Rhesus cytomegalovirus (RhCMV) offers a unique model for studying human cytomegalovirus (HCMV) pathogenesis and vaccine development. RhCMV infection of nonhuman primates greatly broadened the understanding of mechanisms by which CMVs evade or reprogram T cell and natural killer cell responses in vivo. However, the role of humoral immunity and viral modulation of anti-CMV antibodies has not been studied in this model. There is evidence from in vitro studies that HCMVs can evade humoral immunity. By gene mapping and with the help of a novel cell-based reporter assay system we characterized the first RhCMV encoded IgG-Fcγ binding glycoprotein as a potent antagonist of rhesus FcγR activation. We further demonstrate that, unlike evasion of T cell immunity, this viral Fcγ receptor is not required to overcome anti-CMV immunity to establish secondary infections. These findings enable more detailed studies of the in vivo consequences of CMV evasion from IgG responses in nonhuman primate models.
KEYWORDS: Fc receptors, antibody function, cytomegalovirus, immune evasion, immunology, rhesus, virology
ABSTRACT
Receptors recognizing the Fc part of immunoglobulin G (FcγRs) are key determinants in antibody-mediated immune responses. Members of the Herpesviridae interfere with this immune regulatory network by expressing viral FcγRs (vFcγRs). Human cytomegalovirus (HCMV) encodes four distinct vFcγRs that differ with respect to their IgG subtype specificity and their impact on antibody-mediated immune function in vitro. The impact of vFcγRs on HCMV pathogenesis and immunomodulation in vivo is not known. The closest evolutionary animal model of HCMV is rhesus CMV (RhCMV) infection of rhesus macaques. To enable the characterization of vFcγR function in this model, we studied IgG binding by RhCMV. We show that lysates of RhCMV-infected cells contain an IgG-binding protein of 30 kDa encoded by the gene Rh05 that is a predicted type I glycoprotein belonging to the RL11 gene family. Upon deletion of Rh05, IgG-Fc binding by RhCMV strain 68-1 is lost, whereas ectopic expression of Rh05 results in IgG binding to transfected cells consistent with Rh05 being a vFcγR. Using a set of reporter cell lines stably expressing human and rhesus FcγRs, we further demonstrate that Rh05 antagonizes host FcγR activation. Compared to Rh05-intact RhCMV, RhCMVΔRh05 showed an increased activation of host FcγR upon exposure of infected cells to IgG from RhCMV-seropositive animals, suggesting that Rh05 protects infected cells from opsonization and IgG-dependent activation of host FcγRs. However, antagonizing host FcγR activation by Rh05 was not required for the establishment and maintenance of infection of RhCMV, even in a seropositive host, as shown by the induction of T cell responses to heterologous antigens expressed by RhCMV lacking the gene region encoding Rh05. In contrast to viral evasion of natural killer cells or T cell recognition, the evasion of antibody-mediated effects does not seem to be absolutely required for infection or reinfection. The identification of the first vFcγR that efficiently antagonizes host FcγR activation in the RhCMV genome will thus permit more detailed studies of this immunomodulatory mechanism in promoting viral dissemination in the presence of natural or vaccine-induced humoral immunity.
IMPORTANCE Rhesus cytomegalovirus (RhCMV) offers a unique model for studying human cytomegalovirus (HCMV) pathogenesis and vaccine development. RhCMV infection of nonhuman primates greatly broadened the understanding of mechanisms by which CMVs evade or reprogram T cell and natural killer cell responses in vivo. However, the role of humoral immunity and viral modulation of anti-CMV antibodies has not been studied in this model. There is evidence from in vitro studies that HCMVs can evade humoral immunity. By gene mapping and with the help of a novel cell-based reporter assay system we characterized the first RhCMV encoded IgG-Fcγ binding glycoprotein as a potent antagonist of rhesus FcγR activation. We further demonstrate that, unlike evasion of T cell immunity, this viral Fcγ receptor is not required to overcome anti-CMV immunity to establish secondary infections. These findings enable more detailed studies of the in vivo consequences of CMV evasion from IgG responses in nonhuman primate models.
INTRODUCTION
As prototypical members of the β-subgroup of the herpesvirus family, cytomegaloviruses (CMVs) establish lifelong infection characterized by viral latency and reactivation. Human and animal CMVs share sophisticated mechanisms to evade a multitude of antiviral host immune responses, including both innate and adaptive arms of the immune system (1, 2). With respect to cell-mediated immunity, it has been shown that human cytomegalovirus (HCMV) can efficiently evade direct recognition of infected target cells by natural killer (NK) cells, as well as T lymphocytes, using a large repertoire of viral gene products that interfere with antigen presentation, surface receptor transport, or innate receptor signaling (3, 4). Complementing viral evasion of cell-mediated immune responses are strategies for evasion of humoral immunity, such as counteracting IgG-mediated antiviral immunity. Ribosomal profiling identified more than 750 translational products that include many potentially antigenic proteins during the sequential immediate-early (IE), early (E), and late (L) phases of gene expression (5). Despite exposure of these potential viral antigens to the host’s immune system, human and animal CMVs maintain lifelong chronic infections with occasional reactivation. Moreover, CMVs are able to reinfect CMV-immune hosts despite the presence of CMV-specific humoral and cellular immune responses (6, 7). Potentially due to viral immune evasion capabilities, anti-HCMV IgG preparations such as intravenous hyperimmune immunoglobulin (IVIG) or monoclonal antibodies (MAbs) displayed only limited, if any, efficacy in various clinical settings (8–13). In nonhuman primate (NHP) models, prevention of fetal transmission only occurred when IVIG was concentrated from plasma of donors that were preselected for high neutralization activity, whereas IVIG from nonselected plasma was only partially protective, suggesting that RhCMV is able to escape antibody control (14).
Specific viral mechanisms that counteract antibody effector functions might be responsible for limiting the ability of antibodies to control viral infection and dissemination. HCMV evasion from IgG-Fc-mediated effector functions can be attributed to a set of IgG-Fc binding glycoproteins (vFcγRs) encoded by the HCMV genes UL118/119 (gp68) and RL11 (gp34) (15). These vFcγRs were shown to efficiently antagonize host IgG-Fc receptor (FcγR) activation in a cell-based in vitro reporter assay performed on IVIG-opsonized infected cells (16). In addition, RL12 and RL13 have been shown to have vFcγR activity (14). Although HCMV is the only known human betaherpesvirus to encode such glycoproteins, it is not the only herpesvirus for which vFcγRs have been described. Mouse cytomegalovirus (MCMV) encodes the Ig-like glycoprotein fcr-1/m138 (17). Deletion of m138 from the MCMV genome results in drastic attenuation of MCMV in vivo (18). However, since m138 has both Fcγ-related and -unrelated immunoevasive functions (19–21), the role of Fcγ modulation for viral pathogenesis has yet to be established. HSV-1 and VZV glycoproteins E and I (gE/gI) form an IgG-Fc binding heterodimer (22, 23). By clearing antigen/antibody complexes from the infected cell surface (24), the HSV-1 gE/gI complex promotes immune evasion in vivo (25). Interestingly, the VZV gE protein is the major component of the recently developed highly efficient subunit VZV vaccine (26).
Immune responses most prominently governed by host FcγRs include antibody-dependent cell-mediated cytotoxicity, antibody dependent cell-mediated phagocytosis, and the induction of a proinflammatory cytokine profile by various immune cells, including NK cells, macrophages, dendritic cells, B cells, and neutrophils expressing FcγRs (27). FcγRs are further classified by their affinity to IgG-Fc and are highly conserved between humans and nonhuman primates showing strong cross-reactivity (28, 29). There are four known activating receptors comprising the high-affinity receptor CD64/FcγRI, the medium-affinity receptors CD32A/FcγRIIA and CD32C/FcγRIIC, and the low-affinity receptor CD16A/FcγRIIIA. CD32B/FcγRIIB is the only known inhibitory receptor with a medium affinity to IgG-Fc and a single cytosolic ITIM motif (27). Although their affinity to IgG-Fc is also dependent on the IgG subclass, all FcγRs show their highest affinity toward IgG1, while optimal binding in general can only be observed to immune complexed IgG with an intact glycan profile (30). In recent years, FcγR-mediated immune responses have proven to be an essential factor in the antiviral effect of not only nonneutralizing but also neutralizing IgGs specific for important pathogenic viruses such as influenza A (31, 32) and HIV (33, 34).
CMVs are highly species specific, which prevents studying HCMV directly in an animal model. While the closest relative of HCMV is chimpanzee CMV, experimentation in these animals is no longer possible. In contrast, infection of rhesus macaques (RM) (Macaca mulatta) with rhesus cytomegalovirus (RhCMV) is a tractable model and the genomes of NHP CMVs encode homologs of most of the HCMV gene families (35, 36). Therefore, RhCMV infection has emerged as a state of the art model, allowing the study of primate CMV disease infection, immune responses, and pathology in vivo (37), including important aspects of congenital infection (14, 38). While in this model RhCMV genes linked to evasion from CD8+ T lymphocyte and NK cell responses have been extensively investigated (6, 39), little is known about the ability of RhCMV to evade antibody-mediated immunity.
We demonstrate here that the RhCMV RL11 gene family member Rh05 encodes an IgG-Fc binding glycoprotein. Similar to HCMV vFcγRs, this type 1 transmembrane protein is transported to the cell surface, where it efficiently antagonizes FcγR activation triggered by immune IgG. In addition, Rh05 was able to antagonize human FcγRIIIA/CD16A activation by cells opsonized with a rhesusized monoclonal IgG antibody. Interestingly, Rh05 was not required for RhCMV superinfection, suggesting that evasion of preexisting antibodies is not essential for the establishment of secondary infections. These results thus represent the first identification of a vFcγR in RhCMV and highlight the close evolutionary relationship of human and rhesus IgG and FcγRs consistent with the RM/RhCMV model being particularly relevant when studying viral evasion of IgG effector functions in vivo.
RESULTS
RhCMV glycoprotein binding to IgG.
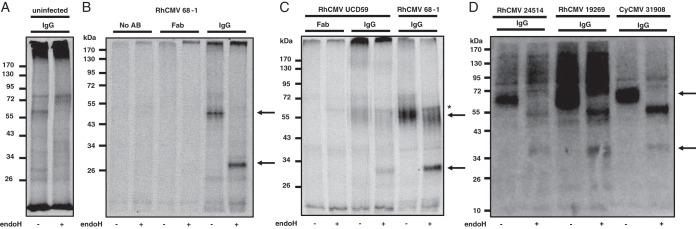
To determine whether RhCMV encodes viral proteins binding to IgG, purified rhesus IgG from RhCMV-seronegative RM was incubated with detergent lysates of [35S]methionine-labeled, RhCMV-infected telomerized rhesus fibroblasts (TRFs). For control, we used Fab fragments generated from rhesus IgG. In addition to the fibroblast-adapted laboratory strain 68-1, which carries a number of gene deletions (36), we also used the primary RhCMV isolate UCD59 (40) and the recently characterized RhCMV isolates 19269 and 24514, as well as the cynomolgus CMV (CyCMV) isolate 31908 (41). Bound proteins were eluted from the protein A/G agarose beads and, where indicated, digested with endoglycosidase H (EndoH) to monitor glycan processing during intracellular transport, followed by separation using SDS-PAGE. As shown in Fig. 1, RhCMV- and CyCMV-infected cell lysates, but not uninfected cell lysates, contained a single protein species of ∼60 kDa bound to IgG. This protein was observed in 68-1-infected cell lysates, as well as in lysates from cells infected with primary NHP CMV isolates. Upon EndoH treatment, the molecular weight of the protein was reduced to ∼30 kDa, suggesting that the protein is highly glycosylated. Both EndoH-sensitive and EndoH-resistant bands were observed consistent with newly synthesized, EndoH-sensitive protein subpopulations in the endoplasmic reticulum that eventually egress to the cell surface.
FIG 1.
RhCMV encodes an IgG binding protein. To detect IgG binding proteins, lysates from metabolically labeled TRFs were incubated with serum from RhCMV-naive RMs, and the total IgG was immunoprecipitated using protein A/G-agarose. Endoglycosidase H (EndoH) was added where indicated. (A) Uninfected cell lysate. (B) TRFs were infected with RhCMV 68-1 (MOI = 3) for 72 h prior to metabolic labeling. Infected cell lysates were either untreated or incubated with purified Fab fragments or whole serum. Immunoprecipitates were separated by SDS-PAGE, and protein bands were visualized by autoradiography. (C) TRFs were infected with RhCMV 68-1 or the low-passage-number isolate UCD59 (MOI = 3) for 72 h prior to metabolic labeling and immunoprecipitation. (D) TRFs were infected with RhCMV 68-1 or the indicated RhCMV and CyCMV low-passage-number isolates (MOI = 3) for 72 h prior to metabolic labeling. IgG immunoprecipitations after incubation with CMV-naive RM serum were performed using protein A/G-agarose. Arrows indicate a single EndoH-sensitive glycoprotein species. *, Nonspecific protein.
Rh05 encodes a viral FcγR.
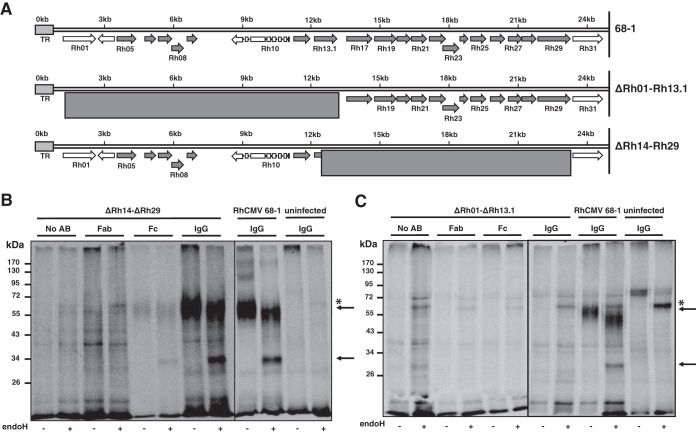
HCMV encodes four vFcγRs: RL11 (gp34), RL12, RL13, and UL119/118 (gp68). RL11, RL12, and RL13 belong to the RL11 gene family, encoding a highly polymorphic glycoprotein family which is also found in RhCMV (36). HCMV gp68 is conserved in RhCMV, including the spliced gene structure, with the putative homologue encoded by Rh152/151 (35). However, the gp68 homologue is truncated in RhCMV 68-1 (36), rendering it possibly nonfunctional. Moreover, the molecular weight of the putative viral Fc receptor was considerably less than predicted for the gp68 homologue of RhCMV. Therefore, we hypothesized that the viral IgG-binding protein was likely a member of the RL11 family. In RhCMV, the RL11 family is encoded in the 5′ end upstream of the open reading frame (ORF) Rh29 (Fig. 2A). To determine whether the putative vFcγR is encoded in this gene region, we generated two deletion mutants lacking Rh01-Rh13.1 and Rh14-Rh29 in RhCMV 68-1 by bacterial artificial chromosome (BAC) recombineering (Fig. 2A). Replacement of the desired genomic regions by a FRT-flanked Kanr cassette was confirmed by restriction digest. Upon electroporation of the BACs, virus was easily recovered, consistent with genes encoded in this genomic region being nonessential for growth in vitro as reported for RhCMV (42) and HCMV (43). To determine whether ΔRh01-13.1 and ΔRh14-29 contained or lacked the putative IgG binding protein, we metabolically labeled infected RF as described above and incubated detergent cell lysates with complete IgG, Fab fragments, or Fc fragments bound to protein A/G-agarose beads or control beads. Upon electrophoretic separation, we observed that lysates of ΔRh14-29-infected cells contained the ∼60 kDa (or 30 kDa upon deglycosylation) protein that was immunoprecipitated with both IgG and Fc, but not with F(ab)2 or beads alone (Fig. 2B). In contrast, the 60-kDa protein was not observed in ΔRh01-13.1-infected cell lysates (Fig. 2C). a finding consistent with the putative vFcγR being encoded in the 5′-terminal region of the genome.
FIG 2.
The IgG-binding protein is encoded in the 5′ end of the RhCMV genome. (A) Schematic overview of the 5′-end genomic region of RhCMV encompassing the RL11 gene family. All RL11 gene family members are highlighted in dark gray. Two deletion mutants, ΔRh01-Rh13.1 and ΔRh14-ΔRh29, that together span the entire RL11 gene family were constructed. The exact region that was deleted in each mutant is indicated by the boxed area. (B and C) TRFs were infected with the indicated deletion mutants or with RhCMV 68-1 WT control at an MOI of 3 for 72 h prior to metabolic labeling. Lysates were either mock incubated or incubated with purified Fab fragments, purified Fc fragments, or whole serum. IgG was immunoprecipitated and treated with EndoH where indicated. Arrows indicate the glycosylated and deglycosylated forms of the RhCMV-encoded protein that coprecipitates with RM IgG or RM IgG Fc fragments from RhCMV 68-1 and from RhCMVΔRh14-29, but not from RhCMVΔRh01-13.1. *, Nonspecific protein. All other unmarked proteins species are also nonspecific.
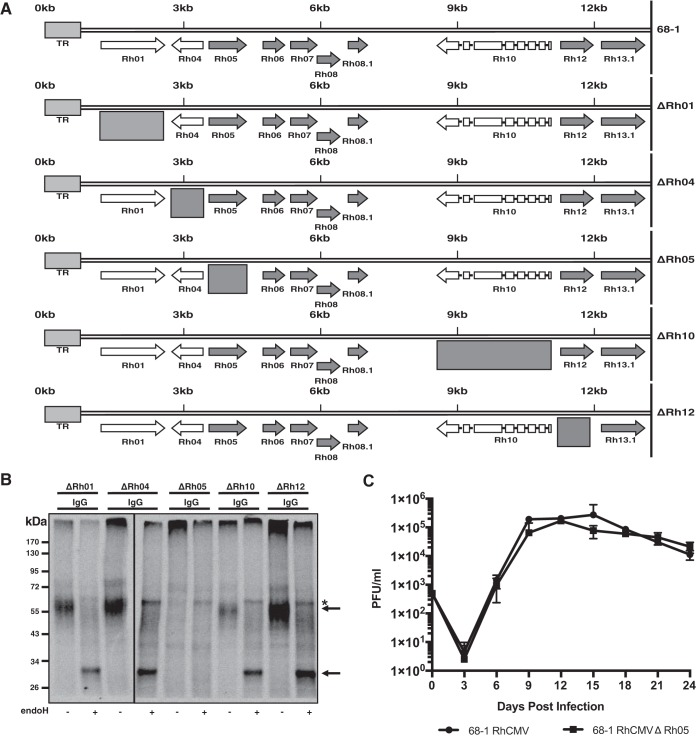
To determine which gene(s) in the Rh01-Rh13.1 region encoded the putative vFcγR, we deleted individual genes in this region from the 68-1 BAC (Fig. 3A). Upon reconstitution of the single deletion constructs, we evaluated IgG binding upon infection of RF. As shown in Fig. 3B, IgG was able to immunoprecipitate the putative vFcγR from all deletions mutants except ΔRh05. To ensure that lack of binding was not due to lack of infection and or gene expression, we also confirmed that ΔRh05 was not essential for infection and growth in vitro (Fig. 3C). These results suggest that the Rh01-Rh13.1 gene region contains a single vFcγR encoded by Rh05.
FIG 3.
Rh05 encodes a viral Fc binding protein. (A) Schematic overview of the RhCMV deletion mutants constructed by BAC recombineering. The entire viral ORF was deleted in each case, as indicated by the boxes. (B) Immunoprecipitations of IgG of RhCMV-naive serum incubated with lysates from TRFs infected with the single deletion mutants (MOI = 3) for 72 h. Half of every sample was EndoH treated, as indicated. Arrows indicate the glycosylated and nonglycosylated form of the IgG binding protein. *, Nonspecific protein. (C) Multistep growth curve of RhCMV 68-1 and RhCMVΔRh05 on primary rhesus fibroblasts. The cells were infected at an MOI of 0.01, samples were harvested every third day, and viral titers were determined by TCID50.
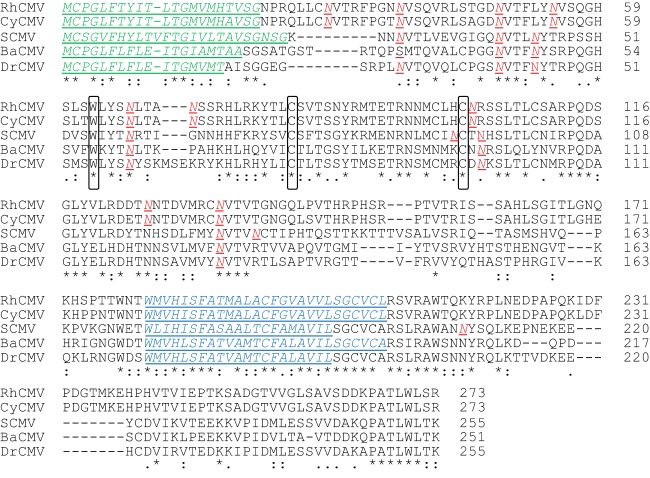
The gene Rh05 encodes an RL11 family protein of 273 amino acids (AA) with a predicted molecular weight of 30.19 kDa. The Rh05 protein displays a type I transmembrane topology with a predicted cleavable amino-terminal signal peptide (AA1-21), a predicted transmembrane domain (AA181-207) and a 65-AA cytoplasmic domain (Fig. 4). Homologous proteins are found in Old World NHP CMVs (Fig. 4). In contrast, none of the RL11-family proteins of human, great ape, or New World NHP CMVs seem be direct homologs of Rh05. The ectodomain is predicted to belong to the immunoglobulin superfamily and contains nine putative N-linked glycosylation sites, several of which being highly conserved, consistent with the protein being highly glycosylated. Also conserved is the C-terminal AA sequence PATLWL[T/S][K/R], which might represent a subcellular sorting signal. The predicted characteristics of this protein are thus, consistent with the observed molecular weight and glycosylation pattern of the Fcγ-binding viral protein.
FIG 4.
RhCMV Rh05 is conserved in Old World monkey CMV species. An alignment of the predicted amino acid sequence of Rh05 with putative homologues of cynomolgus CMV 31908 (CyCMV), simian CMV Colburn (SCMV), Baboon CMV OCOM4-37 (BaCMV), and Drill monkey CMV OCOM6-2 (DrCMV) was generated using the CLUSTAL O (1.2.4) multiple sequence alignment tool. Highlighted are the predicted signal sequence (green, predicted using the SignalP 4.1 server), transmembrane region (blue, predicted using the Phobius server), and potential glycosylation sites (red, using the NetNGlyc 1.0 server). In addition, amino acids that that have been defined as conserved across the RL11 family of proteins are circled in black.
Recombinant Rh05 is an IgG-Fc binding cell surface protein which antagonizes human FcγRIIIA/CD16 activation.
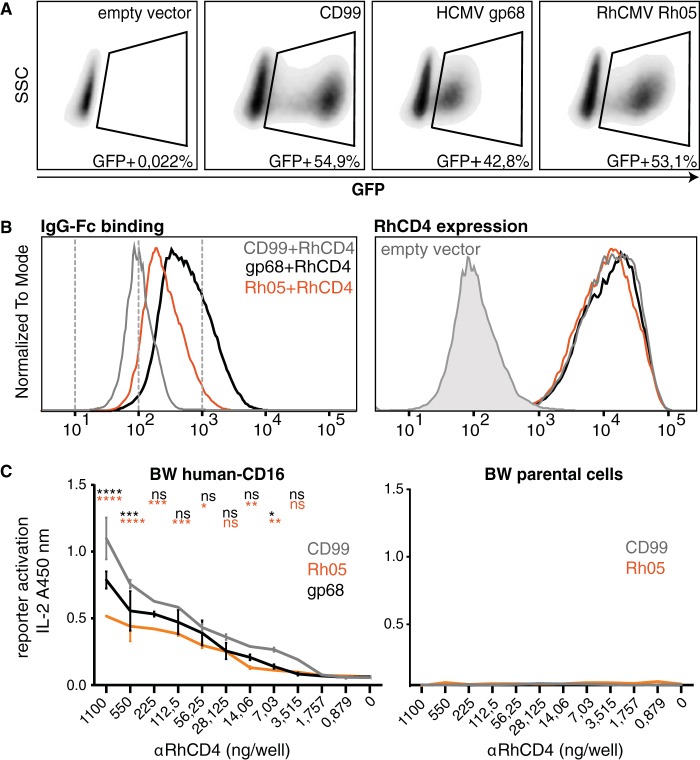
To examine whether Rh05 has the capacity to counteract host Fcγ receptor activation, as reported for the IgG-Fc binding HCMV proteins RL11/gp34 and UL119-118/gp68 (16), we introduced recombinant Rh05 into an established human Fcγ receptor activation assay (44). As a target surface antigen, we chose rhesus CD4 (RhCD4) that can be detected with a recombinant rhesusized IgG1 MAb (αRhCD4 MAb). To this end, we cotransfected HeLa cells with RhCD4 (pCDNA3.1 vector) and a polycistronic pIRES_eGFP vector encoding either recombinant HCMV gp68, RhCMV Rh05, or CD99 control protein, together with green fluorescent protein (GFP) as an expression marker, which allowed us to monitor transfection efficiency (Fig. 5A). As a first step, we wanted to determine whether Rh05 alone would be sufficient to bind to the Fc portion of IgG on the cell surface. By staining the vFcγR/RhCD4-cotransfected HeLa cells with a Texas Red-conjugated human IgG-Fc fragment and gating on the above-mentioned GFP-positive population, we observed that Rh05 is a potent IgG-Fc binding protein compared to HCMV gp68, which served as a positive control (Fig. 5B, left). A human IgG-Fc fragment was used as previous observations already showed high cross-reactivity between human and nonhuman primate IgG-Fc (28, 29). In these experiments, HCMV gp68 was expressed as a fusion protein to the transmembrane domain and cytosolic tail of human CD4 since this fusion protein reaches higher densities on the plasma membrane upon transient expression than wild-type gp68 (P. Kolb and H. Hengel, unpublished observations). Surface expression of cotransfected RhCD4 and binding of αRhCD4 to its antigen in cotransfected HeLa cells was demonstrated by detection of RhCD4 using phycoerythrin (PE)-conjugated αRhCD4 (Fig. 5B, right). Gating on GFP-positive cells allowed us to conclude that cells expressing Rh05, gp68 or CD99 uniformly expressed the target antigen RhCD4 and that surface levels of RhCD4 are not affected by cotransfected genes of interest (Fig. 5B, right).
FIG 5.
Rh05 binds IgG-Fc and antagonizes antibody-dependent FcγR activation. HeLa cells were cotransfected with the target antigen rhesus-CD4 (RhCD4; pcDNA3.1) and either of the indicated genes of interest (CD99, HCMV UL119-118 and RhCMV Rh05; p_IRES-eGFP). (A) GFP-positive cells, gated on live cells using DAPI, were plotted against side scatter. The GFP-positive population, indicated by a gate, demonstrates similar transfection rates for each of the genes of interest. (B, left panel) GFP-positive cells from panel A were analyzed for Fcγ binding by flow cytometry using Texas Red-conjugated human Fcγ fragment. RhCMV Rh05 and HCMV gp68 bound to IgG-Fc, whereas CD99 was negative. (Right panel) The surface expression levels of RhCD4 are not affected by coexpressed genes of interest. RhCD4 was detected in the GFP-positive population from A using a PE-conjugated rhesusized anti-RhCD4 MAb. (C) Rh05 antagonizes antibody-dependent FcγR activation. HeLa cells cotransfected with RhCD4 and the indicated genes of interest were incubated with rhesusized anti-RhCD4 MAb and subsequently cocultured with BW reporter cells expressing the chimeric human receptor CD16ζ (left) or parental BW5147 cells (right). IL-2 levels corresponding to reporter activation were quantified using ELISA. Error bars indicate the standard deviations. Two-way ANOVA (Tukey) was applied: gp68 versus CD99 (black) and Rh05 versus CD99 (orange).
To address the antagonistic potential of Rh05, the cotransfected cells were then cocultured with a reporter cell line expressing the human FcγRIIIA/CD16 ectodomain fused to the CD3-ζ-chain signaling module (BW5147-human-CD16-ζ) after adding graded amounts of αRhCD4. Reporter cell activation was quantified by measuring IL-2 production using a sandwich ELISA as described previously (44). As shown in Fig. 5C, compared to the expression of a non-Fcγ-binding control molecule (CD99), we observed a significant and antibody dose-dependent reduction of CD16-reporter cell activation by target cells expressing Rh05 that exceeded the inhibition mediated by gp68. Control BW cells (i.e., BW5147 mouse thymoma cells) lacking the CD16 FcγR (parental cells) were not activated. Taken together, these data demonstrate that Rh05 represents an IgG-Fc binding glycoprotein with the potential to antagonize the activation of host FcγRs.
Rh05 protects RhCMV-infected cells from FcγR activation by opsonizing IgG.
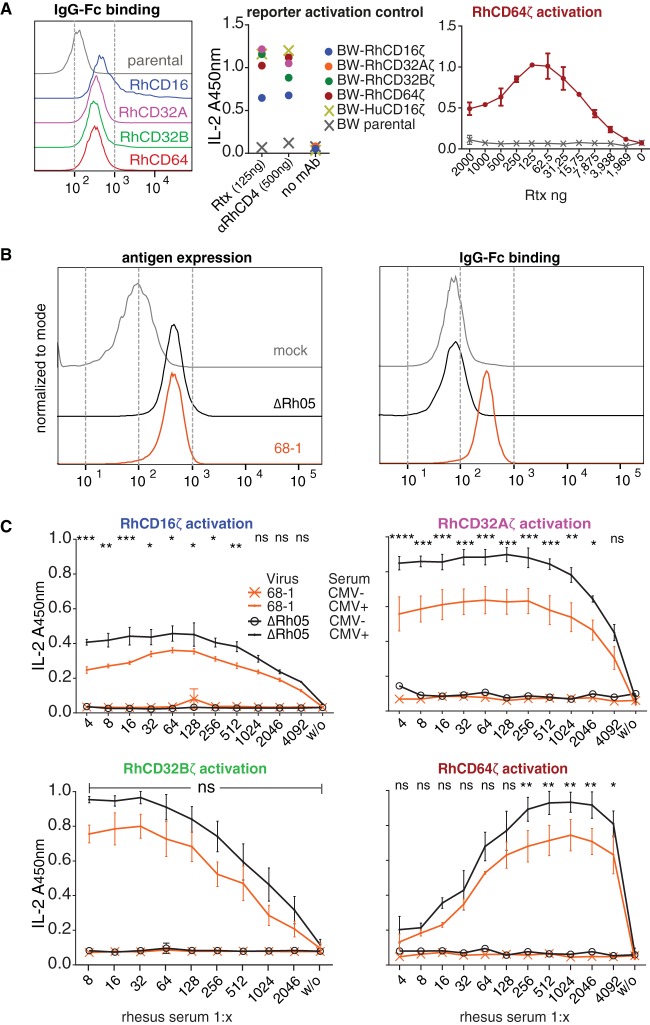
The potent inhibition of human CD16 activation by Rh05 supported our hypothesis that this vFcγR might protect infected cells from Macaca mulatta FcγR-dependent effector mechanisms. To this end, we generated BW reporter cells encoding chimeric rhesus CD16 (RhCD16), RhCD32A, RhCD32B, or RhCD64 consisting of the extracellular FcγR domain fused to the transmembrane and intracellular domains of the mouse CD3ζ chain. FcγR activation can thus be monitored by production of interleukin-2 (IL-2). Surface expression and intact ligand binding of these chimeric Rh-FcγRs was demonstrated by flow cytometry using a Texas Red-conjugated human IgG-Fc fragment (Fig. 6A, left). Next, the ability of these reporter cell lines to generate IL-2 upon FcγR activation was verified by receptor cross-linking by immobilized IgG of human and rhesus origin. All reporter cell lines responded to human IgG1 MAb Rituximab or αRhCD4 (Fig. 6A, middle). Of note, BW-RhCD16ζ yielded lower signals compared to the other cell lines, including BW cells expressing human-CD16ζ. This could be due to the fact that IgG from individual sources can have highly varying affinities to certain isoforms of Rh-FcγRs (29). Interestingly, the dose-response of BW-Rh64ζ cells in this context did not reach an activation plateau that was maintained at high antibody concentrations, but displayed a maximum response at lower antibody concentrations (Fig. 6A, right). In contrast, all other reporter cell lines (including reporter cells expressing hCD64) showed the typical sigmoidal dose-response with plateau activation to the immobilized antibodies above a given antibody concentration (data not shown). While we cannot fully explain this observation, it is possible that RhCD64 reaches suboptimal activation with large amounts of immobilized IgG due to its intrinsic molecular characteristics as a high-affinity FcγR which bind to but are not activated by monomeric IgG (29, 30).
FIG 6.
Rh05 antagonizes FcγR stimulation by infected cells. (A, left) The surface expression of chimeric rhesus FcγRs—RhCD16ζ, RhCD32Aζ, RhCD32Bζ, and RhCD64ζ—on stably transduced BW cells was detected using Texas Red-conjugated human Fcγ fragment. Parental BW cells were used as a control. (Middle) Chimeric rhesus FcγRs are activated upon IgG-Fc binding. The indicated BW reporter cells were assessed for activation by immobilized antibodies (Rtx, rituximab; αRhCD4, recombinant rhesusized anti-rhesus-CD4 MAb). All values are means of technical duplicates and represent plateau activation determined by incubation on titrated amounts of antibody (not shown). (Right) Dose-response upon RhCD64 reporter cell activation by titrated amounts of Rtx. (B) TRF cells were infected with RhCMV 68-1 or RhCMVΔRh05 using centrifugal enhancement at an MOI of 2 for 72 h. (Left) Infected cells were incubated with serum from an RhCMV-seropositive monkey, and overall surface antigen expression was detected via a FITC-conjugated rabbit anti-human IgG polyclonal antibody. (Right) Infected cells were probed with a Texas Red-conjugated human IgG-Fc fragment. (C) Rh05 antagonizes rhesus FcγR activation by antibody bound to infected cells. Infected cells were incubated with serum dilutions of RhCMV-positive or RhCMV-negative monkeys and subsequently cocultured with the indicated BW reporter cells. IL-2 levels corresponding to reporter activation were quantified using ELISA. Error bars indicate the standard errors of the mean. CMV-positive sera, averages of two independent experiments; CMV-negative sera, averages of 1 experiment. Two-way ANOVA (Tukey) was performed. Asterisks indicate statistical comparisons of reporter responses to infected cells opsonized by RhCMV-positive serum.
With these reporter cell lines in hand, we then set out to assess the effect of Rh05 on Rh-FcγR activation. To this end, TRF infected with RhCMV 68-1 or RhCMVΔRh05 were incubated with polyclonal immune serum from RhCMV-positive or -negative animals and then cocultured with the respective reporter cell lines. As expected, surface antigen levels were similar between cells infected with either RhCMV 68-1 or RhCMVΔRh05, as demonstrated by flow cytometry detecting the bound anti-RhCMV serum via a fluorescein isothiocyanate (FITC)-conjugated polyclonal anti-human antibody (Fig. 6B, left). In contrast, IgG-Fc binding was only observed for TRF infected with RhCMV 68-1, but not RhCMVΔRh05, consistent with a complete loss of Fc-binding activity upon deletion of Rh05 (Fig. 6B, right). Applying the FcγR reporter assay, serum from the RhCMV-seropositive animal elicited the typical dose-dependent response in the reporter cell lines, except for RhCD64, which again showed maximal stimulation at lower serum concentrations (Fig. 6C). Serum from the RhCMV-negative animal did not induce IL-2 in any of the reporter cells (Fig. 6C). Importantly, compared to cells infected with RhCMV 68-1, cells infected with RhCMVΔRh05 induced significantly higher reporter cell activation for all examined activating Rh-FcγRs at dilutions of RhCMV-immune serum that elicited maximal stimulation (Fig. 6C). Although there was a similar tendency for the inhibitory RhCD32B receptor, the differences between the RhCMVΔRh05 and 68-1 RhCMV did not reach statistical significance. Based on these results, we conclude that Rh05 limits the ability of IgG antibodies bound to infected cells to activate host FcγRs, thus counteracting opsonization and subsequent FcγR-mediated immune responses.
Reinfection by RL11-family-deleted RhCMV.
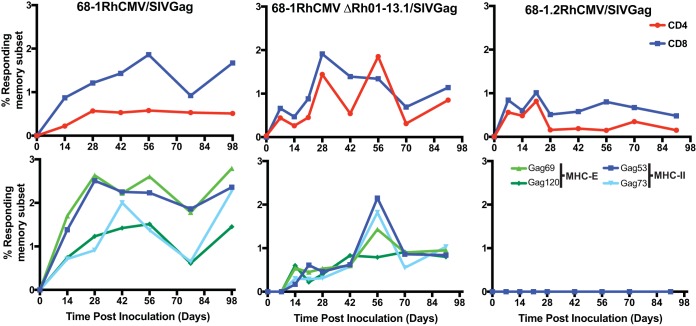
A unique aspect of both RhCMV and HCMV is their ability to establish secondary persistent infections in CMV-immune hosts. We previously demonstrated that viral evasion of CD8+ T cells by US6-family viral inhibitors of MHC-I antigen presentation is necessary for RhCMV to reinfect RhCMV-seropositive animals (6). Furthermore, preventing the activation of NK cells by inhibiting the cell surface expression of ligands for activating NK cell receptors proved to be essential for RhCMV infection in both RhCM-seropositive and -seronegative hosts (39). Therefore, we wondered whether the vFcγR Rh05 would be required for RhCMV to overcome preexisting humoral immunity. T cell responses to heterologous antigens expressed by RhCMV can be used as a surrogate measure for the ability of RhCMV to reinfect seropositive animals (6). Thus, we took advantage of the SIVgag gene inserted during the construction of ΔRh01-13.1 (see Materials and Methods). A total of 5 × 106 PFU of ΔRh01-13.1 was inoculated subcutaneously, and the T cell response to SIVgag was measured biweekly in PBMC by intracellular cytokine staining (ICS) using overlapping peptides spanning the SIVgag sequence. As shown in Fig. 7 (top row), ΔRh01-13.1 elicited robust SIVgag-specific responses for both CD4+ and CD8+ T cells that were comparable to inoculation of 68-1 RhCMV/gag into a different animal. Although these results were only obtained in one animal, they clearly demonstrate that the gene region containing Rh05 is not essential for infection and reinfection.
FIG 7.
Rh05 is not required for superinfection. At day 0, an RhCMV-positive RM was infected subcutaneously with 5 × 106 PFU of the indicated recombinant virus and the SIVgag-specific T cell responses in PBMCs were monitored by ICS for CD69, TNF-α, and IFN-γ using either overlapping SIVgag 15mer peptide mixes to measure total responses (top row) or the indicated MHC-E and MHC-II supertopes to measure epitope-specific responses (bottom row). The results are shown as a percentage of the total memory CD4+ or CD8+ T cells.
We recently reported that recombinant viruses based on strain 68-1, but not the pentamer-intact derivative RhCMV 68-1.2, elicit CD8+ T cells that recognize peptides exclusively in the context of major histocompatibility complex class II (MHC-II) or the nonpolymorphic MHC-E molecule instead of polymorphic MHC-Ia (45, 46). Moreover, some MHC-II- and MHC-E-restricted SIVgag peptide epitopes, termed “supertopes,” are consistently recognized in every animal tested thus far (>100 animals). To determine whether genes encoded in the Rh01-13 region affected this T cell programming we measured the CD8+ T cell responses to two MHC-II and two MHC-E supertopes. Similar to total SIVgag responses, we observed that both 68-1 RhCMV/gag and ΔRh01-13.1 elicited supertope-specific CD8+ T cells, in contrast to 68-1.2 RhCMV/gag that failed to elicit CD8+ T cells to these supertopes (Fig. 7, bottom row). These results suggest that the deletion of Rh05 or any of the other genes encoded in the 5-terminal region of RhCMV does not impact the ability of RhCMV 68-1 to elicit CD8+ T cells to unconventional epitopes.
DISCUSSION
Our results demonstrate that RhCMV Rh05 encodes an IgG-Fc binding glycoprotein that immobilizes antibodies at the cell surface. Using a cell-based assay to measure rhesus IgG-mediated activation of rhesus FcγRs, we further show that Rh05 expressed on the surface of infected cells is a potent antagonist of host FcγR activation by anti-CMV antibodies. Based on these results, we conclude that Rh05 is a vFcγR that counteracts the ability of CMV-specific antibodies to trigger activating host FcγRs, thus supporting viral immune evasion.
Rh05 is the first vFcγR identified in RhCMV. Although Rh05 does not show direct homology to any of the previously identified vFcγRs in HCMV, the protein belongs to the same RL11 gene family as three of the four HCMV vFcγRs: RL11 (gp34), RL12, and RL13 (16, 47). We observed that, similar to gp34, which is able to block all of the activating human FγRs, i.e., FcγRI (CD64), FcγRIIa (CD32a), and FcγRIIIA (CD16), Rh05 reduced the activation of homologous rhesus FcγRs. The diverse RL11 glycoprotein family is characterized by the ∼80-AA RL11 domain containing a conserved tryptophan and two cysteine residues (48, 49). In addition to encoding vFcγRs, members of this gene family have been involved in various immunomodulatory functions (50–54), as well as the viral modulation of angiogenesis, cell differentiation, and reactivation (55, 56). Mutations in the RL13 glycoprotein are rapidly selected in both HCMV and NHP CMVs in tissue culture due to increased growth of RL13-defective variants (41, 57). Due to two frameshift mutations, RhCMV strain 68-1 used in this study is also predicted to lack a functional RL13 homologue (Rh13.1), suggesting that the negative impact of this protein on viral growth in vitro is conserved (36). However, it is presently not known whether intact Rh13.1 also shares the ability to bind Fc with HCMV RL13. Similarly, it is not known whether the RhCMV homologue of HCMV UL118/119 (gp68) is a functional vFcγR. However, given the significant homology, including the spliced gene structure, this is highly likely. Interestingly, the Rh151/152 gene encoding the gp68 homolog is truncated and possibly nonfunctional in RhCMV 68-1 (36). Conceivably, wild-type RhCMV could thus encode additional vFcγRs compared to RhCMV 68-1. However, we observed only a single viral protein band corresponding in size to Rh05 immunoprecipitating with IgG in lysates from cells infected with low-passage-number isolates of RhCMV and CyCMV (Fig. 1). Thus, it is also conceivable that Rh05 is the only vFcγR in NHP CMVs. By studying the homologs of RL13 and gp68 in isolation, we will be able to examine this possibility.
To determine the impact of vFcγR expression on host Fc receptor activation, we introduced Rh-FcγRs into our previously developed FcγR activation assay (44). We showed that this assay delivered reproducible, quantifiable measurements of FcγR activation via immune IgG when applied to infected cells opsonized with polyclonal serum in the context of herpes simplex virus, HCMV, and influenza virus (16, 31, 58). In a mouse influenza virus model, comparative FcγR assay results closely correlated with the protective capacity of antiviral IgGs in vivo (31). By generating Rh-FcγRs fused to mouse CD3ζ, we were able to measure the antibody dose-dependent effect of FcγR activation by antibody binding to RhCMV-infected cells. In doing so, we uncovered an unexpected IgG concentration-dependent optimum of rhesus CD64/FcγRI activation (Fig. 6A and C). In contrast, human FcγRI activation plateaued at high concentrations in this assay system (16). The finding that higher antibody concentrations result in lower FcγR activation could potentially reflect a unique feature, possibly a specific isoform, of the high-affinity rhesus FcγRI.
It is thus possible that the rhesus FcγRI receptors are functionally different from human FcγRI receptors. However, the homologies between RM and human FcγRs are approximately 95, 87, and 91% for FcγRI, RII, and RIII, respectively (29). Some polymorphisms are observed in RM, particularly for FcγRIIA, some of which resulting in impaired antibody binding (29). However, the allotypic variants in this study (FcγRI-3, FcγRIIA-1, FcγRIIB-1, and FcγRIIIA-1) were previously shown to be fully functional but differed with respect to IgG subclass specificity (29). Importantly, Rh05 was able to interfere with the activation of each activating RM FcγR by polyclonal RM serum, suggesting that Rh05 broadly binds IgG subclasses.
Unlike RhCMV lacking the gene region Rh182-189, encoding proteins that prevent MHC-I antigen presentation, or RhCMV lacking NKG2D-ligand-retaining Rh159, deletion of the gene region encompassing Rh05 did not affect the ability of RhCMV to overcome preexisting immunity and establish a secondary infection. If Rh05 is indeed the only vFcγR encoded by RhCMV, this result would indicate that evasion of antibodies is not essential for superinfection. Alternatively, Rh05 is the not the only vFcγRs, and other, yet-to-be-identified vFcγRs support reinfection. In either case, however, these results do not rule out that Rh05 supports viral replication, dissemination, and/or shedding. For instance, although strain 68-1 RhCMV is clearly able to establish secondary persistent infections in RhCMV-seropositive RM, this highly passaged strain is clearly attenuated compared to low-passage-number isolates such as UCD59, resulting in decreased plasma viral titers and decreased shedding during acute infection (59). A more detailed study requiring a larger cohort size will thus be required to quantify the impact of Rh05 on RhCMV infection.
It will also be interesting to study the impact of Rh05 deletion, alone or together with additional putative vFcγRs discussed above, in settings of passive immunization with anti-RhCMV antibodies. The importance of IgG-Fc interaction with host FcγRs for protection by passive immunization against viruses has been illustrated in animal models of influenza and HIV (32, 33, 60). In the case of HIV, it has further been shown that viral antibody escape mutants arise in an Fc-dependent manner (33). However, large DNA viruses such as CMV likely contain multiple epitopes targeted by antibodies, which renders it difficult for the virus to escape immune pressure by mutation. Conceivably, vFcγRs evolved to enable antibody escape by CMVs regardless of the epitope targeted, thus limiting the ability of both neutralizing and nonneutralizing antibodies to prevent viral spread in vivo. This immune evasion mechanism might therefore limit the efficacy of passively administered immunoglobulins to prevent congenital infection by CMV (9). The identification of a vFcγR in a highly relevant animal model of HCMV will contribute to a better understanding of the role of vFcγRs in counteracting immune responses elicited by vaccines and immunotherapies which might be improved by reagents that block vFcγR function.
MATERIALS AND METHODS
Cells.
All cells were cultured in a 5% CO2 atmosphere at 37°C. Telomerized rhesus fibroblasts (TRFs), HEK293T cells, and HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 10% (vol/vol) fetal calf serum (FCS; Biochrom) and antibiotics (1× Pen/Strep; Gibco). TRFs were generated from rhesus fibroblasts (RFs) obtained from animals housed at Oregon National Primate Research Center (ONPRC) and life extended as described previously (61). BW5147 mouse thymoma cells (BW cells; obtained from ATCC TIB-47) were maintained at 3 × 105 to 9 × 105 cells/ml in Roswell Park Memorial Institute medium (RPMI GlutaMAX; Gibco) supplemented with 10% (vol/vol) FCS, antibiotics, sodium pyruvate (1×; Gibco), and β-mercaptoethanol (0.1 mM; Gibco).
Generation of purified Fab and Fc fragments from whole serum.
IgG was isolated from preexisting serum samples of healthy, RhCMV-naive RM at the ONPRC. Fab and Fc fragments were generated using a Pierce Fab preparation kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Protein concentrations of the purified samples were determined using a NanoDrop ND-1000 (Thermo Fisher Scientific), and equal amounts of protein for each sample were separated on a SDS-polyacrylamide gel. To visualize the purified fragments, the gel was fixed with methanol and silver stained using a SilverQuest silver staining kit (Thermo Fisher Scientific).
Metabolic labeling of cells.
TRFs were grown in 60-mm tissue culture dishes (1.5 × 106 cells per dish) and removed using a cell scraper. Cells from two dishes were pooled and transferred into a 50-ml conical tube. The cells were washed twice with phosphate-buffered saline (PBS) and incubated for 1 h in starvation mix (DMEM complete without cysteine or methionine). Afterward, the cells were pelleted, resuspended in 1 ml of starvation mix, and transferred into a 1.5-ml Safe-Lock Eppendorf centrifugation tube, and 300 μCi of 35S was added per sample. The cells were rocked for 30 min at 37°C, pelleted, and washed once with PBS. Finally, the cells were lysed with NP-40 lysis buffer containing protease inhibitors for 45 min at 4°C. Cell debris was removed by centrifugation at 16.100 × g for 20 min. The lysates were stored at –80°C.
Immunoprecipitation of purified Fab, Fc, and IgG from metabolically labeled cells.
Cell lysates were precleared by adding protein A/G-agarose beads, incubated for 1 h at 4°C, followed by pelleting the beads by centrifugation. The supernatant was transferred to a new tube, incubated again with protein A/G-agarose beads at 4°C overnight, and then subjected to centrifugation. The precleared lysates were transferred into a new Eppendorf tube and incubated with 10 μg of either purified Fab, purified Fc, or whole IgG with the addition of protease inhibitors overnight at 4°C. Protein A/G-agarose beads were added to the mixture, and the lysates were incubated for 1 h while rocking at 4°C. The beads were pelleted, the supernatant was discarded, and the beads were washed four times with NET buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40) before resuspension in EndoH buffer. The samples were boiled for 10 min and split in equal parts, with EndoH being added to one part. All samples were incubated at 37°C overnight. 2× Laemmli sample buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue, 0.125 M Tris HCl [pH 6.8]) was added, and the samples were boiled for 5 min and frozen at –80°C.
SDS-PAGE.
We generated 10% SDS-PAGE gels using standard methods. Half of the immunoprecipitate described above was loaded onto the gel, and electrophoresis was performed for 90 min at 100 V. Gels were fixed and dried onto Whatman papers using a slab gel dryer model SGD5040 (Savant). The dried gel was exposed to autoradiography film at –80°C for at least 1 week. The film was developed using an SRX-101A film processor (Konica Minolta, Tokyo, Japan).
Viruses and construction of recombinant mutants.
The primary RhCMV isolate UCD59 was kindly provided by Peter Barry (University of California at Davis) and has been isolated from RM at the CNPRC (59). The primary RhCMV isolates 19269 and 24514, as well as the CyCMV isolate 31908, were isolated from animals at the ONPRC as described previously (40, 41). Isolates 68-1 RhCMV/gag and 68-1.2 RhCMV/gag were also previously described (45, 62). In both constructs, an expression cassette for the simian immunodeficiency virus (SIV) gag gene was inserted into the Rh211 gene. The ΔRh14-Rh29 deletion mutant was generated on the basis of 68-1 RhCMV/gag by homologous Red-mediated recombination (63) using primers with 50-bp homology flanking the desired deletion. In the ΔRh01-Rh13.1 construct, SIVgag replaced the gene Rh01, thus using the endogenous Rh01 promoter for SIVgag expression. Downstream of SIVgag, an aminoglycoside 3′-phosphotransferase (KanR) cassette flanked by FRT sides was inserted, which permits the selection of recombinant clones and subsequent excision of the selection marker using a heat shock inducible flippase (FLP) (64). The constructs were analyzed by restriction digestion with XmaI and Sanger sequencing across the introduced deletion. Recombinant viruses were reconstituted by electroporation of the BAC DNA into primary RFs. Viral cultures were expanded to generate purified viral stocks for experiments.
To generate single ORF deletions in RhCMV, we utilized the en passant method that allows for “scarless” homologous recombination (65). Recombination primers with 100-bp overhangs were designed so that the first 100 bp of the sense primer and the first 50 bp of the antisense primer at the 5′-terminal end corresponded to DNA sequences either directly upstream or downstream of the intended deletion. The 50-bp directly upstream of the intended deletion in the sense primer were repeated in the antisense primer to create a homologous sequence in the intermediate BAC construct. As a template to create the insertion cassette for homologous recombination, we used a plasmid containing the aminoglycoside 3′-phosphotransferase (Kanr) selectable marker with an upstream I-SceI unique restriction site. The primer binding sites for the recombination primers were designed to bind the 5′-end of the I-SceI restriction site and the 3′ end of the KanR selection marker. The KanR cassette was removed by arabinose induced expression of the I-SceI restriction enzyme in Escherichia coli strain GS1783 and by simultaneous induction of the Red recombination genes by heat shock, leading to the homologous recombination of the introduced repeated 50-bp sequences and the “scarless” removal of the targeted ORF. Deletion of the ORF was confirmed by restriction digestion with XmaI and by Sanger sequencing across the deletion. Recombinant viruses were reconstituted and analyzed as described above.
Analysis of RhCMVΔRh05 growth kinetics by using a multistep growth curve.
Primary rhesus fibroblast were seeded out in 24-well plates (5 × 104 cells per well) and infected with either RhCMV 68-1 or RhCMV 68-1 ΔRh05 at a multiplicity of infection (MOI) of 0.01. Supernatants from two wells per sample and time point were harvested every third day starting at day 3, and the supernatants were cleared by centrifugation at 16.100 × g for 5 min before storing them at –80°C. Viral titers of each sample were determined by 50% tissue culture infective dose (TCID50) assays on primary rhesus fibroblasts, and the growth curves were graphed using the arithmetic mean of the two biological repeats per sample.
Molecular cloning, transient transfection, and lentiviral transduction.
Rh05 and rhesus-CD4 (accession no. D63347) were synthesized as gBlock fragments flanked by NheI and BamHI restriction sites (Integrated DNA Technologies [IDT]) and cloned into the pIRES_eGFP expression vector upstream of an internal ribosomal entry site (IRES) and the gene for GFP. Transient expression of recombinant protein was achieved by transfection of HeLa cells using Superfect transfection reagent (Qiagen). BW reporter cells stably expressing chimeric Macaca mulatta FcγR-CD3ζ receptors were generated by lentiviral transduction using HEK293T cells as a packaging cell line. FcγR-CD3ζ chimeric receptors were designed by fusion of the extracellular domain of the respective rhesus FcγRs (RhCD16, accession no. XP_014968661.1; RhCD32a, accession no. XP_014968622.1; RhCD32b, accession no. XP_014968682.1; RhCD64, accession no. NP_001244233.2), with the mouse CD3 signaling module as described previously (44). The Rh-Fcγ receptors were synthesized as gBlock fragments flanked by NheI and BamHI restriction sites (IDT). gBlocks were then cloned into the puc2CL6IPwo lentiviral vector using the above-mentioned restriction sites. For every construct, one 10-cm dish of packaging cell line at roughly 70% density was transfected with the target construct and two supplementing vectors providing the VSV gag/pol and VSV-G-env proteins (6 μg of DNA each) using polyethylenimine (22.5μg/ml) and Polybrene (4 μg/ml; Merck Millipore) in a total volume of 7 ml (2 ml of a 15-min-preincubated transfection mix in serum-free DMEM added to 5 ml of fresh full DMEM). After a medium change, virus supernatant harvested from the packaging cell line 2 days after transfection was then incubated with target BW cells overnight (3.5 ml of supernatant on 106 target cells), followed by expansion and pool selection using 2 μg/ml of puromycin.
Flow cytometry.
BW cells (106) were washed in PBS, equilibrated in staining buffer (PBS, 3% FCS), and sedimented at 1,000 × g and 10°C for 3 min. The cells were resuspended in 100 μl of either primary antibody solution, followed by conjugate antibody solution or conjugate antibody solution alone (1/100 in staining buffer). Every incubation step was carried out at 4°C for 1 h and followed by three washing steps in staining buffer. Dead cells were stained using DAPI (4′,6′-diamidino-2-phenylindole). After the final wash, the cells were resuspended in 400 μl of staining buffer and analyzed on a FACSFortessa instrument (BD Bioscience). Human IgG-Fc-Texas Red (Rockland) and anti-human-IgG-FITC (Miltenyi Biotec) were used as conjugates. PE conjugation was performed using an ab102918 labeling kit (Abcam), as recommended by the supplier.
Fcγ receptor activation assay.
The assay was performed as described earlier (44). Briefly, in a standard assay, target cells were incubated with dilutions of Macaca mulatta sera (RhCMV-infected TRFs) or MAbs (transfected HeLa cells) in DMEM supplemented with 10% (vol/vol) FCS for 30 min at 37°C. The cells were washed before cocultivation with BW reporter cells (effector/target ratio, 20:1) for 16 h at 37°C in a 5% CO2 atmosphere. Cross-link activation of reporter cells was performed by direct coating of target antibody to an enzyme-linked immunosorbent assay (ELISA) plate (Nunc Maxisorp; 96 well, flat transparent), followed by a blocking step and incubation with 2 × 105 reporter cells per well. For all activation assays, mouse IL-2 secretion was quantified by anti-IL-2 ELISA, as described earlier (44). RhCMV-seropositive rhesus macaque serum was provided by the German Primate Center Göttingen from preexisting samples.
Statistical analysis.
Statistical analysis was performed using a two-way analysis of variance (ANOVA), together with Tukey’s range testing. Analyses were performed using the Prism 6 software (GraphPad).
Rhesus macaques.
Adult Macaca mulatta were used at the ONPRC, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. The experiments were conducted in compliance with the Animal Welfare Act in accordance with the Guide for the Care and Use of Laboratory Animals, Institute of Laboratory Animals Resources, National Research Council, and approved by the Institutional Animal Care and Use Committees (IACUC) that adhere to national guidelines established in the Animal Welfare Act (7 U.S.C. Sections 2131 to2159) and the Guide for the Care and Use of Laboratory Animals (8th edition), as mandated by U.S. Public Health Service Policy.
Three purpose-bred, pedigreed, male RMs were used. At assignment, these RMs were positive for RhCMV but free of macacine herpesvirus 1, d-type simian retrovirus, simian T-lymphotrophic virus type 1, simian immunodeficiency virus, and TB. The three RMs were sedated with ketamine HCl or Telazol for the subcutaneous administration of 5 × 106 PFU of either 68-1 RhCMV/gag, RhCMVΔRh01-13.1/gag, or 68-1.2 RhCMVgag, respectively, on day 0.
T cell assays.
SIVgag-specific CD4+ and CD8+ T cell responses were measured bi-weekly in peripheral blood mononuclear cells (PBMCs) by ICS (45, 46, 62, 66). Briefly, PBMCs were incubated with consecutive 15mer peptide mixes (11-AA overlap) comprising SIVgag and the costimulatory molecules CD28 and CD49d (BD Biosciences) for 1 h, followed by the addition of brefeldin A (Sigma-Aldrich) for an additional 8 h. Costimulation without peptides served as a background control. Alternatively, the MHC-E-restricted SIVgag supertope peptides (Gag69276-284 RMYNPTNIL and Gag120482-490 EKQRESREK) or MHC-II-restricted supertope peptides (Gag53211-222 AADWDLQHPQP and Gag73290-301 PKEPFQSYVDRF) were used in this assay.
Stimulated cells were fixed, permeabilized, and stained (45, 46, 62, 66) using combinations of the following fluorochrome-conjugated MAbs: SP34-2 (CD3; Pacific Blue, Alexa700), L200 (CD4; AmCyan, BV510), SK-1 (CD8α; PerCP-Cy5.5), MAB11 (tumor necrosis factor alpha [TNF-α]; FITC, PE), B27 (gamma interferon [IFN-γ]; allophycocyanin [APC]), FN50 (CD69; PE, PE-Texas Red), B56 (Ki-67; FITC), and (in polycytokine analyses) JES6-5H4 (IL-2; PE, PE Cy-7). Data were collected on an LSR-II (BD Biosciences). Analysis was performed using FlowJo software (Tree Star). Lymphocytes were gated for CD3+ and progressive gating on CD4+ and CD8+ T cell subsets. Antigen-responding cells in both CD4+ and CD8+ T cell populations were determined by their intracellular expression of CD69 and one or more cytokines. After subtracting the background, the raw response frequencies were memory corrected (45, 46, 62, 66) using combinations of the following MAbs to define the memory versus naive subsets: SP34-2 (CD3; Alexa700, PerCP-Cy5.5), L200 (CD4; AmCyan), SK-1 (CD8α; APC, PerCP-Cy-5.5), MAb11 (TNF-α; FITC), B27 (IFN-γ; APC), FN50 (CD69; PE), CD28.2 (CD28; PE-Texas Red), DX2 (CD95; PE), 15053 (CCR7; Pacific Blue), and B56 (Ki-67; FITC).
ACKNOWLEDGMENTS
We are grateful to Mike Axthelm, ONPRC, for providing CMV-negative monkey serum and to Andrew Sylwester for help with shipping. We thank Peter Barry for providing UCD59 and Dominique Gütle for support performing the reporter assays.
This study was supported/funded by National Institutes of Health grants RO1AI059457 (to K.F.) and U19AI128741 (to L.J.P.), by Infect-ERA grant BMBF 031L0090 (to H.H.), and by Kompetenznetzwerk Zytomegalie Baden Württemberg (to H.H.). This project was also supported by the National Center for Research Resources and the Office of Research Infrastructure Programs of the NIH through grant P51OD011092. S.S. was supported by a postdoctoral research scholarship from the Belgian American Education Foundation.
REFERENCES
- 1.Powers C, DeFilippis V, Malouli D, Früh K. 2008. Cytomegalovirus immune evasion. Curr Top Microbiol Immunol 325:333–359. [DOI] [PubMed] [Google Scholar]
- 2.Hengel H, Koszinowski UH, Conzelmann KK. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol 26:396–401. doi: 10.1016/j.it.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Halenius A, Gerke C, Hengel H. 2015. Classical and non-classical MHC I molecule manipulation by human cytomegalovirus: so many targets-but how many arrows in the quiver? Cell Mol Immunol 12:139–153. doi: 10.1038/cmi.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson GWG, Tomasec P, Stanton RJ, Armstrong M, Prod’homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, Wang ECY, Griffin CA, Davison AJ. 2008. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS. 2012. Decoding human cytomegalovirus. Science 338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, Axthelm MK, Nelson JA, Jarvis MA, Picker LJ, Früh K. 2010. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328:102–106. doi: 10.1126/science.1185350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross SA, Arora N, Novak Z, Fowler KB, Britt WJ, Boppana SB. 2010. Cytomegalovirus reinfections in healthy seroimmune women. J Infect Dis 201:386–389. doi: 10.1086/649903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida JH, Patel A, Mehta AK, Gatault P, McBride JM, Burgess T, Derby MA, Snydman DR, Emu B, Feierbach B, Fouts AE, Maia M, Deng R, Rosenberger CM, Gennaro LA, Striano NS, Liao XC, Tavel JA. 2017. Phase 2 randomized, double-blind, placebo-controlled trial of RG7667, a combination monoclonal antibody, for prevention of cytomegalovirus infection in high-risk kidney transplant recipients. Antimicrob Agents Chemother 61:e01794-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revello MG, Lazzarotto T, Guerra B, Spinillo A, Ferrazzi E, Kustermann A, Guaschino S, Vergani P, Todros T, Frusca T, Arossa A, Furione M, Rognoni V, Rizzo N, Gabrielli L, Klersy C, Gerna G. 2014. A randomized trial of hyperimmune globulin to prevent congenital cytomegalovirus. N Engl J Med 370:1316–1326. doi: 10.1056/NEJMoa1310214. [DOI] [PubMed] [Google Scholar]
- 10.Nigro G, Adler SP, La Torre R, Best AM. 2005. Passive immunization during pregnancy for congenital cytomegalovirus infection. N Engl J Med 353:1350–1362. doi: 10.1056/NEJMoa043337. [DOI] [PubMed] [Google Scholar]
- 11.Boeckh M, Bowden RA, Storer B, Chao NJ, Spielberger R, Tierney DK, Gallez-Hawkins G, Cunningham T, Blume KG, Levitt D, Zaia JA. 2001. Randomized, placebo-controlled, double-blind study of a cytomegalovirus-specific monoclonal antibody (MSL-109) for prevention of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 7:343–351. doi: 10.1016/S1083-8791(01)80005-7. [DOI] [PubMed] [Google Scholar]
- 12.Hodson EM, Jones CA, Strippoli GF, Webster AC, Craig JC. 2007. Immunoglobulins, vaccines or interferon for preventing cytomegalovirus disease in solid organ transplant recipients. Cochrane Database Syst Rev. [DOI] [PubMed] [Google Scholar]
- 13.Raanani P, Gafter-Gvili A, Paul M, Ben-Bassat I, Leibovici L, Shpilberg O. 2009. Immunoglobulin prophylaxis in hematopoietic stem cell transplantation: systematic review and meta-analysis. J Clin Oncol 27:770–781. doi: 10.1200/JCO.2008.16.8450. [DOI] [PubMed] [Google Scholar]
- 14.Nelson CS, Cruz DV, Tran D, Bialas KM, Stamper L, Wu H, Gilbert M, Blair R, Alvarez X, Itell H, Chen M, Deshpande A, Chiuppesi F, Wussow F, Diamond DJ, Vandergrift N, Walter MR, Barry PA, Cohen-Wolkowiez M, Koelle K, Kaur A, Permar SR. 2017. Preexisting antibodies can protect against congenital cytomegalovirus infection in monkeys. JCI Insight 2:e94002. doi: 10.1172/jci.insight.94002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M, Hengel H. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J Virol 76:8596–8608. doi: 10.1128/JVI.76.17.8596-8608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corrales-Aguilar E, Trilling M, Hunold K, Fiedler M, Le VT, Reinhard H, Ehrhardt K, Merce-Maldonado E, Aliyev E, Zimmermann A, Johnson DC, Hengel H. 2014. Human cytomegalovirus Fcγ binding proteins gp34 and gp68 antagonize Fcγ receptors I, II and III. PLoS Pathog 10:e1004131. doi: 10.1371/journal.ppat.1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thale R, Lucin P, Schneider K, Eggers M, Koszinowski UH. 1994. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J Virol 68:7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crnkovic-Mertens I, Messerle M, Milotic I, Szepan U, Kucic N, Krmpotic A, Jonjic S, Koszinowski UH. 1998. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J Virol 72:1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arapović J, Lenac Rovis T, Reddy AB, Krmpotić A, Jonjić S. 2009. Promiscuity of MCMV immunoevasin of NKG2D: m138/fcr-1 downmodulates RAE-1epsilon in addition to MULT-1 and H60. Mol Immunol 47:114–122. doi: 10.1016/j.molimm.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Lenac T, Budt M, Arapovic J, Hasan M, Zimmermann A, Simic H, Krmpotic A, Messerle M, Ruzsics Z, Koszinowski UH, Hengel H, Jonjic S. 2006. The herpesviral Fc receptor fcr-1 downregulates the NKG2D ligands MULT-1 and H60. J Exp Med 203:1843–1850. doi: 10.1084/jem.20060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mintern JD, Klemm EJ, Wagner M, Paquet ME, Napier MD, Kim YM, Koszinowski UH, Ploegh HL. 2006. Viral interference with B7-1 costimulation: a new role for murine cytomegalovirus fc receptor-1. J Immunol 177:8422–8431. doi: 10.4049/jimmunol.177.12.8422. [DOI] [PubMed] [Google Scholar]
- 22.Johnson DC, Feenstra V. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol 61:2208–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson JK, Grose C. 1998. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor. J Virol 72:1542–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ndjamen B, Farley AH, Lee T, Fraser SE, Bjorkman PJ. 2014. The herpes virus Fc receptor gE-gI mediates antibody bipolar bridging to clear viral antigens from the cell surface. PLoS Pathog 10:e1003961. doi: 10.1371/journal.ppat.1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagashunmugam T, Lubinski J, Wang L, Goldstein LT, Weeks BS, Sundaresan P, Kang EH, Dubin G, Friedman HM. 1998. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol 72:5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham AL. 2016. The herpes zoster subunit vaccine. Expert Opin Biol Ther 16:265–271. doi: 10.1517/14712598.2016.1134481. [DOI] [PubMed] [Google Scholar]
- 27.Nimmerjahn F, Ravetch JV. 2007. Fc-receptors as regulators of immunity. Adv Immunol 96:179–204. doi: 10.1016/S0065-2776(07)96005-8. [DOI] [PubMed] [Google Scholar]
- 28.Rogers KA, Scinicariello F, Attanasio R. 2006. IgG Fc receptor III homologues in nonhuman primate species: genetic characterization and ligand interactions. J Immunol 177:3848–3856. doi: 10.4049/jimmunol.177.6.3848. [DOI] [PubMed] [Google Scholar]
- 29.Chan YN, Boesch AW, Osei-Owusu NY, Emileh A, Crowley AR, Cocklin SL, Finstad SL, Linde CH, Howell RA, Zentner I, Cocklin S, Miles AR, Eckman JW, Alter G, Schmitz JE, Ackerman ME. 2016. IgG binding characteristics of rhesus macaque FcγR. J Immunol 197:2936–2947. doi: 10.4049/jimmunol.1502252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daeron M. 2009. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 31.Van den Hoecke S, Ehrhardt K, Kolpe A, El Bakkouri K, Deng L, Grootaert H, Schoonooghe S, Smet A, Bentahir M, Roose K, Schotsaert M, Schepens B, Callewaert N, Nimmerjahn F, Staeheli P, Hengel H, Saelens X. 2017. Hierarchical and redundant roles of activating FcγRs in protection against influenza disease by M2e-specific IgG1 and IgG2a antibodies. J Virol 91:e02500-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz JA, Bar-On Y, Lu CL, Fera D, Lockhart AAK, Lorenzi JCC, Nogueira L, Golijanin J, Scheid JF, Seaman MS, Gazumyan A, Zolla-Pazner S, Nussenzweig MC. 2017. Non-neutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 170:637–648. doi: 10.1016/j.cell.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, Hoffner M, Streeck H, Ackerman ME, McElrath MJ, Schuitemaker H, Pau MG, Baden LR, Kim JH, Michael NL, Barouch DH, Lauffenburger DA, Alter G. 2015. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell 163:988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol 77:6620–6636. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malouli D, Nakayasu ES, Viswanathan K, Camp DG 2nd, Chang WL, Barry PA, Smith RD, Früh K. 2012. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J Virol 86:8959–8973. doi: 10.1128/JVI.01132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers C, Früh K. 2008. Rhesus CMV: an emerging animal model for human CMV. Med Microbiol Immunol 197:109–115. doi: 10.1007/s00430-007-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bialas KM, Tanaka T, Tran D, Varner V, Cisneros De La Rosa E, Chiuppesi F, Wussow F, Kattenhorn L, Macri S, Kunz EL, Estroff JA, Kirchherr J, Yue Y, Fan Q, Lauck M, O’Connor DH, Hall AHS, Xavier A, Diamond DJ, Barry PA, Kaur A, Permar SR. 2015. Maternal CD4+ T cells protect against severe congenital cytomegalovirus disease in a novel nonhuman primate model of placental cytomegalovirus transmission. Proc Natl Acad Sci U S A 112:13645–13650. doi: 10.1073/pnas.1511526112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturgill ER, Malouli D, Hansen SG, Burwitz BJ, Seo S, Schneider CL, Womack JL, Verweij MC, Ventura AB, Bhusari A, Jeffries KM, Legasse AW, Axthelm MK, Hudson AW, Sacha JB, Picker LJ, Früh K. 2016. Natural killer cell evasion is essential for infection by rhesus cytomegalovirus. PLoS Pathog 12:e1005868. doi: 10.1371/journal.ppat.1005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yue Y, Kaur A, Lilja A, Diamond DJ, Walter MR, Barry PA. 2016. The susceptibility of primary cultured rhesus macaque kidney epithelial cells to rhesus cytomegalovirus strains. J Gen Virol 97:1426–1438. doi: 10.1099/jgv.0.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burwitz BJ, Malouli D, Bimber BN, Reed JS, Ventura AB, Hancock MH, Uebelhoer LS, Bhusari A, Hammond KB, Espinosa Trethewy RG, Klug A, Legasse AW, Axthelm MK, Nelson JA, Park BS, Streblow DN, Hansen SG, Picker LJ, Früh K, Sacha JB. 2016. Cross-species rhesus cytomegalovirus infection of cynomolgus macaques. PLoS Pathog 12:e1006014. doi: 10.1371/journal.ppat.1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lilja AE, Chang WL, Barry PA, Becerra SP, Shenk TE. 2008. Functional genetic analysis of rhesus cytomegalovirus: Rh01 is an epithelial cell tropism factor. J Virol 82:2170–2181. doi: 10.1128/JVI.02316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. 2003. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A 100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corrales-Aguilar E, Trilling M, Reinhard H, Merce-Maldonado E, Widera M, Schaal H, Zimmermann A, Mandelboim O, Hengel H. 2013. A novel assay for detecting virus-specific antibodies triggering activation of Fcγ receptors. J Immunol Methods 387:21–35. doi: 10.1016/j.jim.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, Malouli D, Xu G, Richards R, Whizin N, Reed JS, Hammond KB, Fischer M, Turner JM, Legasse AW, Axthelm MK, Edlefsen PT, Nelson JA, Lifson JD, Früh K, Picker LJ. 2013. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340:1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, Lopez CA, Früh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ. 2016. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351:714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortese M, Calo S, D’Aurizio R, Lilja A, Pacchiani N, Merola M. 2012. Recombinant human cytomegalovirus (HCMV) RL13 binds human immunoglobulin G Fc. PLoS One 7:e50166. doi: 10.1371/journal.pone.0050166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davison AJ, Akter P, Cunningham C, Dolan A, Addison C, Dargan DJ, Hassan-Walker AF, Emery VC, Griffiths PD, Wilkinson GW. 2003. Homology between the human cytomegalovirus RL11 gene family and human adenovirus E3 genes. J Gen Virol 84:657–663. doi: 10.1099/vir.0.18856-0. [DOI] [PubMed] [Google Scholar]
- 49.Sekulin K, Görzer I, Heiss-Czedik D, Puchhammer-Stöckl E. 2007. Analysis of the variability of CMV strains in the RL11D domain of the RL11 multigene family. Virus Genes 35:577–583. doi: 10.1007/s11262-007-0158-0. [DOI] [PubMed] [Google Scholar]
- 50.Perez-Carmona N, Martinez-Vicente P, Farre D, Gabaev I, Messerle M, Engel P, Angulo A. 2018. A prominent role of the human cytomegalovirus UL8 glycoprotein restraining proinflammatory cytokine production by myeloid cells at late times during infection. J Virol 92:e02229–e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruno L, Cortese M, Monda G, Gentile M, Calo S, Schiavetti F, Zedda L, Cattaneo E, Piccioli D, Schaefer M, Notomista E, Maione D, Carfi A, Merola M, Uematsu Y. 2016. Human cytomegalovirus pUL10 interacts with leukocytes and impairs TCR-mediated T-cell activation. Immunol Cell Biol 94:849–860. doi: 10.1038/icb.2016.49. [DOI] [PubMed] [Google Scholar]
- 52.Gabaev I, Steinbruck L, Pokoyski C, Pich A, Stanton RJ, Schwinzer R, Schulz TF, Jacobs R, Messerle M, Kay-Fedorov PC. 2011. The human cytomegalovirus UL11 protein interacts with the receptor tyrosine phosphatase CD45, resulting in functional paralysis of T cells. PLoS Pathog 7:e1002432. doi: 10.1371/journal.ppat.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engel P, Perez-Carmona N, Alba MM, Robertson K, Ghazal P, Angulo A. 2011. Human cytomegalovirus UL7, a homologue of the SLAM-family receptor CD229, impairs cytokine production. Immunol Cell Biol 89:753–766. doi: 10.1038/icb.2011.55. [DOI] [PubMed] [Google Scholar]
- 54.Gabaev I, Elbasani E, Ameres S, Steinbruck L, Stanton R, Doring M, Lenac Rovis T, Kalinke U, Jonjic S, Moosmann A, Messerle M. 2014. Expression of the human cytomegalovirus UL11 glycoprotein in viral infection and evaluation of its effect on virus-specific CD8 T cells. J Virol 88:14326–14339. doi: 10.1128/JVI.01691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crawford LB, Kim JH, Collins-McMillen D, Lee BJ, Landais I, Held C, Nelson JA, Yurochko AD, Caposio P. 2018. Human cytomegalovirus encodes a novel FLT3 receptor ligand necessary for hematopoietic cell differentiation and viral reactivation. mBio 9:e00682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacManiman JD, Meuser A, Botto S, Smith PP, Liu F, Jarvis MA, Nelson JA, Caposio P. 2014. Human cytomegalovirus-encoded pUL7 is a novel CEACAM1-like molecule responsible for promotion of angiogenesis. mBio 5:e02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanton RJ, Baluchova K, Dargan DJ, Cunningham C, Sheehy O, Seirafian S, McSharry BP, Neale ML, Davies JA, Tomasec P, Davison AJ, Wilkinson GW. 2010. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Invest 120:3191–3208. doi: 10.1172/JCI42955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corrales-Aguilar E, Trilling M, Reinhard H, Falcone V, Zimmermann A, Adams O, Santibanez S, Hengel H. 2016. Highly individual patterns of virus-immune IgG effector responses in humans. Med Microbiol Immunol 205:409–424. doi: 10.1007/s00430-016-0457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oxford KL, Strelow L, Yue Y, Chang WL, Schmidt KA, Diamond DJ, Barry PA. 2011. Open reading frames carried on UL/b′ are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J Virol 85:5105–5114. doi: 10.1128/JVI.02631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forthal D, Hope TJ, Alter G. 2013. New paradigms for functional HIV-specific nonneutralizing antibodies. Curr Opin HIV AIDS 8:393–401. doi: 10.1097/COH.0b013e328363d486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang WL, Kirchoff V, Pari GS, Barry PA. 2002. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J Virol Methods 104:135–146. doi: 10.1016/S0166-0934(02)00060-5. [DOI] [PubMed] [Google Scholar]
- 62.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muyrers JP, Zhang Y, Benes V, Testa G, Rientjes JM, Stewart AF. 2004. ET recombination: DNA engineering using homologous recombination in Escherichia coli. Methods Mol Biol 256:107–121. [DOI] [PubMed] [Google Scholar]
- 64.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 65.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 66.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]