Abstract
The isolated type of orofacial cleft, termed non-syndromic cleft lip with or without cleft palate (NSCL/P), is the second most common birth defect in China, with Asians having the highest incidence in the world. NSCL/P involves multiple genes and complex interactions between genetic and environmental factors, imposing difficulty for the genetic assessment of the unborn fetus carrying multiple NSCL/P-susceptible variants. Although genome-wide association studies (GWAS) have uncovered dozens of single nucleotide polymorphism (SNP) loci in different ethnic populations, the genetic diagnostic effectiveness of these SNPs requires further experimental validation in Chinese populations before a diagnostic panel or a predictive model covering multiple SNPs can be built. In this study, we collected blood samples from control and NSCL/P infants in Han and Uyghur Chinese populations to validate the diagnostic effectiveness of 43 candidate SNPs previously detected using GWAS. We then built predictive models with the validated SNPs using different machine learning algorithms and evaluated their prediction performance. Our results showed that logistic regression had the best performance for risk assessment according to the area under curve. Notably, defective variants in MTHFR and RBP4, two genes involved in folic acid and vitamin A biosynthesis, were found to have high contributions to NSCL/P incidence based on feature importance evaluation with logistic regression. This is consistent with the notion that folic acid and vitamin A are both essential nutritional supplements for pregnant women to reduce the risk of conceiving an NSCL/P baby. Moreover, we observed a lower predictive power in Uyghur than in Han cases, likely due to differences in genetic background between these two ethnic populations. Thus, our study highlights the urgency to generate the HapMap for Uyghur population and perform resequencing-based screening of Uyghur-specific NSCL/P markers.
Keywords: Orofacial cleft, Genetic risk, Folic acid, Vitamin A, Nutritional intervention
Introduction
Orofacial cleft is a common birth defect associated with more than 300 recognizable congenital syndromes, and the isolated type of orofacial cleft, termed non-syndromic cleft lip with or without cleft palate (NSCL/P), accounts for ∼70% of affected infants based on world-wide statistics [1]. NSCL/P has been reported to occur in 0.5‰−2‰ of newborns world widely, varying by geographical origin and ethnic groups. In general, East Asians and Native Americans have the highest rates, while Caucasians and Africans exhibited the lowest rates [2]. Notably, the second largest Chinese minority population, Uyghurs, who have a mixed genetic background of both Caucasian and East Asian populations [3], show an incidence of NSCL/P (1.96‰) greater than the average national level (1.42‰) in China [4]. Newborn infants affected by congenital NSCL/P impose great economic burden for the family. Serial therapies for NSCL/P children usually span several years, which include orofacial surgery, orthodontic dentistry, speech correction, and psychological treatment, imposing a substantial financial burden on both families and society [5].
NSCL/P is a multifactorial, partially heritable, congenital disease that involves multiple genes and complex interactions between genetic and environmental cues [1]. During the human embryonic development, maxillary and medial nasal processes derived from neural crest cells fuse to form the upper lip around the first 5–7 weeks [6], and two lateral palatal processes fuse subsequently to form the palate around the 12th week [7]. The rapid proliferative expansion and complex morphogenetic events that control facial development are highly sensitive to environmental influences along with defective gene variants. Among the environmental factors, maternal nutrition has been under intensive investigation for its impact on NSCL/P, as nutrition for fetal development fully depends on food intake and the metabolic efficiency of the maternal body [8]. Several nutrients have been proven to be important in preventing NSCL/P during pregnancy, and among them folic acid and vitamin A are particularly essential [1]. Folic acid is required for the one-carbon metabolism pathway as a donor during the biosynthesis of purines and pyrimidines. It is also an indispensable factor for homocysteine re-methylation and DNA methylation, which in turn play an important role in gene expression regulation [9]. Evidence has shown that an appropriate increase in the intake dosage of folic acid for pregnant women with a familial history of NSCL/P can significantly reduce the chance of having a newborn with NSCL/P [10], [11], [12]. In addition to folic acid, supplementation with vitamin A during pregnancy is also essential for preventing numerous birth defects [13]. A lower vitamin A intake by pregnant women is associated with an increased rate of newborns with NSCL/P [14]. Along the same line, infants with NSCL/P are found to possess significantly lower levels of serum vitamin A than infants without NSCL/P [15]. However, studies using animal models indicate that an excessive vitamin A supplementation might increase the chance of NSCL/P [16]. Therefore, supplementation of vitamin A during the conceptional and peri-conceptional period at the proper dosage is highly recommended to lower the risk of NSCL/P [15].
To discover the NSCL/P-susceptible loci and genetic variants, genome-wide association studies (GWAS) have been carried out in different ethnic populations, leading to the identification of several single nucleotide polymorphism (SNP) sites showing large genetic effects on NSCL/P. For instance, the risk allele forms of the SNPs rs2235371-T and rs861020-A located within the IRF6 gene account for 12% of the genetic contribution to NSCL/P [17]. Recently, Yu et al. performed a GWAS in a Chinese population and identified 26 SNP loci associated with high risk of NSCL/P, though these loci collectively account for only 10.94% of the heritability of NSCL/P [18]. Until now, studies on NSCL/P in China have mostly focused on Han populations, and the genetic and genomic characteristics associated with NSCL/P in other populations like Uyghurs remain largely unknown. To develop a model for genetic risk assessment of NSCL/P, we collected 43 SNP markers and validated their diagnostic ability in 587 Han Chinese or Uyghur Chinese infants with or without NSCL/P recruited for the current study. We found that variations in two nutritional genes, which encode methylenetetrahydrofolate reductase (MTHFR) and retinol binding protein 4 (RBP4), respectively, play important roles in NSCL/P development.
Results
Differential allele frequencies of the 43 SNPs between Caucasian, East Asian, and Uyghur Chinese
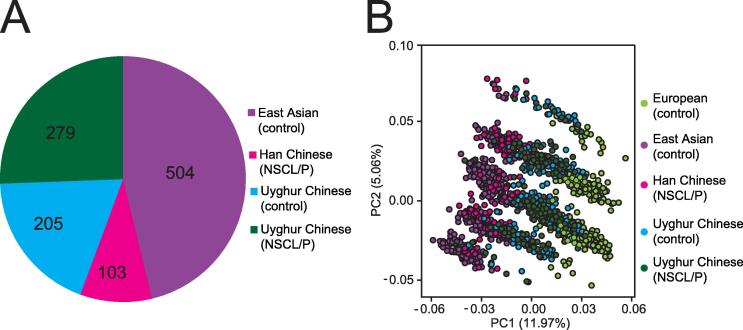
In this study, 43 SNPs associated with NSCL/P were extracted from the GWAScatalog database (https://www.ebi.ac.uk/gwas/) which integrates the significant SNPs from GWAS analyses in multiple ethnic groups. These SNPs were used as the initial candidate SNP set to examine their contributions to the risk of NSCL/P in the Chinese population. The NSCL/P sample set used in this study include 103 Han Chinese infants and 279 Uyghur Chinese infants affected by NSCL/P, with an addition of 205 normal Uyghur infants as control (Figure 1A). First we analyzed their allele frequencies in four ethnic groups, namely, European, East Asian, Han Chinese, and Uyghur Chinese populations. The principal component analysis (PCA) showed nearly identical genotypic frequencies for the 43 SNPs between the 504 East Asian individuals from the 1000-genome database (http://www.internationalgenome.org/) and the 103 Han infants diagnosed with NSCL/P, indicating that the 504 East Asians with normal phenotypes from the public database can be used as background controls for the Han Chinese subjects with NSCL/P (Figure 1B). In contrast, the European population was separated from Asian and Han Chinese populations, indicating allele frequencies of the 43 SNPs are different between European and Asian populations. Interestingly, Uyghur Chinese were present between the Asian and European populations, showing a distribution of the allele frequency between Caucasian and East Asian. Therefore, we use the 504 East Asians with normal phenotypes from the public database as the control group for the Han Chinese cases of NSCL/P, but the normal Uyghur Chinese as the control group for the Uyghur NSCL/P cases in the subsequent analyses.
Figure 1.
PCA classification of ethnic groups
A. The number of NSCL/P cases and controls included in this study. B. PCA classification exhibited different genotypic frequencies based on the 43 SNPs associated with NSCL/P collected from GWAScatalog, among 504 East Asian individuals in 1000 Genomes database, 103 Han Chinese with NSCL/P, 484 Uyghur Chinese, and 503 European individuals in 1 K genome database.
Han and Uyghur ethnic groups showed different ORs for the 43 SNPs
We defined the minor allele of a SNP with a frequency less than 0.5 according to its frequency among the 504 East Asians in the 1000 Genomes database, and then computed the minor allele frequencies (MAFs) of the 43 SNPs in the NSCL/P cases and control samples from the Han and Uyghur populations. We found that most of these SNPs showed different allele frequencies between the two populations. For instance, SNP rs1801133 in the folic acid-related gene MTHFR showed distinct allele frequencies in the control and case groups between Han (44.2%) and Uyghur (32.1%) populations. A same pattern was also seen in rs10882272, a SNP in the vitamin A-related gene RBP4, which shows different allele frequencies of 22.4% in Han Chinese and 33.0% in Uyghur Chinese (Table 1). In addition, all four SNPs in the IRF6 gene showed different frequencies of RAs between the Han and Uyghur populations. For example, the frequency of the RA of rs2235371 was 29.2% in Han Chinese with NSCL/P, while it was only 13.7% in Uyghur Chinese with NSCL/P. Furthermore, some SNPs located in the same genes are highly likely to be linked genetically, such as the rs2235371 and rs10863790 pair, and the rs861020 and rs642961 pair, located in the genic region of IRF6, which exhibits almost the same allele frequencies in both Han and Uyghur populations.
Table 1.
List of the 43 candidate SNPs examined in the current study
| SNP ID | Gene | Major | Minor | RA |
Han Chinese |
Uyghur Chinese |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR |
RA frequency |
OR |
RA frequency |
|||||||
| Control | NSCL/P | Control | NSCL/P | |||||||
| rs742071 | PAX7 | G | T | T | 1.522 | 3.3% | 5.0% | 1.244 | 17.6% | 21.0% |
| rs560426 | ABCA4 | A | G | G | 1.239 | 24.9% | 29.1% | 1.132 | 47.0% | 50.1% |
| rs2235371 | IRF6 | C | T | T | 0.545 | 43.0% | 29.2% | 0.718 | 18.1% | 13.7% |
| rs861020 | IRF6 | G | A | A | 1.509 | 19.0% | 26.2% | 1.024 | 21.9% | 22.3% |
| rs10863790 | IRF6 – DIEXF | A | C | C | 0.574 | 42.9% | 30.1% | 0.682 | 18.3% | 13.3% |
| rs642961 | IRF6 – DIEXF | G | A | A | 2.003 | 19.0% | 32.0% | 1.113 | 24.1% | 25.8% |
| rs4441471 | LOC105373445 | G | A | A | 0.985 | 26.4% | 26.2% | 0.813 | 55.8% | 50.7% |
| rs7590268 | THADA | T | G | G | 2.853 | 2.7% | 8.3% | 0.799 | 23.5% | 19.7% |
| rs3815854 | LOC105373888 | A | G | G | 0.564 | 33.7% | 22.3% | 1.184 | 39.2% | 43.7% |
| rs7632427 | EPHA3 – PROSP | T | C | C | 1.468 | 19.8% | 26.6% | 1.018 | 26.9% | 27.3% |
| rs3733585 | SLC2A9 | T | C | C | 1.198 | 41.7% | 46.2% | 1.130 | 49.3% | 52.4% |
| rs12543318 | LOC105375626 | C | A | A | 0.825 | 42.0% | 37.4% | 0.734 | 51.3% | 43.6% |
| rs987525 | GSDMC – PVT1 | C | A | A | 1.364 | 8.5% | 11.2% | 1.638 | 13.9% | 20.9% |
| rs7078160 | SHTN1 | G | A | A | 1.423 | 41.3% | 50.0% | 1.420 | 28.1% | 35.7% |
| rs9574565 | SPRY2 | C | T | T | 2.126 | 12.5% | 23.3% | 0.829 | 29.3% | 25.9% |
| rs8001641 | LOC105370275 | G | A | A | 1.964 | 13.4% | 23.4% | 1.551 | 24.9% | 34.0% |
| rs1258763 | GREM1 – FMN1 | G | A | A | 1.052 | 7.0% | 7.3% | 0.962 | 45.4% | 44.5% |
| rs1873147 | TPM1 | C | T | T | 1.302 | 14.0% | 17.5% | 0.737 | 44.7% | 37.3% |
| rs8049367 | CREBBP – ADCY9 | C | T | T | 0.98 | 33.9% | 33.4% | 0.764 | 47.0% | 40.8% |
| rs4791774 | NTN1 | A | G | G | 1.323 | 18.3% | 22.9% | 1.157 | 35.2% | 38.6% |
| rs17760296 | NOG | T | G | G | 1.847 | 0.8% | 1.5% | 1.198 | 3.2% | 3.8% |
| rs227731 | NOG | A | C | C | 1.358 | 31.9% | 38.9% | 1.097 | 34.0% | 36.1% |
| rs17085106 | LOC102724913 | G | T | T | NA | 0.0% | 8.0% | 0.624 | 4.9% | 3.2% |
| rs13041247 | LOC102724968 | T | C | C | 0.899 | 41.9% | 39.4% | 0.941 | 43.2% | 41.4% |
| rs2066836 | PTCH1 | C | T | T | 1.248 | 8.4% | 10.2% | 0.918 | 21.5% | 20.1% |
| rs1801133 | MTHFR | C | T | T | 3.391 | 29.6% | 58.8% | 1.075 | 31.3% | 32.8% |
| rs1801131 | MTHFR | A | C | C | 0.967 | 22.0% | 22.4% | 0.795 | 34.1% | 28.8% |
| rs10882272 | FFAR4/RBP4 | T | C | C | 4.189 | 10.6% | 34.1% | 1.390 | 29.2% | 36.8% |
| rs1667255 | TTR | A | C | C | 1.14 | 40.0% | 44.4% | 1.128 | 35.9% | 38.3% |
| rs2236225 | MTHFD1 | C | T | T | 1.399 | 19.9% | 25.8% | 0.941 | 39.7% | 38.2% |
| rs41268753 | GRHL3 | C | T | T | NA | 0.0% | 0.5% | 2.043 | 0.8% | 1.5% |
| rs4460498 | FOXE1 | C | T | T | 1.022 | 11.9% | 13.1% | 1.002 | 36.0% | 36.5% |
| rs13542 | ZIC2 | G | A | A | 1.383 | 26.7% | 33.5% | 0.973 | 30.5% | 30.0% |
| rs1373453 | BEST3 | T | C | C | 1.279 | 22.9% | 28.5% | 0.884 | 17.1% | 15.8% |
| rs1536895 | SMC2 | T | C | C | 1.165 | 13.2% | 16.1% | 1.003 | 10.0% | 10.4% |
| rs2294426 | ADTRP | C | T | T | 2.146 | 28.0% | 44.6% | 0.764 | 58.6% | 51.9% |
| rs4132699 | SEMA4D | A | C | C | 1.094 | 39.5% | 41.7% | 0.997 | 40.8% | 40.4% |
| rs4703516 | ACOT12 | G | T | T | 1.146 | 48.6% | 53.0% | 1.126 | 26.8% | 29.6% |
| rs5765956 | ARHGAP8 | T | C | C | 1.011 | 46.3% | 46.6% | 0.987 | 53.9% | 53.5% |
| rs7820074 | Intergenic | T | C | C | 6.455 | 37.5% | 79.5% | 0.797 | 81.8% | 78.5% |
| rs7950069 | CADM1 | A | G | G | 1.196 | 34.9% | 39.1% | 1.076 | 58.9% | 60.6% |
| rs8076457 | NTN1 | C | T | T | 3.425 | 3.3% | 11.6% | 1.021 | 15.1% | 15.7% |
| rs813218 | CMSS1 | G | A | A | 1.106 | 32.3% | 34.5% | 0.738 | 49.0% | 41.6% |
Note: Major (≥50%) and minor (<50%) alleles were defined according to the genotypic frequencies in the 504 east Asian population in the 1000 Genomes database; RA, risk allele; OR, odds ratio.
Odds ratios (ORs) represent the odds of an incidence in a case/disease group versus that in a control/normal group, with greater OR implicating greater risk of being exposed to the disease. An allele with OR > 1 is defined as a risk allele, whereas an allele with OR < 1 is protective, which may reduce the chance of passing on this disease to an infant. We thus computed the ORs for each SNP site in the two ethnic populations and defined the risk and protective role of the two alleles. Among the 43 SNPs (Table 1), 28 SNPs had ORs consistently above or below one in both the Han and Uyghur populations. For instance, the respective OR values in Han and Uyghur populations was 4.189 and 1.390 for rs10882272, which were 0.545 and 0.718 for rs2235371. Nevertheless, 13 SNPs showed an opposite trend in the two populations. For instance, the OR of rs2294426 was 2.146 in the Han population but 0.764 in the Uyghur population. It is worth noting that the minor allele of SNP rs17085106 was not found in the control Han Chinese group, while it was present in a frequency of 8.0% in Han Chinese with NSCL/P and 3.2% and 4.9% frequencies were observed in Uyghur control and NSCL/P groups, respectively.
Effects of key genes implicated by frequencies of risk alleles
The frequencies of the homozygous and heterozygous forms of the risk alleles may implicate a dominant or recessive genetic effect for each variant (Table S1). As shown in Figure S1, higher percentages subjects with homozygous genotypes for genes IRF6-DIEXF (rs642961), SLC2A9 (rs3733585), and NOG (rs227731) were found in the NSCL/P group than in controls in both Han and Uyghur populations, indicating that these alleles may be recessive to NSCL/P incidence. Conversely, higher percentage of subjects with heterozygous genotypes for gene GSDMC-PVT1 (rs987525) was found in the NSCL/P group than in the control, suggesting its potential dominant effect.
In addition, variants of some nutritional genes also showed differential allele frequencies between the NSCL/P and control groups (Figure S2). Among the Han population, the homozygous genotypes of MTHFR with the risk allele for rs1801133 (TT) and rs1801131 (CC) accounted for 36.8% and 5.8% in the NSCL/P group, respectively, significantly higher than the frequencies of 10.5% and 3.7% in the control group. Nevertheless, these two alleles in the Uyghur population showed a reversed trend, with the percentages of the homozygous genotypes in NSCL/P being lower than those in the control group (Figure S2A and B). For another folic acid metabolism-related gene, MTHFD1, the heterozygous genotypes for the risk allele accounted for 43.8% and 48.5% in the NSCL/P groups of the Han and Uyghur populations, respectively, which was slightly increased compared to 34.7% and 46.6% in the control groups (Figure S2C). Furthermore, the homozygous genotype of the vitamin A-related gene RBP4 accounted for 20.6% and 15.4% in the NSCL/P groups of the Han and Uyghur populations, respectively, in sharp contrast to the 1.5% and 9.3% in the control Han and Uyghur groups, respectively (Figure S2D). Apart from these functionally-known genes, higher percentages of subjects with genotypes homozygous for the risk alleles in the gene ADTRP (rs2294426) and the intergenic SNP rs7820074 were found in the NSCL/P group than in the control group from the Han population but lower in the Uyghur population (Figure S2E and F), similar as shown in MTHFR (rs1801133 and rs1801131).
A relative risk scoring model exhibits better discrimination resolution than an absolute scoring method
An additive linear model is commonly used to assess the genetic risk of a heritable disease by deriving a genetic risk score (GRS) based on the genotypic information of given DNA variants. In our method, the GRS model takes two forms of an OR for each SNP to compute a GRS, with either standardized absolute OR values or relative risk probabilities transformed as the ranks of ORs in the control group.
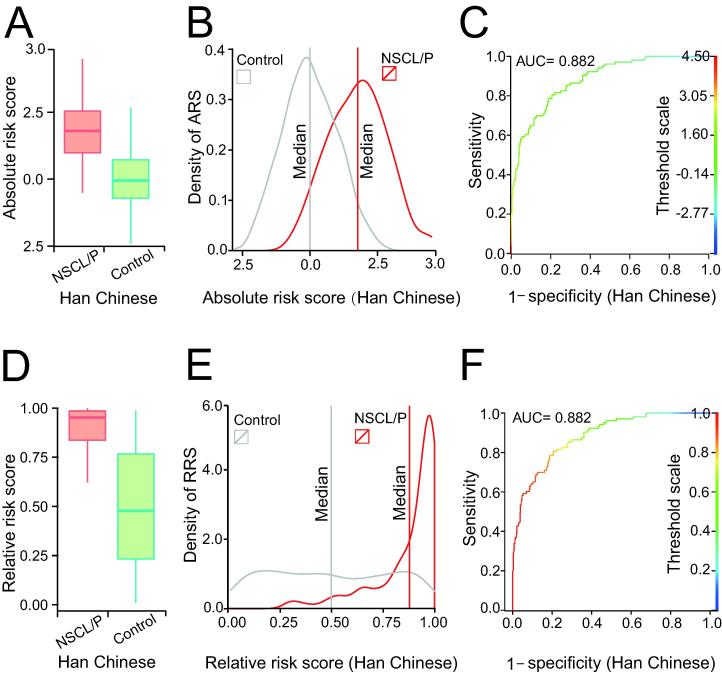
A comparison of the distributions of GRSs computed in the NSCL/P and control groups is shown in Figure 2. For Han Chinese, both absolute OR-based (Figure 2A) and ranking probability-based (Figure 2D) GRSs can clearly distinguish the NSCL/P group from controls, indicating that the model may effectively distinguish the individuals carrying multiple risk alleles that additively increase NSCL/P incidence. While the absolute value-based GRSs of the NSCL/P and control groups partially overlap (Figure 2B), the GRSs based on the relative risk probability (Figure 2E) showed a much clearer discrimination, with a median relative risk probability of 0.94 in NSCL/P versus 0.50 in the control group. In another word, if an individual was assessed with GRS > 0.9, he/she has a much higher chance of being NSCL/P than being normal. The GRS models for NSCL/P risk assessment in Han Chinese population were also evaluated by the receiver operating characteristic curve (ROC) analysis, and both had area under curve (AUC) values of 0.882 (Figure 2C and F).
Figure 2.
The genetic risk scoring models developed for the Han population
A. Boxplot of the ARS in the NSCL/P and control groups. B. Density profile of ARS in the NSCL/P and control groups. The vertical lines represent the median ARS values for the NSCL/P (red) and control (gray) groups, respectively. C. ROC curve for the model based on ARS to distinguish the NSCL/P and control groups. D. Boxplot of RRS in the NSCL/P and control groups. E. Density profile of RRS in the NSCL/P and control groups. The vertical lines represent the median RRS values for the NSCL/P (red) and control (gray) groups, respectively. F. ROC curve for the model based on RRS to distinguish the NSCL/P and control groups. ARS, absolute risk score; RRS, relative risk score; ROC, receiver operating characteristic; AUC, area under curve.
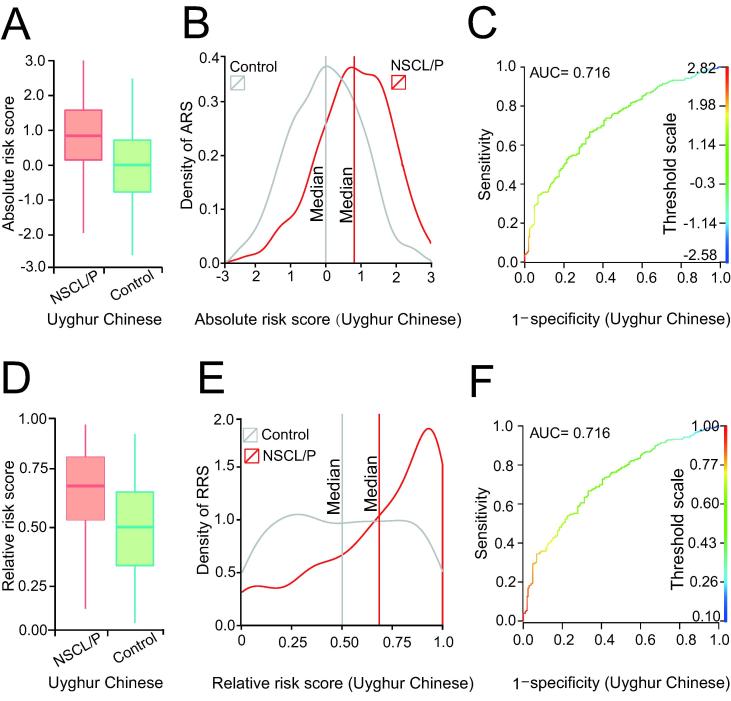
For the Uyghur Chinese population, the predictive power was relatively low (Figure 3). Although the peak value of GRS was 0.89, the lower boundary was approximately 0.7 (Figure 3E). Although AUC values of 0.716 were obtained using GRSs based on the absolute OR or ranking probability, the discrimination resolution in the Uyghur population was lower than that observed in the Chinese population (Figure 3C and F). These results indicate that if an individual was assessed to have GRS > 0.9, the false positive rate would be below 0.1; however, for a subject with GRS of 0.75, there might be a 25% chance of a false positive diagnosis.
Figure 3.
The genetic risk scoring models developed for the Uyghur population
A. Boxplot of the ARS in the NSCL/P and control groups. B. Density profile of the ARS in the NSCL/P and control groups. The vertical line represents the median of the ARS. C. The ROC for the model based on ARS to distinguish between the NSCL/P and control groups. D. Boxplot of the RRS in the NSCL/P and control groups. E. Density profile of the RRS in the NSCL/P and control group. The vertical line represents the median of the RRS. F. The ROC for the model based on RRS to distinguish between the NSCL/P and control groups.
Machine learning methods for genetic risk assessment independent of ORs
The additive models for GRS are dependent on OR values; therefore, a large training set is essential to ensure the accuracy of computed ORs. In addition, when the sampling population changes, the ORs may also change. Moreover, when calculating OR value for each SNP site, all the variants are treated as an additive genetic effect by default, without considering the dominant and recessive effects caused by allele zygosity. The machine learning methods may overcome these shortcomings, as a binary classification of susceptibility or non-susceptibility is purely based on genotypes, without the need to pre-calculate the OR value for each SNP. Therefore, we attempted to build machine learning models for genetic risk assessment.
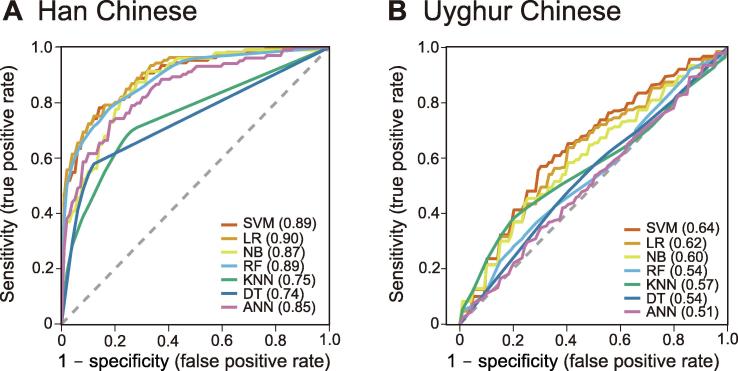
Based on the same panel of 43 SNPs, we used seven types of classical machine learning methods to evaluate their risk assessment accuracy with 10-fold cross-validation in Han and Uyghur populations. These include the support vector machine (SVM), logistic regression (LR), naive Bayesian (NB), random forest (RF), k-nearest neighbor (KNN), decision tree (DT), and artificial neural network (ANN). For the Han population, the best performance was obtained using LR (AUC = 0.90), followed by SVM (AUC = 0.89), and random forest (AUC = 0.89; Figure 4A), which were all slightly higher than the GRS additive model (AUC = 0.88; Figure 2). Similarly, we applied the same seven methods to the Uyghur population, but noticed a poorer performance than that observed in the Han population, with the best method (SVM) only having an AUC of 0.64 (Figure 4B). Although the GRS model for the Uyghur population (AUC = 0.716; Figure 3) showed a higher AUC than the best of the machine learning models.
Figure 4.
Comparison of predictive power by seven machine learning models
Seven NSCL/P risk assessment models were developed based on machine learning methods for the Han population (A) and Uyghur population (B), respectively. SVM, support vector machine; LR, logistic regression; NB, naive bayesian; RF, random forest; KNN, k-nearest neighbor; DT, decision tree; ANN, artificial neural network. The dashed line represents AUC of 0.5, and AUC values obtained using the respective method are included in the parenthesis.
Compilation of a minimum SNP set based on marker effectiveness evaluation
Based on MAFs, ORs, zygosity, discrimination ability and detection quality, we manually selected a panel of 24 SNPs in the Han population (Table 2). We scrutinized SNPs in the genes related to folic acid and vitamin A biosynthesis and metabolism, considering their potential application in nutritional intervention for lowering the risk of NSCL/P incidence.
Table 2.
24 SNP markers for machine learning-based predictive effectiveness evaluation
| No. | SNP ID | Gene | OR | Control | NSCL/P | Genotype | Class |
|---|---|---|---|---|---|---|---|
| 1 | rs1801133 | MTHFR | 3.391 | 10.5% | 36.8% | Homozygous | Risk |
| 2 | rs1801131 | MTHFR | 0.967 | 3.7% | 5.8% | Homozygous | Protective |
| 3 | rs2236225 | MTHFD1 | 1.399 | 34.7% | 43.8% | Heterozygous | Risk |
| 4 | rs10882272 | FFAR4/RBP4 | 4.189 | 1.5% | 20.6% | Homozygous | Risk |
| 5 | rs2294426 | ADTRP | 2.146 | 8.1% | 36.2% | Homozygous | Risk |
| 6 | rs7820074 | intergenic | 6.455 | 12.6% | 63.7% | Homozygous | Risk |
| 7 | rs642961 | IRF6 – DIEXF | 2.003 | 1.9% | 19.4% | Homozygous | Risk |
| 8 | rs7632427 | EPHA3 – PROSP | 1.468 | 5.5% | 17.3% | Homozygous | Risk |
| 9 | rs3733585 | SLC2A9 | 1.198 | 18.0% | 26.2% | Homozygous | Risk |
| 10 | rs7950069 | CADM1 | 1.196 | 12.6% | 16.8% | Homozygous | Risk |
| 11 | rs9574565 | SPRY2 | 2.126 | 21.6% | 40.8% | Heterozygous | Risk |
| 12 | rs8001641 | LOC105370275 | 1.964 | 24.9% | 39.0% | Heterozygous | Risk |
| 13 | rs4791774 | NTN1 | 1.323 | 29.1% | 36.0% | Heterozygous | Risk |
| 14 | rs227731 | NOG | 1.847 | 43.5% | 52.5% | Heterozygous | Risk |
| 15 | rs17085106 | LOC102724913 | 2.146 | 0.0% | 10.0% | Heterozygous | Risk |
| 16 | rs1667255 | TTR | 1.145 | 42.8% | 52.4% | Heterozygous | Risk |
| 17 | rs4703516 | ACOT12 | 1.146 | 50.1% | 61.7% | Heterozygous | Risk |
| 18 | rs7590268 | THADA | 2.853 | 5.4% | 10.7% | Heterozygous | Risk |
| 19 | rs987525 | GSDMC – PVT1 | 1.364 | 15.0% | 20.5% | Heterozygous | Risk |
| 20 | rs7078160 | SHTN1 | 1.423 | 48.1% | 53.4% | Heterozygous | Risk |
| 21 | rs10863790 | IRF6 – DIEXF | 0.574 | 19.0% | 9.7% | Homozygous | Protective |
| 22 | rs3815854 | LOC105373888 | 0.564 | 47.3% | 35.1% | Heterozygous | Protective |
| 23 | rs2235371 | IRF6 | 0.545 | 47.9% | 38.9% | Heterozygous | Protective |
| 24 | rs12543318 | LOC105375626 | 0.825 | 46.7% | 41.8% | Heterozygous | Protective |
Note: The 24 SNPs were manually selected according to a series of criteria, including OR, allele frequency, previous studies, and detection quality of SNP genotyping. An allele with OR > 1 is defined as a risk allele, whereas an allele with OR ≤ 1 is considered protective.
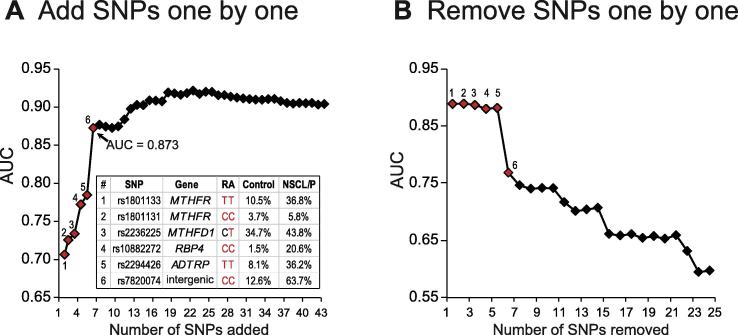
To evaluate the risk assessment efficiency of the 24 selected SNPs, we adopted a strategy of sequentially removing or adding one SNP for each instance of training or prediction using a LR model based on the order shown in Table 2. With the one-by-one adding process, we obtained AUC values of 0.778 and 0.873 using 4 and 6 SNPs, respectively. The highest AUC was obtained (0.925) when 18 SNPs were used to build the model where it then plateaued regardless of the addition of more SNPs (Figure 5A). In the same order, using a one-by-one removal approach, a remarkable decline in the AUC values from 0.87 to 0.76 was observed when the first six SNPs were removed (Figure 5B). Therefore, it appears that the first four SNPs in three genes involved in the folic acid and vitamin A biosynthesis, MTHFR, MTHFD1, and RBP4, play important roles in NSCL/P incidence.
Figure 5.
Evaluation of predictive effectiveness of the 43 SNPs using one-by-one adding and removing SNPs method in the Han population
Alterations in AUC when adding (A) or removing (B) these 24 SNP markers one by one. Numbers above the red dots indicate the ID of the SNPs associated with nutritional genes in the table. Nucleotides marked in red represent the forms of the risk alleles. The genotypic frequencies of the six SNPs are also shown in the table. RA, risk allele.
Discussion
In this study, we first examined the distribution of 43 SNPs in Han and Uyghur populations, which were previously discovered in association with NSCL/P according to GWAScatalog database collection. Then, we developed genetic risk assessment models for NSCL/P in Han Chinese population using traditional risk scoring method and machine learning methods. Compared to scoring-based method, one of the advantages of machine learning method is that it can be directly applied to the genotypes of SNPs without pre-calculation of ORs [19]. This merit of machine learning predictive model may firstly reduce the bias caused by inconsistent OR values of SNPs when populations and samples are changed, and secondly take homozygous and heterozygous forms of risk alleles into account. Among the seven models tested in our study, logistic regression model (LR) showed the best predictive performance in Han population (AUC = 0.903). However, the prediction power of LR was much less robust for the Uyghur population (AUC = 0.627), likely due to the genetic difference between Han and Uyghur Chinese. Therefore, GWAS analyses in Uyghur Chinese population and identification of NSCL/P related variants is highly demanded for risk assessment model development for Uyghur infants.
Among the 43 SNPs, four SNPs are associated with folic acid and vitamin A metabolism-related genes. These include rs1801133 (MTHFR) and rs10882272 (RBP4) whose homozygous risk alleles showing significantly higher enrichment in NSCL/P group compared to control in Han Chinese population. MTHFR encodes an enzyme that catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is the predominant circulatory form of folate and the methyl donor for the re-methylation of homocysteine to regenerate methionine [20]. Previous reports show that TT homozygous genotype of rs1801133, a missense variant causing amino acid alteration (Ala222Val), may reduce folate concentration in serum, plasma, and red blood cells [21], [22], [23], [24], [25]. Variant of rs1801133-T appears to be a risk allele specific to East Asian population, since among the Caucasian infants, the homozygous TT genotypes showed no correlation with NSCL/P [26], [27]. It is of note that homozygous TT genotype showed no correlation with NSCL/P in Uyghur Chinese population. Another missense variant of MTHFR, homozygous CC genotypes of rs1801131 causing amino acid alteration (Glu470Gly), may also reduce enzyme activity to a lesser extent than rs1801133, although it is not associated with lower plasma folate or higher homocysteine levels [28], [29]. This is consistent with our validation result that the OR value of rs1801131 is 0.967 (P = 0.858) and 0.794 (P = 0.103) in Han Chinese and Uyghur Chinese populations.
Another SNP showing high effective discrimination ability is rs10882272, which is located upstream RBP4. RBP4 encodes a plasma transport protein, which acts as a specific carrier of vitamin A and is responsible for transporting retinol from hepatic stores to peripheral target tissues in the body, including the placenta [30]. Previous proteomic studies also show that the RBP4 protein level in serum is significantly lower in infants with NSCL/P than control infants [15]. Our analysis on rs10882272 supports these previous studies, as higher percentage of subjects with homozygous risk genotype CC at rs10882272 are found in the NSCL/P group than in the control group in both Han and Uyghur Chinese populations. Therefore, both proteomics and genomics analyses suggest that appropriate vitamin A supplementation may be helpful to prevent the birth of NSCL/P infants for pregnant women who are carriers of rs10882272 risk alleles.
Addition of two functionally unknown SNPs (rs2294426 and rs7820074) to the four SNPs above can increase the prediction power from AUC = 0.761 to AUC = 0.873. The rs2294426 (homozygous risk genotype TT) and rs7820074 (homozygous risk genotype CC) with OR values of 2.146 and 6.455 indicate that both of them are risk allele in Han Chinese population. However, these two SNPs show opposite trend in Uyghur Chinese population, as the OR values (0.764 for rs2294426 and 0.797 for rs7820074) indicate that both of them are protective alleles in Uyghur population. The SNP rs2294426 at 6p24.1 is located within ADTRP. ADTRP encodes an androgen-dependent tissue factor pathway inhibitor (TFPI) regulating protein, which regulates the expression and function of TFPIs in human endothelial cells under normal conditions and in response to androgens [31], [32]. ADTRP has been reported to be associated with early-onset coronary artery disease in southern Han Chinese populations [31]. Another SNP at 8q21.3, rs7820074, was mapped to an intergenic region, without known function or overlap with any other adjacent genes. Whether this genomic locus harbors any regulatory cis-elements or encodes a non-coding RNA gene related to orofacial cleft development also requires further investigation. More lines of evidence are needed to determine whether these two functionally unknown variants are correlated with the occurrence of NSCL/P.
It’s commonly acknowledged that appropriate supplementation with folic acid and vitamin A during the women’s conception and peri-conception periods is important to reduce the risk of conceiving a NSCL/P baby. Our analysis demonstrates that four SNPs located in the genes involved in folic acid (rs1801133 and rs1801131 in MTHFR, rs2236225 in MTHFD1) and vitamin A (rs10882272 in RBP4) metabolism, account for about 76.1% (AUC = 0.761) prediction ability of the contribution to NSCL/P incidence, indicating high contribution of the variants of MTHFR and RBP4 to NSCL/P occurrence. Thus, our study provides supporting evidence that genetic diagnosis of MTHFR and RBP4 variants may help design nutritional intervention plan to minimize the occurring chance of congenital NSCL/P, especially when both parents carry the same risk alleles. Therefore, these nutritional markers have the application potential to be used as guides for nutritional intervention to reduce NSCL/P incidence in future.
Materials and methods
Subject recruitment
Patients with NSCL/P were recruited from unrelated Chinese infants attending the “Smile Train” in Yantai, Shandong (Han population) and Kashi, Xinjiang (Uyghur population) during 2015–2016. All subjects were interviewed and clinically assessed by at least two experienced clinicians. A full clinical checkup was completed to identify any further anomalies, such as congenital heart disease, hypospadias, or accessory auricles, which would suggest an underlying syndrome. Additional demographic information was obtained through a detailed questionnaire, including gender, age, nationality, and maternal lifestyle during the first trimester of pregnancy. After genetic and phenotypic quality control, 103 and 279 patients with NSCL/P from Han and Uyghur populations, respectively, were included for subsequent analysis.
Healthy individuals without an orofacial cleft or a family history of orofacial cleft were included as controls, comprising 205 Uyghurs and 504 Han Chinese. The 205 Uyghurs were recruited from the local hospitals in Urumqi, China, while the 504 controls for Han population were selected from East Asians (most participants from Beijing, China) in the 1000 Genomes database (http://www.internationalgenome.org).
This study was approved by the Ethics Committee of the Peking University, School and Hospital of Stomatology (approval No. PKUSSIRB-201520012) and was conducted according to the Helsinki Declaration of ethical principles. Written informed consent was obtained from the participants or their guardians for NSCL/P affected and normal individuals in Han and Uyghur Chinese populations.
SNP genotyping
Peripheral blood (3 ml for each person) samples were collected from all subjects. Genomic DNA was extracted from the samples using a standard DNA extraction kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The DNA concentrations were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and normalized to 5 ng/μl for each sample. A total of 43 SNPs (Table 1) that had been previously reported in five well-designed GWAS analyses [18], [33], [34], [35], [36] were selected for subsequent analysis. Genotyping was performed using KASPar chemistry (KBioscience, Hoddesdon, UK), a competitive allele-specific PCR-SNP genotyping system that use ≤ s FRET quencher cassette oligos [37], [38]. The resulting allele call data were viewed graphically as a scatterplot for each assayed marker using SNPViewer (http://www.lgcgenomics.com).
Allele definition
We first re-defined the major allele (≥0.5) and minor allele (<0.5) based on the allele frequencies in the control groups consisting 504 East Asians and 205 Uyghurs, respectively. Then, the frequencies of major and minor alleles were computed in the groups of patients with NSCL/P including 103 Han Chinese and 279 Uyghurs. OR for the two ethnic populations in each groups was calculated as shown below.
| (1) |
| (2) |
| (3) |
An allele is considered as a risk allele, if OR is greater than 1 (OR > 1), and otherwise a protective allele (OR ≤ 1).
Risk scoring
We used two methods to calculate genetic risk scores for a given genotype with an additive model. First, we computed an absolute risk score (ARS) by summing the OR values of SNPs (n = 43) as shown in Equation (4). Each OR of the SNP is multiplied by the assigned digital value of genotypes (0 for the genotype comprising 2 major alleles). Then, the same equation was used to calculate the ARS in the control groups consisting of 504 Asians in order to obtain a distribution of ARS in the control group. Third, we ranked the ARS and assigned the corresponding percentile rank of the ARS value in the control group as the relative risk score (RRS). Finally, the RRS for a given NSCL/P genotype, namely, the percentile rank of the corresponding ARS value in the control group, was used to represent the probability of genetic risk of being an infant with NSCL/P.
| (4) |
Classification accuracy evaluation
We used seven commonly used machine learning models in the Python Scikit-learn package to build the models, including SVM, LR, NB, RF, KNN, DT, and ANN. The ten-fold cross validation method was used during the training process, and then an AUC value was computed to represent the overall performance for each model. 43 risk and protective alleles were collected from GWAScatalog database to evaluate the predictive effectiveness of the 43 SNPs Alleles with OR < 1 were defined as protective alleles, while alleles with OR > 1 were defined as risk alleles. Then, a one-by-one removal strategy and a one-by-one adding strategy were used to test the model performance in which a marker was removed or added from the marker set, respectively.
Authors’ contributions
XXW, FC, XFW, and JL conceived the project; PM, JZ, and PJ collected the samples; XXW and PM conducted the experiments; SZ and XFW analyzed the data. PM, XXW, and XFW wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the open project of Beijing Advanced Innovation Center for Food Nutrition and Human Health, China; the National Natural Science Foundation of China (Grant No. 81860370); Beijing Municipal Natural Science Foundation (Grant No. 7182184); and interdisciplinary medicine Seed Fund of Peking University (Grant No. BMU2017MB006), China. SZ was supported by the National Postdoctoral Program for Innovative Talents (Grant No. BX201600150), China.
Handled by Kai Ye
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2018.07.005.
Contributor Information
Feng Chen, Email: chenfeng2011@hsc.pku.edu.cn.
Xiaoxing Wei, Email: weixiaoxing@tsinghua.org.cn.
Supplementary material
The following are the Supplementary data to this article:
Distribution of genotypes with risk alleles of four known genes in the Han and Uyghur populations Comparison of the frequency of the homozygous genotypes of risk alleles of the SNPs associated with the genes IRF6-DIEXF (A), SLC2A9 (B), GSDMC – PVT1 (C), and NOG (D) between the NSCL/P and control groups. The risk allele of rs642961, rs3733585, rs987525, and rs227731 are A, C, A, and C, respectively. In total 504 and 103 subjects were included in the control and NSCL/P groups in Han Chinese population, respectively; whereas the control and NSCL/P groups in Uyghur Chinese population included 205 and 279 subjects, respectively.
Distribution of genotypes with risk alleles of several genes related to nutritional metabolism in the Han and Uyghur populations Comparison of the frequency of the homozygous genotypes of risk alleles of the SNPs associated with the genes MTHFR (A and B), MTHFD1(C), RBP4 (D), ADTRP (E), and one intergenic region (F) SNP between the NSCL/P and control groups. The risk alleles of rs1801133, rs1801131, rs2236225, rs10882272, rs2294426, and rs7820074 are T, C, T, C, C, and C, respectively. In total 504 and 103 subjects were included in the control and NSCL/P groups in Han Chinese population, respectively; whereas the control and NSCL/P groups in Uyghur Chinese population included 205 and 279 subjects, respectively.
References
- 1.Dixon M.J., Marazita M.L., Beaty T.H., Murray J.C. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mossey P.A., Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski D.F., editor. Cleft lip and palate: from origin to treatment. Oxford University Press; Oxford: 2002. pp. 127–158. [Google Scholar]
- 3.Pei X.X., Zhen C.D., Hui Z.W., Pei X.X., Zhen C.D., Hui Z.W. Serological study on the HLA polymorphism of 8 nationalities in China. Acta Anthropol Sin. 1993;12:158–161. [Google Scholar]
- 4.Zhang R., Xue Z.X. Case-control study of Uyghur babies with cleft lip and palate. Chin J Aesthetic Med. 2003;12:176–179. [Google Scholar]
- 5.Wehby G.L., Cassell C.H. The impact of orofacial clefts on quality of life and healthcare use and costs. Oral Dis. 2010;16:3–10. doi: 10.1111/j.1601-0825.2009.01588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang R., Bush J.O., Lidral A.C. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moxham B.J. The development of the palate — a brief review. Eur J Anat. 2017;7:53–74. [Google Scholar]
- 8.Ackermans M.M., Zhou H., Carels C.E., Wagener F.A., Von den Hoff J.W. Vitamin A and clefting: putative biological mechanisms. Nutr Rev. 2011;69:613–624. doi: 10.1111/j.1753-4887.2011.00425.x. [DOI] [PubMed] [Google Scholar]
- 9.van Rooij I.A., Ocké M.C., Straatman H., Zielhuis G.A., Merkus H.M., Steegers-Theunissen R.P. Periconceptional folate intake by supplement and food reduces the risk of nonsyndromic cleft lip with or without cleft palate. Prev Med. 2004;39:689–694. doi: 10.1016/j.ypmed.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Badovinac R.L., Werler M.M., Williams P.L., Kelsey K.T., Hayes C. Folic acid-containing supplement consumption during pregnancy and risk for oral clefts: a meta-analysis. Birth Defects Res A Clin Mol Teratol. 2007;79:8–15. doi: 10.1002/bdra.20315. [DOI] [PubMed] [Google Scholar]
- 11.Tolarova M., Harris J. Reduced recurrence of orofacial clefts after periconceptional supplementation with high-dose folic acid and multivitamins. Teratology. 1995;51:71–78. doi: 10.1002/tera.1420510205. [DOI] [PubMed] [Google Scholar]
- 12.Wilcox A.J., Lie R.T., Solvoll K., Taylor J., McConnaughey D.R., Abyholm F. Folic acid supplements and risk of facial clefts: national population based case-control study. BMJ. 2007;334:464. doi: 10.1136/bmj.39079.618287.0B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell L.E., Murray J.C., O’Brien S., Christensen K. Retinoic acid receptor alpha gene variants, multivitamin use, and liver intake as risk factors for oral clefts: a population-based case-control study in Denmark, 1991–1994. Am J Epidemiol. 2003;158:69–76. doi: 10.1093/aje/kwg102. [DOI] [PubMed] [Google Scholar]
- 14.Boyles A.L., Wilcox A.J., Taylor J.A., Shi M., Weinberg C.R., Meyer K. Oral facial clefts and gene polymorphisms in metabolism of folate/one-carbon and vitamin A: a pathway-wide association study. Genet Epidemiol. 2009;33:247–255. doi: 10.1002/gepi.20376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Zhou S., Zhang Q., Feng S., Chen Y., Zheng H. Proteomic analysis of RBP4/vitamin A in children with cleft lip and/or palate. J Dent Res. 2014;93:547–552. doi: 10.1177/0022034514530397. [DOI] [PubMed] [Google Scholar]
- 16.Collins M.D., Eckhoff C., Chahoud I., Bochert G., Nau H. 4-Methylpyrazole partially ameliorated the teratogenicity of retinol and reduced the metabolic formation of all-trans-retinoic acid in the mouse. Arch Toxicol. 1992;66:652–659. doi: 10.1007/BF01981505. [DOI] [PubMed] [Google Scholar]
- 17.Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Y., Lameijer E.W., Ye K., Zhang K., Chang S., Wang X. Precision medicine: what challenges are we facing. Genomics Proteomics Bioinformatics. 2016;14:253–261. doi: 10.1016/j.gpb.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rooij I.A., Vermeij-Keers C., Kluijtmans L.A., Ocké M.C., Zielhuis G.A., Goorhuis-Brouwer S.M. Dose the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? Am J Epidemiol. 2003;157:583–591. doi: 10.1093/aje/kwg005. [DOI] [PubMed] [Google Scholar]
- 21.Goyette P., Sumner J.S., Milos R., Duncan A.M., Rosenblatt D.S., Matthews R.G. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet. 1994;7:195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- 22.Frosst P., Blom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 23.Molloy A.M., Daly S., Mills J.L., Kirke P.N., Whitehead A.S., Ramsbottom D. Thermolabile variant of 5,10-methylenetetrahydrofolate reductase associated with low red-cell folates: implications for folate intake recommendations. Lancet. 1997;349:1591–1593. doi: 10.1016/S0140-6736(96)12049-3. [DOI] [PubMed] [Google Scholar]
- 24.Rai V. Strong association of C677T polymorphism of methylenetetrahydrofolate reductase gene with nosyndromic cleft lip/palate (nsCL/P) Indian J Clin Biochem. 2018;33:5–15. doi: 10.1007/s12291-017-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P., Wu T., Schwender H., Wang H., Shi B., Wang Z.Q. Evidence of interaction between genes in the folate/homocysteine metabolic pathway in controlling risk of non-syndromic oral cleft. Oral Dis. 2018;24:820–828. doi: 10.1111/odi.12831. [DOI] [PubMed] [Google Scholar]
- 26.Weisberg I., Tran P., Christensen B., Sibani S., Rozen R. A second genetic poly- morphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 27.van der Put N.M., Gabreëls F., Stevens E.M., Smeitink J.A., Trijbels F.J., Eskes T.K. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolarova M.M., Rooij I.A.L.M.V., Pastor M., Put N.M.J.V.D., Goldberg A.C., Hol F. A common mutation in the MTHFR gene is a risk factor for nonsyndromic cleft lip and palate anomalies. Am J Hum Genet. 1998;63:A27. [Google Scholar]
- 29.Shaw G.M., Rozen R., Finnell R.H., Todoroff K., Lammer E.J. Infant C677T mutation in MTHFR, maternal periconceptional vitamin use, and cleft lip. Am J Med Genet. 1998;80:196–198. doi: 10.1002/(sici)1096-8628(19981116)80:3<196::aid-ajmg2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Blaner W.S. Retinol-binding protein: the serum transport protein for vitamin A. Endocr Rev. 1989;10:308–316. doi: 10.1210/edrv-10-3-308. [DOI] [PubMed] [Google Scholar]
- 31.Huang E.W., Peng L.Y., Zheng J.X., Wang D., Xu Q.Y., Huang L. Common variants in promoter of ADTRP associate with early-onset coronary artery disease in a southern Han Chinese population. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupu C., Zhu H., Popescu N.I., Wren J.D., Lupu F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 2011;118:4463–4471. doi: 10.1182/blood-2011-05-355370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y., Huang Y., Yin A., Pan Y., Wang Y., Wang C. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 2015;6:6414. doi: 10.1038/ncomms7414. [DOI] [PubMed] [Google Scholar]
- 34.Birnbaum S., Ludwig K.U., Reutter H., Herms S., Steffens M., Rubini M. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 35.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Didenko V.V. DNA probes using fluorescence resonance energy transfer (FRET): designs and applications. Biotechniques. 2001;31:1106–1116. doi: 10.2144/01315rv02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuenca J., Aleza P., Navarro L., Ollitrault P. Assignment of SNP allelic configuration in polyploids using competitive allele-specific PCR: application to citrus triploid progeny. Ann Bot. 2013;111:731–742. doi: 10.1093/aob/mct032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of genotypes with risk alleles of four known genes in the Han and Uyghur populations Comparison of the frequency of the homozygous genotypes of risk alleles of the SNPs associated with the genes IRF6-DIEXF (A), SLC2A9 (B), GSDMC – PVT1 (C), and NOG (D) between the NSCL/P and control groups. The risk allele of rs642961, rs3733585, rs987525, and rs227731 are A, C, A, and C, respectively. In total 504 and 103 subjects were included in the control and NSCL/P groups in Han Chinese population, respectively; whereas the control and NSCL/P groups in Uyghur Chinese population included 205 and 279 subjects, respectively.
Distribution of genotypes with risk alleles of several genes related to nutritional metabolism in the Han and Uyghur populations Comparison of the frequency of the homozygous genotypes of risk alleles of the SNPs associated with the genes MTHFR (A and B), MTHFD1(C), RBP4 (D), ADTRP (E), and one intergenic region (F) SNP between the NSCL/P and control groups. The risk alleles of rs1801133, rs1801131, rs2236225, rs10882272, rs2294426, and rs7820074 are T, C, T, C, C, and C, respectively. In total 504 and 103 subjects were included in the control and NSCL/P groups in Han Chinese population, respectively; whereas the control and NSCL/P groups in Uyghur Chinese population included 205 and 279 subjects, respectively.