Hepatitis E virus (HEV) in stools is naked, while that in culture supernatants and patients’ blood is lipid associated. Its life cycle in hepatocytes, polarized cells with a basolateral side communicating with blood and an apical side connected with bile, is incompletely understood. We have developed a polarized hepatocyte model and used the cells to analyze the supernatants bathing the apical and basolateral sides and HEV subcellular distribution. HEV particles from both sides were lipid associated, and most infectious HEV particles left the cell via its apical side. Similar amounts of the open reading frame 2 (ORF2) soluble capsid protein were secreted from both sides of the hepatocytes. This model mimicking physiological conditions should help clarify the HEV cell cycle in polarized hepatocytes.
KEYWORDS: TCID50, bile acid quantification, colocalization, hepatitis E virus, hepatocyte, lipid-associated particle, polarized cell, quasienveloped virus
ABSTRACT
Hepatitis E virus (HEV) is a common cause of acute viral hepatitis worldwide. Most HEV infections are asymptomatic, but immunocompromised patients infected with HEV genotype 3 (HEV3), HEV4, or HEV7 may develop chronic infections. The HEV particles in stools are naked (nHEV), while those in the serum and culture supernatants (eHEV) are associated with lipids. Hepatocytes are polarized epithelial cells that have basolateral (oriented toward the blood) and apical (oriented toward the bile) exosomal pathways. We isolated a subclone, F2, from the human hepatocarcinoma cell line HepG2/C3A that grew as a polarized monolayer culture and had better HEV production than HepG2/C3A cells. F2 cells cultured on semipermeable collagen inserts and infected basolaterally with nHEV3 released 94.6% of virus particles apically, those infected with eHEV3 released 96.8% apically, and eHEV1-infected cells released 99.3% apically. Transcytosis was not involved. Density gradient centrifugation and NP-40 treatment showed that HEV particles released both apically and basolaterally were lipid associated. The apically released HEV3 and HEV1 particles were six and nine times more infectious than those released basolaterally, respectively. Confocal microscopy indicated that the open reading frame 2 (ORF2) capsid protein colocalized apically with ORF3 virus protein, the apical marker DPP4, and the recycling endosome GTPase Rab27a. The amounts of soluble glycosylated ORF2 secreted apically and basolaterally were similar. These polarized-hepatocyte data suggest that infectious HEV particles are mainly released into bile, while the small fraction released into blood could spread HEV throughout the host.
IMPORTANCE Hepatitis E virus (HEV) in stools is naked, while that in culture supernatants and patients’ blood is lipid associated. Its life cycle in hepatocytes, polarized cells with a basolateral side communicating with blood and an apical side connected with bile, is incompletely understood. We have developed a polarized hepatocyte model and used the cells to analyze the supernatants bathing the apical and basolateral sides and HEV subcellular distribution. HEV particles from both sides were lipid associated, and most infectious HEV particles left the cell via its apical side. Similar amounts of the open reading frame 2 (ORF2) soluble capsid protein were secreted from both sides of the hepatocytes. This model mimicking physiological conditions should help clarify the HEV cell cycle in polarized hepatocytes.
INTRODUCTION
Hepatitis E virus (HEV) is one of the most common causes of hepatitis worldwide. HEV is classified within the family Hepeviridae, which contains two genera: Orthohepevirus, comprising four species, named Orthohepevirus A to D, infecting mammals and birds, and Piscihepevirus, infecting trout. The Orthohepevirus A species includes at least eight genotypes, five of which (HEV1 to HEV4 and HEV7) infect humans (1, 2). Genotypes 1 and 2 are restricted to humans and each year cause millions of cases of water-transmitted acute hepatitis in countries with poor sanitation (3). Genotypes 3 and 4 are prevalent in industrialized countries, where they are zoonotic; their reservoirs include pigs, wild boar, deer, and rabbits (4–7). HEV3 and HEV4 can cause acute hepatitis but are also involved in chronic hepatitis in immunocompromised patients; they can worsen chronic liver disorders and can be associated with a range of extrahepatic manifestations (8, 9). HEV7 was detected in dromedary camels and a chronically infected transplant patient who consumed camel milk and meat (10). HEV can also be transmitted by blood transfusion (11).
The 7.2-kb-long HEV RNA genome contains 5′ and 3′ untranslated regions and 3 open reading frames (orf) (12). ORF1 is a nonstructural protein including a methyltransferase, a papain-like cysteine protease, a helicase, and an RNA-dependent RNA polymerase (8). ORF2 is translated from two initiation codons, giving two forms of ORF2. The longer ORF2 form contains a peptide signal and is secreted via the classical secretion pathway, while the shorter ORF2 form becomes part of virus particles (13). These two processes have different kinetics, and only the shorter ORF2 is found in cell lysates (14, 15). ORF3 is a small phosphorylated viroporin that is involved in HEV morphogenesis and release (16–19).
HEV has been cultured in several hepatocarcinoma cell systems (14, 20–23). The HEV secreted from cells is lipid associated (eHEV) (14, 15, 23, 24). This process involves the interaction of ORF3 with the ESCRT (endosome sorting complex required for transport)-associated protein TSG101 (25). Rab27a, a GTPase that takes part in recycling endosomes to the plasma membrane, is also needed for the release of virus particles from infected cells (20). Finally, eHEV is found in a population of vesicles having the characteristics of exosomes (23, 26, 27). The eHEV particles are less infectious than naked HEV (nHEV) but are protected from neutralizing antibodies (15, 23).
Hepatocytes are organized cells that have apical (oriented toward the bile) and basolateral (oriented toward the blood in the spaces of Disse) sides that are delimited by tight junctions. Hepatocyte polarization leads to the segregation of many proteins and functions. For instance, albumin is secreted basolaterally, while bile acids are released apically (28), and dipeptidyl peptidase 4 (DPP4) is a typical apical marker (29). The apical and basolateral endocytosis-exocytosis pathways are different (28), but some proteins are internalized basolaterally and sent to the apical side by transcytosis (28, 30). Studies on unpolarized cells and blood or stool samples suggest that lipid-associated HEV is secreted from both the basolateral and apical sides and that the lipids are removed by bile acids and intestinal proteases (21). However, little is known about the influence of hepatocyte polarity on HEV morphogenesis and egress.
We have found evidence that HEV particles are mainly released from the apical sides of cultured polarized hepatocyte-like cells in a process that does not involve transcytosis. HEV particles are released in a lipid-associated form both apically and basolaterally but are more infectious when released apically. In contrast, similar amounts of soluble ORF2 are secreted on both sides.
RESULTS
HEV production in polarized F2 cells.
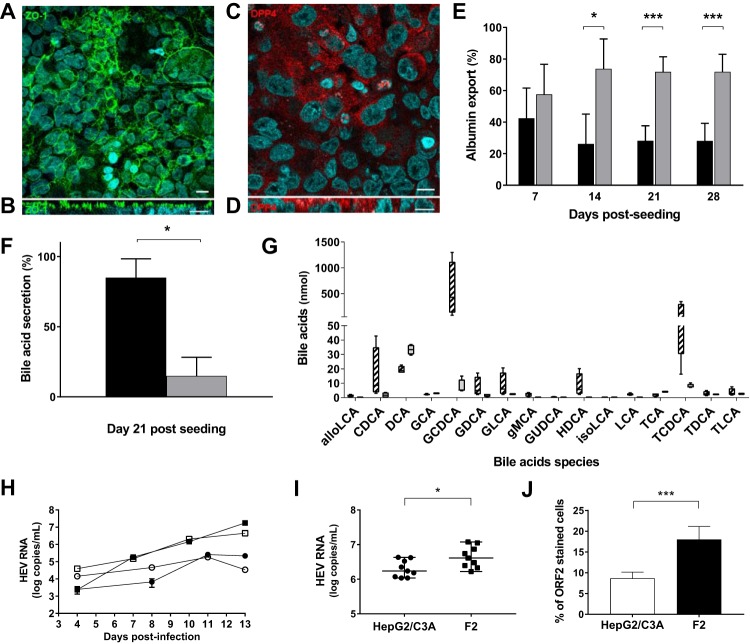
F2 cells, a subclone of HepG2/C3A cells isolated by limiting dilution, were incubated in William’s medium E containing dimethyl sulfoxide (DMSO). The resulting differentiated, polarized hepatocytes were used to analyze the HEV cycle. We collected apical and basolateral culture supernatants from F2 cells grown as monolayers on semipermeable collagen inserts. Confocal microscopy of the F2 cells showed an extensive regular honeycomb pattern of ZO-1 protein staining (Fig. 1A). The (x-z) section of the cell monolayer indicated that ZO-1 was present on the apical sides of the F2 cells (Fig. 1B), which is consistent with a simple columnar epithelium. The cultured F2 cells also contained the apical marker protein DPP4 mainly on their apical sides (Fig. 1C and D). The transepithelial electrical resistance (TEER) was stable throughout the experiment (data not shown), confirming that the apical and basolateral sides of the culture were well distinguished.
FIG 1.
Polarization of HepG2/C3A subclone F2 cells on semipermeable collagen inserts. (A to D) Immunostaining of F2 cells on semipermeable collagen inserts. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Bar =10 μm. (A and B) Maximum-intensity projections of x-y stacks (A) and x-z sections (B) for the tight-junction protein ZO-1. (C and D) x-y (C) and x-z (D) sections for DPP4. (E) Percentages of albumin exported from F2 cells. The albumin in the apical (black bars) and basolateral (gray bars) supernatants was quantified by ELISA. The data shown are means and standard deviations (SD) (n = 12) of the results of four experiments performed in triplicate. *, P < 0.05; ***, P < 0.001. (F) Total bile acids from 21-day cultures of F2 cells quantified by LC-MS. The results are expressed as percentages of exported bile acids in the apical (black bars) and basolateral (gray bars) supernatants. The data shown are means and SD (n = 4). *, P < 0.05. (G) Bile acids were quantified by LC-MS on day 21 postseeding. The amounts of each detected species in the apical (hatched bars) and basolateral (gray bar) supernatants are shown (n = 4). The bile acids quantified were allolithocholic acid (alloLCA), alpha-muricholic acid (aMCA), beta-muricholic acid (bMCA), cholic acid (CA), 3-sulfo-cholic acid (CA-3S), chenodeoxycholic acid (CDCA), 3-sulfo-cheno deoxycholic acid (CDCA-3S), deoxycholic acid (DCA), glycocholic acid (GCA), glycochenodeoxycholic acid (GCDCA), glycodeoxycholic acid (GDCA), glycolithocholic acid (GLCA), gamma-muricholic acid (gMCA), glycoursodeoxycholic acid (GUDCA), hyodeoxycholic acid (HDCA), isodeoxycholic acid (isoDCA), isolithocholic acid (isoLCA), lithocholic acid (LCA), tauroalfamuricholic acid (TaMCA), taurobetamuricholic acid (TbMCA), taurocholic acid (TCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA), taurolithocholic acid (TLCA), 3-sulfotaurolithocholic acid (TLCA-3S), ursodeoxycholic acid (UDCA), and omega muricholic acid (wMCA). For clarity, only detected bile acids are shown. (H) HepG2/C3A (circles) and F2 (squares) cells were infected with nHEV genotype 3 (1.35 × 106 HEV RNA copies/106 cells; white symbols) or eHEV genotype 3 (3.3 × 106 HEV RNA copies/106 cells; black symbols). Supernatants were collected every 2 days, and HEV RNA was quantified by RT-PCR. (I) HepG2/C3A (circles) and F2 (squares) cells were infected with eHEV genotype 3 (2.5 × 108 HEV RNA copies/106 cells). Supernatants were collected on day 15 postinfection, and HEV RNA was quantified by RT-PCR. The data shown are from three independent experiments performed in triplicate. The horizontal bars represent medians. *, P < 0.05. (J) Immunofluorescence of ORF2 protein in HepG2/C3A (white bars) and F2 (black bars) cells 21 to 30 days postinfection. Nuclei were stained with DAPI. The results are expressed as percentages of cells containing ORF2. The data shown are means and SD of the results of three independent experiments. ***, P < 0.001.
The majority of the albumin (over 70%; P < 0.05) was secreted from the basolateral side from the 14th day postseeding (Fig. 1E), and albumin export was maximal in 14- to 28-day-old cultures. The concentrations of bile acids in the culture supernatants were low because they were diluted with medium, but the apical supernatant contained almost all the secreted bile acids (apical, 834 nmol [range, 140 to 1,900 nmol], 92.3%; basolateral, 70 nmol [range, 62 to 87 nmol], 7.7%; P < 0.05) in 21-day cultures (Fig. 1F). The main bile acid was chenodeoxycholic acid, and 94% of the bile acids secreted via the apical side were conjugated to taurine or glycine (glycine/taurine conjugate ratio, ∼3:1) (Fig. 1G). The differential release of albumin and bile acids was consistent with their vectorial export.
We compared the abilities of HEV genotype 3 to infect F2 cells and HepG2/C3A cells by measuring the HEV RNA in their culture supernatants. There was more virus RNA in the supernatants of F2 cells infected with nHEV or eHEV than in the equivalent HepG2/C3A supernatants during days 1 to 13 postinfection (Fig. 1H). The difference was still significant 15 days postinfection (4.9 × 106 [range, 1.66 × 106 to 1.19 × 107] HEV RNA copies/ml in F2 supernatants and 1.72 × 106 [range, 1.08 × 106 to 4.29 × 106] copies/ml in HepG2/C3A supernatants; P < 0.05) (Fig. 1I). Confocal microscopy indicated that significantly more F2 cells (18% ± 3.2%) than HepG2/C3A cells (8.6% ± 1.5%) contained ORF2 (P < 0.001) (Fig. 1J). The inoculum of nHEV required to infect 50% of 105 F2 cells contained 2.9 × 103 copies of HEV RNA, while the inoculum required to infect an equivalent number of HepG2/C3A cells was 6.2 × 103 copies of HEV RNA. Thus, F2 cells were more readily infected by HEV than were HepG2/C3A cells, and this was associated with the release of more HEV RNA into the culture supernatant.
Kinetics of HEV particle production.
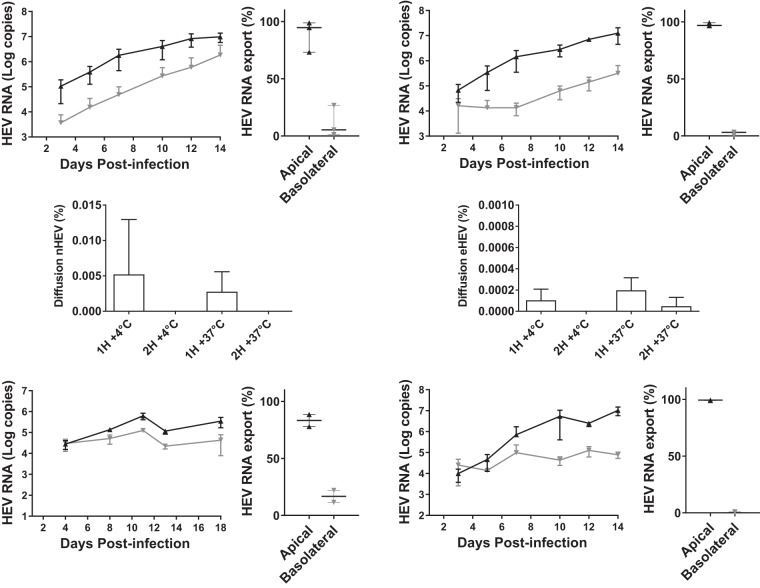
Because we assumed that HEV enters hepatocytes through their basolateral surfaces, we infected 14-day cultures of F2 cells with nHEV or eHEV genotype 3 via their basolateral sides and monitored the HEV RNA secreted for the next 14 days. The inocula contained 6,500 tissue culture infective doses (TCID50) per 106 cells, corresponding to 8.6 × 106 copies of nHEV RNA or 1.7 × 109 copies of eHEV RNA for every 106 cells. HEV RNA was mainly secreted from the apical sides of the cells during the 14 days of infection regardless of the form of the infecting virus particles (Fig. 2A to D). The apical supernatant of nHEV-infected F2 cells contained 94.6% (median, 1.0 × 107 HEV RNA copies; range, 5.6 × 106 to 1.3 × 107), and their basolateral supernatant contained 5.4% (median, 5.8 × 105 HEV RNA copies; range, 7.0 × 104 to 4.9 × 106) of the total HEV RNA secreted (Fig. 2A and B). Similar results were obtained with eHEV-infected F2 cells: the apical supernatant contained 96.8% (median, 1.2 × 107 HEV RNA copies; range, 5.0 × 106 to 2.1 × 107), and the basolateral supernatant contained 3.2% (median, 1.8 × 105 HEV RNA copies; range, 8.3 × 104 to 6.9 × 105) of the total secreted HEV RNA (Fig. 2C and D).
FIG 2.
Polarized release of HEV RNA. (A to D) F2 cells grown on semipermeable inserts for 14 days were infected with nHEV genotype 3 (A and B) or eHEV genotype 3 (C and D) (6,500 TCID50/106 F2 cells) added to the basolateral medium. Apical (black line) and basolateral (gray line) supernatants were collected three times a week, and HEV RNA was quantified by RT-PCR. The data shown are means ± SD of the results of three independent experiments. (B and D) Percentages of total HEV RNA in apical and basolateral compartments infected with nHEV (B) or eHEV (D) for 14 days. (E and F) Basolateral-to-apical transcytosis of nHEV (E) and eHEV (F) 14 days postseeding, HEV RNA (6,500 TCID50/106 cells) was placed in the basolateral compartment and incubated for 2 h at 4°C or 37°C. The results are percentages of the HEV RNA passaged to the apical compartment. The data shown are means and SD of the results of two independent experiments (n = 3). (G to J) HepG2/C3A cells (G and H) or F2 cells (I and J) grown on semipermeable inserts for 14 days were infected with eHEV genotype 3 (5.85 × 108 HEV RNA copies/106 cells) (G and H) or eHEV genotype 1 (2.7 × 107 RNA HEV copies/106 cells) (I and J). Virus in the apical (black lines) and basolateral (gray lines) supernatants was quantified as described for panel A (G and I) and the relative percentages were calculated as for panel B (H and J).
We looked at the role of transcytosis in the apical secretion of HEV by incubating F2 cell monolayers with nHEV or eHEV (6,500 TCID50/106 cells) in the basolateral medium for 1 and 2 h at 4°C or 37.5°C (Fig. 2E and F) and measuring the HEV RNA in the apical and basolateral supernatants. Very little nHEV RNA crossed the cell monolayer at 4°C (5.2 × 10−3% ± 7.7 × 10−3% in 1 h and undetectable after 2 h). Incubating the cells at 37.5°C did not increase these percentages (2.8 × 10−3% ± 2.8 × 10−3% after 1 h and undetectable after 2 h) (Fig. 2E). Even less eHEV RNA crossed the F2 cell monolayer (4°C, 1.0 × 10−4% ± 1.1 × 10−4% after 1 h and undetectable after 2 h; 37.5°C, 1.9 × 10−4% ± 1.2 × 10−4% after 1 h and 4.8 × 10−5% ± 8.3 × 10−4% after 2 h) (Fig. 2F). Hence, transcytosis was not involved in the apical release of HEV from F2 cells.
Control studies with eHEV genotype 3 and HepG2/C3A cells found that the apical supernatant contained 83% (range, 2.2 × 105 to 4.7 × 105 HEV RNA copies) and the basolateral supernatant contained 17% (range, 1.8 × 104 to 6.8 × 104 HEV RNA copies) of the total secreted eHEV RNA (Fig. 2G and H), showing that apical HEV RNA secretion is not specific to F2 cells.
The experiment was repeated with an eHEV genotype 1 strain and F2 cells; the apical supernatant contained 99.3% (median, 7.9 × 106 HEV RNA copies; range, 7.5 × 106 to 1.6 × 107), and the basolateral supernatant contained 0.7% (median, 7.6 × 104 HEV RNA copies; range, 5.3 × 104 to 1.1 × 105) of the total secreted eHEV RNA (Fig. 2I and J). Thus, mainly apical HEV RNA secretion is not specific to HEV genotype 3.
Properties of HEV particles produced by F2 polarized cells.
Density gradient centrifugation of the HEV genotype 3 particles released from F2 cells indicated that the density of particles in the basolateral supernatant was 1.08 while that of particles in the apical supernatant was 1.10 (Fig. 3A). Treating both supernatants with 1% NP-40 resulted in HEV RNA particles with densities of >1.18 (Fig. 3B and C). This pattern was similar to that obtained with virus particles from feces (Fig. 3C), indicating that all the HEV particles were lipid associated. Studies with HEV genotype 1 particles produced similar differences in apical and basolateral densities and the NP-40-triggered density shift (Fig. 3D and E).
FIG 3.
Properties of secreted HEV RNA. (A) Apical (black line) and basolateral (gray line) supernatants (105 copies of HEV RNA) were loaded onto a 7.5 to 40% iodixanol gradient, ultracentrifuged, and fractionated. The percentage of HEV RNA in each fraction (density) is shown. The density gradients are representative of the results of three independent experiments. (B to E) Apical (black line) and basolateral (gray line) supernatants containing 104 copies of HEV RNA genotype 3 (B) or HEV genotype 1 (D) were loaded onto 7.5 to 40% iodixanol minigradients. (C and E) The same samples were treated with 1% NP-40 and ultracentrifuged. Untreated feces from HEV genotype 3 (104 copies of HEV RNA) were added as controls (C, dashed line). (F) The infectivity of apical or basolateral supernatants from F2 cells infected with HEV genotype 3 was evaluated using an endpoint dilution assay, and the TCID50 per million HEV RNA copies was calculated (n = 6). Each symbol represents a pair of apical and basolateral supernatants from the same insert. The bars indicate medians. **, P < 0.01. (G) The infectivity of apical or basolateral supernatants of F2 cells infected with eHEV genotype 1 was evaluated as for panel F (n = 3). The bars represent medians.
Analysis of the infectivity of HEV particles from the apical and basolateral supernatants from 14-day infected polarized hepatocytes by TCID50 gave 5,901 (range, 2,271 to 18,870) TCID50 per million HEV RNA copies for the apical supernatant and 924 (range, 335 to 2,914) for the basolateral supernatant. Thus, approximately 6 times more infectious particles were secreted from the apical side of the cell monolayer than from its basolateral side (Fig. 3F) (P < 0.01).
Similar results were obtained with HEV genotype 1; TCID50 were 2,395 (range, 1,159 to 16,969) per million HEV RNA copies for the apical supernatant and 266 (range, 59 to 1,106) for the basolateral supernatant, corresponding to 9 times more infectious particles secreted apically than basolaterally (Fig. 3G).
Virus ORF2 protein in culture supernatants.
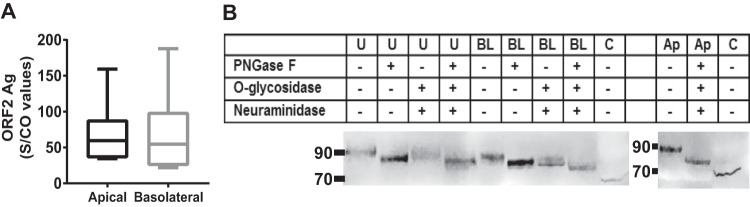
We measured the ORF2 protein in supernatants from HEV genotype 3-infected cells using the Wantaï HEV Ag ELISA (enzyme-linked immunosorbent assay) kit, which detects both the native and glycosylated forms of ORF2. The amounts of ORF2 protein secreted from the apical and basolateral sides were similar (apical median, 62.7 sample/cutoff [S/CO] [range, 58.6 to 159.2]; basolateral median, 47.8 S/CO [range, 22.0 to 187.7]) (Fig. 4A).
FIG 4.
Secretion of ORF2 protein from both sides of F2 cells. (A) The ORF2 protein in HEV3-infected apical or basolateral culture supernatants was quantified with a Wantaï HEV Ag ELISA kit. The results are S/CO values. The box plots show minimal and maximal values, and the horizontal bars represent medians (n = 6). (B) Immunodetection of ORF2 protein in extracts of unpolarized HEV3-infected culture supernatant (U), basolateral supernatant (BL), or apical supernatant (Ap) or cell lysates (C) from HEV3-infected F2 cells. When indicated (+), the same samples were treated with the indicated glycosidases.
Western blotting gave the apparent molecular mass of the ORF2 protein in the apical and basolateral supernatants as 90 kDa (Fig. 4B). Incubating the supernatants with glycosidases to remove N-linked (peptide-N-glycosidase F [PNGase F]) or O-linked sugars (O-glycosidase, when combined with neuraminidase) decreased the apparent molecular mass to 85 kDa, while a combination of all three enzymes decreased the ORF2 apparent molecular mass to 80 kDa, confirming that ORF2 is glycosylated. The cell lysate also contained an ORF2 protein with an apparent molecular mass of 75 kDa.
Distribution of HEV proteins in F2 polarized hepatocytes.
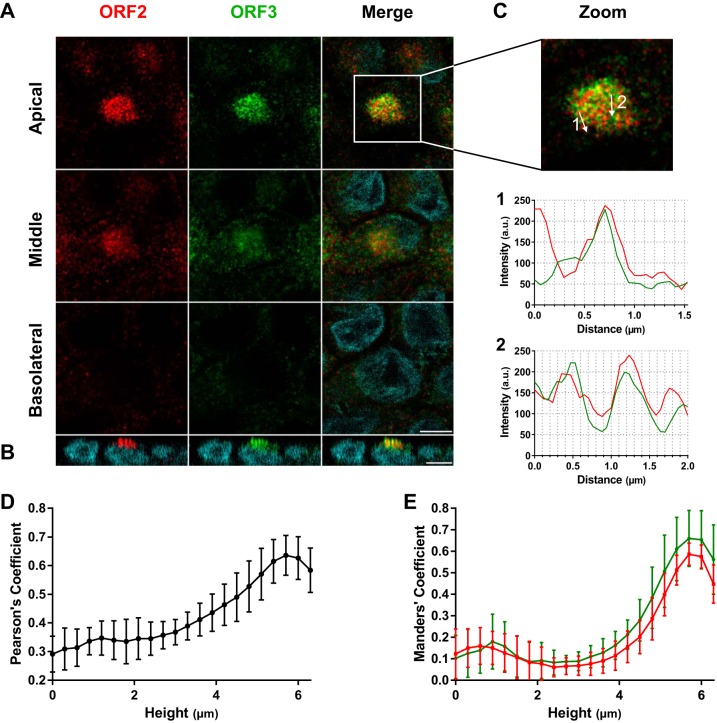
Immunostaining and confocal microscopy indicated that the ORF2 and ORF3 proteins were in vesicle-like structures (Fig. 5A) concentrated at the apical sides of HEV genotype 3-infected cells (Fig. 5A and B). Object-based analysis (OBA) confirmed that the vesicle-like structures contained both ORF2 and ORF3 proteins (Fig. 5C), showing their occasional colocalization. The Pearson correlation coefficient (PCC) increased from 0.29 ± 0.06 at the basolateral slice to 0.64 ± 0.07 at the apical slice (Fig. 5D), indicating that the strength of the linear relation between ORF2 and ORF3 intensities increased toward the apical side. Calculation of Manders coefficients indicated that the proportion of ORF2 in ORF3-labeled voxels increased from 0.12 ± 0.12 to 0.59 ± 0.05 and that the proportion of ORF3 in ORF2-labeled voxels increased from 0.10 ± 0.11 to 0.66 ± 0.13, confirming that the colocalization of both proteins increased from the basolateral to the apical side (Fig. 5E). Similar results were observed in the whole field (see Movie S1 in the supplemental material). Thus, these results confirmed the preferential localization of the ORF2 and ORF3 proteins in the same vesicular structures at the apical side.
FIG 5.
Accumulation of ORF2 and ORF3 proteins at the apical sides of hepatocytes. (A and B) Immunostaining of ORF2 (red) and ORF3 (green) proteins from F2 cells grown for 14 days on inserts, infected with HEV genotype 3, and sampled 14 days postinfection. (A) Z stack of x-y sections of cells acquired by confocal microscopy and three (apical, middle, and basolateral) z sections. Scale bar = 10 μm. (B) x-z section of the cells stained for panel A. Nuclei (cyan) were stained with DAPI. Scale bar = 10 μm. (C) Paths (arrows) of the 2-pixel-wide line scans shown in graphs 1 and 2 below. (D and E) Stacks of 22 (x-y) sections were acquired, slices were separated, and Pearson’s (D) or Manders (E) coefficients were calculated on each section. The mean proportions of ORF2 intensities in ORF3 (red) and of ORF3 intensities in ORF2 (green) are shown. The error bars represent SD; n = 10.
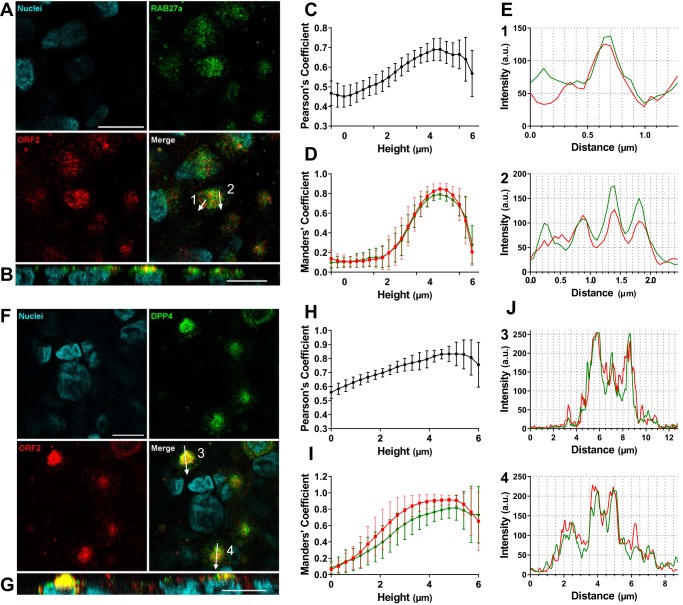
Immunostaining for ORF2 protein and the Rab27a GTPase, involved in endosome recycling at the plasma membrane, showed that Rab27a was also in vesicle-like structures (Fig. 6A) and that there was more Rab27a at the apical sides of HEV genotype 3-infected cells (Fig. 6B; see Movie S2 in the supplemental material). PCC and Manders coefficients increased from the basolateral to the apical sides (PCC, 0.47 ± 0.06 to 0.66 ± 0.07; Manders, ORF2 in Rab27a, 0.13 ± 0.08 to 0.85 ± 0.06, and Rab27a in ORF2, 0.10 ± 0.05 to 0.79 ± 0.05) (Fig. 6C and D; see Movie S2). OBA confirmed their colocalization (Fig. 6E). Similar results were obtained for the canalicular marker DPP4 (PCC, 0.56 ± 0.04 to 0.83 ± 0.08; Manders, ORF2 in DPP4, 0.06 ± 0.07 to 0.91 ± 0.07, and DPP4 in ORF2, 0.08 ± 0.07 to 0.82 ± 0.15) (Fig. 6F to J; see Movie S3 in the supplemental material).
FIG 6.
Colocalization of ORF2 with Rab27A and DPP4 at the apical sides of hepatocytes. (A to E) Immunostaining of ORF2 protein (red) and Rab27a (green) in F2 cells grown on inserts for 14 days, infected with HEV genotype 3, and sampled 14 days postinfection. The arrows across the cells on the merged image indicate the paths of the 2-pixel-wide scans shown in panel E. (C and D) Stacks of 22 (x-y) sections were acquired, slices were separated, and Pearson’s (C) or Manders (D) coefficients were calculated on each section. The mean proportion of ORF2 intensities in Rab27a (red) and of Rab27a intensities in ORF2 (green) are shown. The error bars represent SD; n = 10. (F to J) Immunostaining of ORF2 protein (red) and DPP4 (green) in F2 cells grown on inserts for 14 days, infected with HEV genotype 3, and sampled 14 days postinfection. The arrows across the cells in the merged image indicate the paths of the 2-pixel-wide line scans shown in panel J. (H and I) Stacks of 22 (x-y) sections were acquired, slices were separated, and Pearson’s (H) or Manders (I) coefficients were calculated on each section. The mean proportions of ORF2 intensities in DPP4 (red) and of DPP4 intensities in ORF2 (green) are shown. The error bars represent SD; n = 10. Scale bars = 10 μm.
DISCUSSION
Our studies with polarized hepatocytes in cell culture showed that the HEV particles released from both sides of F2 cells are lipid associated but that most of them leave cells from the apical side by a process that does not seem to involve transcytosis. The virus ORF2 and ORF3 proteins are preferentially located at the apical side, as are Rab27a and DPP4. Ratios of infectious particles to HEV RNA copies were higher at the apical than at the basolateral side, suggesting that the apical pathway is the main release route. In contrast, the amounts of soluble glycosylated ORF2 protein secreted from both sides are similar.
Few cell culture systems support efficient HEV replication and spreading in vitro. Our cells were grown in a medium containing DMSO, which induces and maintains the polarization of many cells in culture, including hepatocytes (31). Yin et al. have shown that media containing DMSO enhance HEV RNA production (15). The cells were cultured on semipermeable collagen-coated inserts to produce confluent monolayers of hepatocyte-derived cells that had the columnar morphology of simple epithelial cells, which enabled us to collect samples of apical and basolateral supernatants (28). We selected a clone, F2, of HepG2/C3A cells that supported better HEV growth than the mother cell line. To produce similar conditions of infection, inocula were adjusted using the TCID50, corresponding to 10 to 100 times more lipid-associated HEV RNA copies than naked HEV. This difference in infectivity has been described in unpolarized systems (20, 23). We found that the kinetics of HEV production by cells infected with eHEV or nHEV were similar. F2 cells grown on inserts had the typical apical proteins ZO-1 and DPP4 and released albumin mainly from their basolateral sides and bile acids from their apical sides. Most of the bile acids were conjugated (glycine-conjugated/taurine-conjugated ratio, ∼3:1), which is characteristic of bile (32). Thus, our system reproduces the main morphological and functional features of polarized hepatocytes and could be a useful tool for studying HEV in polarized cells.
Recent studies indicate that the ORF2 virus capsid protein is also secreted in a soluble glycosylated form involving the classical secretion pathway (13, 14). Both the apical and basolateral supernatants contained a band at 90 kDa that gave an 80-kDa protein after deglycosylation with PNGase F, O-glycosidase, and neuraminidase, confirming that the ORF2 in the culture supernatant was glycosylated. Yin et al. (13) reported a similar difference in the molecular masses of deglycosylated extracellular ORF2 and intracellular ORF2. They also showed that soluble ORF2 does not interfere with the entry of either nHEV or eHEV (13). Soluble ORF2 could alter the ability of IgG and IgM antibodies to neutralize nHEV reaching the portal vein during an infection.
HEV-contaminated food products can transmit HEV genotype 3 (33, 34), but little is known about the form of the virus particles that reach the liver via the portal vein. Regardless of the form of virus (nHEV or eHEV) and whether it is genotype 3 or 1, polarized cells released around 95% of the HEV RNA from their apical sides as lipid-associated particles, supporting the hypothesis that HEV buds off from the apical sides of hepatocytes and loses its quasienvelope due to the detergent action of bile acids (35).
Hepatotrophic viruses like hepatitis A virus (HAV), hepatitis B virus (HBV), and hepatitis C virus (HCV) use hepatocyte polarization for their release (36–39). HAV, like HEV, is a naked virus that is lipid associated in blood (40) and on both sides of polarized cultures (36, 37). Most HAV particles leave hepatocytes grown in a similar system developed from HepG2 cells across their basolateral sides (36), while most HEV particles leave our F2 cells from the apical side. Clearly, the two viruses use different cell mechanisms for their egress. The preferential HEV apical secretion correlates well with the 100-fold-higher concentration of HEV RNA in the bile than in the blood of pigs (41). Although relatively few infectious particles are released basolaterally, they could be sufficient to spread HEV in the host. HEV replication in other organs could then lead to extrahepatic manifestations (9).
We looked for the involvement of transcytosis in HEV transport and accumulation in the apical supernatant, as it is a feature of hepatocytes (28). This was done by measuring the percentages of nHEV and eHEV passing through the cells when active transport was possible (37°C) or not possible (4°C). There appears to be very little transport at either 4°C or 37°C, mainly for eHEV, which is relevant because eHEV is the virus form found in patients’ blood and at the basolateral side of our F2 cultures. Therefore, de novo virus particles reach the apical side of hepatocytes via exocytosis rather than transcytosis.
The pathways of exocytosis from the basolateral and apical sides of hepatocytes are different (28). While all released HEV particles are lipidated, the densities of those released apically were different from those of particles released basolaterally. We have reported that HEV particles are released from cells together with exosomes (23), but the population of exosomes is heterogeneous (42). Studies on polarized colon or kidney cells show that high-density exosomes containing Wnt3 are secreted from the apical side while lower-density exosomes containing CD63 are secreted basolaterally (43, 44). We found that virus particles released from the apical or basolateral side had the same density after incubation with NP-40 as virus particles in feces, suggesting that they differ in their quasienvelope compositions.
Confocal microscopy revealed that ORF2 and ORF3 proteins colocalized and accumulated at the apical side and with the typical apical marker DPP4, a transmembrane protein in the bile canaliculi of hepatocytes in vivo (29). These images per se do not show whether this corresponds to entry or egress, but ORF3 is known to interact with the ESCRT-associated protein Tsg101 via its PSAP motif, which is needed for the release of lipid-associated HEV (17, 25). ORF2 protein is distributed with Rab27a, a major GTPase involved in recycling late endosomes to the plasma membrane (45), which also favors virus egress.
A major finding was that the HEV particles released from the apical sides of cultured cells are more infectious than those released from the basolateral side, indicating that the infectivity of quasienveloped particles can vary. This also indicates that infectious particles are released most efficiently via the apical exocytosis pathway. The polarized targeting of viral proteins is critical for the pathogenesis of several viruses. The influenza A virus M2 protein must be at the apical side of the host cell for the optimal production of infectious particles (46). Similarly, measles virus is released from the apical membranes of polarized epithelial cells by hijacking Rab11A-positive recycling endosomes (47). ORF3 may move toward the ESCRT complex of the apical multivesicular bodies to enable lipid-associated HEV particles to egress, leading to the accumulation of HEV RNA in the apical supernatant.
In summary, we have developed a system of cultured polarized hepatocytes that closely mimics the pathophysiological conditions of HEV infection and found that the majority of infectious HEV particles are released from the bile sides of hepatocytes.
MATERIALS AND METHODS
Culture media and reagents.
Dulbecco's modified Eagle's medium (DMEM), William’s medium E (WME; 32551), fetal bovine serum (FBS), goat serum, 0.05% trypsin-EDTA (1×) containing phenol red, phosphate-buffered saline without calcium and magnesium (PBS), type I collagen (A1048301), rabbit anti-ZO-1 (61-7300), goat anti-mouse 488 (A11029), goat anti-rabbit 555 (A21428), and NucBlue fixed cell stain (R37606) were all purchased from ThermoFisher Scientific, Life Technologies SAS (Saint-Aubin, France). The FBS was heat inactivated and depleted of exosomes by ultracentrifugation (110,000 × g; 14 h; 4°C). Penicillin G (10,000 U/ml), streptomycin (10,000 U/ml), and amphotericin B (25 μg/ml) were supplied as a mixture (PSA) by Bio Whittaker-Lonza (Amboise, France). Bovine serum albumin (BSA), DMSO, Optiprep, Transwell-COL tissue culture inserts (COSTAR; 4.7-cm2 growth area; 3.0-μm pore size), anti-Rab27a (R4655), mouse anti-ORF2 antibodies (clones 1E6 and 4B2), and Fluoromount (F4680) were supplied by Sigma-Aldrich (Saint-Quentin Fallavier, France). The Cytofix/Cytoperm and BDperm/wash were purchased from BD Biosciences (Le Pont de Claix, France). Rabbit anti-ORF3 antibody (bs 0212-R) was from Clinisciences (Nanterre, France). Rabbit anti-CD26/DPP4 (Ab28340) was from Abcam (Paris, France). Odyssey blocking buffer (OBB), IRDye 800 CW donkey anti-mouse IgG (926-32212), and Chameleon Duo prestained protein ladder (928-60000) were supplied by LI-COR Biosciences GmbH (Bad Homburg, Germany).
Cells.
HepG2-derived C3A cells were obtained from the ATCC (HepG2/C3A; ATCC CRL-10741). A subclone, HepG2/F2, was obtained by limiting dilution according to the protocol described by Snooks et al. (36). Briefly, cells were detached by incubation with trypsin-EDTA in Hanks balanced salt solution (Ca2+ and Mg2+ free) containing 3 mM EGTA for 20 min at 37.5°C, passed three times through a 27-gauge needle, diluted, and seeded at 0.5 cells/well in a 96-well plate. HepG2/C3A cells or the F2 clone cells were grown in DMEM supplemented with 10% FBS.
Cells to be infected (106 cells) were seeded on semipermeable inserts of collagen and incubated for 14 days at 37°C in William’s medium E containing 10% FBS, 1% DMSO. Half of the culture medium was replaced three times a week. The results of all experiments using inserts were corrected for the relative volumes of apical (1.5 ml) and basolateral (2.6 ml) culture medium.
Viruses, infection, and transcytosis experiments.
We used clinical samples from patients at the acute phase of infection. Feces from a patient infected with a HEV genotype 3f strain (TLS-09/M0 [48]) were used to prepare suspensions of nHEV. The culture supernatant from the first cell passage of this 3f feces strain on PLC/PRF/5 cells (ATCC; CRL-8024) was used to prepare suspensions of lipid-associated HEV (eHEV). The ORF2 sequences of nHEV and eHEV were identical.
Cells were infected as previously described (23) using a medium containing 96% WME, 2% heat-inactivated exosome-free FBS, 1% PSA, and 1% DMSO. We used an inoculum of nHEV containing 8.6 × 106 HEV RNA copies per 106 cells or eHEV containing 1.7 × 109 HEV RNA copies per 106 cells, corresponding to a TCID50 of 6,500 per 106 cells in each case to infect the cells on inserts. We obtained genotype 1 by growing feces on HepG2/C3A cells and concentrating the supernatants. The resulting concentrated first-passage suspension contained 107 HEV RNA copies/ml.
The nHEV or eHEV inoculum was added to the basolateral side of the F2 cell monolayer (washed three times with PBS). Fresh medium was added to the opposite side, and the plates were incubated for 2 h at 37°C. The inoculum was then removed, the cells were washed three times with PBS, and fresh medium was added. We tested for transcytosis by incubating the cells for 2 h at 4°C and then for 2 h at 37°C with nHEV or eHEV genotype 3 inoculum in the basolateral cell monolayer. The HEV RNA in the basolateral and apical supernatants was quantified by reverse transcription (RT)-PCR at each step. The rate of HEV transcytosis was determined as the percentage of input per hour at 4°C or 37°C.
Albumin and bile acid assays.
Albumin was assayed with the Albumin Human ELISA kit (ab108788; Abcam, Paris, France), following the manufacturer’s instructions. Samples were diluted to lie within the linear range of the assay.
Bile acids were measured (adapted from reference 49) at the Metatoul lipidomic platform (INSERM UMR1048, Toulouse, France) certified to ISO 9001:2015 standards. Supernatants (500 μl) were frozen at −80°C diluted in water (500 μl) containing an internal standard (23NorDCA; Steraloids, Newport, USA). Bile acids were first concentrated on an HLB solid-phase extraction system (Waters SAS, Saint-Quentin-en-Yvelines, France): plates were conditioned (1 ml water, 2 ml methanol), samples were loaded, and the plates were washed with 2 ml water. Finally, bile acids were eluted with 2 ml methanol. The final concentrated extracts were dissolved in 20 μl methanol and stored at −20°C until they were analyzed by high-resolution liquid chromatography-mass spectrometry (LC-MS) on an Ultimate 3000 (ThermoFisher Scientific, Life Technologies SAS, Saint-Aubin, France) equipped with a 2.1- by 100-mm, 1.7-μm Zorbax SB C18 column (Agilent, Santa Clara, CA) at 45°C. The mobile phases were 15 mM ammonium acetate, pH 5.3 (acetic acid) (A); acetonitrile (B); and methanol (C) in gradient mode (flow rate, 0.5 ml/min). The gradient was as follows: 20% B-0% C from 0 min to 2.5 min, 16% B-20% C at 5 min, 1% B-95% C at 23 min, and 20% B-1% C from 23.5 min to 27.5 min. The injection volume was 5 μl. The high-resolution mass spectrometer was an Exactive (ThermoFisher Scientific, Life Technologies SAS, Saint-Aubin, France) running in electrospray ionization (ESI) negative mode with full scan acquisition (200 to 600 m/z). The source parameters were as follows: sheat gas (N2), 30 lb/in2; auxiliary gas flow, 10 lb/in2; sweep flow rate, 0 lb/in2; spray voltage, 2,500 V; capillary temperature, 200°C; capillary voltage, −37.5 V; tube lens voltage, −165 V; heater temperature, 200°C; skimmer voltage, −46 V; maximum injection time; 250 ms. Calibration curves were prepared from standards (from Steraloid and Sigma-Aldrich) in methanol (24, 6, 1.5, 0.375, 0.093, 0.023, and 0.0058 ng/ml). Trace Finder quantitative software (ThermoFisher Scientific, Life Technologies SAS, Saint-Aubin, France) was used to quantify each compound using the resulting ion chromatogram (5-ppm windows). All concentrations were converted to molarity in order to compare amounts of bile acids.
The amounts of apical and basolateral exports of albumin and bile acids per hour were calculated as fractions of the totals (apical plus basolateral).
Density gradient.
HEV-infected supernatants were fractionated on discontinuous gradients of 7.5 to 40% iodixanol prepared from Optiprep as previously described (23). Fractions (0.5 ml) were collected from the top of the tube, and their densities were measured with an Abbe refractometer.
The effect of detergents on HEV particles was analyzed by treating samples with 1% NP-40 for 15 min and loading them on a 1-ml 7.5 to 40% “minigradient” (prepared by adding 100 μl of 40% to 10% iodixanol solutions and the sample diluted in a final 7.5% iodixanol solution). The minigradients were centrifuged at 92,500 × g (T.25 rotor; Beckman-Coulter Optima L80-XP; Beckman Coulter France S.A.S., Paris, France) for 14 h at 10°C. Fractions (90 μl) were collected and analyzed.
HEV RNA quantification.
HEV RNA was extracted from 140-μl samples (QiaAmp viral RNA minikit; Qiagen, Courtaboeuf, France) and quantified by RT-PCR of the orf3 gene. This accredited ISO 15189 method has a limit of detection of 100 HEV RNA copies/ml (50).
Indirect immunofluorescence.
Cells were washed twice with PBS, fixed with Cytofix/Cytoperm (20 min at 4°C), and permeabilized by incubation with BD/PermWash for 10 min at room temperature. Free binding sites were blocked by incubation with 5% goat serum in PBS for 30 min at room temperature. Cells were stained with anti-ORF2 monoclonal antibodies (mouse 1E6 [1/100] plus 4B2 [1/100]) and/or rabbit anti-ORF3 (1/100), anti-ZO-1 (1/50), anti-Rab27a (1/300), or anti-CD26/DPP4 antibodies (1/200) diluted in 5% goat serum in PBS. The cells were washed 5 times in PBS, and immune complexes were detected with Alexa-labeled secondary antibodies (diluted 1/1,000 in 5% goat serum in PBS) and again washed 5 times in PBS. Nuclei were counterstained with NucBlue fixed cell stain following the manufacturer’s instructions and finally mounted in Fluoromount solution.
Confocal microscopy was performed at the Cell Imaging Facility (INSERM UMR1043, Toulouse, France) certified to ISO 9001:2015 and NFX 50 to 900 v2016 standards, using a LEICA SP8 (63× objective; numerical aperture, 1.4; Leica Microsystems, Germany), and the resulting images were analyzed using ImageJ software (authored by Wayne Rasband, Research Services Branch, National Institute of Mental Health, Bethesda, Maryland, USA). Colocalization studies were performed with the JACoP plugin (51) for ImageJ. PCCs were used to evaluate the linear correlation between the intensities of two channels. Manders coefficients sum the intensities of pixels in channel A that find a nonnull counterpart in channel B, which are then divided by the total intensity of pixels in channel A (proportion of A into B), and inversely for the proportion of B into A. Pearson and Manders coefficients were calculated for consecutive cell heights by separating all image slices and calculating both coefficients for each optical slice, applying the same threshold to each optical slice.
ORF2 protein quantification.
The ORF2 protein was quantified with the Wantaï HEV Ag ELISA kit (Wantai Biological Pharmacy Enterprise Co., China). Samples were diluted to lie within the linear range of the assay, and the results are expressed as the ratio of the absorbances of the S/CO and corrected for the dilution and volume of medium on the corresponding side.
Glycosidase digestions.
PNGase F, O-glycosidase, and neuraminidase (New England Biolabs, Ipswich, MA) were used following the manufacturer’s instructions. Briefly, protein extracts were incubated in denaturing buffer for 10 min at 100°C and then with enzyme(s) in 1% NP-40 buffer for 4 h at 37°C. Samples were analyzed by Western blotting.
Western blots.
ORF2 on Western blots was detected (23) by incubation with monoclonal anti-ORF2 antibodies (1E6 [1/2,000] and 4B2 [1/2,000] in OBB) overnight at room temperature and then with IRDye 800CW donkey anti-mouse IgG secondary antibody (diluted 1/15,000 in OBB) for 1 h at room temperature. A Chameleon Duo prestained protein ladder was used as a size marker. Images were acquired using an Odyssey Fc (LI-COR, Bad Homburg, Germany).
Determination of TCID50.
The TCID50, the reciprocal of the virus suspension dilution that infected 50% of the culture wells, was determined by endpoint dilution, using HEV RNA to identify infected wells 10 days postinfection. Briefly, 96-well plates were coated with type I collagen and seeded with 105 F2 cells per well. The cells were inoculated 24 h postseeding by incubation with serial 10-fold dilutions (6 replicates) of virus suspension in infection medium for 6 h at 35.5°C. The cells were then washed 5 times in PBS at 24 h postinfection and maintained in fresh infection medium. Half of each culture supernatant was replaced every 2 days. Half of each culture supernatant was discarded at day 10 postinfection, the cells were lysed by freezing and thawing the plates once, and the HEV RNA in each well was measured. The TCID50 was determined using the Reed and Münch calculation (23).
Statistical analysis.
All statistical analyses were performed with GraphPad Prism 7 software (version 7.03; GraphPad Software, La Jolla, CA). Mann-Whitney and Wilcoxon tests were used to compare quantitative variables and the χ2 test to compare the percentages of infected cells. P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by INSERM UMR1043 (Toulouse, France).
We thank Astrid Canivet at the INSERM UMR 1043 cell imaging facility, Toulouse, France, for technical assistance. The English text was edited by Owen Parkes.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01207-18.
REFERENCES
- 1.Purdy MA, Harrison TJ, Jameel S, Meng X-J, Okamoto H, Van der Poel WHM, Smith DB, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Hepeviridae. J Gen Virol 98:2645–2646. doi: 10.1099/jgv.0.000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DB, Simmonds P. 2018. Classification and genomic diversity of enterically transmitted hepatitis viruses. Cold Spring Harb Perspect Med 8:a031880. doi: 10.1101/cshperspect.a031880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal R, Goel A. 2018. Natural history, clinical manifestations, and pathogenesis of hepatitis E virus genotype 1 and 2 infections. Cold Spring Harb Perspect Med 8:a032136. doi: 10.1101/cshperspect.a032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A 94:9860–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K, Kitajima N, Abe N, Mishiro S. 2004. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330:501–505. doi: 10.1016/j.virol.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, Wang Y. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81:1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- 7.Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guérin J-L. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg Infect Dis 18:1274–1281. doi: 10.3201/eid1808.120057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamar N, Izopet J, Pavio N, Aggarwal R, Labrique A, Wedemeyer H, Dalton HR. 2017. Hepatitis E virus infection. Nat Rev Dis Primers 3:17086. doi: 10.1038/nrdp.2017.86. [DOI] [PubMed] [Google Scholar]
- 9.Kamar N, Marion O, Abravanel F, Izopet J, Dalton HR. 2016. Extrahepatic manifestations of hepatitis E virus. Liver Int 36:467–472. doi: 10.1111/liv.13037. [DOI] [PubMed] [Google Scholar]
- 10.Lee G-H, Tan B-H, Teo EC-Y, Lim S-G, Dan Y-Y, Wee A, Aw PPK, Zhu Y, Hibberd ML, Tan C-K, Purdy MA, Teo C-G. 2016. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150:355–357.e3. doi: 10.1053/j.gastro.2015.10.048. [DOI] [PubMed] [Google Scholar]
- 11.Gallian P, Lhomme S, Piquet Y, Sauné K, Abravanel F, Assal A, Tiberghien P, Izopet J. 2014. Hepatitis E virus infections in blood donors, France. Emerg Infect Dis 20:1914–1917. doi: 10.3201/eid2011.140516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenney SP, Meng X-J. 2018. Hepatitis E virus genome structure and replication strategy. Cold Spring Harb Perspect Med 8:a031724. doi: 10.1101/cshperspect.a031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin X, Ying D, Lhomme S, Tang Z, Walker CM, Xia N, Zheng Z, Feng Z. 2018. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc Natl Acad Sci U S A 115:4773–4778. doi: 10.1073/pnas.1721345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montpellier C, Wychowski C, Sayed IM, Meunier J-C, Saliou J-M, Ankavay M, Bull A, Pillez A, Abravanel F, Helle F, Brochot E, Drobecq H, Farhat R, Aliouat-Denis C-M, Haddad JG, Izopet J, Meuleman P, Goffard A, Dubuisson J, Cocquerel L. 2018. Hepatitis E virus lifecycle and identification of 3 forms of the ORF2 capsid protein. Gastroenterology 154:211–223.e8. doi: 10.1053/j.gastro.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Yin X, Ambardekar C, Lu Y, Feng Z. 2016. Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J Virol 90:4232–4242. doi: 10.1128/JVI.02804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kannan H, Fan S, Patel D, Bossis I, Zhang Y-J. 2009. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol 83:6375–6382. doi: 10.1128/JVI.02571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emerson SU, Nguyen HT, Torian U, Burke D, Engle R, Purcell RH. 2010. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol 84:9059–9069. doi: 10.1128/JVI.00593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagashima S, Takahashi M, Jirintai S, Tanaka T, Yamada K, Nishizawa T, Okamoto H. 2011. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol 92:269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- 19.Ding Q, Heller B, Capuccino JMV, Song B, Nimgaonkar I, Hrebikova G, Contreras JE, Ploss A. 2017. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci U S A 114:1147–1152. doi: 10.1073/pnas.1614955114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108:2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto H. 2011. Hepatitis E virus cell culture models. Virus Res 161:65–77. doi: 10.1016/j.virusres.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Rogée S, Talbot N, Caperna T, Bouquet J, Barnaud E, Pavio N. 2013. New models of hepatitis E virus replication in human and porcine hepatocyte cell lines. J Gen Virol 94:549–558. doi: 10.1099/vir.0.049858-0. [DOI] [PubMed] [Google Scholar]
- 23.Chapuy-Regaud S, Dubois M, Plisson-Chastang C, Bonnefois T, Lhomme S, Bertrand-Michel J, You B, Simoneau S, Gleizes P-E, Flan B, Abravanel F, Izopet J. 2017. Characterization of the lipid envelope of exosome encapsulated HEV particles protected from the immune response. Biochimie 141:70–79. doi: 10.1016/j.biochi.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi M, Tanaka T, Takahashi H, Hoshino Y, Nagashima S, Jirintai S, Mizuo H, Yazaki Y, Takagi T, Azuma M, Kusano E, Isoda N, Sugano K, Okamoto H. 2010. Hepatitis E virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J Clin Microbiol 48:1112–1125. doi: 10.1128/JCM.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagashima S, Takahashi M, Jirintai S, Tanaka T, Nishizawa T, Yasuda J, Okamoto H. 2011. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J Gen Virol 92:2838–2848. doi: 10.1099/vir.0.035378-0. [DOI] [PubMed] [Google Scholar]
- 26.Nagashima S, Jirintai S, Takahashi M, Kobayashi T, Tanggis, Nishizawa T, Kouki T, Yashiro T, Okamoto H. 2014. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol 95:2166–2175. [DOI] [PubMed] [Google Scholar]
- 27.Nagashima S, Takahashi M, Kobayashi T, Tanggis NT, Nishiyama T, Primadharsini PP, Okamoto H. 2017. Characterization of the quasi-enveloped hepatitis E virus particles released by the cellular exosomal pathway. J Virol 91:e00822-17. doi: 10.1128/JVI.00822-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treyer A, Müsch A. 2013. Hepatocyte polarity. Compr Physiol 3:243–287. doi: 10.1002/cphy.c120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaughan GW, Wickson JE, Creswick PF, Gorrell MD. 1990. Identification of the bile canalicular cell surface molecule GP110 as the ectopeptidase dipeptidyl peptidase IV: an analysis by tissue distribution, purification and N-terminal amino acid sequence. Hepatology 11:534–544. doi: 10.1002/hep.1840110403. [DOI] [PubMed] [Google Scholar]
- 30.Gissen P, Arias IM. 2015. Structural and functional hepatocyte polarity and liver disease. J Hepatol 63:1023–1037. doi: 10.1016/j.jhep.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isom HC, Secott T, Georgoff I, Woodworth C, Mummaw J. 1985. Maintenance of differentiated rat hepatocytes in primary culture. Proc Natl Acad Sci U S A 82:3252–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang JYL. 2013. Bile acid metabolism and signaling. Compr Physiol 3:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansuy JM, Gallian P, Dimeglio C, Saune K, Arnaud C, Pelletier B, Morel P, Legrand D, Tiberghien P, Izopet J. 2016. A nationwide survey of hepatitis E viral infection in French blood donors. Hepatol 63:1145–1154. doi: 10.1002/hep.28436. [DOI] [PubMed] [Google Scholar]
- 34.Kamar N, Dalton HR, Abravanel F, Izopet J. 2014. Hepatitis E virus infection. Clin Microbiol Rev 27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto H. 2013. Culture systems for hepatitis E virus. J Gastroenterol 48:147–158. doi: 10.1007/s00535-012-0682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snooks MJ, Bhat P, Mackenzie J, Counihan NA, Vaughan N, Anderson DA. 2008. Vectorial entry and release of hepatitis A virus in polarized human hepatocytes. J Virol 82:8733–8742. doi: 10.1128/JVI.00219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirai-Yuki A, Hensley L, Whitmire JK, Lemon SM. 2016. Biliary secretion of quasi-enveloped human hepatitis A virus. mBio 7:e01998–e01916. doi: 10.1128/mBio.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhat P, Snooks MJ, Anderson DA. 2011. Hepatocytes traffic and export hepatitis B virus basolaterally by polarity-dependent mechanisms. J Virol 85:12474–12481. doi: 10.1128/JVI.05344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belouzard S, Danneels A, Fénéant L, Séron K, Rouillé Y, Dubuisson J. 2017. Entry and release of hepatitis C virus in polarized human hepatocytes. J Virol 91:e00478–e00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong S-H, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiry D, Mauroy A, Pavio N, Purdy MA, Rose N, Thiry E, de Oliveira-Filho EF. 2017. Hepatitis E virus and related viruses in animals. Transbound Emerg Dis 64:37–52. doi: 10.1111/tbed.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colombo M, Raposo G, Théry C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 43.Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ. 2013. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol Cell Proteomics 12:587–598. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Q, Takada R, Noda C, Kobayashi S, Takada S. 2016. Different populations of Wnt-containing vesicles are individually released from polarized epithelial cells. Sci Rep 6:35562. doi: 10.1038/srep35562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 46.Wohlgemuth N, Lane AP, Pekosz A. 2018. Influenza A virus M2 protein apical targeting is required for efficient virus replication. J Virol 92:e01425-18. doi: 10.1128/JVI.01425-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakatsu Y, Ma X, Seki F, Suzuki T, Iwasaki M, Yanagi Y, Komase K, Takeda M. 2013. Intracellular transport of the measles virus ribonucleoprotein complex is mediated by Rab11A-positive recycling endosomes and drives virus release from the apical membrane of polarized epithelial cells. J Virol 87:4683–4693. doi: 10.1128/JVI.02189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lhomme S, Abravanel F, Dubois M, Sandres-Saune K, Mansuy J-M, Rostaing L, Kamar N, Izopet J. 2014. Characterization of the polyproline region of the hepatitis E virus in immunocompromised patients. J Virol 88:12017–12025. doi: 10.1128/JVI.01625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humbert L, Maubert MA, Wolf C, Duboc H, Mahé M, Farabos D, Seksik P, Mallet JM, Trugnan G, Masliah J, Rainteau D. 2012. Bile acid profiling in human biological samples: comparison of extraction procedures and application to normal and cholestatic patients. J Chromatogr B Analyt Technol Biomed Life Sci 899:135–145. doi: 10.1016/j.jchromb.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy J-M, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J Clin Microbiol 50:897–902. doi: 10.1128/JCM.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolte S, Cordelières FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.