RNA viruses mutate rapidly and frequently expand their host ranges to infect novel hosts, leading to serial host shifts. Using an RNA bacteriophage model system (Pseudomonas phage φ6), we studied the impact of preexisting host range mutations on another host range expansion. Results from both clonal Sanger and Illumina sequencing show that extant host range mutations dramatically narrow the neighborhood of potential host range mutations compared to that of wild-type φ6. This research suggests that serial host-shifting viruses may follow a small number of molecular paths to enter additional novel hosts. We also identified new genes involved in φ6 host range expansion, expanding our knowledge of this important model system in experimental evolution.
KEYWORDS: entropy, epistasis, host range mutations, RNA virus
ABSTRACT
RNA viruses are capable of rapid host shifting, typically due to a point mutation that confers expanded host range. As additional point mutations are necessary for further expansions, epistasis among host range mutations can potentially affect the mutational neighborhood and frequency of niche expansion. We mapped the mutational neighborhood of host range expansion using three genotypes of the double-stranded RNA (dsRNA) bacteriophage φ6 (wild type and two isogenic host range mutants) on the novel host Pseudomonas syringae pv. atrofaciens. Both Sanger sequencing of 50 P. syringae pv. atrofaciens mutant clones for each genotype and population Illumina sequencing revealed the same high-frequency mutations allowing infection of P. syringae pv. atrofaciens. Wild-type φ6 had at least nine different ways of mutating to enter the novel host, eight of which are in p3 (host attachment protein gene), and 13/50 clones had unchanged p3 genes. However, the two isogenic mutants had dramatically restricted neighborhoods: only one or two mutations, all in p3. Deep sequencing revealed that wild-type clones without mutations in p3 likely had changes in p12 (morphogenic protein), a region that was not polymorphic for the two isogenic host range mutants. Sanger sequencing confirmed that 10/13 of the wild-type φ6 clones had nonsynonymous mutations in p12, and 2 others had point mutations in p9 and p5. None of these genes had previously been associated with host range expansion in φ6. We demonstrate, for the first time, epistatic constraint in an RNA virus due to host range mutations themselves, which has implications for models of serial host range expansion.
IMPORTANCE RNA viruses mutate rapidly and frequently expand their host ranges to infect novel hosts, leading to serial host shifts. Using an RNA bacteriophage model system (Pseudomonas phage φ6), we studied the impact of preexisting host range mutations on another host range expansion. Results from both clonal Sanger and Illumina sequencing show that extant host range mutations dramatically narrow the neighborhood of potential host range mutations compared to that of wild-type φ6. This research suggests that serial host-shifting viruses may follow a small number of molecular paths to enter additional novel hosts. We also identified new genes involved in φ6 host range expansion, expanding our knowledge of this important model system in experimental evolution.
INTRODUCTION
Emerging and reemerging viruses that shift host to infect new species pose significant economic and health costs to humans, animals, plants, and our ecosystems (1–3). While ecological exposure is an essential part of emergence on a novel host (2), spillover infection of the novel host typically requires a host range mutation, the genetic component of host range expansion (4). These exaptive host range mutations must exist in the viral population prior to contact with the novel host as part of the virus’s standing genetic diversity (5, 6). The exact mutations and mechanisms of host shifting have been intensively studied in emerging zoonotic viruses such as influenza virus, severe acute respiratory syndrome coronavirus (SARS-CoV), and Ebola virus (7, 8).
Given the high mutation rates (9), potentially large population sizes, and fast replication of many emergent RNA viruses (10), these viruses are capable of generating and maintaining substantial genetic variation (11, 12). This variation fuels adaptation, and selective sweeps leave genetic marks of past ecological history in viral genomes. These fixed mutations can alter the fitness landscape and constrain evolutionary trajectories of viruses due to epistatic interactions between mutations (13). Virus evolution is known to be shaped by epistasis, detected by both laboratory experimentation and phylogenetic analysis (14–16), and increased understanding of epistasis promises to improve our predictions of why some viral emergence events are more successful than others (17).
Some emergent viruses experience several hosts, often due to serial emergence events (18). Middle East respiratory syndrome coronavirus (MERS-CoV) is proposed to have jumped hosts from its natural reservoir (bats) into camels, later spilling over to the human population (19). Similarly, canine parvovirus jumped from infecting cats to raccoons and then jumped again to infect dogs (20). Influenza virus strains have also serially shifted hosts (e.g., H3N8 originated from avian hosts infecting horses and then shifted to dogs [21]). This kind of serial emergence allows for the possibility of host range mutations themselves to play a significant role in shaping the landscape of further emergence, one of the legacies of previous host use (7, 22). We used the model RNA virus Pseudomonas double-stranded RNA (dsRNA) bacteriophage φ6 to investigate the role of extant host range mutations on further host range expansion.
Phage φ6 has been a popular model for understanding host range mutations and their fitness effects (5, 23, 24). However, all previous studies have exclusively looked at a wild-type (WT) genotype, replicating in its reservoir host, instead of investigating the interactions of multiple host range mutations during frequent host shifting or serial emergence. In this study, we mapped the host range mutational neighborhoods of wild-type φ6 and two isogenic host range mutants (E8G in P3; G515S in P3) emerging in the novel host Pseudomonas syringae pv. atrofaciens. Significant epistatic constraint was observed with both host range mutants in clonal and Illumina sequencing; only one or two mutations were found that allowed infection of P. syringae pv. atrofaciens. These mutations were a subset of the large mutational neighborhood of P. syringae pv. atrofaciens host range mutations available to wild-type φ6. Additionally, we have identified host range-associated genes at sites other than the canonical site of host range mutations, i.e., three genes on the small segment, not previously implicated in host shifting. Our work supports using deep sequencing to map mutational neighborhoods in future studies though both deep sequencing and the more labor-intensive characterization of clones complemented each other. This work provides a panoramic view of host range mutational neighborhoods in φ6 while demonstrating a significant constraint imposed by host range mutation in a fast-evolving RNA virus.
(This article was submitted to an online preprint archive [25].)
RESULTS
Mapping P3 P. syringae pv. atrofaciens mutational neighborhood.
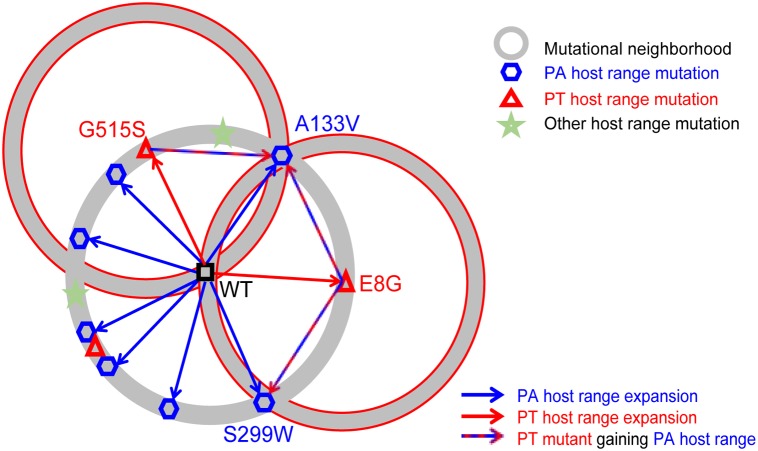
We found that the P. syringae pv. atrofaciens host range mutational neighborhood is highly genotype dependent. Fifty host range mutant plaques were isolated for each of the three genotypes (φ6-WT, φ6-E8G, and φ6-G515S) (Table 1), and their p3 genes for the φ6 attachment protein were Sanger sequenced. We sequenced only the p3 gene because P3 is the only highly accessible protein on the exterior of the virion (26) and the only protein associated with φ6 host range in all previous studies (5, 23, 24). Thirty-five out of the 50 sequenced φ6-WT p3 sequences had single nonsynonymous mutations (seven unique mutations identified), 2 had double mutations, and 13 had no detectable mutations on the p3 gene. Forty-eight of the 50 φ6-E8G host range mutants contained one of the two single mutations present in the φ6-WT clones (A133V and S299W); the remaining two clones had double mutations consisting of A133V and an additional nonsynonymous mutation. All 50 φ6-G515S host range mutants had the A133V mutation, with 43 having a single mutation, 5 having a double mutation, and 2 having a triple mutation. The nonsynonymous mutation A133V was the most frequent in the three tested populations, with 24% in φ6-WT isolates, 96% in φ6-E8G isolates, and 100% in φ6-G515S isolates, making this the most prevalent mutation conferring infection of P. syringae pv. atrofaciens. In addition, there was a noticeable drop in diversity of single host range mutations from the φ6-WT population compared to that of the φ6-E8G and φ6-G515S populations, consistent with epistatic constraint on mutational neighborhood by host range mutations. We have summarized these P3 host range mutational neighborhoods on P. syringae pv. atrofaciens in a two-dimensional schematic (Fig. 1).
TABLE 1.
P. syringae pv. atrofaciens host range mutations detected on P3 with Sanger sequencing
| Mutation type and amino acid position(s) |
WT | E8G | G515S |
|---|---|---|---|
| None | 13 | ||
| Single | |||
| D35A | 3 | ||
| A133V | 12 | 46 | 43 |
| Q140R | 1 | ||
| K144R | 11 | ||
| N146K | 1 | ||
| N146S | 4 | ||
| S299W | 3 | 2 | |
| Double | |||
| A133V K144R | 1 | ||
| A133V A324A | 1 | ||
| A133V E366E | 1 | ||
| A133V Q436Q | 1 | ||
| A133V L461L | 1 | ||
| A133V S515G | 1 | ||
| A133V V606V | 1 | ||
| S299W S628A | 1 | ||
| V326F L147L | 1 | ||
| Triple | |||
| A133V S515G V531A | 1 | ||
| A133V S515G T427T | 1 |
FIG 1.
Two-dimensional schematic representing mutational neighborhoods of φ6 P3. Circles represent P3 mutational neighborhoods of the mutants, which are the centers of circles. The geometric shapes are known P3 mutants. It is assumed that they are distributed on the mutational neighborhood (circles) in a nonrandom way. The arrows are events, such as host range expansion. PA, P. syringae pv. atrofaciens; PT, P. syringae pv. tomato.
Several published works have investigated the mutational neighborhoods of φ6 p3 during expansion of host range (Table 2). Two sites were favored by host range expansion events onto P. pseudoalcaligenes and P. syringae pv. glyclinea: amino acid positions 8 and 554 of attachment protein P3 (66/81 and 18/39 of isolated mutants, respectively [5, 24]). However, the P. syringae pv. atrofaciens mutational neighborhood does not include these frequent sites of mutation to other hosts. There is some overlap of results of previous studies, for instance, N146S (5) and A133V (23), but this suggests that φ6 may interact differently with host P. syringae pv. atrofaciens during attachment than with other Pseudomonas species or P. syringae pathovars. The absence of host range mutations in p3 for 13 of the φ6-WT P. syringae pv. atrofaciens isolates was unexpected since 97% of previously independently isolated host range mutants had nonsynonymous mutations in p3, with no other sites in the φ6 genome identified as causing the expanded host range in the remaining 4/118 (5, 23, 24). This motivated a more in-depth approach: deep sequencing to map the entire mutational neighborhood.
TABLE 2.
Host range mutational neighborhoods inferred from nonsynonymous mutations in P3 from previous publicationsa
| P3 amino acid mutation(s) | Frequency of mutation(s) (no.)b |
||
|---|---|---|---|
| P. syringae pv. atrofaciens (n = 50) | P. syringae pv. glycinea (n = 40) | P. pseudoalcaligenes ERA (n = 69) | |
| G5S | 2 | ||
| E8(K/G/D/A) | 6 | 52 | |
| D35A | 3 | ||
| Q130R | 1 | ||
| A133V | 12 | ||
| Q140R | 1 | ||
| K144R | 11 | ||
| D145G | 3 | ||
| N146(K/S) | 5 | 6 | |
| E178D | 2 | ||
| S299W | 3 | ||
| V326F | 1 | ||
| P339H | 1 | ||
| T516A | 4 | ||
| D533A | 1 | ||
| D535N | 1 | ||
| D554(G/A/V/N) | 11 | 3 | |
| L555F | 1 | ||
| Double/triple | 1 | 1 | 10 |
| None | 13 | 1 | 3 |
P. syringae pv. glycinea data are from Ferris et al. (5), and data for P. pseudoalcaligenes East River isolate A (ERA) were from Ford et al. (24). P. syringae pv. atrofaciens and P. syringae pv. glycinea are closely related to the original host P. syringae pv. phaseolicola while P. pseudoalcaligenes East River isolate A is distantly related to the original host.
n, total number of wild-type isolates studied.
Deep sequencing of φ6 populations.
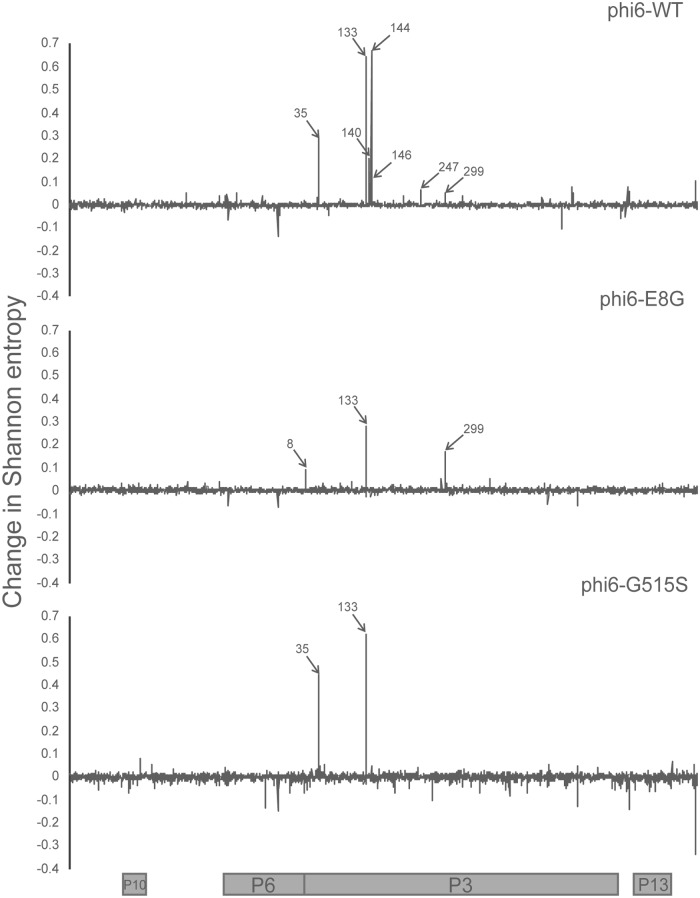
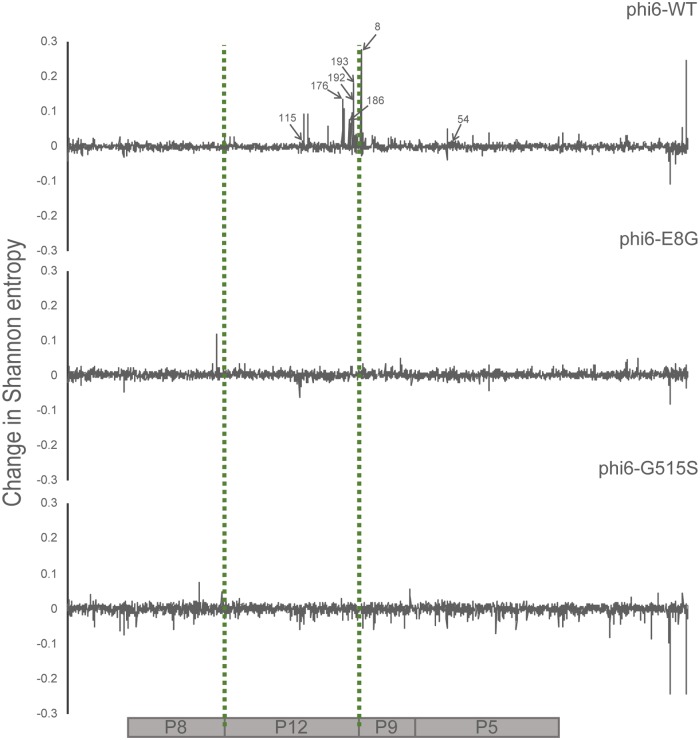
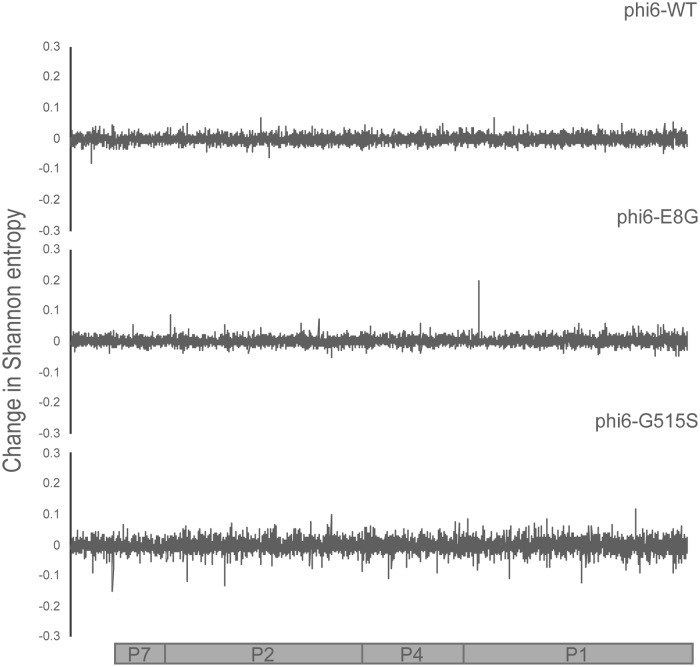
Each φ6 population was raised to high titer on its most recent host (φ6-WT on P. syringae pv. phaseolicola and both φ6-E8G and φ6-G515S on P. syringae pv. tomato) and plated on P. syringae pv. atrofaciens to obtain a lysate made of ∼400 host range mutant plaques. All population lysates from before and after P. syringae pv. atrofaciens host range expansion were sequenced. The change in Shannon entropy was calculated to determine the sites that became more or less variable after overnight growth on P. syringae pv. atrofaciens. The signals of increased variation in the p3 gene matched Sanger sequencing results; all single mutations identified in the three genotypes underwent noticeable entropy changes after gaining the P. syringae pv. atrofaciens host range (Fig. 2). We also found several sites in P3 that may have evaded detection by clonal sampling, including amino acid 247 of φ6-WT and amino acid 35 of φ6-G515S. Deep sequencing also revealed sites of high entropy change in other genes in φ6-WT that could be additional genes controlling host range and suggested targets for sequencing in the φ6-WT clones that did not contain p3 mutations (Fig. 3 and 4). Our results suggested that nonstructural protein genes p12 (encoding the morphogenic protein) and p9 (encoding the major membrane protein) were the most probable sites of additional host range mutations; both genes are involved with viral nucleocapsid vesiculation of the host inner membrane (27, 28). Results from deep sequencing of the p3 gene and of the entire genome further confirmed the constrained neighborhood of host range mutants φ6-E8G and φ6-G515S, revealing fewer possibilities for P. syringae pv. atrofaciens mutations in p3 and none elsewhere in the genome.
FIG 2.
Change in Shannon entropy in the medium segment of φ6-WT, φ6-E8G, and φ6-G515S. Positions labeled correspond to amino acid positions in P3. Coding regions of the medium segment are aligned to the graphs. The x axis corresponds to the nucleotide positions on the medium segment.
FIG 3.
Change in Shannon entropy in the small segment of φ6-WT, φ6-E8G, and φ6-G515S. Coding regions of the small segment are aligned to the graphs. Positions labeled correspond to amino acid positions in aligned genes. The x axis corresponds to the nucleotide positions on the small segment.
FIG 4.
Change in Shannon entropy in large segment of φ6-WT, φ6-E8G, and φ6-G515S. Coding regions of the large segment are aligned to the graphs. The x axis corresponds to the nucleotide positions on the large segment.
Non-p3 φ6-WT mutant sequencing.
We amplified and Sanger sequenced the small segment of all φ6-WT isolates that did not show mutation in p3 (Table 3). Ten of the 13 isolates contained a single nonsynonymous mutation in p12. One contained a single nonsynonymous mutation in p9; another contained a nonsynonymous mutation in p5. The final mutant had a single synonymous mutation in p9. This clone was then fully Sanger sequenced, but no nonsynonymous mutations were identified. These results matched many of the sites with the highest change in Shannon entropy we observed on the small segment of deep-sequenced φ6-WT populations and strongly suggest that mutations in these nonstructural genes can affect φ6 host range.
TABLE 3.
P. syringae pv. atrofaciens host range mutations of φ6-WT detected on the small segment with Sanger sequencing
| Gene and mutation(s) | No. of isolates (n = 13) |
|---|---|
| p12 | |
| K115T | 1 |
| F176L | 3 |
| V186L | 1 |
| K192R | 1 |
| D193G | 3 |
| D193A | 1 |
| p9 | |
| P4P | 1 |
| Q8R | 1 |
| p5 | |
| K54R | 1 |
Estimation of P. syringae pv. atrofaciens mutational neighborhood of φ6-WT.
While 49/50 φ6-WT isolates contained a nonsynonymous mutation that can be correlated with P. syringae pv. atrofaciens expansion, we have not likely exhausted the complete neighborhood of P. syringae pv. atrofaciens mutations available to this genotype. Sanger sequencing detected 16 of these mutations, but the large number of mutations observed only once suggests that there are other P. syringae pv. atrofaciens host range mutations that either occur at low mutation frequency or are of low fitness. Using a jackknife estimator developed for the species number problem (29) and the frequency of the P. syringae pv. atrofaciens mutations identified (Tables 1 and 3), the most likely estimate of the true mutational number is 67 (95% confidence interval, 35 to 99). A similar analysis could not be conducted for the two isogenic mutants because the frequencies of their one or two possible P. syringae pv. atrofaciens mutations is inappropriate for this analysis and suggests that we have well described their limited mutational neighborhoods.
SNPs of host range expanded φ6 populations.
In addition to the sites of highest entropy change, we called single nucleotide polymorphisms (SNPs) present in the deep-sequenced pairs of populations. The counts and details of unique synonymous and nonsynonymous SNPs are summarized in Table 4 and in Tables S1 to S6 in the supplemental material. An increase in detectable polymorphism was observed for φ6-E8G (paired t test, P = 0.001) after host shifting to P. syringae pv. atrofaciens, but no significant change in SNP numbers was observed for φ6-WT and φ6-G515S (paired t test, P = 0.38 and P = 0.40, respectively). The numbers of SNPs detected in the φ6-E8G population grown on P. syringae pv. atrofaciens were also significantly higher than those for the φ6-WT and φ6-G515S populations grown on P. syringae pv. atrofaciens (paired t test, P = 0.03 and P = 0.0001, respectively). Gene p2 on the large segment, coding for the RNA-dependent RNA polymerase, appears to maintain a constant, high level of diversity. The surprisingly large number of low-frequency SNPs for φ6-WT raised on P. syringae pv. phaseolicola demonstrated the potential of a dsRNA virus with a 13-kb-long genome that grows ∼5 generations in overnight plaque growth to generate substantial genetic diversity. This may also be the reason the φ6-WT is more able to readily infect P. syringae pv. atrofaciens with a high P. syringae pv. atrofaciens host range mutation frequency (Fig. 5). The high mutation frequency of φ6-WT was apparent even after an abbreviated 4 h of incubation, which resulted in 100- to 1,000-fold lower population sizes in the lysates (Fig. 5).
TABLE 4.
Pairwise unique SNPs above 0.1% frequency in φ6 populations
| SNP type and/or location | No. of SNPs by organism and typea |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| φ6-WT |
φ6-E8G |
φ6-G515S |
||||||||||
|
P. pseudoalcaligenes |
P. syringae pv. atrofaciens |
P. syringae pv. tomato |
P. syringae pv. atrofaciens |
P. syringae pv. tomato |
P. syringae pv. atrofaciens |
|||||||
| S | NS | S | NS | S | NS | S | NS | S | NS | S | NS | |
| Noncoding | 14 | 20 | 4 | 29 | 7 | 8 | ||||||
| Coding | 23 | 44 | 19 | 54 | 8 | 20 | 39 | 67 | 7 | 24 | 12 | 16 |
| Small segment | ||||||||||||
| P8 | 2 | 4 | 0 | 1 | 0 | 1 | 2 | 3 | 0 | 0 | 2 | 0 |
| P12 | 0 | 1 | 8 | 14 | 0 | 1 | 4 | 5 | 1 | 0 | 1 | 4 |
| P9 | 0 | 3 | 3 | 7 | 0 | 1 | 4 | 5 | 0 | 0 | 1 | 0 |
| P5 | 2 | 4 | 0 | 1 | 0 | 1 | 2 | 3 | 1 | 1 | 0 | 2 |
| Medium segment | ||||||||||||
| P10 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| P6 | 0 | 2 | 1 | 1 | 1 | 0 | 1 | 6 | 1 | 3 | 1 | 1 |
| P3 | 7 | 13 | 5 | 17 | 0 | 2 | 11 | 21 | 0 | 3 | 6 | 6 |
| P13 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| Large segment | ||||||||||||
| P7 | 2 | 3 | 1 | 0 | 2 | 2 | 0 | 1 | 1 | 2 | 0 | 0 |
| P2 | 5 | 7 | 1 | 4 | 4 | 6 | 3 | 10 | 1 | 7 | 0 | 1 |
| P4 | 0 | 3 | 0 | 3 | 1 | 4 | 3 | 5 | 0 | 4 | 0 | 1 |
| P1 | 5 | 2 | 0 | 3 | 0 | 2 | 7 | 6 | 1 | 4 | 1 | 0 |
SNPs were detected by deep sequencing using VarScan. S, synonymous change; NS nonsynonymous change.
FIG 5.
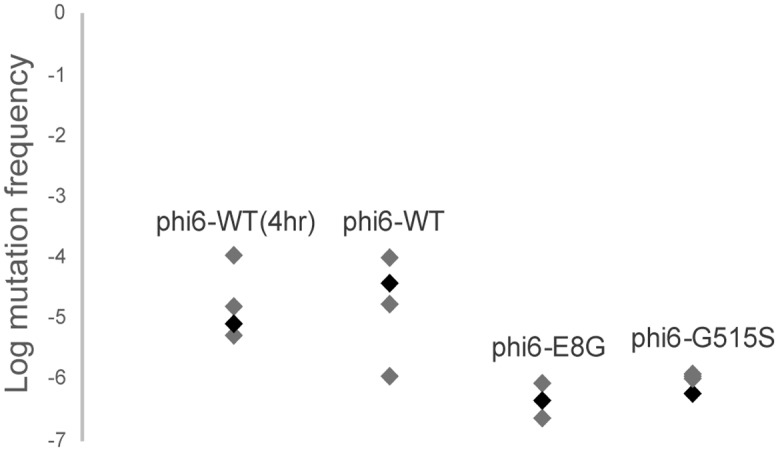
P. syringae pv. atrofaciens host range mutation frequency of φ6-WT (4 h), φ6-WT, φ6-E8G, and φ6-G515S. Values are measured from four purified single plaques. Plaques used for mutational neighborhood mapping are in black.
We compared the SNP results for φ6-WT to the estimated number of P. syringae pv. atrofaciens mutations obtained by jackknifing analysis. The total number of unique SNPs for φ6-WT grown on P. syringae pv. atrofaciens is 54, which is less than the estimated 67, and includes SNPs in genes such as the RNA-dependent RNA polymerase P2, which is unlikely to confer a changed host range. The nonsynonymous SNPs in the four genes now implicated in P. syringae pv. atrofaciens host range expansion (p3 on the medium segment and p12, p9, and p5 on the small segment) total to 39. This is within the 95% confidence interval predicted for the estimate of the complete mutational neighborhood and suggests that some of these detected SNPs may be part of the P. syringae pv. atrofaciens host range mutational neighborhood.
Relative fitness of naturally occurring φ6 P. syringae pv. atrofaciens host range mutants.
A single mutation (A133V) was shared across the three genotypes, which prompted us to look at its fitness effects in the three genetic backgrounds. We used paired growth assays to measure the relative fitness of host range mutants on their shared, original host, P. syringae pv. phaseolicola. Φ6-WT, the ancestor of all the tested strains, was the common competitor for all mutant genotypes and therefore was assigned the relative fitness of 1 (Fig. 6). Relative fitness was not affected when φ6-WT obtained an A133V mutation on p3 (two-tailed one sample t test, P = 0.66). However, when φ6-E8G and φ6-G515S gained the A133V mutation, each significantly increased its fitness on P. syringae pv. phaseolicola (Tukey’s honestly significant differences [HSD] adjusted value, P = 0.000011 and P = 0.011, respectively). K144R, on the other hand, was not beneficial to the two genotypes on the P. syringae pv. phaseolicola host (P < 0.005).
FIG 6.
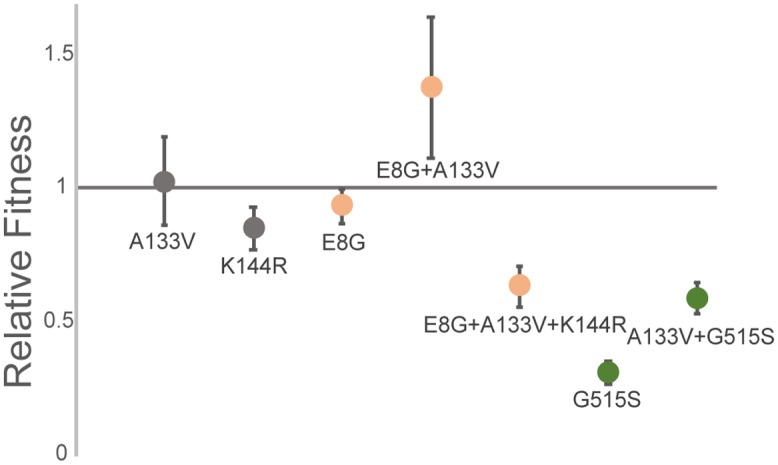
Relative fitness of host range mutants on P. syringae pv. phaseolicola. Same-color genotypes share the same genetic background (φ6-WT, gray; φ6-E8G, yellow; φ6-G515S, green). Values are averages of six replicates each; error bars are standard deviations.
DISCUSSION
Epistasis plays a large role in viral evolution, in part because viral proteins are often highly interactive and multifunctional, and many viral genomes are of limited size (30–32). The constraining effects of larger and smaller beneficial mutational neighborhoods were elegantly demonstrated in φ6 by Burch and Chao, who noted that the high mutation rate of φ6 was not sufficient to allow constrained genotypes to traverse a rugged fitness landscape (33). The ruggedness of viral fitness landscapes has been demonstrated for many viruses, including HIV (31), influenza virus A (34, 35), and Ebola virus (36), and the phenomenon of mutational neighborhoods constraining evolutionary trajectories has been demonstrated in cellular organisms as well (37–39). Many of these studies involved prolonged experimental evolution, whereas our study used a very narrow window of lethal selection: a single night’s selection on a novel host. Nevertheless, we detected strong epistatic constraint from single amino acid changes and in an ecologically realistic scenario for a host-shifting RNA or DNA virus.
Evolvability may vary over evolutionary history (40), and we have characterized only one mutational step (P. syringae pv. atrofaciens mutation frequency) for these three phage genotypes. The genotypes have differences in mutational supply affected more by the size of the P. syringae pv. atrofaciens mutational neighborhood than by overall population size (37). Moderate differences in population size are a concern because φ6-WT was grown on the highly productive host P. syringae pv. phaseolicola, and the two mutants were grown on P. syringae pv. tomato, a host on which they are less productive, but an abbreviated incubation for φ6-WT still produced the same high frequency of P. syringae pv. atrofaciens mutations (P = 0.35). The two mutants could not be grown on P. syringae pv. phaseolicola without risking reversion of their P. syringae pv. tomato host range mutations; G515S is highly deleterious on P. syringae pv. phaseolicola (23), and populations grown on P. syringae pv. phaseolicola quickly revert to glycine at this position (unpublished data from our lab). Nonetheless, the library preparation for Illumina sequencing involved the same amount of RNA, isolated from 24-h growth of twice-purified plaque freezer stocks, enforcing a similar population size of genomes sampled by sequencing. φ6-WT had double the number of SNPs in high-titer lysates in deep sequencing following double-plaque purification, demonstrating that a constrained mutational neighborhood for the two host range mutant genotypes played a large role in the reduced evolvability on P. syringae pv. atrofaciens.
Further, our results do not necessarily mean that φ6-E8G and φ6-G515S are trapped on their fitness landscapes with regard to host range expansion on P. syringae pv. atrofaciens. If these mutants were allowed to evolve further (on the original host or within their novel host range), it is difficult to predict if their evolved descendants would face identical constraints when infecting the P. syringae pv. atrofaciens host, which more accurately reflects the serial host jumping we have observed for mammalian viruses (41). It is easy to imagine that a φ6 P3 protein, somewhat destabilized by the addition of one host range mutation, cannot tolerate further destabilization while retaining its structure and function. The fitness benefits of A133V on the original host (P. syringae pv. phaseolicola) for both φ6-E8G and φ6-G515S may indicate that this is one of few (or the only) P. syringae pv. atrofaciens host range mutations in P3 that improves the mutant’s P3 structure and function. Other compensatory mutations acquired over evolutionary time could stabilize the P3 of descendants of φ6-E8G and φ6-G515S, creating larger mutational neighborhoods for P. syringae pv. atrofaciens host range expansion. However, if growth on P. syringae pv. tomato in and of itself affects P. syringae pv. atrofaciens host shifting, for example, through “maternal effects,” i.e., epigenetic effects due to the host used to generate a high-titer φ6 lysate, which could cause further differences in P. syringae pv. atrofaciens plaquing efficiencies among φ6 strains grown on different hosts (23), then that constraint may continue to affect even derived genotypes of the two isogenic mutants.
Specific mutations found in our study.
The most common means for a mutant to adapt is through additional (potentially compensatory) mutations rather than reversion of mutation (42). We observed three clones in the φ6-G515S population reverting (S515G) while fixing a P. syringae pv. atrofaciens host range mutation. This suggests that the G515S mutation is a relatively deleterious mutation to maintain in the genome when it is not grown on P. syringae pv. tomato, an idea which is bolstered by the low fitness of φ6-G515S on P. syringae pv. phaseolicola (Fig. 6).
While the small segment has not previously been associated with host range, one of our P. syringae pv. atrofaciens host range mutations (P12, F176L) was previously observed in a φ6 evolution experiment on a different novel host (43). Changes in the small segment-encoded protein P5 are known to affect φ6 thermal niche expansion (44), but this is the first time that membrane protein P9, enveloped lytic protein P5, and membrane morphogenic protein P12 (which is not found within the virion) were associated with host range. Across diverse viruses it is not uncommon for a variety of proteins in the envelope (such as P9) to interact with host receptor proteins (6, 45). It is rarer for nonstructural genes that are not on the exterior of the virion to be a host range determinant, but there are examples of this in avian influenza virus (PB2) (46, 47) and in picornaviruses (48, 49). While P5 plays a role in both φ6 entry and egress (50–52), P12 is solely associated with egress from cells (forming membranes from the host cytoplasmic membrane around completed nucleocapsids) and is not detected in φ6 virions (28, 53). P12 was a hot spot of change in entropy for φ6-WT, and 10 of the 13 clones that did not have a mutation in P3 had one of five nonsynonymous mutations in P12, which strongly suggests that this nonstructural protein has a role in host range expansion to P. syringae pv. atrofaciens. Given our current understanding of P12’s role in the φ6 life cycle (27), this would require φ6-WT to be able to attach and infect P. syringae pv. atrofaciens but fail to show its infection through a plaque assay; then mutations in a number of genes could boost the infectivity of φ6 to cause successful plaque formation. As phage host range is a difficult and debated phenotype to measure (54) and as plaque formation is known to be affected by many genetic and environmental factors (55), this could well be the scenario for φ6-WT on hosts closely related to its original host P. syringae pv. phaseolicola, which expresses the same attachment site, the type IV pilus (56). It is known that DNA phage can attach to more hosts than they can productively infect, often due to successful host defense mechanisms such as CRISPR-Cas, restriction endonucleases, and suicide of the infected cell prior to phage maturation (57); our RNA phage, which are not known to trigger abortive infection or be susceptible to these defenses, may still enter hosts they cannot productively exit. This suggests interesting follow-up experiments with φ6-WT and hosts considered outside its current host range. While spot plating is considered a sensitive method for detecting phage host range (58), lack of visible plaques does not definitively mean that phage cannot productively infect a given host at a low level or at a slow pace (59). It further suggests that some of the host range mutations observed here may be better categorized as mutations that aid in spread among novel hosts rather than as attachment mutations that allow for a spillover infections, which are often considered separate steps in the emergence of a virus on a novel host (60). On a practical level, one or more of the P. syringae pv. atrofaciens host range-associated mutations in p12, p9, or p5 may prove useful as a small segment marker for genetic crosses in φ6. The existing mutational markers on the small segment are an easily reverted temperature sensitivity and an unstable genetic insertion (61, 62).
Contrasting clonal sequencing and population deep-sequencing.
Next-generation sequencing (NGS) is now more frequently applied to microbial experimental evolution studies, changing how microbial populations are monitored and analyzed, focusing mostly on relative variant frequencies and their fitness effects (63–66). However, when determining population diversity structure, many studies still use cloning for isolate sequencing and examining chromatograms to describe nucleotide polymorphism (67–69). Increasingly, studies have exclusively used deep sequencing for population SNP detection (70, 71). In this study, both clonal sequencing and population deep sequencing had merits and shortcomings. Clonal mapping of mutational neighborhoods with 50 clones involved relatively small sample sizes but allowed unambiguous identification of single, double, and triple mutant combinations. Illumina sequencing of populations provided a more reassuringly complete picture of the mutational neighborhood, which was highly consistent with that of the clonal sequencing, but this method might recover hitchhiking mutations that might not be responsible for the phenotype of interest and cannot assign combinations of mutations from different segments to a single genome.
Using the change in Shannon entropy provided more accurate data analysis because it allowed us to cancel out a large amount of the noise produced by potential sequencing errors, and this parameter is ideal for our study’s purpose, which is contrasting populations before and after a challenge. However, we found that we could not rely on entropy signals as estimates for mutation frequency or abundance. Although the top three most frequently observed P3 mutations in φ6-WT showed the largest change in entropy, this pattern does not apply to other observed host range mutations (e.g., φ6-WT P3, position 140; φ6-G515S P3, position 35; φ6-WT P9, position 8). Population deep sequencing provides an excellent snapshot of the mutational neighborhood, but it prevents many downstream analyses (including any further experimentation with clones of interest). However, it is less expensive and faster than clonal isolation and will serve the needs of many researchers, especially in studies of host range mutations for emerging disease surveillance.
MATERIALS AND METHODS
Strains and culture conditions.
Wild-type φ6 (ATCC no. 21781-B1) and its standard laboratory host, P. syringae pathovar phaseolicola strain HB10Y (ATCC no. 21781), were originally obtained from the American Type Culture Collection (Bethesda, MD). These, along with novel hosts P. syringae pathovar atrofaciens, P. syringae pathovar tomato, and Pseudomonas pseudoalcaligenes East River isolate A were streaked from glycerol stocks originally obtained from G. Martin (Cornell University, Ithaca, NY), and L. Mindich (Public Health Research Institute, Newark, NJ) as described in previous studies (23, 72). Previously isolated isogenic host range mutants φ6-E8G and φ6-G515S (with E8G and G515S mutations on the host attachment protein P3, respectively) (23) were used to examine genome-wide mutations on host expansion. Both host range mutants can infect P. syringae pathovar phaseolicola, P. syringae pathovar tomato, and P. pseudoalcaligenes. Bacteria were grown in LC medium (LB broth, pH 7.5) at 25°C. Phages were grown with bacteria in 3 ml of 0.7% agar (top layer) on 1.5% agar plates as previously described (23).
Mutational neighborhood mapping.
Twice-plaque-purified φ6-WT, φ6-E8G, and φ6-G515S were raised to high-titer lysates on their respective hosts (i.e., φ6-WT was grown on P. syringae pathovar phaseolicola while φ6-E8G and φ6-G515S were grown on P. syringae pathovar tomato), and titers were determined on their respective hosts. All lysates were tested for the existing P. syringae pv. atrofaciens host range by spot plating approximately 104 to 105 plaque forming units (PFU) on a lawn of P. syringae pv. atrofaciens before plating to select for host range mutants. At least 106 PFU of phage was plated on novel host P. syringae pv. atrofaciens to isolate one host range mutant per lysate. Fifty single plaques were isolated from each lysate by plating on P. syringae pv. atrofaciens. Five of the φ6-G515S P. syringae pv. atrofaciens host range mutant plaques were isolated by the South Brunswick High School spring 2016 Biotechnology class. All 150 plaques were stored in 40% glycerol at −20°C as freezer stock and generated as high-titer lysates again for further analyses.
P. syringae pv. atrofaciens mutation frequency assays.
Four independent clones of twice-plaque-purified φ6-WT, φ6-E8G, and φ6-G515S plaques were raised to high-titer lysates (22 to 24 h overnight incubation) on host P. syringae pathovar phaseolicola (φ6-WT) or P. syringae pathovar tomato (φ6-E8G and φ6-G515S). After titers on these hosts were measured, titers of these high-titer lysates were determined on P. syringae pv. atrofaciens to assess the P. syringae pv. atrofaciens mutation frequency within the population’s standing genetic diversity. The four clones of φ6-WT were also incubated for a shorter length of time (4 h) to assess the P. syringae pv. atrofaciens mutation frequency in smaller populations that had not had as many chances for mutations to accumulate. One of the four clones tested for each genotype was the source of the 50 clones.
Fitness assays.
Twice-purified ancestral plaque freezer stocks (φ6-WT, φ6-E8G, and φ6-G515S), the one E8G A133V K144R plaque, and one representative from the isolated mutant plaques containing mutations A133V, K144R, E8G A133V, and A133V G515S were arbitrarily chosen for paired growth assays (PGAs). Equal amounts of host range mutants were mixed with a common competitor (φ6-WT) in PGAs (73) to test the mutant’s relative fitness on P. syringae pathovar phaseolicola. Ratios of host range mutant to the common competitor (CC) in the mixtures were obtained by counting the PFU of the initial mix (day 0) and after 24 h of growth (day 1). The relative fitness to the common competitor was calculated using the following formula:
To distinguish the different genotypes, two hosts were mixed (20:1, P. syringae pv. phaseolicola to P. syringae pv. atrofaciens) to generate the bacterial lawn as φ6-WT can only infect P. syringae pv. phaseolicola, which creates turbid plaques while the mutants can infect both hosts creating clear plaques. Statistical analyses of fitness data, including analysis of variance (ANOVA) and Tukey’s honestly significant difference (HSD) tests, were performed in R (74).
Sanger sequencing.
One microliter of the host range mutant glycerol stock was plated on a lawn of P. syringae pv. atrofaciens to generate high-titer lysates. Viral RNA was extracted from these lysates using a QiaAmp Viral RNA Mini kit (Qiagen, Valencia, CA) per the manufacturer’s guidelines. Reverse transcription-PCR (RT-PCR) was conducted using SuperScript II Reverse Transcriptase (Invitrogen/Thermo Fisher Scientific, MA) with random hexamers and KAPA Taq DNA polymerase (Kapa Biosystems/Roche) with primers that amplified the regions encoding P3 (host attachment) and P6 (membrane fusion) on the medium segment. Amplified PCR products were cleaned up using EXO-SapIT (US Biological, Swampscott, MA), and Sanger sequencing was performed by Genewiz, Inc. (South Plainfield, NJ). Sequencing results were aligned, and mutations were identified with Sequencher, version 4.10.1.
Mutation estimation.
We used a jackknife algorithm to estimate the true number of P. syringae pv. atrofaciens host range mutations in φ6-WT’s mutational neighborhood (29). As implemented in the R package SPECIES (75), this nonparametric method extrapolates the total number of types from the properties of subsamples of the observed sample (the Sanger sequencing results).
Library preparation.
Φ6-WT, φ6-E8G, and G515S were raised on their most recent hosts to obtain high-titer lysates, as described above. Each high-titer lysate was diluted and plated on P. syringae pv. atrofaciens to obtain a plate comprised of ∼400 plaques each. Each plaque was estimated to contain at least 106 PFU. These plates were harvested to make lysates of φ6-WT, φ6-E8G, and φ6-G515S capable of infecting P. syringae pv. atrofaciens. Viral RNA extracted using a QiaAmp Viral RNA Mini kit (Qiagen, Valencia CA) was purified by 1% low-melting-point agarose gel electrophoresis (IBI Scientific, IA) and GELase digestion according to the manufacturer’s instructions (GELase; Lucigen). Individual RNA samples at a final concentration of ∼15 ng/μl were prepared for Illumina RNA libraries using a TruSeq RNA Library Prep kit (Illumina, CA). Single-ended 150-cycle deep sequencing was performed on an Illumina MiSeq housed in Foran Hall, Rutgers University (SEBS Genome Cooperative).
NGS data analysis.
Raw reads were trimmed and filtered with cutadapt, version 1.12 (Q score cutoff, 30; minimum length cutoff, 75 bp; adapters and terminal Ns of reads removed) (76). Then, the reads were mapped to the Pseudomonas bacteriophage φ6 genomes (reference sequences derived from the Illumina sequencing of φ6-WT, φ6-E8G, and φ6-G515S [also confirmed with Sanger sequencing]), using Burrows-Wheeler Aligner maximal exact match (BWA-MEM) with default settings (77). Although approximately 33.75% to 66.88% of the NGS reads mapped to the Pseudomonas host genome, all virus genome positions had above 1,000× read coverage, with the exception of the large segment of φ6-G515S, which had 322× to 12,058× coverage. Additional file conversion was performed using SAMtools (78). Genome nucleotide counts by position were counted with Integrative Genomics Viewer IGVTools (count options, window size 1 and –bases) (79). Whole-genome variant calling was performed using VarScan (80). Shannon entropy (H) was calculated for each position of the genome with the following equation:
where n = 4 for 4 nucleotides, and P(xi) is the proportion of a single nucleotide over all nucleotides read at that position. Change in Shannon entropy was calculated by subtracting values before growth on P. syringae pv. atrofaciens (Xb) from values after growth on P. syringae pv. atrofaciens (Xa). Shannon entropy shows the polymorphism at each site in the genome through absolute base coverage from deep sequencing. Change in Shannon entropy shows the change in polymorphic base composition at each position in the genome:
When levels of polymorphism were compared directly between populations, the SNPs in each protein-coding gene were considered independent observations for a paired t test (Microsoft Excel, Redmond, WA).
Accession number(s).
Raw sequencing data have been deposited in NCBI under BioProject accession number PRJNA485986.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the National Science Foundation DEB 1453241 (to S.D.).
We thank Nicole Wagner of the SEBS Genome Cooperative and Natasia Jacko for expert technical assistance. German Lagunas Robles and the students of South Brunswick High School’s 2016 Biotechnology class (taught by Asha Sohan and coordinated by Aparna Rajagopal) isolated several of the clones used in this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01385-18.
REFERENCES
- 1.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elena SF, Bedhomme S, Carrasco P, Cuevas JM, de la Iglesia F, Lafforgue G, Lalić J, Pròsper À, Tromas N, Zwart MP. 2011. The evolutionary genetics of emerging plant RNA viruses. Mol Plant Microbe Interact 24:287–293. doi: 10.1094/MPMI-09-10-0214. [DOI] [PubMed] [Google Scholar]
- 3.Marston HD, Folkers GK, Morens DM, Fauci AS. 2014. Emerging viral diseases: confronting threats with new technologies. Sci Transl Med 6:253ps210. [DOI] [PubMed] [Google Scholar]
- 4.Hui EK. 2006. Reasons for the increase in emerging and re-emerging viral infectious diseases. Microbes Infect 8:905–916. doi: 10.1016/j.micinf.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferris MT, Joyce P, Burch CL. 2007. High frequency of mutations that expand the host range of an RNA virus. Genetics 176:1013–1022. doi: 10.1534/genetics.106.064634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lounková A, Kosla J, Přikryl D, Štafl K, Kučerová D, Svoboda J. 2017. Retroviral host range extension is coupled with Env-activating mutations resulting in receptor-independent entry. Proc Natl Acad Sci U S A 114:E5148–E5157. doi: 10.1073/pnas.1704750114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepin KM, Lass S, Pulliam JRC, Read AF, Lloyd-Smith JO. 2010. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat Rev Microbiol 8:802–813. doi: 10.1038/nrmicro2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguas R, Ferguson NM. 2013. Feature selection methods for identifying genetic determinants of host species in RNA viruses. PLoS Comput Biol 9:e1003254. doi: 10.1371/journal.pcbi.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasik BR, Turner PE. 2013. On the biological success of viruses. Annu Rev Microbiol 67:519–541. doi: 10.1146/annurev-micro-090110-102833. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennings PS, Kryazhimskiy S, Wakeley J. 2014. Loss and recovery of genetic diversity in adapting populations of HIV. PLoS Genet 10:e1004000. doi: 10.1371/journal.pgen.1004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elena SF, Solé RV, Sardanyés J. 2010. Simple genomes, complex interactions: epistasis in RNA virus. Chaos 20:026106. doi: 10.1063/1.3449300. [DOI] [PubMed] [Google Scholar]
- 14.Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. 2011. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet 7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalic J, Elena SF. 2012. Magnitude and sign epistasis among deleterious mutations in a positive-sense plant RNA virus. Heredity (Edinb) 109:71–77. doi: 10.1038/hdy.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akand EH, Downard KM. 2018. Identification of epistatic mutations and insights into the evolution of the influenza virus using a mass-based protein phylogenetic approach. Mol Phylogenet Evol 121:132–138. doi: 10.1016/j.ympev.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Holmes EC, Rambaut A. 2004. Viral evolution and the emergence of SARS coronavirus. Philos Trans R Soc Lond B Biol Sci 359:1059–1065. doi: 10.1098/rstb.2004.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, Drexler JF. 2014. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allison AB, Harbison CE, Pagan I, Stucker KM, Kaelber JT, Brown JD, Ruder MG, Keel MK, Dubovi EJ, Holmes EC, Parrish CR. 2012. Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus. J Virol 86:865–872. doi: 10.1128/JVI.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parrish CR, Murcia PR, Holmes EC. 2015. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J Virol 89:2990–2994. doi: 10.1128/JVI.03146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bedhomme S, Hillung J, Elena SF. 2015. Emerging viruses: why they are not jacks of all trades? Curr Opin Virol 10:1–6. doi: 10.1016/j.coviro.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Duffy S, Turner PE, Burch CL. 2006. Pleiotropic costs of niche expansion in the RNA bacteriophage φ6. Genetics 172:751–757. doi: 10.1534/genetics.105.051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford BE, Sun B, Carpino J, Chapler ES, Ching J, Choi Y, Jhun K, Kim JD, Lallos GG, Morgenstern R, Singh S, Theja S, Dennehy JJ. 2014. Frequency and fitness consequences of bacteriophage Φ6 host range mutations. PLoS One 9:e113078. doi: 10.1371/journal.pone.0113078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L, Pasricha MS, Stemate D, Crespo-Bellido A, Gagnon J, Draghi J, Duffy S. 2018. Existing host range mutations constrain further emergence of RNA viruses. bioRxiv 10.1101/394080. [DOI] [PMC free article] [PubMed]
- 26.Stitt BL, Mindich L. 1983. The structure of bacteriophage phi 6: protease digestion of phi 6 virions. Virology 127:459–462. doi: 10.1016/0042-6822(83)90158-7. [DOI] [PubMed] [Google Scholar]
- 27.Johnson MD, Mindich L. 1994. Plasmid-directed assembly of the lipid-containing membrane of bacteriophage phi 6. J Bacteriol 176:4124–4132. doi: 10.1128/jb.176.13.4124-4132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurinavicius S, Kakela R, Bamford DH, Somerharju P. 2004. The origin of phospholipids of the enveloped bacteriophage phi6. Virology 326:182–190. doi: 10.1016/j.virol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 29.Burnham KP, Overton WS. 1979. Robust estimation of population size when capture probabilities vary among animals. Ecology 60:927–936. doi: 10.2307/1936861. [DOI] [Google Scholar]
- 30.Betancourt A. 2010. Lack of evidence for sign epistasis between beneficial mutations in an RNA bacteriophage. J Mol Evol 71:437–443. doi: 10.1007/s00239-010-9397-0. [DOI] [PubMed] [Google Scholar]
- 31.da Silva J, Coetzer M, Nedellec R, Pastore C, Mosier DE. 2010. Fitness epistasis and constraints on adaptation in a human immunodeficiency virus type 1 protein region. Genetics 185:293. doi: 10.1534/genetics.109.112458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalic J, Elena SF. 2015. The impact of high-order epistasis in the within-host fitness of a positive-sense plant RNA virus. J Evol Biol 28:2236–2247. doi: 10.1111/jeb.12748. [DOI] [PubMed] [Google Scholar]
- 33.Burch CL, Chao L. 2000. Evolvability of an RNA virus is determined by its mutational neighborhood. Nature 406:625–628. doi: 10.1038/35020564. [DOI] [PubMed] [Google Scholar]
- 34.Sanjuan R, Moya A, Elena SF. 2004. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci U S A 101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong LI, Suchard MA, Bloom JD. 2013. Stability-mediated epistasis constrains the evolution of an influenza protein. Elife 2:e00631. doi: 10.7554/eLife.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibeh N, Nshogozabahizi JC, Aris-Brosou S. 2016. Both epistasis and diversifying selection drive the structural evolution of the Ebola virus glycoprotein mucin-like domain. J Virol 90:5475–5484. doi: 10.1128/JVI.00322-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall AR, Griffiths VF, MacLean RC, Colegrave N. 2010. Mutational neighbourhood and mutation supply rate constrain adaptation in Pseudomonas aeruginosa. Proceedings of the Royal Society B-Biological Sciences 277:643–650. doi: 10.1098/rspb.2009.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roles AJ, Rutter MT, Dworkin I, Fenster CB, Conner JK. 2016. Field measurements of genotype by environment interaction for fitness caused by spontaneous mutations in Arabidopsis thaliana. Evolution 70:1039–1050. doi: 10.1111/evo.12913. [DOI] [PubMed] [Google Scholar]
- 39.Jerison ER, Kryazhimskiy S, Mitchel JK, Bloom JS, Kruglyak L, Desai MM. 2017. Genetic variation in adaptability and pleiotropy in budding yeast. Elife 6:e27167. doi: 10.7554/eLife.27167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrick JE, Kauth MR, Strelioff CC, Lenski RE. 2010. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol Biol Evol 27:1338–1347. doi: 10.1093/molbev/msq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner PE, Morales NM, Alto BW, Remold SK. 2010. Role of evolved host breadth in the initial emergence of an RNA virus. Evolution 64:3273–3286. doi: 10.1111/j.1558-5646.2010.01051.x. [DOI] [PubMed] [Google Scholar]
- 42.Crill WD, Wichman HA, Bull JJ. 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner PE, McBride RC, Duffy S, Montville R, Wang L-S, Yang YW, Lee SJ, Kim J. 2012. Evolutionary genomics of host-use in bifurcating demes of RNA virus phi-6. BMC Evol Biol 12:153. doi: 10.1186/1471-2148-12-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldhill D, Lee A, Williams ES, Turner PE. 2014. Evolvability and robustness in populations of RNA virus phi6. Front Microbiol 5:35. doi: 10.3389/fmicb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. 1991. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol 65:1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67:1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min JY, Santos C, Fitch A, Twaddle A, Toyoda Y, DePasse JV, Ghedin E, Subbarao K. 2013. Mammalian adaptation in the PB2 gene of avian H5N1 influenza virus. J Virol 87:10884–10888. doi: 10.1128/JVI.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin FH, Lomax NB. 1983. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J Virol 48:410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pacheco JM, Henry TM, O'Donnell VK, Gregory JB, Mason PW. 2003. Role of nonstructural proteins 3A and 3B in host range and pathogenicity of foot-and-mouth disease virus. J Virol 77:13017–13027. doi: 10.1128/JVI.77.24.13017-13027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bamford DH, Romantschuk M, Somerharju PJ. 1987. Membrane fusion in prokaryotes: bacteriophage phi 6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J 6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caldentey J, Bamford DH. 1992. The lytic enzyme of the Pseudomonas phage φ6. Purification and biochemical characterization. Biochim BiophysActa 1159:44–50. doi: 10.1016/0167-4838(92)90073-M. [DOI] [PubMed] [Google Scholar]
- 52.Pei J, Grishin NV. 2005. The P5 protein from bacteriophage phi-6 is a distant homolog of lytic transglycosylases. Protein Sci 14:1370–1374. doi: 10.1110/ps.041250005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinclair JF, Tzagoloff A, Levine D, Mindich L. 1975. Proteins of bacteriophage phi6. J Virol 16:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. doi: 10.1016/s0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- 55.Mullan WMA. 2002. Enumeration of lactococcal bacteriophages. https://www.dairyscience.info/index.php/enumeration-of-lactococcal-bacteriophages.html. Accessed 3 December 2018.
- 56.Roine E, Raineri DM, Romantschuk M, Wilson M, Nunn DN. 1998. Characterization of type IV pilus genes in Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 11:1048–1056. doi: 10.1094/MPMI.1998.11.11.1048. [DOI] [PubMed] [Google Scholar]
- 57.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 58.Xie Y, Wahab L, Gill JJ. 2018. Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses 10:189. doi: 10.3390/v10040189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abedon ST, Yin J. 2009. Bacteriophage plaques: theory and analysis. Methods Mol Biol 501:161–174. doi: 10.1007/978-1-60327-164-6_17. [DOI] [PubMed] [Google Scholar]
- 60.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature 438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mindich L, Lehman J. 1983. Characterization of phi 6 mutants that are temperature sensitive in the morphogenetic protein P12. Virology 127:438–445. doi: 10.1016/0042-6822(83)90156-3. [DOI] [PubMed] [Google Scholar]
- 62.Turner PE, Burch CL, Hanley KA, Chao L. 1999. Hybrid frequencies confirm limit to coinfection in the RNA bacteriophage φ6. J Virol 73:2420–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Acevedo A, Brodsky L, Andino R. 2014. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505:686–690. doi: 10.1038/nature12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS, Sherlock G. 2015. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519:181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, Wu GC, Wielgoss S, Cruveiller S, Medigue C, Schneider D, Lenski RE. 2016. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morley VJ, Turner PE. 2017. Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change. Evolution 71:872–883. doi: 10.1111/evo.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arribas M, Cabanillas L, Kubota K, Lazaro E. 2016. Impact of increased mutagenesis on adaptation to high temperature in bacteriophage Qbeta. Virology 497:163–170. doi: 10.1016/j.virol.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 68.Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, Kishony R. 2016. Spatiotemporal microbial evolution on antibiotic landscapes. Science 353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Presloid JB, Mohammad TF, Lauring AS, Novella IS. 2016. Antigenic diversification is correlated with increased thermostability in a mammalian virus. Virology 496:203–214. doi: 10.1016/j.virol.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cuevas JM, Willemsen A, Hillung J, Zwart MP, Elena SF. 2015. Temporal dynamics of intrahost molecular evolution for a plant RNA virus. Mol Biol Evol 32:1132–1147. doi: 10.1093/molbev/msv028. [DOI] [PubMed] [Google Scholar]
- 71.Pauly MD, Lauring AS. 2015. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J Virol 89:3584–3597. doi: 10.1128/JVI.03483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duffy S, Burch CL, Turner PE. 2007. Evolution of host specificity drives reproductive isolation among RNA viruses. Evolution 61:2614–2622. doi: 10.1111/j.1558-5646.2007.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chao L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 74.R Development Core Team. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 75.Wang J-P. 2011. SPECIES: an R package for species richness estimation. J Stat Soft 40:1–15. [Google Scholar]
- 76.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 77.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv https://arxiv.org/abs/1303.3997.
- 78.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.