The pandemic outbreak of PRRS, which is caused by PRRSV, has greatly affected the swine industry. We still lack an efficient vaccine, and it is an immense challenge to control its infection. An intracellularly expressed Nsp9-specific nanobody, Nb6, has been shown to be able to inhibit PRRSV replication in MARC-145 cells. However, its application is limited, because Nb6 cannot physically enter cells. Here, we demonstrated that the cell-penetrating peptide TAT could deliver Nb6 into cultured cells. In addition, TAT-Nb6 fusion protein could suppress the replication of various PRRSV strains in MARC-145 cells and PAMs. These findings may provide a new approach for drug development to control PRRS.
KEYWORDS: nanobody, PRRSV, antiviral agents
ABSTRACT
Porcine reproductive and respiratory syndrome (PRRS) is of great concern to the swine industry due to pandemic outbreaks of the disease, current ineffective vaccinations, and a lack of efficient antiviral strategies. In our previous study, a PRRSV Nsp9-specific nanobody, Nb6, was successfully isolated, and the intracellularly expressed Nb6 could dramatically inhibit PRRSV replication in MARC-145 cells. However, despite its small size, the application of Nb6 protein in infected cells is greatly limited, as the protein itself cannot enter the cells physically. In this study, a trans-activating transduction (TAT) peptide was fused with Nb6 to promote protein entry into cells. TAT-Nb6 was expressed as an inclusion body in Escherichia coli, and indirect enzyme-linked immunosorbent assays and pulldown assays showed that E. coli-expressed TAT-Nb6 maintained the binding ability to E. coli-expressed or PRRSV-encoded Nsp9. We demonstrated that TAT delivered Nb6 into MARC-145 cells and porcine alveolar macrophages (PAMs) in a dose- and time-dependent manner, and TAT-Nb6 efficiently inhibited the replication of several PRRSV genotype 2 strains as well as a genotype 1 strain. Using a yeast two-hybrid assay, Nb6 recognition sites were identified in the C-terminal part of Nsp9 and spanned two discontinuous regions (Nsp9aa454–551 and Nsp9aa599–646). Taken together, these results suggest that TAT-Nb6 can be developed as an antiviral drug for the inhibition of PRRSV replication and controlling PRRS disease.
IMPORTANCE The pandemic outbreak of PRRS, which is caused by PRRSV, has greatly affected the swine industry. We still lack an efficient vaccine, and it is an immense challenge to control its infection. An intracellularly expressed Nsp9-specific nanobody, Nb6, has been shown to be able to inhibit PRRSV replication in MARC-145 cells. However, its application is limited, because Nb6 cannot physically enter cells. Here, we demonstrated that the cell-penetrating peptide TAT could deliver Nb6 into cultured cells. In addition, TAT-Nb6 fusion protein could suppress the replication of various PRRSV strains in MARC-145 cells and PAMs. These findings may provide a new approach for drug development to control PRRS.
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS), which is caused by PRRS virus (PRRSV), is currently one of the most economically detrimental viral diseases in the swine industry worldwide (1–3). PRRS is characterized by severe reproductive failure in sows and respiratory syndromes and persistent infection in young pigs (4, 5). At present, because of the high antigenic heterogeneity and antibody-dependent enhancement of PRRSV infection, vaccination strategies are obviously ineffective for controlling PRRSV infection (6, 7). A PRRSV-specific treatment for infected herds, or other prevention methods, is urgently needed to inhibit further damage to the industry (8).
PRRSV belongs to the family Arteriviridae within the order Nidovirales (9). It has a single-stranded, positive-sense RNA genome of approximately 15.4 kb, which contains at least 10 open reading frames (ORFs), including ORF1a, ORF1b, ORF2a, ORF2b, ORF3, ORF4, ORF5, ORF5a, ORF6, and ORF7 (10, 11). Viral proteases process polyproteins translated from ORF1a and ORF1b into 16 nonstructural proteins (Nsps): Nsp1α/β, Nsp2-6, Nsp7/7α, Nsp8-12, Nsp2TF, and Nsp2N (12, 13). It has been demonstrated that some of the Nsps are assembled with host cell components to form the viral replication and transcription complex (RTC) (14). In the RTC, Nsp9 possesses RNA-dependent RNA polymerase (RdRp) activity and is essential for genome replication and transcription. Nsp9 is a relatively conserved region in the PRRSV genome, making it a logical antiviral target for the control of PRRSV infection (1).
Nanobodies (Nbs), the variable domain of Camellidae heavy-chain-only antibodies (15), possess attractive advantages over conventional monoclonal antibodies (MAbs), such as small size, ease of genetic manipulation, high specificity, and solubility (16). In addition, nanobodies can be easily produced in prokaryotic or eukaryotic host organisms (17). Considering these characteristics, nanobodies are ideal candidates for drug development (18–20). In our previous study, we isolated a PRRSV Nsp9-specific nanobody, Nb6, which interacted with PRRSV-encoded Nsp9 and blocked PRRSV replication when expressed in MARC-145 cells (1), suggesting it usefulness as an innovative anti-PRRSV agent. However, the impermeability of cell membrane to macromolecules limits the application of such nanobodies in biopharmaceuticals Therefore, it is pertinent to establish an efficient, safe, and nontoxic delivery system to greatly improve the applicability of nanobodies.
Cell-penetrating peptides (CPPs), short peptides with approximately 5 to 30 amino acids, have been widely used for delivering exogenous proteins into living cells and tissues (21–23). It has been reported that CPPs can direct proteins to cross cellular membranes via direct translocation or endocytosis (24, 25). These peptides can enter the cells without causing cytotoxicity and have been used as a tool for the delivery of various cargos into cells, such as plasmid DNA, short interfering RNA (siRNA), therapeutic proteins, viruses, imaging agents, and other nanoparticles (21, 26–28).
The human immunodeficiency virus type 1 (HIV-1) trans-activator TAT protein, which was first discovered in 1988, contains 86 amino acids and can rapidly enter cells. About 10 years after the initial discovery of TAT protein, some studies found that an 11-amino-acid TAT peptide, YGRKKRRQRRR (residues 47 to 57), was sufficient for intracellular transduction and nuclear subcellular localization of exogenous proteins (29–31). For drug delivery, TAT improved the intestinal absorption efficiency of insulin (32), and another study reported that TAT-fused glyoxalase protein inhibited ischemic neuronal cell damage and ameliorates ischemic injury (17). Recently, TAT was also used in the delivery of HIV-1 Nef to enhance the immune response in mice (16). While a TAT-fused nanobody has been reported in recent anticancer research (33), no report has been seen on the use of CPPs as a potential strategy for PRRSV control.
In the present study, cell-penetrating peptide TAT and nanobody Nb6 were expressed as a fused protein, TAT-Nb6, in Escherichia coli. We demonstrated that TAT-Nb6 efficiently entered MARC-145 cells and porcine alveolar macrophages (PAMs), and the nanobody inhibited PRRSV replication in both types of cells. We also identified the Nb6 binding regions on Nsp9 by yeast two-hybrid assay. These data have laid the foundation for future research on the molecular mechanism by which Nb6 inhibits PRRSV replication.
RESULTS
Expression and purification of nanobodies.
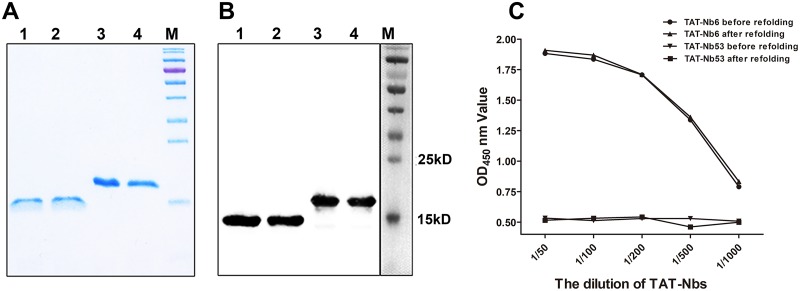
The nanobodies Nb6, Nb53, TAT-Nb6, and TAT-Nb53, each containing a 6×His tag at the C terminus, were successfully expressed in E. coli as inclusion bodies. The four fusion proteins were affinity purified using Ni-nitrilotriacetic acid (NTA) resin under denaturing conditions, refolded and dialyzed, and then examined by SDS-PAGE (Fig. 1A) and Western blot analysis using anti-His monoclonal antibody (Fig. 1B). Analyses show that all four proteins were obtained with high purity. His-tagged Nb6 and Nb53 exhibited the predicted size of 15 kDa, and the two nanobodies fused with TAT leader peptide were approximately 19 kDa, which is consistent with their predicted sizes. Purified Nsp9-His next was used as an indirect enzyme-linked immunosorbent assay (iELISA) coating antigen to test the antigen reactivity of refolded TAT-Nb6. As shown in Fig. 1C, TAT-Nb6 before or after refolding was able to specifically react with Nsp9, unlike TAT-Nb53, and there was no significant difference between these two forms.
FIG 1.
Analysis of purified and refolded nanobodies by SDS-PAGE (A) and Western blotting (B). The predicted sizes of the His-tagged nanobodies are 15 kDa without TAT (NB6 and NB53) and 19 kDa with the TAT leader peptide (TAT-NB6 and TAT-NB53). M, protein marker; lanes 1 to 4, NB6, NB53, TAT-NB6, and TAT-NB53. (C) Determination of the binding activity of TAT-Nb6 to Nsp9 by iELISA. TAT-Nb53 was used as a negative control. Assays were performed in triplicate, and data are presented as means ± standard deviations (SD).
Cellular uptake of TAT-Nb6 and TAT-Nb53.
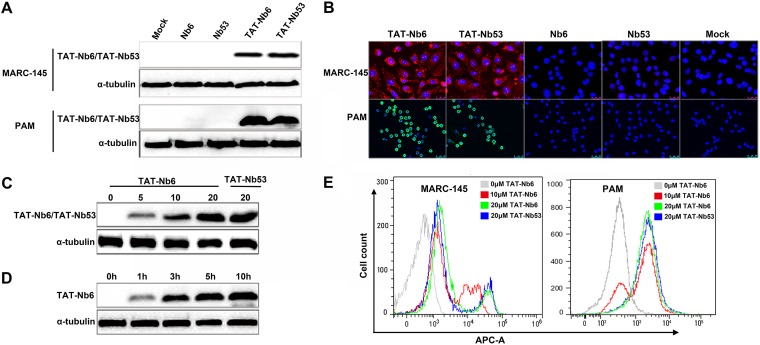
To examine the ability of Nbs to enter cells, the purified proteins with or without TAT were added onto MARC-145 or PAM monolayers at a final concentration of 5 μM. After 5 h of treatment, cells were harvested and the His-tagged nanobodies delivered into the cells were detected by Western blotting and immunofluorescence assay (IFA). As shown in Fig. 2A, TAT-Nb6 and TAT-Nb53 were clearly detected in both MARC-145 cells and PAMs. In contrast, Nbs without TAT were not detectable in the cell lysates. IFA confirmed the Western blotting results, showing that TAT-Nb6 and TAT-Nb53 accumulated in the cytoplasm and also entered the nucleus (Fig. 2B).
FIG 2.
Cellular uptake of TAT-Nbs into MACR-145 and PAMs. (A and B) Western blotting (A) and IFA detection (B) of intracellular TAT-Nbs. MARC-145 and PAMs were treated with 10 μM Nb6, Nb53, TAT-Nb6, and TAT-Nb53 for 5 h. (C and E) Western blot (C) and flow cytometry (E) analyses of the uptake of TAT-NB6 and TAT-NB53 at different concentrations. MARC-145 cells were treated with the nanobodies at the indicated concentrations for 5 h. (D) MARC-145 cells were treated with TAT-NB6 at 20 μM for 0, 1, 3, 5, and 10 h and then examined by Western blotting.
To investigate the effects of protein concentration and incubation time on cellular uptake of the TAT-tagged nanobodies, TAT-Nb6 was incubated with MARC-145 cells at various concentrations for 5 h or at 5 μM for different time periods. The proteins that entered the treated cells were then detected by Western blotting. The data showed that increasing amounts of TAT-Nb6 were delivered into the cells at concentrations ranging from 0 to 20 μM (Fig. 2C). In addition, prolonged incubation time also resulted in more protein accumulation in the cells (Fig. 2D), indicating that the translocation of TAT-Nb6 into cells was dose and time dependent. Flow cytometry assay (FCM) analysis confirmed that TAT-Nb6 internalization was dose dependent in both MARC-145 and PAM cells (Fig. 2E). The delivery efficiency of the control nanobody TAT-Nb53 at 20 μM was similar to that of TAT-Nb6 when detected by Western blotting and FCM (Fig. 2C and E).
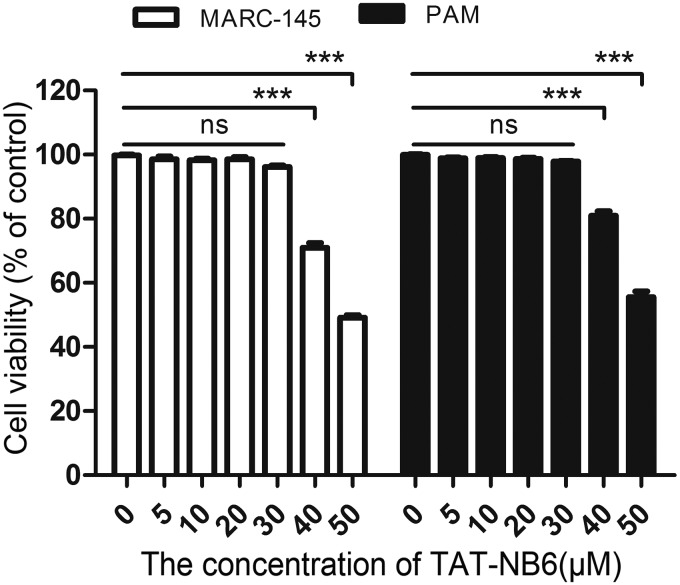
Furthermore, a cell counting kit-8 (CCK-8) assay was performed to test the toxicity of TAT-Nb6 in MARC-145 cells and PAMs. As shown in Fig. 3, the viability of MARC-145 cells and PAMs was close to that of the untreated control cells at concentrations lower than 30 μM, but it rapidly reduced to less than 80% of the control when the concentration of the nanobody was increased to 40 μM. This result suggested that TAT-Nb6, which entered cells in a dose- and time-dependent manner, was not toxic to MARC-145 and PAMs at concentrations lower than 30 μM.
FIG 3.
Detection of TAT-Nb6 toxicity using CCK-8 kits. Data are expressed as means ± SD from three independent experiments. P values were calculated using ANOVA. P values were <0.05 (*), <0.01 (**), and <0.001 (***) compared with the cells infected with PRRSV alone (ns, not significant).
Recognition of Nsp9 in PRRSV-infected cells by TAT-Nb6.
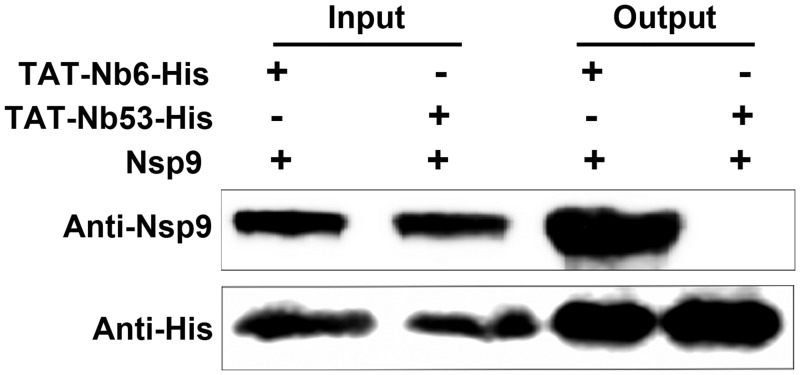
According to the results shown in Fig. 1C, E. coli-expressed TAT-Nb6 could recognize E. coli-expressed Nsp9. To validate the binding ability of TAT-Nb6 to the native Nsp9 produced in virus infection, pulldown assays were performed. MARC-145 cells were lysed after infection with PRRSV strain SD16 for 48 h. The cell lysates were mixed with TAT-Nb6-His or TAT-Nb53-His and incubated with Ni-NTA beads, and the protein complexes bound with the His-tagged proteins were eluted and then detected by Western blotting using anti-His antibody and mouse anti-Nsp9 antiserum. As expected, Nsp9 was pulled down by TAT-Nb6-His but not by the control nanobody, TAT-Nb53-His (Fig. 4). These results demonstrated that TAT-Nb6 maintained the antigen binding ability of the nanobody.
FIG 4.
TAT-Nb6 interacts with PRRSV-encoded Nsp9. MARC-145 cells were infected with SD16 at an MOI of 1 for 48 h, and Nsp9 was pulled down by TAT-Nb6-His or TAT-Nb53-His. The bound proteins were detected by Western blotting using mouse anti-His antibody or mouse anti-Nsp9 antiserum.
TAT-Nb6 inhibits PRRSV replication in MARC-145 cells.
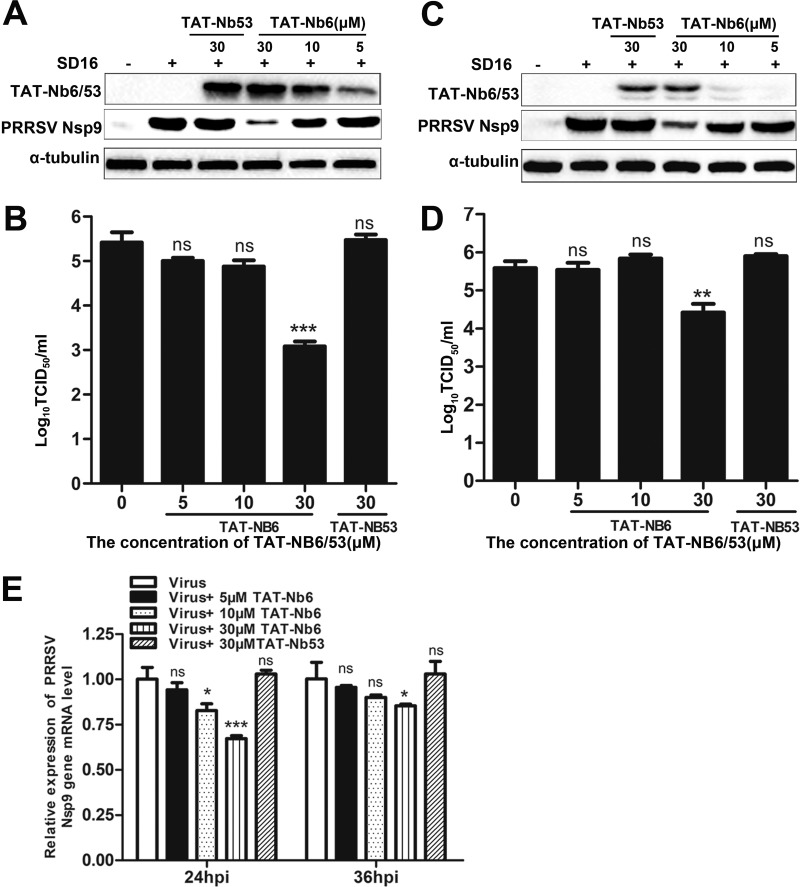
To evaluate the ability of TAT-Nb6 to inhibit PRRSV infection, MARC-145 cells were infected with PRRSV strain SD16 at a multiplicity of infection (MOI) of 0.01. After incubation with the virus for 1 h, TAT-Nbs were added to the infected cells at different concentrations. The treated cells were cultured for 24 h or 36 h, and then the expression levels of PRRSV Nsp9 in the cell lysates were detected by Western blotting. As shown in Fig. 5A and C, Nsp9 expression was significantly suppressed in the cells treated with TAT-Nb6 at 24 hpi and 36 hpi, and the inhibition was dose dependent, suggesting that the replication of PRRSV in MARC-145 cells could be effectively inhibited by TAT-Nb6. We also quantified progeny virus production in cell culture supernatants by TCID50. As shown in Fig. 5B and D, TAT-Nb6 significantly inhibited the propagation of PRRSV SD16 at 30 μM, exhibiting an approximately 100-fold reduction in virus yield at 24 hpi and a 10-fold decrease at 36 hpi compared with the untreated group and TAT-Nb53-treated group. No significant decrease in progeny virus production was detected in the cells treated with TAT-Nb6 at lower concentrations. Consistent with the Nsp9 expression results, titration of progeny viruses in the cell medium also demonstrated that TAT-Nb6 suppressed the virus replication in a dose-dependent manner.
FIG 5.
Inhibition of PRRSV SD16 infection and replication by TAT-NB6 in MARC-145 cells. MARC-145 cells were infected with SD16 at an MOI of 0.01 for 1 h, and then the cell culture media were replaced with fresh DMEM containing 3% FBS and TAT-Nbs at the indicated concentrations. TAT-Nbs and PRRSV Nsp9 were detected at 24 hpi (A) and 36 hpi (C) by Western blotting using anti-His MAb and mouse anti-Nsp9 antiserum, respectively. Progeny virus released in the cell medium was measured by TCID50 at 24 hpi (B) and 36 hpi (D). (E) Relative levels of PRRSV RNA were detected by RT-qPCR using PRRSV Nsp9-specific primers. The GAPDH mRNA level served as an internal reference. Data are expressed as means ± SD from three independent experiments. P values were calculated using ANOVA as <0.05 (*), <0.01 (**), and <0.001 (***) compared with cells infected with PRRSV alone (ns, not significant).
As the RNA-dependent RNA polymerase in PRRSV, Nsp9 plays a critical role in PRRSV RNA synthesis. To detect the effect of TAT-Nb6 on the replication and transcription of PRRSV genome, reverse transcription-quantitative PCR (RT-qPCR) was performed for the detection of the PRRSV nsp9 RNA level. The results reveal that the relative PRRSV RNA level in the cells treated with 30 μM TAT-Nb6 was significantly reduced compared with that of the controls at 24 hpi (Fig. 5E, left). A similar tendency was found at 36 hpi (Fig. 5E, right). Consistent with the virus production and viral protein expression results, the treatment of TAT-Nb6 did not significantly reduce PRRSV RNA synthesis at concentrations lower than 30 μM. These results suggest that TAT-Nb6 efficiently inhibits the RNA synthesis of PRRSV, at least in early infection.
TAT-Nb6 suppresses PRRSV replication in PAMs.
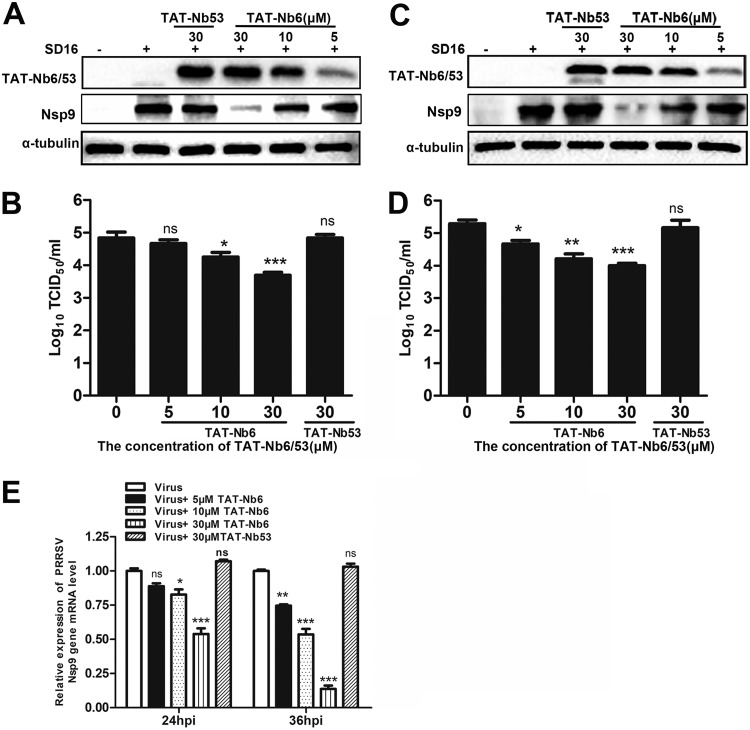
Since TAT-Nb6 exerted significant antiviral activity in PRRSV-infected MARC-145 cells, we further investigated whether TAT-Nb6 could suppress PRRSV replication in PAMs, the primary targets of PRRSV infection in vivo. PAMs were treated with TAT-Nb6 or TAT-Nb53 at different concentrations. The cells and culture supernatants were collected at the indicated time points after infection for Western blotting, qPCR, and titration assays. Consistent with the results detected in MARC-145 cells, TAT-Nb6 treatment significantly inhibited the expression of PRRSV Nsp9, synthesis of PRRSV RNA, and production of progeny viruses in a dose-dependent manner (Fig. 6). Moreover, the data showed that PRRSV replication, especially viral RNA synthesis, was more efficiently inhibited in PAMs than in MARC-145 cells. At 36 hpi, the TAT-Nb6 treatment at a lower concentration of 10 μM also significantly reduced the production of viral RNA, Nsp9, and the progeny viruses.
FIG 6.
Inhibition of PRRSV SD16 infection and replication by TAT-NB6 in PAMs. PAMs were infected with SD16 at an MOI of 0.01 for 1 h, and then the cell culture media were replaced with fresh RPMI 1640 containing 3% FBS and TAT-Nbs at the indicated concentrations. TAT-Nbs and PRRSV Nsp9 were detected at 24 hpi (A) and 36 hpi (C) by Western blotting using anti-His MAb and mouse anti-Nsp9 antiserum, respectively. Progeny virus released in the cell medium was measured by TCID50 at 24 hpi (B) and 36 hpi (D). (E) Relative levels of PRRSV RNA were detected by RT-qPCR using PRRSV Nsp9-specific primers. The GAPDH mRNA level served as an internal reference. Data are expressed as means ± SD from three independent experiments. P values were calculated using ANOVA as <0.05 (*), <0.01 (**), and <0.001 (***) compared with cells infected with PRRSV alone (ns, not significant).
TAT-Nb6 inhibits the replication of multiple PRRSV strains.
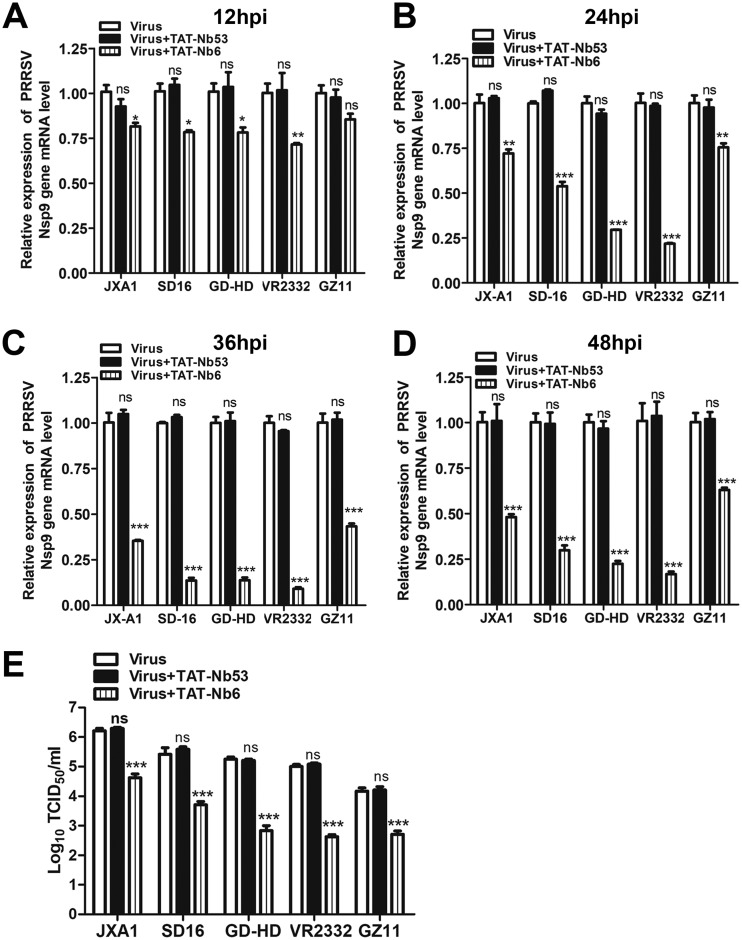
In our previous study, we found that intracellularly expressed Nb6 could significantly inhibit the replication of various PRRSV strains. To determine whether TAT-Nb6 can also suppress the replication of multiple PRRSV strains, three more genotype 2 strains (low-pathogenicity PRRSV strain VR2332 and two high-pathogenicity PRRSV [HP-PRRSV] strains, JXA1 and GD-HD) and genotype 1 strain GZ11-G1 were used to infect PAMs at an MOI of 0.01. At 1 hpi, TAT-Nb6 was added to the infected PAMs at a final concentration of 30 μM. The cells were lysed at the indicated time points, and the transcription levels of N gene were quantified by RT-qPCR. The results showed that the relative RNA levels of all strains were significantly different in all of the groups except GZ11 at 12 hpi (Fig. 7A). After 24 hpi, the RNA synthesis of all strains was predominantly suppressed by TAT-Nb6 (Fig. 7B to D), and the relative RNA levels for strains SD16, GD-HD, and VR2332 were significantly reduced compared to levels for the TAT-Nb53 group and no nanobody group (Fig. 7C and D).
FIG 7.
TAT-NB6 inhibits the replication of multiple PRRSV strains. PAMs were infected with JXA1, SD16, GD-HD, VR2332, or GZ11-G1 at an MOI of 0.01 and incubated with 30 μM TAT-NB6 or TAT-NB53. The transcription levels of PRRSV Nsp9 in the cells were detected via RT-qPCR at 12 (A), 24 (B), 36 (C), and 48 (D) hpi. (E) The titers of progeny virus in the cell culture supernatants were measured by TCID50 at 48 hpi. Data are expressed as means ± SD from three independent experiments. P values were calculated using ANOVA as <0.05 (*), <0.01 (**), and <0.001 (***) compared with cells infected with PRRSV alone (ns, not significant).
Titration of the viruses released in the cell culture revealed that TAT-Nb6 treatment suppressed progeny virus production for all strains at 48 hpi (Fig. 7E). Overall, these results demonstrated that TAT-Nb6 inhibited the replication of PRRSV genotype 1 as well as genotype 2 strains in PAMs, although the inhibition efficiencies for these strains were different.
Nb6 interacts with Nsp9 by binding to the C-terminal end of Nsp9.
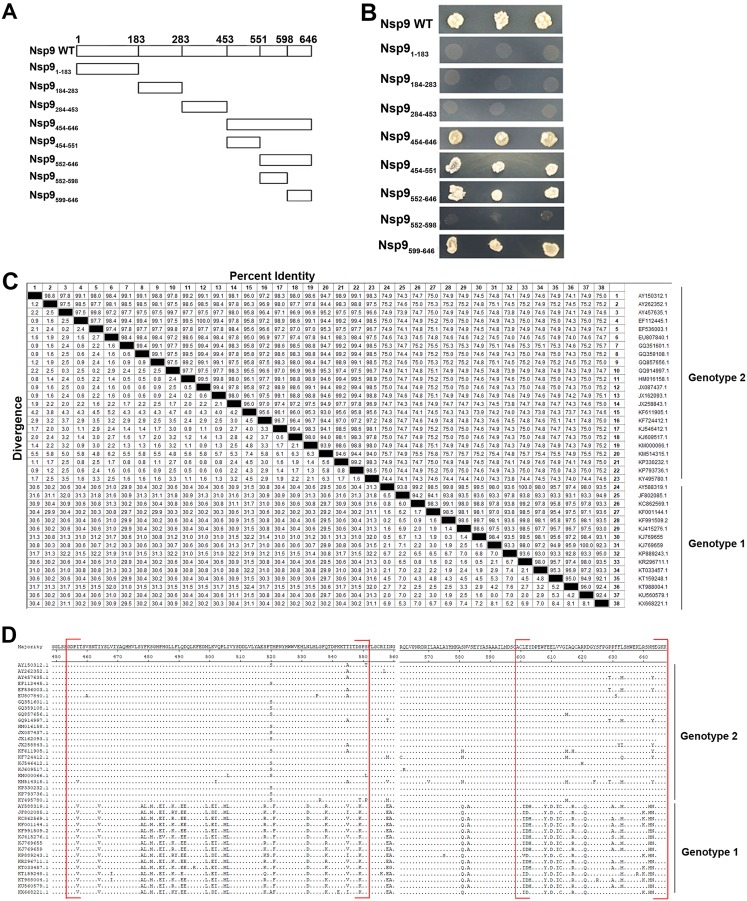
To identify the binding regions within Nsp9 involved in the Nsp9-Nb6 interaction, a yeast two-hybrid assay was performed. According to the three-dimensional (3D) structure of Nsp9 predicted in a previous report (34), the protein was truncated and expressed as four DNA binding domain-fused fragments containing coding sequences Nsp9aa1–183, Nsp9aa184–283, Nsp9aa284–453, and Nsp9aa454–646 (Fig. 8A) for the yeast two-hybrid assay. When coexpressed with the Nb6 activation domain, binding domain-fused full-length Nsp9 and Nsp9aa454–646 activated reporter expression (Fig. 8B), indicating that Nsp9 interacted with Nb6 through the Nsp9aa454–646 region. Subsequently, Nsp9aa454–646 was split into four fragments, including Nsp9aa454–551, Nsp9aa552–598, Nsp9aa599–646, and Nsp9aa552–646. The results revealed that Nb6 could bind to Nsp9aa454–551, Nsp9aa552–646, and Nsp9aa599–646 but not Nsp9aa552–598 (Fig. 8B). These results indicate that Nb6 could recognize two discontinuous regions in the C-terminal end of Nsp9, Nsp9aa454–551 and Nsp9aa599–646.
FIG 8.
Nb6 binds to the C-terminal end of Nsp9. (A) Nsp9 fragments used in this study for the identification of the Nb6-Nsp9 interaction domain. Full-length Nsp9 was truncated into four fragments in the first stage: Nsp1–183, Nsp9184–283, Nsp9284–453, and Nsp454–646. The C-terminal fragment, Nsp9454–646, was subsequently split into fragments Nsp9454–551, Nsp9552–646, Nsp9552–589, and Nsp9599–646. (B) Yeast two-hybrid assay. Vectors expressing the Nb6 protein and the indicated Nsp9 fragments were cotransformed into yeast cells. (C and D) Amino acid sequence comparison and alignment of Nsp9aa454–646. The sequences of Nsp9 from the 38 PRRSV strains in GenBank were analyzed using the Clustal W module of Lasergene 7.1 (DNASTAR, Inc.). Nb6-Nsp9 binding domains are shown in red square brackets, with dots indicating identical amino acid residues.
Based on the results shown in Fig. 7, the suppression efficiencies by TAT-Nb6 were different among the tested PRRSV strains. We speculate that the different binding abilities and, thus, inhibition efficiencies of TAT-Nb6 are attributable to the sequence variations in Nsp9aa454–646 of the PRRSV strains. In order to verify our hypothesis, amino acid sequences of Nsp9aa454–646 from 38 PRRSV strains in GenBank, including 15 genotype 1 and 23 genotype 2 strains, were aligned using the Clustal W module of Lasergene 7.1 (DNASTAR, Inc.). Alignment analysis reveals that the strains among genotype 1 or genotype 2 were highly conserved, and the sequence identity reached 92.2% to 100% and 92.7% to 100%, respectively, whereas the sequence similarity between genotypes 1 and 2 was only about 77.7 to 81.3% (Fig. 8C). Furthermore, the sequences of Nsp9aa454–551 and Nsp9aa599–646 were more variant than those of the rest of the regions on Nsp9454–646 between genotype 1 and genotype 2 (Fig. 8D). Further studies are needed to investigate whether TAT-Nb6 binds to the conserved amino acids in Nsp9aa454–551 and Nsp9aa599–646 and what factors contribute to the different inhibition efficiencies of TAT-Nb6 among the PRRSV strains.
DISCUSSION
To date, PRRS is still of great concern to the swine industry due to the inefficiency of current vaccinations and lack of efficient antiviral strategies. In our previous study, we demonstrated that an intracellularly expressed PRRSV Nsp9-specific nanobody Nb6 could dramatically inhibit PRRSV replication in MARC-145 cells (1). This finding provided motivation and insight for the development of an innovative anti-PRRSV strategy.
Nanobodies are characterized by their unique specificity, affinity, stability, solubility, and small size (35–37). Despite possessing these advantages, nanobodies are unable to physically enter cells, and their application is limited when targeting an intracellular molecule. CPPs provide an ideal means for intracellular delivery of the growing number of pharmaceutically relevant proteins. Previous studies reported that nanobodies fused with CPPs could pass through the cell membrane and interfere with hepatitis C virus replication (38–40).
Previously it was reported that TAT can deliver proteins such as glyoxalase protein and Smar protein, which were expressed as fusion proteins with TAT in prokaryotic expression system, into cells in a time- and dose-dependent manner (31, 41). Our results here demonstrated that TAT-Nb6 and TAT-Nb53 entered MARC-145 cells and PAMs in the same manner (Fig. 2). However, when transfected with various amounts of TAT-Nb6 or 20 μM TAT-Nb53, the MARC-145 cells give two peaks in FCM analysis (Fig. 2E). We speculate that these cells were in different cell cycle stages, and the TAT-Nb transduction efficiency may be influenced by the state of cells.
There are some reports that have shown that 5 amino acids (GRKKR) in TAT peptide act as a potential nuclear localization signal (29), and using a transfection system, mammalian-expressed TAT or TAT-green fluorescent protein (GFP) fusion proteins predominantly localize to the nucleus with accumulation in the nucleolus (31, 42). In addition, it was also demonstrated that E. coli-expressed TAT-GFP was able to enter various cells. However, the subcellular distribution of E. coli-expressed TAT-GFP still remains controversial. Caron et al. (43) and Yang et al. (31) reported that TAT-GFP was mainly located in the cytosol but not in the nucleus, which is in good agreement with the TAT-Nb distribution in PAMs observed in this study (Fig. 2B), whereas Han et al. demonstrated that TAT-GFP was able to be detected in both cytoplasm and nucleus (44), which is inconsistent with our results from TAT-Nb transduction in MARC-145 cells (Fig. 2B). These two different distribution patterns may be influenced by the host cell type, culture medium, or other unknown factors.
Since their discovery, CPPs have attracted the attention of researchers worldwide due to their high efficiency of cellular delivery and low toxicity. In the present study, we tested the cytotoxicity of TAT-Nb6 for MARC-145 cells and PAMs. The results reveal that TAT-Nb6 protein exhibited no toxicity at concentrations up to 30 μM, but the cell toxicity increased rapidly when the concentration was higher than 40 μM (Fig. 3). This indicates that the cytotoxicity of the TAT-tagged protein is related to its concentration, and this side effect in its clinical application should not be underrated.
Through examination of the TAT-Nb6 anti-PRRSV activity, we found that TAT-Nb6 could inhibit the replication of SD16 in both MARC-145 cells and PAMs (Fig. 5 and 6). Interestingly, it seemed that TAT-Nb6 displayed a stronger capability to suppress SD16 replication in PAMs than in MARC-145 cells (Fig. 5 to 7). We observed that the internalized TAT-Nb6 protein level in PAMs was higher than that in MARC-145 cells (Fig. 2A). We postulate that PAMs, as macrophages, are able to take in the exogenous proteins more efficiently than kidney MARC-145 cells. In addition, the internalized nanobody proteins are concentrated in the cytoplasm, where Nsp9 performs its function, and viral RNA syntheses take place in PAMs, while TAT-Nb6 dispersed in the nucleus could be ineffective for the inhibition of Nsp9. These factors may result in the better inhibition of virus replication in PAMs.
In previous studies, PRRSV N gene/protein was used to evaluate virus replication. However, we found that the expression level of N gene/protein was not always consistent with the viral titer in this study. In MARC-145 cells, the reduced ratio in viral titer at 36 hpi was lower than that at 24 hpi, but the N protein production was further reduced at 36 hpi (data not shown). Similarly, the majority of the PRRSV N gene expression was suppressed at 24 hpi and 36 hpi in PAMs treated with 30 μM TAT-Nb6 (data not shown), whereas the virus titer only decreased about 1 and 1.4 log, respectively. These results suggest that N gene or protein level is not a reliable indicator of PRRSV replication in early infection. Given that Nsp9 is the viral RdRp, the core protein of the replication and transcription complex for viral RNA synthesis, we consider that the Nsp9 gene/protein expression level will be more suitable for the reflection of PRRSV replication than N gene/protein. Here, PRRSV Nsp9 instead of N protein was used for the evaluation of the inhibitory effects of TAT-Nb6.
Nsp9, the target of Nb6, is more conserved than other PRRSV proteins (9). In our previous study, we found that intracellularly expressed Nb6 had different antiviral capabilities for different strains (7). Here, we examined the antiviral ability of TAT-Nb6 for four PRRSV type II strains and one type I strain. The results showed that TAT-Nb6 significantly inhibited the replication of all five strains, although the inhibition potency for the type II strains was higher than that for the type I strain (Fig. 7).
Using Nb6 to probe Nsp9 by Western blotting and ELISA, we have found that Nb6 does not bind to the denatured Nsp9 but only interacts with native Nsp9 (data not shown), suggesting that the nanobody recognizes a conformational epitope. Using the yeast two-hybrid assay, we showed that Nb6 bound to the C-terminal part of Nsp9, spanning two discontinuous regions: Nsp9aa454–551 and Nsp9aa599–646 (Fig. 8A and B). This result also implies that Nb6 is a conformational antibody. The amino acids in the two Nsp9 regions involved in forming the antigenic epitope remain to be investigated.
Previous reports propose that PRRSV Nsp9 protein contains a putative RdRp domain in the C-terminal portion and have identified the upstream N-terminal part as a nidovirus RdRp-associated nucleotidyltransferase domain (45, 46). The Nb6 binding regions identified in this study overlap the predicted RdRp catalytic domain (47, 48). As the RdRp of PRRSV, Nsp9 is the key enzyme for viral genome replication and transcription (46). This enzyme is also considered the core component of the transcription and replication complex that forms in viral infections, and it involves both viral and host cellular proteins. Protein interaction studies have revealed that most PRRSV nonstructural proteins (Nsp1α, Nsp1β, Nsp3, Nsp7α, Nsp7β, Nsp8, Nsp11, and Nsp12) can bind directly to Nsp9 (49, 50). PRRSV N protein also interacts with the C-terminal end of Nsp9, and Nsp9-N protein association is involved in the process of viral RNA production (34). Interactions between Nsp9 and cellular cofactors may play a role in the synthesis of viral RNA in an infected cell. For example, several studies demonstrated that the cellular retinoblastoma protein Annexin A2 and DEAD box RNA helicase 5 could bind to Nsp9, and these interactions are beneficial to PRRSV replication (51–53). Therefore, we postulate that Nb6 binding to the C terminus of Nsp9 directly blocks RdRp activity or interferes with the interaction of Nsp9 with other viral or cellular proteins, which resulted in the inhibition of PRRSV replication.
In conclusion, we report in this work that cell-penetrable nanobody TAT-Nb6, which was expressed in a prokaryotic expression system, was able to enter MARC-145 cells and PAMs in a time- and dose-dependent manner and inhibit the replication of both PRRSV genotype 1 and genotype 2 strains. Furthermore, we demonstrated that Nb6 bound to the C-terminal part of Nsp9, which contained two discontinuous regions, Nsp9454–551 and Nsp9599–646, and overlapped the predicted RdRp catalytic domain. These data indicate that TAT-Nb6 exhibits high potential to be further developed as an anti-PRRSV drug. Studies are ongoing to investigate the molecular mechanism by which Nb6 suppresses PRRSV replication and the inhibition efficiency of TAT-Nb6 on PRRSV replication in vivo.
MATERIALS AND METHODS
Cells, viruses, and reagents.
PRRSV-permissive MARC-145 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Corp., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and penicillin-streptomycin (Life Technologies Corp., Grand Island, NY, USA). Porcine alveolar macrophages (PAMs) were obtained from 6-week-old PRRSV-negative pigs using a lung lavage technique as previously described (54) and cultured in RPMI 1640 supplemented with 10% FBS and penicillin-streptomycin. All cells were cultured at 37°C with 5% CO2.
The following viral strains were used in this study: one PRRSV genotype 1 strain, GZ11-G1 (GenBank accession no. KF001144.1), and four genotype 2 strains, including HP-PRRSV strain SD16 (HP-PRRSV/SD16) (JX087437.1), HP-PRRSV/JXA1 (EF112445.1), and HP-PRRSV/GD-HD (KP793736.1), and a low-pathogenicity strain, VR-2332 (EF536003.1). All PRRSV strains were propagated and titrated in MARC-145 cells and stored at −80°C.
Production of recombinant proteins.
The gene fragments for Nb6, Nb53, TAT-Nb6, and TAT-Nb53 were generated by PCR using pEGFP-Nb6 and pEGFP-Nb53 (1) as the template and primers shown in Table 1. Nb53, which did not interact with PRRSV Nsp9 (1), was used as a negative control. The pET-21b(+) plasmid (Novagen, Darmstadt, Germany), containing the sequences for a 6-histidine (6×His) tag downstream of the gene insertion site, was used as the expression vector. The recombinant plasmids, named pET21b-Nb6, pET21b-Nb53, pET21b-TAT-Nb6, and pET21b-TAT-Nb53, were constructed by inserting the corresponding PCR fragments into pET-21b(+) plasmid digested with NdeI and XhoI restriction endonucleases. The recombinant plasmids were transformed into E. coli strain trans5α, single colonies were selected, and the inserted fragments were confirmed by sequencing. For protein expression, the four plasmids were transformed into E. coli BL21(DE3), and protein expression was induced by 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37°C for 6 h. Recombinant proteins in inclusion bodies were dissolved in 8 M urea for denaturation and then purified by Ni-NTA resin (Roche, Mannheim, Germany) under denaturing conditions according to the manufacturer’s instructions. The denatured proteins were refolded by rapid dilution in base refolding buffer (880 mM l-arginine, 55 mM Tris, 21 mM NaCl, 0.88 mM KCl, pH 8.2) with 10 mM EDTA, 150 mM reduced glutathione, and 15 mM oxidized glutathione. For the subsequent cell experiments, the refolded proteins were dialyzed into 0.01 M phosphate-buffered saline (PBS), analyzed by SDS-PAGE and Western blotting, and stored at −80°C until use.
TABLE 1.
Primers used for construction of plasmid pET-21b-TAT-Nbs or pET-21b-Nbs and detecting the level of PRRSV Nsp9 gene by qPCR
| Primer name | Sequencea (5′–3′) | Purpose |
|---|---|---|
| Overlap-TAT-Nb-F1 | CCGCATATGCCTACGGTCGTAAGAAACGTCGCC | pET-21b-TAT-Nbs |
| AGCGTCGCCGTGGAGGCGGTGGCTCGGG | ||
| Overlap-TAT-Nb-F2 | CGGTGGCTCGGGCGGTGGCGGCTCGGGTGGCGG | |
| TGGTTCTCAGGTCCAACTGCAGGAG | pET-21b-Nbs | |
| Nb-F | CCGCATATGCAGGTCCAACTGCA | |
| Nb-R | CCGCTCGAG TGAGGAGACGGTG | |
| Type1-Nsp9-F | GTCTTGAGGCTGACTTGGCT | RT-qPCR |
| Type1-Nsp9-R | TTGGACACACTGGTAACGGG | |
| Type2-Nsp9-F | CCTGCAATTGTCCGCTGGTTTG | |
| Type2-Nsp9-R | GACGACAGGCCACCTCTCTTAG | |
| GAPDH-F | CCTTCCGTGTCCCTACTGCCAAC | |
| GAPDH-R | GACGCCTGCTTCACCACCTTCT |
Underlining indicates restriction sites.
Cellular uptake of TAT-Nbs.
MARC-145 cells or PAMs were seeded in 24-well plates at a density of 1 × 105 or 1 × 106 cells per well, respectively. After confluent monolayers formed, cells were incubated with serum-free culture medium containing TAT-Nb6, TAT-Nb53, Nb6, or Nb53 at concentrations of 0, 5, 10, and 20 μM in a 5% CO2 humidified incubator at 37°C. After a set period of time (0, 1, 3, 5, and 10 h), the cells were washed with PBS three times, and then TAT-Nb6 and TAT-Nb53 taken up by the cells were detected by Western blotting, immunofluorescence assay, and flow cytometry assay.
Cell viability analysis.
Cell viability was evaluated by cell counting kit-8 (CCK-8) assay (Beyotime, Nanjing, China) as described previously (55), with the following modifications. MARC-145 cells or PAMs were seeded at a density of 1 × 104 or 1 × 105 cells per well in 96-well plates and incubated in a 5% CO2 humidified incubator at 37°C. After confluent monolayers formed, cells were washed with PBS and then incubated with different concentrations of TAT-Nb6 and TAT-Nb53 in serum-free medium for 24 h. Ten μl of CCK-8 reagent was added into each well of the 96-well plate containing 100 μl fresh medium and incubated for 2 h at 37°C. The absorbance was measured at 450 nm using an Epoch microplate spectrophotometer (BioTek Instruments, Winooski, VT, USA) for the calculation of cell viability. Results are expressed as the percentage of the optical density of treated cells relative to that of the untreated control cells, which was defined as 100% viability.
Inhibition of PRRSV replication by TAT-Nb6.
MARC-145 cells or PAMs were seeded into 6-well plates at a density of 2 × 105 or 2 × 106 cells per well, respectively. The cells were infected with PRRSV at an MOI of 0.01 for 1 h. The cell culture medium then was replaced with fresh medium supplemented with TAT-Nb6 or TAT-Nb53. At 12, 24, 36, and 48 hpi, supernatants were collected for the progeny virus titration, and the cells were harvested for Western blotting and qPCR analyses.
iELISA.
To detect the function of refolded TAT-Nb6, iELISA was performed as described in previous reports (1, 56). Briefly, the 96-well microtiter plates were coated with Nsp9-His recombinant protein (400 ng/well). After incubation with TAT-Nb6, mouse anti-camel serum (diluted to 1:2,000; prepared in our laboratory) was used to detect TAT-Nb6, and then horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) was added with a dilution of 1:5,000 to detect the bound antibodies. Color was developed using tetramethylbenzidine (TMB; Sigma Chenical Co., St. Louis, MO, USA), and the reaction was stopped with 3 M H2SO4. The optical density at 450 nm was read using an automated ELISA plate reader (Bio-Rad, Hercules, CA, USA).
IFA.
IFA was performed as previously described (57), with the following modifications. The cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 10 min at room temperature, washed three times with PBS, and then permeated with 0.2% Triton X-100 (Sigma-Aldrich) for 10 min. After being blocked with 1% bovine serum albumin (BSA) for 30 min at room temperature, the cells were incubated with mouse anti-His antibody (1:1,000; Tiangen, Beijing, China) for 1 h at room temperature and washed three times with PBS. Incubations with Alexa Fluor 488- or Alexa Fluor 555-conjugated goat anti-mouse IgG (H+L) (1:500; ThermoFisher Scientific, USA) were carried out for 1 h at room temperature, followed by three washes. Prior to mounting, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature. Coverslips were mounted on microscope slides using Fluoro-Gel, and the stained cells were observed under a confocal microscope (AF6000; Leica, Wetzlar, Germany). Mock-infected cells were used as controls to assess background staining.
Flow cytometry assay.
FCM was performed by following the procedure described by the manufacturer (Invitrogen, CA, USA). Briefly, 106 cells were collected by 0.25% trypsin digestion. After three washes with PBS, the cells were incubated with 1 ml fixation/permeabilization working solution at 4°C for 60 min, followed by washing three times with permeabilization buffer. The cells were blocked with 1% BSA for 1 h at 37°C and then incubated with mouse anti-His antibody diluted 1:1,000 for 1.5 h at 37°C. After washing, allophycocyanin-conjugated goat anti-mouse IgG (1:800; ThermoFisher Scientific, USA) was added to the cells and incubated for 1 h at room temperature. Finally, the cells were resuspended in flow cytometry staining buffer and analyzed by flow cytometry.
Quantitative real-time PCR.
Infected MARC-145 and PAMs were washed three times with PBS, and the total RNAs were extracted using TRIzol reagent (Invitrogen, CA, USA). Reverse transcription reactions were performed using a PrimeScript RT master mix kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. qPCR was carried out on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA) using RealStar green fast mixture with ROX (GenStar, Beijing, China) and type 1 or type 2 Nsp9-F/Nsp9-R primer pairs. The reaction was performed in 10 μl, and cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was measured as an internal reference. The primers used for qPCR amplification are listed in Table 1.
Virus titration.
Progeny virus production was detected by titration as previously described (58), with the following modifications. MARC-145 cells were plated on 96-well plates 24 h before viral infection. Virus supernatants were 10-fold serially diluted, and 100 μl of each was added to each well with eight replicates. Six days after infection, the 50% tissue culture infective dose (TCID50) was calculated using the Reed-Muench method (59).
Western blot analysis.
Western blotting was performed as previously described (60), with the following modifications. Briefly, cells were harvested and lysed, and the cellular proteins were separated by 15% SDS-PAGE and blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After being blocked with 2.5% skim milk for 1 h at room temperature, the PVDF membranes were probed with one of the following primary antibodies: mouse anti-6×His tag MAb (1:2,000), mouse anti-PRRSV Nsp9 antiserum (1:5,000; produced in our laboratory), and anti-α-tubulin (1:5,000; Sigma-Aldrich, St. Louis, MO, USA). Membranes were washed three times with PBST, followed by incubation with HRP-conjugated goat anti-mouse IgG at a 1:2,000 dilution as the secondary antibody. The reactions were visualized using an ECL chemiluminescent detection system according to the manufacturer's instructions (Pierce, Rockford, IL, USA).
Pulldown assay.
Pulldown assay was carried out as described previously (34). MARC-145 cells were infected with PRRSV strain SD16 at an MOI of 1 for 48 h, washed with PBS three times, and then lysed with lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.2 mM EDTA, 2 mM EGTA, 10% glycerin, 0.5% Triton X-100) containing a proteinase inhibitor cocktail (Roche, Mannheim, Germany). The cell lysates were collected by centrifugation at 14,000 × g for 15 min at 4°C and mixed with TAT-Nb6 or TAT-Nb53, and then the mixtures were incubated with PureProteome nickel magnetic beads (Millipore, USA). Antigen-bound beads were washed three times with wash buffer (pH 8.0). The bound proteins were eluted with elution buffer (pH 8.0) and analyzed by Western blotting.
Yeast two-hybrid assay.
The yeast two-hybrid assay was performed as described previously (34). The Nsp9 gene of PRRSV, including full-length Nsp9 and truncated fragments of Nsp9, and the Nb6 gene were amplified by PCR using the corresponding primers described in a previous report (34) and then inserted into pGBKT7 and pGADT7, respectively. The pGADT7-Nb6 and pGBKT7-Nsp9 constructs were cotransformed into Saccharomyces cerevisiae (yeast) strain Gold using the Yeastmaker yeast transformation system kit (Clontech, USA) as described by the manufacturer. Each interaction assay was repeated in at least three independent experiments.
Statistical analysis.
All experiments were performed with at least three independent replicates. The results were analyzed using Student's t test for comparisons between two groups or one-way analysis of variance (ANOVA) if more groups were compared. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Key R&D Program of China (grants 2017YFD0501006 and 2016YFD05007) and National Natural Science Foundation of China (grant 31430084).
No animals were used in this study.
REFERENCES
- 1.Liu H, Wang Y, Duan H, Zhang A, Liang C, Gao J, Zhang C, Huang B, Li Q, Li N, Xiao S, Zhou E-M. 2015. An intracellularly expressed Nsp9-specific nanobody in MARC-145 cells inhibits porcine reproductive and respiratory syndrome virus replication. Vet Microbiol 181:252–260. doi: 10.1016/j.vetmic.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Li N, Du T, Yan Y, Zhang A, Gao J, Hou G, Xiao S, Zhou EM. 2016. MicroRNA let-7f-5p inhibits porcine reproductive and respiratory syndrome virus by targeting MYH9. Sci Rep 6:34332. doi: 10.1038/srep34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang A, Zhao L, Li N, Duan H, Liu H, Pu F, Zhang G, Zhou EM, Xiao S. 2017. Carbon monoxide inhibits porcine reproductive and respiratory syndrome virus replication by the cyclic GMP/protein kinase G and NF-kappaB signaling pathway. J Virol 91:e01866-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Y, Du T, Shi Y, Zhang A, Zhang C, Diao Y, Jin G, Zhou E-M. 2016. Synthetic Toll-like receptor 7 ligand inhibits porcine reproductive and respiratory syndrome virus infection in primary porcine alveolar macrophages. Antiviral Res 131:9–18. doi: 10.1016/j.antiviral.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Goyal SM. 1993. Porcine reproductive and respiratory syndrome. J Vet Diagn Investig 5:656–664. doi: 10.1177/104063879300500435. [DOI] [PubMed] [Google Scholar]
- 6.Murtaugh MP, Genzow M. 2011. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 29:8192–8204. doi: 10.1016/j.vaccine.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Bao D, Wang R, Qiao S, Wan B, Wang Y, Liu M, Shi X, Guo J, Zhang G. 2013. Antibody-dependent enhancement of PRRSV infection down-modulates TNF-alpha and IFN-beta transcription in macrophages. Vet Immunol Immunopathol 156:128–134. doi: 10.1016/j.vetimm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Du T, Nan Y, Xiao S, Zhao Q, Zhou EM. 2017. Antiviral strategies against PRRSV infection. Trends Microbiol doi: 10.1016/j.tim.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Darwich L, Gimeno M, Sibila M, Diaz I, de la Torre E, Dotti S, Kuzemtseva L, Martin M, Pujols J, Mateu E. 2011. Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet Microbiol 150:49–62. doi: 10.1016/j.vetmic.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UB, Atkins JF, Snijder EJ, Posthuma CC. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol 92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Shang P, Shyu D, Carrillo C, Naraghi-Arani P, Jaing CJ, Renukaradhya GJ, Firth AE, Snijder EJ, Fang Y. 2018. Nonstructural proteins nsp2TF and nsp2N of porcine reproductive and respiratory syndrome virus (PRRSV) play important roles in suppressing host innate immune responses. Virology 517:164–176. doi: 10.1016/j.virol.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Treffers EE, Napthine S, Tas A, Zhu L, Sun Z, Bell S, Mark BL, van Veelen PA, van Hemert MJ, Firth AE, Brierley I, Snijder EJ, Fang Y. 2014. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc Natl Acad Sci U S A 111:E2172–E2181. doi: 10.1073/pnas.1321930111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 15.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 16.Van Audenhove I, Gettemans J. 2016. Nanobodies as versatile tools to understand, diagnose, visualize and treat cancer. EBioMedicine 8:40–48. doi: 10.1016/j.ebiom.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arezumand R, Alibakhshi A, Ranjbari J, Ramazani A, Muyldermans S. 2017. Nanobodies as novel agents for targeting angiogenesis in solid cancers. Front Immunol 8:1746. doi: 10.3389/fimmu.2017.01746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher D, Helma J, Schneider AFL, Leonhardt H, Hackenberger CPR. 2018. Nanobodies: chemical functionalization strategies and intracellular applications. Angew Chem Int ed Engl 57:2314–2333. doi: 10.1002/anie.201708459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Liu C, Muyldermans S. 2017. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol 8:1442. doi: 10.3389/fimmu.2017.01442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bannas P, Hambach J, Koch-Nolte F. 2017. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol 8:1603. doi: 10.3389/fimmu.2017.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey JD, Flynn NH. 2015. Cell-penetrating peptides transport therapeutics into cells. Pharmacol Ther 154:78–86. doi: 10.1016/j.pharmthera.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Mae M, Langel U. 2006. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr Opin Pharmacol 6:509–514. doi: 10.1016/j.coph.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Borrelli A, Tornesello AL, Tornesello ML, Buonaguro FM. 2018. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules 23:E295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Feng D, Zhang X, Cao D. 2015. Design strategy of cell-penetrating copolymers for high efficient drug delivery. Biomaterials 52:171–179. doi: 10.1016/j.biomaterials.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 25.Vives E, Schmidt J, Pelegrin A. 2008. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta 1786:126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Choi YS, Lee JY, Suh JS, Kwon YM, Lee SJ, Chung JK, Lee DS, Yang VC, Chung CP, Park YJ. 2010. The systemic delivery of siRNAs by a cell penetrating peptide, low molecular weight protamine. Biomaterials 31:1429–1443. doi: 10.1016/j.biomaterials.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Araki D, Takayama K, Inoue M, Watanabe T, Kumon H, Futaki S, Matsui H, Tomizawa K. 2010. Cell-penetrating D-isomer peptides of p53 C-terminus: long-term inhibitory effect on the growth of bladder cancer. Urology 75:813–819. doi: 10.1016/j.urology.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Liu BR, Lin MD, Chiang HJ, Lee HJ. 2012. Arginine-rich cell-penetrating peptides deliver gene into living human cells. Gene 505:37–45. doi: 10.1016/j.gene.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 29.Vives E, Brodin P, Lebleu B. 1997. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem 272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 30.Nagahara H, Vocero-Akbani AM, Snyder EL, Ho A, Latham DG, Lissy NA, Becker-Hapak M, Ezhevsky SA, Dowdy SF. 1998. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med 4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Ma J, Song Z, Wu M. 2002. HIV-1 TAT-mediated protein transduction and subcellular localization using novel expression vectors. FEBS Lett 532:36–44. doi: 10.1016/S0014-5793(02)03624-4. [DOI] [PubMed] [Google Scholar]
- 32.Liang JF, Yang VC. 2005. Insulin-cell penetrating peptide hybrids with improved intestinal absorption efficiency. Biochem Biophys Res Commun 335:734–738. doi: 10.1016/j.bbrc.2005.07.142. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N, Guo H, Zheng W, Wang T, Ma X. 2016. Design and screening of a chimeric survivin-specific nanobody and its anticancer activities in vitro. Anticancer Drugs 27:839–847. doi: 10.1097/CAD.0000000000000394. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Tian J, Nan H, Tian M, Li Y, Xu X, Huang B, Zhou E, Hiscox JA, Chen H. 2016. Porcine reproductive and respiratory syndrome virus nucleocapsid protein interacts with Nsp9 and cellular DHX9 to regulate viral RNA synthesis. J Virol 90:5384–5398. doi: 10.1128/JVI.03216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. 2013. Nanobodies and their potential applications. Nanomedicine (Lond) 8:1013–1026. doi: 10.2217/nnm.13.86. [DOI] [PubMed] [Google Scholar]
- 36.Kijanka M, Dorresteijn B, Oliveira S, van Bergen En Henegouwen PM. 2015. Nanobody-based cancer therapy of solid tumors. Nanomedicine (Lond) 10:161–174. doi: 10.2217/nnm.14.178. [DOI] [PubMed] [Google Scholar]
- 37.Vanlandschoot P, Stortelers C, Beirnaert E, Ibanez LI, Schepens B, Depla E, Saelens X. 2011. Nanobodies: new ammunition to battle viruses. Antiviral Res 92:389–407. doi: 10.1016/j.antiviral.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Jittavisutthikul S, Thanongsaksrikul J, Thueng-In K, Chulanetra M, Srimanote P, Seesuay W, Malik AA, Chaicumpa W. 2015. Humanized-VHH transbodies that inhibit HCV protease and replication. Viruses 7:2030–2056. doi: 10.3390/v7042030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thueng-In K, Thanongsaksrikul J, Jittavisutthikul S, Seesuay W, Chulanetra M, Sakolvaree Y, Srimanote P, Chaicumpa W. 2014. Interference of HCV replication by cell penetrable human monoclonal scFv specific to NS5B polymerase. MAbs 6:1327–1339. doi: 10.4161/mabs.29978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thueng-In K, Thanongsaksrikul J, Srimanote P, Bangphoomi K, Poungpair O, Maneewatch S, Choowongkomon K, Chaicumpa W. 2012. Cell penetrable humanized-VH/V(H)H that inhibit RNA dependent RNA polymerase (NS5B) of HCV. PLoS One 7:e49254. doi: 10.1371/journal.pone.0049254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin MJ, Kim DW, Lee YP, Ahn EH, Jo HS, Kim DS, Kwon OS, Kang TC, Cho YJ, Park J, Eum WS, Choi SY. 2014. Tat-glyoxalase protein inhibits against ischemic neuronal cell damage and ameliorates ischemic injury. Free Radic Biol Med 67:195–210. doi: 10.1016/j.freeradbiomed.2013.10.815. [DOI] [PubMed] [Google Scholar]
- 42.Hauber J, Malim MH, Cullen BR. 1989. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol 63:1181–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caron NJ, Torrente Y, Camirand G, Bujold M, Chapdelaine P, Leriche K, Bresolin N, Tremblay JP. 2001. Intracellular delivery of a Tat-eGFP fusion protein into muscle cells. Mol Ther 3:310–318. doi: 10.1006/mthe.2001.0279. [DOI] [PubMed] [Google Scholar]
- 44.Han K, Jeon MJ, Kim KA, Park J, Choi SY. 2000. Efficient intracellular delivery of GFP by homeodomains of Drosophila Fushi-tarazu and Engrailed proteins. Mol Cells 10:728–732. doi: 10.1007/s100590000036. [DOI] [PubMed] [Google Scholar]
- 45.Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GM, Ruben M, Overkleeft HS, van Veelen PA, Samborskiy DV, Kravchenko AA, Leontovich AM, Sidorov IA, Snijder EJ, Posthuma CC, Gorbalenya AE. 2015. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res 43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beerens N, Selisko B, Ricagno S, Imbert I, van der Zanden L, Snijder EJ, Canard B. 2007. De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J Virol 81:8384–8395. doi: 10.1128/JVI.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie J, Zhou H, Cui J, Chen Y, Zhang M, Deng S, Zhou P, Su S, Zhang G. 2014. Inhibition of porcine reproductive and respiratory syndrome virus by specific siRNA targeting Nsp9 gene. Infect Genet Evol 28:64–70. doi: 10.1016/j.meegid.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Xu X, Tao H, Li Y, Nan H, Wang Y, Tian M, Chen H. 2017. Structural analysis of porcine reproductive and respiratory syndrome virus non-structural protein 7alpha (NSP7alpha) and identification of its interaction with NSP9. Front Microbiol 8:853. doi: 10.3389/fmicb.2017.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nan H, Lan J, Tian M, Dong S, Tian J, Liu L, Xu X, Chen H. 2018. The network of interactions among porcine reproductive and respiratory syndrome virus non-structural proteins. Front Microbiol 9:970. doi: 10.3389/fmicb.2018.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong J, Zhang N, Ge X, Zhou L, Guo X, Yang H. 2014. The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology 464–465:432–440. doi: 10.1016/j.virol.2014.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Guo D, Huang L, Yin M, Liu Q, Wang Y, Yang C, Liu Y, Zhang L, Tian Z, Cai X, Yu L, Weng C. 2014. The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res 189:106–113. doi: 10.1016/j.virusres.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 53.Zhao S, Ge X, Wang X, Liu A, Guo X, Zhou L, Yu K, Yang H. 2015. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res 195:217–224. doi: 10.1016/j.virusres.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 55.Xiao S, Zhang A, Zhang C, Ni H, Gao J, Wang C, Zhao Q, Wang X, Wang X, Ma C, Liu H, Li N, Mu Y, Sun Y, Zhang G, Hiscox JA, Hsu WH, Zhou EM. 2014. Heme oxygenase-1 acts as an antiviral factor for porcine reproductive and respiratory syndrome virus infection and over-expression inhibits virus replication in vitro. Antiviral Res 110:60–69. doi: 10.1016/j.antiviral.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Liu B, Sun Y, Li H, Du T, Nan Y, Hiscox JA, Zhou EM, Zhao Q. 2018. Characterization of three novel linear neutralizing B-cell epitopes in the capsid protein of swine hepatitis E virus. J Virol 92:e00251-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J, Xiao S, Xiao Y, Wang X, Zhang C, Zhao Q, Nan Y, Huang B, Liu H, Liu N, Lv J, Du T, Sun Y, Mu Y, Wang G, Syed SF, Zhang G, Hiscox JA, Goodfellow I, Zhou EM. 2016. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Sci Rep 6:25120. doi: 10.1038/srep25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao S, Wang Q, Gao J, Wang L, He Z, Mo D, Liu X, Chen Y. 2011. Inhibition of highly pathogenic PRRSV replication in MARC-145 cells by artificial microRNAs. Virol J 8:491. doi: 10.1186/1743-422X-8-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pizzi M. 1950. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum Biol 22:151–190. [PubMed] [Google Scholar]
- 60.Zhang A, Duan H, Li N, Zhao L, Pu F, Huang B, Wu C, Nan Y, Du T, Mu Y, Zhao Q, Sun Y, Zhang G, Hiscox JA, Zhou EM, Xiao S. 2017. Heme oxygenase-1 metabolite biliverdin, not iron, inhibits porcine reproductive and respiratory syndrome virus replication. Free Radic Biol Med 102:149–161. doi: 10.1016/j.freeradbiomed.2016.11.044. [DOI] [PubMed] [Google Scholar]