More than 50% of the HIV-1 infections globally are caused by clade C viruses. To date, there is no effective vaccine to prevent HIV-1 infection. Based on the structural information of the currently available HIV-1 bNAbs, attempts are under way to design immunogens that can elicit correlates of protection upon vaccination. Here, we report the isolation and characterization of an HIV-1 N332 supersite-dependent bNAb, AIIMS-P01, from a clade C chronically infected pediatric elite neutralizer. The N332 supersite is an important epitope and is one of the current HIV-1 vaccine targets. AIIMS-P01 potently neutralized the contemporaneous and autologous evolving viruses and exhibited substantial indels despite low somatic hypermutations. Taken together with the information on infant bNAbs, further isolation and characterization of bNAbs contributing to the plasma breadth in HIV-1 chronically infected children may help provide a better understanding of their role in controlling HIV-1 infection.
KEYWORDS: 3D reconstruction, N332 supersite, SOSIP trimer, broadly neutralizing antibodies, clade C, human immunodeficiency virus, negative staining EM, pediatric elite neutralizer, plasma neutralization, single B cells sorting
ABSTRACT
Broadly neutralizing antibodies (bNAbs) have demonstrated protective effects against HIV-1 in primate studies and recent human clinical trials. Elite neutralizers are potential candidates for isolation of HIV-1 bNAbs. The coexistence of bNAbs such as BG18 with neutralization-susceptible autologous viruses in an HIV-1-infected adult elite controller has been suggested to control viremia. Disease progression is faster in HIV-1-infected children than in adults. Plasma bNAbs with multiple epitope specificities are developed in HIV-1 chronically infected children with more potency and breadth than in adults. Therefore, we evaluated the specificity of plasma neutralizing antibodies of an antiretroviral-naive HIV-1 clade C chronically infected pediatric elite neutralizer, AIIMS_330. The plasma antibodies showed broad and potent HIV-1 neutralizing activity with >87% (29/33) breadth, a median inhibitory dilution (ID50) value of 1,246, and presence of N160 and N332 supersite-dependent HIV-1 bNAbs. The sorting of BG505.SOSIP.664.C2 T332N gp140 HIV-1 antigen-specific single B cells of AIIMS_330 resulted in the isolation of an HIV-1 N332 supersite-dependent bNAb, AIIMS-P01. The AIIMS-P01 neutralized 67% of HIV-1 cross-clade viruses, exhibited substantial indels despite limited somatic hypermutations, interacted with native-like HIV-1 trimer as observed in negative stain electron microscopy, and demonstrated high binding affinity. In addition, AIIMS-P01 neutralized the coexisting and evolving autologous viruses, suggesting the coexistence of vulnerable autologous viruses and HIV-1 bNAbs in the AIIMS_330 pediatric elite neutralizer. Such pediatric elite neutralizers can serve as potential candidates for isolation of novel HIV-1 pediatric bNAbs and for understanding the coevolution of virus and host immune response.
IMPORTANCE More than 50% of the HIV-1 infections globally are caused by clade C viruses. To date, there is no effective vaccine to prevent HIV-1 infection. Based on the structural information of the currently available HIV-1 bNAbs, attempts are under way to design immunogens that can elicit correlates of protection upon vaccination. Here, we report the isolation and characterization of an HIV-1 N332 supersite-dependent bNAb, AIIMS-P01, from a clade C chronically infected pediatric elite neutralizer. The N332 supersite is an important epitope and is one of the current HIV-1 vaccine targets. AIIMS-P01 potently neutralized the contemporaneous and autologous evolving viruses and exhibited substantial indels despite low somatic hypermutations. Taken together with the information on infant bNAbs, further isolation and characterization of bNAbs contributing to the plasma breadth in HIV-1 chronically infected children may help provide a better understanding of their role in controlling HIV-1 infection.
TEXT
The promising results of animal challenge studies and human clinical trials of potent HIV-1 broadly neutralizing antibodies (bNAbs) in suppressing viral infection and recent FDA approval of ibalizumab in combination with antiretroviral therapy for individuals with multidrug-resistant HIV-1 have raised hopes for the development of effective HIV-1 therapeutic vaccines (1–4). Based on the structural information of the currently available HIV-1 bNAbs, attempts are under way to design immunogens that can elicit correlates of protection in the vaccinees (5, 6). Studies conducted in chronically HIV-1-infected adult donors suggest that it takes at least 2 to 3 years of infection for the antibodies to undergo affinity maturation in response to viral escape in order to develop potent HIV-1 bNAbs (7–9). The HIV-1 bNAbs evolve in the top 1% of the rare infected individuals classified as elite neutralizers. Further, the plasma antibodies in elite neutralizers show viral neutralizing activity, with a mean inhibitory dilution (ID50) titer of 1:500 against a diverse panel of HIV-1 pseudoviruses. In addition, plasma antibodies of all elite neutralizers neutralize more than one pseudovirus at an ID50 titer of 300 and belong to at least four distinct clades/circulating recombinant forms (CRF) (10). The HIV-1 bNAbs isolated from chronically infected adult elite neutralizers exhibit signature characteristic features, for example, high somatic hypermutations (SHMs), insertions or deletions (indels), a long complementarity-determining region H3 (CDRH3), high potency, and broad viral neutralization breadth (11–16). BG18, an N332 supersite-dependent HIV-1 bNAb with no indels, was isolated from a clade B-infected elite controller, EB354, and is the most potent of this class of bNAbs (12). However, most adult bNAbs, with the exception of BG18, normally fail to neutralize coexisting autologous viruses (7, 12, 17–20), but no such data are available on pediatric elite neutralizer(s).
HIV-1 infection in children is mostly caused by vertical transmission (21). Early treatment with antiretroviral therapy (ART) delays disease progression; however, it cannot prevent HIV-1 infection (22). Without ART, disease progression is faster in infected children than in adults, with a median time to AIDS of 1 year in contrast to 10 years in ART-naive infected adults (23), due to the immaturity and limited diversity of the immune system (24–26). Recently, plasma bNAbs have been shown to evolve in HIV-1-infected infants (27, 28). Longitudinal studies revealed that children infected at birth had high titers of NAbs effective against a few viruses until 3 months postinfection (p.i.), after which their titers decreased. Passively transferred maternal NAbs are probably a source of the NAbs at this stage. The titers of potent NAbs again increased with time, suggesting the development of de novo immune responses in all of the infants at about 1 year p.i. (28). Further, plasma mapping showed that HIV-1 bNAbs can develop early in life and that functional B cells persist in these infants to produce bNAbs, irrespective of high viremia and faster disease progression than that in adults (28, 29). The high viral load, both in infants and adults, possibly promotes the development of NAb breadth (28, 30–32). Moreover, BF520.1, one of the HIV-1 N332 supersite-dependent bNAbs isolated from an infant at 1 year postinfection, has shown broad neutralizing activity, despite limited SHMs (33) and an absence of indels, unlike the bNAbs isolated from adults (13, 16), suggesting that infant bNAbs evolved by different pathways than adult bNAbs.
Few longitudinal studies, including ours, conducted on pediatric HIV-1-directed humoral immune responses showed that plasma bNAbs develop in select chronically infected children and slow progressors, with diverse epitope specificities, (24, 34–37) and with higher breadth and potency than found in infected adults (35). Further, the bNAbs present in the plasma of infected children demonstrated epitope specificities similar to those observed for the adult bNAbs (24). Recently, from pooled peripheral blood mononuclear cells (PBMCs) of select HIV-1 clade C (HIV-1C)-infected pediatric long-term nonprogressors (LTNPs), we constructed an anti-HIV-1 human single-chain recombinant fragment (scFv) phage library, from which one of the CD4-binding site (CD4bs)-targeted scFvs, 2B10, demonstrated broad neutralizing activity with a breadth of 78% (38).
Here, we have characterized the plasma neutralizing activity and epitope specificities of a chronically HIV-1C-infected pediatric elite neutralizer, AIIMS_330. HIV-1-specific single B cells from this donor were sorted using BG505.SOSIP.664.C2 T332N gp140 envelope trimer as antigenic bait, followed by isolation of an N332 supersite-dependent pediatric bNAb AIIMS-P01 that neutralized the autologous coexisting and evolving viruses generated in this study.
(This article was submitted to an online preprint archive [39].)
RESULTS
Identification of a rare pediatric elite neutralizer by longitudinal neutralization activity analysis of the plasma antibodies.
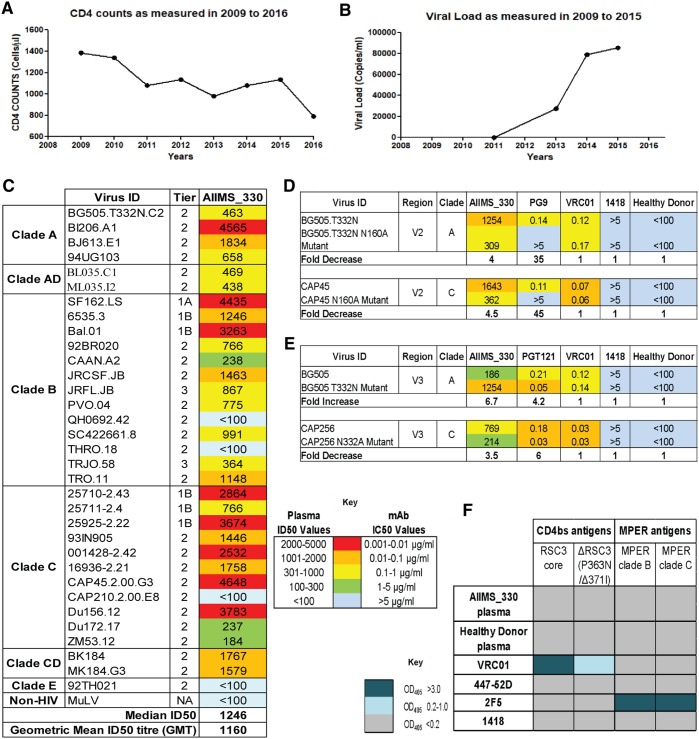
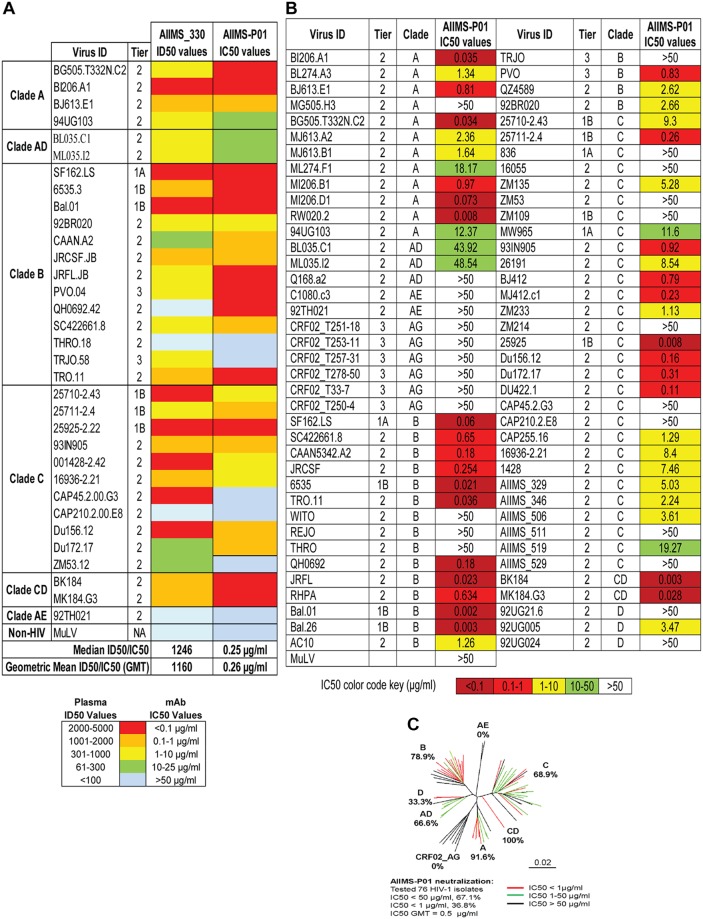
The AIIMS_330 donor is an Indian HIV-1C chronically infected 9-year-old boy who had acquired the infection at birth by vertical transmission. This donor was born in 2006 and was diagnosed to be seropositive for HIV-1 in 2009 at 3 years of age. We first recruited this 3-year-old HIV-1-infected pediatric donor in 2009. The clinical profile of this donor, including the CD4+ T cell counts and viral load, since the time of diagnosis are described in Fig. 1A and B. The plasma neutralizing activity and immune profile of this donor in the year 2009 identified him as an elite controller with CD4+ T cell counts of 1,382 cells/μl and a viral load of <47 RNA copies/ml. At the time of sampling in the present study (2015), the AIIMS_330 donor was antiretroviral (ART) naive; the CD4+ T cell counts and plasma viral loads were 1,033 cells/μl and 85,400 RNA copies/ml, respectively. The plasma antibody neutralization activity of this donor, against a few HIV-1 viruses, has been previously documented (37, 40, 41). In order to assess the evolution of antibody response in this donor, here we further characterized AIIMS_330 (2015) plasma against a large panel of heterologous HIV-1 tier 1, 2, and 3 pseudoviruses (n = 33) (Fig. 1C). The AIIMS_330 plasma antibodies showed broad and potent HIV-1 neutralizing activity, with >87% (29/33) breadth, a median inhibitory dilution (ID50) value of 1,246 (Fig. 1C), and a neutralization score of 2.9 (10), thereby qualifying AIIMS_330 to be an elite neutralizer. The AIIMS_330 plasma showed V1V2 and V3 glycan-directed neutralization specificities, identified using envelope pseudoviruses with point mutations at N160 (CAP45 clade C and BG505 clade A) and N332 (BG505 clade A and CAP256 clade C) glycans and their corresponding wild-type (WT) viruses (Fig. 1D and E). However, no reduction in binding of plasma antibodies was observed in an enzyme-linked immunosorbent assay (ELISA) using a CD4bs probe resurfaced stabilized core 3 (RSC3) mutant protein Δ371I/P363N compared to the level with the wild-type RSC3 core protein. In addition, the plasma antibodies also did not bind to peptides of the membrane-proximal external region (MPER) (Fig. 1F), suggesting the absence of CD4bs- and MPER-directed antibodies in AIIMS_330 plasma of the 2015 time point, as was also seen in its earlier 2013 time point plasma sample (38). Based on the elite neutralizing activity exhibited by AIIMS_330 (2015) plasma, this donor was selected for isolating anti-HIV-1 bNAbs.
FIG 1.
Plasma characterization identified AIIMS_330 as a pediatric elite neutralizer. (A) CD4 counts of the AIIMS_330 pediatric elite neutralizer as measured from 2009 to 2016. (B) Viral load values as measured from 2009 to 2015. (C) Heat map showing the neutralization ID50 titers of AIIMS_330 plasma antibodies against a heterologous panel of HIV-1 viruses (n = 33). MuLV was used as a negative virus control. (D) Heat map showing the N160-dependent analysis of AIIMS_330 plasma using the heterologous clade A virus BG505 and autologous clade C virus CAP45. Values are neutralization ID50 plasma titers or monoclonal antibody IC50 titers. PG9 was a positive control for the V2 region, while VRC01 (CD4bs), 1418 (parvovirus MAb), and healthy donor plasma were used as negative controls. Reduction in the ID50/IC50 titer of >3-fold of the mutant compared to the level of wild-type virus was considered positive dependence. (E) Heat map showing the N332-dependent analysis of AIIMS_330 plasma using the heterologous clade A virus BG505 and autologous clade C virus CAP256. Values are neutralization ID50 plasma titers or monoclonal antibody IC50 titers. PGT121 was a positive control for the V3 glycan N332 region. Reduction in the ID50/IC50 titer of >3-fold of the mutant compared to level of wild-type virus was considered positive dependence. (F) Heat map showing the ELISA results as the optical density (OD) at 405 nm to determine the CD4bs-directed binding of AIIMS_330 plasma antibodies using RSC3 core protein, RSC3 mutant protein Δ371I/P363N, and MPER binding using clades B and C 25-mer MPER peptides. VRC01 and 2F5 were used as positive controls for CD4bs and MPER, respectively, whereas 447-52D (V3 region) and 1418 were used as negative controls. GMT, geometric mean titer.
Pediatric antibody AIIMS-P01 demonstrated broad and potent HIV-1 neutralizing activity.
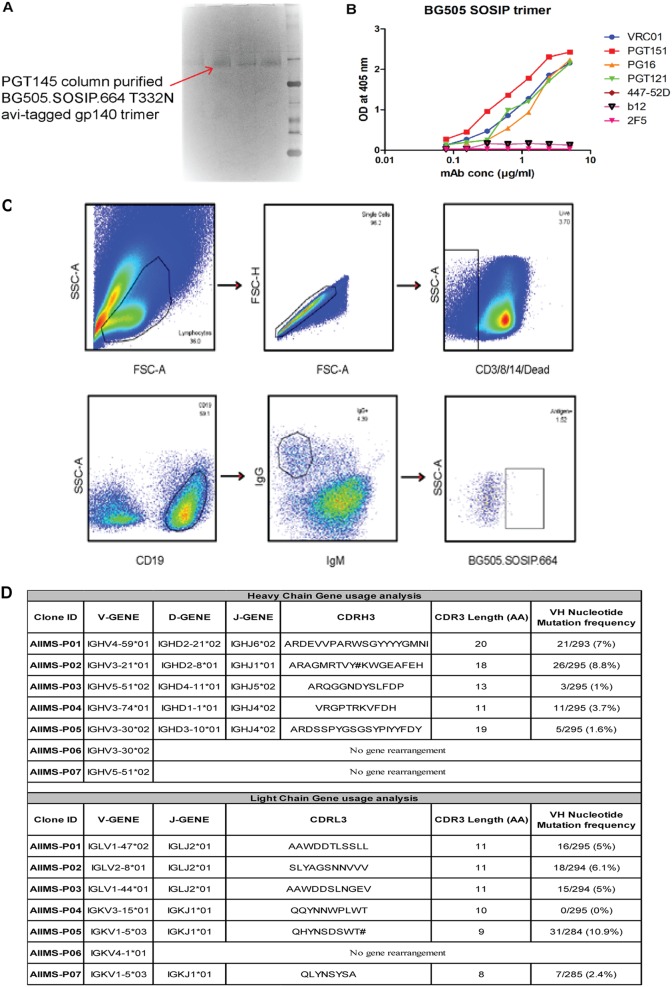
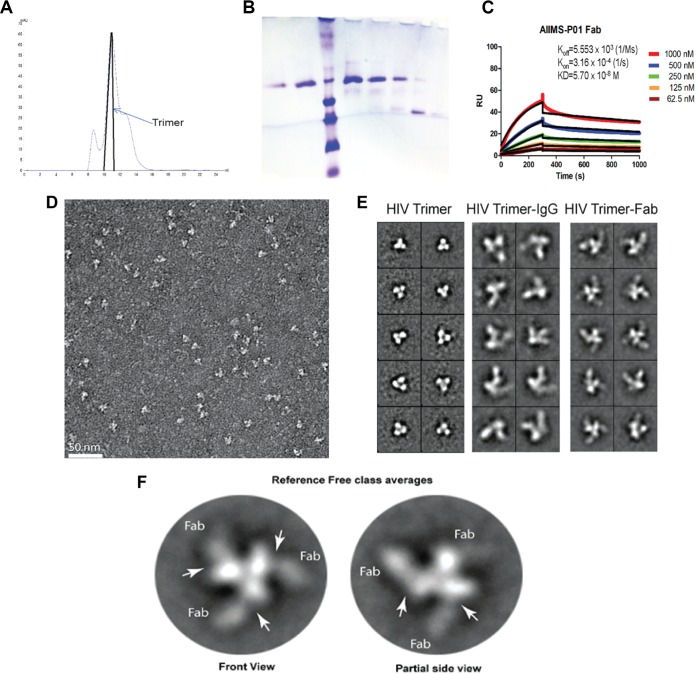
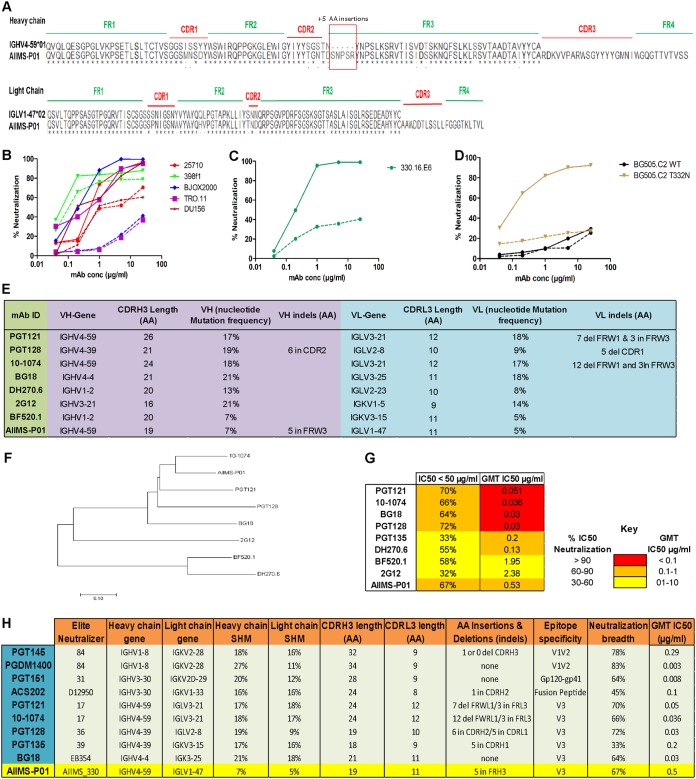
To sort HIV-1 antigen-specific single B cells, initially the antigenic bait, Avi-tagged trimeric BG505.SOSIP.664.C2 T332N gp140 protein, was expressed, purified, and biotinylated, as described previously (14), followed by binding analysis with existing anti-HIV-1 bNAbs to confirm the binding reactivity of the biotinylated trimeric protein with the HIV-1 bNAbs and absence of binding with non-NAbs (Fig. 2A and B). Then, a total of 22 BG505.SOSIP+ single B cells with the phenotype of CD3−, CD8−, CD14−, Dead− cells, CD19+, IgG+, and IgM− were sorted from 2 million stored PBMCs of AIIMS_330 pediatric elite neutralizer (Fig. 2C). Next, DNA was synthesized from single B cells. By nested PCR-based amplification of antibody variable heavy and light chain genes using specific primers (42), we obtained and sequenced seven pairs of heavy- and light-chain genes amplified from single B cells (Fig. 2D). By applying the criteria of having SHMs of >5% (43) in the amplified antibody genes, we identified a singular pediatric HIV-1 bNAb, AIIMS-P01. Therefore, for further experiments, only AIIMS-P01 was characterized.
FIG 2.
Single B cells sorting from the PBMCs of AIIMS_330 pediatric elite neutralizer. (A) BN-PAGE analysis of PGT145 antibody column-purified BG505.SOSIP.664.C2 T332N Avi-tagged trimeric protein. (B) Binding of biotinylated BG505.SOSIP Avi-tagged trimer with HIV-1 bNAbs VRC01, PGT151, PGT121 and PG16. Here, HIV-1 monoclonal antibodies 447-52D, b12, and 2F5 were used as negative controls. (C) Single B cell sorting by FACS of the AIIMS_330 pediatric elite neutralizer using BG505.SOSIP.664.C2 T332N gp140 envelope trimer as antigenic bait. (D) Immunogenetic information of the 7 amplified heavy- and light-chain gene pairs from the HIV-1-infected AIIMS_330 pediatric donor. FSC-A, forward scatter area; FSC-H, forward scatter height; SSC-A, side scatter area.
Next, to determine the neutralization potential of AIIMS-P01, its neutralization activity was tested at concentrations ranging from 50 μg/ml to 0.001 μg/ml against a heterologous panel of HIV-1 viruses, using a TZM-bl-based neutralization assay (44) (Fig. 3). The AIIMS-P01 bNAb effectively neutralized a panel of viruses comprised of susceptible tier 1 to highly resistant tier 3 viruses of different clades (Fig. 3B). Of the viruses tested, AIIMS-P01 neutralized 68% (20/29) clade C viruses and 78% (15/19) clade B viruses. Most of the viruses neutralized by the AIIMS-P01 bNAb were also neutralized by the contemporaneous plasma antibodies, except for the tier 2 clade B virus QH0692 (ID50 titer of 96), as the data from a 1:100 dilution maintain uniformity (Fig. 3A). Three of the viruses, TRJO, CAP45, and ZM53, were not neutralized by the AIIMS-P01 bNAb at the tested concentrations although they were susceptible to neutralization by the contemporaneous plasma antibodies (Fig. 3A). These results demonstrate that the broad viral neutralizing activity of AIIMS-P01 was comparable to that of the contemporaneous plasma antibodies. The pediatric bNAb AIIMS-P01 did not neutralize clade AE viruses (that do not have an Asn glycan at amino acid [aa] residue 332 of the V3 loop), as has also been observed with the existing N332-dependent adult and infant HIV-1 bNAbs, suggesting their N332 glycan dependency (12, 13, 16, 33, 45). The overall breadth of AIIMS-P01 was >67% (neutralized 51/76 HIV-1 viruses), with a geometric mean 50% inhibitory concentration (IC50) titer of 0.5 μg/ml (Fig. 3C).
FIG 3.
AIIMS-P01 demonstrated broad HIV-1 neutralizing activity. (A) Heat map depicting AIIMS_330 plasma ID50 titers and IC50 values of the contemporaneous AIIMS-P01 bNAb tested against viruses of different clades (n = 33). (B) Heat map depicting AIIMS-P01 bNAb neutralization analysis against HIV-1 viruses of different clades (n = 76). Here, MuLV was used as a negative control virus. (C) Dendrogram of neutralization IC50 values of the AIIMS-P01 bNAb against a panel of heterologous HIV-1 viruses (n = 76).
AIIMS-P01 targeted the N332 supersite at the base of the V3 region of HIV-1 envelope.
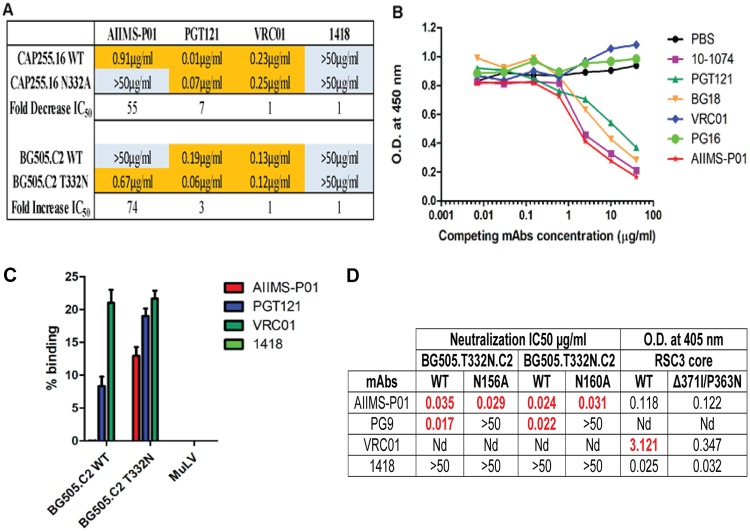
The absence of N332 glycan in clade AE viruses (45), in addition to the inability of AIIMS-P01 to neutralize clade AE viruses, indicates the N332-directed neutralizing dependency of this bNAb. In addition, a 55-fold reduction in the IC50 neutralization activity of AIIMS-P01 against the clade C mutant CAP255.16 N332A, compared to that against the CAP255.16 WT virus, confirmed its N332-directed epitope specificity. Furthermore, when this bNAb was tested against the BG505.C2.T332N mutant that contained the N332 glycan, a 72-fold increase in IC50 was observed compared to that of the BG505.C2 WT that lacked a glycan in this position (Fig. 4A). The N332-dependent neutralizing activity of AIIMS-P01 was further confirmed by competition ELISA using the N332-dependent bNAbs 10-1074, PGT121, and BG18 (Fig. 4B) (12, 13, 16). The binding of AIIMS-P01 with the N332 region on the native-like BG505.SOSIP.664 trimeric envelope was observed in cell surface binding assays (Fig. 4C). The replacement of the Asn332 glycan with Ala resulted in a total reduction in neutralization activity of AIIMS-P01, whereas no reduction was observed with N160A and N156A mutants (Fig. 4D), showing the absence of V1V2-directed neutralization activity of this bNAb. Moreover, no reduction in binding was observed between the CD4bs probes RSC3 core and RSC3 mutant Δ371I/P363N proteins (Fig. 4D), demonstrating that pediatric bNAb AIIMS-P01 was dependent exclusively on HIV-1 V3-glycan N332.
FIG 4.
AIIMS-P01 targets the N332 supersite. (A) Neutralization IC50 values of monoclonal antibodies for their N332 dependence. PGT121 was used as a positive control; VRC01 and 1418 were used as negative controls. (B) Competition ELISA findings. Biotinylated AIIMS-P01 was added at a fixed concentration of 5 μg/ml with increasing concentrations of competing monoclonal antibodies. The black line indicates no competition (PBS) in the absence of competing antibody. (C) Cell surface binding assay results of the percent binding of the monoclonal antibodies. Monoclonal antibodies VRC01 and 1418 and MuLV virus were used as negative controls. PGT121 was used as a positive control. (D) Neutralization IC50 values of AIIMS-P01 for N156 and N160 glycans and CD4bs-dependent analysis. In these neutralization assays, PG9 was used as a positive control for N156 and N160 dependence. In ELISAs VRC01 was used as a positive control for CD4bs-dependent analysis. The 1418 parvovirus antibody was used as a negative control. The optical density (OD) at 405 nm is shown for antibodies tested at a 5-μg/ml concentration. Nd, not determined.
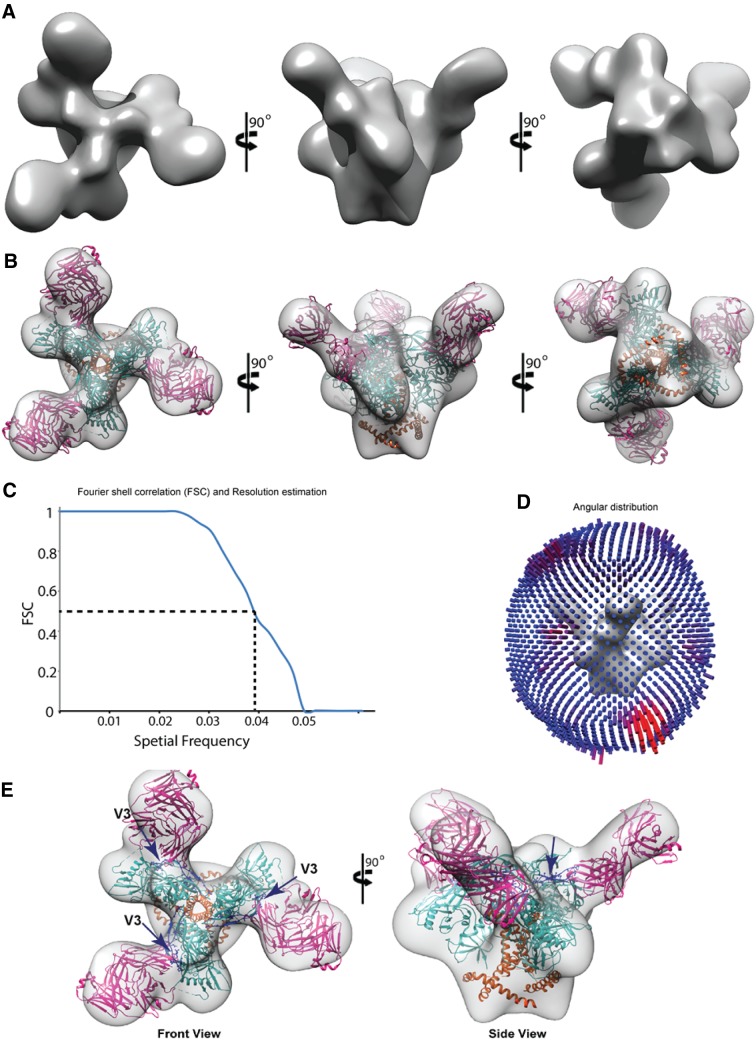
These findings were further substantiated by negative staining electron microscopy (EM) (Fig. 5 and 6). The BG505.SOSIP.664.C2 T332N D7324-tagged trimeric envelope glycoprotein was expressed and purified, and its purity was assessed using blue native PAGE (BN-PAGE) (6) (Fig. 5A and B). AIIMS-P01 Fab was prepared, and its affinity with BG505.SOSIP trimer was assessed by surface plasmon resonance (SPR) (Fig. 5C). Negative staining EM and reference-free classification were performed to visualize the interaction of the native-like HIV-1 trimeric protein with AIIMS-P01 Fab and AIIMS-P01 IgG, respectively (Fig. 5D and E). The density of AIIMS-P01 IgG with HIV-1 trimer in AIIMS-P01 IgG complex is not clearly visible. The plausible explanation for this is that IgG density was averaged out during data processing owing to its inherent flexibility though it binds with HIV trimer. The three Fab fragments were clearly visible in reference-free two-dimensional (2D) class averages (Fig. 5F). The three-dimensional (3D) EM envelope (Fig. 6A and B) suggests the presence of three suitable epitopes in the HIV-1 trimeric protein that has to interact with Fab fragments. The 3D reconstruction of HIV-1 trimer with AIIMS-P01 Fab at 26-Å resolution (Fig. 6C and D) confirmed that the AIIMS-P01 Fab interacts with the amino acid residues at the base of the V3 region on HIV-1 trimeric envelope (Fig. 6E).
FIG 5.
Negative staining EM analysis of AIIMS-P01 Fab and IgG in complex with BG505.SOSIP.664.C2 T332N gp140 trimeric protein. (A) Chromatogram of size exclusion chromatographic purification of lectin affinity column-purified BG505.SOSIP gp140 D7324-tagged trimeric protein. (B) BN-PAGE analysis of size exclusion chromatography-purified BG505.SOSIP.664 T332N D7324-tagged trimeric protein. (C) SPR sensorgram of binding kinetics analysis of AIIMS-P01 Fab with BG505.SOSIP.664 T332N D7324-tagged trimeric protein. (D) Raw EM image of HIV-1 BG505.SOSIP.664 T332N gp140 trimer bound to AIIMS-P01 Fabs embedded in uranyl acetate negative stain. (E) Comparison between representative reference-free 2D class averages of HIV-1 trimer alone, HIV-1 trimer with AIIMS-P01 IgG, and HIV-1 trimer with AIIMS-P01 Fab, as indicated. (F) Reference-free 2D class averages of AIIMS-P01 Fabs bound with HIV-1 BG505.SOSIP.664 T332N gp140 trimer.
FIG 6.
3D reconstruction of AIIMS-P01 Fab in complex with HIV-1 trimer. (A) Solid rendering representation of 3D map of HIV-1 BG505.SOSIP.664 T332N gp140 trimer bound to AIIMS-P01 Fabs. (B) Transparent 3D EM map of HIV-1 BG505.SOSIP.664 T332N gp140 trimer bound to AIIMS-P01 Fabs modelled with the crystal structure. (C) Conventional FSC curve for the 3D reconstruction of HIV-1 BG505.SOSIP.664 T332N gp140 trimer bound to AIIMS-P01 Fabs. The resolution of the map is 26 Å according to a 0.5 cutoff. (D) The plot of angular distribution for the final reconstruction. (E) The left average is the front view, and the right average is the partial side view showing the density of AIIMS-P01 Fabs bound with HIV-1 BG505.SOSIP.664 T332N gp140 trimer. The arrows indicate the V3 region of the HIV-1 trimer, which interacts with AIIMS-P01 Fab.
AIIMS-P01 neutralized autologous contemporaneous and evolving viruses.
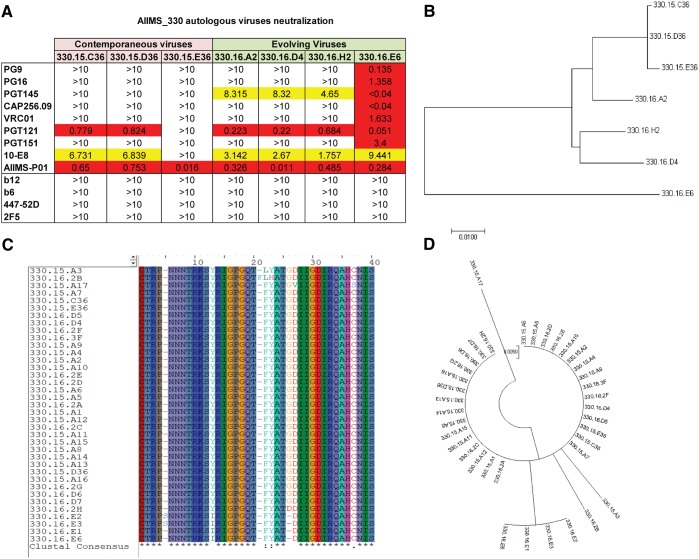
To evaluate the ability of AIIMS-P01 to neutralize the autologous contemporaneous viruses of the 2015 time point, the HIV-1 Rev/Env cassette was amplified by the single-genome amplification (SGA) method, as described previously (46), using cDNA as the template synthesized from plasma viral RNA. A total of 20 autologous viruses were generated from the contemporaneous (2015) time point. The neutralization susceptibility of three HIV-1 enveloped autologous pseudoviruses (2015) to AIIMS-P01 was tested, along with the highly potent second-generation HIV-1 adult bNAbs, first-generation Nabs, and nonneutralizing antibodies (Fig. 7A). The AIIMS-P01 potently neutralized all three autologous contemporaneous viruses (designated series 330.15) (Fig. 7A). Two of three contemporaneous viruses were also neutralized by adult bNAbs PGT121 (N332-directed) and 10E8 (MPER-directed). Further, we generated 17 evolving (2016 time point) autologous viruses (designated series 330.16), and 4 of them were tested for their neutralization susceptibility to AIIMS-P01. We found that all four evolving autologous viruses were neutralized by AIIMS-P01 (Fig. 7A) and also by PGT121, 10E8, and PGT145 (V1V2-directed) adult bNAbs. Six out of the seven AIIMS_330 autologous viruses were resistant to many of the potent adult bNAbs, except 330.16.E6 (Fig. 7A). The phylogenetic analysis (Fig. 7B) depicts the evolution of AIIMS_330 autologous viruses tested in neutralization assays at different time points. Next, we aligned and performed phylogenetic analysis of the V3 region sequences of all the generated autologous viruses, and we found that all the viral sequences had conserved linear V3 epitopes and showed the presence of characteristic residues (N301 and N332 glycans) and motifs (GDIR and NIS motifs in V3 region) required for the susceptibility to neutralization by the V3 glycan N332 supersite-directed HIV-1 bNAbs of PGT121, 10-1074, BG18, BF520, and DH270 class lineages (Fig. 7C and D). These results suggest the coexistence of vulnerable autologous viruses and bNAbs in the AIIMS_330 pediatric elite neutralizer, as has also been observed with BG18, isolated from an adult HIV-1-infected donor EB354 (12).
FIG 7.
AIIMS-P01 neutralized autologous contemporaneous and evolving viruses. (A) Heat map of neutralization IC50 values of pediatric AIIMS-P01 bNAb and adult HIV-1 second-generation bNAbs (PG9, PG16, PGT145, CAP256.09, VRC01, PGT121, PGT151, and 10E8), first-generation NAbs (b12, 2F5, and 447-52D), and non-NAbs (b6) tested at 10 μg/ml against the autologous contemporaneous virus of AIIMS_330 (2015; series 330.15) and a successive time point (2016; series 330.16). (B) Maximum-likelihood phylogenetic tree analysis of AIIMS_330 autologous viruses of two time points, constructed by MEGA7 software. (C) Alignment of V3 region sequences of SGA AIIMS_330 autologous viruses of contemporaneous (2015) (n = 20) and evolving (2016) (n = 17) time points. (D) Phylogenetic tree of V3 region sequences of SGA AIIMS_330 autologous viruses of contemporaneous (2015) (n = 20) and evolving (2016) (n = 17) time points.
AIIMS-P01 showed high affinity with native-like HIV-1 trimeric protein.
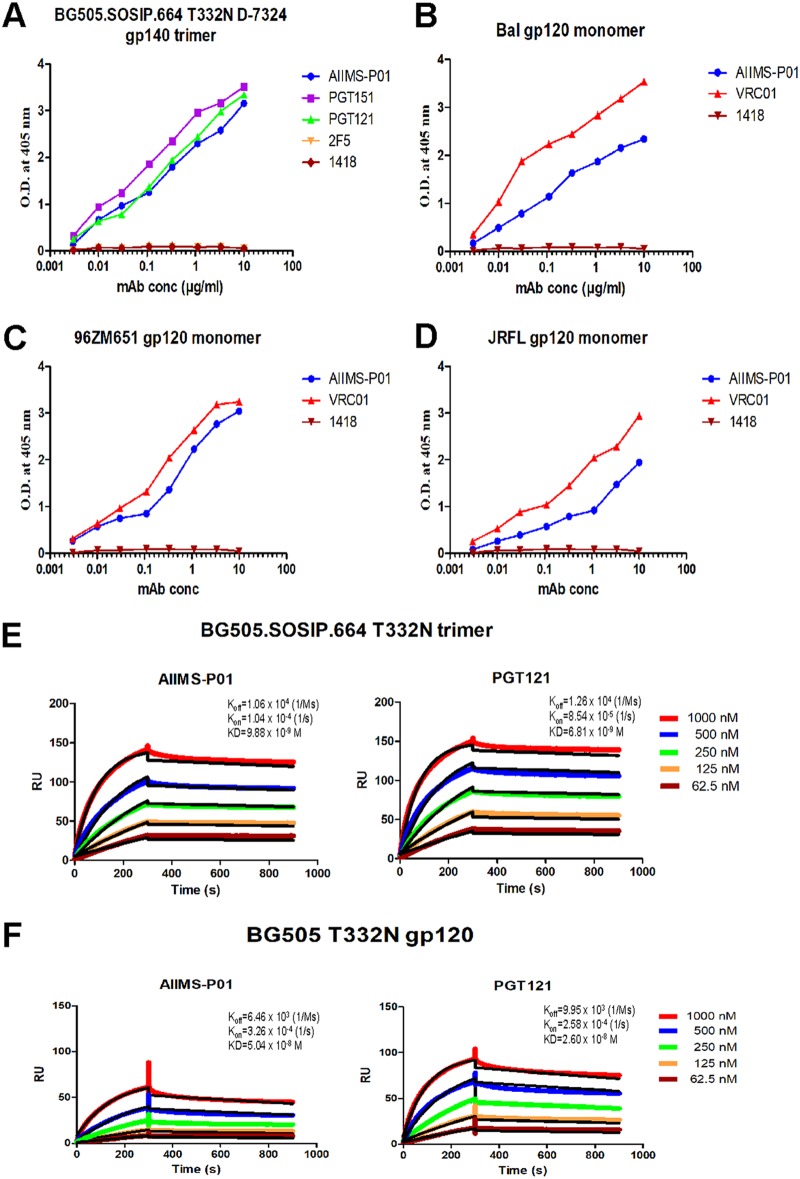
Next, we determined the binding potential of AIIMS-P01 with heterologous HIV-1 monomeric clade C and B envelopes and observed comparable binding efficiencies (Fig. 8B to D). In addition, the binding of AIIMS-P01 with native-like HIV-1 trimeric glycoprotein was comparable to that of PGT121 and PGT151 adult bNAbs (Fig. 8A). ELISA binding analysis showed that AIIMS-P01 displayed high binding with both trimeric HIV-1 antigen and heterologous monomeric antigens (Fig. 8A to D). To further validate these results, the affinity of AIIMS-P01 with trimeric gp140 and monomeric gp120 antigens was assessed using surface plasmon resonance (SPR). AIIMS-P01 displayed low off-rate and higher affinity (equilibrium dissociation constant [KD], = 9.8 nM) with BG505.SOSIP.664 T332N in SPR experiments than a monomeric BG505 gp120 protein (KD = 50 nM) (Fig. 8E and F). These results demonstrate that AIIMS-P01 exhibits high affinity with the native-like HIV-1 trimer.
FIG 8.
AIIMS-P01 showed strong binding with a native-like HIV-1 antigen. (A) Binding curves of AIIMS-P01 with BG505.SOSIP.664 T332N D7324-tagged gp140 trimer. Here, PGT151 trimer-specific HIV-1 bNAb and PG121 (N332) bNAb were used as positive controls; 2F5 (MPER) and 1418 were used as negative controls. (B to D) Binding curves of AIIMS-P01 with monomeric gp120 antigens. Here, VRC01 was used as a positive control; 1418 was used as a negative control. (E) SPR sensorgrams of AIIMS-P01 and PGT121 with BG505.SOSIP gp140 trimer. (F) SPR sensorgrams of AIIMS-P01 and PGT121 with BG505 gp120 monomer. Here, PGT121 was used a positive control.
AIIMS-P01 exhibited the presence of indels with limited somatic hypermutations.
The sequence comparison of pediatric bNAb AIIMS-P01 with its respective germ line heavy- and light-chain genes showed the presence of indels (insertion of 5 amino acids [aa]) in heavy-chain framework region 3 [FRH3]) that have not been observed in the heavy chain of the other N332-directed bNAbs (PGT121 and 10-1074) (Fig. 9A) (13, 16). To gain insight into the effects of the indels on neutralization activity of AIIMS-P01, we tested neutralization breadth and potency of the AIIMS-P01 antibody without the 5-aa insertion in FRH3 on a heterologous panel of five viruses and one autologous virus. Upon removal of the 5-aa insertion in FRH3, we observed a loss of neutralization of AIIMS-P01 in a varied manner in comparison to neutralization by AIIMS-P01 wild-type antibody (Fig. 9B). For example, there was a partial loss of neutralization with 25710, DU156, and 298F1 viruses, while TRO.11 and BJOX2000 viruses became resistant to neutralization by AIIMS-P01 lacking the 5-aa insertion in FRH3 (Fig. 9B). Similarly, deletion of the 5-aa insertion from AIIMS-P01 resulted in resistance to autologous virus neutralization (Fig. 9C). The effect of the 5-aa insertion with respect to AIIMS-P01 was further assessed for its N332 epitope dependence, and we observed that this insertion in AIIMS-P01 is necessary for the neutralization of viruses with an N332 epitope, as observed with BG505.C2 WT (T at N332) and the BG505.C2 T332N mutant (Fig. 9D). These results suggest that the FRH3 insertion is beneficial for neutralization breadth of AIIMS-P01.
FIG 9.
AIIMS-P01 exhibited the presence of indels with low somatic hypermutations. (A) Alignment of heavy- and light-chain amino acid sequences with respective germ line antibody gene sequences. Alignment was done using Clustal X software and BioEdit software. (B) Neutralization activity of AIIMS-P01 antibody without the 5-aa insertion in FRH3 against five heterologous HIV-1 pseudoviruses. (C) Neutralization activity of AIIMS-P01 antibody without the 5-aa insertion in FRH3 with autologous virus 330.16.E6. (D) Neutralization activity of AIIMS-P01 antibody without the 5-aa insertion in FRH3 with BG505.C2 WT and its T332N mutant. (E) Image showing a comparison of AIIMS-P01 with the characteristic features of N332-directed bNAbs of adults and an infant. in, insertion; del, deletion. (F) Maximum-likelihood tree for phylogenetic analysis of N332-directed bNAbs, which was made using MEGA7 software. (G) Heat map showing the comparison of AIIMS-P01 neutralization IC50 values to adult and infant N332-dependent bNAbs. GMT, geometric mean titer. (H) Comparison of the characteristics of HIV-1 bNAbs of adult and pediatric origins identified from all the elite neutralizers reported so far. in, insertion; del, deletion.
A limited frequency of SHMs, 7% in the heavy chain and 5% in the light chain, was found in AIIMS-P01 (Fig. 9E). Further, AIIMS-P01 had similar germ line gene family of IGVH4-59*01 as that of adult bNAbs of 10-1074 and the PGT121 lineage (13, 16), but with a distinct light-chain gene family of IGLV1-47*02 (Fig. 9A and E). Similar to the N332-targeted adult bNAbs (12, 13, 16, 45) and infant bNAbs of the BF520.1 class (33), the CDRH3 region of AIIMS-P01 was 19 amino acids long. A long CDRH3 has been shown to be important for interaction with HIV glycans (47, 48) (Fig. 9E). Phylogenetic analysis of the N332-dependent AIIMS-P01 displayed a close relation with the N332-dependent adult bNAb 10-1074 (Fig. 9F). In addition, a comparison of the neutralization activity of AIIMS-P01 adult and infant bNAbs of N332 epitope specificity revealed that AIIMS-P01 exhibited neutralization breadth comparable to that of the adult N332 bNAbs (Fig. 9G). A comparison of the gene usage, CDR length, the presence of indels, epitope specificities, and neutralizing activity between the N332-directed bNAb AIIMS-P01 and adult bNAbs, along with bNAbs of different specificities, is shown in Fig. 9H.
AIIMS-P01 showed ADCC activity with absence of polyreactivity.
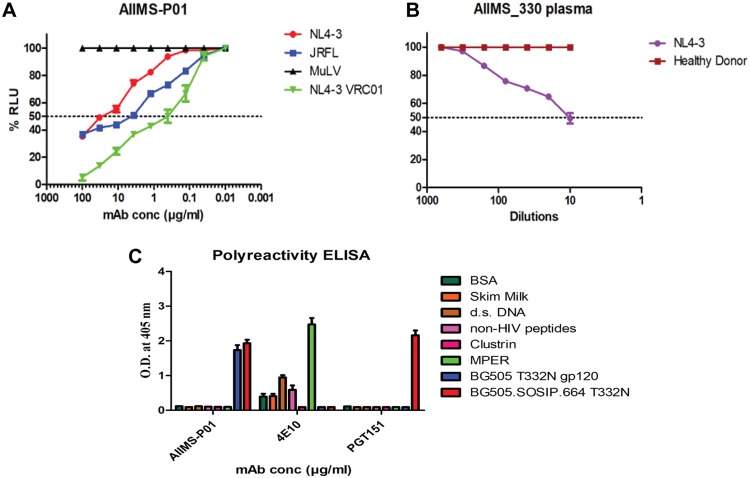
The antibody-dependent cellular cytotoxicity (ADCC) activity of AIIMS-P01 and its contemporaneous plasma antibodies was tested by infecting target cells with NL4-3 and JRFL viruses. The murine leukemia virus (MuLV) virus was used as a negative control. Both AIIMS-P01 bNAb and its contemporaneous plasma antibodies showed ADCC activity (Fig. 10A and B). The AIIMS-P01 bNAb did not show any polyreactivity when tested for binding with multiple antigens, suggesting the lack of binding to non-HIV-1 antigens and cellular self-antigens (Fig. 10C).
FIG 10.
AIIMS-P01 exhibited ADCC activity. (A) ADCC activity analysis of AIIMS-P01 with NL4-3 and JRFL viruses. VRC01 was used as a positive control, and MuLV was used as a negative virus control. (B) The ADCC activity of AIIMS_330 plasma. A healthy donor was used as a negative control. (C) The polyreactivity of AIIMS-P01 was tested by ELISA. The HIV-1 bNAb PGT151 was used as a positive control for trimer binding, and 4E10 was used as a positive control for polyreactivity analysis.
DISCUSSION
Implementation of novel high-throughput techniques such as memory B cell culture and single B cell sorting using antigenic bait led to the successful isolation of second-generation potent HIV-1 human broadly neutralizing antibodies (bNAbs) (49). The epitopes defined by HIV-1 bNAbs form the basis for vaccine design using the Reverse Vaccinology 2.0 strategy (50). The BG18 bNAb, isolated from a clade B HIV-1-infected elite controller, has been shown to coexist with antibody-sensitive viruses, suggesting that similar bNAbs and neutralization-susceptible contemporaneous viruses may be present in viremic controllers (12). Goo et al. for the first time showed the development of bNAbs in infants within 1 year of age (28). In chronically infected children, polyclonal antibodies with multiple specificities have been observed (24, 36), with greater potency and breadth than the adult bNAbs (24, 35), and the pediatric antibodies target similar epitopes as adult bNAbs (24). In addition, we along with others (24) have observed V2 glycan-directed plasma neutralizing activity in infected children, as has also been seen in adults (30, 51–53), suggesting the immunodominance of the V2 glycan epitope on the viral envelope in both HIV-1-infected adults and children. The antigenic stimulus provided by the high viral load in the infected children may contribute to the activation of multiple B cell lineages and the generation of polyclonal antibodies targeting diverse bNAb epitopes. The higher neutralization potency of V1V2 glycan-directed antibodies than that of V3 glycan antibodies (54), and therefore relatively less immune selection pressure on the V3 region of the viral envelope, could be one of the reasons why we observed the autologous viruses to be sensitive to the V3 glycan-directed bNAb AIIMS-P01. Delineating the factors leading to the stimulation in infected children of such bNAb lineages that are effective in neutralizing contemporaneous and evolving viruses will provide information toward the development of multistage HIV-1 envelope-based vaccines.
The recent isolation of the infant bNAb BF520.1 (33) suggested the presence of precursor B cells in infants that can be stimulated by HIV-1. Mapping the specificities of bNAbs at different stages of infection, i.e., those elicited early in infection (28, 33) and ones found in chronically infected children (24, 35, 36), may provide useful information for combinatorial immunization strategies to elicit similar bNAbs in the vaccinees. Recently, we (36) along with others (24, 35) observed the longitudinal evolution of a plasma cross-neutralization antibody response with multiple epitope specificities in select antiretroviral-naive HIV-1C chronically infected children (24). It is pertinent to isolate bNAbs from infected individuals whose plasma antibodies have been extensively mapped for the presence of bNAbs, desirably the LTNPs and elite neutralizers (10, 50).
This study describes for the first time a pediatric bNAb, AIIMS-P01 IgG1, isolated from a single B cell of an ART-naive HIV-1C chronically infected pediatric LTNP, AIIMS_330, who was identified as an elite neutralizer in this study based on plasma mapping analysis. This AIIMS_330 donor belonged to a rare cohort of vertically transmitted ART-naive HIV-1C-infected children that have been followed for more than a decade (37, 38, 40, 41). The AIIMS-P01 bNAb isolated from this elite neutralizer showed comparable neutralization efficiencies against clade C and B HIV-1 viruses, thereby suggesting that this bNAb may neutralize major viral clades responsible for infection globally.
The AIIMS-P01 bNAb is dependent exclusively on the N332 supersite epitope, similar to the HIV-1 bNAbs PGT121, 10-1074, DH270, and BG18 isolated from adult donors (12, 13, 16, 45) and to BF520.1, an infant-derived bNAb (33). The N332 supersite is an important epitope and is one of the current HIV-1 vaccine targets (5, 6). The frequency of somatic hypermutations (7%) observed in AIIMS-P01 was comparable to that found in the infant bNAb BF520.1 (33) and the bNAbs from adults (3.8% to 32.6%) (55, 56). Furthermore, AIIMS-P01 showed 5-amino-acid (aa) insertions in the third framework region of the antibody heavy chain (FRH3), as has been observed to be prevalent in adult bNAbs, with in-frame indels ranging from 0 to 7 aa (13, 15, 16, 57). The high frequency of indels in adult bNAbs has been shown to increase with and is predicted by the frequency of SHMs (58), contributing to their high neutralization potency and breadth (47, 48, 55, 59, 60). However, it is difficult to elicit by immunization similar bNAbs with high SHMs and indels (61) although such antibodies are generated in macaques, with an average SHM of 10% and with the presence of indels, on immunization with trimeric HIV-1 envelopes (62). Our results suggested that AIIMS-P01 has attained high breadth and N332 epitope dependence by virtue of the substantial indels as it has previously been reported for N332- and CD4bs-directed adult bNAbs that indels are critical for making contacts with protein or glycans on HIV (47, 48, 59, 60). The AIIMS-P01 bNAb has acquired indels with limited SHMs; whether such bNAbs can be elicited in vaccinees needs to be addressed. AIIMS-P01 belongs to an IGVH4-59*01 germ line gene family, similar to the existing bNAbs of the PGT121 series and 10-1074, isolated from an adult elite neutralizer (13, 16). The sequence comparison, alignment, and phylogenetic analysis of AIIMS-P01 with existing N332 glycan-dependent bNAbs displayed a close relation to the 10-1074 class of adult bNAbs (13).
The neutralizing activity of AIIMS_330 plasma antibodies mapped to V1V2 and V3 glycan epitopes and corroborates observations of multiple epitope specificities of the plasma antibodies found in chronically infected children (24, 36), supporting the idea that bNAbs from pediatric elite neutralizers can be directed to neutralizing determinants similar to those found in adult elite neutralizers. Likewise, the IGHV1-2 antibody gene usage in infant bNAb BF520.1 (33) is also seen in DH270.6 bNAb from adults (45). Further, the binding interactions of the AIIMS-P01 Fab with the native-like BG505.SOSIP trimeric envelope glycoprotein, as observed in reference-free class averages and negative staining 3D reconstruction, were possibly due to the long CDRH3 of AIIMS-P01 which may promote contact with the envelope glycan residues (47, 48). AIIMS-P01 showed the ability to effectively neutralize not only the autologous contemporaneous viruses but also autologous viral escape mutants of the successive time points, suggesting the coexistence of N332-dependent bNAbs and vulnerable viruses in the AIIMS_330 pediatric elite neutralizer, as has been documented in the EB354 elite controller, with the coexistence of BG18 bNAbs and susceptible contemporaneous autologous viruses (12). Apart from the BG18 bNAb, isolated from an adult elite neutralizer that neutralized autologous viruses (12), the current bNAbs do not neutralize contemporaneous autologous viruses (12, 17–20). The N332 dependence demonstrated by AIIMS-P01 was comparable to that of the contemporaneous plasma antibodies, suggesting that N332-dependent AIIMS-P01-like bNAbs were substantially present in the contemporaneous AIIMS_330 plasma, in addition to antibodies of diverse epitope specificities, as shown by the plasma mapping studies. The pediatric bNAb AIIMS-P01 showed high affinity with native-like soluble trimeric envelope and demonstrated heterologous HIV-1 clade neutralization activity. The inability to neutralize the viruses of the AE clade, which lack the N332 glycan, further confirms the N332 neutralization dependence of AIIMS-P01, as has also been seen earlier with other N332-dependent bNAbs (12, 13, 45). In addition, AIIMS-P01 showed effector functions and the absence of polyreactivity.
To the best of our knowledge, the present study for the first time reports a singular pediatric elite neutralizer followed by isolation and characterization of an HIV-1 N332 supersite-dependent pediatric bNAb, AIIMS-P01. The AIIMS-P01 bNAb contains indels with limited SHMs and belongs to the antibody gene family of the PGT121 class of HIV-1 bNAbs. The AIIMS-P01 bNAb neutralized its coexisting and evolving autologous viruses. However, our observations are based on testing the bNAb against few autologous and evolving viruses generated from two time points, and there are limitations of this study. Further, isolation and testing the neutralization sensitivity of a sizeable number of viruses from each time point of sampling will enable us to show whether the cocirculating viruses remained sensitive to the bNAb in this pediatric donor. As this study was based on a single HIV-1 chronically infected child, our observations encourage further isolation and characterization of the HIV-1 bNAbs contributing to plasma breadth in chronically infected children to better understand their development and evolution.
MATERIALS AND METHODS
Ethics statement and study subject.
Blood (5 ml) of from the HIV-1 clade C-infected AIIMS_330 pediatric donor was drawn after the approval of this study by the institutional ethics committee, All India Institute of Medical Sciences (AIIMS), New Delhi, India (IEC/59/08.01.16 and IEC/NP-536/04.11.2013) and after obtaining written informed consent from the parents/guardian. The peripheral blood mononuclear cells (PBMCs) were isolated by use of a Ficoll density gradient after the plasma was separated by centrifugation at 360 × g for 10 min.
HIV-1 pseudovirus generation.
The HIV-1 pseudoviruses were produced in HEK 293T cells by cotransfecting the full HIV-1 gp160 envelope plasmid and a pSG3ΔEnv backbone plasmid. Briefly, 1 × 105 cells in 2 ml of complete Dulbecco’s modified Eagle’s medium (DMEM) (10% fetal bovine serum [FBS] and 1% penicillin and streptomycin antibiotics) were seeded per well of a six-well cell culture plate (Costar) the day prior to cotransfection for HIV-1 pseudovirus generation. For transfection, the ratio of the envelope (1.25 μg) to PSG3DEnv plasmid (2.50 μg) was 1:2; this complex was made in Opti-MEM (Gibco) with a final volume of 200 μl for each well of a six-well plate and incubated for 5 min at room temperature. Next, 3 μl of polyethylenimine (PEI)-Max transfection reagent (Polysciences) (1 mg/ml) was added to this mixture, mixed well, and further incubated for 15 min at room temperature. This mixture was then added dropwise to HEK 293T cells supplemented with fresh complete DMEM and incubated at 37°C for 48 h. Pseudoviruses were then harvested by filtering cell supernatants with 0.45-mm-pore-size sterile filter (Mdi) and stored frozen at −80°C as aliquots.
HIV-1 neutralization assays.
The HIV-1 neutralization assays of AIIMS_330 plasma antibodies and HIV-1 monoclonal antibodies (MAbs) were performed as described earlier (44). Neutralization was measured as a reduction in luciferase gene expression after a single round of infection of TZM-bl cells (NIH AIDS Reagent Program) with HIV-1 envelope pseudoviruses. The 50% tissue culture infective dose (TCID50) of the HIV-1 pseudovirus was calculated, and 200 TCID50s of the virus were used in neutralization assays by incubation with 1:3 serially diluted heat-inactivated plasma samples at a starting dilution of 1/10 for 1 h at 37°C. After that, TZM-bl cells freshly trypsinized in growth medium (complete DMEM with 10% FBS and 1% penicillin and streptomycin antibiotics) containing 50 μg/ml DEAE-dextran and 1 mM indinavir (in case of primary isolates) at were added 105 cells/well, and plates were incubated at 37°C for 48 h. Virus controls (cells with HIV-1 virus only) and cell controls (cells without virus and antibody or plasma) were included. MuLV was used as a negative control. After the incubation of the plates for 48 h, luciferase activity was measured using a Bright-Glow Luciferase Assay System (Promega). ID50 values of the plasma sample and IC50 values for antibodies were calculated. Values were derived from a dose-response curve fit with a nonlinear function using GraphPad Prism, version 5, software (San Diego, CA).
Binding analysis of AIIMS_330 plasma antibodies.
All proteins (RSC3 core and mutant) and peptides (MPER clades B and C) were coated on ELISA plates at a concentration of 2 μg/ml in 0.1 M NaHCO3 (pH 9.6) and incubated overnight at 4°C. The next day, plates were blocked with 300 μl/well of 15% FBS and 2% bovine serum albumin (BSA) in RPMI medium for 1.5 h, followed by addition of 100 μl/well of heat-inactivated plasma at different dilutions (1:100, 1:300, 1:1,000, 1:3,000, and 1:10,000) or HIV-1 monoclonal antibodies (VRC01, 447-52D, and 2F5) and anti-parvovirus MAb 1418, each at concentrations of 5 to 0.001 μg/ml, and incubated for 1 h at 37°C. Each of the above steps was followed by washing with 1× phosphate-buffered saline (PBS) with 0.1% Tween 20. Then, 100 μl of alkaline phosphatase (AP)-conjugated anti-human IgG Fc antibody (1:2,000 diluted in 1× PBS [Southern Biotech]) was added to the plates; the immune complexes were reacted with AP substrate in 10% diethanolamine (DAE) buffer (1 mg/ml), and absorbance was read at 405 nm.
Expression and purification of BG505.SOSIP.664 T332N trimeric proteins.
The BG505.SOSIP.664.C2 T332N gp140 trimeric proteins with a D7324 tag and Avi tag were expressed in HEK 293F cells and purified by methods described previously (6, 14). Purity was assessed by blue native polyacrylamide gel electrophoresis (BN-PAGE), and binding reactivity with HIV-1 bNAbs was assessed by ELISA. The presence of native-like SOSIP trimers with a D7324 tag was further confirmed by negative staining electron microscopy. The purified Avi-tagged BG505.SOSIP.664.C2 T332N gp140 trimeric protein was first biotinylated using a BirA500 ligase reaction kit (Avidity) according to the manufacturer’s protocol; biotinylation of trimer was assessed by ELISA as described previously (14) and used for the sorting of single B cells of the donor AIIMS_330.
HIV-1 BG505.SOSIP trimer-specific single B cell sorting by FACS.
The frozen PBMCs (2 × 107 cells) of the AIIMS_330 pediatric elite neutralizer were thawed in complete DMEM containing 50 U/ml of Benzonase enzyme (Novagen). Next, cells were stained using a panel of fluorophore-labeled antibodies directed to CD3 (V500), CD8 (V500), CD14 (V500), CD19 (phycoerythrin [PE]-Cy7), human IgM (fluorescein isothiocyanate [FITC]), and human IgG (BV421) and Live/Dead fixable Aqua fluorescent reactive dye (Invitrogen) (to exclude dead cells). All fluorochrome-conjugated antibodies were purchased from BD Biosciences. In addition, to sort antigen-specific single B cells, the HIV-1 BG505.SOSIP.664 gp140 trimer was labeled with streptavidin-allophycocyanin (APC) (Life Technologies) and added to the PBMCs at 4°C for 30 min, followed by two washes with ice-cold 1× PBS. Cells with a phenotype of CD3−/CD8−/CD14−/Dead− cells/CD19+/IgG+/IgM−/APC+ BG505.SOSIP.664 trimer were singly sorted on a BD FACSAria III fluorescence activated cell sorter (FACS) (BD Biosciences) into 96-well PCR plates (Bio-Rad) containing 20 μl/well lysis buffer consisting of 1 U/ml RNase OUT (Thermo Fisher), 0.3125% Igepal CA-630, 1× SuperScript III first-strand buffer, and 6.25 mM dithiothreitol (DTT) provided with the Superscript III reverse transcriptase kit (Thermo Fisher). The sorting data were collected using FACSDiva software (BD Biosciences) and analyzed using FlowJo software (FlowJo, LLC).
Single B cell reverse transcription-PCR (RT-PCR), amplification of antibody genes, and cloning.
The amplification of variable heavy- and light-chain antibody genes was performed as described previously (42, 57, 63) with few modifications. The frozen 96-well plate containing sorted single B cells was first thawed on ice, followed by cDNA synthesis using 1 μl of 200 U/well Superscript III reverse transcriptase (Invitrogen), 1 μl of 10 mM deoxynucleoside triphosphate (dNTP) mix (Thermo Fisher), 0.5 μl of 50 ng/ml random hexamers (Thermo Fisher), and 0.5 μl of oligo(dT) (Thermo Fisher) under the following cycling parameters: 25°C for 10 min, 42°C for 10 min, 50°C for 50 min, 55°C for 10 min, and 85°C for 5 min, with a hold at 4°C. Next, the first-strand cDNA was amplified by two-step nested PCR. In the first step, 0.4 μl of the mixture of 5 μM IgH, Ig(κ), or Ig(λ) chain-specific primers was used for the amplification of antibody variable genes of heavy and light chains in a final reaction volume of 25 μl using 0.3 μl of DreamTaq DNA polymerase (5 U/μl) (Thermo Fisher), 3 μl of first-strand cDNA, and 0.3 μl of 10 mM dNTP mix (Thermo Fisher) under the following cycling parameters: 95°C for 5 min and 35 cycles of 95°C for 30 s, 50 or 52°C for 60 s, 72°C for 1 min, and 72°C for 10 min, with a hold at 4°C. Next, 0.4 μl of the mixture of 5 μM IgH, Ig(κ), or Ig(λ) chain cloning primers was used for the second round of the PCR in a final volume of 25 μl using 0.3 μl of DreamTaq DNA polymerase (5 U/μl) (Thermo Fisher), 0.5 to 2 μl of first-round PCR product, and 0.3 μl of 10 mM dNTP mix (Thermo Fisher) under the following cycling parameters: 95°C for 5 min, followed by 38 cycles of 95°C for 30 s, 55 or 60°C for 60 s, 72°C for 1 min, and 72°C for 10 min, followed by a hold at 4°C. Then, 5 μl of each second-round PCR product was loaded in 1.2% agarose gels and run at 100 V. Gels were visualized under UV light, wells with amplified bands were again amplified using second-round PCR in six tubes of 25 μl, run in 1% agarose gels, purified, digested using restriction enzymes, and cloned into their respective AbVec vectors (63). Four transformed bacterial colonies were randomly picked, checked for the presence of inserts, and sequenced.
Antibody genes sequence analysis.
The sequencing of the antibody genes after their cloning into their corresponding vectors was done commercially by Invitrogen (Thermo Fisher). The sequences were analyzed online through IMGT/V-QUEST (http://www.imgt.org/IMGT_vquest/vquest) and IgBlast (https://www.ncbi.nlm.nih.gov/igblast/). Only selected clones were expressed and purified by the identification of criteria including productive genes, substantial germ line mutations (>5%) indicating a history of affinity maturation, and long CDRH3 loops.
Expression of monoclonal antibodies.
The HIV-1 monoclonal antibodies AIIMS-P01, 10-1074, and BG18 were expressed in HEK 293T (ATCC) or Freestyle 293F cells (Thermo Fisher) by cotransfection of 10 μg each of heavy chain- and light chain-expressing IgG1 plasmids using PEI-Max as the transfection reagent. Following 4 to 6 days of incubation, cells were harvested by centrifugation and filtered through a 0.22-mm-pore-size syringe filter (Mdi). The supernatant was added to a protein A affinity chromatographic column (Pierce). The column was then washed with 1× PBS, and MAbs were eluted with IgG elution buffer (Pierce), immediately neutralized with 1 M Tris (pH 8.0) buffer, and extensively dialyzed against 1× PBS at 4°C. After that, MAbs were concentrated using 10-kDa Amicon Ultra-15 centrifugal filter units (EMD Millipore), filtered through a 0.22-mm-pore-size syringe filter (Mdi), and stored at –80°C for further use. For indel deletion analysis experiments, the minigene lacking the 5-aa insertion in FRH3 of AIIMS-P01 was synthesized by Integrated DNA Technologies (IDT; Iowa) and cloned in the respective monoclonal antibody expression vector, followed by expression and purification as described above.
Binding analysis of monoclonal antibodies by ELISA.
Briefly, 96-well ELISA plates (Costar) were coated with 5 μg/ml recombinant HIV-1 gp120 monomeric proteins overnight at 4°C in 0.1 M NaHCO3 (pH 9.6). The next day, plates were washed three times with 1× PBS (phosphate-buffered saline) and blocked with 15% FBS-RPMI medium and 2% BSA. After 1.5 h of blocking at 37°C, plates were washed three times with 1× PBS. Then, serial dilutions of monoclonal antibodies (MAbs) were added and incubated for 1 h at 37°C. After that, alkaline phosphatase (AP)-labeled anti-Fc secondary antibody (Southern Biotech) at 1:2,000 was added, and plates were incubated at 37°C for 1 h. Plates were then washed three times with 1× PBS; AP substrate tablets (Sigma) dissolved in diethanolamine (DAE) were added and incubated for 30 min at room temperature in the dark, and readout was taken at 405 nm. The BG505.SOSIP.664 gp140 trimeric ELISA was performed as described previously (6). The polyreactivity ELISA was performed as described previously (64). Briefly, all antigens were coated overnight at 37°C in 96-well ELISA plates (Costar). The next morning, after three washes with 1× PBS, the plates were blocked for 1 h at 37°C in 2% BSA and 15% FBS. After the plates were again washed three times with 1× PBS, the anti-HIV-1 antibodies AIIMS-P01, PGT151, and 4E10 as primary antibodies were added at 5 μg/ml diluted in 1× PBS and incubated for 1 h at 37°C. Next, alkaline phosphatase-conjugated anti-human IgG Fc antibody (Southern Biotech) at a 1:2,000 dilution in 1× PBS was added after four washes. Readout was taken at 405 nm by addition of AP substrate (Sigma) in 10% DAE buffer.
AIIMS-P01 Fab fragment preparation.
The Fab fragments were generated from 2 mg of AIIMS-P01 IgG antibody using a Fab fragmentation kit (G Biosciences) according to the manufacturer’s protocol. Purity and size of Fab fragments were assessed by SDS-PAGE, and binding was assessed by surface plasmon resonance (SPR).
SPR analysis.
The affinities and binding kinetics of AIIMS-P01 IgG1 and Fab to BG505.SOSIP.664.C2 soluble gp140 trimer and gp120 monomer were determined by SPR on a Biacore T200 instrument (GE Healthcare) at 25°C with HBS-EP+ buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20). BG505.SOSIP.664 gp120 trimer and gp120 monomer were first immobilized on a CM5 chip at 600 response units (RU) according to the manufacturer’s standard amine coupling kit protocol (GE Healthcare). The HIV-1 bNAbs AIIMS-P01 and PGT121 were injected at a flow rate of 30 μl/min as 2-fold dilutions starting from 1 μM for 300 s, followed by dissociation for 600 s. The cells were regenerated in glycine buffer (pH 2.5). Sensorgrams of the concentration series were corrected with corresponding blank curves and fitted globally with Biacore T200 evaluation software (GE Healthcare) using a 1:1 Langmuir model of binding to calculate the affinity of HIV-1 bNAbs IgG1 and Fab.
Negative staining single-particle EM and 3D reconstruction.
HIV-1 BG505.SOSIP.664 T332N gp140 trimer was incubated with AIIMS-P01 IgG and AIIMS-P01 Fab at concentration ratios of 1:1, 1:2, and 1:3 for 1 h at 4°C. The samples were prepared using conventional negative staining methods (65). Briefly, 3.5 μl of samples was adsorbed on UV-treated carbon-coated copper grids for 1 min. This was followed by washing with three drops of water and staining with 2% uranyl acetate for 20 s. Samples were imaged at room temperature by using a Tecnai T12 electron microscope equipped with an LaB6 filament at 120 kV accelerating voltage. The images were recorded at a magnification of 75,000× on a side-mounted Olympus Velita (2,000- by 2,000-pixel) charge-coupled-device (CCD) camera. A total of 100 tilt paired images were collected at 0° and a 45° tilt angle by tilting the sample stage, and all the data were collected at a defocus range of approximately –1.5 to –1.8 μm with a final pixel size of 2.0 Å at the specimen level. Particles were excised from the untilted images automatically using the “swarm” option of e2boxer.py in the EMAN suite, version 2.12 (66). A total of 11,243 particles were selected from untilted micrographs. The reference-free 2D class averaging was calculated using simple_prime2D in the Simple software package, version 2.1 (67), and the data set was classified into 120 classes. For initial model generation, we employed random conical tilt pair (68) image reconstruction techniques to determine the back-projected 3D model of HIV-1-Fab complex (HIV-1 BG505.SOSIP.664-T332N gp140 with AIIMS-P01 Fab) from the tilted data set using XMIPP (X-windows based microscopy image processing package) (69). Simultaneously, an initial model was calculated from reference-free class averages using e2initialmodel.py in the EMAN suite, version 2.12. The initial models are structurally similar, and both were used as initial models for further data processing. The initial map was filtered to 40 Å using RELION, version 1.4 (70), and used as a reference model for 3D classification. About 7,640 particles were selected from a 3D classification where Fab is clearly visible. These particle sets were used for final auto-refinement in the Relion, version 1.4, software package. The resolution for the map was calculated using a 0.5 cutoff of the Fourier shell correlation (FSC), and the estimated resolution is 26 Å. The three-dimensional EM map was visualized using UCSF Chimera (71). The crystal structure of Fab (PDB accession number 3PIQ) and HIV trimer (PDB accession number 4NCO) (73) were docked into the EM map using the automatic fitting option of the Chimera software package.
Competitive ELISA.
The competitive ELISAs were performed as described previously (12). The ELISA plates were coated overnight at 4°C with 10 μg/ml of anti-C5 (D-7324) antibody (Aalto Biosciences) in 0.1 M NaHCO3 (pH 9.6). The next day, plates were washed three times with 1× PBS and blocked for 1 h at 37°C with 2% BSA and 15% FBS. Then, a 1:4 serial dilution of competing antibodies was added with a starting concentration of 40 μg/ml in the presence of biotinylated AIIMS-P01 at a fixed concentration of 5 μg/ml. Then, plates were washed three times, and streptavidin-horseradish peroxidase (HRP) was added at a 1:1,000 dilution in 1× PBS, followed by addition of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (BioLegend). The reaction was stopped by addition of H2SO4 stop solution before binding was measured at 405 nm. These experiments were performed twice.
Cell surface binding assay.
The binding of the antibodies to cell surface HIV-1 envelopes was assessed using a flow cytometry-based assay as described earlier (64). HEK 293T cells (2 × 105 cells) were transfected with 5 μg of HIV-1 envelope expression plasmid DNA using PEI-Max transfection reagent, followed by harvesting at 48 h posttransfection and incubation at a 5-μg/ml concentration of antibodies for 1 h at 37°C. After cells were washed with 1× PBS, they were next stained with phycoerythrin (PE)-conjugated anti-human IgG Fc antibody (BioLegend) for half an hour at room temperature, followed by acquisition of the data by flow cytometry using a BD FACSCanto II. Data were analyzed using FACSDiva software. Percent binding was calculated as the percentage of PE-positive cells after subtraction of background (antibodies binding to mock-transfected cells) values. Analyses were performed in GraphPad Prism, version 5.0.
HIV-1 envelope amplification and cloning.
The HIV-1 enveloped AIIMS_330 autologous pseudoviruses were generated by single-genome amplification (SGA) as described previously (46). Briefly, viral RNA was extracted from 140 μl of AIIMS_330 plasma of each independent time point by using a QIAamp Viral RNA Mini kit (Qiagen), followed by cDNA synthesis using a Superscript III reverse transcriptase cDNA synthesis kit (Invitrogen) as per the manufacturer’s protocol. The HIV-1 envelope (Rev/Env cassette) was amplified by nested PCR using primers and reaction conditions as described earlier (46). The amplicons were sequenced commercially from Eurofins Genomics. The SGA amplicons with full-length envelope genes were molecularly cloned into a pcDNA3.1 Directional Topo vector (Invitrogen), transformed into TOP10 competent cells, and sequenced.
ADCC assay.
The ADCC assays were done as previously described (72) with few modifications. A total of 5 × 107 target cells (CEM.NKR-CCR5-sLTR-Luc cells) (NIH AIDS reagent program) were infected with HIV-1 NL4-3, HIV-1 JRFL, or MuLV at 200 TCID50s by incubation at 37°C for 1 h in a final volume of 200 μl. At 2 days postinfection, the infected target cells were washed three times with RPMI medium and incubated in round-bottom 96-well plates with effector cells (KHYG-1 cell line) expressing CD16 at a 10:1 effector/target (E/T) ratio in the presence of serial dilutions of AIIMS_330 and healthy donor plasma, AIIMS-P01, and VRC01 HIV-1 bNAb. After an 8-h incubation, cells were lysed with luciferase substrate (BriteGlo; Promega), and luciferase activity in relative light units (RLUs) was measured with a Tecan multiplate mode reader. The ADCC activity of the plasma and antibodies were calculated from the mean RLU value for triplicate wells at each antibody concentration/plasma dilution relative to the mean for background and maximal RLUs from replicate wells containing uninfected and infected cells, respectively, incubated with NK cells but without antibody.
Quantification and statistical analysis.
All statistical analysis was done with GraphPad Prism software, version 5.
Accession number(s).
The sequences of the AIIMS-P01 heavy chain and light chain variable regions have been deposited in GenBank under accession numbers MH267797 and MH267798, respectively. All envelope glycoprotein sequences utilized in this study have been submitted to GenBank under accession numbers MK076687 to MK076724. The accession number for the EM map of AIIMS-P01 Fab complexed with BG505.SOSIP.664.C2 T332N gp140 trimer reported in this study was deposited in the Protein Data Bank in Europe under accession number EMD-6967.
ACKNOWLEDGMENTS
This antibody work was supported by Science and Engineering Research Board (SERB), Department of Science and Technology (DST), India (grant EMR/2015/001276). The HIV-1 pseudoviruses generation work was supported by (BT/PR5066/MED/1582/2012) Department of Biotechnology (DBT), India. The EM work and consumables were supported by (SERB-EMR/2016/000608 & BT/INF/22/SP22844/2017). EM data were collected at the Division of Biological Sciences, IISc, Bangalore. The senior Research Fellowship to SK was supported by SERB, India.
We thank pediatric donor AIIMS_330 for participating. We also thank the following: the NIH AIDS Reagent Program for HIV-1 research reagents; the Neutralizing Antibody Consortium (NAC), International AIDS Vaccine Initiative (IAVI), USA, for HIV-1 neutralizing antibodies; Michel C. Nussenzweig, Rockefeller University, USA, for 10-1074 and BG18 antibody expression plasmids; Dennis R. Burton, The Scripps Research Institute, USA, John P. Moore and Rogier Sanders, Cornell University, USA, and the University of Amsterdam, Netherlands, for BG505.SOSIP.T332N.664 plasmids; Lynn Morris, NICD, South Africa, for N160 and N332 mutant plasmids; Patrick C. Wilson, University of Chicago, USA, for AbVec antibody expression plasmids; Jacob Kopicinski, Imperial College London, United Kingdom, for helping in designing the FACS antibody panel; Dinakar Salunke, Pawan Malhotra, and Naresh Sahoo, ICGEB, New Delhi, India, for ICGEB SPR Facility; Likhesh Sharma (GE Healthcare) for helping with the SPR experiments; and Pradeep Kumar, BD Biosciences, India, for his assistance in sorting of single B cells by FACS.
K.L., S.K., and R.L. have a pending Indian provisional patent application for pediatric bNAb AIIMS-P01 described in the present study. We declare that we have no other competing interests.
S.K. planned and performed the experiments, analyzed the data, and wrote the original manuscript; H.P. helped S.K. in standardizing amplification protocols for antibody genes; M.A.M. and N.M. generated AIIMS_330 autologous pseudoviruses; N.M. analyzed the FACS data and helped S.K. in HIV-1 pseudovirus generation and neutralization assays; H.A. helped S.K. in flow cytometry experiments; R.L. and S.K.K. provided the donor sample, were responsible for clinical care of the AIIMS_330 donor, collected the clinical data from the subject, and reviewed the manuscript; H.C. performed the experiments of AIIMS-P01 indel deleted experiments; E.S.R. helped S.K. in size exclusion chromatography experiments; A.C. provided the scientific inputs for single B cell sorting technique and reviewed the manuscript; S.D. performed and solved the 3D structure from negative stain EM experiments along with H.A.S. and reviewed the manuscript; K.L. conceived, planned, and supervised the experiments and reviewed, edited, and finalized the manuscript.
REFERENCES
- 1.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O'Brien C, Weiland D, Robles A, Kümmerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP, Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F. 2017. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, Zhang L, Ledgerwood J, Grunenberg N, Bailer R, Isaacs A, Seaton K, Mayer KH, Capparelli E, Corey L, Gilbert PB. 2017. Population pharmacokinetics analysis of VRC01, an HIV-1 broadly neutralizing monoclonal antibody, in healthy adults. MAbs 9:792–800. doi: 10.1080/19420862.2017.1311435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobson JM, Kuritzkes DR, Godofsky E, DeJesus E, Larson JA, Weinheimer SP, Lewis ST. 2009. Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrob Agents Chemother 53:450–457. doi: 10.1128/AAC.00942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. 2015. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol 90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. 2013. Antibodies in HIV-1 vaccine development and therapy. Science 341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West AP, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. 2014. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euler Z, Schuitemaker H. 2012. Cross-reactive broadly neutralizing antibodies: timing is everything. Front Immunol 3:215. doi: 10.3389/fimmu.2012.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker L-G, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang C-H, Mcbride R, von Bredow B, Shivatare SS, Wu C-Y, Chan-Hui P-Y, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong C-H, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freund NT, Wang H, Scharf L, Nogueira L, Horwitz JA, Bar-On Y, Golijanin J, Sievers SA, Sok D, Cai H, Cesar Lorenzi JC, Halper-Stromberg A, Toth I, Piechocka-Trocha A, Gristick HB, van Gils MJ, Sanders RW, Wang L-X, Seaman MS, Burton DR, Gazumyan A, Walker BD, West AP, Bjorkman PJ, Nussenzweig MC. 2017. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci Transl Med 9:eaa12144. doi: 10.1126/scitranslmed.aal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sok D, van Gils MJ, Pauthner M, Julien J-P, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, Hua Y, Seaman MS, Moore JP, Ward AB, Wilson IA, Sanders RW, Burton DR. 2014. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A 111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Gils MJ, van den Kerkhof TLGM, Ozorowski G, Cottrell CA, Sok D, Pauthner M, Pallesen J, de Val N, Yasmeen A, de Taeye SW, Schorcht A, Gumbs S, Johanna I, Saye-Francisco K, Liang C-H, Landais E, Nie X, Pritchard LK, Crispin M, Kelsoe G, Wilson IA, Schuitemaker H, Klasse PJ, Moore JP, Burton DR, Ward AB, Sanders RW. 2016. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat Microbiol 2:16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S-l, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang G-Y, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang K-K, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao H-X, Kwong PD, Haynes BF. 2016. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang C-H, Ramos A, Bian CB, Wickramasinghe L, Kong L, Eren K, Wu C-Y, Wong C-H, Kosakovsky Pond SL, Wilson IA, Burton DR, Poignard P, Price MA, Gilmour J, Fast P, Kamali A, Sanders EJ, Anzala O, Allen S, Hunter E, Karita E, Kilembe W, Lakhi S, Inambao M, Edward V, Bekker L-G. 2016. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulterys M, Lepage P. 1998. Mother-to-child transmission of HIV. Curr Opin Pediatr 10:143–150. doi: 10.1097/00008480-199804000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Herout S, Mandorfer M, Breitenecker F, Reiberger T, Grabmeier-Pfistershammer K, Rieger A, Aichelburg MC. 2016. Impact of early initiation of antiretroviral therapy in patients with acute HIV infection in Vienna, Austria. PLoS One 11:e0152910. doi: 10.1371/journal.pone.0152910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder PJ, Lewin SR, Leitman EM. 2016. Paediatric HIV infection: the potential for cure. Nat Rev Immunol 16:259–271. doi: 10.1038/nri.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ditse Z, Muenchhoff M, Adland E, Jooste P, Goulder P, Moore PL, Morris L. 2018. HIV-1 subtype C infected children with exceptional neutralization breadth exhibit polyclonal responses targeting known epitopes. J Virol 93:e00878-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Hoffmann F, He J, He X, Kankasa C, West JT, Mitchell CD, Ruprecht RM, Orti G, Wood C. 2006. Characterization of HIV-1 subtype C envelope glycoproteins from perinatally infected children with different courses of disease. Retrovirology 3:73. doi: 10.1186/1742-4690-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad N. 2005. The vertical transmission of human immunodeficiency virus type 1: molecular and biological properties of the virus. Crit Rev Clin Lab Sci 42:1–34. doi: 10.1080/10408360490512520. [DOI] [PubMed] [Google Scholar]
- 27.Ghulam-Smith M, Olson A, White LF, Chasela CS, Ellington SR, Kourtis AP, Jamieson DJ, Tegha G, Horst CM. v d, Sagar M. 2017. Maternal but not infant anti-HIV-1 neutralizing antibody response associates with enhanced transmission and infant morbidity. mBio 8:e01373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goo L, Chohan V, Nduati R, Overbaugh J. 2014. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat Med 20:655–658. doi: 10.1038/nm.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol 84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, CAPRISA002 Study Team . 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piantadosi A, Panteleeff D, Blish CA, Baeten JM, Jaoko W, McClelland RS, Overbaugh J. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol 83:10269–10274. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. 2009. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS 23:2405–2414. doi: 10.1097/QAD.0b013e32833243e7. [DOI] [PubMed] [Google Scholar]
- 33.Simonich CA, Williams KL, Verkerke HP, Williams JA, Nduati R, Lee KK, Overbaugh J. 2016. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell 166:77–87. doi: 10.1016/j.cell.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraisier C, Van de Perre P, Lepage P, Hitimana DG, Karita E, Desgranges C. 1995. Broadly neutralizing, MN-like PND-directed antibodies in Rwandan children with long-term HIV1 infection. Res Virol 146:201–210. doi: 10.1016/0923-2516(96)80580-2. [DOI] [PubMed] [Google Scholar]
- 35.Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, Leitman E, Moonsamy A, McGregor C, Hurst J, Groll A, Mori M, Sinmyee S, Thobakgale C, Tudor-Williams G, Prendergast AJ, Kloverpris H, Roider J, Leslie A, Shingadia D, Brits T, Daniels S, Frater J, Willberg CB, Walker BD, Ndung’u T, Jooste P, Moore PL, Morris L, Goulder P. 2016. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci Transl Med 8:358ra125. doi: 10.1126/scitranslmed.aag1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makhdoomi MA, Khan L, Kumar S, Aggarwal H, Singh R, Lodha R, Singla M, Das BK, Kabra SK, Luthra K. 2017. Evolution of cross-neutralizing antibodies and mapping epitope specificity in plasma of chronic HIV-1-infected antiretroviral therapy-naïve children from India. J Gen Virol 98:1879–1891. doi: 10.1099/jgv.0.000824. [DOI] [PubMed] [Google Scholar]
- 37.Aggarwal H, Khan L, Chaudhary O, Kumar S, Makhdoomi MA, Singh R, Sharma K, Mishra N, Lodha R, Srinivas M, Das BK, Kabra SK, Luthra K. 2017. Alterations in B cell compartment correlate with poor neutralization response and disease progression in HIV-1 infected children. Front Immunol 8:1697. doi: 10.3389/fimmu.2017.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Kumar R, Khan L, Makhdoomi MA, Thiruvengadam R, Mohata M, Agarwal M, Lodha R, Kabra SK, Sinha S, Luthra K. 2017. CD4-binding site directed cross-neutralizing scFv monoclonals from HIV-1 subtype C-infected Indian children. Front Immunol 8:1568. doi: 10.3389/fimmu.2017.01568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Panda H, Makhdoomi MA, Mishra N, Safdari HA, Chawla H, Aggarwal H, Reddy ES, Lodha R, Kabra SK, Chandele A, Dutta S, Luthra K. 2018. An HIV-1 broadly neutralizing antibody from a clade C-infected pediatric elite neutralizer potently neutralizes the contemporaneous and autologous evolving viruses. bioRxiv 10.1101/403469. [DOI] [PMC free article] [PubMed]
- 40.Prakash SS, Andrabi R, Kumar R, Kabra SK, Lodha R, Vajpayee M, Luthra K. 2011. Binding antibody responses to the immunogenic regions of viral envelope in HIV-1-infected Indian children. Viral Immunol 24:463–469. doi: 10.1089/vim.2011.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prakash SS, Chaudhary AK, Lodha R, Kabra SK, Vajpayee M, Hazarika A, Bagga B, Luthra K. 2011. Efficient neutralization of primary isolates by the plasma from HIV-1 infected Indian children. Viral Immunol 24:409–413. doi: 10.1089/vim.2011.0028. [DOI] [PubMed] [Google Scholar]
- 42.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cale EM, Gorman J, Radakovich NA, Crooks ET, Osawa K, Tong T, Li J, Nagarajan R, Ozorowski G, Ambrozak DR, Asokan M, Bailer RT, Bennici AK, Chen X, Doria-Rose NA, Druz A, Feng Y, Joyce MG, Louder MK, O’Dell S, Oliver C, Pancera M, Connors M, Hope TJ, Kepler TB, Wyatt RT, Ward AB, Georgiev IS, Kwong PD, Mascola JR, Binley JM. 2017. Virus-like particles identify an HIV V1V2 apex-binding neutralizing antibody that lacks a protruding loop. Immunity 46:777–791.e10. doi: 10.1016/j.immuni.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 45.Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang K-K, Saunders KO, Zhang R, Gladden MA, Monroe A, Kumar A, Xia S-M, Cooper M, Louder MK, McKee K, Bailer RT, Pier BW, Jette CA, Kelsoe G, Williams WB, Morris L, Kappes J, Wagh K, Kamanga G, Cohen MS, Hraber PT, Montefiori DC, Trama A, Liao H-X, Kepler TB, Moody MA, Gao F, Danishefsky SJ, Mascola JR, Shaw GM, Hahn BH, Harrison SC, Korber BT, Haynes BF. 2017. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med 9:eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong L, Lee JH, Doores KJ, Murin CD, Julien J-P, McBride R, Liu Y, Marozsan A, Cupo A, Klasse P-J, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong C-H, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burton DR, Hangartner L. 2016. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol 34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burton DR. 2017. What are the most powerful immunogen design vaccine strategies? Reverse Vaccinology 2.0 shows great promise. Cold Spring Harb Perspect Biol 9:a030262. doi: 10.1101/cshperspect.a030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. 2011. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol 85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, Karita E, Kilembe W, Lakhi S, Inambao M, Kamali A, Sanders EJ, Anzala O, Edward V, Bekker L-G, Tang J, Gilmour J, Kosakovsky-Pond SL, Phung P, Wrin T, Crotty S, Godzik A, Poignard P. 2016. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog 12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sok D, Burton DR. 2018. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eroshkin AM, LeBlanc A, Weekes D, Post K, Li Z, Rajput A, Butera ST, Burton DR, Godzik A. 2014. bNAber: database of broadly neutralizing HIV antibodies. Nucleic Acids Res 42:D1133–D1139. doi: 10.1093/nar/gkt1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. 2009. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res 37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kepler TB, Liao H-X, Alam SM, Bhaskarabhatla R, Zhang R, Yandava C, Stewart S, Anasti K, Kelsoe G, Parks R, Lloyd KE, Stolarchuk C, Pritchett J, Solomon E, Friberg E, Morris L, Karim SSA, Cohen MS, Walter E, Moody MA, Wu X, Altae-Tran HR, Georgiev IS, Kwong PD, Boyd SD, Fire AZ, Mascola JR, Haynes BF. 2014. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe 16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien J-P, Briney B, Ramos A, Saye KF, Le K, Mahan A, Wang S, Kardar M, Yaari G, Walker LM, Simen BB, St John EP, Chan-Hui P-Y, Swiderek K, Kleinstein SH, Kleinstein SH, Alter G, Seaman MS, Chakraborty AK, Koller D, Wilson IA, Church GM, Burton DR, Poignard P. 2013. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog 9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Briney BS, Willis JR, Crowe JE. 2012. Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun 13:523–529. doi: 10.1038/gene.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O'Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. 2012. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med 4:142ra96. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, Wilson PC. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohi M, Li Y, Cheng Y, Walz T. 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bell JM, Chen M, Baldwin PR, Ludtke SJ. 2016. High resolution single particle refinement in EMAN2.1. Methods 100:25–34. doi: 10.1016/j.ymeth.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reboul CF, Eager M, Elmlund D, Elmlund H. 2018. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci Publ Protein Soc 27:51–61. doi: 10.1002/pro.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Radermacher M, Wagenknecht T, Verschoor A, Frank J. 1987. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 69.Sorzano COS, Marabini R, Velázquez-Muriel J, Bilbao-Castro JR, Scheres SHW, Carazo JM, Pascual-Montano A. 2004. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 70.Scheres SHW. 2012. A Bayesian view on cryo-EM structure determination. J Mol Biol 415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 72.Alpert MD, Heyer LN, Williams DEJ, Harvey JD, Greenough T, Allhorn M, Evans DT. 2012. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol 86:12039–12052. doi: 10.1128/JVI.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse P-J, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]