Human parechovirus 3 (HPeV3) is receiving increasing attention as a prevalent cause of sepsis-like symptoms in neonates, for which, despite the severity of disease, there are no effective treatments available. Structural and molecular insights into virus neutralization are urgently needed, especially as clinical cases are on the rise. Toward this goal, we present the first structure of HPeV3 in complex with fragments from a neutralizing monoclonal antibody. At high resolution, it was possible to precisely define the epitope that, when targeted, prevents virions from binding to cells. Such an atomic-level description is useful for understanding host-pathogen interactions and viral pathogenesis mechanisms and for finding potential cures for infection and disease.
KEYWORDS: cryo-EM, genome packaging, human parechovirus 3, neutralizing antibodies, picornavirus
ABSTRACT
Human parechovirus 3 (HPeV3) infection is associated with sepsis characterized by significant immune activation and subsequent tissue damage in neonates. Strategies to limit infection have been unsuccessful due to inadequate molecular diagnostic tools for early detection and the lack of a vaccine or specific antiviral therapy. Toward the latter, we present a 2.8-Å-resolution structure of HPeV3 in complex with fragments from a neutralizing human monoclonal antibody, AT12-015, using cryo-electron microscopy (cryo-EM) and image reconstruction. Modeling revealed that the epitope extends across neighboring asymmetric units with contributions from capsid proteins VP0, VP1, and VP3. Antibody decoration was found to block binding of HPeV3 to cultured cells. Additionally, at high resolution, it was possible to model a stretch of RNA inside the virion and, from this, identify the key features that drive and stabilize protein-RNA association during assembly.
IMPORTANCE Human parechovirus 3 (HPeV3) is receiving increasing attention as a prevalent cause of sepsis-like symptoms in neonates, for which, despite the severity of disease, there are no effective treatments available. Structural and molecular insights into virus neutralization are urgently needed, especially as clinical cases are on the rise. Toward this goal, we present the first structure of HPeV3 in complex with fragments from a neutralizing monoclonal antibody. At high resolution, it was possible to precisely define the epitope that, when targeted, prevents virions from binding to cells. Such an atomic-level description is useful for understanding host-pathogen interactions and viral pathogenesis mechanisms and for finding potential cures for infection and disease.
INTRODUCTION
Human parechovirus 3 (HPeV3) is a small, nonenveloped, single-stranded positive-sense RNA virus belonging to the Parechovirus genus of Picornaviridae, which currently includes 19 genotypes most commonly associated with mild gastrointestinal and respiratory illness (1, 2). The increased availability of sequence data in clinical settings has clarified that HPeV3 causes the most virulent infections of the HPeVs, particularly in infants less than 3 months of age where sickness can trigger a sepsis-like dysregulated host response often involving the central nervous system (3–9). In cases of acute meningitis or encephalitis where patients may develop abnormal white matter lesions, neurological sequelae and even death may occur (10–15). To date, no effective treatments for HPeV3 infection are available, highlighting the urgent need for a greater understanding of the structural and molecular basis for HPeV3 neutralization, especially as epidemics are likely to continue (2, 16–18).
The HPeV3 virion is composed of 60 copies of the three structural proteins (VP0, VP1, and VP3) that fit together to form a 28-nm-diameter icosahedral shell around the ∼7.3-kb single-stranded RNA viral genome (19). The genome encodes a single polyprotein that, during infection, is subsequently cleaved into all the essential capsid components and replication proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) (20). Exactly how the HPeV3 particle gets assembled is poorly understood and research is ongoing.
In the case of HPeV1, recent work has shown that newly synthesized viral RNA contains ∼60 spatially defined conserved sequence/structure GXUXUXXU motifs that bind capsid proteins, driving genome encapsidation and efficient capsid self-assembly (21, 22). Assembled capsids lack cleavage of VP0 into VP2 and VP4 products, resulting in a shell made of multiple copies of three proteins rather than the four found in most other picornaviruses. These three proteins that constitute the asymmetric unit are incorporated into pentameric modules, 12 of which form the complete icosahedral capsid of the virus. Around each of the pentamers there is a depression referred to as the canyon. The tips of the 3-fold symmetric propeller-like protrusions are adjacent to this canyon.
The VP1 C termini of several human parechoviruses (e.g., HPeV1, -2, -4, and -5) contain arginine-glycine-aspartic acid (RGD) motifs that can attach to αVβ1, αVβ3, and αVβ6 integrin receptors (23, 24). HPeV3 lacks the RGD motif and thus likely uses a different, as yet unknown, receptor for cell entry (25). Reliance on a different receptor may alter tissue tropism and might explain why HPeV3 infections have different clinical and epidemiological features than other HPeV genotypes.
Human monoclonal antibodies (MAbs) can be exploited to gain valuable insights into the structural basis for neutralizing activity, which in turn can be used for developing effective treatments. High-resolution mapping of MAb binding sites at the HPeV3 capsid surface allows for identification of epitopes recognized by the humoral immune system and may begin to provide mechanistic clues into immune surveillance, evasion, or escape. Here, using high-resolution cryo-electron microscopy (cryo-EM), we define such an epitope, which when targeted by a human monoclonal antibody, blocks attachment of virions to host cells, hence also describing a potential site for receptor binding.
(This article was submitted to an online preprint archive [26].)
RESULTS
Cryo-EM structure of the HPeV3-Fab AT12-015 complex.
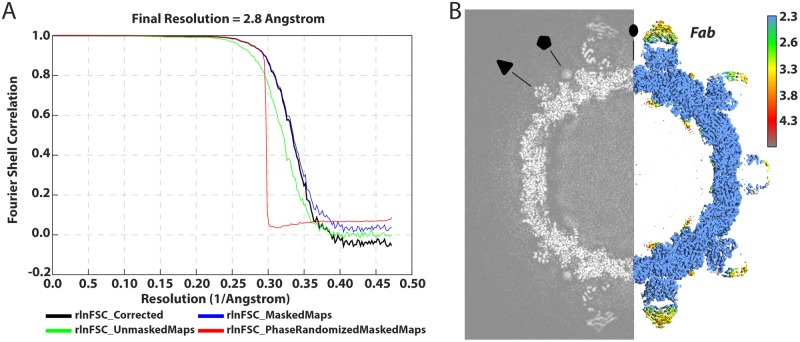
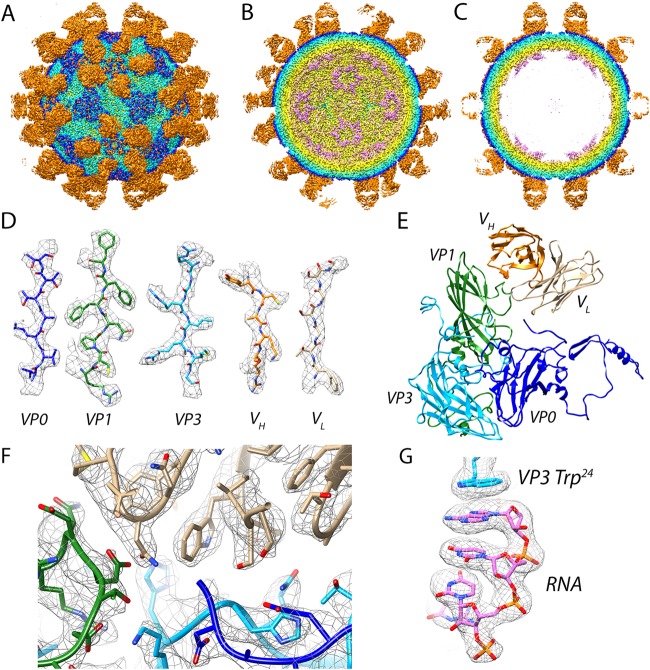
Cryo-grids containing vitrified Fab-labeled virus were imaged, and after data processing in RELION, a total of 74,927 particle projections were selected, which yielded a three-dimensional (3D) reconstruction extending to 2.8-Å resolution according to the gold-standard Fourier shell correlation 0.143 criterion (Table 1 and Fig. 1A) (27). Fab decoration on the capsid surface helped to assign particle orientations during data processing. The capsid was resolved to 2.3 Å, whereas small stretches of capsid-associated RNA and peripheral regions of Fab were defined at a resolution lower than 3.5 Å (Fig. 1B). In the structure, Fab molecules bound to symmetry-related sites at the tips of the propellers on the surface of the virion (Fig. 2A to C). For fitting, large regions of the map showed clear delineation of secondary structural elements, including amino acid and nucleic acid densities (Fig. 2D to G). In this manner, we accurately mapped the antibody footprint on the capsid surface as well as visualized an RNA base-stacking motif that stably anchors the genome to the capsid via interaction with a tryptophan residue (Trp 24) in the HPeV3 VP3 coat protein (Fig. 2F and G).
TABLE 1.
Summary of cryo-EM data collection, refinement, and validation statistics
| HPeV3-Fab complex data categorya | Value |
|---|---|
| Data collection | |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2 · s) | 48 |
| Pixel size (Å) | 1.06 |
| No. of micrographs | 6,541 |
| Reconstruction | |
| No. of particles | 74,927 |
| B factor (Å2) | −70 |
| FSC threshold | 0.143 |
| Resolution (Å) | 2.8 |
| Model building | |
| VP0 (amino acid coverage) | 34–289 |
| VP1 (amino acid coverage) | 25–219 |
| VP3 (amino acid coverage) | 16–256 |
| vRNA (nt) | 8 |
| VH | 2–119 |
| VL | 2–109 |
| Model validation | |
| MolProbity score | 1.55/96th percentile (protein) |
| Ramachandran outliers (%) | 0.93 (protein) |
| Poor rotamers (%) | 3.91 (protein) |
| Clashscore | 0 (protein), 4.35 (vRNA) |
FSC, Fourier shell correlation; nt, nucleotide; VH, Fab heavy chain; VL, Fab light chain.
FIG 1.
Resolution assessment for the HPeV3-Fab complex. (A) “Gold standard” Fourier shell correlation (FSC) curves showing an overall nominal resolution at 2.8 Å. (B) Central cross-section of the three-dimensional density map alongside the structure colored according to local resolution (Å); 2-, 3-, and 5-fold symmetry axes are labeled by the oval, triangle, and pentagon, respectively.
FIG 2.
Visualization of Fab-decorated HPeV3. Virus-Fab reconstruction shown radially color coded (purple, 100 Å; yellow, 120 Å; light blue, 140 Å; blue, 145 Å; and orange, 150 Å). (A) Surface view down a 2-fold axis of symmetry. Propeller regions can be seen as a dark blue triangles on the capsid surface, the Fabs are orange. (B) Cutaway view showing RNA (purple) at 5-fold vertices inside the viral capsid. (C) Central cross-section with well-defined layers of density corresponding to RNA, capsid, and Fab. (D) Side chains for viral coat proteins VP0 (blue), VP1 (green), and VP3 (light blue), as well as Fab heavy and light chains, VH (orange) and VL (light brown), respectively. (E) Modeled asymmetric unit with a Fab molecule bound. (F) High resolution at the Fab AT12-015-HPeV3 interface. (G) RNA anchoring on the inner surface of the virus is mediated by a tryptophan (Trp 24) residue from VP3.
Characterization of the Fab AT12-015 epitope.
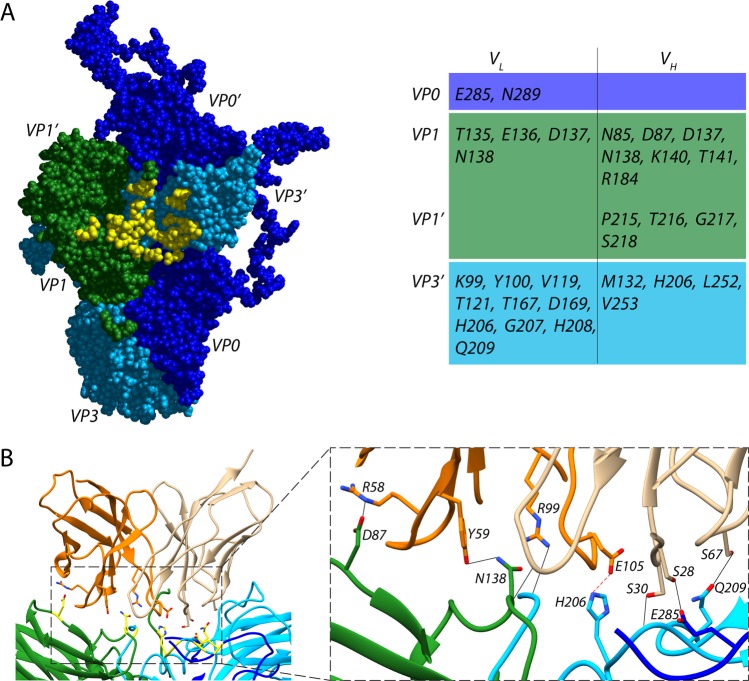
In the reconstruction, the signal related to the Fab heavy and light chains is roughly as strong as that of the viral capsid, indicating 100% occupancy of the 60 available binding sites on the virion (Fig. 1B). The quality of the map was such that we were able to fit atomic coordinates for the Fab, as well as for the three viral coat proteins, and this was followed by molecular dynamics flexible fitting (MDFF) all-atom refinement. Results from flexible fitting revealed that the Fab targets an extended solvent-accessible VP0-VP1-VP1′-VP3′ (′ denotes a neighboring asymmetric unit) conformational epitope. There is no evidence of induced structural changes in any of the capsid proteins upon Fab binding based on a comparison to the crystal structure of HPeV1 (root mean square deviation [RMSD], 0.6) (21). Using a 3.6-Å-distance cutoff, we identified 28 capsid residues forming the epitope and 29 Fab residues that form the paratope (Fig. 3A). Residues from the capsid that are involved in forming the immune complex are conserved among HPeV3 strains but not for HPeV1 or other parechovirus types. Six hydrogen bonds at the interface were identified, including three from the heavy chain (Arg 58 to VP1 residue Asp 87, Tyr 59 to VP1 residue Asn 138, and Arg 99 to VP3 residue Leu 252) and three from the light chain (Ser 28 to VP0 residue Glu 285, Asn 93 to VP1 residue Asp 137, and Ser 30 to VP3 residue Gly 207), that have angles in the range of 138° to 180° and are closely spaced to stably interact with exposed backbone nitrogen and oxygen atoms in the capsid proteins (Fig. 3B). An additional hydrogen bond may form between the Fab light chain residue Ser 67 and VP3 residue Gln 209. However, the density in this area of the map was weak; because of this, it was not possible to assess whether suitable geometric conditions were met for the interaction to occur other than the fact that the residue pair was in close proximity. One salt bridge between heavy-chain-amino-acid Glu 105 and VP3 residue His 206 was inferred by the fact that centroids of the oppositely charged functional groups of the residues were within a 4-Å cutoff, and the Glu carbonyl oxygen atom was within a 4-Å distance from the nitrogen atom of the His side chain (Fig. 3B). This His 206 is centrally located in the footprint for the antibody, and it was recently reported to be critical for binding and neutralization based on experimental selection of an antibody AT12–015-resistant HPeV3 variant (VP3 His 206 to Tyr) (28).
FIG 3.
Interactions between Fab AT12-015 and HPeV3. (A) Fab binds to an epitope extended across neighboring asymmetric units in the assembled virion. Viral capsid residues that participate in Fab heavy (VH) and light (VL) chain binding are highlighted in yellow and are reported in the accompanying table. (B) Stabilizing interactions at the interface. Residues that form hydrogen bonds or a salt bridge are highlighted in yellow on the left, and colored by chain on the enlarged inset on the right. In the inset, hydrogen bonds are shown as black dashed lines along with a centrally located salt bridge highlighted in red.
Human monoclonal antibody AT12-015 prevents HPeV3 entry into cells.
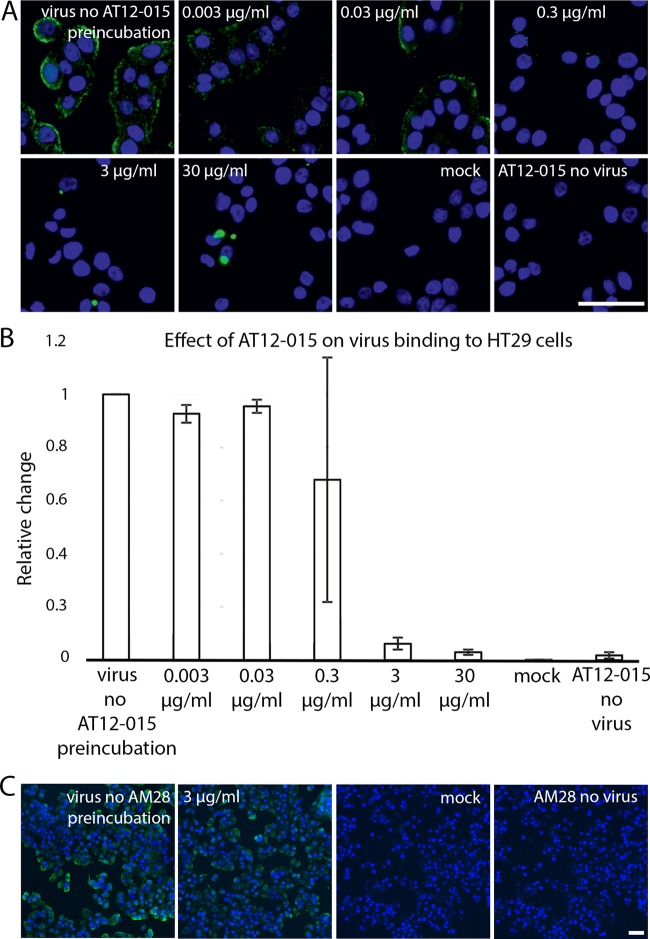
To probe HPeV3 A308/99-specific neutralization, we assayed whether binding of virions to human intestinal HT29 cells was blocked by the presence of human monoclonal antibody AT12-015 (Fig. 4). For antibody-mediated blocking, we preincubated equivalent amounts of virus with antibody at 30, 3, 0.3, 0.03, and 0.003 μg/ml final concentrations for 1 h at 37°C and then added the complexes to cells. Cellular attachment proceeded under ice-cold conditions for 1 h, and afterwards, unbound virions were removed with a series of gentle phosphate-buffered saline (PBS) wash steps. Cold binding ensures that HPeV3 remains at the surface of cells and is not internalized. When preformed antibody-decorated virions were added, either from mixing HPeV3 with the stock or 1:10 dilution of antibody, which is equivalent to 30 or 3 μg/ml of antibody, respectively, no fluorescence signal was observed on the surfaces of HT29 cells, similar to that for the mock infection (no virus) and antibody controls (Fig. 4A, 30 μg/ml, 3 μg/ml, mock, and AT12-015 no virus). Small-sized clusters of virions were observed at excess levels of antibody (Fig. 4A, 30 μg/ml and 3 μg/ml). In contrast, staining was clearly visible on the surfaces of cells incubated with HPeV3 alone as well as with higher dilutions of antibody, and fluorescence intensity was weaker at the 1:100 dilution (0.3 μg/ml), albeit with a high standard deviation (Fig. 4A and B, 0.003 μg/ml, 0.03 μg/ml, and 0.3μg/ml). Virus mixed with 1:100 of antibody showed neutralizing activity without any apparent clumping (Fig. 4B, 0.3 μg/ml). In addition, to be sure that the observed block in virus binding was due to AT12-015 and not because of premixing, we included an unrelated antibody as a control (Fig. 4C). This control antibody AM28 recognizes a conformational epitope in HPeV1 but does not bind to the capsid of HPeV3 (29). Taken together, these results indicate that AT12-015 neutralizes HPeV3 A308/99 infection at least in part extracellularly, but this does not rule out additional downstream (postattachment) inhibitory effects.
FIG 4.
Antibody AT12-015 blocks virus binding to HT29 cells. (A) Representative fluorescence images of HPeV3 incubated in the presence or absence of various amounts of AT12-015 antibody. Cell nuclei were visualized using a Hoechst stain (blue), and bound virus was scored by measuring Alexa Fluor 488 intensity (green). Pictures were acquired using a 20× objective. Scale bar, 50 μm. (B) Effect of preincubation of HPeV3 with different amounts of human monoclonal antibody AT12-015. The results are the averages from three repeats of the cold binding assay. The error bars represent the standard errors of the means (SEMs). (C) AM28 had no effect on HPeV3 binding to HT29 cells. Representative fluorescence images of HPeV3 incubated in the presence (3 μg/ml) or absence (virus no AM28 preincubation) of AM28 antibody and added to the cells for binding. Noninfected cells (mock and AM28 no virus) served as the controls. Stained as in panel A and visualized with a 10× objective. Scale bar, 50 μm.
RNA inside the HPeV3 capsid.
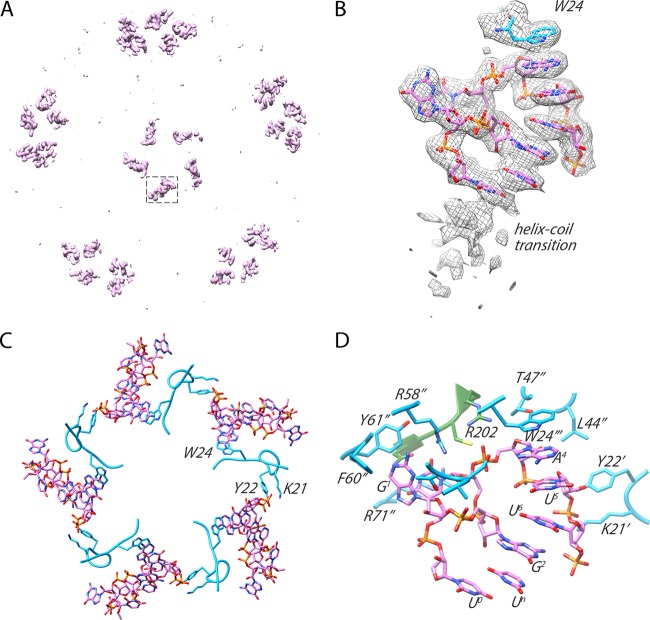
The HPeV3 virion contains ∼7.3 kb of a mostly unstructured single-stranded RNA genome. In the assembled particle, roughly 25% of the RNA adopts a defined conformation near the inner capsid surface, lining symmetry-related sites directly beneath the icosahedral 5-fold vertices (19). For parechoviruses, detailed structural analysis of genome-capsid interactions has only been carried out for HPeV1 (21). Here, we performed a similar analysis for HPeV3. The inside of our 2.8-Å HPeV3 map shows stretches of RNA in the same region as defined for HPeV1. The RNA has a defined tertiary structure, forming a single-stranded loop with a significant portion involved in a rigid base-stacking motif, capped by an aromatic side chain residue (Trp 24 of VP3) with a small conjugated π system (Fig. 5A and B). In this way, the tryptophan has stabilized its π orbitals to a resonance level with the aromatic orbitals of adjacent RNA bases to enable efficient packaging and formation of stable virions (Fig. 2G). EM density for the planar stacking profile is well resolved, on the order of 25 Å in length, before reaching a helix-coil transition (Fig. 5B). The coordinates for the six RNA nucleotides from previous structural work on the HPeV1 virion are in good agreement with those of our modeled stacking motif. We fitted eight bases of RNA beneath the capsid, which included a portion of the packaging sequence described recently for HPeV1 (22). The final sequence docked was U0-G1-G2-U3-A4-U5-U6-Un where the numbering of the last residue is uncertain. Using the RNA motif, we searched the HPeV3 A308/99 genome (GenBank code AB084913) and identified 33 sequences that contained G1-purine2-U3-purine4-U5, 13 of which include the full motif X0-G1-purine2-U3-purine4-U5-X6-X7-U8.
FIG 5.
Ordered RNA inside the HPeV3 virion. (A) RNA density segmented from within the virion seen along an icosahedral 5-fold axis. The boxed segment of the density is enlarged in panel B. (B) Eight nucleotides fit to their corresponding density before reaching the helix-coil transition. (C) VP3 tails bridge two adjacent loops of RNA to promote efficient packaging and assembly. Small portion of the capsid VP3 sequence (amino acids Leu 16 to Arg 26) is shown to clarify the VP3-RNA network on the inner surface of the viral capsid. (D) The binding pocket for RNA on the inside of the capsid involves residues from VP1 and VP3. One RNA loop is stabilized by residues from a single VP1 chain (green) and three VP3 chains (light blue designated by ′, ′′, and ′′′).
In addition to modeling the Trp 24-RNA contact in the cryo-EM structure, two other residues from the same VP3 strand, Lys 21 and Tyr 22, were found to interact with a neighboring RNA loop. These residues, along with Trp 24, in the context of the assembled pentamer, appear to be key for stabilizing the stacked RNA below the capsid vertex (Fig. 5C). In fact, mutation of two of the amino acids, either Tyr 22 or Trp 24 to alanine, is lethal in HPeV1, confirming their essential role in virion stability (22). Other capsid residues that further support the RNA loop under the vertex were identified in VP1 as well as VP3. Specifically, VP1 residues Arg 202, Cys203, and Asn 205 and VP3 residues Ala 18, Ser 19, Thr 20, Leu 44, Thr 47, Arg 58, Phe 60, Tyr 61, and Arg 71, many of which are aromatic or positively charged, form important RNA base and backbone contacts as well as complement the negative charge of the RNA (Fig. 5D). In HPeV1, mutations of VP1 residues Arg 202 and Cys 203 to Ala as well as VP3 residues Thr 44, Arg 55, and Arg 68 to Ala (VP3 residues Thr47, Arg 58, and Arg 71 in our structure) were shown to be lethal, indicating that these residues are important for stability, possibly at the level of RNA packaging and virion assembly (22).
DISCUSSION
Exact knowledge of the structural, antigenic, and immunogenic features of HPeV3 is essential for understanding host-pathogen interactions and viral pathogenesis mechanisms and for finding potential cures for infection and disease, which is pressing, as HPeV3 outbreaks are widespread and may cause severe sepsis-like syndrome in neonates. Recently, we determined a structure of HPeV3 in complex with Fab fragments from a human monoclonal antibody, AT12-015, by cryo-EM (19). However, the strain of virus used in that study, HPeV3 isolate 152037, was not neutralized by addition of AT12-015. Furthermore, the resolution of the reconstruction was only at 15 Å, which prevented atomic characterization of the epitope on the surface of the viral capsid. Here we report a 2.8-Å-resolution cryo-EM structure of an AT12-015 Fab-decorated HPeV3 virion (isolate A308/99). We have shown in a recent study that this particular virus isolate is neutralized by monoclonal antibody AT12-015 (28). In the high-resolution structure, the antigen-antibody interface was well defined, and modeling of viral coat proteins and Fab molecules into densities revealed an extended conformational epitope across the interface of adjacent asymmetric units, involving residues from different parts of neighboring VP0, VP1, and VP3 chains spatially juxtaposed by the structure of the capsid. When bound, monoclonal AT12-015 prevented virus attachment to target HT29 cells except at high dilutions of antibody, indicating that neutralization may occur extracellularly and not by a postattachment mechanism. In this way, we were able to identify the epitope of AT12-015 and determine how the antibody works against HPeV3 A308/99. In addition, the 2.8-Å map provided the most complete picture yet of ordered RNA on the inside of a human parechovirus.
Antibody AT12-015 was first isolated from the immune repertoire of a person with HPeV3 infection using the AIMSelect method, and it broadly recognizes strains of HPeV3 but only neutralizes the prototype A308/99 virus that we used in the study (19, 28, 30). At high resolution, we were able to clearly specify the conserved conformational epitope shared among HPeV3 strains, including solvent-accessible atoms in the interface region, which now includes contributions from VP0 (Glu 285 and Asn 289), VP1 (Asn 85, Asp87, Thr 135 to Asn 138, Lys 140, Thr 141, and Arg 184), VP1′ (Pro 215 to Ser 218), and VP3′ (Lys 99, Tyr 100, Val 119, Thr 121, Met 132, Thr 167, Asp169, His 206 to Gln 209, Leu 252, and Val 253) (Fig. 3A). No structural changes were induced upon Fab-virion complex formation. Atomic-level characterization brings clarity to exactly how Fab AT12-015 binds to the virion. Of particular interest is His 206 of VP3, as it was recently shown that an HPeV3 A308/99 variant mutated at this residue position to Tyr escapes neutralization by AT12-015 (28). Our results provide important context to this observation by showing that the nitrogen atom of VP3 His 206 forms a salt bridge with the carbonyl oxygen of Glu 105 in the Fab heavy chain at the center of the antibody footprint (28). Thus, we can confirm a key role for VP3 His 206 in driving Fab-virion complex formation.
Because antibodies often prevent virus attachment and entry into target cells, we tested whether AT12-015 blocks HPeV3 A308/99 binding to human intestinal HT29 cells. We found that mixing the antibody with virions under saturating conditions efficiently inhibited viral adhesion to cells, and this effect was only reversed at high dilutions of the antibody. At high antibody concentrations, the inhibitory effect may arise in part due to clustering of AT12-015-labeled virions, but specific targeting rather than generalized immune complex formation is supported by the fact that of all HPeV3 strains bound by AT12-015, only isolate A308/99 is neutralized as measured by quantitative reverse transcriptase PCR (RT-PCR) (28). Neutralization thus occurs presumably by either directly or indirectly preventing receptor engagement. To date, little is known about HPeV3 receptor and coreceptor dependencies, as it lacks the RGD motif and hence probably utilizes an uptake mechanism that differs from other parechovirus types. This might account for type-specific neutralization, and in general, usage of a different receptor by HPeV3 likely influences tropism and thus the unique disease severity in the human population.
It has become increasingly appreciated in recent years that for single-stranded RNA viruses, the life cycle is in part regulated by the secondary and tertiary structures of their genomes, hence the current high priority to understand protein-(single-stranded) RNA recognition motifs and RNA sequence-specific folding as it occurs in assembled virions (22, 31–35). Such information may help with efforts to inhibit viral propagation as well as in nanotechnology applications aimed at harnessing either virus or virus-like systems for efficient gene delivery (36). Based on our cryo-EM data, we propose a mechanism for adding HPeV3 RNA to assembling shells that utilizes π electron delocalization from VP3 side chain Trp 24 to assist with nucleotide binding. Here, at the mechanistic level, RNA folding is stabilized by a capsid-locking step where Trp 24, accessible on the inner surface of capsid protein VP3, forms a geometrically favorable short-range stacking interaction with a purine, which is then further strengthened by long-range interactions as a result of electronic resonance through further stacking of adjacent nucleotides. The 14 additional highly conserved capsid residues, namely, VP1 Arg 202, Cys 203, and Asn 205 and VP3 Ala 18, Ser 19, Thr 20, Lys 21, Tyr 22, Leu 44, Thr 47, Arg 58, Phe 60, Tyr 61, and Arg 71, help to position the ordered RNA loop against the inside of the capsid. From an evolutionary perspective, this means that over time, the inner surface of the virus has been fine-tuned to orchestrate the interaction between VP3 Trp 24 and discrete sequences of genomic RNA, and that likewise, the spacing of recognition motifs on the HPeV3 genome has been thermodynamically optimized so as to minimize the free energy of capsid assembly.
MATERIALS AND METHODS
Virus sample preparation.
Human colon adenocarcinoma (HT29) cells were propagated in McCoy’s 5A medium supplemented with 1× nonessential amino acids, 1× antibiotic-antimycotic, and 10% fetal bovine serum (FBS) with the culture condition of 37°C and 5% CO2. Cells were grown to ∼90% confluence before inoculating with human parechovirus 3 (HPeV3) isolate A308/99 at a multiplicity of infection of 0.1. HPeV3 A308/99 was grown in fresh medium as described above except that the medium was slightly modified to contain 1 mM MgCl2, 20 mM HEPES(pH 7.4), and no FBS. Inoculated cells were incubated at 37°C for 3 days. Cells and medium were harvested. At this point, the concentration of HEPES was increased to 40 mM final concentration. The cells were opened by three freeze-thaw cycles, and virus-containing medium was clarified by low-speed centrifugation. Afterwards, the supernatant was carefully removed and concentrated via ultrafiltration using Centricon units with a cutoff at 100 kDa in weight. For purification, we applied a CsCl density gradient (top, 1.2502 g · cm−3; bottom, 1.481 g · cm −3) combined with ultracentrifugation (32,000 rpm, 4°C) for 18 h in a Beckman type SW 41 Ti rotor. The virus band was collected, and the buffer was exchanged into 1× TNM buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2). Ultracentrifugation and buffer exchange were repeated. The concentration was estimated from a Coomassie-blue-stained SDS-PAGE gel, where different concentrations of bovine serum albumin (BSA) solution were used as the standards. Infectivity was measured using a 50% tissue culture infective dose (TCID50) endpoint dilution assay.
Generation of antigen-binding Fab fragment.
The HPeV3 A308/99-specific monoclonal antibody (AT12-015) was obtained from AIMM Therapeutics (the Netherlands). The AT12-015 antibody was digested to produce antigen-binding Fab fragments using the Pierce Fab micro preparation kit (Pierce). The Fab concentration was assessed by a Coomassie-blue-stained SDS-PAGE gel, where different concentrations of bovine serum albumin solution were used as the standards. For complex formation, 30 μl of 0.1 μg/μl virus and 9 μl of 0.1 μg/μl antibody were mixed (1:60 molar ratio) and incubated for 1 h at 37°C.
Cryo-EM data acquisition.
Sample volumes of 3 μl of purified HPeV3 A308/99-Fab complex were applied to glow-discharged ultrathin carbon-coated lacey 400-mesh copper grids (Ted Pella product 01824) and vitrified using a custom-made manual plunger. Cryo-grids were visualized with a FEI Titan Krios electron microscope operating at 300 kV accelerating voltage at a nominal magnification of 75,000× using a FEI Falcon II direct electron detector, corresponding to a pixel size of 1.06 Å on the specimen level. In total, 6,541 images with defocus values in the range of −0.5 to −2.5 μm were recorded in movie mode with 1 s of total acquisition time. Each movie contained 18 frames with an accumulated dose of approximately 48 electrons per Å2.
Image processing and 3D reconstruction.
Dose-fractionated image stacks containing frames from 2 to 17 were subjected to beam-induced motion correction using MotionCor2 (37). Estimation of contrast transfer function parameters for each micrograph was performed using Gctf (38). Particle selection, two-dimensional (2D) classification, and 3D classification were performed on an unbinned data set (1.06 Å/pixel, 480-pixel-box size) using RELION 2.0 (27). In total, 217,212 particle projections were selected. After reference-free 2D classification in RELION, the best classes containing 179,457 particle projections were used for further processing. An ∼10-Å reference map generated in AUTO3DEM from a modest-sized data set of 2,050 particle images collected on a FEI Tecnai TF20 cryo-electron microscope filtered to 60 Å was used for the initial maximum-likelihood-based 3D classification (39). Three classes accounting for 74,927 particles were selected for 3D refinement and reconstruction. During the postprocessing step in RELION, the map was masked with a soft mask and sharpened using a B-factor of −70 Å2. The final refinement resulted in a 2.8-Å map based on the gold-standard Fourier shell correlation 0.143 criterion. Local resolution was determined using ResMap with the unsharpened map as an input (40).
Atomic model building and refinement.
An initial atomic model for the HPeV3 A308/99-Fab complex was generated using I-TASSER and SWISS-MODEL based on the crystal structure of the HPeV1 virion (Protein Data Bank identifier [PDB ID] 4Z92) and Fab fragments of human monoclonal antibody AM28 (PDB ID 4UDF) (21, 29, 41, 42). Docking of atomic coordinates was performed manually using UCSF Chimera, and the fit was further optimized using the “Fit in Map” command (43). Inspection and further refinement were performed using Coot 0.8.8, and this served as the input for MDFF (44). The MDFF program was used together with NAMD and VMD to further enhance the fit of models into cryo-EM density (45–47). A scale factor of 1 was employed to weigh the contribution of the cryo-EM map to the overall potential function used in MDFF. Simulations included 20,000 steps of minimization and 100,000 steps of molecular dynamics under implicit solvent conditions with secondary structure restraints in place. To achieve the best fit of the model to the cryo-EM density, three iterations between Coot and MDFF were performed, with the last step being relaxation of the structure by an energy minimization step using MDFF. For hydrogen bond detection at the virus-antibody interface, we examined the structure in UCSF Chimera using a strict distance cutoff of 3.6 Å, and for geometrical constraints, we only included hydrogen bonds within the range of 138° to 180° (48, 49). The RNA-protein interface was analyzed in UCSF Chimera using the same (3.6 Å) distance cutoff.
Binding assay.
HT29 cells were seeded on 96-well plates at a density of 40,000 cells per well under the same culture conditions as during virus sample preparation. Antibody AT12-015 was incubated as stock (0.5 mg/ml) or as a dilution (1:10, 1:100, 1:1,000, and 1:10,000) with 1× CsCl-gradient-purified HPeV3, giving final antibody concentrations of 30 μg/ml, 3 μg/ml, 0.3 μg/ml, 0.03 μg/ml, and 0.003 μg/ml, respectively. As a control, an HPeV1-specific antibody, AM28, was added to virus similarly as AT12-015. Specifically, 2 μl of antibody was mixed with 2 μl of virus (8 × 105 PFU/ml) for 1 h at 37°C. The incubation took place in McCoy’s 5A medium supplemented with 1× GlutaMAX, 1× nonessential amino acids, 1× antibiotic-antimycotic, 20 mM HEPES, and 30 mM MgCl2. Plates containing cells and tubes containing antibody-virus complexes were placed on ice and allowed to cool. The growth medium of the cells was exchanged to cold-binding medium. Antibody-virus complexes, as well as either viruses or antibodies alone, were then added to cells and incubated for 1 h at an ice-cold temperature. After incubating, the cells were washed 3 times with 0.5% BSA-PBS and incubated for 1 h with primary antibody AT12-015 diluted in 0.5% BSA-PBS. After three additional washing steps and fixing (4% paraformaldehyde, 30 min), a secondary antibody was added for 1 h. The procedure up until staining with secondary antibody was completed on ice. A further series of washes was carried out, and Hoechst (1 μg/ml) was added for the visualization of cell nuclei. Wells were then washed a last time, and plates were sealed for imaging.
High content imaging and analyses.
All experiments were performed in 96-well plates (Perkin Elmer), and images were acquired using the automated fluorescence microscope CellInsight from Thermo Scientific. Image analysis was completed using CellProfiler (http://cellprofiler.org).
Accession number(s).
The final density map has been deposited in the Electron Microscopy Databank (EMDB) with accession code EMD-0069 (https://www.ebi.ac.uk/ebisearch/search.ebi?db=allebi&query=emd-0069&requestFrom=searchBox). The atomic model has been deposited in the Protein Databank (PDB) with accession code 6GV4.
ACKNOWLEDGMENTS
We thank Sergey Guryanov and Shabih Shakeel for helpful discussions. We thank Benita Löflund, Pasi Laurinmäki, Lauri Pulkkinen (University of Helsinki), and Jiri Novacek (Masaryk University) as well as Instruct-FI, the Biocenter Finland National cryo-electron microscopy and light microscopy units, Institute of Biotechnology, and the CSC-IT Center for Science Ltd. for providing technical assistance and facilities to carry out the work. We thank Hiroyuki Shimizu (National Institute of Infectious Diseases) and Katja C. Wolthers (Amsterdam Medical Center) for kindly providing HPeV3 A308/99. We thank Tim Beaumont (AIMM Therapeutics) for kindly providing the AT12-015 antibody.
This work was supported by iNEXT (project number 653706), a Horizon 2020 program of the European Union (iNEXT PID:2141), the CIISB research infrastructure project LM2015043 funded by MEYS CR (CF Cryo-electron Microscopy and Tomography CEITEC Masaryk University, Czech Republic), the Academy of Finland (275199 to S.J.B.), the Sigrid Juselius Foundation (to S.J.B.), the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under an REA grant agreement (PIEF-GA-2013-628150 to A.D.), and the Seventh Framework Programme of the European Union AIPP under contract PIAPP-GA-2013-612308 to S.J.B.
Specific author contributions are as follows: conceptualization, A.D. and S.J.B.; formal analysis, A.D.; data curation, A.D. and S.J.B.; investigation, A.D., J.W.F., and J.J.J.J.; methodology, A.D., J.W.F., J.J.J.J., J.A.G., and S.J.B.; software, J.A.G.; validation, A.D. and J.W.F.; visualization, A.D.; manuscript writing, J.W.F.; manuscript revision, A.D., J.A.G., and S.J.B.; funding acquisition, A.D. and S.J.B.; supervision, A.D. and S.J.B.; project administration, S.J.B.
We declare no competing interests.
REFERENCES
- 1.Ito M, Yamashita T, Tsuzuki H, Takeda N, Sakae K. 2004. Isolation and identification of a novel human parechovirus. J Gen Virol 85:391–398. doi: 10.1099/vir.0.19456-0. [DOI] [PubMed] [Google Scholar]
- 2.Olijve L, Jennings L, Walls T. 2018. Human parechovirus: an increasingly recognized cause of sepsis-like illness in young infants. Clin Microbiol Rev 31:e00047-17. doi: 10.1128/CMR.00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatami A, McMullan BJ, Webber M, Stewart P, Francis S, Timmers KJ, Rodas E, Druce J, Mehta B, Sloggett NA, Cumming G, Papadakis G, Kesson AM. 2015. Sepsis-like disease in infants due to human parechovirus type 3 during an outbreak in Australia. Clin Infect Dis 60:228–236. doi: 10.1093/cid/ciu784. [DOI] [PubMed] [Google Scholar]
- 4.Boivin G, Abed Y, Boucher FD. 2005. Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11:103–105. doi: 10.3201/eid1101.040606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levorson RE, Jantausch BA, Wiedermann BL, Spiegel HM, Campos JM. 2009. Human parechovirus-3 infection: emerging pathogen in neonatal sepsis. Pediatr Infect Dis J 28:545–547. doi: 10.1097/INF.0b013e318194596a. [DOI] [PubMed] [Google Scholar]
- 6.Selvarangan R, Nzabi M, Selvaraju SB, Ketter P, Carpenter C, Harrison CJ. 2011. Human parechovirus 3 causing sepsis-like illness in children from midwestern United States. Pediatr Infect Dis J 30:238–242. doi: 10.1097/INF.0b013e3181fbefc8. [DOI] [PubMed] [Google Scholar]
- 7.Wolthers KC, Benschop KS, Schinkel J, Molenkamp R, Bergevoet RM, Spijkerman IJ, Kraakman HC, Pajkrt D. 2008. Human parechoviruses as an important viral cause of sepsislike illness and meningitis in young children. Clin Infect Dis 47:358–363. doi: 10.1086/589752. [DOI] [PubMed] [Google Scholar]
- 8.Sano K, Hamada H, Hirose S, Sugiura K, Harada S, Koizumi M, Hara M, Nishijima H, Taira M, Ogura A, Ogawa T, Takanashi JI. 2018. Prevalence and characteristics of human parechovirus and enterovirus infection in febrile infants. Pediatr Int 60:142–147. doi: 10.1111/ped.13467. [DOI] [PubMed] [Google Scholar]
- 9.Harvala H, Calvert J, Van Nguyen D, Clasper L, Gadsby N, Molyneaux P, Templeton K, McWilliams Leitch C, Simmonds P. 2014. Comparison of diagnostic clinical samples and environmental sampling for enterovirus and parechovirus surveillance in Scotland, 2010 to 2012. Euro Surveill 19:20772. doi: 10.2807/1560-7917.ES2014.19.15.20772. [DOI] [PubMed] [Google Scholar]
- 10.Britton PN, Dale RC, Nissen MD, Crawford N, Elliott E, Macartney K, Khandaker G, Booy R, Jones CA, Investigators P-A. 2016. Parechovirus encephalitis and neurodevelopmental outcomes. Pediatrics 137:e20152848. doi: 10.1542/peds.2015-2848. [DOI] [PubMed] [Google Scholar]
- 11.Schuffenecker I, Javouhey E, Gillet Y, Kugener B, Billaud G, Floret D, Lina B, Morfin F. 2012. Human parechovirus infections, Lyon, France, 2008–10: evidence for severe cases. J Clin Virol 54:337–341. doi: 10.1016/j.jcv.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Sedmak G, Nix WA, Jentzen J, Haupt TE, Davis JP, Bhattacharyya S, Pallansch MA, Oberste MS. 2010. Infant deaths associated with human parechovirus infection in Wisconsin. Clin Infect Dis 50:357–361. doi: 10.1086/649863. [DOI] [PubMed] [Google Scholar]
- 13.van Zwol AL, Lequin M, Aarts-Tesselaar C, van der Eijk AA, Driessen GA, de Hoog M, Govaert P. 2009. Fatal neonatal parechovirus encephalitis. BMJ Case Rep 2009:bcr05.2009.1883. doi: 10.1136/bcr.05.2009.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verboon-Maciolek MA, Groenendaal F, Hahn CD, Hellmann J, van Loon AM, Boivin G, de Vries LS. 2008. Human parechovirus causes encephalitis with white matter injury in neonates. Ann Neurol 64:266–273. doi: 10.1002/ana.21445. [DOI] [PubMed] [Google Scholar]
- 15.Vergnano S, Kadambari S, Whalley K, Menson EN, Martinez-Alier N, Cooper M, Sanchez E, Heath PT, Lyall H. 2015. Characteristics and outcomes of human parechovirus infection in infants (2008–2012). Eur J Pediatr 174:919–924. doi: 10.1007/s00431-014-2483-3. [DOI] [PubMed] [Google Scholar]
- 16.Aizawa Y, Izumita R, Saitoh A. 2017. Human parechovirus type 3 infection: an emerging infection in neonates and young infants. J Infect Chemother 23:419–426. doi: 10.1016/j.jiac.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Esposito S, Rahamat-Langendoen J, Ascolese B, Senatore L, Castellazzi L, Niesters HG. 2014. Pediatric parechovirus infections. J Clin Virol 60:84–89. doi: 10.1016/j.jcv.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Wildenbeest JG, Harvala H, Pajkrt D, Wolthers KC. 2010. The need for treatment against human parechoviruses: how, why and when? Expert Rev Anti Infect Ther 8:1417–1429. doi: 10.1586/eri.10.130. [DOI] [PubMed] [Google Scholar]
- 19.Shakeel S, Westerhuis BM, Domanska A, Koning RI, Matadeen R, Koster AJ, Bakker AQ, Beaumont T, Wolthers KC, Butcher SJ. 2016. Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis. Nat Commun 7:11387. doi: 10.1038/ncomms11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanway G, Hyypiä T. 1999. Parechoviruses. J Virol 73:5249–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalynych S, Palkova L, Plevka P. 2016. The structure of human parechovirus 1 reveals an association of the RNA genome with the capsid. J Virol 90:1377–1386. doi: 10.1128/JVI.02346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakeel S, Dykeman EC, White SJ, Ora A, Cockburn JJB, Butcher SJ, Stockley PG, Twarock R. 2017. Genomic RNA folding mediates assembly of human parechovirus. Nat Commun 8:5. doi: 10.1038/s41467-016-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boonyakiat Y, Hughes PJ, Ghazi F, Stanway G. 2001. Arginine-glycine-aspartic acid motif is critical for human parechovirus 1 entry. J Virol 75:10000–10004. doi: 10.1128/JVI.75.20.10000-10004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seitsonen J, Susi P, Heikkilä O, Sinkovits RS, Laurinmäki P, Hyypiä T, Butcher SJ. 2010. Interaction of alphaVbeta3 and alphaVbeta6 integrins with human parechovirus 1. J Virol 84:8509–8519. doi: 10.1128/JVI.02176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Sunaidi M, Williams CH, Hughes PJ, Schnurr DP, Stanway G. 2007. Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains. J Virol 81:1013–1021. doi: 10.1128/JVI.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domanska A, Flatt JW, Jukonen JJJ, Geraets JA, Butcher SJ. 2018. 2.8 Å resolution cryo-EM structure of human parechovirus 3 in complex with Fab from a neutralizing antibody. bioRxiv doi: 10.1101/410217. [DOI] [PMC free article] [PubMed]
- 27.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karelehto E, van der Sanden S, Geraets JA, Domanska A, van der Linden L, Hoogendoorn D, Koen G, van Eijk H, Shakeel S, Beaumont T, de Jong M, Pajkrt D, Butcher SJ, Wolthers KC. 2017. Strain-dependent neutralization reveals antigenic variation of human parechovirus 3. Sci Rep 7:12075. doi: 10.1038/s41598-017-12458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shakeel S, Westerhuis BM, Ora A, Koen G, Bakker AQ, Claassen Y, Wagner K, Beaumont T, Wolthers KC, Butcher SJ. 2015. Structural basis of human parechovirus neutralization by human monoclonal antibodies. J Virol 89:9571–9580. doi: 10.1128/JVI.01429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, Radbruch A, Scheeren FA, Spits H, Beaumont T. 2010. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med 16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borodavka A, Dykeman EC, Schrimpf W, Lamb DC. 2017. Protein-mediated RNA folding governs sequence-specific interactions between rotavirus genome segments. Elife 6:e27453. doi: 10.7554/eLife.27453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borodavka A, Tuma R, Stockley PG. 2012. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc Natl Acad Sci U S A 109:15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koning RI, Gomez-Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, Carazo JM, Koster AJ. 2016. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun 7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel N, Wroblewski E, Leonov G, Phillips SEV, Tuma R, Twarock R, Stockley PG. 2017. Rewriting nature's assembly manual for a ssRNA virus. Proc Natl Acad Sci U S A 114:12255–12260. doi: 10.1073/pnas.1706951114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokoloski KJ, Nease LM, May NA, Gebhart NN, Jones CE, Morrison TE, Hardy RW. 2017. Identification of Interactions between Sindbis virus capsid protein and cytoplasmic vRNA as novel virulence determinants. PLoS Pathog 13:e1006473. doi: 10.1371/journal.ppat.1006473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butterfield GL, Lajoie MJ, Gustafson HH, Sellers DL, Nattermann U, Ellis D, Bale JB, Ke S, Lenz GH, Yehdego A, Ravichandran R, Pun SH, King NP, Baker D. 2017. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 552:415–420. doi: 10.1038/nature25157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM–an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. 2008. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker EN, Hubbard RE. 1984. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol 44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 49.Ippolito JA, Alexander RS, Christianson DW. 1990. Hydrogen bond stereochemistry in protein structure and function. J Mol Biol 215:457–471. doi: 10.1016/S0022-2836(05)80364-X. [DOI] [PubMed] [Google Scholar]