doi: 10.1152/ajplung.00007.2005. [DOI] [PubMed] [Google Scholar]
- 51.Weck KE, Dal Canto AJ, Gould JD, O’Guin AK, Roth KA, Saffitz JE, Speck SH, Virgin HW. 1997. Murine gamma-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat Med 3:1346–1353. doi: 10.1038/nm1297-1346. [DOI] [PubMed] [Google Scholar]
- 52.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le TT, Karmouty-Quintana H, Melicoff E, Le TT, Weng T, Chen NY, Pedroza M, Zhou Y, Davies J, Philip K, Molina J, Luo F, George AT, Garcia-Morales LJ, Bunge RR, Bruckner BA, Loebe M, Seethamraju H, Agarwal SK, Blackburn MR. 2014. Blockade of IL-6 trans signaling attenuates pulmonary fibrosis. J Immunol 193:3755–3768. doi: 10.4049/jimmunol.1302470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito F, Tasaka S, Inoue K, Miyamoto K, Nakano Y, Ogawa Y, Yamada W, Shiraishi Y, Hasegawa N, Fujishima S, Takano H, Ishizaka A. 2008. Role of interleukin-6 in bleomycin-induced lung inflammatory changes in mice. Am J Respir Cell Mol Biol 38:566–571. doi: 10.1165/rcmb.2007-0299OC. [DOI] [PubMed] [Google Scholar]
- 55.Biernacka A, Dobaczewski M, Frangogiannis NG. 2011. TGF-beta signaling in fibrosis. Growth Factors 29:196–202. doi: 10.3109/08977194.2011.595714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seo J, Kang J-A, Suh DI, Park E-B, Lee C-R, Choi SA, Kim SY, Kim Y, Park S-H, Ye M, Kwon S-H, Park JD, Lim BC, Lee DH, Kang S-J, Choi M, Park S-G, Chae J-H. 2017. Tofacitinib relieves symptoms of stimulator of interferon genes (STING)–associated vasculopathy with onset in infancy caused by 2 de novo variants in TMEM173. J Allergy Clin Immunol 139:1396–1399.e12. doi: 10.1016/j.jaci.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 57.Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. 1995. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci U S A 92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol 24:468. [PubMed] [Google Scholar]
- 59.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. 2004. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am J Trop Med Hyg 71:493–500. doi: 10.4269/ajtmh.2004.71.493. [DOI] [PubMed] [Google Scholar]
- 60.Miner JJ, Daniels BP, Shrestha B, Proenca-Modena JL, Lew ED, Lazear HM, Gorman MJ, Lemke G, Klein RS, Diamond MS. 2015. The TAM receptor Mertk protects against neuroinvasive viral infection by maintaining blood-brain barrier integrity. Nat Med 21:1464–1472. doi: 10.1038/nm.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee BJ, Santee S, Von Gesjen S, Ware CF, Sarawar SR. 2000. Lymphotoxin-alpha-deficient mice can clear a productive infection with murine gammaherpesvirus 68 but fail to develop splenomegaly or lymphocytosis. J Virol 74:2786–2792. doi: 10.1128/JVI.74.6.2786-2792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coomes SM, Farmen S, Wilke CA, Laouar Y, Moore BB. 2011. Severe gammaherpesvirus-induced pneumonitis and fibrosis in syngeneic bone marrow transplant mice is related to effects of transforming growth factor-beta. Am J Pathol 179:2382–2396. doi: 10.1016/j.ajpath.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krug LT, Collins CM, Gargano LM, Speck SH. 2009. NF-κB p50 plays distinct roles in the establishment and control of murine gammaherpesvirus 68 latency. J Virol 83:4732–4748. doi: 10.1128/JVI.00111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gargano LM, Moser JM, Speck SH. 2008. Role for MyD88 signaling in murine gammaherpesvirus 68 latency. J Virol 82:3853–3863. doi: 10.1128/JVI.02577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freeman ML, Lanzer KG, Cookenham T, Peters B, Sidney J, Wu TT, Sun R, Woodland DL, Sette A, Blackman MA. 2010. Two kinetic patterns of epitope-specific CD8 T-cell responses following murine gammaherpesvirus 68 infection. J Virol 84:2881–2892. doi: 10.1128/JVI.02229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]