Marek’s disease (MD) is a devastating oncogenic disease that affects the poultry industry and is caused by MD alphaherpesvirus (MDV). Current vaccines block induction of disease but do not block chicken-to-chicken transmission. There is a knowledge gap in our understanding of how MDV spreads from chicken to chicken. We studied the Herpesviridae conserved ICP27 regulatory protein in cell culture and during MDV infection in chickens. We determined that MDV ICP27 is important but not required for replication in both cell culture and chickens. In addition, MDV ICP27 was not required for disease induction or oncogenicity but was required for chicken-to-chicken transmission. This study is important because it addresses the role of ICP27 during infection in the natural host and provides important information for the development of therapies to protect chickens against MD.
KEYWORDS: ICP27, herpesvirus, oncogenic virus, replication, transmission
ABSTRACT
The Herpesviridae conserved infected-cell protein 27 (ICP27) is essential for cell culture-based replication of most herpesviruses studied. For members of the Alphaherpesvirinae, ICP27 regulates the expression of many viral genes, including expression of pUL44 (gC), pUL47 (VP13/14), and pUL48 (VP16). These three viral proteins are dysregulated during Marek’s disease alphaherpesvirus (MDV) replication in cell culture. MDV replicates in a highly cell-associated manner in cell culture, producing little to no infectious virus. In contrast, infectious cell-free MDV is produced in specialized feather follicle epithelial (FFE) cells of infected chickens, in which these three genes are abundantly expressed. This led us to hypothesize that MDV ICP27, encoded by gene UL54, is a defining factor for the dysregulation of gC, pUL47, and pUL48 and, ultimately, ineffective virus production in cell culture. To address ICP27’s role in MDV replication, we generated recombinant MDV with ICP27 deleted (vΔ54). Interestingly, vΔ54 replicated, but plaque sizes were significantly reduced compared to those of parental viruses. The reduced cell-to-cell spread was due to ICP27 since plaque sizes were restored in rescued viruses, as well as when vΔ54 was propagated in cells expressing ICP27 in trans. In chickens, vΔ54 replicated, induced disease, and was oncogenic but was unable to transmit from chicken to chicken. To our knowledge, this is the first report showing that the Herpesviridae conserved ICP27 protein is dispensable for replication and disease induction in its natural host.
IMPORTANCE Marek’s disease (MD) is a devastating oncogenic disease that affects the poultry industry and is caused by MD alphaherpesvirus (MDV). Current vaccines block induction of disease but do not block chicken-to-chicken transmission. There is a knowledge gap in our understanding of how MDV spreads from chicken to chicken. We studied the Herpesviridae conserved ICP27 regulatory protein in cell culture and during MDV infection in chickens. We determined that MDV ICP27 is important but not required for replication in both cell culture and chickens. In addition, MDV ICP27 was not required for disease induction or oncogenicity but was required for chicken-to-chicken transmission. This study is important because it addresses the role of ICP27 during infection in the natural host and provides important information for the development of therapies to protect chickens against MD.
INTRODUCTION
Infected-cell protein 27 (ICP27) is an immediate early (IE) multifunctional regulatory protein that is conserved throughout the Herpesviridae. A primary function of ICP27 homologs is regulation of delayed early (DE) and late (L) gene expression. This occurs through several mechanisms, including promotion of transcription through interacting with the C terminus of RNA polymerase II (1, 2), blocking of mRNA splicing through interaction with splicing factors (3–6), and export of viral mRNAs from the nucleus to the cytoplasm through binding to G/C-rich RNA sequences (7). ICP27 is predominately localized to the nucleus but also shuttles to the cytoplasm (8, 9). For most herpesviruses studied to date, ICP27 homologs are essential for growth in tissue culture (reviewed in reference 10); however, this is dependent on the virus. For example, the human herpes simplex virus 1 (HSV-1) ICP27 ortholog, UL54, is necessary for virus replication in cell culture (11, 12). In contrast, deletion mutants of the human cytomegalovirus (HCMV) ICP27 ortholog, UL69, replicate but have severe growth defects (13–15). Likewise, an ICP27-null (vJSΔ54) variant of pseudorabies virus (PRV) is viable but exhibits a slow-growth phenotype in cell culture, depending on the cell type infected (16). PRV vJSΔ54 is attenuated in mice, with mice surviving twice as long as mice inoculated with wild-type PRV. Apart from this work, little is known about the role ICP27 orthologs play during in vivo replication and pathogenesis for other herpesviruses.
Gallid alphaherpesvirus 2 (GaHV-2), better known as Marek’s disease (MD) virus (MDV), is an oncogenic alphaherpesvirus that transforms T cells presenting as solid lymphomas in the viscera and other organs and induces neurological symptoms like ataxia and torticollis. According to our current understanding, infection begins in the respiratory tract by inhalation of MDV shed from infected chickens. MDV initiates infection in macrophages and B cells in the lungs (17, 18) and is then transported to lymphoid organs, where primary cytolytic infection occurs in T cells (17, 18). MDV maintains latency and can induce oncogenic transformation of these cells, ultimately resulting in lymphoma formation and death of the host. To disseminate into the environment, migrating infected immune cells transport MDV to feather follicle (FF) epithelial (FFE) cells in the skin, where infectious virus is shed into the environment, and the virus life cycle is repeated in naive chickens. This process is similar to that of human varicella-zoster virus (19).
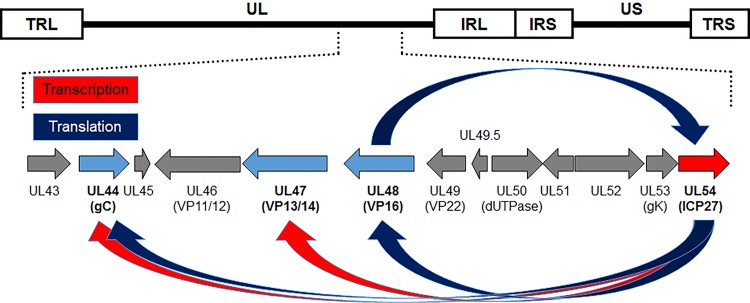
One of the objectives of our laboratory is to identify herpesviral genes required for replication and host-to-host transmission that could be targeted in blocking the spread of herpesviruses in a population. Current vaccines against MD do not block chicken-to-chicken transmission of MDV, resulting in increased MD virulence over the decades (20, 21). Cell culture propagation of MDV does not result in the production of infectious cell-free virus, relying exclusively on cell-to-cell spread (22–24), while fully infectious virus is produced in FFE cells of the skin (25). The generation of infectious cell-free virus is believed to be required for interindividual spread from chicken to chicken (20). Very little is known about the maturation of MD viral particles in cell culture and the shedding of infectious virus from FFE cells. We have identified a number of viral genes that either are expressed at low levels or do not appear to function properly that could explain MDV’s inability to produce infectious cell-free virus in cell culture. Following identification of these genes—which includes the Alphaherpesvirinae conserved pUL44 (glycoprotein C [gC]), pUL47 (VP13/14), and pUL48 (VP16), which are dysregulated in cell culture (26–28)—a common theme evolved that these genes are expected to be regulated by ICP27 (16, 29–31). In particular, the long-known fact that MDV gC mRNA is primarily spliced in cell culture, resulting in secreted gC (26, 32, 33), suggests that ICP27, known to inhibit HSV-1 gC splicing (34, 35), may be linked to this phenomenon. Additionally, expression of both pUL47 and pUL48 is severely deficient in cell culture relative to replication in FFE cells in situ (27, 28), and at least for HSV-1, ICP27 has been shown to be important for transcriptional and translation regulation of these genes (29, 31). Together, our previously published data (16, 29–31) led us to hypothesize that MDV ICP27 is a major factor in the dysregulation of gC, pUL47, and pUL48 and, ultimately, the lack of infectious MD virion production in cell culture (Fig. 1). Since an ICP27 (UL54)-null MDV had not been described in the literature, we began our studies to test the importance of MDV ICP27 for replication in cell culture, in chickens, and on regulation of gC in cell culture.
FIG 1.
ICP27 regulates pUL44 (gC), pUL47, and pUL48 at the transcriptional and translational levels. Schematic representation of the MDV genome depicting the locations of the terminal repeat long (TRL) and short (TRS), internal repeat long (IRL) and short (IRS), and unique long (UL) and short (US) regions. Previous work in other alphaherpesvirus systems showed that ICP27 transcriptionally and translationally regulates pUL44 (gC), pUL47, and pUL48 (16, 29–31). We have shown that these three genes (blue) are dysregulated during MDV replication in cell culture (26–28), leading us to hypothesize that ICP27 is responsible for their dysregulation.
RESULTS
Generation of UL54-null rMDV.
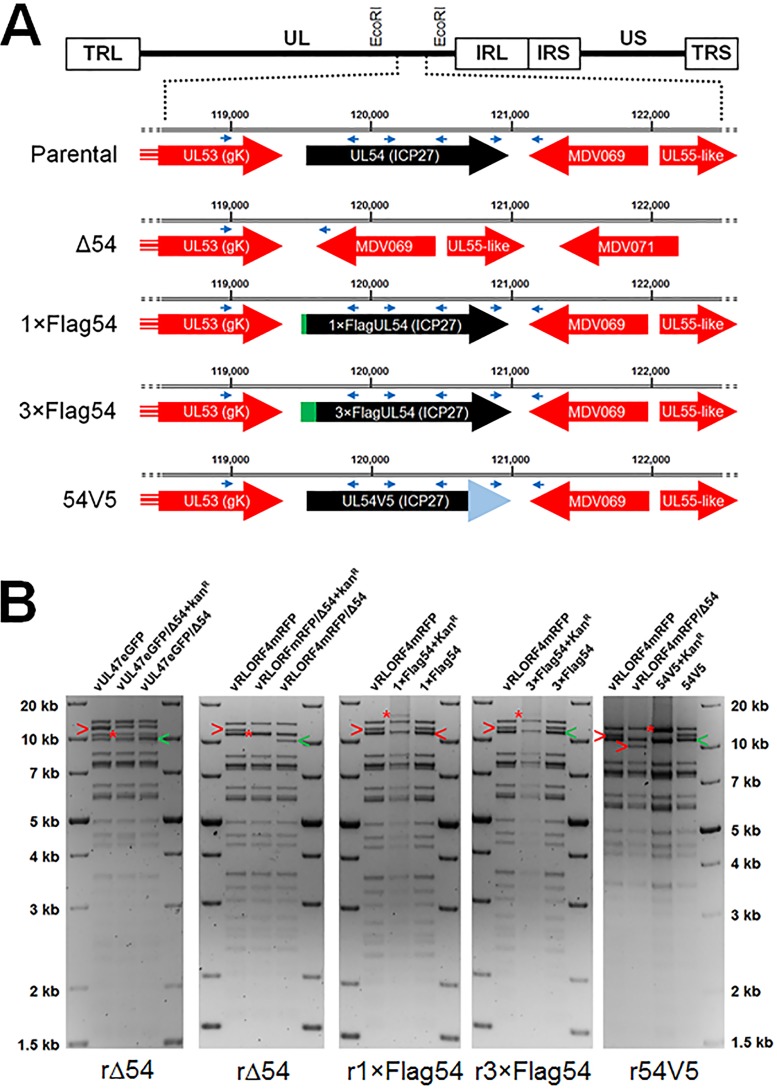
Based on our former work on MDV late genes encoding UL44 (gC), UL47 (VP13/14), and UL48 (VP16), we hypothesized that their dysregulated expression is due to ICP27. However, to date, ICP27’s importance for MDV replication has not been reported. We first generated an ICP27 (UL54)-null MDV in which the entire UL54 gene was removed from a previously published fluorescent MDV bacterial artificial chromosome (BAC) clone (Fig. 2A). We verified the removal of ICP27 from the BAC using restriction fragment length polymorphism (RFLP) analyses of the recombinant MDV (rMDV) clones (Fig. 2B). Moreover, PCR amplification of the regions flanking UL54 confirmed that the UL54 gene was removed without any unexpected changes (data not shown).
FIG 2.
Generation of rMDV. (A) Schematic representation of the MDV genome depicting the locations of the TRL, UL, IRL, IRS, US, and TRS regions. The genes flanking UL54 are shown, including the location within the rMDV BACs used. The locations of PCR primers used for sequencing are shown as blue arrowheads. (B) RFLP analysis of rMDV BAC DNA used in this study. EcoRI restriction digestion maps were used for diagnostic purposes. Integration of the Kanr gene (AphAI) into each locus is shown with a red asterisk, and the originating (red “>”) fragment and final (green “>”) fragments following resolution are also shown. Insertion of the Kanr gene into the UL54 locus resulted in a shift of the 11,294 bp fragment to 10,901 bp. Removal of the Kanr gene resulted in a shift to 9,875 bp, removing 1,419 bp from the original 11,294-bp fragment by deletion of the UL54 open reading frame (ORF). Insertion of 1× and 3×Flag epitopes at the N terminus of UL54 with the Kanr gene resulted in a shift of the 11,294 bp fragment to 12,349- and 12,427-bp fragments, respectively. Removal of the Kanr gene resulted in shifting the respective integrates to 11,318 and 11,360 bp, resulting in total increases of 24 and 66 bp to insert 1×Flag (DYKDDDDK) and 3×Flag (DYKDHDGDYKDHDIDYKDDDDK). Restoration of UL54 with a C-terminal V5 tag plus the Kanr gene into Δ54 resulted in shifting the 9,875-bp fragment to 12,440 bp, followed by reduction to 11,204 bp when resolved. This results in a total change of 108 bp from parental UL54 and UL54V5. No extraneous alterations are evident for all integrates and resolved clones. The molecular weight marker used was the 1-kb Plus DNA ladder from Invitrogen, Inc. (Carlsbad, CA).
In vitro replication of MDV does not require ICP27.
Next, rMDV BAC DNA was transfected into DF-1-Cre cells to reconstitute virus. We initially utilized the rRLORF4mRFP rMDV (36) in order to identify cells transfected with BAC DNA, since transfected cells can be directly visualized by RLORF4mRFP expression. Suprisingly, viral plaques were observed in rΔ54-transfected cells (data not shown). In order to confirm this result, we generated another rMDV using a different viral background, vUL47eGFP (27). Although fluorescence could not be observed following transfection, which is consistent with previous data (27), passage of transfected DF-1-Cre cells with chicken kidney cells (CKCs) resulted in plaques (data not shown). Since rΔ54 in both virus backgrounds resulted in plaque formation, rescued viruses were generated in which the UL54 gene was repaired in vΔ54. The repaired UL54 gene included a V5 epitope tag at the C terminus for downstream identification purposes (Fig. 2A).
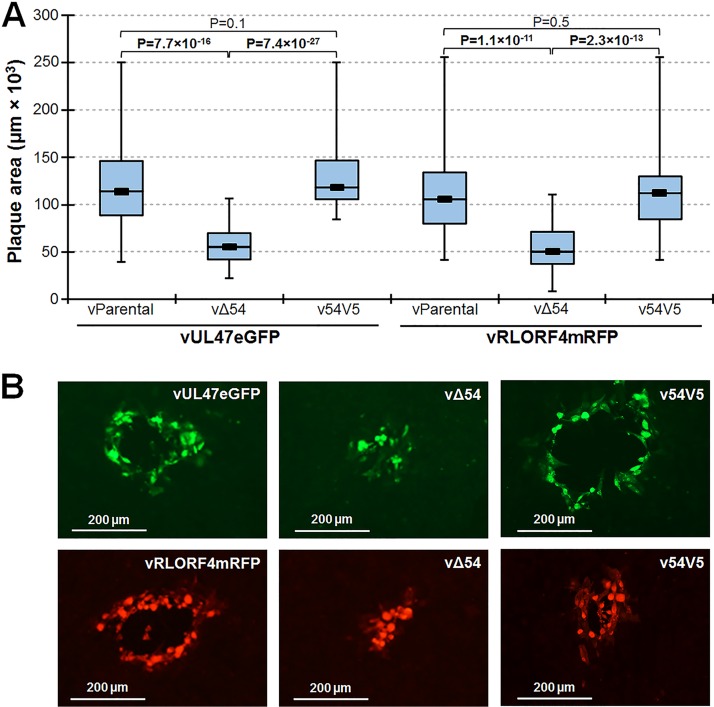
MDV only replicates cell to cell in cell culture such that traditional viral yield growth curves cannot be used to measure virus replication; however, plaque size assays can be used to measure replication based on the virus’s ability to spread from cell to cell. Plaque sizes for vΔ54 were significantly lower than for the respective parental viruses (vParental) (Fig. 3A). Rescuing Δ54 with UL54V5 resulted in plaque sizes similar to those of parental viruses. These data suggests that the plaque formation defect of vΔ54s was due to a lack of ICP27 (UL54) and not extraneous mutations in the genome.
FIG 3.
In vitro replication of Δ54 rMDV. (A) CKCs were seeded in 6-well dishes and infected with 100 PFU of each virus per well. After 5 days, cells were fixed and IFA assays were performed using anti-MDV chicken sera and goat anti-chicken IgY-Alexa Fluor 568 or 488 secondary antibodies. Digital images of 50 individual plaques were taken, and plaque areas were measured using ImageJ. Box plots are shown for each group. Significant differences in mean plaque areas were determined using Student’s t test, and P values are shown. (B) Representative images of the mean plaque areas in panel A are shown for each virus. Plaque areas generated by vΔ54 rMDVs were significantly smaller than parental (vParental) and rescued (v54V5) rMDV in two virus backgrounds.
Inducible expression of MDV ICP27 in trans rescues the cell culture replication defect of rΔ54.
For simplicity, we continued our studies using the RLORF4mRFP rMDV, since expression of mRFP in cell culture allows better visualization of plaques and virus replication (36). To directly test whether the replication defect of vΔ54 was due to a lack of ICP27, we created an MDV ICP27 expression plasmid with a C-terminal V5 epitope tag to rescue the small-plaque phenotype of vΔ54 by transient expression in trans. However, transient transfection of this construct was toxic to cells; therefore, generation of cells stably expressing ICP27 failed.
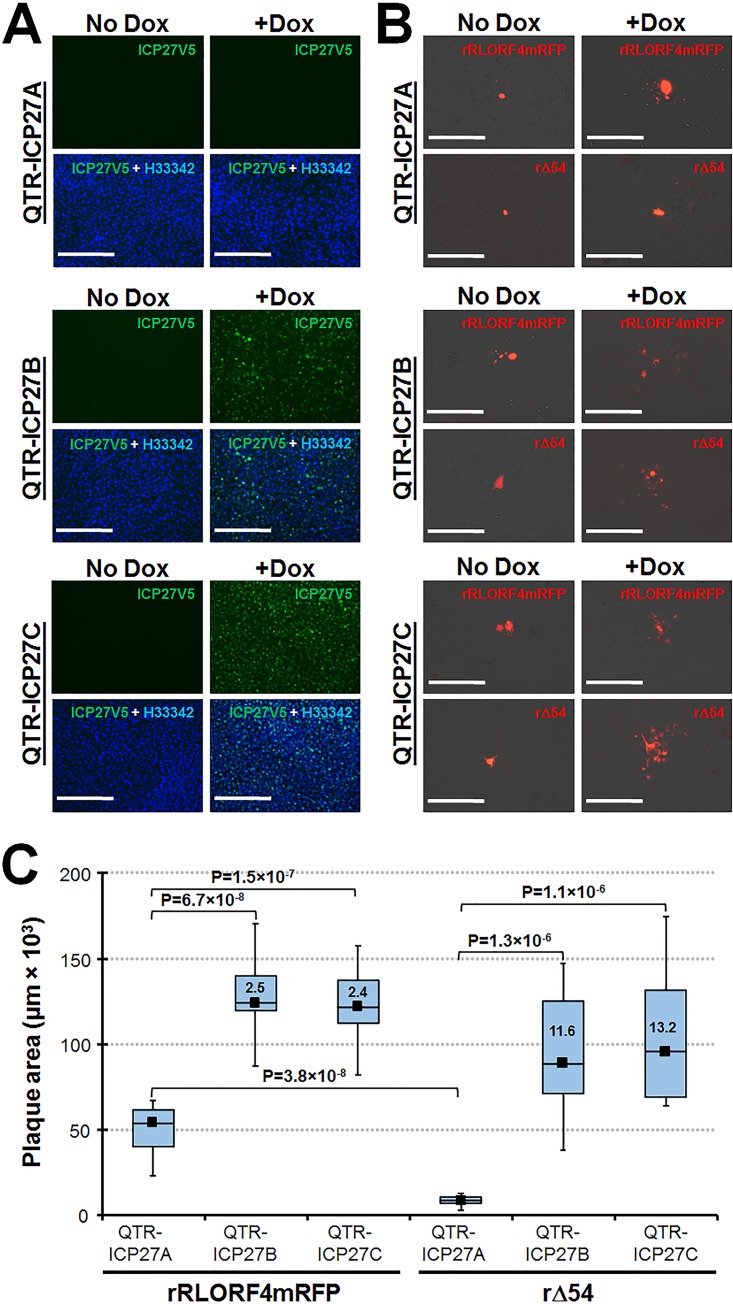
As an alternative approach to express MDV ICP27 in trans, we utilized a previously described system to induce expression of ICP27 in a doxycycline (Dox)-inducible quail cell line that supports MDV replication (37, 38). Multiple clonal cell lines were tested for basal and inducible ICP27 expression, and three clonal cell lines were selected: one with no basal or Dox-inducible expression (QTR-ICP27A) and two in which there was no basal expression but ICP27 was induced (QTR-ICP27B and -ICP27C) by Dox treatment (Fig. 4A). Each cell line was transfected with either rRLORF4mRFP or rΔ54 BAC DNA. The next day, cells were mock treated (no Dox) or treated every other day with Dox, and after 7 days, cells were fixed. Mock-treated QTR-ICP27A, -ICP27B, and -ICP27C supported limited growth of rRLORF4mRFP, while only one or two infected cells could be detected in rΔ54-transfected cells (Fig. 4B). This was consistent with transfection in DF-1-Cre cells described above.
FIG 4.
Inducible expression of MDV ICP27 rescues vΔ54 deficient replication. (A) QTR-ICP27 cell lines were treated with 1 μg/ml Dox (+Dox) or left untreated (no Dox) for 24 h and then fixed. Anti-V5 mAb was used to identify V5-tagged ICP27 protein, and cells were counterstained with Hoechst 33342 (blue). Scale bars represent 200 nm. QTR-ICP27A showed no basal (no Dox) or inducible (+Dox) ICP27 expression, while QTR-ICP27B and -ICP27C had no basal expression but had induced ICP27 expression. (B) QTR-ICP27 cell lines were used to test the ability of inducible expression of ICP27 to rescue cell-to-cell spread of vΔ54. All three cell lines were transfected with rRLORF4mRFP or rΔ54 BAC DNA in duplicate. After 1 day, one well was treated with 1 μg/ml of Dox every other day for 7 days. Cells were fixed and plaques were detected using polyclonal anti-MDV chicken sera plus secondary anti-chicken IgG Alexa Fluor 568. Scale bars represent 200 nm. (C) Plaque size assays showing the mean plaque area produced by rRLORF4mRFP and rΔ54 in Dox-treated QTR-ICP27A, -ICP27B, and -ICP27C at 7 dpi. Ten plaques were measured for each group, and box plots with statistical results are shown. The fold change in plaque sizes generated for rRLORF4mRFP and rΔ54 in QTR-ICP27B and -ICP27C cells compared to QTR-ICP27A is shown in each box.
Following Dox treatment, both rRLORF4mRFP and rΔ54 showed increased plaque sizes in QTR-ICP27B and -ICP27C cells. Figure 4C summarizes plaque sizes generated for both viruses in Dox-treated (+Dox) groups. There was a significant difference between the rRLORF4mRFP and rΔ54 transfected cells in Dox-induced QTR-ICP27A (lacking ICP27 inducible expression) cells, consistent with the importance of ICP27 in replication. However, in QTR-ICP27B and -ICP27C cells induced for ICP27 expression, both rRLORF4mRFP and rΔ54 grew significantly better than in QTR-ICP27A. Induction of ICP27 expression resulted in significantly increased cell-to-cell spread, as evidenced by increased plaque sizes for both vRLORF4mRFP- and vΔ54-transfected cells. Interestingly, enhanced expression of ICP27 increased replication of the parental vRLORF4mRFP 2.5-fold (QTR-ICP27B) and 2.4-fold (QTR-ICP27C) compared to replication in QTR-ICP27A. The increase in replication for vΔ54 was even greater in QTR-ICP27B and -ICP27C, at 11.6-fold and 13.2-fold, respectively, compared to QTR-ICP27A. These results strongly suggest that expression of ICP27 in trans compensates for the lack of endogenous ICP27 expressed from the MDV genome lacking UL54 (ICP27), and therefore, the decreased level of cell-to-cell spread (replication) of vΔ54 is due to a lack of ICP27 expression.
ICP27 regulates MDV gC at the posttranscriptional level in cell culture.
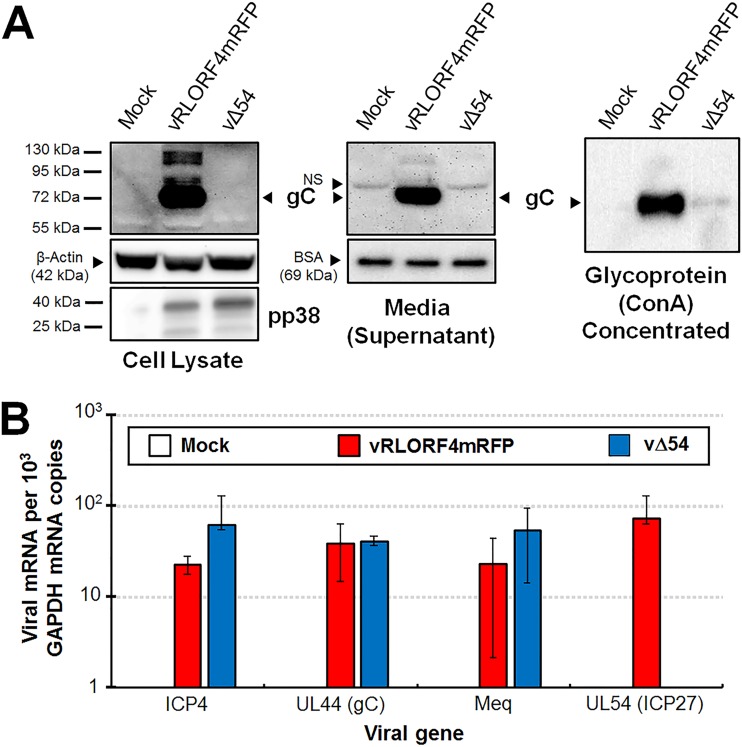
Our original hypothesis for targeting MDV ICP27 centered on its potential role in regulation of late MDV genes, specifically pUL44 (gC), pUL47 (VP13/14), and pUL48 (VP16) (26–28, 36). MDV gC is expressed as a full-length membrane-bound protein and as spliced mRNA variants that are secreted into media (26, 33). One of the many described functions of HSV-1 ICP27 is intron retention of gC mRNA, promoting expression of full-length, membrane-bound HSV-1 gC (34). We hypothesized that the abundance of secreted MDV gC in cell culture was due to ICP27. Therefore, we examined expression of gC in both cellular protein extracts and cell culture media of vRLORF4mRFP- or vΔ54-infected cells using Western blotting analyses. vRLORF4mRFP expressed abundant gC in both cellular and secreted forms, while very little gC protein could be detected in cell lysates or cell culture media of vΔ54-infected cultures (Fig. 5A). Concanavalin A (ConA) concentration of glycoproteins showed that only a small amount of gC could be detected in the vΔ54-infected cell culture media, indicating that MDV gC expression is negligible without ICP27 expression. Thus, we could not conclude whether MDV ICP27 was involved in mRNA splicing and secretion of MDV gC in these studies.
FIG 5.
MDV ICP27 is required for gC protein expression in cell culture. CKCs in 60-mm dishes were infected with 500 PFU of each respective virus. (A) Total cellular protein and cell culture media (supernatant) were collected from mock-, vRLORF4mRFP-, or vΔ54-infected cells at 5 dpi and used in Western blots. Anti-β-actin and -BSA were used for loading controls of cellular and supernatant samples, respectively. Anti-pp38 mAb was used to show comparable infection levels of infected cell cultures. ConA pulldown was used to concentrate glycoproteins. No endogenous control is available for ConA pulldown, but equal amounts of supernatant were used in the pulldown assay. No cellular or secreted MDV gC could be detected in vΔ54-infected CKCs, while abundant gC was present in vRLORF4mRFP-infected cells and supernatant. (B) Total RNA was collected from infected CKCs, cDNA was prepared, and RT-qPCR assays were performed. Specific primers for MDV ICP4, UL44 (gC), Meq (control for gene not expected to be regulated by ICP27), and UL54 (ICP27) were used to quantitate mRNA compared to GAPDH using standard curves. SDs are shown. No amplification of UL54 was detected in vΔ54-infected cells. No significant differences were observed using Student’s t test assuming equal variances at a significance level of P < 0.05. Thus, the lack of gC protein expression in vΔ54 is not due to a lack of mRNA transcription of gC but occurs at the posttranscriptional level.
HSV-1 ICP27 has been shown to regulate HSV-1 gC expression posttranscriptionally (34, 39, 40). Since MDV gC protein levels in vΔ54 were negligible, we next examined MDV gC mRNA expression during cell culture replication. At 5 days postinfection (dpi), MDV gC mRNA transcripts were comparable between vRLORF4mRFP- and vΔ54-infected cells using reverse transcription-quantitative PCR (RT-qPCR) assays (Fig. 5B), suggesting that MDV gC regulation by ICP27 is also at the posttranscriptional level. We also measured expression of two other genes not thought to be regulated by ICP27: the genes for ICP4 (another immediate early) and Meq (oncoprotein not important for replication). There were no significant differences in transcript levels for both ICP4 and Meq in a comparison of vRLORF4mRFP- and vΔ54-infected cells. These results combined with our Western blotting analysis for protein expression show that (i) expression of MDV gC requires ICP27 and (ii) ICP27 most likely plays an essential part in posttranscriptional regulation of gC transcripts, consistent with what has been shown for HSV-1.
Epitope tagging of ICP27 does not negatively affect cell culture replication.
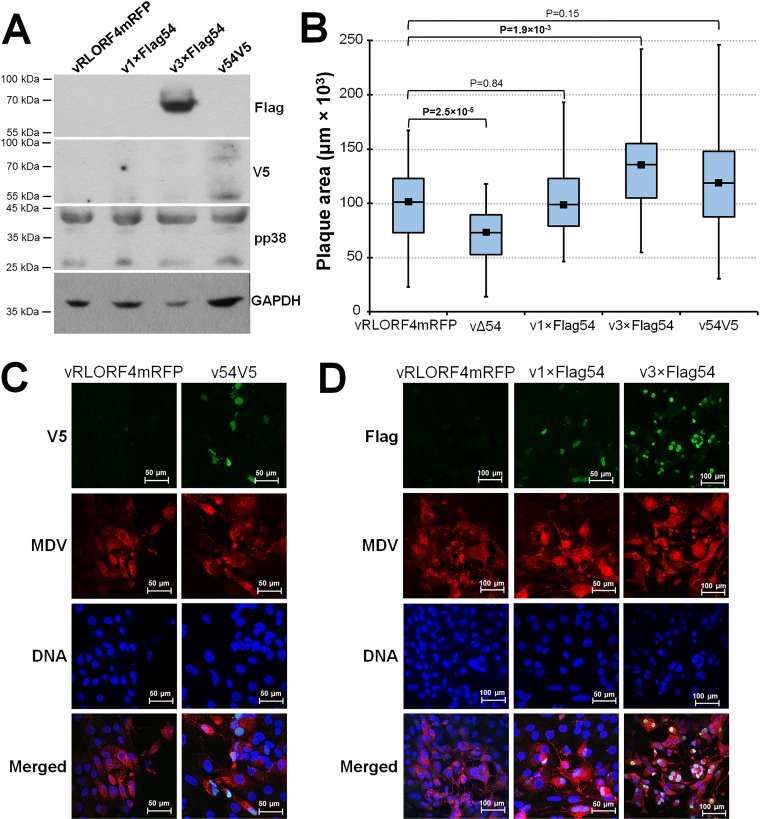
Our ultimate goal was to extend our work to in vivo studies and analyze ICP27 expression in chickens. We generated polyclonal rabbit antibodies against three different ICP27 epitopes but found that their specificity was not optimal (data not shown). Therefore, we used an alternative approach to epitope tag ICP27. Figure 2 shows schematic representation of the constructs used in this study and RFLP analyses. Initially, individual N-terminal 1×Flag and the rescued C-terminal V5 tag viruses were generated; however, detection of ICP27 was difficult using commercial antibodies against Flag and V5, respectively (Fig. 6A). We hypothesized that expression of ICP27 protein would be too low during cell culture replication to detect in Western blotting. To enhance detection, we generated an additional virus in which a 3×Flag tag was added to the N terminus. In contrast to 1×Flag epitope tag, the 3×Flag ICP27 protein was readily detectable by Western blotting of infected cell cultures (Fig. 6A). MDV-specific anti-pp38 MAb was used to compare infection levels of each virus, and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) was used for protein loading controls. Anti-Flag antibody detected the ICP27 protein in v3×Flag54-infected CKCs, while no protein was detected with vRLORF4mRFP, v1×Flag54, or v54V5. Using the anti-V5 antibody, ICP27 protein could be detected in v54V5-infected cells, but the levels were low. Similarly, overexposure of the membrane with Flag antibody showed positive staining with v1×Flag54 (data not shown).
FIG 6.
N- and C-terminally tagged ICP27 rMDV does not negatively affect cell culture replication. CKCs in 60-mm dishes were infected with 500 PFU of each respective virus and at 5 dpi were either used for total protein extraction or fixed for IFA staining. (A) Total protein collected from CKCs infected with each respective virus and anti-Flag, -V5, -pp38, or -GAPDH MAbs were used to detect each respective protein. (B) Box plots of mean plaque areas created by vRLORF4mRFP, vΔ54, v1×Flag54, v3×Flag54, and v54V5 in CKCs. All viruses were used at passage 3. Statistical results are shown. (C and D) Confocal microscopy of CKCs infected with vRLORF4mRFP, v54V5, v1×Flag54, or v3×Flag54. CKCs were infected with 50 PFU/35-mm dish of each respective virus, fixed, and incubated with anti-MDV chicken sera plus goat anti-chicken IgY Alexa Fluor 568 to identify infected cells. (C) V5-tagged ICP27 was detected in v54V5-infected CKCs using anti-V5 MAb plus goat anti-mouse IgG Alexa Fluor 488, and nuclei were stained using Hoechst 33342. (D) Flag-tagged ICP27 was detected in infected CKCs using anti-Flag antibody plus goat anti-mouse IgG Alexa Fluor 488 and counterstained with Hoechst 33342 to identify nuclei. Detection of MDV ICP27 was significantly enhanced by addition of 3×Flag at its N terminus.
To ensure that N- or C-terminal tagging of MDV ICP27 did not have deleterious effects on virus replication, plaque size assays for each virus were performed. Contrary to replication of the vΔ54 rMDVs, all tagged viruses had no defect in cell culture replication (Fig. 6B). Interestingly, addition of the 3×Flag at the N terminus of ICP27 consistently resulted in increased plaque sizes and grew to higher titers compared to parental vRLORF4mRFP in repeated experiments.
Using confocal microscopy, ICP27 proteins tagged with both V5 (Fig. 6C) and 1×Flag or 3×Flag (Fig. 6D) could be detected with their respective epitope antibodies, but consistent with Western blot results, 3×Flag further enhanced detection. MDV ICP27 was predominantly seen in the nucleus regardless of the epitope or terminal tag, similar to what has been observed previously (41). These results show that addition of N- or C-terminal epitope tags to ICP27 did not reduce the ability of MDV to replicate in cell culture and these tags, in particular 3×Flag, can be used for identification of MDV ICP27.
ICP27 is important but not required for MDV eplication in chickens.
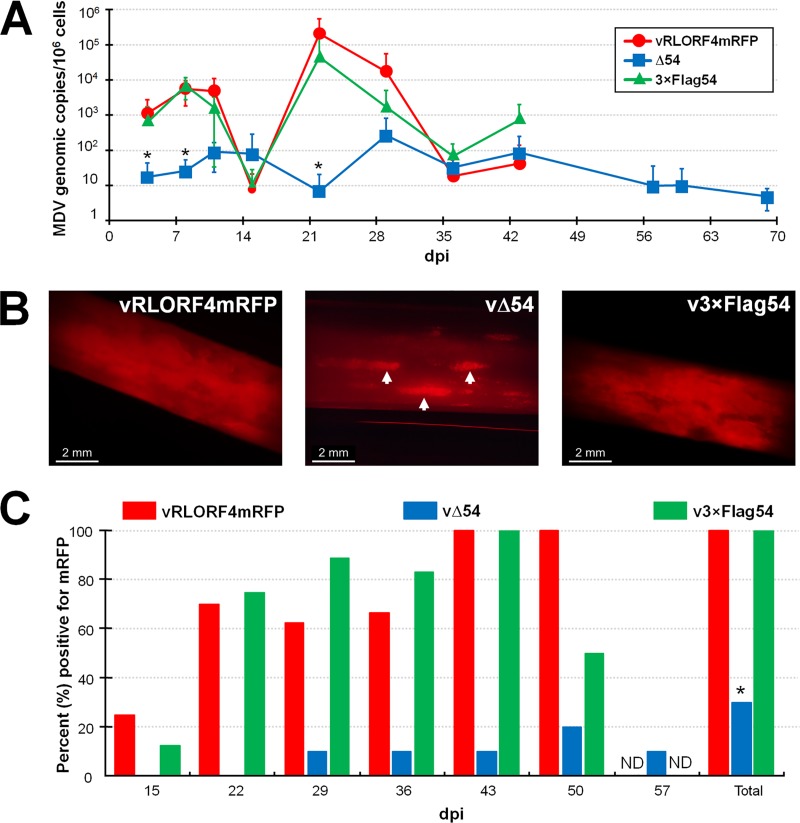
Since MDV ICP27 was important but not required for cell culture replication, we next tested its importance for replication and pathogenesis in chickens. To do this, groups of chickens (n = 8) were inoculated with cell-associated parental virus (vRLORF4mRFP), vΔ54, or v3×Flag54, and viral replication was measured over 10 weeks. Figure 7A shows qPCR analysis of DNA obtained from blood of rMDV-inoculated chickens. MDV genomes were detected in most vΔ54-inoculated birds, although viral DNA copies in the blood were significantly lower at 4, 8, and 21 dpi. vRLORF4mRFP- and v3×Flag54-infected birds had similar replication kinetics throughout the experiment. Although vΔ54 DNA levels were much lower throughout the experiment, viral DNA copies persisted in birds until termination of the experiment at 10 weeks.
FIG 7.
MDV ICP27 is not required for in vivo replication. Pure Columbian chickens were inoculated with cell-associated vRLORF4mRFP, vΔ54, or v3×Flag54 (n = 8/group), and viral DNAs in the blood and virus present in the FFs were examined for 70 days. (A) Mean MDV genomic copies/106 blood cells ± SEMs were determined using qPCR assays. No significant differences were found between the vRLORF4mRFP and v3×Flag54 groups at any of the time points, while the vΔ54 group was significantly lower than both other groups at 4, 7, and 22 dpi. (B) Representative FFs plucked from each group inoculated with virus, showing that all three viruses reach the FFs. Shown are FFs plucked at 36 dpi and visualized with a fluorescent stereoscope. Most of the FFs of vRLORF4mRFP and v3×Flag54 were positive for virus replication, while only spots (white arrowheads) in vΔ54-infected FFs were evident in some birds. (C) Quantitative analysis of the percentage of birds with FFs positive for virus (mRFP) over the course of the experiment, including the total number of birds inoculated with virus that were positive for infection of FFE cells. Fisher’s exact test showed that the vΔ54-infected group was significantly different (P = 0.0031) from the vRLORF4mRFP- and v3×Flag54-infected groups (*), while vRLORF4mRFP- and v3×Flag54-infected groups were not significantly different from each other (P = 1.0). Addition of the 3×Flag on the N terminus of ICP27 did not affect MDV replication in chickens, and vΔ54 was able to replicate and reach the FFs.
Since chickens were infected with fluorescent rMDV, we also monitored the time it took for virus to reach the FF. To do this, wing feathers were plucked every week and examined using a fluorescent stereoscope to visualize the presence of mRFP (Fig. 7B). Our results are summarized in Fig. 7C. Starting at 15 dpi, vRLORF4mRFP and v3×Flag54 were detected in 2 and 1 out of 8 chickens monitored, respectively. Most birds (>50%) were positive in the vRLORF4mRFP and v3×Flag54 groups until termination of those groups at day 60 pi. Interestingly, vΔ54-inoculated birds were intermittently positive in the FF, with one bird positive beginning on day 29 (Fig. 7B) until the bird was euthanized at day 60 due to presenting with neurological symptoms (see below). Two different birds in the vΔ54-inoculated group were also positive at days 50 and 57. Thus, it was clear that vΔ54 was able to replicate and reach the FF in chickens, showing that MDV ICP27 is not required for in vivo replication.
Detection of 3×Flag54 in situ.
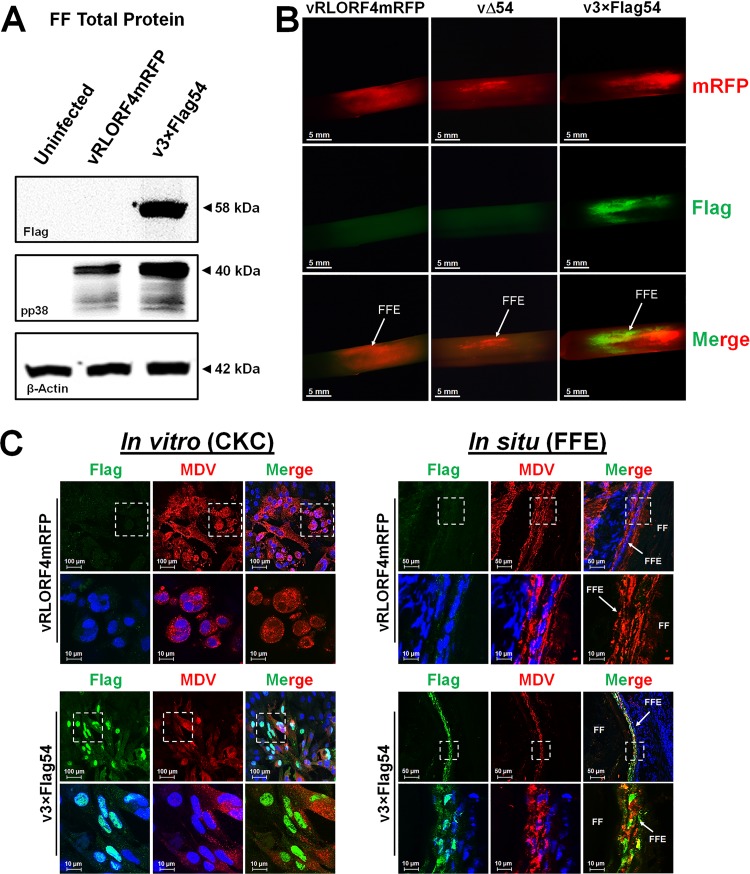
Next we examined MDV ICP27 expression in infected FF and FFE cells using the 3×Flag epitope tag. To detect ICP27 protein in situ, feathers were plucked from vRLORF4mRFP-, vΔ54-, or v3×Flag54-infected birds at 36 dpi and infected FFs were detected by fluorescence. Some feathers were used to collect fluorescent FFE cells using forceps to scrape off the cells, extract total protein, and then use for Western blot analysis. Anti-Flag antibody clearly identified MDV ICP27 in v3×Flag54-infected FFs, while there was no signal in the uninfected or vRLORF4mRFP-infected FFs (Fig. 8A). Anti-pp38 antibody was used as a control for infection levels, and β-actin was used as an endogenous protein control to show that the levels of protein loading for the samples were similar.
FIG 8.
Differential expression of MDV ICP27 in cell culture and FFE cells. (A) Total protein was collected from vRLORF4mRFP, vΔ54, and v3×Flag54 infected FFs at 36 dpi and used in Western blotting assays. Anti-Flag MAb was used to detect ICP27 (3×Flag54), while anti-pp38 MAb was used to show infection levels and anti-β-actin MAb was used for protein loading controls. Only 3×Flag54 was positive for Flag staining. (B) Feathers were plucked from vRLORF4mRFP-, vΔ54-, and v3×Flag54-infected birds at 36 dpi, fixed, and then stained using anti-Flag antibody. There were only limited areas of coexpression of RLOF4mRFP (early gene) and 3×Flag54 (IE gene) in FFs, indicating different phases of replication. Both vRLORF4mRFP- and vΔ54-infected FF were negative for Flag staining. (C) Confocal microscopy of CKCs or FFE cells infected with vRLORF4mRFP or v3×Flag54. White dashed squares are blown up in the images in the bottom row. CKCs infected with infected with v3×Flag54 had strong nuclear staining, consistent with our previous observations (Fig. 6), while FFE cells infected with v3×Flag54 had both nuclear and cytoplasmic staining. This suggests that the localization of MDV ICP27 is different between cell culture (CKC) and in situ (FFE) replication.
Another set of fluorescent feathers were stained with anti-Flag antibody in immunofluorescence (IFA) assays. Figure 8B shows staining of the Flag epitope in v3×Flag54-infected FFs, while both the vRLORF4mRFP- and vΔ54-infected groups were negative for anti-Flag signal. There is a clear difference in expression of MDV ICP27 and RLORF4mRFP, which would be consistent with ICP27 being an IE protein, while RLORF4mRFP is believed to be expressed as an early (E) lytic protein. Together, these data show that addition of the 3×Flag at the N terminus of ICP27 allows the easy detection of MDV ICP27 in FFs that can be used for further studies analyzing the ICP27 protein, in particular to distinguish cells at different stages of lytic viral replication.
Differential localization of ICP27 in cell culture and in situ.
Next, we utilized the 3×Flag to examine localization of MDV ICP27 during cell culture replication in CKCs and in situ replication in FFE cells using frozen skin/feather sections. As was shown earlier (Fig. 6), MDV ICP27 staining was almost exclusively nuclear in cell culture (Fig. 8C). However, in FFE cells, there was clearly more staining throughout the cell than in infected CKCs. These data show that MDV ICP27 localization in cell culture is different from in situ replication.
ICP27 is not required for oncogenicity.
Since ICP27 is conserved among all herpesviruses, but only a few herpesviruses are known to cause cancer, we hypothesized that MDV ICP27 would not be required for oncogenicity. Out of the 10 chickens inoculated with vΔ54, only one developed classical signs of MD over the course of 70 days, while 100% of vRLORF4mRFP- and v3×Flag54-inoculated groups developed MD by 60 dpi (Fig. 9A). Interestingly, the vΔ54-infected chicken showed severe neurological signs presenting as torticollis and ataxia and was the same bird that was positive for virus in the FFE by day 29 (Fig. 7B and C). During necropsy, gross testicle tumors were present and histologically, microscopic tumors in the brain were observed, in which pleomorphic infiltrating lymphocytes positive for MDV Meq oncoprotein were present (Fig. 9B). These results show that MDV ICP27 is not required for oncogenicity of MDV and addition of the 3×Flag at the N terminus of ICP27 does not affect MDV oncogenicity.
FIG 9.
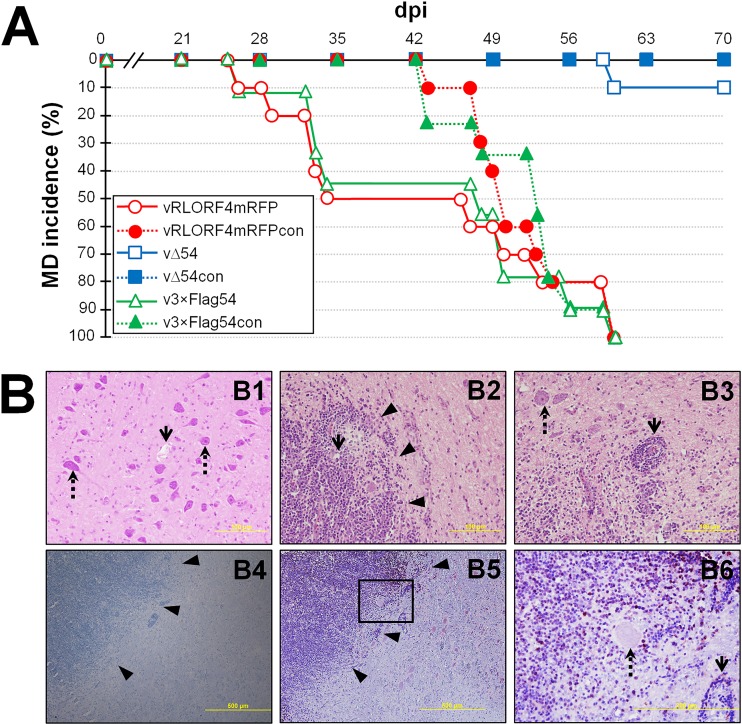
MDV ICP27 is not required for inducing disease or oncogenicity but is required for chicken-to-chicken transmission. Pure Columbian chickens were inoculated with cell-associated vRLORF4mRFP, vΔ54, or v3×Flag54 (n = 10/group) and housed with uninfected contact chickens (n = 10/group) for 70 days. (A) Total MD incidence was determined by identification of gross lesions in dead or euthanized chickens. Fisher’s exact test showed that the vΔ54-infected group was significantly different (P = 0.0001) from the vRLORF4mRFP- and v3×Flag54-infected groups, while the vRLORF4mRFP- and v3×Flag54-infected groups were not significantly different from each other (P = 1.0). (B) Microscopic tumors in cerebrum of vΔ54-infected bird presenting with clinical neurological signs. (B1 to B3) H&E stain. (B1) Age-matched, uninfected control bird. The open arrow shows a blood vessel, and the dotted arrow indicates some of many neurons. (B2) vΔ54-infected bird cerebrum with a high number of neoplastic cells with basophilic round nucleus and small to moderate cytoplasm, consistent with lymphoid cells (arrowheads), surrounding the blood vessel (arrow). (B3) vΔ54-infected cerebrum showing that infiltrating multifocal neoplastic cells invade and efface the normal structure of the cerebrum and expand the Virchow-Robin spaces around the blood vessel (perivascular cuffing [arrow]). Scattered neurons are hypereosinophilic and angular and have pyknotic nuclei (necrosis [dotted arrow]). (B4) vΔ54-infected cerebrum stained with only anti-rabbit secondary antibody and counterstained with hematoxylin as a negative control for anti-Meq staining in B5 and B6. Arrowheads indicate multifocal, invading, and effacing of the normal structure of the cerebrum by infiltrating lymphocytes. (B5) vΔ54-infected cerebrum stained with anti-MDV Meq antibody and counterstained with hematoxylin showing that infiltrating lymphocytes are positive for MDV Meq antigen (brown). (B6) The area in panel B5 is blown up to show neuron (dotted arrow) and blood vessel (arrow) with surrounding invading pleomorphic lymphocytes positive for the MDV Meq antigen (brown).
MDV lacking ICP27 does not spread interindividually.
To test interindividual spread of rMDV, naive contact chickens (n = 10) were housed with each inoculated group (Fig. 9A). Both vRLORF4mRFP and v3×Flag54 were able to transmit and induce disease in 100% of contact chickens (vRLORF4mRFPcon and v3×Flag54con, respectively), while no contact chickens housed with the vΔ54-infected group (vΔ54con) developed disease. As a control for infection, we also tested vΔ54con birds for viral DNA copies and anti-MDV antibodies in the blood, and all were negative for viral genomes and anti-MDV reactive antibodies (data not shown). These data indicate that vΔ54 contact birds were never infected with virus. Overall, these data show that MDV lacking ICP27 (vΔ54) is able to replicate and cause disease in birds experimentally infected with vΔ54 but does not transmit from chicken to chicken.
DISCUSSION
The data presented in this report firmly show that while the Herpesviridae conserved ICP27 of MDV is important for replication, it is not required. It was not completely unexpected that MDV lacking ICP27 (vΔ54) was able to replicate in vitro; however, it was surprising that ICP27-null MDV was able to replicate in chickens and even cause disease. Although a similar observation was observed previously in a study of PRV in mice (16), herpesviral in vivo studies are difficult to interpret when using nonnatural model systems. Here we show that ICP27 is not required for replication of the economically important avian herpesvirus MDV in its natural host species, chickens.
ICP27 performs many functions during cell culture replication, including regulating mRNA splicing (42, 43), promoting mRNA transcription (1, 44), transporting viral mRNAs to the cytoplasm (7, 45), and stimulating translation by interacting with translation initiation factors (31, 46). Very little is known about its role during replication in the natural host. We showed that ICP27 is important for MDV replication in its natural host but is not required for almost all aspects of in vivo replication, including replication in the blood, transformation of cells, and replication in FFE cells of the skin, where virus is shed. The only point at which ICP27 was required in our study was during interindividual transmission. However, it is difficult to confidently conclude its role in interindividual transmission since vΔ54 replication was severely attenuated in chickens. Although vΔ54 reached the skin, we currently do not know whether infectious virus is not produced or is produced, but levels are not sufficient to initiate infection in the new host. In addition, the overall course of infection in chickens by vΔ54 was delayed, such that the amount of time to transmit to naive hosts may also be significantly delayed. It is interesting, though, that gC expression in cell culture was severely affected, suggesting that the inability of vΔ54 to transmit to naive chickens may be due to inefficient gC expression in FFE cells, since gC is required for interindividual spread (26, 47, 48). Nevertheless, ICP27 is most likely involved in multiple aspects of virus replication and gene expression in chickens that could contribute to transmission. We are currently addressing this possibility in more detailed studies on viral gene expression in chickens.
One of the major questions still remaining is whether ICP27 regulates pUL44, pUL47, pUL48, and other viral genes during MDV replication in chickens. We hypothesized that the majority of MDV gC that is normally secreted in cell culture was due to a lack of ICP27 intron retention (34), or possibly ICP27 promoting mRNA splicing, as has been seen for KSHV ICP27 (43). Currently, our data cannot support or refute this hypothesis, since virtually no gC, cellular or secreted, could be detected during vΔ54 replication in CKCs. Former studies showed that MDV ICP27 is able to interact with SR proteins and inhibit mRNA splicing of both cellular telomerase reverse transcriptase (TERT) and viral interleukin-8 (vIL-8) transcripts in cell culture (41); however, UL44 (gC) splicing was never addressed in this study. The data shown in our present report suggest that MDV ICP27 is not required for mRNA transcription, since there was little difference in gC mRNA levels between vRLORF4mRFP and vΔ54 (Fig. 5B), but ICP27 is absolutely required for gC protein production (Fig. 5A). This is consistent with former studies in HSV-1 in which gC is posttranscriptionally regulated by ICP27 (34, 39, 40). Due to the unavailability of antibodies against MDV pUL47 and pUL48 that work in Western blotting, the expression of those genes was not evaluated in this study.
Localization of ICP27 in cell culture was clearly different from that in situ, with ICP27 almost exclusively nuclear in cell culture while present throughout the cell in FFE cells (in situ). This is consistent with our previous results with pUL47 and pUL48 showing that localization of the viral proteins was different between replication in nonproductive CKCs in cell culture and productive FFE cells in situ (27, 28). We surmise that the dysregulation of these genes is directly related to an inability of MDV to produce fully infectious virions in cell culture relative to FFE cells in the skin. Amor et al. (41) showed nuclear localization in transformed T cells reactivating MDV that do not produce infectious cell-free virions, with little to no cytoplasmic staining. This is consistent with our report here. Further studies examining posttranslational modification of ICP27 in cell culture versus in situ may help to explain the differential localization since phosphorylation of HSV-1 ICP27 has been shown to affect its function and localization (49, 50). The use of v3×Flag54 in pulldown assays and proteomics may help to address this possibility.
An unexpected and interesting observation in our in vivo study was the presentation of neurological symptoms and microscopic lesions in the brain of a vΔ54-infected bird (Fig. 9B). Typically, the RB-1B strain of MDV used in our studies is highly oncogenic in the Pure Columbian chicken line, resulting in massive tumors throughout the body, but it rarely induces clinical neurological symptoms. We currently do not know whether ICP27 plays a role in tissue tropism, potentially by dysregulation of late genes. This unexpected result requires further attention.
Our current knowledge of MDV ICP27 is limited, including its importance during cell culture and replication in chickens. We initiated our studies by directly addressing the importance of ICP27 during cell culture and in vivo replication of MDV using UL54-null viruses. Our salient findings are that MDV ICP27 is (i) important but not required for replication in tissue culture cells, (ii) required for expression of gC protein in cell culture, (iii) not required for MDV to complete its replicative cycle within the chicken, (iv) not required for inducing disease, and (v) not required for oncogenicity.
MATERIALS AND METHODS
Cell cultures.
CKCs were prepared from 2- to 4-week-old specific-pathogen-free (SPF) chickens, obtained from the University of Illinois at Urbana-Champaign Poultry Farm, following standard methods (51) and seeded in growth medium consisting of Corning medium 199 (Cellgro, Corning, NY, USA) supplemented with 10% tryptose-phosphate broth (TPB), 0.63% NaHCO3 solution, antibiotics (100 U/ml of penicillin and 100 μg/ml of streptomycin), and 4% fetal bovine serum (FBS). Once CKCs were confluent, medium was replaced with F10.199 maintenance medium consisting of a 1:1 mixture of Ham’s F10 (Cellgro) and medium 199 supplemented with 7.5% TPB, 0.63% NaHCO3, 0.2% FBS, and antibiotics.
The chicken DF-1-Cre fibroblast line (52) was kindly provided by Masahiro Niikura (USDA-ARS-ADOL, Lansing, MI). DF-1-Cre cells were cultivated in a 1:1 mixture of Leibovitz L-15 and McCoy 5A (LM) media (Gibco, Gaithersburg, MD) supplemented with 10% FBS and antibiotics (100 U/ml of penicillin and 100 μg/ml of streptomycin), and maintained in 50 μg/ml of Zeocin (Invitrogen, Carlsbad, CA).
The QT35TR19 quail fibroblastline (37) was propagated in medium 199 supplemented with 10% TPB, 0.63% NaHCO3, antibiotics (100 U/ml of penicillin and 100 μg/ml of streptomycin), 4% FBS, and 5 μg/ml of blasticidin (Invitrogen).
All cells were maintained at 38°C in a humidified atmosphere of 5% CO2.
Expression constructs.
An ICP27 expression construct (pc54V5) was constructed using TOPO cloning of a PCR product using primers shown in Table 1. Briefly, the entire UL54 gene was PCR amplified using BAC DNA from rRB-1B (48) and inserted in frame with the V5 and His epitopes of pcDNA3.1/V5-his construct (Invitrogen Carlsbad, CA) to create pc54V5his.
TABLE 1.
Primers used for generation of expression and transfer plasmids
| Constructa | Directionb | Sequence (5′→3′)c |
|---|---|---|
| pc54v5his | Forward | CACCATGTCTGTAGATGCATT |
| Reverse | CATACCAAACAGAGTATTGCAGT | |
| pc54v5shuttle | Forward | AGATAACCTCAGGCTGAAGCTCGCACCAATAATGATGTGTAAATATGGAACAGAGATAGGGATAACAGGGTAATCGATTT |
| Reverse | GTTAACCCTCAGGGCCAGTGTTACAACCAATTAACC | |
| pcDNA4/TO/54V5 | Forward | GTGGTGGAATTCTGCAGATATCCAGCACAGTGGCGGCCGCTTCACCATGTCTGTAGATGCATTCTCTCGC |
| Reverse | TGATCAGCGGGTTTAAACTCAATGGTGATGGTGATGATGATCAATGGTGATGGTGATGATGAC |
Final construct generated using primers.
Directionality of the primer.
Bold indicates UL54 start codon. Underlining indicates the Bsu361 restriction enzyme sequence used for cloning. Italics indicate the template-binding region of the primers for PCR amplification with pEP-KanS.
A tetracycline-inducible construct was generated using Gibson Assembly cloning (New England BioLabs, Ipswich, MA). Briefly, ICP27V5 from pc54V5 (lacking His) was PCR amplified using primers shown in Table 1 and mixed with pcDNA4/TO/myc-his (Invitrogen) linearized by double digestion with XhoI and AgeI to generate pcDNA4/TO/54V5 in the Gibson Assembly reaction. All clones were confirmed by DNA sequencing using primers shown in Table 2.
TABLE 2.
Primers used for sequencing and diagnostics
| Namea | Directionb | Sequence |
|---|---|---|
| UL54-SeqFor1 | Forward | 5′-AACGATGTTATAGGATCACCTGA-3′ |
| UL54-SeqFor2 | Forward | 5′-CAAGAAGGGCATCACCGAAGAA-3′ |
| UL54-SeqFor3 | Forward | 5′-CCTATCGCAGCAGTTCTTATTAT-3′ |
| UL54-SeqRev1 | Reverse | 5′-TAGACTATGTACCTGGAGAATGT-3′ |
| UL54-SeqRev2 | Reverse | 5′-GACCCGCATGCAATAAATACTC-3′ |
| UL54-SeqRev3 | Reverse | 5′-CCGTGGTATTGCATTGTCA-3′ |
Names of the primers. The function of all the primers was sequencing of the UL54 gene.
Directionality of the primer.
Generation of rΔ54 and rΔ54 rescued viruses.
To generate rΔ54, primers shown in Table 3 were designed to remove the complete UL54 gene, leaving behind only the stop codon, in two rMDV backgrounds (27, 36). Briefly, the aphAI-I-SceI cassette from pEPkan-SII was amplified by PCR using these primers, and two-step Red-mediated recombination was performed in GS1783 Escherichia coli (47, 53). Following integration of the aphAI-I-SceI and RFLP analysis in rMDV clones (Fig. 2B), the aphAI-I-SceI cassette was removed, resulting in rMDV BAC clones lacking the UL54 gene. DNA sequencing and RFLP analyses were used to ensure integrity of the genomes.
TABLE 3.
Primers used for generation of recombinant Marek’s disease viruses
| Modificationa | Directionb | Sequence (5′→3′)c |
|---|---|---|
| 1×Flag54 | Forward | CCTAATGGACTATTATCTATATCAAGATTAAACAAAAAAAATGGATTACAAGGATGACGACGATAAGTAGGGATAACAGGGTAATCGATTT |
| Reverse | GGACTCGCGAGAGAATGCATCTACAGACTTATCGTCGTCATCCTTGTAATCCATTTTTTTTGTTTAATCTGCCAGTGTTACAACCAATTAACC | |
| 3×Flag54 | Forward | TATTATCTATATCAAGATTAAACAAAAAAAATGGATTACAAGGATCATGACGGAGATTACAAGGATCATGACATCGATTACAAGGATGACGACGATAAGTAGGGATAACAGGGTAATCGATTT |
| Reverse | GGACTCGCGAGAGAATGCATCTACAGACTTATCGTCGTCATCCTTGTAATCGATGTCATGATCCTTGTAATCTCCGTCATGATCCTTGTAATCCATTTTTTTTGTTGCCAGTGTTACAACCAATTAACC | |
| 54v5 | Forward | GACTATTATCTATATCAAGATTAAACAAAAAAAATGTCTGTAGATGCATTCTCTCGCGAGTCCGATG |
| Reverse | TATAGGATGTCTTTTAGATAACTATTTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACC |
Modification to UL54 gene or promoter region.
Directionality of the primer.
Underlining indicates Flag tag sequence. Bold indicates the UL54 start or stop codon. Italics indicate the template-binding region of the primers for PCR amplification with pEP-KanS or pcUL54transfer.
To generate rescued Δ54 rMDV, ICP27 with a V5 tag from pc54V5his was introduced into vΔ54 rMDV using a transfer plasmid (53). To generate the transfer plasmid, the aphAI-I-SceI cassette from pEPkan-SII was inserted into pc54V5his using a unique Bsu36I restriction site within UL54 as previously described (53). The UL54V5+aphAI-I-SceI cassette from pcUL54V5transfer was PCR amplified using primers in Table 1 and then used to insert 54V5 where UL54 was previously deleted (Δ54). PCR, DNA sequencing, and RFLP analyses (Fig. 2B) were used to ensure integrity of the genomes and to exclude any fortuitous mutations elsewhere in the genome.
Generation of Flag-tagged UL54 rMDV.
rMDVs were generated using two-step Red-mediated recombination as previously described for GS1783 E. coli (47, 53). Primers used for construction of Flag-tagged UL54 are shown in Table 3. PCR, DNA sequencing, and RFLP analyses (Fig. 2B) were used to ensure integrity of the genomes and to exclude any fortuitous mutations elsewhere in the genome.
Propagation of rMDV.
rMDVs were reconstituted by transfecting DF-1-Cre cells, which efficiently removes the mini-F BAC sequences from the viral genome (47), with BAC DNA plus Lipofectamine 2000 (Invitrogen) using the manufacturer’s instructions. Transfected DF-1-Cre cells were mixed with fresh primary CKCs until plaques formed and then further propagated in CKCs until virus stocks could be stored. All rMDVs were used at ≤5 passages for cell culture and in vivo studies.
IFA assays.
CKCs were infected with rMDVs in 6-well dishes at 100 PFU per well. At 5 dpi, cells were fixed and permeabilized with PFA buffer (2% paraformaldehyde, 0.05% Triton X-100) for 15 min and then washed twice with phosphate-buffered saline (PBS). Infected cells used for antigen detection were blocked in 10% neonatal calf serum and stained with polyclonal anti-MDV chicken sera (47), mouse anti-Flag M2 (Sigma-Aldrich), or mouse anti-V5 E10/V4RR (Thermo Fisher Scientific, Waltham, MA) MAbs. Secondary antibodies used included goat anti-chicken IgY-Alexa Fluor 488 or 568 and goat anti-mouse IgG-Alexa Fluor 488 or 568 (Molecular Probes, Eugene, OR). Hoechst 33342 (2 μg/ml; Molecular Probes) was used to visualize nuclei. Digital images were collected using an EVOS FL cell imaging system (Thermo Fisher Scientific) and compiled using Adobe Photoshop CC 2015 version 7 SP1.
Tetracycline-inducible cell lines.
Tetracycline (Dox)-inducible cell lines were generated based on QT35TR19, a previously described Dox-inducible QT35 cancer cell line (kindly provided by Karel A. Schat, Cornell University), and maintained as described previously (37). Briefly, QT35TR19 cells were transfected with pcDNA4/TO (empty vector) or pcDNA4/TO/54V5 using Lipofectamine 2000 (Invitrogen) by following the manufacturer’s instructions. Monoclonal cell lines were selected with 5 μg/ml of blasticidin and 500 μg/ml of Zeocin. Stable cell lines were tested for basal and inducible expression of UL54V5 (ICP27) using IFA assays. One cell line, designated QTR-ICP27A, had no basal or inducible ICP27 expression, while two cells lines, designed QTR-ICP27B and QTR-ICP27C, had no basal expression of ICP27 but induced expression following treatment with 1 μg/ml of Dox treatment for 24 h.
Measurement of plaque areas.
Plaque areas were measured as previously described (54). Briefly, CKCs were seeded in 6-well dishes and infected with 100 PFU of each virus per well. After 5 days, cells were washed once with PBS, fixed and permeabilized with PFA buffer for 15 min, and then washed twice with PBS. IFA assays were performed using anti-MDV chicken sera and goat anti-chicken IgY-Alexa Fluor 568 or 488 secondary antibody (Molecular Probes, Eugene, OR). Digital images of 10 to 50 individual plaques were obtained using an EVOS FL cell imaging system, plaque areas were measured using ImageJ (55) version 1.51j8 software (https://imagej.nih.gov/ij/), and means were determined for each plaque population. Significant differences in mean plaque areas were determined using Student’s t tests assuming equal variances in Microsoft Excel 2016.
Laser scanning confocal microscopy.
CKCs were infected with rMDV on sterile glass coverslips coated with 40 μg/ml of type I collagen (Sigma-Aldrich, St. Louis, MO) in 24-well dishes at 50 PFU per well. At 4 dpi, cells were fixed and permeabilized with PFA buffer for 15 min and then washed twice with PBS. Skin/feather tissues were collected from MDV-infected chickens and snap-frozen in Tissue Tek optimal cutting temperature (OCT) compound (Sankura Finetek, Torrance, CA) and stored at −80°C until sectioned. Eight- to 10-μm sections were affixed to Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA) and fixed with PFA buffer as described above.
Infected CKCs and cryosectioned tissues used for antigen detection were blocked in 10% FBS and then stained with chicken anti-MDV polyclonal sera (54) and anti-Flag M2 (Sigma-Aldrich) or anti-V5 E10/V4RR (Thermo Fisher Scientific) MAbs. Goat anti-chicken IgY-Alexa Fluor 568 and anti-mouse IgG-Alexa Fluor 488 were used as secondary antibodies. Hoechst 33342 (2 μg/ml; Molecular Probes) was used to visualize nuclei. All images were compiled using Adobe Photoshop CC 2015 version 7 SP1.
Western blot analysis.
Western blot analyses were performed essentially as previously described (32). To detect relative level of MDV infection, MAb H19 (56) was used at a 1:10,000 dilution to detect MDV pp38 protein. Rabbit polyclonal antibodies (pAbs) against MDV ICP27 were generated by GenScript against peptide ERPSPDSKNSQGDDC (pAb ICP27-1), QTKRPHGTGNRKQYC (pAb ICP27-2), or CRRVMHRTSGSKYSY (pAb ICP27-3) and used at a 1:100 dilution. To detect gC, MAb A6 (kindly provided by Jean-Francois Vautherot, INRA, Nouzilly, France) was used at a 1:500 dilution. To detect Flag- or V5-tagged proteins, anti-Flag M2 or anti-V5 E10/V4RR MAbs were used at the manufacturer’s recommended dilutions. For protein loading controls, anti-chicken (ch) GAPDH (GA1R; Thermo Fisher Scientific), anti-β-actin (ACTNO5; Abcam, Cambridge, MA), or anti-bovine serum albumin (anti-BSA; Thermo Fisher Scientific) MAbs were used at their recommended dilutions. Secondary anti-mouse or rabbit IgG-peroxidase conjugate was purchased from GE Healthcare (Piscataway, NJ). Secreted glycoproteins were concentrated from the culture media of MDV-infected CKCs using a concanavalin A (ConA) pulldown method exactly as previously described (32).
RT-qPCR analysis.
Total RNA was collected from 60-mm tissue culture dishes infected with 500 PFU for each virus for 5 days using RNA STAT-60 (Tel-Test, Inc., Friendswood, TX) and Dnase treated using a Turbo DNA-free kit from Thermo Fisher Scientific using the manufacturer’s instructions. RT was performed as a large batch with 5 to 10 μg of DNase-treated total RNA using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific). One-hundred-microliter reactions were carried out according to the manufacturer’s instructions with random primers. The reaction mixture was incubated at 25°C for 10 min, then 37°C for 120 min, followed by 85°C for 5 min.
To amplify cDNA in RT-qPCR assays, 2× Power SYBR green PCR master mix (Thermo Fisher Scientific) was used. Quantification of MDV-specific transcripts were performed using specific primers for each respective MDV transcript and chGAPDH as a normalizing control. Briefly, 3 μl of the RT mixture was used in 20-μl volumes containing 50 μM forward and reverse primers previously published (28). For the generation of standard curves in RT-qPCR assays, a plasmid containing chGAPDH (57) and a BAC clone containing the complete MDV genome were used (47). Serial 10-fold dilutions of each respective plasmid or BAC were used for generating standard curves, starting with approximately 500 pg of DNA. Total copy numbers were determined as previously described (58). The coefficient of regression was always >0.99 for standard curves. Thermal cycling conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All RT-qPCR assays were performed using an Applied Biosystems QuantStudio 3 real-time PCR system (Thermo Fisher Scientific), and the results were analyzed using QuantStudio Design & Analysis Software v1.4.2, supplied by the manufacturer.
Ethics statement.
All animal work was conducted at the University of Illinois at Urbana-Champaign (UIUC) according to national regulations. The animal care facilities and programs of UIUC meet the requirements of the law (89–544, 91–579, 94–276) and NIH regulations on laboratory animals and are in compliance with the Animal Welfare Act, PL 279. The College of Veterinary Medicine at UIUC is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All experimental procedures were in compliance with approval of UIUC’s Institutional Animal Care and Use Committee.
Animal study.
Commercial Pure Columbian chickens were obtained from the University of Illinois Poultry Farm (Urbana, IL) and were from MD-vaccinated parents; therefore, they were considered to be maternal antibody positive. All experimental procedures were conducted in compliance with approved Institutional Animal Care and Use Committee protocols. Water and food were provided ad libitum. Two-day-old chicks were inoculated intra-abdominally with 2,000 PFU of rMDVs in separate rooms (n = 10/group). For each group, another 10 chickens were left uninfected to act as contact controls to determine whether rMDVs were able to transmit to uninfected chickens. Chickens were evaluated daily for symptoms of MD, euthanized when birds showed clinical signs of MD (lethargy, depression, paralysis, torticollis, etc.), and examined for gross MD lesions. Chickens positive for MD included birds succumbing to disease prior to the experimental termination date and birds positive for MD-related lesions at termination of the experiment. Fisher’s exact tests were used to determine statistical differences between groups of chickens for MD incidence at a significance level P < 0.05.
DNA extraction from blood cells and qPCR assays.
Whole blood was collected, as previously described (47), and DNA was extracted using the E.Z. 96 blood DNA kit from Omega Bio-tek, Inc. (Norcross, GA). Blood collection was discontinued for the vRLORF4mRFP and v3×Flag54 groups after 42 days, since only a few inoculated birds remained, while collection of vΔ54 continued until the experiment was terminated. All qPCR assays were performed as described above for the PCR portion of RT-qPCR assays. Standard curves were generated for chGAPDH and MDV ICP4 using previously described templates, and the numbers of MDV genomic copies per 106 cells were determined using the threshold cycle (CT) value for that sample (35). Significant differences in MDV genomic copies at each time point were determined using Student’s t tests assuming equal variances at a significance level of P < 0.05 using Microsoft Excel 2016.
Monitoring rMDV in FFs.
To track the time at which each rMDV reached the FF, two flight feathers were plucked from the right and left wings (4 total) of inoculated birds weekly beginning at 15 dpi, and mRFP expression was examined using a Leica M165FC fluorescent stereomicroscope with a Leica DFC450 digital color microscope camera (Leica Microsystems, Inc., Buffalo Grove, IL). Feather plucking for the vRLORF4mRFP and v3×Flag54 groups was discontinued after 50 days, since only a few inoculated birds remained, while collection of vΔ54 continued until 57 dpi.
Immunohistochemistry.
Cerebrum from vΔ54-infected and age-matched, uninfected control chickens were fixed in 8% PFA and processed for immunohistochemistry. Sections were cut at 8 μm and stained with hematoxylin and eosin (H&E) or rabbit polyclonal anti-Meq antibody plus hematoxylin as a counterstain. The Vectastain Universal Elite ABC, ImmPACT AEC HRP substrate and Vector hematoxylin QS kits were used (Vector Laboratories, Burlingame, CA).
ACKNOWLEDGMENTS
We thank Joanna L. Shisler for her critical evaluation and advice in preparation of the manuscript.
This report was supported by Agriculture and Food Research Initiative competitive grants 2013-67015-26787 and 2016-67015-26777 from the USDA National Institute of Food and Agriculture to K.W.J. Y.-T.T. is supported by a Taiwanese–University of Illinois at Urbana-Champaign scholarship.
REFERENCES
- 1.Zhou C, Knipe DM. 2002. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J Virol 76:5893–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai-Ju JQ, Li L, Johnson LA, Sandri-Goldin RM. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J Virol 80:3567–3581. doi: 10.1128/JVI.80.7.3567-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant HE, Wadd SE, Lamond AI, Silverstein SJ, Clements JB. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J Virol 75:4376–4385. doi: 10.1128/JVI.75.9.4376-4385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. 2010. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology 401:155–164. doi: 10.1016/j.virol.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandri-Goldin RM, Hibbard MK, Hardwicke MA. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol 69:6063–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sciabica KS, Dai QJ, Sandri-Goldin RM. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J 22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbin-Lickfett KA, Chen IH, Cocco MJ, Sandri-Goldin RM. 2009. The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-quartet structures. Nucleic Acids Res 37:7290–7301. doi: 10.1093/nar/gkp793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandri-Goldin RM. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev 12:868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mears WE, Rice SA. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 10.Sandri-Goldin RM. 2011. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol 6:1261–1277. doi: 10.2217/fmb.11.119. [DOI] [PubMed] [Google Scholar]
- 11.Sacks WR, Greene CC, Aschman DP, Schaffer PA. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol 55:796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy AM, McMahan L, Schaffer PA. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol 63:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi ML, Blankenship C, Shenk T. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc Natl Acad Sci U S A 97:2692–2696. doi: 10.1073/pnas.050587597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz JA, Brittle EE, Reynolds AE, Enquist LW, Silverstein SJ. 2006. UL54-null pseudorabies virus is attenuated in mice but productively infects cells in culture. J Virol 80:769–784. doi: 10.1128/JVI.80.2.769-784.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baaten BJ, Staines KA, Smith LP, Skinner H, Davison TF, Butter C. 2009. Early replication in pulmonary B cells after infection with Marek’s disease herpesvirus by the respiratory route. Viral Immunol 22:431–444. doi: 10.1089/vim.2009.0047. [DOI] [PubMed] [Google Scholar]
- 18.Calnek BW. 2001. Pathogenesis of Marek’s disease virus infection. Curr Top Microbiol Immunol 255:25–55. [DOI] [PubMed] [Google Scholar]
- 19.Gershon MD, Gershon AA. 2010. VZV infection of keratinocytes: production of cell-free infectious virions in vivo. Curr Top Microbiol Immunol 342:173–188. doi: 10.1007/82_2010_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarosinski KW. 2017. Interindividual spread of herpesviruses. Adv Anat Embryol Cell Biol 223:195–224. doi: 10.1007/978-3-319-53168-7_9. [DOI] [PubMed] [Google Scholar]
- 21.Read AF, Baigent SJ, Powers C, Kgosana LB, Blackwell L, Smith LP, Kennedy DA, Walkden-Brown SW, Nair VK. 2015. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol 13:e1002198. doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed M, Schidlovsky G. 1968. Electron microscopic localization of herpesvirus-type particles in Marek’s disease. J Virol 2:1443–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Churchill AE, Biggs PM. 1967. Agent of Marek’s disease in tissue culture. Nature 215:528–530. [DOI] [PubMed] [Google Scholar]
- 24.Solomon JJ, Witter RL, Nazerian K, Burmester BR. 1968. Studies on the etiology of Marek’s disease. I. Propagation of the agent in cell culture. Proc Soc Exp Biol Med 127:173–177. [DOI] [PubMed] [Google Scholar]
- 25.Calnek BW, Adldinger HK, Kahn DE. 1970. Feather follicle epithelium: a source of enveloped and infectious cell-free herpesvirus from Marek’s disease. Avian Dis 14:219–233. [PubMed] [Google Scholar]
- 26.Jarosinski KW, Osterrieder N. 2012. Marek’s disease virus expresses multiple UL44 (gC) variants through mRNA splicing that are all required for efficient horizontal transmission. J Virol 86:7896–7906. doi: 10.1128/JVI.00908-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarosinski KW, Arndt S, Kaufer BB, Osterrieder N. 2012. Fluorescently tagged pUL47 of Marek’s disease virus reveals differential tissue expression of the tegument protein in vivo. J Virol 86:2428–2436. doi: 10.1128/JVI.06719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarosinski KW, Vautherot JF. 2015. Differential expression of Marek’s disease virus (MDV) late proteins during in vitro and in situ replication: role for pUL47 in regulation of the MDV UL46-UL49 gene locus. Virology 484:213–226. doi: 10.1016/j.virol.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Ellison KS, Maranchuk RA, Mottet KL, Smiley JR. 2005. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J Virol 79:4120–4131. doi: 10.1128/JVI.79.7.4120-4131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jean S, LeVan KM, Song B, Levine M, Knipe DM. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma 2) genes in infected cells. Virology 283:273–284. doi: 10.1006/viro.2001.0902. [DOI] [PubMed] [Google Scholar]
- 31.Fontaine-Rodriguez EC, Knipe DM. 2008. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J Virol 82:3538–3545. doi: 10.1128/JVI.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tischer BK, Schumacher D, Chabanne-Vautherot D, Zelnik V, Vautherot JF, Osterrieder N. 2005. High-level expression of Marek’s disease virus glycoprotein C is detrimental to virus growth in vitro. J Virol 79:5889–5899. doi: 10.1128/JVI.79.10.5889-5899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isfort RJ, Stringer RA, Kung HJ, Velicer LF. 1986. Synthesis, processing, and secretion of the Marek’s disease herpesvirus A antigen glycoprotein. J Virol 57:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedlackova L, Perkins KD, Lengyel J, Strain AK, van Santen VL, Rice SA. 2008. Herpes simplex virus type 1 ICP27 regulates expression of a variant, secreted form of glycoprotein C by an intron retention mechanism. J Virol 82:7443–7455. doi: 10.1128/JVI.00388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedlackova L, Perkins KD, Meyer J, Strain AK, Goldman O, Rice SA. 2010. Identification of an ICP27-responsive element in the coding region of a herpes simplex virus type 1 late gene. J Virol 84:2707–2718. doi: 10.1128/JVI.02005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarosinski KW, Donovan KM, Du G. 2015. Expression of fluorescent proteins within the repeat long region of the Marek’s disease virus genome allows direct identification of infected cells while retaining full pathogenicity. Virus Res 201:50–60. doi: 10.1016/j.virusres.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Jarosinski KW, Schat KA. 2006. Expression of Marek’s disease virus phosphorylated polypeptide pp38 produces splice variants and enhances metabolic activity. Vet Microbiol 117:154–168. doi: 10.1016/j.vetmic.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 38.Kaufer BB, Arndt S, Trapp S, Osterrieder N, Jarosinski KW. 2011. Herpesvirus telomerase RNA (vTR) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog 7:e1002333. doi: 10.1371/journal.ppat.1002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice SA, Knipe DM. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J Virol 64:1704–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith IL, Hardwicke MA, Sandri-Goldin RM. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 186:74–86. [DOI] [PubMed] [Google Scholar]
- 41.Amor S, Strassheim S, Dambrine G, Remy S, Rasschaert D, Laurent S. 2011. ICP27 protein of Marek’s disease virus interacts with SR proteins and inhibits the splicing of cellular telomerase chTERT and viral vIL8 transcripts. J Gen Virol 92:1273–1278. doi: 10.1099/vir.0.028969-0. [DOI] [PubMed] [Google Scholar]
- 42.Hardy WR, Sandri-Goldin RM. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol 68:7790–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. 2008. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol 82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen IH, Li L, Silva L, Sandri-Goldin RM. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J Virol 79:3949–3961. doi: 10.1128/JVI.79.7.3949-3961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen IH, Sciabica KS, Sandri-Goldin RM. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J Virol 76:12877–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM. 2004. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330:487–492. doi: 10.1016/j.virol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Jarosinski KW, Margulis NG, Kamil JP, Spatz SJ, Nair VK, Osterrieder N. 2007. Horizontal transmission of Marek’s disease virus requires US2, the UL13 protein kinase, and gC. J Virol 81:10575–10587. doi: 10.1128/JVI.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarosinski KW, Osterrieder N. 2010. Further analysis of Marek’s disease virus horizontal transmission confirms that U(L)44 (gC) and U(L)13 protein kinase activity are essential, while U(S)2 is nonessential. J Virol 84:7911–7916. doi: 10.1128/JVI.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas S, Corbin-Lickfett KA, Escudero-Paunetto L, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants are defective in herpes simplex virus 1 replication and gene expression. J Virol 84:2200–2211. doi: 10.1128/JVI.00917-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin-Lickfett KA, Rojas S, Li L, Cocco MJ, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants display altered functional interactions with cellular export factors Aly/REF and TAP/NXF1 but are able to bind herpes simplex virus 1 RNA. J Virol 84:2212–2222. doi: 10.1128/JVI.01388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schat KA, Sellers HS. 2008. Cell-culture methods, p 195–203. In Dufour-Zavala L, Swayne DE, Glisson JR, Pearson JE, Reed WM, Jackwood MW, Woolcock PR (ed), A laboratory manual for the identification and characterization of avian pathogens, 5th ed. American Association of Avian Pathologists, Jacksonville, FL. [Google Scholar]
- 52.Niikura M, Kim T, Silva RF, Dodgson J, Cheng HH. 2011. Virulent Marek’s disease virus generated from infectious bacterial artificial chromosome clones with complete DNA sequence and the implication of viral genetic homogeneity in pathogenesis. J Gen Virol 92:598–607. doi: 10.1099/vir.0.026864-0. [DOI] [PubMed] [Google Scholar]
- 53.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 54.Jarosinski KW, Osterrieder N, Nair VK, Schat KA. 2005. Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J Virol 79:11647–11659. doi: 10.1128/JVI.79.18.11647-11659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
- 56.Lee LF, Liu X, Witter RL. 1983. Monoclonal antibodies with specificity for three different serotypes of Marek’s disease viruses in chickens. J Immunol 130:1003–1006. [PubMed] [Google Scholar]
- 57.Jarosinski KW, O'Connell PH, Schat KA. 2003. Impact of deletions within the Bam HI-L fragment of attenuated Marek’s disease virus on vIL-8 expression and the newly identified transcript of open reading frame LORF4. Virus Genes 26:255–269. [DOI] [PubMed] [Google Scholar]
- 58.Jarosinski KW, Yunis R, O'Connell PH, Markowski-Grimsrud CJ, Schat KA. 2002. Influence of genetic resistance of the chicken and virulence of Marek’s disease virus (MDV) on nitric oxide responses after MDV infection. Avian Dis 46:636–649. doi: 10.1637/0005-2086(2002)046[0636:IOGROT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]