Abstract
Butyrate is a major short-chain fatty acid (SCFA) produced by microbial fermentation of dietary fiber in the gastrointestinal tract. Butyrate is also a well-known broad-spectrum histone deacetylase (HDAC) inhibitor. Butyrate has been reported to improve energy metabolism in rodents, which is associated with its beneficial effects on skeletal muscle, brown fat tissue and pancreatic β-cells. The present study investigated the direct effect of butyrate on hepatic gluconeogenesis in mouse primary hepatocytes and the underlying mechanism. Isolated mouse primary hepatocytes were incubated with sodium butyrate, other HDAC inhibitors and other SCFAs. Hepatic glucose production was measured and gluconeogenic gene expression was detected by polymerase chain reaction analysis. The phosphorylation of cyclic adenosine monophosphate (cAMP) response element binding protein (CREB) was assessed by western blot analysis. The results revealed that sodium butyrate dose-dependently increased hepatic glucose production and gluconeogenic gene expression in isolated mouse primary hepatocytes. Trichostatin A, a potent broad-spectrum HDAC inhibitor, had the opposite effect. Similar to sodium butyrate, propionate, which is another SCFA, promoted hepatic glucose production and gluconeogenic gene expression in the presence or absence of gluconeogenic substrates, which were further enhanced by cAMP. Furthermore, sodium butyrate also increased the accumulation of intracellular ATP and induced the phosphorylation of CREB in mouse hepatocytes. In conclusion, the present study suggested that butyrate stimulates hepatic gluconeogenesis and induces gluconeogenic gene expression as a substrate and cAMP/CREB signaling activator.
Keywords: butyrate, gluconeogenesis, short chain fatty acid, histone deacetylase, cyclic adenosine monophosphate response element binding protein
Introduction
Metabolic syndrome is a cluster of risk factors, including obesity, type 2 diabetes, dyslipidemia, hypertension and cardiovascular disease (1). Dietary intervention is a potential strategy to prevent and treat metabolic syndrome. It has been revealed that fiber-enriched diets improve obesity, insulin sensitivity and glucose tolerance, which is attributed to the production of short-chain fatty acid (SCFA) mediated by the gut microbiota (2–5). The most abundant SCFAs in the gut include acetate, propionate and butyrate (6). Although the major source of butyrate is the fermentation of dietary fibers, it is also contained in butter and cheese (7). Butyrate supplementation has been demonstrated to ameliorate diet-induced obesity, dyslipidemia, insulin resistance and glucose intolerance in mice (8). However, the mechanisms underlying the beneficial effect of butyrate has remained largely elusive.
It is well accepted that butyrate acts not only as a signaling molecule for the G-protein-coupled-receptor 41 (GPR41) and GPR43, but also as a wide-spectrum histone deacetylase (HDAC) inhibitor (9,10). Histone deacetylases regulate gene transcription by deacetylation of proteins, including histone proteins and transcription factors. In recent years, HDAC has emerged as a novel molecular target in the treatment of type 2 diabetes. It has been demonstrated that HDACs are crucial regulators of pancreatic cell fate determination (11). Butyrate treatment was reported to improve β-cell proliferation, function and glucose homeostasis as well as to reduce β-cell apoptosis through HDAC inhibition and histone acetylation in diabetic rats (12). Li et al (13) observed that sodium butyrate-stimulated fibroblast growth factor 21 expression and enhanced fatty acid oxidation in the liver by inhibition of HDAC3. In addition, sodium butyrate has been demonstrated to improve systemic insulin sensitivity and increase energy expenditure in mice via upregulating mitochondrial function in skeletal muscle and brown fat through peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) induction and elevation of adenosine monophosphate-activated protein kinase activity (8). However, the direct effect of butyrate on hepatic gluconeogenesis has remained to be elucidated.
Gluconeogenesis is an important pathological contributing factor in diabetic subjects and abnormally high during the progression of diabetes (14). In the liver, class IIa HDACs mediate glucogan-induced gluconeogenic gene expression through promoting deacetylation and activation of forkhead box O (FoxO) transcription factors. Hepatic knockdown of Class IIa HDACs in vivo resulted in lowered blood glucose in mice (15). Therefore, the present study investigated the direct effect of butyrate on gluconeogenesis in isolated mouse primary hepatocytes. Unexpectedly, butyrate significantly increased hepatic gluconeogenesis and the expression of gluconeogenic genes, which was different from the actions of other HDAC inhibitors.
Materials and methods
Materials
Hepatocyte medium was purchased from ScienCell (Carlsbad, CA, USA). Dulbecco's modified Eagle's medium (DMEM), Hank's balanced salt solution (HBSS), PBS and collagenase type IV (200 units/mg) were from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Sodium pyruvate, sodium L-lactate, dexamethasone, bovine serum albumin (BSA), 8-bromo-cyclic adenosine monophosphate (8-bromo-cAMP), acetate, propionate, sodium butyrate, Trichostatin A (TSA), CI994, Entinostat (MS-275), PCI-34051 and tubacin were obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). All primers used for reverse-transcription quantitative polymerase chain reaction (RT-qPCR) were synthesized by Shanghai Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). Anti-cAMP response element-binding protein (CREB; cat no. 9197S) and anti-phosphorylated (p)-CREB (Ser133; cat no. 9198) as well as anti-mouse immunoglobulin (Ig)G (cat no. 14709) and anti-rabbit IgG conjugated with horseradish peroxidase (cat no. 7074) were from Cell Signaling Technology Inc. (Beverly, MA, USA). The antibody to GAPDH (cat no. sc25778) was purchased from Santa Cruz Biotechnology, Inc. (Danvers, MA, USA).
Isolation and culture of mouse primary hepatocytes
A total of 20 Male C57BL/6 mice (age, 6–8 weeks; weight, 16–18 g) were purchased from Shanghai Slack Experimental Center (Shanghai, China). Primary hepatocytes were isolated from C57BL/6 mice by a modified version of the collagenase method. In a biosafety cabinet, mouse livers were perfused with 10 ml calcium-free HBSS through the portal vein under anesthesia with 10% chloral hydrate (Sigma-Aldrich; Merck, KGaA). The liver was excised by careful dissection and transferred to a 10-cm tissue culture dish. Mice were then sacrificed via cervical dislocation. The isolated liver was perfused with 0.05% collagenase IV (Gibco; Thermo Fisher Scientific, Inc.) dissolved in 20 ml calcium-containing HBSS in a recirculating manner for 15 min. Hepatocytes extracted from digested livers were filtered through a 100-µm cell strainer, washed 3 times with PBS and re-suspended in hepatocyte medium containing 100 U/ml penicillin, 100 µg/ml streptomycin, 0.1% BSA and hepatocyte growth factor (all from ScienCell Research Laboratories, Inc., San Diego, CA, USA). The cells were seeded on 6-well or 24-well plates and incubated in a tissue culture incubator at 37°C (95% air and 5% CO2). After 24 h, the cells had already attached to the plates and the medium was replaced with DMEM containing 5.5 mM glucose and 0.25% BSA, followed by drug treatment. All mice used in the present study for isolation of hepatocytes were fed a normal diet under a regular schedule and were not fasted. All of the experimental procedures involving the use of animals were approved by the Animal Use and Care Committee of Shanghai Jiaotong University School of Medicine (Shanghai, China).
Cell culture and treatments
Mouse primary hepatocytes were cultured in DMEM supplemented with 0.25% BSA overnight at 37°C with 5% CO2. To evaluate the effect of butyrate on hepatic gluconeogenesis, mouse primary hepatocytes were incubated with sodium butyrate for 8 h at various concentrations (0, 0.1, 1, 5, or 10 mmol/l). To investigate the effect of HDAC inhibitors on hepatic gluconeogenesis, cells were incubated in glucose-free DMEM containing gluconeogenic substrates (10 mmol/l sodium lactate and 1 mmol/l sodium pyruvate), 8-bromo-cAMP (100 µmol/l) and with the following HDAC inhibitors: TSA (100 nmol/l), CI994 (10 µmol/l), MS-275 (10 µmol/l), PCI-34051 (10 µmol/l), Tubacin (10 µmol/l) or sodium butyrate (5 mmol/l). Following 8 h incubation at 37°C, cell culture supernatants were collected for measuring the glucose content. Mouse primary hepatocytes were then treated with sodium butyrate (5 mmol/l) and TSA (100 nmol/l) in the presence of 8-bromo-cAMP (100 µmol/l) for 8 h at 37°C, the mRNA expression of gluconeogenic genes were detected using RT-qPCR. To compare the effects of three SCFAs on hepatic glucose production, mouse hepatocytes were treated with acetate (5 mmol/l), propionate (5 mmol/l) or sodium butyrate (5 mmol/l) for 8 h at 37°C in the presence of gluconeogenic substrates (10 mmol/l sodium lactate and 1 mmol/l sodium pyruvate). To test whether SCFAs affect hepatic gluconeogenesis as substrates, mouse hepatocytes were incubated with one of three different SCFAs (acetate, propionate, or sodium butyrate; 5 mmol/l each) and 100 µM 8-bromo-cAMP in the absence of gluconeogenic substrates (10 mmol/l sodium lactate and 1 mmol/l sodium pyruvate) for 8 h at 37°C. To assess the phosphorylation levels of CREB, mouse hepatocytes were treated with butyrate (5 mmol/l) or 8-bromo-cAMP (100 µmol/l) for 1 h at 37°C.
In vitro glucose production assay
Glucose production was assayed as described previously (16). In brief, hepatocytes were seeded into 24-well plates at 2.5×105 cells/well. After 24 h, these hepatocytes were pre-stimulated with DMEM containing 5.5 mM glucose, 0.25% BSA and 100 nM dexamethasone for 16 h. Cells were then washed three times with PBS and incubated in glucose production buffer (DMEM without glucose, serum or phenol red, and supplemented with 1 mM sodium pyruvate and 10 mM sodium lactate). After 8 h, the cell culture supernatants were collected for measuring the glucose content using a glucose oxidase kit (Applygen Co., Beijing, China).
RNA extraction and RT-qPCR
Total RNA was extracted from mouse primary hepatocytes using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Complementary DNA was synthesized by RT using Random Primers (Promega Corp., Madison, WI, USA) according to the manufacturer's protocol. RT-qPCR was performed in a Roche LightCycler 480 system (Roche Diagnostics, Basel, Switzerland) using SYBR Premix EX Taq (Takara, Tokyo, Japan). The PCR conditions were as follows: Denaturation at 95°C for 10 sec, followed by 40 cycles of 95°C for 5 sec. The annealing temperature was then reduced to 60°C for 31 sec with a final elongation step at 72°C for 300 sec. At the end of the amplification, a melting curve was generated in the temperature range of 60–95°C. The sequences of primers used are listed in Table I. Relative gene expression levels were quantified based on the cycle threshold (Cq) values and normalized to the reference gene β-actin. The relative gene expression was calculated using the 2−ΔΔCq method (17).
Table I.
Sequences of primers used for polymerase chain reaction.
| Gene | Sequence no. | Primer sequence (5′-3′) | Product length (bp) |
|---|---|---|---|
| PEPCK | NM_011044.2 | F, GTGCTGGAGTGGATGTTCGG | 258 |
| R, CTGGCTGATTCTCTGTTTCAGG | |||
| G6pase | NM_008061.4 | F, ACTGTGGGCATCAATCTCCTC | 344 |
| R, CGGGACAGACAGACGTTCAGC | |||
| FoxO1 | NM_019739.3 | F, AAGAGCGTGCCCTACTTCAA | 157 |
| R, CTCCCTCTGGATTGAGCATC | |||
| PGC-1α | NM_008904.2 | F, ATACCGCAAAGAGCACGAGAAG | 253 |
| R, CTCAAGAGCAGCGAAAGCGTCACAG | |||
| HNF4α | NM_008261.3 | F, ATGCGACTCTCTAAAACCCTTG | 135 |
| R, ACCTTCAGATGGGGACGTGT | |||
| β-actin | NM_007393.5 | F, GGCTGTATTCCCCTCCATCG | 154 |
| R, CCAGTTGGTAACAATGCCATGT |
FOX, forkhead box; PEPCK, phosphoenolpyruvate carboxykinase; G6pase, glucose 6-phosphatase; PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; HNF4α, hepatocyte nuclear factor 4α; F, forward; R, reverse.
Western blot analysis
Primary hepatocytes were treated with radioimmunoprecipitation lysis buffer containing protease and phosphatase inhibitors (Merck KGaA) and centrifuged at a speed of 6,000 × g for 10 min at 4°C. The supernatant was then used to analyze the expression levels of specific proteins. Protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). A total of 10 µg protein was loaded per lane and separated in 15% SDS-PAGE. Samples were then transferred onto polyvinylidene fluoride membranes (ECL Advance; Cell Signaling Technology, Inc). Prior to western blot analysis, membranes were blocked with 5% skimmed milk for 2 h at 37°C and subsequently incubated at 4°C overnight with rabbit anti-mouse primary antibodies sourced from Cell Signaling Technology Inc. These included cAMP response element-binding protein (CREB; cat. no. 9197S; 1:1,000) and phosphorylated (p)-CREB (Ser133; cat. no. 9198; 1:1,000). GAPDH antibodies (cat. no. sc25778; 1:1,000) were purchased from Santa Cruz Biotechnology, Inc. Membranes were then incubated with mouse anti-rabbit horseradish peroxidase-labeled IgG secondary antibodies (1:2,000; cat. no. 7074; Cell Signaling Technology Inc.,) overnight at 4°C. The results were visualized using an immobilon ECL ultra western HRP substrate (cat. no. WBULS0100; Merck KGaA) and images were captured using an LAS-4000 Super CCD Remote Control Science Imaging System (Fuji, Tokyo, Japan).
Adenosine triphosphate (ATP) assay
The amount of ATP was measured by the luciferin-luciferase method according to the protocol of the ATP detection kit (cat. no. S0026; Beyotime Institute of Biotechnology, Inc., Haimen, China). Primary hepatocytes were seeded onto 24-well plates at 2.5×105 cells/well. After 24 h, these cells received the same dexamethasone pre-stimulation as described in the glucose production assay. The cells were then treated with propionate (5 mmol/l) and different concentrations of sodium butyrate (0, 0.1, 1, 5 and 10 mmol/l) for 1 h prior to lysis with lysis buffer (200 µl/well) from the ATP detection kit. After centrifugation at 12,000 × g for 5 min at 4°C, the supernatant was transferred to a fresh tube for the ATP test. The luminescence of a 20-µl sample was assayed in a luminometer (Perkin Elmer, Inc., Waltham, MA, USA) together with 100 µl ATP detection buffer from the ATP detection kit. The protein concentration was determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). The concentration of ATP was normalized to that of protein in the same cell lysate.
Statistical analysis
All values are expressed as the mean ± standard error of the mean from at least three independent experiments. Two group comparisons were performed using a Student's t-test. All statistical analyses were performed using SPSS 19.0 (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Sodium butyrate stimulates hepatic glucose production and gluconeogenic gene expression
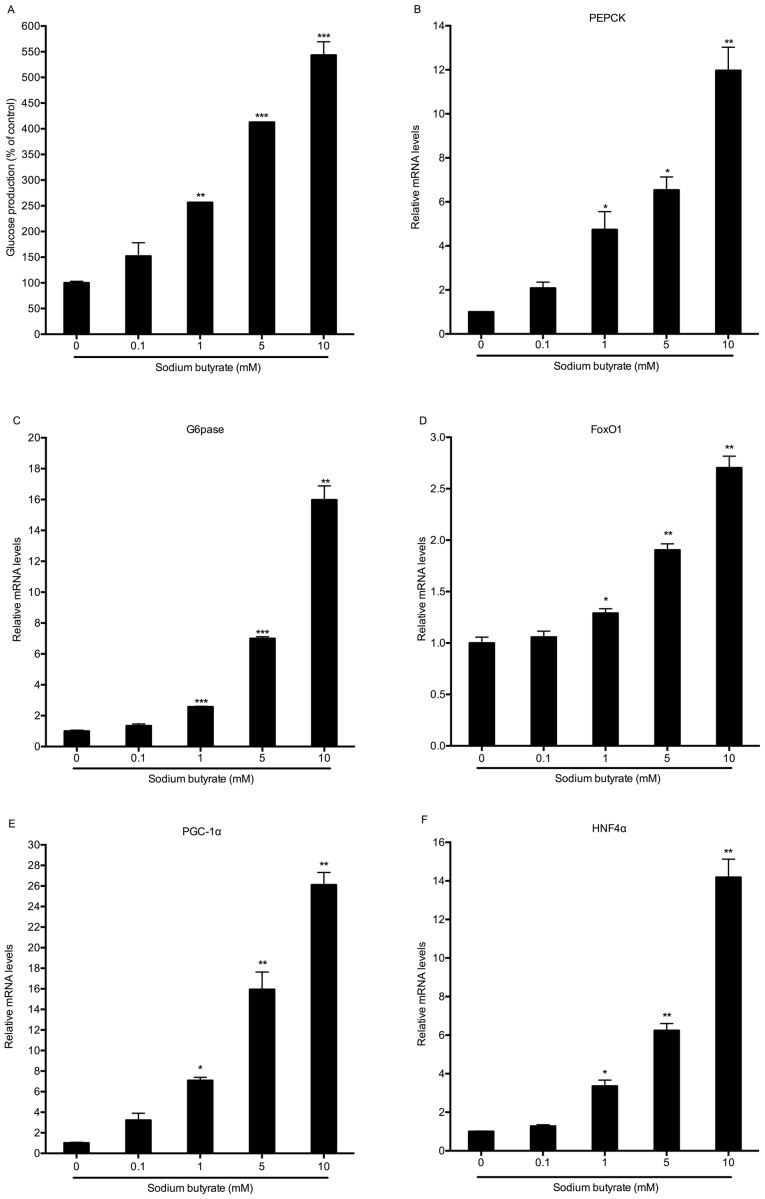
To determine the effects of butyrate on hepatic gluconeogenesis, mouse primary hepatocytes were incubated with various concentrations of sodium butyrate for 8 h. Unexpectedly, sodium butyrate stimulated hepatic glucose production in a dose-dependent manner, causing a significant increase at the concentration of 1 mM (P<0.01; Fig. 1A). Under the same conditions, the expression of genes associated with hepatic gluconeogenesis, including phosphoenolpyruvate carboxykinase (PEPCK), glucose-6-phosphatase (G6Pase), FoxO1, PGC-1α and hepatocyte nuclear factor 4α (HNF4α) was also increased. Consistent with the results on gluconeogenesis, sodium butyrate dose-dependently increased the mRNA expression of these gluconeogenic genes, with the maximum efficacy at the concentration of 10 mM (Fig. 1B-F).
Figure 1.
Sodium butyrate dose-dependently increases hepatic glucose production and gluconeogenic gene expression. (A) Mouse primary hepatocytes were incubated with the indicated concentrations of sodium butyrate for 8 h. The cell culture supernatants were subsequently collected for measuring the glucose content. (B-F) The mRNA expression of gluconeogenic genes in mouse hepatocytes treated with sodium butyrate for 8 h was detected by reverse-transcription quantitative polymerase chain reaction analysis. Values are expressed as the mean ± standard error of the mean of three separate experiments. *P<0.05, **P<0.01, ***P<0.001 compared with control group. FOX, forkhead box; PEPCK, phosphoenolpyruvate carboxykinase; G6pase, glucose 6-phosphatase; PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; HNF4α, hepatocyte nuclear factor 4α.
Effects of various HDAC inhibitors on hepatic gluconeogenesis
To investigate whether sodium butyrate-stimulated gluconeogenesis is involved in the inhibition of HDACs, the effects of other HDAC inhibitors, including TSA, CI994, MS-275, PCI-34051 and tubacin, on hepatic glucose production in mouse primary hepatocytes were detected. After mouse hepatocytes were incubated with 100 µM 8-bromo-cAMP for 8 h, glucose production was markedly increased. In the presence of 100 nM TSA, 10 µM CI994 or 10 µM PCI-34051, 8-bromo-cAMP-stimulated gluconeogenesis was significantly decreased. However, sodium butyrate treatment further enhanced glucose production induced by 8-bromo-cAMP. The other two HDAC inhibitors had no significant effect (Fig. 2A). TSA is a potent inhibitor of class I and II HDAC (18). The present study further compared the effects of TSA and sodium butyrate on the expression of the five gluconeogenic genes. As expected, the expression of all of these gluconeogenic genes was strongly induced by 8-bromo-cAMP. Contrary to the action of sodium butyrate, TSA suppressed cAMP-stimulated gluconeogenic gene expression (Fig. 2B-F). It is therefore unlikely that sodium butyrate promotes gluconeogenesis via inhibiting HDAC activity.
Figure 2.
Effect of various HDAC inhibitors on hepatic glucose production. (A) Mouse hepatocytes were incubated with different HDAC inhibitors (100 nM TSA, 10 µM CI994, 10 µM MS-275, 10 µM PCI-34051, 10 µM Tubacin or 5 mM SB) in glucose-free Dulbecco's modified Eagle's medium containing gluconeogenic substrates (10 mM sodium lactate and 1 mM sodium pyruvate) and 100 µM 8-bromo-cAMP for 8 h. The cell culture supernatants were collected for measuring the glucose content. (B-F) After mouse hepatocytes were treated with 5 mM sodium butyrate and 100 nM TSA in the presence of 100 µM 8-bromo-cAMP for 8 h, the mRNA expressions of gluconeogenic genes were detected by reverse-transcription quantitative polymerase chain reaction analysis. Values are expressed as the mean ± standard error of the mean of three separate experiments. *P<0.05, **P<0.01 compared with control group; #P<0.05, ##P<0.01, ###P<0.001 compared with 8-bromo-cAMP group. 8-bromo-cAMP, 8-bromo-cyclic adenosine monophosphate; TSA, Trichostatin A; MS-275, Entinostat; HDAC, histone deacetylase; SB, sodium butyrate; FOX, forkhead box; PEPCK, phosphoenolpyruvate carboxykinase; G6pase, glucose 6-phosphatase; PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; HNF4α, hepatocyte nuclear factor 4α.
Effects of various SCFAs on hepatic glucose production and gluconeogenic gene expression
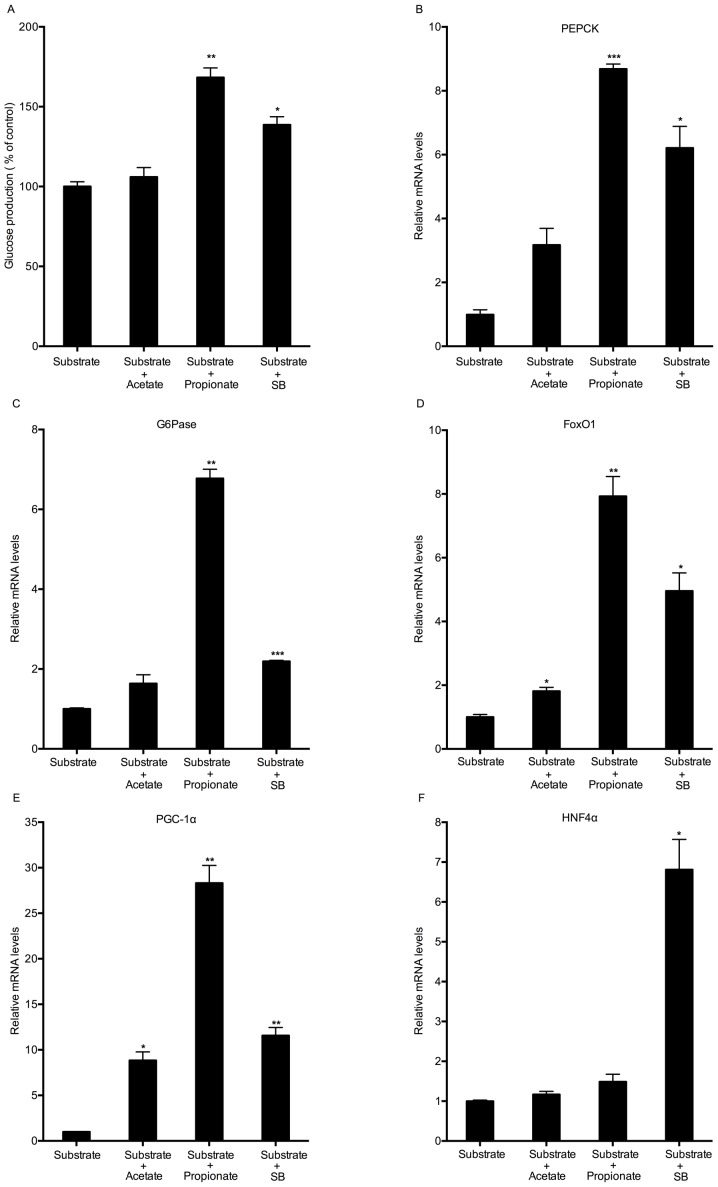
To investigate whether sodium butyrate stimulates gluconeogenesis as an energy substrate, the present study compared the effects of three SCFAs on hepatic glucose production. Mouse hepatocytes were treated with 5 mM acetate, propionate or sodium butyrate for 8 h in the presence of gluconeogenic substrates (sodium lactate and sodium pyruvate). Similar to sodium butyrate, propionate also promoted hepatic glucose production (Fig. 3A). Sodium butyrate supplementation led to increases in the expression of all of the selected gluconeogenic genes. Propionate significantly triggered the mRNA expression of PEPCK, G6Pase, FoxO1 and PGC-1α, but not that of HNF4α (Fig. 3B-F). Although acetate had no significant effects on glucose production, it significantly increased the expression of FoxO1 and PGC-1α (Fig. 3A, D and E).
Figure 3.
Effect of various short-chain fatty acids on hepatic glucose production and gluconeogenic gene expression. (A) Mouse hepatocytes were incubated with 5 mM acetate, propionate or SB in glucose-free Dulbecco's modified Eagle's medium containing gluconeogenic substrates (10 mM sodium lactate and 1 mM sodium pyruvate) for 8 h. The cell culture supernatants were collected for measuring glucose content. (B-F) Under the same conditions, the mRNA expression of gluconeogenic genes was detected by reverse-transcription quantitative polymerase chain reaction analysis. Values are expressed as the mean ± standard error of the mean of three separate experiments. *P<0.05, **P<0.01, ***P<0.001 compared with substrate group. SB, sodium butyrate; FOX, forkhead box; PEPCK, phosphoenolpyruvate carboxykinase; G6pase, glucose 6-phosphatase; PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; HNF4α, hepatocyte nuclear factor 4α.
Sodium butyrate stimulates hepatic glucose production as a substrate
To test whether sodium butyrate induces glucose production as a type of gluconeogenic substrate, mouse primary hepatocytes were incubated with one of three SCFAs in the absence of gluconeogenic substrates. Acetate, propionate and sodium butyrate all increased hepatic gluconeogenesis. Propionate had the most noticeable effect. In the presence of three SCFAs, addition of 8-bromo-cAMP further enhanced the glucose production (Fig. 4A). In agreement with the results on gluconeogenesis, 8-bromo-cAMP significantly enhanced butyrate and propionate-stimulated expression of the five gluconeogenic genes (Fig. 4B-F). Acetate supplementation only led to mild increases in PEPCK and PGC-1α expression (Fig. 4B and E). It is well accepted that propionate is a substrate for hepatic glucose production (19). Therefore, it is reasonable to presume that butyrate also stimulates hepatic gluconeogenesis as a substrate.
Figure 4.
Sodium butyrate stimulates hepatic glucose production as a substrate. (A) Mouse hepatocytes were incubated with different short-chain fatty acids (5 mM) and 100 µM 8-bromo-cAMP in the absence of gluconeogenic substrates (sodium lactate and sodium pyruvate) for 8 h. The cell culture supernatants were collected for measuring the glucose content. (B-F) Under the same conditions, the mRNA expression of gluconeogenic genes was detected by reverse-transcription quantitative polymerase chain reaction analysis. Values are expressed as the mean ± standard error of the mean of three separate experiments. *P<0.05, ***P<0.001 compared with control group; #P<0.05, ##P<0.01, ###P<0.001 compared with CON group (without 8-bromo-cAMP). CON, control; 8-bromo-cAMP, 8-bromo-cyclic adenosine monophosphate; SB, sodium butyrate; FOX, forkhead box; PEPCK, phosphoenolpyruvate carboxykinase; G6pase, glucose 6-phosphatase; PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; HNF4α, hepatocyte nuclear factor 4α.
Sodium butyrate activates CREB in mouse hepatocytes
It is well-known that cAMP/CREB is an important signaling pathway for hepatic gluconeogenesis (20,21). The present study investigated whether the cAMP/CREB signaling pathway is involved in butyrate-mediated induction of gluconeogenesis. As ATP is the substrate for cAMP production, the present study first detected the intracellular ATP concentration after mouse primary hepatocytes were incubated with various concentrations of sodium butyrate for 1 h. In parallel with glucose production, sodium butyrate treatment resulted in the accumulation of intracellular ATP in a dose-dependent manner, exhibiting a significant effect at the concentration of 1 mM (P<0.05; Fig. 5A). However, gluconeogenic substrates (sodium lactate and sodium pyruvate) as well as propionate did not alter the intracellular ATP concentration (Fig. 5B). Next, western blot analysis was performed to evaluate the phosphorylation state of CREB in butyrate-treated hepatocytes. Similar to 8-bromo-cAMP, sodium butyrate also stimulated the phosphorylation of CREB (Fig. 5C). Overall, these results indicated that butyrate activates cAMP/CREB signaling and stimulates the expression of hepatic gluconeogenic genes, at least in part via increasing the accumulation of intracellular ATP.
Figure 5.
Effects of butyrate on intracellular ATP content and CREB phosphorylation. (A) Mouse hepatocytes were incubated with the indicated concentrations of sodium butyrate for 1 h. Intracellular ATP concentration was measured by the luciferin-luciferase method. (B) Mouse hepatocytes were incubated with gluconeogenic substrates (10 mM sodium lactate and 1 mM sodium pyruvate), 5 mM proprionate, and 5 mM butyrate for 1 h. Intracellular ATP concentration was measured. (C) Mouse hepatocytes were treated with 5 mM butyrate or 100 µM 8-bromo-cAMP for 60 min. The phosphorylation level of CREB was detected by western blot analysis. Values are expressed as the mean ± standard error of the mean. *P<0.05, **P<0.01, ***P<0.001 compared with control group. ATP, adenosine triphosphate; p/t-CREB, phosphorylated/total cAMP response binding protein; 8-bromo-cAMP, 8-bromo-cyclic adenosine monophosphate.
Discussion
The liver is the major site of gluconeogenesis from red blood cell-derived pyruvate and lactate and from amino acid precursors. Hepatic metabolism has a key role in the regulation of the energy status of the whole body. Increased glucose production through abnormally elevated hepatic gluconeogenesis is central to the manifestation of hyperglycaemia in type 2 diabetes (22). The present study demonstrated that butyrate promoted gluconeogenesis in mouse primary hepatocytes as a substrate and signaling molecule, which appears paradoxical to its beneficial effect on the whole-body energy metabolism in rodent animals. To the best of our knowledge, there is little evidence demonstrating that butyrate directly improves blood glucose levels in human studies.
Gluconeogenesis is tightly controlled through the transcriptional regulation of PEPCK and G6Pase. Numerous transcription factors and co-activators, such as FoxO1, PGC-1α and HNF4α, are involved in the induction of the two hepatic gluconeogenic genes (23). Class IIa HDAC, together with HDAC3, was reported to regulate hepatic gluconeogenesis via deacetylation of FoxO1 (15). Butyrate is a well-characterized HDAC inhibitor. Similar to TSA, butyrate was reported to inhibit HDAC enzymes and increase the overall level of histone H3 acetylation (24). A previous study demonstrated that butyrate functioned as an HDAC inhibitor to stimulate the proliferation of colonocytes through changing their glucose metabolism (25). Therefore, the present study first investigated whether butyrate affected hepatic glucose production by acting as an HDAC inhibitor. Among six HDAC inhibitors, only sodium butyrate increased gluconeogenesis and TSA exerted the opposite effect. TSA is typically considered a broad-spectrum HDAC inhibitor. The present study further detected the expression of gluconeogenic genes in isolated mouse primary hepatocytes treated with sodium butyrate and TSA. In consistency with the results on gluconeogenesis, sodium butyrate enhanced 8-bromo-cAMP-stimulated hepatic gluconeogenic gene expression, while TSA had the opposite effect. These results indicated that butyrate stimulates gluconeogenesis and upregulates gluconeogenic gene expression independent of HDAC.
Besides being a broad-spectrum HDAC inhibitor, butyrate is an SCFA (6). In the present study, the effects of three SCFAs on hepatic glucose production and gluconeogenic gene expression were compared at the same concentration. Similar to sodium butyrate, propionate also stimulated gluconeogenesis and the expression of associated genes in mouse primary hepatocytes. Propionate has long been described as a hepatic gluconeogenic substrate (26). Infused propionate as a tracer was reported to markedly increase hepatic tricarboxylic acid metabolism and drive hepatic glucose production in a dose-dependent manner (19). Dietary supplementation with SCFAs, including propionate and butyrate, induced intestinal glucose production. Butyrate directly stimulated PEPCK and G6Pase expression in enterocytes in vitro (6). Therefore, it is likely that butyrate, as propionate, is also a gluconeogenic substrate. As expected, the present study demonstrated that sodium butyrate alone induced hepatic glucose production and gluconeogenic gene expression in mouse hepatocytes cultured with media void of other gluconeogenic substrates (sodium lactate and pyruvate).
cAMP has a pivotal role in the signaling pathways of hepatic gluconeogenesis (20). Since intracellular ATP is the substrate of cAMP production, the present study detected the intracellular ATP content in mouse hepatocytes incubated with butyrate and propionate. In agreement with the results of a previous study (27), propionate had no effect on the ATP concentration in the present study. However, butyrate dose-dependently induced the accumulation of intracellular ATP, demonstrating a similar increase to that during gluconeogenesis. Another mechanism underlying butyrate-stimulated gluconeogenesis except as a substrate has been reported; butyrate increased cAMP levels and protein kinase A activity in T-cells (28) and regulated the proliferation of porcine peripheral blood mononuclear cells in a cAMP-dependent manner (29). The present study revealed that butyrate stimulated the phosphorylation of CREB in mouse primary hepatocytes. These results suggested that butyrate stimulates gluconeogenesis via activating cAMP/CREB signaling.
In conclusion, the present study demonstrated that butyrate has a dual role in stimulating hepatic glucose production as a substrate and signaling molecule. cAMP/CREB signaling is involved in butyrate-induced hepatic gluconeogenic gene expression. Apparently, the beneficial effects of butyrate on other tissues counterbalances its action on hepatic gluconeogenesis, eventually improving the whole-body energy metabolism in rodent animals. However, additional studies should be performed before sodium butyrate is used in the treatment of type 2 diabetes mellitus in the clinic.
Acknowledgements
Not applicable.
Funding
This work was funded by grants from the National Natural Science Foundation of China (grant nos. 81270910, 81370876, 81471030, 81570693 and 30600294), the Natural Science Foundation of Shanghai (grant no. 15ZR1434000), and the Academic Leaders Training Program of Pudong Health Bureau of Shanghai (grant no. PWRd2011-01).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XJ and LZ designed the study. XJ and FZ conducted the experiments. YZ and WX analysed the data. XJ wrote the manuscript. LZ had primary responsibility for final content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal procedures were reviewed and approved by the Animal Use and Care Committee of Shanghai Jiao Tong University (Shanghai, China) and the China Experimental Animal Protection Association. All efforts were made to minimize the suffering of the experimental mice.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ning G. Decade in review-type 2 diabetes mellitus: At the centre of things. Nat Rev Endocrinol. 2015;11:636–638. doi: 10.1038/nrendo.2015.147. [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 3.Papathanasopoulos A, Camilleri M. Dietary fiber supplements: Effects in obesity and metabolic syndrome and relationship to gastrointestinal functions. Gastroenterology. 2010;138:65–72.e1-e2. doi: 10.1053/j.gastro.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson MD, Currie JM, Morgan LM, Jewell DP, Frayn KN. Prior short-term consumption of resistant starch enhances postprandial insulin sensitivity in healthy subjects. Diabetologia. 2003;46:659–665. doi: 10.1007/s00125-003-1081-0. [DOI] [PubMed] [Google Scholar]
- 5.Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr. 2005;82:559–567. doi: 10.1093/ajcn/82.3.559. [DOI] [PubMed] [Google Scholar]
- 6.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Robertson MD, Currie JM, Morgan LM, Jewell DP, Frayn KN. Prior short-term consumption of resistant starch enhances postprandial insulin sensitivity in healthy subjects. Diabetologia. 2003;46:659–665. doi: 10.1007/s00125-003-1081-0. [DOI] [PubMed] [Google Scholar]
- 8.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin HV, Frassetto A, Kowalik EJ, Jr, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ. Butyrate and propionate protectagainst diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis L, Hammers H, Pili R. Targeting tumor angiogenesis with histone deacetylase inhibitors. Cancer Lett. 2009;280:145–153. doi: 10.1016/j.canlet.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haumaitre C, Lenoir O, Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. 2008;28:6373–6383. doi: 10.1128/MCB.00413-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan S, Jena GB. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: Study in juvenile diabetic rat. Chem Biol Interact. 2014;213:1–12. doi: 10.1016/j.cbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Gao Z, Zhang J, Ye X, Xu A, Ye J, Jia W. Sodium butyrate stimulates expression of fibroblast growth factor 21 in liver by inhibition of histone deacetylase 3. Diabetes. 2012;61:797–806. doi: 10.2337/db11-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hundal RS, Krssak M, Dufour S, Laurent D, Lebon V, Chandramouli V, Inzucchi SE, Schumann WC, Petersen KF, Landau BR, Shulman GI. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes. 2000;49:2063–2069. doi: 10.2337/diabetes.49.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ip W, Shao W, Chiang YT, Jin T. The Wnt signaling pathway effector TCF7L2 is upregulated by insulin and represses hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2012;303:E1166–E1176. doi: 10.1152/ajpendo.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Taddei A, Roche D, Bickmore WA, Almouzni G. The effects of histone deacetylase inhibitors on heterochromatin: Implications for anticancer therapy? EMBO Rep. 2005;6:520–524. doi: 10.1038/sj.embor.7400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry RJ, Borders CB, Cline GW, Zhang XM, Alves TC, Petersen KF, Rothman DL, Kibbey RG, Shulman GI. Propionate increases hepatic pyruvate cycling, anaplerosis and alters mitochondrial metabolism. J Biol Chem. 2016;291:12161–12170. doi: 10.1074/jbc.M116.720631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR III, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Perriello G, Pampanelli S, Del Sindaco P, Lalli C, Ciofetta M, Volpi E, Santeusanio F, Brunetti P, Bolli GB. Evidence of increased systemic glucose production and gluconeogenesis in an early stage of NIDDM. Diabetes. 1997;46:1010–1016. doi: 10.2337/diabetes.46.6.1010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, et al. FoxO1 regulates multiple metabolic pathways in the liver: Effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 24.Xiong H, Guo B, Gan Z, Song D, Lu Z, Yi H, Wu Y, Wang Y, Du H. Butyrate upregulates endogenous host defense peptides to enhance disease resistance in piglets via histone deacetylase inhibition. Sci Rep. 2016;6:27070. doi: 10.1038/srep27070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson JW, Bridges SR. Short-chain fatty acid fermentation products of plant fiber affect glucose metabolism of isolated rat hepatocytes. Proc Soc Exp Biol Med. 1984;177:372–376. doi: 10.3181/00379727-177-41958. [DOI] [PubMed] [Google Scholar]
- 27.Massillon D, Arinze IJ, Xu C, Bone F. Regulation of glucose-6-phosphatase gene expression in cultured hepatocytes and H4IIE cells by short-chain fatty acids: Role of hepatic nuclear factor-4alpha. J Biol Chem. 2003;278:40694–40701. doi: 10.1074/jbc.M303182200. [DOI] [PubMed] [Google Scholar]
- 28.Diakos C, Prieschl EE, Saemann M, Novotny V, Bohmig G, Csonga R, Baumruker T, Zlabinger GJ. Novel mode of interference with nuclear factor of activated T-cells regulation in T-cells by the bacterial metabolite n-butyrate. J Biol Chem. 2002;277:24243–24251. doi: 10.1074/jbc.M200191200. [DOI] [PubMed] [Google Scholar]
- 29.Weber TE, Kerr BJ. Butyrate differentially regulates cytokines and proliferation in porcine peripheral blood mononuclear cells. Vet Immunol Immunopathol. 2006;113:139–147. doi: 10.1016/j.vetimm.2006.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.