Abstract
Circular RNAs (circRNAs) have a great potential as clinical biomarkers; however, specific circRNAs with a diagnostic value for essential hypertension (EH) largely remain to be identified. In the present study, the potential application of Homo sapiens (hsa)_circ_0014243, which was identified to be significantly upregulated in whole blood samples of EH patients in a previous microarray profiling study by our group, in the diagnosis of EH was evaluated. Reverse transcription-quantitative polymerase chain reaction analysis was performed to determine the expression levels of hsa_circ_0014243 and hsa-microRNA (miR)-10a-5p in a total of 178 blood samples collected from 89 healthy controls and 89 patients diagnosed with EH. Divergent primers were designed for circRNAs, while conventional primers were used for miRs. Independent-samples t-tests and bivariate correlation analyses were performed to analyze the association between clinical factors influencing EH and hsa_circ_0014243 expression levels. A receiver operating characteristics (ROC) curve was generated to estimate the diagnostic value of hsa_circ_0014243 for EH. Finally, the expression levels of circRNAs and miRNAs were combined to propose a possible prediction model for EH. The results indicated that hsa_circ_0014243 was upregulated in whole blood samples of EH patients compared with that in the controls (P<0.001). Furthermore, the relative expression levels of hsa_circ_0014243 (Δ quantification cycle) were identified to be significantly correlated with age (r=−0.259, P<0.001), high-density lipoprotein levels (r=0.196, P=0.009) and glucose levels (r=−0.204, P=0.006). The area under the ROC curve (AUC) of the model using hsa_circ_0014243 as a predictor was 0.732. Of note, the AUC increased to 0.781 when hsa_circ_0014243 levels were combined with hsa-miR-10a-5p levels as predictors. The present results suggest that hsa_circ_0014243 has a crucial role in the genesis and development of EH, and presents a certain diagnostic capability for EH.
Keywords: circular RNA, hsa_circ_0014243, essential hypertension, biomarker, diagnosis
Introduction
Essential hypertension (EH) is a multi-factorial chronic disease that is characterized by elevated arterial pressure and may be accompanied by pathological changes in organs including the heart, blood vessels, kidney and brain. Environmental as well as hereditary factors have prominent roles in the initiation and development of EH (1). EH affects all income groups in the world and the World Health Organization estimated that 29.2% of the world's population will suffer from hypertension by 2025 (2). Studies have demonstrated that low-grade inflammation in vascular tissues causes endothelial dysfunction and promotes oxidative stress, and is therefore a major contributor to the physiopathology of EH (3,4). However, the molecular mechanisms underlying the pathology of EH, including the roles of circular RNAs (circRNAs), remain to be fully elucidated.
CircRNAs are widely expressed in mammalian cells and constitute a class of endogenous non-coding RNAs without 5′ caps or 3′ tails (5,6). In contrast to linear RNAs, circRNAs are synthesized from transcripts derived from exons and/or introns by back-splicing (7,8). Several studies have demonstrated that circRNAs regulate gene expression and act as specific microRNA (miRNA/miR) sponges by weakening the inhibitory effects of miRNAs on their associated target mRNAs (9). Certain circRNAs are important genetic material of polygenic inheritance and have the ability to inhibit the degradation by RNase R or RNA exonuclease (10). Furthermore, an RNA sequencing analysis using human whole blood samples identified circRNAs as a novel class of putative biomarkers (11). In view of all of these traits, future studies should focus on investigating circRNAs to establish a stable biomarker system that may improve the diagnosis and treatment of associated diseases.
A present, microRNAs are more extensively studied than circRNAs. Certain miRNAs have been reported to be involved in cardiovascular diseases and were demonstrated to be closely associated with EH (12,13). Zheng et al (14) suggested that miR-155 levels are negatively correlated with blood pressure. Recent studies indicated that Homo sapiens (hsa)-miR-10a-5p represses the inflammatory response by inhibiting nuclear factor (NF)-κB activation (5,15). Therefore, it may be speculated that hsa-miR-10a-5p is negatively correlated with EH. However, the function and regulatory mechanisms underlying the roles of circRNAs and miRNAs in EH progression remain to be elucidated.
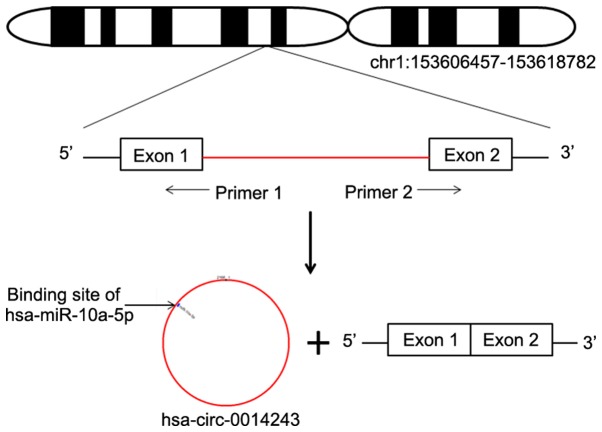
Hsa_circ_0014243 is a circRNA with a spliced length of 2,166 nt. As presented in Fig. 1, hsa_circ_0014243 is a type of circular intronic RNA that is formed by the circularization of introns. Hsa_circ_0014243 contains multiple miRNA-binding sites on its surface, including a hsa-miR-10a-5p-binding site. Hsa_circ_0014243 is located on chromosome 1:153606457-153618782 and its encoding gene is chromatin target of protein arginine methyltransferase 1. In the present study, the association between hsa_circ_0014243 and EH was investigated based on previous results indicating that hsa_circ_0014243 expression is upregulated in whole blood samples of EH patients (16). Furthermore, bioinformatics prediction indicated the presence of a potential hsa-miR-10a-5p-binding site on the surface of hsa_circ_0014243.
Figure 1.
Schematic illustrating the generation of the circular RNA hsa_circ_0014243. Chr, chromosome; miR, microRNA; circ, circular RNA; hsa, Homo sapiens.
Materials and methods
Patients and samples
A total of 178 subjects, comprising 89 healthy controls and 89 patients diagnosed with EH, were recruited at Ningbo Seventh Hospital and Community Health Service Center of Baizhang and Dongjiao Street (Ningbo, China). Following the guidelines established by the American Heart Association and American College of Cardiology in 2017, EH patients were defined as individuals whose three consecutive measurements of systolic blood pressure (SBP) were ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg (17). The controls had a SBP and DBP of <130 and <85 mmHg, respectively, and had never been diagnosed with hypertension by reliable health facilities. All controls and EH patients were recruited from Han Chinese families who resided in Ningbo City for at least three generations. All participants had similar occupations as office staff and their age ranged from 35 to 75 years and their median age was 64 years old. In addition, none of the individuals had genetic hypertension or any other serious condition, including secondary hypertension, diabetes mellitus, heart disease, kidney disease, malignant cancer, cerebrovascular disease, thyroid disorder or drug abuse. Whole blood samples collected from the participants were immediately stored at −80°C.
The participants filled in a standard questionnaire assessing demographic features and environmental factors. Physical examination and laboratory measurements were performed by experienced medical staff. Demographic features and environmental factors assessed included sex, age, history of smoking and drinking, family history and medication history. Physical examination comprised the measurement of routine parameters, including height, weight and blood pressure, while the laboratory examination mainly included the measurement of blood glucose and blood lipid levels.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of hsa_circ_0014243 and hsa-miR-10a-5p
Total RNA was extracted from whole blood samples with TRIzol reagent (BioTeke, Beijing, China) according to the manufacturer's instructions. Agarose gel electrophoresis was performed to evaluate RNA integrity. Total RNA quantity and purity was determined using the Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
CircRNA from total RNA was reverse-transcribed to complementary (c)DNA using the GoScript™ RT system (Promega Corporation, Madison, WI, USA). RT of the circRNAs was performed using the following conditions: 42°C for 15 min and 70°C for 15 min, followed by storage at −20°C. miRNAs from total RNA were reverse transcribed into cDNA using the miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China) following the manufacturer's protocol. RT of miRNAs was performed using the following conditions: Incubation at 42°C for 60 min, 95°C for 3 min and storage at −20°C. RT was performed in a Mastercycler pro PCR System (Eppendorf, Hamburg, Germany).
Real-time qPCR was performed using the miRcute Plus miRNA qPCR Detection Kit (Tiangen) and the GoTaq qPCR Master Mix (Promega) according to the manufacturer's protocols on a Roche LightCycler 480 Real Time PCR instrument (Roche Diagnostics, Basel, Switzerland). Commonly used references were selected as controls based on the experimental methods used by previous studies. GAPDH was amplified as the internal control for analysis of circRNA levels (18,19), while Caenorhabditis elegans (cel)-miR-39 was amplified as the external control for miRNA levels (20). Instead of the commonly used convergent primers, divergent primers were designed for circRNA amplification, while primers for miRNAs were designed using the conventional method. The reason why circRNAs primers differ from linear RNA is that circRNA is derived from reverse splicing of RNA transcript and the specific expression of the splicing site can be demonstrated by the reverse primer that covers the splicing site in the amplification region as shown in Fig. 1. The authors of the current study evaluated putative interactions between the miRNA sequences and hsa_circ_0014243 using Circular RNA Interactome (https://circinteractome.nia.nih.gov/). The primers were synthesized by Invitrogen (Shanghai, China) and their sequences are presented in Table I. The reaction conditions for circRNA amplification were as follows: 95°C for 2 min and 45 cycles of 95°C for 5 sec, 58°C for 30 sec and 72°C for 30 sec. The miRNAs were amplified using the following profile: Denaturation at 95°C for 15 min; five cycles of 94°C for 20 sec, 65°C for 30 sec and 72°C for 34 sec; and extension with 45 cycles of 94°C for 20 sec and 60°C for 34 sec. Expression levels were determined by using the ΔCq method with GAPDH or cel-miR-39 as a reference (21). All experimental data are expressed as the mean ± standard deviation of three measurements.
Table I.
Sequences of polymerase chain reaction primers used for analysis of circRNA and miRNA levels.
| Primer name | Primer sequence |
|---|---|
| Hsa_circ_0014243 F | 5′-ACGAACTGCTGGAAGAGGTC-3′ |
| Hsa_circ_0014243 R | 5′-GCAATTACGTCAGGCGTCAG-3′ |
| GAPDH F | 5′-ACCCACTCCTCCACCTTTGAC-3′ |
| GAPDH R | 5′-TGTTGCTGTAGCCAAATTCGTT-3′ |
| Hsa-miR-10a-5p | 5′-GTACCCTGTAGATCCGAATTTGTG-3′ |
| Cel-miR-39 | 5′-TCACCGGGTGTAAATCAGCTT-3′ |
miR, microRNA; hsa, Homo sapiens; circ, circular RNA; F, forward; R, reverse; Cel, Caenorhabditis elegans.
Statistical analysis
Experimental data were analyzed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA, USA) was used to generate graphs. Continuous variables, including ΔCq value, age, body mass index (BMI), high-density lipoprotein (HDL), low-density lipoprotein (LDL) levels, triglyceride levels, total cholesterol (TC) levels, serum creatinine levels and neutrophil granulocyte counts were compared between controls and EH patients using Student's t-test. Pearson's correlation analysis or Spearman's rank correlation analysis was performed to assess the correlation between hsa_circ_0014243 expression and patient characteristics. Pearson's χ2 test was performed to analyse the association between categorical variables (smoking history, drinking history and sex) in the two patient groups. Logistic regression was performed with the continuous variable method to identify confounding factors (22). Receiver operating characteristic (ROC) curves were generated to evaluate the diagnostic value of the predictive variables. A two-sided P<0.05 was considered to indicate a statistically significant difference.
Results
Hsa_circ_0014243 expression is upregulated in whole blood samples of EH patients
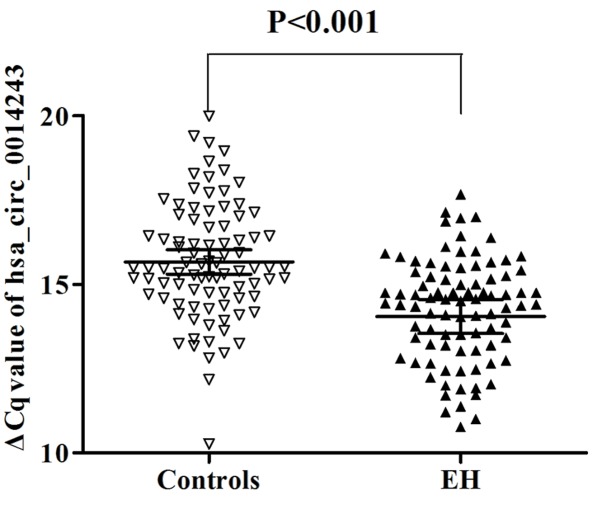
Table II presents the comparison of characteristics between the 89 healthy controls and 89 EH patients. The controls and EH patients exhibited no significant differences in sex and age. However, the subjects in the control and EH groups had significant differences in smoking (t=11.38, P<0.001), drinking (t=4.93, P=0.026), BMI (t=3.54, P<0.001), HDL levels (t=−2.23, P=0.027), blood uric acid levels (t=2.29, P=0.023), TG (t=2.40, P=0.018), homocysteine levels (t=2.48, P=0.014) and glucose levels (t=2.61, P=0.032). RT-qPCR was performed to determine whether hsa_circ_0014243 expression levels were associated with hsa-miR-10a-5p levels, which have been recognized to be correlated with EH (23). Patients diagnosed with EH had higher ΔCq values for hsa-miR-10a-5p (4.04±2.20 vs. 2.55±2.75; P<0.001) and lower ΔCq values for hsa_circ_0014243 (14.05±2.40 vs. 15.66±1.71; P<0.001) relative to the controls. As presented in Fig. 2, the expression levels of hsa_circ_0014243 were upregulated in whole blood samples of EH patients relative to those in the controls.
Table II.
Comparison of characteristics and parameters between controls and EH group.
| Characteristic/parameter | Normal ranges | Controls | EH | t/χ2 | P-value |
|---|---|---|---|---|---|
| Age (years) | N/A | 59.78±11.03 | 62.21±9.75 | 1.56 | 0.120 |
| Sex (M/F) | N/A | 57/32 | 57/32 | 0.000 | 1.000 |
| Smoking (Y/N) | N/A | 18/71 | 39/50 | 11.38 | <0.001 |
| Drinking (Y/N) | N/A | 23/66 | 37/52 | 4.93 | 0.026 |
| BMI (kg m2) | 18.5–23.9 | 23.38±2.85 | 25.13±3.67 | 3.54 | <0.001 |
| SBP (mmHg) | 90–139 | 128.82±14.18 | 144.84±15.98 | 7.07 | <0.001 |
| DBP (mmHg) | 60–89 | 79.88±9.12 | 86.27±11.97 | 4.01 | <0.001 |
| HDL (mmol/l) | >1.04 | 1.48±0.37 | 1.38±0.22 | −2.23 | 0.027 |
| LDL (mmol/l) | 0–3.12 | 2.99±0.54 | 3.17±0.75 | 1.85 | 0.067 |
| ALT (IU/l) | 9–50 | 28.23±14.92 | 28.57±21.51 | 0.12 | 0.903 |
| AST (IU/l) | 15–40 | 30.45±10.10 | 29.27±7.16 | −0.903 | 0.368 |
| BUN (mmol/l) | 3.10–8.00 | 5.35±1.06 | 5.25±1.04 | −0.654 | 0.514 |
| BUA (µmol/l) | 208–428 | 314.68±59.75 | 339.48±82.97 | 2.29 | 0.023 |
| TC (mmol/l) | 0–5.20 | 4.95±0.66 | 5.14±0.69 | 1.93 | 0.055 |
| TG (mmol/l) | 0–1.70 | 1.28±0.49 | 1.55±0.91 | 2.40 | 0.018 |
| RBC (1012/l) | 4.30–5.80 | 4.57±0.35 | 4.61±0.40 | 0.68 | 0.499 |
| HCY (µmol/l) | 4.0–15.4 | 14.50±2.98 | 16.38±6.48 | 2.48 | 0.014 |
| HB (g/l) | 130–175 | 142±10.75 | 144.13±12.67 | 1.21 | 0.228 |
| Scr (µmol/l) | 57–97 | 86.44±13.81 | 90.68±15.80 | 1.91 | 0.058 |
| Glu (mmol/l) | 3.89–6.11 | 5.58±1.00 | 5.97±1.39 | 2.16 | 0.032 |
| WBC (109/l) | 4.0–10.0 | 5.77±1.01 | 6.13±1.45 | 1.93 | 0.055 |
| Lym (109/l) | 1.10–3.20 | 1.97±0.48 | 2.02±0.51 | 0.73 | 0.469 |
| Neut (109/l) | 1.80–6.30 | 3.33±0.76 | 3.68±1.10 | 2.49 | 0.014 |
| ΔCq hsa-miR-10a-5p | N/A | 2.55±2.75 | 4.04±2.20 | 4.01 | <0.001 |
| ΔCq hsa_circ_0014243 | N/A | 15.66±1.71 | 14.05±2.40 | −5.17 | <0.001 |
Values are expressed as the mean ± standard deviation or n. ALT, alanine transaminase; AST, aspartate transaminase; BMI, body mass index; BUA, blood uric acid; BUN, blood urea nitrogen; DBP, diastolic blood pressure; EH, essential hypertension; F, female; Glu, glucose; HCY, homocysteine; HDL, high-density lipoprotein; LDL, low-density lipoprotein; Lym, lymphocyte; M, male; N, no; Neut, neutrophile granulocyte; RBC, red blood cells; SBP, systolic blood pressure; Scr, serum creatinine; TG, triglyceride; TC, total cholesterol; WBC, white blood cells; Y, yes; miR, microRNA; hsa, Homo sapiens; circ, circular RNA; Cq, quantification cycle.
Figure 2.
Hsa_circ_0014243 expression levels in controls and patients with EH. A lower ΔCq value indicates higher expression. Hsa_circ_0014243 expression in controls was lower compared with that in patients with EH (n=89, P<0.001). EH, essential hypertension; Cq, quantification cycle; circ, circular RNA.
Association between hsa_circ_0014243 levels and various factors potentially associated with EH
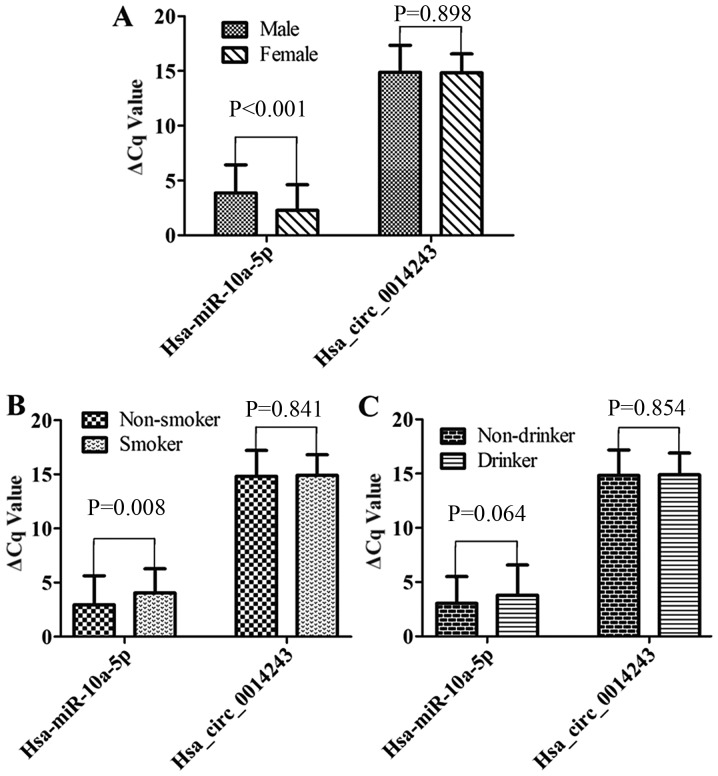
As presented in Fig. 3, males and females exhibited statistically significant differences in ΔCq values for hsa-miR-10a-5p (P<0.001; Fig. 3A), indicating that males had significantly lower expression levels when compared with females. Non-smokers had significantly higher ΔCq values for hsa-miR-10a-5p relative to smokers (P=0.008; Fig. 3B). However, hsa_circ_0014243 was not significantly associated with sex (P=0.898; Fig. 3A), smoking history (P=0.841; Fig. 3B) and drinking history (P=0.854; Fig. 3C).
Figure 3.
Comparison of hsa_circ_0014243 and hsa-miR-10a-5p levels between (A) males and females, (B) smokers and non-smokers and (C) drinkers and non-drinkers. A lower ΔCq value indicates higher expression. miR, microRNA; hsa, Homo sapiens; circ, circular RNA; Cq, quantification cycle.
The correlation between hsa_circ_0014243 expression levels and various indicators of physiological, biochemical and behavioral characteristics of the subjects, which comprised 89 healthy controls and 89 EH patients, was then assessed. Pearson correlation analysis was applied to binary normal distribution data, while Spearman's rank correlation was used for non-normally distributed data and ranked data. As presented in Table III, the expression levels of hsa_circ_0014243 were significantly associated with age (r=−0.259, P<0.001), HDL levels (r=0.196, P=0.009) and glucose levels (r=−0.204, P=0.006), but not with any of the other characteristics assessed, including BMI, sex, and smoking and drinking history.
Table III.
Pearson or Spearman's rank correlation analysis of the correlation between the expression levels of Homo sapiens circular RNA-0014243 (Δ quantification cycle) and laboratory parameters.
| Parameter | r | P-value |
|---|---|---|
| Age (years) | −0.259 | <0.001 |
| BMI (kg/m2) | −0.050 | 0.510a |
| ALT (IU/l) | 0.115 | 0.126 |
| AST (IU/l) | 0.123 | 0.101 |
| BUN (mmol/l) | 0.107 | 0.155 |
| BUA (µmol/l) | −0.057 | 0.446 |
| HDL (mmol/l) | 0.196 | 0.009 |
| LDL (mmol/l) | −0.053 | 0.481 |
| Scr (µmol/l) | −0.034 | 0.655 |
| Glu (mmol/l) | −0.204 | 0.006 |
| HCY (µmol/l) | −0.112 | 0.136 |
Pearson correlation analysis. ALT, alanine transaminase; AST, aspartate transaminase; BMI, body mass index; BUA, blood uric acid; BUN, blood urea nitrogen; Glu, glucose; HCY, homocysteine; HDL, high-density lipoprotein; LDL, low-density lipoprotein; Scr, serum creatinine.
Potential value of hsa_circ_0014243 as a biomarker for EH
The results of the logistic regression analysis with P-value adjustment indicated that the ΔCq values of hsa-miR-10a-5p [odds ratio=1.344, 95% confidence interval (CI): 1.094–1.652, P=0.005] and hsa_circ_0014243 (odds ratio=0.546, 95% CI: 0.414–0.720, P<0.001) were significant predictors of EH (Table IV). The odds ratio above/below 1 indicates that hsa_circ_0014243 is a risk factor and hsa-miR-10a-5p a protective factor regarding EH. Furthermore, sex (odds ratio=0.218, 95% CI: 0.070–0.684, adjusted P=0.009) smoking (odds ratio=5.534, 95% CI: 1.485–20.626, adjusted P=0.011) and BMI (odds ratio=1.175, 95% CI: 1.034–1.334, adjusted P=0.013) were identified as potential factors influencing EH.
Table IV.
The effect of predictors for essential hypertension.
| Variable | OR (95% CI) | Wald | P-value |
|---|---|---|---|
| Sex (M/F) | 0.218 (0.070,0.684) | 6.826 | 0.009 |
| Age (years) | 1.004 (0.958,1.053) | 0.033 | 0.855 |
| BMI (kg/m2) | 1.175 (1.034,1.334) | 6.103 | 0.013 |
| Smoking (Y vs. N) | 5.534 (1.485,20.626) | 6.496 | 0.011 |
| Drinking (Y vs. N) | 1.074 (0.307,3.755) | 0.012 | 0.911 |
| ALT (IU/l) | 1.006 (0.983,1.029) | 0.234 | 0.628 |
| AST (IU/l) | 0.966 (0.917,1.018) | 1.642 | 0.200 |
| Scr (µmol/l) | 1.025 (0.991,1.059) | 2.102 | 0.147 |
| BUN (mmol/l) | 0.785 (0.532,1.157) | 1.497 | 0.221 |
| BUA (mmol/l) | 1.001 (0.995,1.007) | 0.159 | 0.690 |
| HCY (µmol/l) | 1.027 (0.915,1.152) | 0.204 | 0.651 |
| LDL (mmol/l) | 0.995 (0.413,2.399) | 0.001 | 0.991 |
| TC (mmol/l) | 1.752 (0.950,3.231) | 3.227 | 0.072 |
| HDL (mmol/l) | 0.601 (0.140,2.585) | 0.469 | 0.494 |
| TG (mmol/l) | 1.465 (0.640,3.356) | 0.817 | 0.366 |
| ΔCq hsa-miR-10a-5p | 1.344 (1.094,1.652) | 7.900 | 0.005 |
| ΔCq hsa_circ_0014243 | 0.546 (0.414,0.720) | 18.414 | <0.001 |
Values were analysed using the continuous variables method in a logistic regression analysis. CI, confidence interval; OR, odds ratio; ALT, alanine transaminase; AST, aspartate transaminase; BMI, body mass index; BUA, blood uric acid; BUN, blood urea nitrogen; HCY, homocysteine; HDL, high-density lipoprotein; LDL, low-density lipoprotein; TG, triglyceride; TC, total cholesterol; Scr, serum creatinine; hsa, Homo sapiens; circ, circular RNA; miR, microRNA; Y, yes; N, no; M, males; F, females.
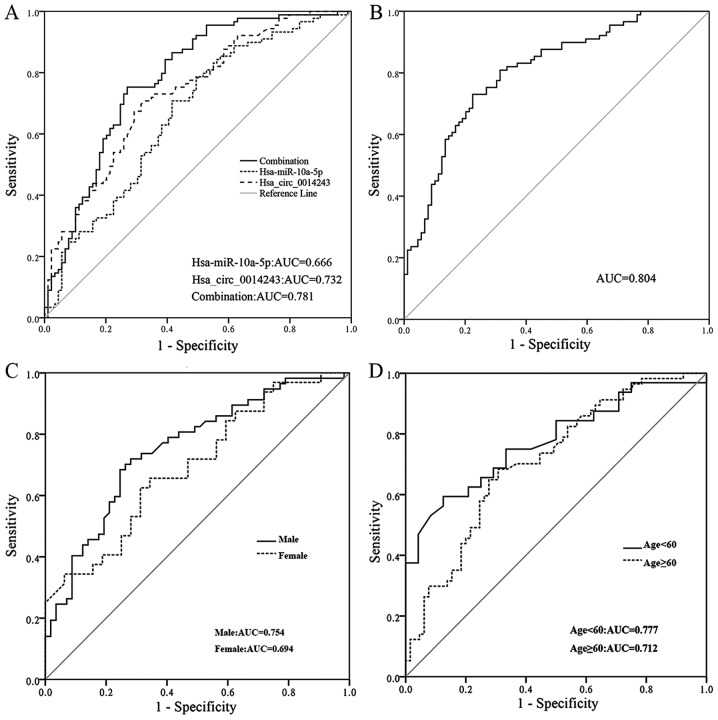
To further explore the potential value and evaluate the accuracy of hsa_circ_0014243 in the diagnosis of EH, the ROC curve was generated using hsa_circ_0014243 as the predictor. A larger value of the area under the ROC curve (AUC) indicates a greater diagnostic value and a better prediction performance. As presented in Fig. 4A, an AUC of 0.732 was obtained using hsa_circ_0014243 as the predictor. The sensitivity and specificity were 0.708 and 0.674, respectively; the cut-off value was 15.010. The AUC of 0.666 was obtained using hsa-miR-10a-5p as predictor, and the sensitivity and specificity were 0.708 and 0.584, respectively; the cut-off value was 4.247. When the expression levels of hsa_circ_0014243 were combined with those of hsa-miR-10a-5p, the AUC increased to 0.781, with corresponding sensitivity and specificity values of 0.865 and 0.584, respectively; the cut-off value was 0.377. As presented in Fig. 4B, the AUC of the model increased to 0.804 when hsa_circ_0014243 was combined with other risk factors, including smoking history, drinking history and BMI, for EH as predictor variables; the cut-off value of this combination was 0.516. Furthermore, to determine the corresponding diagnostic value of the model in population, ROC curves were generated in population sub-groups based on sex and age. Of note, the AUC of the ROC for hsa_circ_0014243 for predicting EH in males and individuals aged <60 years was 0.754 and 0.777, respectively (Fig. 4C and D); the cut-off values were 15.145 and 16.180, respectively. For men aged <60 years, the AUC obtained for using hsa_circ_0014243 as a predictor was 0.820 and the cutoff value was 14.605.
Figure 4.
Receiver operating characteristic curve analysis. Utility of (A) hsa_circ_0014243, hsa-miR-10a-5p and their combination and (B) hsa_circ_0014243 + risk factors (smoking history, drinking history and BMI) in the diagnosis of EH. Comparison of the diagnostic value of hsa_circ_0014243 between (C) males and females and (D) patients aged <60 and ≥60 years for EH. EH, essential hypertension; miR, microRNA; hsa, Homo sapiens; circ, circular RNA; AUC area under the curve.
Discussion
circRNAs are covalently closed RNA molecules that are widely expressed in human cells (11). circRNAs are abundant, and circRNA levels may exceed those of their linear isomers by >10-fold in certain cases (24). In addition, compared to other non-coding linear RNAs, the most important properties of circRNAs are the highly conserved sequences that exhibit tissue-specific and stable expression in mammalian cells (24,25). These characteristics make circRNAs ideal biomarkers for certain diseases. An increasing number of circRNAs have been suggested as biomarkers for cancer and cardiovascular diseases. Previous studies have demonstrated the potential use of hsa_circ_0001895 and hsa_circ_0074362 as biomarkers for gastric cancer (18,26). Jiang et al (27) reported that the circRNA cerebellar degeneration related protein 1 antisense, may serve as a prognostic biomarker for cholangiocarcinoma. However, to date, only few circRNAs have been reported as ideal biomarkers for EH.
In the present study, hsa_circ_0014243 was revealed to be significantly upregulated in EH patients relative to controls and logistic regression analysis identified it as an important risk factor for EH. The diagnostic value of this circRNA was then evaluated by ROC curve analysis. Compared with hsa-miR-10a-5p, hsa_circ_0014243 was upregulated in EH and had a higher AUC and a higher specificity. Previous research has indicated that circRNAs may serve as potential risk factors of EH and the reported AUC for hsa_circ_0037911 and hsa_circ_0002161 was 0.627 and 0.573, respectively (16). In the current study, hsa_circ_0014243 had a higher diagnostic value compared with hsa_circ_0037911 and hsa_circ_0002161. These results indicated that hsa_circ_0014243 has a high potential as a biomarker for EH.
Although the function and regulatory mechanisms underlying the effects of circRNAs in the formation and development of cardiovascular disease remain to be fully elucidated, studies have demonstrated that circRNAs regulate gene expression by acting as specific miRNAs sponges. Wang et al (28) reported that heart-related circRNA directly binds to miR-223 and acts as a specific miRNA sponge to inhibit heart failure and cardiac hypertrophy. Thus, circRNA-miRNA interactions are likely to be involved in intricate regulatory networks by regulating the expression levels of target genes. The bioinformatics prediction research indicated an interaction between hsa-miR-10a-5p and hsa_circ_0014243. Previous studies indicated that hsa-miR-10a-5p may be transferred from endothelial cells to monocytic cells and suppress inflammation by inhibiting NF-κB activation, which targets the β-transducin-repeat-containing gene and the 3′-untranslated region of the gene encoding mitogen-activated protein kinase-7 (5,15). Therefore, the present study hypothesized that hsa-miR-10a-5p is negatively associated with EH. The results revealed that hsa_circ_0014243 was upregulated (P<0.001) and hsa-miR-10a-5p was downregulated (P<0.001) in patients diagnosed with EH relative to those in control subjects. It is likely that hsa_circ_0014243 acts as a specific sponge targeting hsa-miR-10a-5p, possibly to participate in the genesis and development of EH.
Multiple studies have demonstrated the potential value of certain circRNAs as biomarkers for various types of cancer and cardiovascular disease. To the best of our knowledge, the present study was the first to demonstrate that hsa_circ_0014243 expression is upregulated in EH (P<0.001 vs. controls) and thus exhibits a marked diagnostic value.
Of note, sex and lifestyle factors, including smoking and drinking, were identified as crucial factors involved in the development and progression of EH. Male sex and smoking were identified to be associated with lower hsa-miR-10a-5p expression levels, which were identified as a risk factor for EH based on logistic regression analysis. The above results were consistent with the known increased risk of smokers to develop EH. However, logistic regression results demonstrated that males were less likely to have an occurrence of EH compared with females. A national hypertension survey from China revealed that, in 18–44-year-olds, the prevalence rate of EH in males was higher than that in females (20.6/14.3%); in 45–59-year-olds, the prevalence rate of EH in males was close to that in females (40.4/39.8%); and in >60-year-olds, the prevalence rate of EH in males was lower than that in females (57.4/59.0%) (29). Given that the median age of male and female individual in the current study was >60 years old, it was hypothesized that females are at greater risk of developing hypertension than males due to low estrogen level after menopause. Willinger et al (30) reported six miRNAs detected in whole blood samples that were significantly associated with the three-level smoking status. Another study indicated that cigarette smoke exposure may induce pulmonary hypertension by downregulating the mRNA expression of the potassium channels Kv1.5 and Kv2.1 mRNA in rats (31). However, the roles of miRNAs or circRNAs in smoking-induced EH remain largely elusive.
Epidemiology studies on hypertension have reported that the prevalence of hypertension increases with advancing age and is associated with various alterations in the autonomic regulation of the cardiovascular system (32). An estimated 90% of individuals who are non-hypertensive at 55 years of age will develop hypertension in their lifetime (33). Hence, the present study investigated the effects of aging on hsa_circ_0014243 expression levels. The results suggested that the expression levels of hsa_circ_0014243 are correlated with age and increase with advancing age (P<0.001). Given that circRNAs are upregulated on a genome-wide scale with advancing age in Drosophila, mice and Cel (33–35), aging-associated circRNA accumulation appears to be a universal phenomenon among different species. Cheng et al (36) reported that circRNA_104816 and circRNA_103827 levels in human granulosa cells are positively associated with maternal age. Another study reported that hsa_circ_0000069 levels were upregulated in colorectal cancer tissues when compared with adjacent noncancerous tissues that the levels correlated positively with the patients' age (37). Therefore, it is required to identify factors that potentially contribute to aging-associated accumulation of circRNAs. Recent studies proposed that the high stability of circRNAs and alterations in alternative splicing of circRNAs contribute to cardiovascular diseases (38). The enhanced stability of circRNAs may gradually accumulate over time as a result of uninterrupted transcription, which may explain for the observed increases in circRNA levels with increasing age (35). In addition to the potential role of the high stability of circRNA in aging-associated circRNA accumulation, back-splicing may also explain for the aging-associated accumulation of circRNAs (38). Thus, it may be speculated that hsa_circ_0014243 accumulates in individuals with age and then induces EH once it reaches a high concentration.
Previous studies have indicated that certain clinical characteristics and lifestyle habits may be associated with circRNA expression levels. In the present study, Pearson or Spearman's rank correlation analysis was performed to evaluate the correlation between the levels of hsa_circ_0014243 and various factors that potentially contribute to EH. Considering that circRNAs may affect some hypertension-associated factors and lead to blood pressure changes, the authors have provided an explanation for the changes in blood pressure caused by aberrant expression of this circRNA. The levels of hsa_circ_0014243 were indicated to be negatively correlated with HDL levels. It has been proven that HDL is one of risk factors for high blood pressure and the blood pressure may increase with the levels of HDL. It may be hypothesized that hsa_circ_0014243 affects the blood pressure by changing the concentration of HDL.
Of note, the present study has several limitations. First, the levels of hsa-miR-10a-5p and hsa_circ_0014243 were analyzed in only 89 healthy controls and 89 EH patients, and more samples should be analyzed in future studies. Furthermore, the present study did not include any cell culture or animal model studies that may have increased the credibility of the causality indicated by the current results. Third, no Luciferase assay was performed to confirm the direct binding interaction of hsa-miR-10a-5p and hsa_circ_0014243. These limitations will be addressed in future studies by our group.
In conclusion, the results of the present study indicated that the levels of hsa_circ_0014243 were upregulated in patients with EH. It may be hypothesized that hsa_circ_0014243 acts as a specific sponge of hsa-miR-10a-5p, whose expression levels are known to be downregulated in EH. Hsa_circ_0014243 levels were identified to be significantly associated with age, HDL levels and glucose levels. The present results suggest a crucial role of hsa_circ_0014243 in the genesis and development of EH and its potential use as a biomarker for the diagnosis of EH.
Acknowledgements
The authors would like to thank Dr Han Cen of the Department of Preventative Medicine, Medical School of Ningbo University for reading the manuscript and providing valuable suggestions.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81773528), Ningbo Scientific Innovation Team for Environmental Hazardous Factor Control and Prevention (grant no. 2016C51001), Zhejiang Province Social Development Research Project (grant no. 2016C33178) and the K.C. Wong Magna Fund at Ningbo University.
Availability of data and materials
All data generated or analyzed during the current study are included in this published article.
Authors' contributions
SZ, TZ and LZ designed the experiments, and SZ, TG and XB performed them. JS and JZ collected and analyzed the data. SZ wrote the manuscript.
Ethical approval and consent to participate
The present study, including the recruitment of patients from affiliated hospitals, was approved by the Human Research Ethics Committee of Ningbo University (Ningbo, China). Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Staessen JA, Wang J, Bianchi G, Birkenhäger WH. Essential hypertension. Lancet. 2003;361:1629–1641. doi: 10.1016/S0140-6736(03)13302-8. [DOI] [PubMed] [Google Scholar]
- 2.Mao S, Gu T, Zhong F, Fan R, Zhu F, Ren P, Yin F, Zhang L. Hypomethylation of the Toll-like receptor-2 gene increases the risk of essential hypertension. Mol Med Rep. 2017;16:964–970. doi: 10.3892/mmr.2017.6653. [DOI] [PubMed] [Google Scholar]
- 3.Savoia C, Sada L, Zezza L, Pucci L, Lauri FM, Befani A, Alonzo A, Volpe M. Vascular inflammation and endothelial dysfunction in experimental hypertension. Int J Hypertens. 2011;2011:281240. doi: 10.4061/2011/281240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao SQ, Sun JH, Gu TL, Zhu FB, Yin FY, Zhang LN. Hypomethylation of interleukin-6 (IL-6) gene increases the risk of essential hypertension: A matched case-control study. J Hum Hypertens. 2017;31:530–536. doi: 10.1038/jhh.2017.7. [DOI] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, Zhou J, Tang ZY, Huang XY. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Li C, Tan C, Liu X. Circular RNAs: A new frontier in the study of human diseases. J Med Genet. 2016;53:359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 9.Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michell DL, Vickers KC. HDL and microRNA therapeutics in cardiovascular disease. Pharmacol Ther. 2016;168:43–52. doi: 10.1016/j.pharmthera.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, Liao J, Liu B, Zeng F, Zhang L. Mechanisms and therapeutic potential of microRNAs in hypertension. Drug Discov Today. 2015;20:1188–1204. doi: 10.1016/j.drudis.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, Xu CC, Chen WD, Shen WL, Ruan CC, Zhu LM, Zhu DL, Gao PJ. MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts. Biochem Biophys Res Commun. 2010;400:483–488. doi: 10.1016/j.bbrc.2010.08.067. [DOI] [PubMed] [Google Scholar]
- 15.Njock MS, Cheng HS, Dang LT, Nazari-Jahantigh M, Lau AC, Boudreau E, Roufaiel M, Cybulsky MI, Schober A, Fish JE. Endothelial cells suppress monocyte activation through secretion of extracellular vesicles containing antiinflammatory microRNAs. Blood. 2015;125:3202–3212. doi: 10.1182/blood-2014-11-611046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, Zhang L. A potential risk factor of essential hypertension in case-control study: Circular RNA hsa_circ_0037911. Biochem Biophys Res Commun. 2018;498:789–794. doi: 10.1016/j.bbrc.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 17.Burnier M, Oparil S, Narkiewicz K, Kjeldsen SE. New 2017 American Heart Association and American College of Cardiology guideline for hypertension in the adults: Major paradigm shifts, but will they help to fight against the hypertension disease burden? Blood Press. 2018;27:62–65. doi: 10.1080/08037051.2018.1430504. [DOI] [PubMed] [Google Scholar]
- 18.Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, Guo J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317699125. doi: 10.1177/1010428317699125. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijmans JG, Diehl KJ, Bammert TD, Kavlich PJ, Lincenberg GM, Greiner JJ, Stauffer BL, DeSouza CA. Association between hypertension and circulating vascular-related microRNAs. J Hum Hypertens. 2018;32:440–447. doi: 10.1038/s41371-018-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 22.Schellingerhout JM, Heymans MW, de Vet HC, Koes BW, Verhagen AP. Categorizing continuous variables resulted in different predictors in a prognostic model for nonspecific neck pain. J Clin Epidemiol. 2009;62:868–874. doi: 10.1016/j.jclinepi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345–8354. doi: 10.3748/wjg.v23.i47.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao B, Guo J. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11–20. doi: 10.2217/bmm-2017-0114. [DOI] [PubMed] [Google Scholar]
- 27.Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui YF, Sun DJ. A novel prognostic biomarker for cholangiocarcinoma: CircRNA Cdr1as. Eur Rev Med Pharmacol Sci. 2018;22:365–371. doi: 10.26355/eurrev_201801_14182. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Zhang L, Wang F, Liu L, Wang H. China National Survey of Chronic Kidney Disease Working Group: Prevalence, awareness, treatment, and control of hypertension in China: Results from a national survey. Am J Hypertens. 2014;27:1355–1361. doi: 10.1093/ajh/hpu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willinger CM, Rong J, Tanriverdi K, Courchesne PL, Huan T, Wasserman GA, Lin H, Dupuis J, Joehanes R, Jones MR, et al. MicroRNA signature of cigarette smoking and evidence for a putative causal role of MicroRNAs in smoking-related inflammation and target organ damage. Circ Cardiovasc Genet. 2017;10:e001678. doi: 10.1161/CIRCGENETICS.116.001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin CY, Wan LM, Chen YQ, Ou HT, Zhao L, Liang YT, Ma R, Lu WJ, Wang J. Cigarette smoke exposure induced pulmonary artery pressure increase through inhibiting Kv1.5 and Kv2.1 mRNA expression in rat pulmonary artery smooth muscles. Zhonghua Jie He He Hu Xi Za Zhi. 2012;35:695–699. (In Chinese) [PubMed] [Google Scholar]
- 32.Arnold AC, Gallagher PE, Diz DI. Brain renin-angiotensin system in the nexus of hypertension and aging. Hypertens Res. 2013;36:5–13. doi: 10.1038/hr.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, et al. The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure: The JNC 7 report. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 34.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortés-López M, Gruner MR, Cooper DA, Gruner HN, Voda AI, van der Linden AM, Miura P. Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genomics. 2018;19:8. doi: 10.1186/s12864-017-4386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng J, Huang J, Yuan S, Zhou S, Yan W, Shen W, Chen Y, Xia X, Luo A, Zhu D, Wang S. Circular RNA expression profiling of human granulosa cells during maternal aging reveals novel transcripts associated with assisted reproductive technology outcomes. PLoS One. 2017;12:e0177888. doi: 10.1371/journal.pone.0177888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, Huang MD, He JD. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the current study are included in this published article.